Abstract
Ovarian follicular development and ovulation are complex and tightly regulated processes that involve regulation by microRNAs (miRNAs). We previously identified differentially expressed mRNAs between human cumulus granulosa cells (CGCs) from immature early antral follicles (germinal vesicle - GV) and mature preovulatory follicles (metaphase II - M2). In this study, we performed an integrated analysis of the transcriptome and miRNome in CGCs obtained from the GV cumulus-oocyte complex (COC) obtained from IVM and M2 COC obtained from IVF. A total of 43 differentially expressed miRNAs were identified. Using Ingenuity IPA analysis, we identified 7288 potential miRNA-regulated target genes. Two hundred thirty-four of these target genes were also found in our previously generated ovulatory gene library while exhibiting anti-correlated expression to the identified miRNAs. IPA pathway analysis suggested that miR-21 and FOXM1 cooperatively inhibit CDC25A, TOP2A and PRC1. We identified a mechanism for the temporary inhibition of VEGF during ovulation by TGFB1, miR-16-5p and miR-34a-5p. The linkage bioinformatics analysis between the libraries of the coding genes from our preliminary study with the newly generated library of regulatory miRNAs provides us a comprehensive, integrated overview of the miRNA-mRNA co-regulatory networks that may play a key role in controlling post-transcriptomic regulation of the ovulatory process.
Introduction
Ovarian follicular development and ovulation in mammals are highly complex and tightly regulated processes that involve the selection of a dominant follicle, reactivation of oocyte meiosis, rupture of the follicle wall, cumulus-oocyte expansion and tissue remodeling to form the corpus luteum. Oocyte growth and maturation are linked to the developmental properties of somatic cell populations in the expanding follicle as bi-directional signaling that occurs between oocytes and granulosa cells1. The genes involved in follicle maturation and ovulation are the subject of growing interest and have been well-studied by several laboratories, including ours2,3.
MicroRNAs (miRNAs) are small (~22-nucleotide), non-coding regulatory RNA molecules encoded by plants, animals and some viruses that regulate a variety of developmental and physiological processes (reviewed in4,5).
In the last few years, by virtue of the application of high-throughput sequencing techniques, it became clear that eukaryotes transcribe up to 90% of their genomic DNA. However, only 1-2% of these transcripts are protein-coding genes6.
Several studies have found specific miRNAs that target important ovarian molecules such as progesterone receptor (PGR)7, aromatase (CYP19A1), follicle stimulating hormone receptor (FSHR)8 and the FYN pathway9. miRNAs were suggested to function as RNA-based paracrine factors in the crosstalk between oocytes and support cells of the follicle10. Furthermore, the expression of a subset of miRNAs has been shown to vary during follicle/luteal development11–13. Effects on granulosa cell function and/or gene expression, largely in mice, have been reported for a handful of miRNAs, and these include effects on cell survival (miR-21 and miR-23a), proliferation (miR-145, miR-503, and miR-224), estradiol production (miR-224, miR-383, and miR-378) and terminal differentiation (miR-132 and miR-212) of cultured cells (reviewed in14,15).
In humans, miRNAs have already been profiled in cumulus versus mural granulosa cells8, Polycystic Ovary Syndrome (PCOS) patients16 and cumulus cells according to oocyte nuclear maturity17. miRNAs have been suggested as potential biomarkers in IVF18.
In the past several years, miRNA and mRNA integrated analysis (MMIA) has become a tool for examining the biological functions of miRNA expression. As the biological effects of miRNAs are due to their modulation of target RNA expression, accurate miRNA target prediction is essential to any study of miRNA function.
To understand the molecular and regulatory mechanisms necessary for follicular maturation and ovulation, we aimed to uncover the miRNA transcriptome involved in these processes. Recent advances in genomics allow a systematic approach to identify critical genes and regulated pathways involved in oocyte maturation and ovulation. Using whole transcriptome sequencing, we recently identified mRNAs that are differentially expressed between immature mid-antral follicles and mature pre-ovulatory follicles3. The resulting database provides unprecedented insight into the processes and pathways involved in follicular maturation and ovulation. To complete the identification of factors involved in the ovulatory process, the aim of this work was to generate a library of global miRNAs involved in this process and to link the new ovulatory miRNA library with the previously described mRNA library. This enabled us to identify new regulatory mechanisms responsible for the final follicular maturation and ovulatory processes.
Results
miRNA profile differences in compact and expanded cumulus cells
To elucidate the roles of miRNAs in human folliculogenesis and ovulation, we generated a library of regulated miRNAs during the final stage of follicular maturation and ovulation. We used NanoString on RNA extracted from two groups of cumulus granulosa cells: (1) Compact cumulus cells surrounding GV oocytes that were acquired during IVM treatment (CCGV n = 4) and (2) Expanded cumulus cells enclosing M2 oocytes that were acquired during IVF treatment (CCM2, n = 3). One CCGV sample was an outlier and was removed from further downstream analysis. Thus, the final number of samples in each group was three.
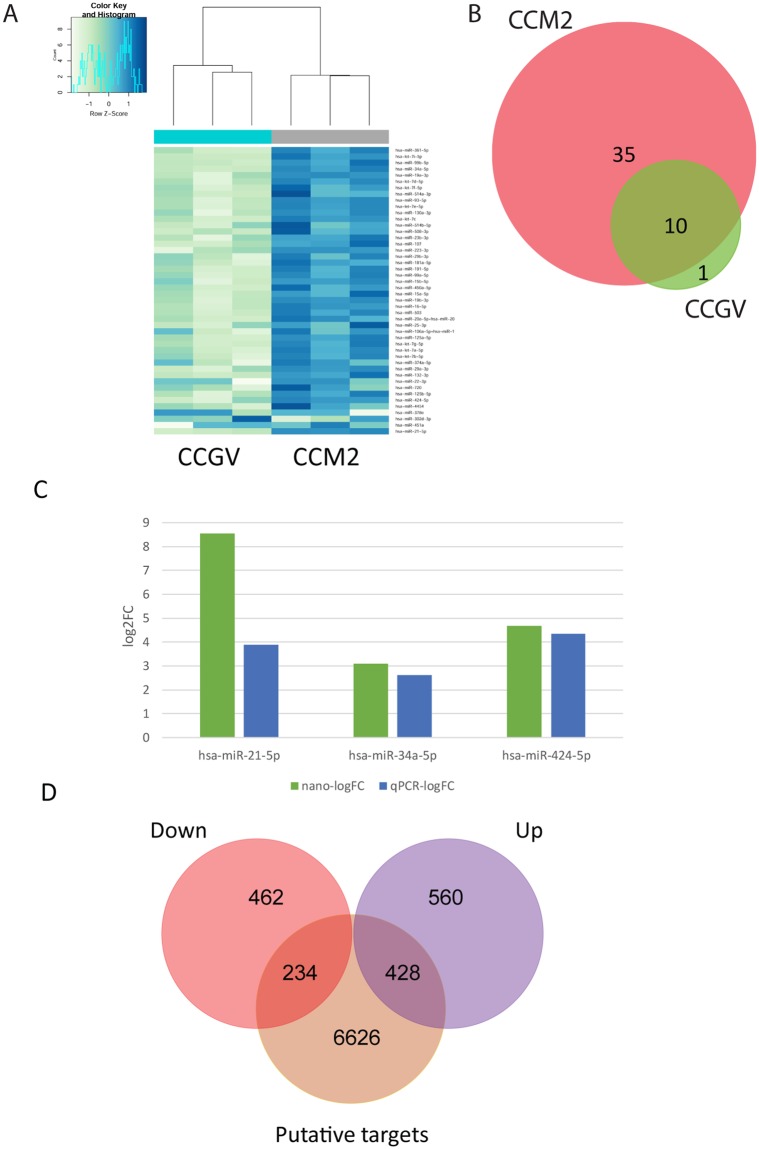
A Venn diagram of these groups is shown in Fig. 1B, illustrating the numbers of selective and co-expressed miRNAs in the two conditions, CCGV and CCM2. A total of 45 miRNAs were detected in CCM2 and 11 miRNAs were detected in CCGV. Most notably, 35 miRNAs were exclusively detected in the CCM2 group; only 1 was exclusively detected in the CCGV group and 10 miRNAs were shared between the two conditions (Table 1). hsa-miR-378e was exclusively expressed in the CCGV group, although the differential expression level was not statistically significant.
Figure 1.
(A) Hierarchical clustering of CCGV (n = 3) and CCM2 (n = 3) samples based on miRNA expression levels. Each column represents a sample and each row represents a transcript. Expression level of each miRNA in a single sample is depicted according to the color scale. (B) Venn diagram of selective and co-expressed miRs in CCGV and CCM2 samples. A total of 45 miRs were expressed in CCM2 (pink) and 11 miRs were expressed in CCGV (green). Most notably, 35 miRs were exclusively expressed in the IVF group and 10 miRs were shared by both samples. (C) Total mRNA was purified from CCs denuded from GV COC aspirated during IVM procedures (CCGV) and CCs denuded from M2 COC (CCM2) aspirated during IVF procedures. The miRNAs were subjected to qPCR in duplicate with the examined genes and RNU6B primers. Gene expression was calculated relative to the RNU6B level in the same sample and expression levels were compared using Student’s t-test. The difference reached a level of significance (P < 0.05) for all tested genes. NanoString (green) and qPCR (blue) results are presented as log2-fold change between CCM2 and CCGV samples. (D) Venn diagram showing the relationship between the putative miRNA targets (brown) and experimentally differentially expressed mRNAs (from Yerushaslmi et al.3, upregulated mRNA (purple), downregulated mRNA (red)). Of all the putative miRNA targets, 234 were negatively correlated.
Table 1.
miRNAs identified by the NanoString nCounter miRNA expression assay in human cumulus granulosa cells. Forty-three of the miRNAs were differentially expressed with a fold change >2 and an FDR < 5%.
| miR | logFC CCM2 vs. CCGV | Average Expression CCGV | Average Expression CCM2 | p-value | Adj. p-value | Putative granulosa cell function | References |
|---|---|---|---|---|---|---|---|
| hsa-let-7a-5p* | 3.81363 | 3642.69 | 43742.2 | 0.000154 | 0.000256 | Apoptosis, cumulus vs. mural | 8, 50, 63 |
| hsa-let-7b-5p | 3.30288 | 2591.97 | 20276.4 | 0.000497 | 0.000693 | ||
| hsa-let-7c | 3.87105 | 506.408 | 6725.24 | 3.53E-06 | 2.91E-05 | ||
| hsa-let-7d-5p | 3.10472 | 376.498 | 2855.3 | 3.91E-05 | 0.000106 | ||
| hsa-let-7e-5p | 3.54614 | 491.637 | 5066.29 | 1.15E-05 | 4.41E-05 | ||
| hsa-let-7f-5p* | 2.66036 | 348.055 | 2230.98 | 0.000156 | 0.000256 | ||
| hsa-let-7g-5p* | 4.17388 | 1762.22 | 31204.5 | 1.99E-05 | 6.11E-05 | ||
| hsa-let-7i-5p | 3.2881 | 279.589 | 2871.92 | 7.02E-06 | 0.000036 | ||
| hsa-miR-106a-5p/ miR-17-5p | 1.89582 | 1418.37 | 4268.29 | 0.017636 | 0.019316 | Bovine follicle development | 64 |
| hsa-miR-107 | 2.20654 | 289.935 | 1226.41 | 0.000389 | 0.00056 | Bovine follicle development, Steroidogenesis | 65, 66 |
| hsa-miR-125a-5p | 3.81061 | 2536.53 | 30936.3 | 9.05E-05 | 0.000189 | Apoptosis | 67 |
| hsa-miR-125b-5p | 5.00927 | 2516.64 | 70782.3 | 3.79E-06 | 2.91E-05 | Bovine follicle development, Steroidogenesis, cumulus vs. mural | 8, 65, 66 |
| hsa-miR-130a-3p | 3.27081 | 559.043 | 4177.33 | 5.68E-05 | 0.000131 | Bovine follicle development, PCOS | 66, 68 |
| hsa-miR-132-3p | 5.59724 | 290.378 | 15338.4 | 3.9E-09 | 8.96E-08 | cumulus vs. mural, Steroidogenesis, PCOS, murine follicle development | 8, 11, 68 |
| hsa-miR-15a-5p | 3.15798 | 1280.84 | 10868 | 0.000342 | 0.000507 | Steroidogenesis | 65 |
| hsa-miR-15b-5p | 2.42639 | 1952.83 | 8482.74 | 0.004694 | 0.005682 | Steroidogenesis | 65 |
| hsa-miR-16-5p | 3.5448 | 1493.38 | 16847.5 | 0.000103 | 0.000206 | Bovine follicle development | 8 |
| hsa-miR-181a-5p | 3.82643 | 404.396 | 5005.58 | 7.08E-06 | 0.000036 | cumulus vs. mural, proliferation | 8, 69 |
| hsa-miR-191-5p* | 2.80069 | 1557.66 | 10258.1 | 0.001191 | 0.001565 | Bovine follicle development, cumulus vs. mural | 8, 70 |
| hsa-miR-19a-3p | 3.09878 | 381.085 | 2980.11 | 4.71E-05 | 0.000114 | Bovine follicle development, Steroidogenesis, PCOS | 64– 66, 68 |
| hsa-miR-19b-3p | 3.33449 | 1177.47 | 10517.6 | 0.000143 | 0.000253 | Ovarian hyperstimulation | 71 |
| hsa-miR-20a-5p/ miR-20b-5p | 3.85536 | 1094.21 | 16458.7 | 2.87E-05 | 8.26E-05 | cumulus vs. mural | 8 |
| hsa-miR-21-5p* | 8.54162 | 21.5677 | 6740.8 | 1.69E-11 | 7.79E-10 | cumulus vs. mural, murine follicle development, apoptosis | 8, 11, 40 |
| hsa-miR-223-3p | 2.58013 | 254.152 | 1404.38 | 0.000114 | 0.000216 | cumulus vs. mural, Steroidogenesis, PCOS | 8, 72 |
| hsa-miR-22-3p* | 1.75262 | 1807.13 | 4565.75 | 0.041854 | 0.044774 | cumulus vs. mural, Steroidogenesis, PCOS | 8, 72 |
| hsa-miR-23b-3p | 2.59206 | 281.984 | 1384.31 | 0.000117 | 0.000216 | Bovine ovary | 73 |
| hsa-miR-25-3p | 2.06898 | 1683.34 | 7156.33 | 0.013105 | 0.014703 | Steroidogenesis | 65 |
| hsa-miR-29a-3p | 4.94371 | 536.265 | 13587.1 | 5.05E-07 | 7.74E-06 | Steroidogenesis | 74 |
| hsa-miR-29b-3p | 3.91754 | 672.73 | 7187.08 | 0.000016 | 5.26E-05 | Steroidogenesis, cumulus vs. mural | 8, 65 |
| hsa-miR-302d-3p | −0.25 | 1660.7 | 1305.76 | 0.710 | 0.710** | n/a | |
| hsa-miR-34a-5p | 3.09029 | 245.28 | 2018.34 | 8.49E-06 | 0.000036 | Apoptosis | 55, 75 |
| hsa-miR-361-5p | 3.15727 | 329.908 | 2675.28 | 1.59E-05 | 5.26E-05 | Murine follicle development | 11 |
| hsa-miR-374a-5p | 2.05355 | 841.464 | 2757.13 | 0.007376 | 0.008482 | cumulus vs. mural, | 8 |
| hsa-miR-378e | −1.3021 | 3643.64 | 2397.74 | 0.13083 | 0.13678** | Steroidogenesis | 76 |
| hsa-miR-424-5p | 4.68931 | 1405.72 | 27294.5 | 7.48E-06 | 0.000036 | Age | 17 |
| hsa-miR-4454 | 3.79594 | 2852.29 | 62869.6 | 0.000325 | 0.000498 | n/a | |
| hsa-miR-450a-5p | 2.70382 | 1349.62 | 8663.35 | 0.001472 | 0.001881 | Bovine follicle development | 66 |
| hsa-miR-451a* | 1.218 | 1669.72 | 3424.96 | 0.176 | 0.180** | cumulus vs. mural | 8 |
| hsa-miR-503 | 4.2726 | 750.23 | 13810.1 | 2.66E-06 | 2.91E-05 | Bovine follicle development, | 12, 68, 77 |
| hsa-miR-508-3p | 3.27969 | 208.958 | 2839.37 | 8.61E-06 | 0.000036 | PCOS | 16 |
| hsa-miR-514a-3p | 2.63133 | 282.897 | 2477.97 | 0.000185 | 0.000294 | PCOS | 16 |
| hsa-miR-514b-5p | 1.8413 | 431.937 | 1844.75 | 0.005467 | 0.006449 | PCOS | 72 |
| hsa-miR-720 | 3.10848 | 1958.04 | 22615.3 | 0.001832 | 0.002277 | PCOS, Age | 16, 17 |
| hsa-miR-93-5p | 3.30979 | 620.555 | 5549.2 | 0.000045 | 0.000114 | Proliferation, PCOS | 78, 79 |
| hsa-miR-99a-5p* | 2.89148 | 1585.53 | 10215.6 | 0.000935 | 0.001266 | cumulus vs. mural, Bovine follicle development | 8, 70, 80 |
| hsa-miR-99b-5p | 2.74138 | 357.19 | 2252.67 | 7.72E-05 | 0.000169 | Age, Bovine follicle development, PCOS | 64, 67, 81 |
*miRNA detected as one of the 10 most abundant in human cumulus cells8.
**Differential expression between CCGV and CCM2 samples was not significant.
With NanoString data inspected, all quality control parameters were within the acceptable ranges. Unsupervised hierarchical cluster analysis based on the expression of all detected miRNAs shows that the biological replicates cluster to their corresponding groups (Fig. 1A).
Almost all of the expressed miRNAs were significantly differentially expressed (43 of the 46 identified miRNAs) between CCGV and CCM2, with a fold change >2 and a false discovery rate (FDR) < 5%. Of these, 25 represent unique seed sequences. Our results indicate that all differentially expressed miRNAs were up-regulated in CCM2 cells.
Validation of selected miRNA expression levels as estimated by qPCR
We used qPCR analysis to validate the results obtained by NanoString. We selected three miRNAs that were identified as differentially expressed by NanoString. qPCR was performed in three different experiments, as shown as fold induction in Fig. 1C, and demonstrated agreement with NanoString relative expression data.
Identification of differentially expressed miRNA target genes – in silico analysis
Because miRNAs act by regulating the expression of target genes, precise miRNA target prediction is important for elucidating miRNA function. The identification of miRNA target genes was performed using the microRNA target filter tool of QIAGEN’s Ingenuity Pathway Analysis software, taking into account only experimentally validated and highly confident predictions of miRNA target interactions. We found 7288 putative miRNA target genes of which 727 (294 unique) were experimentally validated (Supp. Tables S3 and S4).
Identification of differentially expressed miRNAs that negatively correlated with ovulatory target genes - experimental analysis
To further dissect the putative ovulatory miRNA targets, we combined our previously generated mRNA library3 and miRNA expression data using the microRNA target filter tool of QIAGEN’s Ingenuity Pathway Analysis19. A Venn diagram depicting the relationships between the putative miRNA targets and experimentally differentially expressed mRNAs is depicted in Fig. 1D.
We chose to focus on negatively correlated targets. Since all of the differentially expressed CCM2 miRNAs were upregulated, all chosen paired mRNA targets were downregulated. The 696 downregulated mRNAs in the CCM2 group (compared to CCGV group) were paired with the 7288 putative miRNA targets. Of the 696 downregulated mRNAs, 234 were targets of 22 differentially expressed miRNAs (of the 25 having unique seed sequences). In total, these 234 genes formed 479 miRNA-target gene pairs with an inverse correlation of expression (Table 2).
Table 2.
The 234 miRNA target genes that negatively correlated with the expression of 22 differentially expressed miRNAs.
| ID | Symbol | Count of Targets | Target mRNA |
|---|---|---|---|
| hsa-let-7g-5p | let-7a-5p (and other miRNAs w/seed GAGGUAG) | 40 | ADAMTS15, ANGPTL2, AURKB, B3GAT1, C15orf39, CDC25A, CHRD, CMTM6, DAPK1, DSP, ESPL1, ESR2, ETNK2, EZH2, FANCD2, FRMD4B, GAS7, GATM, IFNLR1, KIF21B, LINGO1, MMP11, MYO5B, MYRIP, NEMP1, NOS1, PAG1, PARM1, PLXNC1, PPT2, PRIM1, RBM38, RIMS3, RRM2, SLC1A4, SLC37A4, THBS1, TMPO, TYMS, UNC5A |
| hsa-miR-107 | miR-103-3p (and other miRNAs w/seed GCAGCAU) | 26 | ADGRB3, AJUBA, BCL11A, CDCA4, CLSPN, ESR1, HOXD10, HSDL1, IGSF3, IHH, LRP1, MBOAT1, NAV1, NEIL1, NOS1, NRP2, OLFM1, PAG1, RIMS3, RNF19A, RTKN2, S1PR3, SOWAHC, SYNDIG1, TMEM35, WHSC1 |
| hsa-miR-125b-5p | miR-125b-5p (and other miRNAs w/seed CCCUGAG) | 29 | ADAMTS15, AJUBA, C15orf39, CACNB2, CDC25A, ENTPD1, FAM78A, GLB1L2, HAPLN1, HCN1, HCN4, HOXD9, KCNH3, KCNIP3, LOXL1, MMP11, NEMP1, NUP210, OPALIN, PARM1, PPT2, PRSS35, RBM38, SLC4A8, ST6GAL1, STMN3, TMCC2, TNFSF4, VEGFA |
| hsa-miR-130a-3p | miR-130a-3p (and other miRNAs w/seed AGUGCAA) | 32 | ADAMTS18, ADCY2, ADGRB3, ARX, BCL11A, CEP55, CHST1, DEPDC1, DIAPH3, ESCO2, ESR1, FAM78A, GSE1, HES1, IGSF3, INHBB, ITPKB, KLHDC8A, SIK1, MB21D2, MTCL1, MYO1D, NRP2, PLCL2, PLLP, PRR15, RALGAPA2, SHANK2, SOX4, SOX5, ST8SIA5, ZEB1 |
| hsa-miR-132-3p | miR-132-3p (and other miRNAs w/seed AACAGUC) | 12 | ARX, COL4A4, HAPLN1, HUNK, ITPKB, KIF21B, OLFM1, PALM2, RAP2B, SHANK2, SOX4, SOX5 |
| hsa-miR-424-5p | miR-16-5p (and other miRNAs w/seed AGCAGCA) | 39 | ADAMTS18, C14orf37, CASR, CD47, CDC25A, CDCA4, CHEK1, CLSPN, CRHBP, DPH5, GLCE, GSE1, HCN1, HERC6, IHH, LY6E, MARCH4, MCF2L, MSH2, MYO5B, MYRIP, NAV1, NOS1, NRP2, NUP210, PAG1, PARM1, PLXNA2, PPIF, PPT2, PRIM1, RIMS3, SLC4A8, SOWAHC, SOX5, SYNDIG1, VEGFA, WHSC1, ZNF423 |
| hsa-miR-20a-5p | miR-17-5p (and other miRNAs w/seed AAAGUGC) | 36 | AJUBA, CDC25A, CEP128, CHAF1A, CNNM3, DDIAS, DPF3, E2F1, ELAVL2, ESR1, FRMD4B, GUCY1A3, HAUS8, HCN4, ITPKB, SIK1, MARCH4, MCF2L, MCM3, MYLIP, MYO1D, MYO5B, NR4A3, NRP2, PBK, POLQ, PRR15, RASL11B, RRM2, SEMA7A, SHANK2, SORL1, SOWAHC, SOX4, VEGFA, WHSC1 |
| hsa-miR-181a-5p | miR-181a-5p (and other miRNAs w/seed ACAUUCA) | 35 | ACAN, ADAMTS18, ADGRB3, ATP1B1, ELAVL2, ERG, ESR1, FAM19A2, GAL3ST3, GAS7, GREM1, GSE1, HAPLN1, HCN1, HEY2, HMGB2, MB21D2, MBOAT1, MTCL1, NR4A3, PAG1, PALM2, PARM1, PEG3, PLCL2, POLQ, PPP1R12B, RALGAPA2, RFTN2, RTKN2, SFRP4, SOX5, THBS2, UNC5A, WHSC1 |
| hsa-miR-191-5p | miR-191-5p (and other miRNAs w/seed AACGGAA) | 2 | BCL11A, SOX4 |
| hsa-miR-19b-3p | miR-19b-3p (and other miRNAs w/seed GUGCAAA) | 36 | ARHGAP11A, CEP55, CHST1, DAAM1, DAG1, EDARADD, ELAVL2, ENC1, ESR1, IFI44L, IGSF3, INHBB, ITPKB, PCLAF, MATN2, MATN3, MB21D2, MTCL1, MYLIP, NAV1, NRP2, PARM1, PLCL2, PLXNC1, PRC1, RAP2B, RNF19A, SHANK2, SOGA1, SORL1, SOX4, SOX5, SPTSSB, SYNPO2, THBS1 |
| hsa-miR-21-5p | miR-21-5p (and other miRNAs w/seed AGCUUAU) | 9 | BCL11A, CDC25A, DAG1, ERG, MATN2, MSH2, PAG1, SOX5, ST6GAL1 |
| hsa-miR-22-3p | miR-22-3p (miRNAs w/seed AGCUGCC) | 10 | ADCK2, CHGA, DERL3, EMILIN3, ESR1, GATM, HUNK, NUSAP1, RAPGEF3, VIT |
| hsa-miR-223-3p | miR-223-3p (miRNAs w/seed GUCAGUU) | 14 | ADGRB3, ATP1B1, CSPG5, CTSV, E2F1, ECT2, GALNT18, LAYN, MYO5B, NUP210, OLFM1, RIMS3, STMN1, ZEB1 |
| hsa-miR-23b-3p | miR-23a-3p (and other miRNAs w/seed UCACAUU) | 41 | ARHGEF6, BCL11A, CDC6, COL4A4, DAPK1, DDAH1, DEPDC1, DTL, EDARADD, ENC1, EXOC3L4, FANCI, FSHR, GALNT12, GLCE, GREM1, HAPLN1, HES1, HMGB2, HOXD10, IHH, KCNIP4, MAP7D2, MKX, NDC1, NEMP1, PDGFA, PLXNC1, PPIF, PRSS35, RAD51AP1, RAP2B, SFRP4, SOWAHC, SPTSSB, SYNPO2, TMPO, TOP2A, WHSC1, ZEB1, ZNF423 |
| hsa-miR-29a-3p | miR-29b-3p (and other miRNAs w/seed AGCACCA) | 29 | ADAMTS18, ADAMTS2, ADCYAP1R1, AGPAT4, ATP1B1, AUNIP, BCL11A, CLEC2L, COL4A4, CSPG4, GAS7, HAPLN1, KIF24, LPL, MAP2K6, MFAP2, MYBL2, NASP, NAV1, NLGN3, PAG1, PALM2, PDGFC, RNF19A, RTKN2, SLC16A1, SOWAHC, TMEM132A, VEGFA |
| hsa-miR-34a-5p | miR-34a-5p (and other miRNAs w/seed GGCAGUG) | 25 | ABLIM1, ADCY5, ANK2, CD47, CDC25A, COL4A4, DAAM1, FAM107A, GABRA3, GLCE, INHBB, MYRIP, NAV1, NOS1, PAG1, PALM2, PTGIS, RIMS3, SDK2, SLCO3A1, SOGA1, SOX4, TMEM35, VEGFA, WHSC1 |
| hsa-miR-361-5p | miR-361-5p (miRNAs w/seed UAUCAGA) | 7 | ADCY2, ERG, FOXM1, GCOM1, PEG3, VEGFA, VWDE |
| hsa-miR-374a-5p | miR-374b-5p (and other miRNAs w/seed UAUAAUA) | 21 | CD47, FAM169A, FAM19A2, GAS7, HAPLN1, HES1, HUNK, INHBB, MAP2K6, MKX, MMRN1, NR4A3, PLXNA2, RTKN2, SHANK2, SNTB1, SOX4, SPTSSB, UST, VEGFA, ZNF423 |
| hsa-miR-503 | miR-503-5p (miRNAs w/seed AGCAGCG) | 14 | CASR, CDC25A, CDCA4, CHEK1, CNNM3, DNAAF3, GLCE, IHH, NAV1, NOS1, PARM1, SOX5, VEGFA, ZNF423 |
| hsa-miR-514b-5p | miR-513c-5p (and other miRNAs w/seed UCUCAAG) | 2 | BRCA2, PDE6A |
| hsa-miR-514a-3p | miR-514a-3p (and other miRNAs w/seed UUGACAC) | 1 | PEG3 |
| hsa-miR-25-3p | miR-92a-3p (and other miRNAs w/seed AUUGCAC) | 19 | ACAN, ADGRB3, ANGPTL2, BCL11A, CHGA, CHST1, DAG1, GLCE, GRIA1, HAPLN1, HOXD10, SIK1, MARCH4, MYLIP, NR4A3, PALM2, SORL1, SOX4, SYNDIG1 |
To understand the putative roles of these miRNA target genes, two lists of putative regulated genes were generated. The first list included all of the putative miRNA targets (in silico analysis) and the second list included only the negatively correlated differentially expressed targets (experimental analysis). This contributes to both a broad and targeted view of the ontologies and pathways related to the roles of miRNAs in ovulation. Gene Ontology (GO) and pathway analyses were performed using the GeneAnalytics tool20.
Gene Ontology and pathway classification of the in silico analysis of miRNA target genes
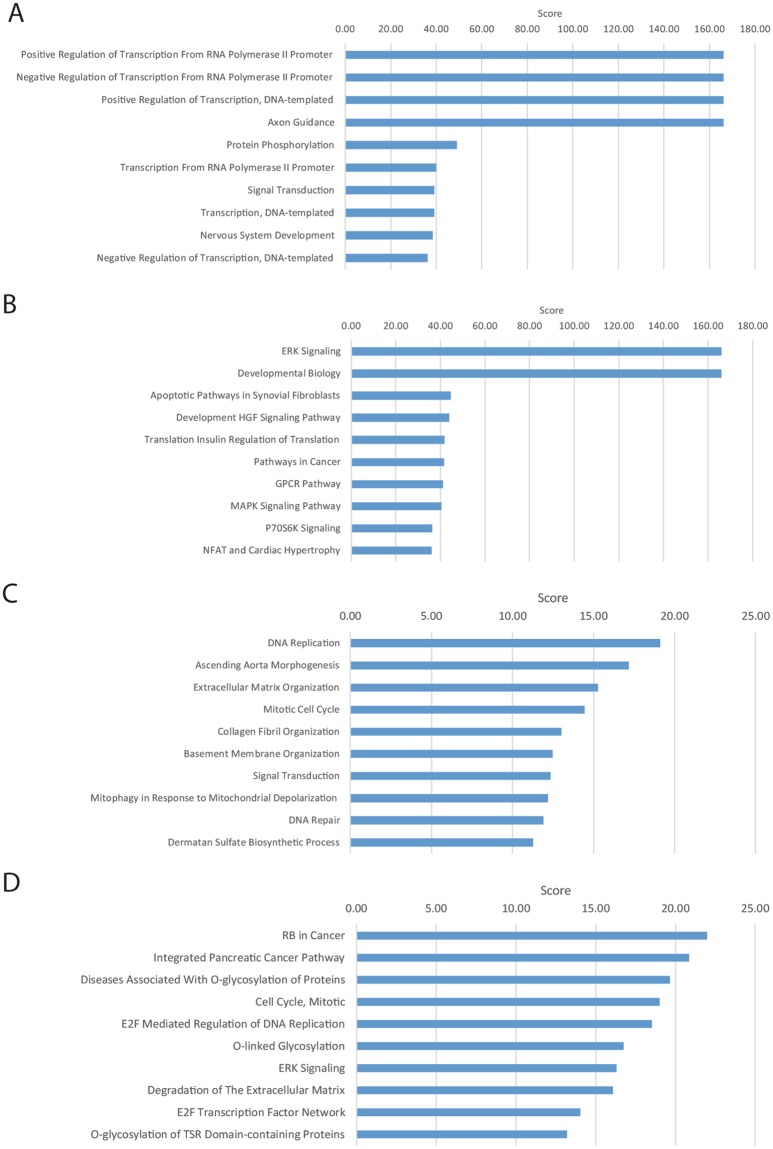
For the unfiltered miRNA targets (7288 genes), we identified 78 high-score GO terms (Fig. 2A and Supp. Table S5). The GO terms included 9 “transcription regulation” ontologies and 15 “signaling” ontologies, including neurotropin TRK signaling, FGF receptor signaling, EGF receptor signaling, TGFB receptor signaling, WNT signaling, VEGF signaling, insulin receptor signaling and RAS signaling. Other important processes included axon guidance, nervous system development, negative regulation of cell proliferation, in utero embryonic development and angiogenesis and cell migration.
Figure 2.
GeneAnalytics analysis of all 7288 putative miRNA targets (A,B) and of the 234 negatively correlated differentially expressed miRNA targets (C,D). Presented are the top 10 enriched GO terms (A,C) and the top 10 enriched pathways (B,D). The presented GeneAnalytics score is a transformation (−log2) of the p-value; hence, higher scores represent stronger enrichment.
We identified 144 high-score pathways that included 67 signaling pathways (Fig. 2B and Supp. Table S6). The top scoring signaling pathways were ERK signaling, HGF signaling, GPCR signaling, MAPK signaling, P70-S6K signaling and RAS signaling. Other pathways included NFAT and cardiac hypertrophy, proteoglycans in cancer, focal adhesion, endocytosis and axon guidance.
Gene Ontology and pathway classification of the negatively correlated miRNA target genes
For the subset of negatively correlated miRNA targets (234 genes), only four high scoring and four medium scoring GO terms were identified. The GO terms included DNA replication, ascending aorta morphogenesis, extracellular matrix organization and mitotic cell cycle. The top medium scoring GO terms included collagen fibril, basement membrane organization and signal transduction (Fig. 2C and Supp. Table S7). The nine high scoring pathways included RB in cancer, integrated pancreatic cancer pathway, diseases associated with O-glycosylation, cell cycle, E2F-mediated DNA replication, degradation of extracellular matrix and ERK signaling (Fig. 2D and Supp. Table S8).
Upstream regulator analysis of negatively correlated miRNA target genes
We used Ingenuity’s IPA upstream regulator analytic feature to identify a cascade of upstream transcriptional regulators that can explain the observed gene expression changes of the negatively correlated genes beyond the identified miRNAs. These regulators can be transcription factors (TFs) and any gene or small molecule that has been observed experimentally to affect gene expression. TF and miRNA may mutually regulate each another or regulate a shared target gene and, as such, enhance the robustness of gene regulation. Hence, TF-miRNA regulatory network analysis will be helpful to decipher gene expression regulatory mechanisms of the ovulatory process.
A total of 68 significant upstream regulators were identified (Z-score > ±2, Supp. Table S9). The top ten inhibited regulators and activated regulators are presented in Tables 3 and 4, respectively.
Table 3.
Top 10 inhibited upstream regulators of negatively correlated miRNAs and targets identified by ingenuity IPA analysis19.
| Upstream Regulator | Exp Log Ratio | Molecule Type | Activation z-score | p-value of overlap | Target molecules in dataset |
|---|---|---|---|---|---|
| TGFB1 | growth factor | −4.118 | 5.51E-07 | ACAN, ADAMTS2, CDC25A, CSPG4, DAAM1, DAPK1, DSP, E2F1, ESPL1, ESR2, FAM107A, GAS7, GATM, GLCE, GREM1, GRIA1, GSE1, HES1, INHBB, LOXL1, LPL, MFAP2, MMP11, MYBL2, NR4A3, PDGFA, PLXNC1, PRC1, PRIM1, RAD51AP1, RAPGEF3, RASL11B, S1PR3, SEMA7A, SOX4, THBS1, TOP2A, TYMS, UST, VEGFA, ZEB1 | |
| MITF | transcription regulator | −3.606 | 3.85E-07 | ACAN, AURKB, CEP55, CHAF1A, DAPK1, ECT2, ESPL1, FRMD4B, HAPLN1, HAUS8, HES1, ITPKB, SOX5, TMCC2 | |
| CSF2 | cytokine | −3.448 | 1.48E-03 | CHAF1A, FOXM1, HAUS8, IFNLR1, MCM3, NUSAP1, PPIF, PRC1, RRM2, SNTB1, STMN1, THBS1, TOP2A | |
| MYC | transcription regulator | −3.281 | 9.61E-04 | ACAN, AURKB, CD47, CDC25A, CHEK1, CSPG4, CTSV, DSP, E2F1, EZH2, FOXM1, HAPLN1, HES1, MSH2, PEG3, RRM2, SLC16A1, SOX5, STMN1, THBS1, THBS2, TYMS, VEGFA | |
| FOXM1 | −4.219 | transcription regulator | −3.088 | 6.30E-07 | AURKB, CDC25A, ESR1, FOXM1, PDGFA, PRC1, STMN1, TOP2A, VEGFA, ZEB1 |
| HGF | growth factor | −2.925 | 1.12E-03 | ANGPTL2, AURKB, CDC25A, CDC6, ENTPD1, FOXM1, HES1, NR4A3, PDGFA, PLXNA2, PRC1, THBS1, VEGFA, ZEB1 | |
| SP1 | transcription regulator | −2.918 | 6.72E-03 | CHGA, E2F1, ESR1, FOXM1, GRIA1, LPL, MMP11, MYBL2, NOS1, PDGFA, PDGFC, TYMS, VEGFA | |
| CCND1 | transcription regulator | −2.884 | 2.97E-07 | CDC6, CEP55, CLSPN, DDIAS, DEPDC1, DTL, E2F1, ESCO2, FOXM1, KIAA0101, MYRIP, RRM2, SOX4, TYMS, ZNF423 | |
| Vegf | group | −2.834 | 3.46E-05 | ACAN, ANGPTL2, AURKB, CDC25A, CDC6, DPF3, ENTPD1, FOXM1, HES1, IHH, INHBB, NR4A3, PDGFA, PLXNA2, PRC1, VEGFA, ZEB1 | |
| PTGER2 | g-protein coupled receptor | −2.828 | 1.97E-05 | CEP55, DEPDC1, ECT2, NUSAP1, PBK, PRC1, THBS1, VEGFA |
Table 4.
Top 10 activated upstream regulators of negatively correlated miRNAs and targets identified by ingenuity IPA analysis19.
| Upstream Regulator | Exp Log Ratio | Molecule Type | Activation z-score | p-value of overlap | Target molecules in dataset |
|---|---|---|---|---|---|
| let-7 | microrna | 3.246 | 1.97E-05 | AURKB,BRCA2, CDC25A, CDC6, CHEK1, EZH2, FANCD2, MCM3, PPP1R12B, RRM2, THBS1 | |
| mir-21 | 8.541619 | microrna | 3.093 | 3.46E-05 | CDC25A, DDAH1, ECT2, MSH2, NUSAP1, PBK, PRC1, RAD51AP1, STMN1, TOP2A |
| RBL1 | transcription regulator | 2.923 | 3.79E-10 | AURKB, CDC25A, CDC6, E2F1, HES1, MCM3, MYBL2, RRM2, THBS1, TYMS, ZEB1 | |
| CDKN2A | transcription regulator | 2.837 | 1.04E-07 | AURKB, CDC25A, CDCA4, CHAF1A, E2F1, EZH2, GAS7, HMGB2, HUNK, MYBL2, PDGFA, PEG3, RAD51AP1, RRM2, TMPO, VEGFA | |
| BNIP3L | other | 2.828 | 1.48E-07 | CD47, CHEK1, E2F1, FANCD2, MYBL2, PRIM1, RRM2, TOP2A | |
| miR-16-5p | 4.689 | mature microrna | 2.777 | 1.71E-03 | CDC25A, CHEK1, CRHBP, HERC6, MSH2, PPIF, PRIM1, VEGFA |
| let-7a-5p | 4.174 | mature microrna | 2.768 | 2.61E-04 | AURKB, CDC25A, DSP, FANCD2, PRIM1, SLC1A4, THBS1, TYMS |
| LY294002 | chemical - kinase inhibitor | 2.73 | 1.25E-03 | ACAN, BRCA2, ESR1, HAPLN1, HES1, HMGB2, INHBB, LOC102724428/SIK1, NR4A3, THBS1, TOP2A, TYMS, VEGFA, ZEB1 | |
| CDKN1A | kinase | 2.605 | 4.66E-12 | AURKB, CDC25A, CDC6, CEP55, CHEK1, DTL, FANCI, FOXM1, HMGB2, KIAA0101, MCM3, MYBL2, NUSAP1, PBK, PRC1, STMN1, TOP2A, TYMS, VEGFA | |
| sirolimus | chemical drug | 2.527 | 2.84E-02 | CDC25A, CHEK1, E2F1, GRIA1, NR4A3, STMN1, TMPO, TOP2A, TYMS, VEGFA |
As expected, among the activated regulators, we identified 5 miRNAs, 3 of which were also differentially expressed in our analysis (Supp. Table S9).
We plotted several of the upstream regulators and their targets, as shown in Fig. 3. We identified miR-21 to be involved in the regulation of CDC25A, DDAH1, ECT2, MSH2, NUSAP1, PBK, PRC1, RAD51AP1, STMN1 and TOP2A (Fig. 3A). Interestingly, most genes are involved in reproductive disease (highlighted in pink).
Figure 3.
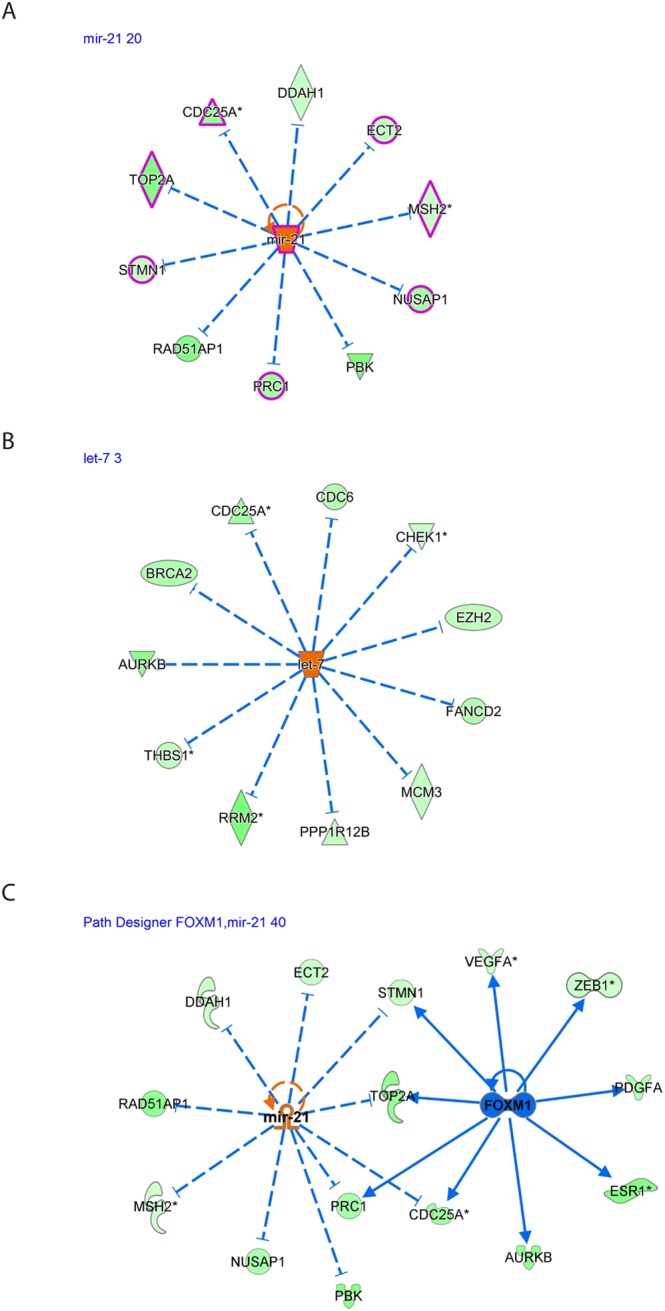
The resultant DE miRNAs were analyzed through the use of IPA (Ingenuity Pathway Analysis, QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis)19 to explore the inversely correlated miRNA-regulated pathways and mRNA expression in preovulatory granulosa cells. This analysis revealed that both (A) miR-21 and (B) let-7 are involved in the downregulation of several important genes. (C) Putative crosstalk between the FOXM1 signaling pathway and miR-21 signaling. A number of genes and pathways are reciprocally regulated by these negatively correlated transcription regulators. mir-21 expression is upregulated, whereas FOXM1 expression is downregulated during the ovulatory process.
We identified let-7 to be involved in the regulation of AURKB, BRCA2, CDC25A, CDC6, CHEK1, EZH2, FANCD2, MCM3, PPP1R12B, RRM2 and THBS1 (Fig. 3B).
Among the notable interactions found was the reciprocal crosstalk between miR-21 (activated) and FOXM1 (inhibited) signaling. Both transcriptional regulators inhibit STMN1, TOP2A, CDC25A and PRC1 (Fig. 3C).
Also noted was the reciprocal crosstalk between TGFB1 (inhibited), miR-16-5p and miR-34a-5p (both activated) signaling (Fig. 4). All three transcription regulators inhibit VEGFA expression observed during the ovulatory process. TGFB1 (inhibited) activates E2F1, while miR-34a (activated) inhibits E2F1 signaling to the net effect of E2F1 inhibition. TGFB1 also inhibits mir-34a.
Figure 4.
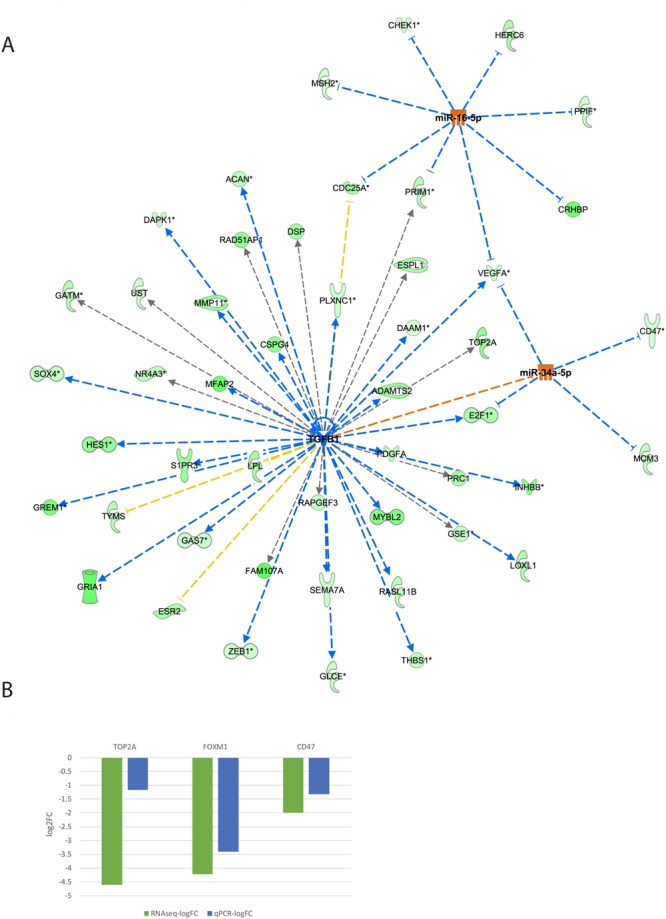
(A) The resultant DE miRNAs were analyzed through the use of IPA (Ingenuity Pathway Analysis, QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis)19 to examine the crosstalk between the TGFB1 signaling pathway and miR-34a and miR-16a-5p (seed of miR-424-5p). We revealed a number of genes and pathways, most notably VEGFA, that are reciprocally regulated by these negatively correlated transcription regulators. (B) Total mRNA was purified from CCs denuded from GV COC and M2 COC aspirated during IVF procedures. The mRNAs were subjected to qPCR in duplicate with the examined genes and ACTB primers. Gene expression was calculated relative to the ACTB level in the same sample and expression levels were compared using Student’s t-test. The difference reached p = 0.006 for FOXM1, p = 0.01 for TOP2A and p = 0.08 for CD47. RNAseq (green) and qPCR (blue) results are presented as log2-fold change between COC M2 and COC GV samples.
Interestingly, both upstream regulators, miR-16-5p (seed sequence of miR-424-5p) and miR-34a-5p, are also upregulated in granulosa cells during ovulation (Table 2 and Fig. 1C).
Functional validation of upstream regulator analysis results
We chose several of the putative miRNA target genes and regulators that were identified by the upstream regulator analysis and performed qPCR analysis. In CCM2 cells, FOXM1 and TOP2A were downregulated, whereas miR-21 was upregulated (Figs 3C and 4B, fold induction). Additionally, miR-34a-5p was upregulated, while TOP2A and CD47 were downregulated (Fig. 4, fold induction).
Discussion
A number of previous studies have examined mRNA and/or miRNA expression in cumulus granulosa cells. However, no study has examined global miRNA expression in human cumulus granulosa cells during final follicular maturation and ovulation. Moreover, this is the first study to link the miRNA-target gene pairs with an inverse correlation of expression. This approach allows the identification of the regulated mRNA-miRNA networks during final follicular maturation and ovulation.
Differential expression analysis revealed dramatic changes in miRNA expression during the CC maturation process; all differentially expressed miRNAs were up-regulated in CCM2 cells.
Compared to published miRNA expression in CCs obtained from M2 COC during IVF treatment8, we observed that 8 of the 10 most abundant miRNAs were also present in the CCM2 sample and 43 of the 46 expressed miRNAs were identified by Velthut-Meikas et al.
Forty-two of the differentially expressed miRNAs were previously identified in granulosa cells. Among them, 14 are involved in steroidogenesis or PCOS, 14 are involved in apoptosis (eight of which belong to the let-7 family), 12 have been implicated in bovine follicular development and 20 were described as differentially expressed between human cumulus and mural granulosa cells. miR-4454 was not previously described in granulosa cells and was recently implicated in the pathogenesis of cartilage degeneration by promoting inflammatory, catabolic, and cell death activity in chondrocytes21. We believe that miR-4454 is implicated in the control of final follicular maturation perhaps by promoting similar proinflammatory-like activities in granulosa cells.
The differentially expressed miRNAs were predicted to regulate 696 target genes that were differentially expressed in our previously generated ovulatory cDNA library, of which, 234 displayed an inverse correlation.
During the final stages of ovulation, several upregulated processes were previously identified by the analysis of global mRNA expression, such as cellular movement, inflammatory response, immune cell trafficking, tissue development and lipid metabolism3,22,23. However, several processes, such as tissue morphology, DNA replication and cell cycle, are downregulated3,22,23. These downregulated processes were also represented in our analyses of miRNA targets. We observed that a large number of the putative miRNA target genes are related to cell cycle and DNA replication processes. These are probably downstream to the FSH blocking effect following the LH surge, thus leading to decreased proliferation24. Interestingly, all of the differentially expressed miRNAs were upregulated in our analysis. This result suggests that the main role of miRNA in the late ovulatory process is more toward control and inhibition of granulosa cell proliferation and less toward other processes found when analyzing gene, rather than miRNA, expression.
The top identified pathways among the miRNA negatively correlated targets include several pathways that pertain to cellular proliferation and survival, including “RB in cancer”, “E2F regulation of DNA replication” and “cell cycle”. Among them, the retinoblastoma protein (RB) was identified as an important factor in ovarian physiology. Conditional deletion of the RB gene in murine ovarian granulosa cells leads to increased follicular recruitment and premature ovarian failure25.
We also identified the O-glycosylation of proteins pathway, which might pertain to cumulus matrix maintenance since cumulus complexes from O-glycan-deficient oocytes were smaller and contained fewer CCs, although fertility was not impaired26.
Several pathways involved in ovulation were identified in our analyses, including the ERK signaling pathway27, HGF signaling28, MAPK pathway27, P70-S6K signaling29, neurotropin TRK signaling30, FGF receptor signaling31, EGF receptor signaling31, TGFB receptor signaling32, WNT signaling33, VEGF signaling34, insulin receptor signaling35, and RAS signaling36. These findings strengthen and validate our miRNA results.
As expected, more potential novel pathways, including NFAT (nuclear factor of activated T-cells) and PAK (p21-activated kinase) signaling pathways, were found in the broader analysis, which included all miRNA targets (and not only the anti-correlated ones).
NFAT has not been described in granulosa cells, although it has been implicated in GnRH signaling37. NFAT was also described as a regulator of COX2 in endometrial stromal cells38 and other tissues and may play a role in prostaglandin regulation during ovulation.
PAK signaling was implicated in cell migration by altering actin cytoskeletal dynamics downstream to Rac/Cdc42, although this activity was not described in granulosa cells and may be important during cumulus expansion and ovulation39.
miR-21 is highly expressed and upregulated by LH in murine granulosa cells40 and ovine follicles12. miR-21 suppression results in granulosa cell apoptosis40. Our results further corroborate miR-21 upregulation in humans during the ovulatory process and, by combining our negatively correlated ovulatory gene expression data, we suggest a mechanism of action for miR-21 function in granulosa cells. We found 10 different transcripts regulated by miR-21 that were downregulated in our library, all of which are involved in the cell cycle and apoptosis. These findings expand our knowledge of the processes that lead to the observed inhibition of proliferation on granulosa cells during the final stages of the ovulatory process. Some of the targets were already identified in granulosa cells, including CDC25a (apoptosis, cell cycle41), MSH2 (apoptosis42), PRC1 (apoptosis43) and STMN1 (cell migration, apoptosis44). Other targets that were not previously described in granulosa cells include TOP2A (apoptosis, cell cycle), DDAH1 (apoptosis), ECT2 (cell cycle), RAD51AP1 (cell cycle), PBK (apoptosis) and NUSAP1 (cell migration, apoptosis). The known and previously unknown targets provide insight into miR-21 activity in granulosa cells. The negative correlation between TOP2A and miR-21 was experimentally validated (Figs 1C and 4B).
Forkhead box M1 (FOXM1), a member of the large family of Forkhead box transcription factors, is highly expressed in proliferating cells and plays pivotal roles in embryonic and fetal development, DNA replication and mitosis45. The role of FOXM1 in ovulation and granulosa cells has not yet been established. FOXM1 is downregulated in granulosa cells obtained from obese patients undergoing IVF compared to lean patients46 and was also found as an inhibited upstream regulator of negatively correlated miRNA target genes in our analysis. Our observation suggests that the reciprocal activation of miR-21 and suppression of FOXM1 synergize to cause the observed inhibition of granulosa cell proliferation during the final follicular maturation just prior to ovulation. Interestingly, high expression of miR-21 is implicated in increased proliferation in cancer and inflammation47. However, the inhibition of breast cancer cell growth by 3, 3′-diindolylmethane is mediated by the upregulation of miR-21 and CDC25A and FOXM1 suppression48, consistent with that observed in cumulus cells during ovulation3. These in silico findings between FOXM1, TOP2A, and miR-21 were experimentally validated (Figs 1C and 4B).
The let-7 family of miRNAs is believed to act as tumor suppressors49 and has been implicated in granulosa cell atresia in porcine50. This family was among the most upregulated miRNAs during final follicular maturation. Several of its negatively correlated targets were previously identified in granulosa cells, including AURKB (cell cycle51), CDC25A (apoptosis, cell cycle41), EZH2 (histone methylation52), and THBS1 (adhesion, angiogenesis53). Other targets that were not described in granulosa cells include BRCA2 (DNA repair), CDC6 (DNA replication), FANCD2 (DNA repair), MCM3 (DNA replication), PPP1R12B (myosin phosphatase), RRM2 (DNA synthesis) and CHEK1 (cell cycle). Most negatively correlated let-7 targets are related to DNA replication/repair and cell cycle progression. Thus, the putative role of let-7 upregulation during final follicular maturation may be in mediating the observed inhibition of granulosa cell proliferation.
We determined that miR-34a is upregulated in CCM2 compared to CCGV. miR-34a is a tumor suppressor gene that suppresses ovarian cancer proliferation and motility by targeting AXL receptor tyrosine kinase54. miR-34a levels are increased in sheep granulosa cells during the follicular to luteal transition12. In addition, miR-34a may be involved in granulosa cell apoptosis in pig ovaries by targeting the inhibin B gene55. The integrated mRNA-miRNA analysis suggests that miR-34a acts through the regulation of E2F1 (transcription), CD47 (adhesion), MCM3 (proliferation), VEGFA (angiogenesis) and TGFB1 signaling during final follicular maturation and luteinization.
Furthermore, the roles of the TGFB1 signaling pathways in folliculogenesis have been extensively studied56. The Ingenuity’s IPA upstream regulator analytic identified 38 inhibited TGFB1 targets of the 234 putative miRNA negatively correlated targets, making it the most significant miRNA-regulated pathway. When TGFB1 targets were plotted along with miR-16-5p (seed of miRNA-424-3p) and miR-34a targets, we determined that VEGFA expression was affected by all of these regulators. It was previously shown by us3 and others2 that VEGFA expression is reduced just prior to ovulation and increased in the luteal phase. This identified regulatory network suggests a novel cooperative mechanism for VEGF transient downregulation.
In conclusion, integrated analysis between the expression of coding genes from our preliminary study with the newly generated library of regulatory miRNAs enabled us to better understand the regulation of coding ovulatory genes by non-coding transcripts and their function from the single-molecule level to whole pathways.
Methods
Ethical approval
The current study was approved by the Sheba Medical Center Institutional Review Board (IRB) Committee (ethical approval number 8707-11-SMC), and written informed consent was obtained from each patient.
All experiments were performed in accordance with relevant guidelines and regulations.
IVF protocol
Ovarian stimulation was carried out as previously described57,58 according to the “short antagonist” protocol. The protocol consisted of controlled ovarian hyperstimulation with recombinant FSH (rFSH), either Gonal-F; Merck Serono or Puregon Pen; Schering Plough) and human menopausal gonadotropin (HMG; Menopur; Ferring) followed by the addition of ovarian suppression with GnRH antagonists (0.25 mg/day, Cetrorelix, Cetrotide; Serono International, SR) when the leading follicle was more than 12 mm in diameter. When three or more follicles exceeded 18 mm in diameter, 250 µg of hCG (Ovitrelle; Merck Serono) was administered to trigger ovulation. Transvaginal follicular aspiration was performed 35 hours later with ultrasound guidance.
For NanoString analysis, three women (ages 25–35 years) undergoing IVF donated cumulus cells from one M2 COC (see Supp. Table S1 for details).
For the miRNA NanoString validation experiments by qPCR, cumulus cells were obtained from 3–4 different women and pooled to generate a single replicate. Each woman donated CGCs from one M2 COC. The women’s ages and infertility etiologies were similar to those described in Table S1.
For functional validation of upstream regulator analysis, cumulus cells were obtained from 3–4 different women and pooled to generate a single replicate. Each woman (total −21 women) donated CGCs of one M2 or one GV COC. Due to the limited availability of GV COC obtained from IVM, we used cumulus cells of GV COC obtained during IVF. The age of the women was 28–40 years (average 36.5 years). The etiology of infertility included male factor and PGD.
IVM protocol
Four women (ages 28–40 years) undergoing IVM were selected for this study (see Supp. Table S1 for details). In the final analysis, only 3 women were included; one woman (Patient code 59 A in Table S1) was excluded because she was an outlier. Each woman donated one single COC. IVM cycles were carried out as previously described59. Briefly, sonographic assessment of the antral follicle count and of endometrial thickness was carried out on day 3 of a spontaneous menstrual cycle. The serum concentrations of estradiol and progesterone were also determined. Next, 150 IU/day rFSH were administered to the patients for 3 days. A second evaluation was performed on day 6. An injection of 10000 IU hCG (Pregnyl; Organon, Oss, Holland) was administered subcutaneously when the endometrial thickness was ≥5 mm and the leading follicle was at least 12 mm. Oocyte retrieval was carried out 36 hours later. Compact CCs were obtained from surplus germinal vesicle (GV) oocytes that were acquired during IVM treatment (CCGV group, provided by Dr. Ruben Fadini from the Biogenesi Reproductive Medicine Center, Istituti Clinici Zucchi, Monza, Italy).
For NanoString analysis, four women (ages 28–40 years) undergoing IVM were selected (see Supp. Table S1 for details). In the final analysis, only 3 women were included; one woman (Patient code 59 A in Table S1) was excluded because she was an outlier. Each woman donated one single COC. For the miRNA NanoString validation experiments, cumulus cells were obtained from 3–4 different women and pooled to generate a single replicate. Each woman donated CGCs from one GV COC. The women’s ages and infertility etiologies were similar to those described in Table S1.
Cumulus granulosa cell collection
CGCs were obtained through oocyte denudation in the course of intracytoplasmic sperm injection (ICSI) procedures. After oocyte retrieval, CGCs of each oocyte were removed using hyaluronidase (SAGE) and a glass denudation pipette (Swemed). The CGCs were washed in PBS and centrifuged at 5000 × g for 5 minutes at room temperature. The resulting pellets were stored at −80 °C until RNA isolation.
RNA extraction
Total RNA was extracted from seven independent CCs samples of seven different women obtained from compact GV (4 samples) and expanded M2 of single COC (3 samples) using a Micro RNA Isolation Kit (Zymo Research Corp., CA USA) according to the manufacturer’s instructions. RNA purity and concentration were assessed using a NanoDrop spectrophotometer (NanoDrop 2000C, Thermo Fisher Scientific).
Quantitative real-time PCR
cDNA synthesis for the detection of mature miRNAs was performed with the miScript Reverse Transcription Kit (QIAGEN) using a blend of oligo(dT) and random primers (iScript™ cDNA Synthesis Kit). cDNA synthesis for the quantification of mRNA expression was performed with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). miRNA expression was determined using the miScript SYBR Green Kit (QIAGEN) and mRNA expression was determined using Fast SYBR Green Master Mix (Applied Biosystems). The StepOnePlus Real-Time PCR System (Applied Biosystems) was used to detect amplification.
Expression levels were normalized to ACTB (for mRNA) and RNU6B (for miRNA). qPCR results were analyzed with StepOne software. Relative gene expression was calculated using the delta-delta Ct method60. Details of the primers are shown in Table S2.
NanoString and bioinformatics sample analysis
Evaluation of miRNA expression was carried out using NanoString technology, which is not based on sequencing but rather based on a digital molecular barcoding system. Each barcode is attached to a single target-specific probe corresponding to a specific miRNA. The output from this technology is the digital count of 800 tested miRNAs.
A total of 3 μl (20–120 ng) of each RNA sample was prepared according to the manufacturer’s instructions. Mature miRNAs were ligated to a species-specific tag sequence (miRtag) via a thermally controlled splinted ligation. miRNA assays were performed according to the NanoString miRNA Assay Manual. Hybridizations were carried out by combining 5 µl of each miRNA assay with 20 µl of nCounter Reporter probes in hybridization buffer and 5 µl of nCounter Capture probes for a total reaction volume of 30 µl. miRNA expression was assessed using the NanoString nCounter system (NanoString Technologies, Seattle, WA, USA), which enables multiplexed direct digital counting of 800 human miRNA molecules. The raw count data from the 6 samples were analyzed using the edgeR61 and limma R62 packages. First, TMM normalization was applied to the data followed by voom transformation. To assess the number of miRNAs expressed under each condition, we required that probes be detectable in all replicates of the tested condition. As detectable, we defined probes having a normalized expression level that exceeded that of the “highest expressed” negative control probe. This resulted in 54 expressed miRNAs. Next, linear models were used to assess differential expression using the limma method. Pairwise comparisons were performed using moderated t statistics. To perform multiple group comparisons, one-way ANOVA was applied, except the residual mean squares were moderated between genes (see the limma package for details62). miRNAs were considered differentially expressed if their FDR was <5%.
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Bioinformatics analysis of the library
The resultant differentially expressed miRNAs were analyzed through the use of IPA (Ingenuity Pathway Analysis QIAGEN Inc. https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis)19. Putative miRNA targets found using Ingenuity miRNA target filter for experimentally validated and putative predicted targets (high confidence level) – in silico analysis.
Using whole transcriptome sequencing, we previously generated an mRNA ovulatory genes library3. The library was comprised of the same groups as in the current study (CCGV and CCM2), encompassing all differentially expressed mRNAs between immature mid-antral follicles and mature pre-ovulatory follicles.
Our previously generated mRNA library and the miRNA putative targets were combined using the microRNA target filter to generate a subset of the negatively correlated miRNA targets – experimental analysis.
Both mRNA target lists (in silico analysis target list and experimental analysis target list) were analyzed for GO terms and pathways using the GeneAnalytics tool20.
The DE miRNAs and negatively correlated targets were also analyzed using the Ingenuity Upstream Regulator Analysis tool to predict upstream molecules, including miRNA and transcription factors, that may have caused the observed gene expression changes.
Statistics
Each experiment was performed at least three times. Data are expressed as the mean ± standard error of the mean (SEM) and were evaluated using Student’s t-test with a two-tailed distribution, with two samples equaling variance, or with ANOVA (ANalysis Of VAriance) for more than two variances using the post hoc Tukey test assuming equal variance, or the Games–Howell test for unequal variance. When appropriate, the Kruskal–Wallis non-parametric comparison test was used. For all statistical analyses, SPSS 22 software (IBM, Armonk, NY, USA) was used. P-value < 0.05 was considered statistically significant.
Electronic supplementary material
Acknowledgements
This study was supported by grants from the Ministry of Health, Israel (3-00000-7410) and from the Chaim Sheba Medical Center (R&D Sheba grant 5/2014).
Author Contributions
A.H., G.M.Y., E.M. and M.D. designed the study, performed the analysis and interpretation of the data, prepared figures and tables, and wrote and finalized the manuscript. A.H., G.M.Y., M.D. and L.S. prepared figures and tables. Y.Y., M.B., R.M. and L.O. contributed to the interpretation of the data and manuscript preparation. L.O., Y.Y. and M.D.C. were in charge of tissue and oocyte processing. A.H., G.C., R.F., M.M.R. and M.D.C. were involved in patient recruitment. All of the authors contributed to data analysis and reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33807-y.
References
- 1.Li Q, McKenzie LJ, Matzuk MM. Revisiting oocyte-somatic cell interactions: in search of novel intrafollicular predictors and regulators of oocyte developmental competence. Mol Hum Reprod. 2008;14:673–678. doi: 10.1093/molehr/gan064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wissing ML, et al. Identification of new ovulation-related genes in humans by comparing the transcriptome of granulosa cells before and after ovulation triggering in the same controlled ovarian stimulation cycle. Human reproduction (Oxford, England) 2014;29:997–1010. doi: 10.1093/humrep/deu008. [DOI] [PubMed] [Google Scholar]
- 3.Yerushalmi GM, et al. Characterization of the human cumulus cell transcriptome during final follicular maturation and ovulation. Mol Hum Reprod. 2014 doi: 10.1093/molehr/gau031. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 6.Kaikkonen MU, Lam MTY, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovascular Research. 2011;90:430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toms D, Xu S, Pan B, Wu D, Li J. Progesterone receptor expression in granulosa cells is suppressed by microRNA-378-3p. Mol Cell Endocrinol. 2015;399:95–102. doi: 10.1016/j.mce.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 8.Velthut-Meikas A, et al. Research resource: small RNA-seq of human granulosa cells reveals miRNAs in FSHR and aromatase genes. Molecular endocrinology. 2013;27:1128–1141. doi: 10.1210/me.2013-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ninio-Many L, Grossman H, Shomron N, Chuderland D, Shalgi R. microRNA-125a-3p reduces cell proliferation and migration by targeting Fyn. J Cell Sci. 2013;126:2867–2876. doi: 10.1242/jcs.123414. [DOI] [PubMed] [Google Scholar]
- 10.Hawkins SM, Matzuk MM. Oocyte-somatic cell communication and microRNA function in the ovary. Ann Endocrinol (Paris) 2010;71:144–148. doi: 10.1016/j.ando.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiedler SD, Carletti MZ, Hong X, Christenson LK. Hormonal regulation of MicroRNA expression in periovulatory mouse mural granulosa cells. Biol Reprod. 2008;79:1030–1037. doi: 10.1095/biolreprod.108.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride D, et al. Identification of miRNAs associated with the follicular-luteal transition in the ruminant ovary. Reproduction (Cambridge, England) 2012;144:221–233. doi: 10.1530/rep-12-0025. [DOI] [PubMed] [Google Scholar]
- 13.Schauer SN, Sontakke SD, Watson ED, Esteves CL, Donadeu FX. Involvement of miRNAs in equine follicle development. Reproduction (Cambridge, England) 2013;146:273–282. doi: 10.1530/rep-13-0107. [DOI] [PubMed] [Google Scholar]
- 14.Donadeu FX, Schauer SN, Sontakke SD. Involvement of miRNAs in ovarian follicular and luteal development. The Journal of endocrinology. 2012;215:323–334. doi: 10.1530/JOE-12-0252. [DOI] [PubMed] [Google Scholar]
- 15.Imbar T, Eisenberg I. Regulatory role of microRNAs in ovarian function. Fertility and sterility. 2014;101:1524–1530. doi: 10.1016/j.fertnstert.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 16.Liu S, et al. Altered microRNAs expression profiling in cumulus cells from patients with polycystic ovary syndrome. J Transl Med. 2015;13:238. doi: 10.1186/s12967-015-0605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno Juan Manuel, Núñez María José, Quiñonero Alicia, Martínez Sebastian, de la Orden Marina, Simón Carlos, Pellicer Antonio, Díaz-García César, Domínguez Francisco. Follicular fluid and mural granulosa cells microRNA profiles vary in in vitro fertilization patients depending on their age and oocyte maturation stage. Fertility and Sterility. 2015;104(4):1037-1046.e1. doi: 10.1016/j.fertnstert.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Scalici E, et al. Circulating microRNAs in follicular fluid, powerful tools to explore in vitro fertilization process. Scientific reports. 2016;6:24976. doi: 10.1038/srep24976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Ari Fuchs S, et al. GeneAnalytics: An Integrative Gene Set Analysis Tool for Next Generation Sequencing, RNAseq and Microarray Data. OMICS. 2016;20:139–151. doi: 10.1089/omi.2015.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura, A. et al. Identification of microRNA-181a-5p and microRNA-4454 as mediators of facet cartilage degeneration. JCI Insight1, 10.1172/jci.insight.86820 (2016). [DOI] [PMC free article] [PubMed]
- 22.Lee YS, et al. Extensive effects of in vitro oocyte maturation on rhesus monkey cumulus cell transcriptome. Am J Physiol Endocrinol Metab. 2011;301:E196–209. doi: 10.1152/ajpendo.00686.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouandaogo Zamalou Gisèle, Frydman Nelly, Hesters Laetitia, Assou Said, Haouzi Delphine, Dechaud Hervé, Frydman René, Hamamah Samir. Differences in transcriptomic profiles of human cumulus cells isolated from oocytes at GV, MI and MII stages after in vivo and in vitro oocyte maturation. Human Reproduction. 2012;27(8):2438–2447. doi: 10.1093/humrep/des172. [DOI] [PubMed] [Google Scholar]
- 24.Yong EL, Baird DT, Hillier SG. Mediation of gonadotrophin-stimulated growth and differentiation of human granulosa cells by adenosine-3′,5′-monophosphate: one molecule, two messages. Clin Endocrinol (Oxf) 1992;37:51–58. doi: 10.1111/j.1365-2265.1992.tb02283.x. [DOI] [PubMed] [Google Scholar]
- 25.Andreu-Vieyra C, Chen R, Matzuk MM. Conditional deletion of the retinoblastoma (Rb) gene in ovarian granulosa cells leads to premature ovarian failure. Molecular endocrinology. 2008;22:2141–2161. doi: 10.1210/me.2008-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ploutarchou P, Melo P, Day AJ, Milner CM, Williams SA. Molecular analysis of the cumulus matrix: insights from mice with O-glycan-deficient oocytes. Reproduction (Cambridge, England) 2015;149:533–543. doi: 10.1530/REP-14-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan HY, et al. MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science. 2009;324:938–941. doi: 10.1126/science.1171396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zachow R, Uzumcu M. The hepatocyte growth factor system as a regulator of female and male gonadal function. The Journal of endocrinology. 2007;195:359–371. doi: 10.1677/JOE-07-0466. [DOI] [PubMed] [Google Scholar]
- 29.Alam H, et al. Follicle-stimulating hormone activation of hypoxia-inducible factor-1 by the phosphatidylinositol 3-kinase/AKT/Ras homolog enriched in brain (Rheb)/mammalian target of rapamycin (mTOR) pathway is necessary for induction of select protein markers of follicular differentiation. The Journal of biological chemistry. 2004;279:19431–19440. doi: 10.1074/jbc.M401235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vera C, Tapia V, Vega M, Romero C. Role of nerve growth factor and its TRKA receptor in normal ovarian and epithelial ovarian cancer angiogenesis. Journal of ovarian research. 2014;7:82. doi: 10.1186/s13048-014-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaves RN, de Matos MH, Buratini J, Jr, de Figueiredo JR. The fibroblast growth factor family: involvement in the regulation of folliculogenesis. Reprod Fertil Dev. 2012;24:905–915. doi: 10.1071/rd11318. [DOI] [PubMed] [Google Scholar]
- 32.Juengel JL, McNatty KP. The role of proteins of the transforming growth factor-beta superfamily in the intraovarian regulation of follicular development. Hum Reprod Update. 2005;11:143–160. doi: 10.1093/humupd/dmh061. [DOI] [PubMed] [Google Scholar]
- 33.Fan HY, et al. Beta-catenin (CTNNB1) promotes preovulatory follicular development but represses LH-mediated ovulation and luteinization. Molecular endocrinology. 2010;24:1529–1542. doi: 10.1210/me.2010-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otani N, et al. The vascular endothelial growth factor/fms-like tyrosine kinase system in human ovary during the menstrual cycle and early pregnancy. The Journal of clinical endocrinology and metabolism. 1999;84:3845–3851. doi: 10.1210/jcem.84.10.6025. [DOI] [PubMed] [Google Scholar]
- 35.Brogan RS, Mix S, Puttabyatappa M, VandeVoort CA, Chaffin CL. Expression of the insulin-like growth factor and insulin systems in the luteinizing macaque ovarian follicle. Fertility and sterility. 2010;93:1421–1429. doi: 10.1016/j.fertnstert.2008.12.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wayne CM, Fan HY, Cheng X, Richards JS. Follicle-stimulating hormone induces multiple signaling cascades: evidence that activation of Rous sarcoma oncogene, RAS, and the epidermal growth factor receptor are critical for granulosa cell differentiation. Molecular endocrinology. 2007;21:1940–1957. doi: 10.1210/me.2007-0020. [DOI] [PubMed] [Google Scholar]
- 37.Binder AK, Grammer JC, Herndon MK, Stanton JD, Nilson JH. GnRH regulation of Jun and Atf3 requires calcium, calcineurin, and NFAT. Molecular endocrinology. 2012;26:873–886. doi: 10.1210/me.2012-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abraham F, Sacerdoti F, De León R, Gentile T, Canellada A. Angiotensin II Activates the Calcineurin/NFAT Signaling Pathway and Induces Cyclooxygenase-2 Expression in Rat Endometrial Stromal Cells. PLoS ONE. 2012;7:e37750. doi: 10.1371/journal.pone.0037750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nature cell biology. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 40.Carletti MZ, Fiedler SD, Christenson LK. MicroRNA 21 blocks apoptosis in mouse periovulatory granulosa cells. Biol Reprod. 2010;83:286–295. doi: 10.1095/biolreprod.109.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan HA, et al. CDC25 protein expression and interaction with DAZL in human corpus luteum. Fertility and sterility. 2009;92:1997–2003. doi: 10.1016/j.fertnstert.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 42.Donadeu FX, Mohammed BT, Ioannidis J. A miRNA target network putatively involved in follicular atresia. Domest Anim Endocrinol. 2017;58:76–83. doi: 10.1016/j.domaniend.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ndiaye K, Fayad T, Silversides DW, Sirois J, Lussier JG. Identification of downregulated messenger RNAs in bovine granulosa cells of dominant follicles following stimulation with human chorionic gonadotropin. Biol Reprod. 2005;73:324–333. doi: 10.1095/biolreprod.104.038026. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Rudaz C, et al. Excessive ovarian production of nerve growth factor elicits granulosa cell apoptosis by setting in motion a tumor necrosis factor alpha/stathmin-mediated death signaling pathway. Reproduction (Cambridge, England) 2011;142:319–331. doi: 10.1530/rep-11-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bella L, Zona S, Nestal de Moraes G, Lam EW. FOXM1: A key oncofoetal transcription factor in health and disease. Semin Cancer Biol. 2014;29:32–39. doi: 10.1016/j.semcancer.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 46.Merhi Z, et al. Adiposity Alters Genes Important in Inflammation and Cell Cycle Division in Human Cumulus Granulosa Cell. Reprod Sci. 2015;22:1220–1228. doi: 10.1177/1933719115572484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheedy FJ. Turning 21: Induction of miR-21 as a Key Switch in the Inflammatory Response. Front Immunol. 2015;6:19. doi: 10.3389/fimmu.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin Y. 3,3′-Diindolylmethane inhibits breast cancer cell growth via miR-21-mediated Cdc25A degradation. Mol Cell Biochem. 2011;358:345–354. doi: 10.1007/s11010-011-0985-0. [DOI] [PubMed] [Google Scholar]
- 49.Shimizu S, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52:698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 50.Cao R, et al. Expression and preliminary functional profiling of the let-7 family during porcine ovary follicle atresia. Mol Cells. 2015;38:304–311. doi: 10.14348/molcells.2015.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oktay K, Buyuk E, Oktem O, Oktay M, Giancotti FG. The c-Jun N-terminal kinase JNK functions upstream of Aurora B to promote entry into mitosis. Cell Cycle. 2008;7:533–541. doi: 10.4161/cc.7.4.5660. [DOI] [PubMed] [Google Scholar]
- 52.Maekawa R, et al. Changes in gene expression of histone modification enzymes in rat granulosa cells undergoing luteinization during ovulation. Journal of ovarian research. 2016;9:15. doi: 10.1186/s13048-016-0225-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berisha B, Schams D, Rodler D, Sinowatz F, Pfaffl MW. Expression and localization of members of the thrombospondin family during final follicle maturation and corpus luteum formation and function in the bovine ovary. J Reprod Dev. 2016;62:501–510. doi: 10.1262/jrd.2016-056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li R, et al. MiR-34a suppresses ovarian cancer proliferation and motility by targeting AXL. Tumour Biol. 2015;36:7277–7283. doi: 10.1007/s13277-015-3445-8. [DOI] [PubMed] [Google Scholar]
- 55.Tu F, et al. miR-34a targets the inhibin beta B gene, promoting granulosa cell apoptosis in the porcine ovary. Genet Mol Res. 2014;13:2504–2512. doi: 10.4238/2014.January.14.6. [DOI] [PubMed] [Google Scholar]
- 56.Myers M, Pangas SA. Regulatory roles of transforming growth factor beta family members in folliculogenesis. Wiley Interdiscip Rev Syst Biol Med. 2010;2:117–125. doi: 10.1002/wsbm.21. [DOI] [PubMed] [Google Scholar]
- 57.Elizur SE, et al. Factors predicting IVF treatment outcome: a multivariate analysis of 5310 cycles. Reprod Biomed Online. 2005;10:645–649. doi: 10.1016/S1472-6483(10)61673-2. [DOI] [PubMed] [Google Scholar]
- 58.Hourvitz A, et al. Role of embryo quality in predicting early pregnancy loss following assisted reproductive technology. Reprod Biomed Online. 2006;13:504–509. doi: 10.1016/S1472-6483(10)60637-2. [DOI] [PubMed] [Google Scholar]
- 59.Fadini R, et al. Effect of different gonadotrophin priming on IVM of oocytes from women with normal ovaries: a prospective randomized study. Reprod Biomed Online. 2009;19:343–351. doi: 10.1016/S1472-6483(10)60168-X. [DOI] [PubMed] [Google Scholar]
- 60.Yuan JS, Wang D, Stewart CN., Jr. Statistical methods for efficiency adjusted real-time PCR quantification. Biotechnology journal. 2008;3:112–123. doi: 10.1002/biot.200700169. [DOI] [PubMed] [Google Scholar]
- 61.Robinson MD, McCarthy DJ, Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou J, et al. The let-7g microRNA promotes follicular granulosa cell apoptosis by targeting transforming growth factor-beta type 1 receptor. Mol Cell Endocrinol. 2015;409:103–112. doi: 10.1016/j.mce.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 64.Gebremedhn S, et al. MicroRNA Expression Profile in Bovine Granulosa Cells of Preovulatory Dominant and Subordinate Follicles during the Late Follicular Phase of the Estrous Cycle. PLoS One. 2015;10:e0125912. doi: 10.1371/journal.pone.0125912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sirotkin AV, Ovcharenko D, Grossmann R, Laukova M, Mlyncek M. Identification of microRNAs controlling human ovarian cell steroidogenesis via a genome-scale screen. J Cell Physiol. 2009;219:415–420. doi: 10.1002/jcp.21689. [DOI] [PubMed] [Google Scholar]
- 66.Navakanitworakul R, et al. Characterization and Small RNA Content of Extracellular Vesicles in Follicular Fluid of Developing Bovine Antral Follicles. Scientific reports. 2016;6:25486. doi: 10.1038/srep25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang C, et al. MicroRNA-125a-5p induces mouse granulosa cell apoptosis by targeting signal transducer and activator of transcription 3. Menopause. 2016;23:100–107. doi: 10.1097/GME.0000000000000507. [DOI] [PubMed] [Google Scholar]
- 68.Sang Q, et al. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. The Journal of clinical endocrinology and metabolism. 2013;98:3068–3079. doi: 10.1210/jc.2013-1715. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Q, et al. MicroRNA-181a suppresses mouse granulosa cell proliferation by targeting activin receptor IIA. PLoS One. 2013;8:e59667. doi: 10.1371/journal.pone.0059667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salilew-Wondim D, et al. The expression pattern of microRNAs in granulosa cells of subordinate and dominant follicles during the early luteal phase of the bovine estrous cycle. PLoS One. 2014;9:e106795. doi: 10.1371/journal.pone.0106795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie S, Batnasan E, Zhang Q, Li Y. MicroRNA Expression is Altered in Granulosa Cells of Ovarian Hyperresponders. Reproductive Sciences. 2016;23:1001–1010. doi: 10.1177/1933719115625849. [DOI] [PubMed] [Google Scholar]
- 72.Xu B, Zhang YW, Tong XH, Liu YS. Characterization of microRNA profile in human cumulus granulosa cells: Identification of microRNAs that regulate Notch signaling and are associated with PCOS. Mol Cell Endocrinol. 2015;404:26–36. doi: 10.1016/j.mce.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 73.Hossain MM, et al. Identification and characterization of miRNAs expressed in the bovine ovary. BMC Genomics. 2009;10:443. doi: 10.1186/1471-2164-10-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yao N, et al. Follicle-stimulating hormone regulation of microRNA expression on progesterone production in cultured rat granulosa cells. Endocrine. 2010;38:158–166. doi: 10.1007/s12020-010-9345-1. [DOI] [PubMed] [Google Scholar]
- 75.Sontakke SD, Mohammed BT, McNeilly AS, Donadeu FX. Characterization of microRNAs differentially expressed during bovine follicle development. Reproduction (Cambridge, England) 2014;148:271–283. doi: 10.1530/REP-14-0140. [DOI] [PubMed] [Google Scholar]
- 76.Xu S, Linher-Melville K, Yang BB, Wu D, Li J. Micro-RNA378 (miR-378) regulates ovarian estradiol production by targeting aromatase. Endocrinology. 2011;152:3941–3951. doi: 10.1210/en.2011-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lei L, Jin S, Gonzalez G, Behringer RR, Woodruff TK. The regulatory role of Dicer in folliculogenesis in mice. Mol Cell Endocrinol. 2010;315:63–73. doi: 10.1016/j.mce.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sathyapalan T, David R, Gooderham NJ, Atkin SL. Increased expression of circulating miRNA-93 in women with polycystic ovary syndrome may represent a novel, non-invasive biomarker for diagnosis. Scientific reports. 2015;5:16890. doi: 10.1038/srep16890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang L, et al. MicroRNA-93 promotes ovarian granulosa cells proliferation through targeting CDKN1A in polycystic ovarian syndrome. The Journal of clinical endocrinology and metabolism. 2015;100:E729–738. doi: 10.1210/jc.2014-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim YJ, et al. MicroRNA Profile of Granulosa Cells after Ovarian Stimulation Differs According to Maturity of Retrieved Oocytes. Geburtshilfe Frauenheilkd. 2016;76:704–708. doi: 10.1055/s-0041-111173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Diez-Fraile A, et al. Age-associated differential microRNA levels in human follicular fluid reveal pathways potentially determining fertility and success of in vitro fertilization. Hum Fertil (Camb) 2014;17:90–98. doi: 10.3109/14647273.2014.897006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.