Abstract
Intrinsically disordered proteins (IDPs) are highly unorthodox proteins that do not form three-dimensional structures under physiological conditions. The discovery of IDPs has destroyed the classical structure-function paradigm in protein science, 3-D structure = function, because IDPs even without well-folded 3-D structures are still capable of performing important biological functions and furthermore are associated with fatal diseases such as cancers, neurodegenerative diseases and viral pandemics. Pre-structured motifs (PreSMos) refer to transient local secondary structural elements present in the target-unbound state of IDPs. During the last two decades PreSMos have been steadily acknowledged as the critical determinants for target binding in dozens of IDPs. To date, the PreSMo concept provides the most convincing structural rationale explaining the IDP-target binding behavior at an atomic resolution. Here we present a brief developmental history of PreSMos and describe their common characteristics. We also provide a list of newly discovered PreSMos along with their functional relevance.
Keywords: IDPs, IDR (Intrinsically Disordered Region), NMR, IUPs (Intrinsically Unfolded Proteins), PreSMos (Pre-Structured Motifs)
INTRODUCTION
Intrinsically Disordered Proteins
The central dogma in protein science, established over the last half-century, states that “a well-folded 3-D structure is a prerequisite for protein function”. The 3-D structure in this statement refers to the one that is observed under near-physiological conditions, (i.e., ~ pH 7, ambient temperature, and aqueous buffer, etc.). Intrinsically unstructured/unfolded proteins (IUPs), now more commonly known as intrinsically disordered proteins (IDPs) (Dunker et al., 2013), are very peculiar proteins that do not form well-folded 3-D structures even under non-denaturing conditions. Naturally, IDPs are of great importance from a protein folding perspective. More intriguing are the observations that IDPs are functional or active without 3-D structures, for example, being involved in transcription (Lee et al., 2000; Sherr, 2004; Kim et al., 2017a; 2017b), translation (Fletcher and Wagner, 1998; Kim et al., 2015), cell cycle regulation (Pavletich, 1999), chaperoning (Hong et al., 2005), and membrane-binding (Atwal et al., 2007; Eliezer et al., 2001). The discovery of many, as much as half of the entire human proteome (Dunker et al., 2000), such highly unorthodox proteins has strongly suggested that the classical structure-function relationship of proteins needs to be reexamined. Cleary, the golden paradigm in structural biology, 3-D structure = protein function, is no longer valid. Several reviews dealing with general aspects of IDPs are available for further reading (Chavali et al., 2017; Dunker et al., 2013; Lee et al., 2012; Uversky and Dunker, 2010; Uversky, 2015).
Not only because of a basic scientific point of view are our interests in IDPs keen but also because of the fact that these proteins are involved in many fatal diseases. For example, ~80% of human cancers are associated with IDPs (Galea et al., 2008) such as eIF4E-binding proteins (4EBPs) (Fletcher and Wagner, 1998; Kim et al., 2015), Bcl-XL (Xu et al., 2009], human glucocorticoid receptors (Kim et al., 2017b), E7 (Lee et al., 2016), hypoxia inducible factors (Semenza, 2003; Kim et al., 2009a) and p53 all of which are so-called “hybrid-type” IDPs where intrinsically disordered regions (IDRs) coexist with globular domains (Lee et al., 2000; Wells et al., 2008). The causative agents of mad cow disease or Creutzfeldt-Jakob disease (CJD) in humans are prions that are also IDPs where a C-terminal globular domain coexists with a long intrinsically disordered region (IDR) at the N-terminus encompassing ~120 amino acid residues (James et al., 1997; Liu et al., 1999). Alpha-synuclein (Eliezer et al., 2001) and tau (Bibow et al., 2011; Künze et al., 2012), implicated in PD (Parkinson’s diseases) and AD (Alzheimer’s disease) respectively, are also IDPs. Furthermore, several viral strains including the well-known AIDS-causing HIV-1 produce IDPs (Chi et al., 2007; Feuerstein et al., 2012; Kim et al., 2009b; Lee et al., 2016; Liang et al., 2007; Reingewertz et al., 2009; To et al., 2016). Clearly, there is an immediate and strong need to acquire very thorough knowledge not only on the normal functionality of IDPs but also on their pathologic connection to above diseases since it has become apparent that the classical globular protein based approach is unlikely to provide us with sufficient information that can be used for developing effective weaponry against IDP-associated diseases.
PreSMos: Pre-Structured Motifs, a Historical Perspective
The most obvious characteristic of IDPs is that they do not possess spatially-disposed active pockets, a fact that brings us to a simple but profound question of how then these long malleable stretches of amino acids (sometimes hundreds of amino acids) can bind their targets. Targets of IDPs are not just proteins, but can be nucleic acids (Thapar et al., 2004; To et al., 2016; Wells et al., 2008), lipids, metals, and small molecules (Follis et al., 2008; Metallo, 2010). Efforts were made recently to classify IDPs into several subfamilies (van der Lee et al., 2014). While intuitive, such a classification fails to provide detailed insights into how all these different subfamilies bind their targets. The well-cited expression “coupled folding and binding” (Dyson and Wright, 2002) is a useful term, but only as far as one tries to depict the rather easily-predictable topological change that IDPs need experience upon binding to their partners. This generic description therefore fails to provide any atomistic details associated with IDP-target binding that, if available, would be highly valuable for IDP-based drug design. As the axiom “the devil is in the details” dictates, the question one must answer is rather specific. It has been amply demonstrated that only certain segments or residues of IDPs/IDRs are involved in direct physical contact with target. Do we then have a clear answer on what specific features in these segments or residues make target binding possible? Why does mutating just a few (often just one) sparsely-disposed hydrophobic residues in acidic transactivation domains (TADs) drastically affect the transcriptional activity whereas mutating several of the abundant acidic residues has only a marginal effect on the activity? (Chang et al., 1995; Drysdale et al., 1995) An early investigation attempted to address this question by employing wild type GAL4 and its scrambled mutant with no transcriptional activity (Giniger and Ptashne, 1987) and concluded that the mutant was inactive because its helix-forming propensity was compromised. This study triggered a huge controversy over whether target-free acidic TADs should form an amphipathic helix as the specificity determinant for activity (Van Hoy et al., 1993).
Direct and quantitative evidence that some sort of a secondary structural element, e.g., helix, is needed for transcriptional activity came from an NMR study on p53 TAD (Lee et al., 2000). The 73-residue long p53 TAD in its unbound form was found “unstructured” in a tertiary sense, yet contained a transient (~25% populated only) amphipathic helix whose residues formed a stable amphipathic helix when complexed with the N-terminal p53-binding domain (residues 3–109) of mdm2 (Kussie et al., 1996). This pioneering NMR study heralded the birth of the PreSMo concept. Subsequent NMR reports confirmed that pre-existing, pre-formed, or pre-ordered residual secondary structures, no matter what they may be called, do exist in unbound IDPs and are important for target binding (Lee et al., 2012). In the early days of IDP research, another line of thought prevailed advocating a notion of induced fit (IF), arguing that no pre-existing secondary structures were needed for target binding based upon the conclusion that IDPs are fully unstructured. A well-known example is the 4EBP1, a 118-residue translational inhibitor, which was reported to have “no regions of local order in the absence of eIF4E” (Fletcher et al., 1998). For the last two decades, this IDP has been known as the symbol of the completely unstructured (CU) nature of IDPs; however, a recent NMR study revealed that this IDP also contains a pre-structured helix which mediates its binding to eIF4E (Kim et al., 2015). Another well-known IDP is the kinase-inducible domain (KID) of CREB the NMR results on which supported the concept that IDPs must be in the CU state so that they must undergo “a coil -> helix folding transition” via IF (Radhakrishnan et al., 1997). It is unclear how the authors of this particular report reached the conclusion that “the population of helix in free pKID is extremely small.” when their NMR data indicated presence of two transient helices (one ~60% and the other ~10% populated). Another group which worked on the same system concluded that two helix PreSMos were present (Table 1; Hua et al., 1998; Lee et al., 2012).
Table 1.
A list of MU-type IDPs/IDRs containing PreSMos
| Name | Number of residues | P/Rb | Location of PreSMo residuesc | Populationd (%) | Role/Binding | References |
|---|---|---|---|---|---|---|
| FlgM | 97 | P | 60–73 | 50±10 | σ28 | Daughdrill et al., 1997 |
| 83–90 | 50±10 | |||||
| 42–50 | 20 | |||||
| KID | 60 | R | 119–129 | >50 | KIX | Radhakrishnan et al., 1998 |
| 134–143 | ~10 | Hua et al., 1998 | ||||
| GBD/CRIB in WASP W7 | 68 | R | 252–264 | ~14 | Cdc42/Rac | Rudolph et al., 1998 |
| (201–268) | ||||||
| HIV-1 Nef | 56 | R | 14–22 : helix I | 18 | Geyer et al., 1999 | |
| (2–57) | 35–41 : helix II (Hα only) | |||||
| Synaptobrevin-2 | 96 | R | 78–91 | 45 | core complex forming | Hazzard et al., 1999 |
| APPC | 47 | R | 20–23 | 30 | X11 | Ramelot et al., 2000 |
| (649–695) | 27–35 | 20 | ||||
| 37–45 (Hα only) | 30 | |||||
| p53 TAD | 73 | R | 18–26 : helix | 20 | Mdm2 | Lee et al., 2000 |
| 40–44 : turn I | 5 | RPA, TFEII | ||||
| 48–53 : turn II | 15 | |||||
| RPS4 | 200 | P | 12–15 | 8 | rRNA, ribosomal proteins | Sayers et al., 2000 |
| 30–33: β? | 23 | |||||
| α-Synuclein | 140 | P | 18–31 | ~10 | amyloid-forming | Eliezer et al., 2001 |
| Murrali et al., 2018 | ||||||
| N-term. Tmod 1 | 92 | R | 24–35 | NA | tropomyosin | Greenfield et al., 2005 |
| VP16 TAD | 79 | 443–447 | 25 | hTAFII31 PC4 | Jonker et al., 2005 | |
| (412–490) | R | 469–483 | 15 | |||
| VP16 TAD | 79 | R | 424–433/442–446, 465–467/472–479 (Hα only) | 60/40 | hTAFII31 PC4 | Kim et al., 2009 |
| (412–490) | 10/20 | |||||
| Dynein interm. chain | 40 | R | 223–228 | NA | light chains | Benison et al., 2006 |
| (198–237) | Benison et al., 2007 | |||||
| γ-Synuclein | 127 | P | 49–99 | ~15 | Marsh et al., 2006 | |
| HMGA1 | 107 | P | 3–9 | 8 | 20 different proteins | Buchko et al., 2007 |
| 64–67 | ||||||
| CFTR | 185 | R | α-helix | interaction between R region and NT-binding domain 1 | Baker et al., 2007 | |
| (654–838) | 654–668, 759–764, 766–776, 801–817 | >5 | ||||
| >5 | ||||||
| β-strand | ||||||
| 744–753 | >5 | |||||
| NS5A-D2 (HCV) | 93 | R | L48-V57 | 20 | - | Liang et al., 2007 |
| (250–342) | L86-E96 (Hα only) | 25 | ||||
| preS1 of HBV | 119 | R | 32–36, 41–45 | ~10 | hepatocyte receptor-binding | Chi et al., 2007 |
| 11–18, 22–25, 37–40, 46–50. (Hα only) | ~10 | |||||
| ~10 | ||||||
| β-synuclein | 134 | P | NA | ~20 | - | Sung et al., 2007 |
| Securin | 202 | P | 150–159 : helix | 45 | - | Csizmok et al., 2008 |
| 113–127 (β) | 15 | |||||
| 174–178 | 20 | |||||
| C-XPCe | 126 | R | 818–843: helix | ~30 | Centri2 | Miron et al., 2008 |
| (815–940) | 847–860: helix | ~30 | TFIIH | |||
| 891–901: helix | NA | |||||
| 908–915: helix | NA | |||||
| 923–930: helix | NA | |||||
| MSP2 | 237 | P | 14–21 | 35 | - | Zhang et al., 2008 |
| 140–150 | 35 | |||||
| 197–211 | 20 | |||||
| DARPP-32 | 118 | R | 22–29 | 50 | PP1 | Dancheck et al., 2008 |
| 103–114 | 25 | |||||
| I-2 | 156 | R | 36–42 | 30 | PP1 | Dancheck et al., 2008 |
| (9–164) | 96–106 | 48 (70) | ||||
| 127–154 | 67 (90) | |||||
| 132–138 | >98 | |||||
| ENSA | 121 | P | 32–36 | 40 | - | Boettcher et al., 2008 |
| 48–50 | 10 | Boettcher et al., 2007 | ||||
| 65–70 | 30 | |||||
| ODD/HIF-1α | 74 | R | 438–440 | ~10 | - | Kim et al., 2009 |
| (404–477) | 467–477 | |||||
| Sml1 | 104 | P | 4–14: helix | ~20 | RNR binding | Zhao et al., 2000 |
| (1–104) | 61–80: helix | ~70 | Dimer forming | |||
| Myb25 | 25 | R | 295–309 : helix | 25~30 | KIX | Zor et al., 2002 |
| (291–315) | ||||||
| N tail | 125 | R | 488–499 : helix | NA | phosphoprotein P | Bourhis et al., 2004 |
| Measles virus nucleoprotein | (401–525) | |||||
| dSLBP | 92 | R | 28–45 : helix | NA | mRNA | Thapar et al., 2004 |
| (17–108) | 50–57 : helix | stem-loop | ||||
| 66–75 : helix | ||||||
| 91–96 : helix | ||||||
| Tβ-4 | 43 | P | 5–16 : helix | NA | Ca ATP | Domanski et al., 2004 |
| (1–43) | G-actin | |||||
| N tail | 82 | R | 479–484 | 36 | phosphoprotein P | Jensen et al., 2008 |
| Sendai Virus nucleoprotein | (443–524) | 476–488 | 38 | |||
| 478–492 | 11 | |||||
| Sic1 | 90 | R | 20–30 | 20 | Cdc4 | Mittag et al., 2008 |
| (1–90) | 63–68 | |||||
| c-Myc | 88 | R | 26–34 : helix | 40 | Bin-SH3 domain | Andresen et al., 2012 |
| (1–88) | 47–52 : helix | 25 | 24–31(TRRAP binding) | |||
| 20–23 : β-turn | ||||||
| ExsE | 88 | P | 42–51: helix | NA | ExsC | Zheng et al., 2012 |
| (1–88) | 61–65: helix | |||||
| NS5A | 415 | R | 401–412 : helix | NA | Bin1-SH3 | Braeuning, 2013 |
| HCV | (33–447) | 427–445 : helix | ||||
| NS5A | 179 | R | 205–221 : helix I | 38 | Bin1-SH3 | Feuerstein et al., 2012 |
| HCV | (191–369) | 251–266 : helix II | 38 | Solyom et al., 2015 | ||
| 292–306 : helix III | 51 | |||||
| 4EBP2 | 120 | P | 1–5 | 15~37 | eIF4E | Lukhele et al., 2013 |
| (1–120) | 33–37 | |||||
| 50–64 | ||||||
| 86–89 | ||||||
| 96–105 | ||||||
| E7 | 40 | R | 8–13 : helix | NA | E2 | Noval et al., 2013 |
| HPV | (1–40) | 17–29 : helix | ||||
| 33–38 : PPII | ||||||
| 4EBP1 | 70 | R | 56–63 : helix | 20 | eIF4E | Kim et al., 2015 |
| (49–118) | ||||||
| Myb32 | 32 | R | 290–310 : helix | ~70 | KIX | Arai et al., 2015 |
| (284–315) | ||||||
| E7 | 46 | R | 7–14 : helix | 10 | E2 | Lee et al., 2016 |
| HPV | (1–46) | 20–26 : helix | 20 | |||
| CBP-ID4 | 207 | R | 1852–1875: helix | ~60 | - | Piai et al., 2016 |
| (1851–2057) | 1951–1978: helix | |||||
| HIV-1 Tat | 121 | P | 27–32: helix | ~20 | Fab’ | To et al., 2016 |
| (1–121)a | 41–59: helix | ~30 | P-TEFb | |||
| 70–81: β sheet | ~25 | TAR-cyclin T1 | ||||
| 93–99: β sheet | ~25 | |||||
| 105–112: β sheet | ~10 | |||||
| SUSP4 | 100 | R | 263–291 : helix | ~30 | mdm2 | Kim et al., 2017 |
| (201–300) | 265–270 : helix | ~10 | ||||
| 281–291 : helix | ||||||
| hGRtau1c | 64 | R | 185–202: helix | 20~30 | TAZ2 | Kim et al., 2017 |
| (181–244) | 206–225: helix | |||||
| 232–244: helix | ||||||
| Huntingtin Httex1 25Q | 95 | P | 18–42: helix | NA | Cytotoxic | Newcombe et al., 2018 |
| (1–95) | Membrane binding | |||||
| Aggregation |
The numbering includes a 20-residue N-terminal tag.
An IDP (P) versus an IDR (R).
Residue numbers are taken from the original report.
Population of PreSMos are read from the mid-point of the SSP scores that are calculated from chemical shifts in BMRB or literature. Shown in bold are the populations described in the original report. When the populations described in the original report without SSP scores differed significantly from the calculated SSP scores, the SSP scores are provided in parenthesis.
NA = not available.
Determined by SAXS.
While the conceptual development on PreSMos has been somewhat delayed due to previous misconceptions that IDPs were completely unstructured, the presence of local residual secondary structures in isolated IDPs has been increasingly detected by many NMR investigations including a few critical NMR reports published at the turn of the century. The first key report found that p53 TAD has local structural elements (a helix and two turns) in the unbound state, as described above (Lee et al., 2000). The second report made by Ramelot et al. demonstrated that the cytoplasmic tail of the amyloid precursor protein forms a transient structure and such a pre-ordered structure is important for its binding to cytosolic factors (Ramelot et al., 2000). Sayers et al. also reported that structural preordering important for target binding was detected in the N-terminal region of ribosomal protein S4 (Sayers et al., 2000). Zhao et al. reported local structural elements in the overall loosely folded Sml1 (Zhao et al., 2000). Zitzewitz et al. published an article in 2000 with a title of “Preformed secondary structure drives the association reaction of GCN4-p1, a model coiled-coil system” (Zitzewitz et al., 2000). Another report by Bienkiewicz et al. described the functional consequences of pre-organized helical structure in the intrinsically disordered cell-cycle inhibitor p27 (Kip1) (Bienkiewicz et al., 2002). All these early NMR studies contributed to the foundation of the PreSMo concept, the idea that IDPs are not completely unstructured, but mostly unstructured (MU), and contain PreSMos. Following these NMR reports, bioinformatics studies proposed similar concepts such as PSE (Pre-formed Structural Element) (Fuxreiter et al., 2004), MoRF (Molecular Recognition Element) (Mohan et al., 2006; Oldfield et al., 2005), or primary contact sites a few years later. All these results, NMR experimental or predicted, point in unison to the idea that IDPs possess local secondary structural elements that are “hot spots” for target-binding.
In 2012 we published the first comprehensive review on PreSMos (Lee et al., 2012) because no explicit articles on the subject were available, despite the fact that PreSMos (whatever they may be called) have been recognized for more than a decade as very important (perhaps the most significant) features explaining IDP-target binding on a per-residue basis. Several additional pieces of evidence have recently been published, demonstrating the functional significance of PreSMos (Kim et al., 2017b; Iešmantavičius et al., 2014; Mohan et al., 2014; Salamanova et al., 2018). In the first review, we presented 27 IDPs/IDRs containing PreSMos which constitute ~56% of all IDPs characterized by then. Most critically, we introduced the term pre-structured motifs (PreSMos) in order to unambiguously point out the importance of the pre-structured nature of target-binding segments in free IDPs and to provide a convenient term that can replace various names “transient, nascent, residual, minimally-structured, non-negligible, pre-existing, pre-formed, or pre-ordered secondary structures”. These terms were used mainly by NMR structural biologists who did not hasten to generalize the concept with a particular name realizing that PreSMos had only been observed in a handful of IDPs until 2005. This review is a follow-up to our 2012 review. Because we have found 20 more PreSMos since our first review here we provide an updated list of PreSMos and a brief description on their functional significance; however, we acknowledge that the list may still be incomplete. In addition, we describe differences between the PreSMos that are detected experimentally and the terms derived from bioinformatics predictions. With this review we now have 47 IDPs/IDRs containing PreSMos, strongly suggesting that PreSMos are general signatures in most IDPs.
DISCUSSION
Definition of a PreSMo
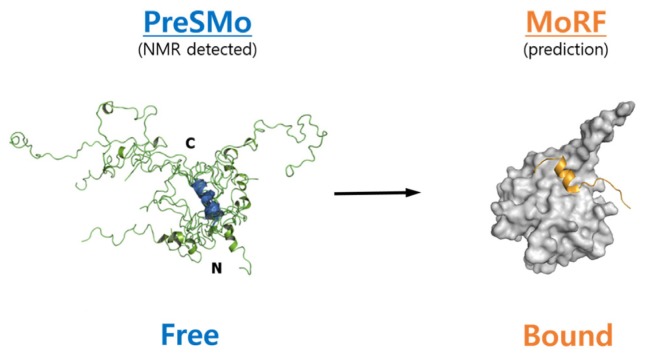
The definition of a PreSMo was given in our 2012 review (Lee et al., 2012); PreSMos are NMR-detected transient secondary structural elements within long (minimally 40 residues) and functionally-active IDRs of IDPs. We underline the fact that PreSMos are the experimentally observable entitites in NMR analyses or other atomic-resolution experiments no matter how minimally it might be pre-populated; it is a measured quantity, not predicted notions. This contrasts with MoRF (Mohan et al., 2006), which is a theoretical concept derived from the target-bound conformations of short segments (peptides) of IDRs (Fig. 1). IDPs exist as an ensemble of many different conformers separated by small energy differences. A conformer with a PreSMo would be one in the ensemble that is populated to an NMR-detectable degree. The lowest population of a PreSMo-containing conformer observed to date is ~10% (Lee et al., 2012).
Fig. 1. PreSMo vs. MoRF. A schematic diagram of the main differences between a PreSMo and a MoRF.
A PreSMo is observed mostly by NMR experiments in the target-free state of IDPs. Since free-state IDPs exist as an ensemble of many conformers separated by small energy differences, structural superposition among different conformers along the backbone atoms is not possible. Nevertheless, a structural zsuperposition along a PreSMo is possible as shown in the left panel for the helix PreSMo of 4EBP1 (Kim et al., 2015). A PreSMo may become a MoRF upon target binding as illustrated for this helix PreSMo in 4EBP1 which becomes an α-MoRF when bound to eIF4E.
Table 1 is an updated list of PreSMos found in 47 IDPs/IDRs. The total number of IDPs studied in detail by NMR (with an exception of C-XPC studied by SAXS) is 70 even though the number of reports are more than 70 reports because some IDPs were investigated more than once. Notably, several IDPs (4EBP1, HIV-1 Tat, VP16 TAD, securin, and p21Waf1/Cipl/Sdil) that were originally reported as CU types with no PreSMo turned out to be MU types in later studies. For convenience, we added the 20 newly-identified PreSMos (starting from Myb25) at the end of Table 1, including a few PreSMos that were actually reported before 2012, but were not included in our 2102 review. Although the number of investigated IDPs is small compared to the possible number of IDPs/IDRs predicted by bioinformatics (thousands or more) it is sufficient to provide an overview on PreSMos. In 2012, the number of IDPs/IDRs with PreSMos was 27 (out of 48 studied) it is now 48 out of 70; the proportion of MU type IDPs/IDRs increased from 56% to 69%. The proportion is likely to increase if more IDPs/IDRs are characterized. One immediate feature noted in Table 1 is that in most cases we essentially study IDRs rather than IDPs (only 15 are IDPs), although we speak of IDPs. Note that all IDPs/IDRs in Table 1 are composed of more than 40 residues except for Myb25/Myb32. IDPs by definition consist of a minimal 40 residues and are distinct from the short flexible linkers and loops typically composed of fewer than 20 residues. The other feature shown in Table 1 is that most PreSMos are helices even though some are turns, β-strands and poly-proline type II helices. A high percentage of helices is also noted in MoRFs where α-MoRFs are the majority (Mohan et al., 2006; Oldfield et al., 2005).
NMR is the main tool that enables quantitative definition of a PreSMo (Chi et al., 2007; Eliezer et al., 2001; Kim et al., 2009a; 2009b; 2015; 2017b; Lee et al., 2000; 2012; 2016; Liu et al., 1999; Xu et al., 2009). The beauty of NMR technique is that the presence of a PreSMo is reflected in several independent NMR parameters. In the early days, one needed to provide all of these NMR parameters (chemical shifts, inter-proton NOEs, J-couplings, T1 and T2 relaxation times, heteronuclear NOEs, temperature coefficients of backbone amide protons, etc.) to prove the existence of a PreSMo (Lee et al., 2000), whereas it usually is sufficient in recent years to just provide SSP (secondary structure propensity) scores (Marsh et al., 2006) as the concept of PreSMos has become more and more widely accepted. The SSP scores derived from CSIs (chemical shift indices) reveal an actual percentile value of a PreSMo population whereas CSIs can only indicate whether or not a PreSMo is present. A very important feature of a PreSMo is that it is never 100% populated. On the average, they are ~30% pre-populated, i.e., transient (Lee et al., 2012). This transient nature of PreSMos probably is the main cause that made several NMR investigators fail to detect them in the early days (Fletcher and Wagner, 1998; O’Hare and Williams, 1992; Radhakrishnan et al., 1997).
PreSMo vs. MoRF
The most common bioinformatics term used interchangeably with PreSMos is MoRFs (Mohan et al., 2006). For example, the mdm2-binding helix PreSMo detected by NMR in free p53 TAD is reported as an α-MoRF, a MoRF seen as an alpha helix in the target-bound state (Oldfield et al., 2005). Although there are a few more (out of more than a hundred) MoRFs that overlap with PreSMos fundamental differences exist between MoRFs and PreSMos. By definition MoRFs were identified in the x-ray structures of complexes between target proteins and short fragments of IDPs/IDRs that were predicted to be disordered by bioinformatics disorder prediction algorithms. The concept of the MoRF implicitly acknowledges the idea that the structured, bound-conformation is induced only upon target binding which is based on the early-day idea that IDPs have no pre-structured secondary structures. On the other hand, the definition of a PreSMo is not associated with the target-bound structure at all. In this regard, stating that a MoRF is found by NMR experiments is inaccurate (Bourhis et al., 2004) since one cannot tell if a MoRF would exist within an isolated IDP. One has to obtain a complex structure between a target and a PreSMo/MoRF in order to conclude that the putative MoRF (which is actually a PreSMo) is indeed a MoRF. Thus, a helix PreSMo may become an α-MoRF, but the opposite may not necessarily be true. With PreSMos we get the realistic percentage of the pre-structuredness whereas MoRFs do not provide such information. The term PreSMo was introduced as late as in 2012, but we underline that the PreSMos mentioned here refer to all the pre-existing or pre-formed residual secondary structures detected by NMR years before the term MoRF was introduced. It will be interesting to see how many of MoRFs may indeed coincide with PreSMos. One has to use a MoRF fragment, or preferably a longer IDR that encompasses such a MoRF fragment, to answer this question. An active pocket is a property of a globular protein that exists before binding to its target. In this regard, PreSMos qualify as the “active sites”, albeit not pockets, of IDPs since they are present before target binding. The same cannot be said for MoRFs. In Fig. 1, we show a conceptual scheme depicting what we have just described.
Characteristics of PreSMos
PreSMos are the “active sites” of IDPs
As is evident from Table 1 the PreSMos are the target-binding hot spots already present in free IDPs/IDRs; PreSMos are primed in a conformation similar to the target-bound conformation. Such pre-structuring is certainly advantageous for avoiding an entropic penalty that has to be paid when malleable IDPs/IDRs bind globular targets. Recent mutation studies demonstrated that the degree of pre-population of PreSMos is subtly controlled for efficient target binding (Borcherds et al., 2014; Iešmantavičius et al., 2014; Kim et al., 2017b; Salamanova et al., 2018). In many globular proteins a single mutation in the active site completely nullifies protein function by disabling the binding of ligands. PreSMos are often found in tandem within sufficiently long transcription factor IDPs/IDRs separated by ~30 residues (Chi et al., 2005). One PreSMo may be a high-affinity binding site to a target whereas the other is a low-affinity site to the same target. A synergistic effect of multiple PreSMos for efficient target binding has been discussed previously (Lee et al., 2000).
Shape complementarity in IDPs
Since it was believed that any secondary structure in IDPs should be induced only upon target binding many implicitly concluded that IDPs would totally lie outside of the classical structure-function paradigm, not obeying the rules established by structural biology such as shape complementarity. However, PreSMos reveal to us that IDPs abide by the shape complementarity extremely well via binding to targets (see Fig. 3 in Lee et al., 2012). In other words, when the secondary structural aspects for IDP-target binding are considered IDPs are not unorthodox at all. The genuine novelty of IDPs is the absence of 3-D structures only, not the absence of secondary structures. Structure (or PreSMos) does dictate function in the case of IDPs.
Practical tips for NMR detection of PreSMos
The NMR spectral quality of hybrid-type IDPs is often not good enough for a full resonance assignment since a globular domain and an IDR will tumble around in different time scales. Consequently, a reductionist approach of using an IDR instead of a whole IDP is often necessary. One precaution when using such an approach is that one should use a sufficiently long region, not a short fragment since PreSMos may exist in the outside of the region covered by a short peptide (Botuyan et al., 1997; Uesugi et al., 1997). A longer IDR often contains a more populated PreSMo due to a tertiary effect that stabilizes the transient secondary structures, as was demonstrated in the case of p53 TAD and its short helical peptide (Botuyan et al., 1997; Lee et al., 2000). Another case demonstrating the significance of using a fragment of appropriate length is Myb 25/Myb32 (Table 1; Arai et al., 2015). The populations of a helix PreSMo in Myb25 and in Myb32 are ~30% and ~70%, respectively, demonstrating that having just 7 more residues in Myb32 drastically increases the PreSMo population by ~40%. Using bioinformatics disorder prediction programs may keep one from choosing an inappropriate IDR for NMR experiments. The inappropriate choice of an IDR for NMR investigation might be another reason why some NMR studies failed to detect PreSMos.
CONCLUSION & PERSPECTIVE
Because IDPs are relatively a new field several new (sometimes rather vague) terms and expressions were introduced in order to describe novel concepts or phenomena associated with IDPs (van der Lee et al., 2014). Aside from bioinformatics terms (PSEs, MoRFs) other numerous expressions basically with the same meaning as PreSMos were proposed such as “only partly structured” (Zor et al., 2002), “small islands of secondary structures” (Laptenko and Prives, 2006), “weakly structured” (Chumakov, 2007), “limited structure” (Lavery and McEwan, 2008), “minimal ordering of short linear motifs” (Mittag et al., 2008), “residual secondary structural elements” (Kim et al., 2009b), “transient order” (Feuerstein et al., 2012), “transiently ordered regions”, “localized structurally ordered regions” (Zheng et al., 2012), and dynamic local structure (Lum et al., 2012) just to name a few.
Being flooded with so many terms that are intended to denote PreSMos is not unique for PreSMos. For example, it took more than a decade for the IDP research community to come up with a more or less consensus term for IDPs in 2013 (Dunker et al., 2013). Yet overly creative names not precisely in line with the classical concepts and terms in structural biology or protein science created a certain degree of confusion that led to a situation where the importance of IDPs was not duly appreciated for some time (Uversky and Dunker, 2010). Here, we present again an easy-to-use term of PreSMos to designate what has been described by several generic names realizing that the existence and functional significance of PreSMos will be appreciated more and more (now in ~70% of IDPs). Most importantly, the statement that IDPs would adopt structure only upon target binding is misleading because it implies that IDPs are structureless down to the level of secondary structures. On the contrary, target binding only tightens (some structural induction) a PreSMo into a more stable conformation, but does not let a random-coil turn into a structure. In hindsight, the presence of PreSMos is in excellent agreement with the observations that a protein cannot exist in a fully random-coil state; denatured globular proteins are not random coils (Baldwin and Zimm, 2000; Bernadó et al., 2005; Neri et al., 1992).
Approximately 20 years have passed since IDPs emerged in protein science and structural biology communities. With more than ~5,000 papers on the subject no one would deny that IDPs have brought a critical paradigm shift to protein research, undoubtedly requiring that biochemistry textbooks be revised to include IDPs. There has been a tendency to put excessive emphasis on the disordered nature per se of IDPs with subsequent attempts trying to relate it to function due to an early-day misconception. For example, some reports on PreSMos were interpreted simply as evidence for disorder itself rather than as evidence for the existence of PreSMos (Cheng et al., 2006; Midic et al., 2009; Radivojac et al., 2007). It is important for the protein science community to learn a non-traditional view on proteins and their structures in two aspects. First, it is now well-known fact that long regions (40 residues and up) of proteins can be intrinsically disordered beyond the level of short disordered loops (Dunker et al., 2000). Proteins exist as dynamic conformational ensembles, not as snap-short entities that the PDB structures (both x-ray and NMR) have depicted for a long time. Second, in the absence of a well-defined 3D structure, the minimal residual secondary structures embedded into the flexible long IDR play key roles in target binding and govern the function of IDPs. Even in globular proteins, an important role of tertiary structure is to place the interacting (or active) secondary structures in a proper orientation relative to target proteins.
A discussion of PreSMos naturally brings us to the question of whether the mechanism of IDP-target binding follows IF (induced fit) or CS (conformational selection). In the case of KID-KIX binding IF (Sugase et al., 2007) was shown to be dominant whereas in the N-tail of viral nucleoproteins CS appeared prevalent (Jensen et al., 2008). In recent years, it is believed that these two mechanisms would work in concert; CS at the start of binding and IF at the final stage of binding (tightening). The existence of PreSMos itself is not an evidence for CS and one need to use a kinetics approach in order to determine if faster binding (kon increased) can be achieved with more pre-structuring of the PreSMo segments. Future works employing PreSMo mutants should provide a more concreate answer on this aspect. No matter whether PreSMos are pre-structured or not, i.e., even if a PreSMo may become unstructured and re-structured for binding as one may envision in the IF model (To et al., 2016) it still does not change the fact that the fragment forming a PreSMo per se is important for target binding.
It is possible that PreSMos are also important for aggregation via oligomerization (Atwal et al., 2007; Eliezer et al., 2001). Both oligomerization and IDP-target binding are protein-protein interactions; the former is homogenous IDP-IDP self-binding while the latter is heterogeneous binding. Even though the PreSMo concept is broadly (~70%) applicable we do not expect that it should be applicable to all IDPs since there are IDPs/IDRs that are composed of simple dipeptide repeats (Lee et al., 2016). The PreSMo concept is also unlikely to be applicable to highly charged polyvalent IDPs which maintain unfolded topology even after target binding (Borgia et al., 2018). Due to strong attractive electrostatic interactions these IDPs have a very high affinity (pM) towards each other, unlike MU-type IDPs that bind their targets via PreSMos typically with μM affinities. However, it is noteworthy that even polyglutamine and polyproline were shown to form α-helical and PPII helix type secondary structures, respectively (Mukrasch et al., 2009; Newcombe et al., 2018). Recent reports showed that IDP studies may lead to the development of new pharmaceuticals. For example, some PreSMo-antagonists against target proteins could serve as anti-cancer compounds (Kim et al., 2017a) and certain small molecule inhibitors can directly inhibit IDPs themselves (Follis et al., 2008; Metallo, 2010).
ACKNOWLEDGMENTS
This work was supported by a Korea-Hungary and Pan EU collaborative project from National Research Council of Science and Technology (NST) (NTM2231712) to KH
REFERENCES
- Andresen C., Helander S., Lemak A., Farès C., Csizmok V., Carlsson J., Penn L.Z., Forman-Kay J.D., Arrowsmith C.H., Lundström P., et al. Transient structure and dynamics in the disordered c-Myc transactivation domain affect Bin1 binding. Nucleic Acids Res. 2012;40:6353–6366. doi: 10.1093/nar/gks263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai M., Sugase K., Dyson H.J., Wright P.E. Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding. Proc Natl Acad Sci USA. 2015;112:9614–9619. doi: 10.1073/pnas.1512799112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal R.S., Xia J., Pinchev D., Taylor J., Epand R.M., Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- Baker J.M., Hudson R.P., Kanelis V., Choy W.Y., Thibodeau P.H., Thomas P.J., Forman-Kay J.D. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin R.L., Zimm B.H. Are denatured proteins ever random coils? Proc Natl Acad Sci U S A. 2000;97:12391–12392. doi: 10.1073/pnas.97.23.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benison G., Nyarko A., Barbar E. Heteronuclear NMR identifies a nascent helix in intrinsically disordered dynein intermediate chain: implications for folding and dimerization. J Mol Biol. 2006;362:1082–1093. doi: 10.1016/j.jmb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Benison G., Berkholz D.S., Barbar E. Protein assignments without peak lists using higher-order spectra. J Magn Reson. 2007;189:173–181. doi: 10.1016/j.jmr.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Bernadó P., Bertoncini C.W., Griesinger C., Zweckstetter M., Blackledge M. Defining long-range order and local disorder in native alpha-synuclein using residual dipolar couplings. J Am Chem Soc. 2005;127:17968–17969. doi: 10.1021/ja055538p. [DOI] [PubMed] [Google Scholar]
- Bibow S., Mukrasch M.D., Chinnathambi S., Biernat J., Griesinger C., Mandelkow E., Zweckstetter M. The Dynamic Structure of Filamentous Tau. Angew Chem Int Ed Engl. 2011;50:11520–11524. doi: 10.1002/anie.201105493. [DOI] [PubMed] [Google Scholar]
- Bienkiewicz E.A., Adkins J.N., Lumb K.J. Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27(Kip1) Biochemistry. 2002;41:752–759. doi: 10.1021/bi015763t. [DOI] [PubMed] [Google Scholar]
- Boettcher J.M., Hartman K.L., Ladror D.T., Qi Z., Woods W.S., George J.M., Rienstra C.M. Membrane-induced folding of the cAMP-regulated phosphoprotein endosulfine-alpha. Biochemistry. 2008;47:12357–12364. doi: 10.1021/bi801450t. [DOI] [PubMed] [Google Scholar]
- Boettcher J.M., Hartman K.L., Ladror D.T., Qi Z., Woods W.S., George J.M., Rienstra C.M. (1)H, (13)C, and (15)N resonance assignment of the cAMP-regulated phosphoprotein endosulfine-alpha in free and micelle-bound states. Biomol NMR Assign. 2007;1:167–169. doi: 10.1007/s12104-007-9063-7. [DOI] [PubMed] [Google Scholar]
- Borcherds W., Theillet F.X., Katzer A., Finzel A., Mishall K.M., Powell A.T., Wu H., Manieri W., Dieterich C., Selenko P., Loewer A., Daughdrill G.W. Disorder and residual helicity alter p53-Mdm2 binding affinity and signaling in cells. Nat Chem Biol. 2014;10:1000–1002. doi: 10.1038/nchembio.1668. [DOI] [PubMed] [Google Scholar]
- Borgia A., Borgia M.B., Bugge K., Kissling V.M., Heidarsson P.O., Fernandes C.B., Sottini A., Soranno A., Buholzer K.J., Nettels D., Kragelund B.B., Best R.B., Schuler B. Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018;555:61–66. doi: 10.1038/nature25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhis J.M., Johansson K., Receveur-Bréchot V., Oldfield C.J., Dunker K.A., Canard B., Longhi S. The C-terminal domain of measles virus nucleoprotein belongs to the class of intrinsically disordered proteins that fold upon bind to their physiological partner. Virus Res. 2004;99:157–167. doi: 10.1016/j.virusres.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Botuyan M.V., Momand J., Chen Y. Solution conformation of an essential region of the p53 transactivation domain. Fold Des. 1997;2:331–342. doi: 10.1016/S1359-0278(97)00047-3. [DOI] [PubMed] [Google Scholar]
- Braeuning A. The connection of β-catenin and phenobarbital in murine hepatocarcinogenesis: a critical discussion of Awuah et al., PLoS ONE 7, e39771. Arch Toxicol. 2013;87:401–402. doi: 10.1007/s00204-012-1002-4. [DOI] [PubMed] [Google Scholar]
- Buchko G.W., Ni S., Lourette N.M., Reeves R., Kennedy M.A. NMR resonance assignments of the human high mobility group protein HMGA1. J Biomol NMR. 2007;38:185. doi: 10.1007/s10858-006-9116-8. [DOI] [PubMed] [Google Scholar]
- Chang J., Kim D.H., Lee S.W., Choi K.Y., Sung Y.C. Transactivation ability of p53 transcriptional activation domain is directly related to the binding affinity to TATA-binding protein. J Biol Chem. 1995;270:25014–25019. doi: 10.1074/jbc.270.42.25014. [DOI] [PubMed] [Google Scholar]
- Chavali S., Gunnarsson A., Babu M.M. Intrinsically disordered proteins adaptively reorganize cellular matter during stress. Trends Biochem Sci. 2017;42:410–412. doi: 10.1016/j.tibs.2017.04.007. [DOI] [PubMed] [Google Scholar]
- Cheng Y., LeGall T., Oldfield C.J., Dunker A.K., Uversky V.N. Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry. 2006;45:10448–10460. doi: 10.1021/bi060981d. [DOI] [PubMed] [Google Scholar]
- Chi S.W., Lee S.H., Kim D.H., Ahn M.J., Kim J.S., Woo J.Y., Torizawa T., Kainosho M., Han K.H. Structural details on mdm2-p53 interaction. J Biol Chem. 2005;280:38795–38802. doi: 10.1074/jbc.M508578200. [DOI] [PubMed] [Google Scholar]
- Chi S.W., Kim D.H., Lee S.H., Chang I., Han K.H. Pre-structured motifs in the natively unstructured preS1 surface antigen of hepatitis B virus. Protein Sci. 2007;10:2108–2117. doi: 10.1110/ps.072983507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumakov P.M. Versatile functions of p53 protein in multicellular organisms. Biochemistry. 2007;72:1399–1421. doi: 10.1134/s0006297907130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csizmok V., Felli I.C., Tompa P., Banci L., Bertini I. Structural and dynamic characterization of intrinsically disordered human securin by NMR spectroscopy. J Am Chem Soc. 2008;130:16873–16879. doi: 10.1021/ja805510b. [DOI] [PubMed] [Google Scholar]
- Dancheck B., Nairn A.C., Peti W. Detailed structural characterization of unbound protein phosphatase 1 inhibitors. Biochemistry. 2008;47:12346–12356. doi: 10.1021/bi801308y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughdrill G.W., Chadsey M.S., Karlinsey J.E., Hughes K.T., Dahlquist F.W. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- Domanski M., Hertzog M., Coutant J., Gutsche-Perelroizen I., Bontems F., Carlier M.F., Guittet E., van Heijenoort C. Coupling of folding and binding of thymosin beta4 upon interaction with monomeric actin monitored by nuclear magnetic resonance. J Biol Chem. 2004;279:23637–23645. doi: 10.1074/jbc.M311413200. [DOI] [PubMed] [Google Scholar]
- Drysdale C.M., Dueñas E., Jackson B.M., Reusser U., Braus G.H., Hinnebusch A.G. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol. 1995;15:1220–1233. doi: 10.1128/mcb.15.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A.K., Babu M.M., Barbar E., Blackledge M., Bondosm S.E., Dosztányi Z., Dyson H.J., Forman-Kay J., Fuxreiter M., Gsponer J., Han K.H., Jones D.T., Longhi S., Metallo S.J., Nishikawa K., Nussinov R., Obradovic Z., Pappu R.V., Rost B., Selenko P., Subramaniam V., Sussman J.L., Tompa P., Uversky V.N. What’s in a name? Why these proteins are intrinsically disordered. Why these proteins are intrinsically disordered. Intrinsically Disordered Proteins. 2013;1:e24157. doi: 10.4161/idp.24157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A.K., Obradovic Z., Romero P., Garner E.C., Brown C.J. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- Dyson H.J., Wright P.E. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Eliezer D., Kutluay E., Bussell R., Jr, Browne G. Conformational properties of a-synuclein in its free and lipid-associated states. J Mol Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- Feuerstein S., Solyom Z., Aladag A., Favier A., Schwarten M., Hoffmann S., Willbold D., Brutscher B. Transient structure and SH3 interaction sites in an intrinsically disordered fragment of the hepatitis C virus protein NS5A. J Mol Biol. 2012;420:310–323. doi: 10.1016/j.jmb.2012.04.023. [DOI] [PubMed] [Google Scholar]
- Fletcher C.M., Wagner G. The interaction of eIF4E with 4E-BP1 is an induced fit to a completely disordered protein. Protein Sci. 1998;7:1639–1642. doi: 10.1002/pro.5560070720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follis A.V., Hammoudeh D.I., Wang H., Prochownik E.V., Metallo S.J. Structural rationale for the coupled binding and unfolding of the c-myc oncoprotein by small molecules. Chem Biol. 2008;15:1149–1155. doi: 10.1016/j.chembiol.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M., Simon I., Friedrich P., Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol. 2004;338:1015–1026. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Galea C.A., Wang Y., Sivakolundu S.G., Kriwacki R.W. Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits. Biochemistry. 2008;47:7598–7609. doi: 10.1021/bi8006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer M., Munte C.E., Schorr J., Kellner R., Kalbitzer H.R. Structure of the anchor-domain of myristoylated and non-myristoylated HIV-1 Nef protein. J Mol Biol. 1999;289:123–138. doi: 10.1006/jmbi.1999.2740. [DOI] [PubMed] [Google Scholar]
- Giniger E., Ptashne M. Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature. 1987;330:670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- Greenfield N.J., Kostyukova A.S., Hitchcock-DeGregori S.E. Structure and tropomyosin binding properties of the N-terminal capping domain of tropomodulin 1. Biophys J. 2005;88:372–383. doi: 10.1529/biophysj.104.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzard J., Südhof T.C., Rizo J. NMR analysis of the structure of synaptobrevin and of its interaction with syntaxin. J Biomol NMR. 1999;14:203–207. doi: 10.1023/a:1008382027065. [DOI] [PubMed] [Google Scholar]
- Hong W., Jiao W., Hu J., Zhang J., Liu C., Fu X., Shen D., Xia B., Chang Z. Periplasmic protein HdeA exhibits chaperone-like activity exclusively within stomach pH range by transforming into disordered conformation. J Biol Chem. 2005;280:27029–27034. doi: 10.1074/jbc.M503934200. [DOI] [PubMed] [Google Scholar]
- Hua Q.X., Jia W.H., Bullock B.P., Habener J.F., Weiss M.A. Transcriptional activator-coactivator recognition: nascent folding of kinase-inducible transcativation domain predicts its structure on coactivator binding. Biochemistry. 1998;37:5858–5866. doi: 10.1021/bi9800808. [DOI] [PubMed] [Google Scholar]
- Iešmantavičius V., Dogan J., Jemth P., Teilum K., Kjaergaard M. Helical propensity in an intrinsically disordered protein accelerates ligand binding. Angew Chem Int Ed Engl. 2014;53:1548–1551. doi: 10.1002/anie.201307712. [DOI] [PubMed] [Google Scholar]
- James T.L., Liu H., Ulyanov N.B., Farr-Jones S., Zhang H., Donne D.G., Kaneko K., Groth D., Mehlhorn I., Prusiner S.B., Cohen F.E. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.R., Houben K., Lescop E., Blanchard L., Ruigrok R.W., Blackledge M. Quantitative conformational analysis of partially folded proteins from residual dipolar couplings: application to the molecular recognition element of Sendai virus nucleoprotein. J Am Chem Soc. 2008;130:8055–8061. doi: 10.1021/ja801332d. [DOI] [PubMed] [Google Scholar]
- Jonker H.R., Wechselberger R.W., Boelens R., Folkers G.E., Kaptein R. Structural properties of the promiscuous VP16 activation domain. Biochemistry. 2005;25:827–839. doi: 10.1021/bi0482912. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Lee S.H., Chi S.W., Nam K.H., Han K.H. Backbone resonance assignment of a proteolysis-resistant fragment in the oxygen-dependent degradation domain of the hypoxia inducible factor 1α. Mol Cells. 2009a;27:493–496. doi: 10.1007/s10059-009-0065-4. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Lee C., Lee S.H., Kim K.T., Han J.J., Cha E.J., Lim J.E., Cho Y.J., Hong S.H., Han K.H. The Mechanism of p53 Rescue by SUSP4. Angew Chem Int Ed Engl. 2017a;56:1278–1282. doi: 10.1002/anie.201607819. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Lee S.H., Nam K.H., Chi S.W., Chang I., Han K.H. Multiple hTAF(II)31-binding motifs in the intrinsically unfolded transcriptional activation domain of VP16. BMB Rep. 2009b;42:411–417. doi: 10.5483/bmbrep.2009.42.7.411. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Lee C., Cho Y.J., Lee S.H., Cha E.J., Lim J.E., Sabo T.M., Griesinger C., Lee D., Han K.H. A pre-structured helix in the intrinsically disordered 4EBP1. Mol BioSyst. 2015;11:366–369. doi: 10.1039/c4mb00532e. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Wright A., Han K.H. An NMR study on the intrinsically disordered core transactivation domain of human glucocorticoid receptor. BMB Rep. 2017b;10:522–527. doi: 10.5483/BMBRep.2017.50.10.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künze G., Barré P., Scheidt H.A., Thomas L., Eliezer D., Huster D. Binding of the three-repeat domain of tau to phospholipid membranes induces an aggregated-like state of the protein. Biochim Biophys Acta. 2012;1818:2302–2313. doi: 10.1016/j.bbamem.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussie P.H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A.J., Pavletich N.P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Laptenki O., Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- Lavery D.N., McEwan I.J. Structural characterization of the native NH2-terminal transactivation domain of the human androgen receptor: a collapsed disordered conformation underlies structural plasticity and protein-induced folding. Biochemistry. 2008;47:3360–3369. doi: 10.1021/bi702221e. [DOI] [PubMed] [Google Scholar]
- Lee C., Kim D.H., Lee S.H., Su J., Han K.H. Structural investigation on the intrinsically disordered N-terminal region of HPV16 E7 protein. BMB Rep. 2016;49:431–436. doi: 10.5483/BMBRep.2016.49.8.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Mok K.H., Muhandiram R., Park K.H., Suk J.E., Kim D.H., Chang J., Sung Y.C., Choi K.Y., Han K.H. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- Lee K.H., Zhang P., Kim H.J., Mitrea D.M., Sarkar M., Freibaum B.D., Cika J., Coughlin M., Messing J., Molliex A., Maxwell B.A., Kim N.C., Temirov J., Moore J., Kolaitis R.M., Shaw T.I., Bai B., Peng J., Kriwacki R.W., Taylor J.P. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell. 2016;167:774–788. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Kim D.H., Han J.J., Cha E.J., Lim J.E., Cho Y.J., Lee C., Han K.H. Understanding pre-structured motifs (PreSMos) in intrinsically unfolded proteins. Curr Protein Pept Sci. 2012;13:34–54. doi: 10.2174/138920312799277974. [DOI] [PubMed] [Google Scholar]
- Liu H., Farr-Jones S., Ulyanov N.B., Llinas M., Marqusee S., Groth D., Cohen F.E., Prusiner S.B., James T.L. Solution Structure of Syrian Hamster Prion Protein rPrP(90–231) Biochemistry. 1999;38:5362–5377. doi: 10.1021/bi982878x. [DOI] [PubMed] [Google Scholar]
- Liang Y., Ye H., Kang C.B., Yoon H.S. Domain 2 of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfolded. Biochemistry. 2007;46:11550–11558. doi: 10.1021/bi700776e. [DOI] [PubMed] [Google Scholar]
- Lukhele S., Bah A., Lin H., Sonenberg N., Forman-Kay J.D. Interaction of the eukaryotic initiation factor 4E with 4E-BP2 at a dynamic bipartite interface. Structure. 2013;21:2186–2196. doi: 10.1016/j.str.2013.08.030. [DOI] [PubMed] [Google Scholar]
- Lum J.K., Neuweiler H., Fersht A.R. Long-range modulation of chain motions within the intrinsically disordered transavctivation of tumor supressor p53. J Am Chem Soc. 2012;124:1617–1622. doi: 10.1021/ja2078619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J.A., Singh V.K., Jia Z., Forman-Kay J.D. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: Implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo S.J. Intrinsically disordered proteins are potential drug targets. Curr Opin Chem Biol. 2010;14:481–488. doi: 10.1016/j.cbpa.2010.06.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midic U., Oldfield C.J., Dunker A.K., Obradovic Z., Uversky V.N. Protein disorder in the human diseasome: unfoldomics of human genetic diseases. BMC Genomics. 2009;10:S12. doi: 10.1186/1471-2164-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron S., Duchambon P., Blouquit Y., Durand D., Craescu C.T. The carboxy-terminal domain of xeroderma pigmentosum complementation group C protein, involved in TFIIH and centrin binding, is highly disordered. Biochemistry. 2008;47:1403–1413. doi: 10.1021/bi701863u. [DOI] [PubMed] [Google Scholar]
- Mittag T., Orlicky S., Choy W.Y., Tang X., Lin H., Sicheri F., Kay L.E., Tyers M., Forman-Kay J.D. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci USA. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan A., Oldfield C.J., Radivojac P., Vacic V., Cortese M.S., Dunker A.K., Uversky V.N. Analysis of Molecular Recognition Features (MoRFs) J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- Mukrasch M.D., Bibow S., Korukottu J., Jeganathan S., Biernat J., Griesinger C., Mandelkow E., Zweckstetter M. Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol. 2009;7:e34. doi: 10.1371/journal.pbio.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrali M.G., Schiavina M., Sainati V., Bermel W., Pierattelli R., Felli I.C. 13C APSY-NMR for sequential assignment of intrinsically disordered proteins. J Biomol NMR. 2018;70:167–175. doi: 10.1007/s10858-018-0167-4. [DOI] [PubMed] [Google Scholar]
- Neri D., Billeter M., Wider G., Wüthrich K. NMR determination of residual structure in a urea-denatured protein, the 434-repressor. Science. 1992;257:1559–1563. doi: 10.1126/science.1523410. [DOI] [PubMed] [Google Scholar]
- Newcombe E.A., Ruff K.M., Sethi A., Ormsby A.R., Ramdzan Y.M., Fox A., Purcell A.W., Gooley P.R., Pappu R.V., Hatters D.M. Tadpole-like conformations of huntingtin exon 1 are characterized by conformational heterogeneity that persists regardless of polyglutamine length. J Mol Biol. 2018;430:1442–1458. doi: 10.1016/j.jmb.2018.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noval M.G., Gallo M., Perrone S., Salvay A.G., Chemes L.B., de Prat-Gay G. Conformational dissection of a viral intrinsically disordered domain involved in cellular transformation. PLoS One. 2013;8:e72760. doi: 10.1371/journal.pone.0072760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hare P., Williams G. Structural studies of the acidic transactivation domain of the Vmw65 protein of herpes simplex virus using 1H NMR. Biochemistry. 1992;31:4150–4156. doi: 10.1021/bi00131a035. [DOI] [PubMed] [Google Scholar]
- Oldfield C.J., Cheng Y., Cortese M.S., Romero P., Uversky V.N., Dunker A.K. Coupled folding and binding with alpha-helix forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- Pavletich N.P. Mechanisms of cyclin-dependent kinase regulation: structures of cdks, their cyclin activators, and cip and INK4 inhibitors, J. Mol Biol. 1999;287:821–828. doi: 10.1006/jmbi.1999.2640. [DOI] [PubMed] [Google Scholar]
- Piai A., Calçada E.O., Tarenzi T., Grande A.D., Varadi M., Tompa P., Felli I.C., Pierattelli R. Just a Flexible Linker? The structural and dynamic properties of CBP-ID4 revealed by NMR spectroscopy. Biophys J. 2016;110:372–381. doi: 10.1016/j.bpj.2015.11.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P., Iakoucheva L.M., Oldfield C.J., Obradovic A., Uversky V.N., Dunker A.K. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan I., Pérez-Alvarado G.C., Parker D., Dyson H.J., Montminy M.R., Wright P.E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Pérez-Alvarado G.C., Dyson H.J., Wright P.E. Conformational preferences in the Ser133-phosphorylated and non-phosphorylated forms of the kinase inducible transactivation domain of CREB. FEBS Lett. 1998;430:317–322. doi: 10.1016/s0014-5793(98)00680-2. [DOI] [PubMed] [Google Scholar]
- Ramelot T.A., Gentile L.N., Nicholson L.K. Transient structure of the amyloid precursor protein cytoplasmic tail indicates preordering of structure for binding to cytosolic factors. Biochemistry. 2000;39:2714–2725. doi: 10.1021/bi992580m. [DOI] [PubMed] [Google Scholar]
- Reingewertz T.H., Benyamini H., Lebendiker M., Shalev D.E., Friedler A. The C-terminal domain of the HIV-1 Vif protein is natively unfolded in its unbound state. Protein Eng Des Sel. 2009;22:281–287. doi: 10.1093/protein/gzp004. [DOI] [PubMed] [Google Scholar]
- Rudolph M.G., Bayer P., Abo A., Kuhlmann J., Vetter I.R., Wittinghofer A. The Cdc42/Rac interactive binding region motif of the Wiskott Aldrich syndrome protein (WASP) is necessary but not sufficient for tight binding to Cdc42 and structure formation. J Biol Chem. 1998;273:18067–18076. doi: 10.1074/jbc.273.29.18067. [DOI] [PubMed] [Google Scholar]
- Salamanova E., Costeira-Paulo J., Han K.H., Kim D.H., Nilsson L., Wright A.P.H. A subset of functional adaptation mutations alter propensity for α-helical conformation in the intrinsically disordered glucocorticoid receptor tau1core activation domain. Biochim Biophys Acta. 2018;1862:1452–1461. doi: 10.1016/j.bbagen.2018.03.015. [DOI] [PubMed] [Google Scholar]
- Sayers E.W., Gerstner R.B., Draper D.E., Torchia D.A. Structural preordering in the N-terminal region of ribosomal protein S4 revealed by heteronuclear NMR spectroscopy. Biochemistry. 2000;39:13602–13613. doi: 10.1021/bi0013391. [DOI] [PubMed] [Google Scholar]
- Semenza G.L. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. Principles of tumor suppression. Cell. 2004;116:235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- Sólyom Z., Ma P., Schwarten M., Bosco M., Polidori A., Durand G., Willbold D., Brutscher B. The Disordered Region of the HCV Protein NS5A: Conformational Dynamics, SH3 Binding, and Phosphorylation. Biophys J. 2015;109:1483–1496. doi: 10.1016/j.bpj.2015.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugase K., Dyson H.J., Wright P.E. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- Sung Y.H., Eliezer D. Residual structure, backbone dynamics, and interactions within the synuclein family. J Mol Biol. 2007;372:689–707. doi: 10.1016/j.jmb.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapar R., Mueller G.A., Marzluff W.F. The N-terminal domain of the Drosophila histone mRNA binding protein, SLBP, is intrinsically disordered with nascent helical structure. Biochemistry. 2004;43:9390–9400. doi: 10.1021/bi036314r. [DOI] [PubMed] [Google Scholar]
- To V., Dzananovic E., McKenna S.A., O’Neil J. The Dynamic Landscape of the Full-Length HIV-1 Transactivator of Transcription. Biochemistry. 2016;55:1314–1325. doi: 10.1021/acs.biochem.5b01178. [DOI] [PubMed] [Google Scholar]
- Uesugi M., Nyanguile O., Lu H., Levine A.J., Verdine G.L. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- Uversky V.N. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015;282:1182–1189. doi: 10.1111/febs.13202. [DOI] [PubMed] [Google Scholar]
- Uversky V.N., Dunker A.K. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K., Fuxreiter M., Gough J., Gsponer J., Jones D.T., Kim P.M., Kriwacki R.W., Oldfield C.J., Pappu R.V., Tompa P., Uversky V.N., Wright P.E., Babu M.M. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoy M., Leuther K.K., Kodadek T., Johnston S.A. The acidic activation domains of the GCN4 and GAL4 proteins are not alpha helical but form beta sheets. Cell. 1993;72:587–594. doi: 10.1016/0092-8674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- Wells M., Tidow H., Rutherford T.J., Markwick P., Jensen M.R., Mylonas E., Svergun D.I., Blackledge M., Fersht A.R. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc Natl Acad Sci USA. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Ye H., Osman N.E., Sadler K., Won E.Y., Chi S.W., Yoon H.S. The MDM2-binding region in the transactivation domain of p53 also acts as a Bcl-X(L)-binding motif. Biochemistry. 2009;48:12159–12168. doi: 10.1021/bi901188s. [DOI] [PubMed] [Google Scholar]
- Zhang X., Perugini M.A., Yao S., Adda C.G., Murphy V.J., Low A., Anders R.F., Norton R.S. Solution conformation, backbone dynamics and lipid interactions of the intrinsically unstructured malaria surface protein MSP2. J Mol Biol. 2008;379:105–121. doi: 10.1016/j.jmb.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Georgieva B., Chabes A., Domkin V., Ippel J.H., Schleucher J., Wijmenga S., Thelander L., Rothstein R. Mutational and structural analyses of the ribonucleotide reductase inhibitor Sml1 define its Rnr1 interaction domain whose inactivation allows suppression of mec1 and rad53 lethality. Mol Cell Biol. 2000;23:9076–9083. doi: 10.1128/mcb.20.23.9076-9083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Ma D., Yahr T.L., Chen L. The transiently ordered regions in intrinsically disordered ExsE are correlated with structural elements involved in chaperone vinding. Biochem Biophys Res Commun. 2012;417:129–134. doi: 10.1016/j.bbrc.2011.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitzewitz J.A., Ibarra-Molero B., Fishel D.R., Terry K.L., Matthews C.R. Preformed secondary structure drives the association reaction of GCN4-p1, a model coiled-coil system. J Mol Biol. 2000;296:1105–1116. doi: 10.1006/jmbi.2000.3507. [DOI] [PubMed] [Google Scholar]
- Zor T., Mayr B.M., Dyson H.J., Montminy M.R., Wright P.E. Roles of phosphorylation and helix propensity in the binding of the KIX domain of CREB-binding protein by constitutive (c-Myb) and inducible (CREB) activators. J Biol Chem. 2002;277:42241–42248. doi: 10.1074/jbc.M207361200. [DOI] [PubMed] [Google Scholar]