Abstract
Mandelic acid is an important aromatic fine chemical and is currently mainly produced via chemical synthesis. Recently, mandelic acid production was achieved by microbial fermentations using engineered Escherichia coli and Saccharomyces cerevisiae expressing heterologous hydroxymandelate synthases (hmaS). The best-performing strains carried a deletion of the gene encoding the first enzyme of the tyrosine biosynthetic pathway and therefore were auxotrophic for tyrosine. This was necessary to avoid formation of the competing intermediate hydroxyphenylpyruvate, the preferred substrate for HmaS, which would have resulted in the predominant production of hydroxymandelic acid. However, feeding tyrosine to the medium would increase fermentation costs. In order to engineer a tyrosine prototrophic mandelic acid-producing S. cerevisiae strain, we tested three strategies: (1) rational engineering of the HmaS active site for reduced binding of hydroxyphenylpyruvate, (2) compartmentalization of the mandelic acid biosynthesis pathway by relocating HmaS together with the two upstream enzymes chorismate mutase Aro7 and prephenate dehydratase Pha2 into mitochondria or peroxisomes, and (3) utilizing a feedback-resistant version of the bifunctional E. coli enzyme PheA (PheAfbr) in an aro7 deletion strain. PheA has both chorismate mutase and prephenate dehydratase activity. Whereas the enzyme engineering approaches were only successful in respect to reducing the preference of HmaS for hydroxyphenylpyruvate but not in increasing mandelic acid titers, we could show that strategies (2) and (3) significantly reduced hydroxymandelic acid production in favor of increased mandelic acid production, without causing tyrosine auxotrophy. Using the bifunctional enzyme PheAfbr turned out to be the most promising strategy, and mandelic acid production could be increased 12-fold, yielding titers up to 120 mg/L. Moreover, our results indicate that utilizing PheAfbr also shows promise for other industrial applications with S. cerevisiae that depend on a strong flux into the phenylalanine biosynthetic pathway.
Abbreviations: HmaS, Hydroxymandelate synthase; MA, mandelic acid; HMA, 4-hydroxymandelic acid
Keywords: Mandelic acid, Hydroxymandelate synthase, Chorismate mutase-prephenate dehydratase, Compartmentalization, Tyrosine prototrophy
Graphical abstract
Highlights
-
•
Increased mandelic acid production in tyrosine prototrophic S. cerevisiae.
-
•
Bifunctional E. coli enzyme PheA increases flux into yeast phenylalanine branch.
-
•
PheA allows mandelic acid production in prototrophic S. cerevisiae.
-
•
Compartmentalized mandelic acid production in yeast mitochondria/peroxisomes.
1. Introduction
Metabolic engineering can serve to convert microorganisms into microbial cell factories to biotechnologically produce desired compounds like chemicals, pharmaceuticals and food ingredients. The yeast Saccharomyces cerevisiae can be easily modified by a large amount of available molecular biology tools. Although higher titers and yields are often reached with bacterial hosts (Averesch and Krömer, 2018), the robustness of S. cerevisiae and its stress tolerance in fermentative processes (Gibson et al., 2007) represent distinct advantages of this yeast (Weber et al., 2010).
We recently published a metabolic engineering approach allowing for the production of the aromatic fine chemicals mandelic acid (MA) and hydroxymandelic acid (HMA) in S. cerevisiae (Reifenrath and Boles, 2018). Prior to that, three studies focused on the production of MA or MA derivatives in Escherichia coli (Liu et al., 2014, Müller et al., 2006, Sun et al., 2011). MA finds use in the cosmetic industry and serves as a precursor for the production of pharmaceutically active compounds (Chang et al., 2007, Furlenmeier et al., 1976, Mill et al., 1983, Nishizawa et al., 1985, Saravanan and Singh, 1998, Ward et al., 2007). MA production in E. coli and S. cerevisiae was achieved by introducing heterologous hydroxymandelate synthases (HmaS), which convert phenylpyruvate, the precursor of phenylalanine, to MA (Liu et al., 2014, Müller et al., 2006, Reifenrath and Boles, 2018, Sun et al., 2011). However, hydroxymandelate synthases have a much higher affinity towards their native substrate hydroxyphenylpyruvate, the precursor of tyrosine, which they convert to hydroxymandelic acid (HMA) (Reifenrath and Boles, 2018, Sun et al., 2011). Consequently, in fermentations with strains exhibiting an intact tyrosine biosynthetic pathway mainly HMA will be formed. Therefore, all MA production strains published so far, carried disruptions in the tyrosine biosynthetic pathway (Liu et al., 2014, Müller et al., 2006, Reifenrath and Boles, 2018, Sun et al., 2011). As feeding of tyrosine to the medium would increase production costs, our goal was to construct an S. cerevisiae MA producer strain that does not require supplementation of tyrosine. After our attempts to rationally engineer the active site of HmaS to increase affinity for phenylpyruvate and decrease the conversion of hydroxyphenylpyruvate did not yield satisfactory results (Reifenrath and Boles, 2018, and this study), we shifted our focus to metabolic engineering strategies to improve MA production. We aimed to increase flux into the phenylalanine pathway and decrease flux into the tyrosine pathway without inhibiting tyrosine synthesis completely.
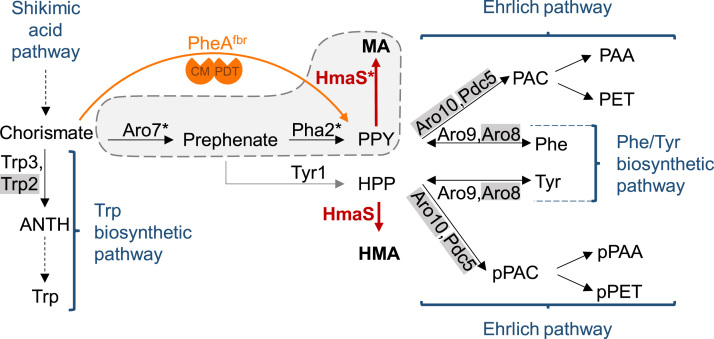
The core of the aromatic amino acid biosynthesis pathway in S. cerevisiae is the shikimic acid pathway (Braus, 1991). Chorismate, which originates from the shikimic acid pathway, represents the last common precursor of the three aromatic amino acids tryptophan, phenylalanine and tyrosine (see Fig. 1). Chorismate is either converted by Trp2/Trp3 to anthranilate, thereby entering the tryptophan biosynthetic pathway, or by the chorismate mutase Aro7 to prephenate. Prephenate, the last common intermediate of the biosynthetic pathways of phenylalanine and tyrosine, is either converted by the prephenate dehydratase Pha2 to phenylpyruvate or by the prephenate dehydrogenase Tyr1 to hydroxyphenylpyruvate (Fig. 1). Phenylpyruvate and hydroxyphenylpyruvate are both substrates of the transaminases Aro8 and Aro9, which convert them to phenylalanine and tyrosine, respectively (Braus, 1991). The degradation of phenylalanine and tyrosine is carried out via the Ehrlich pathway and results in the production of fusel alcohols and the corresponding acids (Fig. 1)(Hazelwood et al., 2008).
Fig. 1.
Applied modifications of the aromatic amino acid pathway of S. cerevisiae for mandelic acid production. The heterologous hydroxymandelate synthase (HmaS, red) converts the intermediates of the aromatic amino acid pathway phenylpyruvate (PPY) and hydroxyphenylpyruvate (HPP) to mandelic acid (MA) and hydroxymandelic acid (HMA), respectively. Enzymes that were targeted to mitochondria/peroxisomes in our compartmentalization approach are marked with a star. The compartment (mitochondrion or peroxisome) containing the three consecutive enzymes Aro7fbr, Pha2 and HmaS is depicted as a grey area surrounded by a dashed line. The bifunctional E. coli enzyme PheAfbr is indicated in orange (CM, chorismate mutase; PDT, prephenate dehydratase). Dashed arrows indicate multiple enzymatic steps. Enzymes whose genes were deleted in a part of the strains used in this work are labeled in grey. ANTH, anthranilate; Trp, tryptophan; PAC, phenylacetaldehyde; pPAC, p-hydroxyphenylacetaldehyde; Phe, phenylalanine; Tyr, tyrosine; PAA, phenylacetic acid; pPAA, p-hydroxyphenylacetic acid; PET, phenylethanol; pPET, p-hydroxyphenylethanol.
Aiming to construct a tyrosine prototrophic S. cerevisiae MA producer strain we tested two metabolic engineering approaches – using pathway compartmentalization and using the heterologous, bifunctional E. coli enzyme PheA.
Compartmentalization of metabolic pathways in yeast organelles has been successfully used to increase yields of various industrially relevant products (reviewed by Hammer and Avalos, 2017). Compartmentalizing pathways can be advantageous due to the special environment inside of an organelle (e.g. substrate or cofactor availability, pH, etc.), because of shorter diffusion distances, high protein content or the spatial separation from enzymes present in the cytosol. In our approach, we mainly aimed to spatially separate the MA production pathway (Aro7, Pha2, HmaS) from competing enzymes like Tyr1, from the Ehrlich pathway enzymes and from the competing substrate hydroxyphenylpyruvate (compare Fig. 1). We chose to test compartmentalization of the pathway in mitochondria and peroxisomes – two organelles which substantially differ in their physicochemical environment (Deloache and Dueber, 2013, Hu et al., 2008, Orij et al., 2009, Roermund et al., 2004) and their permeability (Cooper, 2000, DeLoache et al., 2016) but have both been successfully used for pathway compartmentalization approaches in S. cerevisiae (Hammer and Avalos, 2017).
In our second metabolic engineering approach for the production of MA in a tyrosine prototrophic S. cerevisiae strain we used the bifunctional E. coli enzyme PheA in a feedback resistant version (PheAfbr). In contrast to the aromatic amino acid pathway of S. cerevisiae, in E. coli phenylpyruvate and hydroxyphenylpyruvate are formed by bifunctional enzymes, PheA and TyrA, respectively (Ikeda, 2006, Rodriguez et al., 2014). Both enzymes exhibit chorismate mutase activity, and PheA additionally exhibits prephenate dehydratase activity whereas TyrA additionally exhibits prephenate dehydrogenase activity. By using PheAfbr in an aro7Δ S. cerevisiae strain we aimed to increase flux into the phenylalanine biosynthetic pathway and decrease levels of hydroxyphenylpyruvate. We assumed that there might be a channeling effect caused by a transfer of the intermediate prephenate formed by the chorismate mutase domain to the prephenate dehydratase domain of PheAfbr, thereby favoring entry into the phenylalanine branch (compare Fig. 1).
Whereas the introduced mutations in the active site of HmaS did not yield the desired positive effect on MA production, we demonstrate that the two metabolic engineering strategies – compartmentalization and pheAfbr expression – successfully resulted in a shift from HMA to MA production without causing tyrosine auxotrophy. Of these two strategies, using the bifunctional PheA enzyme led to much higher MA titers compared to the mitochondrial and peroxisomal compartmentalization strategy.
2. Material and methods
2.1. Plasmid and strain construction
Plasmids and S. cerevisiae strains used in this study are listed in Supplementary tables S1 and S2, respectively. For subcloning electrocompetent E. coli DH10β (Gibco BRL, Gaithersburg, MD) were used according to Dower et al. (1988). E. coli transformants were selected on lysogeny broth (LB) agar plates (Sambrook et al., 1989) containing 100 mg/L ampicillin. Plasmids were constructed with Gibson assembly (Gibson et al., 2009) or homologous recombination. The respective empty vectors were linearized with SacI and BamHI and then used for Gibson assembly or transformation together with the according PCR-fragments. S. cerevisiae was transformed according to the protocol of Gietz and Schiestl (2007). All primers used in this study are listed in Supplementary table S3.
To receive a feedback-resistant variant of PheA (Uniprot accession number P0A9J8) the mutations ΔT304, G305K and ΔQ306 (Nelms et al., 1992) were introduced. The gene was codon-optimized according to Wiedemann and Boles (2008) and ordered as a String DNA fragment from Thermo Fischer Scientific Germany with 30 base pairs overhangs to both the HXT7 promoter and the CYC1 terminator. The enhanced peroxisomal targeting sequence (ePTS1, DeLoache et al., 2016) was codon-optimized (Wiedemann and Boles, 2008) and introduced by the primers MRP232–234. The mitochondrial targeting sequence (MTS, amino acids 1–69 of Neurospora crassa F0 ATP synthase subunit 9, GenBank accession number CAA24230) was codon-optimized with the DNA 2.0 software and ordered from the company DNA 2.0. The DNA sequences of codon-optimized pheAfbr, MTS and ePTS1 can be found in Supplementary table S4. To receive a feedback resistant form of Aro7 (Aro7fbr, Schnappauf et al., 1998) the mutation G141S was introduced by the primers MGP11 and MGP12. All constructs were confirmed by sequencing (GATC Biotech, Germany).
Strains constructed in this study and their parent strains are listed in Supplementary table S2. Shik↑ indicates the genotype aro4Δ::HXT7pr-ARO1 ura3Δ::TPI1pr-ARO4fbr_HXT7pr-ARO3fbr resulting in an overexpression of ARO1 and of the feedback resistant versions of ARO4 and ARO3 as published in Reifenrath and Boles (2018). Genomic deletions were performed with the CRISPR/Cas9 system as described in Generoso et al. (2016). Cells were selected on YPD medium containing antibiotics G418 (200 mg/L) or clonNAT/Nourseothricin (100 mg/L). The gRNA sequences chosen for the deletions are listed in Supplementary table S5. The genomic reintegration of TRP2 into strain MRY31 (CEN.PK2-1C TRP1 Shik↑ aro10Δ aro8Δ pdc5Δ trp2Δ aro7Δ) to yield the tryptophan prototrophic strain MRY36 was achieved by simply transforming the TRP2-PCR product with overhang to TRP2 promoter and terminator and selecting the cells on plates lacking tryptophan.
All experiments were done using the codon-optimized hmaS from Nocardia uniformis as this enzyme performed best in S. cerevisiae in our recent study (Reifenrath and Boles, 2018).
2.2. Strain cultivation
S. cerevisiae strains were always freshly transformed and plated onto synthetic complete medium (SCD, 20 g/L glucose, as described in Bruder et al. (2016) omitting leucine, histidine and/or uracil according to auxotrophic requirements.
For fermentations, precultures (75 ml in 300 ml flasks) and main cultures (20 ml in 100 ml flasks) were grown in synthetic minimal medium (SMD, 1.7 g/L Difco yeast nitrogen base without amino acids and 5 g/L ammonium sulfate, 20 g/L glucose, 20 mM KH2PO4, pH 6.3), and uracil (0.171 mM) and histidine (0.124 mM) were added when required. The precultures of the compartmentalization fermentation (results in Fig. 3C and D) were supplemented with additional aromatic amino acids (0.160 mM tyrosine, 0.582 mM phenylalanine, 0.2 mM anthranilate as precursor for tryptophan) to ensure comparable growth of all strains. Precultures were harvested in the exponential phase, washed and resuspended to an optical density (λ = 600 nm) of 5. The optical density at 600 nm (OD600) was analyzed with a spectrophotometer (Ultrospec 2100 pro spectrophotometer, GE Healthcare, USA). All fermentations were performed in biological duplicates.
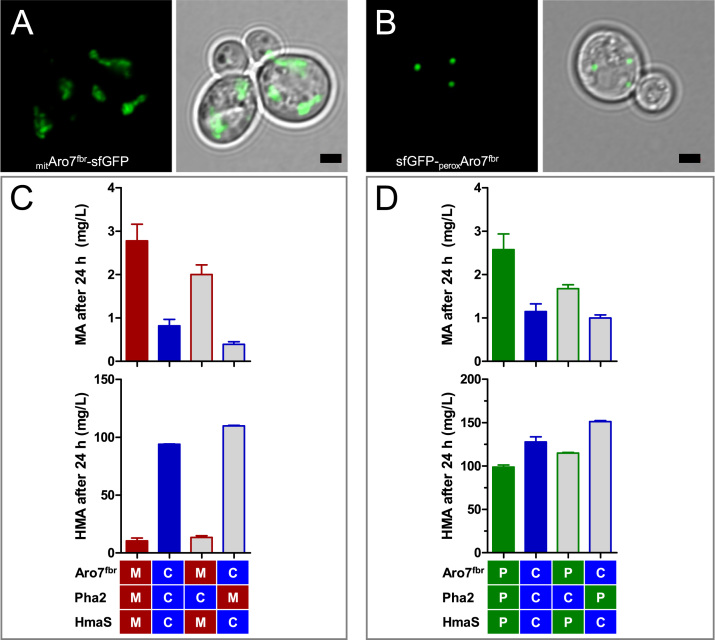
Fig. 3.
Pathway compartmentalization in mitochondria and peroxisomes of S. cerevisiae for mandelic acid production. A) and B) Intracellular localization of mitAro7fbr-sfGFP and sfGFP-peroxAro7fbr, respectively, visualized by fluorescence microscopy. CEN.PK2-1C cells expressing MTS-ARO7fbr-sfGFP (A) and sfGFP-ARO7fbr-ePTS1 (B) from plasmids were grown until the exponential phase in lf-SCD medium. The scale bars indicate 2 µm. C) and D) mandelic acid (MA, top) and hydroxymandelic acid (HMA, bottom) titers after 24 h of fermentation with MRY25 (CEN.PK2-1C TRP1 Shik↑ aro7Δ trp2Δ) with Aro7fbr, Pha2 and N. uniformis HmaS targeted to mitochondria (M, red), cytosol (C, blue) or peroxisomes (P, green). Plain-colored bars present samples in which all three consecutive enzymes were localized in one compartment (blue, cytosol; red, mitochondria; green, peroxisomes). The fermentation was performed in SMD (20 g/L glucose) with anthranilate supplementation and a starting OD600 of 5. Error bars indicate standard deviation of biological duplicates (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
For the growth tests on agar plates cells were resuspended in H2O, washed once, brought to an OD600 of 1, and dilution series were generated (OD600 of 0.1, 0.01 and 0.001). 4 µl of the cell suspensions were then dropped onto SCD or SMD agar plates containing different combinations of the aromatic amino acids tyrosine, phenylalanine and tryptophan. Pictures of the plates were taken after 3 days of incubation at 30 °C.
2.3. Metabolite analysis
Samples of the supernatant for metabolite analysis were obtained by centrifugation and stored at −20 °C until analyzed. Samples for glucose and ethanol analysis were treated and analyzed as described previously (Reifenrath and Boles, 2018).
For the analysis of MA and HMA, acetonitrile was added to the samples to a final concentration of 20%. Quantification was performed using an HPLC (Dionex) equipped with an Agilent Zorbax SB-C8 column (4.6 × 150 mm, 3.5 µm) at 40 °C. The samples were analyzed under the following gradient of eluent A (0.1% (v/v) formic acid) and eluent B (acetonitrile with 0.1% (v/v) formic acid): 2 min 0% B, 13 min linear gradient to 10% B, 5 min linear gradient to 25% B, 4 min 25% B, 1 min linear gradient to 0% B, 5 min 0% B. The flow rate was 1 ml/min. MA was detected at 190 nm (compartmentalization approach, Fig. 3C and D) or 200 nm (PheA approach, Fig. 4B, C and D), HMA was detected at 200 nm and phenylacetic acid and phenylethanol eluted together and were detected at 215 nm (Dionex UlitMate 3000 Variable Wavelength Detector). The standard solutions for phenylacetic acid and phenylethanol calibration contained the same amount of phenylacetic acid and phenylethanol (w/v) as described previously (Reifenrath and Boles, 2018). MA, phenylethanol and phenylacetic acid were purchased from Sigma Aldrich (778052, 77861 and P16621, respectively) and HMA was purchased from Tokyo Chemical Industry Co. (H0060). Data analysis and graphing were performed with the software Prism 5 (GraphPad, USA).
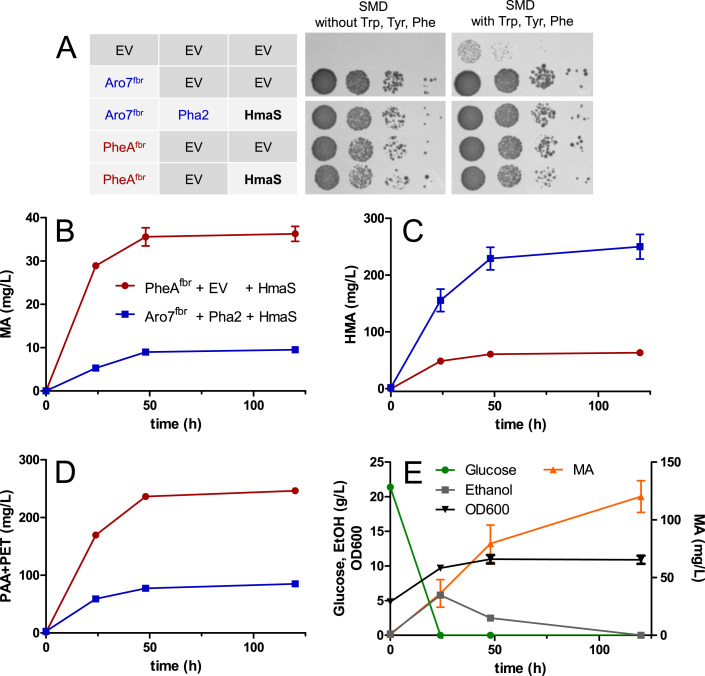
Fig. 4.
Using the bifunctional E. coli enzyme PheAfbr for mandelic acid production in an aro7Δ S. cerevisiae strain. A) Growth test with MRY36 (CEN.PK2-1C TRP1 Shik↑ aro10Δ aro8Δ pdc5Δ aro7Δ) cells harboring different plasmid combinations on SMD with and without supplementation of aromatic amino acids. Cell dilutions of OD600 1, 10−1, 10−2 and 10−3 (from left to right, respectively) were used. The picture was taken after 3d at 30 °C. EV, empty vector. B), C) and D) Production of mandelic acid (MA, B), hydroxymandelic acid (HMA, C) and phenylacetic acid + phenylethanol (PAA+PET, D) by the aro7Δ strain MRY36 expressing either pheAfbr and hmaS (red) or ARO7fbr, PHA2 and hmaS (blue). E) Fermentation with the aro7Δ strain MRY36 harboring the hmaS-pheAfbr plasmid MRV144. MA, glucose and ethanol titers and OD600 in SMD without supplementation of aromatic amino acids. The fermentations were performed in SMD (20 g/L glucose) without supplementation aromatic amino acids and with a starting OD600 of 5. Error bars indicate standard deviation of biological duplicates (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
2.4. Fluorescence microscopy
For fluorescence microscopy CEN.PK2-1C harboring the MTS-ARO7fbr-sfGFP or the sfGFP-ARO7fbr-ePTS1 plasmid were grown in low fluorescence SCD medium (lf-SCD) containing 6.9 g/L yeast nitrogen base (YNB) with ammonium sulfate, without amino acids, without folic acid and without riboflavin (MP Biomedicals), 20 g/L glucose and amino acids as stated in Bruder et al. (2016), omitting uracil. The medium was filter-sterilized. Cells were grown to the exponential phase, then diluted 1:1 with fresh lf-SCD containing 0.6% low melting agarose (Roth) and applied to the glass microscope slide. Samples were analyzed with the confocal laser scanning microscope ZEISS LSM 780 with objective plan-apochromat 63 × /1.40 Oil DIC M27 and excitation at 488 nm and an emission bandwidth of 493–598 nm.
3. Results and discussion
3.1. Engineering hydroxymandelate synthase HmaS for mandelic acid production
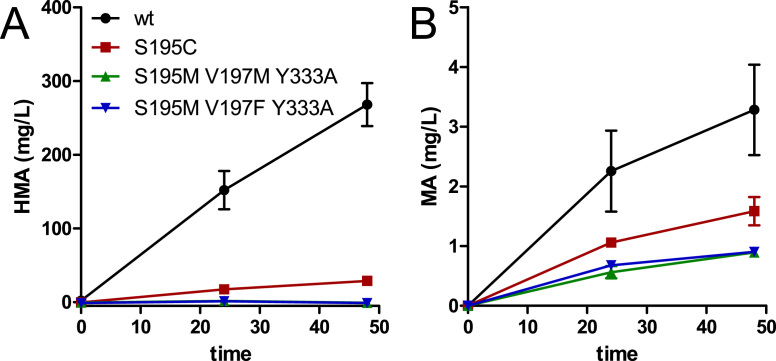
HmaS converts hydroxyphenylpyruvate to HMA and, less preferred, phenylpyruvate to MA (Reifenrath and Boles, 2018, Sun et al., 2011). Apart from completely blocking the tyrosine biosynthetic pathway, one approach to specifically produce MA would be to prevent hydroxyphenylpyruvate from binding to HmaS and to simultaneously increase the conversion of phenylpyruvate. A serine in the active site of HmaS forms a hydrogen bond to the para-hydroxy group of its favored substrate hydroxyphenylpyruvate (Brownlee et al., 2008). In our previous work on MA and HMA production in S. cerevisiae, we mutated this serine to hydrophobic amino acids to decrease affinity for hydroxyphenylpyruvate and optimize the HmaS active site for phenylpyruvate binding with the goal of improving specific MA production. The most promising exchange (S201V, A. orientalis HmaS) increased MA production but only to relatively low MA titers (4 mg/L). This suggested that the low turnover of phenylpyruvate by the HmaSS201V variant cannot be attributed solely to the competition with hydroxyphenylpyruvate for the active site, but rather to mechanistic constraints or a severe alteration of the active site by the substitution of serine to valine. In the present work, we therefore tested a more moderate substitution (serine to cysteine). We chose the N. uniformis HmaS as this enzyme had been shown to promote higher production of MA/HMA in yeast (Reifenrath and Boles, 2018). With the serine to cysteine mutation (HmaSS195C) we solely replaced an OH hydroxy group with a slightly less electronegative SH thiol group. We hypothesized that this exchange would lead to a more pronounced shift in the preference of phenylpyruvate compared to hydroxyphenylpyruvate, thereby increasing production of MA. However, the expression of N. uniformis HmaSS195C from plasmid MRV140 in strain Y4 (CEN.PK2-1C TRP1 Shik↑ aro10Δ aro8Δ pdc5Δ, Reifenrath and Boles, 2018) grown in SMD medium only led to a severe decrease in HMA titers and no increase in MA titers (Fig. 2A and B).
Fig. 2.
In vivo hydroxymandelic acid and mandelic acid production by engineered HmaS variants. A) Hydroxymandelic acid (HMA) titers and B) mandelic acid (MA) titers of Y4 (CEN.PK2-1C TRP1 Shik↑ aro10Δ aro8Δ pdc5Δ, Reifenrath and Boles, 2018) expressing the different variants of N. uniformis hmaS. The fermentations were performed in SMD (20 g/L glucose) without supplementation of aromatic amino acids and with a starting OD600 of 5. Error bars indicate standard deviation of biological duplicates.
Additionally, we tested active site mutations as published by Pratter et al. (2013) who discovered Streptomyces coelicolor HmaS mutants with improved steady state kinetics (higher kcatapp and higher kcatapp/KMapp) for phenylpyruvate conversion. Actually, Pratter et al. (2013) mainly aimed for an inversion of the enantioselectivity of the enzyme for the production of R-MA instead of S-MA. Additionally, they observed that the two mutant versions with highest selectivity for R-MA production (S221M+V223M+Y359A and S221M+V223F+Y359A) also showed improved steady state characteristics for MA production. They did not analyze steady state kinetics with hydroxyphenylpyruvate as a substrate but as the serine forming the hydrogen bond with the para-hydroxy group of hydroxyphenylpyruvate (S221) was replaced by a methionine in both mutants, a negative effect on HMA production was most likely. Aiming for increased MA production, we therefore transferred the corresponding mutations to the HmaS of N. uniformis (S195M+V197M+Y333A and S195M+V197F+Y333A) and tested the variants in S. cerevisiae. However, after expressing the hmaS variants (plasmids MRV141 and MRV142) in strain Y4 (CEN.PK2-1C TRP1 Shik↑ aro10Δ aro8Δ pdc5Δ, Reifenrath and Boles, 2018) grown in SMD medium, no positive effect on MA production could be detected although the HMA titers dropped as expected (Fig. 2A and B).
To conclude, all mutations that we introduced into the active site of HmaS did not yield the desired effects of increased MA production documenting the difficulties of rational enzyme engineering. One solution could be a sensor system allowing to screen for improved MA production to make the identification of mutants with increased activity towards phenylpyruvate possible. However, to our knowledge such a sensor system, which would require sensitivity to MA and should not sense HMA, is not yet available.
3.2. Compartmentalizing the mandelic acid biosynthetic pathway in peroxisomes and mitochondria
As the amino acid replacements introduced into the HmaS active site did not improve MA production, we shifted our strategy towards pathway compartmentalization. We aimed to increase flux into the phenylalanine branch and decrease flux into the tyrosine branch by locating the three consecutive enzymatic reactions catalyzed by Aro7, Pha2 and HmaS into mitochondria or peroxisomes. Localizing these three enzymes into one compartment should favor production of phenylpyruvate and MA due to spatial separation of the MA production pathway from the competing enzyme Tyr1 and the competing substrate hydroxyphenylpyruvate (Fig. 1). During yeast fermentations, intermediates of the aromatic amino acid pathway can be found in culture supernatants (Luttik et al., 2008). Even though no specific transporters for these hydrophilic molecules are known, transport via unspecific or unknown transporters might allow their release into the medium (Generoso et al., 2017, Ramos et al., 2016). In the same way, unspecific transport across mitochondrial membranes might occur and peroxisomal membranes have been shown to be highly permeable to small hydrophilic molecules (DeLoache et al., 2016). Therefore, we reasoned that uptake of chorismate into and release of MA out of the organelles should not be a major problem. Moreover, a minor fraction of prephenate must be transported back into the cytosol to be available for tyrosine production via Tyr1.
We chose a feedback resistant version of Aro7 (Aro7fbr, G141S, according to Schnappauf et al., 1998) to avoid the inhibiting influence of aromatic amino acid levels on chorismate mutase activity. We constructed plasmids from which ARO7fbr, PHA2 and hmaS from N. uniformis were expressed – either with or without a mitochondrial or a peroxisomal targeting sequence, MTS or ePTS1, respectively (for codon-optimized sequences see Supplementary table S4). We chose the mitochondrial targeting sequence of N. crassa ATP synthase subunit 9, which was previously shown to efficiently localize proteins to the mitochondria of S. cerevisiae (Benisch and Boles, 2014, Weber et al., 2017, Westermann and Neupert, 2000). For peroxisomal localization we chose the enhanced peroxisomal targeting sequence ePTS1 that was identified by DeLoache et al. (2016) and shown to cause efficient protein relocation into peroxisomes. Fluorescence microscopy performed with CEN.PK2-1C (Entian and Kötter, 2007) expressing MTS-ARO7fbr or ARO7fbr-ePTS1 tagged with superfolder GFP (Pédelacq et al., 2006), MTS- ARO7fbr-sfGFP and sfGFP-ARO7fbr-ePTS1, respectively, indicated that MTS and ePTS1 directed the proteins to the respective organelles (Fig. 3A and B).
Next, we tested whether Aro7fbr and Pha2 remain active when they are targeted to mitochondria or peroxisomes. A growth test with aro7 or pha2 deletion strains showed that both the mitochondrial and peroxisomal versions of Aro7fbr and Pha2 could replace the missing cytosolic reactions and enabled growth on media without aromatic amino acid supplementation (see Supplementary Fig. S1A and B). Only in case of MTS-ARO7fbr expression in an aro7Δ strain, the growth test showed a slight reduction in growth in comparison to cytosolic ARO7fbr expression (see Supplementary Fig. S1A). Overall, the growth tests suggested that transporters or pores in the organellar membranes (DeLoache et al., 2016, Palmieri and Pierri, 2010) allow passage of chorismate, prephenate and phenylpyruvate. However, we cannot exclude that a minor fraction of the MTS of ePTS1 fusion proteins remained in the cytosol.
Having shown that Aro7fbr and Pha2 targeted to mitochondria (mitAro7fbr and mitPha2, respectively) and peroxisomes (peroxAro7fbr and peroxPha2, respectively) are active, we fermented with the aro7Δ strain MRY25 (CEN.PK2-1C TRP1 Shik↑ aro7Δ trp2Δ) expressing from plasmids ARO7fbr, PHA2 and hmaS with or without added targeting sequences and analyzed MA and HMA titers (Fig. 3C and D). Besides the expression of PHA2 variants from a plasmid, PHA2 was additionally expressed from the genome as MRY25 only carried an aro7 deletion but no pha2 deletion. The presence of additional cytosolically localized Pha2 needs to be considered when interpreting the fermentation results.
When mitAro7fbr, mitPha2 and mitHmaS were used, the titer of MA increased threefold whereas the HMA titer decreased 9-fold in comparison to the cytosolically localized enzymes (Fig. 3C, red vs. blue bars). A similar but weaker tendency was seen for peroxAro7fbr, peroxPha2 and peroxHmaS. Here, the MA titer increased 1.6-fold whereas the HMA titer decreased only slightly in comparison to the cytosolically localized enzymes (Fig. 3D, green vs. blue bars). In both cases, peroxisomal or mitochondrial targeting of Aro7fbr, Pha2 and HmaS was beneficial for MA production.
For the controls where solely Pha2 was localized in mitochondria or peroxisomes, we expected an increased conversion of prephenate by Tyr1 resulting in enhanced HMA production. The results for Aro7fbr+mit/peroxPha2+HmaS confirmed this hypothesis ‒ the MA concentration dropped and the HMA concentration increased slightly in comparison to the cytosolic control (Fig. 3C and D, grey bars with blue border vs. blue bars).
Interestingly, when Aro7fbr and HmaS were localized in mitochondria (mitAro7fbr, (mit)Pha2 and mitHmaS, Fig. 3C) the HMA titers decreased strongly from 94 mg/L to less than 20 mg/L. This might suggest a limited permeability of the mitochondrial membrane for hydroxyphenylpyruvate or HMA. For peroxisomal localization of Aro7 and HmaS we detected a much weaker effect on HMA levels (peroxAro7fbr, (perox)Pha2 and peroxHmaS, Fig. 3D), which can be explained by the high permeability of the peroxisomal membrane to small hydrophilic molecules (DeLoache et al., 2016).
Unexpectedly, we detected increased MA for the controls where only the second enzyme Pha2 was localized in the cytosol and the two other enzymes were localized in the organelles (mit/peroxAro7fbr, Pha2 and mit/peroxHmaS, Fig. 3C and D, grey bars with red/green border). As the second out of the three enzymes was not localized in the respective organelle we had expected a decrease in MA production. Furthermore, the titers of the Ehrlich pathway products phenylacetic acid and phenylethanol remained inconclusive (see Supplementary Fig. S2). We detected a strong decrease in phenylacetic acid and phenylethanol (PAA+PET) titers when Aro7fbr, Pha2 and HmaS were localized in mitochondria, but also when the second enzyme Pha2 was localized in the cytosol (Supplementary Fig. S2A). For peroxisomal localization of the enzymes the phenylacetic acid and phenylethanol (PAA+PET) titers remained nearly unchanged (Supplementary Fig. S2B).
The results described were unexpected - from pathway compartmentalization we had expected advantages for MA production due to the following circumstances: (1) production of prephenate inside the organelle (catalyzed by mit/peroxAro7fbr) whereas the competing enzyme Tyr1 is localized in the cytosol; (2) production of phenylpyruvate inside the organelle (catalyzed by mit/peroxPha2) whereas the competing Ehrlich pathway enzymes are localized in the cytosol; (3) cytosolic production of the competing substrate hydroxyphenylpyruvate (catalyzed by Tyr1) whereas mit/peroxPha2 produces phenylpyruvate within the organelle where also mit/peroxHmaS is localized and (4) clustering of enzymes leading to a channeling effect (Castellana et al., 2014) (see Fig. 1).
However, our results – especially the controls where only the second enzyme Pha2 was localized in the cytosol – did not entirely support the above-mentioned hypotheses. We therefore conclude that our observations are the result of an interplay of several factors like (restricted) permeability of the organellar membranes, regulation on expressional level, varying enzyme activities depending on their localization (organelle vs. cytosol) together with the desired pathway compartmentalization effects.
Altogether, we could show that pathway compartmentalization had a beneficial effect on the shift from HMA to MA production. However, regarding the ambiguous results and the low MA titers (2.7 mg/L in mitochondria and 2.6 mg/L in peroxisomes) we conclude that even though peroxisomal and mitochondrial pathway compartmentalization positively influenced MA production, it is not a promising option for future applications.
3.3. Using the bifunctional enzyme PheA leads to increased mandelic acid production
As an alternative approach to shift production from HMA to MA in S. cerevisiae, we used the bifunctional E. coli enzyme PheA in an aro7Δ strain. PheA has both chorismate mutase and prephenate dehydratase activity and converts chorismate to phenylpyruvate via prephenate (see Fig. 1). We assumed that in PheA some kind of substrate channeling might occur (Castellana et al., 2014), thereby favoring the flux of prephenate towards phenylpyruvate over the flux towards hydroxyphenylpyruvate. We used a feedback resistant version of PheA (PheAfbr, ΔT304, G305K and ΔQ306; according to Nelms et al. (1992); codon-optimized sequence in Supplementary Table S4).
First, we tested in a growth test whether the chorismate mutase domain of PheAfbr is active in S. cerevisiae and whether pheAfbr expression impairs growth on medium without tyrosine. We therefore grew the aro7Δ strain MRY36 expressing pheAfbr (and hmaS) on SMD medium without supplementation of aromatic amino acids (Fig. 4A). MRY36 expressing pheAfbr (± hmaS) grew comparable to MRY36 expressing ARO7fbr (± PHA2, ± hmaS) confirming that the chorismate mutase domain of PheAfbr is active in S. cerevisiae. Additionally, the results showed that the intermediate prephenate is also still available for prephenate dehydrogenase Tyr1 to be further converted to tyrosine.
Furthermore, we also found that PheAfbr can replace the prephenate dehydratase Pha2 of S. cerevisiae as expression of pheAfbr in a pha2Δ strain (MRY15, CEN.PK2-1C TRP1 Shik↑ pha2Δ) enabled growth on medium without phenylalanine (see growth test in Supplementary Fig. S4).
Having confirmed that both the chorismate mutase and the prephenate dehydratase domain of PheAfbr are active in S. cerevisiae, we tested PheAfbr together with the HmaS of N. uniformis in a fermentation using the aro7Δ strain MRY36. The fermentation was performed in SMD medium without amino acid supplementation. We compared cells expressing pheAfbr and hmaS with cells expressing ARO7fbr, PHA2 and hmaS (see Fig. 4B-D). Upon expression of ARO7fbr, PHA2 and hmaS 9.5 mg/L MA was produced after 120 h (Fig. 4B). When hmaS was expressed together with pheAfbr instead, the MA titer increased to 36 mg/L (Fig. 4B). Additionally, a strong increase of the Ehrlich pathway products phenylacetic acid and phenylethanol confirmed that PheAfbr caused an increased flux into the phenylalanine biosynthesis branch (Fig. 4D). Furthermore, HMA titers decreased 4-fold when pheAfbr was expressed instead of ARO7fbr and PHA2 (Fig. 4C).
The increased flux into the phenylalanine branch upon expression of pheAfbr could be caused by a higher prephenate dehydratase activity of PheAfbr in comparison to Pha2 or by a substrate channeling effect. PheA is a well-studied enzyme and its catalytic efficiency was analyzed in several in vitro studies (Dorpheide et al., 1972, Lee et al., 1995, Liu et al., 1996, Zhang et al., 2000). However, these studies were performed under different conditions and reported varying KM and kcat values. Additionally, we did not find in vitro studies analyzing the catalytic efficiency of Pha2 which would allow comparing the prephenate dehydratase activities of Pha2 and PheA. Several studies on PheA suggest that there is no channeling or at least no tight channeling of prephenate from the chorismate mutase domain to the prephenate dehydratase domain (Duggleby et al., 1978, Zhang et al., 2000, Zhang et al., 1998). This is in accordance with our results which show that at least a fraction of prephenate diffuses from PheA to enter the tyrosine branch (growth test Fig. 4A and fermentation Fig. 4C). However, this does not exclude that partial channeling between the two domains or channeling due to enzyme clustering (Castellana et al., 2014) could occur.
To conclude, there are several possible explanations for the effect seen for pheAfbr expression in S. cerevisiae. Either way, using PheAfbr in S. cerevisiae enabled us to significantly increase MA production in comparison to the same strain overexpressing the individual yeast genes ARO7fbr and PHA2.
3.4. Expressing pheAfbr and hmaS from a single plasmid further increases mandelic acid production
In the experiments described in chapter 3.3, every gene was expressed from an individual plasmid so that each cell contained three plasmids. As this might cause heterogeneous expression of the genes on single cell level (Lee et al., 2015) and/or plasmid burden effects (Karim et al., 2013), we constructed a single plasmid from which both pheAfbr and hmaS were expressed. We first created a new hmaS construct where we replaced the methionine repressible MET25prom promoter by the strong constitutive ZEO1prom promoter (Tochigi et al., 2010) and the CYC1ter by the expression-enhancing PRM9ter terminator (Curran et al., 2013) (plasmid MRV143). The new plasmid MRV143 did not lead to higher MA and HMA production when compared to the original hmaS plasmid MRV104 (see Supplementary Fig. S4).
Nevertheless, we used ZEO1prom-hmaS-PRM9ter to construct the hmaS-pheAfbr plasmid MRV144 (ZEO1prom-hmaS-PRM9ter + HXT7prom-pheAfbr-CYC1ter, see Supplementary Table S1). Intriguingly, the aro7Δ strain MRY36 harboring the new hmaS-pheAfbr plasmid MRV144 reached MA titers up to 120 mg/L after 120 h (Fig. 4E), which is a threefold increase compared to MRY36 expressing the genes from individual plasmids (Fig. 4B). The yield of MA after 120 h was 5.6 (± 0.5) mg/g glucose. The results outline the strong impact of heterogeneity on single cell level (Lee et al., 2015) and/or plasmid burden effects (Karim et al., 2013) when using several 2 µ plasmids for enzyme expression. A genomic integration of multiple copies of pheAfbr and hmaS could further reduce plasmid burden effects and might yield even higher MA levels.
Overall, using PheAfbr increased MA titers 12-fold from 10 mg/L to 120 mg/L. This titer is approximately half of what we achieved in our previous study (236 mg/L MA, Reifenrath and Boles, 2018). However, the strain used in that previous work was auxotrophic for all aromatic amino acids, barely grew within 120 h (from OD600 = 5 to OD600 = 6) and did not fully consume glucose and ethanol after 120 h in medium supplemented with aromatic amino acids (Reifenrath and Boles, 2018). The MA producer strain constructed in this study (aro7Δ strain MRY36 harboring the hmaS-pheAfbr plasmid MRV144) did not need supplementation of any aromatic amino acid, grew well during the fermentation (from OD600 = 5 to OD600 = 11) and completely consumed glucose and ethanol (Fig. 4E).
3.5. Conclusion and outlook
We tested three different strategies to increase mandelic acid production in tyrosine prototrophic S. cerevisiae strains: (1) engineering the active site of HmaS, (2) compartmentalizing the MA pathway and (3) using a feedback resistant form of the bifunctional chorismate mutase-prephenate dehydratase PheA from E. coli (scheme in Fig. 1). The third approach was most promising. Expression of pheAfbr together with hmaS from a single plasmid in the aro7Δ S. cerevisiae strain MRY36 increased MA production 12-fold (comp. Fig. 4A and E). With PheAfbr, we achieved our goal of demonstrating a promising strategy to create a prototrophic MA producer strain. Moreover, our results suggest that using PheAfbr in S. cerevisiae is very interesting also for the production of other phenylalanine branch derived compounds such as phenylethanol or heterologous trans-cinnamic acid derivatives like styrene, pinocembrin dihydrochalcone, cinnamaldehyde and cinnamyl alcohol (Gottardi et al., 2017, Suástegui and Shao, 2016).
Acknowledgements
We thank Jun-yong Choe for helpful suggestions regarding protein engineering. Financial support by the German Federal Ministry of Education and Research following a decision of the German Bundestag (Grants 031A542 and 031B0218) is gratefully acknowledged.
Acknowledgments
Conflict of interest
None.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.mec.2018.e00079.
Contributor Information
Mara Reifenrath, Email: reifenrath@bio.uni-frankfurt.de.
Mislav Oreb, Email: m.oreb@bio.uni-frankfurt.de.
Eckhard Boles, Email: e.boles@bio.uni-frankfurt.de.
Appendix A. Supplementary material
Supplementary material
References
- Averesch N.J.H., Krömer J.O. Metabolic engineering of the shikimate pathway for production of aromatics and derived compounds—present and future strain construction strategies. Front. Bioeng. Biotechnol. 2018;6 doi: 10.3389/fbioe.2018.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benisch F., Boles E. The bacterial Entner-Doudoroff pathway does not replace glycolysis in Saccharomyces cerevisiae due to the lack of activity of iron-sulfur cluster enzyme 6-phosphogluconate dehydratase. J. Biotechnol. 2014;171:45–55. doi: 10.1016/j.jbiotec.2013.11.025. [DOI] [PubMed] [Google Scholar]
- Braus G.H. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol. Rev. 1991;55:349–370. doi: 10.1128/mr.55.3.349-370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee J., He P., Moran G.R., Harrison D.H.T. Two roads diverged: the structure of hydroxymandelate synthase from Amycolatopsis orientalis in complex with 4-hydroxymandelate. Biochemistry. 2008;47:2002–2013. doi: 10.1021/bi701438r. [DOI] [PubMed] [Google Scholar]
- Bruder S., Reifenrath M., Thomik T., Boles E., Herzog K. Parallelised online biomass monitoring in shake flasks enables efficient strain and carbon source dependent growth characterisation of Saccharomyces cerevisiae. Microb. Cell Fact. 2016;15:127. doi: 10.1186/s12934-016-0526-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana M., Wilson M.Z., Xu Y., Joshi P., Cristea I.M., Rabinowitz J.D., Gitai Z., Wingreen N.S. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat. Biotechnol. 2014;32:1011–1018. doi: 10.1038/nbt.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.L., Teleshova N., Rapista A., Paluch M., Anderson R.A., Waller D.P., Zaneveld L.J.D., Granelli-Piperno A., Klotman M.E. SAMMA, a mandelic acid condensation polymer, inhibits dendritic cell-mediated HIV transmission. FEBS Lett. 2007;581:4596–4602. doi: 10.1016/j.febslet.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G.M. 2nd ed. Sinauer Associates; Sunderland (MA): 2000. The Cell: A Molecular Approach. [Google Scholar]
- Curran K.A ., Karim A.S., Gupta A., Alper H.S. Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications. Metab. Eng. 2013;19:88–97. doi: 10.1016/j.ymben.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloache W.C., Dueber J.E. Compartmentalizing metabolic pathways in organelles. Nat. Biotechnol. 2013 doi: 10.1038/nbt.2549. [DOI] [PubMed] [Google Scholar]
- DeLoache W.C., Russ Z.N., Dueber J.E. Towards repurposing the yeast peroxisome for compartmentalizing heterologous metabolic pathways. Nat. Commun. 2016;7:11152. doi: 10.1038/ncomms11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorpheide T.A.A., Crewther P., Davidson B.E. Chorismate from Escherichia coli K-12 dehydratase. J. Biol. Chem. 1972;247:4447–4452. [PubMed] [Google Scholar]
- Dower W.J., Miller J.F., Ragsdale C.W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggleby R.G., Sneddon M.K., Morrison J.F. Chorismate mutase-prephenate dehydratase from Escherichia coli: active sites of a bifunctional enzyme. Biochemistry. 1978;17:1548–1554. doi: 10.1021/bi00601a030. [DOI] [PubMed] [Google Scholar]
- Entian K.D., Kötter P. Yeast genetic strain and plasmid collections. Methods Microbiol. 2007 [Google Scholar]
- Furlenmeier A., Quitt P., Vogler K., Lanz P., 1976. 6-Acyl derivatives of aminopenicillanic acid. US Patent 3900464.
- Generoso W.C., Gottardi M., Oreb M., Boles E. Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J. Microbiol. Methods. 2016;127:203–205. doi: 10.1016/j.mimet.2016.06.020. [DOI] [PubMed] [Google Scholar]
- Generoso W.C., Brinek M., Dietz H., Oreb M., Boles E. Secretion of 2,3-dihydroxyisovalerate as a limiting factor for isobutanol production in Saccharomyces cerevisiae. FEMS Yeast Res. 2017 doi: 10.1093/femsyr/fox029. [DOI] [PubMed] [Google Scholar]
- Gibson B.R., Lawrence S.J., Leclaire J.P.R., Powell C.D., Smart K.A. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol. Rev. 2007 doi: 10.1111/j.1574-6976.2007.00076.x. [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O., Iii C.A.H., America N. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:1–4. doi: 10.1038/nprot.2007.17. [DOI] [PubMed] [Google Scholar]
- Gottardi M., Reifenrath M., Boles E., Tripp J. Pathway engineering for the production of heterologous aromatic chemicals and their derivatives in Saccharomyces cerevisiae: bioconversion from glucose. FEMS Yeast Res. 2017;17:1–22. doi: 10.1093/femsyr/fox035. [DOI] [PubMed] [Google Scholar]
- Hammer S.K., Avalos J.L. Harnessing yeast organelles for metabolic engineering. Nat. Chem. Biol. 2017;13:823–832. doi: 10.1038/nchembio.2429. [DOI] [PubMed] [Google Scholar]
- Hazelwood L.A., Daran J.M., Van Maris A.J.A., Pronk J.T., Dickinson J.R. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 2008;74:3920. doi: 10.1128/AEM.02625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Dong L., Outten C.E. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J. Biol. Chem. 2008;283:29126–29134. doi: 10.1074/jbc.M803028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M. Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 2006 doi: 10.1007/s00253-005-0252-y. [DOI] [PubMed] [Google Scholar]
- Karim A.S., Curran K.A., Alper H.S. Characterization of plasmid burden and copy number in Saccharomyces cerevisiae for optimization of metabolic engineering applications. FEMS Yeast Res. 2013;13:107–116. doi: 10.1111/1567-1364.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.Y., Karplus P.A., Ganem B., Clardy J. Atomic structure of the buried catalytic pocket of Escherichia coli Chorismate Mutase. J. Am. Chem. Soc. 1995;117:3627–3628. [Google Scholar]
- Lee M.E., DeLoache W.C., Cervantes B., Dueber J.E. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol. 2015 doi: 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- Liu D.R., Cload S.T., Pastor R.M., Schultz P.G. Analysis of active site residues in Escherichia coli chorismate mutase by site-directed mutagenesis. J. Am. Chem. Soc. 1996;118:1789–1790. [Google Scholar]
- Liu S.P., Liu R.X., El-Rotail A.A.M.M., Ding Z.Y., Gu Z.H., Zhang L., Shi G.Y. Heterologous pathway for the production of l-phenylglycine from glucose by E. coli. J. Biotechnol. 2014;186:91–97. doi: 10.1016/j.jbiotec.2014.06.033. [DOI] [PubMed] [Google Scholar]
- Luttik M.A.H., Vuralhan Z., Suir E., Braus G.H., Pronk J.T., Daran J.M. Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: quantification of metabolic impact. Metab. Eng. 2008;10:141–153. doi: 10.1016/j.ymben.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Mill J., Schmiegel K.K., Shaw W.N., 1983. Phenethanolamines, compositions containing the same, and method for effecting weight control. U.S. Patent 4391826.
- Müller U., van Assema F., Gunsior M., Orf S., Kremer S., Schipper D., Wagemans A., Townsend C.A., Sonke T., Bovenberg R., Wubbolts M. Metabolic engineering of the E. coli L-phenylalanine pathway for the production of d-phenylglycine (d-Phg) Metab. Eng. 2006;8:196–208. doi: 10.1016/j.ymben.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Nelms J., Edwards R.M., Warwick J., Fotheringham I. Novel mutations in the pheA gene of Escherichia coli K-12 which result in highly feedback inhibition-resistant variants of chorismate mutase/prephenate dehydratase. Appl. Environ. Microbiol. 1992;58:2592–2598. doi: 10.1128/aem.58.8.2592-2598.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa, S., Tamaki, S., Kawamura, T., Kakeya, N., Kitao, K., 1985. Mandelic acid derivatives. WO Patent 1985001046.
- Orij R., Postmus J., Beek A., Ter, Brul S., Smits G.J. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology. 2009;155:268–278. doi: 10.1099/mic.0.022038-0. [DOI] [PubMed] [Google Scholar]
- Palmieri F., Pierri C.L. Mitochondrial metabolite transport. Essays Biochem. 2010;47:37–52. doi: 10.1042/bse0470037. [DOI] [PubMed] [Google Scholar]
- Pédelacq J.-D., Cabantous S., Tran T., Terwilliger T.C., Waldo G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Pratter S.M., Konstantinovics C., Di Giuro C.M.L., Leitner E., Kumar D., De Visser S.P., Grogan G., Straganz G.D. Inversion of enantioselectivity of a mononuclear non-heme iron(II)-dependent hydroxylase by tuning the interplay of metal-center geometry and protein structure. Angew. Chem. Int. Ed. 2013;52:9677–9681. doi: 10.1002/anie.201304633. [DOI] [PubMed] [Google Scholar]
- Ramos J., Sychrová H., Kschischo M. Yeast Membrane Transport. 2016. [PubMed] [Google Scholar]
- Reifenrath M., Boles E. Engineering of hydroxymandelate synthases and the aromatic amino pathway enables de novo biosynthesis of mandelic and 4-hydroxymandelic acid with Saccharomyces cerevisiae. Metab. Eng. 2018;45:246–254. doi: 10.1016/j.ymben.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Martínez J.A., Flores N., Escalante A., Gosset G., Bolivar F. Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds. Microb. Cell Fact. 2014 doi: 10.1186/s12934-014-0126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harb. Lab. Press. 1989 [Google Scholar]
- Saravanan P., Singh V.K. An efficient synthesis of chiral nonracemic diamines: application in asymmetric synthesis. Tetrahedron Lett. 1998;39:167–170. [Google Scholar]
- Schnappauf G., Krappmann S., Braus G.H. Tyrosine and tryptophan act through the same binding site at the dimer interface of yeast chorismate mutase. J. Biol. Chem. 1998;273:17012–17017. doi: 10.1074/jbc.273.27.17012. [DOI] [PubMed] [Google Scholar]
- Suástegui M., Shao Z. Yeast factories for the production of aromatic compounds: from building blocks to plant secondary metabolites. J. Ind. Microbiol. Biotechnol. 2016;43:1611–1624. doi: 10.1007/s10295-016-1824-9. [DOI] [PubMed] [Google Scholar]
- Sun Z., Ning Y., Liu L., Liu Y., Sun B., Jiang W., Yang C., Yang S. Metabolic engineering of the L-phenylalanine pathway in Escherichia coli for the production of S- or R-mandelic acid. Microb. Cell Fact. 2011;10:71. doi: 10.1186/1475-2859-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochigi Y., Sato N., Sahara T., Wu C., Saito S., Irie T., Fujibuchi W., Goda T., Yamaji R., Ogawa M., Ohmiya Y., Ohgiya S. Sensitive and convenient yeast reporter assay for high-throughput analysis by using a secretory luciferase from Cypridina noctiluca. Anal. Chem. 2010;82:5768–5776. doi: 10.1021/ac100832b. [DOI] [PubMed] [Google Scholar]
- Roermund C.W.T, de Jong M., IJlst L., van Marle J., Dansen T.B., Wanders R.J.A., Waterham H.R. The peroxisomal lumen in Saccharomyces cerevisiae is alkaline. J. Cell Sci. 2004;117:4231–4237. doi: 10.1242/jcs.01305. [DOI] [PubMed] [Google Scholar]
- Ward M., Yu B., Wyatt V., Griffith J., Craft T., Neurath A.R., Strick N., Li Y.Y., Wertz D.L., Pojman J.A., Lowe A.B. Anti-HIV-1 activity of poly(mandelic acid) derivatives. Biomacromolecules. 2007;8:3308–3316. doi: 10.1021/bm070221y. [DOI] [PubMed] [Google Scholar]
- Weber C., Farwick A., Benisch F., Brat D., Dietz H., Subtil T., Boles E. Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Appl. Microbiol. Biotechnol. 2010;87:1303–1315. doi: 10.1007/s00253-010-2707-z. [DOI] [PubMed] [Google Scholar]
- Weber H.E., Gottardi M., Brückner C., Oreb M., Boles E., Tripp J. Requirement of a functional flavin mononucleotide prenyltransferase for the activity of a bacterial decarboxylase in a heterologous muconic acid pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2017;83:1–13. doi: 10.1128/AEM.03472-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B., Neupert W. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000 doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Wiedemann B., Boles E. Codon-optimized bacterial genes improve L-arabinose fermentation in recombinant Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2008;74:2043–2050. doi: 10.1128/AEM.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Pohnert G., Kongsaeree P., Wilson D.B., Clardy J., Ganem B. Chorismate mutase/prephenate dehydratase from Escherichia coli. J. Biol. Chem. 1998;273:6248–6253. doi: 10.1074/jbc.273.11.6248. [DOI] [PubMed] [Google Scholar]
- Zhang S., Wilson D.B., Ganem B. Probing the catalytic mechanism of prephenate dehydratase by site- directed mutagenesis of the Escherichia coli P-protein dehydratase domain. Biochemistry. 2000;39:4722–4728. doi: 10.1021/bi9926680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material