Abstract
The inclusion of introgressive processes in evolutionary studies induces a less constrained view of evolution. Network-based methods (like large-scale similarity networks) allow to include in comparative genomics all extrachromosomic carriers (like viruses, the most abundant biological entities on the planet) with their cellular hosts. The integration of several levels of biological organization (genes, genomes, communities, environments) enables more comprehensive analyses of gene sharing and improved sequence-based classifications. However, the algorithmic tools for the analysis of such networks are usually restricted to people with high programming skills. We present an integrated suite of software tools named MultiTwin, aimed at the construction, structuring, and analysis of multipartite graphs for evolutionary biology. Typically, this kind of graph is useful for the comparative analysis of the gene content of genomes in microbial communities from the environment and for exploring patterns of gene sharing, for example between distantly related cellular genomes, pangenomes, or between cellular genomes and their mobile genetic elements. We illustrate the use of this tool with an application of the bipartite approach (using gene family–genome graphs) for the analysis of pathogenicity traits in prokaryotes.
Keywords: multipartite networks, genome evolution, twins, articulation points, gene sharing, pathogenicity
Introduction
The network paradigm is increasingly used as a complement for the invaluable phylogenetic tree reconstruction for biological evolutionary studies (Halary et al. 2010; Kloesges et al. 2011; Leigh et al. 2011; Tamminen et al. 2012; Corel et al. 2016, 2018; Iranzo, Krupovic, et al. 2016). Recently, multipartite graph analysis is in particular starting to receive an increased attention in evolutionary studies (Lanza et al. 2015; Iranzo, Koonin, et al. 2016; Corel et al. 2018). These graphs encompass several levels of biological organization. Bipartite graphs have been up to now most commonly used for comparative genomics (Ahn et al. 2011; Himmelstein et al. 2015; Lanza et al. 2017), and particularly gene family–genome bipartite graphs have already demonstrated their usefulness, like uncovering membrane-related genes shared between recently discovered ultrasmall bacteria (CPR) and archaea (Jaffe et al. 2016), proposing finer classifications of archaeal or ds-DNA viruses (Iranzo, Koonin, et al. 2016; Iranzo, Krupovic, et al. 2016), or analyzing the transmission of antibiotic resistance through Firmicute plasmids (Lanza et al. 2015). Moreover detecting structural features in genome-based graphs informs on the degree of redundancy of genomic data, and gives a summarization of the genomes under study, with possible applications to the detection of functional modules (bio-bricks). Higher level applications of multipartite graph analysis would also be of considerable interest, like the study of environmental adaptive traits with tripartite gene–genome–environment graphs or quadri-partite domain–gene–genome–environment graphs, and are starting to gain attention both from the computational (Murata 2010) and applied point of view (Alaimo et al. 2014). Here, we introduce a general framework and dedicated tools developed in Python for the basic construction, structuring, and analysis of multipartite graphs.
Our tool implements the exhaustive search for two types of structures in multipartite graphs: twin nodes and articulation points. Twin nodes are useful since they describe entities having exclusively identical distributions, such as gene families that are shared by the same sets of genomes (Corel et al. 2018). As well as being interesting from a biological perspective as sets of genes that are codistributed, these sets of exclusive genes can be used to reduce the size of the graph, which is useful when analyzing large data sets. Articulation points, which are points whose removal disconnects the graph, represent in contrast the unique bridge between otherwise completely unrelated nodes in the graph, and hint at the existence of communities in the graph, without having to rely on an explicit clustering algorithm. In the example of a gene family–genome bipartite graph these points may be a bridging gene family or set of gene families that are shared by otherwise very different genomes, indicative of long-distance gene sharing (Corel et al. 2018). Furthermore, the MultiTwin suite can generate a bipartite gene family–genome graph directly from genomic or proteome sequence data (i.e., either from sequences themselves or the output file of a BLAST all-against-all run on the set of sequences) to summarize all gene sharing between that set of genomes.
Materials and Methods
A graph is k-partite if there exists a partition of the set of nodes into k subsets, such that an edge only connects nodes from two different subsets of the partition. For example, a gene family–genome graph has two types of nodes (gene families and genomes). An edge only connects gene families and genomes, specifically when one member of a gene family is found in a genome. Nodes related by an edge are said to be neighbors. In k-partite graphs, neighbors are necessarily in different subsets of the partition. MultiTwin contains programs of two kinds: analysis of k-partite graphs, and modifications of k-partite graphs.
Analysis of k-Partite Graphs
MultiTwin includes a program named detect_twins.py for the detection of exclusively shared nodes, that is, nodes having exactly the same neighbors, and of their support (i.e., their common neighborhood). It also includes a dedicated tool to construct gene families (familydetector). Finally, it features a module (description.py) to annotate the content of a k-partite graph (and of its possible intermediate modifications). For instance, figure 1 represents a bipartite graph that is subjected to three successive modifications: intermediate modifications are construction of gene families (level 1) and detection of twins (level 2).
Fig. 1.
—Outline of the bipartite graph generation and analysis. At the root level, the bipartite graph only consists in disjoint star graphs. Level 1 and level 2 are constructed by two successive runs of factorgraph.py using the maps described in blue. The first factoring is based is the gene family clustering produced by our script familydetector. Different similarity thresholds can be used, resulting in differently structured graph (assuming a molecular clock, these graphs can be seen as time slices of evolution). The second factoring corresponds to the identification of twins by detect_twins.py. The change of identifiers in the graph is recorded in the trail files as indicated on the bottom line. At level 3, the operation is a terminal one, since it produces overlapping clusters. The analysis of the resulting components is performed by the description.py script, and is based on the annotations (at the root level) and the specified trail files.
Modifications of k-Partite Graphs
MultiTwin operates via the following basic operations on graphs: subgraphs, factoring, and (overlapping) clustering. We distinguish between iterable and terminal operations, according to whether the operation on the graph preserves or destroys the graph’s structure.
A subgraph is defined by a subset of the graph’s edges. It can be used, for example, to restrict the graph to nodes having certain properties (say, gene families with particular functions), or to edges between nodes of some kind (say, genomes having the same taxonomy). It is iterable, since it preserves the k-partiteness of the graph. It is implemented in MultiTwin’s subgraph.py script.
-
Factoring is defined by clustering nonoverlapping subsets of nodes. It results in a factor graph, where each node represents a cluster of nodes of the original graph. This operation can be used to find properties that are common to some subsets of nodes (for example, to construct gene family–genome bipartite graphs, one replaces all members of a gene family by a single node). This operation (implemented in factorgraph.py) is iterable as long as one does not cluster nodes of different types together. It also reduces the number of nodes and edges of the graph, and therefore usually implies to rename the nodes (encoded in a trail file).
To this end, an initial graph has to be defined (the root graph), whose node identifiers will serve as reference. The graph can be iteratively modified, and all intermediate steps can be considered in the analysis. This is achieved by the use of a trail file, which maintains the correspondence between the original node identifiers (in the root graph) and those of the increasingly compressed graph (cf. fig. 1).
The rationale behind the choice to maintain the reference to the identifiers of the root graph is that some biological annotations are most likely available for the entities forming the nodes of the root graph. In our example, the root graph is the gene–genome bipartite graph. Functional and taxonomic annotations are thus available for individual genes and for genomes, respectively.
Overlapping clustering happens when a single node can be assigned to multiple clusters. This operation can be used for instance when assessing taxonomic consistency of genomes containing given functional gene categories (a function performed by a cluster of gene families is present in several taxonomical categories). This is a terminal graph operation, since the overlap of clusters forces the merging of several clusters into a same supernode, and thus destroys the graph’s structure in some extent.
File Formats and Types
All files generically follow the same syntax XTABY. MultiTwin works with the following basic files.
Node type files where X is a node ID and Y is the node type (e.g., type 1 corresponding to genes and type 2 to genomes). Node type files can be omitted for unipartite graphs, and also for bipartite graphs, provided that the first column of the edge file only contains type 1 nodes, and the second column type 2 nodes.
Edge files where X denotes the head and Y the tail of an edge.
Community files where X is a node ID and Y is a community ID. These files encode overlapping and nonoverlapping clusterings (depending on whether node IDs are repeated or not). Community files are generated by MultiTwin but can also be supplied by the user. For example, the decomposition of the graph into connected components can be encoded as a community file. Clusters can be any kind of node subsets: groups of gene families or genomes, nodes forming a connected component, communities returned by an external clustering algorithm, and so on.
Trail files, where X refers to the node ID in the root graph, Y to the node ID in the compressed graph. These files start with a two-line header recalling the operation that has produced the compressed graph. Trail files allow to track consistently successive modifications of multipartite graphs and are therefore exclusively generated by the MultiTwin suite.
Only the annotation file, provided by the user, and which contains the biological information has a different format. It consists of a tab-separated file, whose first row contains the attribute names corresponding to the column below, and whose remaining rows start with the identifier used in the root graph, like in the following example:
| UniqID | Species | Project ID | Pathogen |
|---|---|---|---|
| 57955 | Rhizobium leguminosarum | PRJNA57955 | No |
Implementation and Availability
The implementation of the framework was carried out in Python (version 3.5) with some additional original code in C++. The Python code includes efficient implementations of graph algorithms from the igraph package (Csárdi and Nepusz 2006), that can moreover be accessed through the python-igraph wrapper.
The source code available at https://github.com/TeamAIRE/MultiTwin, last accessed October 6, 2018; accepts different kinds of inputs, depending on the user’s objectives. A detailed file with installation and usage information is provided. A user-friendly graphical interface is available for the main scripts of the MultiTwin suite. The data used in the application is also included as supplementary material online with a dedicated guide file that allows to replicate our analysis.
Standalone Generation and Analysis of Bipartite Gene Family–Genome Graphs
The standalone program bitwin.py performs the construction and the analysis of bipartite gene family–genome graphs (fig. 2).
Fig. 2.

—Overall structure of the bitwin.py program. The green boxes denote the user input files (optional when connected with a dotted line). The oval boxes represent the programs called by the bitwin.py script, and the in and outgoing arrows represent the input and output files. The blue boxes represented the output files generated overall by the bitwin.py script.
Construction of gene families. By default, MultiTwin assumes that the sequences have been subjected to an all-against-all BLAST or Diamond run (that can optionally be performed if the raw sequences are supplied in FASTA format). Only the reciprocal best hit is kept, and the sequence similarities are filtered for ≥80% mutual coverage and E-value ≤10−5 Then MultiTwin constructs a sequence similarity network (SSN) by filtering the remaining similarities above a user-defined similarity threshold (≥30% sequence identity by default) (Bittner et al. 2010). The sequences are then grouped into gene families. There are two ways to construct gene families (like in the software program CompositeSearch; Pathmanathan et al. 2018). With the option 1, each gene family is a connected component of the SSN (i.e., all sequences are directly related, or indirectly by a path in the graph). With the option 2, which gives a finer-grained definition of gene families, the gene families are the clusters of the SSN produced by the community multilevel algorithm (a.k.a. Louvain algorithm) (Blondel et al. 2008).
Construction of the gene family – genome bipartite graph. The user-supplied genome-sequence file is seen as the edge file of a bipartite graph, and the previously constructed gene families are stored as a community file. The bipartite graph is obtained by factoring the graph by this community file.
All the resulting bipartite graphs produced by the pipeline are stored in a hierarchy of directories below the current working directory.
Custom Usage
The MultiTwin code can also be used as a framework for the analysis of user-supplied multipartite graphs, such as a tripartite graph of gene families within different genomes, within different environments (fig. 3).
Fig. 3.

—Twin nodes in a toy example of tripartite graph. Twin classes are formed by all the nodes having exactly the same neighborhood. In this example, we highlighted in the same color the nodes forming the graph’s three twin classes containing more than one node. All nodes in black have a different set of neighbors (and form thus each their own twin class). In a multipartite graph, twins can be homogeneous, like twin 1 (in yellow) or heterogeneous, like twins 2 and 3. The detect_twins.py script implements an option to detect only homogeneous twins (possibly even of a given type). In a tripartite graph where nodes of respective types 1, 2, and 3 are gene families, genomes, and environments, it may be interesting to detect patterns like twin 2, where a gene family is found in the strict subset of those genomes that thrive in the same environment. Twin 3 is likely less informative, since the environment is nondiscriminating (core genes are nevertheless detected on the lower layer).
Results
We have applied the MultiTwin suite to the study of gene sharing in the microbial world (Corel et al. 2018). Here, we propose a small application for the study of pathogenicity traits in prokaryotes. We assembled a data set of 20 pairs of genomes with comparable sizes, coming from phylogenetically closely related pathogen and nonpathogen organisms (supplementary table 1, Supplementary Material online), to test the existence of genes exclusively associated with pathogenicity or nonpathogenicity. According to (Merhej et al. 2009; Georgiades and Raoult 2011), there are good reasons to expect finding such genes. Organisms were assigned as “pathogens” or “nonpathogens” based on metadata from the GOLD (Mukherjee et al. 2017) and PATRIC (Wattam et al. 2017) databases. Protein sequences from these genomes were used in an all-against-all BLAST search with parameters as described in (Bittner et al. 2010), and the bipartite network was generated using bitwin.py, with a minimum of 30% identity and 80% mutual coverage between sequences and gene family direction set to assemble connected components (with the option 1). COG annotations were assigned to gene families using RPS-BLAST (Marchler-Bauer et al. 2002). The detect_twins.py function was used to identify sets of genomes sharing exclusive gene families. These exclusive gene families were used as a community file for factorgraph.py, collapsing gene-families that have an identical species distribution into a single node in the factored bipartite graph. Should the gene content of prokaryotic genomes have evolved largely in a tree-like fashion, one would expect to find mostly exclusive gene families in host genomes having the same taxonomy. However, our study uncovered many sets of genes shared exclusively by polyphyletic groups of genomes.
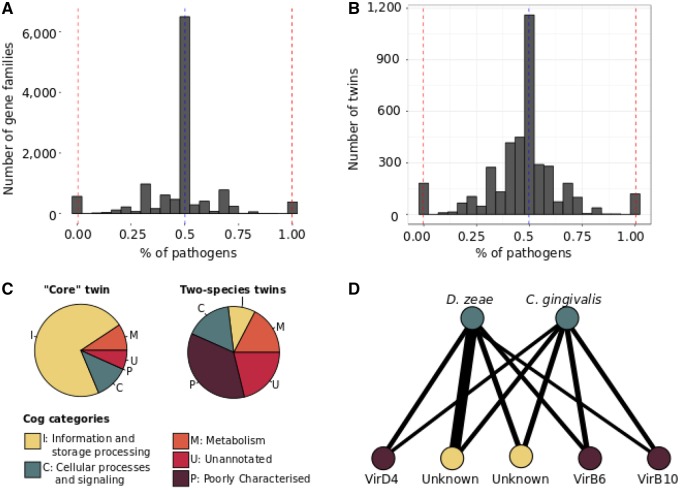
In total, 26,228 gene families were compressed to 3,982 groups of exclusively shared gene families, of which 3,197 consisted of a single gene family with a unique taxonomic distribution (80.29%), and 785 of multiple gene families with a unique taxonomic distribution (19.71%) (fig. 4). The latter include a “core” bacterial group, composed of 50 gene families (comprising 4,371 genes) that are universally conserved in all 40 genomes included in the analysis (fig. 4).
Fig. 4.
—Summary of the bipartite graph analysis of forty prokaryotic genomes. (A) The majority of gene families contained an equal proportion of pathogen and nonpathogen genes. Comparatively few are enriched in either pathogens or nonpathogens, with an extreme drop off from the peak at 0.5. A subset of gene families are exclusive to pathogens or to nonpathogens, indicated by peaks at 0 and 1, however the majority of these are only found in one genome. (B) Most groups of exclusively shared gene families also contain an equal proportion of pathogens and nonpathogens, however the peak at 0.5 is less extreme in comparison to the surrounding distribution. There is a more gradual decline in number of exclusively shared gene families from this peak toward the extremities at 0 and 1 than in the distribution at the gene family level. (C) Functional analysis revealed that the group of exclusively shared gene families containing all “core” gene families was predominantly composed of gene families involved in information and storage processing. This contrasts the groups of exclusively shared gene families containing gene families found in only two species, where informational genes are the least represented COG. Gene families found in two species are predominantly either associated with poorly characterized COGs or unannotated. (D) An example of group of exclusively shared gene families of four gene families (bottom nodes) codistributing in two relatively distantly related pathogen genomes (top nodes) from Dickeya zeae (Gamma-proteobacteria) and Capnocytophaga gingivalis (Flavobacteria). Two gene families (purple) contain components of the type IV secretion system, while two (yellow) have no known COG annotations. Their codistribution with components of the type IV secretion system in distantly related taxa suggests that these may play a role in pathogenicity.
Additionally, 119 pathogen-specific groups of exclusively shared gene families were identified that included sequences from more than one pathogen species (58 single gene family groups and 61 containing multiple gene families) (supplementary table 2, Supplementary Material online), as well as 20 species-specific groups of exclusively shared gene families. The strongest cases for pathogen-specific traits identified by bipartite analysis are the pathogen-specific groups of exclusively shared gene families that are most broadly distributed across multiple pathogenic genomes. Indeed, the majority of pathogen-specific groups of exclusively shared gene families (84) only included sequences from two different pathogen genomes, and none included sequences exclusive to all pathogen genomes, meaning that there is no “core” pool of gene families exclusively shared by pathogens. Two strategies were used to screen for exclusively shared gene families enriched in pathogens but also present in nonpathogens—either a coarse cutoff value of >80% of genes within a given group being pathogen-derived (42 groups of exclusive gene families in total) or a hypergeometric test followed by FDR correction to identify groups of exclusive gene families significantly enriched in pathogen-derived genes (five groups) (supplementary table 2, Supplementary Material online).
Both pathogen-specific and pathogen-enriched groups of exclusive gene families identified in this analysis include gene families with known roles in pathogenicity. One of the most broadly distributed pathogen-specific group of exclusive gene families, ADP-heptose: LPS heptosyltransferase (COG0859), is a part of the core machinery for LPS biosynthesis which, as an endotoxin, is a characterized factor in the pathogenesis of a broad range of Gram-negative bacteria (Raetz and Whitfield 2002). Another pathogenicity factor, haemolysin coregulated protein 1, was enriched in pathogens (based on the >80% cutoff). This is part of the type 6 secretion machinery, and has been proposed as a chaperone for effector protein secretion (Silverman et al. 2013). In addition to pathogenicity factors, a chloramphenicol-O-acetyl transferase and a beta-lactamase class D were enriched in pathogen genomes, enzymes conferring antibiotic resistance (Schwarz et al. 2004).
The identification of these known pathogen gene families within our set of groups of exclusive gene families can be seen as a proof of concept. It demonstrates the effectiveness of the bipartite graph approach for gene rediscovery. Moreover, this approach could be applied to identify novel genes associated with a particular feature. For example, in this data set many groups of exclusive gene families unique to pathogens and enriched in pathogen genomes compared with nonpathogens are either annotated by COG as conserved proteins of unknown function, or unannotated in COG. Their enrichment in pathogen genomes compared with other groups of exclusive gene families suggests a potential role in pathogenicity for these thus far uncharacterized genes.
Likewise, 181 nonpathogen-specific groups of exclusive gene families (98 groups containing a single gene family and 83 containing multiple gene families) were identified in this analysis, and none were featured in all nonpathogen genomes. Additionally, we identified 96 groups of exclusive gene families in which >80% of genes were from nonpathogens, and 29 groups of exclusive gene families enriched in nonpathogens, using the hypergeometric test. The nonpathogen-specific and enriched groups of exclusive gene families are more abundant and generally have broader distribution than those found in pathogens. This greater abundance is consistent with the findings of a broader analysis on 317 genomes (Merhej et al. 2009), which suggested that gene loss, in opposed to acquisition of virulence factors, has driven the evolution of parasites in their adaptation to their host cell. This included the loss of rRNA genes and transcriptional regulators, a result which is mirrored in our analysis. Another five broadly distributed nonpathogen-enriched groups of exclusive gene families (two of them containing several gene families) are associated with loss of transcriptional regulation, supporting the idea that the evolution of pathogenesis could be related to the loss of regulation. Our approach independently found a correlation between nutrient acquisition (Merhej et al. 2009), and specifically a nitrogen fixation ability and a nonpathogenic lifestyle. Two large and broadly distributed groups of several exclusive gene families enriched in nonpathogens are made up entirely of ABC-transport protein gene families, with predicted substrates including sugars and amino acids and oxoions. Four different groups of exclusive gene families also each include different components of the TRAP-type C4-dicarboxylate transport system, with substrates including succinate, malate and fumarate. This transport system is required for nitrogen fixation (Finan et al. 1983). Another more broadly distributed group of several exclusive gene families present only in nonpathogens includes two gene families, a predicted Fe-S oxidoreductase and nitrogenase molybdenum-iron protein (alpha and beta chains). These are central components of the pathway for nitrogen fixation (Dixon and Kahn 2004). Moreover, one exclusive gene family, unique to nonpathogens, is annotated as a Sec-independent protein secretion pathway component. This secretion system has a broad range of functions, one of which is its requirement for nitrogen oxide reduction in the nitrogen cycle (Natale et al. 2008). Finally, two groups of exclusive gene families containing >80% nonpathogen genes include additional parts of the pathway for nitrogen fixation: nitrogenase subunit NifH and Nitrate/Nitrite transport proteins. While elements of the nitrogen fixation pathway are shared between pathogens and nonpathogens (Carvalho et al. 2010), our bipartite graph analysis reinforces the argument that nitrogen fixation is a predominantly a feature of nonpathogenic bacteria.
Discussion
This relatively small scale bipartite graph analysis identified known signatures of pathogenesis and antibiotic resistance that were exclusive to or enriched in pathogen genomes, as well as genes of thus far unknown function which may play similar roles in pathogen biology, highlighting the potential of the approach for gene discovery. A larger number of groups of exclusive gene families were associated with nonpathogen genomes, consistent with the idea that pathogens undergo reductive evolution during their adaptation to the host environment including deregulation of gene expression (Merhej et al. 2009; Georgiades and Raoult 2011). Nonpathogen-enriched groups of exclusive gene families associated were also associated with nitrogen fixation. Nitrogen fixation within a prokaryotic community can be viewed as an example of the production of a “public good”—it is a pathway that produces an important commodity that can be shared by an entire community, but its phylogenetic distribution within that community is patchy. Though some pathogens are known to encode genes involved in the production of public goods, it would be interesting to explore whether there is a broad trend toward the production of public goods by nonpathogens. MultiTwin would allow to test this hypothesis on a larger scale data set, as well as extend the analysis to additional levels of organization: investigating the subgenic level, by using tripartite domain–gene–genome graphs or, at the other end of the scale, the environmental conditions, by using tripartite gene–genome–environments graphs (or even considering 4-partite domain–gene–genome–environments graphs). Another likely direction would be to use MultiTwin to define core and shell genes of pangenomes.
Supplementary Material
Acknowledgments
We thank J. O. McInerney, M. Habib, F. de Montgolfier, and T. Hujsa for critical discussions, and R. Lannes for help with the Python code. This work has been supported by the European Research Council (grant FP7/2007-2013 Grant Agreement #615274 to J.S.P., A.K.W., E.C., and E.B.) and a grant from Région Ile-de-France (DIM Malinf 2011-2013) to S.K.
Literature Cited
- Ahn Y-Y, Ahnert SE, Bagrow JP, Barabási A-L.. 2011. Flavor network and the principles of food pairing. Sci Rep. 1:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaimo S, Giugno R, Pulvirenti A.. 2014. ncPred: ncRNA-disease association prediction through tripartite network-based inference. Front Bioeng Biotechnol. 2(71). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner L, et al. 2010. Some considerations for analyzing biodiversity using integrative metagenomics and gene networks. Biol Direct 5(1):47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E.. 2008. Fast unfolding of communities in large networks. J Stat Mech Theory Exp. 2008(10):P10008–P10008. [Google Scholar]
- Carvalho FM, Souza RC, Barcellos FG, Hungria M, Vasconcelos ATR.. 2010. Genomic and evolutionary comparisons of diazotrophic and pathogenic bacteria of the order Rhizobiales. BMC Microbiol. 10(1):37.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corel E, et al. 2018. Bipartite network analysis of gene sharings in the microbial world. Mol Biol Evol. 34(4):899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corel E, Lopez P, Méheust R, Bapteste E.. 2016. Network-thinking: graphs to analyze microbial complexity and evolution. Trends Microbiol. 24(3):224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csárdi G, Nepusz T.. 2006. The igraph software package for complex network research. InterJournal Complex Syst. 1695:1695. [Google Scholar]
- Dixon R, Kahn D.. 2004. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol. 2(8):621–631. [DOI] [PubMed] [Google Scholar]
- Finan TM, Wood JM, Jordan DC.. 1983. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J Bacteriol. 154:1403–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades K, Raoult D.. 2011. Defining pathogenic bacterial species in the genomic era. Front Microbiol. 1:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E.. 2010. Network analyses structure genetic diversity in independent genetic worlds. Proc Natl Acad Sci U S A. 107(1):127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelstein DS, et al. 2015. Heterogeneous network edge prediction: a data integration approach to prioritize disease-associated genes. PLoS Comput. Biol. 11(7):e1004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranzo J, Koonin EV, Prangishvili D, Krupovic M.. 2016. Bipartite network analysis of the archaeal virosphere: evolutionary connections between viruses and capsid-less mobile elements. J Virol. 90(24):11043–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranzo J, Krupovic M, Koonin EV.. 2016. The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. MBio 7:e00978–e00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AL, Corel E, Pathmanathan JS, Lopez P, Bapteste E.. 2016. Bipartite graph analyses reveal interdomain LGT involving ultrasmall prokaryotes and their divergent, membrane-related proteins. Environ Microbiol. 18(12):5072–5081. [DOI] [PubMed] [Google Scholar]
- Kloesges T, Popa O, Martin W, Dagan T.. 2011. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol. 28(2):1057–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza VF, Baquero F, de la Cruz F, Coque TM.. 2017. AcCNET (Accessory Genome Constellation Network): comparative genomics software for accessory genome analysis using bipartite networks. Bioinformatics 33(2):283–285. [DOI] [PubMed] [Google Scholar]
- Lanza VF, et al. 2015. The plasmidome of firmicutes: impact on the emergence and the spread of resistance to antimicrobials. Microbiol Spectr. 3:PLAS-0039-2014. [DOI] [PubMed] [Google Scholar]
- Leigh JW, Schliep K, Lopez P, Bapteste E.. 2011. Let them fall where they may: congruence analysis in massive phylogenetically messy data sets. Mol Biol Evol. 28(10):2773–2785. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2002. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30(1):281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D.. 2009. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol Direct 4(1):13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, et al. 2017. Genomes OnLine Database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 45(D1):D446–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T. 2010. Detecting communities from tripartite networks. Proceedings of the 19th international conference on World wide web – WWW ’10. New York (NY): ACM Press. p. 1159.
- Natale P, Brüser T, Driessen AJM.. 2008. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—distinct translocases and mechanisms. Biochim Biophys Acta Biomembr. 1778(9):1735–1756. [DOI] [PubMed] [Google Scholar]
- Pathmanathan JS, Lopez P, Lapointe F-J, Bapteste E.. 2018. CompositeSearch: a generalized network approach for composite gene families detection. Mol Biol Evol. 35(1):252–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CRH, Whitfield C.. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem. 71:635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A.. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev. 28(5):519–542. [DOI] [PubMed] [Google Scholar]
- Silverman JM, et al. 2013. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell 51(5):584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamminen M, Virta M, Fani R, Fondi M.. 2012. Large-scale analysis of plasmid relationships through gene-sharing networks. Mol Biol Evol. 29(4):1225–1240. [DOI] [PubMed] [Google Scholar]
- Wattam AR, et al. 2017. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 45(D1):D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.