Abstract
Background
The purpose of this study is to discuss whether genetic variants (rs2230199, rs1047286, rs2230205, and rs2250656) in the C3 gene account for a significant risk of advanced AMD.
Methods
We performed a meta-analysis using electronic databases to search relevant articles. A total of 40 case-control studies from 38 available articles (20,673 cases and 20,025 controls) were included in our study.
Results
In our meta-analysis, the pooled results showed that the carriage of G allele for rs2230199 and the T allele for rs1047286 had a tendency to the risk of advanced AMD (OR = 1.49, 95% CI = 1.39–1.59, P < 0.001; OR = 1.45, 95% CI = 1.37–1.54, P < 0.001). Moreover, in the subgroup analysis based on ethnicity, rs2230199 and rs1047286 polymorphisms were more likely to be a predictor of response for Caucasian region (OR = 1.48, 95% CI = 1.38–1.59, P < 0.001; OR = 1.45, 95% CI = 1.37–1.54, P < 0.001). Besides, pooled results suggested that the G allele of rs2230199 could confer susceptibility to advanced AMD in Middle East (OR = 1.62, 95% CI = 1.33–1.97, P < 0.001).
Conclusion
In our meta-analysis, C3 genetic polymorphisms unveiled a positive effect on the risk of advanced AMD, especially in Caucasians. Furthermore, numerous well-designed studies with large sample-size are required to validate this conclusion.
Electronic supplementary material
The online version of this article (10.1186/s12886-018-0945-5) contains supplementary material, which is available to authorized users.
Keywords: Age-related macular degeneration, C3 gene, Polymorphism, Meta-analysis
Background
Age-related macular degeneration (AMD) is a complex and progressive retinal disorder influenced by family history, aging, race, smoking and diet, which caused irreversible visual impairment in a growing number of elderly persons [1, 2]. The early stage of AMD is characterized by pigmentary abnormalities of the retinal pigment epithelium (RPE) and extracellular deposits called drusen under the retina [3]. As the condition progresses, two advanced forms of this disease are developed: extensive pigment epithelium atrophy (geographic atrophy or dry AMD) or subretinal choroidal neovascular membrane (exudative or wet AMD). Although constituting only 10–15% of all AMD cases, advanced forms account for nearly 80% of AMD-related blindness in western countries [4]. It has been reported that the prevalence of advanced AMD is estimated at 3% in people aged > 65 years old, rising to 11% in those > 85 years old in developed world [5]. While, the pooled prevalence of advanced AMD is 0.56% among aged 40–79 years in Asian countries [6].
Advanced AMD has been implicated with important risk factors listed above, it is a multifactorial disease which influenced by a combination of environmental and genetic susceptibility [1, 3, 7, 8]. Although the well-defined pathogeny of advanced AMD remains to be unresolved, genetic association studies have provided consequential insights into the molecular basis of advanced AMD. Several genes at chromosomal loci 1q32 and 10q26, involving in inflammation and complement activation pathway, have been plausible candidate, as supported by the laboratory research in vitro and vivo that inflammation and immune response related proteins were found in drusen [9–11]. So far, the strongest genetic association has been identified on 1q32 with single nucleotide polymorphisms (SNPs) in complement factor H (CFH) gene by candidate region and whole genome association analyses [12, 13].
Apart from CFH, the central element of the complement cascade, complement component C3a has been interconnected with the vascular endothelial growth factor expression, geographic atrophy, retinal pigment epithelium deterioration, and progression to choroidal neovascularization [11, 14, 15]. These studies strongly indicated that aberrant regulation or activation of the complement pathway confer susceptibility to the main mechanism of advanced AMD. As the main regulator of the alternative complement pathway, several genetic variants in C3 gene have been investigated with advanced AMD in different ethnic groups, the pooled results are incompatible and ambiguous. According to the International HapMap Project database, the human C3 gene is located on chromosome 19 and exhibits nine common genetic SNPs (rs2230199, rs1047286, rs2241394, rs2250656, rs344542, rs2230205, rs339392, rs3745565, and rs11569536). Used for screening the electronic database and manual searching, the most widely condidate polymorphisms of the C3 gene which at least has been surveyed in three pertinent studies are rs2230199, rs1047286, rs2230205 and rs2250656. In order to better understand the genetic risk of C3 gene in the relationship with exudative AMD, we performed a meta-analysis to illuminate this association and determine whether the genetic variants of C3 gene conferred susceptibility to advanced AMD.
Materials and methods
Literature search
A systematic search of electronic database such as PubMed, Embase, CNKI, Cochrane library and Web of Science was conducted with the following keywords: (“AMD” or “maculopathy” or “macular degeneration” or “age-related maculopathy” or “age-related macular degeneration”) and (“complement 3” or “complement C3” or “C3” or “complement component 3”) and (“variant” or “mutation” or “genetic” or “SNP” or “polymorphism” or “genetic polymorphism” or “genetic variant” or “single nucleotide polymorphism”). Each database was thoroughly scanned and was up to date as of September 1 2018. Our meta-analysis was mainly focused on case-control studies, without any language limitation imposed in the literature searching.
Study selection
Retrieved articles were considered eligible for our meta-analysis when they met the following inclusion criteria: (1) investigating the disease risk of C3 polymorphism with advanced AMD; (2) detailed genetyping data for each site could be acquired to estimate the odds ratio (OR) and 95% confidence interval (CI) based on genetic model contrast; (3) individual for all selected samples met the modified version of the age-related eye disease study (AREDS) grading system as described elsewhere. Major exclusion criteria were limited to several items as follow: (1) overlapping subjects in several articles for the same research group; (2) only focused on families’ individuals rather than sporadic advanced AMD patients; (3) abstract from conferences, letters, review articles and case reports. When several articles included some of the same samples, the one with largest individuals and thorough genotype information would be winnowed for our meta-analysis.
Data extraction
Data from the retrieved studies were extracted independently by two reviewers (J.Z. and S.L.). The following items obtained from each eligible articles included: the first author, the year of publication, country and ethnicity of subjects, information on study design, sample size, genotyping methods and distribution in case and control groups. Two authors carefully inspected the raw statistics and reached a consensus in all aspects. If any disagreement still existed, the third author (S.H.) would be invited to chew over current controversy and resolve the dispute.
Quality assessment
Quality assessment of the screened studies was also independently conducted by two reviewers (J.Z. and S.L.) in the basis of the HuGENet Handbook [16]. A total of six bias assessment items were refined to investigate the relationship between genes and diseases from this handbook, including bias in selection of cases, bias in selection of controls, bias in genotyping cases, bias in genotyping controls, bias in population stratification, confounding bias, multiple tests, and selective outcome reports. The quality evaluation of every items for extracted articles was defined as “Yes” or“No”. Separately, “Unclear” was designated if there was not enough information to make a decision. A series of corrections and judgements were performed independently by another coauthor (S.H.) if debate still lasted in the assessment. Consensus referring all items was achieved after discussion.
Statistical analysis
Allele and genotype frequency of each C3 polymorphic site were counted between cases and healthy controls. The genetic strength association including pooled ORs and 95% CIs was assessed using different genetic models, including allele model (A vs. a), homozygote model (AA vs. aa), heterozygote model (Aa vs. aa), dominant (AA+Aa vs. aa), recessive (AA vs. Aa+aa). The heterogeneity assumption between studies was estimated and evaluated by Cochran’s Q statistic as well as the I2 statistic. The result that our P value of Q statistic was less than 0.05 or the I2 value was greater than 50% suggested apparent heterogeneity, thus a random-effect model was utilized in our model analysis. Otherwise, the fixed-effect model was performed [17]. Sensitivity analyses was conducted to assess the effect of each study and the stability of the pooled ORs by removing included study in turn from the compiled list. Begg’s funnel plots [18] and Egger’s regression test [19] were furthered to detect the potential publication bias. All statistical analysis using two-sided P values was executed by STATA 12.0 software (StataCorp LP, College Station, Texas, USA). A significant difference was estimated under the level of 0.05. The final results needed to be tested and verified by two authors (J.Z. and S.L.) respectively.
Results
Overall characteristics of selected studies and quality assessment
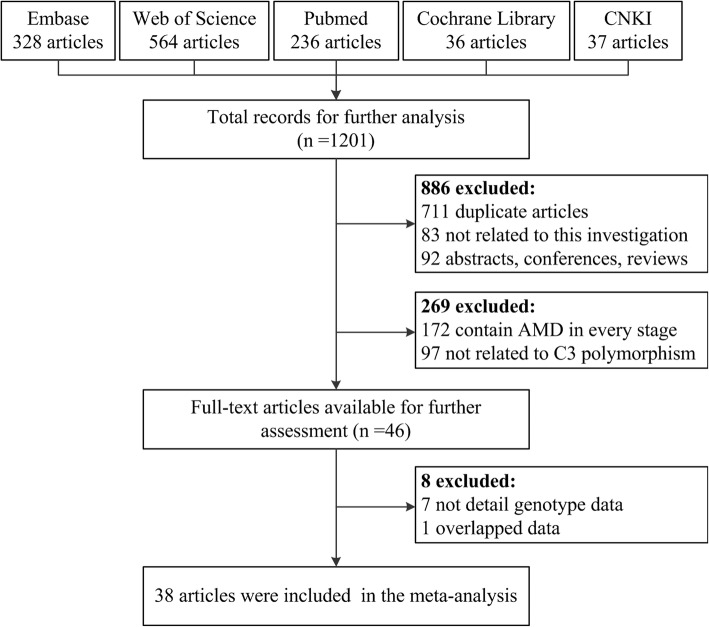
The flow diagram for literature searching is summarized in Fig. 1. A total of 1201 articles from the five databases (Additional file 1) were filtered by our search method. Of which, 886 studies were excluded for the three aspects: (1) 711 duplicated articles; (2) 83 articles not related to the theme of this investigation; (3) 92 articles mainly referred to abstract, conference, review, and case report. Through our rigorous inspection, 269 ones in the rest of articles were stroke out. Of them, 172 articles were focused on the every stage of AMD but not advanced AMD, 97 articles were not concerned with the association between advanced AMD and C3 genetic polymorphism. The other 46 full-text articles were left in our meta-analysis. Seven of them did not have detailed genotype data after cautiously reading the included literatures.Besides, two papers were investigated by the same author and the same batch of patients from Iran [20, 21]. We decided to choose the one which had larger samples size and more comprehensive directions. Finally, 40 case-control studies regarding the association of C3 gene with advanced AMD from 38 available publications were generally contained in our current meta-analysis [4, 20, 22–57]. The common characteristics of each article are generally showed in Table 1. As listed in the table, 30 studies from Caucasian region, 7 studies from East Asian group and 3 studies from Middle East have been chosen in our meta-analysis. The genotyping methods for our whole sample are distinct and the results could be validated in different ways.
Fig. 1.
Flow diagram presenting the result of literature searching process in meta-analysis
Table 1.
The General Characteristics of All Studies Included in our Meta-Analysis
| Refs | Year | Country | Ethnicity | Case/Control | Mean age of AMD | Mean age of control | Typing teaching | Study design |
|---|---|---|---|---|---|---|---|---|
| Yate et al. [54] | 2007 | UK | Caucasian | 603/350 | 79.4 ± 7.2 | 75.3 ± 7.8 | SNaPshot | Sex-,age-,ethnic-,matched |
| Yate et al. [54] | 2007 | Scotland | Caucasian | 244/351 | 77.8 ± 9.2 | 78.0 ± 8.5 | TaqMan | Sex-,age-,ethnic-,matched |
| Maller et al. [41] | 2007 | America | Caucasian | 1238/934 | NA | NA | MALDI-TOF MS | Age-,ethnic-, matched |
| Edwards et al. [31] | 2008 | America | Caucasian | 444/300 | NA | NA | Illumina GoldenGate | Age-,ethnic-, matched |
| Spencer et al. [50] | 2008 | America | Caucasian | 286/701 | 76.5 ± 7.7 | 66.9 ± 8.3 | TaqMan | Sex-,age-,ethnic-,matched |
| Scholl et al. [4] | 2009 | German | Caucasian | 99/612 | 71.8 ± 7.4 | 76.2 ± 5.3 | MALDI-TOF MS | Sex-,age-,ethnic-,matched |
| Francis et al. [32] | 2009 | America | Caucasian | 211/187 | 79 | 74 | Sequencing | Age-,ethnic-, matched |
| Park et al. [44] | 2009 | America | Caucasian | 898/599 | 80.6 ± 5.0 | 77.6 ± 4.3 | Illumina GoldenGate | Sex-,age-,ethnic-,matched |
| Despriet et al. [30] | 2009 | Netherlands | Caucasian | 268/173 | 78.7 ± 7.7 | 74.1 ± 6.3 | TaqMan | Sex-,age-,ethnic-,matched |
| Bergeron et al. [22] | 2009 | America | Caucasian | 421/215 | 64.8 | 66.5 | TaqMan | Sex-,age-,ethnic-,matched |
| Reynolds et al. [47] | 2009 | America | Caucasian | 120/60 | 82.0 ± 6.9 | 79.0 ± 4.4 | MALDI-TOF MS | Sex-,age-,ethnic-,matched |
| Pei et al.[45] | 2009 | China | East Asian | 123/130 | 70.6 ± 8.2 | 69.2 ± 10.1 | MALDI-TOF MS | Sex-,age-,ethnic-, matched |
| Cui et al. [29] | 2010 | China | East Asian | 150/161 | 66.6 ± 8.4 | 65.7 ± 7.8 | PCR-RFLP/ Sequencing | Sex-,age-,ethnic-, matched |
| Zerbib et al. [56] | 2010 | France | Caucasian | 1080/406 | 79.0 ± 7.4 | 67.8 ± 7.7 | TaqMan | Sex-,age-,ethnic-, matched |
| McKay et al. [43] | 2010 | Northern Ireland | Caucasian | 437/436 | 77.6 | 74.9 | SNaPshot | Sex-,age-,ethnic-, matched |
| Chen et al. [25] | 2010 | America | Caucasian | 2157/1150 | 78.6 | 74.1 | Illumina GoldenGate | Sex-,age-,ethnic-, matched |
| Kopplin et al. [37] | 2010 | America | Caucasian | 377/161 | NA | NA | Affymetrix GeneChip | Age-,ethnic-, matched |
| Liu et al. [39] | 2010 | China | East Asian | 158/220 | 64.0 ± 6.6 | 63.0 ± 7.8 | SNaPshot | Sex-,age-,ethnic-, matched |
| Yu et al. [55] | 2011 | America | Caucasian | 1082/221 | 79.5 ± 5.5 | 77.0 ± 4.6 | MALDI-TOF MS | Sex-,age-,ethnic-, matched |
| Chen et al. [26] | 2011 | America | Caucasian | 1335/509 | 70.2 ± 5.1 | 67.0 ± 4.3 | SNaPshot | Sex-,age-,ethnic-, matched |
| Hageman et al. [33] | 2011 | America | Caucasian | 1132/822 | 76.5 ± 7.1 | 76.4 ± 7.3 | NA | Sex-,age-,ethnic-, matched |
| Peter et al. [46] | 2011 | America | Caucasian | 48/1260 | NA | NA | TaqMan | Age-,ethnic-, matched |
| Yanagisawa et al. [53] | 2011 | Japan | East Asian | 420/197 | 74.0 ± 7.5 | 72.0 ± 6.0 | TaqMan | Sex-,age-,ethnic-, matched |
| Martinez et al. [42] | 2012 | Spain | Caucasian | 259/191 | NA | NA | SNaPshot | Age-,ethnic-, matched |
| Smailhodzic et al. [49] | 2012 | Netherlands | Caucasian | 197/150 | NA | NA | Sequencing | Sex-,age-,ethnic-, matched |
| Buentello et al. [23] | 2012 | Mexico | Caucasian | 159/152 | 76.4 ± 8.1 | 73.5 ± 6.8 | PCR-RFLP | Sex-,age-,ethnic-, matched |
| Tian et al. [51] | 2012 | China | East Asian | 535/469 | NA | NA | MALDI-TOF MS | Age-,ethnic-, matched |
| Losonczy et al. [40] | 2012 | Hungary | Caucasian | 275/106 | 76.0 ± 7.3 | 79.1 ± 6.1 | PCR-RFLP | Sex-,age-,ethnic-, matched |
| Cipriani et al. [27] | 2012 | UK | Caucasian | 893/2199 | 78.6 ± 7.5 | NA | Illumina BeadChip | Sex-,age-,ethnic-, matched |
| Jaouni et al. [36] | 2012 | Israel | Middle East | 317/159 | 78.1 ± 7.6 | 70.8 ± 8.2 | PCR-RFLP | Sex-,age-,ethnic-, matched |
| Wu et al. [52] | 2013 | China | East Asian | 165/216 | 69.4 ± 10 | 64.5 ± 8.0 | TaqMan | Sex-,age-,ethnic-, matched |
| Helgason et al. [35] | 2013 | Iceland | Caucasian | 1107/2869 | NA | NA | Illumina BeadChip | Age-,ethnic-, matched |
| Helgason et al. [35] | 2013 | America | Caucasian | 1525/1288 | NA | NA | Illumina BeadChip | Age-,ethnic-, matched |
| Contreras et al. [28] | 2014 | Mexico | Caucasian | 273/201 | 76.0 ± 8.0 | 65.5 ± 9.8 | TaqMan | Sex-,age-,ethnic-, matched |
| Caire et al. [24] | 2014 | Spain | Caucasian | 154/141 | 75.4 ± 7.2 | 78.5 ± 7.2 | SNaPshot | Sex-,age-,ethnic-, matched |
| Liu et al. [38] | 2014 | China | East Asian | 200/275 | 75.3 ± 7.7 | 74.3 ± 7.6 | TaqMan | Sex-,age-,ethnic-, matched |
| Hautamaki et al. [34] | 2015 | Finland | Caucasian | 301/119 | NA | NA | Sequencing | Age-,ethnic-, matched |
| Saksens et al. [48] | 2016 | Netherlands | Caucasian | 571/900 | 76.6 ± 8.5 | 71.3 ± 6.7 | KASP | Sex-,age-,ethnic-, matched |
| Bonyadi et al. [20, 21] | 2017 | Iran | Middle East | 266/228 | 76.4 ± 7.6 | 72.7 ± 6.8 | PCR-RFLP | Sex-,age-,ethnic-, matched |
| Habibi et al. [57] | 2017 | Tunisia | Middle East | 145/207 | 73.1 ± 8.1 | NA | PCR-SSP | Age-,ethnic-, matched |
Bias assessment of the included studies
Overall results in Table 2 primarily expound the evaluation of potential sources of bias in our included studies. Overall, the quality of the included studies was consistently absolute. Of the studies, there was no obvious bias in the selection of cases and controls, genotyping controls, population stratification, confounding bias, multiple tests, or selective outcome reports.
Table 2.
Assessment of potential bias in included studies
| Year | First author | Bias in selection of cases | Bias in selection of controls | Bias in genotyping controls | Bias in population stratification | Confounding bias | Multiple test and Selective outcome reports |
|---|---|---|---|---|---|---|---|
| 2007 | Yate et al. [54] | NO | NO | NO | NO | NO | NO |
| 2007 | Maller et al. [41] | NO | NO | NO | NO | NO | NO |
| 2008 | Edwards et al. [31] | NO | NO | NO | NO | NO | NO |
| 2008 | Spencer et al. [50] | NO | NO | NO | NO | NO | NO |
| 2009 | Scholl et al. [4] | NO | NO | NO | NO | NO | NO |
| 2009 | Francis et al. [32] | NO | NO | NO | NO | NO | NO |
| 2009 | Park et al. [44] | NO | NO | NO | NO | NO | NO |
| 2009 | Despriet et al. [30] | NO | NO | NO | NO | NO | NO |
| 2009 | Bergeron et al. [22] | NO | NO | NO | NO | NO | NO |
| 2009 | Reynolds et al. [47] | NO | NO | NO | NO | NO | NO |
| 2009 | Pei et al. [45] | NO | NO | NO | NO | NO | NO |
| 2010 | Cui et al. [29] | NO | NO | NO | NO | NO | NO |
| 2010 | Zerbib et al. [56] | NO | NO | NO | NO | NO | NO |
| 2010 | McKay et al. [43] | NO | NO | NO | NO | NO | NO |
| 2010 | Chen et al. [25] | NO | NO | NO | NO | NO | NO |
| 2010 | Kopplin et al. [37] | NO | NO | NO | Unclear | NO | NO |
| 2010 | Liu et al. [39] | NO | NO | NO | NO | NO | NO |
| 2011 | Yu et al. [55] | NO | NO | NO | NO | NO | NO |
| 2011 | Chen et al. [26] | NO | NO | NO | NO | NO | NO |
| 2011 | Hageman et al. [33] | NO | NO | NO | Unclear | NO | NO |
| 2011 | Peter et al. [46] | Yes | NO | NO | Unclear | NO | NO |
| 2011 | Yanagisawa et al. [53] | NO | NO | NO | NO | NO | NO |
| 2012 | Martinez et al. [42] | NO | NO | NO | NO | NO | NO |
| 2012 | Smailhodzic et al. [49] | NO | NO | NO | NO | NO | NO |
| 2012 | Buentello et al. [23] | NO | NO | NO | NO | NO | NO |
| 2012 | Tian et al. [51] | NO | NO | NO | NO | NO | NO |
| 2012 | Losonczy et al. [40] | NO | NO | NO | NO | NO | NO |
| 2012 | Cipriani et al. [27] | NO | Yes | NO | NO | NO | NO |
| 2012 | Jaouni et al. [36] | NO | NO | NO | NO | NO | NO |
| 2013 | Wu et al. [52] | NO | NO | NO | NO | NO | NO |
| 2013 | Helgason et al. [35] | NO | NO | NO | NO | NO | NO |
| 2014 | Contreras et al. [28] | NO | NO | NO | NO | NO | NO |
| 2014 | Caire et al. [24] | NO | NO | NO | NO | NO | NO |
| 2014 | Liu et al. [38] | NO | NO | NO | NO | NO | NO |
| 2015 | Hautamaki et al. [34] | NO | NO | NO | NO | NO | NO |
| 2016 | Saksens et al. [48] | NO | NO | NO | NO | NO | NO |
| 2017 | Bonyadi et al. [20, 21] | NO | NO | NO | NO | NO | NO |
| 2017 | Habibi et al. [57] | NO | NO | NO | NO | NO | NO |
Relationship of C3 gene polymorphisms with advanced AMD susceptibility
Several genetic models for C3 polymorphisms including rs2230199, rs1047286, rs2230205, rs2250656 were used in our meta-analysis and the combined results are presented in Table 3. Briefly, 36 studies discussed the association of rs2230199 with advanced AMD, 13 studies investigated the relationship between rs1047286 and advanced AMD, 5 studies referred to rs2230205, rs2250656, respectively.
Table 3.
Main Results of Pooled ORs and Analysis of C3 gene polymorphism with advanced AMD in our Meta-Analysis
| Subgroup | No. of studies | No. of patients | Allele model | Homozygote model | Heterozygote model | Dominant model | Recessive model | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Control | OR(95% CI) | P | OR(95% CI) | P | OR(95% CI) | P | OR(95% CI) | P | OR(95% CI) | P | ||
| C3 rs2230199 (Associated allele vs. Reference allele: G vs. C) | |||||||||||||
| Overall | 36 | 34,805 | 29,499 | 1.49 (1.39,1.59) | < 0.001 | 2.33 (1.98,2.74) | < 0.001 | 1.53 (1.41,1.64) | < 0.001 | 1.62 (1.51,1.74) | < 0.001 | 1.99 (1.70,2.34) | < 0.001 |
| Caucasian | 28 | 31,372 | 26,130 | 1.48 (1.38,1.59) | < 0.001 | 2.20 (1.87,2.60) | < 0.001 | 1.55 (1.43,1.67) | < 0.001 | 1.63 (1.51,1.75) | < 0.001 | 1.88 (1.59,2.21) | < 0.001 |
| East Asian | 5 | 2122 | 2388 | 1.11 (0.56,2.20) | 0.76 | – | – | 1.32 (0.91,1.93) | 0.144 | 1.49 (1.04,2.15) | 0.032 | 5.60 (1.57,19.9) | – |
| Middle East | 3 | 1311 | 981 | 1.62 (1.33,1.97) | < 0.001 | – | – | 1.07 (0.66,1.73) | 0.798 | 1.49 (0.95,2.34) | 0.085 | 25.5 (3.35,194) | – |
| C3 rs1047286 (Associated allele vs. Reference allele: T vs. C) | |||||||||||||
| Overall | 13 | 16,232 | 16,222 | 1.45 (1.37,1.54) | < 0.001 | 2.06 (1.56,2.72) | < 0.001 | 1.72 (1.51,1.96) | < 0.001 | 1.76 (1.56,2.00) | < 0.001 | 1.71 (1.30,2.24) | < 0.001 |
| Caucasian | 10 | 14,688 | 14,548 | 1.45 (1.37,1.54) | < 0.001 | 2.06 (1.56,2.72) | < 0.001 | 1.72 (1.50,1.96) | < 0.001 | 1.76 (1.55,2.00) | < 0.001 | 1.71 (1.30,2.24) | < 0.001 |
| East Asian | 3 | 1544 | 1674 | 1.75 (0.49,6.29) | 0.388 | – | – | 2.06 (0.38,11.3) | 0.404 | 2.06 (0.38,11.3) | 0.404 | – | – |
| C3 rs2230205 (Associated allele vs. Reference allele: A vs. G) | |||||||||||||
| Overall | 5 | 3302 | 2732 | 0.99 (0.89,1.11) | 0.903 | 1.04 (0.77,1.42) | 0.780 | 1.00 (0.80,1.23) | 0.967 | 1.00 (0.81,1.22) | 0.992 | 1.06 (0.81,1.37) | 0.687 |
| Caucasian | 1 | 880 | 598 | 0.90 (0.66,1.23) | 0.507 | 0.45 (0.12,1.60) | 0.215 | 0.98 (0.69,1.39) | 0.902 | 0.93 (0.66,1.32) | 0.699 | 0.45 (0.13,1.60) | 0.217 |
| East Asian | 4 | 2422 | 2134 | 1.01 (0.89,1.14) | 0.903 | 1.10 (0.80,1.51) | 0.546 | 1.01 (0.77,1.32) | 0.967 | 1.04 (0.80,1.33) | 0.787 | 1.10 (0.84,1.43) | 0.497 |
| C3 rs2250656 (Associated allele vs. Reference allele: G vs. A) | |||||||||||||
| Overall | 5 | 3278 | 2632 | 0.90 (0.75,1.08) | 0.257 | 0.76 (0.49,1.16) | 0.207 | 0.78 (0.65,0.95) | 0.014 | 0.78 (0.65,0.94) | 0.010 | 0.83 (0.55,1.27) | 0.391 |
| Caucasian | 1 | 874 | 512 | 0.82 (0.64,1.05) | 0.117 | 0.77 (0.42,1.42) | 0.407 | 0.76 (0.55,1.05) | 0.097 | 0.76 (0.56,1.04) | 0.085 | 0.87 (0.48,1.57) | 0.642 |
| East Asian | 4 | 2404 | 2120 | 0.92 (0.73,1.16) | 0.486 | 0.74 (0.41,1.37) | 0.340 | 0.80 (0.63,1.02) | 0.068 | 0.79 (0.63,1.00) | 0.052 | 0.80 (0.44,1.45) | 0.456 |
Association between SNP rs2230199 of C3 gene and advanced AMD
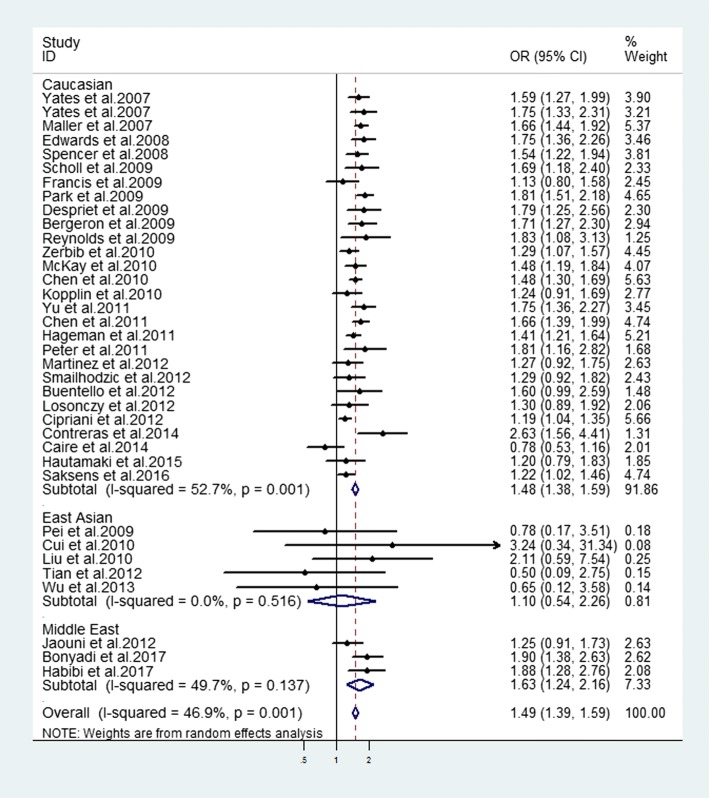
As shown in Table 3, there was a significant association between the rs2230199 SNP and advanced AMD susceptibility in the overall populations (allelic model: OR = 1.49, 95% CI = 1.39–1.59, P < 0.001; homozygote model: OR = 2.33, 95% CI = 1.98–2.74, P < 0.001; heterozygote model: OR = 1.53, 95% CI = 1.41–1.64, P < 0.001; dominant model: OR = 1.62, 95% CI = 1.51–1.74, P < 0.001; recessive model: OR = 1.99, 95% CI = 1.70–2.34, P < 0.001). Moreover, the subgroup analysis straitified by ethnicity indicated that rs2230199 conferred obvious susceptibility to advanced AMD in the group of Caucasian in allelic (OR = 1.48, 95% CI = 1.38–1.59, P < 0.001) (Fig. 2), homozygote (OR = 2.20, 95% CI = 1.87–2.60, P < 0.001), heterozygote (OR = 1.55, 95% CI = 1.43–1.67, P < 0.001), dominant (OR = 1.63, 95% CI = 1.51–1.75, P < 0.001), recessive (OR = 1.88, 95% CI = 1.59–2.21, P < 0.001) models (Table 3). Besides, the allelic comparison yielded a positive correlation in Middle East group (OR = 1.62, 95% CI = 1.33–1.97, P < 0.001). However, this relationship was not significant in East Asian group for any genetic models (Table 3).
Fig. 2.
Evaluation of the association between C3 genetic polymorphism (rs2230199) with advanced AMD
Association between SNP rs1047286 of C3 gene and advanced AMD
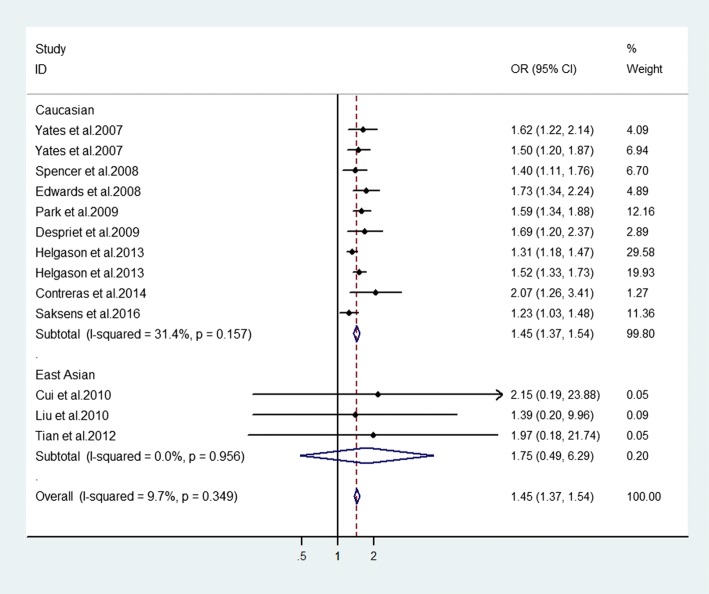
Significant association between this SNP and advanced AMD was confirmed in the overall populations (allelic model: OR = 1.45, 95% CI = 1.37–1.54, P < 0.001; homozygote model: OR = 2.06, 95% CI = 1.56–2.72, P < 0.001; heterozygote model: OR = 1.72, 95% CI = 1.51–1.96, P < 0.001; dominant model: OR = 1.76, 95% CI = 1.56–2.00, P < 0.001; recessive model: OR = 1.71, 95% CI = 1.30–2.24, P < 0.001). In subgroup analysis stratified by ethnicity, our meta-analysis indicated significant correlation of rs1047286 with advanced AMD in the five genetic models (allelic model: OR = 1.45, 95% CI = 1.37–1.54, P < 0.001 (Fig. 3); homozygote model: OR = 2.06, 95% CI = 1.56–2.72, P < 0.001; heterozygote model: OR = 1.72, 95% CI = 1.50–1.96, P < 0.001; dominant model: OR = 1.76, 95% CI = 1.55–2.00, P < 0.001; recessive model: OR = 1.71, 95% CI = 1.30–2.24, P < 0.001) (Table 3). This association could not be found in East Asian group for any genetic model (Table 3).
Fig. 3.
Assessment of the association between C3 genetic polymorphism (rs1047286) with advanced AMD
Association between SNP rs2230205 of C3 gene and advanced AMD
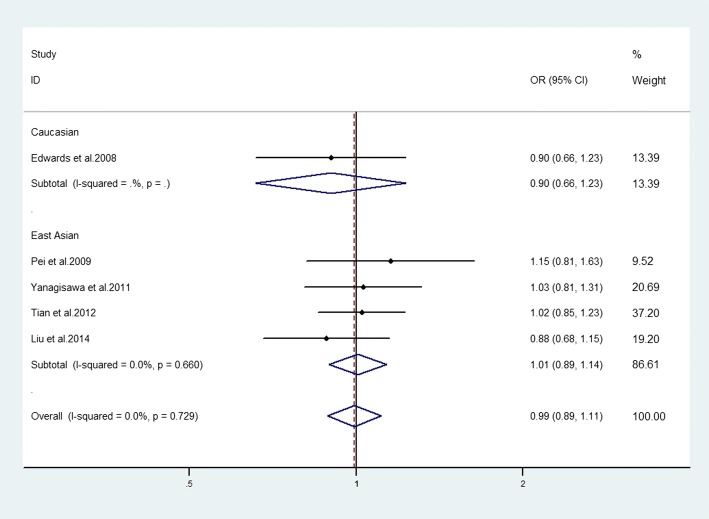
No association between this SNP and advanced AMD was achieved in the overall populations (allelic model: OR = 0.99, 95% CI = 0.89–1.11, P = 0.903; homozygote model: OR = 1.04, 95% CI = 0.77–1.42, P = 0.780; heterozygote model: OR = 1.00, 95% CI = 0.80–1.23, P = 0.967; dominant model: OR = 1.00, 95% CI = 0.81–1.22, P = 0.992; recessive model: OR = 1.06, 95% CI = 0.81–1.37, P = 0.687). Subgroup analysis of Caucasian and East Asian group showed that there was a lack of relationship in any of the genetic models (Fig. 4, Table 3).
Fig. 4.
Estimation of the association between C3 genetic polymorphism (rs2230205) with advanced AMD
Association between SNP rs2250656 of C3 gene and advanced AMD
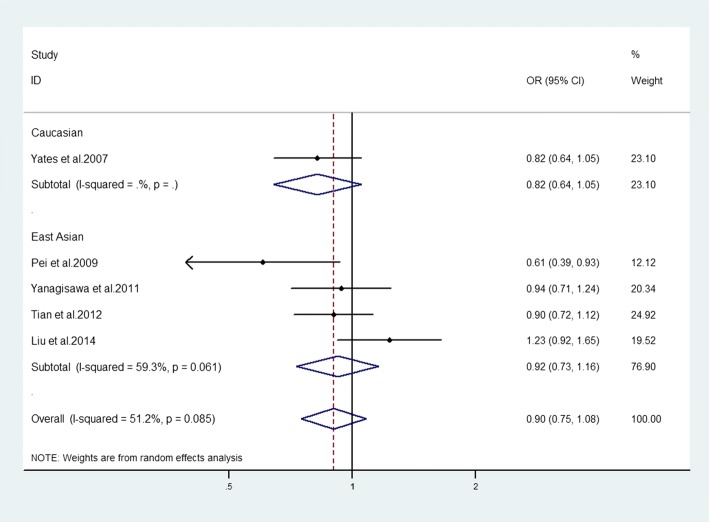
The results of meta-analysis showed that there was not a positive association between this SNP and advanced AMD in the overall populations (allelic model: OR = 0.90, 95% CI = 0.75–1.08, P = 0.257; homozygote model: OR = 0.76, 95% CI = 0.49–1.16, P = 0.207; recessive model: OR = 0.83, 95% CI = 0.55–1.27, P = 0.391). But a weakly protective risk between this SNP and advanced AMD was observed in heterozygote model and dominant model (OR = 0.78, 95% CI = 0.65–0.95, P = 0.014; OR = 0.78, 95% CI = 0.65–0.94, P = 0.010, respectively). In the stratified analysis by ethnicity, there was no association in any of the genetic models. (Fig. 5, Table 3).
Fig. 5.
Evaluation of the association between C3 genetic polymorphism (rs225065) with advanced AMD
Heterogeneity test and sensitivity analysis
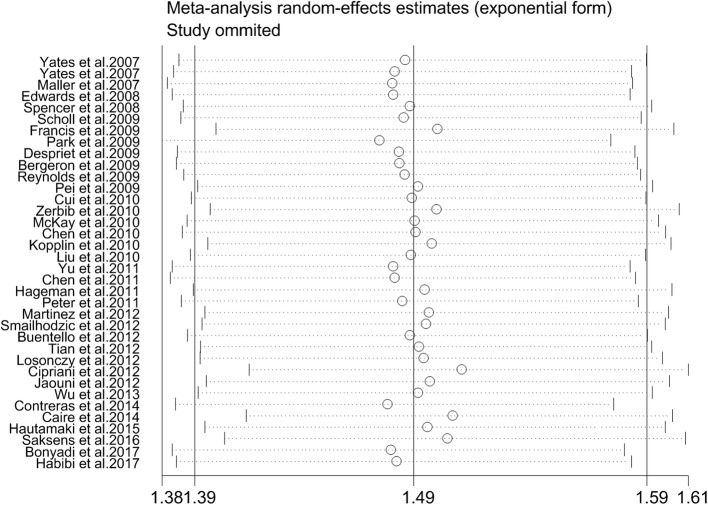
Significant heterogeneity between these studies was observed among two SNPs (rs2230199 and rs2250656) (P < 0.1) (Figs. 2, 5). The results of our subgroup analysis confirmed that ethnicity was the primary sources of heterogeneity. Additionally, sensitivity analysis was conducted to evaluate the effect of individual study on the pooled ORs by sequentially omitting each study. The pooled ORs were not affected by removing any study (Fig. 6, the sensitivity analysis of rs2230199; others see Additional file 2: Figures S1-S3).
Fig. 6.
Evaluation of the sensitivity analysis between C3 genetic polymorphism (rs2230199) with advanced AMD
Publication bias
Publication bias is a potential problem, thus Begg’s funnel plots and Egger’s regression tests were applied to investigate the publication bias for C3 genetic polymorphism. Four symmetrical funnel plots suggested that both tests had no evidence of significant bias (data not shown). Furthermore, as emerged in Table 4, the pooled P values for both tests are more than 0.05.
Table 4.
Bias between C3 genetic polymorphism with advanced AMD in our Meta-Analysis
| Polymorphism | Number of publication | Publication bias | |
|---|---|---|---|
| Begg’s test | Egger’s test | ||
| rs2230199 | 36 | 0.653 | 0.790 |
| rs1047286 | 13 | 0.428 | 0.124 |
| rs2230205 | 5 | 1.000 | 0.905 |
| rs2250656 | 5 | 1.000 | 0.594 |
Discussion
AMD is a multifactorial disease, in which complement system mediated inflammation plays a pivotal role. Several pathways including the alternative complement component have been described to be implicated in the development of AMD [54]. As the central element of the complement cascade, C3 has been a plausible candidate gene since its cleavage product C3a was confirmed in drusen. In our current meta-analysis, 20,673 patients and 20,025 controls from 38 articles were combined to detect the association of C3 genetic polymorphisms with advanced AMD. We came to the conclusion that two nonsynonymous SNPs rs2230199 and rs1047286 were demonstrated an increased pathogenic effect on advanced AMD (rs2230199: allelic model: OR = 1.49, 95% CI = 1.39–1.59, P < 0.001; homozygote model: OR = 2.33, 95% CI = 1.98–2.74, P < 0.001; rs1047286: allelic model: OR = 1.45, 95% CI = 1.37–1.54, P < 0.001; homozygote model: OR = 2.06, 95% CI = 1.56–2.72, P < 0.001). Moreover, our meta-analysis discovered that SNP rs2250656 decreased the risk of advanced AMD susceptibility, which a protective association was acquired in heterozygote model and dominant model. Obviously, the results of SNP rs2250656 with advanced AMD needed to be validated with larger samples and studies in different ethnicity.
Being consistent with previous studies, the G allele of rs2230199 conferred susceptibility to advanced AMD in Caucasian group. In our meta-analysis, we first confirmed that the G allele of rs2230199 could be linked with AMD in Middle East but not East Asian region, though rather larger population needed to be validated in the future. Besides, our meta-analysis found a novel association between the T allele of rs1047286 and advanced AMD in Caucasian but not East Asian group.
The common polymorphisms rs2230199 and rs1047286 in the C3 gene have been identified as genetic risk factors for advanced AMD in Caucasian populations. However, the allele frequencies of rs2230199 vary widely among different ethnicities. Frequencies of the risk G allele at rs2230199 were 25% to 31% in AMD cases and 19% to 21% in controls in Caucasians [29]. Besides, the frequencies of G allele was 14% to 25% in both cases and controls in Middle East region [20, 57]. While, the risk allele were absent in Japanese and rare (< 1%) in Chinese populations [53]. For rs1047286, frequencies of the risk T allele were 27% to 29% in AMD and 20% to 22% in controls in Caucasians. Cui et al. [29] also foud that rs1047286 was only 0.3% to 1% in both cases and controls and was not significantly associated with advanced AMD in Chinese population. The facts that rs2230199 and rs1047286 did not show tendency to risk of advanced AMD in East East but in Caucasian and Middle East could be explained by the lower minor allele frequencies (< 5%) of this two SNPs, suggesting that the susceptibility to advanced AMD by the variants of rs2230199 and rs1047286 did not transcend ethnic lines. In other words, this difference in the association between different ethnicities may result from other influence factors such as geography, the level of socioeconomic development or race.
The gene for C3 is located on the short arm of chromosome 19 and consists of 41 exons, which forms 13 functional domains. C3 is the most abundant complement component and significant C3 messenger RNA is detected in the neural retina, choroid, RPE, and cultured RPE cells [58]. Cleavage of C3 into C3a and C3b is the central step in complement activation, which amplifies the complement response, resulting in the formation of lytic pores in the cell membrane. Janssen et al. [59] argued that cleaved native C3 undergoes important structural rearrangements which causes conformational changes exposing binding sites for complement components and drusen including C3 and its activation products was confirmed in the finding that local inflammation and activation of the complement cascade can contribute to the pathogenesis of AMD. Notably, animal studies conducted by Bora et al. [60, 61] have indicated that C3 deficiency in C3−/− mice prevented the formation of choroidal neovascularization in advanced AMD (wet AMD), indicating that C3 is a pivotal element of this activation process.
In our meta-analysis, four SNPs including rs2230199, rs1047286, rs223205 and rs2250656 were analyzed in the pooled data. Among them, rs2230199 and rs1047286 are located in the first ring of macroglobulin domains, which conduct a prominent function for correct orientation of the thioester-containing domain. The amino acid changes induced by the genetic mutations may alter the configuration of the macroglobulin ring [62]. With evidence supporting a biologic functional effect through the formation of two electrophoretic allotypes in rs2230199 genetic site (C3F and C3S), the two alleles showed a differential capacity to bind monocyte- complement receptor. Helgason et al. [35] noted that the G allele in rs2230199 (C3F) was associated with the reduction of C3 gene binding to CFH, which leads to an increase in complement activation. Additionally, rs2230199 variant may alter the net charge of the molecule and influence the position of the thioester-containing domain. Except for advanced AMD, the risk variant of rs2230199 has been previously considered as associated with other immune-mediated conditions, such as IgA nephropathy, systemic vasculitis.
In the current meta-analysis, rs1047286 variant showed significant association with advanced AMD in Caucasian populations. Despriet et al. [30] argued that rs2230199 and rs1047286 variants were in high linkage disequilibrium (LD) (D’ = 0.90, r2 = 0.80), which haplotype analyses suggested that the effect of the C3 alleles was independent from the established genetic and environmental risk factors. Furthermore, our pooled analysis of neighboring SNPs of rs2230199 indicated that the allele frequency of the variant rs2230205 and rs2250656 was not significantly different between the advanced AMD cases and controls. Pei et al. [45] confirmed that the G allele of rs2250656 variant may be a protective factor for the development of AMD in East Asian. Given that the site of rs2250656 lies near the junction of intron 2 and exon 3, which contain short sequences and regulate the expression of gene and neighboring genes, it may contribute to the low risk for advanced AMD. Obviously, our pooled results were inconsistent with Pei’s report, owing to the relative small sample size and distinct environmental elements.
In a previous meta-analysis where a total of 15 independent studies with 5593 cases and 5181 controls were included, Zhang et al. [63] indicated that rs2230199 C > G SNP increased the risk of AMD development and the G allele was a risk factor for AMD in Caucasian but not Asians. Moreover, Yu et al. [64] have implemented a systemic meta-analysis and the overall results suggested a positive association between rs2230199, rs1047286 and AMD susceptibility. Additionally, Despriet et al. [30] have clarified these positive associations for only four available studies. In comparison to previous meta-analyses, our analysis mainly focused on the major form of AMD (advanced AMD) and was involved with a greater number of studies and larger sample size. These would make our pooled ORs more believable, stable, and accurate than before, especially in the association with advanced AMD. Moreover, our present meta-analysis encompassed an acceptable quality evaluation system, minimizing the potential bias.
Considerable efforts have been paid to discuss the potential relationship between C3 genetic polymorphisms and advanced AMD, some limitations for our present meta-analysis need to be declared. First, heterogeneity among the ethnic groups was discovered when investigating the association of C3 genetic variants with advanced AMD. However, based on the results of the sensitivity analysis, it is clear that the overall effect was not affected by heterogeneity. Additionally, there was no obvious publication bias detected in the contrast of C3 gene with advanced AMD. Second, the number of patients and controls was relatively small in each included study; therefore, a great number of samples from different ethnic regions are required for further analysis. Third, the effects of common confounding factors, including sex, age, body mass index, smoking, and diet were not evaluated in the present study because of insufficient data. Fourth limitation is that only three ethnic backgrounds with relatively few studies were taken into consideration, thus further efforts to reduce the incidence of ethnic bias will be needed once raw data become available. Finally, the electronic databases from which we selected eligible studies were listed in English and Chinese; therefore, a language bias may be existed in our meta-analysis.
Conclusion
The present meta-analysis provided a series of evidence-based pooled data for a significant association between rs2230199, rs1047286 and susceptibility to advanced AMD, especially in Caucasians. Additional well-designed work with a larger number of studies in which incorporate different ethnicities together with gene-gene and gene-environment is recommended to better confirm the functional role of the two nonsynonymous polymorphisms.
Additional files
The full details of databases searching terms. (DOC 38 kb)
Figures S1-S3. The sensitivity analysis of C3 genetic polymorphisms (rs1047286,rs2230205,rs2250656). (DOC 106 kb)
Acknowledgments
None.
Funding
This work was supported by National Natural Science Foundation Project (81300761) and Key Project of Health and Family Planning Commission of Wuhan Municipality (WX18A08).
Availability of data and materials
All data has been shared in the Figures and Tables.
Authors’ contributions
YX designed the study. JZ and SL collected and checked the available information from eligible articles in this meta-analysis. SH analyzed the data. JY prepared the Fig. 1-6, Additional file 1: Fig. S1-S3 and Table 1-4. JZ and SL wrote the main manuscript text. YX reviewed and revised the manuscript. All authors censored and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kokotas H, Grigoriadou M, Petersen MB. Age-related macular degeneration: genetic and clinical findings. Clin Chem Lab Med. 2011;49(4):601–616. doi: 10.1515/CCLM.2011.091. [DOI] [PubMed] [Google Scholar]
- 2.Klein R, Peto T, Bird A, Vannewkirk MR. The epidemiology of age-related macular degeneration. Am J Ophthalmol. 2004;137(3):486–495. doi: 10.1016/j.ajo.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 3.de Jong PT. Age-related macular degeneration. N Engl J Med. 2006;355(14):1474–1485. doi: 10.1056/NEJMra062326. [DOI] [PubMed] [Google Scholar]
- 4.Scholl HP, Fleckenstein M, Fritsche LG, Schmitz-Valckenberg S, Gobel A, Adrion C, Herold C, Keilhauer CN, Mackensen F, Mossner A, et al. CFH, C3 and ARMS2 are significant risk loci for susceptibility but not for disease progression of geographic atrophy due to AMD. PLoS One. 2009;4(10):e7418. doi: 10.1371/journal.pone.0007418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vingerling JR, Klaver CC, Hofman A, de Jong PT. Epidemiology of age-related maculopathy. Epidemiol Rev. 1995;17(2):347–360. doi: 10.1093/oxfordjournals.epirev.a036198. [DOI] [PubMed] [Google Scholar]
- 6.Kawasaki R, Yasuda M, Song SJ, Chen SJ, Jonas JB, Wang JJ, Mitchell P, Wong TY. The prevalence of age-related macular degeneration in Asians: a systematic review and meta-analysis. Ophthalmology. 2010;117(5):921–927. doi: 10.1016/j.ophtha.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Haddad S, Chen CA, Santangelo SL, Seddon JM. The genetics of age-related macular degeneration: a review of progress to date. Surv Ophthalmol. 2006;51(4):316–363. doi: 10.1016/j.survophthal.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Cackett P, Wong TY, Aung T, Saw SM, Tay WT, Rochtchina E, Mitchell P, Wang JJ. Smoking, cardiovascular risk factors, and age-related macular degeneration in Asians: the Singapore Malay eye study. Am J Ophthalmol. 2008;146(6):960–967. doi: 10.1016/j.ajo.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 9.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102(20):7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson PT, Betts KE, Radeke MJ, Hageman GS, Anderson DH, Johnson LV. Individuals homozygous for the age-related macular degeneration risk-conferring variant of complement factor H have elevated levels of CRP in the choroid. Proc Natl Acad Sci U S A. 2006;103(46):17456–17461. doi: 10.1073/pnas.0606234103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nozaki M, Raisler BJ, Sakurai E, Sarma JV, Barnum SR, Lambris JD, Chen Y, Zhang K, Ambati BK, Baffi JZ, et al. Drusen complement components C3a and C5a promote choroidal neovascularization. Proc Natl Acad Sci U S A. 2006;103(7):2328–2333. doi: 10.1073/pnas.0408835103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308(5720):419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 13.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hageman GS, Luthert PJ, Victor Chong NH, Johnson LV, Anderson DH, Mullins RF. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch's membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res. 2001;20(6):705–732. doi: 10.1016/S1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- 15.Johnson LV, Leitner WP, Staples MK, Anderson DH. Complement activation and inflammatory processes in Drusen formation and age related macular degeneration. Exp Eye Res. 2001;73(6):887–896. doi: 10.1006/exer.2001.1094. [DOI] [PubMed] [Google Scholar]
- 16.Little J, Higgins J, Bray M, Ioannidis J, Khoury M, Manolio T, Smeeth L, Sterne J. The HuGENet™ HuGE Review Handbook, version 1.0. 2006. [Google Scholar]
- 17.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 19.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonyadi M, Mohammadian T, Jabbarpoor Bonyadi MH, Fotouhi N, Soheilian M, Javadzadeh A, Moein H, Yaseri M. Association of polymorphisms in complement component 3 with age-related macular degeneration in an Iranian population. Ophthalmic Genet. 2017;38(1):61–66. doi: 10.3109/13816810.2015.1126612. [DOI] [PubMed] [Google Scholar]
- 21.Bonyadi M, Jabbarpoor Bonyadi MH, Yaseri M, Mohammadian T, Fotouhi N, Javadzadeh A, Soheilian M. Joint association of complement component 3 and CC-cytokine ligand2 (CCL2) or complement component 3 and CFH polymorphisms in age-related macular degeneration. Ophthalmic Genet. 2017;38:1–6. doi: 10.1080/13816810.2016.1275195. [DOI] [PubMed] [Google Scholar]
- 22.Bergeron-Sawitzke J, Gold B, Olsh A, Schlotterbeck S, Lemon K, Visvanathan K, Allikmets R, Dean M. Multilocus analysis of age-related macular degeneration. European journal of human genetics : EJHG. 2009;17(9):1190–1199. doi: 10.1038/ejhg.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buentello-Volante B, Rodriguez-Ruiz G, Miranda-Duarte A, Pompa-Mera EN, Graue-Wiechers F, Bekker-Mendez C, Ayala-Ramirez R, Quezada C, Rodriguez-Loaiza JL, Zenteno JC. Susceptibility to advanced age-related macular degeneration and alleles of complement factor H, complement factor B, complement component 2, complement component 3, and age-related maculopathy susceptibility 2 genes in a Mexican population. Mol Vis. 2012;18:2518–2525. [PMC free article] [PubMed] [Google Scholar]
- 24.Caire J, Recalde S, Velazquez-Villoria A, Garcia-Garcia L, Reiter N, Anter J, Fernandez-Robredo P, Alfredo G-L. Growth of geographic atrophy on fundus autofluorescence and polymorphisms of CFH, CFB, C3, FHR1-3, and ARMS2 in age-related macular degeneration. JAMA ophthalmology. 2014;132(5):528–534. doi: 10.1001/jamaophthalmol.2013.8175. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, Jakobsdottir J, Tosakulwong N, Pericak-Vance MA, Campochiaro PA, Klein ML, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107(16):7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Zeng J, Zhao C, Wang K, Trood E, Buehler J, Weed M, Kasuga D, Bernstein PS, Hughes G, et al. Assessing susceptibility to age-related macular degeneration with genetic markers and environmental factors. Arch Ophthalmol (Chicago, Ill : 1960) 2011;129(3):344–351. doi: 10.1001/archophthalmol.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cipriani V, Leung HT, Plagnol V, Bunce C, Khan JC, Shahid H, Moore AT, Harding SP, Bishop PN, Hayward C, et al. Genome-wide association study of age-related macular degeneration identifies associated variants in the TNXB-FKBPL-NOTCH4 region of chromosome 6p21.3. Hum Mol Genet. 2012;21(18):4138–4150. doi: 10.1093/hmg/dds225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Contreras AV, Zenteno JC, Fernandez-Lopez JC, Rodriguez-Corona U, Falfan-Valencia R, Sebastian L, Morales F, Ochoa-Contreras D, Carnevale A, Silva-Zolezzi I. CFH haplotypes and ARMS2, C2, C3, and CFB alleles show association with susceptibility to age-related macular degeneration in Mexicans. Mol Vis. 2014;20:105–116. [PMC free article] [PubMed] [Google Scholar]
- 29.Cui L, Zhou H, Yu J, Sun E, Zhang Y, Jia W, Jiao Y, Snellingen T, Liu X, Lim A, et al. Noncoding variant in the complement factor H gene and risk of exudative age-related macular degeneration in a Chinese population. Invest Ophthalmol Vis Sci. 2010;51(2):1116–1120. doi: 10.1167/iovs.09-4265. [DOI] [PubMed] [Google Scholar]
- 30.Despriet DD, van Duijn CM, Oostra BA, Uitterlinden AG, Hofman A, Wright AF, ten Brink JB, Bakker A, de Jong PT, Vingerling JR, et al. Complement component C3 and risk of age-related macular degeneration. Ophthalmology. 2009;116(3):474–480. doi: 10.1016/j.ophtha.2008.09.055. [DOI] [PubMed] [Google Scholar]
- 31.Edwards AO, Fridley BL, James KM, Sharma AK, Cunningham JM, Tosakulwong N. Evaluation of clustering and genotype distribution for replication in genome wide association studies: the age-related eye disease study. PLoS One. 2008;3(11):e3813. doi: 10.1371/journal.pone.0003813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis PJ, Hamon SC, Ott J, Weleber RG, Klein ML. Polymorphisms in C2, CFB and C3 are associated with progression to advanced age related macular degeneration associated with visual loss. J Med Genet. 2009;46(5):300–307. doi: 10.1136/jmg.2008.062737. [DOI] [PubMed] [Google Scholar]
- 33.Hageman GS, Gehrs K, Lejnine S, Bansal AT, Deangelis MM, Guymer RH, Baird PN, Allikmets R, Deciu C, Oeth P, et al. Clinical validation of a genetic model to estimate the risk of developing choroidal neovascular age-related macular degeneration. Human genomics. 2011;5(5):420–440. doi: 10.1186/1479-7364-5-5-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hautamaki A, Seitsonen S, Holopainen JM, Moilanen JA, Kivioja J, Onkamo P, Jarvela I, Immonen I. The genetic variant rs4073 A-->T of the Interleukin-8 promoter region is associated with the earlier onset of exudative age-related macular degeneration. Acta Ophthalmol. 2015;93(8):726–733. doi: 10.1111/aos.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helgason H, Sulem P, Duvvari MR, Luo H, Thorleifsson G, Stefansson H, Jonsdottir I, Masson G, Gudbjartsson DF, Walters GB, et al. A rare nonsynonymous sequence variant in C3 is associated with high risk of age-related macular degeneration. Nat Genet. 2013;45(11):1371–1374. doi: 10.1038/ng.2740. [DOI] [PubMed] [Google Scholar]
- 36.Jaouni T, Averbukh E, Burstyn-Cohen T, Grunin M, Banin E, Sharon D, Chowers I. Association of pattern dystrophy with an HTRA1 single-nucleotide polymorphism. Arch Ophthalmol (Chicago, Ill : 1960) 2012;130(8):987–991. doi: 10.1001/archophthalmol.2012.1483. [DOI] [PubMed] [Google Scholar]
- 37.Kopplin LJ, Igo RP, Jr, Wang Y, Sivakumaran TA, Hagstrom SA, Peachey NS, Francis PJ, Klein ML, SanGiovanni JP, Chew EY, et al. Genome-wide association identifies SKIV2L and MYRIP as protective factors for age-related macular degeneration. Genes Immun. 2010;11(8):609–621. doi: 10.1038/gene.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu K, Lai TY, Chiang SW, Chan VC, Young AL, Tam PO, Pang CP, Chen LJ. Gender specific association of a complement component 3 polymorphism with polypoidal choroidal vasculopathy. Sci Rep. 2014;4:7018. doi: 10.1038/srep07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Zhao P, Tang S, Lu F, Hu J, Lei C, Yang X, Lin Y, Ma S, Yang J, et al. Association study of complement factor H, C2, CFB, and C3 and age-related macular degeneration in a Han Chinese population. Retina (Philadelphia, Pa) 2010;30(8):1177–1184. doi: 10.1097/IAE.0b013e3181cea676. [DOI] [PubMed] [Google Scholar]
- 40.Losonczy G, Vajas A, Takacs L, Dzsudzsak E, Fekete A, Marhoffer E, Kardos L, Ajzner E, Hurtado B, de Frutos PG, et al. Effect of the Gas6 c.834+7G>a polymorphism and the interaction of known risk factors on AMD pathogenesis in Hungarian patients. PLoS One. 2012;7(11):e50181. doi: 10.1371/journal.pone.0050181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39(10):1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Barricarte R, Recalde S, Fernandez-Robredo P, Millan I, Olavarrieta L, Vinuela A, Perez-Perez J, Garcia-Layana A, Rodriguez de Cordoba S. Relevance of complement factor H-related 1 (CFHR1) genotypes in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2012;53(3):1087–1094. doi: 10.1167/iovs.11-8709. [DOI] [PubMed] [Google Scholar]
- 43.McKay GJ, Dasari S, Patterson CC, Chakravarthy U, Silvestri G. Complement component 3: an assessment of association with AMD and analysis of gene-gene and gene-environment interactions in a northern Irish cohort. Mol Vis. 2010;16:194–199. [PMC free article] [PubMed] [Google Scholar]
- 44.Park KH, Fridley BL, Ryu E, Tosakulwong N, Edwards AO. Complement component 3 (C3) haplotypes and risk of advanced age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50(7):3386–3393. doi: 10.1167/iovs.08-3231. [DOI] [PubMed] [Google Scholar]
- 45.Pei XT, Li XX, Bao YZ, Yu WZ, Yan Z, Qi HJ, Qian T, Xiao HX. Association of c3 gene polymorphisms with neovascular age-related macular degeneration in a chinese population. Curr Eye Res. 2009;34(8):615–622. doi: 10.1080/02713680903003484. [DOI] [PubMed] [Google Scholar]
- 46.Peter I, Huggins GS, Ordovas JM, Haan M, Seddon JM. Evaluation of new and established age-related macular degeneration susceptibility genes in the Women's Health Initiative sight exam (WHI-SE) study. Am J Ophthalmol. 2011;152(6):1005–1013. doi: 10.1016/j.ajo.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reynolds R, Hartnett ME, Atkinson JP, Giclas PC, Rosner B, Seddon JM. Plasma complement components and activation fragments: associations with age-related macular degeneration genotypes and phenotypes. Invest Ophthalmol Vis Sci. 2009;50(12):5818–5827. doi: 10.1167/iovs.09-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saksens NT, Lechanteur YT, Verbakel SK, Groenewoud JM, Daha MR, Schick T, Fauser S, Boon CJ, Hoyng CB, den Hollander AI. Analysis of risk alleles and complement activation levels in familial and non-familial age-related macular degeneration. PLoS One. 2016;11(6):e0144367. doi: 10.1371/journal.pone.0144367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smailhodzic D, Klaver CC, Klevering BJ, Boon CJ, Groenewoud JM, Kirchhof B, Daha MR, den Hollander AI, Hoyng CB. Risk alleles in CFH and ARMS2 are independently associated with systemic complement activation in age-related macular degeneration. Ophthalmology. 2012;119(2):339–346. doi: 10.1016/j.ophtha.2011.07.056. [DOI] [PubMed] [Google Scholar]
- 50.Spencer KL, Olson LM, Anderson BM, Schnetz-Boutaud N, Scott WK, Gallins P, Agarwal A, Postel EA, Pericak-Vance MA, Haines JL. C3 R102G polymorphism increases risk of age-related macular degeneration. Hum Mol Genet. 2008;17(12):1821–1824. doi: 10.1093/hmg/ddn075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian J, Yu W, Qin X, Fang K, Chen Q, Hou J, Li J, Chen D, Hu Y, Li X. Association of genetic polymorphisms and age-related macular degeneration in Chinese population. Invest Ophthalmol Vis Sci. 2012;53(7):4262–4269. doi: 10.1167/iovs.11-8542. [DOI] [PubMed] [Google Scholar]
- 52.Wu L, Tao Q, Chen W, Wang Z, Song Y, Sheng S, Li P, Zhou J. Association between polymorphisms of complement pathway genes and age-related macular degeneration in a Chinese population. Invest Ophthalmol Vis Sci. 2013;54(1):170–174. doi: 10.1167/iovs.12-10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanagisawa S, Kondo N, Miki A, Matsumiya W, Kusuhara S, Tsukahara Y, Honda S, Negi A. A common complement C3 variant is associated with protection against wet age-related macular degeneration in a Japanese population. PLoS One. 2011;6(12):e28847. doi: 10.1371/journal.pone.0028847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yates JR, Sepp T, Matharu BK, Khan JC, Thurlby DA, Shahid H, Clayton DG, Hayward C, Morgan J, Wright AF, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357(6):553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 55.Yu Y, Reynolds R, Fagerness J, Rosner B, Daly MJ, Seddon JM. Association of variants in the LIPC and ABCA1 genes with intermediate and large drusen and advanced age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52(7):4663–4670. doi: 10.1167/iovs.10-7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zerbib J, Richard F, Puche N, Leveziel N, Cohen SY, Korobelnik JF, Sahel J, Munnich A, Kaplan J, Rozet JM, et al. R102G polymorphism of the C3 gene associated with exudative age-related macular degeneration in a French population. Mol Vis. 2010;16:1324–1330. [PMC free article] [PubMed] [Google Scholar]
- 57.Habibi I, Sfar I, Kort F, Bouraoui R, Chebil A, Limaiem R, Ayed S, Ben Abdallah T, El Matri L, Gorgi Y. Complement component C3 variant (R102G) and the risk of Neovascular age-related macular degeneration in a Tunisian population. Klin Monatsbl Augenheilkd. 2017;234(4):478–482. doi: 10.1055/s-0043-104419. [DOI] [PubMed] [Google Scholar]
- 58.Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J : official publication of the Federation of American Societies for Experimental Biology. 2000;14(7):835–846. doi: 10.1096/fasebj.14.7.835. [DOI] [PubMed] [Google Scholar]
- 59.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444(7116):213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 60.Bora PS, Hu Z, Tezel TH, Sohn JH, Kang SG, Cruz JM, Bora NS, Garen A, Kaplan HJ. Immunotherapy for choroidal neovascularization in a laser-induced mouse model simulating exudative (wet) macular degeneration. Proc Natl Acad Sci U S A. 2003;100(5):2679–2684. doi: 10.1073/pnas.0438014100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bora PS, Sohn JH, Cruz JM, Jha P, Nishihori H, Wang Y, Kaliappan S, Kaplan HJ, Bora NS. Role of complement and complement membrane attack complex in laser-induced choroidal neovascularization. J Immunol (Baltimore, Md : 1950) 2005;174(1):491–497. doi: 10.4049/jimmunol.174.1.491. [DOI] [PubMed] [Google Scholar]
- 62.Nishida N, Walz T, Springer TA. Structural transitions of complement component C3 and its activation products. Proc Natl Acad Sci U S A. 2006;103(52):19737–19742. doi: 10.1073/pnas.0609791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang MX, Zhao XF, Ren YC, Geng TT, Yang H, Feng T, Jin TB, Chen C. Association between a functional genetic polymorphism (rs2230199) and age-related macular degeneration risk: a meta-analysis. Gen Mol Res : GMR. 2015;14(4):12567–12576. doi: 10.4238/2015.October.16.24. [DOI] [PubMed] [Google Scholar]
- 64.Qian-Qian Y, Yong Y, Jing Z, Xin B, Tian-Hua X, Chao S, Jia C. Nonsynonymous single nucleotide polymorphisms in the complement component 3 gene are associated with risk of age-related macular degeneration: a meta-analysis. Gene. 2015;561(2):249–255. doi: 10.1016/j.gene.2015.02.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The full details of databases searching terms. (DOC 38 kb)
Figures S1-S3. The sensitivity analysis of C3 genetic polymorphisms (rs1047286,rs2230205,rs2250656). (DOC 106 kb)
Data Availability Statement
All data has been shared in the Figures and Tables.