Abstract
We report a case of meningitis in a neonate in China, which was caused by a novel multidrug-resistant Cronobacter sakazakii strain, sequence type 256, capsular profile K1:CA1. We identified genetic factors associated with bacterial pathogenicity and antimicrobial drug resistance in the genome and plasmids. Enhanced surveillance of this organism is warranted.
Cronobacter sakazakii is a foodborne pathogen associated with outbreaks of life-threatening necrotizing enterocolitis, meningitis, and sepsis in neonates and infants. Although the incidence of C. sakazakii infection is low, fatality rates range from 40% to 80% (1). Infections are usually limited to specific sequence types (STs) and complex clonal complexes (2–4). C. sakazakii ST4 is predominantly associated with meningitis in neonates; C. sakazakii ST12, with necrotizing enterocolitis in neonates (2,3). C. sakazakii is usually resistant to cephalothin and penicillin but is more sensitive to antimicrobial drugs than are other members of the family Enterobacteriaceae. Few reports describe drug-resistance patterns in C. sakazakii isolates (4–6). We report 1 multidrug-resistant (MDR) C. sakazakii ST256 strain that caused meningitis in a neonate in China.
On September 29, 2015, a 26-day-old boy, who was born after 38 weeks’ gestation and had abdominal distention, fever, and jaundice, was hospitalized in a children’s hospital in Guangzhou, China. He was fed breast milk; however, it could not be determined whether he had been exclusively breast-fed or whether the breast milk had been expressed by use of a pump. His cerebrospinal fluid contained numerous leukocytes, and his cerebrum contained abscesses. After a series of symptomatic treatments, including initial intravenous ceftazidime followed by meropenem, his clinical signs gradually improved. However, when discharged from the hospital after 3 weeks, his mental and physical development were remarkably impaired.
Cronobacter, isolated from brain abscess fluid, was identified by using an automated VITEK 2 Compact system (bioMérieux, Marcy l’Etoile, France). An isolate, GZcsf-1, was determined to be C. sakazakii ST256 with serotype O1. The Cronobacter PubMLST (https://pubmlst.org/cronobacter/) contains 2 ST256 isolates: MOD1-Ls15 g, isolated from the alimentary canal of the green bottle fly, Lucilia sericata, in the United States; and 2061 (no source information), detected in France. This ST had not been reported to cause meningitis in neonates. Susceptibility to 15 antimicrobials was tested by using the broth dilution method; MICs are shown in the Technical Appendix. GZcsf-1 was resistant to 8 antimicrobials: ampicillin, cefazolin, ceftriaxone, aztreonam, gentamicin, tetracycline, chloramphenicol, and trimethoprim/sulfamethoxazole.
The Beijing Genomics Institute performed genomic and plasmid DNA sequencing by using the PacBio RS II (Pacific Biosciences, Menlo Park, CA, USA) and HiSeq (Illumina, San Diego, CA, USA) platforms. The annotations were performed as previously described (7). C. sakazakii GZcsf-1 had 1 circular chromosome, 4.43 Mb long, containing 56.87% GC and 2 plasmids (denoted pGW1, 340,723 bp, 57.2% GC; pGW2, 135,306 bp, 54.0% GC) (GenBank accession nos. CP028974–6). On the basis of the characteristics of K antigen and colanic acid biosynthesis encoding genes in GZcsf-1, we determined its capsular profile to be K1:CA1, which differs from the capsular profile K2:CA2, proposed to be strongly associated with C. sakazakii isolated from neonates with severe infection (3). The subtype I-E CRISPR-Cas system and CRISPR1 to CRISPR3 arrays were detected (7).
We identified mobile elements and different types of secretion systems by using VRprofile (http://202.120.12.134/STEP/STEP_VR.html) (Technical Appendix). Although virulence genes in C. sakazakii have not yet been clarified, T4SS, T6SS, and prophages may contribute to the pathogenicity of GZcsf-1. More importantly, full-plasmid comparison by blastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi) revealed pGW2 to be closely related to IncFIB-type plasmid pESA3, which has been widely identified as a virulence plasmid in pathogenic strains of C. sakazakii (8).
We identified drug-resistance genes by using ResFinder 2.1 (https://cge.cbs.dtu.dk/services/ResFinder-2.1/). Except for 2 drug-resistance genes in the genome, 19 were integrated in pGW1, indicating that the MDR phenotype of C. sakazakii GZcsf-1 resulted from plasmid-mediated resistance (Technical Appendix). By performing plasmid multilocus sequence typing (https://pubmlst.org/bigsdb?db=pubmlst_plasmid_seqdef&page=sequenceQuery), we determined that pGW1 is IncHI2-ST1 and is mainly found in other countries (9). Full-plasmid comparison revealed that pGW1 is closely related to p505108-MDR (5).
The drug-resistance gene blaDHA-1, encoding an AmpC β-lactamase and conferring cephalosporin resistance, was found in pGW1 (Figure, panel A). According to a previous report, this gene is associated with ISCR1 (10). The difference between the 2 plasmids pGW1and p505108-MDR was in the insertion of a 7-kb fragment, associated with mercury resistance, between sul1 and IS26 in pGW1 (Figure, panel A). This fragment was also detected in plasmid unitig_2, implying that there might be a transposition or homologous recombination. Of 19 drug-resistance genes, 9 (47.4%) were integrated in the MDR region of pGW1 (Figure, panel B). The fragment from Tn3 remnant to mrr gene in pGW1 was identical to that in p505108-MDR. Compared with the Tn2 remnant in p505108-MDR, only blaTEM-1B in reverse orientation was conserved in pGW1. We found 5 new drug-resistance genes in pGW1—mph(A), sul1, aadA2, dfrA12, and aac(3)-IId—adjacent to IS26, IS5075, and ISCfr1, constituting new accessory modules. Mobilization of these accessory resistance modules into plasmid backbones may be promoted by transposition and homologous recombination.
Figure.
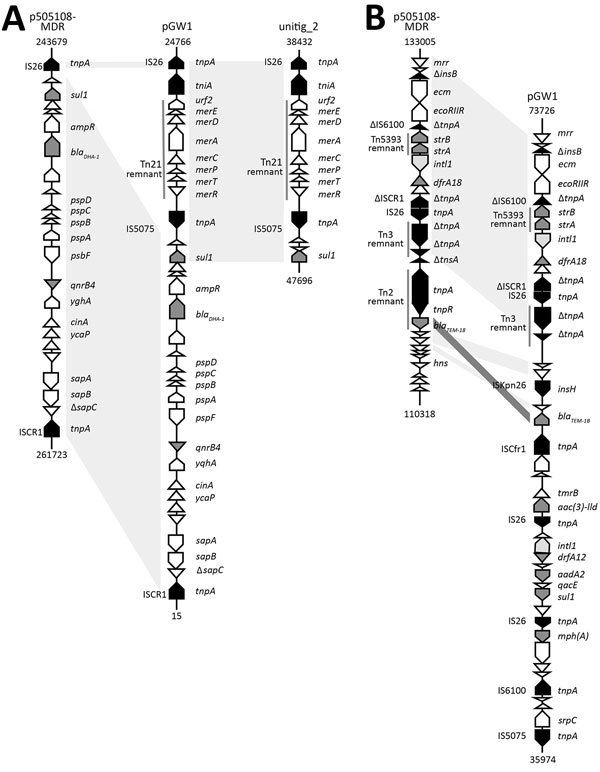
Analysis of resistance plasmid pGW1 in Cronobacter sakazakii sequence type 256, isolated from neonate with meningitis, China, 2015, showing genetic organization of major drug-resistance determinants in pGW1, along with its structural comparison with that of p505108-MDR. The blaDHA-1 region (A) and multidrug-resistance region (B) of plasmid pGW1 are compared with those of plasmid p505108-MDR, unitig_2, or both. Black indicates genes encoding transposase, light gray indicates genes encoding integrase, dark gray indicates genes encoding drug resistance, and white indicates other genes. A few hypothetical genes with nucleotide sequences <200 bp are not shown. Shading denotes regions of homology (>95% nt identity); light gray shading indicates same orientation, and dark gray shading indicates reverse orientation. Numbers indicate nucleotide positions within the corresponding plasmids.
This study provides a new insight into C. sakazakii pathovars and raises concern that plasmid-mediated MDR C. sakazakii maybe a threat to infant health. Enhanced surveillance of antimicrobial drug–resistant Cronobacter is warranted.
Antimicrobial drug susceptibility profiles and large mobile elements and secretion systems regions and drug resistance genes identified in Chronobacter sakazakii strain GZcsf-1, isolated from a neonate in China, 2015.
Acknowledgments
This work was supported by grants from the National Key R&D Program of China (2017YFC1601200), the National Natural Science Foundation of China (31601571), Pearl River S&T Nova Program of Guangzhou (201806010062), and the Guangdong Academy of Sciences Special Project of Science and Technology Development (2017GDASCX-0201).
Biography
Dr. Zeng is an assistant professor at Guangdong Institute of Microbiology, Guangzhou, Guangdong Province, China. Her research is focused on adaptive evolution, drug resistance, and pathogenesis of Cronobacter.
Footnotes
Suggested citation for this article: Zeng H, Lei T, He W, Zhang J, Liang B, Li C, et al. Novel multidrug-resistant Cronobacter sakazakii causing meningitis in neonate, China, 2015. Emerg Infect Dis. 2018 Nov [date cited]. https://doi.org/10.3201/eid2411.180718
These authors contributed equally to this article.
References
- 1.Bowen AB, Braden CR. Invasive Enterobacter sakazakii disease in infants. Emerg Infect Dis. 2006;12:1185–9. 10.3201/eid1208.051509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin A, Loughlin M, Caubilla-Barron J, Kucerova E, Manning G, Dowson C, et al. Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance which do not correlate with biotypes. BMC Microbiol. 2009;9:223. 10.1186/1471-2180-9-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogrodzki P, Forsythe SJ. DNA-sequence based typing of the Cronobacter genus using MLST, CRISPR-cas array and capsular profiling. Front Microbiol. 2017;8:1875. 10.3389/fmicb.2017.01875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui JH, Yu B, Xiang Y, Zhang Z, Zhang T, Zeng YC, et al. Two cases of multi-antibiotic resistant Cronobacter spp. infections of infants in China. Biomed Environ Sci. 2017;30:601–5. [DOI] [PubMed] [Google Scholar]
- 5.Shi L, Liang Q, Zhan Z, Feng J, Zhao Y, Chen Y, et al. Co-occurrence of 3 different resistance plasmids in a multi-drug resistant Cronobacter sakazakii isolate causing neonatal infections. Virulence. 2018;9:110–20. 10.1080/21505594.2017.1356537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu BT, Song FJ, Zou M, Hao ZH, Shan H. Emergence of colistin resistance gene mcr-1 in Cronobacter sakazakii producing NDM-9 and Escherichia coli from the same animal. Antimicrob Agents Chemother. 2017;61:e01444–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng H, Zhang J, Li C, Xie T, Ling N, Wu Q, et al. The driving force of prophages and CRISPR-Cas system in the evolution of Cronobacter sakazakii. Sci Rep. 2017;7:40206. 10.1038/srep40206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joseph S, Desai P, Ji Y, Cummings CA, Shih R, Degoricija L, et al. Comparative analysis of genome sequences covering the seven cronobacter species. PLoS One. 2012;7:e49455. 10.1371/journal.pone.0049455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haenni M, Saras E, Ponsin C, Dahmen S, Petitjean M, Hocquet D, et al. High prevalence of international ESBL CTX-M-15-producing Enterobacter cloacae ST114 clone in animals. J Antimicrob Chemother. 2016;71:1497–500. 10.1093/jac/dkw006 [DOI] [PubMed] [Google Scholar]
- 10.Guo Q, Spychala CN, McElheny CL, Doi Y. Comparative analysis of an IncR plasmid carrying armA, blaDHA-1 and qnrB4 from Klebsiella pneumoniae ST37 isolates. J Antimicrob Chemother. 2016;71:882–6. 10.1093/jac/dkv444 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Antimicrobial drug susceptibility profiles and large mobile elements and secretion systems regions and drug resistance genes identified in Chronobacter sakazakii strain GZcsf-1, isolated from a neonate in China, 2015.


