Abstract
The Hfq protein is a global small RNA chaperone that interacts with regulatory bacterial small RNAs (sRNA) and plays a role in the post-transcriptional regulation of gene expression. The roles of Hfq in the virulence and pathogenicity of several infectious bacteria have been reported. This study was conducted to elucidate the functions of two hfq genes in Burkholderia glumae, a causal agent of rice grain rot. Therefore, mutant strains of the rice-pathogenic B. glumae BGR1, targeting each of the two hfq genes, as well as the double defective mutant were constructed and tested for several phenotypic characteristics. Bacterial swarming motility, toxoflavin production, virulence in rice, siderophore production, sensitivity to H2O2, and lipase production assays were conducted to compare the mutant strains with the wild-type B. glumae BGR1 and complementation strains. The hfq1 gene showed more influence on bacterial motility and toxoflavin production than the hfq2 gene. Both genes were involved in the full virulence of B. glumae in rice plants. Other biochemical characteristics such as siderophore production and sensitivity to H2O2 induced oxidative stress were also found to be regulated by the hfq1 gene. However, lipase activity was shown to be unassociated with both tested genes. To the best of our knowledge, this is the first study to elucidate the functions of two hfq genes in B. glumae. Identification of virulence-related factors in B. glumae will facilitate the development of efficient control measures.
Keywords: Burkholderia glumae, gene functions, hfq gene, RNA chaperone
The Hfq RNA chaperone is a ubiquitous protein found in the majority of sequenced bacterial species. It was first identified as a bacterial host factor required for bacteriophage Qβ RNA replication in Escherichia coli (De Fernandez et al., 1968). Hfq plays several roles, such as binding bacterial small RNA (sRNA) and messenger RNA (mRNAs), and facilitating RNA-RNA interactions (Hajnsdorf and Regnier, 2000). Moreover, it has been reported that Hfq can influence polyadenylation of mRNAs and act independently as a regulator of gene expression (Valentin-Hansen et al., 2004). In addition to RNAs binding, Hfq regulates the expression of various bacterial genes at the post-transcriptional level, and directly or indirectly affects a number of proteins and controls the ribo-regulation of the expression of multiple genes (Mohanty et al., 2004; Morita et al., 2005).
A number of bacterial species, such as Staphylococcus aureus and Bacillus subtilus, have only one hfq gene, while some species, such as Burkholderia spp. and Novosphingobium aromaticivorans have two hfq genes (Sun et al., 2002). In Bacillus anthracis, three hfq gene sequences were identified, and their functional properties were investigated (Panda et al., 2015). The construction of an E. coli mutant by hfq insertion mutations demonstrated the importance of Hfq. The mutant exhibited pleiotropic phenotypic changes, including reduced growth rates, altered cellular morphology, and improved cellular sensitivity to stress (Tsui et al., 1994). Furthermore, Hfq in E. coli was shown to be an essential element in the translation of RpoS, which encodes the σS subunit of the RNA polymerase, the master regulator for many stress-regulated genes (Muffler et al., 1997). Hfq was also found to contribute greatly to the translational regulation of several other mRNAs, by assisting sRNA binding (Wroblewska and Olejniczak, 2016).
Following the study of the null hfq mutant in E. coli, the functional regulatory roles of Hfq in several other bacterial species have been studied (Chao and Vogel, 2010). Hfq was found to regulate genes encoding diverse traits, due to its nature as a multifunctional global sRNA chaperone. Traits such as protein composition of the cell membrane, tolerance to different forms of cellular stress, motility, and the metabolism of various nutrients, have been found to be regulated by Hfq (Chao and Vogel, 2010). Moreover, Hfq has also been found to be responsible for the regulation of virulence in several animal-pathogenic bacterial species such as Vibrio cholerae, Yersinia pestis, and Pseudomonas aeruginosa (Ding et al., 2004; Geng et al., 2009; Sonnleitner et al., 2003). In Burkholderia cepacia, the hfq mutant is more susceptible than the wild type to stress conditions resembling those found during infection, and exhibits reduced motility, extra polysaccharide production and host colonization ability. Such results suggest that Hfq plays a major role in the increased virulence of B. cepacia (Sousa et al., 2010). Although numerous studies have been conducted on the roles of Hfq in animal pathogenic bacteria, only few of them have described its roles in plantpathogenic species. In Agrobacterium tumefaciens, Hfq influences the sRNA AbcR1 and its corresponding mRNA target; an hfq mutant was reported to overproduce around eight proteins, and to exhibit altered cell morphology and reduced growth, motility and virulence (Wilms et al., 2012). Similarly, Hfq was found to regulate the virulence of the fire blight pathogen Erwinia amylovora (Zeng et al., 2013).
Burkholderia glumae is a bacterial pathogen of rice that causes panicle blight on infected plants. It was first reported in Japan causing seedling, sheath, and grain rot (Goto and Ohata, 1956; Goto et al., 1987). Since then, B. glumae has been described in several other regions including East Asia, Latin America, and the USA (Jeong et al., 2003; Nandakumar et al., 2007, 2009; Trung et al., 1993). The disease is considered a potentially major threat to rice cultivation worldwide (Ham et al., 2011). B. glumae has been reported to produce toxoflavin, which is essential for its virulence and highly toxic to plant cells (Sato et al., 1989). The seedborne nature of the pathogen and its effect of grain weight reduction, floret sterility, and seed germination inhibition may cause up to 75% yield loss in heavily infested fields (Trung et al., 1993). Understanding the factors contributing to the pathogenicity and virulence of B. glumae might be useful to facilitate the development of efficient control measures against this devastating pathogen. To our knowledge, no previous studies have investigated the roles of hfq genes in B. glumae. Therefore, this study aimed to elucidate the roles of two hfq genes on B. glumae through the construction of hfq mutants of B. glumae BGR1, and to investigate the phenotypic characteristics related to virulence, stress tolerance and other biochemical activities.
Materials and Methods
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. To prepare the bacterial cultures, strains were grown on Luria-Bertani (LB) agar plates; for liquid cultures, single colonies from the pure cultures on agar plates were transferred to LB broth and incubated in a shaking incubator at 200 rpm at 37°C. Culture media were supplemented with antibiotics (rifampicin, 50 μg/ml; kanamycin, 30 or 50 μg/ml; tetracycline, 10 μg/ml; ampicillin, 100 μg/ml), when necessary.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | References |
|---|---|---|
| B. glumae | ||
| BGR1 | Wild-type, Rif | (Jeong et al., 2003) |
| BGS2 | BGR1 tofI::Ω | (Kim et al., 2004) |
| HFQ1 | BGR1 hfq1::Ω | This study |
| HFQ2 | BGR1 Δhfq2 | This study |
| HFQ12 | BGR1 hfq1::Ω Δhfq2 | This study |
| HFQ1C | HFQ1 harboring pRK_H1 | This study |
| HFQ2C | HFQ2 harboring pRK_H2 | This study |
| HFQ12C | HFQ12 harboring pRK_H12 | This study |
| E. coli | ||
| DH5α λpir | For cloning and replication of plasmids | Lab collection |
| S17-1 λpir | For transferring cloning plasmids to recipient cells | Lab collection |
| Plasmids | ||
| pK18mobsacB | Allelic exchange suicide vector, sacB, Km | |
| pVIK112 | R6K suicide vector, lacZY for transcriptional fusions, Km | |
| pRK415 | Broad host range expression vector, Tet | |
| pVIK_Hfq1 | pVIK112 vector cloning internal region of hfq1 | This study |
| pK18_Hfq2 | pK18mobsacB vector cloning LR fragment of hfq2 | This study |
| pRK_H1 | pRK415 vector including hfq1 gene | This study |
| pRK_H2 | pRK415 vector including hfq2 gene | This study |
| pRK_H12 | pRK415 vector including hfq1 and hfq2 gene | This study |
Bacterial genomics DNA extraction and polymerase chain reactions
Bacterial genomic DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA) and following the manufacturer’s instructions. Plasmid DNA was separated with the Dokdo-PrepTM Plasmid Mini-Prep Kit (Elpis-Biotech, Daejeon, South Korea). The concentration and the quality of the extracted DNA were assessed using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Polymerase chain reaction (PCR) was performed using Pfu-X DNA Polymerase (Solgent, Daejeon, South Korea) or Taq polymerase (TaKaRa, Kusatsushi, Japan). The PCR mixture and reaction conditions were maintained according to the manufacturer’s instructions using a Sure Cycler 8800 Thermal cycler (Agilent Technologies, Santa Clara, CA, USA). The list of primers and their targets used in this study are shown in Table 2.
Table 2.
Primers used in this study
| Purpose | Primer name | Sequence (5’ to 3’) |
|---|---|---|
| Amplifying internal region of hfq1 | Hfq1_F | AAAAGAATTCGGAGTACGCCATGAGCAACA |
| Amplifying internal region of hfq1 | Hfq1_R | AAAAGGTACCGGAGCAACACGACGTACTGG |
| Confirming vectors and mutants | Lacfuse_R | GGGGATGTGCTGCAAGGCG |
| Confirming for mutants | Hfq1_Up_F | TAGTGTAGCGATTGACGGCG |
| Amplifying L fragment of hfq2 | Hfq2 _LF | AAAAGAATTCTTTTCTCGGGTCACATCCGC |
| Amplifying L fragment of hfq2 | Hfq2 _LR | AAAGGATCCCATTGAGAAGCGGCTTGAGC |
| Amplifying R fragment of hfq2 | Hfq2 _RF | AAAGGATCCAATCGCACGAAGGCGTCTC |
| Amplifying R fragment of hfq2 | Hfq2 _RR | AAAAAGCTTGAAACGCGGATGTGAAGACG |
| Confirming for mutants | Hfq2_Gene_Up_F | GACCCGAAGAACCCCGACTA |
| Confirming for mutants | Hfq2_Gene_Down_R | CCAGAAGGCACAGACGGACC |
| Confirming for mutants | pk18_Down_R | GTGAAGCTAGCTTATCGCCAT |
| RT-PCR | RT_Hfq1_F3 | CGATTGACGGCGGATCTCTT |
| RT-PCR | RT_ Hfq1_R2 | TTAGGAGGACGCGCCTTCCG |
| RT-PCR | RT_ Hfq2_F2 | ATGGCCAATCCTGCAGAATCC |
| RT-PCR | RT_ Hfq2_R2 | TTACTCGTTGCTGCCGCCGG |
| Amplifying hfq1 to add in pRK415 | C_ P2-F | AAAAAGCTTCGGCAATCAGGATTCGCATG |
| Amplifying hfq1 to add in pRK415 | C_ P2-R | AAGGTACCGAATCGTCCTTTGCTATGATGA |
| Amplifying hfq1 to add in pRK415 | C_kpn1_Hfq1_F | AAGGTACCATGGAGTACGCCATGAGCAA |
| Amplifying hfq1 to add in pRK415 | C_BamH1_Hfq1_R | AAAGGATCCGATAAGGCGGCTGGCGTGGA |
| Amplifying hfq2 to add in pRK415 | C_EcoR1_Hfq2_F | AAAAGAATTCGTTTTCTCGGGTCACATCCG |
| Amplifying hfq2 to add in pRK415 | C_BamH1_Hfq2_R | AAAGGATCCAGATGCAGGTCCAGTCGCT |
Generation of mutant and complementation strains
For the construction of the mutant strains, standard general procedures for recombinant DNA construction were followed (Sambrook et al., 1989). To generate the insertion mutant strain HFQ1, the target hfq1-like gene of B. glumae BGR1 was disrupted by insertion mutagenesis using the recombinant suicide vector pVIK112 (Kalogeraki and Winans, 1997).
The target gene was amplified by PCR using the corresponding primers (Table 2). To ligate the amplified fragment into the suicide vector pVIK112, the active sites of restriction enzymes (EcoRI and KpnI) were inserted in the interior region of PCR primers of the target genes. PCR fragments and pVIK112 were digested using restriction enzymes, and ligated by T4 DNA ligase. Escherichia coli DH5α λpir competent cells were transformed with pVIK112 plasmid, and grown in LB medium containing kanamycin (50 μg/ml). Plasmid DNA was extracted from the transformants, and restriction enzyme digestion followed by gel electrophoresis were performed to confirm the correct ligation. Following confirmation, the assembled recombinant plasmids were again introduced into E. coli S17-1 λpir competent cells. The recombinant plasmid was introduced into B. glumae BGR1 by biparental mating. To properly select transformant B. glumae BGR1 cells, mated cells were grown on a plate with two antibiotics (kanamycin, 100 μg/ml; rifampicin, 100 μg/ml), and the selected cells were verified by PCR using an internal sequence of the pVIIK112 vector and an upstream sequence of the target gene as primers (Table 2).
For the construction of the deletion mutant HFQ2 strain, two DNA fragments of the target hfq2-like gene upstream region (L fragment) and downstream region (R fragment) were amplified by PCR using the corresponding primers (Table 2). The two fragments were then cleaved by BamHI, then ligated and amplified. The amplified PCR product and the pK18mobsacB suicide vector were digested by restriction enzymes (EcoRI and HindIII) and ligated. The recombinant plasmids were introduced into E. coli DH5α λpir competent cells and the transformants were cultured in LB medium with 30 μg/ml of kanamycin. To introduce the recombinant plasmids into B. glumae, the wild-type BGR1 (recipient strain) and E. coli S17-1 (donor strain) harboring identical recombinant plasmids were conjugated by biparental mating. Transformant cells were selected after incubation in LB broth containing 60 μg/ml of kanamycin and 100 μg/ml of rifampicin; transformation was confirmed by PCR using the corresponding primers (Table 2). Transformant cells were subsequently subcultured in fresh LB medium with 50 μg/ml of rifampicin and then incubated in LB medium containing 30% sucrose (w/v). As a result of double crossover recombination events, an in-frame deletion mutant strain was obtained.
For the construction of the double mutant defective in two hfq genes, HFQ12, the same methodology as for the construction of the HFQ2 deletion mutant was followed, along with the procedures of disruptive insertion within the hfq1-like gene on the resultant strain.
Construction of complementation strains
To generate constructs for complementation of the HFQ1, HFQ2, and HFQ12 strains, the whole open reading frame (ORF) of each gene was amplified by PCR using the corresponding primers listed in Table 2. The obtained fragments were cloned into the expression vector pRK415, and the cloned vectors were transformed by conjugation as explained above. The resultant strains from these steps were verified by PCR.
Total RNA extraction and RT-PCR
The wild-type B. glumae BGR1 and mutant strains were cultured overnight in 2 ml of μB broth at 37°C with shaking, and then subcultured for 10 or 24 h under the same conditions. Total bacterial RNA from each strain was extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Residual genomic DNA was removed by performing on-column RNase-free DNase digestion following the manufacturer’s instructions (Qiagen, Hilden, Germany). Concentration and quality of the extracted RNA were assessed using a NanoDrop ND-2000 spectrophotometer. To confirm the gene expression, 500 ng of extracted RNA was reverse transcribed with random hexamer primers using the SuperiorScript III cDNA synthesis kit (Enzynomics, Daejeon, South Korea). The cDNA was used as template for RT-PCR analysis using the primers listed in Table 2.
Phenotypic analysis
Growth rate, swarming motility and toxoflavin production
To compare the growth rate of the tested mutants (HFQ1, HFQ2 and HFQ12), complemented strains (HFQ1C, HFQ2C and HFQ12C) and wild-type B. glumae BGR1, strains were cultured as described above and after inoculation into fresh LB broth, the growth rate was monitored by measuring optical density (OD600) values on intervals.
Phenotypic characteristics associated with virulence were compared between the resultant mutant strains and the wild-type B. glumae BGR1. Swarming motility was previously confirmed to be absent in the quorum sensing (QS)-defective mutant, B. glumae BGS2 (tofI::Ω mutant strain), and therefore, it served as negative control in this assay (Kim et al., 2007). Swarming motility assays were conducted on LB supplemented with 0.4% (w/v) agar plates. Bacterial strains were cultured on LB broth, as explained above, and cells were harvested from 1 ml of overnight culture by centrifugation at 13,500 rpm for 1 min. Harvested cells were washed twice and suspended in 100 μl of fresh LB broth. From the suspension, 5 μl were spotted on the assay plates and incubated for 24 h at 37°C.
Production of toxoflavin was detected using thin-layer chromatography (TLC) assay (Lee et al., 2016). Briefly, 1 ml from the cultures of each strain was centrifuged at 13,500 rpm for 1 min, and 500 μl from the supernatant were transferred into new tubes and mixed thoroughly with same volume of chloroform. The mixture was centrifuged at 13,500 rpm for 10 min; 450 μl of infranatant were transferred into a new tube and concentrated with a centrifugal vaporizer (EYELA, Tokyo, Japan). The solutes were then dissolved in 10 μl of 80% methanol, and 1 μl was spotted onto TLC silica gel plate (Merck Millipore, Darmstadt, Germany). The latter step was repeated 10 times and the plates were placed into the developing chamber with 95:5 chloroform:methanol solvent system. Produced toxoflavin was observed as visible bands on the TLC plates using 3UV™ Multi-Wavelength lamps (UVP, Upland, CA, USA).
Pathogenicity test for the assessment of virulence in rice plants
Rice (Oryza sativa L. var. glutinosa Blanco) grown under greenhouse conditions at flowering stage was used for testing the virulence of the wild-type B. glumae (BGR1) and related mutant strains (HFQ1, HFQ2, HFQ12, HFQ1C, HFQ2C, and HFQ12C). Briefly, rice heads were dipped for 1 min into suspensions (OD600 = 1.0) of the tested strains, prepared as explained previously and diluted in distilled water (DW). One week after inoculation, disease severity was assessed on rice panicles according to the following scale: 0, clear (no discoloration); 1, 0–20% discolored; 2, 20–40% discolored; 3, 40–60% discolored; 4, 40–80% discolored; 5, 80–100% discolored. Disease severity for each replicate was calculated based on the number of diseased grains per rice head using the following formula: Disease severity = Σ (number of seeds at each ratio × ratio value) / the total number of seeds. Three replicates for each treatment were used, and rice treated with DW served as negative control.
Siderophore production, H2O2 sensitivity, and lipase activity assays
Siderophore production by the tested strains was assessed on Chrome azurol S (CAS)-blue agar plate (Ames-Gottfred et al., 1989; Schwyn and Neilands, 1987). Briefly, 10 μl of bacterial culture (OD600 = 1.0) were inoculated into holes punched on CAS-blue plates and incubated at 37°C for 2 days. Siderophore production was detected by a color change from blue to orange around the boundaries of bacterial spots. Three replicates were used per treatment and a non-siderophore producing Bacillus sp. was used as negative control.
The assay for hydrogen peroxide (H2O2) sensitivity was conducted using an H2O2 disk diffusion assay as described by King et al. (2000). Briefly, overnight cultures of tested strains grown on LB broth were seeded into autoclaved and cooled LB agar for a final OD600 = 0.5, and then they were evenly spread in petri dishes. Sterile disks were soaked in 30% H2O2 solution and placed on top of the bacterial agar plates. The treated plates were incubated at 37°C for 1 day, and clear zones reflecting the sensitivity of the bacterial strains to H2O2 were photographed and measured.
The lipase activity assay was performed using precipitation test on T80 agar plates according to the method described by Rai et al. (2014). First, 10 μl of bacterial culture (OD600 = 1.0) were inoculated onto holes punched on LB agar plates containing 1% tween 80 (v/v) and 0.01% CaCl2 (w/v), and incubated for 2 days at 37°C. Lipase activity was then assessed by observing the white zones surrounding bacterial spots, which are indicative of lipase production capacity.
Statistical analysis
Data statistical analysis was performed using the Statistical Analysis Systems (SAS Institute, Cary, NC). All experiments were conducted twice with three replicates. In order to perform comparisons on disease severity and biochemical tests results, analysis of variance was performed using the GLM procedures, and the means were separated using Tukey’s honest significant difference test at P < 0.05.
Results
Location and sequence analysis of two distinct hfq genes in B. glumae BGR1
The two target hfq genes in B. glumae BGR1 were localized and analyzed before conducting the experiments. The two copies of hfq genes present in B. glumae were located in chromosome 1, bglu_1g14550 (hfq1 gene) and bglu_1g19700 (hfq2 gene); the length of hfq1 gene was 255 nt, compared with the 555 nt sequence length of the hfq2 gene (Supplementary Fig. 1). The hfq1 gene in B. glumae was found to be highly conserved in comparison with other bacterial species such as E coli, Pseudomonas aeruginosa, Salmonella typhimurium, Bacillus cereus, and Burkholderia cenocepacia. In contrast, the hfq2 gene was found to be completely different in length. Moreover, the C-terminal part of the Hfq protein was much longer in B. glumae than in other bacteria (Supplementary Fig. 1).
Construction and verification of B. glumae BGR1 mutant strains
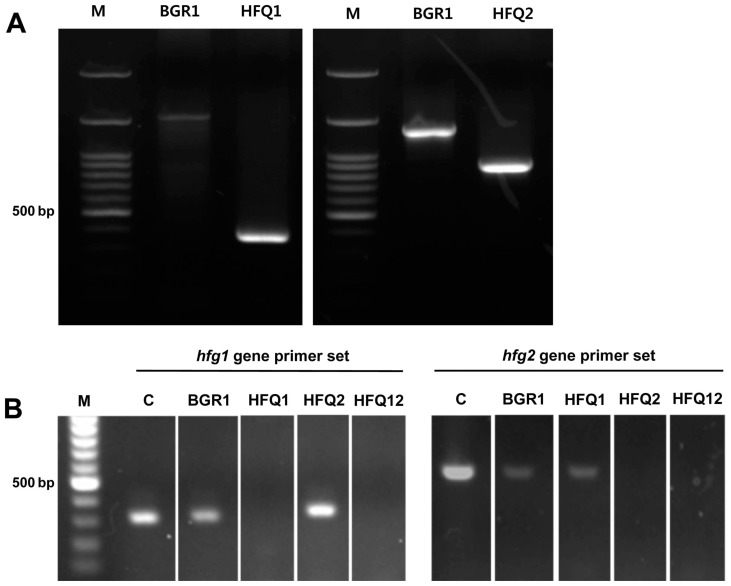
To study the functions of two hfq genes in B. glumae BGR1, a mutant for each of the two hfq genes and a double defective mutant were generated. The defective hfq1 gene (bglu_1g14550) was verified by PCR, targeting the upstream sequence of the target gene, and an inner sequence of the pVIK112 vector. As a result, a clear band was observed in the HFQ1 mutant, but not in B. glumae BGR1 (Fig. 1A). The in-frame mutation of the hfq2 gene (bglu_1g19700) was verified through PCR, targeting the specific upstream and downstream sequences of the target genes. The results indicate that 445 bp, within the target gene were deleted from HFQ2 compared with the wild-type B. glumae BGR1 (Fig. 1A). Moreover, the mutations of the respective gene in each mutant, and of both genes in the double defective mutant strain were verified by the RT-PCR (Fig. 1B).
Fig. 1.
Construction and confirmation of the mutant strains deficient in two hfq genes. (A) Agarose gel electrophoresis from the PCR assays for confirmation of the transformation of the two hfq genes. The transformed pVIK112 vector with the insertion of an hfq1-like gene was missing in the wild-type Burkholderia glumae BGR1, and confirmed in the HFQ1 mutant strain, and the 445 bp within the hfq2-like gene was absent in the HFQ2 deletion strain, in comparison with wild-type B. glumae BGR1. M, 100 bp DNA ladder (B) Agarose gel electrophoresis from RT-PCR was performed for confirmation of the two hfq gene-expression in wild-type B. glumae BGR1 and deficient mutant strains HFQ1, HFQ2, and HFQ12. M, 100 bp DNA ladder; C, B. glumae BGR1 genomic DNA.
Comparative assessment of bacterial growth and major virulence factors in B. glumae BGR1 mutant strains
By observing the growth rates of the tested strains, the hfq1 defective mutant (HFQ1) and the double defective mutant (HFQ12) showed clear retarded growth in comparison to the wild-type B. glumae BGR1. The hfq2 defective mutant (HFQ2) growth rate did not differ from the wild-type B. glumae BGR1. The complemented strains evidenced partial recovery of normal bacterial growth rate (Supplementary Fig. 2).
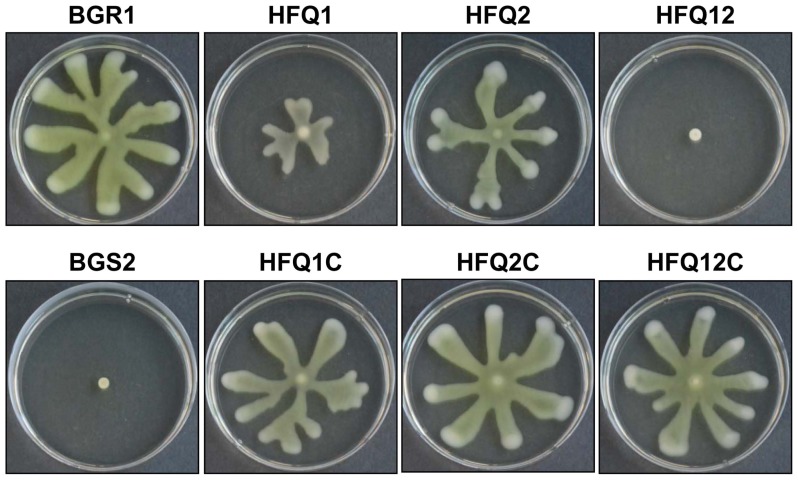
Compared with the wild-type B. glumae BGR1; swarming activity was clearly reduced in the mutant strains HFQ1, HFQ2 and HFQ12. In particular, the double defective mutant HFQ12 (hfq1::Ω Δhfq2) showed no swarming motility, similar to the negative control B. glumae BGS2 (tofI::Ω). The HFQ1 mutant strain showed more reduction in swarming motility compared with the HFQ2 mutant. Furthermore, swarming motility was almost fully recovered in the complementation mutant strains HFQ1C, HFQ2C, and HFQ12C (Fig. 2). The clear reduction in swarming motility in hfq1 deficient mutants suggests a major role of this gene in the regulation of swarming motility in B. glumae.
Fig. 2.
Photographs of the swarming motility of wild-type Burkholderia glumae (BGR1), as well as of mutant strains deficient in one or two hfq genes (HFQ1, HFQ2, and HFQ12), and hfq complementation strains (HFQ1C, HFQ2C, and HFQ12C). Quorum sensing (QS)-deficient strains (BGS2) were used as negative controls, Bacterial strains were spotted on Luria-Bertani supplemented with 0.4% agar and incubated for 1 day at 37°C.
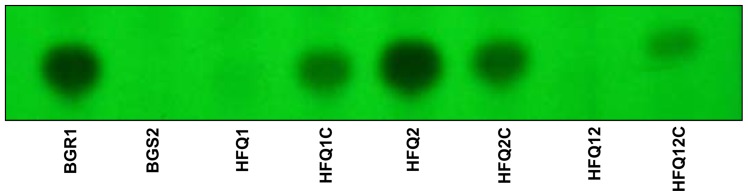
The other main virulence factor tested was the ability of the strains to produce toxoflavin. When the toxoflavin production by mutant strains was compared with the wild-type BGR1 using the TLC assay, no spots were observed either in the HFQ1 mutant or in the double defective mutant HFQ12, indicating the loss of toxoflavin production. However, clear spots were observed in the wild-type and in the HFQ2 mutant. Pale spots were observed in the complementation strains, indicating a partial recovery of toxoflavin production. No spots were observed in the negative control QS-defective mutant BGS2 (Fig. 3). This result suggests that hfq1, but not hfq2, plays a major role in the regulation of toxoflavin production.
Fig. 3.
Thin-layer chromatography (TLC) assay was performed to assess toxoflavin production in the wild-type Burkholderia glumae (BGR1), as well as in the strains deficient in one or two hfq genes, and the complementation strains (HFQ1, HFQ1C, HFQ2, HFQ2C, HFQ12, and HFQ12C). Quorum sensing (QS)-deficient strains (BGS2) were used as negative control. Produced toxoflavin was observed as black dots on a TLC silica gel plate examined under UV light at 254 nm.
In vivo virulence of B. glumae BGR1 mutant strains in rice
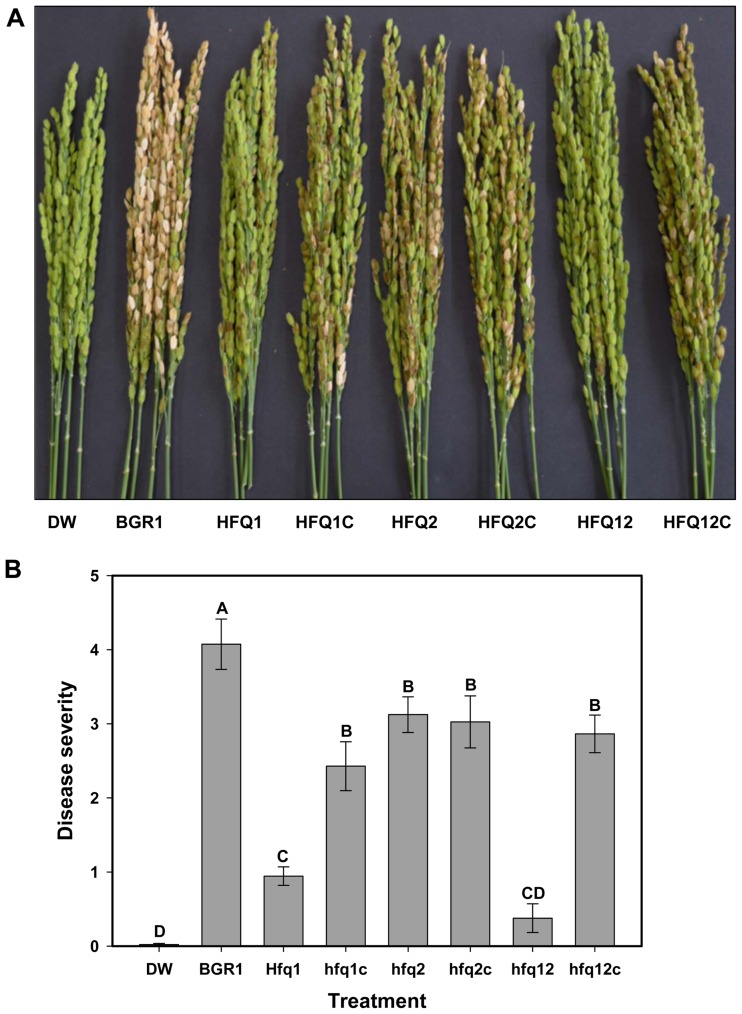
The above-mentioned results clearly indicate that hfq1 gene is important for the regulation of swarming motility sand toxoflavin production, which are the main virulence factors in B. glumae. In consistence with such results, rice plants inoculated with HFQ1 and HFQ12 exhibited a significant reduction in disease severity compared with the wild-type BGR1 (Fig. 4). While rice inoculated with HFQ2 strain exhibited only a partial reduction in disease severity in comparison with the wild-type BGR1, it was also evident that the complementation strains HFQ1C and HFG12C recovered significantly more virulence than the mutant strains (Fig. 4).
Fig. 4.
Disease severity in rice plants 7 days after inoculation by wild-type Burkholderia glumae (BGR1) and by strains deficient in one or two hfq genes, and the complementation strains (HFQ1, HFQ1C, HFQ2, HFQ2C, HFQ12, and HFQ12C). Distilled water (DW) was used as negative control. (A) Photograph showing the difference in virulence between the treatments. (B) Histogram showing the disease severity results in inoculated rice plants. Different letters on the error bars (Standard deviations, n = 3) indicate significant differences between the treatments, according to Tukey’s honest significant difference test at P < 0.05.
Evaluation of various biochemical activities of B. glumae BGR1 mutant strains
To study the effect of the two targeted hfq genes on various biochemical activities that are crucial for survival and stress resistance, the mutant strains were compared with the wild type B. glumae BGR1 in their ability to produce siderophores and in their sensitivity to H2O2. Lipase activity was also a parameter for comparison.
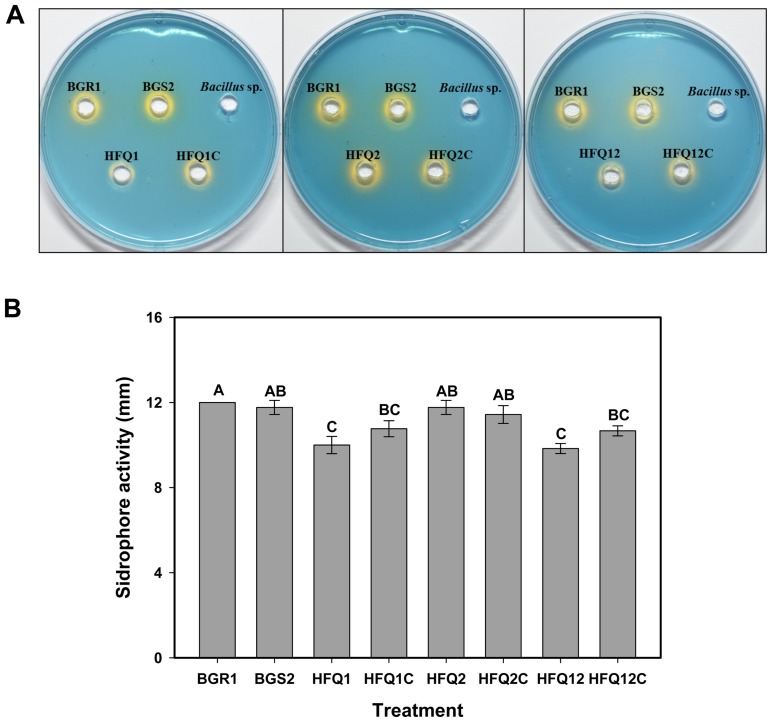
When the bacterial strains were tested using the siderophore assay, the wild-type B. glumae BGR1, as well as the QS-defective mutant BGS2, evidenced the production of siderophore, as indicated by the change in the color of CAS-blue agar plates, from blue to orange, around the bacterial growth (Fig. 5A). The hfq1 gene defective mutant HFQ1 as well as the double defective strain HFQ12 exhibited a clear reduction in the ability to produce siderophore, as observed in (Fig. 5A), and this was further confirmed by assessment of the siderophore activity zones, which were found to be significantly reduced in comparison with other treatments (Fig. 5B). The complementation strains partially recovered the siderophore production ability. No color change was observed in the negative control Bacillus sp. These results indicate the importance of the hfq1 gene in the regulation of siderophore production by B. glumae.
Fig. 5.
Siderophore production by the wild-type Burkholderia glumae (BGR1), and by strains deficient in one or two hfq genes, the complementation strains (HFQ1, HFQ1C, HFQ2, HFQ2C, HFQ12, and HFQ12C), and the quorum sensing (QS)-deficient strains (BGS2). Bacillus sp. was used as negative control. Bacterial strains were grown on chrome azurol S (CAS)-blue medium for 2 days at 37°C, and siderophore production was confirmed by the color change indicative of siderophores binding with ferric ion (A) Photographs showing the change in color of CAS plates. (B) Histogram showing the results of the siderophore detection assay. Measured diameters of the color change induced in the CAS medium by each of the strains used in the study. Different letters on the error bars (Standard deviations, n = 3) indicate significant difference between the treatments, according to Tukey’s honest significant difference test at P < 0.05.
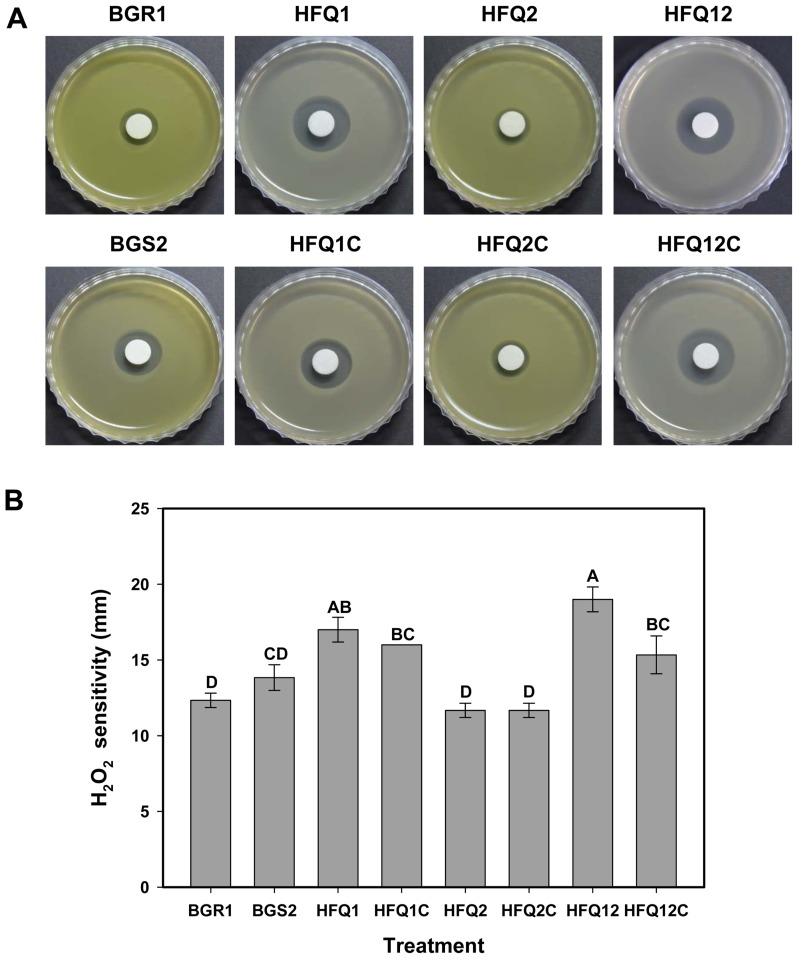
The sensitivity of the mutant strains and the wild-type B. glumae BGR1 to H2O2 treatment was tested using an H2O2 disk diffusion assay. Observation of the resulting clear zones around the H2O2 disks (Fig. 6A) indicated that the hfq1 defective mutant HFQ1 and the double defective mutant HFQ12 were less tolerant to H2O2 oxidative stress than the wild-type B. glumae BGR1. Assessment of the clear zone around the H2O2 disk confirmed the earlier observation that the HFQ1 and HFQ12, but not HFQ2, strains were more sensitive to H2O2 than the wild-type BGR1 or the QS-deficient mutant BGS2 (Fig. 6B). Similar to the previous assays, the complemented strains were found to partially recover H2O2 resistance, which indicates the role of the hfq1-like gene in the regulation of catalase and/or peroxidase activity and in tolerance to H2O2.
Fig. 6.
Hydrogen peroxide (H2O2) sensitivity assay. (A) Photographs of the bacterial culture plates 1 day after incubation at 37°C showing a clear halo corresponding to inhibition zones of bacterial growth around the H2O2 disks, indicating bacterial sensitivity to H2O2. (B) Histogram showing the results of the H2O2 disk assay. Measured diameters of the inhibition zones around the H2O2 disks. Different letters on the error bars (Standard deviations, n = 3) indicate significant differences between the treatments, according to Tukey’s honest significant difference test at P < 0.05.
As for the lipase activity assay, white precipitate zones were observed along the boundaries in all tested strains except for the QS-defective mutant BGS2. There were no apparent differences in the lipase activity between the mutant strains constructed in this study and the wild-type B. glumae BGR1, which suggests that the two hfq genes tested have no influence on lipase production in B. glumae BGR1 (Supplementary Fig. 3).
Discussion
Noncoding regulatory sRNAs play a key role in facilitating the response of bacteria to environmental fluctuations by modulating the stability of the post-transcriptional regulation of specific target mRNA transcripts, and by adjusting translational efficiency (Fröhlich and Vogel, 2009). The activation and stability of sRNAs are mainly controlled by the RNA chaperone Hfq protein (Link et al., 2009). The roles and functions of Hfq have been studied in several bacterial species, and particularly in gram-negative bacteria such as E. coli (Tsui et al., 1994), S. typhimurium (Monteiro et al., 2012; Sittka et al., 2007), and P. aeruginosa (Sonnleitner et al., 2002; Sonnleitner and Blasi, 2014). The functions of the hfq gene have been evaluated through phenomenological experiments; knock-out of the hfq gene and by testing the mutants using basic phenotypic assays such as estimation of growth rate, observation of cellular morphology, changes in virulence, and resistance to stress conditions (Bibova et al., 2013; Fantappiè et al., 2009; Sousa et al., 2010; Wilms et al., 2012). In some bacterial species such as Magnetospirillum magnetotacticum, Novosphingobium aromaticivorans, and Burkholderia genus, more than one copy of the hfq gene exist (Salgado-Garrido et al., 1999). However, only few studies were conducted on the functions and characterization of the two distinct hfq genes. In B. cepacia, the two Hfq proteins, Hfq and Hfq2, were reported to be essential for optimal survival in stress conditions and for full virulence (Ramos et al., 2011; Sousa et al., 2010). In the current study, the functions of two hfq genes in B. glumae were studied, and their roles in the regulation of virulence-related and other biochemical factors were investigated.
Two mutants targeting both the hfq1 and hfq2 genes, as well as a double defective mutant, were constructed and tested in the presence of the wild type B. glumae BGR1, and the complementation strains. Retarded growth rate was observed in the hfq1 mutant (HFQ1), which was consistent with several other studies suggesting that Hfq is an essential component for cell growth (Sobrero and Valverde, 2012). However, unlike HFQ1, the hfq2 defective mutant (HFQ2) didn’t show alteration in the bacterial growth compared the wild-type B. glumae BGR1. As observed with other phenotypic characteristics in this study, the double mutant HFQ12 has shown further reduction in the bacterial growth rate, indicating possible accumulative effect of both genes. The hfq1 gene was also shown to be more important than hfq2 for the regulation of virulence-related characters such as swarming motility and toxoflavin production. This was confirmed by the observation of a significant reduction of disease severity in rice plants inoculated with the HFQ1 mutant compared with HFQ2. Swarming motility with polar flagella and toxoflavin production are crucial virulence factors regulated by quorum sensing in B. glumae (Kim et al., 2004; 2007). Both studied genes exhibited an accumulative effect, as observed from the phenotypic assays in which the double defective mutant clearly evidenced more alterations of the phenotype. The hfq2 may have a role in assisting hfq1 in the display of full virulence, since the double defective mutant in both genes showed an almost complete loss of motility, toxoflavin production, and bacterial virulence in rice. In a previous study, a differential expression was found between the hfq and hfq2 genes. In that study, the maximal transcription of hfq was found at the early exponential phase, while for hfq2 the maximal transcription of hfq was found at the stationary phase of bacterial growth. Both genes were found to be involved in full virulence and stress tolerance (Ramos et al., 2011). A recent study pointed out that the functionality of the hfq2 gene is affected by the sRNA h2cR in B. cenocepacia (Ramos et al., 2012). The involvement of the two hfq genes in virulence reported in this study is consistent with other previous studies reporting reduced virulence of hfq mutant bacterial strains. Deletion of hfq in E. amylovora significantly reduced bacterial virulence in apple and pear fruits via the regulatory roles of Hfq on amylovoran exopolysaccharide production, biofilm formation, motility, and the type III secretion system (Zeng et al., 2013). Unlike most of the reported results on the effect of hfq on bacterial virulence and various altered phenotypes, a contrary finding was reported in Staphylococcus aureus strains, in which hfq mutations did not result in any detected phenotypic alteration (Bohn et al., 2007).
Similar to the previous assays, the siderophore detection assay and the H2O2 sensitivity test showed a similar trend indicating that hfq1 is more influential than hfq2. Likewise, in a previous study, the Hfq protein was found to be essential for normal siderophore production and to contribute to the tolerance to oxidative stress by H2O2 (Sobrero et al., 2012). The altered phenotypes of the mutant strains could be related to the ribo-regulation activity of Hfq in association with sRNA, thus controlling the gene expression involved in motility, production of toxoflavin and siderophore, and tolerance to H2O2 oxidative stress. Previously, three copies of Hfq RNA chaperones were identified in B. anthracis, and their functional and regulatory properties were characterized. From this, an RNA chaperoning activity was proposed for two Hfq proteins (Panda et al., 2015) Contrary to the phenotypic activity that seemed to be affected by mutations of the hfq genes, our results indicate no apparent roles of the two tested hfq genes on lipase production activity.
Although several aspects on the molecular mechanism remain not well-understood, previous studies have identified various sRNAs that associate with Hfq and their regulatory functions on target genes (Sobrero and Valverde, 2012). Using electrophoretic mobility shift assays, rpoS mRNA was found to form complexes with DsrA sRNA and Hfq (Soper and Woodson, 2008). Salim and Feig (2010) reported that the possibility of simultaneous binding of the mRNA fhlA and the sRNA OxyS to Hfq. The OxyS is a regulatory RNA that affects the expression of multiple genes and is expressed in response to oxidative stress. The Hfq was also found to promote the binding of several other sRNAs such as LhrA, LhrB, LhrC, RprA, Spot42, and RyhB to specific mRNAs (Sobrero and Valverde, 2012; Christiansen et al., 2006). Further future work is needed for characterization of the mechanisms and identification of the functional sRNAs involved in the gene regulation responsible for the pleiotropic phenotypic alterations related to virulence and stress tolerance observed in the current study.
Taken together, the present study revealed the functions of two hfq genes in B glumae that had not been studied previously. The results indicate that the two studied hfq genes are required for full virulence of B. glumae BGR1 in rice, and that the hfq1 gene has a more apparent influence on bacterial motility and toxoflavin production than hfq2. Additionally, the hfq1 gene was shown to be involved in siderophore production and in the tolerance to H2O2 oxidative stress. Finally, the two hfq genes are most likely unrelated to lipase production in B. glumae. Results of the current study and other similar studies will advance our understanding of the essential factors involved in the regulation of B. glumae virulence, paving the way for implementing successful disease control measures.
Supplementary data
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1D-1A1B03930678), and by grants from the Strategic Initiative for Microbiomes in Agriculture and Food, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea (No. 916009021SB010).
References
- Ames-Gottfred NP, Christie BR, Jordan DC. Use of the Chrome Azurol-S Agar Plate Technique to Differentiate Strains and Field Isolates of Rhizobium-Leguminosarum Biovar Trifolii. Appl Environ Microbiol. 1989;55:707–710. doi: 10.1128/aem.55.3.707-710.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibova I, Skopova K, Masin J, Cerny O, Hot D, Sebo P, Vecerek B. The RNA Chaperone Hfq Is Required for Virulence of Bordetella pertussis. Infect Immun. 2013;81:4081–4090. doi: 10.1128/IAI.00345-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn C, Rigoulay C, Bouloc P. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 2007;7:10. doi: 10.1186/1471-2180-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Søgaard-Andersen L, Kallipolitis BH. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA. 2006;12:1383–1396. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fernandez MTF, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qβ-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- Fantappiè L, Metruccio MM, Seib KL, Oriente F, Cartocci E, Ferlicca F, Giuliani MM, Scarlato V, Delany I. The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect Immun. 2009;77:1842–1853. doi: 10.1128/IAI.01216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Geng J, Song Y, Yang L, Feng Y, Qiu Y, Li G, Guo J, Bi Y, Qu Y, Wang W, Wang X, Guo Z, Yang R, Han Y. Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PLoS One. 2009;4:e6213. doi: 10.1371/journal.pone.0006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Ohata K. New bacterial diseases of rice (brown stripe and grain rot) Ann Phytopathol Soc Jpn. 1956;21:46–47. [Google Scholar]
- Goto T, Nishiyama K, Ohata K. Bacteria causing grain rot of rice. Japanese J Phytopathol. 1987;53:141–149. doi: 10.3186/jjphytopath.53.141. [DOI] [Google Scholar]
- Hajnsdorf E, Regnier P. Host factor Hfq of Escherichia coli stimulates elongation of poly(a) tails by poly(a) polymerase I. Proc Natl Acad Sci U S A. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham JH, Melanson RA, Rush MC. Burkholderia glumae: next major pathogen of rice? Mol Plant Pathol. 2011;12:329–339. doi: 10.1111/j.1364-3703.2010.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, Kim J, Kim S, Kang Y, Nagamatsu T, Hwang I. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis. 2003;87:890–895. doi: 10.1094/PDIS.2003.87.8.890. [DOI] [PubMed] [Google Scholar]
- Kalogeraki VS, Winans SC. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene. 1997;188:69–75. doi: 10.1016/S0378-1119(96)00778-0. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim JG, Kang Y, Jang JY, Jog GJ, Lim JY, Kim S, Suga H, Nagamatsu T, Hwang I. Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol Microbiol. 2004;54:921–934. doi: 10.1111/j.1365-2958.2004.04338.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Kang Y, Choi O, Jeong Y, Jeong JE, Lim JY, Kim M, Moon JS, Suga H, Hwang I. Regulation of polar flagellum genes is mediated by quorum sensing and FlhDC in Burkholderia glumae. Mol Microbiol. 2007;64:165–179. doi: 10.1111/j.1365-2958.2007.05646.x. [DOI] [PubMed] [Google Scholar]
- King KY, Horenstein JA, Caparon MG. Aerotolerance and peroxide resistance in peroxidase and PerR mutants of Streptococcus pyogenes. J Bacteriol. 2000;182:5290–5299. doi: 10.1128/JB.182.19.5290-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Park J, Kim S, Park I, Seo YS. Differential regulation of toxoflavin production and its role in the enhanced virulence of Burkholderia gladioli. Mol Plant Pathol. 2016;17:65–76. doi: 10.1111/mpp.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci U S A. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Maples VF, Kushner SR. The Smlike protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- Monteiro C, Papenfort K, Hentrich K, Ahmad I, Le Guyon S, Reimann R, Grantcharova N, Römling U. Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol. 2012;9:489–502. doi: 10.4161/rna.19682. [DOI] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffler A, Traulsen DD, Fischer D, Lange R, Hengge-Aronis R. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the sigmaS subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1997;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar R, Rush MC, Correa F. Association of Burkholderia glumae and B. gladioli with panicle blight symptoms on rice in Panama. Plant Dis. 2007;91:767. doi: 10.1094/PDIS-91-6-0767C. [DOI] [PubMed] [Google Scholar]
- Nandakumar R, Shahjahan AKM, Yuan XL, Dickstein ER, Groth DE, Clark CA, Cartwright RD, Rush MC. Burkholderia glumae and B. gladioli cause bacterial panicle blight in rice in the southern United States. Plant Dis. 2009;93:896–905. doi: 10.1094/PDIS-93-9-0896. [DOI] [PubMed] [Google Scholar]
- Panda G, Tanwer P, Ansari S, Khare D, Bhatnagar R. Regulation and RNA-binding properties of Hfq RNA chaperones in Bacillus anthracis. Biochim Biophys Acta. 20151850:1661–1668. doi: 10.1016/j.bbagen.2015.03.016. [DOI] [PubMed] [Google Scholar]
- Rai B, Shrestha A, Sharma S, Joshi J. Screening, optimization and process scale up for pilot scale production of lipase by Aspergillus niger. Biomed Biotechnol. 2014;2:54–59. [Google Scholar]
- Ramos CG, Sousa SA, Grilo AM, Feliciano JR, Leitão JH. The second RNA chaperone, Hfq2, is also required for survival under stress and full virulence of Burkholderia cenocepacia J2315. J Bacteriol. 2011;193:1515–1526. doi: 10.1128/JB.01375-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ramos CG, da Costa PJP, Döring G, Leitão JH. The novel cis-encoded small RNA h2cR is a negative regulator of hfq2 in Burkholderia cenocepacia. PloS one. 2012;7:e47896. doi: 10.1371/journal.pone.0047896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Seraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim NN, Feig AL. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PloS one. 2010;5:e13028. doi: 10.1371/journal.pone.0013028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press; NY, USA: 1989. p. 1546. [Google Scholar]
- Sato Z, Koiso Y, Iwasaki S, Matsuda I, Shirata A. Toxins produced by Pseudomonas glumae. Japanese J Phytopathol. 1989;55:353–356. doi: 10.3186/jjphytopath.55.353. [DOI] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrero P, Valverde C. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit Rev Microbiol. 2012;38:276–299. doi: 10.3109/1040841X.2012.664540. [DOI] [PubMed] [Google Scholar]
- Sobrero P, Schlüter JP, Lanner U, Schlosser A, Becker A, Valverde C. Quantitative proteomic analysis of the Hfq-regulon in Sinorhizobium meliloti 2011. PLoS One. 2012;7:e48494. doi: 10.1371/journal.pone.0048494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E, Moll I, Bläsi U. Functional replacement of the Escherichia coli hfq gene by the homologue of Pseudomonas aeruginosa. Microbiology. 2002;148:883–891. doi: 10.1099/00221287-148-3-883. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jäger KE, Bläsi U. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb Pathog. 2003;35:217–228. doi: 10.1016/S0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E, Bläsi U. Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa Carbon Catabolite Repression. PLoS Genet. 2014;10:e1004440. doi: 10.1371/journal.pgen.1004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa SA, Ramos CG, Moreira LM, Leitão JH. The hfq gene is required for stress resistance and full virulence of Burkholderia cepacia to the nematode Caenorhabditis elegans. Microbiology. 2010;156:896–908. doi: 10.1099/mic.0.035139-0. [DOI] [PubMed] [Google Scholar]
- Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trung HM, Van NV, Vien NV, Lam DT, Lien M. Occurrence of rice grain rot disease in Vietnam. Int Rice Res Notes. 1993;18:30. [Google Scholar]
- Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Wilms I, Möller P, Stock AM, Gurski R, Lai EM, Narberhaus F. Hfq influences multiple transport systems and virulence in the plant pathogen Agrobacterium tumefaciens. J Bacteriol. 2012;194:5209–5217. doi: 10.1128/JB.00510-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewska Z, Olejniczak M. Hfq assists small RNAs in binding to the coding sequence of ompD mRNA and in rearranging its structure. RNA. 2016;22:979–994. doi: 10.1261/rna.055251.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, McNally RR, Sundin GW. Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora. J Bacteriol. 2013;195:1706–1717. doi: 10.1128/JB.02056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.