Abstract
Receptor-like proteins (RLPs) are involved in plant development and disease resistance. Only some of the RLPs in tomato (Solanum lycopersicum L.) have been functionally characterized though 176 genes encoding RLPs, which have been identified in the tomato genome. To further understand the role of RLPs in tomato, we performed genome-guided classification and transcriptome analysis of these genes. Phylogenic comparisons revealed that the tomato RLP members could be divided into eight subgroups and that the genes evolved independently compared to similar genes in Arabidopsis. Based on location and physical clustering analyses, we conclude that tomato RLPs likely expanded primarily through tandem duplication events. According to tissue specific RNA-seq data, 71 RLPs were expressed in at least one of the following tissues: root, leaf, bud, flower, or fruit. Several genes had expression patterns that were tissue specific. In addition, tomato RLP expression profiles after infection with different pathogens showed distinguish gene regulations according to disease induction and resistance response as well as infection by bacteria and virus. Notably, Some RLPs were highly and/or unique expressed in susceptible tomato to pathogen, suggesting that the RLP could be involved in disease response, possibly as a host-susceptibility factor. Our study could provide an important clues for further investigations into the function of tomato RLPs involved in developmental and response to pathogens.
Keywords: gene expression profiling, host-susceptibility factor, plant-microbe interaction, receptor-like protein (RLP), Solanum lycopersicum
Plants have evolved various immune mechanisms to recognize and respond to pathogens (Hulbert et al., 2001; Kruijt et al., 2005; Yeom et al., 2011; Yeom et al., 2012). Among these mechanisms are two innate immune systems based on receptors located on the cell surface or within the cell (Lee and Yeom, 2015; Tör et al., 2009). The cell surface receptors are referred to as pattern recognition receptors and include two major classes: receptor-like kinases and receptor-like proteins (RLP) (Kruijt et al., 2005; Monaghan and Zipfel, 2012; Tör et al., 2009).
The two groups of pattern recognition receptors have leucine-rich repeat (LRR) structural elements, which are extracellular domains that interact with ligands. Receptor-like kinases contain extracellular LRRs, a transmembrane spanning domain, and a cytoplasmic kinase domain; while RLPs contain extracellular LRRs and transmembrane spanning domains, but also have a signal peptide and lack the cytoplasmic kinase domain (Monaghan and Zipfel, 2012). RLPs are composed of several conserved domains designated A through G based on their amino acid sequences. Domain A is a putative signal peptide and domain B is a cys-rich domain. Domain C is annotated as an LRR and consists of three domains: C1, C2 (a non-LRR region), and C3. Domains D, E, F, and G are a spacer, an acidic domain, a transmembrane domain, and a short cytoplasmic domain, respectively (Fritz-Laylin et al., 2005).
RLPs comprise a large gene family in plants that is involved in defense and development. To date, most RLPs with assigned functions are involved in disease resistance. The first RLP gene identified was the tomato gene Cf-9 that is involved in disease resistance against the fungal pathogen Cladosporium fulvum (Jones et al., 1994). Since its discovery, several other Cf genes involved in plant resistance to C. fulvum have been cloned and characterized from tomato (Dixon et al., 1996; Dixon et al., 1998; Takken et al., 1999; Thomas et al., 1997). Other RLPs have been identified that regulate resistance to pathogens in tomato, Arabidopsis, and apple. In tomato, Ve genes confer race-specific resistance against Verticillium (Kawchuk et al., 2001). Also in tomato, LeEIX1 and LeEIX2 mediate recognition of the ethylene-inducing xylanase of Trichoderma viride (Ron and Avni, 2004). HcrVf-2 in apple confers resistance against Venturia inequalis (Belfanti et al., 2004). In Arabidopsis, ReMAX may regulate recognition of the microbe-associated molecular pattern eMAX of Xanthomonas (Jehle et al., 2013) and RFO2 mediates resistance to Fusarium oxysporum forma specialis matthioli (Shen and Diener, 2013). Screens of T-DNA insertion lines (Wang et al., 2008) indicated that AtRLP30 influences susceptibility of Pseudomonas syringae pv. phaseolicola to non-host resistance. RBPG1 in Arabidopsis is a microbe-associated molecular pattern conferring resistance to fungal endo-polygalacturonases (Zhang et al., 2014). Although most RLPs are involved in resistance, they can also regulate development. CLAVATA2 (CLV2) in Arabidopsis is crucial in meristem development (Jeong et al., 1999), and its orthologue in maize, FASCINATED EAR2 (FEA2), also regulates development of the shoot meristem (Taguchi-Shiobara et al., 2001). The Arabidopsis TOO MANY MOUTHS (TMM) is involved in stomatal distribution on the epidermis (Nadeau and Sack, 2002).
Genome sequences can provide valuable information for genome-based investigation, such as gene cloning and genome-wide analysis of a target gene family (Kim et al., 2017; Seo et al., 2016; Tomato Genome, 2012). RLPs have also been investigated using global identification techniques and genome analysis. To date, 57 RLPs in Arabidopsis (Wang et al., 2008), 90 RLPs in rice (Fritz-Laylin et al., 2005), 82 RLPs in poplar (Petre et al., 2014), and 144 RLPs in cotton (Chen et al., 2015) have been identified. However, there is little data from genome-wide investigations of RLPs in tomato. Consequently, we performed genome-wide identification and classification of RLP family members in tomato and identified their locations on chromosomes and their physical clustering. To assume their function, we investigated the gene expression profiles using tissue specific and pathogen-infected RNA-seq data. These results will provide useful information on the function of RLP gene family members of tomato and relatives, Solanaceous crops, and will assist in their further functional characterization.
Materials and Methods
Identification of RLP genes in the tomato genome
One hundred seventy-six tomato RLP genes have been identified from ITAG2.4 gene models in the tomato database (https://solgenomics.net/; (Andolfo et al., 2013; Tomato Genome, 2012). To verify these genes, we created Pfam domains using C3-D amino acid sequences extracted from known RLP proteins. The identified genes were screened using the Pfam protein domain database (http://pfam.sanger.ac.uk/) based on the C3-D domains of RLP genes using a Hidden Markov Model (HMMERv.3, http://hmmer.org) search. The RLP genes were subjected to a Basic Local Alignment Search Tool (BLAST) search in the National Center for Biology Information (NCBI) database to validate the genes. The putative RLP genes in tomato were analyzed by functional annotation with SMART (http://smart.embl-heidelberg.de) to find RLP domains, such as the signal peptide, transmembrane domain, and LRRs. The 57 Arabidopsis RLP genes were obtained from a study by (Wang et al., 2008).
Phylogenetic analysis and classification
Amino acid sequences of the C3-D domains for each tomato and Arabidopsis RLP were extracted. Among them, C3-D domains from 54 Arabidopsis and 109 tomato proteins (> 70% of the full-length of the C3-D domains) were selected for multiple sequence alignment and phylogenetic analysis. Multiple sequence alignments were performed with MUSCLE (Edgar, 2004) using MEGA software (http://www.megasoftware.net/). The phylogenetic tree was constructed with multiple alignment results using PhyML (http://www.phylogeny.fr) with the default values.
Classification of RLPs was performed based on sequence similarity using the phylogenetic tree and OrthoMCL according to a previously reported method (Kang et al., 2016; Seo et al., 2016). RLPs were classified according to their clade with characterized RLP genes in the phylogenetic tree. Orthologues were clustered by comparison with the full-length amino acid sequences from all identified tomato and Arabidopsis RLPs using OrthoMCL (http://orthomcl.cbil.upenn.edu). RLPs belonging to the same cluster were classified as within the same phylogenetic groups. Singletons, which were not classified in the phylogenetic tree or OrthoMCL, were further compared using accelerated protein-protein BLAST (BLASTP) against all RLPs and were assigned to subgroups.
Chromosome locations and physical clustering of tomato RLPs
Tomato RLP genes were mapped onto tomato chromosomes based on the physical position of each gene from ITAG2.4. Visualization of chromosome locations was performed using the MapChart program (Voorrips, 2002). Physical clustering among RLPs was analyzed and calculated with the following conditions: clusters contain less than 200 kb between RLPs and there are less than eight genes between RLPs (Seo et al., 2016).
Motif analysis of RLPs in tomato
The conserved motifs among tomato RLPs were identified using the MEME suite program (http://meme-suite.org/tools/meme). The parameters were as follows: maximum number of motifs, 20; minimum width of motifs, 6; maximum width of motifs, 50. Other conditions were set to default values.
Expression profiling of tomato RLPs
Tomato genes from RNA-seq data from various tissues (Tomato Genome, 2012), including root, leaf, bud, flower, and fruit (six developmental stages), were analyzed. We also obtained public RNA-seq data from pathogen-infected tomato samples. For Tomato yellow leaf curl virus (TYLCV) infected tomato samples, leaf samples were collected at 0, 3, 5, 7 days post inoculation (dpi) and then, leaf samples at 3, 5, 7 dpi were pooled together for RNA isolation and library preparation (Chen et al., 2013). For tomato samples with Pseudomonas infection, the third true leaf of 4-week-old Rio Grande prf3 tomato plants were syringe-infiltrated with bacterial strains and tissue was collected 6 h after infiltration, then total RNA was isolated and libraries were constructed (Rosli et al., 2013). Transcript data from tomato plants infected with TYLCV (Chen et al., 2013) or Pseudomonas (Rosli et al., 2013) were trimmed and normalized using in-house pipelines. RNA-seq data were normalized using log2 [reads per kilobase of per million mapped reads (RPKM)] values. Visualization of expression profiling was performed using R (http://bioconductor.org/) to develop heatmaps.
Results and Discussion
Identification and structure analysis of RLP genes in tomato
To identify the RLP genes in tomato, we used previously published RLP gene information as guide genes (Andolfo et al., 2013). A total of 176 RLP-encoding genes, including ten known RLPs, were identified in the tomato genome. One hundred seventy-six is approximately two to three times more than in Arabidopsis (57 genes) or rice (90 genes), which is consistent with the extensive expansion in the tomato genome. The RLPs consist of several domains, designated A to G (Fritz-Laylin et al., 2005). Domain C3 (a LRR domain) and D (a linker domain) are conserved sequences, so we used these to confirm identification of RLPs and to conduct the phylogenetic analysis (Supplementary Fig. 1).
We further examined the RLPs that we had identified by conducting HMMER program (Finn et al., 2011) comparing the C3-D domain against the 176 putative RLP genes. All of the identified 176 genes contained a C3-D domain, so we defined them all as the RLP gene set (Supplementary Table 1). The RLP genes were annotated to find putative functional domains, such as signal peptides, transmembrane domains, and LRRs (Table 1). Fifty-four (30.7%) genes were considered full-type RLPs as they encompass all three defined domains. Seventy-five (42.6%) genes included two domains. These genes were subdivided into 42 genes containing the signal peptide and LRRs and 33 genes with LRRs and the transmembrane. The remaining 47 (26.7%) genes contained only a single domain with LRRs. The high proportion of partial-type genes could be due to annotation problems, such as early stops, truncated domains, and assembly errors.
Table 1.
The number of predicted receptor-like protein (RLP)-encoding genes in the tomato genome
| RLP type | No. of genes |
|---|---|
| Signal peptide/LRR/transmembrane domain | 54 |
| Signal peptide/LRR | 42 |
| LRR/transmembrane domain | 33 |
| LRR | 47 |
|
| |
| Total | 176 |
Phylogenetic analysis and classification of tomato RLPs
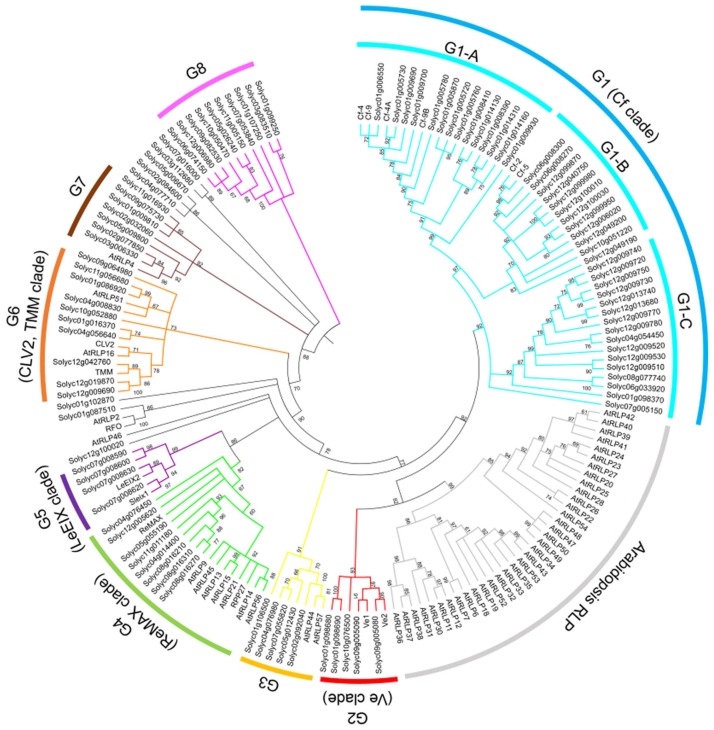
To analyze the phylogenetic relationship of tomato and Arabidopsis RLPs, the amino acid sequences of the C3-D domains of tomato and Arabidopsis RLP genes with sequences containing more than 70% of the full-length sequence were used for multiple sequence alignment using MEGA software (http://www.megasoftware.net/). We obtained 54 RLPs from Arabidopsis and 109 RLPs from tomato for further analysis. The phylogenetic tree was constructed using the multiple sequence alignment of the 54 Arabidopsis genes and 109 tomato genes (Fig. 1). A few clades included RLP genes from both species. However, most tomato RLPs formed a well-separated clade from that of Arabidopsis RLPs indicating that most RLP gene family members may have evolved and diversified independently after speciation. The expansion of RLPs along a specific lineage could lead to functional divergence among plant species and their relatives.
Fig. 1.
Phylogenetic relationship of Arabidopsis and tomato receptor-like proteins (RLPs). The phylogenetic tree was constructed using amino acid sequences of the C3-D domains using PhyML. Tomato RLPs were classified into eight groups (G1 to G8). Bootstrap values > 60% are indicated above branches. Colors indicate individual groups.
A total of 132 tomato RLPs were classified into eight groups designated G1 to G8 according to the phylogenetic relationship and sequence similarity clustering methods using OrthoMCL described in previous study (Seo et al., 2016). Forty-four RLP genes did not belong to any group (Supplementary Table 1). G1 is the largest group that includes the known Cf genes and 47 (26.7%) tomato RLPs. G1 is further divided into three subgroups, G1-A, G1-B, and G1-C, based on branching and clustering with known Cf genes. Even though G1 contained the largest number of RLPs among the RLP groups, G1 was comprised of only tomato RLP genes. Thus, the Cf genes and their orthologues may have been created by extensive expansion in tomato after speciation. Similarly, G2, G5, and G8 contained only tomato RLP genes and may have expanded independently after speciation. The RLPs in G3, G4, G6, and G7 contained both tomato and Arabidopsis RLPs suggesting that these members might have evolved from a common ancestor.
Chromosomal location and physical clustering within the tomato genome
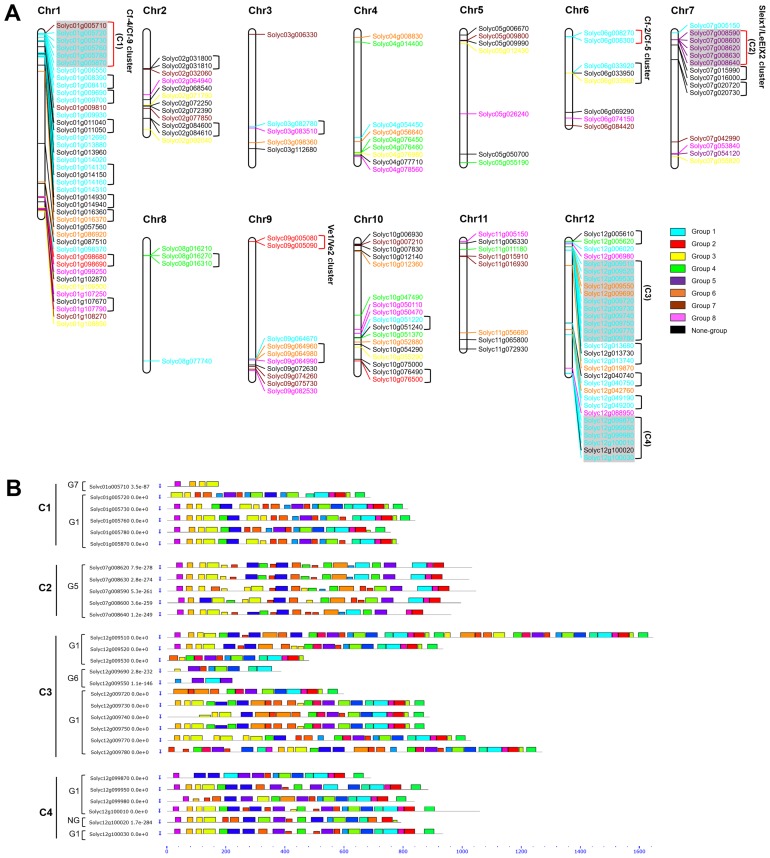
Genomic positions of tomato RLPs were obtained from the Solanaceae lycopersicum gene model ITAG2.4 and were mapped onto the corresponding tomato chromosome locations. The 166 RLPs were distributed across all tomato chromosomes. The twelve chromosomes contained various numbers of RLP genes and the distribution was uneven (Fig. 2A). Chromosomes 1 and 12 had the largest number of RLPs with 41 and 31 genes, respectively. Chromosomes 3 and 8 contained only five and four RLPs, respectively. Most RLPs made physical clusters with members of the same group at specific positions. For example, many RLPs in G1 were located on chromosomes 1 and 12 and RLPs in G5 were located on chromosome 7, suggesting that the formation of these physical clusters and the expansion of tomato RLPs may be due to tandem duplication.
Fig. 2.
Physical genome distribution and conserved motif analysis of tomato receptor-like protein (RLP) genes on chromosomes. (A) Chromosomal locations and physical clusters of SlRLPs. Tomato chromosomes are represented as white bars. Colors represent the phylogenetic groups. Square brackets to the right of the gene IDs indicate tandem arrays and physical gene clusters. Physical clusters with previously cloned genes are denoted with red square brackets and the cloned gene names. (B) Conserved motif analysis of SlRLPs within physical clusters. Representative four physical clusters contained more than three genes (C1, C2, C3, and C4) were analyzed conserved motifs. Black solid line represents corresponding RLPs and their size. Various colored boxes indicate different motifs.
To analyze the duplication history of tomato RLPs, physical clusters were established based on previously published determining methods (Jupe et al., 2012). Twenty-eight physical clusters including 82 genes (49.4%) on eleven chromosomes were identified (Fig. 2A). Chromosome 1 contained the largest number of clusters (9 clusters), whereas chromosome 11 contained no physical clusters. Most of the physical clusters contained two or three genes, while four clusters contained more than three genes. Specifically, most of the physical clusters were composed of RLPs belonging to the same group, such as a Cf-2/Cf-5 cluster and a Sleix1/LeEIX2 cluster. To figure out the structure similarity of RLPs in the same physical cluster, we conducted the conserved motifs analysis in tomato RLP amino acid sequences. The motif analysis in four physical clustering including more than three genes showed that the motifs of closely related genes within each physical cluster shared common sequences and positions (Fig. 2B and Supplementary Fig. 1). Thus, clustered RLPs were likely duplicated from a recent common ancestor via tandem duplication events. This distribution and physical clustering pattern is consistent with RLP gene families and nucleotide-binding site leucine-rich repeat (NBS-LRR) gene families in other plant species (Andolfo et al., 2013; Seo et al., 2016).
Tissue specific expression of tomato RLPs
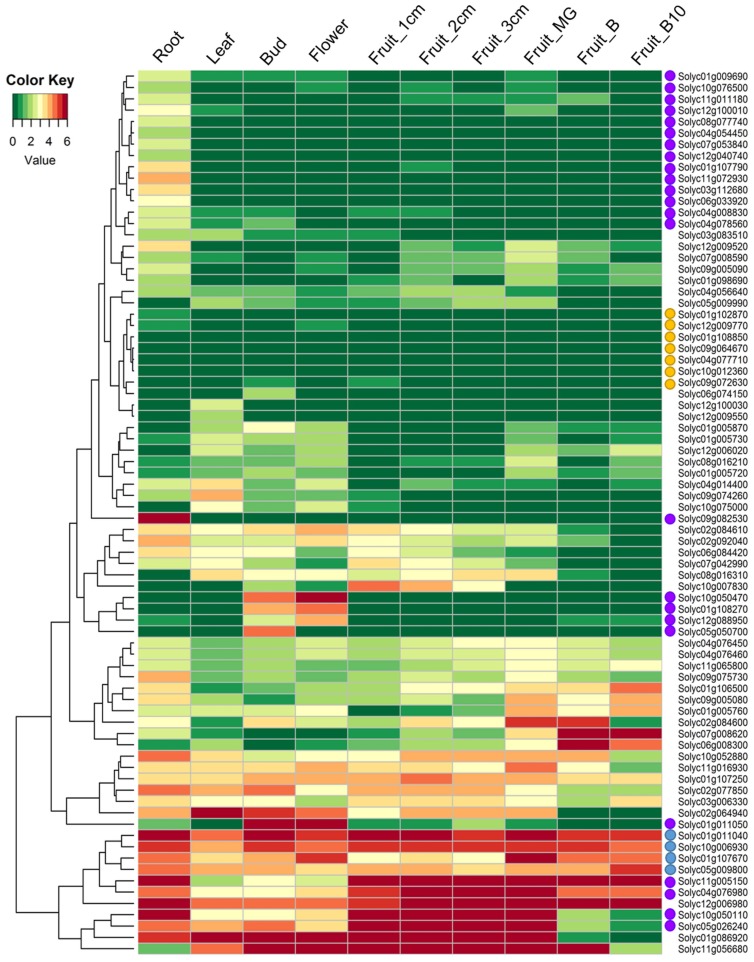
RLPs can play a role in plant development, such as TMM and CLV2 in Arabidopsis (Jeong et al., 1999; Nadeau and Sack, 2002). To understand the putative function of RLP genes, transcriptional profiles of tomato RLP genes were analyzed using RNA-seq data from five tissues (root, leaf, bud, flower, and fruit) and six fruit developmental stages (Tomato Genome, 2012) (Fig. 3). The tomato RLP gene expression profiles varied between different tissue and developmental stages. Among tomato RLPs, 29 RLPs showed zero reads in any tissues or stages, and 66 RLPs transcripts were at extremely low levels in all tested tissues. Therefore, these 95 gene were excluded from further analysis. Four genes (Solyc01g011040, Solyc10g006930, Solyc01g107690, and Solyc05g009800; blue circles on the left side of gene names in Fig. 3) showed high transcript levels in all tested tissues, whereas seven genes were expressed at low levels in all tested tissues (yellow circles in Fig. 3). Some gene transcripts were expressed only in specific vegetative organs or reproductive organs. For example, 15 genes (purple circles in Fig. 3) were expressed at relatively higher levels in root than in other tissues. Specifically, one RLP (Solyc09g082530) among the root-specific RLPs had a very high level of expression, indicating it may be function as a root-specific regulator in tomato. Several genes revealed specific expression patterns in reproductive tissues compared to other tested tissues. High transcript levels of three RLPs (Solyc10g050470, Solyc01g108270, and Solyc01g011050; purple circles in Fig. 3) were detected in buds and flowers. Solyc12g088950 and Solyc05g050700 were solely expressed in flowers and buds, respectively, indicating that these genes may be involved in bud and flower developmental processes. Four genes (Solyc11g005150, Solyc04g076980, Solyc10g050110, and Solyc05g026240; purple circles in Fig. 3) showed specific expression patterns during fruit developmental stages.
Fig. 3.
Expression profiles of tomato receptor-like protein (RLP) genes across different tissues. The tomato transcriptome data were generated in-house pipeline. Expression data for each gene were normalized to log2[reads per kilobase of per million mapped reads (RPKM)] values and hierarchically clustered. Colors represent relative expression levels. MG, mature green stage; B, breaker stage; B10, 10 days post-breaker. Circles (blue, yellow, and purple) on the left side of gene names indicate tissue specific expressed genes.
Development-related genes have evolved while maintaining function and conserved sequences with orthologues (Dolinski and Botstein, 2007). The Arabidopsis CLV2 gene has a function in controlling the size of stem population in the shoot apical meristem (Wang et al., 2010). The CLV2 gene mutants result in phenotypic alterations in gynoecia, flower pedicels, stamens and rosette leaves (Kayes and Clark, 1998; Pan et al., 2016). The expression profile of CLV2 during development stages in Arabidopsis showed constitutive expression patterns in developmental stages such as root, stem, leaf, apex, flowers, flower organs, seed, and whole plant in AtGenExpress (www.arabidopsis.org). The Arabidopsis TMM gene is known to play a role in stomatal distribution on the epidermis (Nadeau and Sack, 2002). However, expressions of TMM are also detected in various tissues in Arabidopsis (www.arabidopsis.org). To investigate whether tomato RLPs classified with CLV2 and TMM genes in G6 play a role in plant development, we analyzed the expression of tomato RLPs in G6. Five genes had detectable expression levels on the heatmap (Fig. 3), while nine genes showed very low expression levels and were excluded from the heatmap. Three genes (Solyc11g056680, Solyc01g086920, and Solyc10g052880) were expressed in most of the tested tissues and two genes (Solyc04g008830 and Solyc04g056640) were expressed at higher levels in roots than in the other tissues. Taken together, these results suggest that tomato RLP genes are involved in biological functions during plant development in vegetative and reproductive organs.
Global gene expression of tomato RLPs during pathogen infection
RLPs, such as Cf and Ve proteins in tomato and HcrVf2 protein in apple, can mediate resistance responses against pathogens such as Cladosporium fulvum, Verticillium spp., and Venturia inequalis, respectively (Belfanti et al., 2004; Dixon et al., 1996; Dixon et al., 1998; Jones et al., 1994; Kawchuk et al., 2001; Takken et al., 1999; Thomas et al., 1997). In addition, RLPs are associated with susceptibility to pathogens. The T-DNA mutant line of AtRLP in Arabidopsis shows enhanced susceptibility against the non-host bacterial pathogen Pseudomonas syringae pv phaseolicola strain 1448A (Wang et al., 2008).
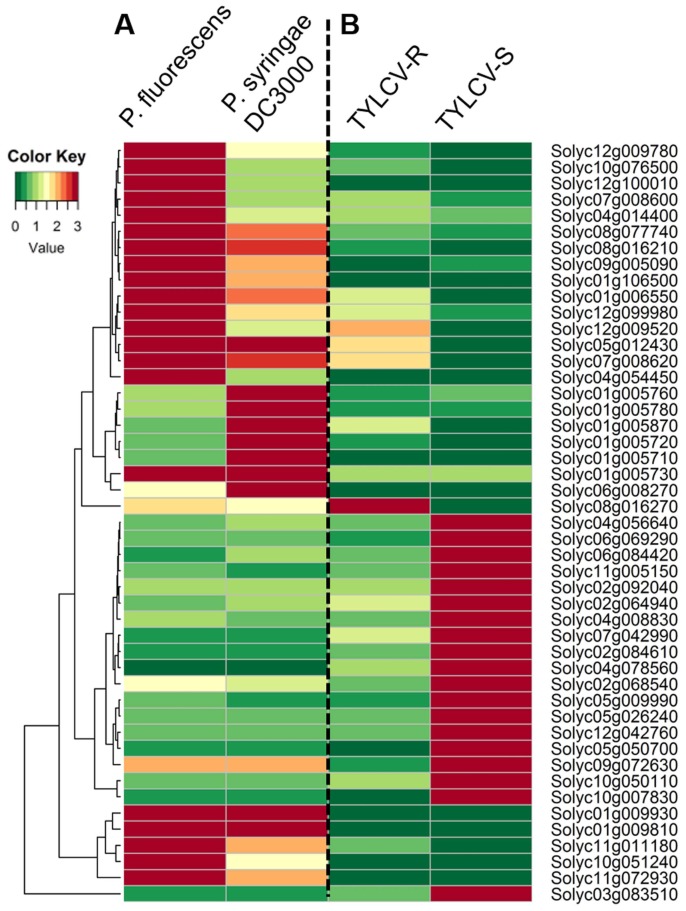
Transcriptome-based expression analysis of tomato RLPs during biotic stresses revealed various expression patterns during different pathogen infection conditions. We obtained previously published RNA-seq data from tomatoes challenged with Pseudomonas and TYLCV (Chen et al., 2013; Rosli et al., 2013). A large number of RLPs showed very low transcript levels under both biotic stresses (Pseudomonas and TYLCV challenges), therefore 47 tomato RLPs were selected for heatmap construction to analyze expression profiles.
We further analyzed RNA-seq data from Rio Grande prf3 tomato plants infected with the wild-type bacterial strains Pseudomonas syringae pv. tomato (Pst) DC3000 or Pseudomonas fluorescens 55 (Rosli et al., 2013) (Fig. 4). The Rio Grande prf3 tomato plants showed different responses to infection with the two different Pseudomonas strains, such as disease induction (Abramovitch et al., 2003) and non-hypersensitive response (HR) (Kim et al., 2002). The RNA-seq data were generated and analyzed via an in-house pipeline. The RLP genes had varying expression patterns between the P. fluorescens- and P. syringae DC300-infected plants. Five RLP genes (Solyc01g009930, Solyc01g009810, Solyc01g005730, Solyc05g012430, and Solyc07g008620) were upregulated in response to both infections. Expression of sixteen genes was higher in P. fluorescens-infected samples than in those challenged with P. syringae DC3000. Conversely, six genes had higher transcript levels in P. syringae DC3000-infected plants than in P. fluorescens-infected plants. These six genes may be involved in host susceptibility to P. syringae DC3000.
Fig. 4.
Expression profiling of tomato receptor-like proteins (RLPs) during pathogen infection. (A) Tomato RLP gene expression patterns during challenge with Pseudomonas fluorescens and P. syringae DC3000. Expression values were normalized to a mock control of each gene. (B) Expression profiles of RLP genes in resistant (TYLCV-R) and susceptible (TYLCV-S) tomato cultivars against TYLCV. RLP expression values were normalized to a mock control of each gene. Relative expression values were transformed to log2. Colors represent relative expression levels.
To assess tomato RLP expression in response to TYLCV infection, we obtained and analyzed RNA-seq data from a TYLCV-resistant tomato breeding line (R) and a TYLCV-susceptible tomato breeding line (S) challenged with TYLCV (Chen et al., 2013). A great number of RLP genes were undetected or barely expressed during TYLCV infection in either line. Nineteen RLPs were highly upregulated in S plants, while the expression levels of these genes in R plants were lower (Fig. 4). In contrast, four genes (Solyc12g009520, Solyc05g012430, Solyc07g008620, and Solyc08g016270) were expressed at higher levels in R plants than in S plants. The Solyc08g016270 gene exhibited the highest expression level in R plants indicating that it may be associated with defense-related functions against TYLCV. Taken together, our findings suggest that tomato RLPs may not only be involved in resistance against, but also in susceptibility to pathogens.
A few of the tomato RLPs have been functionally characterized based on past genetic and molecular approaches. In this study, we identified 176 RLPs divided into 8 subgroups. We investigated the physical location with gene clustering composition in tomato genome, indicating some RLP expansions primarily through tandem duplication events. To help uncovering the role of RLPs in tomato, we performed expression profile of tomato RLP genes upon plant tissues and different response to pathogens, showing that some are unique expressed and may are involved in host-susceptibility. Our results could provide valuable resources for future investigations in the biological function of tomato RLP genes and other species.
Supplementary data
Acknowledgement
This work was supported by the National Research Foundation of Korea Grant funded (NRF-2015R1A6A1A03031413) and Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ010939062018) from Rural Development Administration by the Korean Government.
References
- Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 2003;22:60–69. doi: 10.1093/emboj/cdg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfo G, Sanseverino W, Rombauts S, Van de Peer Y, Bradeen JM, Carputo D, Frusciante L, Ercolano MR. Overview of tomato (Solanum lycopersicum) candidate pathogen recognition genes reveals important Solanum R locus dynamics. New phytol. 2013;197:223–237. doi: 10.1111/j.1469-8137.2012.04380.x. [DOI] [PubMed] [Google Scholar]
- Belfanti E, Silfverberg-Dilworth E, Tartarini S, Patocchi A, Barbieri M, Zhu J, Vinatzer BA, Gianfranceschi L, Gessler C, Sansavini S. The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety. Proc Natl Acad Sci USA. 2004;101:886–890. doi: 10.1073/pnas.0304808101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Huang JQ, Li NY, Ma XF, Wang JL, Liu C, Liu YF, Liang Y, Bao YM, Dai XF. Genome-wide analysis of the gene families of resistance gene analogues in cotton and their response to Verticillium wilt. BMC Plant Biol. 2015;15:148. doi: 10.1186/s12870-015-0508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Lv Y, Zhao T, Li N, Yang Y, Yu W, He X, Liu T, Zhang B. Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by tomato yellow leaf curl virus. PloS one. 2013;8:e80816. doi: 10.1371/journal.pone.0080816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JD. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell. 1996;84:451–459. doi: 10.1016/S0092-8674(00)81290-8. [DOI] [PubMed] [Google Scholar]
- Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JD. The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell. 1998;10:1915–1925. doi: 10.1105/tpc.10.11.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski K, Botstein D. Orthology and functional conservation in eukaryotes. Annu Rev Genet. 2007;41:465–507. doi: 10.1146/annurev.genet.40.110405.090439. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Krishnamurthy N, Tor M, Sjolander KV, Jones JD. Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 2005;138:611–623. doi: 10.1104/pp.104.054452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert SH, Webb CA, Smith SM, Sun Q. Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- Jehle AK, Lipschis M, Albert M, Fallahzadeh-Mamaghani V, Furst U, Mueller K, Felix G. The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas. Plant Cell. 2013;25:2330–2340. doi: 10.1105/tpc.113.110833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell. 1999;11:1925–1934. doi: 10.1105/tpc.11.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JD. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science. 1994;266:789–793. doi: 10.1126/science.7973631. [DOI] [PubMed] [Google Scholar]
- Jupe F, Pritchard L, Etherington GJ, Mackenzie K, Cock PJ, Wright F, Sharma SK, Bolser D, Bryan GJ, Jones JD, Hein I. Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genomics. 2012;13:75. doi: 10.1186/1471-2164-13-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang WH, Kim S, Lee HA, Choi D, Yeom SI. Genome-wide analysis of Dof transcription factors reveals functional characteristics during development and response to biotic stresses in pepper. Sci Rep. 2016;6:33332. doi: 10.1038/srep33332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, Fischer R, Prufer D. Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci USA. 2001;98:6511–6515. doi: 10.1073/pnas.091114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayes JM, Clark SE. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development. 1998;125:3843–3851. doi: 10.1242/dev.125.19.3843. [DOI] [PubMed] [Google Scholar]
- Kim SB, Kang WH, Huy HN, Yeom SI, An JT, Kim S, Kang MY, Kim HJ, Jo YD, Ha Y, Choi D, Kang BC. Divergent evolution of multiple virus-resistance genes from a progenitor in Capsicum spp. New Phytol. 2017;213:886–899. doi: 10.1111/nph.14177. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/S0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Kruijt M, De Kock MJ, de Wit PJ. Receptor-like proteins involved in plant disease resistance. Mol Plant Pathol. 2005;6:85–97. doi: 10.1111/j.1364-3703.2004.00264.x. [DOI] [PubMed] [Google Scholar]
- Lee HA, Yeom SI. Plant NB-LRR proteins: tightly regulated sensors in a complex manner. Brief Funct Genomics. 2015;14:233–242. doi: 10.1093/bfgp/elv012. [DOI] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C. Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol. 2012;15:349–357. doi: 10.1016/j.pbi.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD. Control of stomatal distribution on the Arabidopsis leaf surface. Science. 2002;296:1697–1700. doi: 10.1126/science.1069596. [DOI] [PubMed] [Google Scholar]
- Pan L, Lv S, Yang N, Lv Y, Liu Z, Wu J, Wang G. The Multifunction of CLAVATA2 in Plant Development and Immunity. Front Plant Sci. 2016;7:1573. doi: 10.3389/fpls.2016.01573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petre B, Hacquard S, Duplessis S, Rouhier N. Genome analysis of poplar LRR-RLP gene clusters reveals RISP, a defense-related gene coding a candidate endogenous peptide elicitor. Front Plant Sci. 2014;5:111. doi: 10.3389/fpls.2014.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Avni A. The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell. 2004;16:1604–1615. doi: 10.1105/tpc.022475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosli HG, Zheng Y, Pombo MA, Zhong S, Bombarely A, Fei Z, Collmer A, Martin GB. Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biol. 2013;14:R139. doi: 10.1186/gb-2013-14-12-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E, Kim S, Yeom SI, Choi D. Genome-Wide Comparative Analyses Reveal the Dynamic Evolution of Nucleotide-Binding Leucine-Rich Repeat Gene Family among Solanaceae Plants. Front Plant Sci. 2016;7:1205. doi: 10.3389/fpls.2016.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Diener AC. Arabidopsis thaliana resistance to fusarium oxysporum 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection. PLoS Genet. 2013;9:e1003525. doi: 10.1371/journal.pgen.1003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 2001;15:2755–2766. doi: 10.1101/gad.208501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FL, Thomas CM, Joosten MH, Golstein C, Westerink N, Hille J, Nijkamp HJ, De Wit PJ, Jones JD. A second gene at the tomato Cf-4 locus confers resistance to cladosporium fulvum through recognition of a novel avirulence determinant. Plant J. 1999;20:279–288. doi: 10.1046/j.1365-313X.1999.t01-1-00601.x. [DOI] [PubMed] [Google Scholar]
- Thomas CM, Jones DA, Parniske M, Harrison K, Balint-Kurti PJ, Hatzixanthis K, Jones JD. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell. 1997;9:2209–2224. doi: 10.1105/tpc.9.12.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome C. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tör M, Lotze MT, Holton N. Receptor-mediated signalling in plants: molecular patterns and programmes. J Exp Bot. 2009;60:3645–3654. doi: 10.1093/jxb/erp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wang G, Long Y, Thomma BPJ, de Wit PJ, Angenent GC, Fiers M. Functional analyses of the CLAVATA2-like proteins and their domains that contribute to CLAVATA2 specificity. Plant Physiol. 2010;152:320–331. doi: 10.1104/pp.109.148197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ellendorff U, Kemp B, Mansfield JW, Forsyth A, Mitchell K, Bastas K, Liu CM, Woods-Tor A, Zipfel C, de Wit PJ, Jones JD, Tor M, Thomma B. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 2008;147:503–517. doi: 10.1104/pp.108.119487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom SI, Seo E, Oh SK, Kim KW, Choi D. A common plant cell-wall protein HyPRP1 has dual roles as a positive regulator of cell death and a negative regulator of basal defense against pathogens. Plant J. 2012;69:755–768. doi: 10.1111/j.1365-313X.2011.04828.x. [DOI] [PubMed] [Google Scholar]
- Yeom SI, Baek HK, Oh SK, Kang WH, Lee SJ, Lee JM, Seo E, Rose JK, Kim BD, Choi D. Use of a secretion trap screen in pepper following Phytophthora capsici infection reveals novel functions of secreted plant proteins in modulating cell death. Mol Plant-Microbe Interact. 2011;24:671–684. doi: 10.1094/MPMI-08-10-0183. [DOI] [PubMed] [Google Scholar]
- Zhang L, Kars I, Essenstam B, Liebrand TW, Wagemakers L, Elberse J, Tagkalaki P, Tjoitang D, van den Ackerveken G, van Kan JA. Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the arabidopsis receptor-like protein Responsiveness to Botrytis Polygalacturonases 1. Plant Physiol. 2014;164:352–364. doi: 10.1104/pp.113.230698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.