Antigenic variation is a strategy used by many pathogens to escape host immune surveillance and establish persistent infections. This study successfully applies next-generation sequencing to study pilin antigenic variation in the human pathogen Neisseria meningitidis. This assay provides an affordable and efficient solution for quantifying antigenic variation frequency in mutant strains and for defining the recombination products of the process. We determined that there is a nonuniformity of silent donor copies used during meningococcus antigenic variation, and by the analysis of selected mutants deficient for specific recombination pathways, we show for the first time that the processes are conserved between N. meningitidis and Neisseria gonorrhoeae.
KEYWORDS: pilus, gene conversion, antigenic variation, 454 sequencing, diversity generation
ABSTRACT
Many pathogenic microbes evade host immune surveillance by varying the surface antigens, a process termed antigenic variation. While the process of pilin antigenic variation has been extensively studied in the human pathogen Neisseria gonorrhoeae (gonococcus [Gc]), relatively few studies of pilin antigenic variation have been conducted with Neisseria meningitidis (meningococcus [Mc]). Mc is usually a commensal organism that colonizes the human nasopharynx, but when it translocates to the bloodstream or meninges, it results in the severe and often deadly meningococcal disease. The type IV pili of Mc isolates play a critical role in host surface adherence, and its major pilin component (PilE) can undergo antigenic variation. In this study, Roche 454 pyrosequencing was used to examine the pilin antigenic variation of Mc strain 8013, as well as 8013 recA, recX, recQ, rep, and recJ mutants, Gc orthologues which have been shown to play a role in pilin antigenic variation. This study confirms that the Mc recA, rep, and recJ genes are essential for pilin antigenic variation. While the Mc recQ and recX gene products contribute to normal frequencies of antigenic variation, the loss of these factors does not alter the types of pilin variants produced. Overall, this study shows that the mechanisms of pilin antigenic variation are conserved between Gc and Mc.
IMPORTANCE Antigenic variation is a strategy used by many pathogens to escape host immune surveillance and establish persistent infections. This study successfully applies next-generation sequencing to study pilin antigenic variation in the human pathogen Neisseria meningitidis. This assay provides an affordable and efficient solution for quantifying antigenic variation frequency in mutant strains and for defining the recombination products of the process. We determined that there is a nonuniformity of silent donor copies used during meningococcus antigenic variation, and by the analysis of selected mutants deficient for specific recombination pathways, we show for the first time that the processes are conserved between N. meningitidis and Neisseria gonorrhoeae.
INTRODUCTION
Many viruses, bacteria, and parasites can alter the expression or composition of their surface-exposed proteins to escape host immune surveillance, a process defined as antigenic variation (1). Antigenic variation is one of the most prevalent strategies that pathogens employ to establish persistent infections in immunocompetent hosts (2). A common mechanism of diversity generation is gene conversion, which involves a unidirectional recombination event that incorporates donor sequences into a homologous locus, while the donor sequence remains unchanged (3).
One of the best-studied antigenic variation systems is the pilin antigenic variation system of the pathogenic Neisseria, Neisseria gonorrhoeae (gonococcus [Gc]) and Neisseria meningitidis (meningococcus [Mc]) (4). Gc is an obligate human pathogen that colonizes the urogenital tract and is the sole causative agent of the sexually transmitted infection, gonorrhea. Mc is an obligate human resident that usually colonizes the nasopharynx asymptomatically but occasionally causes life-threating meningitis and sepsis. The emerging antibiotic resistance in Gc and the lethality of disseminated Mc infection make them serious public health threats (5).
Type IV pili (Tfp) are crucial virulence factors for pathogenic Neisseria and are important for adhesion, twitching motility, resistance to neutrophil killing, and DNA transformation (3, 6–8). Tfp are filamentous appendages expressed on the bacterial surface that are composed of thousands of copies of PilE subunits encoded by the pilE gene. Pilin antigenic variation occurs when variant DNA sequences from a silent pilS copy transfer to the expressed pilE locus, generating a pilin variant (9, 10). Gc has one expressed pilE gene and up to 19 silent copies distributed in four or five silent loci across the chromosome. Most Mc isolates have fewer of these pilS copies compared to Gc isolates, usually, four to eight copies contained in one pilS locus (11). These pilS copies lack a promoter and ribosome binding site, serving as reservoirs for the genetic information used for pilin antigenic variation (4). A subset of Mc isolates do not undergo pilin antigenic variation but instead have pilE genes that are more similar to those of commensal Neisseria (10, 11) and have altered glycosylation (12).
Many different approaches have been applied to study pilin antigenic variation in Gc isolates. One common approach used to estimate pilin antigenic variation in Gc isolates is the pilus-dependent colony morphology change (PDCMC) assay (13, 14). Some Gc pilin antigenic variation variants produce a nonpiliated (P−) colony phenotype due to poor expression of the variant or loss of the pilin protein (13, 15). In a Gc piliated (P+) colony, P− blebs emerge at the edges of P+ colonies over time, and their appearance can be used as a reproducible measure of pilin antigenic variation frequency (13). As long as the starting pilE sequences and the growth rates between the strains are the same, the frequencies of pilin antigenic variation can be compared between isogenic strains (4). In contrast, PDCMC cannot be used to estimate Mc pilin antigenic variation, as no P− blebs are observed arising in Mc colonies from pilin antigenic variation for unknown reasons. Traditional DNA sequencing has also been used to quantify the frequency and rate of Gc and Mc pilin antigenic variation (10, 16). Generally, these studies have utilized an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible recA allele that was engineered at the endogenous recA to control when pilin antigenic variation can occur. To analyze pilin variation, individual colonies are grown on solid medium with IPTG to allow for pilin antigenic variation, and individual progeny from those progenitors are subsequently grown on solid medium without IPTG to lock in the resultant changes in the pilE genes of individual progeny. Sequencing of pilE in progeny colonies provides a measure of the relative level of antigenic variation and the types of recombinants produced (16). The traditional sequencing assay has the advantage of analyzing P+ or P− variants, but the assay is labor intensive and expensive and is not sensitive enough to measure pilin variation in mutants with a reduced frequency. Therefore, next-generation sequencing technology (NGS) has been applied to measure the frequency of pilin antigenic variation in populations of Gc and Mc isolates (17, 18).
Gc pilin antigenic variation is mediated via a RecF-like homologous recombination pathway, which can repair single-strand gaps or double-strand breaks (19). Proteins that affect the frequency of Gc pilin antigenic variation include enzymes involved in general DNA recombination or DNA repair pathways. RecA, a recombinase that generally mediates strand exchange, coprotease activity, and G4 structure binding, is required for pilin antigenic variation (20). RecOR is a recombinase that assists in RecA-mediated strand exchange when gapped substrates are used (21) and is also essential for pilin antigenic variation (13, 19). Other factors involved in pilin antigenic variation include RecX, RdgC, and RecG. RecG is a helicase that processes Holliday junctions. RecX has been shown to antagonize RecA and mediates efficient recombination. RdgC has been shown to bind DNA and inhibit RecA strand exchange (13, 14, 19, 22–25). While RuvA, RuVB, and RuvC were reported to be involved in mediating Gc pilin antigenic variation, we have shown that this phenotype was due to the slower growth of mutants and not a direct reduction in pilin antigenic variation (18). In this study, we tested whether the pathways used by Mc isolates were the same as by Gc by analyzing pilin antigenic variation in Mc strain 8013 recA, recX, rep, recJ, and recQ loss-of-function mutants.
An analysis of the pilE variants produced in Mc8013 showed that mutations that lowered the frequency (recX and recQ) produced similar patterns of variants to the parental strain. These analyses also show that silent pilS copy 3 is the dominant donor sequence and silent copy 5 is used infrequently. This work shows for the first time that Gc and Mc isolates use similar recombination pathways for pilin antigenic variation and demonstrates the utility of NGS to analyze Mc pilin antigenic variation.
RESULTS
Phenotypic analysis of Mc8013 recA, recJ, rep, recX, and recQ mutants.
We picked the serogroup C strain Mc8013 for this study as it has been sequenced, it is relatively easy to transform with Gc DNA, and our group has been vaccinated against serogroup C Mc. To test whether Mc uses similar recombination pathways as Gc for pilin antigenic variation, the orthologues of recA, recJ, rep, recX, and recQ were identified in Mc8013 via NCBI blast, and the genes with the highest homology were selected for further analysis. Mc8013 genes NMV_0941, NMV_0907, NMV_0300, NMV_1567, and NMV_0939 each share more than 95% protein sequence identity with the corresponding Gc genes and were designated Mc recA, recX, recQ, recJ, and rep, respectively.
To study the function of Mc recA, recX, recQ, recJ, and rep genes, loss-of-function mutants were constructed in strain Mc8013 recA6. DNA transformation assays using spectinomycin (Spc)-resistant chromosomal DNA revealed that the recX, recJ, recQ, and rep mutants exhibited levels of DNA transformation efficiency similar to that of the parental strain, while the recA mutant exhibited an approximately 100-fold decrease in DNA transformation efficiency (Fig. 1A). The few CFU detected in the recA mutant are likely due to the spontaneous antibiotic resistance, as no DNA control for this mutant showed a similar background level (data not shown). The UV survival assay was performed to measure the DNA repair capabilities of the mutants. The Mc recA and recJ mutants were the most sensitive to the UV exposure, while the recQ mutant had an intermediate phenotype (Fig. 1B). The recX and rep mutants, on the other hand, exhibited wild-type level sensitivities to UV treatment (Fig. 1B). The nalidixic acid (Nal) survival assay was also performed to measure the sensitivity of the mutants to double-strand breaks induced by the inhibition of gyrase (22). The recJ, rep, and recA mutants showed decreased survival to Nal. The recX and recQ mutants exhibited slightly reduced survival to Nal exposure that was not statistically significant (Fig. 1C).
FIG 1.
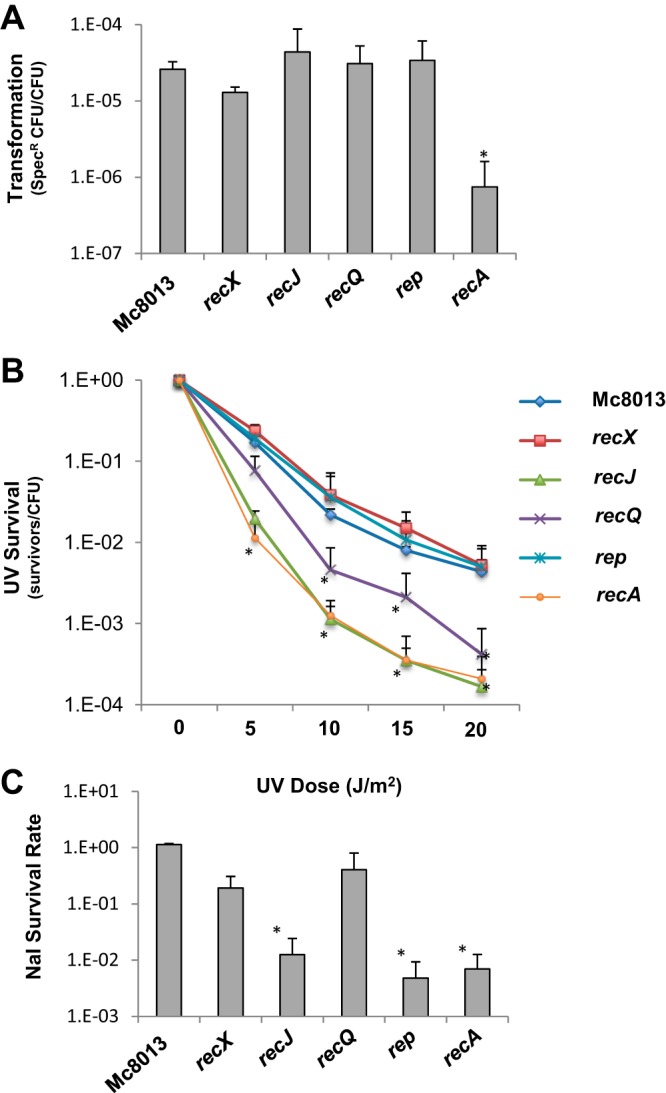
DNA transformation and DNA repair of Mc8013 mutants. (A) DNA transformation efficiency of Mc8013, recX, recJ, recQ, rep, and recA mutants. (B) UV survival rates of Mc8013, recX, recJ, recQ, rep, and recA mutants. (C) Nalidixic acid survival rates of Mc8013, recX, recJ, recQ, rep, and recA mutants. Error bars represent the standard deviations from three independent experiments. *, P < 0.05 by the Student t test relative to parental Mc8013.
Growth of the strains for analysis of pilin antigenic variation.
As the growth rates might affect the frequency of pilin antigenic variation, a time course was conducted measuring CFU per colonies of the Mc8013 mutants growing on solid medium. For the Mc8013 parent strain, recQ, recX, rep, and recJ mutants, IPTG was added to the solid medium to induce the expression of recA. For the recA-deficient strain, no IPTG was added to the solid medium (26). Each Mc strain was grown to 1 × 107 CFU/colony on solid medium for the following times: Mc8013 parent strain, 11 h; recA mutant, 15 h; recQ mutant, 11 h; recX mutant, 13 h; rep mutant, 15 h; and recJ mutant, 13 h. For 1 × 108 CFU/colony, strains were grown for 17 h (Mc8013, rep) and 21 h (recA, recQ, recX, and recJ mutants).
Measuring the frequency of pilin antigenic variation by next-generation sequencing.
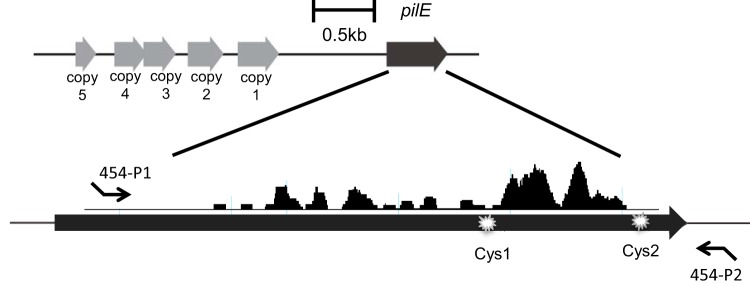
Roche 454 pyrosequencing was used to analyze the frequency of pilE antigenic variation of strain Mc8013 and the recJ, rep, recX, and recQ mutants. The reason that 454 sequencing was chosen over other NGS approaches is that the optimal size range to obtain high-quality reads for 454 sequencing is 200 to 600 bp (it covers the length of the pilE PCR product), while the optimal size range for ion torrent sequencing and Illumina sequencing is significant shorter. Mc8013 has one expressed pilE gene and five tandemly arranged variant silent copies upstream of pilE (Fig. 2). To amplify the majority of pilE variants, the forward PCR primer was designed to bind to the conserved pilE region and the reverse primer was designed to bind to the sequence 33 bp after the stop codon of pilE (Fig. 2). Different multiplex identifiers (MIDs) were designed into the forward primers to identify each strain. The resultant PCR products had an appropriate length of 554 bp (including the primers), excluding any changes in length mediated by variation of the donor silent copy.
FIG 2.
Gene organization of pilE and upstream silent copies in Mc8013. The five gray arrows indicate pilin silent copies 1 to 5. The black arrow represents the expressed pilE gene in Mc8013. The silent copies and pilE share significant DNA homology and their level of discrepancy is indicated by black peaks when aligned (generated by DNAstar SeqMan Pro). Cys1 and Cys2 represent the two conserved cysteine amino acids in the pilE gene. The binding sites of the 454 primers are represented by small black arrows, with the 454-P1 primer binding to the conserved region of pilE and the 454-P2 primer binding to the region 33 bp after the stop codon of pilE.
The 454 sequencing reads were analyzed to determine the pilin antigenic variation frequency for each strain. The sequence reads were first aligned to the pilE starting sequence by the software DNAstar SeqMan_Ngen_12 module, and all pilE sequence variants were identified. While the inherent error rate of 454 pyrosequencing resulted in a low level of sequencing errors, including nucleotide mutations, deletions, and additions, pilE antigenic variants were identified when the altered sequence matched one or more silent copy sequence. The frequency of pilin antigenic variation was calculated by dividing the number of sequence variants by the total number of pilE sequences. The pilin antigenic variation frequency for the Mc8013 parent strain was 2.74% when grown on solid plates for 17 h (1 × 108 CFU per colony) and 1.39% when grown for 11 h (1 × 107 CFU per colony) (Table 1). In both cases, the antigenic variation frequency of recX mutants was reduced to approximately 20% and the recQ mutant to approximately 4% of the parental strain, showing that as in Gc, these genes are not essential for the recombination process but aid in the efficiency. In contrast, the recA, rep, and recJ mutants were totally deficient in pilin antigenic variation (Table 1). These data confirm that the recX and recQ genes are important for Mc pilin antigenic variation, while recA, rep, and recJ genes are required for Mc pilin antigenic variation, which are similar phenotypes to the same mutants in Gc.
TABLE 1.
Pilin antigenic variation frequencies of Mc8013 strains
| Growth (CFU/colony)a | Antigenic variation frequency (%)b of strain: |
|||||
|---|---|---|---|---|---|---|
| Mc8013 | recA | recX | recQ | rep | recJ | |
| 107c | 1.39 | NDd | 0.22 | 0.04 | ND | ND |
| 108e | 2.74 | 0.02 | 0.54 | 0.12 | 0.01 | 0.03 |
Strains were grown on solid medium.
Antigenic variation frequency was calculated by the number of pilE variant reads divided by the total read number of each strain. All mutants were statistically different from Mc8013 parental strain (P < 0.05) using the two-tailed test of population proportion.
Mc8013 and recQ strains were grown for 11 h, recX and recJ strains were grown for 13 h, and the recA and rep strains were grown for 15 h.
ND, no variant reads detected.
Mc8013 and rep strains were grown for 17 h, and recA, recQ, recX, and recJ strains were grown for 21 h.
Determination of the donor silent-copy contribution to pilin antigenic variation.
During Gc pilin antigenic variation, silent copies at the same or different loci are not equally incorporated into the expressed pilE gene (16, 27). We examined whether the use of silent-copy donors in Mc isolates is also nonrandom by analyzing the donor silent-copy profiles in Mc8013 pilin variants. We also tested if RecX or RecQ contributed to silent-copy incorporation in the mutant variants. Among the 132 pilE variants that were identified in the Mc8013 parent strain, silent copy 3 was the most frequent donor (63 variants), followed by silent copy 4 (24 variants), silent copy 2 (21 variants), silent copy 1 (14 variants), and silent copy 5 (1 variant) (Fig. 3). Silent copy 3 shares the longest region of continuous homology with the pilE sequence (∼531 bp), while silent copy 5 is the shortest copy (∼210 bp) (Fig. 4). Shared homology may contribute to the selection of silent copies as donors. Eight pilE variants were identified with a single nucleotide change that could have originated from different donor silent copies that we presume are minimal antigenic variation events (Fig. 3). One pilE variant was identified as a hybrid with donor sequences that originated from both silent copy 3 and copy 4 (Fig. 3). The silent-copy profiles were also analyzed in recX and recQ mutants, and while the numbers are small, a similar pattern of donor preference was also observed in these mutants with reduced frequencies of variation (Fig. 3). These results are consistent with the conclusions that silent-copy incorporation into pilE is not random and that RecX and RecQ are not involved in the process of antigenic variation in Mc isolates.
FIG 3.
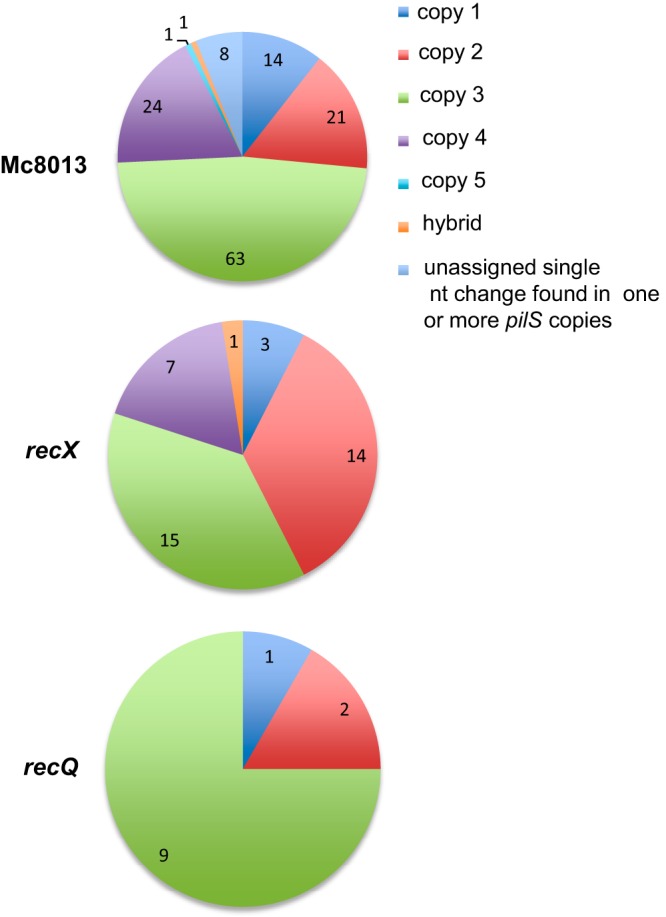
The profile of donor silent copies incorporated into the pilE gene. The pie chart represents the numbers of pilE variants with donor sequences from a specific silent copy, hybrid silent copies, or an unassigned single nucleotide change. The three pie charts show the donor silent copy profile for Mc8013, the recX mutant, and the recQ mutant.
FIG 4.
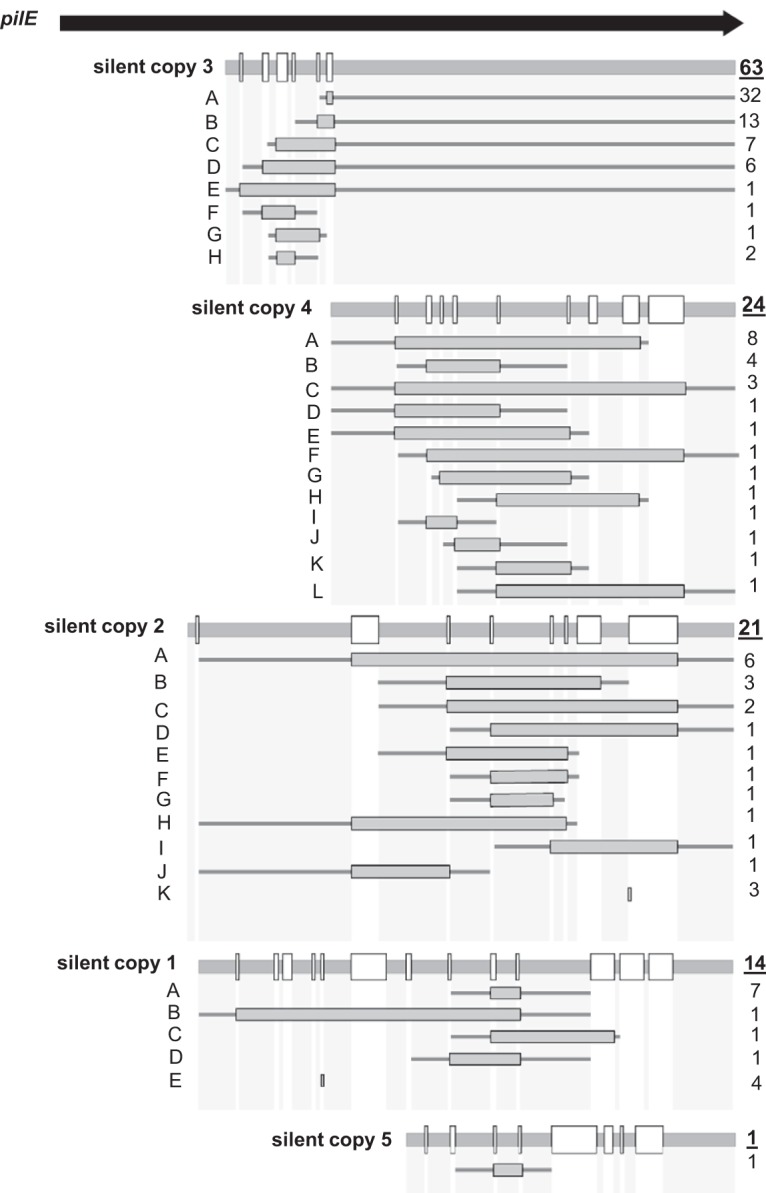
Detailed analysis of pilE variants in Mc8013. The pilE gene is represented by the arrow at the top. For each silent donor copy, the level of sequence conservation with the starting pilE sequence is shown with sequences that are identical with pilE by the thick gray horizontal line and sequences that differ between the pilS copy and pilE represented by white boxes. The frequency of each recombination event is indicated on the right of each cartoon where the underlined number on top indicates the total number of events using the same silent copy donor. For each recombination event, the frequency of individual recombination events is indicated on the right of each cartoon, consisting of a gray box that indicates the extent of the transferred silent copy sequence in pilE and thin gray lines that indicate flanking sequences shared by pilE and the silent copy for each product.
To characterize the products of Mc pilin antigenic variation, each individual pilE recombination event from the Mc8013 parental strain was defined (Fig. 4). The number of times each specific recombination event was found and, for each recombination event, the flanking regions shared by pilE and the silent copy, as well as the region that recombined, were identified and mapped (Fig. 4). The most frequent antigenic variation event (26% of all variants) was a three-nucleotide (CCA) insertion into pilE with the donor sequence originating from silent copy 3 (Fig. 4, silent copy 3). Multiple recombination products were detected that received sequences from each of the five donor loci, showing that even with only five silent loci, the possible pilin diversity generated in Mc isolates is much more than just the number of donor silent copies. Interestingly, none of the recombination events introduced presumptive premature stop codons or a pilE truncation, as has been reported previously for Gc isolates (16, 18).
DISCUSSION
Pilin antigenic variation is an important strategy for pathogenic Neisseria species to escape host immune surveillance and alter the expression and possibly functions of the type IV pilus. While Gc pilin antigenic variation can be measured using the surrogate assay of colony phase variation (13), this approach is not applicable to Mc, as Mc isolates do not have distinguishable, pilus-dependent colony variation phenotypes. Traditional Sanger sequencing assays for Gc or Mc pilin antigenic variation involves the amplification of the pilE genes from hundreds of individual colonies (10, 28). This assay is labor intensive and not ideal for measuring Mc mutants with decreased pilin antigenic variation frequency. An alternate approach used Illumina sequencing to investigate the pilin antigenic variation in Neisseria species (17). However, due to the short reads of the Illumina platform, this approach does not identify individual pilE variants but only characterizes the overall population of the recombinants. Roche 454 targeted sequencing of the Mc pilE enabled us to identify the pilE variants through one strand sequence analysis that covered the entire pilE variable region. Moreover, the pilE PCR products of different strains were barcoded with unique MIDs and pooled in a library, thus enabling the measurement of the pilin antigenic variation frequencies of multiple strains at the same time. This procedure enables the detection of Mc pilE variation when the frequency is lower than 1%. This study shows it is feasible to measure Mc pilin antigenic variation in a way that determines both the frequency and the characteristics of the recombination reactions. Unfortunately, Roche 454 sequencing technology is no longer being supported, and its availability as a sequencing platform is rapidly disappearing. To continue to use this type of deep sequencing, one would have to use Illumina sequencing, which does not provide individual variants (17), or adopt PacBio or Nanopore sequencing in a way that overcomes the intrinsic error rate of these platforms.
There are many factors that have been shown to be involved in Gc pilin variation, including RecA, RecX, RecJ, RecQ, Rep, RecO, and RecR (reviewed in references 4 and 25). Inactivation of the genes encoding these proteins resulted in different Gc frequencies of pilin antigenic variation. This study showed that the inactivation of recA lowered the transformation efficiency, highlighting a role for RecA in DNA uptake and incorporation. Mutations of recA, recJ, and recQ all lowered survival after UV radiation, implicating these genes in pyrimidine dimer repair. RecA, RecJ, and Rep are involved in double-strand break repair. This study confirms that the orthologues of recA, recX, recJ, recQ, and rep gene products also are involved in Mc pilin antigenic variation, showing that the mechanism is conserved between these related species.
During the course of pilin antigenic variation of Neisseria species, it is important to know whether each silent copy has an equal chance to be incorporated into the pilE gene. During pilin antigenic variation of Gc isolates, silent copies at the same or different loci are not equally incorporated into the expressed pilE gene (16). Our data show that the use of donor silent copies is also nonuniform in Mc strain 8013; in the variants analyzed, silent copy 3 was the dominant donor sequence, while silent copy 5 was represented the least. A further examination of silent copies revealed that silent copy 3 shares the longest homologous region with the parental pilE sequence (∼531 bp on one side only), and silent copy 5 is the shortest silent copy (∼210 bp) (Fig. 4). These factors might explain the frequent use of copy 3 as a donor and the low use of copy 5. However, stretches of sequence homology and the size of the silent copy do not correlate with the relative donor frequency of the other silent copies (Fig. 4). Neither relative homology nor size explains the nonuniform use of Gc silent donor copies (16). Therefore, there may be mechanisms of silent copy usage that pertain to specific silent copies and are not generalizable to other silent copies in Mc isolates. Taken together, our study successfully applied next-generation sequencing to investigate Mc pilin antigenic variation. It confirmed the critical roles of recA, recX, recQ, recJ, and rep in Mc pilin antigenic variation and revealed the nonuniformity of silent copy incorporation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains, plasmids, and oligonucleotides used in this study are described in Tables 2 and 3. Escherichia coli TOP10 competent cells (Invitrogen) were grown in Luria-Bertani (LB) broth or on LB solid medium containing 15 g/liter agar at 37°C and used to propagate plasmids. The Mc strains used in this study were primarily derivatives of Mc strain 8013 (Mc8013) (Table 2). All meningococcal strains used in this study contained the IPTG-inducible recA6 allele that replaced the original recA gene to enable control of pilE antigenic variation (26). All Mc strains contained the same starting pilE sequence as the published Mc8013 pilE gene (identification [ID] NMV_0019).
TABLE 2.
Strain and plasmid descriptions and sources
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| N. meningitidis strains | ||
| 8013 | Mc serogroup C parental strain | 33 |
| 8013 recA6 | Contains an IPTG-inducible recA gene | This study |
| 8013 recQ::erm recA6 | The recQ gene is replaced by the erm antibiotic resistant cassette | This study |
| 8013 recJ::kan recA6 | The recJ gene is replaced by the kan antibiotic resistant cassette | This study |
| 8013 recX::erm recA6 | The recX gene is replaced by the erm antibiotic resistant cassette | This study |
| 8013 rep::kan recA6 | The rep gene is replaced by the kan antibiotic resistant cassette | This study |
| 8013 Spr recA6 | Spontaneous Spc-resistant mutant, used for DNA transformation assay | This study |
| Gc FA1090 recQ::erm | The Gc recQ gene is insertionally disrupted by Erm | 14 |
| Plasmids | ||
| precXKOerm | Plasmid to knock out recX gene, Ermr | 31 |
| precJ::Kan | Plasmid to knock out recJ gene, Kanr | This study |
| prep::Kan | Plasmid to knock out rep gene, Kanr | This study |
| precQ::erm | Plasmid to knock out recQ gene, Ermr | This study |
TABLE 3.
Oligonucleotide sequences and descriptions
| Oligonucleotide name | Sequencea | Description |
|---|---|---|
| ΔrecJ-P1 | GGTTCGTCGTCAATCCTGAA | Primer to interrupt Nm8013 recJ by replacing recJ with kan |
| ΔrecJ-P2 | GCAGGTCGACTCTAGAGGATCAACGTGGAACTTTACAGTGCGGGT | Primer to interrupt Nm8013 recJ by replacing recJ with kan |
| ΔrecJ-P3(Kan) | ACCCGCACTGTAAAGTTCCACGTTGATCCTCTAGAGTCGACCTGC | Primer to interrupt Nm8013 recJ by replacing recJ with kan |
| ΔrecJ-P4(Kan) | CGCTTCCCAGTAGTCGATATAAAGTCAGAAGAACTCGTCAAGAAGGCG | Primer to interrupt Nm8013 recJ by replacing recJ with kan |
| ΔrecJ-P5 | CGCCTTCTTGACGAGTTCTTCTGACTTTATATCGACTACTGGGAAGCG | Primer to interrupt Nm8013 recJ by replacing recJ with kan |
| ΔrecJ-P6 | CCGACCACATAAGCCTGAAA | Primer to interrupt Nm8013 recJ by replacing recJ with kan |
| Δrep-P1 | CCGTGTTCGGACGGTTATTA | Primer to interrupt Nm8013 rep by replacing recJ with kan |
| Δrep-P2 | GCAGGTCGACTCTAGAGGATCGTTTTGATGCCGTCTGAAATC | Primer to interrupt Nm8013 rep by replacing recJ with kan |
| Δrep-P3(Kan) | GATTTCAGACGGCATCAAAACGATCCTCTAGAGTCGACCTGC | Primer to interrupt Nm8013 rep by replacing recJ with kan |
| Δrep-P4(Kan) | CATTGCGGCTCCGGTTTAATCTCAGAAGAACTCGTCAAGAAGGCG | Primer to interrupt Nm8013 rep by replacing recJ with kan |
| Δrep-P5 | CGCCTTCTTGACGAGTTCTTCTGAGATTAAACCGGAGCCGCAATG | Primer to interrupt Nm8013 rep by replacing recJ with kan |
| Δrep-P6 | CGGCGAACTCTTAAACCATTTC | Primer to interrupt Nm8013 rep by replacing recJ with kan |
| ΔrecQ-P1 | GGGACAATTGGAACGCTTTG | Forward primer to amplify the recQ::erm region from the FA1090 recQ mutant |
| ΔrecQ-P2 | GCAACACCCAATCGTTCAAG | Reverse primer to amplify the recQ::erm region from the FA1090 recQ mutant |
| ΔrecJ-colony PCR1 | ACCCGCACTGTAAAGTTCCACGTT | Forward primer to verify recJ interruption |
| ΔrecJ-colony PCR2 | CGCTTCCCAGTAGTCGATATAAAG | Reverse primer to verify recJ interruption |
| Δrep-colony PCR1 | GATTTCAGACGGCATCAAAAC | Forward primer to verify rep interruption |
| Δrep-colony PCR2 | CATTGCGGCTCCGGTTTAATC | Reverse primer to verify rep interruption |
| 454-8013-P1 | CCATCTCATCCCTGCGTGTCTCCGACTCAGACGAGTGCGTTCGCCCTTCCTGCTTATCAA | Forward primer to amplify the pilE region for 454 sequencing, with MID for Nm8013 parent strain grown on solid medium for 11 h |
| 454-P2 | CCTATCCCCTGTGTGCCTTGGCAGTCTCAGCGTGGAAAATCACTTACCGC | Reverse primer to amplify the pilE region for 454 sequencing |
| 454-recA-P1 | CCATCTCATCCCTGCGTGTCTCCGACTCAGAGTATACATATCGCCCTTCCTGCTTATCAA | Forward primer to amplify the pilE region for 454 sequencing, with MID for Nm8013 recA mutant grown on solid medium for 15 h |
| 454-recX-P1 | CCATCTCATCCCTGCGTGTCTCCGACTCAGCGTCTAGTACTCGCCCTTCCTGCTTATCAA | Forward primer to amplify the pilE region for 454 sequencing, with MID for Nm8013 recX mutant grown on solid medium for 13 h |
| 454-recQ-P1 | CCATCTCATCCCTGCGTGTCTCCGACTCAGAGCACTGTAGTCGCCCTTCCTGCTTATCAA | Forward primer to amplify the pilE region for 454 sequencing, with MID for Nm8013 recQ mutant grown on solid medium for 11 h |
| 454-recJ-P1 | CCATCTCATCCCTGCGTGTCTCCGACTCAGTGTACTACTCTCGCCCTTCCTGCTTATCAA | Forward primer to amplify the pilE region for 454 sequencing, with MID for Nm8013 recJ mutant grown on solid medium for 13 h |
| 454-rep-P1 | CCATCTCATCCCTGCGTGTCTCCGACTCAGTACGAGTATGTCGCCCTTCCTGCTTATCAA | Forward primer to amplify the pilE region for 454 sequencing, with MID for Nm8013 rep mutant grown on solid medium for 15 h |
| 454-8013-P1-17h | CCATCTCATCCCTGCGTGTCTCCGACTCAGACGACAGCTCTCGCCCTTCCTGCTTATCAA | Forward primer to amplify the pilE region for 454 sequencing, with MID for Nm8013 parent strain grown on solid media for 17 h |
| 454-recA-P1-21h | CCATCTCATCCCTGCGTGTCTCCGACTCAGTAGCTCTATCTCGCCCTTCCTGCTTATCAA | Forward primer to amplify the pilE region for 454 sequencing, with MID for Nm8013 recA mutant grown on solid medium for 21 h |
| 454-recX-P1-21h | CCATCTCATCCCTGCGTGTCTCCGACTCAGACTCATCTACTCGCCCTTCCTGCTTATCAA | Forward primer to amplify the pilE region for 454 sequencing, with MID for Nm8013 recX mutant grown on solid medium for 21 h |
| 454-recQ-P1-21h | CCATCTCATCCCTGCGTGTCTCCGACTCAGTGATGTGTACTCGCCCTTCCTGCTTATCAA | Forward primer to amplify the pilE region for 454 sequencing, with MID for Nm8013 recQ mutant grown on solid medium for 21 h |
| 454-recJ-P1-21h | CCATCTCATCCCTGCGTGTCTCCGACTCAGAGAGCGTCACTCGCCCTTCCTGCTTATCAA | Forward primer to amplify the pilE region for 454 sequencing, with MID for Nm8013 recJ mutant grown on solid medium for 21 h |
| 454-rep-P1-17h | CCATCTCATCCCTGCGTGTCTCCGACTCAGAGCGACTAGCTCGCCCTTCCTGCTTATCAA | Forward primer to amplify the pilE region for 454 sequencing, with MID for Nm8013 rep mutant grown on solid medium for 17 h |
The MID for each sample is underlined.
Mc strains were grown on GC medium base (Difco) plus Kellogg supplements [22.2 mM glucose, 0.68 mM glutamine, 0.45 mM cocarboxylase, 1.23 mM Fe(NO3)3; all from Sigma] (7) at 37°C with 5% CO2 for 11 to 21 h. For the transformation assays, Mc strains were grown in Gc liquid medium (GCBL; 1.5% protease peptone no. 3 [Difco], 0.4% K2HPO4 [Fisher], 0.1% KH2PO4 [Fisher], 0.1% NaCl [Fisher]) with Kellogg supplements and 0.042% sodium bicarbonate (Sigma) at 37°C. Antibiotics were added at the following concentrations for Mc: kanamycin (Kan), 50 μg/ml; erythromycin (Erm), 2 μg/ml; tetracycline (Tet), 0.2 μg/ml; and spectinomycin (Spc), 50 μg/ml. For E. coli, the concentrations were Kan, 50 μg/ml; Erm, 250 μg/ml; and Tet, 20 μg/ml. IPTG (1 mM; Diagnostic Chemicals) was added to solid medium when necessary.
Construction of an IPTG-inducible recA6 strain in Mc strain 8013.
The native recA gene of Mc8013 was replaced with the Gc FA1090 recA6 construct by transforming with Gc chromosomal DNA selecting for the Tet marker associated with the recA6 allele (26). The recA6 strains are RecA+ when IPTG is provided in the medium and are RecA deficient when grown without IPTG (26).
Insertional inactivation of recJ, rep, recX, and recQ in Mc8013 recA6.
To mutate recJ and rep, a Kan cassette was used to replace the majority of each gene using the SOEing PCR approach (29). The primers listed in Table 3 and Phusion Hot Start Flex DNA polymerase (NEB) was used for all PCRs in this study; a standard PCR protocol was followed. Two sets of primers were used to amplify the regions adjacent to the genes from the Mc8013 chromosomal DNA template. Complementary primers were used to amplify the kanamycin cassette from plasmid pBSL86 (ATCC). The three PCR products were gel purified and used as the template in another PCR using the two flanking gene-specific primers, resulting in the Kan marker flanked by sequences adjacent to the deleted gene. The correct-sized PCR product was isolated and cloned into a pSMART vector (Lucigen). After confirmation by DNA sequence analysis, each mutation was transformed into Mc8013 recA6 by liquid transformation as described previously (22), selected on medium containing Kan and verified by colony PCR and sequencing.
To mutate the Mc8013 recQ and recX genes, flanking primers (Table 3) were used to amplify the mutated gene from Gc FA1090 (recQ::erm [30] and recX::erm [31]). Each PCR product was cloned into a pSMART vector (Lucigen), and the proper construct was confirmed by DNA sequence analysis and transformed into Mc8013 recA6 by liquid transformation as described previously (22). The final strain was selected on medium containing Erm and verified by colony PCR and sequencing.
Determination of growth on solid medium.
Measurements of growth were performed on solid medium as previously described (22). Briefly, a piece of sterile filter paper was used to pick five colonies after growth on GCB agar containing IPTG for 11 to 21 h, the cells were released from the filter paper into GCBL by vortexing, and serial dilutions were plated to determine CFU/colony.
Liquid transformation assay.
Mc transformation efficiency was measured as previously described (32). Briefly, Mc strains were grown for 18 h on GCB plates and resuspended in liquid transformation medium (LTM; 1 mM IPTG, 1.5% protease peptone no. 3 [Difco], 0.1% NaCl, 200 mM HEPES [Sigma], 5 mM MgSO4, and Kellogg supplements I and II [pH 7.2]) to an optical density at 600 nm of approximately 1.5. Thirty microliters of the cell suspension was added to tubes containing 150 ng Spcr Mc chromosomal DNA and 200 μl LTM. Following incubation at 37°C for 20 min, transformation solutions were added to prewarmed 2 ml LTM and incubated at 37°C in the presence of 5% CO2 for 4 h. The mixtures were serially diluted 10-fold in GCBL, and 20-μl serial 10-fold dilutions were spotted on GCB plates in the presence and absence of Spcr. The transformation efficiencies are reported as antibiotic-resistant CFU divided by total CFU and are the means from three replicates.
UV survival assay.
UV survival assays were performed as described previously (22). Briefly, Mc bacterial lawns were grown for 24 h on GCB agar supplemented with IPTG when appropriate, collected with a Dacron swab, and resuspended in 1 ml of GCBL. The cells were serially diluted and spotted onto GCB agar with IPTG when appropriate. Plates were exposed to 0, 5, 10, 15, or 20 mJ/m2 UV light (UV Stratalinker 1800; Stratagene) and incubated at 37°C in the presence of 5% CO2 overnight. The UV survival rate was calculated by dividing the number of CFU in the irradiated sample by the number of CFU in the nonirradiated sample. The assay was performed in duplicates from at least three different experiments.
Nalidixic acid survival assay.
Nal survival assays were performed as previously described (28). Bacterial lawns were grown for 18 h on GCB plates, swabbed, and resuspended in 1 ml of liquid medium. Cells were serially diluted and spotted onto GCB agar in the presence or absence of 0.5 μg/ml nalidixic acid (Nal). The survival rate was calculated by dividing the number of CFU surviving Nal treatment by the total number of CFU. The assay was performed in duplicates within at least three independent experiments.
Roche 454 pyrosequencing.
Mc strains were grown on fresh GCB plates for 18 h. For each strain, one colony was picked with a sterilized filter disk and dispersed in 500 μl GCBL. A 1-μl culture was diluted in 500 μl GCBL, and 30-, 40-, and 50-μl cultures were spread on fresh GCB plates with or without IPTG. Bacteria were grown to a cell density of 1 × 107 or 1 × 108 CFU per colonies as indicated. Approximately 300 colonies per strain were collected in 1.5-ml GCBL by Dacron swabs. The QIAmp DNA minikit (Qiagen) was used to isolate genomic DNA, according to the manufacturer's instructions. The pilE region for each strain was amplified using Phusion Hot Start Flex DNA polymerase (NEB) and 10 ng genomic DNA with 454-P1 primer and 454-P2 primer (Table 3). The primer design was based on 454 sequencing application brief no. 001-2009. Each 454-P1 primer contains a multiplex identifier (MID) as a unique identifier for samples, and MIDs were designed according to 454 sequencing technical bulletin no. 005-2009. DNA was quantified with a Nanodrop (ND-1000) and by gel densitometry (Bio-Rad Quantity One software). The final genomic DNA concentration for each sample was adjusted to approximately 20 ng/μl, and 2 μl per sample was pooled as a sequencing library. The 454 pyrosequencing was performed by the Microbiome core facility at the University of North Carolina. The library was sequenced on a Roche 454 Genome Sequencer FLX Titanium instrument using standard GS FLX Titanium XLR70 sequencing reagents and protocols. One-eighth of the plate was used for the sequencing, and 86,765 raw sequence reads were obtained.
ACKNOWLEDGMENTS
This work was supported by Wellcome Trust grant R24378/CN001 and NIH grant R37 AI033493 to H.S.S.
Christoph Tang and Vladimir Pelecic helped provide the basis for this work. We thank Ella Rotman, Kyle Obergfell, Lauren Priniski, and Linda Hu for critical reading of the manuscript.
REFERENCES
- 1.Palmer GH, Bankhead T, Seifert HS. 2016. Antigenic variation in bacterial pathogens. Microbiol Spectr 4:VMBF-0005-2015. doi: 10.1128/microbiolspec.VMBF-0005-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang FT, Yan J, Mbow ML, Sviat SL, Gilmore RD, Mamula M, Fikrig E. 2004. Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect Immun 72:5759–5767. doi: 10.1128/IAI.72.10.5759-5767.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obergfell KP, Seifert HS. 2015. Mobile DNA in the pathogenic Neisseria. Microbiol Spectr 3:MDNA3-0015-2014. doi: 10.1128/microbiolspec.MDNA3-0015-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rotman E, Seifert HS. 2014. The genetics of Neisseria species. Annu Rev Genet 48:405–431. doi: 10.1146/annurev-genet-120213-092007. [DOI] [PubMed] [Google Scholar]
- 5.Quillin SJ, Seifert HS. 2018. Neisseria gonorrhoeae host adaptation and pathogenesis. Nat Rev Microbiol 16:226–240. doi: 10.1038/nrmicro.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig L, Pique ME, Tainer JA. 2004. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 7.Kellogg DS Jr, Peacock WL Jr, Deacon WE, Brown L, Pirkle DI. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J Bacteriol 85:1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stohl EA, Dale EM, Criss AK, Seifert HS. 2013. Neisseria gonorrhoeae metalloprotease NGO1686 is required for full piliation, and piliation is required for resistance to H2O2- and neutrophil-mediated killing. mBio 4:e00399-13. doi: 10.1128/mBio.00399-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seifert HS. 1996. Questions about gonococcal pilus phase- and antigenic variation. Mol Microbiol 21:433–440. doi: 10.1111/j.1365-2958.1996.tb02552.x. [DOI] [PubMed] [Google Scholar]
- 10.Helm RA, Seifert HS. 2010. Frequency and rate of pilin antigenic variation of Neisseria meningitidis. J Bacteriol 192:3822–3823. doi: 10.1128/JB.00280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wörmann ME, Horien CL, Bennett JS, Jolley KA, Maiden MC, Tang CM, Aho EL, Exley RM. 2014. Sequence, distribution and chromosomal context of class I and class II pilin genes of Neisseria meningitidis identified in whole genome sequences. BMC Genomics 15:253. doi: 10.1186/1471-2164-15-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gault J, Ferber M, Machata S, Imhaus AF, Malosse C, Charles-Orszag A, Millien C, Bouvier G, Bardiaux B, Pehau-Arnaudet G, Klinge K, Podglajen I, Ploy MC, Seifert HS, Nilges M, Chamot-Rooke J, Dumenil G. 2015. Neisseria meningitidis type IV pili composed of sequence invariable pilins are masked by multisite glycosylation. PLoS Pathog 11:e1005162. doi: 10.1371/journal.ppat.1005162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sechman EV, Rohrer MS, Seifert HS. 2005. A genetic screen identifies genes and sites involved in pilin antigenic variation in Neisseria gonorrhoeae. Mol Microbiol 57:468–483. doi: 10.1111/j.1365-2958.2005.04657.x. [DOI] [PubMed] [Google Scholar]
- 14.Sechman EV, Kline KA, Seifert HS. 2006. Loss of both Holliday junction processing pathways is synthetically lethal in the presence of gonococcal pilin antigenic variation. Mol Microbiol 61:185–193. doi: 10.1111/j.1365-2958.2006.05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson J, Bergström S, Barrera O, Robbins K, Corwin D. 1985. Pilus-gonococcal variants. Evidence for multiple forms of piliation control. J Exp Med 162:729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Criss AK, Kline KA, Seifert HS. 2005. The frequency and rate of pilin antigenic variation in Neisseria gonorrhoeae. Mol Microbiol 58:510–519. doi: 10.1111/j.1365-2958.2005.04838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies JK, Harrison PF, Lin YH, Bartley S, Khoo CA, Seemann T, Ryan CS, Kahler CM, Hill SA. 2014. The use of high-throughput DNA sequencing in the investigation of antigenic variation: application to Neisseria species. PLoS One 9:e86704. doi: 10.1371/journal.pone.0086704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rotman E, Webber DM, Seifert HS. 2016. Analyzing Neisseria gonorrhoeae pilin antigenic variation using 454 sequencing technology. J Bacteriol 198:2470–2482. doi: 10.1128/JB.00330-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehr IJ, Seifert HS. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation, and DNA repair. Mol Microbiol 30:697–710. doi: 10.1046/j.1365-2958.1998.01089.x. [DOI] [PubMed] [Google Scholar]
- 20.Koomey M, Gotschlich EC, Robbins K, Bergstrom S, Swanson J. 1987. Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics 117:391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morimatsu K, Kowalczykowski SC. 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell 11:1337–1347. [DOI] [PubMed] [Google Scholar]
- 22.Stohl EA, Seifert HS. 2001. The recX gene potentiates homologous recombination in Neisseria gonorrhoeae. Mol Microbiol 40:1301–1310. doi: 10.1046/j.1365-2958.2001.02463.x. [DOI] [PubMed] [Google Scholar]
- 23.Mehr IJ, Long CD, Serkin CD, Seifert HS. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154:523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skaar EP, Lazio MP, Seifert HS. 2002. Roles of the recJ and recN genes in homologous recombination and DNA repair pathways of Neisseria gonorrhoeae. J Bacteriol 184:919–927. doi: 10.1128/jb.184.4.919-927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kline KA, Seifert HS. 2005. Role of the Rep helicase gene in homologous recombination in Neisseria gonorrhoeae. J Bacteriol 187:2903–2907. doi: 10.1128/JB.187.8.2903-2907.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seifert HS. 1997. Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene 188:215–220. doi: 10.1016/S0378-1119(96)00810-4. [DOI] [PubMed] [Google Scholar]
- 27.Cahoon LA, Seifert HS. 2009. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helm RA, Seifert HS. 2009. Pilin antigenic variation occurs independently of the RecBCD pathway in Neisseria gonorrhoeae. J Bacteriol 191:5613–5621. doi: 10.1128/JB.00535-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallejo AN, Pogulis RJ, Pease LR. 2008. PCR mutagenesis by overlap extension and gene SOE. CSH Protoc 2008:pdb prot4861. doi: 10.1101/pdb.prot4861. [DOI] [PubMed] [Google Scholar]
- 30.Cahoon LA, Manthei KA, Rotman E, Keck JL, Seifert HS. 2013. Neisseria gonorrhoeae RecQ helicase HRDC domains are essential for efficient binding and unwinding of the pilE guanine quartet structure required for pilin antigenic variation. J Bacteriol 195:2255–2261. doi: 10.1128/JB.02217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stohl EA, Brockman JP, Burkle KL, Morimatsu K, Kowalczykowski SC, Seifert HS. 2003. Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J Biol Chem 278:2278–2285. doi: 10.1074/jbc.M210496200. [DOI] [PubMed] [Google Scholar]
- 32.Duffin PM, Seifert HS. 2010. DNA uptake sequence-mediated enhancement of transformation in Neisseria gonorrhoeae is strain dependent. J Bacteriol 192:4436–4444. doi: 10.1128/JB.00442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nassif X, Puaoi D, So M. 1991. Transposition of Tn1545-Δ3 in the pathogenic neisseriae: a genetic tool for mutagenesis. J Bacteriol 173:2147–2154. doi: 10.1128/jb.173.7.2147-2154.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]