Abstract
Background
The eukaryotic transcription factor NF-Y (which consists of NF-YA, NF-YB and NF-YC subunits) is involved in many important plant development processes. There are many reports about the NF-Y family in Arabidopsis and other plant species. However, there are no reports about the NF-Y family in walnut (Juglans regia L.).
Results
Thirty-three walnut NF-Y genes (JrNF-Ys) were identified and mapped on the walnut genome. The JrNF-Y gene family consisted of 17 NF-YA genes, 9 NF-YB genes, and 7 NF-YC genes. The structural features of the JrNF-Y genes were investigated by comparing their evolutionary relationship and motif distributions. The comparisons indicated the NF-Y gene structure was both conserved and altered during evolution. Functional prediction and protein interaction analysis were performed by comparing the JrNF-Y protein structure with that in Arabidopsis. Two differentially expressed JrNF-Y genes were identified. Their expression was compared with that of three JrCOs and two JrFTs using quantitative real-time PCR (qPCR). The results revealed that the expression of JrCO2 was positively correlated with the expression of JrNF-YA11 and JrNF-YA12. In contrast, JrNF-CO1 and JrNF-YA12 were negatively correlated.
Conclusions
Thirty-three JrNF-Ys were identified and their evolutionary, structure, biological function and expression pattern were analyzed. Two of the JrNF-Ys were screened out, their expression was differentially expressed in different development periods of female flower buds, and in different tissues (female flower buds and leaf buds). Based on prediction and experimental data, JrNF-Ys may be involved in flowering regulation by co-regulate the expression of flowering genes with other transcription factors (TFs). The results of this study may make contribution to the further investigation of JrNF-Y family.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1459-2) contains supplementary material, which is available to authorized users.
Keywords: Walnut (Juglans regia), NF-Y transcription factor, Phylogenetic analysis, Expression profiles, Transcriptome sequencing
Background
Nuclear factor Y (NF-Y), which was previously known as heme activator protein (HAP) or CCAAT binding factor (CBF), is a trimeric transcription factor that is present in nearly all eukaryotes. A conserved NF-Y transcription factor has three subunits, NF-YA/B/C (also called HAP2/3/5 or CBF-B/A/C) [1], which can specifically bind to a cis-element, CCAAT-box, in eukaryotic promoters [2]. A single NF-Y subunit cannot regulate transcription. The subunits can only function in the form of a dimer or trimer [3–7]. Initially, NF-YB and NF-YC form a dimer in the cytoplasm. They then bind with NF-YA protein to form a trimer in the nucleus [8, 9]. A recent study suggests that some transcription factors can combine with the NF-YB-YC dimer to form a NF-YB-YC-TF trimer instead of the traditional NF-YB-YC-YA trimer. Both trimers can bind to the promoter region of the target gene and regulate its expression [3].
In many mammals and yeast, a single NF-Y gene encodes each NF-Y subfamily [10]. For example, each NF-Y subfamily in mice and humans is encoded by only one NF-Y gene. In plants, however, many NF-Y genes encode each NF-Y subfamily. For example, it has been reported that in Arabidopsis thaliana, the NF-YA subfamily is encoded by ten NF-YA genes, the NF-YB subfamily is encoded by thirteen NF-YB genes, and the NF-YC subfamily is encoded by thirteen NF-YC genes [11]. Other studies indicated that each NF-Y subfamily in Arabidopsis thaliana is encoded by ten genes [3, 12].
In recent years, more and more plant NF-Y genes have been isolated and identified, including Triticum aestivum L. [13], Arabidopsis thaliana (L.) Heynh. [11], populus euphratica Olivier. [14], Glycine max (L.) Merr. [15], Brassica napus L. [16], Phaseolus vulgaris L. [17], Physcomitrella patens (Hedw.) Bruch & Schimp. [18], Vitis vinifera L. [19], Solanum lycopersicum L. [20], Citrullus lanatus (Thunb.) Matsum. & Nakai. [21], Citrus sinensis (L.) Osbeck and citrus clementina Hort ex Tan [22]. These NF-Y genes are involved in many plant developmental processes, such as flowering time regulation [3, 11, 23–33], root growth [34, 35], embryo development [36–42], seed germination [43], meristem formation [44] and fruit maturation [20]. The NF-Y genes also participate in plant physiological processes including photosynthesis [45–48] and stress response of endoplasmic reticulum (ER) [49, 50]. In addition, NF-Y genes are also involved in plant responses to abiotic stresses [14, 15, 51–58] and in processes related to plant-microbe interactions [59].
The wood and fruit of walnut (J. regia L.) are highly valuable, and the research of walnut focus on molecular breeding and flowering in recent years [60–65]. However, less attention were paid to walnut compare with other plants, because it must grow for many years before it becomes productive. The walnut genome was only published recently [66]. The purpose of this study was to identify NF-Y gene family in walnut (JrNF-Y) and to characterize their structure and function. Flower transition is an important time in plant growth [30], therefore, we focused on this period. Reverse genetic analysis makes it easier to predict the function of the same structural proteins among different species by constructing phylogenetic trees [67] and by analyzing gene expression patterns [28, 68]. The NF-Y family in Arabidopsis has been well characterized and annotated [11]. Therefore, sequencing results from walnut flower buds and leaf buds were searched with Arabidopsis NF-Y protein sequences to identify candidate NF-Y transcription factors in walnut. These candidate NF-Y members were then aligned with the published walnut genome. The NF-Y proteins sequences of walnut and Arabidopsis were aligned and a phylogenetic tree was constructed. The conserved domains of the walnut NF-Y protein sequences were aligned with mouse NF-Y protein sequences to further analyze the evolutionary relationships. The motifs of the walnut NF-Y proteins were predicted to analyze their structural features. The functions of walnut NF-Y members were annotated and their interactions were analyzed based on corresponding NF-Ys in Arabidopsis. Microarray data from transcriptome sequencing was used to construct the expression patterns of JrNF-Ys at different stages and in different tissues. Differentially expressed NF-Y members and the annotated FLOWER LOCUS T (FT) and CONSTANS (CO) genes were identified in the walnut transcriptome, and their relative expression levels were measured using real-time quantitative PCR (qRT-PCR) method. The relative expression levels were used to investigate possible associations among NF-Y, CO, and FT. Published data about walnut protein and cDNA data is limited. Therefore, some walnut NF-Ys were probably not included in our retrieval results. However, the results of this experiment provide a beginning point for further study about the NF-Y gene family in walnut.
Results
Identification and genomic localization of NF-Ys in walnut
The full-length protein sequences of Arabidopsis NF-Ys [11] were used to search the walnut transcriptome database using BLAST (version 2.60) [69] and HMMER (version 3.0) software [70]. Eighty-eight candidate NF-Y genes were identified in walnut by BLAST. Forty-four candidate genes were identified by HMMER. The results of the two search methods were merged resulting in 104 candidate NF-Y genes. Some of the candidate genes were discarded because they were too long or too short or because they had improper domains. Some sequences were considered to be the same gene because their similarity was > 98%. Finally, 33 candidate NF-Y genes were identified and translated into amino acid sequences according to the code frame shown in CD-Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The 33 candidate genes included 17 NF-YA genes, 9 NF-YB genes, and 7 NF-YC genes. These genes were named JrNY-F for J. regia. The number after gene name indicated the numerical order of the local gene ID. Each JrNF-Y protein was matched against one NF-Y protein in Arabidopsis (Table 1; priority: Query Cover>Ident>E value).
Table 1.
NF-Y genes in walnut
| Family | Name | Gene ID | Best match in Juglans regia Genome | ||||
|---|---|---|---|---|---|---|---|
| Query Cover | E value | Ident | Accession | Alignments Ranges | |||
| NF-YA Subfamily | JrNF-YA1 | Cluster-14,922.20995 | 97% | 0.0 | 98% | NW_017389264.1 | 43,746–44,361,44,488-44,976,45,840-46,105,46,556–46,737 |
| JrNF-YA2 | Cluster-14,922.27809 | 100% | 0.0 | 99% | NW_017388893.1 | 293,307–293,587,293,372-294,101,294,234–294,305, 294,531–294,693,295,892-296,322,296,422–296,951 |
|
| JrNF-YA3 | Cluster-14,922.32667 | 100% | 0.0 | 84% | NW_017441761.1 | 116,121–117,095,117,697-117,838,118,791–118,861, 118,959–119,069,119,978-120,307,120,912–121,157 |
|
| JrNF-YA4 | Cluster-14,922.32809 | 100% | 0.0 | 99% | NW_017388893.1 | 291,714–291,800,292,443-292,699,293,985-294,101,294,234- 294,305,294,531-294,693,295,892-296,322,296,422–296,951 |
|
| JrNF-YA5 | Cluster-14,922.40699 | 89% | 0.0 | 98% | NW_017442734.1 | 7508–8233,8334-8501,8723-8792,8858–8973, 9835–10,224,12,530–12,634 |
|
| JrNF-YA6 | Cluster-14,922.46822 | 98% | 0.0 | 99% | NW_017436745.1 | 2743–3638,4155-4319,6065-6137,6280-6398,6591–6912, 7558–7868 |
|
| JrNF-YA7 | Cluster-14,922.46846 | 89% | 0.0 | 98% | NW_017442734.1 | 7508–8233,8334-8501,8723-8973,9835–10,224 12,530–12,634 |
|
| JrNF-YA8 | Cluster-14,922.46847 | 89% | 0.0 | 98% | NW_017442734.1 | 7508–8233,8334-8501,8723-8792,8858–8973 9835–10,233,12,279–12,443 |
|
| JrNF-YA9 | Cluster-14,922.50230 | 99% | 0.0 | 99% | NW_017388973.1 | 706,166–707,095,707,522-707,639,707,717–707,791, 708,295–708,465,708,579–709,333 |
|
| JrNF-YA10 | Cluster-14,922.52088 | 83% | 0.0 | 99% | NW_017443622.1 | 162,218–163,195,163,717-163,881,165,641–165,713, 165,852–165,981,166,205–166,527 |
|
| JrNF-YA11 | Cluster-14,922.60069 | 96% | 0.0 | 99% | NW_017388854.1 | 1,241,389–1,241,687,1,244,904-1,245,304,1,246,166–1,246,427 1,246,636–1,246,803,1,247,015–1,247,884 |
|
| JrNF-YA12 | Cluster-14,922.69778 | 99% | 0.0 | 99% | NW_017438713.1 | 17,627–17,902,19,198-19,481,20,521-21,677,23,066–23,146 22,468–23,146 |
|
| JrNF-YA13 | Cluster-14,922.72325 | 90% | 0.0 | 99% | NW_017436745.1 | 2743–3638,4155–4703 | |
| JrNF-YA14 | Cluster-14,922.79874 | 100% | 0.0 | 99% | NW_017441761.1 | 115,996–117,095,117,697-117,838,118,791–118,861, 118,959–119,069,119,978-120,307,120,912–121,157 |
|
| JrNF-YA15 | Cluster-14,922.82902 | 100% | 0.0 | 99% | NW_017388893.1 | 291,714–291,800,292,443-292,720,293,981–294,101, 294,234–294,305,294,531-294,693,295,892–296,951 |
|
| JrNF-YA16 | Cluster-14,922.84458 | 100% | 0.0 | 99% | NW_017441761.1 | 115,996–117,095,117,697-117,838,118,791–118,861, 118,959–119,069,119,978–120,934 |
|
| JrNF-YA17 | Cluster-14,922.95251 | 100% | 0.0 | 99% | NW_017389365.1 | 22–832,959-1206,1227-1715,1842–2448 | |
| NF-YB Subfamily | JrNF-YB1 | Cluster-14,922.21265 | 100% | 0.0 | 99% | NW_017443611.1 | 188,307–189,170,197,160-197,297,197,295–197,376 |
| JrNF-YB2 | Cluster-14,922.29372 | 99% | 8e-144 | 100% | NW_017440443.1 | 158,098–158,332,158,425–158,702 | |
| JrNF-YB3 | Cluster-14,922.32115 | 99% | 0.0 | 100% | NW_017440443.1 | 157,907–158,332,158,425-158,554,159,022–159,104, 159,272–159,324,161,023–161,350 |
|
| JrNF-YB4 | Cluster-14,922.39672 | 100% | 0.0 | 99% | NW_017388887.1 | 1,546,260–1,547,648,1,547,636–1,547,998 | |
| JrNF-YB5 | Cluster-14,922.54864 | 99% | 6e-165 | 99% | NW_017440443.1 | 157,770–157,906,158,098-158,332,158,425–158,554, 159,022–159,104,159,272–159,324,161,023–161,350 |
|
| JrNF-YB6 | Cluster-14,922.57314 | 100% | 0.0 | 99% | NW_017441173.1 | 105,472–107,835 | |
| JrNF-YB7 | Cluster-14,922.58354 | 99% | 0.0 | 99% | NW_017389061.1 | 97,582–98,028,99,008-99,091,99,750-99,791,99,916–100,435 101,311–101,443,101,707-101,956,102,119–102,407 |
|
| JrNF-YB8 | Cluster-14,922.64236 | 98% | 0.0 | 99% | NW_017436168.1 | 34,788–35,884 | |
| JrNF-YB9 | Cluster-14,922.72337 | 96% | 0.0 | 99% | NW_017443591.1 | 1,605,638–1,606,317 | |
| NF-YC Subfamily | JrNF-YC1 | Cluster-14,922.36868 | 100% | 0.0 | 99% | NW_017443565.1 | 579,861–578,990,574,608–574,273 |
| JrNF-YC2 | Cluster-14,922.44401 | 99% | 0.0 | 100% | NW_017389361.1 | 1,038,089–1,039,233,1,035,966-1,036,022,1,036,219–1,036,261 | |
| JrNF-YC3 | Cluster-14,922.50411 | 74% | 0.0 | 97% | NW_017389324.1 | 983,265–984,133 | |
| JrNF-YC4 | Cluster-14,922.50413 | 100% | 0.0 | 100% | NW_017389324.1 | 983,265–984,857 | |
| JrNF-YC5 | Cluster-14,922.55754 | 76% | 0.0 | 99% | NW_017389752.1 | 432,148–433,061,431,991–432,052 | |
| JrNF-YC6 | Cluster-14,922.62047 | 99% | 0.0 | 99% | NW_017389324.1 | 983,265–984,362,985,618–985,795 | |
| JrNF-YC7 | Cluster-14,922.71538 | 71% | 0.0 | 97% | NW_017389361.1 | 1,038,255–1,038,849 | |
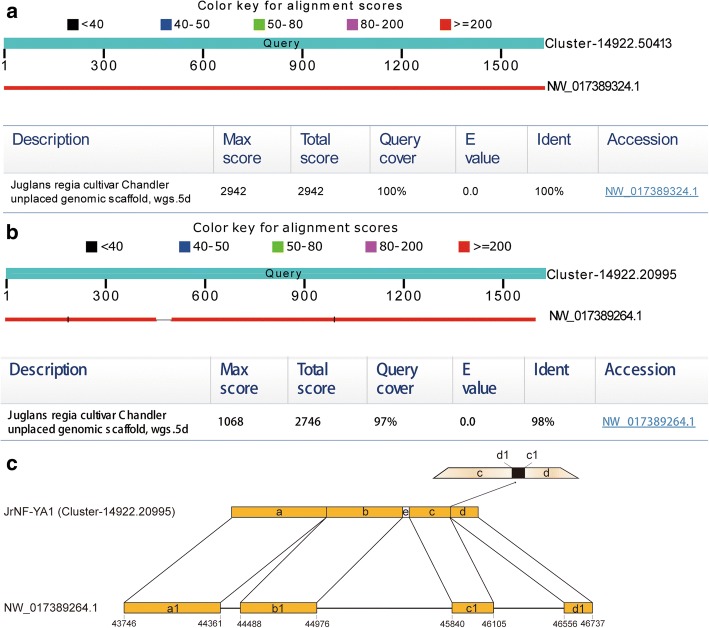
Walnut genome data was uploaded to NCBI (https://www.ncbi.nlm.nih.gov/bioproject/291087) in 2015. This was an enormous contribution even though the data was spliced at the level of scaffold. We attempted to map the cDNA sequences of the candidate NF-Ys on the published walnut genome (GCA 001411555.1wgs.5d scaffolds). In general, cDNA acquired by transcriptome sequencing does not match well against a single scaffold due to post-transcriptional processing. Most of the other NF-Ys partially matched the published data [e.g., Cluster-14,922.20995 (JrNF-YA1) partially matched NW_017389264.1] (Fig. 1). However, Cluster-14,922.50413 (JrNF-YC4) completely matched NW_017389324.1. Optimal matching results of walnut genomic scaffolds for each JrNF-Y are shown in Table 1. The table also shows the initiation and termination sites as references for further study.
Fig. 1.
Blast results of the cDNA sequences with the genomic scaffold in walnut. a cDNA (Cluster-14,922.50413) was 100% homologous with the genomic scaffold (NW_017389324.1). b cDNA (Cluster-14,922.20995) was 98% homologous with the genomic scaffold (NW_017389264.1). c Correspondence of different zones between cDNA and the genomic scaffold in walnut; a/b/c/d/e, different segments in cDNA (Cluster-14,922.20995); a1/b1/c1/d1, different segments in the genomic scaffold (NW_017389264.1). The a, b, c, and d segments were respectively matched with a1, b1, c1, and d1. The enlarged part in the upper right shows that (i) the last position of c1 matched the first position of d, (ii) the first position of d1 matched the last position of c, and (iii) c was adjacent to d. The numbers below the scaffold indicate the site number
Multiple alignments and phylogenetic analyses of the JrNF-Ys
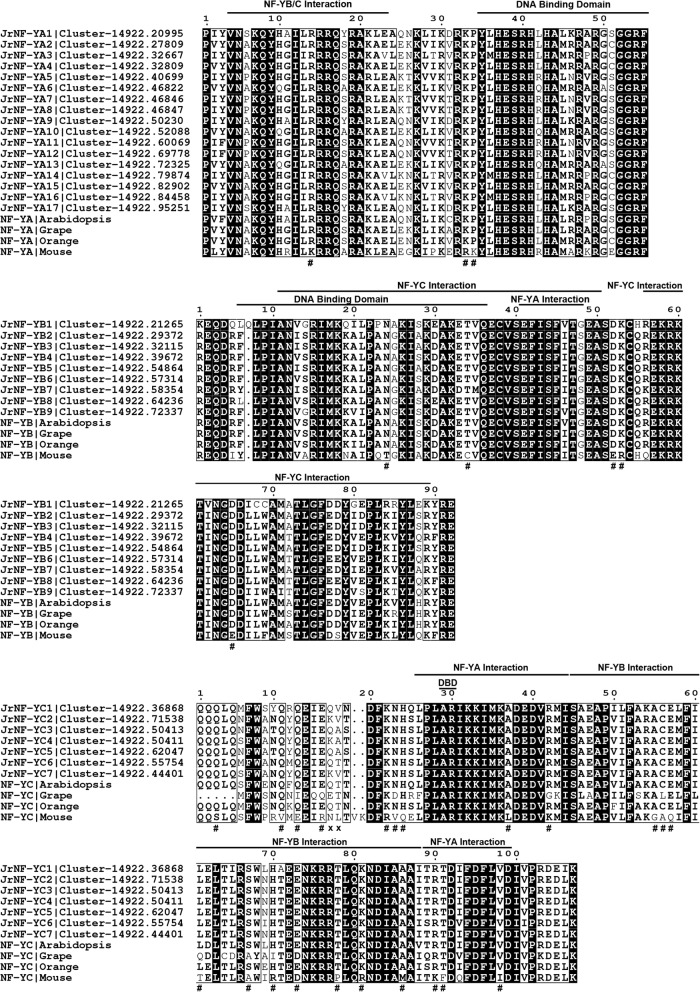
Protein sequences of the three NF-Y subfamilies (i.e., 33 candidate NF-Y genes) were aligned using clustalX software [71]. The results showed that each member of the JrNF-Y family member contains an interaction domain for interacting with other NF-Y subunits and a DNA binding domain for recognizing CCAAT binding sites. The three NF-Y subunits of grape, orange and mouse were included as an out-group to root the phylogenetic trees and for comparison. The interaction domain and the DNA binding domain were well conserved between plants and animal. The core conserved regions of the JrNF-YA, JrNF-YB and JrNF-YC proteins were 55, 92, and 107 amino acids long, respectively.
In most eukaryotes there is a clear boundary between the conserved and non-conserved regions of NF-Y proteins [11, 28]. Studies in yeast have found that CBF-B (NF-YA) and CBF-C (NF-YC) subfamilies are both often accompanied by large amounts of glutamine and some hydrophobic residues, which are involved in transcriptional activation [72]. The conserved regions in the JrNF-Ys were similar; however, there were obvious differences among them (Fig. 2, Additional file 1). Even within the same subfamily, the NF-Y protein sequences showed variability. Therefore, the transcriptional activities of the transcription factors also need to be verified.
Fig. 2.
Multiple alignment of the JrNF-Y family was performed by Clustal X. Conserved regions of all JrNF-Y proteins and Mus musculus NF-Y proteins (NF-YMouse) are shown. NF-YAMouse, NF-YBMouse, NF-YCMouse were respectively compared with the JrNF-YA, JrNF-YB, JrNF-YC subfamilies. Regions required for DNA binding or interacted with NF-YA, NF-YB and NF-YC subfamilies were previously defined in mammals and yeast. Amino acids in black boxes/white letters were identical in 100% of all aligned sequences. Amino acid sequences with the symbol “#” were conserved in walnut but divided when compared with mouse. Amino acid sequences with the symbol “x” were unique in mouse, and only in the NF-YC subfamily
Multiple sequence alignment of the conserved regions in the three subfamilies showed that 31 of 55 amino acid residues in the conserved region of NF-YA were absolutely conserved compared with 58 of 92 residues in NF-YB and 86 of 107 residues in NF-YC. These values were much greater than those in Arabidopsis, which had 24 of 53 residues absolutely conserved in NF-YA, 9 of 100 residues absolutely conserved in NF-YB, and 4 of 90 residues absolutely conserved in NF-YC [11]. The conserved regions of the JrNF-YA and JrNF-YB subfamilies were remarkably consistent with the NF-YA and NF-YB subfamilies in grape, orange and mouse. Comparing the conserved regions in walnut with those in mouse, 22 of 86 residues were different in NF-YC, 3 of 31 residues were different in NF-YA, and 5 of 58 residues were different in NF-YB.
The above results indicate that a less proportion of residues were conserved in JrNF-YC than in JrNF-YA and JrNF-YB. The valine(V) and lysine(K) between the nineteenth and twentieth amino acids were unique in mouse and were not observed in seven JrNF-YC sequences and other plant NF-YC sequences (V and K were not numbered and were marked with an “X” at the bottom of NF-YC sequences in Fig. 2). This may reflect the difference between animal and plant. It is worth noting that the initial position of the NF-YC domain reported in Arabidopsis was identified at the L locus whereas the initial position of the JrNF-YC domain was 25 amino acids before the L locus [11].
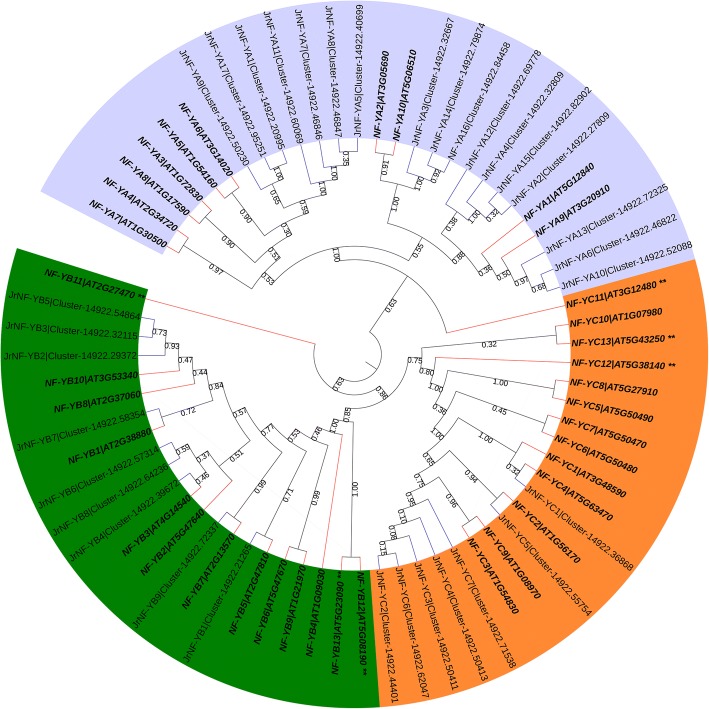
To investigate the evolutionary relationship between the walnut NF-Y family and the Arabidopsis NF-Y family, an un-rooted phylogenetic tree was constructed using the NF-Y protein sequences of Arabidopsis and walnut (Fig. 3). The phylogenetic tree showed close relationships among the candidate NF-Ys within each of the three subfamilies. The exceptions were JrNF-YC1 and JrNF-YB11. The close evolutionary relationships indicated that the NF-Y protein family in Arabidopsis has similar structure and function to that in walnut.
Fig. 3.
Phylogenetic analysis of NF-Y proteins in walnut and Arabidopsis. The phylogenetic tree was constructed by protein sequences of thirty-three NF-Ys in walnut (JrNF-Ys) and thirty-six NF-Ys in Arabidopsis [11]. Some reports indicate only 30 NF-Y proteins in Arabidopsis [3, 12]. ** indicate NF-Ys reported by Siefers et al. [11] but not reported by either Zhao et al. [3] or Petroni et al. [12]. Purple, green and orange indicate the NF-YA, NF-YB, and NF-YC subfamilies, respectively. The nodes of walnut and Arabidopsis are indicated by blue and red lines, respectively. The bootstrap values are shown on branches
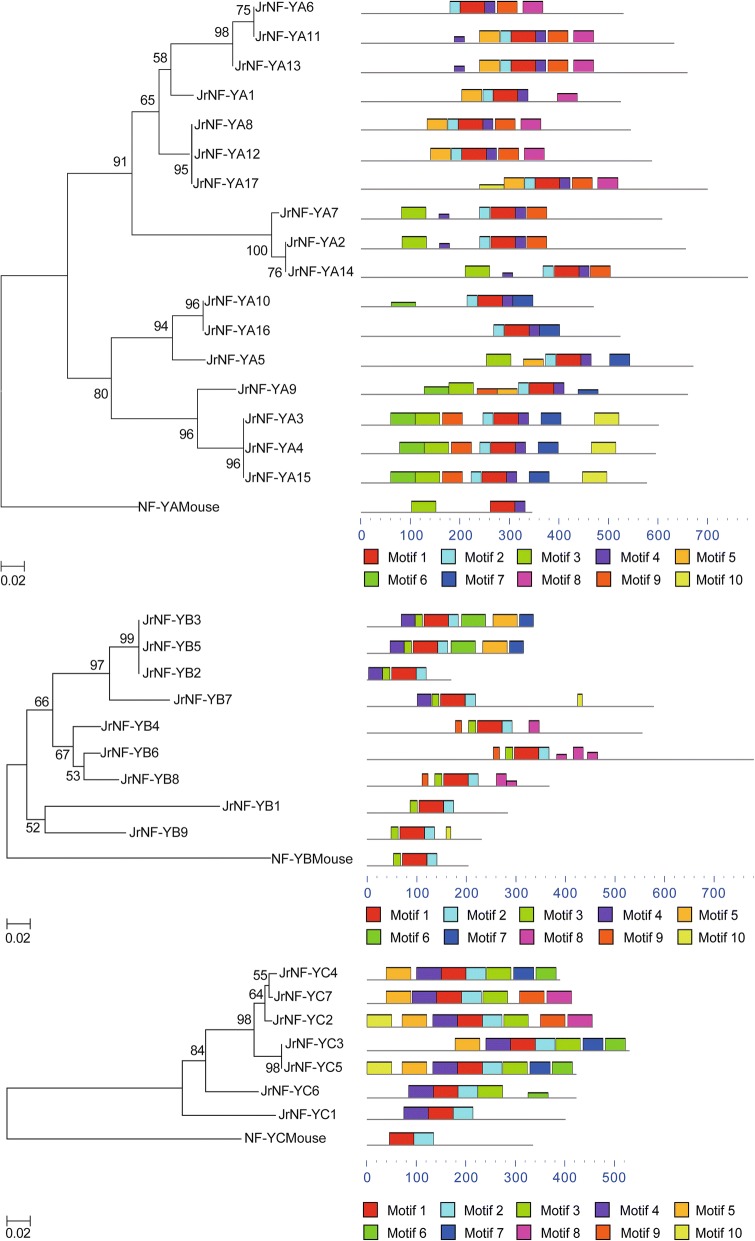
A rooted phylogenetic tree of each JrNF-Y subfamily was generated with the conserved domain sequences (Fig. 4). The sequences of the three NF-Y subfamilies in mouse were used as the roots of the phylogenetic trees. The multiple sequence alignment results of each JrNF-Y domain were then used to construct an adjacent evolutionary tree. MEME software (http://meme-suite.org/tools/meme) was used to predict the motif distributions with the full-length protein sequences of the three JrNF-Y subfamilies (Fig. 4) [73]. Initially, the construction of the adjacent evolutionary tree and the prediction of the motif distributions were done separately. Then we observed that the two parts showed some important relationships. For example, the evolutionary relationships indicated close genetic relationships among JrNF-YA3, JrNF-YA4, and JrNF-YA15. Furthermore, their motif distributions were similar. Although the motif distributions were predicted by the JrNF-Y sequence (full-length) and the phylogenetic tree was constructed using domain sequences (fragments), the two results were in good agreement. This phenomenon was also observed in Arabidopsis [11].
Fig. 4.
Phylogenetic trees and motif distributions of NF-YA, NF-YB, NF-YC subfamilies in walnut proteins and mouse proteins. Phylogenetic trees were constructed with the conserved regions shown in Fig. 2. The bootstrap values (> 50%) are shown at each branch (1000 replicates). Motif distributions were predicted with the full-length protein sequences of walnut and mouse. The motifs are distinguished by different colors as shown in the legend
Function prediction and protein interaction
Because of the lack of relevant data about walnut proteins, we predicted the function of JrNF-Y proteins based on corresponding NF-Y proteins in Arabidopsis [68]. We used the Blastp program to align the 33 walnut NF-Y proteins with 36 Arabidopsis NF-Y proteins [69]. Each JrNF-Y protein was closely aligned to at least one Arabidopsis NF-Y protein. Some of the JrNF-Y proteins were closely aligned to the same Arabidopsis NF-Y protein. Overall, the 33 JrNF-Y proteins were most closely aligned to 11 Arabidopsis NF-Y proteins (NF-YA1/NF-YA3/NF-YA9/NF-YA10/NF-YB3/NF-YB5/NF-YB7/NF-YB8/NF-YC1/NF-YC2/NF-YC9) (Table 2).
Table 2.
Annotation of the JrNF-Y gene family
| Gene name | Putative Arabidopsis Orthologs | Function in Arabidopsis |
|---|---|---|
| JrNF-YA1 | NF-YA1 | Flowering time regulation [30]; Salt stress response [54]; Male gametogenesis, embryogenesis, and seed development [39]. |
| JrNF-YA8 | ||
| JrNF-YA11 | ||
| JrNF-YA12 | ||
| JrNF-YA13 | ||
| JrNF-YA17 | ||
| JrNF-YA3 | NF-YA3 | Early embryogenesis [41]; Abiotic stress tolerance [55]. |
| JrNF-YA4 | ||
| JrNF-YA5 | ||
| JrNF-YA9 | ||
| JrNF-YA10 | ||
| JrNF-YA15 | ||
| JrNF-YA16 | ||
| JrNF-YA6 | NF-YA9 | Male gametogenesis, embryogenesis, and seed development [39] [43]. |
| JrNF-YA2 | NF-YA10 | Root growth [34]; Seed germination [43]. |
| JrNF-YA7 | ||
| JrNF-YA14 | ||
| JrNF-YB4 | NF-YB3 | Flowering time regulation [25]; ER stress response [49]; Heat stress response [58]. |
| JrNF-YB6 | ||
| JrNF-YB8 | ||
| JrNF-YB1 | NF-YB5 | Unknown |
| JrNF-YB9 | NF-YB7 | embryo development [36, 42] |
| JrNF-YB2 | NF-YB8 | Unknown |
| JrNF-YB3 | ||
| JrNF-YB5 | ||
| JrNF-YB7 | ||
| JrNF-YC1 | NF-YC1 | Flowering time regulation [8]; Freezing stress resistance [56]. |
| JrNF-YC3 | ||
| JrNF-YC6 | NF-YC2 | Photooxidative stress response and flowering time regulation [31]; ER stress response [49]. |
| JrNF-YC2 | NF-YC9 | Flowering time regulation [26, 28]. |
| JrNF-YC4 | ||
| JrNF-YC5 | ||
| JrNF-YC7 |
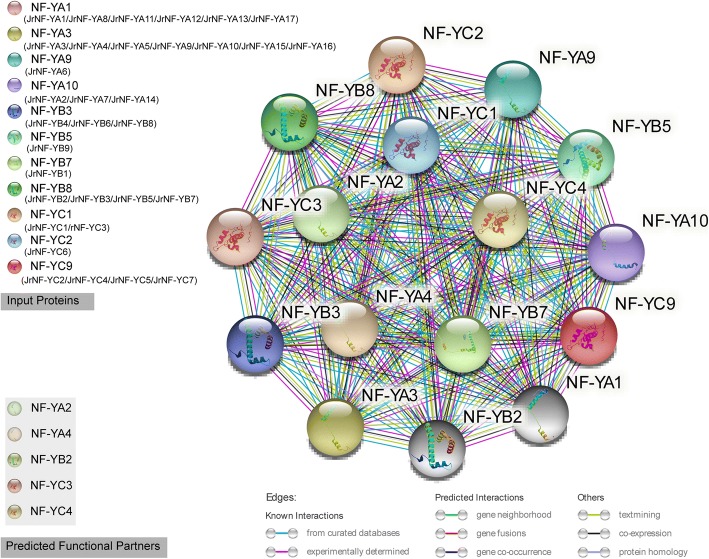
In order to investigate the interaction between the 33 JrNF-Ys, we uploaded the 11 Arabidopsis NF-Y proteins which represented the 33 JrNF-Y proteins to the String website [74]. The interaction networks were mapped out according to the 11 input proteins and their 5 predicted functional partners (Fig. 5). The 11 input proteins were annotated to the common function of stimulating the transcription of various genes by recognizing and binding to a CCAAT motif in promoters. Besides, other functions were annotated to these proteins, such as regulation of timing of transition from vegetative to reproductive phase (NF-YA1), embryo development (NF-YA9), long-day photoperiodism and flowering (NF-YB2), positive regulation of transcription (NF-YA3/ NF-YA3/NF-YA10/NF-YB3/NF-YB5/NF-YB7/NF-YB8/NF-YC1/NF-YC2), abscisic acid-activated signaling pathway (NF-YB6/NF-YC9). In addition, the interaction in NF-YA1&NF-YC3, NF-YA1&NF-YC9, NF-YC2&NF-YB3, NF-YC3&NF-YB2, NF-YC3&NF-YB3, NF-YC9&NF-YB2, NF-YC9&NF-YB3 have been validated by lab experiments (https://string-db.org/).
Fig. 5.
Interaction network of JrNF-Y proteins. The interaction network of the JrNF-Y proteins was constructed using homologous NF-Y proteins in Arabidopsis. The correspondence between the walnut NF-Y proteins (JrNF-Ys) and the Arabidopsis NF-Y proteins (identified in Table 2) are listed in the upper left part of the figure. The network was constructed using the input proteins and six predicted functional partners of the input proteins. The network nodes represent proteins. The 3D structure of the proteins is shown inside the nodes. Edges represent protein-protein associations. The associations are meant to be specific and meaningful, i.e. proteins jointly contribute to a shared function; this does not necessarily mean they are physically bound to each other. The colors of the line indicate different data sources (https://string-db.org/) [74]
Expression patterns of JrNF-Ys in female flower buds and leaf buds
We compared the relative expression (FPKM) of the 33 JrNF-Y genes in F_1, F_2, F_3 and JRL, heat maps were constructed and cluster analysis were conducted to compare the expression (FPKM) patterns of the 33 JrNF-Y genes in F_1, F_2, F_3 and JRL.
Cluster analysis showed that the leaf buds (JRL) have distant relationship with female flower buds (F_1, F_2, and F_3). The relative expressions of seven JrNF-Y genes (i.e., A10/A13/B6/B8/C1/C5/C6) were clustered together for their high expression in F_1, F_2, F_3 and JRL. In contrast, twelve JrNF-Y genes (i.e., A1/A2/A3/A4/A7/A8/A16/B1/B4/B9/C3/C7) were clustered together for their low expression in F_1, F_2, F_3 and JRL (Fig. 6).
Fig. 6.
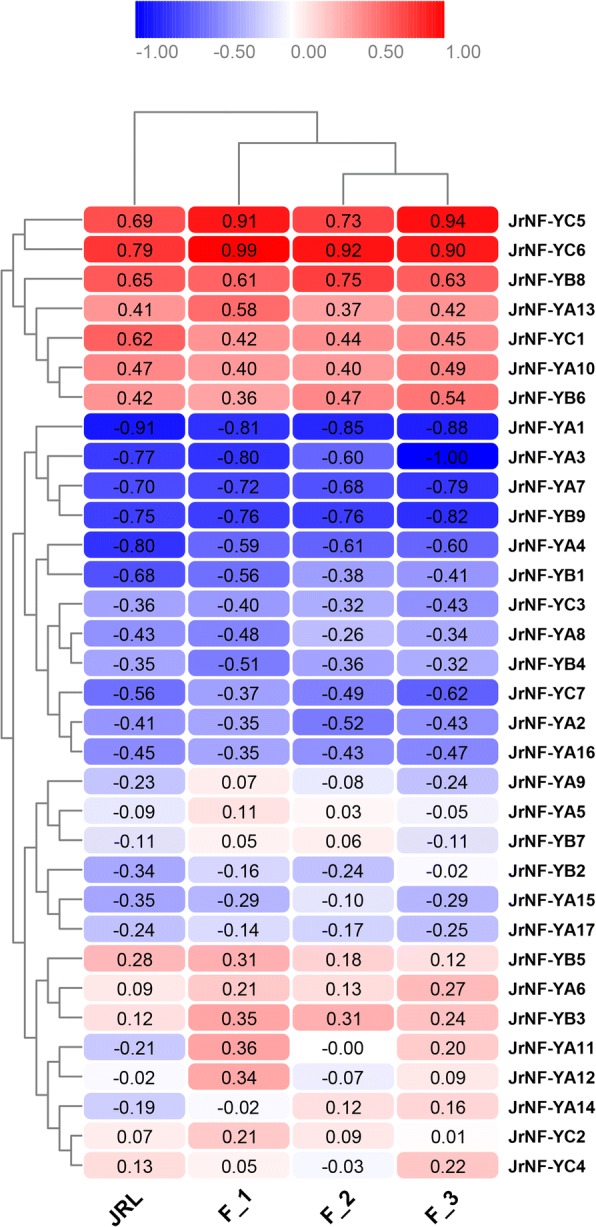
Expression patterns of JrNF-Ys at different development stages and in different walnut tissues. The FPKM values obtained from transcriptome sequencing were used to represent the relative expression level of JrNF-Ys [76]
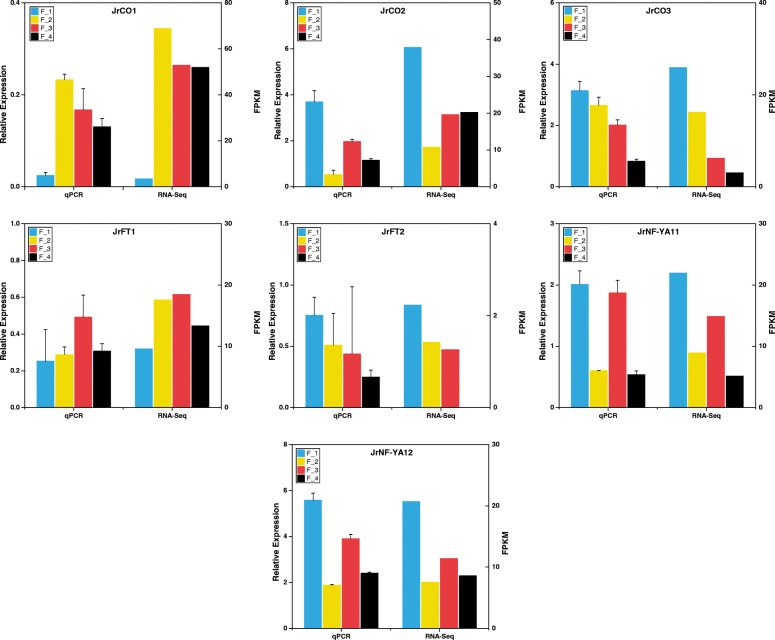
JrNF-YA11 (q valueF_1vsJRL = 0.001, log2ratioF_1vsJRL = 2.02) and JrNF-YA12 (q valueF_1vsJRL = 0.046, log2ratioF_1vsJRL = 1.21; q valueF_1vsF_2 = 0.023, log2ratioF_1vsF_2 = 1.47) were screened out for their differential expression patterns. The expression of JrNF-YA11 was up-regulated in female flower buds before flower transition (F_1) compared with that in leaf buds during flower transition (JRL) (Fig. 6). The expression of JrNF-YA12 in female flower buds before flower transition (F_1) was upregulated compared with that in (i) female flower buds during flower transition (F_2) and (ii) leaf buds during flower transition (JRL).
Some studies indicate that the transcription factor CO competes with other transcription factors (TFs) to regulate the expression of the FT gene [3]. We selected two walnut FTs (JrFT1 and JrFT2) and three walnut COs (JrCO1, JrCO2, and JrCO3) from the transcriptome sequencing data (Additional file 2). The relative expressions of the JrFTs, the JrCOs, and the differentially expressed JrNF-Ys were determined by qPCR (Fig. 7). The expression pattern of JrCO2 was similar to that of JrNF-YA11 and JrNF-YA12, and their expression trend were down-up-down in F_1, F_2, F_3 and JRL. The similarities also exist between the expression pattern of JrCO3 and JrFT2, and their expression trend were continuous decline in F_1, F_2, F_3 and JRL.
Fig. 7.
Expression profile of JrCOs, JrFTs and differential expressed JrNF-Ys. Relative expression levels of two differential expressed JrNF-Ys (JrNF-YA11, JrNF-YA12), three JrCOs (JrCO1, JrCO2, JrCO3), and two JrFTs (JrFT1, JrFT2) were obtained from qPCR experiments and were used to construct a heatmap. The data normalization was performed with the computing method of log10 + 2
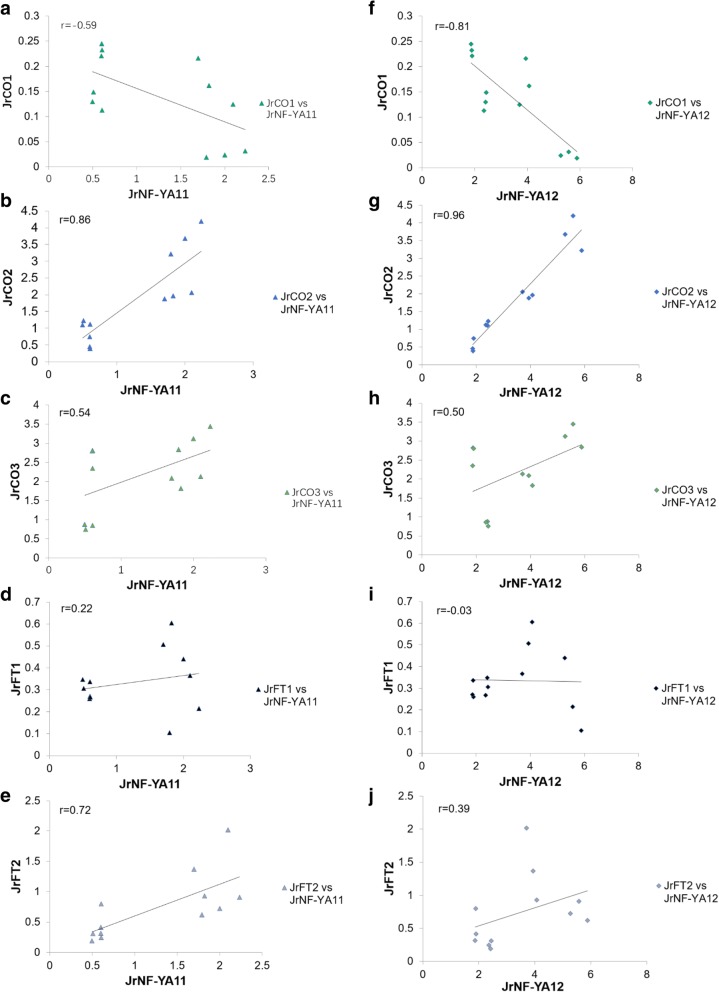
The Pearson Correlation Coefficients among these genes is shown in Fig. 8. JrCO2 showed good correlation with JrNF-YA11 (r = 0.86) and JrNF-YA12 (r = 0.96). JrNF-CO1 was negatively correlated with JrNF-YA12 (r = − 0.81). P-value analysis (Additional file 3) showed that P JrCO2 vs JrNF-YA11 = 0.239 > 0.05, which indicated there were no significant difference between JrCO2 and JrNF-YA11 and validated the correlation between JrCO2 and JrNF-YA11. However, P JrCO2 vs JrNF-YA12 = 0.003 < 0.05, P JrNF-CO1 vs JrNF-YA12 = 0.032 < 0.05, which cannot support the correlation between JrCO2 and JrNF-YA12, and the correlation between JrCO1 and JrNF-YA12.
Fig. 8.
Correlations in expression levels between JrCOs, JrFTs with JrNF-Ys at different periods and in different tissues of walnut. Relative expression levels were used for linear regression analysis. Panels a, b, c, d, e, are for the correlation about JrCO1 vs JrNF-YA11, JrCO2 vs JrNF-YA11, JrCO3 vs JrNF-YA11, JrFT1 vs JrNF-YA11, and JrFT2 vs JrNF-YA11, respectively. Panels f, g, h, i, j, are for the correlation about JrCO1 vs JrNF-YA12, JrCO2 vs JrNF-YA12, JrCO3 vs JrNF-YA12, JrFT1 vs JrNF-YA12, and JrFT2 vs JrNF-YA12, respectively. The correlation coefficient are shown in the figure
Discussion
The cDNA sequences of the JrNF-Y genes were aligned with the walnut genome. The cDNAs that were mapped to the genome segments were considered as the exon regions (e.g., the section between a1 and b1, Fig. 1c). Considering the post- transcriptional processing, we did not judge the adjacent regions (e.g., NW_017389264.1 44,361 to 44,488; Fig. 1c) to be intron regions even though the possibility exists. We only recorded the information about the start and end sites where cDNA matched the genomic scaffold segments (Table 1).
Thirty-six NF-Y protein sequences of Arabidopsis were used to construct a phylogenetic tree with thirty-three NF-Y protein sequences of walnut. Some studies suggest that six NF-Ys (i.e., NF-YB11/12/13 and NF-YC10/11/13) should not be included in the Arabidopsis NF-Y family because they do not include the proper structure [3]. Our phylogenetic tree seems to support this view (Fig. 3). The six NF-Ys of Arabidopsis have a distant evolutionary relationship with the three clusters of NF-YA/B/C. None of the 33 JrNF-Ys was included in the same sub-cluster with the six Arabidopsis NF-Ys mentioned above.
There are obvious differences between NF-Y proteins in animals and plants (Fig. 2). Two amino acids were observed in mouse NF-Y sequences but not in plants NF-YC sequences. However, the evolutionary conservation of NF-Y proteins in mouse and plants was also demonstrated. The conserved region of NF-Y proteins in mouse and plants showed high similarity in all three subfamilies. In previous report [12], NF-YC conserved regions in Arabidopsis strart form the leucine (L) at the twenty-sixth site (Fig. 2). Sequence alignment indictaed that the 25 amino acid residues before the NF-YC conserved regions in previous report were consistent between mouse, Arabidopsis, walnut and orange.
Conservation and differentiation also exists between plants. Absolutely conserved sequences in black boxes/white letters were shared by Arabidopsis, walnut, grape and orange. However, the first five amino acids (QQQLQ) of NF-YC (Fig. 2) were missing in NF-YC sequences of grape, and this situation did not exist in Arabidopsis, walnut and orange.
An interaction network among the 33 JrNF-Y proteins was established based on the 11 correlated Arabidopsis NF-Y proteins. The 11 input proteins were annotated to the function of regulation of timing of transition from vegetative to reproductive phase, embryo development, long-day photoperiodism and flowering, positive regulation of transcription, abscisic acid-activated signaling pathway. In addition, seven protein-protein interactions have been validated by lab experiments and other interaction relationships were predicted (https://string-db.org/). There is no doubt that the network provides valuable information for further research.
With the exception of JrNF-YA10, the expression of the JrNF-Y genes in female flower buds varied among development stages (i.e., before, during, and after flower transition, Fig. 6). This suggests that the NF-Y genes directly or indirectly participate in the process of flower bud development. Previous studies have confirmed or predicted that most NF-Ys are involved in the regulation of flowering time [3, 11, 23–33]. This is also supported by our observation that the expression of 24 of 33 NF-Ys was greater in female flower buds (F_2) than in leaf buds (JRL) (Fig. 2). The expression of nine NF-Ys was greater in leaf buds than in female flower buds. It is possible that these NF-Ys inhibit flowering during the vegetative stage and this need more experimental evidence.
Previous studies have indicated that CO and NF-Y compete to regulate FT expression in Arabidopsis [3]. The NF-YA and CO proteins both can combine with an NF-YB-YC dimer to form either (i) an NF-YA-YB-YC trimer which inhibits FT expression or (ii) an NF-YA-YB-CO trimer which promotes FT expression. In the photoperiodic pathway of Arabidopsis, NF-YA expression is greatest during the day, whereas CO expression is greatest at night. The expression of FT reflects this diurnal pattern. Specifically, expression of FT is low during the day (when NF-YA expression is high) and high during the night (when CO expression is high). We observed that JrCO1 expression was negatively correlated with the expression of both JrNF-YA11 and JrNF-YA12. However, JrFT2 was positively rather than negatively correlated with JrNF-YA11 and JrNF-YA12 in female flower buds (i.e., F_1, F_2, F_3) and in leaf buds (i.e., JRL). JrNF-YA11 and JrNF-YA12 had greater expression in female flower buds during flower transition (F_2) and leaf buds (JRL) than in flower buds before flower transition (F_1) or after flower transition (F_3). A complex network is involved in the regulation of flowering. The expression of FT is regulated by many transcription factors. However, the results suggest that JrCO or other TFs compete with JrNF-YA proteins to combine with the JrNF-YB-YC dimer and promote the expression of JrFT2. This hypothesis needs to be tested in future research work.
Conclusions
Thirty-three JrNF-Ys were identified and their evolutionary, structural, and biological functions were analyzed. The biological function of the JrNF-Y proteins was predicted by comparative analysis with Arabidopsis NF-Y proteins, and this provided a rudimentary understood for the less-studied JrNF-Ys. Further more, Two JrNF-Ys were differentially expressed during the process of flower transition, which revealed that JrNF-Ys might play a role in flower transition. The results of this study may contribute to the future studies about the JrNF-Y family.
Methods
Plant materials
Walnut (J. regia L.) trees were grown under natural conditions in the southern part of the Xinjiang Uyghur Autonomous Region, China. Leaf buds were collected during the flower transition period (JRL) and female flower buds were collected before, during, after the flower transition period (F_1, F_2 and F_3). The leaf buds(JRL) were collected at the same period during the flower transition(F_2). The samples were immediately frozen in liquid N and stored at − 80 °C.
Transcriptome sequencing and de novo assembly
Solexa/Illumina sequencing was carried out by Novogene, Beijing, China. Total RNA was extracted from three female flower buds at each stage (i.e., F_1, F_2, and F_3). Total RNA was extracted from 18 leaf buds (JRL). Total RNA was extracted using RNAout 1.0 (Tianenze, Beijing, China). A total of 1.5 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext ® Ultra™ RNA Library Prep Kit for Illumina ® (NEB, USA). The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia). After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2000 platform and paired-end reads were generated. For the assembly library, clean data(clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. Clean reads were de novo assembled using Trinity [75], and the transcriptome reference database was obtained. FPKM was used to obtain the relative expression levels [76].
Identification of JrNF-Ys
The protein sequences of 36 NF-Y genes (10 NF-YA genes, 13 NF-YB genes, and 13 NF-YC genes) in Arabidopsis were downloaded from TAIR (http://www.arabidopsis.org/) (Additional file 4) [11]. These sequences were used to search our walnut transcriptome database (unpublished) with the tblastn program in BLAST (blast-2.60) [69]. The screening threshold was set as 1e-10. The protein sequences of 10 NF-YA genes, 13 NF-YB genes, and 13 NF-YC genes were used to establish three Hidden Markov Models (HMMs) (Additional file 5) [70]. The three models were used as the query to search the transcriptome database with the screening threshold set at 1e-10. The results of the BLAST and HMMER searchers were merged, resulting in 104 candidate NF-Y genes in walnut.
All 104 candidates were uploaded to the NCBI to verify the existence of the core domain using Conserved Domain Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) Some candidates were abandoned because they lacked the core domain. Other candidates were abandoned because their sequences were either too long or too short. Finally, 33 unigenes were identified and translated into amino acid sequences (Additional file 6).
Multiple alignments and phylogenetic analyses
Clustal X 2.1 [71] was used to align the protein sequences of the JrNF-Y genes. The conserved regions of the three subfamilies were identified using Arabidopsis as a reference. The conserved domains of three subfamilies in Arabidopsis, walnut, grape, orange and mouse were uploaded to the ESPript website (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) for editing [77]. The three subfamily sequences of Arabidopsis, grape and orange were download from the website of PlantTFDB (http://planttfdb.cbi.pku.edu.cn/) [78], and then HMM model of NF-YA, NF-YB, NF-YC of Arabidopsis, grape and orange were built based on these sequences (Additional file 5).
Protein sequences of NF-Y genes in Arabidopsis and walnut were used to construct a neighbor-joining tree with 1000 bootstrap replications using MEGA 6 software [79]. The phylogenetic tree constructed by MEGA was uploaded to iTOL (http://itol.embl.de/) for further editing. Motifs were predicted using MEME software (http://meme-suite.org/tools/meme). A protein interaction network was constructed with String software (https://string-db.org/) [74].
Quantitative real-time PCR
Total RNA was extracted using RNAout 1.0 (Tianenze, Beijing, China) by Novogene, Beijing, China. The synthesis of cDNA was performed using a PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). Real-time quantification was performed using a CFX manager (Bio-Rad, USA) with the SYBR Green Realtime PCR Master Mix (Toyobo, Osaka, Japan). The protocol of the real-time PCR was as follows: initiation with 95 °C for 5 min, followed by 40 cycles for 30 s at 94 °C, 30 s at 55 °C, and 30 s at 72 °C. A melting curve was included from 65 to 95 °C to verify the specificity of the amplified product. Each reaction was repeated three times. Walnut actin gene (forward: 5′-CCATCCAGGCTGTTCTCTC-3′, and reverse: 5′-GCAAGGTCCAGACGAAGG -3′) and walnut gadph gene (forward: 5′-ATTTGGAATCGTTGAGGGTCTTATG-3′ and reverse: 5′- AATGATGTTGAAGGAAGCAGCAC-3′) were used as the normalizer (Additional file 7). The results were evaluated by the method of the 2 -ΔCt [80].
Differential expression analysis
Prior to differential gene expression analysis, the read counts for each sequenced library were adjusted with EdgeR software. Differential expression analysis of two samples was performed using the DEGseq (2010) R package. The thresholds for significant differential expression were qvalue < 0.05 and |log2(foldchange)| > 1.
Additional files
Figure S1. The conserved regions in the full length of the JrNF-Ys. (DOC 2960 kb)
Unigene sequences of JrCOs and JrFTs. (DOC 36.0 kb)
Table S1. Correlation and P-value in Expression Level. (DOC 40.0 kb)
Full length and conserved sequences of the Arabidopsis and mouse NF-Ys. (DOC 44.0 kb)
The HMM models and domain sequences of NF-YA, NF-YB and NF-YC of Arabidopsis, grape and orange. (DOC 44.0 kb)
Unigene sequences and translated amino acid sequences of 33 walnut NF-Ys. (DOC 116 kb)
Primers involved in this article. (DOC 32.0 kb)
Acknowledgments
We thank Dr. Gale (Shihezi University) for careful editing of this manuscript.
Funding
This work was supported by the important National Science and Technology Specific projects of Xinjiang (No. 201130102–1-4) and the National Natural Science Foundation of China (No. 30560090).
Availability of data and materials
Data generated or analyzed during this study are included in this article and its supplementary information files.
Abbreviations
- CBF
CCAAT binding factor
- CO
CONSTANS
- ER
Endoplasmic reticulum
- F_1
Female flower buds before flower transition
- F_2
Female flower buds during flower transition
- F_3
Female flower buds after flower transition
- FT
FLOWERING LOCUS T
- HAP
Heme activator protein
- HMMs
Hidden Markov Models
- JRL
Leaf buds during flower transition
- qRT-PCR
Real-time quantitative PCR
- TFs
Transcription factors
Authors’ contributions
JXN led and coordinated the project, JXN and SWQ designed the study, SWQ, LZ, HX, LM and YQ collected the plant materials and isolated the RNA. SWQ and LZ conducted the real-time quantitative PCR. SWQ conducted the bioinformatics analysis and wrote the paper. All authors have read and agree with the final manuscript. JXN is the corresponding author and is responsible for all contact and correspondence. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shaowen Quan, Email: 731982806@qq.com.
Jianxin Niu, Email: njx105@163.com.
Li Zhou, Email: 125492367@qq.com.
Hang Xu, Email: 254567382@qq.com.
Li Ma, Email: 2606516390@qq.com.
Yang Qin, Email: 742621732@qq.com.
References
- 1.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239(1):15–27. doi: 10.1016/S0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 2.Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, Vonrhein C, Moras D, Romier C, Bolognesi M, et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell. 2013;152(1–2):132–143. doi: 10.1016/j.cell.2012.11.047. [DOI] [PubMed] [Google Scholar]
- 3.Zhao H, Wu D, Kong F, Lin K, Zhang H, Li G. The Arabidopsis thaliana nuclear factor Y transcription factors. Front Plant Sci. 2016;7:2045. doi: 10.3389/fpls.2016.02045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romier C, Cocchiarella F, Mantovani R, Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J Biol Chem. 2003;278(2):1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- 5.Sinha S, Maity SN, Lu J, de Crombrugghe B. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci U S A. 1995;92(5):1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinha S, Kim IS, Sohn KY, de Crombrugghe B, Maity SN. Three classes of mutations in the a subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol Cell Biol. 1996;16(1):328–337. doi: 10.1128/MCB.16.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maity SN, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23(5):174–178. doi: 10.1016/S0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 8.Hackenberg D, Wu Y, Voigt A, Adams R, Schramm P, Grimm B. Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol Plant. 2012;5(4):876–888. doi: 10.1093/mp/ssr107. [DOI] [PubMed] [Google Scholar]
- 9.Laloum Tom, De Mita Stéphane, Gamas Pascal, Baudin Maël, Niebel Andreas. CCAAT-box binding transcription factors in plants: Y so many? Trends in Plant Science. 2013;18(3):157–166. doi: 10.1016/j.tplants.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Li XY, HvHR MR, Benoist C, Mathis D. Intron-exon organization of the NF-Y genes. Tissue-specific splicing modifies an activation domain. J Biol Chem. 1992;267:8984–8990. [PubMed] [Google Scholar]
- 11.Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G, Holt BF., 3rd Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009;149(2):625–641. doi: 10.1104/pp.108.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF, 3rd, Mantovani R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell. 2012;24(12):4777–4792. doi: 10.1105/tpc.112.105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephenson TJ, McIntyre CL, Collet C, Xue GP. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol Biol. 2007;65(1–2):77–92. doi: 10.1007/s11103-007-9200-9. [DOI] [PubMed] [Google Scholar]
- 14.Yan DHXX, Yin WL. NF-YB family genes identified in a poplar genome-wide analysis and expressed in Populus euphratica are responsive to drought stress. Plant Mol Biol Rep. 2013;31:363–370. doi: 10.1007/s11105-012-0508-5. [DOI] [Google Scholar]
- 15.Quach TN, Nguyen HT, Valliyodan B, Joshi T, Xu D, Nguyen HT. Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol Gen Genomics. 2015;290(3):1095–1115. doi: 10.1007/s00438-014-0978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang M, Yin X, Lin Z, Zheng Q, Liu G, Zhao G. Identification and characterization of NF-Y transcription factor families in canola (Brassica napus L.) Planta. 2014;239(1):107–126. doi: 10.1007/s00425-013-1964-3. [DOI] [PubMed] [Google Scholar]
- 17.Ripodas C, Castaingts M, Clua J, Blanco F, Zanetti ME. Annotation, phylogeny and expression analysis of the nuclear factor Y gene families in common bean (Phaseolus vulgaris) Front Plant Sci. 2014;5:761. doi: 10.3389/fpls.2014.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F, Han M, Lv Q, Bao F, He Y. Identification and expression profile analysis of NUCLEAR FACTOR-Y families in Physcomitrella patens. Front Plant Sci. 2015;6:642. doi: 10.3389/fpls.2015.00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren C, Zhang Z, Wang Y, Li S, Liang Z. Genome-wide identification and characterization of the NF-Y gene family in grape (vitis vinifera L.) BMC Genomics. 2016;17(1):605. doi: 10.1186/s12864-016-2989-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Li K, Ju Z, Cao D, Fu D, Zhu H, Zhu B, Luo Y. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genomics. 2016;17(1):36. doi: 10.1186/s12864-015-2334-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Zhu JH, Yang YX. Genome-wide identification and expression analysis of NF-Y transcription factor families in watermelon (Citrullus lanatus) J Plant Growth Regul. 2017;36(3):590–607. doi: 10.1007/s00344-017-9670-1. [DOI] [Google Scholar]
- 22.Pereira Suzam L. S., Martins Cristina P. S., Sousa Aurizangela O., Camillo Luciana R., Araújo Caroline P., Alcantara Grazielle M., Camargo Danielle S., Cidade Luciana C., de Almeida Alex-Alan F., Costa Marcio G. C. Genome-wide characterization and expression analysis of citrus NUCLEAR FACTOR-Y (NF-Y) transcription factors identified a novel NF-YA gene involved in drought-stress response and tolerance. PLOS ONE. 2018;13(6):e0199187. doi: 10.1371/journal.pone.0199187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006;46(3):462–476. doi: 10.1111/j.1365-313X.2006.02706.x. [DOI] [PubMed] [Google Scholar]
- 24.Cai X, Ballif J, Endo S, Davis E, Liang M, Chen D, DeWald D, Kreps J, Zhu T, Wu Y. A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 2007;145(1):98–105. doi: 10.1104/pp.107.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen NZ, Zhang XQ, Wei PC, Chen QJ, Ren F, Chen J, Wang XC. AtHAP3b plays a crucial role in the regulation of flowering time in Arabidopsis during osmotic stress. J Biochem Mol Biol. 2007;40(6):1083–1089. doi: 10.5483/bmbrep.2007.40.6.1083. [DOI] [PubMed] [Google Scholar]
- 26.Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, Marion CM, Hempel FD, Ratcliffe OJ. The nuclear factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta. 2008;228(5):709–723. doi: 10.1007/s00425-008-0773-6. [DOI] [PubMed] [Google Scholar]
- 27.Kumimoto RW, Zhang Y, Siefers N, Holt BF., 3rd NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 2010;63(3):379–391. doi: 10.1111/j.1365-313X.2010.04247.x. [DOI] [PubMed] [Google Scholar]
- 28.Cao S, Kumimoto RW, Siriwardana CL, Risinger JR, Holt BF., 3rd Identification and characterization of NF-Y transcription factor families in the monocot model plant Brachypodium distachyon. PLoS One. 2011;6(6):e21805. doi: 10.1371/journal.pone.0021805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao S, Kumimoto RW, Gnesutta N, Calogero AM, Mantovani R, Holt BF., 3rd A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell. 2014;26(3):1009–1017. doi: 10.1105/tpc.113.120352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan MR, Ai XY, Zhang JZ. Genetic regulation of flowering time in annual and perennial plants. Wiley interdisciplinary reviews RNA. 2014;5(3):347–359. doi: 10.1002/wrna.1215. [DOI] [PubMed] [Google Scholar]
- 31.Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18(11):2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hackenberg D, Keetman U, Grimm B. Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int J Mol Sci. 2012;13(3):3458–3477. doi: 10.3390/ijms13033458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brambilla V, Fornara F. Y flowering? Regulation and activity of CONSTANS and CCT-domain proteins in Arabidopsis and crop species. Biochim Biophys Acta. 2017;1860(5):655–660. doi: 10.1016/j.bbagrm.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 34.Ballif J, Endo S, Kotani M, MacAdam J, Wu Y. Over-expression of HAP3b enhances primary root elongation in Arabidopsis. Plant Physiol Biochem. 2011;49(6):579–583. doi: 10.1016/j.plaphy.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 35.Sorin C, Declerck M, Christ A, Blein T, Ma L, Lelandais-Briere C, Njo MF, Beeckman T, Crespi M, Hartmann C. A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 2014;202(4):1197–1211. doi: 10.1111/nph.12735. [DOI] [PubMed] [Google Scholar]
- 36.Lotan TOM, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/S0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 37.Kwong RW. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. The Plant Cell Online. 2002;15(1):5–18. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.West M, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and Cotyledon identity in Arabidopsis. Plant Cell. 1994;6(12):1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee H, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci U S A. 2003;100(4):2152–2156. doi: 10.1073/pnas.0437909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mu J, Tan H, Hong S, Liang Y, Zuo J. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant. 2013;6(1):188–201. doi: 10.1093/mp/sss061. [DOI] [PubMed] [Google Scholar]
- 41.Huang M, Hu Y, Liu X, Li Y, Hou X. Arabidopsis LEAFY COTYLEDON1 controls cell fate determination during post-embryonic development. Front Plant Sci. 2015;6:955. doi: 10.3389/fpls.2015.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fornari M, Calvenzani V, Masiero S, Tonelli C, Petroni K. The Arabidopsis NF-YA3 and NF-YA8 genes are functionally redundant and are required in early embryogenesis. PLoS One. 2013;8(11):e82043. doi: 10.1371/journal.pone.0082043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siriwardana CL, Kumimoto RW, Jones DS, Holt BF., 3rd Gene family analysis of the Arabidopsis NF-YA transcription factors reveals opposing Abscisic acid responses during seed germination. Plant Mol Biol Report. 2014;32(5):971–986. doi: 10.1007/s11105-014-0704-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernie T, Ott T, Gamas P, Crespi M, et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006;20(22):3084–3088. doi: 10.1101/gad.402806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephenson TJMC, Collet C, Xue GP. TaNF-YC11, one of the light-upregulated NF-YC members in Triticum aestivum, is co-regulated with photosynthesis-related genes. Funct Integr Genomics. 2010;10:265–276. doi: 10.1007/s10142-010-0158-3. [DOI] [PubMed] [Google Scholar]
- 46.Kusnetsov VLM, Meurer J, Oelmuller R. The assembly of the CAAT-box binding complex at a photosynthesis gene promoter is regulated by light, cytokinin, and the stage of the plastids. J Biol Chem. 1999;274:36009–36014. doi: 10.1074/jbc.274.50.36009. [DOI] [PubMed] [Google Scholar]
- 47.Stephenson TJ, McIntyre CL, Collet C, Xue GP. TaNF-YB3 is involved in the regulation of photosynthesis genes in Triticum aestivum. Funct Integr Genomics. 2011;11(2):327–340. doi: 10.1007/s10142-011-0212-9. [DOI] [PubMed] [Google Scholar]
- 48.Alam MM, Tanaka T, Nakamura H, Ichikawa H, Kobayashi K, Yaeno T, Yamaoka N, Shimomoto K, Takayama K, Nishina H, et al. Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnol J. 2015;13(1):85–96. doi: 10.1111/pbi.12239. [DOI] [PubMed] [Google Scholar]
- 49.Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22(3):782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6alpha and 6beta that activates the mammalian unfolded protein response. Mol Cell Biol. 2001;21(4):1239–1248. doi: 10.1128/MCB.21.4.1239-1248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci U S A. 2007;104(42):16450–16455. doi: 10.1073/pnas.0707193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20(8):2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang M, Zhao Y, Shi S, Du X, Gu J, Xiao K. Wheat nuclear factor Y (NF-Y) B subfamily gene TaNF-YB3;l confers critical drought tolerance through modulation of the ABA-associated signaling pathway. Plant Cell, Tissue and Organ Culture (PCTOC) 2016;128(1):97–111. doi: 10.1007/s11240-016-1088-0. [DOI] [Google Scholar]
- 54.Li Yan-Jie, Fang Yi, Fu Ya-Ru, Huang Jin-Guang, Wu Chang-Ai, Zheng Cheng-Chao. NFYA1 Is Involved in Regulation of Postgermination Growth Arrest Under Salt Stress in Arabidopsis. PLoS ONE. 2013;8(4):e61289. doi: 10.1371/journal.pone.0061289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leyva-Gonzalez MA, Ibarra-Laclette E, Cruz-Ramirez A, Herrera-Estrella L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS One. 2012;7(10):e48138. doi: 10.1371/journal.pone.0048138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi H, Ye T, Zhong B, Liu X, Jin R, Chan Z. AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol. 2014;203(2):554–567. doi: 10.1111/nph.12812. [DOI] [PubMed] [Google Scholar]
- 57.Zhao M, Ding H, Zhu JK, Zhang F, Li WX. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011;190(4):906–915. doi: 10.1111/j.1469-8137.2011.03647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato H, Mizoi J, Tanaka H, Maruyama K, Qin F, Osakabe Y, Morimoto K, Ohori T, Kusakabe K, Nagata M, et al. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell. 2014;26(12):4954–4973. doi: 10.1105/tpc.114.132928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zanetti ME, Ripodas C, Niebel A. Plant NF-Y transcription factors: key players in plant-microbe interactions, root development and adaptation to stress. Biochim Biophys Acta. 2017;1860(5):645–654. doi: 10.1016/j.bbagrm.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 60.Karimi REA, Vahdati K, Woeste K. Molecular characterization of Persian walnut populations in Iran with microsatellite markers. Hortscience. 2010:45(9):1403–1406.
- 61.Amiri R, Vahdati K, Mohsenipoor S, Mozaffari MR, Leslie C. Correlations between some horticultural traits in walnut. Hortscience. 2010;45(11):1690–1694. [Google Scholar]
- 62.Pop IF, Vicol AC, Botu M, Raica PA, Vahdati K, Pamfil D. Relationships of walnut cultivars in a germplasm collection: comparative analysis of phenotypic and molecular data. Sci Hortic. 2013;153(Issue):124–135. doi: 10.1016/j.scienta.2013.02.013. [DOI] [Google Scholar]
- 63.Avanzato D, McGranahan G, Vahdati K, Botu MIL, VA J. Following walnut footprints (Juglans regia L.). cultivation and culture, folklore and history, traditions and uses. Belgium: ISHS; 2014. [Google Scholar]
- 64.Vahdati K, Pourtaklu SM, Karimi R, Barzehkar R, Amiri R, Mozaffari M, Woeste K. Genetic diversity and gene flow of some Persian walnut populations in southeast of Iran revealed by SSR markers. Plant Systematics Evolution. 2015;301(2):691–699. doi: 10.1007/s00606-014-1107-8. [DOI] [Google Scholar]
- 65.Hassankhah A, Rahemi M, Mozafari MR, Vahdati K. Flower development in walnut: altering the flowering pattern by Gibberellic acid application. Notulae Botanicae Horti Agrobotanici Cluj-Napoca. 2018;46(2):700. doi: 10.15835/nbha46211183. [DOI] [Google Scholar]
- 66.Martinez-Garcia PJ, Crepeau MW, Puiu D, Gonzalez-Ibeas D, Whalen J, Stevens KA, Paul R, Butterfield TS, Britton MT, Reagan RL, et al. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. 2016;87(5):507–532. doi: 10.1111/tpj.13207. [DOI] [PubMed] [Google Scholar]
- 67.Barker D, Pagel M. Predicting functional gene links from phylogenetic-statistical analyses of whole genomes. PLoS Comput Biol. 2005;1(1):e3. doi: 10.1371/journal.pcbi.0010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aoki K, Ogata Y, Shibata D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant & cell physiology. 2007;48(3):381–390. doi: 10.1093/pcp/pcm013. [DOI] [PubMed] [Google Scholar]
- 69.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 71.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 72.Coustry F, Maity SN, Sinha S, deCrombrugghe B. The transcriptional activity of the CCAAT-binding factor CBF is mediated by two distinct activation domains, one in the CBF-B subunit and the other in the CBF-C subunit. J Biol Chem. 1996;271(24):14485–14491. doi: 10.1074/jbc.271.24.14485. [DOI] [PubMed] [Google Scholar]
- 73.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(Web Server issue):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45(Database issue):D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods (San Diego, Calif) 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The conserved regions in the full length of the JrNF-Ys. (DOC 2960 kb)
Unigene sequences of JrCOs and JrFTs. (DOC 36.0 kb)
Table S1. Correlation and P-value in Expression Level. (DOC 40.0 kb)
Full length and conserved sequences of the Arabidopsis and mouse NF-Ys. (DOC 44.0 kb)
The HMM models and domain sequences of NF-YA, NF-YB and NF-YC of Arabidopsis, grape and orange. (DOC 44.0 kb)
Unigene sequences and translated amino acid sequences of 33 walnut NF-Ys. (DOC 116 kb)
Primers involved in this article. (DOC 32.0 kb)
Data Availability Statement
Data generated or analyzed during this study are included in this article and its supplementary information files.