Abstract
Bacillus thuringiensis is a well-known biopesticide that has been used for more than 80 years. This spore-forming bacterium belongs to the group of Bacillus cereus that also includes, among others, emetic and diarrheic pathotypes of B. cereus, the animal pathogen Bacillus anthracis and the psychrotolerant Bacillus weihenstephanensis. Bacillus thuringiensis is rather unique since it has adapted its lifestyle as an efficient pathogen of specific insect larvae. One of the peculiarities of B. thuringiensis strains is the extent of their extrachromosomal pool, with strains harbouring more than 10 distinct plasmid molecules. Among the numerous serovars of B. thuringiensis, ‘israelensis’ is certainly emblematic since its host spectrum is apparently restricted to dipteran insects like mosquitoes and black flies, vectors of human and animal diseases such as malaria, yellow fever, or river blindness. In this review, the putative role of the mobile gene pool of B. thuringiensis serovar israelensis in its pathogenicity and dedicated lifestyle is reviewed, with specific emphasis on the nature, diversity, and potential mobility of its constituents. Variations among the few related strains of B. thuringiensis serovar israelensis will also be reported and discussed in the scope of this specialised insect pathogen, whose lifestyle in the environment remains largely unknown.
Keywords: biopesticide, Bacillus thuringiensis serovar israelensis; mosquitoes, plasmids, phages, transposons
This review describes the potential roles of the mobile gene pool of the entomopathogenic Bacillus thuringiensis serovar israelensis to its pathogenicity and dedicated lifestyle.
BACILLUS THURINGIENSIS SV. ISRAELENSIS, A NEAT BIOPESTICIDE FOR THE CONTROL OF MOSQUITO-TRANSMITTED DISEASES
Discovery
In 1976, Goldberg and Margalit discovered a bacterial strain demonstrating high insecticidal activity against mosquito larvae (Goldberg and Margalit 1977). The corresponding spore-forming bacteria was subsequently identified as Bacillus thuringiensis, which is a Gram-positive, aerobic, endospore-forming saprophyte bacterium, naturally occurring in various soil and aquatic habitats (Lacey and Goettel 1995). Huguette de Barjac later established that this bacteria was a new B. thuringiensis serovar (sv.), namely H-14, and it was given the name ‘israelensis’ indicating that it was found in Israel (de Barjac 1978). Thanks to its high entomopathogenic efficiency and specificity against dipteran larvae, as well as its safety for mammals and other non-target organisms (Lacey 2007; Roh et al.2007), B. thuringiensis sv. israelensis was rapidly commercialised and opened a gateway to all B. thuringiensis-based products, which represented 10% of the total insecticide and 97% of the bioinsecticide markets in 2006 (Brar et al.2006). However, with increasing knowledge, studies and formulation techniques, the mosquito biocontrol market witnessed substantial growth in the last years, with a current estimation of an 8 million dollars market, mainly concentrated in the USA, Brazil and Europe (Forrest Innovations LTD). The majority of biolarvicidal products are B. thuringiensis sv. israelensis based (e.g. VectoBac®, MosquitoBits®).
Mosquito-borne diseases
Mosquitoes, with over 3500 described species, belong to the order Diptera. These insects can be phytopathogens that attack plants and cause massive loss of crops and a decrease in the global food production. For example, Mayetiola destructor, also known as the Hessian fly, causes serious damage to wheat, barley and rye in Europe and North America. Mosquitoes can also be disease carrying vectors with over 18 described illnesses caused by a bacteria, virus or parasite transmitted by a mosquito. Every year, there are more than 700,000 deaths from vector-borne diseases that account for 17% of all infectious diseases (WHO, October 2017). For instance, malaria, the most significant human disease caused by a parasite, continues to spread in endemic regions, with 216 million cases in 91 countries and 445,000 deaths recorded in 2016 (WHO, November 2017). Malaria is caused by several Plasmodium species, transmitted by the bite of Anopheles female mosquitoes (Aponte et al.2007; Tolle 2009). Encephalitis, caused by the Japanese Encephalitis Virus (JEV), whose primary vectors are Culex spp. mosquitoes (Mackenzie, Gubler and Petersen 2004), is another example of mosquito-transmitted diseases. JEV is endemic in 24 countries of the south East Asian and western pacific regions with 1 in 250 infections resulting in severe clinical illness and possibly death (WHO, December 2015). Other mosquito-borne diseases are dengue and yellow fever, and are still prevalent in forested areas of tropical West Africa and Asia. Although similar in epidemiology and human-mosquito-human life cycles, dengue viruses, unlike the yellow fever virus, have undergone significant genetic variation allowing their full adaptation to Aedes aegypti, thus insuring a more efficient life cycle maintenance (Gubler 2004). This constitutes a major problem since almost half of the world population lives in dengue endemic regions with an estimated 390 millions dengue infection cases per year in 2013 (WHO, December 2016). Massive use of chemical and synthetic pesticides has led to the emergence and spread of many resistant vector strains as well as other drawbacks such as the spreading of environmental pollutants and the safety risks for humans and domestic animals. Therefore, vector control can no longer depend on the use of chemicals and major efforts are employed for the development of environmentally friendly biocontrol agents such as B. thuringiensis formulations (Margalith and Ben-Dov 2000).
Bacillus thuringiensis sv. israelensis and its mosquitocidal parasporal crystal
Bacillus thuringiensis owes its insecticidal potential to several factors. However, the main factor is a parasporal crystal that contains a variety of δ-endotoxins (Palma et al.2014). These toxins are pore-forming Cry (crystal) and/or Cyt (cytolytic) proteins that are solubilised in the insect alkaline midgut upon ingestion. Cry toxins currently constitute the largest group of insecticidal proteins produced by a Bacillus species, with 801 described proteins divided into 75 families (http://www.btnomenclature.info/, on 18 January 2018). This current classification depends on the evolutionary degree of divergence between Cry proteins, estimated by algorithms and represented by a phylogenetic tree. In B. thuringiensis sv. israelensis, the genes coding for these proteins are carried on a 128-kb megaplasmid, which will be discussed in a later section.
Each Cry protein family shows specificity to a certain insect group: e.g. Cry1 and 9 families are toxic to lepidopteran larvae whereas Cry4, 10 and 11 are toxic for dipteran larvae (Van Frankenhuyzen 2009, 2013; El Khoury et al.2014). Besides, some Cry proteins can be toxic to more than one group of insects, as is the case of Cry2, which is active against both lepidopteran and dipteran larvae (Höfte and Whiteley 1989). Nonetheless, a comparison between the amino acid sequences of the various families showed the presence of several conserved sequence blocks, which delimitate three main protein domains (Schnepf et al.1998). X-ray crystallography also reinforced this three-domain globular structure (Bravo, Gill and Soberon 2007; Pardo-López, Soberón and Bravo 2013). Each domain plays a specific role in the toxicity. Domain I, or perforating domain, consists of eight α-helices connected by loops and is located towards the N-terminus of the protein. Its main function is membrane insertion and pore formation through the hydrophobic helices (Ben-Dov 2014; Xu et al.2014). Domain II, or central domain, consists of three antiparallel β-sheets and is responsible for toxin-receptor interactions. Finally, domain III, or galactose-binding domain, is a sandwich formed by two antiparallel β-sheets and is involved in both receptor binding and pore formation (Xu et al.2014). The solubilised Cry toxins are activated in the insect midgut by several proteases, and then form a pore in the intestinal membrane once they are bound to their cognate receptors (Zhang, Hua and Adang 2017).
The parasporal crystal also contains Cyt toxins that constitute another relevant insecticidal factor. These toxins are cytolytic (haemolytic) and exhibit a predominant dipteran specific activity in vivo (Guerchicoff, Delécluse and Rubinstein 2001; Butko 2003; de Maagd et al.2003). Three Cyt families have been characterised so far, with structural studies revealing that Cyt1 and 2 are single domain, three-layer alpha-beta proteins (Cohen et al. 2008, 2011). In addition to being toxic on their own, Cyt toxins play an important role in B. thuringiensis sv. israelensis toxicity against dipteran larvae due to their synergistic effects with other δ-endotoxins, which not only increases the toxicity per se, but also help overcoming field resistance (Wirth, Georghiou and Federici 1997; Wirth et al.2005; Ben-Dov 2014). Several studies have in fact highlighted this synergy between Cyt and various Cry toxins, such as Cry11A and Cry4 (Pérez et al.2005; Elleuch et al.2015). For B. thuringiensis sv. israelensis Cyt1A constitutes a massive 60% in volume of the parasporal crystal, thus playing a key role in anti-dipteran activity. Cyt2Ba and Cyt1C however accumulate in small amounts in the parasporal body (Ben-Dov 2014; Palma et al.2014). In contrast to Cry toxins, the Cyt ones do not recognise a specific receptor, but target instead membrane phospholipids with a marked affinity to some specific unsaturated fatty acids (Cohen et al.2011).
Mosquito resistance to B. thuringiensis sv. israelensis toxins
Despite their high toxicity, any alteration in Cry toxin–receptor interaction may lead to resistance to the corresponding toxin (de Maagd et al.2003). However, as mentioned above, B. thuringiensis sv. israelensis toxicity is due to a cocktail of toxins forming the parasporal crystal, including the cytolytic toxins. Therefore, resistance to this bacterium is a more complex phenotype involving environmental, behavioural and genomic factors. For instance, leaf litter in mosquito breeding sites is capable of decreasing overall B. thuringiensis sv. israelensis toxicity by 70% either by damaging the bioviability of the toxins in the environment, thus the chance of their ingestion by insect larvae, or by hindering the synergistic effect of Cyt toxins (Tetreau et al.2012b). In addition, this contact may cause the crystal to sediment more rapidly to the bottom of the water in the breeding sites, lessening the probability of their ingestion by dipteran larvae that only feed from the surface of the water. On a genomic level, as shown by transcriptomic and proteomic approaches, modifications affecting the receptors at different levels in the larval midgut conform with B. thuringiensis resistance mechanisms previously described in Lepidoptera (Tetreau et al.2012a). Other methods such as combined AFLP and sequencing analysis and differential expression analysis on a whole genome level combined with SNP detection have confirmed the high genomic complexity of field mosquito resistance to B. thuringiensis sv. israelensis, and a more recent study has shown that six chromosomal loci in A. aegypti are involved in its resistance to B. thuringiensis sv. israelensis toxins (Paris and Després 2012; Després et al.2014; Bonin et al.2015).
Other mosquitocidal factors
Other insecticidal factors involved in B. thuringiensis biocontrol potential have been described. First, the β-exotoxin (also named thuringiensin) is a small thermostable oligosaccharide secreted by B. thuringiensis during its vegetative growth, independently of parasporal crystal and spore formation. It is active against several insect groups including Diptera (Liu et al.2014). Second, some B. thuringiensis strains secrete, during their vegetative growth, Vip (for vegetative insecticidal proteins), which have no known activity against Diptera, and Sip (for secreted insecticidal protein), a 41-kDa protein active against coleopteran larvae. However, the N-terminal part of the Sip toxin is very similar to Mtx3, a mosquitocidal toxin from Lysinibacillus sphaericus (formerly known as Bacillus sphaericus) (Palma et al.2014). Lysinibacillus sphaericus is known for its activity against mosquito larvae, which is comparable to that of B. thuringiensis sv. israelensis (Berry 2012). Lysinibacillus sphaericus was also shown to be able to acquire plasmids from B. thuringiensis sv. israelensis by conjugation (Gammon et al.2006). Interestingly, L. sphaericus toxins can act in synergy with B. thuringiensis sv. israelensis Cry4A, Cry4B, Cry11A and Cyt1A toxins (Wirth, Federici and Walton 2000; Wirth et al.2004, 2007, 2014).
BACILLUS THURINGIENSIS SV. ISRAELENSIS AND KIN
Based on a dataset of 45 genome sequences, Zwick et al. (2012) have reported that the core genome of B. cereus sensu lato (s.l.) contains ca. 1750 genes, and its pan-genome includes more than 23 000 genes, recently extended up to 60 000 (Bazinet 2017) indicating a vast potential of adaptability to environmental and host-related niches. What have we learned on the genomics of the group known as B. thuringiensis sv. israelensis, also known as serotype H-14?
Originally, the B. thuringiensis sv. israelensis strains were empirically defined as those revealing a particular serotype H-14, able to synthesise major toxins killing the Dipteran insects and, in some cases, possessing characteristic plasmid profiles (Margalit and Dean 1985). At present, 453 genomic sequences in GenBank are labelled as or very closely related to the B. thuringiensis cluster. This raises the question of recognition, among all these sequences, those that can be assigned as B. thuringiensis sv. israelensis. Apparently, the B. thuringiensis sv. israelensis cluster should contain strains sharing significant similarity of their chromosomes to those that were defined as reference B. thuringiensis sv. israelensis strains (Ankarloo et al.2000). To be pertinent, it should also be statistically separated from other particular clusters of B. thuringiensis strains. At the same time, since there are several common plasmids in the reference strains, especially those essential for mosquito-killing phenotype, they should also be considered. A MLST (multi-locus sequence typing) study of random B. thuringiensis isolates, including B. thuringiensis sv. israelensis ATCC 35646 and 4Q7 (also known as 4Q2–81) as references, revealed that about 10% (6 of 55) of the isolates could be regarded as B. thuringiensis sv. israelensis since they fell into the same clonal complex (Sorokin et al.2006). A similar percentage can be expected if the set of 453 B. thuringiensis strains in GenBank is non-biased.
In a recent publication, the homology of the sequences applied for MLST studies was used to identify the genomes from GenBank corresponding to the B. thuringiensis sv. israelensis strains (Bolotin et al.2017). This simple protocol allowed us to identify several strains that were not regarded as sv. israelensis when deposited, including B. thuringiensis HD 1002 that appeared to be rather important in our study since it was the only GenBank sample containing a completely assembled pBtic360 plasmid, not identified earlier (see below). Notably, this strain does not contain the pBtoxis plasmid, as is the case for ATCC 35646, the first B. thuringiensis sv. israelensis strain sequenced (Anderson et al.2005). The ‘israelensis’ serovar was also assigned to B. thuringiensis IBL 200 that contained in its sequence a counterpart of pBtic360. In fact, we show below that IBL 200 should not be formally considered as B. thuringiensis sv. israelensis. In the cited study (Bolotin et al.2017), it was also observed that the presence of small plasmids fluctuates among strains in the B. thuringiensis sv. israelensis cluster. They should not therefore be used as additional features for the assignment of strains to this cluster. By contrary, large plasmids, of 100 kb and higher, are more stable and therefore their presence could be regarded as additional property of relatedness of the strains to this cluster.
An extensive analysis of genomic phylogeny for 140 B. thuringiensis strains, based on comparison of concatenated single-copy protein sequences, was recently published (Zheng et al.2017). The proposed tree placed together the strains of serovars novosibirsk BGSC 4AX1, israelensis BGSC 4Q1, IBL 4222, israelensis HD-789 and israelensis 4Q7. It places strain IBL 200 into a separate cluster, earlier assigned as sv. israelensis based on MLST sequences (Bolotin et al.2017). Although Zheng et al. did not indicate a quantitative parameter that can be used for strain separation, all strains that could be regarded as pertaining to the israelensis serovar were quite well distinct of others on their analyses.
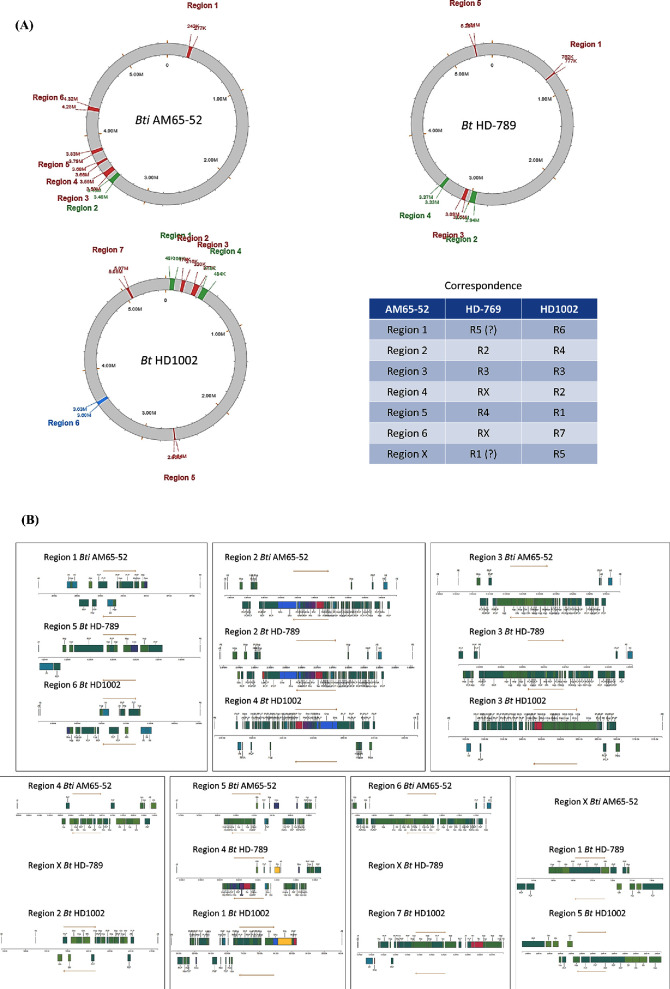
Alignments of 453 available B. thuringiensis genomic sequences (November 2017) were used to further clarify the situation with the presence of B. thuringiensis sv. israelensis strains in this set. The purpose was also to verify if a quantitative parameter, such as the indicator of similarity between genomes, could be used for the definition of B. thuringiensis sv. israelensis and what are the limitations of its use. First, ANI (average nucleotide identity) parameter (Richter and Rossello-Mora 2009) and tools implemented as pyani Python3 module (http://widdowquinn.github.io/pyani/) were used to construct a phylogenetic tree of the 453 B. thuringiensis strains. This analysis identified a cluster of 13 strains (highlighted with pale orange colour in Fig. 1) closely related to three B. thuringiensis sv. israelensis strains, AM65–52, 4Q7 and HD-789, which can be considered as references.
Figure 1.
(A) Circular and unrooted tree representing the phylogeny of genomic sequences of the B. thuringiensis strains. The strain names correspond to their labelling in NCBI genomic database. The colours are arbitrary assigned to clearly distinct formal clusters. Pale orange colour corresponds to the cluster closely related to three B. thuringiensis sv. israelensis reference strains, AM65–52, 4Q7 and HD-789. (B) Unrooted tree is drawn for better presentation of differences between clades. See the text for more detailed comments.
The observation that another B. thuringiensis sv. israelensis reference strain, ATCC 35646, was placed apart from this cluster is noteworthy. This unexpected result might however be due to insufficient quality of the ATCC 35646 sequence, the only reference strain done using the Sanger technique with low coverage (Anderson et al.2005). Incidentally, the strain ATCC 35646 was not present on the tree reported by Zheng et al. (2017). From these data, one could conclude that there is a cluster of potential B. thuringiensis sv. israelensis strains with genomic sequence identity between them and reference strains of 99.95% or higher. The most relevant data of this analysis are presented in Table 1.
Table 1.
Genome-wide identity between sequenced strains of B. thuringiensis closely related to the B. thuringiensis sv. israelensis cluster.
| B. thuringiensis strains | NCBI Bio-Project | Assembly size (kb) | No. of contigs | Identity to AM65–52 | Identity to HD-789 | Identity to 4Q7 | Status as B. thuringiensis sv. israelensis |
|---|---|---|---|---|---|---|---|
| sv. israelensis AM65–52 | PRJNA303961 | 6715 | 10 | 1 | 0.99982 | 0.99985 | Yes |
| HD-789 | PRJNA171844 | 6335 | 7 | 0.99982 | 1 | 0.99987 | Yes |
| sv. israelensis 4Q7 | PRJNA238495 | 5040 | 52 | 0.99985 | 0.99987 | 1 | Yes |
| sv. israelensis BGSC 4Q1 | PRJNA349211 | 6394 | 85 | 0.99965 | 0.9997 | 0.99983 | Yes |
| sv. israelensis BR58 | PRJNA292320 | 6364 | 261 | 0.99949 | 0.9997 | 0.99959 | Yes |
| sv. israelensis ATCC 35646 | PRJNA15522 | 5880 | 867 | 0.99626 | 0.99593 | 0.99619 | Yes |
| IBL 4222 | PRJNA29735 | 6612 | 383 | 0.99994 | 0.99989 | 0.99991 | Yes |
| 147 | PRJNA288912 | 6168 | 94 | 0.99986 | 0.99987 | 0.99987 | Yes |
| HD 1002 | PRJNA236049 | 6573 | 8 | 0.99974 | 0.99973 | 0.99987 | Yes |
| sv. novosibirsk BGSC 4AX1 | PRJNA349211 | 6737 | 112 | 0.99968 | 0.99964 | 0.99983 | Yes |
| ‘Bacillus sp’ Root11 | PRJNA297942 | 6622 | 63 | 0.99964 | 0.99971 | 0.99985 | Yes |
| ‘Bacillus sp’ Root131 | PRJNA297942 | 6524 | 61 | 0.9996 | 0.9997 | 0.99984 | Yes |
| UBA3967 | PRJNA348753 | 5215 | 95 | 0.99992 | 0.99988 | 0.99987 | No |
| Lr7/2 | PRJNA260736 | 5610 | 94 | 0.9934 | 0.99349 | 0.99355 | No |
| IBL 200 | PRJNA29733 | 6732 | 244 | 0.9715 | 0.97209 | 0.9731 | No |
| sv. morissoni BGSC 4AA1 | PRJNA271502 | 6180 | 7 | 0.99035 | 0.99059 | 0.99114 | No |
| sv.kurstaki HD73 | PRJNA185468 | 5909 | 8 | 0.95955 | 0.95972 | 0.96014 | No |
The strains labelled as ‘Yes’ in the ‘Status as B. thuringiensis sv. israelensis’ column are regarded as serovar israelensis due to criteria of genome proximity and plasmid content. Strains sv. morissoni BGSC 4AA1 and sv. kurstaki HD73 are presented as an out-group. For strain IBL 200, see comments in the text.
To make deeper scrutiny of these results, the presence of plasmids identified in three reference B. thuringiensis sv. israelensis strains with completed genomes (HD-789, AM65–52 and HD 1002) was searched for in all 453 B. thuringiensis strains. The results are illustrated in Table 2. Three of the above 13 potential B. thuringiensis sv. israelensis strains (4Q7, UBA3967 and Lr7/2) showed significant differences in plasmid content from the references. Strain 4Q7 is a laboratory strain artificially cured from plasmids. It contains only the 235-kb plasmid pBtic235 (Bolotin et al.2017; Gillis et al.2017b), as our analysis confirmed (see also below). The two other strains are presumably close relatives of the sv. israelensis cluster but contain a completely different set of plasmids.
Table 2.
Lengths of identical regions in genomic assemblies of different B. thuringiensis strains with large plasmids from B. thuringiensis sv. israelensis strain AM65–52.
| Plasmid names in AM65–52 | 1–360K | 2–350K | 3–235K | 4–128K | 5–100K | |
|---|---|---|---|---|---|---|
| Alternative plasmid names | pBtic360 | pXO16 | pBtic235 | pBtoxis | pBtic100 | Status as B. thuringiensis |
| sv. israelensis | ||||||
| B. thuringiensis strains | Length of identity to AM65–52 plasmids (bp) | |||||
| sv. israelensis AM65–52 | 359 560 | 349 600 | 235 424 | 127 938 | 99 993 | Yes |
| HD-789 | 38 552 | 349 600 | 235 424 | 127 526 | 101 695 | Yes |
| sv. israelensis 4Q7 | 18 927 | 0 | 235 472 | 7 064 | 6 779 | Yes |
| sv. israelensis BGSC 4Q1 | 36 877 | 470 675 | 235 424 | 27 737 | 101 282 | Yes |
| sv. israelensis BR58 | 405 796 | 0 | 237 806 | 140 548 | 104 442 | Yes |
| sv. israelensis ATCC 35646 | 360 161 | 328 014 | 230 106 | 27 991 | 100 639 | Yes |
| IBL 4222 | 358 462 | 349 600 | 235 354 | 115 130 | 97 052 | Yes |
| 147 | 359 606 | 0 | 235 523 | 121 171 | 98 048 | Yes |
| HD 1002 | 360 258 | 349 600 | 235 424 | 29 034 | 100 003 | Yes |
| sv. novosibirsk BGSC 4AX1 | 367 164 | 369 542 | 235 424 | 133 440 | 102 999 | Yes |
| ‘Bacillus sp’ Root11 | 362 213 | 349 698 | 235 424 | 118 825 | 98 931 | Yes |
| ‘Bacillus sp’ Root131 | ND | ND | ND | ND | ND | Yes |
| UBA3967 | 30 302 | 0 | 0 | 7 064 | 6 578 | No |
| Lr7/2 | 32 503 | 0 | 1 337 | 7 969 | 2 956 | No |
| IBL 200 | 226 809 | 0 | 1 912 | 23 458 | 10 595 | No |
| sv. morissoni BGSC 4AA1 | 46 544 | 0 | 174 | 19 230 | 6 520 | No |
| sv. kurstaki HD73 | 49 001 | 0 | 174 | 8 996 | 2 776 | No |
Reference plasmids of B. thuringiensis sv. israelensis AM65–52 strain are labelled as they are deposited in GenBank and with alternative names used in this review. Lengths of identity in the B. thuringiensis sv. israelensis AM65–52 strain are equal to the sizes of the plasmids in this strain. Large differences between identity length and the size of plasmid in B. thuringiensis sv. israelensis AM65–52 indicate the absence of corresponding counterpart plasmid in the query strain. The column ‘Status as B. thuringiensis sv. israelensis’ refers to the same column in Table A1. ND: Not Determined.
In this respect, strain B. thuringiensis UBA3967 was the most interesting since none of the plasmids from the sv. israelensis reference set could be detected, and the assembly size of about 5.2 Mb suggests that it is a natural plasmidless B. thuringiensis sv. israelensis strain. However, this sample originated from assembly and filtering for individual bacterial genomes of more than 1500 sets of GenBank metagenomic data (Parks et al.2017). Such filtrated assemblies are also known as MAGs (metagenome-assembled genomes). Apparently, the filtration protocols are limited in considering plasmid sequences. The proposed labelling of this genome as UBA (uncultivated bacteria and archaea) (Parks et al.2017) is also somewhat misleading since B. thuringiensis sv. israelensis strains are easily cultivable. It would be more pertinent to label it as MAG. It is surprising why the authors did not filter such sequences out before submitting the whole set of their assemblies into the GenBank. The origin of strain Lr7/2 was not further analysed since there is not much of its description available. However, the assembly has the total size of 5.6 Mb, suggesting the presence of some plasmids absent in the reference set of sv. israelensis strains (Tables 1 and 2). It should be noted that the levels of identity in the related parts of the present plasmids are 99.9% or higher, and for the non-present plasmids it is usually below 97%. It can therefore be concluded that 12 strains, indicated in Tables 1 and 2, constitute at present, by both criteria, chromosomal and plasmid content closeness, the B. thuringiensis sv. israelensis cluster with available genomic sequences.
Some other interesting observations of this analysis are also worth to be noted. Two of the 12 B. thuringiensis sv. israelensis cluster strains, 147 and BR58, do not contain counterparts of the 350-kb conjugative plasmid pXO16. Interestingly, both strains were isolated in Brazil (Barbosa et al.2015; Zorzetti et al.2015). In addition, strain 147 is the only natural strain from this group that lacks the pGIL01 prophage (see below and Gillis and Mahillon 2014c). By contrast, the strains B. thuringiensis sv. yunnanensis BGSC 4AM1, sv. pondicheriensis BGSC 4BA1 and B. thuringiensis HD 1011 contain counterparts of pXO16. It is surprising that this plasmid is so rare in natural isolates (3/440 non-B. thuringiensis sv. israelensis strains) since the conjugation competent recipient spectrum seemed not to be limited to the israelensis serovar (Jensen et al.1996; Makart et al.2018). It is worth to note that prophage GIL16, a pGIL01 closely related plasmidial prophage, is well represented in clusters of B. thuringiensis sv. aizawai and sv. kurstaki (Verheust, Fornelos and Mahillon 2005; Gillis and Mahillon 2014c).
Many of the analysed B. thuringiensis strains contain more than 200 kb of similarity with the 360-kb plasmid pBtic360 (see below). Similarity with this enigmatic plasmid, not known before the NGS era, was mostly detected with the anonymous AFS strains, massively sequenced recently (NCBI BioProject PRJNA400804). Presumably a smaller counterpart of ca. 250 kb is present in many strains outside the B. thuringiensis sv. israelensis group, exemplified by B. thuringiensis IBL 200 in Table 2.
Almost no homology to the pBtic235 plasmid was detected outside of the B. thuringiensis sv. israelensis cluster, which appeared to be a prophage (see below and Gillis et al.2017b). This is presumably related to its phage nature, displaying a narrow host range. The insecticidal toxin carrying plasmids pBtoxis and pBtic100 do not reveal extensively similar counterparts outside of the B. thuringiensis sv. israelensis group. Moreover, it is known that some B. thuringiensis sv. israelensis strains, like ATCC 35646 and B. thuringiensis HD 1002, do not carry pBtoxis, but only pBtic100. Our analysis adds the strain B. thuringiensis sv. israelensis BGSC 4Q1 (whose original name was HD-567) to this list.
THE EXTRACHROMOSOMAL ZOO
Bacillus thuringiensis strains have long been recognised to carry numerous extrachromosomal molecules (González and Carlton 1980), whose sizes vary from 2 to more than 500 kb (Sheppard et al.2013) and numbers reaching up to 17 distinct molecules (Reyes-Ramírez and Ibarra 2008). As already indicated, the main focus brought to these elements relates to the entomopathogenesis of their host strains and the plasmid-borne genetic determinants of the entomotoxins: Cry (González and Carlton 1984), Cyt (Faust et al.1983; Berry et al.2002) and VIP (Hollensteiner et al.2017a). In addition, most of the entomotoxin genes are located inside or adjacent to mobile entities (see below) such as insertion sequences (IS), composite and complex transposons (Tn) (Léonard, Chen and Mahillon 1997; Rosso, Mahillon and Delécluse 2000; Murawska, Fiedoruk and Swiecicka 2014).
For strain 4Q2 (also known as HD-500), the total size of the extrachromosomal DNA is 747 771 bp for seven molecules, excluding the genome of the potential phage-like particle reported by Tam and Fitz-James (1986) (see below). For strain HD-789, the extrachromosomal genome was reported to totalise 839 352 bp, as compared to the 5495 278 bp of its chromosome, which represents ca. 13% of the whole genome.
No specific direct experiments have been performed to precisely determine the copy number of the various plasmid molecules. Analysis of plasmid pattern ran in agarose gels suggested a much higher copy number for the three small plasmids than for pGIL01 and, even more so, for the large molecules. In addition, as detailed below, at least one molecule (pBtic235) is particularly difficult to see on a gel (Jensen et al.1996; Gillis et al.2017b). Recent genome sequencing data have suggested that the large plasmids are present in low copy number (< 3), while those of the small ones vary from 10 to 40 copies, depending on the strain. For pGIL01, a variation from 10 to 15 copies was estimated, with the median value of 12–13 (Bolotin et al.2017).
As is generally the case for bacterial plasmids, most B. thuringiensis sv. israelensis extrachromosomal molecules are circular, with the notable exception of the temperate tectiviruses (i.e. pGIL01, see below) that reside as 15-kb linear prophage molecules. The following sections give further details on the main features and, when appropriate, the potential role(s) of the plasmids encountered in the B. thuringiensis sv. israelensis strains, using strain 4Q2 as reference.
The three small circular pTX14 plasmids
The first small circular plasmids of B. thuringiensis sv. israelensis were isolated and characterised from strain 4Q2 and were named pTX14–1 (5 415 bp), pTX14–2 (6 829 bp) and pTX14–3 (7 649 bp) (Clark et al.1985; Andrup et al.1994, 2003; Andrup, Bendixen and Jensen 1995). Their maps are illustrated in Fig. 2. As detailed below, they all harbour at least two functional units, one for replication and another for mobilisation. Since these plasmids replicate using a rolling circle replication (RCR) mechanism, they contain, at minima, a replication (Rep) gene, a double-strand origin (dso or leading-strand origin) and a single-strand origin (sso or lagging-strand origin) (for some recent reviews, see Ruiz-Masó et al.2015; Wawrzyniak, Płucienniczak and Bartosik 2017). Their mobilisation relies on Mob proteins that recognise their cognate oriT, which are then nicked at the initiation step of mobilisation. The actual transfer is then taken over by the transfer machinery of a co-resident conjugative plasmid (Andrup et al.2003).
Figure 2.
Circular maps of the three small plasmids pTX14–1, pTX14–2 and pTX14–3 from B. thuringiensis sv. israelensis. The block arrows in the outer circles indicate the predicted CDSs in their direction of transcription. The black circle represents the GC content plotted using a sliding window, as the deviation from the average GC content of the entire sequence. The green/magenta circles represent the GC-skew calculated using a sliding window, as (G-C)/(G+C), and plotted as the deviation from the average GC skew of the entire sequence. Double-strand origins (dso) and single-strand origins (sso) are presented as rose block arrows; origins of transfer (oriT) are indicated by the orange bar. CDSs encoding for putative ‘Bacillus-collagen-like’, Rep and Mob proteins are highlighted by blue, grey and purple block arrows, respectively. CDSs encoding for hypothetical proteins are indicated in light green. Other relevant loci are indicated. Maps were generated by CGView (Grant and Stothard 2008) using the sequences of B. thuringiensis subsp. israelensis plasmids pTX14–1 (Acc. #: NC_002091), pTX14–2 (Acc. #: NC_004334) and pTX14–3 (Acc. #: NC_001446).
A striking feature of these pTX14 plasmids is the presence, in each plasmid, of a distinct ‘bcol’ (Bacillus-collagen-like) gene coding for a protein containing a central domain with a repetitive triplet motive analogous to those found in eukaryotic collagen (Gly-X-Y). So far, no specific function has been attributed to these putative peptides.
Also, contrary to what was observed for pGI1 from strain H1.1 of B. thuringiensis sv. thuringiensis (Fico and Mahillon 2006), no obvious toxin–antitoxin (TA) modules were noticed among the pTX14 elements. These TA systems, also known as addiction systems, have been suggested to participate to the segregational stability of their host plasmids during the cell partitioning.
pTX14-1
pTX14–1 is the smallest of the three pTX-14 plasmids and its relatives are also the rarest among B. thuringiensis strains. pTX14–1 rep14–1 and mob14–1 genes have homologues in a few other plasmids such as pBTm019A (12.3 kb) from B. thuringiensis (Nagamatsu et al.2010), p12509 (12.5 kb) from B. thuringiensis Bt18247 (Hollensteiner et al.2017b) or pBTZ_5 (4.9 kb) from B. mycoides strain BTZ (Johnson, Daligault and Davenport 2015). As recently reported, the pTX14–1 counterparts are missing from the B. thuringiensis sv. israelensis strains ATCC 35646, HD-789 and HD 1002, but they are present in strains AM65–52, IPS 82 and BMP144 (Bolotin et al.2017).
pTX14–1 replication protein belongs to the pC194/pUB110 family of RCR Rep proteins that is widespread among Gram-positive bacteria, in particular among Firmicutes such as Bacillus spp., Listeria spp. and Staphylococcus spp. (Ruiz-Masó et al.2015). Similarly, its 420-residue Mob14–1 displays homologies (mainly in its N-terminal domain) to Mob proteins encoded by plasmids as diverse as pPL1 from Marinococcus halophilus DSM 20408T (Louis and Galinski 1997) or pAP3.9 from an alpha-proteobacterial endosymbiont of Amoeba proteus (Park et al.2009).
As indicated above, the third CDS (coding DNA sequence(s)) of pTX14–1 encodes a putative ‘Bacillus-collagen-like’ protein. Although the bcol14–1 gene sequence does not share similarity with the corresponding genes of the other two pTX14 plasmids, the Bcol14–1 protein is related to putative proteins, generally larger (from 500 to more than 700 residues), found among members of the B. cereus group, as well as among Clostridium species. The actual role of each such protein is elusive. The most studied glycoprotein of this family, BclA, synthesised during sporulation, was demonstrated to be an important component of the exosporium and thus plays multiple roles in surface properties of mature B. cereus spores (Terry et al.2017). The particular repeat patterns are however very variable and a particular biological function of each paralogous protein can be unique. It is noteworthy that the high variability of repeats in genes encoding such proteins was used in developing of the most sensitive strain identification and discrimination methods, like VNTR (variable-number tandem repeat) or MLVA (multiple locus VNTR analysis) (Keim et al.2000; Castanha et al.2006).
pTX14-2
pTX14–2 and its close relatives are present in most if not all the strains of the B. thuringiensis sv. israelensis group. Besides the rep14–2, mob14–2 and bcol14–2 genes, this plasmid bears at least two other CDS, including one coding for a potential transcriptional regulator from the Xre (xenobiotic response element) family (McDonnell et al.1994). pTX14–2 replicon has been shown to reside in a 2.2 kb fragment that contains the rep14–2 gene and its cognate dso site (Andrup et al.2003). Mob14–2 has homologues in both close and distantly related plasmids such as pTA1015 and pTA1060 from Bacillus subtilis (Meijer et al.1998, 41% identity) to pSD853_7.9 from Salmonella enterica subsp. enterica serovar Dublin plasmid (YP_004376195.1, 37% identity). Interestingly, as previously shown, the Rep14–2 and Mob14–2 belong to families distinct from those of Rep14–1 and Mob14–1, respectively, of pTX14–1. Yet, Rep14–2 belongs to the same group VII of RCR replication proteins as Rep14–3 from the co-resident plasmid pTX14–3 (Andrup et al.2003).
pTX14-3
pTX14–3, the largest of the small resident plasmids of strain 4Q2, is absent from the other strains of the B. thuringiensis sv. israelensis group, with the exception of strain ATCC 35646 (Bolotin et al.2017). It shares however important analogy (rep, mob or both) with plasmids from other B. cereus s.l. strains such as pBCT8 (7 971 bp from Bacillus toyonensis) (Jimenez et al.2013), pSin9.7 b (9 698 bp from B. mycoides Sinv) (Di Franco et al.2005) or pFR12 (12 095 bp from B. thuringiensis INTA-FR7–4) (Amadio, Benintende and Zandomeni 2009). As for pTX14–2, its replication region was cloned and contained rep14–3 and both the sso and dso sites (Madsen, Andrup and Boe 1993; Andrup et al.2003).
In addition to the replication, mobilisation and ‘Collagen-like’ modules, this plasmid codes for at least four other putative proteins including a well-conserved hypothetical 133-residue protein, annotated as ‘transporter’ also found outside the B. cereus group (e.g. Bacillus flexus or Paenibacillus terrigena), and another 133-aa protein belonging to the Xre family of transcriptional regulator.
pGIL01, the plasmidial tectivirus prophage
The fourth B. thuringiensis sv. israelensis plasmid (in increasing size), namely pGIL01, is a linear molecule of 14 931 bp delimited by imperfect 73-bp terminal repeats (Verheust, Jensen and Mahillon 2003). Remarkably, this molecule is also a lysogenic phage, known as GIL01 (Fig. 3), which can reside and replicate independently as a linear plasmidial (non-integrated) prophage inside the host cell and, after induction by DNA-damaging treatments, is able to produce viable phage particles (Verheust, Jensen and Mahillon 2003; Gillis and Mahillon 2014b,c). This phage belongs to the genus Betatectivirus within the family Tectiviridae (Gillis et al.2017a; Adriaenssens et al.2018). The type species for this genus is Bacillus virus Bam35 owing to all the biological, genomic, proteomic and structural data available for this virus (Gillis et al.2017a). Even though most studies covering the biology of this type of phages refer to the archetype tectivirus PRD1 (genus Alphatectivirus), which infects Gram-negative bacteria, several studies have emphasised the structural and functional properties shared by PRD1 and Bam35 (Benson et al.2004; Fuller 2005). Consequently, many of the morphological characteristics described for PRD1 are also assumed to be similar for tectiviruses in Gram-positive bacteria. All possess an isometric protein coat surrounding a lipid membrane, which encloses the linear double-stranded DNA with covalently linked terminal proteins (TP). Both groups are tail-less phages, but the lipid membrane can act as a tail-like structure for genome delivery upon adsorption on a susceptible host (Oksanen and Bamford 2012; Gillis and Mahillon 2014b). Comparison of cryo-electron microscopy and three-dimensional image reconstructions of Bam35 with the ones of PRD1 showed that the capsids of the two phages are identical in size (64.5 nm between opposite faces), with spikes that protrude from the vertices of the virions, and that the structures associated with the assembly and fold of Bam35 major capsid protein closely resembles that of PRD1 (Benson et al.2004; Fuller 2005). Also, despite the absence of any sequence homology, the general organisation of the genomes in both groups of phages is maintained (Ravantti et al.2003). Yet, the major biological difference between PRD1- and Bam35-like phages resides in their respective propagation styles: PRD1-like phages are exclusively lytic phages, whereas Bam35-like ones are temperate, establishing a prophage state (Strömsten et al.2003; Verheust, Jensen and Mahillon 2003; Fornelos, Bamford and Mahillon 2011; Gillis and Mahillon 2014c).
Figure 3.
Functional map of B. thuringiensis sv. israelensis linear plasmidial prophage pGIL01. Predicted CDSs and their direction of transcription are represented as block arrows and are divided into two lines for figure display. The graphs below the block arrows represent the %GC content plotted using a sliding window, as the deviation from the average GC content (red) of the entire sequence [GC(blue)/AT(green)]. CDS numbers are indicated above the block arrows. CDSs previously shown or suggested function (Fornelos et al.2011; Jalasvuori et al.2013; Gillis and Mahillon 2014c; Berjón-Otero et al.2017) are colour-coded: rose, regulatory element; grey, unknown function; blue, replication; purple, DNA packaging and assembly; orange, capsid structural component; yellow, membrane structural component; green, lysis. Inverted terminal repeats (ITR) are highlighted by the red block arrows at both ends of the genome. Promoters P1, P2 and P3 are indicated by angled arrows, whereas putative Pho-independent transcription terminators are depicted as open stem loops downstream CDSs 8 and 30. Three gene modules based on functional grouping are indicated. The rulers represent base pairs. Map was generated using Geneious 11.1.2 (http://www.geneious.com; Kearse et al.2012) and the sequence of Bacillus phage pGIL01 (Acc. #: AJ536073) (Verheust et al.2003).
The genomes of phages Bam35 (genome length: 14 935 bp) and GIL01 were sequenced independently in 2003 (Ravantti et al.2003; Verheust, Jensen and Mahillon 2003), revealing that they have almost identical genomes. Based on the literature, these two phages were isolated from different hosts: Bam35 was isolated from B. thuringiensis sv. alesti, while GIL01 was found in B. thuringiensis sv. israelensis, as previously pointed out (Ackermann et al.1978; Verheust, Jensen and Mahillon 2003). Nevertheless, subsequent studies that focused on analysing the diversity among the tectiviruses infecting B. cereus s.l. have highlighted that particular phage subclusters contained GIL01-like phages all derived from B. thuringiensis sv. israelensis strains, except for Bam35 (Gillis and Mahillon 2014c). Due to the homogeneity of GIL01-like subclusters, it is plausible that the strain originally harbouring Bam35 was wrongly identified as B. thuringiensis sv. alesti, as the classification of B. thuringiensis serovars was developed on the basis of H-flagellar antigens (de Barjac and Frachon 1990). Thus, for purposes of this review, we will further consider Bam35 and GIL01 as essentially the same phage.
GIL01 modular genome
GIL01 and other tectiviruses infecting B. cereus s.l. possess a modular genome divided into a ‘replication-regulation region’ that encodes all proteins involved in phage genome replication and regulation, ensuring the replication of the linear molecule inside the host cell, and a ‘packaging-lysis region’ harbouring a module encoding for virion structural and DNA packaging proteins and another module encoding for host recognition and lytic proteins (Fig. 3) (Gillis and Mahillon 2014c).
GIL01 genome delivery and replication
The entry process of Bam35/GIL01 into its hosts has been dissected into three distinct steps: (i) receptor binding, (ii) peptidoglycan digestion, and (iii) interaction with the host cytoplasmic membrane (Gaidelytė et al.2006). Bam35/GIL01 adsorbs to the N-acetyl-muramic acid that constitutes the peptidoglycan of the host cell (Gaidelytė et al.2006). However, the specific molecules acting as receptor(s) in susceptible hosts are still unknown. Lytic activities of recombinant endolysins, Mur1 and Mur2 (CDSs 26 and 30 in GIL01, respectively), have been demonstrated (Verheust, Fornelos and Mahillon 2004), but which of these two enzymes is involved in peptidoglycan digestion for phage entry is still not resolved. After genome is delivered into the host cell, Bam35/GIL01 can either establish a carrier state and reside inside the cell as a linear plasmid or lyse the infected cells leading to the release of the virion progeny (Strömsten et al.2003; Verheust, Jensen and Mahillon 2003; Fornelos, Bamford and Mahillon 2011).
Recently, it has been demonstrated that Bam35/GIL01 replicates by means of a protein-primed mechanism that involves a subgroup of family B DNA polymerases (DNAPs) (Berjón-Otero et al.2015). In the protein-primed mechanism, replication starts from both ends of the TP-containing DNA (TP-DNA) by the formation of a phosphoester bond of the first nucleotide to an OH group of a serine, a threonine or, as in the case of PRD1 and Bam35/GIL01, a tyrosine of the TP and progresses asymmetrically from both ends (Berjón-Otero et al.2016; Lujan, Williams and Kunkel 2016). Protein-primed genome replication usually involves a terminal repetition that allows the recovery of the 3΄-end information in the template strand after initiation in an internal position, by means of a backward translocation of the primed TP-DNA complex (Berjón-Otero et al.2016; Lujan, Williams and Kunkel 2016). In the case of Bam35, the priming reaction is directed by the third base of the template strand and the genetic information of the genome end is recovered by a single-nucleotide jumping-back mechanism (Berjón-Otero et al.2016). DNAPs involved in protein-primed DNA replication are characterised by high processive polymerisation and strand displacement capacity, and the one from Bam35/GIL01 is no exception. However, despite its fidelity and proofreading activity, Bam35/GIL01 DNAP is also able to perform abasic site translesion synthesis. This latter activity might safeguard the phage genome integrity, when exposed to genotoxic agents (Berjón-Otero et al.2015).
GIL01 lytic switch and lysogeny
Bam35/GIL01 lytic switch is regulated by a multicomponent system that relies on three promotor regions (P1, P2 and P3), which guide the transcription of all genes in the same direction (Fig. 3) (Fornelos, Bamford and Mahillon 2011; Fornelos et al.2015, 2018). The induction of the phage is SOS-dependent, but unlike phage lambda, it does not depend on a CI-like repressor. Instead, stable lysogeny relies on the host transcription factor LexA, as it represses the phage transcription by binding to a set of boxes (dinBox sequences) located in the lysogenic promoter P1 (Fig. 3), which controls the expression of genes involved in phage DNA replication and transcription regulation (Fornelos, Bamford and Mahillon 2011). However, LexA is unable to efficiently repress Bam35/GIL01 transcription unless a 50-amino acid phage-encoded polypeptide, named gp7 (encoded by CDS 7), is also present. Gp7 enhances LexA binding to operator DNA by forming stable complexes with free or DNA-bound LexA and, most importantly, impairs RecA-mediated autocleavage of LexA (Fornelos et al.2015). Moreover, it has been recently shown that promoter P3, which controls the expression of late genes (i.e. structural and lytic genes), is also repressed by the LexA-gp7 complex by binding to tandem LexA boxes located within the promoter. Both promoters P1 and P3 are switch on by DNA damage, but P3 expression is delayed in comparison with P1 and requires a second small phage-encoded protein, gp6 (encoded by CDS 6) that is a LexA homologous (Fornelos et al.2018). These recent findings indicate that upon DNA damage gp6 can directly activate transcription at P3, resulting in the expression of late genes required for virion assembly and lysis of the host. Another protein, gp1 (encoded by CDS 1), has also been reported as required for Bam35/GIL01 lysogeny. This protein displays a DNA-binding domain similar to the MerR (repressor of mercury Resistance operons) transcriptional regulator, and, thus, it might be acting as a modulator of DNA structure regulating the transcription of the phage (Fornelos, Bamford and Mahillon 2011).
As several other temperate phages, Bam35/GIL01 has established a stable relationship with its host bacteria being able to modify some of the B. thuringiensis survival/ecological traits when present in lysogenic state. For example, it was found that Bam35/GIL01 lysogeny modifies the bacterial growth (i.e. larger colonies), decreases sporulation rates, has a negative impact on biofilm formation and enhances swarming motility, all by not yet identified mechanisms (Gillis and Mahillon 2014a).
When the lytic pathway is induced, the progeny virions are assembled within the host cytoplasm around 40 min post-infection (p.i.). Also, a strong K+ efflux has been registered approximately 40 min p.i. and was proposed to be associated with the insertion of holin molecules into the host cytoplasmic membrane. Cell lysis begins around 45 min p.i. and is completed in 15 min (Gaidelytė et al.2006; Daugelavičius et al.2007).
pBtoxis, the mosquito nightmare
As discussed earlier, B. thuringiensis sv. israelensis strains are key players in worldwide mosquito biocontrol. Their entomopathogenic activity mostly depends on toxins produced as a parasporal crystal during the sporulation phase (Lacey 2007; Palma et al.2014). In 1984, González and Carlton demonstrated that a large 75-MDa plasmid is required for crystal toxin production in B. thuringiensis sv. israelensis (González and Carlton 1984). This plasmid can undergo genomic rearrangements (i.e. recombination) with a 110-kb plasmid to create two new plasmids of 105 and 135 kb. This plasmid was later on named pBtoxis, the 128-kb toxin-coding plasmid. Given its importance in mosquitocidal activity and other cellular functions, pBtoxis was first mapped (Ben-Dov et al.1999) and then completely sequenced and annotated (Acc. #: AL731825) (Berry et al.2002). The predicted 125 CDSs were associated to the production of insecticidal factors, sporulation and germination, transcription regulation, production of antimicrobial peptides, amino acid metabolism and plasmid replication and partition. A circular map of pBtoxis is shown in Fig. 4. Bolotin et al. (2017) have recently compared the extrachromosomal pool of different B. thuringiensis sv. israelensis genomes. Concerning the plasmids carrying insecticidal crystal protein genes, this study showed that strain HD-789 contains a 225 kb plasmid, one part highly similar to pBtoxis and the other to a 100 kb plasmid (Doggett et al.2013). This might be due either to faulty artificial contig joining or to plasmid co-integration. As for strain AM65–52, it carries a 128-kb plasmid identical to pBtoxis, with the exception of a symmetrical inversion of almost 60 kb, presumably due to a single recombination event (Bolotin et al.2017).
Figure 4.
Circular map of B. thuringiensis sv. israelensis plasmid pBtoxis. The block arrows in the outer circles indicate the predicted CDSs in their direction of transcription, with or without functional annotation or relevant homologues. The black circle represents the GC content plotted using a sliding window, as the deviation from the average GC content of the entire sequence. The green/magenta circles represent the GC-skew calculated using a sliding window, as (G-C)/(G+C), and plotted as the deviation from the average GC skew of the entire sequence. CDSs encoding for mosquitocidal toxins (Cry and Cyt) and other accessory proteins (P19 and P20) with relevant roles in promoting pesticidal crystal formation are indicated by green block arrows. MGE including IS and transposons are highlighted by rose block arrows. CDSs potentially involved in peptide antibiotic production and export are indicated in blue. Remaining predicted CDSs are indicated in purple. Map was generated by CGView (Grant and Stothard 2008) using the sequence of B. thuringiensis subsp. israelensis plasmid pBtoxis (Acc. #: AL731825) (Berry et al.2002).
The following sections discuss some of the various functions of this plasmid and the genes it carries, based on the annotation given by Berry et al. (2002).
Mosquitocidal toxins
Five cry genes are located on pBtoxis: cry4Aa, two cry4Ba, cry10Aa and cry11Aa, as well as three cyt genes cyt1Aa, cyt2Ba and cyt1Ca and one haemagglutinin-related protein coding gene. The cry and cyt genes code for the parasporal crystal proteins which are produced abundantly during the sporulation phase (Stein et al.2006). However, in some cases, pBtoxis can undergo DNA rearrangements leading to deletion of certain cry genes. This was the case for strain BUPM97 isolated in Tunisia, in which a DNA fragment containing cry4A and cry10A genes was deleted (Zghal and Jaoua 2006). In other cases, such as strains BGSC 4Q1, ATCC 35646 and HD 1002, most of the pBtoxis plasmid is absent (Table 2). This correlates with the fact that strain ATCC 35646 did not show larvicidal activity when tested against A. aegypti larvae (Bolotin et al.2017). Apart from these strains, a divergence from the classical plasmid content of B. thuringiensis sv. israelensis is the one of strain HD-789 mentioned above (pBTHD789–3, Acc. #: CP003766) (Doggett et al.2013). This plasmid carries seven parasporal crystal coding genes consisting of cry4Aa3, cry4Ba5 (two genes), cry10Aa3, cry11Aa3, cry60Ba3 and cry60Aa3, in addition to three cyt and one haemagglutinin gene. It is also important to note three non-toxic key elements, coded by genes located on pBtoxis and necessary for full B. thuringiensis sv. israelensis insecticidal activity. One is a 54-kDa protein, containing metallophosphatase and ricin-like domains. Although this protein is not a toxin, it was shown to be specific to the parasporal body and important for its structural integrity (Diaz-Mendoza, Bideshi and Federici 2012). The other two are the accessory proteins P19 and P20, indispensable for correct folding of Cry and Cyt proteins (Manasherob et al.2001; Shi et al.2006).
Sporulation and germination genes
Another role played by pBtoxis is its potential ability to facilitate spore formation and/or germination. CDSs pBt031 and pBt145 are the putative proteins that may be associated with sporulation (Stein et al.2006). The former is similar to a peptidoglycan hydrolase, known to have an effect on sporulation in B. subtilis (Kuroda, Asami and Sekiguchi 1993). The other is a putative cell-coat associated protein. Another three CDSs organised in a single operon (pBt084, pBt085 and pBt086) are co-transcribed and encode putative spore germination proteins similar to GerAC, GerBB and GerKA of B. subtilis, respectively (Stein et al.2006). Two CDSs, pBt060 and pBt063, similar to pBt086 and pBt085, respectively, are apparent pseudogenes. The ger operon (pBt084, pBt085 and pBt086) was cloned and expressed in an acrystalliferous strain, and evaluated for its involvement in alkaline spore germination (Abdoarrahem et al.2009). The presence of the ger genes clearly increases spore germination capacity in an alkaline environment, resembling that of the insect midgut. This study also suggested that germination proteins encoded by the ger operon and the parasporal crystal toxins create a multifactorial influence, resulting in an improved germination in alkaline conditions in the midgut of the target insects (Abdoarrahem et al.2009).
Plasmid replication
For any given plasmid, replication initiation depends on a sequence known as the origin of replication (ori). pBtoxis ori is located in a 2.2 kb region spanning from the nucleotide position 124 407 to 126 636 as determined in a study conducted in 2006. This area of pBtoxis contains the ori and an operon coding for two proteins, CDS 157 and CDS 156 (Tang et al.2006). It also contains two cis-elements: a putative DnaA box motif responsible for the binding of initiator DnaA protein (Leonard and Grimwade 2010) and iterons implicated in the formation of iteron–initiator complexes necessary for conformational change and subsequent plasmid replication start (Chattoraj 2002). In pBtoxis, these iterons are small A+T rich imperfect direct repeats (DRs), mainly acting as binding sites for plasmid-encoded replication proteins (Tang et al.2007). Further analysis showed that CDS 157 is an iteron-binding protein, which forms a protein–DNA complex with iterons present upstream of its sequence. CDS 156 is a FstZ-like protein: a Mg2+-dependent GTPase that interacts with this complex and thus serves as a replication initiator protein (Tang et al.2007). In general, circular plasmid replication can occur in one of three different modes: theta replication, rolling circle or strand displacement. The replication elements present in pBtoxis suggest a theta replication mode (Tang et al.2007; Lilly and Camps 2015).
Other functions
Several other important functions are encoded by genes located on pBtoxis. For instance, CDSs pBt136 to pBt140 are transcribed (Stein et al.2006) and take part in the synthesis of a bacteriocin similar to AS48 produced by Enterococcus faecalis (Shehata et al.2017). Proteins encoded by CDSs pBt130 to pBt134 are ABC transporter components, reportedly involved in the bacteriocin secretion (Berry et al.2002). Five CDSs code for transcription regulators, four of which are transcribed (pBt014, 094, 148 and 149) (Stein et al.2006). Several transposases, which will be detailed in a later section, are also located on pBtoxis.
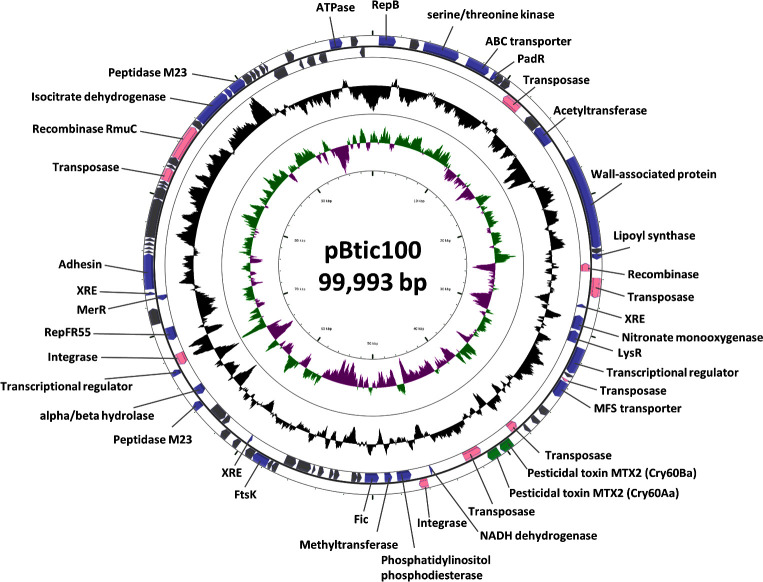
pBtic100, the 100-kb element
At present, the consensus knowledge about the insecticidal plasmid of the B. thuringiensis sv. israelensis group is that the active strains contain the 128 kb plasmid pBtoxis (Berry et al.2002) and this plasmid directs the synthesis of four different detectable Cry toxins and two Cyt toxins (Ben-Dov 2014). Three of the Cry toxins (Cry4Aa, Cry4Ba and Cry11Aa) represent the main set, toxic against A. aegypti, and Cyt1Aa acts synergistically and improves their efficacy, as previously explained (Ben-Dov 2014). Although on the level of biochemistry it appears to be correct, the recent analysis of genomic data indicates that the picture might be more complex, especially if one takes into account targets other than A. aegypti (Doggett et al.2013; Monnerat et al.2014; Bolotin et al.2017). The picture is further complicated by the fact that another plasmid, designated here as pBtic100 (Fig. 5), is also present in all B. thuringiensis sv. israelensis strains and this plasmid carries two additional Cry genes, encoding Cry60Aa and Cry60Ba toxins (Bolotin et al.2017). These proteins, sometimes designated as Cry15Aa, are classified as Etx/Mtx3 beta-sheet toxins (Berry and Crickmore 2017). They were originally described in B. thuringiensis sv. jegathesan, are identical to those of B. thuringiensis sv. israelensis ATCC 35646 and are active against the mosquito Culex quinquefasciatus (Sun et al.2013). The same authors mentioned that an identical toxin is produced by B. thuringiensis sv. malayensis BGSC 4AV1 and the corresponding protein can be found in GenBank under the accession number GU810818. We did not however identify a contig corresponding to this gene in the genomic assembly of this strain (Acc. ##: NFCR0100001 to NFCR01000104).
Figure 5.
Circular map of B. thuringiensis sv. israelensis plasmid pBtic100. The block arrows in the outer circles indicate the predicted CDSs in their direction of transcription. The black circle represents the GC content plotted using a sliding window, as the deviation from the average GC content of the entire sequence. The green/magenta circles represent the GC-skew calculated using a sliding window, as (G-C)/(G+C), and plotted as the deviation from the average GC skew of the entire sequence. CDSs with functional annotation are indicated in purple block arrows. CDSs in grey represent genes coding for hypothetical proteins. CDSs encoding for pesticidal toxins are indicated by green block arrows. Transposases, recombinases and integrases are highlighted by rose block arrows. Map was generated by CGView (Grant and Stothard 2008) using the sequence of B. thuringiensis subsp. israelensis AM65–52 plasmid pAM65–52-5–100K (Acc. #: CP013280) (Bolotin et al.2017).
Thus, two large plasmids, pBtoxis and pBtic100, out of five found in B. thuringiensis sv. israelensis strains, encode active insecticidal toxins. As already indicated above, of the 11 GenBank genomic sequences that were assigned to B. thuringiensis sv. israelensis, three (BGSC 4Q1, ATCC 35646 and HD 1002) contain only a small part of pBtoxis (Table 2). In one strain, HD-789, the two plasmids were reported as parts of pBTHD789–3, of 225 kb (Doggett et al.2013). We can therefore consider that either there are two relatively stable lineages of B. thuringiensis sv. israelensis strain possessing or not pBtoxis or that this plasmid can be easily lost and again acquired. It is also possible that the two plasmids can co-integrate, presumably due to a transposon of the Tn552 family, recently identified in both plasmids and only as one copy in their co-integrate pBTHD789–3 (Doggett et al.2013; Bolotin et al.2017).
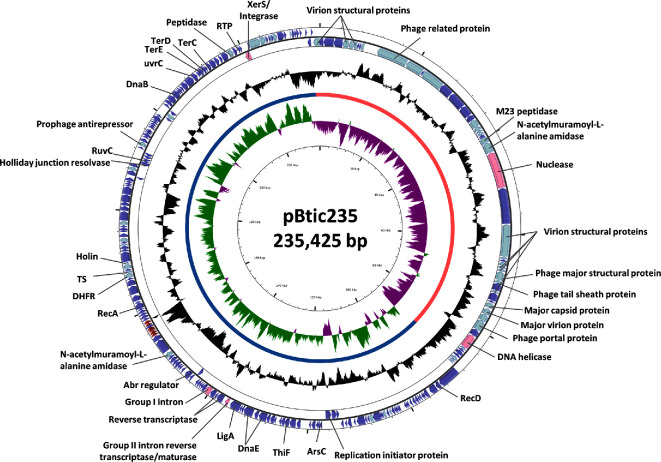
pBtic235, the cryptic molecule
In 1996, starting from strain B. thuringiensis sv. israelensis 4Q2, Jensen et al. obtained a plasmid-cured derivative they named 4Q7. This strain was mainly used to demonstrate the implication of pXO16 in the spectacular aggregation phenotype associated with conjugation (see the next section). However, in their paper, Jensen et al. (1996) noticed ‘the presence of a faint band in some of their plasmid preparations’. Recently, using several complementary approaches, it was shown that this ‘elusive’ band corresponded to a prophage-like molecule that was named pBtic235 in reference to its genome size of 235 kb (Gillis et al.2017b) (Fig. 6). This hybrid molecule has been detected in the 12 strains that constitute the B. thuringiensis sv. israelensis cluster (see Table 2 and Doggett et al.2013; Jeong, Park and Choi 2014; Johnson, Daligault and Davenport 2015; Bolotin et al.2017; Gillis et al.2017b), being almost a hallmark for this serovar.
Figure 6.
Circular map B. thuringiensis sv. israelensis plasmid pBtic235. The block arrows in the outer circles indicate the predicted CDSs in their direction of transcription, with or without functional annotation or relevant homologues. The black circle represents the GC content plotted using a sliding window, as the deviation from the average GC content of the entire sequence. The green/magenta circles represent the GC-skew calculated using a sliding window, as (G-C)/(G+C), and plotted as the deviation from the average GC skew of the entire sequence. The orange and blue semi-circles indicate the phage- and plasmid-like modules, respectively. CDSs in light blue represent genes coding for proteins found in phages. Enzymes commonly associated with excision of MGE are highlighted by rose block arrows. tRNAs are presented as red block arrows. Remaining predicted CDSs are indicated in purple. Map was generated by CGView (Grant and Stothard 2008) using the sequence of B. thuringiensis subsp. israelensis plasmid pBTHD789–2 (Acc. #: NC_018509) (Doggett et al.2013) and the functional annotation described by Gillis et al. (2017b).
The genome of pBtic235 displays potential plasmid- and phage-like modules transcribed mainly in opposite senses from each other (Gillis et al.2017b), suggesting that this molecule could be the result of the fusion between a plasmid and a phage. It possesses 17 tRNAs and 240 putative CDSs, many of them having no homologues in the databases. Nevertheless, CDSs coding for potential proteins involved in replication, genome packaging and virion structure, cell lysis, regulation of lytic-lysogenic cycles, metabolite transporters, stress and metal resistance have been predicted (Fig. 6) (Gillis et al.2017b). Of particular interest is the phage-like module, since many of the hypothetical phage structural proteins share a strong homology with their counterparts present in 0305phi8–36, a jumbo-myovirus that might have also originated from different type of molecules as it displays two replicative systems (Hardies, Thomas and Serwer 2007).
As a plasmid, pBtic235 is not capable of self-transfer to a recipient bacterium by conjugation, albeit it seems able to take advantage of co-resident conjugative plasmid pXO16 to promote its own mobilisation at frequencies of 10−5 transconjugants per recipient cell (Gillis et al.2017b). However, one of the most interesting features of this large cryptic molecule is its capacity to be induced by DNA-damaging treatments and occasionally produce turbid phage plaques in lawns of susceptible B. thuringiensis hosts, therefore displaying a dual plasmid-prophage nature. This striking feature provides new clues about gene transfer inside the B. cereus group. It has been proposed that pBtic235 might correspond to the previously reported prophage SU-11 induced also from a B. thuringiensis sv. israelensis strain (Kanda et al.1999; Gillis et al.2017b).
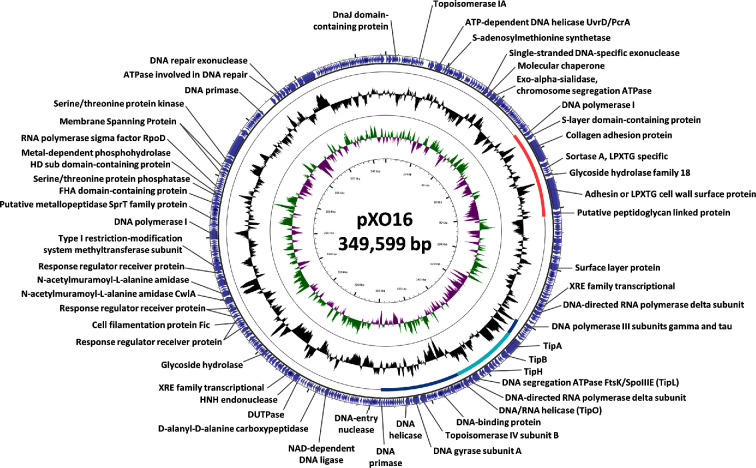
pXO16, the giant conjugative device
Conjugative capabilities of B. thuringiensis sv. israelensis are associated with pXO16, a very large and low-copy conjugative plasmid (350 kb) that has been studied for its peculiar transfer mechanism (Fig. 7). Originally discovered by Reddy, Battisti and Thorne (1987) for its ability to mobilise pBC16, a tetracycline-resistant (TetR) plasmid from B. cereus (Bernhard, Schrempf and Goebel 1978), the interest in pXO16 was renewed when its transfer was correlated to a characteristic cell aggregation phenomenon (Jensen et al.1995).
Figure 7.
Circular map of pXO16, the conjugative plasmid of B. thuringiensis sv. israelensis. The purple block arrows in the outer circle indicate the predicted CDSs in their direction of transcription, with or without functional annotation or relevant homologues. The black circle represents the GC content plotted using a sliding window, as the deviation from the average GC content of the entire sequence. The green/magenta circles represent the GC-skew calculated using a sliding window, as (G-C)/(G+C), and plotted as the deviation from the average GC skew of the entire sequence. The orange and blue bars represent the aggregation and replication regions, respectively. The ‘transfer israelensis plasmid’ (tip) region (Makart et al.2018) is highlighted by the cyan bar inside the replication region. Map was generated by CGView (Grant and Stothard 2008) by using the reverse complement sequence of B. thuringiensis subsp. israelensis plasmid pBTHD789–1 (Acc. #: NC_018516) (Doggett et al.2013) and submitting it to RAST server based on SEED subsystems for CDSs prediction and functional (re-)annotation (Aziz et al.2008; Overbeek et al.2014). Then, CDSs (re-) annotations were manually verified and compared with those of plasmid pBTHD789–1.
pXO16 aggregation
pXO16 conjugative aggregation phenotype can be witnessed, in ‘liquid mating’, by the appearance of macroscopic aggregates between two compatible strains, Agr+ and Agr− (Andrup, Damgaard and Wassermann 1993). The Agr+ phenotype and its transfer capabilities were unambiguously associated to the presence of pXO16, while the Agr− phenotype corresponded to pXO16-cured strains of B. thuringiensis sv. israelensis (Jensen et al.1995). The pXO16-encoded aggregation is not pheromone-induced since no aggregation-generating signals were detected in filtrates of donors, recipients and mating mixes (Andrup, Damgaard and Wassermann 1993). It therefore distinguishes itself from the conjugative aggregations observed in E. faecalis and Lactobacillus plantarum, which are induced by secreted pheromone molecules (Clewell and Weaver 1989; Ahn et al.1992).
It is generally considered that Gram-positive conjugation systems differ from the Gram-negative ones by the absence of sexual pilus, and that they rather rely on aggregating substances to form mating pairs (reviewed in Grohmann, Muth and Espinosa 2003). The addition of proteases during pXO16 mating abolished co-aggregation and decreased plasmid mobilisation (see below), suggesting the involvement of a protein as an aggregating modulator. Protease-treated Agr+ cells were still able to normally produce aggregates, whereas protease-treated Agr− cells delayed the appearance of aggregates. This was interpreted as the implication of a protein on the surface of Agr− cells and a non-proteinaceous molecule on the surface of Agr+ cells (Andrup, Damgaard and Wassermann 1993). Later, a large S-layer protein (> 200 kDa) located in the outer layer of the B. thuringiensis sv. israelensis strain 4Q2 was also reported to be absent in its plasmid-cured derivatives. Besides, anti-S-layer antibodies were shown to inhibit plasmid mobilisation. It was therefore suggested that pXO16 encodes this S-layer protein, which could be involved in the transfer mechanism (Wiwat et al.1995).
Interestingly, scanning electron micrographs of mating mixtures showed ‘connections’ between aggregating cells, one cell often attached to multiple partner cells (Andrup et al.1996). It is yet unknown whether these structures are used to get the cells in close contact or to actually convey the DNA from the donor to the recipient cells. Recently, however, pXO16 aggregation phenotype proved to greatly benefit during conjugative transfer while not being mandatory (Makart et al.2018).
pXO16 conjugation properties and host range
pXO16 conjugative transfer reaches 100% of the recipients in about 40 to 50 min in B. thuringiensis sv. israelensis matings, proving to be very fast and very efficient in this system (Andrup, Damgaard and Wassermann 1993; Timmery et al.2009). Nonetheless, it apparently displays a rather narrow host range, since only 10 B. thuringiensis serovars out of 21 tested were Agr− (i.e. able to produce aggregation when mixed with pXO16-containing strains). Three tested B. cereus strains were also Agr− (and therefore potentially acting as recipients), but the Bacillus megaterium, L. sphaericus and B. subtilis strains were not. All Agr− strains were able to receive pXO16 and their corresponding transconjugants effectively became Agr+ pXO16 donors (Jensen et al.1996). No taxonomic relationship could be found between Agr− strains. It is also worth noting that Wiwat, Panbangred and Bhumiratana (1990) successfully transferred pBC16 and pC194 (a chloramphenicol-resistant plasmid originating from Staphylococcus aureus; Ehrlich 1977) from B. thuringiensis sv. israelensis donors to 25 B. thuringiensis strains, as well as chromosomic markers to 12 subspecies of B. thuringiensis. It is yet unknown if this could be fully attributed to pXO16, as the implication of other conjugative plasmids and transposons in the recipients has not been ruled out. Notably, it was recently shown that pXO16 displays a broader host range than previously thought since it could actually be transferred to several strains of the B. cereus group that were not Agr−, i.e. not producing aggregates in the presence of Agr+ donor cells (Makart et al.2018).
pXO16 transfer kinetics and mechanism
Conjugative transfer kinetics were accurately described by Andrup et al. (1998) using the Michaelis-Menten model for enzyme kinetics, most notably because donor saturation was demonstrated. The minimal time for a conjugative transfer is about 3.5–4 min, which includes the formation of the mating pair and the expression of a TetR marker gene. A donor cell needs a recovery time of about 10 min before it can re-donate the plasmid. The maturation time needed for a transconjugant to become a donor is ca. 40 min. This fits the average time for the expression of an S-layer protein to cover the whole cell surface in Bacillus species (reviewed in Sidhu and Olsen, 1997). These kinetics studies also gave evidences in favour of a ‘mating pair’ model of transfer (one donor—one recipient pair) since donors did not seem to transfer pXO16 to more than one recipient cell at a time, even though aggregates are made of thousands of cells clumped together (Jensen et al.1996).
The sequence of pXO16 has been identified as corresponding to the pBTHD789–1 and pAM65–52-2–350K plasmids in the sequenced genomes of B. thuringiensis sv. israelensis HD-789 (Doggett et al.2013; Makart, Gillis and Mahillon 2015) and AM65–52 (Bolotin et al.2017) (Table 2). Sequence analysis showed that pXO16 sequence is highly coding but only few of its CDS have homologues in the databases. Nonetheless, hypothetical regions responsible for the aggregation phenotype and the plasmid replication have been highlighted (Makart, Gillis and Mahillon 2015). The common orientation of all CDS and the presence of a high number of potential paralogues have also suggested a phage-like nature, even though phage plaques have not yet been isolated. Concerning conjugative functions, no significant homologues to known type IV secretion systems (T4SS) were initially observed, suggesting that pXO16 possessed an unforeseen conjugative system. However, pXO16 conjugative system was recently proved to transfer single-stranded DNA through a mechanism distantly related to T4SS, like what is observed for the conjugative plasmid pCW3 from Clostridium perfringens (Makart et al.2018).
pXO16 mobilisation and retromobilisation
pXO16 can mobilise rolling-circle and theta-replicating plasmids, including the so called ‘non-mobilisable’ ones lacking the mob gene and the oriT site. The mob gene and the oriT site favour mobilisation but are not necessary for high-frequency mobilisation (Reddy, Battisti and Thorne 1987; Andrup, Damgaard and Wassermann 1993; Andrup et al.1996; Timmery et al.2009). However, an intact single-strand origin of replication seems to be crucial, with its orientation also having a role. Neither recombination nor deletion was detected upon plasmid mobilisation, suggesting that the mobilisation is processed by ‘donation’, namely without any physical recombination between the conjugative plasmid and the mobilised plasmid (Andrup et al.1996). Following the observation of cell connections in aggregates by electron scanning microscopy, it was hypothesised that transfer of non-mobilisable plasmids might result from random migration through these structures rather than actively conducted by the conjugation machinery.
Retromobilisation, the ‘capture’ of plasmid DNA from a recipient cell by a donor cell, was also observed for both mobilisable and non-mobilisable plasmids, albeit at lower frequencies for the latter (Timmery et al.2009). Retromobilisation was shown to follow a successive model, i.e. the conjugative transfer of pXO16 to the recipient is followed by mobilisation of the recipient plasmid to the donor. This model was supported by the observation of a delay in retromobilisation compared to pXO16 own transfer (Timmery et al.2009). Retromobilisation was also affected by the type of plasmid, since the kanamycin-resistant plasmid pUB110 was retromobilised faster and at higher frequencies than the erythromycin-resistant plasmid pE194 even though they both have similar size and originate from S. aureus (Lacey, Lewis and Rosdahl 1974; Horinouchi et al.1982; Timmery et al.2009).
Chromosomal marker mobilisation by pXO16
Recently, pXO16 was shown to mediate conjugative transfer of chromosomal loci at frequencies of ca. 10−6–10−5 transconjugants/donor cell (Makart et al.2017). In these experiments, mobilisation of chromosomal regions of at least 46 kb was demonstrated. As these frequencies coincided with those of non-mobilisable plasmid transfer, both phenomena are believed to occur by the same mechanism. Whereas most bacterial chromosomal transfer systems occur via the integration of conjugative elements into the chromosome prior to its transfer, pXO16 appears to transfer the chromosomal markers in the absence of physical integration, but rather through a ‘donation-type’ mobilisation. The use of variant calling sequencing has also allowed to fully map the chromosomal regions obtained by the transconjugants, and these transferred regions were shown to range from 2.5 kb to up to 791 kb (Makart et al.2018).
pXO16 ecology
pXO16 conjugative transfer and pBC16 mobilisation were detected in a broad range of temperature, pH values and salinity (Thomas et al.2001). This leads to believe that pXO16 conjugative system is rather insensible to environmental conditions. However, pBC16 mobilisation was not detected in river water and dead mosquito larvae whereas pXO16 self-transfer was observed but at lower frequencies. These effects might have been due to the sporulation of introduced cells and/or to the competition of indigenous bacteria, especially in dead larvae (Thomas et al.2001). An increase in the number of transconjugants in river water after 7 days coincided with sporulation, indicating a possible link between sporulation and pXO16 conjugation events (Thomas et al.2001). Concerning pXO16 transfer in food-related environments, it was proved to be very effective in cow milk, soymilk, rice milk and rice pudding (Van der Auwera et al.2007; Timmery et al.2009). On the other hand, mobilisation frequencies were reduced in these foodstuffs (Timmery et al.2009). This illustrates an increased sensibility of the mobilisation mechanism to the medium compared to the conjugative transfer. Finally, pXO16 conjugative transfer has also been detected in the intestine of gnotobiotic rats, but the cells bearing pXO16 were unexpectedly not stably maintained in this environment, while it is very stable in culture media (Wilcks et al.2008).
pBtic360, an elusive XXL plasmid
The plasmid designated as pBtic360 (Table 2) has never been detected yet, physically or functionally, as a plasmid entity. The only evidence for its presence in B. thuringiensis sv. israelensis strains is based on genomic sequencing. The reasons are that the plasmid possesses a very large size of 359.6 kb (Fig. 8), a low copy number of 1.0±0.2 per chromosome (Bolotin et al.2017) and has no apparent or easily detectable biological function as a whole.
Figure 8.
Circular map B. thuringiensis sv. israelensis plasmid pBtic360. The block arrows in the outer circles indicate the predicted CDSs in their direction of transcription. The black circle represents the GC content plotted using a sliding window, as the deviation from the average GC content of the entire sequence. The green/magenta circles represent the GC-skew calculated using a sliding window, as (G-C)/(G+C), and plotted as the deviation from the average GC skew of the entire sequence. CDSs with functional annotation are indicated in purple block arrows, and CDSs putatively involved in replication and/or regulation are highlighted by gray block arrows. CDS encoding for a putative mosquitocidal toxin is indicated in green. Transposases, recombinases and integrases enzymes are highlighted by rose block arrows. Map was generated by CGView (Grant and Stothard 2008) using the sequence of B. thuringiensis subsp. israelensis AM65–52 plasmid pAM65–52-5–360K (Acc. #: CP013276) (Bolotin et al.2017).
The first completed assembly of the plasmid was reported in the genomic sequence of the B. thuringiensis HD 1002 as a contig designated as plasmid 1 and deposited under the Acc. # CP009349, although the authors reported the strain as belonging to the thuringiensis serovar (Johnson, Daligault and Davenport 2015). Recently, the plasmid was independently assembled de novo using the reads generated for strain AM65–52 (Bolotin et al.2017). Its alignment with the contig of strain B. thuringiensis HD 1002 indicated almost complete identity. Co-linear contigs were also detected in assemblies of other strains, including that of BGSC 4AJ1, a remote strain B. thuringiensis sv. monterrey, where the counterpart plasmid appears to be rather different from those of the B. thuringiensis sv. israelensis strains. It is thus reasonable to assume that these elements represent a bona fide plasmid family among the B. thuringiensis strains. The reported complete assembly of B. thuringiensis HD-789 does not contain similar contigs (Doggett et al.2013). However, one can speculate that the plasmid should be present in this strain also, since it was detected in all genomic sequences of B. thuringiensis sv. israelensis strains, excluding only the artificially plasmid-cured strain 4Q7 (Table 2, see also Jeong, Park and Choi 2014; Bolotin et al.2017).
It is interesting to mention that there is some divergence between the counterpart plasmids detected in different strains. Whereas the pBtic360 molecules in strains HD 1002, AM65–52, IBL 4222 and ATCC 35646 differ only in few nucleotides, the corresponding contigs from strains B. thuringiensis IBL 200 and B. thuringiensis subsp monterrey BGSC 4JA1 share 95 and 93% of identity, respectively.
At present, the biological function of pBtic360 can only be judged by putative functions based on similarities of detected genes. Since many transporters were detected in the plasmid, it was postulated that the main advantage pBtic360 provides is to facilitate the transport of multiple metabolites (Bolotin et al.2017). The plasmid also contains more than 10 transposons and IS elements, suggesting a possibility for extensive gene exchange. Since the strains of B. thuringiensis sv. israelensis and sv. monterrey BGSC 4AJ1 target different insects, Diptera and Coleoptera, respectively (Zheng et al.2017), it can be suggested that pBtic360 and relatives do not specify adaptation to the insect host, but rather to general survival. An interesting question is also how similar plasmids were transferred to such different bacterial hosts. Since pBtic360 codes for the putative FtsK-like DNA transport protein and T4SS VirD4 protein, it was suggested that the plasmid was able of conjugative self-transfer (Bolotin et al.2017).
THE MOBILE GENETIC POOL
Although the extrachromosomal gene pool of the B. thuringiensis sv. israelensis strains represents a major adaptation potential, the extent of their intracellular mobile genetic elements (MGE) grants them additional opportunities to redesign and reshuffle their genomes, to haul individual genes and to modulate gene expression. MGE include IS, class II transposons (Tn, also named complex or replicative transposons), mobile insertion cassettes (MIC, Chen et al.1999) and miniature inverted repeat transposable elements (MITE) as well as integrons, type II introns, B. cereus repeats (bcr) and prophages (Craig et al.2002). In the following section, the MGE found in the genomes of B. thuringiensis sv. israelensis strains will be discussed, with the focus on the fully assembled genomes (strains AM65–52, HD-789 and HD 1002).
IS and MIC
Insertion sequences are the most common prokaryotic transposable elements (TE) with a very simple organisation, given they only carry what is necessary for their transposition: two terminal inverted repeats (IR) flanking a DNA transposase gene. They are omnipresent in prokaryotic genomes and can occur in very high numbers (Mahillon 2002; Siguier, Gourbeyre and Chandler 2014). IS can also generate short DR flanking the target site on insertion (Siguier et al.2015). The transposase can be coded by one or more open reading frames, depending on the absence or presence of a programmed ribosomal frameshifting (Nagy and Chandler 2004; Fayet and Prère 2010). In fact, a recent study has found that transposase coding genes are the most prevalent genes in nature (Aziz, Breitbart and Edwards 2010). Their density in the case of bacterial chromosomes is generally below 3%. However, in the case of plasmids, this density varies from 5 to 20% reaching an extreme of 40% in the case of pW100, the 221-kb megaplasmid of Shigella flexneri (Siguier, Filée and Chandler 2006).
IS are classified into different families using a variety of characteristics, not only depending on the transposase itself, but also including several structural features (Mahillon and Chandler 1998). The ISFinder database includes more than 4000 different IS, classified in 26 families (https://www-is.biotoul.fr/) (Siguier, Filée and Chandler 2006; Siguier et al.2006). A simple search in this database will reveal only seven reported IS for B. thuringiensis sv. israelensis, belonging to the IS4 and IS6 families. IS belonging to these families as well as other TEs have been extensively studied and reviewed for their structural association with genes coding for δ-endotoxins (Mahillon et al.1994) with IS231 being the first IS isolated from B. thuringiensis (Mahillon et al. 1985, 1987; Rezsöhazy, Hallet and Delcour 1992; Rezsöhazy, Hallet and Mahillon 1993). However, a more in-depth analysis of B. thuringiensis sv. israelensis IS content has not been conducted.
The complete genomes of the B. thuringiensis sv. israelensis strains HD-789 (Doggett et al.2013), HD 1002 (Johnson, Daligault and Davenport 2015) and AM65–52 (Bolotin et al.2017) were analysed for the presence of IS elements, using the online tool ISsaga (http://issaga.biotoul.fr/ISsaga2/). The results are shown in Fig. 9 and a summary is given in Table 3. A quick scan of these IS shows their proximity to toxin coding genes (data not shown). In fact, the plasmid carrying toxin coding genes pBtoxis has several IS, as shown in Fig. 4, such as IS240 from the IS6 family, first described in 1989, in B. thuringiensis sv. israelensis (Delécluse et al.1989). Two IS240 copies flank the 134-kDa protein-coding gene. This forms a composite class II TE, which supports the hypothesis linking this IS to B. thuringiensis sv. israelensis toxicity against dipteran larvae. Other IS found in B. thuringiensis sv. israelensis genomes belong to IS3 (Duval-Valentin, Marty-Cointin and Chandler 2004), IS4 (Hallet, Rezsohazy and Delcour 1991; Rezsohazy et al.1993; Hallet et al.1994; Léonard and Mahillon 1998; De Palmenaer, Siguier and Mahillon 2008), IS6, IS200/IS605 (Barabas et al.2008; He et al.2015), IS607 (Boocock and Rice 2013) and IS1182 families, as shown in Fig. 9. A search using tBlastN (Altschul et al.1997; Camacho et al.2009) of transposase sequences from different families, in the non-completed B. thuringiensis sv. israelensis genomes, confirmed the data obtained by analysing the complete genomes for each family (data not shown). However, the distribution of these IS between the chromosome and the plasmids differs. The most notable difference is for elements belonging to the IS3 and IS607 families that showed a preference to be located on chromosomes. As for IS from the IS4, IS6 and IS1182 families, they have a clear preference for plasmids (Table 3).
Figure 9.

Distribution of complete IS family members among the three complete genomes of B. thuringiensis sv. israelensis strains. The ISsaga web tool (Varani et al.2011) was used for IS annotation. The number of complete IS was detected in each strain genome. Only the families with at least one complete IS located on either the chromosome or the plasmids are shown in this graphic representation.
Table 3.
Distribution of IS family members among the three fully annotated sequences of B. thuringiensis sv. israelensis strains.
| Family* | Transposase chemistry | Transposition mechanism | Chromosomal copies | Plasmidial copies | Total |
|---|---|---|---|---|---|
| IS3 | DDE: 2 ORFs | Copy-paste | 30 | 5 | 35 |
| IS4 | DDE: 1 ORF | Cut-paste | 9 | 18 | 27 |
| IS6 | DDE: 1 ORF | Co-integrate | 0 | 15 | 15 |
| IS200 | HuH: 1 or 2 ORF | Peel-paste | 4 | 6 | 10 |
| IS607/IS605 | Serine TnpA | Cut-paste | 5 | 0 | 5 |
| IS1182 | DDE: 1 ORF | Unknown | 0 | 4 | 4 |
*Details on the IS families can be obtained at https://www-is.biotoul.fr/index.php. See also Siguier et al. (Siguier, Filée and Chandler 2006; Siguier et al.2006, 2015) for references.
MIC are a group of IS related TE. They consist of two IR lacking a transposase-coding gene, flanking one or more CDS (Chen et al.1999). Their sizes can vary from ca. 700 bp to 3 kb. The IS231 subgroup from the IS4 family includes many examples of MIC: 10 MIC231 were described by De Palmenaer, Vermeiren and Mahillon (2004). This study revealed that the various MIC231s, despite carrying different passenger genes, share a conserved region with IS231 elements next to the left IR, named the CA region (several Cs followed by a number of As). This study also proved that trans-transposition is possible in strains with a functional IS231 element. This is because the IS231 transposase recognises and acts on IS231-related IRs (De Palmenaer, Vermeiren and Mahillon 2004). Although several IS231 elements have been noted in B. thuringiensis sv. israelensis, no MIC has been characterised to date. Given their structure, variability and distribution, they will not be further analysed in this review.
Class II transposable elements (Tn)
Class II transposable elements (Tn) are divided into two groups: composite and non-composite or unit transposons. Composite transposons are two IS flanking one or more passenger genes such as antibiotic resistance or metabolic genes. This is the case for Tn5 and Tn10 families (Obukowicz et al.1986; Lawley, Burland and Taylor 2000). These composite elements, illustrated in the previous section, will not be further discussed in this review given that their mechanism and distribution resemble those of classic ISs. The other group of transposons, or unit transposons, are represented in B. thuringiensis sv. israelensis by elements belonging to the Tn3 family or that are labelled Tn3-like elements. These elements carry a DDE transposase operating by a copy-paste mechanism, which depends on the formation of a co-integrate structure that is resolved later on by a resolvase or invertase (Nicolas et al. 2015, 2017).
Tn4430 was the first class II transposable element discovered in B. thuringiensis (Lereclus, Menou and Lecadet 1983; Mahillon and Lereclus 1988). Even though it was isolated in several serotypes, it was not found in B. thuringiensis sv. israelensis strains (Mahillon et al.1994). Another class II transposon, Tn5401 (Baum 1994), was isolated from a sporulation deficient B. thuringiensis sv. morissoni and was associated with cry genes (Malvar and Baum 1994). TnBth3 was the first class II transposon identified in B. thuringiensis sv. israelensis, strain HD-789 (Doggett et al.2013). Table 4 shows the class II transposable elements found in B. thuringiensis sv. israelensis strains using ISsaga. Several class II transposable elements related to Tn3 were also identified by BlastN in non-completed assemblies. They include TnBth1, TnBth2, TnBth3, Tn4430 and Tn5401, with their copy numbers varying from 0 to 5 (Data not shown).
Table 4.
Distribution of Class II transposons (Tn3 family) and related MITES among the three complete B. thuringiensis sv. israelensis genome sequences.
| Elements | Transposase chemistry | Transposition mechanism | HD-789 | AM65–52 | HD 1002 |
|---|---|---|---|---|---|
| TnBth1 | DDE | Replicative transposition | 1 | 2 | 2 |
| TnBth2 | 0 | 0 | 0 | ||
| TnBth3 | 1 | 1 | 1 | ||
| Tn4430 | 0 | 0 | 0 | ||
| Tn5401 | 1 | 1 | 1 | ||
| MITEBth3 | 2 | 2 | 2 |
In addition, MITEBth3 related to TnBth1 was identified in all scanned B. thuringiensis sv. israelensis strains (Table 4). MITEs or TIMEs (Tn3 derived inverted-repeat miniature elements) are a group of short non-autonomous class II transposable elements (Fattash et al.2013; Szuplewska et al.2014). They are small multicopy elements, consisting of conserved terminal IR sequences, typical for the Tn3-family, as well as short stretches of conserved sequences in their core regions (Szuplewska et al.2014). TIMEs, like autonomous Tn3 family members, are present in low copy number in bacterial chromosomes and plasmids (Grindley 2002), which is also the case for B. thuringiensis sv. israelensis strains.
Bacillus cereus repeats (bcr)
During analysis of B. cereus genomes, small repeated DNA elements were characterised and named bcr1, for B. cereus repeat 1 (Økstad et al.1999). This element exhibits features of mobile DNA elements and is present in all strains of the B. cereus s.l. group but not in Bacillus strains outside of this group (Økstad et al.2004). Beside bcr1, bcr2–18 elements were identified and characterised by Tourasse et al. (2006). These elements, although somewhat similar in sequence, differ in their genomic distribution. Their mobility is associated with ‘TTTAT’ sequences that could be duplicated upon insertion. This sequence may be implicated in a site-specific recombination-like mechanism, which allows bcr integration, yet to be investigated (Klevan et al.2007). Bcr1–18 are organised in secondary structure, and divided into three groups (bcr1–3, bcr4–6 and bcr7–18). Each group may be implicated in important genome evolution, either through modulating gene expression levels or through involvement in genomic rearrangements and acquisition of new genes (Kristoffersen et al.2011).
Figure 10 shows the distribution of all the B. cereus repeats in the complete genomes of B. thuringiensis sv. israelensis HD-789, AM65–52 and HD 1002 strains. The numbers of complete and partial B. cereus repeats in these genomes are shown in Table 5. Bcr1 is the most abundant repeat, with around 12 complete repeats per genome, followed by bcr3 (up to 8 complete copies). The other repeats are mostly present as one or two copies, which raises the question of their functionality. bcr7 and bcr18 are absent from all analysed genomes.
Figure 10.
Distribution of all B. cereus repeats in the complete genomes of B. thuringiensis sv. israelensis HD-789, AM65–52 and HD 1002 strains. Note that the number of complete and partial B. cereus repeats in these genomes is shown in Table 5.
Table 5.
Distribution of the bcr copies among the three fully sequenced B. thuringiensis sv. israelensis genomes.
| HD-789 | AM65–52 | HD 1002 | ||||
|---|---|---|---|---|---|---|
| Repeats | Complete copies | Partial copies | Complete copies | Partial copies | Complete copies | Partial copies |
| bcr1 | 12 | 70 | 12 | 71 | 12 | 71 |
| bcr2 | 2 | 23 | 2 | 23 | 2 | 22 |
| bcr3 | 6 | 14 | 7 | 14 | 8 | 14 |
| bcr4 | 0 | 0 | 5 | 0 | 6 | 0 |
| bcr5 | 2 | 0 | 2 | 0 | 2 | 0 |
| bcr6 | 2 | 0 | 2 | 0 | 2 | 0 |
| bcr7 | 0 | 0 | 0 | 0 | 0 | 0 |
| bcr8 | 1 | 0 | 1 | 0 | 1 | 0 |
| bcr9 | 1 | 1 | 1 | 1 | 1 | 1 |
| bcr10 | 1 | 1 | 1 | 1 | 1 | 1 |
| bcr11 | 1 | 0 | 1 | 0 | 1 | 0 |
| bcr12 | 1 | 0 | 2 | 0 | 2 | 0 |
| bcr13 | 2 | 0 | 2 | 0 | 2 | 0 |
| bcr14 | 1 | 2 | 1 | 2 | 1 | 2 |
| bcr15 | 1 | 1 | 1 | 1 | 1 | 1 |
| bcr16 | 1 | 1 | 1 | 1 | 1 | 1 |
| bcr17 | 1 | 1 | 1 | 1 | 1 | 1 |
| bcr18 | 0 | 0 | 0 | 0 | 0 | 0 |
Group II introns
Group I and II introns, first discovered more than 20 years ago, are self-splicing catalytic RNAs. These elements are considered mobile elements since they encode proteins that allow their reverse transcription and mobility from one location to another in the genome (Lehmann and Schmidt 2003; Tourasse and Kolstø 2008). Introns can be mobilised either by intron homing, by retro-homing (i.e. inserting into homologous intronless DNA sites) or by transposition (i.e. inserting into novel genomic locations) (Tourasse and Kolstø 2008). Both groups can be found in bacterial genomes; however, the main differences reside in their size, secondary structure, location and mobility mechanism (Ko, Choi and Park 2002; Robart and Zimmerly 2005). Group II introns are genetic retro-elements and are often found in non-coding regions or even in mobile DNA elements such as plasmids, Tn, IS or pathogenicity islands (Dai 2002; Klein and Dunny 2002; Robart and Zimmerly 2005). Their size can vary between 0.7 and 3 kb, and they encode mainly a target-primed reverse transcriptase (RT), a maturase and an endonuclease (Dai et al.2003; Lambowitz and Zimmerly 2004).
These elements are widespread in all bacterial genomes, including those of the B. cereus group. B.th.I1 and B.th.I2 are group II introns located on pAW63, a conjugative plasmid from B. thuringiensis sv. kurstaki (Van der Auwera and Mahillon 2008). These elements are active and are located within conjugation genes as well as a regulatory control circuit. B.th.I3 was however found in B. thuringiensis sv. israelensis strain ATCC 35646, within a gene encoding a RT, and the 5’ intron binding site (IBS) is also present in the RT active site (Tourasse and Kolstø 2008). Detailed analysis of B. thuringiensis sv. israelensis genomes revealed one full-length B.th.I3 element in each strain. In all cases, this element was inserted within a gene coding a RT, with the 5’ IBS being the RT active site.
Integrons
Integrons are DNA assembly platforms capable of incorporating, through site-specific recombination, exogenous DNA containing CDS. These elements then ensure the expression of the CDS, turning it into a functional gene (Mazel 2006). All integrons contain three core elements necessary for the capture of exogenous CDS: an integrase coding gene, a primary recombination site (attI) composed of two integrase binding sites (L and R) and an outward oriented promoter responsible for the transcription of the integrated cassettes (Hall and Collis 1995). Another fundamental component of integron cassettes is the attC site. This sequence, although not conserved between integrons, possesses a conserved palindromic organisation, and is composed of two imperfect IRs with a variable central region (Escudero et al.2014). Integrons are classified into two groups: the very large chromosomal superintegrons, which can carry more than 20 cassettes, and the mobile integrons, physically associated to mobile DNA elements such as IS or Tn and primarily involved in the spread of antibiotic-resistance genes. These elements are divided into five classes based on their integrase coding gene (Mazel 2006; Escudero et al.2014; Deng et al.2015). To date, no study has investigated the presence of integron cassettes, mobile or superintegrons, in B. thuringiensis strains. Therefore, the complete B. thuringiensis sv. israelensis genomes (strains AM65–52, HD-789 and HD 1002) were scanned using IntegronFinder v1.5 (Cury et al.2016). Although several integrase coding genes are present in the studied genomes and were detected by BlastN search (eight complete and one partial chromosomal copies, one or two plasmidial copies), the lack of the recombination sites (attI or attC) suggested the absence of any functional integron.
Integrative and conjugative elements
Conjugative transposons or integrative and conjugative elements (ICEs) are self-transmissible MGE, ranging from ca. 20 kb to more than 500 kb in size (Wozniak and Waldor 2010; Johnson and Grossman 2015). ICEs carry the components necessary for bacterial conjugation and for their integration and excision from the host chromosome. An integrase (int) and an excisionase (xis) recognise and operate on a specific recombination site (attP on the circularised ICE and attB on the target chromosome). The T4SS is responsible for the DNA transfer from the donor to the recipient cell (Lee et al.2007; Smillie et al.2010; Auchtung et al.2016). These elements are sometimes capable of facilitating the transfer or mobilisation of other genetic elements (Menard and Grossman 2013). ICEs can also carry several passenger genes coding for antibiotic resistance or metabolic function (Wozniak and Waldor 2010). While conjugative plasmids have been extensively studied and their mechanisms and distribution unravelled, such analysis is still in the early stages for ICEs. Very few ICEs have in fact been experimentally studied in terms of conjugation and borders. However, bioinformatic analyses have allowed the finding of a large number of these elements (Wozniak et al.2009; Guglielmini et al.2011; Cury, Touchon and Rocha 2017), that have now been regrouped in a dedicated database named ICEberg (http://db-mml.sjtu.edu.cn/ICEberg/) (Bi et al.2012). ICEs were found in less than 30% of the Firmicutes genomes and none were in a B. thuringiensis genome. A BlastN of B. thuringiensis sv. israelensis complete genomes against ICE elements present in the ICEberg database gave no positive results.
Prophages
Generally, three types of temperate phages can be considered. The most common phages are the ones able to integrate into the bacterial chromosome, thus completely relying on the bacterial chromosome replication system in their lysogenic state. Those able to maintain low copy number replication mode and thus existing as plasmids without massive cell lysis (also referred to as ‘plasmidial prophages’; Gillis and Mahillon 2014b) are rarer. The third and rarest type are phages capable of integrating into self-replicative plasmids. The rarity of the last type is probably due to the inability of most plasmids to evolve in association with a prophage due to restrictions on the sizes of packaged DNA. As already indicated, almost all B. thuringiensis sv. israelensis strains contain pGIL01-related plasmids able to develop into active GIL01 phage (Gillis and Mahillon 2014c), which is maintained at about 10–15 copies in the lysogenic state (Bolotin et al.2017). It was also recently demonstrated that the plasmid pBtic235 could develop as phage infecting susceptible B. thuringiensis cells, as previously indicated (Gillis et al.2017b).
The only experimental work, where the induction of a chromosomal prophage was demonstrated, showed that a prophage called phIS3501 could form circular replicative forms (Moumen, Nguen-The and Sorokin 2012). It is interesting that this prophage is integrated into a gene encoding haemolysin II and thus able to form the potentially active toxin upon phage excision. The detection of other chromosomal prophages is possible with the relevant computer tools based either on detecting local clusters of genes coding for proteins similar to those occurring in phages (Lima-Mendez et al.2008) or less direct and more sophisticated on detecting of phage sequence related DNA properties (Akhter, Aziz and Edwards 2012). The best experimental method to predict the inducible phage lysogens in bacterial genome would probably be one of the NGS technologies applied to DNA extracted from cells treated with sublethal doses of DNA damage agents. Such treatment induces phage replication and thus leads to an increase of their DNA. However, this approach has not yet been used for the B. thuringiensis sv. israelensis strains. Since none of the available bioinformatics tools is optimal for prophage prediction, the user-friendly WEB tool PHASTER (Zhou et al.2011) was used together with a manual scrutiny of the results, using different databases.
A short summary of PHASTER results obtained for the chromosomes of completely assembled genomes of B. thuringiensis sv. israelensis AM65–52, HD-789 and HD 1002 is shown in Fig. 11. Manual scrutiny of gene functions detected in potential prophage regions indicated that only Region 2, 3, 5 and 6 of strain AM65–52 and their counterparts from the other two strains could be regarded as the ones encoding for potentially inducible prophages. Notably, Region 2 corresponds to the above mentioned prophage phIS3501. The capacity for induction of the other regions still has to be experimentally verified. Although Region 4 (around 3665 kb in AM65–52 genome) contains a potential replication protein, all genes necessary for the phage envelope formation seem to be lost.
Figure 11.
Detection of prophage-like regions in the completed genomes of B. thuringiensis sv. israelensis strains. The FASTER web tool (Zhou et al.2011) was used for prophage region detection. (A) Direct outputs of PHASTER tool representing circular chromosomes of analysed genomes. Coordinate numbering corresponds to GenBank entries (Acc. ##: CP013275 for AM65–52, NC_018508 for HD-789 and NZ_CP009351 for HD 1002). The detected regions were automatically numbered by the PHASTER tool and the colours correspond to reliability of prediction (green > blue > brown). The estimated region correspondence is indicated in the bottom right corner. Region X indicates that no corresponding prediction was provided by the tool for the given strain, question mark indicates that predicted regions cannot be unambiguously counter parted. (B) Each panel represents CDS maps of counter parted regions as provided and coloured by the PHASTER tool. Pale and dark green colours correspond to hypothetical and phage-related functions. Other colours indicate identified and annotated phage functions. The maps for strain HD 1002 are reverse complemented in relation to other two genomes in concordance with the GenBank entry, as the corresponding circular map in A. See additional explanations in the text.
To further verify the results for unfinished B. thuringiensis sv. israelensis genomes from our list (Tables 1 and 2), CDS comparisons using Prokka (Seemann 2014) genome annotation and Roary (Page et al.2015) pan-genome construction tools were applied. Interestingly, although this analysis confirmed the presence of Regions 2 and 6 in all strains containing the pXO16 conjugative plasmid, it showed their absence in the three strains that are lacking pXO16: B. thuringiensis 147 and Br58 and sv. israelensis 4Q7 (not shown). The latter was artificially cured from its plasmids and presumably has also lost some of its prophages. The 147 and Br58 strains are the natural isolates (Barbosa et al.2015; Zorzetti et al.2015) containing all other B. thuringiensis sv. israelensis-specific plasmids, except for pXO16, as previously indicated (Table 2). Region 3 is only absent in the 4Q7 strain (not shown).
Putative prophage Region 3, the most complete after phIS3501 (Region 2), interrupts the formate dehydrogenase accessory protein FdhD gene (at 3507 kb in AM65–52 genome, Acc. # AND25365). It is not clear if the prophage Region 5 can be induced. It is integrated into one of dipicolinate synthase subunits (3828 kb in AM65–52, Acc. #: AND25687). The induction ability is also unclear for Region 6, integrated close to the superoxide dismutase gene (at 4321 kb in AM65–52 genome, Acc. # AND26206). These two insertion elements, Region 5 and 6, rather represent remnant prophages than the inducible ones, although it is noteworthy that Region 6 is lost in the 147, Br58 and 4Q7 strains, all missing pXO16.
CONCLUDING REMARKS
In 1976, several spore-forming bacteria were isolated from soil samples of known mosquito larval breeding sites in Israel. One bacterium however demonstrated rapid larvicidal activity against dipteran larvae from various species. This bacterium was identified as B. thuringiensis and later on serotyped as H-14 and given the name ‘israelensis’. It distinguished itself from the other B. thuringiensis serotypes by the presence of Cyt toxins that work synergistically with the classic Cry δ-endotoxins. Some 40 years later, bioinformatic approaches allowed us to define a cluster of 12 B. thuringiensis sv. israelensis strains among the several hundred genomes of B. thuringiensis strains registered in the database.
A multitude of B. thuringiensis plasmids, variable in number and size, can form up to 13% of the whole genome. In this review, plasmids usually present in B. thuringiensis sv. israelensis genomes were extensively analysed and detailed. The nine described plasmids are: pTX14–1, 14–2 and 14–3, pGIL01, pBtoxis, pBtic100, pBtic235, pXO16 and pBtic360. Each plasmid is distinguished by its role in horizontal DNA transfer and/or by the genes it carries, whose function and role mostly remain cryptic. Although these plasmids are key players in gene exchange, allowing bacteria to adapt to their host and their environment, intracellular mobile genetic elements also play a vital role in genome dynamics and gene transfer. In this review, we highlighted the presence of different types of MGE in available B. thuringiensis sv. israelensis genomes. A variety of IS, class II transposons, MITEs and TIMEs, group II introns, bcr, as well as prophages were found in both chromosomes and plasmids of the analysed strains. However, mobile insertion cassettes, integrons and integrative and conjugative elements were not detected using our approach. Because most entomotoxin genes are located within or adjacent to mobile entities (Fiedoruk et al. 2017) and because B. thuringiensis sv. israelensis strains carry numerous extrachromosomal molecules, they will always be able to adapt and survive by overcoming environmental changes and difficulties.
After listing the members of the extrachromosomal zoo of B. thuringiensis sv. israelensis, and given its high importance as bioinsecticide, a one by one study of these elements should take place in order to decipher their true impact on the anti-diptera activity of B. thuringiensis sv. israelensis. Diptera, especially mosquitoes, are insects that cause a number of problems worldwide as vectors of important human diseases. Therefore, the world needs an ecofriendly biocontrol method, as efficient as synthetic insecticides, but much safer and less probe to insect resistance. Better understanding the biology and life cycle of B. thuringiensis sv. israelensis will ensure the appropriate and sustainable use of this effective bioinsecticide.
However, the possibility of moving genes among plasmids or between plasmids and chromosome, as well as plasmid mobilisation from one strain to another of the same or of different species, shows the power of genome dynamics in the adaptation, evolution and survival of bacteria.
Acknowledgements
Lars Andrup and Gert Jensen for their extensive characterisation of B. thuringiensis sv. israelensis plasmid bonanza and for their seminal description of the conjugative behaviour of pXO16.
FUNDING
This study was funded by the FNRS (National Fund for Scientific Research) through grants to A. Gillis and J. Mahillon, the Université catholique de Louvain, the Research Department of the Communauté française de Belgique (Concerted Research Action, ARC12–17/046) through a grant to L. Makart, the National Council for Scientific Research in Lebanon (CNRS-L) through a grant to N. Fayad and the PCSI of the AUF-BMO for researcher mobility.
Conflict of interest. None declared.
REFERENCES
- Abdoarrahem MM, Gammon K, Dancer BN et al. Genetic basis for alkaline activation of germination in Bacillus thuringiensis subsp. israelensis. Appl Environ Microb 2009;75:6410–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriaenssens EM, Wittmann J, Kuhn JH et al. Taxonomy of prokaryotic viruses: 2017 update from the ICTV Bacterial and archaeal viruses subcommittee. Arch Virol 2018;163:1125–9. [DOI] [PubMed] [Google Scholar]
- Ackermann HW, Roy R, Martin M et al. Partial characterization of a cubic Bacillus phage. Can J Microbiol 1978;24:986–93. [DOI] [PubMed] [Google Scholar]
- Ahn C, Collins-Thompson D, Duncan C et al. Mobilization and location of the genetic determinant of chloramphenicol resistance from Lactobacillus plantarum caTC2R. Plasmid 1992;27:169–76. [DOI] [PubMed] [Google Scholar]
- Akhter S, Aziz RK, Edwards RA. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res 2012;40:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadio AF, Benintende GB, Zandomeni RO. Complete sequence of three plasmids from Bacillus thuringiensis INTA-FR7-4 environmental isolate and comparison with related plasmids from the Bacillus cereus group. Plasmid 2009;62:172–82. [DOI] [PubMed] [Google Scholar]
- Anderson I, Sorokin A, Kapatral V et al. Comparative genome analysis of Bacillus cereus group genomes with Bacillus subtilis. FEMS Microbiol Lett 2005;250:175–84. [DOI] [PubMed] [Google Scholar]
- Andrup L, Bendixen HH, Jensen GB. Mobilization of Bacillus thuringiensis plasmid pTX14-3. Plasmid 1995;33:159–67. [DOI] [PubMed] [Google Scholar]
- Andrup L, Damgaard J, Wassermann K. Mobilization of small plasmids in Bacillus thuringiensis subsp. israelensis is accompanied by specific aggregation. J Bacteriol 1993;175:6530–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrup L, Damgaard J, Wassermann K et al. Complete nucleotide sequence of the Bacillus thuringiensis subsp. israelensis plasmid pTX14-3 and its correlation with biological properties. Plasmid 1994;31:72–88. [DOI] [PubMed] [Google Scholar]
- Andrup L, Jensen GB, Wilcks A et al. The patchwork nature of rolling-circle plasmids: comparison of six plasmids from two distinct Bacillus thuringiensis serotypes. Plasmid 2003;49:205–32. [DOI] [PubMed] [Google Scholar]
- Andrup L, Jørgensen O, Wilcks A et al. Mobilization of “nonmobilizable” plasmids by the aggregation-mediated conjugation system of Bacillus thuringiensis. Plasmid 1996;36:75–85. [DOI] [PubMed] [Google Scholar]
- Andrup L, Smidt L, Andersen K et al. Kinetics of conjugative transfer: a study of the plasmid pXO16 from Bacillus thuringiensis subsp. israelensis. Plasmid 1998;40:30–43. [DOI] [PubMed] [Google Scholar]
- Ankarloo J, Caugant DA, Hansen BM et al. Genome stability of Bacillus thuringiensis subsp. israelensis isolates. Curr Microbiol 2000;40:51–6. [DOI] [PubMed] [Google Scholar]
- Aponte JJ, Aide P, Renom M et al. Is malaria eradication possible? Lancet North Am Ed 2007;370:1459. [Google Scholar]
- Auchtung JM, Aleksanyan N, Bulku A et al. Biology of ICE Bs1, an integrative and conjugative element in Bacillus subtilis. Plasmid 2016;86:14–25. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 2008;9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res 2010;38:4207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabas O, Ronning DR, Guynet C et al. Mechanism of IS200/IS605 family DNA transposases: activation and transposon-directed target site selection. Cell 2008;132:208–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa LC, Farias DL, Silva Ide M et al. Draft genome sequence of Bacillus thuringiensis 147, a Brazilian strain with high insecticidal activity. Mem Inst Oswaldo Cruz 2015;110:822–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum JA. Tn5401, a new class II transposable element from Bacillus thuringiensis. J Bacteriol 1994;176:2835–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazinet AL. Pan-genome and phylogeny of Bacillus cereus sensu lato. BMC Evol Biol 2017;17:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Dov E. Bacillus thuringiensis subsp. israelensis and its dipteran-specific toxins. Toxins 2014;6:1222–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Dov E, Nissan G, Pelleg N et al. Refined, circular restriction map of the Bacillus thuringiensis subsp. israelensis plasmid carrying the mosquito larvicidal genes. Plasmid 1999;42:186–91. [DOI] [PubMed] [Google Scholar]
- Benson SD, Bamford JK, Bamford DH et al. Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell 2004;5:673–85. [DOI] [PubMed] [Google Scholar]
- Berjón-Otero M, Lechuga A, Mehla J et al. Bam35 tectivirus intraviral interaction map unveils new function and localization of phage orfan proteins. J Virol 2017;91:e00870–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berjón-Otero M, Villar L, de Vega M et al. DNA polymerase from temperate phage Bam35 is endowed with processive polymerization and abasic sites translesion synthesis capacity. Proc Natl Acad Sci USA 2015;112:E3476–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berjón-Otero M, Villar L, Salas M et al. Disclosing early steps of protein-primed genome replication of the Gram-positive tectivirus Bam35. Nucleic Acids Res 2016;44:9733–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard K, Schrempf H, Goebel W. Bacteriocin and antibiotic resistance plasmids in Bacillus cereus and Bacillus subtilis. J Bacteriol 1978;133:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C. The bacterium, Lysinibacillus sphaericus, as an insect pathogen. J Invertebr Pathol 2012;109:1–10. [DOI] [PubMed] [Google Scholar]
- Berry C, Crickmore N. Structural classification of insecticidal proteins—towards an in silico characterization of novel toxins. J Invertebr Pathol 2017;142:16–22. [DOI] [PubMed] [Google Scholar]
- Berry C, O’Neil S, Ben-Dov E et al. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl Environ Microb 2002;68:5082–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi D, Xu Z, Harrison EM et al. ICEberg: A web-based resource for integrative and conjugative elements found in bacteria. Nucleic Acids Res 2012;40:D621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Gillis A, Sanchis V et al. Comparative genomics of extrachromosomal elements in Bacillus thuringiensis subsp. israelensis. Res Microbiol 2017;168:331–44. [DOI] [PubMed] [Google Scholar]
- Bonin A, Paris M, Frérot H et al. The genetic architecture of a complex trait: resistance to multiple toxins produced by Bacillus thuringiensis israelensis in the dengue and yellow fever vector, the mosquito Aedes aegypti. Infect Genet Evol 2015;35:204–13. [DOI] [PubMed] [Google Scholar]
- Boocock MR, Rice PA. A proposed mechanism for IS607-family serine transposases. Mobile DNA 2013;4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar SK, Verma M, Tyagi RD et al. Recent advances in downstream processing and formulations of Bacillus thuringiensis based biopesticides. Process Biochem 2006;41:323–42. [Google Scholar]
- Bravo A, Gill S, Soberon M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 2007;49:423–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butko P. Cytolytic toxin Cyt1A and its mechanism of membrane damage: data and hypotheses. Appl Environ Microb 2003;69:2415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V et al. BLAST+: architecture and applications. BMC Bioinformatics 2009;10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanha ER, Swiger RR, Senior B et al. Strain discrimination among B. anthracis and related organisms by characterization of bclA polymorphisms using PCR coupled with agarose gel or microchannel fluidics electrophoresis. J Microbiol Methods 2006;64:27–45. [DOI] [PubMed] [Google Scholar]
- Chattoraj DK. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol Microbiol 2002;37:467–76. [DOI] [PubMed] [Google Scholar]
- Chen Y, Braathen P, Léonard C et al. MIC231, a naturally occurring mobile insertion cassette from Bacillus cereus. Mol Microbiol 1999;32:657–68. [DOI] [PubMed] [Google Scholar]
- Clark BD, Boyle TM, Chu CY et al. Restriction endonuclease mapping of three plasmids from Bacillus thuringiensis var. israelensis. Gene 1985;36:169–71. [DOI] [PubMed] [Google Scholar]
- Clewell DB, Weaver KE. Sex pheromones and plasmid transfer in Enterococcus faecalis. Plasmid 1989;21:175–84. [DOI] [PubMed] [Google Scholar]
- Cohen S, Albeck S, Ben-Dov E et al. Cyt1Aa toxin: crystal structure reveals implications for its membrane-perforating function. J Mol Biol 2011;413:804–14. [DOI] [PubMed] [Google Scholar]
- Cohen S, Dym O, Albeck S et al. High-Resolution crystal structure of activated Cyt2Ba monomer from Bacillus thuringiensis subsp. israelensis. J Mol Biol 2008;380:820–7. [DOI] [PubMed] [Google Scholar]
- Craig NL, Gragie R, Gellert M et al. Mobile DNA II. Washington, DC: ASM Press, 2002. [Google Scholar]
- Cury J, Jove T, Touchon M et al. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res 2016;44:4539–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cury J, Touchon M, Rocha EPC. Integrative and conjugative elements and their hosts: composition, distribution and organization. Nucleic Acids Res 2017;45:8943–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L. Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res 2002;30:1091–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Toor N, Olson R et al. Database for mobile group II introns. Nucleic Acids Res 2003;31:424–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugelavičius R, Gaidelytė A, Cvirkaitė-Krupovič V et al. On-line monitoring of changes in host cell physiology during the one-step growth cycle of Bacillus phage Bam35. J Microbiol Methods 2007;69:174–9. [DOI] [PubMed] [Google Scholar]
- de Barjac H. A new variety of Bacillus thuringiensis very toxic to mosquitoes: B. thuringiensis var. israelensis serotype 14. C R Acad Sci Hebd Séances Acad Sci D 1978;286:797–800. [PubMed] [Google Scholar]
- de Barjac H, Frachon E. Classification of Bacillus thuringiensis strains. Entomophaga 1990;35:233–40. [Google Scholar]
- Delécluse A, Bourgouin C, Klier A et al. Nucleotide sequence and characterization of a new insertion element, IS240, from Bacillus thuringiensis israelensis. Plasmid 1989;21:71–8. [DOI] [PubMed] [Google Scholar]
- de Maagd RA, Bravo A, Berry C et al. Structure, diversity, and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu Rev Genet 2003;37:409–33. [DOI] [PubMed] [Google Scholar]
- Deng Y, Bao X, Ji L et al. Resistance integrons: class 1, 2 and 3 integrons. Ann Clin Microb Anti 2015;14:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palmenaer D, Siguier P, Mahillon J. IS4 family goes genomic. BMC Evol Biol 2008;8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palmenaer D, Vermeiren C, Mahillon J. IS231-MIC231 elements from Bacillus cereus sensu lato are modular. Mol Microbiol 2004;53:457–67. [DOI] [PubMed] [Google Scholar]
- Després L, Stalinski R, Tetreau G et al. Gene expression patterns and sequence polymorphisms associated with mosquito resistance to Bacillus thuringiensis israelensis toxins. BMC Genomics 2014;15:926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Mendoza M, Bideshi DK, Federici BA. A 54-kilodalton protein encoded by pBtoxis is required for parasporal body structural integrity in Bacillus thuringiensis subsp. israelensis. J Bacteriol 2012;194:1562–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Franco C, Santini T, Pisaneschi G et al. Insights into the genetic organization of the Bacillus mycoides cryptic plasmids pDx14.2 and pSin9.7 deduced from their complete nucleotide sequence. Plasmid 2005;54:288–93. [DOI] [PubMed] [Google Scholar]
- Doggett NA, Stubben CJ, Chertkov O et al. Complete Genome Sequence of Bacillus thuringiensis serovar israelensis strain HD-789. Genome Announc 2013;1:e01023–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval-Valentin G, Marty-Cointin B, Chandler M. Requirement of IS911 replication before integration defines a new bacterial transposition pathway. EMBO J 2004;23:3897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich SD. Replication and expression of plasmids from Staphylococcus aureus in Bacillus subtilis. Proc Natl Acad Sci USA 1977;74:1680–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleuch J, Jaoua S, Darriet F et al. Cry4Ba and Cyt1Aa proteins from Bacillus thuringiensis israelensis: Interactions and toxicity mechanism against Aedes aegypti. Toxicon 2015;104:83–90. [DOI] [PubMed] [Google Scholar]
- El Khoury M, Azzouz H, Chavanieu A et al. Isolation and characterization of a new Bacillus thuringiensis strain Lip harboring a new cry1Aa gene highly toxic to Ephestia kuehniella (Lepidoptera: Pyralidae) larvae. Arch Microbiol 2014;196:435–44. [DOI] [PubMed] [Google Scholar]
- Escudero JA, Loot C, Nivina A et al. The integron: adaptation on demand. Microbiol Spectr 2014;3:MDNA3–0019–2014. [DOI] [PubMed] [Google Scholar]
- Fattash I, Rooke R, Wong A et al. Miniature inverted-repeat transposable elements: discovery, distribution, and activity. Genome 2013;56:475–86. [DOI] [PubMed] [Google Scholar]
- Faust RM, Abe K, Held GA et al. Evidence for plasmid-associated crystal toxin production in Bacillus thuringiensis subsp. israelensis. Plasmid 1983;9:98–103. [DOI] [PubMed] [Google Scholar]
- Fayet O, Prère M-F. Programmed Ribosomal −1 Frameshifting as a tradition: the bacterial transposable elements of the IS3 Family. In: Atkins J, Gesteland R (eds.). Recoding: Expansion of Decoding Rules Enriches Gene Expression: Nucleic Acids and Molecular Biology, Vol 24, Springer, New York, NY, 2010, 259–80. [Google Scholar]
- Fico S, Mahillon J. TasA - tasB, a new putative toxin-antitoxin (TA) system from Bacillus thuringiensis pGI1 plasmid is a widely distributed composite mazE-doc TA system. BMC Genomics 2006;7:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedoruk K, Daniluk T, Mahillon J et al. Genetic environment of cry1 genes indicates their common origin. Genome Biol Evol 2017;9:2265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornelos N, Bamford JKH, Mahillon J. Phage-borne factors and host LexA regulate the lytic switch in phage GIL01. J Bacteriol 2011;193:6008–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornelos N, Browning DF, Pavlin A et al. Lytic gene expression in the temperate bacteriophage GIL01 is activated by a phage-encoded LexA homologue. Nucleic Acids Res 2018; DOI: 10.1093/nar/gky646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornelos N, Butala M, Hodnik V et al. Bacteriophage GIL01 gp7 interacts with host LexA repressor to enhance DNA binding and inhibit RecA-mediated auto-cleavage. Nucleic Acids Res 2015;43:7315–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S. A PRD1 by another name? Structure 2005;13:1738–40. [DOI] [PubMed] [Google Scholar]
- Gaidelytė A, Cvirkaitė-Krupovic V, Daugelavicius R et al. The entry mechanism of membrane-containing phage Bam35 infecting Bacillus thuringiensis. J Bacteriol 2006;188:5925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammon K, Jones GW, Hope SJ et al. Conjugal transfer of a toxin-coding megaplasmid from Bacillus thuringiensis subsp. israelensis to mosquitocidal strains of Bacillus sphaericus. Appl Environ Microb 2006;72:1766–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis A, Bin Jang H, Mahillon J et al. Taxonomic Proposal 2017.013B.A.v2. To rename the existing genus Tectivirus (proposed new name Alphatectivirus) in the family Tectiviridae, create a new genus, Betatectivirus, and include three (3) new species. International Committee on Taxonomy of Viruses (ICTV) 2017a. https://talk.ictvonline.org/files/ictv_official_taxonomy_updates_since_the_8th_report/m/prokaryote-official/7109 (assessed on March 17, 2018).
- Gillis A, Guo S, Bolotin A et al. Detection of the cryptic prophage-like molecule pBtic235 in Bacillus thuringiensis subsp. israelensis. Res Microbiol 2017b;168:319–30. [DOI] [PubMed] [Google Scholar]
- Gillis A, Mahillon J. Influence of lysogeny of tectiviruses GIL01 and GIL16 on Bacillus thuringiensis growth, biofilm formation, and swarming motility. Appl Environ Microb 2014a;80:7620–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis A, Mahillon J. Phages preying on Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: past, present and future. Viruses 2014b;6:2623–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis A, Mahillon J. Prevalence, genetic diversity, and host range of tectiviruses among members of the bacillus cereus group. Appl Environ Microb 2014c;80:4138–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg LJ, Margalit J. A bacterial spore demonstrating rapid larvicidal activity against Anopheles sergentii, Uranotaenia unguiculata, Culex univitattus, Aedes aegypti and Culex pipiens. Mosq News 1977;37:355–8. [Google Scholar]
- González JM Jr, Carlton BC. Patterns of plasmid DNA in crystalliferous and acrystalliferous strains of Bacillus thuringiensis. Plasmid 1980;3:92–8. [DOI] [PubMed] [Google Scholar]
- González JM Jr, Carlton BC. A large transmissible plasmid is required for crystal toxin production in Bacillus thuringiensis variety israelensis. Plasmid 1984;11:28–38. [DOI] [PubMed] [Google Scholar]
- Grant JR, Stothard P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res 2008;36:W181–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. The movement of Tn3-like elements: transposition and cointegrate resolution. In: Craig NL, Gragie R, Gellert M et al. (eds). Mobile DNA II. Washington, DC: ASM Press, 2002, 272–302. [Google Scholar]
- Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol R 2003;67:277–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ. The changing epidemiology of yellow fever and dengue, 1900 to 2003: full circle? Comp Immunol Microb 2004;27:319–30. [DOI] [PubMed] [Google Scholar]
- Guerchicoff A, Delécluse A, Rubinstein CP. The Bacillus thuringiensis cyt genes for hemolytic endotoxins constitute a gene family. Appl Environ Microb 2001;67:1090–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmini J, Quintais L, Garcillán-Barcia MP et al. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 2011;8:e1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol 1995;15:593–600. [DOI] [PubMed] [Google Scholar]
- Hallet B, Rezsohazy R, Delcour J. IS231A from Bacillus thuringiensis is functional in Escherichia coli: transposition and insertion specificity. J Bacteriol 1991;173:4526–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallet B, Rezsöhazy R, Mahillon J et al. IS231A insertion specificity: consensus sequence and DNA bending at the target site. Mol Microbiol 1994;14:131–9. [DOI] [PubMed] [Google Scholar]
- Hardies SC, Thomas JA, Serwer P. Comparative genomics of Bacillus thuringiensis phage 0305?8-36: defining patterns of descent in a novel ancient phage lineage. Virol J 2007;4:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Corneloup A, Guynet C et al. The IS200/IS605 family and ‘Peel and Paste’ single-strand transposition mechanism. Microbiol Spectr 2015;3:1–21. [DOI] [PubMed] [Google Scholar]
- Höfte H, Whiteley HR. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev 1989;53:242–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollensteiner J, Poehlein A, Spröer C et al. Complete genome sequence of the nematicidal Bacillus thuringiensis MYBT18246. Stand Genomic Sci 2017a;12:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollensteiner J, Poehlein A, Spröer C et al. Complete genome sequence of the nematicidal Bacillus thuringiensis MYBT18247. J Biotechnol 2017b;260:48–52. [DOI] [PubMed] [Google Scholar]
- Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol 1982;150:804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalasvuori M, Palmu S, Gillis A et al. Identification of five novel tectiviruses in Bacillus strains: analysis of a highly variable region generating genetic diversity. Res Microbiol 2013;164:118–26. [DOI] [PubMed] [Google Scholar]
- Jensen GB, Andrup L, Wilcks A et al. The aggregation-mediated conjugation system of Bacillus thuringiensis subsp. israelensis: host range and kinetics of transfer. Curr Microbiol 1996;33:228–36. [DOI] [PubMed] [Google Scholar]
- Jensen GB, Wilcks A, Petersen SS et al. The genetic basis of the aggregation system in Bacillus thuringiensis subsp. israelensis is located on the large conjugative plasmid pXO16. J Bacteriol 1995;177:2914–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Park SH, Choi SK. Genome sequence of the acrystalliferous Bacillus thuringiensis serovar israelensis strain 4Q7, widely used as a recombination host. Genome Announc 2014;2:e00231–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez G, Blanch AR, Tamames J et al. Complete genome sequence of Bacillus toyonensis BCT-7112T, the active ingredient of the feed additive preparation Toyocerin. Genome Announc 2013;1:e01080–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Grossman AD. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet 2015;49:577–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Daligault HE, Davenport KW. Complete genome sequences for 35 biothreat assay-relevant Bacillus species. Genome Announc 2015;3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda K, Ohderaotoshi T, Shimojyo A et al. An extrachromosomal prophage naturally associated with Bacillus thuringiensis serovar israelensis. Lett Appl Microbiol 1999;28:305–8. [Google Scholar]
- Kearse M, Moir R, Wilson A et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012;28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P, Price LB, Klevytska AM et al. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J Bacteriol 2000;182:2928–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J, Dunny G. Bacterial group II introns and their association with mobile genetic elements. Front Biosci 2002;7:1843–56. [DOI] [PubMed] [Google Scholar]
- Klevan A, Tourasse NJ, Stabell FB et al. Exploring the evolution of the Bacillus cereus group repeat element bcr1 by comparative genome analysis of closely related strains. Microbiology 2007;153:3894–908. [DOI] [PubMed] [Google Scholar]
- Ko M, Choi H, Park C. Group I self-splicing intron in the recA gene of Bacillus anthracis. J Bacteriol 2002;184:3917–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristoffersen SM, Tourasse NJ, Kolstø AB et al. Interspersed DNA repeats bcr1-bcr18 of Bacillus cereus group bacteria form three distinct groups with different evolutionary and functional patterns. Mol Biol Evol 2011;28:963–83. [DOI] [PubMed] [Google Scholar]
- Kuroda A, Asami Y, Sekiguchi J. Molecular cloning of a sporulation-specific cell wall hydrolase gene of Bacillus subtilis. J Bacteriol 1993;175:6260–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey LA. Bacillus thuringiensis serovariety israelensis and Bacillus sphaericus for mosquito control. J Am Mosq Control Assoc 2007;23:133–63. [DOI] [PubMed] [Google Scholar]
- Lacey LA, Goettel M. Current developments in microbial control of insect pests and prospects for the early 21st century. Entomophaga 1995;40:3–27. [Google Scholar]
- Lacey RW, Lewis E, Rosdahl VT. Evolution of plasmids in vivo in a strain of Staphylococcus aureus. J Med Microbiol 1974;7:117–25. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. Mobile Group II Introns. Annu Rev Genet 2004;38:1–35. [DOI] [PubMed] [Google Scholar]
- Lawley TD, Burland V, Taylor DE. Analysis of the complete nucleotide sequence of the tetracycline-resistance transposon Tn10. Plasmid 2000;43:235–9. [DOI] [PubMed] [Google Scholar]
- Lee CA, Auchtung JM, Monson RE et al. Identification and characterization of int (integrase), xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICEBs1 of Bacillus subtilis. Mol Microbiol 2007;66:1356–69. [DOI] [PubMed] [Google Scholar]
- Lehmann K, Schmidt U. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol 2003;38:249–303. [DOI] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE. Regulating DnaA complex assembly: it is time to fill the gaps. Curr Opin Microbiol 2010;13:766–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léonard C, Chen Y, Mahillon J. Diversity and differential distribution of IS231, IS232 and IS240 among Bacillus cereus, Bacillus thuringiensis and Bacillus mycoides. Microbiology 1997;143:2537–47. [DOI] [PubMed] [Google Scholar]
- Léonard C, Mahillon J. IS231A transposition: conservative versus replicative pathway. Res Microbiol 1998;149:549–55. [DOI] [PubMed] [Google Scholar]
- Lereclus D, Menou G, Lecadet MM. Isolation of a DNA sequence related to several plasmids from Bacillus thuringiensis after a mating involving the Streptococcus faecalis plasmid pAMBeta1. Mol Gen Genet 1983;191:307–13. [DOI] [PubMed] [Google Scholar]
- Lilly J, Camps M. Mechanisms of theta plasmid replication. Microbiol Spectr 2015;3:PLAS-0029–2014. [DOI] [PubMed] [Google Scholar]
- Lima-Mendez G, Van Helden J, Toussaint A et al. Prophinder: A computational tool for prophage prediction in prokaryotic genomes. Bioinformatics 2008;24:863–5. [DOI] [PubMed] [Google Scholar]
- Liu X, Ruan L, Peng D et al. Thuringiensin: A thermostable secondary metabolite from Bacillus thuringiensis with insecticidal activity against a wide range of insects. Toxins 2014;6:2229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Galinski EA. Identification of plasmids in the genus Marinococcus and complete nucleotide sequence of plasmid pPL1 from Marinococcus halophilus. Plasmid 1997;38:107–14. [DOI] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Kunkel TA. DNA polymerases divide the labor of genome replication. Trends Cell Biol 2016;26:640–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell GE, Wood H, Devine KM et al. Genetic control of bacterial suicide: regulation of the induction of PBSX in Bacillus subtilis. J Bacteriol 1994;176:5820–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: The spread and resurgence of Japanese Encephalitis, West Nile and Dengue viruses. Nat Med 2004;10:S98–S109. [DOI] [PubMed] [Google Scholar]
- Madsen SM, Andrup L, Boe L. Fine mapping and DNA sequence of replication functions of Bacillus thuringiensis plasmid pTX14-3. Plasmid 1993;30:119–30. [DOI] [PubMed] [Google Scholar]
- Mahillon J, Rezsöhazy R, Hallet B et al. IS231 and other Bacillus thuringiensis transposable elements: a review. Genetica 1994;93:13–26. [DOI] [PubMed] [Google Scholar]
- Mahillon J. Insertion Sequence elements and transposons in Bacillus. In: Berkeley R, Heyndrickx M, Logan N et al. (eds). Applications and Systematics of Bacillus and Relatives. London: Blackwell Science, 2002:236–53. [Google Scholar]
- Mahillon J, Chandler M. Insertion Sequences. Microbiol Mol Biol R 1998;62:725–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J, Lereclus D. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J 1988;7:1515–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J, Seurinck J, Delcour J et al. Cloning and nucleotide sequence of different iso-IS231 elements and their structural association with the Tn4430 transposon in Bacillus thuringiensis. Gene 1987;51:187–96. [DOI] [PubMed] [Google Scholar]
- Mahillon J, Seurinck J, Van Rompuy L et al. Nucleotide sequence and structural organization of an insertion sequence element (IS231) from Bacillus thuringiensis strain berliner 1715. EMBO J 1985;4:3895–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makart L, Commans F, Gillis A et al. Horizontal transfer of chromosomal markers mediated by the large conjugative plasmid pXO16 from Bacillus thuringiensis serovar israelensis. Plasmid 2017;91:76–81. [DOI] [PubMed] [Google Scholar]
- Makart L, Gillis A, Hinnekens P et al. A novel T4SS-mediated DNA transfer used by pXO16, a conjugative plasmid from Bacillus thuringiensis serovar israelensis. Environ Microbiol 2018;20:1550–61. [DOI] [PubMed] [Google Scholar]
- Makart L, Gillis A, Mahillon J. pXO16 from Bacillus thuringiensis serovar israelensis: Almost 350 kb of terra incognita. Plasmid 2015;80:8–15. [DOI] [PubMed] [Google Scholar]
- Malvar T, Baum JA. Tn5401 disruption of the spo0F gene, identified by direct chromosomal sequencing, results in CryIIIA overproduction in Bacillus thuringiensis. J Bacteriol 1994;176:4750–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manasherob R, Zaritsky A, Ben-Dov E et al. Effect of accessory proteins P19 and P20 on cytolytic activity of Cyt1Aa from Bacillus thuringiensis subsp. israelensis in Escherichia coli. Curr Microbiol 2001;43:355–64. [DOI] [PubMed] [Google Scholar]
- Margalit J, Dean D. The story of Bacillus thuringiensis var. israelensis (B.t.i.). J Am Mosq Control Assoc 1985;1:1–7. [PubMed] [Google Scholar]
- Margalith Y, Ben-Dov YM. Biological control by Bacillus thuringiensis subsp. israelensis. Insect Pest Manag Tech Environ Prot 2000;200:243–301. [Google Scholar]
- Mazel D. Integrons: Agents of bacterial evolution. Nat Rev Microbiol 2006;4:608–20. [DOI] [PubMed] [Google Scholar]
- Meijer WJ, Wisman GA, Terpstra P et al. Rolling-circle plasmids from Bacillus subtilis: complete nucleotide sequences and analyses of genes of pTA1015, pTA1040, pTA1050 and pTA1060, and comparisons with related plasmids from Gram-positive bacteria. FEMS Microbiol Rev 1998;21:337–68. [DOI] [PubMed] [Google Scholar]
- Menard KL, Grossman AD. Selective pressures to maintain attachment site specificity of integrative and conjugative elements. PLoS Genet 2013;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnerat R, Pereira E, Teles B et al. Synergistic activity of Bacillus thuringiensis toxins against Simulium spp. larvae. J Invertebr Pathol 2014;121:70–3. [DOI] [PubMed] [Google Scholar]
- Moumen B, Nguen-The C, Sorokin A. Sequence analysis of inducible prophage phIS3501 integrated into the haemolysin II gene of Bacillus thuringiensis var israelensis ATCC 35646. Genet Res Int 2012;2012:543286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawska E, Fiedoruk K, Swiecicka I. Modular genetic architecture of the toxigenic plasmid pIS56-63 harboring cry1Ab21 in Bacillus thuringiensis subsp. thuringiensis strain IS5056. Pol J Microbiol 2014;63:147–56. [PubMed] [Google Scholar]
- Nagamatsu Y, Okamura S, Saitou H et al. Three cry toxins in two types from Bacillus thuringiensis strain M019 preferentially kill human hepatocyte cancer and uterus cervix cancer cells. Biosci Biotech Bioch 2010;74:494–8. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Chandler M. Regulation of transposition in bacteria. Res Microbiol 2004;155:387–98. [DOI] [PubMed] [Google Scholar]
- Nicolas E, Lambin M, Dandoy D et al. The Tn3 -family of replicative transposons. Microbiol Spectr 2015;1:1–32. [DOI] [PubMed] [Google Scholar]
- Nicolas E, Oger CA, Nguyen N et al. Unlocking Tn3-family transposase activity in vitro unveils an asymetric pathway for transposome assembly. Proc Natl Acad Sci USA 2017;114:E669–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obukowicz MG, Perlak FJ, Kusano-Kretzmer K et al. Integration of the delta-endotoxin gene of Bacillus thuringiensis into the chromosome of root-colonizing strains of pseudomonads using Tn5. Gene 1986;45:327–31. [DOI] [PubMed] [Google Scholar]
- Oksanen HM, Bamford DH. Family Tectiviridae. In: King AMQ, Adams MJ, Carstens EB. et al (eds). Virus Taxonomy Ninth Report of the International Committee on Taxonomy of Viruses. London: Academic Press, 2012, 317–21. [Google Scholar]
- Økstad OA, Hegna I, Lindba T et al. Genome organization is not conserved between Bacillus cereus and Bacillus subtilis. Microbiology 1999;145:621–31. [DOI] [PubMed] [Google Scholar]
- Økstad OA, Tourasse NJ, Stabell FB et al. The bcr1 DNA repeat element is specific to the Bacillus cereus group and exhibits mobile element characteristics. J Bacteriol 2004;186:7714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Olson R, Pusch GD et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 2014;42:D206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AJ, Cummins CA, Hunt M et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015;31:3691–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma L, Muñoz D, Berry C et al. Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins 2014;6:3296–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-López L, Soberón M, Bravo A. Bacillus thuringiensis insecticidal three-domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS Microbiol Rev 2013;37:3–22. [DOI] [PubMed] [Google Scholar]
- Paris M, Després L. Identifying insecticide resistance genes in mosquito by combining AFLP genome scans and 454 pyrosequencing. Mol Ecol 2012;21:1672–86. [DOI] [PubMed] [Google Scholar]
- Park M, Kim MS, Lee KM et al. Characterization of a cryptic plasmid from an alpha-proteobacterial endosymbiont of Amoeba proteus. Plasmid 2009;61:78–87. [DOI] [PubMed] [Google Scholar]
- Parks DH, Rinke C, Chuvochina M et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol 2017;2:1533–42. [DOI] [PubMed] [Google Scholar]
- Pérez C, Fernandez LE, Sun J et al. Bacillus thuringiensis subsp. israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. Proc Natl Acad Sci USA 2005;102:18303–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravantti JJ, Gaidelyte A, Bamford DH et al. Comparative analysis of bacterial viruses Bam35, infecting a gram-positive host, and PRD1, infecting gram-negative hosts, demonstrates a viral lineage. Virology 2003;313:401–14. [DOI] [PubMed] [Google Scholar]
- Reddy A, Battisti L, Thorne CB. Identification of self-transmissible plasmids in four Bacillus thuringiensis subspecies. J Bacteriol 1987;169:5263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Ramírez A, Ibarra JE. Plasmid patterns of Bacillus thuringiensis type strains. Appl Environ Microb 2008;74:125–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezsöhazy R, Hallet B, Delcour J. IS231 D, E and F, three new insertion sequences in Bacillus thuringiensis: extension of the IS231 family. Mol Microbiol 1992;6:1959–67. [DOI] [PubMed] [Google Scholar]
- Rezsöhazy R, Hallet B, Mahillon J. IS231V and W from Bacillus thuringiensis subsp. israelensis, two distant members of the IS231 family of insertion sequences. Plasmid 1993;30:141–9. [DOI] [PubMed] [Google Scholar]
- Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 2009;106:19126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, Zimmerly S. Group II intron retroelements: function and diversity. Cytogenet Genome Res 2005;110:589–97. [DOI] [PubMed] [Google Scholar]
- Roh JY, Choi JY, Li MS et al. Bacillus thuringiensis as a specific, safe, and effective tool for insect pest control. J Microbiol Biotechn 2007;17:547–59. [PubMed] [Google Scholar]
- Rosso M-L, Mahillon J, Delécluse A. Genetic and genomic contexts of toxin genes. In: Charles J-F, Delécluse A, Nielsen-LeRoux C (eds). Entomopathogenic Bacteria: from Laboratory to Field Application. The Netherlands: Kluwer Academic Publishers, 2000:143–66. [Google Scholar]
- Ruiz-Masó JA, MachóN C, Bordanaba-Ruiseco L et al. Plasmid rolling-circle replication. Microbiol Spectr 2015;3:PLAS-0035–2014. [DOI] [PubMed] [Google Scholar]
- Schnepf E, Crickmore N, Van Rie J et al. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol R 1998;62:775–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014;30:2068–9. [DOI] [PubMed] [Google Scholar]
- Shehata AA, Tarabees R, Basiouni S et al. Phenotypic and genotypic characterization of bacteriocinogenic enterococci against Clostridium botulinum. Probiotics Antimicro 2017;9:182–8. [DOI] [PubMed] [Google Scholar]
- Sheppard AE, Poehlein A, Rosenstiel P et al. Complete genome sequence of Bacillus thuringiensis Strain 407 Cry-. Genome Announc 2013;1:e00158–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y-X, Zeng S-L, Yuan M-J et al. Influence of accessory protein P19 from Bacillus thuringiensis on insecticidal crystal protein Cry11Aa. Wei Sheng Wu Xue Bao 2006;46:353–7. [PubMed] [Google Scholar]
- Sidhu MS, Olsen I. S-layers of Bacillus species. Microbiology 1997;143:1039–52. [DOI] [PubMed] [Google Scholar]
- Siguier P, Filée J, Chandler M. Insertion sequences in prokaryotic genomes. Curr Opin Microbiol 2006;9:526–31. [DOI] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L et al. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 2006;34:D32–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Gourbeyre E, Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev 2014;38:865–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Gourbeyre E, Varani A et al. Everyman's guide to bacterial insertion sequences. Microbiol Spectr 2015;3:1–35. [DOI] [PubMed] [Google Scholar]
- Smillie C, Garcillan-Barcia MP, Francia MV et al. Mobility of plasmids. Microbiol Mol Biol R 2010;74:434–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin A, Candelon B, Guilloux K et al. Multiple-locus sequence typing analysis of Bacillus cereus and Bacillus thuringiensis reveals separate clustering and a distinct population structure of psychrotrophic strains. Appl Environ Microb 2006;72:1569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein C, Jones GW, Chalmers T et al. Transcriptional analysis of the toxin-coding plasmid pBtoxis from Bacillus thuringiensis subsp. israelensis. Appl Environ Microb 2006;72:1771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strömsten NJ, Benson SD, Burnett RM et al. The Bacillus thuringiensis linear double-stranded DNA phage Bam35, which is highly similar to the Bacillus cereus linear plasmid pBClin15, has a prophage state. J Bacteriol 2003;185:6985–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhao Q, Xia L et al. Identification and characterization of three previously undescribed crystal proteins from Bacillus thuringiensis subsp. jegathesan. Appl Environ Microb 2013;79:3364–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuplewska M, Ludwiczak M, Lyzwa K et al. Mobility and generation of mosaic non-autonomous transposons by Tn3-derived Inverted-repeat Miniature Elements (TIMEs). PLoS One 2014;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam A, Fitz-James P. Plasmids associated with a phagelike particle and with a satellite inclusion in Bacillus thuringiensis ssp. israelensis. Can J Microbiol 1986;32:382–8. [DOI] [PubMed] [Google Scholar]
- Tang M, Bideshi DK, Park HW et al. Minireplicon from pBtoxis of Bacillus thuringiensis subsp. israelensis. Appl Environ Microb 2006;72:6948–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Bideshi DK, Park HW et al. Iteron-binding ORF157 and FtsZ-like ORF156 proteins encoded by pBtoxis play a role in its replication in Bacillus thuringiensis subsp. israelensis. J Bacteriol 2007;189:8053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry C, Jiang S, Radford DS et al. Molecular tiling on the surface of a bacterial spore - the exosporium of the Bacillus anthracis/cereus/thuringiensis group. Mol Microbiol 2017;104:539–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetreau G, Bayyareddy K, Jones CM et al. Larval midgut modifications associated with Bti resistance in the yellow fever mosquito using proteomic and transcriptomic approaches. BMC Genomics 2012a;13:248–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetreau G, Stalinski R, Kersusan D et al. Decreased toxicity of Bacillus thuringiensis subsp. israelensis to mosquito larvae after contact with leaf litter. Appl Environ Microb 2012b;78:5189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DJ, Morgan JA, Whipps JM et al. Plasmid transfer between Bacillus thuringiensis subsp. israelensis strains in laboratory culture, river water, and dipteran larvae. Appl Environ Microb 2001;67:330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmery S, Modrie P, Minet O et al. Plasmid capture by the Bacillus thuringiensis conjugative plasmid pXO16. J Bacteriol 2009;191:2197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolle MA. Mosquito-borne diseases. Curr Probl Pediatr Adolesc Health Care 2009;39:97–140. [DOI] [PubMed] [Google Scholar]
- Tourasse NJ, Helgason E, Økstad OA et al. The Bacillus cereus group: novel aspects of population structure and genome dynamics. J Appl Microbiol 2006;101:579–93. [DOI] [PubMed] [Google Scholar]
- Tourasse NJ, Kolstø AB. Survey of group I and group II introns in 29 sequenced genomes of the Bacillus cereus group: insights into their spread and evolution. Nucleic Acids Res 2008;36:4529–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G, Mahillon J. Transcriptional analysis of the conjugative plasmid pAW63 from Bacillus thuringiensis. Plasmid 2008;60:190–9. [DOI] [PubMed] [Google Scholar]
- Van der Auwera G, Timmery S, Hoton F et al. Plasmid exchanges among members of the Bacillus cereus group in foodstuffs. Int J Food Microbiol 2007;113:164–72. [DOI] [PubMed] [Google Scholar]
- Van Frankenhuyzen K. Insecticidal activity of Bacillus thuringiensis crystal proteins. J Invertebr Pathol 2009;101:1–16. [DOI] [PubMed] [Google Scholar]
- Van Frankenhuyzen K. Cross-order and cross-phylum activity of Bacillus thuringiensis pesticidal proteins. J Invertebr Pathol 2013;114:76–85. [DOI] [PubMed] [Google Scholar]
- Varani AM, Siguier P, Gourbeyre E et al. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol 2011;12:R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheust C, Fornelos N, Mahillon J. The Bacillus thuringiensis phage GIL01 encodes two enzymes with peptidoglycan hydrolase activity. FEMS Microbiol Lett 2004;237:289–95. [DOI] [PubMed] [Google Scholar]
- Verheust C, Fornelos N, Mahillon J. GIL16, a new Gram-positive tectiviral phage related to the Bacillus thuringiensis GIL01 and the Bacillus cereus pBClin15 elements. J Bacteriol 2005;187:1966–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheust C, Jensen GB, Mahillon J. pGIL01, a linear tectiviral plasmid prophage originating from Bacillus thuringiensis serovar israelensis. Microbiology 2003;149:2083–92. [DOI] [PubMed] [Google Scholar]
- Wawrzyniak P, Płucienniczak G, Bartosik D. The different faces of rolling-circle replication and its multifunctional initiator proteins. Front Microbiol 2017;8:2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcks A, Smidt L, Bahl MI et al. Germination and conjugation of Bacillus thuringiensis subsp. israelensis in the intestine of gnotobiotic rats. J Appl Microbiol 2008;104:1252–9. [DOI] [PubMed] [Google Scholar]
- Wirth MC, Berry C, Walton WE et al. Mtx toxins from Lysinibacillus sphaericus enhance mosquitocidal cry-toxin activity and suppress cry-resistance in Culex quinquefasciatus. J Invertebr Pathol 2014;115:62–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth MC, Federici BA, Walton WE. Cyt1A from Bacillus thuringiensis synergizes activity of Bacillus sphaericus against Aedes aegypti (Diptera: Culicidae). Appl Environ Microb 2000;66:1093–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth MC, Georghiou GP, Federici BA. CytA enables CryIV endotoxins of Bacillus thuringiensis to overcome high levels of CryIV resistance in the mosquito, Culex quinquefasciatus. Proc Natl Acad Sci USA 1997;94:10536–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth MC, Jiannino JA, Federici BA et al. Synergy between toxins of Bacillus thuringiensis subsp. israelensis and Bacillus sphaericus. J Med Entomol 2004;41:935–41. [DOI] [PubMed] [Google Scholar]
- Wirth MC, Park H, Walton WE et al. Cyt1A of Bacillus thuringiensis delays evolution of resistance to Cry11A in the mosquito Culex quinquefasciatus. Appl Environ Microb 2005;71:185–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth MC, Yang K, Walton WE et al. Mtx toxins synergize Bacillus sphaericus and Cry11Aa against susceptible and insecticide-resistant Culex quinquefasciatus larvae. Appl Environ Microb 2007;73:6066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiwat C, Panbangred W, Bhumiratana A. Transfer of plasmids and chromosomal genes amongst subspecies of Bacillus thuringiensis. J Ind Microbiol 1990;6:19–27. [Google Scholar]
- Wiwat C, Panbangred W, Mongkolsuk S et al. Inhibition of a conjugation-like gene transfer process in Bacillus thuringiensis subsp. Israelensis by the anti-S-layer protein antibody. Curr Microbiol 1995;30:69–75. [Google Scholar]
- Wozniak RAF, Fouts DE, Spagnoletti M et al. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet 2009;5:e1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RAF, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Micro 2010;8:552–63. [DOI] [PubMed] [Google Scholar]
- Xu C, Wang BC, Yu Z et al. Structural insights into Bacillus thuringiensis Cry, Cyt and parasporin toxins. Toxins 2014;6:2732–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Hua G, Adang MJ. Effects and mechanisms of Bacillus thuringiensis crystal toxins for mosquito larvae. Insect Sci 2017;24:714–29. [DOI] [PubMed] [Google Scholar]
- Zheng J, Gao Q, Liu L et al. Comparative genomics of Bacillus thuringiensis reveals a path to specialized exploitation of multiple invertebrate hosts. mBio 2017;8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zghal RZ, Jaoua S. Evidence of DNA rearrangements in the 128-kilobase pBtoxis plasmid of Bacillus thuringiensis israelensis. Mol Biotechnol 2006;33:191–8. [DOI] [PubMed] [Google Scholar]
- Zorzetti J, Ricietto AP, da Silva CR et al. Genome sequence of the mosquitocidal Bacillus thuringiensis strain BR58, a biopesticide product effective against the coffee berry borer (Hypothenemus hampei). Genome Announc 2015;3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH et al. PHAST: A fast phage search tool. Nucleic Acids Res 2011;39:W347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick ME, Joseph SJ, Didelot X et al. Genomic characterization of the Bacillus cereus sensu lato species: backdrop to the evolution of Bacillus anthracis. Genome Res 2012;22:1512–24. [DOI] [PMC free article] [PubMed] [Google Scholar]