Staphylococcus aureus is a major cause of life-threatening infections. The bacterium expresses alpha-toxin, a hemolysin and cytotoxin responsible for many of the pathologies of S. aureus. Alpha-toxin production is enhanced by subinhibitory concentrations of antibiotics. Here, we show that this process is dependent on the long noncoding RNA, SSR42. Further, SSR42 itself is regulated by several global regulators, thereby integrating environmental and nutritional signals that modulate hemolysis of the pathogen.
KEYWORDS: Staphylococcus aureus, β-lactams, hemolysin gene regulation, noncoding RNA
ABSTRACT
Staphylococcus aureus is a human pathogen causing a variety of diseases by versatile expression of a large set of virulence factors that most prominently features the cytotoxic and hemolytic pore-forming alpha-toxin. Expression of alpha-toxin is regulated by an intricate network of transcription factors. These include two-component systems sensing quorum and environmental signals as well as regulators reacting to the nutritional status of the pathogen. We previously identified the repressor of surface proteins (Rsp) as a virulence regulator. Acute cytotoxicity and hemolysis are strongly decreased in rsp mutants, which are characterized by decreased transcription of toxin genes as well as loss of transcription of a 1,232-nucleotide (nt)-long noncoding RNA (ncRNA), SSR42. Here, we show that SSR42 is the effector of Rsp in transcription regulation of the alpha-toxin gene, hla. SSR42 transcription is enhanced after exposure of S. aureus to subinhibitory concentrations of oxacillin which thus leads to an SSR42-dependent increase in hemolysis. Aside from Rsp, SSR42 transcription is under the control of additional global regulators, such as CodY, AgrA, CcpE, and σB, but is positioned upstream of the two-component system SaeRS in the regulatory cascade leading to alpha-toxin production. Thus, alpha-toxin expression depends on two long ncRNAs, SSR42 and RNAIII, which control production of the cytolytic toxin on the transcriptional and translational levels, respectively, with SSR42 as an important regulator of SaeRS-dependent S. aureus toxin production in response to environmental and metabolic signals.
IMPORTANCE Staphylococcus aureus is a major cause of life-threatening infections. The bacterium expresses alpha-toxin, a hemolysin and cytotoxin responsible for many of the pathologies of S. aureus. Alpha-toxin production is enhanced by subinhibitory concentrations of antibiotics. Here, we show that this process is dependent on the long noncoding RNA, SSR42. Further, SSR42 itself is regulated by several global regulators, thereby integrating environmental and nutritional signals that modulate hemolysis of the pathogen.
INTRODUCTION
Community-acquired and health care-associated methicillin-resistant Staphylococcus aureus (MRSA) strains are major causes for a variety of diseases (1, 2) ranging from superficial skin infections and skin and deep abscesses to severe conditions with high morbidity, such as osteomyelitis, endocarditis, and sepsis (3). S. aureus adapts to different host environments by coordinated expression of certain virulence factors (4–6). A major virulence factor of S. aureus is the pore-forming alpha-toxin (encoded by hla). In addition to the hemolytic, cytolytic, and dermonecrotic properties of alpha-toxin (7, 8), the generation of alpha-toxin pores within target cell membranes triggers release of cytokines and host cell apoptosis (9, 10). Alpha-toxin thereby acts on a multitude of different cell types, including macrophages, monocytes, lymphocytes, epithelial cells, and fibroblasts (7, 9, 11), thus having a major impact on the pathogenesis of infections such as pneumonia (12), corneal infections (13), and sepsis (14). Alpha-toxin production in S. aureus is tightly regulated in a time-dependent manner by two-component systems, such as the quorum sensing system accessory gene regulator (agr) (15, 16) and SaeRS (17), MarR-type DNA-binding proteins belonging to the staphylococcal accessory regulator (sar) family, as well as other regulators (reviewed in reference 18).
SaeRS is essential for S. aureus virulence, regulating the transcription of toxin-encoding genes, such as hla, and surface proteins as well as capsular biosynthesis (17, 19–21). Targets of the SaeRS two-component system are classified into two groups. Class I genes, e.g., coa, fnbA, eap, efb, saeP, fib, and emp, depend on the histidine kinase activity of SaeS, whereas for transcriptional activation of class II genes such as hla and hlb, a low basal phosphorylation level of the response regulator SaeR is sufficient (22; reviewed in references 17 and 23). sae is transcribed from two promoters, P1 and P3. Transcription from promoter P1 results in transcript T1 encoding a four-gene operon, saePQRS. saeP and saeQ are auxiliary genes (19, 24). Both resulting proteins, SaeP and SaeQ, were described to form a ternary complex activating phosphatase activity within SaeS while being dispensable for its kinase activity (22). The T1 transcript is unstable. It is processed by the endoribonuclease RNase Y to create the more stable transcript T2, consisting of saeQRS (25, 26). Transcript T3 is driven by the constitutively active promoter P3 and contains saeRS (25). A fourth transcript harboring only saeP, T4, is initiated at P1 and results either from processing of T1 or by premature termination (24, 27).
The agr quorum and diffusion sensing system constitutes another major virulence regulator in S. aureus. An auto-inducing peptide pheromone (AIP) derived from the precursor AgrD is secreted and modified by the membrane protein, AgrB. At high densities of bacteria, the AIP sensor histidine kinase AgrC phosphorylates the response regulator AgrA, which in turn leads to transcription of the main agr effector, RNAIII. Whereas some virulence factor genes are directly dependent on AgrA, a majority are regulated by the RNA (28) on either the transcriptional or translational level (29, 30). Noncoding RNAs (ncRNAs) have been shown to regulate gene expression acting in cis or trans, thereby modulating transcription, translation, and mRNA stability (31), and especially in bacteria ncRNAs play major roles in gene and virulence regulation (32). S. aureus produces an array of small ncRNAs whose full regulatory properties have not yet been completely deciphered (33–35). RNAIII regulates staphylococcal alpha-toxin on a translational level by base pairing to hla mRNA, thereby liberating a Shine-Dalgarno sequence which otherwise is sequestered within a secondary structure and inaccessible to the ribosomes (30).
In a transposon mutant screen we previously identified repressor of surface proteins(Rsp) as another virulence regulator. We demonstrated that deletion of rsp resulted in reduced hemolysis and cytotoxicity of S. aureus as well as a shift in pathotype (36). We identified the long ncRNA SSR42 as a major target of Rsp since SSR42 transcription was completely lost in insertional as well as deletion mutants of rsp (36). Prior to that study, SSR42 had been identified as an 891-nucleotide (nt) ncRNA that was stable during the stationary growth phase and thus was designated a small stable RNA (SSR) (37, 38). SSRs were implicated in bacterial adaption to detrimental conditions, thereby enhancing survival (38). SSR42 further regulates the transcription of 80 mRNA species in S. aureus strains UAMS-1 and LAC. Among the regulated genes were alpha-toxin, Panton-Valentine leukocidin (PVL), and protein A as well as genes for capsule biosynthesis (37). SSR42 has also been identified in other studies, where it has been designated Teg27 (39), sRNA363 (4), or srn_4470_RsaX28 (40), although with slightly different length estimates. However, transcriptome sequencing (RNA-seq) revealed a primary SSR42 transcript of 1,232 nt in the methicillin-resistant S. aureus (MRSA) strain JE2 (36). Another study demonstrated cleavage of SSR42 (there termed RsaX28) by the endoribonuclease RNase Y in a so-called EMOTE (exact mapping of transcriptome ends) assay (40).
In the present study, we show that SSR42 is the effector of Rsp and that SSR42 is required for efficient transcription of hla message in an SaeRS-dependent manner. We further show that hemolysis is enhanced upon exposure of S. aureus to subinhibitory concentrations of antibiotics and that this process is dependent on SSR42. In order to elucidate the signal transduction cascade involved in this process, we demonstrated that SSR42 transcription is influenced by global regulators and thus serves as an integrator for various stimuli, which results in increased hemolysis.
RESULTS
SSR42 is the effector of Rsp-mediated hemolysis in S. aureus.
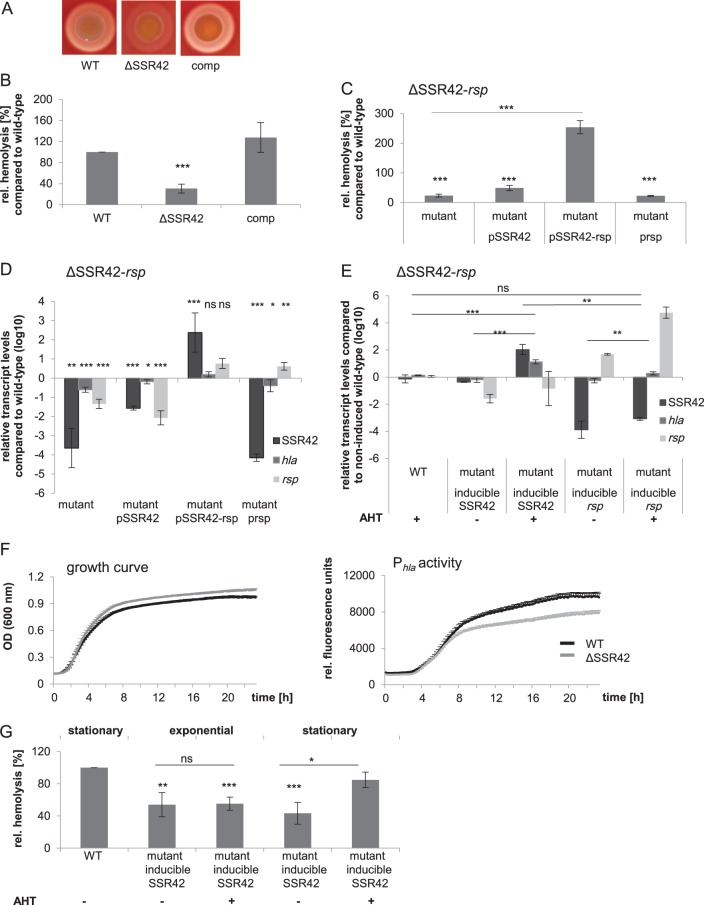
We recently identified the AraC-type transcriptional regulator Rsp as a regulator of various virulence traits of S. aureus ranging from hemolysis to cytotoxicity. However, loss of rsp did not influence staphylococcal survival in whole blood or deep-abscess formation, suggesting that Rsp deficiency altered the pathotype of S. aureus. Rsp-dependently transcribed genes were determined by RNA-seq and included a set of virulence factors as well as the long noncoding RNA (ncRNA) SSR42. SSR42 is located directly upstream of rsp in an antiparallel orientation, and its transcription is strongly dependent on functional Rsp (36). Deletion of rsp strongly reduced hemolysis of S. aureus (36). Since SSR42 was previously described to influence hemolysis (37), we tested whether the hemolysis defect of an rsp mutant resulted from the loss of SSR42 and thus determined the potential role of SSR42 as a potential effector of Rsp. We generated a markerless SSR42 knockout mutant (ΔSSR42) by allelic replacement in the cytolytic strain S. aureus 6850, which encompassed the complete primary ncRNA transcript. Whereas deletion of SSR42 did not alter bacterial growth dynamics (see Fig. S1 in the supplemental material), hemolysis on sheep blood agar was strongly reduced (Fig. 1A). Similarly, hemolysis of erythrocyte suspensions in samples treated with culture supernatants of S. aureus ΔSSR42 were reduced to 30% of the wild-type level. Hemolysis was readily complemented in trans by expression of SSR42 from a plasmid, illustrating that the regulatory cascade of alpha-toxin expression was still intact in the deletion strain (Fig. 1B). We further excluded potential polar effects of the gene deletion by using various complementation plasmids (Fig. S1B and C).
FIG 1.
SSR42 is required for hemolysis in S. aureus and is Rsp dependent. S. aureus 6850 wild-type (WT) hemolysis is lost in a ΔSSR42 mutant but is complemented in trans (comp) on sheep blood agar (A) or upon exposure of sheep erythrocytes to stationary-growth-phase culture supernatants (B) rel, relative. (C) Episomal complementation with only SSR42 (pSSR42) or rsp (prsp) is not sufficient to complement hemolysis in an SSR42-rsp double-deletion mutant, whereas reintroduction of the locus rsp-SSR42 in trans readily complements hemolysis (pSSR42-rsp). (D) Quantitative real-time PCR analysis of wild-type S. aureus, a ΔSSR42-rsp double-knockout mutant, and complemented mutants for comparison of transcript levels of SSR42, hla, and rsp. The transcription levels of SSR42, hla, and rsp were significantly reduced in the double-knockout mutant. SSR42 and hla transcription could be restored only when complementation plasmid pSSR42-rsp (harboring both SSR42 and rsp) was used. hla transcription could not be restored to wild-type levels when only rsp (prsp) or SSR42 (pSSR42) was complemented in trans. Introduction of SSR42 (pSSR42) in trans could not successfully complement SSR42 transcription. (E) ncRNA SSR42 is required for efficient hla transcription. Quantitative real-time PCR analysis was performed of hla levels in wild-type S. aureus 6850, a ΔSSR42-rsp double-knockout mutant, and a mutant complemented with AHT-inducible transcription of either SSR42 (pAHT-SSR42) or rsp (pAHT-rsp). Only when SSR42 transcription was induced via treatment with AHT (+) did hla transcript levels show a significant increase and reach wild-type levels. (F) Growth curve and hla promoter (Phla) activity profile in wild-type S. aureus 6850 (WT) and an isogenic ΔSSR42 mutant. hla promoter activity is significantly reduced in ΔSSR42 at 9 h postinoculation (P = 7.34 × 10−5 normalized to optical density). (G) Hemolysis of S. aureus ΔSSR42 culture supernatants is complemented by SSR42 (pAHT-SSR42) upon AHT-induced (+ AHT) expression only in stationary growth phase and not in the exponential phase. For quantification of hemolysis, sheep erythrocytes were exposed to stationary-growth-phase culture supernatants, and absorbance of liberated heme at 405 nm was determined. Hemolytic level of wild-type supernatants was set to 100%. Statistical analysis was performed using Student's t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Northern blotting using two different probes against SSR42 (Fig. S2A) demonstrated a transcript of an approximate length of 1,200 nt (Fig. S2A to C), confirming the RNA-seq data (36). Next to the predominant full-length SSR42 we identified a smaller RNA, which suggested processing of the primary transcript. Global mapping of potential RNase processing sites demonstrated an RNase Y cleavage site within SSR42 (Fig. S2H) (40). Indeed smaller transcript versions of SSR42 were not detectable in a Δrny mutant of S. aureus (Fig. 2D). We thus analyzed SSR42 transcript stability in both wild-type and rny-negative backgrounds. However, these results did not reveal a role for RNase Y in destabilizing the SSR42 transcript (Fig. S2E and F).
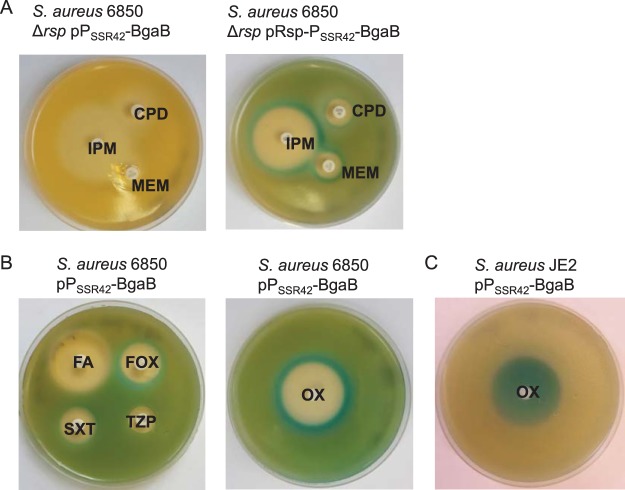
FIG 2.
Promoter PSSR42 is rsp-dependently activated upon exposure to subinhibitory concentrations of antibiotics. (A and B) Disk diffusion assays with S. aureus 6850 Δrsp harboring a transcriptional fusion of PSSR42 with β-galactosidase BgaB in the presence or absence of rsp (pPSSR42-BgaB or pRsp-PSSR42-BgaB) demonstrate activation of the promoter by production of blue dye on X-Gal agar plates. IPM, imipenem; CPD, cefpodoxime; MEM, meropenem; OX, oxacillin; FA, fusidic acid; FOX, cefoxitin; SXT, trimethoprim-sulfamethoxazole; TZP, piperacillin-tazobactam. agar plates. (C) Induction of SSR42 promoter activity with oxacillin in the MRSA strain JE2 harboring plasmid pPSSR42-BgaB.
Next, we generated a double-knockout S. aureus strain, ΔSSR42-rsp, in which we deleted the chromosomal region encoding both SSR42 and rsp (Fig. 1C and S1). We further transformed complementation plasmids into S. aureus ΔSSR42-rsp that reintroduced either SSR42, rsp, or both of the genes (pSSR42, prsp, or pSSR42-rsp, respectively) (Fig. 1C to F) and tested the functional complementation of hemolysis (Fig. 1C). S. aureus ΔSSR42-rsp displayed strongly reduced hemolysis which was not complemented by either pSSR42 or prsp (Fig. 1C). In contrast, reintroduction of the complete region encompassing SSR42 and rsp in trans (pSSR42-rsp) completely restored wild-type hemolysis levels.
We then isolated RNA from wild-type, mutant, and complemented mutant strains and analyzed transcript levels of SSR42, rsp, and hla message RNAs by quantitative PCR (qPCR). hla transcription was restored only in S. aureus ΔSSR42-rsp carrying pSSR42-rsp (Fig. 1D), corroborating that Rsp is required for SSR42 transcription. We therefore generated a plasmid which encoded either SSR42 (pAHT-SSR42) or rsp (pAHT-rsp) under the control of an anhydrous tetracycline (AHT)-inducible promoter. Only upon induction of SSR42 transcription did we also observe a significant increase in hla transcript levels, suggesting that SSR42 transcription is involved in hla transcription, whereas Rsp mainly serves as a regulator for SSR42 transcription (Fig. 1E). Thus, effects of Rsp on S. aureus hemolysis are mediated via SSR42.
Next, we investigated if the complete SSR42 region was required for regulation of hemolysis. We therefore deleted various fragments of approximately 70 nt from the molecule in pSSR42 (deletions Δ1 through Δ8) and tested hemolysis of S. aureus ΔSSR42 complemented with the resulting plasmids (Fig. S3A to D). A 65-nt deletion (deletion Δ7; bp 2352858 to 2352794 according to the genome sequence of S. aureus 6850; NCBI RefSeq accession number NC_022222) spanning the region previously identified as the potential 5′ end of SSR42 (37) resulted in significantly reduced hemolysis and accumulation of RNA degradation products, as evidenced by Northern blotting. Another 63-nt region (Δ6; bp 2352688 to 2352742) was also important for SSR42 stability since its deletion resulted in reduction of RNA abundance. Deletion of 62 nucleotides upstream of a predicted RNase Y cleavage site (Δ8; bp 2352917 to 2352978) had no impact on hemolysis, whereas deletion of a 76-nt region (Δ1; bp 2352023 to 2352099) had a strong impact on hemolysis of S. aureus but not on stability of SSR42 (Fig. S3A to D).
Introducing small mutations in SSR42 revealed a broad sequence range which was not implicated in the hemolysis regulation. A total of 437 nt encompassing this region (bp 2351972 to 2352410) were deleted from the SSR42 complementation plasmid, creating minimal SSR42 version 1. By deletion of another 128 nt (bp 2353043 to 2352915) of the 5′ end of SSR42, a complementation plasmid for minimal SSR42 version 2 was created. Secondary structure prediction showed the conservation of previous observed structural motifs (Fig. S3E and F). Due to the length of the molecule, the secondary structure predictions are only for visualization and rather incorrect. Only when a ΔSSR42 mutant was complemented with minimal version 1 were the hemolytic activity deficiencies partially complemented (75.64%, P > 0.001). Expression of minimal version 2 did not restore the hemolytic activity of a ΔSSR42 mutant (Fig. S3G). Analysis of transcript levels via Northern blotting revealed a complete lack of SSR42 transcripts, indicating the requirement of the deleted sequence in stabilizing SSR42 (Fig. S3H).
Sequence Δ1 (bp 2352023 to 2352099) was thought to regulate hemolysis since its absence did not affect the stability of the molecule as did the absence of sequences Δ6 and Δ7. To investigate whether region Δ1 would be sufficient to restore hemolysis in a ΔSSR42 mutant, the stem-loop structure encompassing this region was introduced in trans (pstemloop). Transcript levels of both SSR42 and hla were still significantly reduced compared to the wild-type level after introduction of pstemloop (Fig. S3I).
We next tested effects of SSR42 deletion on promoter activity of the hla transcript (Phla). We generated a transcriptional fusion of Phla and green fluorescent protein (GFP) (Fig. 1F and S4A) and introduced the reporter in the wild-type and ΔSSR42 backgrounds. As expected, hla promoter activity in wild-type bacteria was strongest in the stationary growth phase of wild-type S. aureus when transcription of hla is induced and stabilized by factors such as SaeR and SarA (37, 41). In contrast, in the ΔSSR42 mutant, Phla promoter activity was significantly reduced during stationary growth phase, thereby corroborating that transcriptional activation of hla requires SSR42 (37). By monitoring Phla activity over time in both the wild-type and ΔSSR42 backgrounds, we further show that hla transcription begins during transition from the exponential to the stationary growth phase and is highest during late stationary growth of the bacteria. Whereas we observed overexpression of hla message in S. aureus ΔSSR42 harboring pAHT-SSR42 after AHT addition already during exponential growth of the bacteria (Fig. S4B), hemolysis was restored only upon induction of SSR42 transcription during the stationary growth phase (Fig. 1G). Since the quorum sensing-controlled RNAIII is required for translation initiation of hla mRNA (30) yet only is transcribed during stationary phase, functional alpha-toxin can be formed only whenever RNAIII is present.
SSR42 transcription is modulated in response to subinhibitory concentrations of antibiotics.
Subinhibitory concentrations of antibiotics have been shown to alter the toxin expression of S. aureus (42–45); however, the molecular details of this regulation are not known. Since SSR42 is required for transcript abundance of the alpha-toxin mRNA, we generated S. aureus reporter strains. These harbored a reporter plasmid in which the promoter PSSR42 was transcriptionally fused to the open reading frame bgaB encoding a β-galactosidase. We tested the effect of antibiotics on SSR42 transcription using disk diffusion assays, and activation of PSSR42 was detected by production of blue indigo dye on agar plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal).
PSSR42 promoter activity was induced by antibiotics such as oxacillin, imipenem, meropenem, or cefpodoxime, and also by the DNA-damaging agent mitomycin C (Fig. 2 and S5; Table 1). This induction again was strongly dependent on functional Rsp (Fig. 2A). Further, we observed by disk diffusion as well as MIC strips (Fig. S5C) that the β-galactosidase activity was active in distinct zones beyond the zones of growth inhibition in which subinhibitory concentrations of the substances were present. We also tested PSSR42 promoter activation by a GFP reporter plasmid in S. aureus JE2. Planktonic growth in tryptic soy broth (TSB) of S. aureus JE2 was inhibited by 10 and 64 μg/ml oxacillin, and accordingly PSSR42 promoter activity was not detected. Upon exposure of the bacteria to 0.05 μg/ml oxacillin, which is only slightly inhibitory to growth of S. aureus JE2 in TSB, PSSR42 activity decreased initially, rose after 11 h, and eventually exceeded activity of untreated bacteria (Fig. S6B). In the methicillin-susceptible S. aureus (MSSA) strain 6850, subinhibitory concentrations of oxacillin (0.025 μg/ml) led to an overall increase in PSSR42 activity (Fig. S6C). In contrast, treatment of S. aureus JE2 with colistin reduced PSSR42 promoter activity in a GFP promoter activity assay (Fig. S6D).
TABLE 1.
The SSR42 promoter PSSR42 is induced by various antibiotics
| Drug (amt) | Description and/or effect(s) | Influence on PSSR42a |
|---|---|---|
| Cefpodoxime (10 μg) | β-Lactam antibiotic; inhibition of transpeptidation, cell wall disruption | + |
| Cefoxitin (30 μg) | + | |
| Oxacillin (10 μg) | + | |
| Ampicillin (10 μg) | NA | |
| Piperacillin-tazobactam (30 μg/60 μg) | NA | |
| Imipenem (10 μg) | + | |
| Meropenem (10 μg) | + | |
| Amoxicillin-clavulanate (20 μg/10μg) | NA | |
| Mitomycin C (10 μg) | DNA double-strand breaks, inhibition of DNA synthesis | + |
| Fusidic acid (10 μg) | Inhibition of protein synthesis, prevents turnover of elongation factor G from the ribosome | NA |
| Colistin (600 μg) | Disruption of cell membrane, amphiphilic interaction with cell membrane | − |
An increase (+) or decrease (−) in promoter activity is indicated. NA, not applicable (activity was not observed).
Since antibiotics as well as the DNA-damaging agent mitomycin C induced the promoter of SSR42 (Fig. S5D) and since the substances previously had been shown to mediate the S. aureus SOS response (4, 46, 47), we tested if PSSR42 activation was part of the SOS response in S. aureus. However, mitomycin C-induced activation of PSSR42 was still observed at wild-type levels in an S. aureus lexA-G94E strain, a mutant in which the autoproteolytic cleavage of LexA and thus induction of the SOS-response are prevented (Fig. S5D) (48). Thus, SSR42 transcription is not affected by lack of functional LexA. However, involvement of the SOS response in regulation of transcriptional control cannot be completely excluded and needs further investigation.
hla transcription is modulated in response to subinhibitory concentrations of antibiotics in an SSR42-dependent manner.
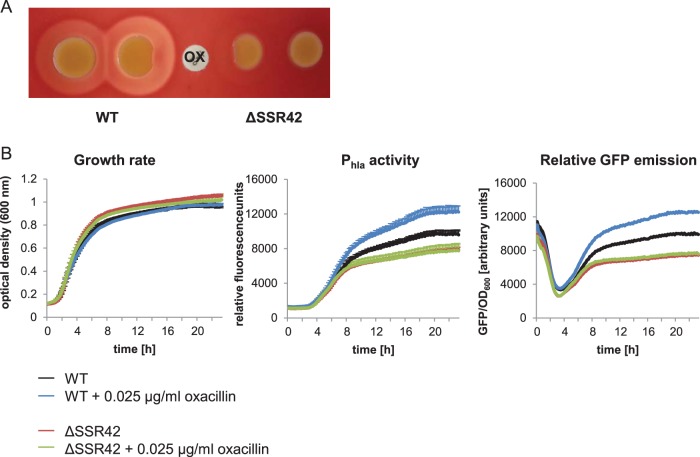
We next investigated if oxacillin would induce upregulation of hla transcription in an SSR42-dependent manner. We therefore spotted bacterial cultures of wild-type S. aureus and an isogenic ΔSSR42 mutant on sheep blood agar and placed an oxacillin-containing disk (1 μg) between the two spotted strains (Fig. 3A). Wild-type bacteria close to the oxacillin disk displayed stronger hemolysis than distant bacterial colonies. We also recorded bacterial growth and GFP fluorescence in wild-type S. aureus as well as in an isogenic ΔSSR42 mutant equipped with a Phla-GFP transcriptional fusion. Again, upregulation of hla promoter activity in response to oxacillin was observed only in wild-type S. aureus and not in the ΔSSR42 mutant (Fig. 3B). Our results thereby indicated that upregulation of hemolysin transcription in response to subinhibitory concentrations of oxacillin required the noncoding RNA SSR42.
FIG 3.
Oxacillin enhances hemolysis in an SSR42-dependent fashion. (A) Hemolysis of wild-type S. aureus and the ΔSSR42 mutant on sheep blood agar in the presence of oxacillin (OX). Wild-type bacteria display enhanced hemolysis when grown in close proximity to the 1-μg oxacillin disk. (B) Promoter Phla shows strong activity in the presence of 0.025 μg/ml oxacillin in wild-type S. aureus 6850 but not in an isogenic SSR42 knockout mutant. Promoter activity was recorded over 23 h using a strain harboring a transcriptional fusion of Phla with GFP (pPhla-GFP) and measuring fluorescence as well as the OD600. Depicted is GFP emission normalized to the OD (GFP/OD600).
SSR42 transcription is controlled by multiple global regulators and is required upstream of SaeRS.
Since SSR42 was required for oxacillin-induced hemolysis of S. aureus, we sought to identify factors involved in transcription of the ncRNA. For that purpose, we used a transcriptional fusion of the SSR42 promoter (PSSR42) and GFP and transduced the resulting reporter plasmid (pPSSR42-GFP) into several different S. aureus strains and recorded PSSR42 activity over time courses of 23 h. We found two distinct promoter activity curves in all of the tested strains. Whereas the MRSA strains JE2 and MW2 as well as MSSA strain Newman exhibited sigmoidal activity profiles, the strains 6850, RN4220, COL, Cowan I, and HG003 displayed only one peak of PSSR42 activity (Fig. S6A).
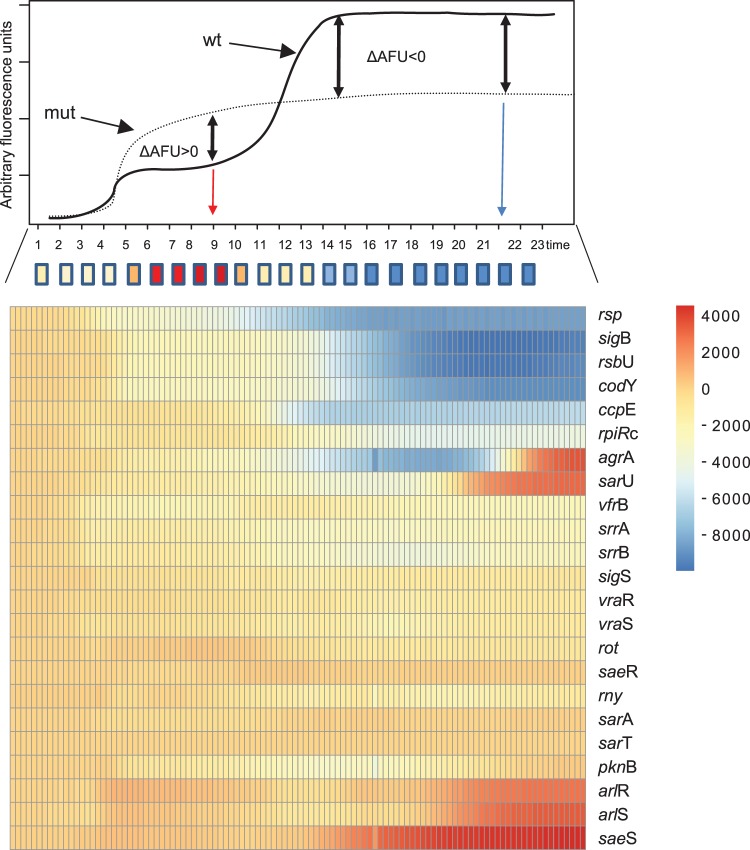
We next transduced the reporter plasmid in a variety of insertional mutants of S. aureus JE2 (49) (Fig. 4 and S7). Figure 4 depicts a heat map of the differences in GFP fluorescence levels between the wild type and the respective mutant strains. Activity of PSSR42 was completely lost in an rsp mutant (Network on Antimicrobial Resistance in Staphylococcus aureus [NARSA] Strain Repository number NE1304) (Fig. 4 and S7A), corroborating our findings on the strict Rsp dependency of SSR42 transcription (36). We also observed strongly reduced PSSR42 activity in mutants of the alternative sigma factor σB (sigB or rpoF; strain NE1109) and the σB-regulatory protein RsbU (NE1607) (Fig. S7B), whereas inactivation of the alternative sigma factor σS by gene deletion resulted in only a minute decrease in PSSR42 activity (Fig. 4 and S7I)
FIG 4.
Dependency of SSR42 promoter activity on global regulators. Activity of the SSR42 promoter (PSSR42) was measured by a transcriptional fusion with GFP in S. aureus JE2 and insertional mutants from NTML. GFP emission was recorded over 23 h. Shown are the promoter activity curve of wild-type bacteria (upper panel) and a heat map of differential activities of PSSR42 in mutants from the NTML (lower panel) compared to PSSR42 activity in wild-type S. aureus JE2. AFU, arbitrary fluorescence units.
Insertional transposon mutants within genes for the carbon catabolite repressor CcpE (strain NE1560) as well as the global repressor CodY (NE1555) and a knockout of RpiRc (50) also demonstrated reduced PSSR42 promoter activity (Fig. S7C and D). Interestingly, strains carrying mutations in the CodY and CcpE genes exhibited strongly reduced PSSR42 activity completely lacking the second peak of PSSR42 activation (Fig. S7C).
Insertional inactivation of the gene encoding the quorum sensing response regulator AgrA (strain NE1532) and inactivation of a positive regulator of the agr system, SarU (NE96), drastically altered the activation profile by delaying the second peak of promoter activity from 9 h to approximately 17 h after inoculation. At these later time points, however, GFP fluorescence levels exceeded the level of isogenic wild-type bacteria (Fig. S7E and F). SarU is a MarR-type transcriptional regulator and part of the multigene sarA family of regulators in S. aureus. However, mutations within sarA and sarT did not significantly alter the expression profile of PSSR42, and disruption of the gene encoding repressor of toxins, rot (strain NE386), led only to a slightly reduced activity of PSSR42 starting approximately at 11 h after inoculation (Fig. S7J). Insertional disruption of histidine kinase gene saeS (NE1296) led to stronger promoter activation starting 11 h after inoculation, whereas disruption of saeR (NE1622) did not change the activity of PSSR42 (Fig. S7P). Further, mutations in the two-component system ArlRS (strains NE1684 and NE1183) (Fig. S7N) led to overall higher activity of PSSR42, whereas strains carrying mutations within SrrAB (NE1309 and NE588) (Fig. S7L) as well as VraRS (NE554 and NE823) (Fig. S7M) displayed only slightly reduced GFP fluorescence levels. Although insertional inactivation of sigS, pknB, srrAB, and vraR resulted in only minor effects on PSSR42 activity, SSR42 transcript levels were significantly decreased, whereas the arlR mutant demonstrated significantly enhanced SSR42 levels, as observed by quantitative reverse transcription-PCR (qRT-PCR), thereby corroborating the promoter activity data (Fig. S7R).
We next tested rsp promoter activities (Prsp) in selected insertional mutants within S. aureus JE2 and recorded time courses of GFP fluorescence as well as bacterial growth. Strains carrying mutations within each of the genes agrA, rpiRc, codY, and arlR demonstrated Prsp promoter activity profiles similar to the ones recorded for PSSR42. This suggests that PSSR42 activities in these mutants indirectly resulted from altered expression of Rsp. However, this was not the case for the other regulators tested, ccpE, rsbU, and sigB (Fig. S8).
Thus, expression of the Rsp/SSR42 system is dependent on several global regulators, two-component systems, and alternative σ factors, thereby highlighting the central role of the molecules in the virulence regulatory circuit of S. aureus.
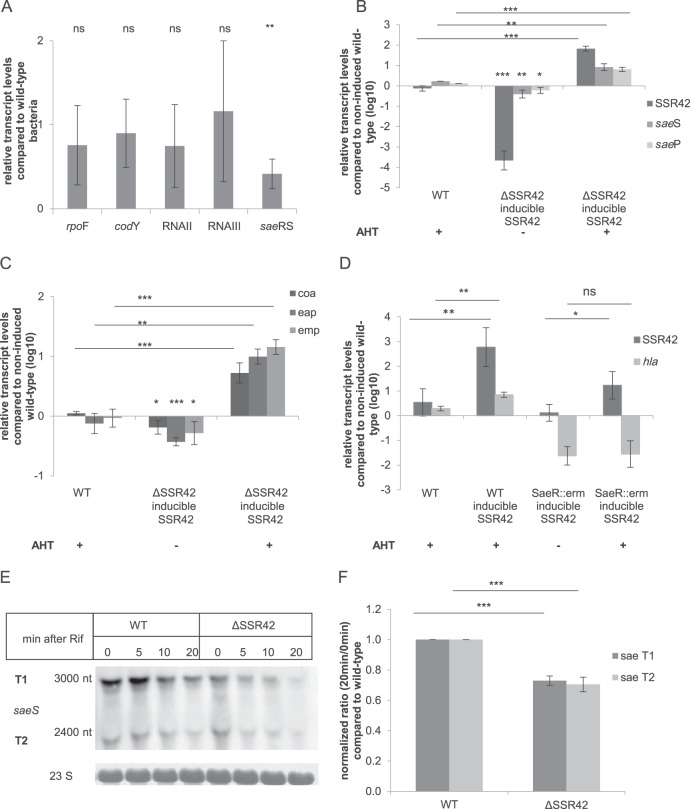
We next tested transcript levels of key regulators of PSSR42. Whereas mRNA levels of sigB, codY, RNAII (agrB), and RNAIII were unaltered in S. aureus ΔSSR42 compared to the wild-type levels, saeS transcript levels were significantly reduced in the ΔSSR42 background (Fig. 5A). We further found saeS and saeP mRNAs at significantly elevated levels upon AHT induction of SSR42 transcription in a ΔSSR42 mutant, whereas the genes were significantly reduced in noninduced samples (Fig. 5B). Since we observed a significant increase in hla transcription upon inducible transcription of SSR42 (Fig. 1F and S4B), we also tested the transcription of class I SaeRS target genes for their dependency on SSR42. Whereas significantly elevated mRNA levels of coa, eap, and emp were detected upon induction of SSR42 transcription during exponential growth, mRNA levels in noninduced controls were significantly reduced compared to those of wild-type bacteria (Fig. 5C).
FIG 5.
SSR42 regulates hla transcription upstream of SaeRS. (A) In S. aureus ΔSSR42, saeRS is transcribed at significantly decreased levels, whereas other genes of global regulators such as rpoF (sigB), codY, agrB, and RNAIII are unaltered. (B) Upon induction of SSR42 transcription by AHT in S. aureus ΔSSR42 pAHT-SSR42, transcriptional upregulation of saeP and saeS is observed in contrast to levels in noninduced samples. Without induction saeP and saeS are present at significantly reduced levels compared to the level of S. aureus wild type. (C) AHT induction of SSR42 transcription results in significantly elevated transcript levels of the class I targets coa, eap, and emp compared to levels in the noninduced complemented mutant and AHT-treated wild-type bacteria (WT). (D) SSR42-dependent upregulation of hla transcription is dependent on a functional SaeRS system. hla transcript levels are elevated upon AHT induction of SSR42 transcription in S. aureus JE2 (WT) yet are absent in an isogenic saeR insertional mutant strain. (E and F) sae T1 and T2 transcripts are less stable in a ΔSSR42 mutant of S. aureus 6850. Stability was assayed by addition of rifampin and assaying transcripts by Northern blotting. Chemiluminescence signals were quantified using ImageJ and normalized to transcript levels of wild-type bacteria. Statistical analysis was performed using Student's t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Our data thus suggested that SSR42 regulates the SaeRS two-component system. In order to test if SSR42 is functional upstream of the SaeRS two-component system, we analyzed transcription of hla in a genetic background deficient in the response regulator SaeR (Fig. 5D). We therefore introduced pAHT-SSR42 in wild-type S. aureus JE2 as well as an insertional saeR mutant (NE1622) (49). Whereas induction of SSR42 transcription resulted in significantly elevated hla mRNA levels in wild-type bacteria, hla mRNA levels were unaltered in the saeR mutant despite overexpression of SSR42 (Fig. 5D). Similar results were found for class I target genes of the SaeRS system, coa, eap, and emp (Fig. S9). We next monitored sae transcript stability by a rifampin assay. Our data show that the stability of sae transcripts T1 and T2 was significantly lower in a ΔSSR4 mutant background than the level in wild-type bacteria at 20 min after addition of rifampin (Fig. 5E).
DISCUSSION
SSR42 serves as an effector of Rsp in expression of alpha-toxin.
We previously identified repressor of surface proteins (Rsp) as a global regulator of S. aureus hemolysis, cytotoxicity, and virulence and found that Rsp is required for transcription of SSR42, which is located directly upstream of Rsp in an antiparallel orientation (36). Loss of hemolysis has been described for mutants of rsp (36, 49, 51), and functional Rsp is a requirement for transcription of SSR42 (36). SSR42 is essential for wild-type hemolysis in S. aureus (Fig. 1). Upon deletion of arbitrarily selected regions of the ncRNA, SSR42 RNA stability was found diminished, and accordingly hemolysis of bacterial culture supernatants was reduced (see Fig. S2E to H in the supplemental material). This illustrated the requirement of a full-length SSR42 molecule for RNA stability as well as phenotypic hemolysis. Since we observed both hemolysis and SSR42 transcription only in the presence of the rsp gene, this demonstrates that Rsp-dependent hemolysis is regulated by way of the SSR42 transcript. Therefore, the ncRNA is the effector of the transcriptional regulator (Fig. 1). A previous study demonstrated SSR42 involvement in hemolysis yet determined SSR42 to encompass 891 nt in strain UAMS-1 (37). Incidentally, the construct used to complement the SSR42 deletion in the previous study contained a genomic fragment, which consisted of the entire SSR42 transcript, thereby explaining the efficient complementation of the mutant phenotype (37). We can exclude the notion that UAMS-1 produces a truncated version of the RNA since our Northern blots demonstrate an RNA of a size similar to that of JE2 but at low abundance (Fig. S2C).
By inducible expression of SSR42 in a ΔSSR42 mutant, we produced the ncRNA either during stationary phase or ectopically during exponential growth phase. Whereas high hla mRNA levels were obtained after induction of SSR42 transcription during exponential phase, S. aureus hemolysis was restored only upon SSR42 induction during the stationary growth phase (Fig. 1G and S4B). Since the agr quorum sensing effector RNAIII is required for translation of the hla message (30) and is expressed only during stationary growth (29, 52–54), our data thus show that alpha-toxin production involves two ncRNAs: the 1,232-nt SSR42 is required for transcription of the hla message, whereas the 514-nt RNAIII renders the Shine-Dalgarno sequence of the mRNA accessible to ribosomes.
Hemolysis induction by antibiotics is dependent on the ncRNA SSR42.
Enhanced alpha-toxin production upon exposure of S. aureus to subinhibitory concentrations of antibiotics has been previously described (44, 55). Especially β-lactam antibiotics such as oxacillin significantly altered toxin expression profiles of the bacteria (45) and enhanced hla transcription in S. aureus (44) or altered disease progression (56). During the course of treatment of bacterial infections, the pathogens may encounter subinhibitory concentrations of antibiotics, e.g., during low-dosage therapy, at the beginning and end of a treatment (57), or within biofilms (58). However, the underlying molecular mechanisms by which antibiotics would increase S. aureus hemolysis are only incompletely understood. The saeRS (55, 59), alternative σ factor σS (60), and VraRS (59) were found involved; however, the last two are not directly linked to hemolysin production. We show that SSR42 promoter activity of PSSR42 as well as transcription of the ncRNA is increased by various antibiotics at subinhibitory concentrations (Fig. S5 and 6B to D; Table 1). Unexpectedly, PSSR42 was also activated when S. aureus reporter strains were exposed to mitomycin C (Fig. S5D). Both β-lactam antibiotics and mitomycin C were implicated in activation of the S. aureus SOS response (4). However, in a lexA mutant, the PSSR42 promoter was still activated by mitomycin C exposure, thus demonstrating that PSSR42 induction is independent of LexA derepression. Similarly, subinhibitory concentrations of oxacillin induced the hla promoter (Phla) in a process that depended on SSR42 (Fig. 3). Thus, β-lactam-enhanced S. aureus hemolysis is regulated via the Rsp/SSR42 axis, thereby identifying a hitherto undescribed role for both factors.
Multiple global regulators integrate SSR42 transcription.
hla expression is regulated by a variety of factors and conditions (29, 61–65). In order to assess the position of SSR42 in this complex regulatory network, we assayed the activity of PSSR42 promoter in a variety of regulator mutants. Aside from rsp (Fig. 2A, 4, and S7A), mutation of agrA, sarU, codY, σB, saeS, arlRS, ccpE, and rpiRc significantly altered PSSR42 activity, whereas the effect of saeR, σS, vraRS, or srrAB mutation was negligible (Fig. 4 and S7). Insertional disruption of agr and sarU, an activator of agr, resulted in similar PSSR42 profiles with a long phase of inactivity and a late stationary-phase boost eventually exceeding PSSR42 activity of the wild type (Fig. 4 and S7E and F). In contrast, insertional mutations within sarA, sarT, and rot did not drastically alter PSSR42 activity over time. ArlRS was shown to positively regulate expression of agr (66); however, its influence on hemolysis is controversial (66–70). Here, we observed increased PSSR42 activity in insertional arlR and arlS mutants. In contrast, two other two-component systems implicated in altered β-lactam susceptibility, srrAB and vraRS (71), did not significantly contribute to PSSR42 activity.
In the codY mutant, PSSR42 activity was strongly reduced, completely lacking the second peak of activation. The second phase of promoter activation commenced about 9 h after inoculation and thus far into the stationary growth phase. The resulting sigmoidal PSSR42 activity was strongly dependent on the strain background and was observed in S. aureus JE2, MW2, and Newman strains but not in the remainder of the strains (6850, COL, Cowan I, HG003, and RN4220) (Fig. S6A), indicating that the peculiar profile does not correlate with methicillin resistance, virulence, or hemolysis. Since the SSR42 promoter lacks a typical CodY-binding motif (data not shown) and is not bound by CodY (72), regulation of SSR42 by CodY likely is indirect. For instance, rsbU as well as rpiRc is activated by CodY (72). Consistently, inactivation of sigB, rsbU, and rpiRc led to PSSR42 inactivation (Fig. 4 and S7) (73, 74). Our data hence suggest that CodY indirectly regulates SSR42 by way of σB and RpiRc activity. σB itself positively regulates approximately 120 genes in response to various conditions (75, 76). However, a consensus σB-binding site is absent in PSSR42 (data not shown). RpiRc reacts to the metabolic state of the bacteria and was reported to control pentose phosphate pathway genes as well as RNAIII, likely via σB and SarA (50). However, in our analyses sarA did not influence PSSR42 activity.
CcpE, another virulence regulator monitoring metabolic levels, represses not only tricarboxylic acid (TCA) cycle genes but also RNAIII, hla, psmα, and capA in the presence of glucose (77, 78). The ccpE mutant demonstrated a drastically altered PSSR42 profile in which the second activation peak was lost.
Since alpha-toxin transcription depended on SSR42, we also investigated SaeRS, the major transcriptional regulator of hla. SaeR induces hla transcription predominantly in the postexponential growth phase (19, 61), and SSR42-dependent promoter activity of Phla begins at the transition to the stationary growth phase (Fig. 1F). Deletion of SSR42 resulted in a significant decrease in expression of sae as well as class I and class II target genes (Fig. 5). Together, these data suggested that SSR42 is required upstream of SaeRS. We thus found that SSR42 modulates the stability of the sae transcripts T1 and T2 (Fig. 5E). However, it has been shown previously that small amounts of SaeRS are sufficient to initiate transcription of the class II SaeR target hla (17) and that only a complete lack of SaeR activity results in loss of Phla activity (79). The mechanism by which SSR42 stabilizes sae mRNA is currently unknown. Since both transcripts, SSR42 (40) and saePQRS (26), are processed by the endoribonuclease RNase Y, SSR42 interaction with RNase Y thereby may affect the stability of sae mRNA by influencing sae T1 cleavage. Whereas the stability of sae is affected by RNase Y cleavage (26), SSR42 stability remains unaltered in the absence of rny (Fig. 2E). We also observed that transcriptional activation of SSR42 levels is attenuated by SaeS at time points of high SSR42 levels (Fig. S7H). This interdependency may illustrate a negative-feedback loop between the two molecules. Most strikingly, the effects were not observed in an saeR response regulator mutant. It is questionable if the observed loss of saeRS transcript stability in the SSR42 mutant is sufficient for the strong phenotypic decrease in hemolysis (79). In addition, knockout of Rsp, the direct regulator of SSR42, resulted in an increase in transcription of sae (36). The precise role of the interplay of SSR42, saePQRS, and RNase Y (Fig. 6) therefore remains elusive.
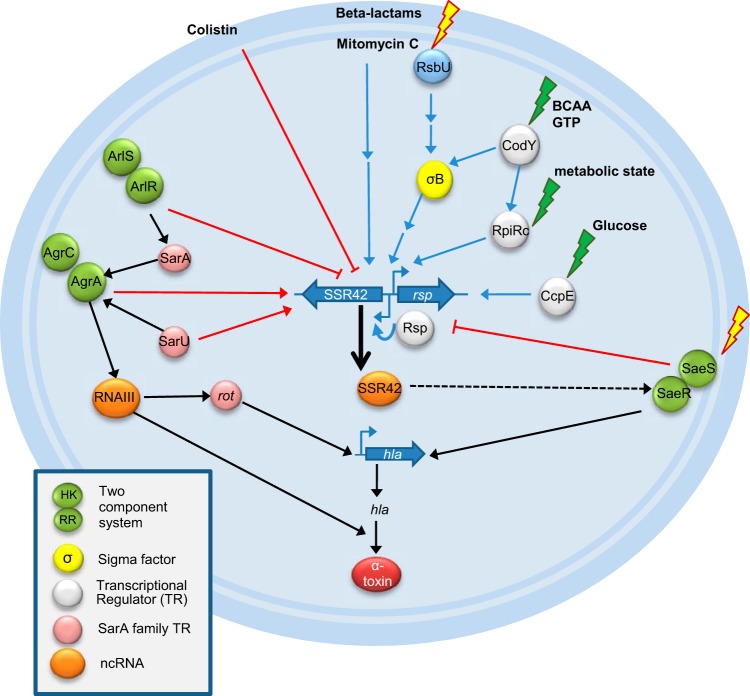
FIG 6.
Overview of SSR42-dependent alpha-toxin expression. Disruption of the agr quorum sensing system results in complete lack of hemolysis due to the absence of RNAIII as well as reduced sae mRNA levels and also strongly delays SSR42 transcription with a dynamic similar to that of an sarU insertional mutant strain. In codY, σB, and rsbU mutants, PSSR42 activity was drastically reduced, illustrating positive regulation of SSR42 transcription by these factors. CodY acts presumably via RsbU and RpiRc. Further, in a ccpE mutant the second peak of the PSSR42 activity profile activation was lost. In contrast, loss of the ArlRS two-component system led to induction of PSSR42 activity. Similarly, this also was observed in an saeS mutant, illustrating feedback between SSR42 and sae, since SSR42 acts via SaeRS and the effects of ectopic ncRNA expression are lost in an saeR mutant. The exact mode of action of SSR42 on sae is unknown. However, both the SSR42 and sae transcripts are processed by RNase Y, which therefore suggests that RNA stability is involved. The β-lactam-dependent induction of hla transcription thereby is dependent on SSR42, whereas colistin reduces PSSR42 promoter activity. For details, refer to the text. Black arrows, known interactions; red arrows, downregulation; blue arrows, upregulation of SSR42 by factors shown in this study.
Staphylococcal alpha-toxin is controlled by a variety of virulence regulators on the transcriptional as well as posttranscriptional level. We show that SSR42 contributes to hla transcription in a process upstream of SaeRS. SSR42 transcription itself is modulated by global regulators such as Rsp, AgrA, SarU, ArlRS, SaeS, CodY, σB, CcpE, and RpiRc, thereby demonstrating that the ncRNA is involved in integrating nutritional as well as environmental signals during cytolysin production and virulence. By this pathway, S. aureus hemolysis is enhanced SSR42 dependently upon exposure to subinhibitory β-lactam concentrations. Thus, exposure to antibiotics eventually can alter S. aureus virulence.
MATERIALS AND METHODS
Bacterial culture conditions.
Staphylococcus aureus strains were grown on tryptic soy agar (TSA) or in tryptic soy broth (TSB) (lot number BCBP7262V; Sigma) supplemented with 0.25% glucose and appropriate antibiotics. Escherichia coli strains were grown on LB using appropriate antibiotics. Broth cultures were grown aerobically at 37°C overnight at 180 rpm.
Bacterial growth curves.
S. aureus strains were grown in TSB at 37°C at 180 rpm in air. Triplicates of the cultures were diluted to an optical density at 600 nm (OD600) of 0.1 in 400 μl of fresh TSB and were grown for 23 h in a 48-well microwell plate. Absorbance was measured automatically at 600 nm every 10 min using a Tecan Infinite M200 plate reader.
Construction of bacterial strains and plasmids.
For all strains, plasmids, and oligonucleotides used, see Table S1 in the supplemental material. All S. aureus insertional transposon mutants available through the Nebraska Transposon Mutant Library (NTML) Library were transduced via phage 11 into the erythromycin-sensitive genetic background of wild-type S. aureus JE2 in order to avoid secondary-site mutations. Markerless targeted gene deletions of SSR42 and SSR42-rsp were generated using the vectors pBASE6-SSR42 and pBASE6-SSR42-rsp. Gene deletions were performed as described previously (80). For inducible complementation of SSR42 or Rsp, plasmids pAHT-SSR42 and pAHT-rsp were used, respectively. For complementation of ΔSSR42 and ΔSSR42-rsp mutants, plasmids pSSR42, prsp, pSSR42-rsp, and p2216-2218 were used. Small deletions in SSR42 were investigated using complementation plasmids pSSR42Δ1, pSSR42Δ2, pSSR42Δ3, pSSR42Δ4, pSSR42Δ5, pSSR42Δ6, pSSR42Δ7, and pSSR42Δ8. Promoter activities of PSSR42 and Phla were monitored using reporter plasmid pRsp-PSSR42-BgaB, p-PSSR42-BgaB, pPSSR42-GFP, or pPhla-GFP. The construction of all strains and plasmids is described in Text S1 in the supplemental material.
RNA isolation.
Bacterial RNA was extracted using a previously described TRIzol method (81) and treated with DNase I.
qRT-PCR.
Reverse transcription of total isolated RNA was performed using RevertAID reverse transcriptase (Thermo Scientific). A 10-ng sample of cDNA was used to perform qRT-PCR in a one-step reaction using Sybr Green master mix (2×; Genaxxon) on a StepOne Plus real-time PCR system (Applied Biosystems). For primers used for qRT-PCR see Table S1 in the supplemental material. Analysis was performed using the 2−ΔΔCT (where CT is threshold cycle) method. Relative gene expression was normalized to expression of the housekeeping gene of gyrase subunit B (gyrB) and to the corresponding expression in wild-type cells.
Northern blotting.
Northern blotting of RNA was performed as previously described (82) using digoxigenin-labeled probes. Primers for the generation of probes are listed in Table S1. For detailed information on RNA methods, see Text S1 in the supplemental material.
Rifampin assay.
S. aureus strains were grown in TSB at 37°C in air until cultures reached stationary growth phase. Rifampin was added to 500 μg/ml, and bacteria were harvested at time points indicated within the figures by flash freezing. RNA was isolated and analyzed by Northern blotting. ImageJ was used for quantification of signals (83).
Hemolysis assay.
Bacteria were grown overnight in TSB at 37°C. Hemolysis of S. aureus was determined by spotting 10 μl of 100-fold-diluted culture on Columbia agar (BD Biosciences) supplemented with 5% defibrinated sheep blood (Fiebig Nährstofftechnik, Germany). For quantitative analysis, sheep erythrocytes (Fiebig Nährstofftechnik, Germany) were washed with 0.9% NaCl and then diluted to a final concentration of 1% in the same buffer. Bacteria were grown overnight in TSB at 37°C and harvested, and supernatant was collected and sterile filtered (0.45-μm pore size). Sterile-filtered supernatant (5%, vol/vol) of S. aureus was added to a 1% erythrocyte solution and incubated for 1 h at 37°C. Thereafter, the suspension was centrifuged, and supernatants were analyzed in technical replicates for heme release by measuring absorbance at 405 ± 9 nm using a Tecan Infinite M200 plate reader.
β-Galactosidase assay.
SSR42 promoter activity upon treatment with chemicals and antibiotics was analyzed using a β-galactosidase reporter construct. Strains harboring this reporter plasmid were grown overnight at 37°C in TSB with 10 μg/ml chloramphenicol. One hundred microliters of bacteria was added to 5 ml of soft agar containing 40 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and plated on agar plates containing 40 mg/ml X-Gal. Bacteria were either streaked on the agar or plated within soft agar. Diffusion disks containing different antibiotics were placed upon the agar plates, which were incubated overnight for 37°C.
Fluorescence-based promoter activity assay.
Promoter activities during bacterial growth were assessed by monitoring GFP fluorescence (excitation, 488 ± 9 nm; emission, 518 ± 20 nm) as well as optical density (600 nm) using a Tecan Infinite M200 multiplate reader. For this, bacteria were grown in TSB overnight at 37°C at 180 rpm in air. The cultures were diluted in 400 μl of fresh TSB to an OD600 of 0.1 in triplicates. Concentrations of supplemented antibiotics are indicated within the figures. Bacteria were grown for 23 h in a 48-well microwell plate. Absorbance and GFP fluorescence were measured automatically every 10 min using a Tecan Infinite M200 multiplate reader.
Heat maps.
Heat maps were generated by calculating the difference in fluorescence units between each mutant and respective wild-type strain for each time point. The resulting matrix was visualized using the R library application pheatmat.
Statistics.
If not stated otherwise, statistical analyses were performed using Student's t test.
Supplementary Material
ACKNOWLEDGMENTS
We thank the German Science Foundation (http://www.dfg.de) for funding this project within the Transregional Research Collaborative TRR34, project C11 (M.F. and T.R.). The Helmholtz Institute for RNA-based Infection Research (HIRI) supported this work with a seed grant through funds from the Bavarian Ministry of Economic Affairs and Media, Energy and Technology (grant allocation no. 0703/68674/5/2017 and 0703/89374/3/2017). S. aureus JE2 mutants were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) Program supported under NIAID/NIH contract number HHSN272200700055C.
We further are indebted to Stefanie Feuerbaum and Tim Teufel for initial cloning of constructs and experimentation, to Rosemarie Gaupp and Markus Bischoff for the ΔrpiRc mutant, and to Lisa Münzenmayer for critically reading the manuscript.
We declare that we have no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00252-18.
REFERENCES
- 1.Dantes R, Mu Y, Belflower R, Aragon D, Dumyati G, Harrison LH, Lessa FC, Lynfield R, Nadle J, Petit S, Ray SM, Schaffner W, Townes J, Fridkin S, Emerging Infections Program-Active Bacterial Core Surveillance MRSA Surveillance Investigators. 2013. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med 173:1970–1978. doi: 10.1001/jamainternmed.2013.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee BY, Singh A, David MZ, Bartsch SM, Slayton RB, Huang SS, Zimmer SM, Potter MA, Macal CM, Lauderdale DS, Miller LG, Daum RS. 2013. The economic burden of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA). Clin Microbiol Infect 19:528–536. doi: 10.1111/j.1469-0691.2012.03914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 4.Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol 188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clauditz A, Resch A, Wieland KP, Peschel A, Gotz F. 2006. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun 74:4950–4953. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soutourina O, Dubrac S, Poupel O, Msadek T, Martin-Verstraete I. 2010. The pleiotropic CymR regulator of Staphylococcus aureus plays an important role in virulence and stress response. PLoS Pathog 6:e1000894. doi: 10.1371/journal.ppat.1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhakdi S, Tranum-Jensen J. 1991. Alpha-toxin of Staphylococcus aureus. Microbiol Rev 55:733–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. 1996. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 9.Bhakdi S, Muhly M, Korom S, Hugo F. 1989. Release of interleukin-1 beta associated with potent cytocidal action of staphylococcal alpha-toxin on human monocytes. Infect Immun 57:3512–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menzies BE, Kourteva I. 2000. Staphylococcus aureus alpha-toxin induces apoptosis in endothelial cells. FEMS Immunol Med Microbiol 29:39–45. doi: 10.1111/j.1574-695X.2000.tb01503.x. [DOI] [PubMed] [Google Scholar]
- 11.Walev I, Palmer M, Martin E, Jonas D, Weller U, Hohn-Bentz H, Husmann M, Bhakdi S. 1994. Recovery of human fibroblasts from attack by the pore-forming alpha-toxin of Staphylococcus aureus. Microb Pathog 17:187–201. doi: 10.1006/mpat.1994.1065. [DOI] [PubMed] [Google Scholar]
- 12.Bubeck Wardenburg J, Schneewind O. 2008. Vaccine protection against Staphylococcus aureus pneumonia. J Exp Med 205:287–294. doi: 10.1084/jem.20072208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Callaghan RJ, Callegan MC, Moreau JM, Green LC, Foster TJ, Hartford OM, Engel LS, Hill JM. 1997. Specific roles of alpha-toxin and beta-toxin during Staphylococcus aureus corneal infection. Infect Immun 65:1571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powers ME, Becker REN, Sailer A, Turner JR, Wardenburg JB. 2015. Synergistic action of Staphylococcus aureus α-toxin on platelets and myeloid lineage cells contributes to lethal sepsis. Cell Host Microbe 17:775–787. doi: 10.1016/j.chom.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bronner S, Monteil H, Prévost G. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol Rev 28:183–200. doi: 10.1016/j.femsre.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 17.Mainiero M, Goerke C, Geiger T, Gonser C, Herbert S, Wolz C. 2010. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J Bacteriol 192:613–623. doi: 10.1128/JB.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Junecko JM, Zielinska AK, Mrak LN, Ryan DC, Graham JW, Smeltzer MS, Lee CY. 2012. Transcribing virulence in Staphylococcus aureus. World J Clin Infect Dis 2:63–76. doi: 10.5495/wjcid.v2.i4.63. [DOI] [Google Scholar]
- 19.Novick RP, Jiang D. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149:2709–2717. doi: 10.1099/mic.0.26575-0. [DOI] [PubMed] [Google Scholar]
- 20.Sun F, Li C, Jeong D, Sohn C, He C, Bae T. 2010. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol 192:2111–2127. doi: 10.1128/JB.01524-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voyich JM, Vuong C, DeWald M, Nygaard TK, Kocianova S, Griffith S, Jones J, Iverson C, Sturdevant DE, Braughton KR, Whitney AR, Otto M, DeLeo FR. 2009. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J Infect Dis 199:1698–1706. doi: 10.1086/598967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong D-W, Cho H, Jones MB, Shatzkes K, Sun F, Ji Q, Liu Q, Peterson SN, He C, Bae T. 2012. The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol Microbiol 86:331–348. doi: 10.1111/j.1365-2958.2012.08198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q, Yeo W-S, Bae T. 2016. The SaeRS two-component system of Staphylococcus aureus. Genes (Basel) 7:81. doi: 10.3390/genes7100081. [DOI] [Google Scholar]
- 24.Steinhuber A, Goerke C, Bayer MG, Döring G, Wolz C. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J Bacteriol 185:6278–6286. doi: 10.1128/JB.185.21.6278-6286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geiger T, Goerke C, Mainiero M, Kraus D, Wolz C. 2008. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J Bacteriol 190:3419–3428. doi: 10.1128/JB.01927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marincola G, Schafer T, Behler J, Bernhardt J, Ohlsen K, Goerke C, Wolz C. 2012. RNase Y of Staphylococcus aureus and its role in the activation of virulence genes. Mol Microbiol 85:817–832. doi: 10.1111/j.1365-2958.2012.08144.x. [DOI] [PubMed] [Google Scholar]
- 27.Adhikari RP, Novick RP. 2008. Regulatory organization of the staphylococcal sae locus. Microbiology 154:949–959. doi: 10.1099/mic.0.2007/012245-0. [DOI] [PubMed] [Google Scholar]
- 28.Queck SY, Jameson-Lee M, Villaruz AE, Bach T-HL, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell 32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morfeldt E, Taylor D, von Gabain A, Arvidson S. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J 14:4569–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storz G, Altuvia S, Wassarman KM. 2005. An abundance of RNA regulators. Annu Rev Biochem 74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 32.Han Y, Liu L, Fang N, Yang R, Zhou D. 2013. Regulation of pathogenicity by noncoding RNAs in bacteria. Future Microbiol 8:579–591. doi: 10.2217/fmb.13.20. [DOI] [PubMed] [Google Scholar]
- 33.Geissmann T, Chevalier C, Cros M-J, Boisset S, Fechter P, Noirot C, Schrenzel J, François P, Vandenesch F, Gaspin C, Romby P. 2009. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res 37:7239–7257. doi: 10.1093/nar/gkp668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livny J, Teonadi H, Livny M, Waldor MK. 2008. High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS One 3:e3197. doi: 10.1371/journal.pone.0003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romilly C, Lays C, Tomasini A, Caldelari I, Benito Y, Hammann P, Geissmann T, Boisset S, Romby P, Vandenesch F. 2014. A non-coding RNA promotes bacterial persistence and decreases virulence by regulating a regulator in Staphylococcus aureus. PLoS Pathog 10:e1003979. doi: 10.1371/journal.ppat.1003979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das S, Lindemann C, Young BC, Muller J, Österreich B, Ternette N, Winkler AC, Paprotka K, Reinhardt R, Förstner KU, Allen E, Flaxman A, Yamaguchi Y, Rollier CS, van Diemen P, Blättner S, Remmele CW, Selle M, Dittrich M, Müller T, Vogel J, Ohlsen K, Crook DW, Massey R, Wilson DJ, Rudel T, Wyllie DH, Fraunholz MJ. 2016. Natural mutations in a Staphylococcus aureus virulence regulator attenuate cytotoxicity but permit bacteremia and abscess formation. Proc Natl Acad Sci U S A 113:E3101–E3110. doi: 10.1073/pnas.1520255113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrison JM, Miller EW, Benson MA, Alonzo F III, Yoong P, Torres VJ, Hinrichs SH, Dunman PM. 2012. Characterization of SSR42, a novel virulence factor regulatory RNA that contributes to the pathogenesis of a Staphylococcus aureus USA300 representative. J Bacteriol 194:2924–2938. doi: 10.1128/JB.06708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olson PD, Kuechenmeister LJ, Anderson KL, Daily S, Beenken KE, Roux CM, Reniere ML, Lewis TL, Weiss WJ, Pulse M, Nguyen P, Simecka JW, Morrison JM, Sayood K, Asojo OA, Smeltzer MS, Skaar EP, Dunman PM. 2011. Small molecule inhibitors of Staphylococcus aureus RnpA alter cellular mRNA turnover, exhibit antimicrobial activity, and attenuate pathogenesis. PLoS Pathog 7:e1001287. doi: 10.1371/journal.ppat.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beaume M, Hernandez D, Farinelli L, Deluen C, Linder P, Gaspin C, Romby P, Schrenzel J, Francois P. 2010. Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS One 5:e10725. doi: 10.1371/journal.pone.0010725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khemici V, Prados J, Linder P, Redder P. 2015. Decay-initiating endoribonucleolytic cleavage by RNase Y is kept under tight control via sequence preference and sub-cellular localisation. PLoS Genet 11:e1005577. doi: 10.1371/journal.pgen.1005577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nygaard TK, Pallister KB, Ruzevich P, Griffith S, Vuong C, Voyich JM. 2010. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J Infect Dis 201:241–254. doi: 10.1086/649570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dickgiesser N, Wallach U. 1987. Toxic shock syndrome toxin-1 (TSST-1): influence of its production by subinhibitory antibiotic concentrations. Infection 15:351–353. doi: 10.1007/BF01647737. [DOI] [PubMed] [Google Scholar]
- 43.Dumitrescu O, Boisset S, Badiou C, Bes M, Benito Y, Reverdy M-E, Vandenesch F, Etienne J, Lina G. 2007. Effect of antibiotics on Staphylococcus aureus producing Panton-Valentine leukocidin. Antimicrob Agents Chemother 51:1515–1519. doi: 10.1128/AAC.01201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohlsen K, Ziebuhr W, Koller K-P, Hell W, Wichelhaus TA, Hacker J. 1998. Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother 42:2817–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudkin JK, Laabei M, Edwards AM, Joo HS, Otto M, Lennon KL, O'Gara JP, Waterfield NR, Massey RC. 2014. Oxacillin alters the toxin expression profile of community-associated methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 58:1100–1107. doi: 10.1128/AAC.01618-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cirz RT, Jones MB, Gingles NA, Minogue TD, Jarrahi B, Peterson SN, Romesberg FE. 2007. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J Bacteriol 189:531–539. doi: 10.1128/JB.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maiques E, Úbeda C, Campoy S, Salvador N, Lasa Í, Novick RP, Barbé J, Penadés JR. 2006. β-Lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol 188:2726–2729. doi: 10.1128/JB.188.7.2726-2729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schroder W, Goerke C, Wolz C. 2013. Opposing effects of aminocoumarins and fluoroquinolones on the SOS response and adaptability in Staphylococcus aureus. J Antimicrob Chemother 68:529–538. doi: 10.1093/jac/dks456. [DOI] [PubMed] [Google Scholar]
- 49.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaupp R, Wirf J, Wonnenberg B, Biegel T, Eisenbeis J, Graham J, Herrmann M, Lee CY, Beisswenger C, Wolz C, Tschernig T, Bischoff M, Somerville GA. 2016. RpiRc is a pleiotropic effector of virulence determinant synthesis and attenuates pathogenicity in Staphylococcus aureus. Infect Immun 84:2031–2041. doi: 10.1128/IAI.00285-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li T, He L, Song Y, Villaruz AE, Joo HS, Liu Q, Zhu Y, Wang Y, Qin J, Otto M, Li M. 2015. AraC-Type regulator Rsp adapts Staphylococcus aureus gene expression to acute infection. Infect Immun 84:723–734. doi: 10.1128/IAI.01088-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chabelskaya S, Bordeau V, Felden B. 2014. Dual RNA regulatory control of a Staphylococcus aureus virulence factor. Nucleic Acids Res 42:4847–4858. doi: 10.1093/nar/gku119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong YQ, Van Wamel W, Nast CC, Yeaman MR, Cheung AL, Bayer AS. 2002. Activation and transcriptional interaction between agr RNAII and RNAIII in Staphylococcus aureus in vitro and in an experimental endocarditis model. J Infect Dis 186:668–677. doi: 10.1086/342046. [DOI] [PubMed] [Google Scholar]
- 54.Yarwood JM, McCormick JK, Paustian ML, Kapur V, Schlievert PM. 2002. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J Bacteriol 184:1095–1101. doi: 10.1128/jb.184.4.1095-1101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuroda H, Kuroda M, Cui L, Hiramatsu K. 2007. Subinhibitory concentrations of beta-lactam induce haemolytic activity in Staphylococcus aureus through the SaeRS two-component system. FEMS Microbiol Lett 268:98–105. doi: 10.1111/j.1574-6968.2006.00568.x. [DOI] [PubMed] [Google Scholar]
- 56.Kernodle DS, McGraw PA, Barg NL, Menzies BE, Voladri RK, Harshman S. 1995. Growth of Staphylococcus aureus with nafcillin in vitro induces alpha-toxin production and increases the lethal activity of sterile broth filtrates in a murine model. J Infect Dis 172:410–419. doi: 10.1093/infdis/172.2.410. [DOI] [PubMed] [Google Scholar]
- 57.Odenholt I. 2001. Pharmacodynamic effects of subinhibitory antibiotic concentrations. Int J Antimicrob Agents 17:1–8. doi: 10.1016/S0924-8579(00)00243-0. [DOI] [PubMed] [Google Scholar]
- 58.Singh R, Ray P, Das A, Sharma M. 2010. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J Antimicrob Chemother 65:1955–1958. doi: 10.1093/jac/dkq257. [DOI] [PubMed] [Google Scholar]
- 59.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49:807–821. doi: 10.1046/j.1365-2958.2003.03599.x. [DOI] [PubMed] [Google Scholar]
- 60.Miller HK, Carroll RK, Burda WN, Krute CN, Davenport JE, Shaw LN. 2012. The extracytoplasmic function sigma factor σS protects against both intracellular and extracytoplasmic stresses in Staphylococcus aureus. J Bacteriol 194:4342–4354. doi: 10.1128/JB.00484-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giraudo AT, Cheung AL, Nagel R. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch Microbiol 168:53–58. doi: 10.1007/s002030050469. [DOI] [PubMed] [Google Scholar]
- 62.McNamara PJ, Milligan-Monroe KC, Khalili S, Proctor RA. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J Bacteriol 182:3197–3203. doi: 10.1128/JB.182.11.3197-3203.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohlsen K, Koller KP, Hacker J. 1997. Analysis of expression of the alpha-toxin gene (hla) of Staphylococcus aureus by using a chromosomally encoded hla::lacZ gene fusion. Infect Immun 65:3606–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saïd-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, McNamara PJ, Arvidson S, Foster TJ, Projan SJ, Kreiswirth BN. 2003. Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol 185:610–619. doi: 10.1128/JB.185.2.610-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmidt KA, Manna AC, Gill S, Cheung AL. 2001. SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect Immun 69:4749–4758. doi: 10.1128/IAI.69.8.4749-4758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang X, Zheng L, Landwehr C, Lunsford D, Holmes D, Ji Y. 2005. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J Bacteriol 187:5486–5492. doi: 10.1128/JB.187.15.5486-5492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fournier B, Klier A, Rapoport G. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol 41:247–261. doi: 10.1046/j.1365-2958.2001.02515.x. [DOI] [PubMed] [Google Scholar]
- 68.Harper L, Balasubramanian D, Ohneck EA, Sause WE, Chapman J, Mejia-Sosa B, Lhakhang T, Heguy A, Tsirigos A, Ueberheide B, Boyd JM, Lun DS, Torres VJ. 2018. Staphylococcus aureus responds to the central metabolite pyruvate to regulate virulence. mBio 9:e02272-17. doi: 10.1128/mBio.02272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Radin JN, Kelliher JL, Parraga Solorzano PK, Kehl-Fie TE. 2016. The two-component system ArlRS and alterations in metabolism enable Staphylococcus aureus to resist calprotectin-induced manganese starvation. PLoS Pathog 12:e1006040. doi: 10.1371/journal.ppat.1006040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker JN, Crosby HA, Spaulding AR, Salgado-Pabon W, Malone CL, Rosenthal CB, Schlievert PM, Boyd JM, Horswill AR. 2013. The Staphylococcus aureus ArlRS two-component system is a novel regulator of agglutination and pathogenesis. PLoS Pathog 9:e1003819. doi: 10.1371/journal.ppat.1003819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsuo M, Kato F, Oogai Y, Kawai T, Sugai M, Komatsuzawa H. 2010. Distinct two-component systems in methicillin-resistant Staphylococcus aureus can change the susceptibility to antimicrobial agents. J Antimicrob Chemother 65:1536–1537. doi: 10.1093/jac/dkq141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. 2010. Direct targets of CodY in Staphylococcus aureus. J Bacteriol 192:2861–2877. doi: 10.1128/JB.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giachino P, Engelmann S, Bischoff M. 2001. σB Activity depends on RsbU in Staphylococcus aureus. J Bacteriol 183:1843–1852. doi: 10.1128/JB.183.6.1843-1852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palma M, Cheung AL. 2001. σB activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect Immun 69:7858–7865. doi: 10.1128/IAI.69.12.7858-7865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rogasch K, Rühmling V, Pané-Farré J, Höper D, Weinberg C, Fuchs S, Schmudde M, Bröker BM, Wolz C, Hecker M, Engelmann S. 2006. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J Bacteriol 188:7742–7758. doi: 10.1128/JB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seidl K, Stucki M, Ruegg M, Goerke C, Wolz C, Harris L, Berger-Bachi B, Bischoff M. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Chemother 50:1183–1194. doi: 10.1128/AAC.50.4.1183-1194.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ding Y, Liu X, Chen F, Di H, Xu B, Zhou L, Deng X, Wu M, Yang CG, Lan L. 2014. Metabolic sensor governing bacterial virulence in Staphylococcus aureus. Proc Natl Acad Sci U S A 111:E4981–E4990. doi: 10.1073/pnas.1411077111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hartmann T, Baronian G, Nippe N, Voss M, Schulthess B, Wolz C, Eisenbeis J, Schmidt-Hohagen K, Gaupp R, Sunderkotter C, Beisswenger C, Bals R, Somerville GA, Herrmann M, Molle V, Bischoff M. 2014. The catabolite control protein E (CcpE) affects virulence determinant production and pathogenesis of Staphylococcus aureus. J Biol Chem 289:29701–29711. doi: 10.1074/jbc.M114.584979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiong YQ, Willard J, Yeaman MR, Cheung AL, Bayer AS. 2006. Regulation of Staphylococcus aureus alpha-toxin gene (hla) expression by agr, sarA, and sae in vitro and in experimental infective endocarditis. J Infect Dis 194:1267–1275. doi: 10.1086/508210. [DOI] [PubMed] [Google Scholar]
- 80.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 81.Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penades JR, Valle J, Solano C, Gingeras TR. 2011. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci U S A 108:20172–20177. doi: 10.1073/pnas.1113521108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goerke C, Campana S, Bayer MG, Doring G, Botzenhart K, Wolz C. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect Immun 68:1304–1311. doi: 10.1128/IAI.68.3.1304-1311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abramoff MD, Magelhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int 11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.