Abstract
Antimicrobial resistance (AMR) is a global problem hindering treatment of bacterial infections, rendering many aspects of modern medicine less effective. AMR genes (ARGs) are frequently located on plasmids, which are self-replicating elements of DNA. They are often transmissible between bacteria, and some have spread globally. Novel strategies to combat AMR are needed, and plasmid curing and anti-plasmid approaches could reduce ARG prevalence, and sensitise bacteria to antibiotics. We discuss the use of curing agents as laboratory tools including chemicals (e.g. detergents and intercalating agents), drugs used in medicine including ascorbic acid, psychotropic drugs (e.g. chlorpromazine), antibiotics (e.g. aminocoumarins, quinolones and rifampicin) and plant-derived compounds. Novel strategies are examined; these include conjugation inhibitors (e.g. TraE inhibitors, linoleic, oleic, 2-hexadecynoic and tanzawaic acids), systems designed around plasmid incompatibility, phages and CRISPR/Cas-based approaches. Currently, there is a general lack of in vivo curing options. This review highlights this important shortfall, which if filled could provide a promising mechanism to reduce ARG prevalence in humans and animals. Plasmid curing mechanisms which are not suitable for in vivo use could still prove important for reducing the global burden of AMR, as high levels of ARGs exist in the environment.
Keywords: antimicrobial resistance, plasmid, plasmid curing, CRISPR/Cas, antibiotics, conjugation inhibitors
Removing plasmids from bacteria in different ecosystems could be an important aspect of fighting antimicrobial resistance.
INTRODUCTION
One of the major threats facing society is the rise in number of antimicrobial-resistant (AMR) bacteria (O’Neill 2016). Antimicrobials underpin modern medicine; they are used to treat infections, to prevent infections (prophylaxis) during medical procedures (e.g. surgery) and they are crucial for patients with compromised immune function (Holmes et al.2016; Laxminarayan et al.2016). Between 2000 and 2010, global human use of antibiotics increased by 36%, and the use of two last-resort antibiotics, carbapenems and polymyxins, increased by 45% and 13%, respectively (Van Boeckel et al.2014). Antimicrobials have many non-human uses including in animals for growth promotion, veterinary treatment and aquaculture (Cabello 2006; Meek, Vyas and Piddock 2015; Van Boeckel et al.2015). In 2013, an estimated 131 109 tons of antimicrobials were used globally in food animals; by 2030 this is expected to increase to 200 235 tons (Van Boeckel et al.2017). However, there is a growing trend to improve antimicrobial stewardship in many countries. For example, in Switzerland veterinary antimicrobial sales increased between 2006 and 2008, but then steadily decreased, reaching a 26.2% reduction in 2013 (Carmo et al.2017). In addition to human and animal use, many cleaning and personal hygiene products contain biocides, such as triclosan, which can select for mutants resistant to biocides, and in some cases to antibiotics used in medicine (Meek, Vyas and Piddock 2015; Webber et al.2015, 2017).
A key factor that has led to the rise and global dissemination of multidrug-resistant (MDR) bacteria are mobile antimicrobial resistance genes (ARGs). These are frequently located on plasmids, which are pieces of usually circular, self-replicating DNA which can code for a variety of different functional gene groups. Aspects of plasmid biology have been extensively reviewed elsewhere, but, in brief, plasmids often include partitioning systems, toxin–antitoxin (TA) systems and conjugative/transmission systems (Van Melderen and Saavedra De Bast 2009; Pinto, Pappas and Winans 2012; Carattoli 2013; Baxter and Funnell 2014; Goessweiner-Mohr et al.2014; Kado 2014; MacLean and San Millan 2015; Ruiz-Maso et al.2015; Cabezon et al.2015; Ilangovan, Connery and Waksman 2015; Chan, Espinosa and Yeo 2016; Banuelos-Vazquez, Torres Tejerizo and Brom 2017; Hall et al.2017; Hulter et al.2017). Conjugation is mediated by type IV secretion coupled with a relaxosome complex to mediate DNA movement from one cell to another (Ilangovan, Connery and Waksman 2015).
Plasmids are frequently categorised based on incompatibility groups (Inc), defined as the inability of two related plasmids to be propagated stably in the same cell and may be due to competition for the same replication or segregation sites, or caused by repression of replication initiation (Novick 1987; Carattoli 2009). Reviews on incompatibility groups and plasmid classification can be found elsewhere (Novick 1987; Carattoli 2011; Shintani, Sanchez and Kimbara 2015; Orlek et al.2017). Plasmids that share the same mechanisms for replication or partitioning are placed in the same incompatibility groups. Plasmid incompatibility has been used to follow the movement and evolution of plasmids conferring AMR (Carattoli et al.2005).
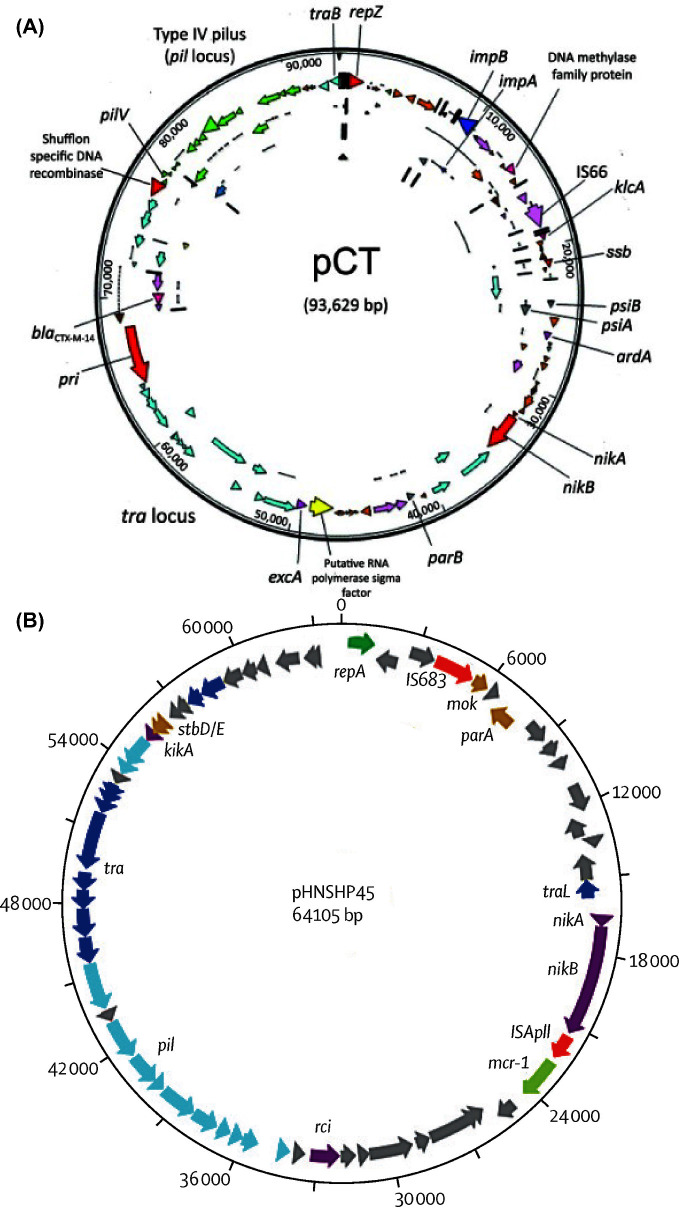
ARGs that pose a serious threat to human medicine are typically found in Gram-negative bacteria. These include genes coding for extended spectrum β-lactamases (ESBL) (e.g. CTX-M), carbapenemases (e.g. KPC, NDM and OXA-58) (Holmes et al.2016) and colistin resistance (e.g. MCR-1) (Liu et al.2016). The issues surrounding AMR plasmids are derived in part by their substantial complexity. Plasmids often display a high degree of plasticity, with frequent insertions, deletions and rearrangements of DNA including changes to specific ARGs (Kado 2014). For example, the blaCTX-M gene is highly variable, and the CTX-M family of ESBLs are commonly coded for by multiple different plasmids, such as pCT (Fig. 1A) (Cottell et al.2011, Bevan, Jones and Hawkey 2017). According to the Beta-Lactamase DataBase, 207 variants of blaCTX-M have been identified (accessed on 11 May 2018) (Naas et al.2017). Another example of a plasmid-mediated ARG is the mcr-1 gene, first identified on a transmissible plasmid, pHNSHP45, in 2016 (Fig. 1b) (Liu et al.2016). Since then mcr-1 and variants of this gene have been identified on multiple plasmid backbones and host strains. Of concern are isolates carrying colistin and carbapenem ARGs, as few treatment options would remain for infections caused by such bacteria (Lai et al.2017; Wang et al.2017; Zhou et al.2017). In addition to these examples, plasmids can carry a variety of other resistance genes, including qnr variants, aac(6΄)-lb-cr and plasmid-mediated efflux pump genes such as oqxAB and qepA, which confer low levels of resistance to quinolone antimicrobials (Jacoby, Strahilevitz and Hooper 2014). Increasingly, research should focus on ARGs which are frequently mobilised and transmit between bacteria (Crofts, Gasparrini and Dantas 2017).
Figure 1.
Organisation of two antibiotic resistance plasmids. (A) pCTCTX-M (IncK). Brown, pseudogenes; orange, hypothetic proteins; light pink, insertion sequences; light blue, tra locus; green, pil locus; dark pink, antimicrobial drug resistance gene; yellow, putative sigma factor; red, replication-associated genes. Arrows show the direction of transcription. Reproduced with permission from Cottell et al. (2011). (B) pHNSHP45mcr-1. Light blue, type IV pilus; dark blue, transfer region; yellow, plasmid stability; dark green, plasmid replication; red, insertion sequence; light green, antimicrobial resistance; purple, other proteins; grey, hypothetical proteins. Reproduced with permission from Liu et al. (2016).
In the European Union, resistance to carbapenem antibiotics in invasive Klebsiella pneumoniae isolates ranges from 66.9% (Greece), 33.9% (Italy), 2.1% (Spain) to <5% (Northern Europe) (ECDC 2016). For invasive E. coli infections, resistance to third-generation cephalosporins ranges from 5% in Iceland to 50% in Italy, Slovakia and Bulgaria, while carbapenem resistance in E. coli is <1% for most of the EU and between 1–5% for Romania (ECDC 2016). A study of travellers returning to the Netherlands found 30.5% of participants had ESBLs in their bacterial flora, while only 8.6% had ESBLs before their trip (Paltansing et al.2013). A large prospective study of 2001 Dutch travellers found 34.7% with no ESBL producing Enterobacteriaceae prior to international travel returned with ESBL producing strains (Arcilla et al.2017). A similar study of 188 Swedish travellers found 32% returned from regions associated with high levels of ESBL producing Enterobacteriaceae carrying these antibiotic-resistant bacteria (Vading et al.2016). One isolate contained both blaCTX-M and mcr-1 (Vading et al.2016). Indeed, mcr-1 was detected by metagenomics in 4.9% of faecal samples from 122 healthy Dutch travellers upon return from travel to South/East Asia and/or Southern Africa undertaken between 2011 and 2012 (von Wintersdorff et al.2016). However, in this study little is known about the index isolate in which the mcr-1 gene originated, including the isolate's susceptibility profiles. Therefore, it is possible that the isolates were susceptible to other antimicrobials. In the majority of studies travellers who obtained ESBL-producing bacteria eventually lost the ESBL genes upon return. Of 15 Swiss volunteers, 3 were colonised by ESBL-resistant Enterobacteriaceae before their trip, all were colonised upon return and 6 were still colonised 6 months post-travel (Pires et al.2016). Of the resistant isolates 80% contained IncF family plasmids, and in some of the participants who were colonised 6 months after travel, the plasmids had moved into new host bacteria (Pires et al.2016). blaCTX-M-15 was the most prevalent ESBL, comprising 92% of the ESBL producers immediately after travel (Pires et al.2016). Together, this highlights the need to reduce the prevalence of ARGs on a global scale.
Could plasmid curing be a strategy to reduce AMR?
Plasmid curing is the process by which plasmids are removed from bacterial populations. This is an attractive strategy to combat AMR as it has the potential to remove ARGs from a population while leaving the bacterial community intact. This means, for example, that the structure of the gastrointestinal microbiome of a chicken treated with a plasmid curing agent might remain largely unchanged, but potentially pathogenic bacteria which may unfortunately be transmitted into the food chain would be susceptible to antibiotics. Alternatively, a plasmid curing agent could be given to a patient prior to surgery, to reduce the likelihood of a resistant hospital acquired infection. Plasmid curing agents could also be taken by international travellers to reduce the global spread of AMR. Unfortunately, at the moment no such treatment options are in use. In fact, there are very few curing mechanisms that have been tested in vivo, even in experimental models. Therefore, research in this area is urgently needed. Recently, it was shown that 24% of non-antibacterial drugs impact growth of members of the human microbiome (Maier et al.2018). Studies such as this would be important for determining any impact of anti-plasmid compounds on the microbiome.
The ‘One Health’ approach to tacking AMR is based around the notion that AMR does not abide by human, animal or political boundaries, and therefore a multisectoral and multifaceted approach is required. Likewise, anti-plasmid strategies should also adopt a One Health strategy, and not be focused on human medicine alone. Indeed some anti-plasmid strategies are unsuitable or unviable for human use. Furthermore, anti-plasmid strategies alone will never ‘solve’ AMR; nonetheless, they could play an important role in reducing global resistance levels. Removing drug-resistance plasmids is a strategy for all sectors to reduce the overall burden of AMR. For example, plasmid curing could be used to remove ARGs from bacteria in sewage before release into the environment. Human and animal waste is often recycled and used to fertilise agricultural land; this can contain high concentrations and varieties of ARGs which can be passed on to people (Meek, Vyas and Piddock 2015; Rahube et al.2016). One study performed in Canada found in the first year vegetables grown above, on and below the surface of soil treated with sewage contained significantly more ARGs than non-treated soil (Rahube et al.2016). An abundance of ARGs were detected in plasmid metagenome libraries constructed from the influent, activated sludge and digested sludge from two wastewater treatment plants in Hong Kong, demonstrating that these were important reservoirs of ARGs (Li, Li and Zhang 2015). River samples taken upstream and downstream of a tertiary waste water treatment plant in the UK in 2009 and 2011 were examined for third-generation cephalosporin-resistant Enterobacteriaceae (Amos et al.2014). Significantly higher amounts of blaCTX-M-15 were found downstream of the plant, and 10 novel genetic contexts were identified (Amos et al.2014). The plasmids containing blaCTX-M-15 were conjugative, and were in pathogens such as the highly successful extraintestinal E. coli ST131 (Stoesser et al.2016), and other species never before reported to carry blaCTX-M-15 (Amos et al.2014). IncP-1ε plasmids were detected in manure and arable soil in Germany, and a correlation was found between the presence of IncP-1ε plasmids and antibiotic use (Heuer et al.2012). A waste water treatment plant in Brazil found 34% of E. coli and 27% of K. pneumoniae were resistant to cephalosporins and/or quinolones, and 5.4% of Klebsiella species were carbapenem resistant in raw as well as treated water (Conte et al.2016). Analysis of these ARGs showed a high prevalence of blaCTX-M and blaSHV (Conte et al.2016). Recent work from our group examined wastewater used for irrigation of urban agriculture plots in Burkina Faso. This wastewater contained multiple ARGs including ESBLs, 10 different Enterobacteriaceae-associated plasmid incompatibility groups and 30 Gram-positive replicons associated with ARGs (Bougnom et al., submitted). Together, these studies demonstrate that a treatment such as plasmid curing agents to remove ARGs from manure, sewage and waste water are needed.
The search for plasmid curing compounds began decades ago, and gained momentum in the 1970s (Table 1). The number of publications peaked in the 1980s (based on searches for publications relating to plasmid curing performed on NCBI PubMed). However, most compounds were toxic, and would produce adverse or unwanted side effects and thus had little use in human medicine. This was followed by a decline in interest and publications. Generally, plasmid curing properties have been evaluated by culturing strains in the presence of a compound or extract at subgrowth inhibitory concentrations. Curing effects are then confirmed by the reversal of plasmid-mediated antibiotic resistance and/or by physical loss of the plasmid(s). Therefore, many of the older publications only refer to the loss of an AMR phenotype.
Table 1.
Plasmid curing compounds.
| Curing Agent | Species | Plasmid Cured | Key Findings | Reference |
|---|---|---|---|---|
| Acridine orange | E. coli | Small plasmids (UTI isolates) | 75 μg/mL: 11.76% CF for plasmids ≤2.7 mDa | Zaman, Pasha and Akhter (2010) |
| pBR322 | 100 μg/mL: 35% CF | Keyhani et al. (2006) | ||
| pBR325 | 100 μg/mL: 15% CF | Keyhani et al. (2006) | ||
| pUK657 | 375 μg/mL: 14.28% CF | Beg and Ahmad (2000) | ||
| V. parahaemolyticus | AMR plasmid | 0.2 mg/mL cured 6/13 plasmids from isolates (1.2–10kb). | Letchumanan et al. (2015) | |
| L. plantarum | Raffinose & lactose metabolising plasmid | 0.1 mg/mL cured 10/12 plasmids | Adeyemo and Onilude (2015) | |
| S. aureus | Staphyloccocin plasmid | 15 μg/mL: 12.1% CF | Jetten and Vogels (1973) | |
| pED503 | 15 μg/mL: 3.4% CF | Ersfeld-Dressen, Sahl and Brandis (1984) | ||
| B. fragilis | AMR plasmid | 16 μg/mL cured resistance to Ery and Clin | Rotimi, Duerden and Hafiz (1981) | |
| B. thetaiotaomicron | AMR plasmid | 16 μg/mL cured resistance to Ery and Clin | Rotimi, Duerden and Hafiz (1981) | |
| Acriflavine | S. enterica | AMR plasmids | Of plasmids with five resistance phenotypes, 35% CF of S. Oranienburg, 5% CF of S. Panama. Of plasmids with one resistance phenotype, 98% CF of S. Panama and S. paratyphi B | Bouanchaud and Chabbert (1971) |
| E. coli | AMR plasmid | Three plasmids cured at 5, 12 and 22% CF | Bouanchaud and Chabbert (1971) | |
| Haemolysin producing plasmids | 24 h incubation with 10 μg/mL resulted in low CF | Mitchell and Kenworthy (1977) | ||
| Group A Streptococci | AMR plasmid | 0.2 μg/mL for 18 h: 2.1%–4.3% CF of three plasmids | Nakae, Inoue and Mitsuhashi (1975) | |
| L. casei | pDR101 | 10 μg/mL for 48 h: 7.2% CF | Chassy, Gibson and Guiffrida (1978) | |
| L. reuteri | pLUL631 (lactose fermenting) | 2 μg/mL: 1%–10% CF | Axelsson et al. (1988) | |
| S. aureus | Staphyloccocin plasmids | 2 μg/mL: 25% CF | Jetten and Vogels (1973) | |
| B. fragilis | AMR plasmid | 16 μg/mL, 18–21 days: loss of Ery, Clin and Tet resistance plasmid | Rotimi, Duerden and Hafiz (1981) | |
| B. thetaiotaomicron | AMR plasmid | 16 μg/mL, 18–21 days: loss of Ery, Clin and Tet resistance plasmid | Rotimi, Duerden and Hafiz (1981) | |
| O. oeni | pRS1, pRS2, pRS3 | 2.5–10 μg/Ml, CF of: 18.7% (pRS1), 6.2% (pRS2), 62.5% (pRS3), 31.2% (pRS2 & pRS3 simultaneously) | Mesas, Rodriguez and Alegre (2004) | |
| E. faecium | AMR plasmids | Sub-MIC levels resulted in cured isolates | Coleri et al. (2004) | |
| E. faecalis | AMR plasmids | Sub-MIC levels resulted in cured isolates | Coleri et al. (2004) | |
| Ascorbic Acid | S. aureus | Penicillinase plasmid | 1 mM for 6 h: 12%–35% CF | Amábile Cuevas (1988) |
| Aminoglycoside resistance plasmid | 1 mM for 6 h: 4 of six strains cured, with 10%–48% CF | Amábile-Cuevas, Piña-Zentella and Wah-Laborde (1991) | ||
| pI55cI | 1 mM for 6 h: 48% CF | Amábile-Cuevas, Piña-Zentella and Wah-Laborde (1991) | ||
| P. acidilactici | Pediocin producing plasmid | 1 mM: 35% CF of 7.8 kb plasmid | Ramesh, Halami and Chandrashekar (2000) | |
| Bile | S. enterica Typhimurium | pSLT | 15% ox bile: 10−6 frequency of plasmid loss in wild type. In ccdB mutant frequency was 10−4 | García-Quintanilla et al. (2006) |
| S. enterica Infantis | pESI | 1%–4% bile: reduced CF | Aviv, Rahav and Gal-mor (2016) | |
| Chlorpromazine | E. coli | F’lac plasmid | 20–60 μg/mL: 5%–20% CF, most efficient at pH 7.6 | Mandi et al. (1975) |
| R-factor | 50 μg/mL: plasmid curing was observed | Molnar, Mandi and Kiraly (1976) | ||
| R114 plasmid | Enhanced curing activity with methylene blue | Molnar et al. (1980) | ||
| S. aureus | QacA encoding plasmid | Successive passaging in 2–20 mg/mL resulted in curing | Costa et al. (2010) | |
| Ethidium bromide | S. aureus | Penicillinase carrying plasmids | 8 × 10−6M at pH 7.2: CF of 50% (maximum). 6 × 10−6M: CF average of 20%, ranging from 0.21%–58% depending on plasmid/strain. Curing peaked at 10–12 h, became refractory to additional curing | Bouanchaud, Scavizzi and Chabbert (1969); Rubin and Rosenblum (1971) |
| Staphyloccocin producing plasmid | 1.25 μg/mL: 94% CF | Jetten and Vogels (1973) | ||
| pED503 | 3.6 μg/mL: 4.4% CF | Ersfeld-Dressen, Sahl and Brandis (1984) | ||
| AMR plasmids | 32% and 60% CF for Pen and mercury resistance plasmids | Bouanchaud and Chabbert (1971) | ||
| E. aerogenes | pKpQIL-like (blaTEM-1 and blaKPC-3) | 400–600 μg/mL: 85% CF | Pulcrano et al. (2016) | |
| Salmonella | AMR plasmids | 100–2000 μg/mL for 1–7 days cured 2/17 strains | Poppe and Gyles (1988) | |
| E. coli | F’-lac plasmids | 6–250 × 10−5M: 20% CF | Bouanchaud, Scavizzi and Chabbert (1969) | |
| p424 | 0.52 mM cured plasmid, four cured variants had altered colony morphology and biochemical modifications | Rosas et al. (1983) | ||
| pUK651 | 200 μg/mL: 36.6% CF | Beg and Ahmad (2000) | ||
| Haemolysin producing plasmids | 50 μg/mL: low frequency of plasmid loss at 24 h | Mitchell and Kenworthy (1977) | ||
| AMR plasmids | 7.5 × 10−5 and 1.3 × 10−3M: CF of 71% and 32%, respectively | Bouanchaud, Scavizzi and Chabbert (1969) | ||
| AMR plasmids | 32% curing of resistance to five antibiotics | Bouanchaud and Chabbert (1971) | ||
| UTI plasmids | 125 μg/mL: 17.65% CF | Zaman, Pasha and Akhter (2010) | ||
| B. cereus | Hydrocarbon degrading plasmid | 100 μg/mL: cured isolates enabling testing of plasmid properties | Borah and Yadav (2015) | |
| B. fragilis | AMR plasmid | 16 μg/mL cured Ery and Clin resistance. Curing of Tet resistance required 18–21 days | Rotimi, Duerden and Hafiz (1981) | |
| B. thetaiotaomicron | AMR plasmid | 16 μg/mL cured Ery and Clin resistance. Curing of Tet resistance required 18–21 days | Rotimi, Duerden and Hafiz (1981) | |
| Irgasan (Triclosan) | E. coli | pMIB4 | 100× below MIC cured plasmid. Effective in broth and embedded in silicone hydrogels | Riber et al. (2016) |
| Lawsone | S. aureus | Van resistance plasmid | 200 μg/mL: 20% CF (1/2 MIC) | Jahagirdar, Patwardhan and Dhakephalkar (2008) |
| Plumbagin | E. coli | R6K | 200 μg/mL: 42% CF of 2/6 resistance markers | Lakhmi, Padma and Polasa (1987) |
| TP181 | 100 μg/mL: 100% CF | Lakhmi, Padma and Polasa (1987) | ||
| R162 | 100 μg/mL: 100% CF | Lakhmi, Padma and Polasa (1987) | ||
| TP154 | 100 μg/mL: 45% CF of 3/6 resistance markers | Lakhmi, Padma and Polasa (1987) | ||
| RP4 | 12.5 μg/mL: 32% CF | Bharathi and Polasa (1991) | ||
| pKT231 | 12.5 μg/mL: 10% CF | Bharathi and Polasa (1991) | ||
| pTP181-derivatives | 25 μg/mL: 11%–47% CFCaused by interference with plasmid replication and maintenance | Lakshmi and Thomas (1996) | ||
| pUK651 | 7000 μg/mL: 14% CF (sub-MIC) | (Beg and Ahmad (2000) | ||
| R plasmid | 1000 μg/mL: 15% CF. | Patwardhan et al. (2015) | ||
| S. aureus | Van resistance plasmid | 25 μg/mL: 4% CF, 50 μg/mL inhibited growth | Jahagirdar, Patwardhan and Dhakephalkar (2008) | |
| P. aeruginosa | R plasmid | 1000 μg/mL: 13% CF | Patwardhan et al. (2015) | |
| P. vulgaris | R plasmid | 500 μg/mL: 32% CF | Patwardhan et al. (2015) | |
| K. pneumoniae | R plasmid | 500 μg/mL: 30% CF | Patwardhan et al. (2015) | |
| Promethazine | E. coli | AMR plasmid | Plasmids eliminated | Spengler et al. (2003) |
| F’lac plasmid | At 37°C, 80 μg/mL: 79.6% CF At 39°C, 80 μg/mL: 88% CF Multi-species co-cultures reduced promethazine concentration required for curing | Molnár, Amaral and Molnár (2003); Spengler et al. (2003) | ||
| pBR322 | TF-14 (a potential proton pump inhibitor) increased promethazine CF | Wolfart et al. (2006) | ||
| Rifampicin | E. coli | Haemolysin plasmids | 2 μg/mL, 24 h incubation led to high CF | Mitchell and Kenworthy (1977) |
| F’lac | 3–7.5 μg/mL resulted in curing. Rif/RNA polymerase interaction required for curing | Bazzicalupo and Tocchini-Valentini (1972) | ||
| S. aureus | Penicillinase plasmid | 0.1 μg/mL: 20% CF, 0.05 μg/mL: 5% CF | Johnston and Richmond (1970); Wood, Carter and Best (1977) | |
| Sodium dodecyl sulphate (SDS) | E. coli | R and F factors | 24 h of 10% SDS: 5.3%–22% CF, 72 h resulted in 95%–100% CF | Tomoeda et al. (1968) |
| p424 | 10% cured variants had altered colony morphology and biochemical modifications | Rosas et al. (1983) | ||
| pR4 | 100 μg/mL: 12.5% CF | Bharathi and Polasa (1991) | ||
| pKT231 | 200 μg/mL: 7.5% CF | Bharathi and Polasa (1991) | ||
| pBR322 | 0.25%–1%: 27%–35% CF | Keyhani et al. (2006) | ||
| UTI plasmids | 10% w/v: 7.4% CF | Zaman, Pasha and Akhter (2010) | ||
| K. pneumoniae | Large indigenous plasmid (96 kb) | 4% resulted in 1/8 colonies successfully cured | El-Mansi et al. (2000) | |
| Lactobacillus isolates (milk) | AMR plasmids | 1% cured 5 of 7 isolates | Lavanya et al. (2011) | |
| P. aeruginosa | pBC15 | 10% was effective | Raja and Selvam (2009) | |
| S. aureus | Staphyloccocin producing plasmid | 30 μg/mL: 100% CF | Jetten and Vogels (1973) | |
| Thioridazine | E. coli | AMR plasmid | 75% MIC eliminated resistance | Radhakrishnan et al. (1999) |
| S. flexneri | AMR plasmid | 75% MIC eliminated resistance | Radhakrishnan et al. (1999) | |
| V. cholera | AMR plasmid | 75% MIC eliminated resistance | Radhakrishnan et al. (1999) | |
| Trifluoperazine | E. coli | AMR plasmid | Reviewed in detail by | Spengler et al. (2003) |
| 1΄-acetoxychavicol acetate | E. coli | pAR1813 | 400 μg/mL: 32% CF | Latha et al. (2009) |
| RP4 | 400 μg/mL: 7% CF | Latha et al. (2009) | ||
| S. Typhi | pAR1814 | 800 μg/mL: 75% CF | Latha et al. (2009) | |
| P. aeruginosa | pAR1816 | 800 μg/mL: 75% CF | Latha et al. (2009) | |
| E. faecalis | pAR1812 | 400 μg/mL: 66% CF | Latha et al. (2009) | |
| B. cereus | pAR1817 | 400 μg/mL: 6% CF | Latha et al. (2009) | |
| 8-epidiosbulbin E acetate | E. coli | RP4 | 25 μg/mL: 44% CF | Shriram et al. (2008) |
| pARI813 | 25 μg/mL: 44% CF | Shriram et al. (2008) | ||
| B. subtilis | pUB110 | 100 μg/mL: 48% CF | Shriram et al. (2008) | |
| P. aeruginosa | RMS163 | 200 μg/mL: 30% CF | Shriram et al. (2008) | |
| RIP64 | 100 μg/mL: 64% CF | Shriram et al. (2008) | ||
| E. faecalis | pARI812 | 200 μg/mL: 48% CF | Shriram et al. (2008) | |
| S. sonnei | pARI815 | 25 μg/mL: 32% CF | Shriram et al. (2008) |
CF—Curing Frequency: the proportion of colonies which were cured of the plasmid compared to non-cured colonies. Ery—erythromycin, Clin—clindamycin, Tet—tetracycline, Pen—penicillin, Van—vancomycin, Rif—rifampicin.
The rise in AMR, specifically plasmid-mediated resistance, combined with the dwindling pipeline of new drugs in development has resulted in a resurgence of interest in plasmid curing. Strategies of plasmid curing vary greatly, such as the use of chemicals, drugs, natural products, phage therapies, other plasmids and even CRISPR/Cas. A recent study demonstrated that inhibiting plasmid conjugation was an effective means to remove a plasmid from a bacterial population over time (Lopatkin et al.2017). The authors concluded that strategies to prevent plasmid conjugation should be explored as a means to reduce AMR plasmid prevalence (Lopatkin et al.2017). Plasmid curing of a population can also occur when plasmid replication is prevented or reduced, or if plasmid segregation is disrupted, resulting in gradual reduction in plasmid carrying cells. Plasmid curing can also be achieved by increasing the fitness cost associated with plasmid carriage. We anticipate over the next decade that these mechanisms will be studied, streamlined and new practical ways to reduce global AMR plasmid carriage, and hence presence of ARGs, will be developed.
PLASMID CURING COMPOUNDS
Many compounds have shown some plasmid curing activity. These include detergents, biocides, DNA intercalating agents, antibiotics (e.g. aminocoumarins, quinolones, rifampicin), ascorbic acid, psychotropic drugs (e.g. chlorpromazine) and plant-derived compounds (Table 1). The effectiveness of these compounds varies greatly and depends on bacterial strain, plasmid and growth conditions. Plasmid curing compounds can act through different mechanisms. In many cases, the compound disrupts plasmid replication by integrating into the DNA (e.g. intercalating agents and chlorpromazine), causing breaks in DNA (e.g. ascorbic acid) or by influencing plasmid supercoiling (e.g. aminocoumarins and quinolones). Plasmid curing compounds can also act by preventing conjugation (e.g. unsaturated fatty acids and TraE inhibitors). Each of these can result in reduced plasmid prevalence within the population over time. The mechanism of action of some curing agents remains to be fully elucidated. One could hypothesise that plasmid curing compounds could also target plasmid segregation, by preventing equal distribution among daughter cells, or increase the fitness burden associated with plasmid carriage.
Detergents
The detergents bile and sodium dodecyl sulphate (SDS) are able to cure some plasmids from some bacterial strains (Table 1). Four notable examples include a study where bile salts dose-dependently caused the loss of the Salmonella enterica serovar Typhimurium virulence plasmid, pSLT (García-Quintanilla et al.2006). However, the level of bile required was 10%–15%, which is significantly higher than that found normally within the small intestine (0.2%–2%) (García-Quintanilla et al.2006; Kristoffersen et al.2007). The Salmonella virulence plasmid can be transmitted to new hosts in the mouse intestine, but transmission is unlikely to occur in areas with high levels of bile (García-Quintanilla, Ramos-Morales and Casadesús 2008). Bile (>1%) reduced expression of conjugative pilus genes pilV and pilT, and decreased conjugation of S. enterica Infantis mega plasmid pESI (280 kb), encoding resistance to tetracycline, sulfamethoxazole and trimethoprim as well as virulence traits (Aviv et al.2014; Aviv, Rahav and Gal-mor 2016). The relevance of bile-mediated plasmid curing during human Salmonella infections remains unclear. In addition, the levels of bile required for plasmid curing or to reduce plasmid transmission may result in diarrhoea, and therefore bile is unlikely to be used as a treatment.
SDS-based plasmid curing methods have been used as a laboratory tool for decades. In 1968, SDS was shown to reduce carriage of fertility and resistance factors (F and R factors/plasmids) (Tomoeda et al.1968). Over the years, SDS has been used to cure plasmids from E. coli (Rosas et al.1983; Bharathi and Polasa 1991; Keyhani et al.2006; Zaman, Pasha and Akhter 2010), K. pneumoniae (El-Mansi et al.2000), Pseudomonas aeruginosa (Raja and Selvam 2009), Lactobacillus species (Lavanya et al.2011) and Staphylococcus aureus (Jetten and Vogels 1973) (Table 1). SDS also had other effects on bacteria; these included changes in the peptidoglycan layer, bacterial cell size, septation and loss of outer membrane components (Rosas et al.1983).
In summary, detergents are unlikely to be used in humans or animals to reduce AMR plasmids, mainly due to the high concentrations needed, and the associated unwanted gastrointestinal side effects, such as SDS-induced colitis. However, detergents continue to be used in the laboratory setting as a tool to study plasmid biology.
Biocides
Recently, it was shown that concentrations well below the MIC of triclosan (also called irgasan) increased the loss of a GFP reporter plasmid pMIB4 from E. coli (Riber et al.2016). A key finding of this paper was that triclosan embedded in interpenetrating polymer networks of silicone hydrogels was effective at reducing plasmid carriage. The use of such technology as a drug delivery system is appealing, especially for items such as indwelling medical devices (e.g. catheters). However, exposure to triclosan can select for MDR bacterial mutants, largely due to overexpression of bacterial efflux pumps (Chuanchuen et al.2001; Webber et al., 2008, 2015; Hernandez et al.2011; Fernando et al.2014; Rensch et al.2014; Gantzhorn, Olsen and Thomsen 2015). Therefore, caution should be used implementing such a strategy.
DNA intercalating agents
The DNA intercalating agents acridine orange, ethidium bromide and acriflavine also have plasmid curing properties. Acridine orange cured E. coli (Keyhani et al.2006; Zaman, Pasha and Akhter 2010), Vibrio parahaemolyticus (Letchumanan et al.2015), Lactobacillus plantarum (Adeyemo and Onilude 2015), S. aureus (Jetten and Vogels 1973; Ersfeld-Dressen, Sahl and Brandis 1984), Bacteroides fragilis and B. thetaiotaomicron (Rotimi, Duerden and Hafiz 1981) (Table 1). In the late 1960s and early 1970s, ethidium bromide was found to eliminate plasmids from various strains of S. aureus and Escherichia coli (Table 1) (Bouanchaud, Scavizzi and Chabbert 1969; Rubin and Rosenblum 1971). More recently, it has been used to cure other plasmids from E. coli (Rosas et al.1983), Bacillus cereus (Borah and RNS 2015), clinical Enterobacter aerogenes isolates of a blaTEM-1 and blaKPC-3 pKpQIL-like plasmid (Pulcrano et al.2016), and cured plasmids from two avian Salmonella strains (Poppe and Gyles 1988) (Table 1).
Acriflavine cured some resistance plasmids from Salmonella Oranienburg, S. Panama and E. coli K12 in vitro and in a murine in vivo model (Bouanchaud and Chabbert 1971). Acriflavine also cured E. coli of haemolysin production (Mitchell and Kenworthy 1977). However, it is more commonly associated with curing of Gram-positive bacteria. It cured plasmids from Group A Streptococci (Nakae, Inoue and Mitsuhashi 1975), Lactobacillus casei (Chassy, Gibson and Guiffrida 1978), L. reuteri (Axelsson et al.1988) and Oenococcus oeni (used in wine production) (Mesas, Rodriguez and Alegre 2004) (Table 1). Acriflavine was effective at curing resistance from antibiotic-resistant Enterococcus faecium and E. faecalis (Coleri et al.2004).
Acriflavine, ethidium bromide and acridine orange caused loss of a plasmid-encoded staphylococcin production in Staphylococcus species (Jetten and Vogels 1973; Ersfeld-Dressen, Sahl and Brandis 1984); however, as strains became resistant to acriflavine they also became resistant to its curing effects (Jetten and Vogels 1973). Acriflavine, acridine orange and ethidium bromide cured resistance to antimicrobials from both donor and transconjugants B. fragilis and B. thetaiotaomicron (Rotimi, Duerden and Hafiz 1981).
The practical applications of DNA intercalating agents are few, due to their activity as powerful mutagens, associated with significant toxicity and the carcinogenic nature of these molecules. The harm of using such compounds vastly outweighs any potential benefit derived from plasmid curing. In addition, as many intercalating agents are substrates of bacterial efflux pumps, the use of such compounds could select for overexpression of efflux pumps which can lead to MDR (Piddock 2006). However, these compounds can still be useful in a laboratory setting to cure strains of plasmids (Coleri et al.2004; Mesas, Rodriguez and Alegre 2004; Chin et al.2005; Raja and Selvam 2009; Zaman, Pasha and Akhter 2010; Adeyemo and Onilude 2015; Pulcrano et al.2016).
Plant-derived compounds
Many well-studied plant extracts come from traditional medicine. Plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone) is a yellow dye derived from the root of the tropical/subtropical Plumbago species (Patwardhan et al.2015). Plumbagin is reported to have anticancer, antifungal and antimicrobial activity (Padhye et al.2012; Tyagi and Menghani 2014). In E. coli, plumbagin effectively eliminated a conjugative, MDR plasmid (Lakhmi, Padma and Polasa 1987) and the RP4 plasmid (Bharathi and Polasa 1991). Plumbagin eliminated plasmids from E. coli, by decreasing plasmid copy number and reducing the toxic effect of plasmid loss (Lakshmi and Thomas 1996) (Table 1).
Subinhibitory concentrations of Plumbago zeylanica root extract were tested on MDR clinical isolates of S. Paratyphi, S. aureus, E. coli and Shigella dysenteriae, as well as E. coli containing pUK651, but the extract only cured 14% of E. coli of pUK651 (Beg and Ahmad 2000). Subinhibitory concentrations of P. auriculata root extracts cured drug-resistance plasmids from P. aeruginosa, E. coli, Proteus vulgaris and K. pneumoniae, which were slightly higher than pure plumbagin (Patwardhan et al.2015) (Table 1).
8-epidiosbulbin E acetate is isolated from the bulbs of Dioscorea bulbifera, a plant known in Ayurvedic alternative medicine (Shriram et al.2008). 8-epidiosbulbin E acetate belongs to the clerodane class of diterpenes. Its antibacterial and curing activity was evaluated, and it cured reference strains of E. coli, B. subtilis, P. aeruginosa, and clinical isolates of E. coli, E. faecalis and S. sonnei with an average efficiency of 34% (Shriram et al.2008) (Table 1).
The curing activity of the crude extract of Alpinia galanga (L.) Swartz, a medicinal plant indigenous to Southeast Asian countries, was tested (Latha et al.2009). The bioactive fraction containing 1΄-acetoxychavicol acetate was tested on nine bacterial reference strains carrying antibiotic-resistance plasmids. A subinhibitory concentration of crude extract cured plasmids from S. Typhi, E. coli and E. faecalis. Purified 1΄-acetoxychavicol acetate cured MDR plasmids from S. Typhi, P. aeruginosa, E. faecalis, E. coli and B. cereus (Latha et al.2009) (Table 1).
Taken together, plant-derived compounds can be effective at curing plasmids in vitro; however, more research is needed to confirm spectrum of activity, identify the active components and to determine any toxicity and in vivo efficacy.
Conjugation inhibiting compounds
Unsaturated fatty acids
Work from de la Cruz and colleagues has focused on performing high-throughput screens of compounds to search for inhibitors of conjugation (Fernandez-Lopez et al.2005; Getino et al. 2015, 2016; Ripoll-Rozada et al.2016). Their high-throughput screening method used a lux reporter under the control of the lac promoter, on a simple conjugative plasmid derived from R388 in E. coli (Fernandez-Lopez et al.2005). The donor carried the lacI repressor; thus, luminescence was only produced after conjugation (Fernandez-Lopez et al.2005). They tested a library of microbial extracts, and showed that unsaturated fatty acids, including dehydrocrypenynic acid, linoleic acid and oleic acid, inhibited conjugation (Fernandez-Lopez et al.2005). Recently, Lopatkin et al. (2017) used linoleic acid to determine the impact of reduced conjugation on plasmid persistence within a population. Indeed, 3.5 μM linoleic acid was sufficient to destabilise a plasmid with low conjugation efficiency from a population; however, it was ineffective for plasmids with higher conjugation efficiencies or which carried a fitness benefit (Lopatkin et al.2017).
A study of synthetic fatty acids demonstrated that 2-alynoic fatty acids inhibited conjugation; of these, 2-hexadecynoic acid was the most potent, followed by 2-octadecynoic acid (Getino et al.2015). At concentrations of 0.4 mM, 2-hexadecynoic acid reduced conjugation frequencies of IncW, IncH and IncF plasmids by 100 times, while concentrations of 1 mM were required to reduce conjugation of IncI, IncL/M and IncX plasmids. Conjugation of IncP and IncN plasmids was not affected by 2-hexadecynoic acid. Using molecules with similar structures, they determined that the carboxylic group, a 16-carbon chain and one unsaturated bond were optimal for conjugation inhibition. They showed that 2-hexadecanoic acid acted on the donor, and inhibited conjugation in E. coli, S. enterica, P. putida and Acinetobacter baumannii (Getino et al.2015).
Four unsaturated fatty acids (linoleic, oleic, 2-hexadecynoic and 2-ocatadecynoic acid) inhibited the activity of the plasmid encoded TrwD ATPase (VirB11 homologue) (Ripoll-Rozada et al.2016). TrwD acts as a traffic ATPase, regulating switching between pilus biogenesis and DNA translocation through the conjugation machinery (Ripoll-Rozada et al.2013). Fatty acids which did not inhibit conjugation had no impact on TrwD activity (Ripoll-Rozada et al.2016). The authors suggested that the mechanism for the conjugation inhibiting activity of unsaturated fatty acids was due to their binding to the N-terminal domain and linker region of TrwD, inhibiting the movement of the N-terminal domain over the C-terminal domain, thus preventing ATPase activity of the enzyme (Ripoll-Rozada et al.2016).
One of the concerns about any clinical use of synthetic fatty acids, such as 2-hexadecanoic acid, is toxicity in people or animals. Recent work focused on finding less-toxic molecules by screening a natural compound library produced by aquatic microbes (Getino et al.2016). Tanzawaic acid A and B, polyketides produced by Penicillium species, were identified as effective conjugation inhibitors of IncW and IncFII plasmids. Tanzawaic acid B (0.4 mM) reduced conjugation by 100-fold for IncW and IncFII, as compared to untreated controls. However, they were only moderately effective on IncFI, IncI, IncL/M, IncX and IncH plasmids, reducing conjugation by between 10% and 50% compared to untreated cells. In addition, they did not inhibit conjugation of IncN and IncP plasmids (Getino et al.2016). Importantly, oleic acid, linoleic acid and tanzawaic acids A and B were less toxic on bacteria, fungi and tissue culture cells than 2-hexadecynoic and 2-oxydecynoic acid (Getino et al.2016).
Unsaturated fatty acids have been shown to be effective conjugation inhibitors in many laboratory settings, and on a variety of plasmids. Furthermore, they are associated with reduced toxicity on tissue culture cells. Further studies are needed to determine the in vivo safety and efficacy of unsaturated fatty acids, but they are promising candidates for future plasmid curing work.
TraE inhibitors
Using a targeted approach, Baron and colleagues have identified small molecules which bind to and inhibit the dimerisation of TraE, an essential component of the type IV secretion system involved in a variety of functions including conjugation of pKM101 (Paschos et al.2011; Casu et al. 2016, 2017). Structural studies of the pKM101 encoded TraE dimerisation (VirB8 homologue) were used as a basis for uncovering small molecules which inhibited dimerisation, four of which (molecules B8I-16, BAR-072, BAR-073 and UM-024) also inhibited transmission of pKM101 (Casu et al.2016). None of these molecules impacted upon transmission of RP4, highlighting their specificity for pKM101 TraE (Casu et al.2016). In a follow-up study, Casu et al. (2017) screened a fragment library for compounds which bound to TraE. They used this information to design two molecules which bound with high affinity to TraE and were able to reduce transmission of pKM101 (molecules 105055 and 239852) (Casu et al.2017). Together, this work demonstrates the feasibility and specificity of structure-based design of anti-plasmid compounds.
Drugs used in human medicine
DNA gyrase/topoisomerase inhibitors
DNA gyrase is essential in bacteria as it introduces supercoiling into DNA molecules; it is comprised of two GyrA and two GyrB monomers (Andriole 2005). Multiple antibiotics target DNA gyrase. Aminocoumarin antibiotics, such as novobiocin and coumermycin A, inhibit GyrB (Gellert et al.1976). These and the related compounds clorobiocin and isobutyryl novenamine were effective at plasmid curing (Hooper et al.1984). The GyrB inhibiting activities of aminocoumarins are responsible for their plasmid curing properties (Taylor and Levine 1979), and the E. coli gyrase B subunit is required for plasmid maintenance, and curing activity of coumermycin A1 (Wolfson et al.1982). Novobiocin interfered with plasmid maintenance, rather than selecting plasmid-free isolates (Hooper et al.1984). Furthermore, bacteria with a mutation in gyrB conferring resistance to coumermycin required higher levels of the antibiotic to produce the curing effect (Hooper et al.1984).
Novobiocin was effective at curing plasmids from many Gram-positive bacteria including L. plantarum, Lactobacillus strains isolated from chickens, L. acidophilus isolated from molasses, E. faecalis, clinical isolates of enterococci, B. subtilis and S. aureus (Table 2) (McHugh and Swartz 1977; Ruiz-Barba, Piard and Jiménez-Díaz 1991; Chin et al.2005; Karthikeyan and Santosh 2010). Escherichia coli and other Gram-negative Enterobacteriaceae were cured of a variety of plasmids by novobiocin (Michel-briand et al.1986). Novobiocin eliminated the Salmonella virulence plasmid from S. Typhimurium, resistance plasmids from Serratia marcescens and a cryptic plasmid from Chlamydia muridarum (Gulig and Curtiss III 1987; Llanes et al.1990; O’Connell and Nicks 2006). Coumermycin eliminated some plasmids from E. coli, but not RP4 (Danilevskaya and Gragerov 1980; Wolfson et al.1982; Bharathi and Polasa 1991).
Table 2.
Quinolone and aminocoumarin antimicrobials with plasmid curing properties.
| Quinolone | Species | Plasmid cured | Key findings | Reference |
|---|---|---|---|---|
| Ciprofloxacin | E. coli | R446b | 1/2 MIC: no curing, 0.06 μg/mL (sub-MIC): 30% CF | Weisser and Wiedemann (1985); Michel-briand et al. (1986) |
| R386 | 0.07 μg/mL (sub-MIC): 2% CF | Michel-briand et al. (1986) | ||
| F’lac | 1/2 MIC: 50% CF | Weisser and Wiedemann (1985) | ||
| R16 | 1/2 MIC: 1% CF | Weisser and Wiedemann (1985) | ||
| Rts1 | 1/2 MIC: 32% CF | Weisser and Wiedemann (1985) | ||
| 5 large plasmids | Sub-MIC: 10%–90% CF. Small high copy plasmids not cured | Platt and Black (1987) | ||
| S. sonnei | pWR105 | 0.05 μg/mL (sub-MIC): 50% CF | Michel-briand et al. (1986) | |
| Coumermycin A | E. coli | pBR322 | 5 μg/mL: 90% CF, 7 μg/mL: 45% CF. Mechanism involves antagonism of DNA gyrase | Danilevskaya and Gragerov (1980); Wolfson et al. (1982) |
| pMG110 | 7 μg/mL: 70% CF and mechanism involves antagonism of DNA gyrase. | Wolfson et al. (1982) | ||
| pMB9 | 5 μg/mL: 64.7% CF. Cou resistant mutant had 5% CF at 10 μg/mL | Danilevskaya and Gragerov (1980) | ||
| pOD162 | 5 μg/mL: 64.5% CF | Danilevskaya and Gragerov (1980) | ||
| pSC101 | 2 μg/mL: 32.5% CF | Danilevskaya and Gragerov (1980) | ||
| pKT231 | 3.15 μg/mL: 90% CF | Bharathi and Polasa (1991) | ||
| pRK2013 | 3.15 μg/mL: 35.5% CF | Bharathi and Polasa (1991) | ||
| Enoxacin | E. coli | R446b | 1/2 MIC: 24% CF, 0.5 μg/mL (sub-MIC): 2% CF | Weisser and Wiedemann (1985); Michel-briand et al. (1986) |
| R386 | 0.05 μg/mL (sub-MIC): 2% CF | Michel-briand et al. (1986) | ||
| S-a | 0.5 μg/mL (sub-MIC): 1% CF | Michel-briand et al. (1986) | ||
| F’lac | 1/2 MIC: 66% CF | Weisser and Wiedemann (1985) | ||
| R16 | 1/2 MIC: 11% CF | Weisser and Wiedemann (1985) | ||
| Rts1 | Sub-MIC concentrations: 98% CF | Weisser and Wiedemann (1985) | ||
| pORF2 | Sub-MIC concentrations: 43% CF | Fu et al. (1988) | ||
| S. sonnei | pWR105 | 0.12 μg/mL (sub-MIC): 11% CF | Michel-briand et al. (1986) | |
| Flumequine | E. coli | R446b | 8 μg/mL (sub-MIC): 2% CF | Michel-briand et al. (1986) |
| S-a | 4 μg/mL (sub-MIC): 1% CF | Michel-briand et al. (1986) | ||
| S. sonnei | pWR105 | 0.25 μg/mL (sub-MIC): <1% CF | Michel-briand et al. (1986) | |
| S. dysenteriae | pWR24 | 0.12 μg/mL (sub-MIC): 2% CF | Michel-briand et al. (1986) | |
| S. flexneri | PWR110 | 0.12 μg/mL (sub-MIC): <1% CF | Michel-briand et al. (1986) | |
| Nalidixic Acid | E. coli | pMG110 | 4.3 μM (sub-MIC): 1% CF | Hooper et al. (1984) |
| R446b | 1/2 MIC: 8% CF, 64 μg/mL (sub-MIC): 4% CF | Weisser and Wiedemann (1985); Michel-briand et al. (1986) | ||
| F’lac | 1/2 MIC: 18% CF | Weisser and Wiedemann (1985) | ||
| R16 | 1/2 MIC: 41% CF | Weisser and Wiedemann (1985) | ||
| Rts1 | 1/2 MIC: 4% CF | Weisser and Wiedemann (1985) | ||
| pMC1314 | Sub-MIC concentrations of 0.3 μg/mL: 9.6% CF; 0.6 μg/mL: 17% CF; 1.2 μg/mL: 36% CF | Courtright, Turowski and Sonstein (1988) | ||
| S-a | 32 μg/mL (sub-MIC): 1.5% CF | Michel-briand et al. (1986) | ||
| S. sonnei | pWR105 | 8 μg/mL (sub-MIC): 1% CF | Michel-briand et al. (1986) | |
| S. enterica Typhimurium | R1 plasmids | 6.25 μM eliminated resistance with CFs of: 70% Kan, 56% Chl, 60% Str, 64% Amp | Hahn and Ciak (1976) | |
| Norfloxacin | E. coli | R446b | 1/2 MIC: 18% CF, 0.1 μg/mL (sub-MIC): 1% CF | Weisser and Wiedemann (1985); Michel-briand et al. (1986) |
| S-a | 0.25 μg/mL (sub-MIC): 3% CF | Michel-briand et al. (1986) | ||
| F’lac | 1/2 MIC: 19% CF | Weisser and Wiedemann (1985) | ||
| R16 | 1/2 MIC: 25% CF | Weisser and Wiedemann (1985) | ||
| Rts1 | 1/4 MIC: 52% CF | Weisser and Wiedemann (1985) | ||
| S. sonnei | pWR105 | 0.5 μg/mL (sub-MIC): <1% CF | Michel-briand et al. (1986) | |
| Novobiocin | E. coli | pDT4 | Novobiocin-sensitive strain was cured, but isogenic resistant strain was not | Taylor and Levine (1979) |
| pMG110 | 22 μM: 99% CF in wild-type strain, in gyrB resistant strain 990 μM: 33.3% CF | Hooper et al. (1984) | ||
| R386 | 200 μg/mL: 15% (IncFI) CF | McHugh and Swartz (1977) | ||
| R1–16 | 175 μg/mL: 34% (IncFII) CF | McHugh and Swartz (1977) | ||
| R726 | 175 μg/mL: 16.1% (IncH) CF | McHugh and Swartz (1977) | ||
| pMG102 | 50 μg/mL: 20.3%, 100 μg/mL: 14.7% CF | McHugh and Swartz (1977) | ||
| S. enterica | Virulence plasmid (100 kb) | 200–250 μg/mL used to cure virulence plasmid | Gulig and Curtiss III (1987) | |
| Enterobacter | pMG150 | 225 μg/mL: 52.5% CF | McHugh and Swartz (1977) | |
| E. faecalis | pJH1 | 8 μg/mL: 34% CF | McHugh and Swartz (1977) | |
| Enterococcus | pDR1 | 10 μg/mL: 28% CF | McHugh and Swartz (1977) | |
| L. plantarum | Multiple unidentified plasmids (2–68 kb) | 0.125–0.25 μg/mL: 94%–100% CF for four isolates | Ruiz-Barba, Piard and Jiménez-Díaz (1991) | |
| L. fermentum | Ery resistance plasmid | 1.8–40 μg/mL (sub-MIC): 64% CF, and 2.1% CF for two strains | Chin et al. (2005) | |
| L. acidophilus | Ery resistance plasmids (4.4–11.5 kb) | 1.8–40 μg/mL (sub-MIC): 3.3%–9.0% CF | Chin et al. (2005) | |
| Chl resistance plasmid (20.3 kb) | 2.4 μg/mL: 4.6% CF, peaked at 18 h | Karthikeyan and Santosh (2010) | ||
| C. muridarum | Cryptic plasmid (7.5 kb) | 4%–30% effective, but optimal concentration inhibited 99% of bacterial growth | O’Connell and Nicks (2006) | |
| Ofloxacin | E. coli | R446b | 1/2 MIC: 10% CF | Weisser and Wiedemann (1985); Michel-briand et al. (1986) |
| F’lac | 1/2 MIC: 39% CF | Weisser and Wiedemann (1985) | ||
| R16 | 1/2 MIC: 19% CF | Weisser and Wiedemann (1985) | ||
| Rts1 | 1/4 MIC: 32% CF | Weisser and Wiedemann (1985) | ||
| Oxolinic acid | E. coli | pMC1314 | Sub-MIC concentrations of 0.06 μg/mL: 24% CF; 0.12 μg/mL: 36% CF; 0.25 μg/mL: 100% CF | Courtright, Turowski and Sonstein (1988) |
| Pefloxacin | E. coli | R446b | 1/2 MIC: 21% CF, 0.1 μg/mL(sub-MIC): 1% CF | Weisser and Wiedemann (1985); Michel-briand et al. (1986) |
| F’lac | 1/2 MIC: 6% CF | Weisser and Wiedemann (1985); Selan et al. (1988) | ||
| R16 | 1/2 MIC: 16% CF | Weisser and Wiedemann (1985) | ||
| Rts1 | 1/2 MIC: 27% CF | Weisser and Wiedemann (1985) | ||
| S. sonnei | pWR105 | 1 μg/mL (sub-MIC): 2% CF | Michel-briand et al. (1986) | |
| S. dysenteriae | pWR24 | 1 μg/mL (sub-MIC): 4% CF | Michel-briand et al. (1986) | |
| S. flexneri | PWR110 | 1 μg/mL (sub-MIC): 4% CF | Michel-briand et al. (1986) | |
| Pipemidic acid | E. coli | R446b | 1/2 MIC: 4% CF4 μg/mL (sub-MIC): 6% CF | Weisser and Wiedemann (1985); Michel-briand et al. (1986) |
| F’lac | 1/2 MIC: 35% CF | Weisser and Wiedemann (1985) | ||
| R16 | 1/2 MIC: 31% CF | Weisser and Wiedemann (1985) | ||
| Rts1 | 1/2 MIC: 47% CF | Weisser and Wiedemann (1985) | ||
| R386 | 2 μg/mL (sub-MIC): 0.5% CF | Michel-briand et al. (1986) | ||
| S-a | 4 μg/mL (sub-MIC): 1% CF | Michel-briand et al. (1986) | ||
| S. sonnei | pWR105 | 1 μg/mL (sub-MIC): no curing | Michel-briand et al. (1986) | |
| Trovafloxacin | E. coli | pT713 (partial) | MIC: 50% CF | Brandi, Falconi and Ripa (2000) |
| pJEL144 (partial) | ⅓ MIC: 50% CF | Brandi, Falconi and Ripa (2000) | ||
| pRK2 (partial) | 1/2 MIC: 30% CF. Also reduced copy number | Brandi, Falconi and Ripa (2000) | ||
| Other Quinolones | E.coli | R446b | Rosoxacin: 2 μg/mL (sub-MIC): 1% CF β-Hydroxypiromydic acid: 32 μg/mL (sub-MIC): 3% CF Cinoxacin: 4 μg/mL (sub-MIC): 1% CF | Michel-briand et al. (1986) |
| R386 | Rosoxacin: 0.05 μg/mL (sub-MIC): 0.5% CF β-Hydroxypiromydic acid: 4 μg/ml (sub-MIC): 0.5% CF Cinoxacin: 4 μg/mL (sub-MIC): 0.5% CF | Michel-briand et al. (1986) | ||
| S-a | Rosoxacin: 2 μg/mL (sub-MIC): 1% CF β-Hydroxypiromydic acid: 64 μg/mL (sub-MIC): no curing Cinoxacin: 4 μg/mL (sub-MIC): 1% CF | Michel-briand et al. (1986) | ||
| S. sonnei | pWR105 | Rosoxacin: 0.12 μg/mL (sub-MIC): <1% CF β-Hydroxypiromydic acid: 0.25 μg/mL (sub-MIC): <1% CF Cinoxacin: 1 μg/mL (sub-MIC): 12% CF | Michel-briand et al. (1986) |
CF—Curing Frequency: the proportion of colonies which were cured of the plasmid compared to non-cured colonies. Kan—kanamycin, Chl—chloramphenicol, Str—streptomycin, Amp—ampicillin, Cou—coumermycin.
Quinolone antimicrobials also target DNA gyrase. There have been numerous reports of plasmids cured from various bacterial species by different quinolone antibiotics (Table 2). The majority of studies have been done using E. coli. For example, five fluoroquinolones and two quinolones cured four plasmids (Weisser and Wiedemann 1985), and subinhibitory levels of quinolones cured E. coli of various plasmids including large clinical plasmids (Table 2) (Oliva et al.1985; Platt and Black 1987; Courtright, Turowski and Sonstein 1988; Selan et al.1988). However, quinolones have variable curing activity on some plasmids (Weisser and Wiedemann 1985, 1986). For example, quinolones resulted in incomplete curing and reduced copy number of several plasmids (Table 2) (Phillips and Towner 1990; Brandi, Falconi and Ripa 2000), and were ineffective at curing E. coli of other plasmids (pBP1, R391, R27 or three small, high-copy plasmids from a clinical E. coli isolate) (Weisser and Wiedemann 1985; Platt and Black 1987). In line with this, one study demonstrated in E. coli that quinolones cured pORF2 with high efficiency, three plasmids were poorly cured and three plasmids were unaffected (Table 2) (Fu et al.1988). Interestingly, this study also examined quinolone efficacy at curing pORF2 from E. coli in vivo. They found quinolone treatment of mice infected with E. coli/pORF2 led to significant reduction in plasmid carriage (Fu et al.1988).
In a large study, 12 quinolones were tested for their ability to cure 11 plasmids of different incompatibility groups from E. coli, and virulence plasmids in five other species of Enterobacteriaceae (Table 2) (Michel-briand et al.1986). The authors concluded that non-fluorinated quinolones had slightly higher curing activity, but that novobiocin cured better than quinolones (Michel-briand et al.1986). Other studies examining a range of bacteria showed subinhibitory concentrations of quinolones reduced resistance and virulence plasmids in S. aureus, S. Typhimurium, E. coli, P. aeruginosa and Yersinia pseudotuberculosis (Hahn and Ciak 1976; Sonstein and Burnham 1993).
In summary, aminocoumarin-mediated curing appears to be more effective on Gram-positive bacteria than Gram-negative bacteria. Quinolone-mediated plasmid curing is effective on some plasmids in Gram-negative bacteria such as E. coli. However, this is complicated by the presence of plasmid-mediated quinolone-resistance genes, such as qnr, aac(6΄)-1b-cr, qepA and oqxAB (Jacoby, Strahilevitz and Hooper 2014; Rodriguez-Martinez et al.2016). Attempting to use quinolones to cure plasmids carrying quinolone-resistance genes could provide a fitness advantage to plasmid-containing cells, and would therefore select for plasmid maintenance. Furthermore, plasmid-mediated quinolone-resistance genes are frequently coded for by plasmids which carry other resistance genes conferring resistance to antimicrobials including beta-lactams, extended spectrum beta-lactams, carbapenems, aminoglycosides, trimethoprim and chloramphenicol (Rodriguez-Martinez et al.2016). Taken together, it is unlikely that antibiotics will be used to cure AMR plasmids in humans, animals or the environment as this will provide selection pressure for resistance to arise or be maintained within bacteria. Therefore, aminocoumarin and quinolone antibiotics are an effective laboratory tool, but are unlikely to be used elsewhere for plasmid curing.
Rifampicin
The antibiotic rifampicin inhibits RNA polymerase and is used to treat tuberculosis. Subinhibitory concentrations of rifampicin cured a penicillin-resistance plasmid from S. aureus (Johnston and Richmond 1970) and the F’lac plasmid from E. coli (Bazzicalupo and Tocchini-Valentini 1972). However, a rifampicin-resistant strain was not susceptible to curing, suggesting that the mechanism of curing was dependent upon the interaction of rifampicin with RNA polymerase (Bazzicalupo and Tocchini-Valentini 1972). In E. coli, haemolysin production was effectively cured by rifampicin (Mitchell and Kenworthy 1977). A gentamicin-resistance plasmid was not cured from two S. aureus strains using rifampicin, but it did cure one strain of a penicillin-resistance plasmid (Wood, Carter and Best 1977). Multiple studies have found rifampicin to be less effective than other curing agents (Rubin and Rosenblum 1971; Poppe and Gyles 1988), and given the importance of rifampicin in treating infections such as tuberculosis, it is unlikely to be used as a general plasmid curing agent.
Ascorbic acid
Research on the bioactive compound ascorbic acid (vitamin C) dates back to the first half of the 20th century. In aerobic conditions, ascorbic acid converts circular covalently closed DNA into open circular DNA (Morgan, Cone and Elgert 1975). To investigate the mechanism of action, fragments of pBR322 with radio-labelled 3΄ ends were used to demonstrate that efficient cleavage occurred preferentially at purine-rich regions (Chiou et al.1985). Studies on DNA extracted from E. coli demonstrated that ascorbic acid specificity was linked to negative torsion of the DNA, and this was influenced by ionic strength, salt concentration and pH (Wang and Ness 1989). In vitro studies on plasmid pBR322 DNA showed that ascorbic acid increased the damaging effects of dimethylarsinous acid and human liver ferritin (Ahmad, Kitchin and Cullen 2002). Synthesised ascorbic acid variants with protected (non-reactive) hydroxyl groups were tested for their ability to relax pUC19, which demonstrated that the hydroxyl groups at position C2 and C3 were essential for DNA damage (Liu et al.2006).
In S. aureus 1 mM ascorbic acid resulted in loss of penicillin and aminoglycoside resistance encoding plasmids (Table 1) (Amábile Cuevas 1988; Amábile-Cuevas, Piña-Zentella and Wah-Laborde 1991). Two plasmids, pI258 (penicillin resistance) and pT181 (tetracycline resistance), were not cured by ascorbic acid. However, there was a significant decrease in the MIC of tetracycline, which the authors hypothesised was due to reduction in plasmid copy number (Amábile-Cuevas, Piña-Zentella and Wah-Laborde 1991).
Ascorbic acid (1 mM) cured the lactic acid bacterium Pediococcus acidilactici of a plasmid coding for the production of pediocin, a metabolite which inhibits growth of some pathogenic bacteria, thus minimising food spoilage (Ramesh, Halami and Chandrashekar 2000). Ascorbic acid is non-toxic and is associated with human health benefits. This makes it an attractive curing agent, although it seems to be more effective at curing plasmids from Gram-positive rather than Gram-negative bacteria. Furthermore, after vitamin C supplementation concentrations of ascorbic acid in the plasma are relatively low (0.07 mM), but concentrations in lymphocytes can be much higher (3.5 mM), and concentrations in duodenal biopsies were around 1.2 mmol/kg (Levine et al.1996; Waring et al.1996). Conversely, Maier et al. (2018) estimated ascorbic acid concentrations in the intestine to be around 0.379 mM. Together this shows that while plasma concentrations after supplementation would not reach sufficient levels to have anti-plasmid activity, ascorbic acid is concentrated in the intestine, where it could potentially affect plasmids within intestinal bacteria. However this remains to be demonstrated in vivo.
Psychotropic drugs
The phenothiazines have been widely used in human medicine, originally as anti-helminthics, but now this class of drugs comprises the largest of five classes of anti-psychotic drugs (Ohlow and Moosmann 2011). The impact of these molecules on bacteria has been reviewed elsewhere (Amaral, Viveiros and Molnar 2004; Spengler et al.2006; Varga et al.2017). Plasmid curing properties have also been attributed to phenothiazines (Table 1) (Amaral, Viveiros and Molnar 2004; Spengler et al.2006; Dastidar et al.2013). In addition, a recent study found that chlorpromazine significantly impacted the growth of diverse members of the human microbiome, including Akkermansia muciniphila, Bacteroides uniformis, B. vulgatus, Clostridium perfringens, Parabacteroides distasonis and P. merdae (Maier et al.2018). Phenothiazines, including chlorpromazine, cured plasmids from E. coli (Table 1) (Mandi et al.1975; Molnar, Mandi and Kiraly 1976), and the curing activity was enhanced by methylene blue (Molnar et al.1980). Thioridazine cured the AMR phenotype from E. coli, S. flexneri and V. cholerae isolates, but not from S. aureus (Table 1) (Radhakrishnan et al.1999), while promethazine and trifluoperazine were tested on clinical isolates of E. coli, Citrobacter freundii and E. cloacae, but only one E. coli isolate was cured, despite E. coli K12 being readily cured of a lac-reporter plasmid (Spengler et al.2003). However, trifluoroketone 18 or trifluoromethyl-ketone 14 (proton pump inhibitors) enhanced curing activity of the phenothiazines, suggesting the compounds may be effluxed (Spengler et al.2003; Wolfart et al.2006). In mixed cultures of E. coli, B. cereus and S. epidermidis, promethazine cured F’lac from E. coli (Molnár, Amaral and Molnár 2003). Chlorpromazine cured the MRSA Iberian clone strain HPV107 of a plasmid encoding the QacA efflux pump (Costa et al.2010).
Together, this shows phenothiazines have in vitro curing activity on some bacteria and plasmid combinations. However, their in vivo efficacy as plasmid curing compounds remains unclear. Any potential connection between anti-plasmid and the anti-commensal activity of chlorpromazine remains to be elucidated. In patients being treated for psychosis with chlorpromazine, serum concentrations are around 0.1–0.3 μg/mL, and toxic side effects occur at 0.75 μg/mL (Sanofi-Aventis 2016). However, Maier et al. (2018) estimate intestinal concentration of chlorpromazine to be around 46 μM (14.67 μg/mL). The concentrations used for plasmid curing are generally around 10–100 μg/mL (Mandi et al.1975; Spengler et al.2003). Therefore, concentrations resulting in curing may be reached in the intestines of individuals being treated with chlorpromazine. Novel approaches involving targeted drug delivery or preventing uptake of orally administered phenothiazines may help to improve curing efficacy and reduce toxicity. Until such obstacles are overcome, the use of phenothiazines for in vivo plasmid curing is unlikely.
INCOMPATIBILITY-BASED PLASMID CURING SYSTEMS
Curing based upon the principle of plasmid incompatibility is an alternative method to chemical or drug-based strategies to remove plasmids from bacteria. Plasmid curing using an incompatible plasmid vector has been widely used in plasmid characterisation of Gram-positive and Gram-negative species. Introducing a smaller high-copy-number plasmid from the same incompatibility group may specifically eliminate a resident plasmid (Bringel, Frey and Hubert 1989). Incompatibility-based curing has been useful for investigating incompatibility mechanisms, plasmid–host interactions and for the construction of gene transfer systems (Uraji, Suzuki and Yoshida 2002). The main advantage of this method is the reduced risk of chromosomal mutations and toxicity sometimes associated with chemical curing agents (Hovi et al.1988; Poppe and Gyles 1988). In addition, incompatibility-based curing is specific to plasmids of the targeted incompatibility group. One major drawback of incompatibility-based curing methods is the extensive cloning required for set up, and the detailed knowledge of the target plasmid. Ni et al. (2008) reported the main difficulty in constructing incompatibility plasmids for curing is the replication control and/or partition region of the plasmid must be identified before curing (Ni et al.2008). Additional plasmid genes (e.g. antitoxin from a TA system) may need to be included (Ni et al.2008; Hale et al.2010).
Incompatibility-based curing has been used in a variety of bacteria and plasmids (Table 3). In particular, when chemical curing methods have proven less effective, e.g. Lactobacillus, and Y. pestis (Ruiz-Barba, Piard and Jiménez-Díaz 1991; Chin et al.2005; Ni et al.2008; Karthikeyan and Santosh 2010). Incompatibility-based curing systems were designed and used in L. acidophilus, L. plantarum and L. pentosus (Table 3) (Bringel, Frey and Hubert 1989; Posno et al.1991). Incompatibility has been used to study the contribution of plasmids to bacterial pathogenesis, including a systematic investigation of the role of plasmids in Y. pestis pathogenesis (Table 3) (Ni et al.2008). Incompatibility was used to cure vaccine and wild-type strains of B. anthracis of two large pathogenicity-related plasmids (Table 3), allowing study of their contribution to capsule and anthrax toxin production (Wang et al.2011; Liu et al.2012). Incompatibility has been used not only in human pathogens, but also to remove tumour inducing (Ti) plasmids from Agrobacterium tumefaciens, a dicotyledonous plant pathogen, in which Ti plasmids are responsible for inducing vegetable tumours (Table 3) (Uraji, Suzuki and Yoshida 2002).
Table 3.
Incompatibility based curing plasmids.
| Species | Curing plasmid details | Cured Plasmid Details | Key Findings | Delivery | Reference |
|---|---|---|---|---|---|
| L. plantarum | pULP8 and pULP9 6.6 kb, Amp and Ery resistance. Constructed by inserting the Ery-resistance gene from pVA891 into a pUC19-pLP1 construct | 2.1 kb pLp1 endogenous plasmid | Maintained in 5% of bacteria after 20 generations (selection free media). TE: 2 × 10−7 CFU/μg DNA | Electroporation | Bringel, Frey and Hubert (1989) |
| L. pentosus | pLP3537, 6.3 kb, Ery resistance. Constructed by inserting 2.3 kb endogenous plasmid into a screening vector, pEI2. Contained lactobacillus replicon | 2.3 kb endogenous plasmid | Maintained in 8% of bacteria after 100 generations (selection free media). TE: 102–103 CFU/μg DNA | Electroporation | Posno et al. (1991) |
| pLPE323, 3.6 kb, Ery resistance. Constructed by inserting 2.3 kb endogenous plasmid into pE194 vector. Contained lactobacillus replicon | 2.3 kb endogenous plasmid | Maintained in 100% of bacteria after 100 generations (selection free media). TE: 102–103 CFU/μg DNA | Electroporation | Posno et al. (1991) | |
| pGK12, 4.4 kb, broad Gram-positive host range plasmid | 1.7 kb endogenous plasmid | Maintained in <1% of bacteria after 100 generations (selection-free media). TE: 103 | Electroporation | Posno et al. (1991) | |
| A. tumefaciens | pMGTrep1, contained pTi repABC genes and sacB (sucrose sensitivity gene) to select for pMGTrep1 loss | pTi-SAKURA (206kb) pTiC58 (214kb) | Between 32% (pTi-SAKURA) and 99% (pTiC58) of transconjugants were cured of pTi | Conjugation | Uraji, Suzuki and Yoshida (2002) |
| Y. pestis | pEX18-PCP- pPCP1 replicon, sacB | pPCP1 virulence plasmid (ColE1) | 64% of colonies cured | Electroporation | Ni et al. (2008) |
| pEX18-MT- pMT1 replicon, sacB | pMT1 virulence plasmid (repA) | 30% of colonies cured | Electroporation | Ni et al. (2008) | |
| pEX18-CD- pCD1 replicon, sacB | pCD1 virulence plasmid (IncFIIA) | 98% of colonies cured | Electroporation | Ni et al. (2008) | |
| pEX18-CRY- pCRY replicon, sacB | pCRY (21.7 kb) cryptic plasmid | 70% of colonies cured | Electroporation | Ni et al. (2008) | |
| B. anthracis | pKS5K, contains ORF43–46, temperature sensitive | pXO1 (181.6 kb) encodes anthrax toxin/regulatory genes (pagA, lef, cya,atxA, pagR) | Isolate was successfully cured. CF not determined | Electroporation | Liu et al. (2012) |
| pKSV7-oriIV, contains pXO2 repS, repB, ori sequences, temperature sensitive | pXO2 (93.5 kb) encodes capsule synthesis and degradation genes (capABCD). | Isolate was successfully cured. CF not determined | Electroporation | Wang et al. (2011) | |
| pKORT, derived from pKSV7, contains pXO1 and pXO2 origins of replication, temperature sensitive | pXO1 and pXO2 | Isolate was successfully cured. CF not determined | Electroporation | Wang et al. (2015) | |
| E. coli | pCURE1, anti-pO157, pMB1 replicon, oriTRK2, sacB, Amp and Kan resistance | pO157 (F-like plasmid) | Isolate was successfully cured. CF not determined | Transformation or mobilisation by IncP-1 transfer system (due to oriTRK2) | Hale et al. (2010) |
| pCURE2, anti-IncF pMB1 replicon, oriTRK2, sacB, Amp and Kan resistance | IncF-like plasmids including p1658/97, pKDSC50 (RepFIB and RepFIIA), F and F’ plasmids (RepFIA) | Highly effective on IncF plasmids, CF up to 100%. Inclusion of anti-toxin genes on pCURE2 increased efficacy | Transformation or mobilisation by IncP-1 transfer system (due to oriTRK2) | Hale et al. (2010) | |
| pCURE11, anti-IncP-1α, pMB1 replicon, oriTRK2, sacB, Amp and Kan resistance | pRK24 (IncP-1α), derivative of RK2 | CF: 100% of tested colonies | Transformation or mobilisation by IncP-1 transfer system (due to oriTRK2) | Hale et al. (2010) | |
| pJIMK3, pemI anti-toxin gene, no incompatibility genes included | pEI1573 (IncL/M), carries blaIMP-4 isolated from E. cloacae | 30% CF of pEI1573 in E. coli | Transformation | Kamruzzaman et al. (2017) | |
| pJIMK25, pJIMK3 with addition of IncL/M replication fragments | pEI1573 | 100% CF pEI1573 in E. coli | Transformation | Kamruzzaman et al. (2017) | |
| E. coli, K. pneumoniae, C. freundii, M. morganii | pJIMK46, pemI anti-toxin gene, fosA3, IncL/M and IncI1 replicons | pEI1573, pJIE512b (conjugative IncI1 plasmid with blaCMY-2 isolated from E. coli) | Cured when curing plasmid was selected for using antibiotics. Cured in vitro in E. coli, K. pneumoniae, C. freundii, M. morganii. Cured pEI1573 in vivo, from E. coli, but required antibiotic selection for pJIMK46 | Conjugation | Kamruzzaman et al. (2017) |
TE—Transformation efficiency of curing plasmid, CF—curing frequency of plasmid, Ery—erythromycin, Amp—ampicillin, Kan—kanamycin.
Incompatibility-based plasmids called pCURE were constructed for curing pO157 (a typical F-like plasmid), other F-like and IncP-1α plasmids from E. coli (Table 3) (Hale et al.2010). To create the pCURE constructs, elements expected to interfere with specific functions were chosen, such as repressing vital components (e.g. transcriptional repressor, antisense RNA or other translational regulators) and competition for vital steps (e.g. replication origin) (Hale et al.2010). To control the TA system, either the putative antitoxin or antisense RNA repressor was included (Hale et al.2010).
In a recent study, ‘interference plasmids’ were designed which combined an antitoxin gene and replicon genes to cure blaIMP-4 and blaCMY-2 encoding plasmids both in vitro and in vivo (Table 3) (Kamruzzaman et al.2017). In the presence of the antibiotic selecting for the interference plasmid, target plasmids were effectively removed from E. coli, K. pneumoniae, C. freundii and Morganella morganii in vitro, and from E. coli colonising the mouse intestine. Interference plasmids were lost from the mouse intestine after cessation of antibiotic treatment.
One targeted approach sought small molecules which mimic the incompatibility system of IncB plasmids (Denap et al.2004). They found the aminoglycoside apramycin binds to the SLI region of the RepA mRNA, preventing translation of RepA, which is necessary for plasmid replication (Denap et al.2004). Treatment of E. coli harbouring pMU2403 (IncB) with apramycin resulted in almost complete plasmid elimination (Denap et al.2004).
An important question regarding use outside the laboratory of incompatibility-based curing systems is how to apply the curing plasmids to people, animals or the environment. Plasmids could be delivered via bacteria or phage. However, the potential requirement for antibiotic treatment to select for the curing plasmids (Kamruzzaman et al.2017) would be a significant drawback. Another concern regarding curing plasmids is the potential for acquisition of ARG(s) onto the curing backbone. More research is needed in increasingly complex plasmid systems to study the dynamics between curing plasmids and AMR plasmids, including research focused on minimising the need for antibiotic selection.
PHAGE-BASED ANTI-PLASMID SYSTEMS
For the past 50 years, bacteriophages which specifically target the pili of plasmid conjugation systems have been studied (Caro and Schnös 1966). More recently, this has been studied in the context of AMR plasmids. Phages which target the conjugation pilus preferentially kill bacteria with high pilus expression (Dionisio 2005). Low pilus expression results in reduced susceptibility to phage, but also reduced conjugation rates. Therefore, diversity in pilus expression within a bacterial population improves the chances of plasmid survival (Dionisio 2005). Another example of bacteriophages specifically targeting AMR plasmids involved the phage PRD1 which targeted the mating pair complex of plasmids RP4 and RN3 (Jalasvuori et al.2011). PRD1 reduced plasmid carriage within E. coli and Salmonella populations from 100% to 5% after 10 days. Furthermore, the 5% which retained plasmid had lost the ability to conjugate (Jalasvuori et al.2011). PRD1 significantly reduced the number of E. coli K12 RP4 transconjugants, and even reduced transconjugants when single, sub-MIC antibiotic selection was applied (Ojala, Laitalainen and Jalasvuori 2013). However, when double selection for transconjugants was applied phage-resistant mutants arose, but 65% had lost the ability to conjugate (Ojala, Laitalainen and Jalasvuori 2013). Together, this demonstrates the use of phage to produce an evolutionary pressure which results in either plasmid loss or evolution of a non-conjugative plasmid. This fits with the other research focused on using phage-mediated directed evolution to select for antibiotic sensitive bacteria (Chan et al.2016).
The M13 filamentous phage minor coat protein g3p was necessary and sufficient to inhibit F-plasmid conjugation in E. coli (Lin et al.2011). Another study modelled the dynamics of the F-plasmid and M13 phage in E. coli (Wan and Goddard 2012). They found M13 infection reduced cell growth rate, and the conjugation rate was only one order of magnitude faster than the rate of phage infection. This implies that a high concentration of phage would be required to effectively prevent conjugation, and they showed that conjugation continues even with phage (Wan and Goddard 2012). Recently, the evolutionary and ecological implications of lytic bacteriophage predation on plasmid maintenance in a population of P. fluorescens were examined (Harrison et al.2015). They concluded that phage accelerates plasmid loss in the absence of selective pressure (Harrison et al.2015).
In summary, these studies show that phage can be a highly effective tool for reducing plasmid prevalence within a population. Another advantage of bacteriophage approaches is their status as ‘generally regarded as safe’, which streamlines downstream applications such as use of phage to decolonise surfaces, as a probiotic or use on farms. However, unclear regulatory pathways for use of phage as medication still pose a problem. Another problem associated with phage therapy is bacterial evolution of resistance to phage. By understanding the evolutionary pressures applied to bacteria by phage predation, this evolution can be harnessed to increase susceptibility to antibiotics (Jalasvuori et al.2011; Ojala, Laitalainen and Jalasvuori 2013; Chan et al.2016). Future research is needed to further our understanding of the phage-plasmid-host dynamics, to improve upon evolution-optimised approaches and to test these approaches in increasingly complex models.
CRISPR/CAS-BASED PLASMID CURING SYSTEMS
CRISRP/Cas is a bacterial ‘adaptive immune system’ which allows recognition, degradation and memory of foreign DNA sequences. CRISPR/Cas works as a result of spacer DNA segments coded by the bacteria that are transcribed into crRNA. The crRNA is bound by the Cas protein complex which cleaves nucleic acid sequences matching the crRNA, resulting in double-stranded breaks (Sternberg and Doudna 2015). DNA repair mechanisms can be used to insert a desired sequence into the break (Sternberg and Doudna 2015). In bacteria, double-stranded breaks are often fatal, but combination with traditional recombineering systems such as λ-red can allow for effective genome editing (Peters et al.2015). The highly specific CRISPR/Cas system has been extensively described and reviewed elsewhere (Jiang and Doudna 2015; Sternberg and Doudna 2015; Wright, Nuñez and Doudna 2016). In a seminal paper, Garneau et al. (2010) showed that Streptococcus thermophiles isolates which had lost the plasmid pNT1 had acquired new spacer sequences which targeted pNT1. This work demonstrated that CRISPR/Cas acted to remove plasmid DNA from bacteria.
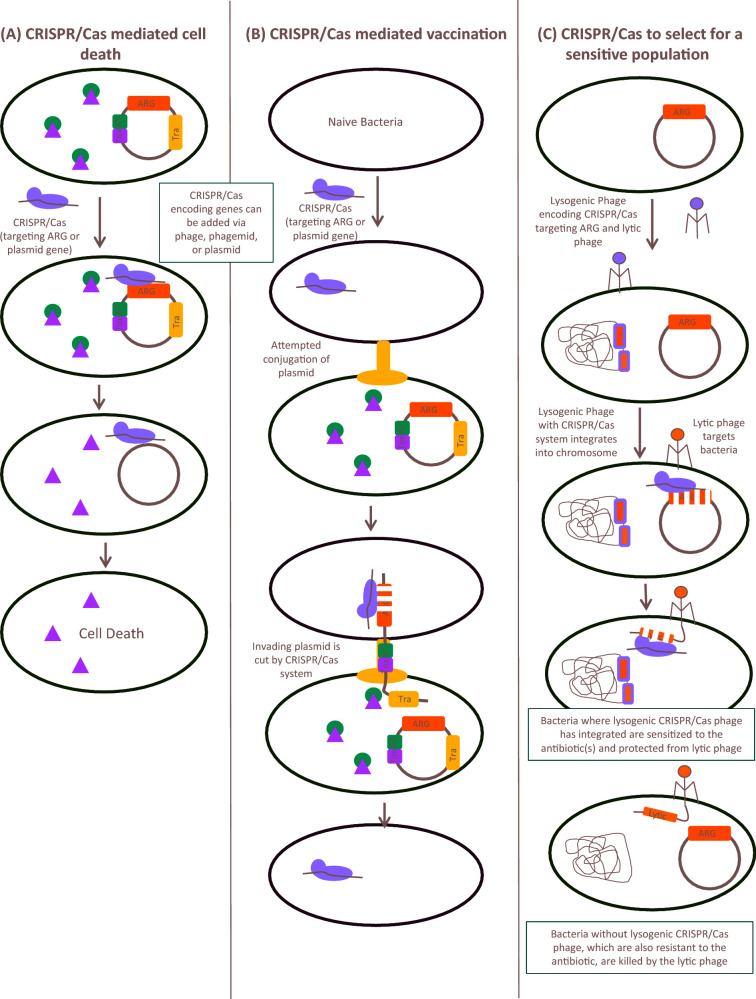
Recently, the CRISPR/Cas system has been explored as a method for plasmid curing. Firstly, it can be designed to target specific plasmid genes, including ARGs. The double-stranded breaks introduced in the process can reduce the stability of the plasmid, and in some cases result in plasmid loss (Fig. 2a) (Kim et al.2016; Lin et al.2016). Plasmids in isolates from man, animals or the environment frequently carry TA systems. TA systems, sometimes called addiction systems, are comprised of a toxin and an antitoxin gene (Van Melderen and Saavedra De Bast 2009; Chan, Espinosa and Yeo 2016). Generally, the activity of the stable toxin is mitigated by a less stable antitoxin. Therefore, as long as the antitoxin is produced, the toxin cannot act (Van Melderen and Saavedra De Bast 2009; Chan, Espinosa and Yeo 2016). When encoded on plasmids, the TA system functions by killing daughter cells which do not contain a copy of the plasmid coding for the antitoxin gene, a process termed postsegregational killing (Chan, Espinosa and Yeo 2016). Therefore, targeting plasmids with TA systems resulted in bacterial cell death (Fig. 2a) (Citorik, Mimee and Lu 2014). Toxin-mediated cell death could be complemented by antitoxin-encoding phage (Citorik, Mimee and Lu 2014). Specific ARGs can also be targeted by CRISPR/Cas systems (Citorik, Mimee and Lu 2014; Kim et al.2016). For example, homologous regions in TEM and SHV beta-lactamases were targeted (Kim et al.2016). CRISPR/Cas systems can also target plasmid backbone genes such as replicase genes (Cao et al.2017). CRISPR/Cas systems targeted and removed multiple AMR plasmids simultaneously (Yosef et al.2015).
Figure 2.
CRISPR/Cas as an anti-plasmid strategy. (A) CRISPR/Cas systems (purple) which target plasmid encoded genes cause double-stranded breaks in the AMR plasmid, leading to plasmid degradation. In plasmids with toxin (Tox, blue) antitoxin (AT, green) systems, loss of plasmid leads to active toxin. The toxin then mediates cell death, resulting in removal of AMR plasmid carrying bacteria from a population. (B) CRISPR/Cas system prevents uptake of plasmid DNA. Bacteria encoding CRISPR/Cas system that targets plasmid genes degrade incoming DNA, including conjugative (Tra, orange) AMR plasmids, thus preventing spread of AMR palsmids. (C) CRISPR/Cas system combined with lysogenic and lytic phages selects for an antimicrobial sensitive population. Lysogenic phages encoding CRISPR/Cas systems which target both AMR plasmid and lytic phage are administered to bacteria. Production of the CRISPR/Cas system results in degradation of the AMR plasmid, and protection from lytic phages. Administration of lytic phages kills all non-sensitised bacteria, which do not encode the lytic phage resistance, thus producing evolutionary pressure for an antimicrobial sensitive population.
Secondly, CRISPR/Cas is an attractive strategy because it can be used to prevent plasmid transmission by ‘vaccination’ (Fig. 2b). Methicillin-sensitive S. aureus was vaccinated against pUSA02, the plasmid responsible for methicillin resistance in the epidemic MRSA strain USA300 (Bikard et al.2014). Likewise, E. coli containing CRISPR/Cas targeting blaCTX-M-15 and blaNDM-1 were less efficiently transformed with an AMR plasmid carrying these genes (Yosef et al.2015). In E. faecalis the CRISPR3-Cas locus was deleted, resulting in significantly higher acquisition of the pAD1 plasmid (target sequence located in CRISPR3), while acquisition of pCF10 was unaffected, as it is not targeted by CRISPR3 (Price et al.2016). In line with this, carbapenem-resistant K. pneumoniae were less likely to have active CRISPR/Cas systems than carbapenem-sensitive strains (Lin et al.2016). Similarly, type I-F CRISPR/Cas systems were more common in E. coli isolates that were antimicrobial sensitive (Aydin et al.2017). Some of these CRISPR spacers aligned to sequences commonly found in IncFII and IncI1 plasmids, which are associated with clinical resistance (Aydin et al.2017). This strongly suggests that antimicrobial-sensitive isolates can use CRISPR/Cas systems to degrade incoming AMR plasmids.
The benefits of using CRISPR/Cas systems to cure bacterial plasmids are clear. However, there are significant drawbacks associated with this strategy. One of the primary problems is delivery. A variety of delivery methods including plasmid transformation, conjugation, phagemid and bacteriophages have been used predominantly in vitro, with limited studies using in vivo models (Bikard et al.2014; Yosef et al.2015; Kim et al.2016). Despite this, so far the practical use of these systems is limited. One study used bacteria sensitised to antibiotics by a lysogenic CRISPR phage which also carried resistance to a lytic phage, thus combining use of the lytic and lysogenic phages to put evolutionary pressure on bacteria to become drug sensitive (Fig. 2c) (Yosef et al.2015). A recently devised system termed GOTraP enhances DNA transduction to a variety of bacterial species, including E. coli, S. sonnei and K. pneumoniae (Yosef et al.2017). Such a strategy could be employed to improve the delivery of anti-ARG or anti-plasmid CRISPR/Cas systems. In another study, a phagemid was effective for treating S. aureus skin lesions in mice (Bikard et al.2014). CRISPR/Cas may be an effective treatment for curing plasmids from surface wounds and burns, but advances in systemic treatment are still required.
The specificity of CRISPR/Cas is both a benefit and a drawback. For example, the lack of common sequences among variants of blaOXA and blaCTX-M beta-lactamases restricted CRISPR design (Kim et al.2016). Frequently, plasmid-mediated antibiotic resistance genes have multiple sequence variants, so if CRISPR/Cas sequences were used to kill bacteria there could be selective pressure for mutations in the regions targeted by CRISPR to give a CRISPR-resistant plasmid (Gomaa et al.2014). Similarly, in a phagemid system, resistance occurred due to deletions of the cas9 gene on the phagemid (Bikard et al.2014). Future research must consider the evolutionary pressure, and as some have done (Yosef et al.2015), design strategies to reduce development of resistance.
FUTURE OF PLASMID CURING AND ANTI-PLASMID APPROACHES
We and others anticipate that future research will continue in this area, driven in large part by the need to prevent and treat resistant infections (Getino and de la Cruz 2018). Methods to effectively and safely cure plasmids have the potential to diminish the severity of the impact of drug-resistant infections. Currently, few studies have examined curing methods in vivo. These studies along with future in vivo plasmid curing studies will be crucial in developing methods to sensitise bacteria to existing antibiotics. In the future, it may be that doctors prescribe a plasmid curing agent to help ensure that the antibiotics taken by the patient are effective. Alternatively, a plasmid curing agent could be taken by an individual (e.g. on return from an area where plasmid-mediated drug-resistance is common) as a way of restoring drug-susceptible bacteria to the gastrointestinal microbiome.
The use of plasmid curing strategies in settings other than in humans and animals should not be under appreciated. Reducing the global burden of AMR will require a multifaceted One-Health approach, and curing AMR plasmids from ARG hot spots such as waste water, manure and downstream of pharmaceutical (antibiotic) factories is a viable strategy. Some of the approaches described may be more suited to an environmental or agricultural setting and not for human use. For instance, another potential use of curing strategies could be on farms where livestock are often exposed to antibiotics, and harbour multiple MDR plasmids. Soil, waste water treatment and aquaculture could all be treated with plasmid curing agents to reduce drug resistance.
One concern surrounding plasmid curing is the potential for the development of resistance to anti-plasmid approaches, and the impact of these approaches on the bacterial community structure. Bacteria are constantly evolving, which makes developing ‘evolution-proof’ drugs extremely challenging, but by striving for this gold standard it may be possible to delay resistance (Bell and MacLean 2018). While these ideas apply to antimicrobials, similar principles can applied to plasmid curing strategies. If an anti-plasmid approach kills or produces a fitness defect in the bacteria, it seems likely that resistance will occur. For example, some of the CRISPR/Cas approaches selectively kill resistant bacteria by targeting specific sequences. Mutations in these sequences would then result in survival of the mutants, and expansion due to the resistance phenotypes. Thus, methods to provide selective advantages for the sensitive strains or to guide evolution towards antimicrobial susceptible strains can be utilised to help overcome these challenges. Antibiotics which have curing properties (e.g. quinolones and rifampicin) are obvious examples where resistance would be selected, and therefore should not be used as curing compounds outside the laboratory. Another concern is cell permeability. If plasmids provide a beneficial trait, mutations which reduce permeability or increase efflux may be selected by plasmid curing compounds. Indeed bacteria may increase expression of efflux to remove potentially detrimental plasmid curing compounds from the cells. Such mutations could also produce reduced susceptibility to clinically important antimicrobials. Research should consider and address these concerns.
Altogether, plasmid curing has come a long way, from the use of toxic compounds to novel designer curing methods based on incompatibility or CRISPR/Cas. Further research is now needed to uncover safe and effective means to cure plasmids, particularly in the face of the global AMR crisis.
Acknowledgements
We thank Finbarr Hayes for critically reading this manuscript and providing constructive feedback. MMCB was funded in part by a fellowship from the AXA Research Fund [14-AXA-PDOC-269] and the Wellcome Trust ISSF. MMCB and MLC are funded by a grant to LJVP from the Medical Research Council, UK [MR/N012933/1]. The funders had no role in this work or the decision to submit the work for publication.
FUNDING
This work was supported by the Medical Research Council [MR/N012933/1], the AXA Research Fund [14-AXA-PDOC-269 to MMCB], and the Wellcome Trust ISSF [to MMCB].
Conflicts of interest. None declared.
REFERENCES
- Adeyemo SM, Onilude AA. Plasmid curing and its effect on the growth and physiological characteristics of Lactobacillus plantarum isolated from fermented cereals. J Microbiol Res 2015;5:11–22. [Google Scholar]
- Ahmad S, Kitchin KT, Cullen WR. Plasmid DNA damage caused by methylated arsenicals, ascorbic acid and human liver ferritin. Toxicol Lett 2002;133:47–57. [DOI] [PubMed] [Google Scholar]
- Amábile Cuevas CF. Loss of penicillinase plasmids of Staphylococcus aureus after treatment with L-ascorbic acid. Mutat Res Lett 1988;207:107–9. [DOI] [PubMed] [Google Scholar]
- Amábile-Cuevas CF, Piña-Zentella RM, Wah-Laborde ME. Decreased reistance to antibiotics and plasmid loss in plasmid-carrying strains of Staphylocccus aureus treated with ascorbic acid. Mutat Res Lett 1991;264:119–25. [DOI] [PubMed] [Google Scholar]
- Amaral L, Viveiros M, Molnar J. Antimicrobial activity of phenothiazines. In Vivo (Brooklyn) 2004;18:725–32. [PubMed] [Google Scholar]
- Amos GCA, Hawkey PM, Gaze WH et al. Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J Antimicrob Chemoth 2014;69:1785–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriole VT. The quinolones: past, present, and future. Clin Infect Dis 2005;41:S113–9. [DOI] [PubMed] [Google Scholar]
- Arcilla MS, van Hattem JM, Haverkate MR et al. Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect Dis 2017;17:78–85. [DOI] [PubMed] [Google Scholar]
- Aviv G, Rahav G, Gal-mor O. Horizontal transfer of the Salmonella enterica serovar infantis resistance and virulence plasmid pesi to the gut microbiota of warm-blooded hosts. mBio 2016;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv G, Tsyba K, Steck N et al. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ Microbiol 2014;16:977–94. [DOI] [PubMed] [Google Scholar]
- Axelsson LT, Ahrné SEI, Andersson MC et al. Identification and cloning of a plasmid-encoded erythromycin resistance determinant from Lactobacillus reuteri. Plasmid 1988;20:171–4. [DOI] [PubMed] [Google Scholar]
- Aydin S, Personne Y, Newire E et al. Presence of Type I-F CRISPR/Cas systems is associated with antimicrobial susceptibility in Escherichia coli. J Antimicrob Chemoth 2017;72:2213–8. [DOI] [PubMed] [Google Scholar]
- Banuelos-Vazquez LA, Torres Tejerizo G, Brom S. Regulation of conjugative transfer of plasmids and integrative conjugative elements. Plasmid 2017;91:82–89. [DOI] [PubMed] [Google Scholar]
- Baxter JC, Funnell BE. Plasmid partition mechanisms. Microbiol Spectr 2014;2:1–20. [DOI] [PubMed] [Google Scholar]
- Bazzicalupo P, Tocchini-Valentini GP. Curing of an Escherichia coli episome by rifampicin. P Natl Acad Sci USA 1972;69:298–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg AZ, Ahmad I. Effect of Plumbago zeylanica extract and certain curing agents on multidrug resistant bacteria of clinical origin. World J Microb Biot 2000;16:841–4. [Google Scholar]
- Bell G, MacLean C. The search for “evolution-proof” antibiotics. Trends Microbiol 2018;26:471–83. [DOI] [PubMed] [Google Scholar]
- Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M beta-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemoth 2017;72:2145–55. [DOI] [PubMed] [Google Scholar]
- Bharathi A, Polasa H. Elimination of broad-host range plasmid vectors in Escherichia coli by curring agents. FEMS Microbiol Lett 1991;84:37–40. [DOI] [PubMed] [Google Scholar]
- Bikard D, Euler CW, Jiang W et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol 2014;32:1146–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah D, Yadav, RNS. Plasmid curing of a novel hydrocarbon degrading Bacillus cereus strain DRDU1 revealed its involvement in petroleum oil degradation. J Pet Env Biotechnol 2015;6:1–4. [Google Scholar]
- Bouanchaud DH, Chabbert YA. Practical effectiveness of agents curing R factors and plasmids. Ann NY Acad Sci 1971;65:305–11. [DOI] [PubMed] [Google Scholar]
- Bouanchaud DH, Scavizzi MR, Chabbert YA. Elimination by ethidium bromide of antibiotic resistance in Enterobacteria and Staphylococci. J Gen Microbiol 1968;54:417–25. [DOI] [PubMed] [Google Scholar]
- Brandi L, Falconi M, Ripa S. Plasmid curing effect of trovafloxacin. FEMS Microbiol Lett 2000;184:297–302. [DOI] [PubMed] [Google Scholar]
- Bringel F, Frey L, Hubert JC. Characterization, cloning, curing, and distribution in lactic acid bacteria of pLP1, a plasmid from Lactobacillus plantarum CCM 1904 and its use in shuttle vector construction. Plasmid 1989;22:193–202. [DOI] [PubMed] [Google Scholar]
- Cabello FC. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol 2006;8:1137–44. [DOI] [PubMed] [Google Scholar]
- Cabezon E, Ripoll-Rozada J, Pena A et al. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev 2015;39:81–95. [DOI] [PubMed] [Google Scholar]
- Cao Q-H, Shao H-H, Qiu H et al. Using the CRISPR/Cas9 system to eliminate native plasmids of Zymomonas mobilis ZM4. Biosci Biotechnol Biochem 2017;81:453–9. [DOI] [PubMed] [Google Scholar]
- Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Ch 2009;53:2227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. Plasmids in Gram negatives: Molecular typing of resistance plasmids. Inte J Med Microbiol 2011;301:654–8. [DOI] [PubMed] [Google Scholar]
- Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol 2013;303:298–304. [DOI] [PubMed] [Google Scholar]
- Carattoli A, Bertini A, Villa L et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Meth 2005;63:219–28. [DOI] [PubMed] [Google Scholar]
- Carmo LP, Schüpbach-Regula G, Müntener C et al. Approaches for quantifying antimicrobial consumption per animal species based on national sales data: a Swiss example, 2006 to 2013. Euro Surveill 2017;22:30458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro LG, Schnös M. The attachment of the male-specific bacteriophage F1 to sensitive strains of Escherichia coli. P Natl Acad Sci USA 1966;56:126–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casu B, Arya T, Bessette B et al. Fragment-based screening identifies novel targets for inhibitors of conjugative transfer of antimicrobial resistance by plasmid pKM101. Sci Rep 2017;7:14907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casu B, Smart J, Hancock MA et al. Structural analysis and inhibition of TraE from the pKM101 type IV secretion system. J Biol Chem 2016;291:23817–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan BK, Sistrom M, Wertz JE et al. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep 2016;6:26717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WT, Espinosa M, Yeo CC. Keeping the wolves at bay: antitoxins of prokaryotic type II toxin-antitoxin systems. Front Mol Biosci 2016;3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassy BM, Gibson EM, Guiffrida A. Evidence for plasmid-associated lactose metabolism in Lactobacillus casei subsp.casei. Curr Microbiol 1978;1:141–4. [DOI] [PubMed] [Google Scholar]
- Chin SC, Abdullah N, Siang TW et al. Plasmid profiling and curing of Lactobacillus strains isolated from the gastrointestinal tract of chicken. J Microbiol 2005;43:251–6. [PubMed] [Google Scholar]
- Chiou SH, Chang WC, Jou YS et al. Specific cleavages of DNA by ascorbate in the presence of copper ion or copper chelates. J Biochem 1985;98:1723–6. [DOI] [PubMed] [Google Scholar]
- Chuanchuen R, Beinlich K, Hoang TT et al. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob Age Ch 2001;45:428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol 2014;32:1141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleri A, Cokmus C, Ozcan B et al. Determination of antibiotic resistance and resistance plasmids of clinical Enterococcus species. J Gen Appl Microbiol 2004;50:213–9. [DOI] [PubMed] [Google Scholar]
- Conte D, Palmeiro JK, da Silva Nogueira K et al. Characterization of CTX-M enzymes, quinolone resistance determinants, and antimicrobial residues from hospital sewage, wastewater treatment plant, and river water. Ecotoxicol Environ Safe 2017;136:62–69. [DOI] [PubMed] [Google Scholar]
- Costa SS, Ntokou E, Martins A et al. Identification of the plasmid-encoded qacA efflux pump gene in meticillin-resistant Staphylococcus aureus (MRSA) strain HPV107, a representative of the MRSA Iberian clone. Int J Antimicrob Ag 2010;36:557–61. [DOI] [PubMed] [Google Scholar]
- Cottell JL, Webber MA, Coldham NG et al. Complete sequence and molecular epidemiology of IncK epidemic plasmid encoding bla CTX-M-14. Emerg Infect Dis 2011;17:645–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtright JB, Turowski DA, Sonstein SA. Alteration of bacterial DNA structure, gene expression, and plasmid encoded antibiotic resistance following exposure to enoxacin. J Antimicrob Chemoth 1988;21:1–18. [DOI] [PubMed] [Google Scholar]
- Crofts TS, Gasparrini AJ, Dantas G. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol 2017;15:422–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Gragerov AI. Curing of Escherichia coli K12 plasmids by coumermycin. Mol Gen Genet 1980;178:233–5. [DOI] [PubMed] [Google Scholar]
- Dastidar S, Kristiansen J, Molnar J et al. Role of phenothiazines and structurally similar compounds of plant origin in the fight against infections by drug resistant bacteria. Antibiotics 2013;2:58–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denap JCB, Thomas JR, Musk DJ et al. Combating drug-resistant bacteria: small molecule mimics of plasmid incompatibility as antiplasmid compounds. J Am Chem Soc 2004;126:15402–4. [DOI] [PubMed] [Google Scholar]
- Dionisio F. Plasmids surive despite their cost and male-specific phages due to heterogeneity of bacterial populations. Evol Ecol Res 2005;7:1089–107. [Google Scholar]
- ECDC Surveillance of antimicrobial resistance in Europe 2016. Annual Report of the European Antimicrobial Resistance Surveillance Network 2016:1–100. [Google Scholar]
- El-Mansi M, Anderson KJ, Inche CA et al. Isolation and curing of the Klebsiella pneumoniae large indigenous plasmid using sodium dodecyl sulphate. Res Microbiol 2000;151:201–8. [DOI] [PubMed] [Google Scholar]
- Ersfeld-Dressen H, Sahl H-G, Brandis H. Plasmid involvement in production of and immunity to the staphylococcin-like peptide Pep 5. Microbiology 1984;130:3029–35. [DOI] [PubMed] [Google Scholar]
- Fernandez-Lopez R, Machon C, Longshaw CM et al. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology 2005;151:3517–26. [DOI] [PubMed] [Google Scholar]
- Fernando DM, Xu W, Loewen PC et al. Triclosan can select for an AdeIJK-overexpressing mutant of Acinetobacter baumannii ATCC 17978 that displays reduced susceptibility to multiple antibiotics. Antimicrob Agents Ch 2014;58:6424–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu KP, Grace ME, Hsiao CL et al. Elimination of antibiotic-resistant plasmids by quinolone antibiotics. Chemotherapy 1988;34:415–8. [DOI] [PubMed] [Google Scholar]
- Gantzhorn MR, Olsen JE, Thomsen LE. Importance of sigma factor mutations in increased triclosan resistance in Salmonella Typhimurium. BMC Microbiol 2015;15:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Quintanilla M, Prieto AI, Barnes L et al. Bile-induced curing of the virulence plasmid in Salmonella enterica serovar typhimurium. J Bacteriol 2006;188:7963–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Quintanilla M, Ramos-Morales F, Casadesús J. Conjugal transfer of the Salmonella enterica virulence plasmid in the mouse intestine. J Bacteriol 2008;190:1922–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010;468:67–71. [DOI] [PubMed] [Google Scholar]
- Gellert M, O’Dea MH, Itoh T et al. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. P Natl Acad Sci USA 1976;73:4474–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getino M, de la Cruz F. Natural and artificial strategies to control the conjugative transmission of plasmids. Microbiol Spectr 2018;6:MTBP-0015-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getino M, Fernandez-Lopez R, Palencia-Gandara C et al. Tanzawaic acids, a chemically novel set of bacterial conjugation inhibitors. PLoS ONE 2016;11:e0148098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getino M, Sanabria-Rios DJ, Fernandez-Lopez R et al. Synthetic fatty acids prevent plasmid-mediated horizontal gene transfer. mBio 2015;6:e01032–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessweiner-Mohr N, Arends K, Keller W et al. Conjugation in gram-positive bacteria. Microbiol Spectr 2014;2:PLAS-0004-2013. [DOI] [PubMed] [Google Scholar]
- Gomaa AA, Klumpe HE, Luo ML et al. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio 2014;5:e00928–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig PA, Curtiss R III. Plasmid- associated virulence of Salmonella typhimurium. Infect Immun 1987;55:2891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn FE, Ciak J. Elimination of resistance determinants from R-factor R1 by intercalative compounds. Antimicrob Agents Ch 1976;9:77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale L, Lazos O, Haines A et al. An efficient stress-free strategy to displace stable bacterial plasmids. BioTechniques 2010;48:223–8. [DOI] [PubMed] [Google Scholar]
- Hall JPJ, Brockhurst MA, Dytham C et al. The evolution of plasmid stability: Are infectious transmission and compensatory evolution competing evolutionary trajectories? Plasmid 2017;91:90–95. [DOI] [PubMed] [Google Scholar]
- Harrison E, Wood AJ, Dytham C et al. Bacteriophages limit the existence conditions for conjugative plasmids. mBio 2015;6:e00586–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A, Ruiz FM, Romero A et al. The binding of triclosan to SmeT, the repressor of the multidrug efflux pump SmeDEF, induces antibiotic resistance in Stenotrophomonas maltophilia. PLoS Pathog 2011;7:e1002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer H, Binh CTT, Jechalke S et al. IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: diversification driven by class 1 integron gene cassettes. Front Microbio 2012;3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AH, Moore LS, Sundsfjord A et al. Understanding the mechanisms and drivers of antimicrobial resistance. The Lancet 2016;387:176–87. [DOI] [PubMed] [Google Scholar]
- Hooper DC, Wolfson JS, McHugh GL et al. Elimination of plasmid pMG110 from Escherichia coli by novobiocin and other inhibitors of DNA gyrase. Antimicrob Agents Ch 1984;25:586–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovi M, Sukupolvi S, Edwards MF et al. Plasmid-associated virulence of Salmonella enteritidis. Microbial Pathog 1988;4:385–91. [DOI] [PubMed] [Google Scholar]
- Hulter N, Ilhan J, Wein T et al. An evolutionary perspective on plasmid lifestyle modes. Curr Opin Microbiol 2017;38:74–80. [DOI] [PubMed] [Google Scholar]
- Ilangovan A, Connery S, Waksman G. Structural biology of the Gram-negative bacterial conjugation systems. Trends Microbiol 2015;23:301–10. [DOI] [PubMed] [Google Scholar]
- Jacoby GA, Strahilevitz J, Hooper DC. Plasmid-mediated quinolone resistance. Microbiol Spectr 2014;2:PLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahagirdar S, Patwardhan R, Dhakephalkar PK. Curing plasmid-mediated vancomycin resistance in Staphylococcus aureus using herbal naphthoquinones. J Hosp Infect 2008;70:289–91. [DOI] [PubMed] [Google Scholar]
- Jalasvuori M, Friman V-P, Nieminen A et al. Bacteriophage selection against a plasmid-encoded sex apparatus leads to the loss of antibiotic-resistance plasmids. Biol Lett 2011;7:902–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten AM, Vogels GD. Characterization and extrachromosomal control of bacteriocin production in Staphylococcus aureus. Antimicrob Agents Ch 1973;4:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Doudna JA. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol 2015;30:100–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JH, Richmond MH. The increased rate of loss of penicillinase plasmids from Staphylococcus aureus in the presence of rifampicin. J Gen Microbiol 1970;60:137–9. [DOI] [PubMed] [Google Scholar]
- Kado CI. Historical events that spawned the field of plasmid biology. Microbiol Spectr 2014;2:PLAS-0019-2013. [DOI] [PubMed] [Google Scholar]
- Kamruzzaman M, Shoma S, Thomas CM et al. Plasmid interference for curing antibiotic resistance plasmids in vivo. PLoS ONE 2017;12:e0172913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan V, Santosh SW. Comparing the efficacy of plasmid curing agents in Lactobacillus acidophilus. Benef Microbes 2010;1:155–8. [DOI] [PubMed] [Google Scholar]
- Keyhani J, Keyhani E, Attar F et al. Sensitivity to detergents and plasmid curing in Enterococcus faecalis. J Ind Microbiol Biot 2006;33:238–42. [DOI] [PubMed] [Google Scholar]
- Kim JS, Cho DH, Park M et al. CRISPR/Cas9-mediated re-sensitization of antibiotic-resistant Escherichia coli harboring extended-spectrum beta-lactamases. J Microbiol Biot 2016;26:394–401. [DOI] [PubMed] [Google Scholar]
- Kristoffersen SM, Ravnum S, Tourasse NJ et al. Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14570. J Bacteriol 2007;189:5302–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C-C, Chuang Y-C, Chen C-C et al. Coexistence of MCR-1 and NDM-9 in a clinical carbapenem-resistant Escherichia coli isolate. Int J Antimicrob Ag 2017;49:517–8. [DOI] [PubMed] [Google Scholar]
- Lakhmi VV, Padma S, Polasa H. Elimination of multidrug-resistant plasmid in bacteria by plumbagin, a compound derived from a plant. Curr Microbiol 1987;16:159–61. [Google Scholar]
- Lakshmi VV, Thomas CM. Curing of F-like plasmid TP181 by plumbagin is due to interference with both replication and maintenance functions. Microbiology 1996;142:2399–406. [Google Scholar]
- Latha C, Shriram VD, Jahagirdar SS et al. Antiplasmid activity of 1΄-acetoxychavicol acetate from Alpinia galanga against multi-drug resistant bacteria. J Ethnopharmacol 2009;123:522–5. [DOI] [PubMed] [Google Scholar]
- Lavanya B, Sowmiya S, Balaji S et al. Plasmid profiling and curing of Lactobacillus strains isolated from fermented milk for probiotic applications. Adv J Food Sci Technol 2011;3:95–101. [Google Scholar]
- Laxminarayan R, Matsoso P, Pant S et al. Access to effective antimicrobials: a worldwide challenge. Lancet 2016;387:168–75. [DOI] [PubMed] [Google Scholar]
- Letchumanan V, Pusparajah P, Tan LTH et al. Occurrence and antibiotic resistance of Vibrio parahaemolyticus from Shellfish in Selangor, Malaysia. Front Microbiol 2015;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Conry-Cantilena C, Wang Y et al. Vitamin C pharmacokinetics in healthy volunteers: evidence for a recommended dietary allowance. P Natl Acad Sci USA 1996;93:3704–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A-D, Li L-G, Zhang T. Exploring antibiotic resistance genes and metal resistance genes in plasmid metagenomes from wastewater treatment plants. Front Microbiol 2015;6:1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Jimenez J, Derr J et al. Inhibition of bacterial conjugation by phage M13 and its protein g3p: quantitative analysis and model. PLoS ONE 2011;6:e19991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TL, Pan YJ, Hsieh PF et al. Imipenem represses CRISPR-Cas interference of DNA acquisition through H-NS stimulation in Klebsiella pneumoniae. Sci Rep 2016;6:31644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P-Y, Jiang N, Zhang J et al. The oxidative damage of plasmid DNA by ascorbic acid derivatives in vitro: the first research on the relationship between the structure of ascorbic acid and the oxidative damage of plasmid DNA. Chem Biodivers 2006;3:958–66. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang D, Wang H et al. Curing of plasmid pXO1 from Bacillus anthracis using plasmid incompatibility. PLoS ONE 2012;7:e29875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-Y, Wang Y, Walsh TR et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016;16:161–8. [DOI] [PubMed] [Google Scholar]
- Llanes C, Michel-Briand Y, Thouverez M et al. Stability of conjugative and non-conjugative R-plasmids from Serratia marcescens to gyrase inhibitors. Microbiologica 1990;13:157–9. [PubMed] [Google Scholar]
- Lopatkin AJ, Meredith HR, Srimani JK et al. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat Commun 2017;8:1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh GL, Swartz MN. Elimination of plasmids from several bacterial species by novobiocin. Antimicrob Agents Ch 1977;12:423–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean RC, San Millan A. Microbial evolution: towards resolving the plasmid paradox. Curr Biol 2015;25:R764–7. [DOI] [PubMed] [Google Scholar]
- Maier L, Pruteanu M, Kuhn M et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018;555:623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandi TY, Molnar J, Holland IB et al. Efficient curing of an Escherichia coli F-prime plasmid by phenothiazines. Genet Res 1975;26:109–11. [DOI] [PubMed] [Google Scholar]
- Meek RW, Vyas H, Piddock LJ. Nonmedical uses of antibiotics: time to restrict their use? PLoS Biol 2015;13:e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesas JM, Rodriguez MC, Alegre MT. Plasmid curing of Oenococcus oeni. Plasmid 2004;51:37–40. [DOI] [PubMed] [Google Scholar]
- Michel-briand Y, Uccedi V, Laporte J et al. Elimination of plasmids from Enterobacteriaceae by 4-quinolone derivatives. J Antimicrob Chemoth 1986;18:667–74. [DOI] [PubMed] [Google Scholar]
- Mitchell I, Kenworthy R. Attempted elimination of plasmid-determined haemolysin, k88 antigen and enterotoxin from Escherichia coli pathogenic for pigs. J Appl Bacteriol 1977;42:207–12. [DOI] [PubMed] [Google Scholar]
- Molnár A, Amaral L, Molnár J. Antiplasmid effect of promethazine in mixed bacterial cultures. Int J Antimicrob Ag 2003;22:217–22. [DOI] [PubMed] [Google Scholar]
- Molnar J, Mandi Y, Kiraly J. Antibacterial effect of some phenothiazine compounds and R-factor elimination by chlorpromazine. Acta Microbiol Acad Sci Hung 1976;23:45–54. [PubMed] [Google Scholar]
- Molnar J, Schneider B, Mandi Y et al. New mechanism of plasmid curing by psychotropic drugs. Acta Microbiol Acad Sci Hung 1980;27:309–15. [PubMed] [Google Scholar]
- Morgan AR, Cone RL, Elgert TM. The mechanism of DNA strand breakage by vitamin C and superoxide and the protective roles of catalase and superoxide dismutase. Nucleic Acids Res 1976;3:1139–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T, Oueslati S, Bonnin RA et al. Beta-lactamase database (BLDB) – structure and function. J Enzym Inhib Med Ch 2017;32:917–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae M, Inoue M, Mitsuhashi S. Artificial elimination of drug resistance from group A beta-hemolytic Streptococci. Antimicrob Agents Ch 1975;7:719–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B, Du Z, Guo Z et al. Curing of four different plasmids in Yersinia pestis using plasmid incompatibility. Lett Appl Microbiol 2008;47:235–40. [DOI] [PubMed] [Google Scholar]
- Novick RP. Plasmid incompatibility. Microbiol Rev 1987;51:381–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell CM, Nicks KM. A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology 2006;152:1601–7. [DOI] [PubMed] [Google Scholar]
- O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations the Review on Antimicrobial Resistance 2016, https://arm-review.org/publications.html.
- Ohlow MJ, Moosmann B. Phenothiazine: the seven lives of pharmacology's first lead structure. Drug Discov Today 2011;16:119–31. [DOI] [PubMed] [Google Scholar]
- Ojala V, Laitalainen J, Jalasvuori M. Fight evolution with evolution: plasmid-dependent phages with a wide host range prevent the spread of antibiotic resistance. Evol Appl 2013;6:925–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva B, Selan L, Ravagnan G et al. Inhibition of conjugal transfer by new quinolinic compounds. Chemioterapia 1985;4:199–201. [PubMed] [Google Scholar]
- Orlek A, Stoesser N, Anjum MF et al. Plasmid classification in an era of whole-genome sequencing: application in studies of antibiotic resistance epidemiology. Front Microbiol 2017;8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhye S, Dandawate P, Yusufi M et al. Perspectives on medicinal properties of plumbagin and its analogs. Med Res Rev 2012;32:1131–58. [DOI] [PubMed] [Google Scholar]
- Paltansing S, Vlot JA, Kraakman ME et al. Extended-spectrum beta-lactamase-producing Enterobacteriaceae among travelers from the Netherlands. Emerg Infect Dis 2013;19:1206–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos A, den Hartigh A, Smith MA et al. An in vivo high-throughput screening approach targeting the type IV secretion system component VirB8 identified inhibitors of Brucella abortus 2308 proliferation. Infect Immun 2011;79:1033–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan RB, Shinde PS, Chavan KR et al. Reversal of plasmid encoded antibiotic resistance from nosocomial pathogens by using Plumbago auriculata root extracts. Int J Curr Microbiol Appl Sci 2015;2:187–98. [Google Scholar]
- Peters JM, Silvis MR, Zhao D et al. Bacterial CRISPR: Accomplishments and prospects. Curr Opin Microbiol 2015;27:121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AC, Towner KJ. Use of a non-radioactive DNA hybridization technique to study the effect of quinolone antibiotics on plasmid replication and caring. J Antimicrob Chemoth 1990;25:745–50. [DOI] [PubMed] [Google Scholar]
- Piddock LJV. Multidrug-resistance efflux pumps - not just for resistance. Nat Rev Microbiol 2006;4:629–36. [DOI] [PubMed] [Google Scholar]
- Pinto UM, Pappas KM, Winans SC. The ABCs of plasmid replication and segregation. Nat Rev Microbiol 2012;10:755–65. [DOI] [PubMed] [Google Scholar]
- Pires J, Kuenzli E, Kasraian S et al. Polyclonal intestinal colonization with extended-spectrum cephalosporin-resistant Enterobacteriaceae upon traveling to India. Front Microbiol 2016;7:1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt DJ, Black AC. Plasmid ecology and tbe elimination of plasmids by 4-quinolones. J Antimicrob Chemoth 1987;20:137–8. [DOI] [PubMed] [Google Scholar]
- Poppe C, Gyles CL. Tagging and elimination of plasmids in Salmonella of avian origin. Vet Microbiol 1988;18:73–87. [DOI] [PubMed] [Google Scholar]
- Posno M, Leer RJ, van Luijk N et al. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic lactobacillus plasmids and segregational instability of the introduced vectors. Appl Environ Microb 1991;57:1822–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price VJ, Huo W, Sharifi A et al. CRISPR-Cas and restriction-modification act additively against conjugative antibiotic resistance plasmid transfer in Enterococcus faecalis. mSphere 2016;1:e00064–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulcrano G, Pignanelli S, Vollaro A et al. Isolation of Enterobacter aerogenes carrying bla TEM-1 and bla KPC-3 genes recovered from a hospital Intensive Care Unit. APMIS 2016;124:516–21. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan V, Ganguly K, Ganguly M et al. Potentiality of tricyclic compound thioridazine as an effective antibacterial and antiplasmid agent. Indian J Exp Biol 1999;37:671–5. [PubMed] [Google Scholar]
- Rahube TO, Marti R, Scott A et al. Persistence of antibiotic resistance and plasmid-associated genes in soil following application of sewage sludge and abundance on vegetables at harvest. Can J Microbiol 2016;62:600–7. [DOI] [PubMed] [Google Scholar]
- Raja CE, Selvam GS. Plasmid profile and curing analysis of Pseudomonas aeruginosa as metal resistant. Int J Environ Sci Technol 2009;6:259–66. [Google Scholar]
- Ramesh A, Halami PM, Chandrashekar A. Ascorbic acid-induced loss of a pediocin-encoding plasmid in Pediococcus acidilactici CFR K7. World J Microb Biot 2000;16:695–7. [Google Scholar]
- Rensch U, Nishino K, Klein G et al. Salmonella enterica serovar Typhimurium multidrug efflux pumps EmrAB and AcrEF support the major efflux system AcrAB in decreased susceptibility to triclosan. Int J Antimicrob Ag 2014;44:179–80. [DOI] [PubMed] [Google Scholar]
- Riber L, Burmolle M, Alm M et al. Enhanced plasmid loss in bacterial populations exposed to the antimicrobial compound irgasan delivered from interpenetrating polymer network silicone hydrogels. Plasmid 2016;87–88:72–78. [DOI] [PubMed] [Google Scholar]
- Ripoll-Rozada J, Garcia-Cazorla Y, Getino M et al. Type IV traffic ATPase TrwD as molecular target to inhibit bacterial conjugation. Mol Microbiol 2016;100:912–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll-Rozada J, Zunzunegui S, de la Cruz F et al. Functional interactions of VirB11 traffic ATPases with VirB4 and VirD4 molecular motors in type IV secretion systems. J Bacteriol 2013;195:4195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Martinez JM, Machuca J, Cano ME et al. Plasmid-mediated quinolone resistance: two decades on. Drug Resist Updates 2016;29:13–29. [DOI] [PubMed] [Google Scholar]
- Rosas SB, Calzolari A, La Torre JL et al. Involvement of a plasmid in Escherichia coli envelope alterations. J Bacteriol 1983;155:402–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotimi VO, Duerden BI, Hafiz S. Transferable plasmid-mediated antibiotic resistance in Bacteroides. J Med Microbiol 1981;14:359–70. [DOI] [PubMed] [Google Scholar]
- Rubin SJ, Rosenblum ED. Effects of ethidium bromide on growth and on loss of the penicillinase plasmid of Staphylococcus aureus. J Bacteriol 1971;108:1200–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Barba JL, Piard JC, Jiménez-Díaz R. Plasmid profiles and curing of plasmids in Lactobacillus plantarum strains isolated from green olive fermentations. J Appl Bacteriol 1991;71:417–21. [DOI] [PubMed] [Google Scholar]
- Ruiz-Maso JA, MachoN C, Bordanaba-Ruiseco L et al. Plasmid rolling-circle replication. Microbiol Spectr 2015;3:PLAS-0035-2014. [DOI] [PubMed] [Google Scholar]
- Sanofi-Aventis NZ. Data Sheet Largactil. http://www.medsafe.govt.nz/profs/Datasheet/l/largactiltabinjsusp.pdf2016:largactil-ccdsv1-dsv8-05aug16.
- Selan L, Scazzocchio F, Oliva B et al. Plasmid “curing” by some recently synthetized 4-quinolone compounds. Chemioterapia 1988;7:292–4. [PubMed] [Google Scholar]
- Shintani M, Sanchez ZK, Kimbara K. Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front Microbiol 2015;6:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriram V, Jahagirdar S, Latha C et al. A potential plasmid-curing agent, 8-epidiosbulbin E acetate, from Dioscorea bulbifera L. against multidrug-resistant bacteria. Int J Antimicrob Ag 2008;32:405–10. [DOI] [PubMed] [Google Scholar]
- Sonstein SA, Burnham JC. Effect of low concentrations of quinolone antibiotics on bacterial virulence mechanisms. Diagn Microbiol Infect Dis 1993;16:277–89. [DOI] [PubMed] [Google Scholar]
- Spengler G, Miczák A, Hajdú E et al. Enhancement of plasmid curing by 9-aminoacridine and two phenothiazines in the presence of proton pump inhibitor 1-(2-benzoxazolyl)-3,3,3-trifluoro-2-propanone. Int J Antimicrob Ag 2003;22:223–7. [DOI] [PubMed] [Google Scholar]
- Spengler G, Molnár A, Schelz Z et al. The mechanism of plasmid curing in bacteria. Curr Drug Targets 2006;7:823–41. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, Doudna JA. Expanding the Biologist's Toolkit with CRISPR-Cas9. Mol Cell 2015;58:568–74. [DOI] [PubMed] [Google Scholar]
- Stoesser N, Sheppard AE, Pankhurst L et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 2016;7:e02162–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DE, Levine JG. Characterization of a plasmid mutation affecting maintenance, transfer and elimination by novobiocin. Mol Gen Genet 1979;174:127–33. [DOI] [PubMed] [Google Scholar]
- Tomoeda M, Inuzuka M, Kubo N et al. Effective elimination of drug resistance and sex factors in Escherichia coli by sodium dodecyl sulfate. J Bacteriol 1968;95:1078–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi R, Menghani E. A Review on Plumbago zeylanica: a compelling herb. Int J Pharma Sci Res 2014;5:119–26. [Google Scholar]
- Uraji M, Suzuki K, Yoshida K. A novel plasmid curing method using incompatibility of plant pathogenic Ti plasmids in Agrobacterium tumefaciens. Genes Genet Syst 2002;77:1–9. [DOI] [PubMed] [Google Scholar]
- Vading M, Kabir MH, Kalin M et al. Frequent acquisition of low-virulence strains of ESBL-producing Escherichia coli in travellers. J Antimicrob Chemoth 2016;71:3548–55. [DOI] [PubMed] [Google Scholar]
- Van Boeckel TP, Brower C, Gilbert M et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA 2015;112:5649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Boeckel TP, Gandra S, Ashok A et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis 2014;14:742–50. [DOI] [PubMed] [Google Scholar]
- Van Boeckel TP, Glennon EE, Chen D et al. Reducing antimicrobial use in food animals. Science 2017;357:1350–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen L, Saavedra De Bast M. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet 2009;5:e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga B, Csonka Á, Csonka A et al. Possible biological and clinical applications of phenothiazines. Anticancer Res 2017;37:5983–93. [DOI] [PubMed] [Google Scholar]
- von Wintersdorff CJH, Wolffs PFG, van Niekerk JM et al. Detection of the plasmid-mediated colistin-resistance gene mcr-1 in faecal metagenomes of Dutch travellers. J Antimicrob Chemoth 2016;71:3416–9. [DOI] [PubMed] [Google Scholar]
- Wan Z, Goddard NL. Competition between conjugation and M13 phage infection in Escherichia coli in the absence of selection pressure: a kinetic study. G3 2012;2:1137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Gao Z, Wang H et al. Curing both virulent mega-plasmids from Bacillus anthracis wild-type strain A16 simultaneously using plasmid incompatibility. J Microbiol Biotechnol 2015;25:1614–20. [DOI] [PubMed] [Google Scholar]
- Wang H, Liu X, Feng E et al. Curing the plasmid pXO2 from Bacillus anthracis A16 using plasmid incompatibility. Curr Microbiol 2011;62:703–9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ness B Van. Site-specific cleavage of supercoiled DNA by ascorbate/Cu(II). Nucleic Acids Res 1989;17:6915–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang R, Li J et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol 2017;2:16260. [DOI] [PubMed] [Google Scholar]
- Waring AJ, Drake IM, Schorah CJ et al. Ascorbic acid and total vitamin C concentrations in plasma, gastric juice, and gastrointestinal mucosa: effects of gastritis and oral supplementation. Gut 1996;38:171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber MA, Buckner MMC, Redgrave LS et al. Quinolone-resistant gyrase mutants demonstrate decreased susceptibility to triclosan. J Antimicrob Chemoth 2017;72:2755–63. [DOI] [PubMed] [Google Scholar]
- Webber MA, Randall LP, Cooles S et al. Triclosan resistance in Salmonella enterica serovar Typhimurium. J Antimicrob Chemoth 2008;62:83–91. [DOI] [PubMed] [Google Scholar]
- Webber MA, Whitehead RN, Mount M et al. Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J Antimicrob Chemoth 2015;70:2241–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser J, Wiedemann B. Elimination of plasmids by new 4-quinolones. Antimicrob Agents Ch 1985;28:700–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser J, Wiedemann B. Elimination of plasmids by enoxacin and ofloxacin at near inhibitory concentrations. J Antimicrob Chemoth 1986;18:575–83. [DOI] [PubMed] [Google Scholar]
- Wolfart K, Spengler G, Kawase M et al. Synergistic interaction between proton pump inhibitors and resistance modifiers: promoting effects of antibiotics and plasmid curing. In Vivo (Brooklyn) 2006;20:367–72. [PubMed] [Google Scholar]
- Wolfson JS, Hooper DC, Swartz MN et al. Antagonism of the B subunit of DNA gyrase eliminates plasmids pBR322 and pMG110 from Escherichia coli. J Bacteriol 1982;152:338–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DO, Carter MJ, Best GK. Plasmid-mediated resistance to gentamicin in Staphylococcus aureus. Antimicrob Agents Chemoth 1977;12:513–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell 2016;164:29–44. [DOI] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Globus R et al. Extending the host range of bacteriophage particles for DNA transduction. Mol Cell 2017;66:721–728.e3.e3. [DOI] [PubMed] [Google Scholar]
- Yosef I, Manor M, Kiro R et al. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci USA 2015;112:7267–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman MA, Pasha MH, Akhter MZ. Plasmid curing of Escherichia coli cells with ethidium bromide, sodium dodecyl sulfate and acridine orange. Bangladesh J Microbiol 2010;27:28–31. [Google Scholar]
- Zhou Y-F, Tao M-T, Feng Y et al. Increased activity of colistin in combination with amikacin against Escherichia coli co-producing NDM-5 and MCR-1. J Antimicrob Chemoth 2017;72:1723–30. [DOI] [PubMed] [Google Scholar]