Keywords: peripheral nerve injury, aging, Sprague-Dawley rat, transcriptome, sequencing, mRNA, rat age, Schwann cells, neural regeneration
Abstract
An aging-induced decrease in Schwann cell viability can affect regeneration following peripheral nerve injury in mammals. It is therefore necessary to investigate possible age-related changes in gene expression that may affect the biological function of peripheral nerves. Ten 1-week-old and ten 12-month-old healthy male Sprague-Dawley rats were divided into young (1 week old) and adult (12 months old) groups according to their ages. mRNA expression in the sciatic nerve was compared between young and adult rats using next-generation sequencing (NGS) and bioinformatics (n = 4/group). The 18 groups of differentially expressed mRNA (DEmRNAs) were also tested by quantitative reverse transcription polymerase chain reaction (n = 6/group). Results revealed that (1) compared with young rats, adult rats had 3608 groups of DEmRNAs. Of these, 2684 were groups of upregulated genes, and 924 were groups of downregulated genes. Their functions mainly involved cell viability, proliferation, differentiation, regeneration, and myelination. (2) The gene with the most obvious increase of all DEmRNAs in adult rats was Thrsp (log2 FC = 9.01, P < 0.05), and the gene with the most obvious reduction was Col2a1 (log2FC = –8.89, P < 0.05). (3) Gene Ontology analysis showed that DEmRNAs were mainly concentrated in oligosaccharide binding, nucleotide-binding oligomerization domain containing one signaling pathway, and peptide-transporting ATPase activity. (4) Analysis using the Kyoto Encyclopedia of Genes and Genomes showed that, with increased age, DEmRNAs were mainly enriched in steroid biosynthesis, Staphylococcus aureus infection, and graft-versus-host disease. (5) Spearman's correlation coefficient method for evaluating NGS accuracy showed that the NGS results and quantitative reverse transcription polymerase chain reaction results were positively correlated (rs = 0.74, P < 0.05). These findings confirm a difference in sciatic nerve gene expression between adult and young rats, suggesting that, in peripheral nerves, cells and the microenvironment change with age, thus influencing the function and repair of peripheral nerves.
Introduction
The treatment of limb paralysis and dysfunction caused by peripheral nerve injury is a major challenge in medical science. In the USA alone, the number of peripheral nerve injury surgeries performed each year has reached 50,000 cases, and the economic burden is over 7 billion US dollars. Therefore, the question of how to improve treatment and effectively repair peripheral nerve injury has become a major public health issue that needs to be addressed by the global healthcare community (Wang et al., 2010; Yang et al., 2011; Chen et al., 2018). Thus, studies of peripheral nerve regeneration have significance in both scientific research and clinical practice (Zheng et al., 2014; He et al., 2015; Hung et al., 2015; Qiu et al., 2015; Pan et al., 2017; Zou et al., 2018).
The reduction in nerve repair ability in older animals is a research hotspot in the field of peripheral nerve injury repair (Verdu et al., 2000; Amer et al., 2014). For a long time, the majority of researchers have considered that a decline in neuronal regeneration ability is the major reason that axonal regeneration is slow in old animals (Graciarena et al., 2014; Moldovan et al., 2016; Lim et al., 2017). For example, Zou et al. (2013) showed that intrinsic changes in older neurons influence neuronal regeneration ability in a Caenorhabditis elegans model. They found that in older neurons, let-7 down-regulating LIN-41, which contributes to a developmental decline in axonal regeneration. However, in the younger neurons, LIN-41 inhibits let-7 expression via Argonatute ALG-1 to ensure that axonal regeneration could only be inhibited in older neurons. Some other scholars focused on cells other than neurons. For example, Painter et al. (2014) demonstrated that the peripheral nerve regeneration ability of 24-month-old mice is worse than that of 2-month-old mice; although in vitro gene detection revealed that only 14 genes in the dorsal root ganglion cells of old mice are significantly different from those in young mice. In addition, genes that are directly associated with regeneration ability (such as ATF3, GAP and prr1a) did not markedly differ between old and young mice. Therefore, some researchers have suggested that the major factor influencing peripheral nerve regeneration in mammals such as mice, rats, and humans is not the neurons themselves, but instead the decline of Schwann cell viability in peripheral nerves (Michio et al., 2014; Couve et al., 2017; Xu et al., 2017).
This study therefore aimed to compare mRNA expression in the sciatic nerves of Sprague-Dawley rats at different ages using next-generation, high-throughput whole RNA sequencing and bioinformatics. Using these techniques, the aim was to discover age-related molecules that influence the biological functions of peripheral nerves, and to investigate any underlying mechanisms, to provide theoretical bases for improving the treatment of aging peripheral nerves after injury (Canta et al., 2016; Sakita et al., 2016; Zhou. et al., 2017).
Materials and Methods
Animals
Ten 1-week-old and ten 12-month-old healthy male Sprague-Dawley rats, bought from the Animal Center of Medical School of Sun Yat-Sen University of China, were randomly selected as experimental animals. This study was approved by the Experimental Animal Administration Committee of Sun Yat-Sen University (approval number: (2013)A-055). Efforts were taken to minimize animal suffering during the experiment.
Collection of sciatic nerves
Four rats in the young or adult group were randomly selected for the next-generation RNA high-throughput sequencing detection of sciatic nerves. Rats in the adult group were deeply anesthetized using an intraperitoneal injection of 10% chloral hydrate (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China; 0.3 mL/100 g body weight). Rats in the young group were sacrificed by cervical dislocation. The specific operational procedures have been previously published (Zhu et al., 2015, 2017). Under aseptic conditions, the skin of the left leg was cut parallel to the femur, and the sciatic nerve was exposed by splitting the superficial gluteus muscle. The bilateral sciatic nerves (≥ 15 mm in length) were excised from the lower edge of piriform muscle from the rats and then external fat and connective tissue were removed.
Library preparation and RNA sequencing
Comparison of the senescence pathway genes was performed by next-generation sequencing (NGS). All sequencing programs were performed by Shanghai Biotechnology Corporation (Shanghai, China). Briefly, RNA extraction was performed using the mir Vana™ miRNA isolation kit (Cat# AM1561, Ambion, Austin, TX, USA) following the standard operating procedures provided by the manufacturer. The quality of the extracted RNA was detected using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Total RNA was then purified using the RNAClean XP kit (Cat A63987, Beckman Coulter, Inc., CA, USA) and the RNase-Free DNase Set (Cat#79254, Qiagen, Germany). The quality of the initial total RNA sample for the sequencing experiment was detected using a NanoDrop ND-2000 spectrophotometer and an Agilent Bioanalyzer 2100. Total RNA that passed the quality control was used in subsequent sequencing experiments (Pertea et al., 2015).
For transcriptome sequencing, we used the number of reads mapping to the gene region to estimate the gene expression level (Pertea et al., 2016). However, in addition to being proportional to the gene expression level, the number of reads was also associated with the length of genes and the sequencing data volume. To compare gene expression levels between different genes and different samples, reads were converted into fragments per kilobase of exon model per million mapped reads (FPKM) for normalization of gene expression levels (Mortazavi et al., 2008; Yu et al., 2017). First, the number of fragments of each gene after Hisat2 comparison was counted using Stringtie v1.3.0 (The Center for Computational Biology at Johns Hopkins University, Baltimore, MD, USA). Next, normalization was performed using the trimmed mean of M values method (Robinson et al., 2010). Finally, the FPKM value of each gene was calculated using the Perl program.
Quantitative real time polymerase chain reaction (RT-PCR)
Quantitative analysis of targeted mRNA expression was performed using RT-PCR and the relative standard curve method. Six rats in each group were randomly selected. The bilateral sciatic nerves were collected using the previously described method, and total RNA was extracted. The RNA concentration and purity were detected using a Nanodrop 2000. RNA samples with high concentrations were diluted to a final concentration of 200 ng/μL. RNA (1 μg) was used for RNA reverse transcription using the RevertAid first strand cDNA synthesis kit (Thermo Fisher, Waltham, MA, USA). An appropriate amount of cDNA was amplified using FastStart Universal SYBR Green Master Mix (Roche, Basel, Switzerland) in a fluorescence quantitative PCR machine (Stepone Plus, Thermo Fisher). The specific procedures were performed according to the manufacturer's product manual. Each sample was run in triplicate. GAPDH expression was used to normalize mRNA expression. Gene information and primers are shown in Table 1. The specificity of RT-PCR was confirmed via routine agarose gel electrophoresis and melting curve analysis. The 2-ΔΔCt method was used to calculate relative gene expression (Huang et al., 2017).
Table 1.
Primer information for the genes that were detected

Bioinformatics and statistical analyses
Analyses of differential genes among samples were performed using edgeR (Robinson et al., 2010). After the P-value was obtained, multiple testing was performed for correction. The threshold value of the P-value was determined by controlling for false discovery rate (Benjamini et al., 1995, 2001). The corrected P-value was the Q-value. In addition, the differential expression fold was calculated based on the FPKM value, which was the fold change. The screening condition of differential genes was limited to Q-value ≤ 0.05 and fold change ≥ 2. Both gene ontology (GO) analysis (http://www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (http://www.genome.jp/kegg/) were performed to determine the roles of the differentially expressed mRNAs. Fisher's exact test was used to test the significance of GO terms and pathway identifiers enriched in the differentially expressed gene list. RNA sequencing and bioinformatics analyses were performed by Shanghai Biotechnology Corporation (Shanghai, China).
The gene data of RT-PCR conform to normal distribution and were presented as the mean ± SD. SPSS 13.0 software (SPSS, Chicago, IL, USA) was used to analyze the accuracy of NGS. Quantitative RT-PCR results were compared with sequencing results using Spearman's correlation coefficient method. P < 0.05 was considered statistically significant.
Results
Analyses of mRNA expression levels in the sciatic nerve of young and adult rats
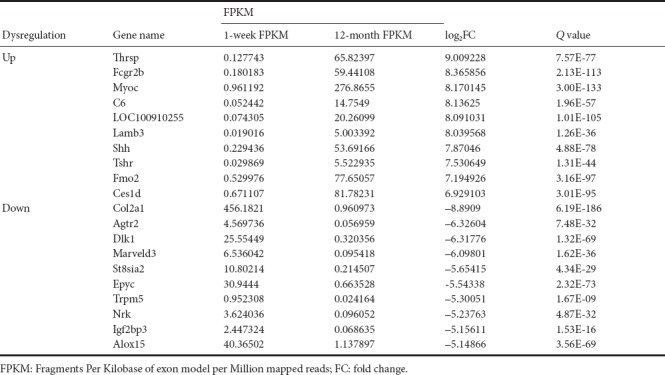
Transcriptome gene expression of sciatic nerves in young and adult rats was analyzed using NGS. A scatter map and a heat map were used to present the distribution of differentially expressed mRNAs (DEmRNAs; Figure 1). A total of 30,957 mRNA groups were detected using RNA sequencing (RNA-seq), and the DEmRNA screening was performed using fold change ≥ 2.0 and P < 0.05 as the criteria. Compared with young rats, adult rats had 3608 groups of DEmRNAs. Of these, 2684 groups were upregulated genes, and 924 groups were downregulated genes. The gene with the greatest mRNA increase in adult rats was Thrsp (log2FC = 9.01, P < 0.05), and the gene with the greatest reduction was Col2a1 (log2FC = –8.89, P < 0.05). The 10 groups of genes with the most obvious DEmRNAs are shown in Table 2. Additionally, 208 mRNAs were only expressed in adult rats and 50 mRNAs were only expressed in young rats. The gene with the greatest mRNA increase in adult rats was Rn60_X_0752.3 (FPKM = 227.23, P < 0.05), and the gene with the most obvious increase in young rats was Ddx3y (FPKM = 32.69, P < 0.05). The top 10 most highly expressed mRNAs which were not expressed by rats in the opposite group are shown in Table 3.
Figure 1.
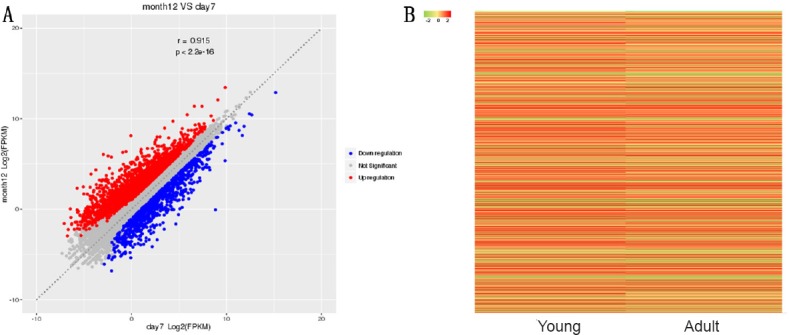
Scatter map and heat map of mRNA expression in the sciatic nerve in young (1 week old, n = 4) and adult (12 months old, n = 4) Sprague-Dawley rats.
(A) Scatter map of mRNA expression. Red indicates upregulated genes in adult rats compared with young rats, while blue indicates downregulated genes. (B) Heat map of mRNA expression. Green in the heat map indicates a fold change ≤ 2, and red indicates a fold change > 2.
Table 2.
Top 10 mRNA groups that were only expressed in the sciatic nerve of rats in young (1 week old, n = 4) and adult (12 months old, n = 4) Sprague-Dawley rats
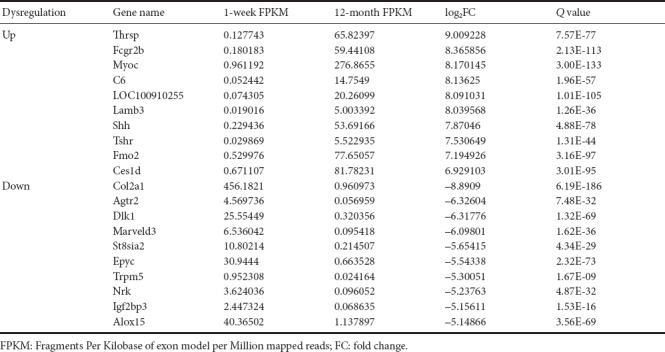
Table 3.
Top 10 differentially expressed mRNA groups in sciatic nerves that were upregulated or downregulated in young (1 week old, n = 4) and adult (12 months old, n = 4) Sprague-Dawley rats
GO and KEGG analyses of DEmRNAs
Clustering analysis of DEmRNAs can help to identify the function of unknown transcripts, or the unknown function of known transcripts, by gathering similar expression patterns or similar genes to a class (Yao et al., 2012). Functional classification by GO is an internationally standardized classification system for gene function that offers a dynamic, updated, and controlled vocabulary, employing strictly defined concepts to comprehensively describe the properties of genes and their products in any organism. GO classification encompasses three domains, as follows: molecular function, cellular component, and biological process (Yi et al., 2006). GO analysis revealed that DEmRNAs were mainly concentrated in genes related to oligosaccharide-binding, nucleotide-binding oligomerization domain containing one (NOD1) signaling pathway, peptide-transporting ATPase activity, fibrinogen binding, and marginal zone B cell differentiation (Figure 2 and Table 4).
Figure 2.
Schematic diagram of the Gene Ontology (GO) category and gene enrichment of differentially expressed mRNAs (DEmRNAs) in the sciatic nerve of 12-month-old and 1-week-old Sprague-Dawley rats.
(A) GO classification: Red columns represent biological process; green columns represent cellular component; blue columns represent molecular function. (B) GO enrichment: Different shapes indicate different types of function. Circles represent biological process; triangles represent cellular component, and squares represent molecular function. The area of the circles represents the number of DEmRNAs involved in the terms. Different colors indicate different Q-values of DEmRNAs in the GO terms between adult rats and young rats. Green represents Q > 0.10; yellow represents Q > 0.05 and Q < 0.075; orange represents Q < 0.05 and Q > 0.025; and red represents Q < 0.025. The standard of the test was set as 0.05 (n = 4 rats per group).
Table 4.
Categories in Gene Ontology (GO) analyses that corresponded to the top 20 groups of differentially expressed mRNAs (DEmRNAs) in the sciatic nerve of Sprague-Dawley rats at the ages of 1 week and 12 months

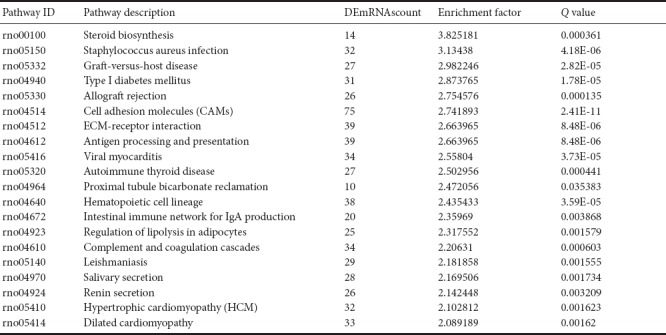
The pathway enrichment of DEmRNAs was analyzed using KEGG. The KEGG pathway database comprises information about networks of molecular interactions for numerous organisms, thus permitting functional classification. In the adult rats, DEmRNAs were mainly enriched in steroid biosynthesis, Staphylococcus aureus infection, graft-versus-host disease, type 1 diabetes mellitus, and allograft rejection categories (Figure 3 and Table 5).
Figure 3.
Schematic diagram of the pathway classification and gene enrichment of differentially expressed mRNAs (DEmRNAs) in the sciatic nerve of 12-month-old and 1-week-old Sprague Dawley rats.
(A) Kyoto Encyclopedia of Genes and Genomes (KEGG) classification: Orange columns represent cellular process; brown columns represent environmental information processing; green columns represent genetic information processing; blue columns represent metabolism; and purple columns represent organismal systems. (B) KEGG pathway enrichment: The area of the circles represents the number of DEmRNAs involved in the pathway. Different colors indicate different Q-values of DEmRNAs in the pathway between adult rats and young rats. Green represents Q > 0.15; yellow represents Q > 0.10 and Q < 0.15; orange represents Q < 0.10 and Q > 0.05; and red represents Q < 0.05. The standard of the test was set as 0.05 (n = 4 per group).
Table 5.
Pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses that corresponded to the top 20 groups of enriched differentially expressed mRNAs (DEmRNAs, age of 12 months vs. age of 1 week)
Validation of microarray sequencing accuracy using quantitative RT-PCR
Five groups of upregulated mRNAs, namely nerve growth factor receptor (NGFR), betacellulin (BTC), activin A receptor type 1C (ACVR1C), disks large homolog 2 (DLG2), and Jun proto-oncogene (JUN), and five groups of downregulated mRNAs, namely noncompact myelin associated protein (NCMAP), myelin basic protein (MBP), PDZ binding kinase (PBK), baculoviral IAP repeat containing 5 (BIRC5), and early growth response 2 (EGR2), were selected for quantitative RT-PCR analyses in rats with different genotypes. The housekeeping gene GAPDH was used as the internal control to compare the sequencing results. Among the 10 groups of DEmRNAs, only BTC expression differed from the sequencing results. The sequencing results showed that BTC expression was upregulated, whereas the quantitative RT-PCR results demonstrated that BTC expression was downregulated. In contrast, the fold changes of expression in the quantitative RT-PCR results of other DEmRNAs were similar to those of the RNA microarray sequencing results (Figure 4).
Figure 4.

Comparison of quantitative RT-PCR (n = 6/group) and RNA sequencing (RNA-seq; n = 4/group) results of differentially expressed mRNAs in the sciatic nerve of 12-month-old and 1-week-old Sprague-Dawley rats.
To achieve baseline consistency, the quantitative RT-PCR fold change data were expressed as 2-ΔΔCt, and RNA-seq data are expressed as log2FC. NGFR: Nerve growth factor receptor; ACVR1C: activin A receptor type 1C; DLG2: discs large MAGUK scaffold protein 2; JUN: Jun proto-oncogene; BTC: betacellulin; NCMAP: noncompact myelin associated protein; MBP: myelin basic protein; BIRC5: baculoviral IAP repeat containing 5; EGR2: early growth response 2; PBK: PDZ binding kinase; FC: fold change; RT-PCR: real time polymerase chain reaction.
A total of 18 groups of DEmRNAs were randomly selected, including the above 10 groups of genes, as well as KIF2C, MAP3K, PDGFB, MPZ, PMP22, MAL, WWC1 and CDC20. To evaluate the accuracy of NGS sequencing, the correlation between RNA sequencing results and quantitative RT-PCR results was evaluated using Spearman correlation coefficient analyses. The correlation coefficient rs was 0.74 and P < 0.05, indicating that the NGS results and quantitative RT-PCR results were positively correlated (Figure 5).
Figure 5.
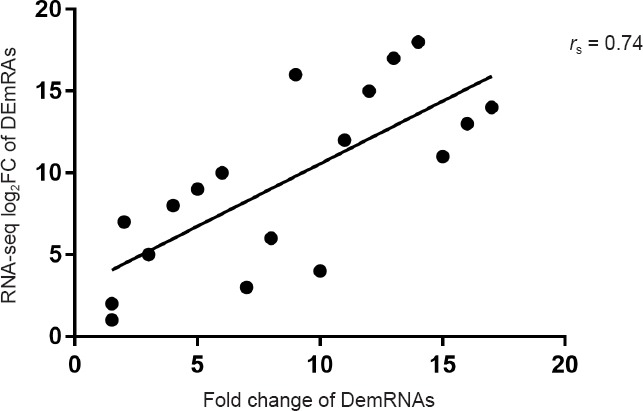
Scatter map of Spearman correlation coefficient analyses between quantitative real time polymerase chain reaction and sequencing data (Seq) of differentially expressed mRNAs (DEmRNAs) in the sciatic nerve of 12-month-old (adult) and 1-week-old (young) Sprague-Dawley rats.
The correlation coefficient rs was 0.74, and P < 0.05 (n = 6/group).
Discussion
After peripheral nerve injury, neurons initiate axonal regeneration. Distal axons, cut from the neuronal cell body, develop Wallerian degeneration (Szepanowski and Kieseier, 2016; Xiao et al., 2016; Roberts et al., 2017; Cervellini et al., 2018). Proximal axons form growth cones and extend to distal ends, which grow into the target organ to achieve functional recovery (Scheib et al., 2013; Kızılay et al., 2016; Ma et al., 2016; Wang et al., 2016; Tallon and Farah, 2017). However, with increasing age, the speed of nerve regeneration and functional recovery greatly decreases (Kawabuchi et al., 2011). We previously reviewed literature on the repair efficacy of peripheral nerve injury in the upper limb from a 20-year period, to perform a univariate meta-analysis. The results indicated that a 1-year increase in age had an odds ratio of 0.97–0.98 for excellent recovery (He et al., 2014). For example, if the regeneration ability following nerve injury in a newborn is 100%, the nerve regeneration ability after injury during old age (e.g., in a 60-year-old male) is only 16–30% of that at birth. These results suggest that the “aging” factor has a relatively large influence on nerve injury repair efficacy (Esquisatto et al., 2014; Ugrenović et al., 2016; Scheib et al., 2016; Castelnovo et al., 2017).
Some researchers suggested that the major factor influencing peripheral nerve regeneration in mammals, such as mice, rats, and humans, is not neurons themselves but instead the decline of Schwann cell viability in peripheral nerves (Belin et al., 2017; Plaza-Zabala et al., 2017). Kang et al. (2013) used phase-contrast microscopy and immunohistochemistry to detect the rate of nerve regeneration in mice at different ages. The results showed that the axonal regeneration rate of old animals does not markedly differ from that of young animals. However, endoneurial tubes in old animals have more nerve and myelin sheath debris to block the growth of axons. Therefore, these researchers considered that the “dedifferentiation-proliferation-differentiation” timing of Schwann cells, and degree of Schwann cells impact the time course of Wallerian degeneration to further influence the rate of axonal regeneration. Painter et al. (2014) compared the differences in gene expression in Schwann cells between old and young mice. They demonstrated that the expression levels of growth-factor-related coding genes (BTC, NGFR, and BDNF) and mitosis-related genes (KIF2C, PBK, BIRC5, and CDC20) markedly decrease in old mice, while expression levels of myelination-associated genes (PMP2, MPZ, MAL, and EGR2) increase in old mice. These results suggest that the dedifferentiation ability of Schwann cells declines in old mice, thereby reducing the rate of nerve regeneration.
In the current study, we compared transcriptome gene expression in the sciatic nerve of rats at different ages and found that 3608 groups of mRNAs were differentially expressed between adult rats (12 months old) and young rats (1 week old). The upregulated genes were mainly concentrated in changes in development, anatomical morphology, cell differentiation, and the extracellular matrix. To validate the accuracy of microarray sequencing, we selected 10 groups of genes associated with proliferation, differentiation, myelination, injury, and synaptic transmission for quantitative RT-PCR. The results were compared with sequencing results to validate their accuracy. The proliferation- and differentiation-related genes included NGFR, ACVR1C, BTC, survivin (BIRC5) and EGR2. The injury-related genes included JUN, and the myelination-related genes included MBP and NCMAP. The synaptic transmission-related genes included DLG2. The PCR results demonstrated that the expression trends of all groups of genes, except for BTC, were consistent with the microarray results. Spearman correlation coefficient analysis showed that the results of these two groups showed a positive correlation, which further indicated the accuracy of the microarray sequencing. The growth, development, conduction, and repair of peripheral nerves is a complex process. Although more than 1000 groups of differential mRNA sequences were obtained from the NGS array of sciatic nerves of rats at different ages, further in-depth experimental studies, as well as the function and regulatory method of deep mining of differential sequences, are required to fill the gap between age and peripheral nerve structure and function.
Collectively, this study showed that gene expression in sciatic nerves differs between adult and young rats, and these differences occur in genes involved in cell activity, proliferation, differentiation, regeneration, myelination, and injury. These results suggest that, in peripheral nerves, both cells and the microenvironment change with age to influence the function and repair of peripheral nerves. These results thus provide important theoretical bases for further studies on the relationship between peripheral nerves, regeneration, and age.
Additional file: Open peer review reports 1 (10.4KB, pdf) and 2 (8.1KB, pdf) .
Footnotes
Conflicts of interest: The authors declare that there are no conflicts of interest associated with this manuscript.
Financial support: This study was supported by the National Natural Science Foundation of China, No. 81201546 (to YXL); the Doctoral Start-up Program of Natural Science Foundation of Guangdong Province of China, No. 2017A030310302 (to ZWZ); the Medical Scientific Research Foundation of Guangdong Province of China, No. A2016018 (to BH); the Science and Technology Project of Guangdong Province of China, No. 2016A010103012 (to JHL). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: This study was approved by the Experimental Animal Administration Committee of Sun Yat-sen University of China (approval number: (2013)A-055). All experimental procedures described here were in accordance with the National Institutes of Health (NIH) guidelines for the Care and Use of Laboratory Animals.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewers: Ben Christensen, University of Utah, Salt Lake City, USA; Dumitru Andrei Iacobas, Prairie View A&M University, USA.
Funding: This study was supported by the National Natural Science Foundation of China, No. 81201546 (to YXL); the Doctoral Start-up Program of Natural Science Foundation of Guangdong Province of China, No. 2017A030310302 (to ZWZ); the Medical Scientific Research Foundation of Guangdong Province of China, No. A2016018 (to BH); the Science and Technology Project of Guangdong Province of China, No. 2016A010103012 (to JHL).
P-Reviewer: Christensen B, Iacobas DA; C-Editor: Zhao M; S-Editor: Wang J, Li CH; L-Editor: Qiu Y, Song LP; T-Editor: Liu XL
References
- 1.Amer MG, Mazen NF, Mohamed NM. Role of calorie restriction in alleviation of age-related morphological and biochemical changes in sciatic nerve. Tissue Cell. 2014;46:497–504. doi: 10.1016/j.tice.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Belin S, Zuloaga KL, Poitelon Y. Influence of mechanical stimuli on Schwann Cell Biology. Front Cell Neurosci. 2017;11:347. doi: 10.3389/fncel.2017.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- 4.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Statist. 2001;29:1165–1188. [Google Scholar]
- 5.Canta A, Chiorazzi A, Carozzi VA, Meregalli C, Oggioni N, Bossi M, Rodriguez-Menendez V, Avezza F, Crippa L, Lombardi R, de Vito G, Piazza V, Cavaletti G, Marmiroli P. Age-related changes in the function and structure of the peripheral sensory pathway in mice. Neurobiol Aging. 2016;45:136–148. doi: 10.1016/j.neurobiolaging.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Castelnovo LF, Bonalume V, Melfi S, Ballabio M, Colleoni D, Magnaghi V. Schwann cell development, maturation and regeneration: a focus on classic and emerging intracellular signaling pathways. Neural Regen Res. 2017;12:1013–1023. doi: 10.4103/1673-5374.211172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cervellini I, Galino J, Zhu N, Allen S, Birchmeier C, Bennett DL. Sustained MAPK/ERK activation in adult Schwann cells impairs nerve repair. J Neurosci. 2018;38:679–690. doi: 10.1523/JNEUROSCI.2255-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Xiang J, Wu J, He B, Lin T, Zhu Q, Liu X, Zheng C. Expression patterns and role of PTEN in rat peripheral nerve development and injury. Neurosci Lett. 2018;676:78–84. doi: 10.1016/j.neulet.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 9.Couve E, Lovera M, Suzuki K, Schmachtenberg O. Schwann cell phenotype changes in aging human dental pulp. J Dent Res. 2017;97:347–355. doi: 10.1177/0022034517733967. [DOI] [PubMed] [Google Scholar]
- 10.Esquisatto MAM, de Aro AA, Fêo HB, Gomes L. Changes in the connective tissue sheath of Wistar rat nerve with aging. Ann Anat. 2014;196:441–448. doi: 10.1016/j.aanat.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Graciarena M, Dambly-Chaudière C, Ghysen A. Dynamics of axonal regeneration in adult and aging zebrafish reveal the promoting effect of a first lesion. Proc Natl Acad Sci U S A. 2014;111:1610–1615. doi: 10.1073/pnas.1319405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He B, Zhu Q, Chai Y, Ding X, Tang J, Gu L, Xiang J, Yang Y, Zhu J, Liu X. Safety and efficacy evaluation of a human acellular nerve graft as a digital nerve scaffold: a prospective, multicentre controlled clinical trial. J Tissue Eng Regen Med. 2015;9:286–295. doi: 10.1002/term.1707. [DOI] [PubMed] [Google Scholar]
- 13.He B, Zhu Z, Zhu Q, Zhou X, Zheng C, Li P, Zhu S, Liu X, Zhu J. Factors predicting sensory and motor recovery after the repair of upper limb peripheral nerve injuries. Neural Regen Res. 2014;9:661–672. doi: 10.4103/1673-5374.130094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang G, Kang Y, Huang Z, Zhang Z, Meng F, Chen W, Fu M, Liao W, Zhang Z. Identification and characterization of long non-coding rnas in osteogenic differentiation of human adipose-derived stem cells. Cell Physiol Biochem. 2017;42:1037–1050. doi: 10.1159/000478751. [DOI] [PubMed] [Google Scholar]
- 15.Hung HA, Sun G, Keles S, Svaren J. Dynamic regulation of Schwann cell enhancers after peripheral nerve injury. J Biol Chem. 2015;290:6937–6950. doi: 10.1074/jbc.M114.622878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang H, Lichtman JW. Motor axon regeneration and muscle reinnervation in young adult and aged animals. J Neurosci. 2013;33:19480–19491. doi: 10.1523/JNEUROSCI.4067-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawabuchi M, Tan H, Wang S. Age affects reciprocal cellular interactions in neuromuscular synapses following peripheral nerve injury. Ageing Res Rev. 2011;10:43–53. doi: 10.1016/j.arr.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Kızılay Z, Erken HA, Çetin NK, Aktaş S, Abas Bİ, Yılmaz A. Boric acid reduces axonal and myelin damage in experimental sciatic nerve injury. Neural Regen Res. 2016;11:1660–1665. doi: 10.4103/1673-5374.193247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim EF, Musa A, Frederick A, Ousman SS. AlphaB-crystallin expression correlates with aging deficits in the peripheral nervous system. Neurobiol Aging. 2017;53:138–149. doi: 10.1016/j.neurobiolaging.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Ma KH, Hung HA, Svaren J. Epigenomic regulation of Schwann cell reprogramming in peripheral nerve injury. J Neurosci. 2016;36:9135–9147. doi: 10.1523/JNEUROSCI.1370-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moldovan M, Rosberg MR, Alvarez S, Klein D, Martini R, Krarup C. Aging-associated changes in motor axon voltage-gated Na+ channel function in mice. Neurobiol Aging. 2016;39:128–139. doi: 10.1016/j.neurobiolaging.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 23.Painter MW, Brosius Lutz A, Cheng YC, Latremoliere A, Duong K, Miller CM, Posada S, Cobos EJ, Zhang AX, Wagers AJ, Havton LA, Barres B, Omura T, Woolf CJ. Diminished Schwann cell repair responses underlie age-associated impaired axonal regeneration. Neuron. 2014;83:331–343. doi: 10.1016/j.neuron.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan B, Zhou HX, Liu Y, Yan JY, Wang Y, Yao X, Deng YQ, Chen SY, Lu L, Wei ZJ, Kong XH, Feng SQ. Time-dependent differential expression of long non-coding RNAs following peripheral nerve injury. Int J Mol Med. 2017;39:1381–1392. doi: 10.3892/ijmm.2017.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, stringtie and ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plaza-Zabala A, Sierra-Torre V, Sierra A. Autophagy and microglia: novel partners in neurodegeneration and aging. Int J Mol Sci. 2017;18:598. doi: 10.3390/ijms18030598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu L, He B, Hu J, Zhu Z, Liu X, Zhu J. Cartilage oligomeric matrix protein angiopoeitin-1 provides benefits during nerve regeneration in vivo and in vitro. Ann Biomed Eng. 2015;43:2924–2940. doi: 10.1007/s10439-015-1342-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts SL, Dun X, Doddrell RDS, Mindos T, Drake LK, Onaitis MW, Florio F, Quattrini A, Lloyd AC, D’Antonio M, Parkinson DB. Sox2 expression in Schwann cells inhibits myelinationin vivo and induces influx of macrophages to the nerve. Development. 2017;144:3114–3125. doi: 10.1242/dev.150656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakita M, Murakami S, Fujino H. Age-related morphological regression of myelinated fibers and capillary architecture of distal peripheral nerves in rats. BMC Neurosci. 2016;17:39. doi: 10.1186/s12868-016-0277-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheib J, Hoke A. Advances in peripheral nerve regeneration. Nat Rev Neurol. 2013;9:668–676. doi: 10.1038/nrneurol.2013.227. [DOI] [PubMed] [Google Scholar]
- 34.Scheib JL, Höke A. An attenuated immune response by Schwann cells and macrophages inhibits nerve regeneration in aged rats. Neurobiol Aging. 2016;45:1–9. doi: 10.1016/j.neurobiolaging.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Szepanowski F, Kieseier BC. Targeting lysophospholipid signaling as a therapeutic approach towards improved peripheral nerve regeneration. Neural Regen Res. 2016;11:1754–1755. doi: 10.4103/1673-5374.194720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tallon C, Farah MH. Beta secretase activity in peripheral nerve regeneration. Neural Regen Res. 2017;12:1565–1574. doi: 10.4103/1673-5374.217319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ugrenović S, Jovanović I, Vasović L, Kundalić B, Čukuranović R, Stefanović V. Morphometric analysis of the diameter and g-ratio of the myelinated nerve fibers of the human sciatic nerve during the aging process. Anat Sci Int. 2016;91:238–245. doi: 10.1007/s12565-015-0287-9. [DOI] [PubMed] [Google Scholar]
- 38.Verdu E, Ceballos D, Vilches JJ, Navarro X. Influence of aging on peripheral nerve function and regeneration. J Peripher Nerv Syst. 2000;5:191–208. doi: 10.1046/j.1529-8027.2000.00026.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang D, Liu X, Zhu J, Hu J, Jiang L, Zhang Y, Yang L, Wang H, Zhu Q, Yi J, Xi T. Repairing large radial nerve defects by acellular nerve allografts seeded with autologous bone marrow stromal cells in a monkey model. J Neurotraum. 2010;27:1935–1943. doi: 10.1089/neu.2010.1352. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Zhao Y, Sun C, Hu W, Zhao J, Li G, Zhang L, Liu M, Liu Y, Ding F, Yang Y, Gu X. Chitosan degradation products promote nerve regeneration by stimulating Schwann cell proliferation via miR-27a/FOXO1 axis. Mol Neurobiol. 2016;53:28–39. doi: 10.1007/s12035-014-8968-2. [DOI] [PubMed] [Google Scholar]
- 41.Xiao B, Rao F, Guo ZY, Sun X, Wang YG, Liu SY, Wang AY, Guo QY, Meng HY, Zhao Q, Peng J, Wang Y, Lu SB. Extracellular matrix from human umbilical cord-derived mesenchymal stem cells as a scaffold for peripheral nerve regeneration. Neural Regen Res. 2016;11:1172–1179. doi: 10.4103/1673-5374.187061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu S, Ao J, Gu H, Wang X, Xie C, Meng D, Wang L, Liu M. IL-22 impedes the proliferation of schwann cells: transcriptome sequencing and bioinformatics analysis. Mol Neurobiol. 2017;54:2395–2405. doi: 10.1007/s12035-016-9699-3. [DOI] [PubMed] [Google Scholar]
- 43.Yang LM, Liu XL, Zhu QT, Zhang Y, Xi TF, Hu J, He CF, Jiang L. Human peripheral nerve-derived scaffold for tissue-engineered nerve grafts: histology and biocompatibility analysis. J Biomed Mater Res B Appl Biomater. 2011;96:25–33. doi: 10.1002/jbm.b.31719. [DOI] [PubMed] [Google Scholar]
- 44.Yao D, Li M, Shen D, Ding F, Lu S, Zhao Q, Gu X. Gene expression profiling of the rat sciatic nerve in early Wallerian degeneration after injury. Neural Regen Res. 2012;7:1285–1292. doi: 10.3969/j.issn.1673-5374.2012.17.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi M, Horton JD, Cohen JC, Hobbs HH, Stephens RM. Whole pathway scope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006;7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu D, Du Z, Pian L, Li T, Wen X, Li W, Kim S, Xiao J, Cohen P, Cui J, Hoffman AR, Hu J. Mitochondrial DNA hypomethylation is a biomarker associated with induced senescence in human fetal heart mesenchymal stem cells. Stem Cells Int. 2017;2017:1–12. doi: 10.1155/2017/1764549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng C, Zhu Q, Liu X, Huang X, He C, Jiang L, Quan D. Improved peripheral nerve regeneration using acellular nerve allografts loaded with platelet-rich plasma. Tissue Eng Part A. 2014;20:3228–3240. doi: 10.1089/ten.tea.2013.0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J, Fa n Y, Chen H. Analyses of long non-coding RNA and mRNA profiles in the spinal cord of rats using RNA sequencing during the progression of neuropathic pain in an SNI model. RNA Biol. 2017;14:1810–1826. doi: 10.1080/15476286.2017.1371400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Z, Huang Y, Zou X, Zheng C, Liu J, Qiu L, He B, Zhu Q, Liu X. The vascularization pattern of acellular nerve allografts after nerve repair in Sprague-Dawley rats. Neurol Res. 2017;39:1014–1021. doi: 10.1080/01616412.2017.1365423. [DOI] [PubMed] [Google Scholar]
- 50.Zhu Z, Zhou X, He B, Dai T, Zheng C, Yang C, Zhu S, Zhu J, Zhu Q, Liu X. Ginkgo biloba extract (EGb 761) promotes peripheral nerve regeneration and neovascularization after acellular nerve allografts in a rat model. Cell Mol Neurobiol. 2015;35:273–282. doi: 10.1007/s10571-014-0122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou J, Liu S, Sun J, Yang W, Xu Y, Rao Z, Jiang B, Zhu Q, Liu X, Wu J, Chang C, Mao H, Ling E, Quan D, Zeng Y. Peripheral nerve-derived matrix hydrogel promotes remyelination and inhibits synapse formation. Adv Funct Mater. 2018;28:1705739. [Google Scholar]
- 52.Zou Y, Chiu H, Zinovyeva A, Ambros V, Chuang CF, Chang C. Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science. 2013;340:372–376. doi: 10.1126/science.1231321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




