Abstract
Background
Abasic sites are formed spontaneously and by nucleobase chemical modifications and base excision repair. A chemically stable abasic site analog was site-specifically introduced into replicable plasmid DNAs, which were transfected into human U2OS cells. The amplified DNAs were recovered from the cells and used for the transformation of a bacterial indicator strain.
Results
Large deletion mutations were induced by the analog, in addition to point mutations at the modified site. No apparent sequence homology at the deletion junctions was found.
Conclusion
These results suggested that the large deletions induced by the abasic site analog are formed by homology-independent events.
Keywords: Abasic site, Abasic site analog, Large deletion
Background
Chemical modifications of nucleic acids have severe effects on organisms because they disturb replication, transcription, and translation, resulting in mutagenesis, carcinogenesis, and cell death. Various DNA lesions are formed spontaneously and by mutagens, such as reactive oxygen species and ultraviolet light [1–5]. An abasic (apurinic/apyrimidinic, AP) site is formed by the spontaneous hydrolysis of the N-glycosyl bonds of DNA, and this hydrolysis is accelerated by the chemical modifications of bases [6]. Moreover, this DNA lesion is produced as an intermediate of base excision repair when a monofunctional DNA glycosylase, such as uracil DNA glycosylase, acts on a damaged base. Approximately 10,000–50,000 abasic sites are reportedly present per mammalian cell under normal physiological conditions [7, 8]. Thus, the abasic site is one of the important DNA lesions causing mutagenesis/carcinogenesis, due to its abundance and loss-of-base character.
In general, the incorporation of a 2′-deoxyribonucleotide opposite the abasic site and its analogs by mammalian replicative DNA polymerases (pols) is inefficient. For example, in vitro DNA synthesis by human DNA pol ε is severely blocked at the nucleotide preceding an abasic site analog [9]. Instead, translesion synthesis DNA pols are considered to conduct the bypass of the DNA lesion [10]. Due to the lack of a nucleobase, DNA pols “misincorporate” nucleotides opposite the DNA lesion. In living mammalian cells, various point mutations are induced by the abasic site (analog), depending on its opposite bases, flanking sequences, and cell type [11–15].
Previously, large deletion mutations were found when shuttle vectors containing ultraviolet-induced DNA lesions, cyclobutane and 6–4 thymine-thymine dimers, were replicated in simian cells [16]. Since these dimers block nucleotide incorporation by replicative DNA pols as with the abasic site [17], it may also cause large deletion mutations in addition to point mutations. In this study, we focused on deletion mutations upon the transfection of a shuttle plasmid DNA bearing a chemically stable, frequently used tetrahydrofuran analog (THF) of the abasic site. The analog actually caused large deletion mutations in living human cells, without any apparent homology at the deletion junctions.
Methods
Materials
The oligodeoxyribonucleotides (ODNs) used for plasmid construction and sequence analysis are listed in Table 1. ODN-1 and ODN-2 were previously synthesized [18, 19]. ODN-3 and ODN-5 containing THF, and ODN-4 containing G instead of THF, were synthesized in this study. These ODNs were chemically phosphorylated on the support and purified by HPLC, as described previously [20]. PCR primers were purchased from Hokkaido System Science (Sapporo, Japan) and Eurofins Genomics (Tokyo, Japan) in purified forms. Escherichia coli KS40/pOF105, used as the indicator strain of the supF mutants, was provided by Professor Tatsuo Nunoshiba of the International Christian University [21].
Table 1.
Oligodeoxyribonucleotides used in this study
| Name | Sequence (5′- > 3′)a |
|---|---|
| ODN-1 | P-CGACTTCGAAGGTTCGAATCC |
| ODN-2 | P-CGACTTCGAAGOTTCGAATCC |
| ODN-3 | P-CGACTTCGAAGFTTCGAATCC |
| ODN-4 | P-TCCGAAAGAATTGAGCGTCAGA |
| ODN-5 | P-TCCGAAAGAATTFAGCGTCAGA |
| ODN-seq1 | GGCGGTGCTACAGAGTTCTT |
| ODN-seq2 | GCACCCAACTGATCTTCAGC |
aF, O, and P represent THF, GO, and the phosphate, respectively
Plasmid DNA containing THF
The double-stranded shuttle vectors containing THF were prepared from the single-stranded forms of pZ189-T_E107K/D402E (formerly pZ189-107 K/402E) and ODN-3 or ODN-5 by enzymatic reactions catalyzed by T4 DNA pol and T4 DNA ligase, as described [22, 23]. To obtain highly pure DNAs, they were treated with Plasmid-Safe ATP-Dependent DNase (Epicentre, Madison, WI, USA). The DNAs were purified with a PureLink PCR Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA) to remove proteins, ODNs, and mononucleotides before and after the DNase treatment. The plasmid DNAs bearing G or 8-oxo-7,8-dihydroguanine (GO) were prepared by the same procedures.
Transfection of plasmid DNAs and mutant frequency determination
U2OS cells (5.0 × 104 cells) were cultured in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum. After 24 h, 100 ng (29 fmol) of the plasmid DNA bearing THF was transfected with Lipofectamine (Thermo Fisher Scientific), and the cells were cultured for 48 h. The plasmid molecules propagated in the U2OS cells was isolated by the Hirt procedure [24]. The DNA was extensively digested by DpnI to break down the unreplicated plasmid. E. coli KS40/pOF105 cells were electroporated with the plasmid, and the supF mutant frequency was calculated [21, 25]. The plasmids bearing G and GO, instead of THF, were transfected as controls. The sequence of the mutant supF plasmid was analyzed, using the ODN-seq1 and ODN-seq2 primers (Table 1).
Results
Induction of large deletions by an abasic site analog in the supF gene (experiment 1)
First, we incorporated THF into position 122 of the supF gene. The mutational properties of other DNA lesions, GO and O6-methylguanine, in this position have previously been examined [18, 19, 26]. The plasmid bearing the unique THF was prepared by enzyme reactions using ODN-3 (Table 1). The DNA was transfected into human U2OS cells, recovered from the cells, and introduced into E. coli KS40/pOF105 cells. The numbers of colonies on the titer plate and the selection plates containing nalidixic acid, streptomycin, and X-gal were counted [21, 25]. Mutations in the supF gene confer resistance to the two antibiotics and the lacZ− phenotype to E. coli. The plasmid DNAs containing G and GO, instead of THF, were used as controls.
The numbers of colonies on the titer plates, which semi-quantitatively reflect the amounts of plasmid DNA amplified in the U2OS cells, were comparable for all progeny plasmids, indicating the similar replication efficiencies of the G-, GO-, and THF-plasmids and THF removal by DNA repair. The supF mutant cells formed white or pale blue colonies on the selection plates. The supF mutant frequency was calculated by dividing the numbers of supF mutant colonies on the selection plates by those of the total colonies on the titer plates.
The supF mutant frequency was 1.1 × 10− 3 when the control plasmid containing G, instead of THF, was transfected (Fig. 1A). This value was similar to the expected one calculated by the error frequency of the DNA pol used in the plasmid preparation and the length of the supF gene, suggesting that a significant portion of the mutations was due to nucleotide misincorporation during the plasmid preparation [27]. The replacement of the G with THF at position 122 greatly enhanced the supF mutant frequency (1.2 × 10− 2). This value was much higher than the mutant frequency for another major DNA lesion, GO.
Fig. 1.
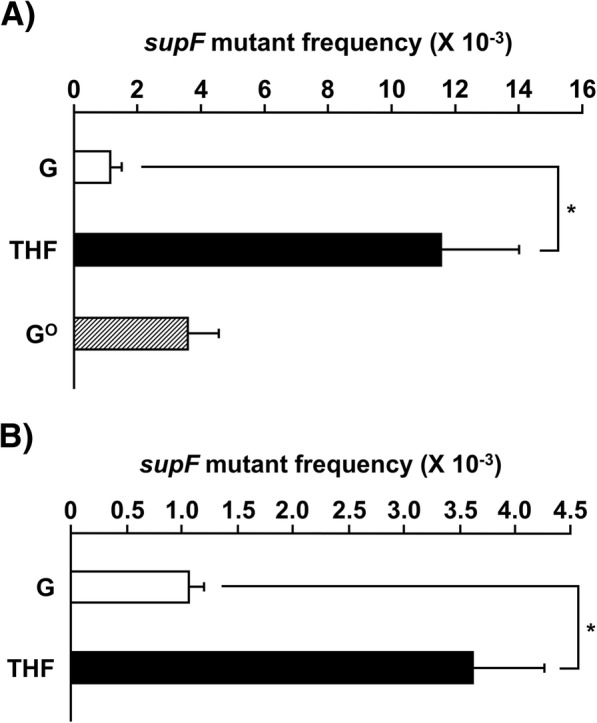
The supF mutant frequency in U2OS cells transfected with plasmid DNA containing THF (a) at position 122 of the supF gene and (b) outside of the gene (at “position 171”). Transfection experiments were performed four times. Data are expressed as the means + standard errors. *P < 0.05 vs. control plasmid (Student’s t-test)
We next analyzed the supF plasmids in the mutant colonies (Tables 2 and 3). Previously, THF has been shown to induce base-substitution mutations [15]. These types of mutations were also found in the experiment. THF➔C and THF➔T mutations were observed as targeted substitutions in this study. In addition, a targeted − 1 deletion was detected. Moreover, as we expected, large deletion mutations were present among the mutants. The frequencies of the large deletion mutations were calculated as the products of the total supF mutant frequencies and the ratios of the large deletions. The frequencies were 4.8 × 10− 4 and 2.5 × 10− 3 in the G and THF experimental groups, respectively. In the case of GO, the frequency was 6.4 × 10− 4, indicating that not every DNA lesion induces large deletion mutations. Thus, the abasic site analog, but not GO, caused large deletions in human cells.
Table 2.
Mutations detected in the supF gene (experiment 1)a
| THF | |
|---|---|
| 11C- > A, 95C- > A, 101C- > T | 1 |
| 39G- > GG, 101C- > T, 111C- > T | 1 |
| 70C- > T, 71C- > T | 1 |
| 73A- > C, 74G- > A | 1 |
| 73G- > T, 118G- > A | 2 |
| 74A- > C, 75G- > A | 2 |
| 86G- > T, 91G- > A | 2 |
| 86G- > T, 95C- > A, 101C- > T, 138C- > T, 147C- > T, 161C- > A | 1 |
| 95C- > G | 2 |
| 101C- > T, 125C- > G | 1 |
| 121G- > C, 122G- > T | 1 |
| 122G- > T | 2 |
| large insetion | 1 |
| large deletion | 6 |
| large deletion, 99A- > G | 1 |
| large deletion, large insertion | 5 |
| total colonies analyzed | 30 |
| GO | |
| 5G- > C, 85G- > T | 2 |
| 5G- > A, 19C- > T, 27G- > A | 1 |
| 5G- > A, 19C- > T, 27G- > A, 118G- > T | 1 |
| 5G- > A, 27G- > T, 126G- > C | 1 |
| 91G- > GG, 118G- > T, 126G- > A | 1 |
| 95C- > G | 1 |
| 96 T- > G | 2 |
| 121G- > T | 1 |
| 122G- > T | 23 |
| 122G- > C | 8 |
| 122 deletion | 1 |
| large deletion | 7 |
| large deletion, large insertion | 1 |
| total colonies analyzed | 50 |
| THF | |
| 5G- > C, 27G- > C, 33G- > A, 40G- > A, 73G- > C, 126G- > C | 2 |
| 5G- > C, 27G- > A, 91G- > T | 1 |
| 5G- > C, 26C- > A, 27G- > A, 112G- > T, 121G- > T | 1 |
| 27G- > A, 73G- > A, 118G- > A | 1 |
| 27G- > C, 65G- > T, 73G- > T | 1 |
| 27G- > T, 73G- > C, 78G- > T, 118G- > A | 1 |
| 73G- > C, 106G- > A | 1 |
| 91 G- > A 112 G- > A | 1 |
| 91G- > C, 112G- > A | 1 |
| 91G- > A, 112G- > A, 126G- > C | 1 |
| 95C- > A, 101C- > T | 1 |
| 117C- > G, 126G- > T | 2 |
| 118G- > C | 2 |
| 121GG- > T | 1 |
| 122G- > AAGA | 1 |
| 122G- > T | 7 |
| 122G- > C | 11 |
| 122 deletion | 11 |
| 122G- > T, 127 deletion | 1 |
| large deletion | 10 |
| large deletion, 122G- > T | 1 |
| large deletion, large insetion | 1 |
| total colonies analyzed | 60 |
aMutations detected in single colonies are represented. The sequence of the upper strand is shown. The numbers of colonies are shown on the right side
Table 3.
Spectra of mutations detected in the supF gene (experiment 1)a
| G | GO | THF | ||
|---|---|---|---|---|
| point mutations at position 122 | ||||
| G:C - > A:Tb | 0 (0) | 0 (0) | 0 (0) | |
| G:C - > T:Ac | 3 (10) | 23 (46) | 9 (15) | |
| G:C - > C:Gd | 0 (0) | 8 (16) | 11 (18) | |
| deletion | 0 (0) | 1 (2) | 11 (18) | |
| mutations at other positions | ||||
| transition | ||||
| A:T - > G:C | 1 (3) | 0 (0) | 0 (0) | |
| G:C - > A:T | 16 (53) | 8 (16) | 17 (28) | |
| transversion | ||||
| A:T - > T:A | 0 (0) | 0 (0) | 0 (0) | |
| A:T - > C:G | 3 (10) | 2 (4) | 0 (0) | |
| G:C - > T:A | 9 (30) | 6 (12) | 11 (18) | |
| G:C - > C:G | 4 (13) | 4 (8) | 19 (32) | |
| small insertion (1–2 bp) | 1 (3) | 1 (2) | 0 (0) | |
| large insertion (> 2 bp) | 6 (20) | 1 (2) | 1 (2) | |
| small deletion (1–2 bp) | 0 (0) | 0 (0) | 1 (2) | |
| large deletion (> 2 bp) | 12 (40) | 8 (16) | 12 (20) | |
| others | 0 (0) | 0 (0) | 2 (3)e | |
| total mutations | 55 | 62 | 94 | |
| total colonies analyzed | 30 (100) | 50 (100) | 60 (100) | |
aAll data are represented as cases found (%)
bThe mutation corresponds to incorporation of TMP opposite G/GO/THF
cThe mutation corresponds to incorporation of dAMP opposite G/GO/THF
dThe mutation corresponds to incorporation of dGMP opposite G/GO/THF
eGG - > T at positions 121–122 and G - > AAGA at position 122
Induction of large deletions including the supF gene by an abasic site analog located outside the gene (experiment 2)
Next, we incorporated THF outside the supF gene, to detect large deletion mutations more easily. The analog was introduced into the site located 9 “bases” downstream of the gene, which has a length of 162 bases. We named this site “position 171”, although it is outside of the gene. Point mutations at this site alone do not inactivate the supF gene, but large deletions containing a part of the gene produce supF mutants. ODN-5 was used for plasmid construction (Table 1).
The supF mutant frequencies were 1.1 and 3.6 × 10− 3 when the plasmid DNAs without and with THF, respectively, were transfected (Fig. 1B). Sequence analyses (Tables 4 and 5) and calculations of the large deletion frequency revealed that THF also induced large deletions in this case: the frequencies were 2.5 × 10− 4 and 2.8 × 10− 3 for the control and THF plasmid DNAs, respectively (P < 0.01, Student’s t-test). Thus, we confirmed that the presence of the abasic site analog induces large deletions.
Table 4.
Mutations detected in the supF gene (experiment 2)a
| G | |
| 5G- > C, 126G- > C | 1 |
| 65G- > GAA, 71C- > G, 73GAGC- > CCTT, 78GC- > AA, 81A- > G, 85GGA- > ACG | 1 |
| 70C- > A, 91G- > C | 2 |
| 70C- > A, 95C- > G | 2 |
| 91G- > T, 126G- > T | 2 |
| 95C- > A | 2 |
| 95C- > G | 4 |
| 121G- > C | 3 |
| 122G- > A | 1 |
| 131C- > A | 1 |
| 131C- > G | 2 |
| large deletion | 7 |
| total colonies analyzed | 28 |
| THF | |
| 5G- > A, 91G- > C | 2 |
| 5G- > C, 73G- > C, 112G- > A | 1 |
| 5G- > C, 27G- > T, 73G- > T, 91G- > T | 1 |
| 63 deletion | 1 |
| 73G- > C, 91G- > C | 2 |
| 95C- > A | 1 |
| 98A- > T | 1 |
| 118G- > C | 2 |
| large insertion | 2 |
| large deletion | 33 |
| large deletion, small insertion | 8 |
| total colonies analyzed | 54 |
aMutations detected in single colonies are represented. The sequence of the upper strand is shown. The numbers of colonies are shown on the right side
Table 5.
Spectra of mutations detected in the supF gene (experiment 2)a
| G | THF | |
|---|---|---|
| transition | ||
| A:T - > G:C | 2 (7) | 0 (0) |
| G:C - > A:T | 4 (14) | 3 (6) |
| transversion | ||
| A:T - > T:A | 0 (0) | 1 (2) |
| A:T - > C:G | 1 (4) | 0 (0) |
| G:C - > T:A | 13 (46) | 4 (7) |
| G:C - > C:G | 18 (64) | 11 (20) |
| small insertion (1–2 bp) | 1 (4) | 8 (15) |
| large insertion (> 2 bp) | 0 (0) | 2 (4) |
| small deletion (1–2 bp) | 0 (0) | 1 (2) |
| large deletion (> 2 bp) | 7 (25) | 41 (76) |
| others | 0 (0) | 0 (0) |
| total mutations | 46 | 71 |
| total colonies analyzed | 28 (100) | 54 (100) |
aAll data are represented as cases found (%)
Analysis of deleted sequences and junctions
Finally, we analyzed the deleted sequences and junctions. Sequence homology at the junctions of deleted regions should be found, if the deletions occurred in a homology-dependent manner. However, we did not find apparent similarity in any case (Table 6). Thus, homology-mediated events were not involved in the deletion formation.
Table 6.
Sequences of deletion junction
aThe sequence of the upper strand is shown. Deleted portion is shown in red
bA TACTGGCCTGCTCCCT sequence is present instead of the deleted sequence
cSubstitution at position 99 is shown in bold
dAn ATGCAGCGA sequence is present instead of the deleted sequence
eAn ATCAT sequence is present instead of the deleted sequence
fAn AG sequence is present instead of the deleted sequence
gAn AT sequence is present instead of the deleted sequence
hA CT sequence is present instead of the deleted sequence
Twelve mutants obtained in the THF (position 122) experiment contained large deletions. Among them, position 122 was included in the deleted portions for 10 mutants (Table 6). The other two lacked regions upstream of the modified position. No large deletions downstream of the supF gene retaining position 122 were found. Meanwhile, position 171 was included in the deleted regions for 26 among the 41 deletion mutants derived from the THF-plasmid (position 171). The other deletion mutants lost the upstream portion of the modified position.
Discussion
In this study, we examined the large deletion mutations induced by the abasic site analog THF in human cells. Unsurprisingly, substantial fractions of the progenies of THF-bearing vectors contained large deletions (Tables 3 and 5). In the case of THF at position 122 (experiment 1), most of the deleted regions were upstream of the modified site, although the downstream deletion produces supF mutants and thus is detectable. Since THF was introduced into the upper strand, the region corresponding to the 5′-side of the lesion was lost. A similar tendency was observed for THF at position 171 (experiment 2), although the lesion was located downstream of the supF gene and thus the deletion of the upstream region is essential to inactivate the gene.
When we consider replication, the 5′-side of the lesion is equivalent to the downstream region of the opposite site in the nascent strand. As described in the Introduction section, large deletion mutations were also induced by cyclobutane and 6–4 thymine-thymine dimers in mammalian cells [16]. Since the photodimers and THF are replication blocking lesions, we hypothesize that the inefficient bypass of THF causes the large deletion generated during replication.
Alternatively, the large deletion might be formed as a byproduct of DNA repair. APE1 (APEX, HAP, Ref-1) is the major apurinic/apyrimidinic endonuclease in human cells [28–31]. THF is a substrate of APE1 [32] and the enzyme incises DNA 5′ to THF, resulting in single-strand break (SSB) formation. Op het Veld et al. reported that large deletions were frequently observed in XRCC1-deficient Chinese hamster cells, after a treatment with methyl methanesulfonate [33]. Since the protein is involved in SSB repair and the compound induces the N-methylation of nucleobases, they hypothesized that the deletions could be caused by the accumulation of SSBs during abasic site formation and the subsequent incision of the lesions.
Simonelli et al. examined the mutagenic properties of the abasic site in African green monkey COS-7 cells [34]. Although they also found that large deletion mutations (8–168 bases) were induced by the DNA lesion, no information about the deletion junctions was reported. We detected the absence of homology in the deletion junctions we analyzed (Table 6) and concluded that the deletions were due to homology-independent events, such as DNA replication block and/or SSB formation, as described above.
A deaminated base, hypoxanthine, was previously shown to cause an A➔G mutation in mammalian cells [35]. However, DeVito et al. recently reported that the DNA lesion induced large deletions in human HEK293 and HCT116 cells [36]. N-methylpurine DNA glycosylase excises this modified base, leaving an abasic site [37]. Since hypoxanthine does not seem to be a replication blocking lesion due to its structural similarity to G, the abasic sites formed by the DNA glycosylase possibly contributed to the deletion mutations, as DeVito et al. discussed. In contrast, GO did not induce large deletions in U2OS cells (experiment 1) although the major repair enzyme OGG1 produces an abasic site. This discrepancy might be due to balance of DNA glycosylase and apurinic/apyrimidinic endonuclease activities. The amounts of the repair proteins would depend on cell types and the abasic sites formed by N-methylpurine DNA glycosylase might not be efficiently removed in HEK293 and HCT116 cells. In line with this possibility, GO induces large deletions in HCT116 cells [36]. The repair by monofunctional DNA glycosylases possibly acts as a double-edged sword in cells.
In this study, we found THF➔C and THF➔T mutations at position 122 (Table 3). These substitution mutations seemed to be caused by dGMP and dAMP incorporations, respectively, opposite the lesion. In contrast to E. coli, no clear rule of nucleotide incorporation was present in mammalian cells [11, 15]. Interestingly, the − 1 deletion at the THF site (5′-G-THF-T-3′) was observed, in contrast to the previous reports. This mutation might be triggered by dCMP incorporation opposite THF, followed by mispairing of the 5′-flanking G and the incorporated C after looping-out of THF. Alternatively, it might be initiated by dCMP incorporation opposite the 5′-flanking G after looping-out of THF, followed by the extension of the G:C pair.
The detection and quantitation of abasic sites are important research, and useful chemical probes have been developed [38, 39]. Investigations of the mutational properties of abasic sites, as well as their quantitation, are necessary for understanding the contributions of this important DNA lesion to mutagenesis, carcinogenesis, aging, and neurodegeneration.
Conclusions
In conclusion, the abasic site analog THF induced large deletion mutations in human cells. The lack of homology at their junction sites indicated that the deletions are caused by homology-independent events. Investigations of their molecular mechanism are quite pivotal, and further experiments are currently in progress.
Funding
This work was supported in part by Japan Society for the Promotion of Science (JSPS) KAKENHI grant number JP 16H02956 (HK).
Abbreviations
- GO
8-oxo-7,8-dihydroguanine
- ODN
oligodeoxyribonucleotide
- pol
polymerase
- SSB
single-strand break
- THF
tetrahydrofuran analog
Authors’ contributions
TS and HK conceived of the study, participated in its design and coordination, and drafted the manuscript. YKa performed the mutagenesis studies, the sequence analysis, and the data analysis. YKo synthesized and purified ODNs containing THF. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tetsuya Suzuki, Email: suzukite@hiroshima-u.ac.jp.
Yuri Katayama, Email: b134018@hiroshima-u.ac.jp.
Yasuo Komatsu, Email: komatsu-yasuo@aist.go.jp.
Hiroyuki Kamiya, Phone: +81-82-257-5300, Email: hirokam@hiroshima-u.ac.jp.
References
- 1.Kamiya H. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: approaches using synthetic oligonucleotides and nucleotides: survey and summary. Nucleic Acids Res. 2003;31:517–531. doi: 10.1093/nar/gkg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki T, Kamiya H. Mutations induced by 8-hydroxyguanine (8-oxo-7,8-dihydroguanine), a representative oxidized base, in mammalian cells. Genes Environ. 2016;39:2. doi: 10.1186/s41021-016-0051-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kino K, Hirao-Suzuki M, Morikawa M, Sakaga A, Miyazawa H. Generation, repair and replication of guanine oxidation products. Genes Environ. 2017;39:21. doi: 10.1186/s41021-017-0081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeifer GP, You Y-H, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res Fund Mol Mech Mutagen. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 5.Ikehata H, Ono T. The mechanisms of UV mutagenesis. J Radiat Res. 2011;52:115–125. doi: 10.1269/jrr.10175. [DOI] [PubMed] [Google Scholar]
- 6.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 7.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 9.Locatelli GA, Pospiech H, Tanguy Le Gac N, van Loon B, Hubscher U, Parkkinen S, Syväoja JE, Villani G. Effect of 8-oxoguanine and abasic site DNA lesions on in vitro elongation by human DNA polymerase ε in the presence of replication protein a and proliferating-cell nuclear antigen. Biochem J. 2010;429:573–582. doi: 10.1042/BJ20100405. [DOI] [PubMed] [Google Scholar]
- 10.Choi J-Y, Lim S, Kim E-J, Jo A, Guengerich FP. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases α, δ, η, ι, κ, and REV1. J Mol Biol. 2010;404:34–44. doi: 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamiya H, Suzuki M, Komatsu Y, Miura H, Kikuchi K, Sakaguchi T, Murata N, Masutani C, Hanaoka F, Ohtsuka E. An abasic site analogue activates a c-ha-ras gene by a point mutation at modified and adjacent positions. Nucleic Acids Res. 1992;20:4409–4415. doi: 10.1093/nar/20.17.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamiya H, Suzuki M, Ohtsuka E. Mutation-spectrum of a true abasic site in codon 12 of a c-ha-ras gene in mammalian cells. FEBS Lett. 1993;328:125–129. doi: 10.1016/0014-5793(93)80979-5. [DOI] [PubMed] [Google Scholar]
- 13.Gentil A, Cabral-Neto JB, Mariage-Samson R, Margot A, Imbach JL, Rayner B, Sarasin A. Mutagenicity of a unique apurinic/apyrimidinic site in mammalian cells. J Mol Biol. 1992;227:981–984. doi: 10.1016/0022-2836(92)90513-J. [DOI] [PubMed] [Google Scholar]
- 14.Cabral Neto JB, Cabral RE, Margot A, Le Page F, Sarasin A, Gentil A. Coding properties of a unique apurinic/apyrimidinic site replicated in mammalian cells. J Mol Biol. 1994;240:416–420. doi: 10.1006/jmbi.1994.1457. [DOI] [PubMed] [Google Scholar]
- 15.Weerasooriya S, Jasti VP, Basu AK. Replicative bypass of abasic site in Escherichia coli and human cells: similarities and differences. PLoS One. 2014;9:e107915. doi: 10.1371/journal.pone.0107915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamiya H, Iwai S, Kasai H. The (6-4) photoproduct of thymine-thymine induces targeted substitution mutations in mammalian cells. Nucleic Acids Res. 1998;26:2611–2617. doi: 10.1093/nar/26.11.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaufmann WK, Cleaver JE. Mechanisms of inhibition of DNA replication by ultraviolet light in normal human and xeroderma pigmentosum fibroblasts. J Mol Biol. 1981;149:171–187. doi: 10.1016/0022-2836(81)90297-7. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki T, Harashima H, Kamiya H. Effects of base excision repair proteins on mutagenesis by 8-oxo-7,8-dihydroguanine (8-hydroxyguanine) paired with cytosine and adenine. DNA Repair (Amst) 2010;9:542–550. doi: 10.1016/j.dnarep.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Kamiya H, Yamaguchi A, Suzuki T, Harashima H. Roles of specialized DNA polymerases in mutagenesis by 8-hydroxyguanine in human cells. Mutat Res Fund Mol Mech Mutagen. 2010;686:90–95. doi: 10.1016/j.mrfmmm.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Kamiya H, Kasai H. Substitution and deletion mutations induced by 2-hydroxyadenine in Escherichia coli: effects of sequence contexts in leading and lagging strands. Nucleic Acids Res. 1997;25:304–310. doi: 10.1093/nar/25.2.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obata F, Nunoshiba T, Hashimoto-Gotoh T, Yamamoto K. An improved system for selection of forward mutations in an Escherichia coli supF gene carried by plasmids. J Radiat Res. 1998;39:263–270. doi: 10.1269/jrr.39.263. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki T, Harashima H, Kamiya H. Unexpectedly weak impacts of decreased p53 and retinoblastoma protein levels on mutagenesis by 8-oxo-7,8-dihydroguanine (8-hydroxyguanine) Genes Environ. 2011;33:103–108. doi: 10.3123/jemsge.33.103. [DOI] [Google Scholar]
- 23.Tornaletti S, Maeda LS, Kolodner RD, Hanawalt PC. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair (Amst) 2004;3:483–494. doi: 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Stary A, Sarasin A. Simian virus 40 (SV40) large T antigen-dependent amplification of an Epstein-Barr virus-SV40 hybrid shuttle vector integrated into the human HeLa cell genome. J Gen Virol. 1992;73:1679–1685. doi: 10.1099/0022-1317-73-7-1679. [DOI] [PubMed] [Google Scholar]
- 25.Satou K, Hori M, Kawai K, Kasai H, Harashima H, Kamiya H. Involvement of specialized DNA polymerases in mutagenesis by 8-hydroxy-dGTP in human cells. DNA Repair (Amst) 2009;8:637–642. doi: 10.1016/j.dnarep.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki T, Kuramoto Y, Kamiya H. Reduction of Werner syndrome protein enhances G:C➔a:T transition by O6-methylguanine in human cells. Chem Res Toxicol. 2018;31:319–324. doi: 10.1021/acs.chemrestox.8b00009. [DOI] [PubMed] [Google Scholar]
- 27.Eckert KA, Kunkel TA. DNA polymerase fidelity and the polymerase chain reaction. Genome Res. 1991;1:17–24. doi: 10.1101/gr.1.1.17. [DOI] [PubMed] [Google Scholar]
- 28.Robson CN, Hickson ID. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res. 1991;19:5519–5523. doi: 10.1093/nar/19.20.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci U S A. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seki S, Hatsushika M, Watanabe S, Akiyama K, Nagao K, Tsutsui K. cDNA cloning, sequencing, expression and possible domain structure of human APEX nuclease homologous to Escherichia coli exonuclease III. Biochim Biophys Acta. 1992;1131:287–299. doi: 10.1016/0167-4781(92)90027-W. [DOI] [PubMed] [Google Scholar]
- 31.Xanthoudakis S, Miao G, Wang F, Pan Y-CE, Curran T. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson DM, III, Takeshita M, Grollman AP, Demple B. Incision activity of human apurinic endonuclease (ape) at abasic site analogs in DNA. J Biol Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 33.Op het Veld CW, Jansen J, Zdzienicke MZ, Vrieling H, van Zeeland AA. Methyl methanesulfonate-induced hprt mutation spectra in the Chinese hamster cell line CHO9 and its xrcc1-deficient derivative EM-C11. Mutat Res. 1998;398:83–92. doi: 10.1016/S0027-5107(97)00243-1. [DOI] [PubMed] [Google Scholar]
- 34.Simonelli V, Narciso L, Dogliotti E, Fortini P. Base excision repair intermediates are mutagenic in mammalian cells. Nucleic Acids Res. 2005;33:4404–4411. doi: 10.1093/nar/gki749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamiya H, Miura H, Kato H, Nishimura S, Ohtsuka E. Induction of mutation of a synthetic c-ha-ras gene containing hypoxanthine. Cancer Res. 1992;52:1836–1839. [PubMed] [Google Scholar]
- 36.DeVito S, Woodrick J, Song L, Roy R. Mutagenic potential of hypoxanthine in live human cells. Mutat Res Fund Mol Mech Mutagen. 2017;803–805:9–16. doi: 10.1016/j.mrfmmm.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy R, Kennel SJ, Mitra S. Distinct substrate preference of human and mouse N-methylpurine-DNA glycosylases. Carcinogenesis. 1996;17:2177–2182. doi: 10.1093/carcin/17.10.2177. [DOI] [PubMed] [Google Scholar]
- 38.Ide H, Akamatsu K, Kimura Y, Michiue K, Makino K, Asaeda A, Takamori Y, Kubo K. Synthesis and damage specificity of a novel probe for the detection of abasic sites in DNA. Biochemistry. 1993;32:8276–8283. doi: 10.1021/bi00083a031. [DOI] [PubMed] [Google Scholar]
- 39.Kojima N, Takebayashi T, Mikami A, Ohtsuka E, Komatsu Y. Construction of highly reactive probes for abasic site detection by introduction of an aromatic and a guanidine residue into an aminooxy group. J Am Chem Soc. 2009;131:13208–13209. doi: 10.1021/ja904767k. [DOI] [PubMed] [Google Scholar]



