Abstract
There are two main types of hair follicle in Inner Mongolia Cashmere goats, the primary hair follicle (PHF) producing hair fibers and the secondary hair follicle (SHF) producing cashmere fibers. Of both fibers from cashmere-bearing goats in Aerbasi, Inner Mongolia, the timing of cyclical phases for the cashmere have been well clarified but hair fibers have been less noticeable. Herein, we evaluated transcriptome of PHF and SHF from the same three goats in Aerbasi at the catagen- and telogen phase of cashmere growth. We totally found 1977 DEGs between PHFs at the telogen and catagen phases of SHF, 1199 DEGs between telogen- and catagen SHF, 2629 DEGs between PHF at the catagen phase of SHF and catagen SHF, and 755 DEGs between PHF at the telogen phase of SHF and telogen SHF. By analyzing gene functions based on GO and KEGG database, we found that the DEGs have functions in muscle contraction and muscle filament sliding between catagen- and telogen SHF, indicating that arrector pilli muscles might play a role on the transition from catagen to telogen. Moreover, considering that the enriched GO and KEGG categories of the DEGs between PHF and SHF, we suggested that part of PHF might rest in their own anagen phase when SHF are at catagen, but PHF might enter into the telogen phase at SHF’s telogen. Finally, we highly recommended the several potential genes acting as the regulators of the transition between growth phases including IL17RB and eight members of ZNF. These results provide insight into molecular mechanisms on the transition of SHF from catagen to telogen together with PHF’s growth situation at SHF’s catagen and telogen in Inner Mongolia Cashmere goats.
Introduction
Hair follicles on animal skin execute many useful biology function except for producing hair, such as immune defense and grease secretion, etc. There are two main types of hair follicle in goats, the primary hair follicle (PHF) producing hair fibers and the secondary hair follicle (SHF) producing cashmere fibers. Cashmere is finer, stronger, lighter, softer, and more insulating than sheep wool, surely it looks as a luxury fiber[1]. As a major province of raw cashmere in China, Inner Mongolia bears three types of Cashmere goats—Erlangshan, Aerbasi and Alashan; these Cashmere goats are mainly distributed in the northwest of Inner Mongolia Plateau (Fig 1). They are superior in the production of cashmere with diameter less than 16 μm.
Fig 1.
(a) is Inner Mongolia Autonomous Region map and the location is Otog Banner where Inner Mongolia Cashmere goat strain line of Aerbasi mainly distributed. (b) is a female goat of this breed.
Hair follicles is a unique, highly regenerative organ with a normal developmental cycle, which is mainly determined by the interaction of epithelial cells and dermal papilla that are generally composed of dermal mesenchymal cells [2]. A growth cycle consists mainly of three distinct stages—anagen, catagen and telogen [2,3,4]. In Cashmere goat, SHF takes approximately one year to complete a growth cycle. For Inner Mongolia Cashmere goats, the anagen phase of SHF begins from April until November, the catagen phase is from December to January, and the telogen phase is from February to March[5]. HFs help cashmere to grow at the anagen phase, turn into apoptosis at the catagen phase and finally release cashmere at the telogen phase. When the next anagen phase comes, HFs launch a new regenerative cycle. To increase the harvest accompanying with the thinner cashmere, researchers try to find major genes and pathways, which may affect the of development and growth the cashmere by molecular biology methods.
RNA-seq is usual and convenient for researchers to study different animal phenotypes due to differentially expressed genes (DEGs), because we could get enough transcriptome data from small quantities of tissue samples[6,7]. In previous studies of hair follicle in Cashmere goats, the scientists have already found many important factors that may affect hair follicle cycle in Wnt signal transduction pathway, fibroblast growth factor (FGF) family, bone morphogenetic protein (BMP) family, Sonichedgehog (Shh) signal transduction pathway, transforming growth factor (TGF) family, Notch signal transduction pathway and so on [8,9,10]. Some genes play an activator role while other genes play roles in these pathways as the inhibitors. β-catenin has been confirmed as an important accelerator in Wnt signal transduction pathway, but more importantly, its activity is simultaneous with the apoptosis of hair follicle stem cells [11]. Maksim V. Plikus found that BMPs may be the long-sought inhibitors of hair growth postulated by classical experiments[12].
In this study, we used RNA-seq technology to find some important factors in SHF of Inner Mongolia Cashmere goats by comparing catagen SHF with telogen SHF. Furthermore, functional annotation analysis also helps us find some important genes that influence cashmere growth. By comparing the DEGs of PHF at the catagen and telogen phase of SHF, the growth condition of PHF became relatively clear. Our data might take insight into the molecular mechanism on the growth of SHF and PHF at the catagen- and telogen phase of SHF.
Material and methods
1. Ethics statement
In this study, hair follicles were collected in accordance with the International Guiding Principles for Biomedical Research Involving Animals and was approved by the Animal Ethics Committee of the Inner Mongolia Academy of Agriculture and Animal Husbandry Sciences that is responsible for Animal Care and Use in the Inner Mongolia Autonomous Region of China. In our study, no specific permissions were required for these activities and the animals did not involve endangered or protected species.
2. Hair follicle samples preparation for RNA-seq and qRT-PCR validation
Three 3-years-old female Aerbasi Inner Mongolia Cashmere goats from goat stud farm (Aerbasi White Cashmere Goat Breeding Farm, Erdos, Inner Mongolia) were used in this study for RNA-seq. At the dorsal side of goats, we collected PHF and SHF samples by pulled out hair and cashmere from the root of HFs in SHF’s catagen (in mid-January) and its telogen (in mid-March), respectively; and then put them in liquid nitrogen as soon as possible and finally stored in -80°C refrigerator for a long term. About 50 to 100 hair follicles of three cashmere-bearing goats were collected as a sequencing sample. Six female goats, which come from the same group at same age as the RNA-seq goats, were used in qRT-PCR.
3. Construction of RNA library, sequencing and quality control
Total RNA was extracted and test the concentration. Each RNA sample needs over than 2 μg and RIN (the integrity of RNA) value larger than six. We mixed three cashmere goats total RNA and then constructed a library. In hence, four libraries were constructed. After high throughput sequencing, we used Fast-QC to measure the quality of sequence data, including base quality distribution, GC%, content of PCR duplication and frequency of kmer.
4. Mapping reads to reference genome and gene structure analyze
After filtration, we mapped the clean reads to goat gene sequences (http://goat.kiz.ac.cn/GGD/download.htm) and goat reference genome sequences (http://goat.kiz.ac.cn/GGD/download.htm) by MapSplice[4,13]. Then calculated the gene structure, exons, introns and intergenic regions.
5. Expression level analysis
RPKM means the genes expression and calculating bases on the reads counts mapped to this gene and gene length[14]. The different expression genes between catagen and telogen was calculated by EBSeq (Log2FC>1 or Log2FC<-1, FDR<0.05)[15].
6. GO and KEGG pathway analysis
We analyzed genes function by Gene Ontology (GO), an international gene functional classification system, on three categories including biological process, molecular function and cellular component[4,16]. We used Fisher formula to exam the P-Value of each GO analysis and blasted result onto human and mouse reference genomes (P-Value<0.01 represent gene function is significance). Different expression genes (DEGs) were annotated by Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg) to identify the pathway enrichments[17,18,19].
7. qRT-PCR to validate RNA-seq data
To validate the veracity of RNA-seq data and genes expression level, we used quantitative real time PCR in this experiment. First, we extracted total RNAfrom catagen’s- and telogen’s SHF from six goats. Second, we synthesized cDNA from mRNA. Then, the primers we used was designed and synthesized depending on the mRNA sequences published on NCBI database. QRT-PCR was used SYBR GreenⅡ. In the qRT-PCR, β-actin was acted as internal reference and DEGs expression level was calculated by 2-ΔΔct. The primer sequences information was shown in Table 1.
Table 1. The genes primer sequences information.
| gene name | Reference Sequence | primer sequence | TM (°C) |
|---|---|---|---|
| IL17RB | XM_005695840.1 | F: CTCCCAAGACCTGTTCCACC | 61 |
| R: GCGCACTGTAGCTGTCTTTG | |||
| β-actin | NM_001314342.1 |
F: GGCAGGTCATCACCATCGG R: CGTGTTGGCGTAGAGGTCTTT |
60 |
Reaction system of qRT-PCR was totally 20μl, including SYBR Green Ⅱ 10μl, primer F 0.5μl, primer R 0.5μl, ROX 0.4μl, cDNA 2μl and H2O 6.6μl. The condition of the reaction was departed into amplification stage and dissociation stage. Amplification stage includes 95°C 30s for 1 cycle, 95°C 5s and annealing temperature (TM) 34s for 40 cycles, dissociation stage includes 95°C 15s, 60°C 60s and 95°C 15s. Result were analyzed by SPSS 17.0.
Result
To gain insight into the growth of hair follicles at SHF’s cyclic phases, we quantified the whole-genome transcriptomes of primary- or secondary hair follicles at the catagen and telogen phases of SHF in 3 female Inner Mongolia Cashmere goats. In total, we obtained over 25 million clean reads from each library after trimming, adaptor sequences, low quality reads and multiple mapped reads (Table 2). Gene structure was showed in Fig 2. The clean reads were mapped to 22175 genes annotated in the goat reference genome (Capra Hircus 1.0). By comparing the RNA-seq data, we then identified DEGs in the following four groups including (i) PHF at the telogen and catagen phases of SHF, (ii) telogen- and catagen SHF, (iii) PHF at the catagen phase of SHF and catagen SHF, and (v) PHF at the telogen phase of SHF and telogen SHF (Table 3). Using the thresholds of false discovery rate (FDR) < 0.05 and difference ratio of RPKM (reads per kilobase of exon per million fragments mapped) > 2, we defined 1977 DEGs between PHF at the telogen and catagen phases of SHF, 1199 DEGs between telogen and catagen SHF, 2629 DEGs between PHF at the catagen phase of SHF and catagen SHF, and 755 DEGs between PHF at the telogen phase of SHF and telogen SHF. When focusing on the characterized transcripts, the number of DEGs among the four groups arrays in ascending order as follows: 1783 DEGs between PHF at the catagen phase of SHF and catagen SHF, 1092 between PHF at the telogen and catagen phases of SHF, 552 between telogen and catagen SHF, and 422 between PHF at the telogen phase of SHF and telogen SHF. It is noteworthy that, based on the number of DEGs, the largest distinction at transcription level should be between PHF at SHF’s catagen phase and catagen SHF in sharp contrast to the smallest difference between PHF at SHF’s telogen phase and telogen SHF. Interestingly, the transcriptomic changes seem to be relatively small between telogen- and catagen SHF compared with those between PHF at the catagen- and telogen phase of SHF.
Table 2. Mapping statistics of SHF and PHF in both catagen and telogen.
| Statistics Item | SHF in catagen | SHF in telogen | PHF in catagen | PHF in telogen |
|---|---|---|---|---|
| All reads | 39905590 | 38656183 | 40472235 | 38929776 |
| Unmapped | 13543655 | 8353891 | 6029692 | 9019823 |
| Mapped | 26361935 | 30302292 | 34442543 | 29909953 |
| Mapped rate | 0.661 | 0.784 | 0.851 | 0.768 |
| Unique mapped | 25111435 | 28929192 | 32528469 | 28709741 |
| Unique mapped rate | 0.629 | 0.748 | 0.804 | 0.737 |
| Repeat mapped | 1250515 | 1373099 | 1914088 | 1200212 |
| Junction all mapped | 5307898 | 1338926 | 7746160 | 3207050 |
| Junction unique mapped | 5304888 | 1337307 | 7742669 | 3205570 |
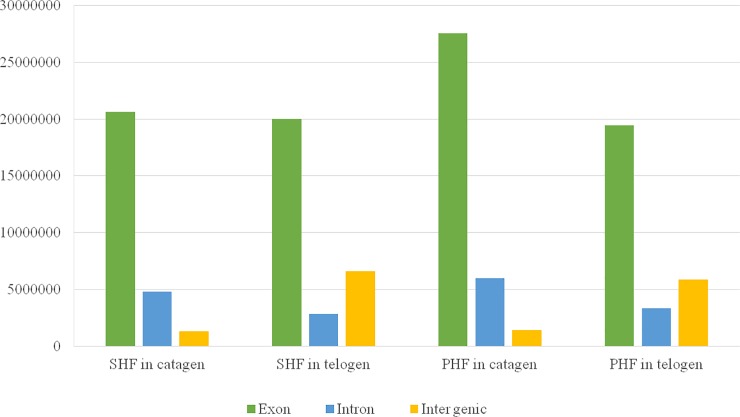
Fig 2. Gene structure in catagen and telogen of SHF and PHF.
The number of exons of SHF in catagen and telogen respectively are 20617193 and 19999609, introns are 4822842 and 2832110, intergenic regions are 1309521 and 6618361. However, the number of exons of PHF in catagen and telogen respectively are 27541761 and 19455711, introns are 5950502 and 3337148, intergenic regions are 1387033 and 5853974. From the figure, we can find that in these four samples, the largest number is exon. But for intron and intergenic regions, there are some differences between catagen and telogen. In catagen, the number of intron is higher than intergenic regions, while in telogen is opposite.
Table 3. Different expression genes identified by RNA-seq.
| PHF_telogen vs PHF_catagen | SHF_telogen vs SHF_catagen | SHF_ catagen vs PHF_ catagen |
SHF_telogen vs PHF_telogen | |||||
|---|---|---|---|---|---|---|---|---|
| upregulated | downregulated | upregulated | downregulated | upregulated | downregulated | upregulated | downregulated | |
| Characterized genes | 591 | 501 | 360 | 192 | 939 | 844 | 302 | 120 |
| Uncharacterized genes | 667 | 218 | 593 | 54 | 601 | 245 | 290 | 43 |
Next, we performed a GO annotation analysis for these characterized DEGs in the four groups. The biology processes were considered to be significant with the thresholds of false discovery rate (FDR) < 0.05 as showed in Table 4. Hairs detach dermal papilla and stop growing since catagen, and then they become dull and lifeless at telogen because of no blood supply. For the only muscle in hair follicles, the arrector pilli muscles is not just an attacher to hair follicle–as had been reported earlier- but is an essential part of the hair follicle cycle by interacting with the follicle mesenchyme[20]. As expected, between catagen SHF and telogen SHF, the DEGs were related to muscle contraction (GO:0006936) and muscle filament sliding (GO:0030049) instead of growth and metabolism. Our results indicated that arrector pilli muscles, which attached to hair follicles, might have functions on helping cashmere stay in the hair follicles at catagen. Interestingly, the DEGs of PHF between the two phases of SHF were involved in those biology processes including translation (GO:0006412, 0006413, 0006414 and 0006415), RNA metabolism (GO:0016070 and 0016071), protein metabolism (GO:0044267), and viral growth (GO:0019083, 0019058 and 0016032). There is not any enriched GO category of the DEGs between PHF and SHF at the telogen phase of SHF, which is satisfied with the threshold at the telogen phase of SHF. In contrast, we found 22 enriched GO categories for the DEGs between the PHF at the catagen of SHF and catagen SHF, such as extracellular matrix organization (GO:0030198), muscle contraction (GO:0030049 and 0006936), translation (GO:0006413, 0006414, and 0006415), and metabolism (GO:0044281, 0006749, 0005975, and 0006805). All in all, these results might hint that the catagen phase of PHF might overlap with the catagen phase of SHF.
Table 4. GO annotation analysis.
| GO ID | GO Term | DifGene | P-Value | FDR |
|---|---|---|---|---|
| PHF_catagen/PHF_telogen | ||||
| GO:0006414 | translational elongation | 82 | 7.16×10−21 | 1.76×10−17 |
| GO:0019083 | viral transcription | 73 | 5.03×10−20 | 6.17×10−17 |
| GO:0006415 | translational termination | 73 | 1.25×10−19 | 1.02×10−16 |
| GO:0019058 | viral life cycle | 73 | 6.23×10−19 | 3.82×10−16 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 73 | 2.54×10−18 | 1.06×10−15 |
| GO:0006412 | translation | 101 | 2.60×10−18 | 1.06×10−15 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 75 | 3.02×10−18 | 1.06×10−15 |
| GO:0006413 | translational initiation | 78 | 7.87×10−18 | 2.41×10−15 |
| GO:0016071 | mRNA metabolic process | 80 | 9.53×10−15 | 2.60×10−12 |
| GO:0016070 | RNA metabolic process | 81 | 4.19×10−14 | 1.03×10−11 |
| GO:0016032 | viral process | 87 | 4.09×10−13 | 9.12×10−11 |
| GO:0044267 | cellular protein metabolic process | 104 | 3.53×10−12 | 7.23×10−10 |
| GO:0010467 | gene expression | 120 | 3.85×10−11 | 7.26×10−9 |
| GO:0042273 | ribosomal large subunit biogenesis | 11 | 3.64×10−5 | 6.39×10−3 |
| SHF_catagen/SHF_telogen | ||||
| GO:0006936 | muscle contraction | 21 | 6.84×10−11 | 1.30×10−7 |
| GO:0030049 | muscle filament sliding | 14 | 3.23×10−10 | 3.07×10−7 |
| GO:0060048 | cardiac muscle contraction | 9 | 1.40×10−5 | 7.64×10−3 |
| GO:0045214 | sarcomere organization | 8 | 1.61×10−5 | 7.64×10−3 |
| SHF_catagen/PHF_catagen | ||||
| GO:0030198 | extracellular matrix organization | 64 | 1.32×10−9 | 3.69×10−6 |
| GO:0030049 | muscle filament sliding | 23 | 1.88×10−9 | 3.69×10−6 |
| GO:0006936 | muscle contraction | 32 | 1.73×10−7 | 2.27×10−4 |
| GO:0006414 | translational elongation | 72 | 5.57×10−7 | 5.47×10−4 |
| GO:0019083 | viral transcription | 62 | 1.55×10−6 | 1.22×10−3 |
| GO:0019058 | viral life cycle | 64 | 2.00×10−6 | 1.31×10−3 |
| GO:0006415 | translational termination | 62 | 2.64×10−6 | 1.48×10−3 |
| GO:0030199 | collagen fibril organization | 17 | 4.45×10−6 | 2.19×10−3 |
| GO:0044281 | small molecule metabolic process | 207 | 6.15×10−6 | 2.69×10−3 |
| GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 65 | 1.08×10−5 | 4.26×10−3 |
| GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 62 | 1.48×10−5 | 5.29×10−3 |
| GO:0006749 | glutathione metabolic process | 14 | 2.94×10−5 | 9.62×10−3 |
| GO:0060048 | cardiac muscle contraction | 16 | 3.19×10−5 | 9.64×10−3 |
| GO:0006937 | regulation of muscle contraction | 11 | 5.96×10−5 | 1.67×10−2 |
| GO:0006413 | translational initiation | 66 | 6.60×10−5 | 1.72×10−2 |
| GO:0005975 | carbohydrate metabolic process | 73 | 6.99×10−5 | 1.72×10−2 |
| GO:0001666 | response to hypoxia | 33 | 1.46×10−4 | 3.37×10−2 |
| GO:1901687 | glutathione derivative biosynthetic process | 12 | 1.63×10−4 | 3.56×10−2 |
| GO:0042060 | wound healing | 20 | 1.97×10−4 | 4.08×10−2 |
| GO:0016071 | mRNA metabolic process | 74 | 2.11×10−4 | 4.14×10−2 |
| GO:0007155 | cell adhesion | 92 | 2.35×10−4 | 4.27×10−2 |
| GO:0006805 | xenobiotic metabolic process | 32 | 2.39×10−4 | 4.27×10−2 |
| SHF_telogen/PHF_telogen | ||||
Besides the GO analysis, we also enriched the KEGG pathways for DEGs with FDR < 0.05 in order to collect the molecular interaction, reaction and relation among DEGs. As same as the enriched biological processes in the GO analysis, the pathways for the DEGs between catagen SHF and telogen SHF are also associated with the function of muscle (Table 5). Furthermore, we also observed that the most pathways were observed for the DEGs between the PHF at the catagen of SHF and catagen SHF, including extracellular matrix interaction (PATH:04512 and 04510), metabolism (PATH:04974, 0006749, 00480, 00980 and 05240) and ribosome (PATH:03010). Against that several pathways for the comparisons as above described, the only enriched pathway for the DEGs of PHF between the two phases of SHF was ribosome (PATH:03010); besides, none can be enriched for the DEGs between PHF and SHF at the telogen phase of SHF.
Table 5. KEGG pathways analysis.
| Pathway ID | Pathway Term | DifGene | P-Value | FDR |
|---|---|---|---|---|
| PHF_catagen/PHF_telogen | ||||
| PATH:03010 | Ribosome | 79 | 3.29×10−18 | 8.00×10−16 |
| SHF_catagen/SHF_telogen | ||||
| PATH:04530 | Tight junction | 16 | 5.62×10−5 | 1.20×10−2 |
| PATH:05410 | Hypertrophic cardiomyopathy (HCM) | 11 | 4.35×10−4 | 4.51×10−2 |
| PATH:05414 | Dilated cardiomyopathy | 11 | 6.63×10−4 | 4.51×10−2 |
| PATH:05416 | Viral myocarditis | 9 | 8.47×10−4 | 4.51×10−2 |
| SHF_catagen/PHF_catagen | ||||
| PATH:04512 | ECM-receptor interaction | 29 | 2.27×10−6 | 6.09×10−4 |
| PATH:04510 | Focal adhesion | 47 | 1.28×10−5 | 1.71×10−3 |
| PATH:04974 | Protein digestion and absorption | 27 | 2.71×10−5 | 2.42×10−3 |
| PATH:00480 | Glutathione metabolism | 18 | 1.19×10−4 | 6.92×10−3 |
| PATH:03010 | Ribosome | 66 | 1.29×10−4 | 6.92×10−3 |
| PATH:00980 | Metabolism of xenobiotics by cytochrome P450 | 21 | 5.37×10−4 | 2.40×10−2 |
| PATH:05134 | Legionellosis | 19 | 1.19×10−3 | 4.55×10−2 |
| PATH:05204 | Chemical carcinogenesis | 18 | 1.36×10−3 | 4.55×10−2 |
| SHF_telogen/PHF_telogen | ||||
According to GO terms, we made an examination using qRT-PCR for the DEGs, such as IL17RB (Interleukin 17 receptor B), HPS6(biogenesis of lysosomal organelles complex 2 subunit 3) and ALPL(alkaline phosphatase), which are functionally related to hair follicle growth Fig 3). The result showed that these three genes were upregulated in catagen SHF compared with telogen SHF, which is consistent with the result of RNA-seq. We also found that the DEGs between catagen SHF and telogen included eight members of zinc finger protein family (Table 6), of which seven members were upregulated except for ZNF347.
Fig 3. IL17RB, HPS6 and ALPL was detected by the qRT-PCR, and the trend was similar to RNA-seq.
** p-value < 0.01, * p-value < 0.05.
Table 6. DEGs from zinc finger protein family in SHF during catagen to telogen.
| AccID | expression counts in catagen | expression counts in telogen | Blast Human AccID | gene name |
|---|---|---|---|---|
| GOAT_ENSP00000395277 | 5647 | 52510 | NM_015037.3 | ZSWIM8 |
| GOAT_ENSP00000331462 | 5 | 70 | NM_032651.1 | ZNF704 |
| GOAT_ENSBTAP00000001064 | 1547 | 44890 | NM_022103.3 | ZNF667 |
| goat_GLEAN_10007887 | 10 | 18 | NM_182594.2 | ZNF454 |
| goat_GLEAN_10012700 | 0 | 6 | NM_017757.2 | ZNF407 |
| GOAT_ENSBTAP00000052171-D2 | 164 | 1 | NM_032584.2 | ZNF347 |
| GOAT_ENSBTAP00000018112 | 20 | 26 | NM_003417.4 | ZNF264 |
| goat_GLEAN_10011985 | 0 | 3 | NM_024721.4 | ZFHX4 |
Discussion
In this study, we chose hair follicle as an alternative to skin in order to reveal the molecular mechanism of the growth cycle of Inner Mongolia Cashmere goat hair follicle. Compared with skin, hair follicle is beneficial to remove the signals of hair stem cells in skin and to eliminate some unexpected influence. To date, we are acquainted with the growth law of SHF in Inner Mongolia Cashmere goat but are completely at a loss for the time-based growth pattern of PHF yet. This study might be helpful to find the relationship between these two hair follicles by collecting PHF at the catagen/telogen phase of SHF. We uncovered that the distinction of gene expression are mainly associated with muscle movement between catagen SHF and telogen one. However, the growth cycle of PHF is not as same as SHF’s; at SHF’s catagen phase, PHF looks like stay in an active state because many metabolism-related genes were enriched(S1 Table). But, interestingly, PHF might be of the same inactive state as SHF at SHF’ telogen phase because no one GO category was identified for DEGs between PHF and SHF.
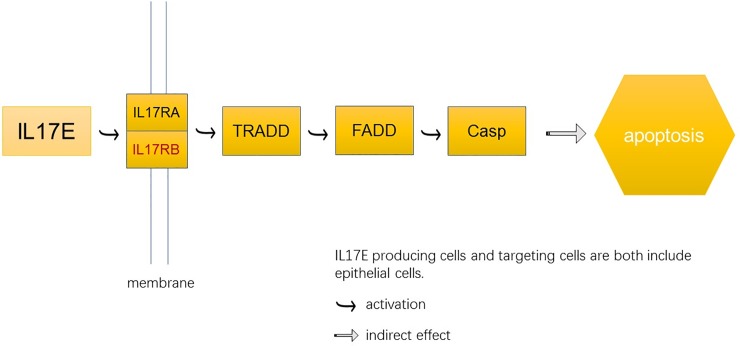
In addition, we analyzed the expression pattern of several critical genes in SHF at the catagen- and telogen. We found that a critical gene IL17RB was upregulated in catagen SHF compared with telogen SHF. IL17 superfamily is a relatively new family of cytokines, which consists of 6 ligands and can bind 5 receptor subtypes[21]. According to recent researches and our transcriptome data, we predicted that IL17RB might play a role in the development of SHF because it acts as a mediate signaling in many other physiological functions[22,23]. A branch of IL-17 pathway, IL17E pathway, targets epithelial cell. The compound of IL17RB and IL17RA worked as membrane receptor signals of IL17E which would activate TRADD, FADD and Casp in order. Finally, this pathway may indirectly affect the apoptosis of epithelial cells (Fig 4). It might indicate that IL17RB plays a positive role in cell apoptosis, and then launches the transition from catagen to telogen. Also, two IL17RB can also form a compound which acted as a membrane receptor of IL17B in IL17RB-IL17B pathway. Vahideh Alinejad et al. found that the IL17RB-IL17B pathway affected breast cancer together with the upstream and downstream cytokines[24]. They summarized IL17 family includes six protein members—IL17A, IL17B, IL17C, IL17D, IL17E and IL17F, of which IL17B and its receptor(IL17RB) are least known genes[25,26].
Fig 4. IL17RB-IL17E pathway may due to epithelial cells apoptosis.
Then, we identified that the eight members of zinc finger protein family were the DEGs between catagen SHF and telogen one (Table 6), of which seven members were upregulated except for ZNF347. Zinc finger proteins are important structural elements in many nucleic acid binding proteins [27]. The researchers further found that zinc finger proteins might affect the growth cycle of hair follicle [28,29]. A zinc finger transcription factor Trps1 mainly expressed at the nuclei of mesenchymal cells during hair follicle morphogenesis; its expression level increased in dermis but decreased in epidermis during early skin morphogenesis [28]. In 2013, Zfp157, a member of KRAB zinc finger protein family, expressed in both epithelial cells of ducts and sebaceous glands of hair follicles in mice [29]. Notably, ZNF667 can inhibit the expression and promoter activity of the rat proapoptotic gene Bax [30]. ZNF667 was a DEG from catagen to telogen of both SHF and PHF in our study, this may indicate that ZNF667 might be an important impact factor for the transition of SHF and PHF from catagen to telogen.
In our previous study, we found that STC2 was downregulated from anagen to catagen[31]. Teng Xu et al. identified approximate 7,000 transcripts that were differentially expressed between anagen and telogen of Inner Mongolia Cashmere goat. In addition, the genes were mainly enriched in ECM receptor interaction, focal adhesion and gap junction from the KEGG pathway database[32]. Rongqing Geng et al. totally identified 1,332 DEGs in Shaanbei Cashmere goat among anagen, catagen and telogen and these genes were mainly playing important roles in Wnt, Shh, TGF-β, and Notch signaling pathways[8]. Xiao-yang Ji et al. found that ubiquitin-mediated proteolysis pathway is a prominent signaling pathway that can distinguish SHF from PHF in cashmere goats [33]. Jianping Li et al. found ten microRNAs have influence in hair follicle growth in Liaoning Cashmere goat and fine-wool sheep telogen skins [34].
The researchers also kept on excavating inhibitors that may affect the growth of hair follicle from model animals to cashmere goats. Plikus et al. indicated BMPs might be a potential signal that played as an inhibitor in the propagation of hair stem cells in mice[12]. Wen L. Bai et al. found that BMP4 is generally upregulated from anagen to telogen in Liaoning Cashmere goat skin tissue, a higher BMP4 methylation level in skin coincides with a lower expression of BMP4 mRNA[35]. They also found seven downregulated miRNAs could joined some signal pathways through their target genes directly or indirectly from anagen to telogen[36]. Mathieu Castela et al. suggested IGF1R would together with BMP4 to control hair follicle goes into anagen early and into catagen late using K15-IGF1RKO mice [37]. Xianghui Ma et al. found MSI2, a RNA-binding protein of Musashi, would keep hair follicle resting in process of telogen-to-anagen transition in mice[38]. In recent study, Guangxian Zhou et al. revealed ncRNA might play a regulatory role in skin of cashmere goat through whole transcriptome[39]. Our study might improve the understanding on the molecular mechanism of the growth cycle of SHF and PHF in Cashmere goats.
Conclusion
In this study, we aim to find out about some interesting factors and signaling pathways, using transcriptome sequencing, that have functions on the growth of two kinds of hair in Inner Mongolia Cashmere goats at the catagen- and telogen phase of SHF. We totally found 1977 DEGs between PHF at the telogen and catagen phases of SHF, 1199 DEGs between telogen and catagen SHF, 2629 DEGs between catagen SHF and PHF at the catagen phase of SHF, and 755 DEGs between telogen SHF and PHF at the telogen phase of SHF. Between catagen- and telogen SHF, the DEGs were related to muscle contraction (GO:0006936) and muscle filament sliding (GO:0030049). Moreover, according to GO category of the DEGs between PHF and SHF, part of PHF might be in anagen phase at SHF’s catagen, but PHF might enter into telogen phase at SHF’s telogen. Among DEGs, IL17RB and eight members of ZNF might play important roles in the growth at the transition of hair follicles from catagen to telogen. These results would benefit the studies of the growth of two kinds of hair follicles in Inner Mongolia Cashmere goats.
Supporting information
(DOCX)
Data Availability
The sequencing reads of each sequencing libraries have been deposited under NCBI with Project ID SUB3555047.
Funding Statement
This work was financially supported by China Agriculture Research System (CARS-39-06), National Natural Science Foundation of China (31660639, 31402052).
References
- 1.Ye G, Wang X, Yan H, Jie Z, Ma S, Niu Y, et al. Comparative Transcriptome Analysis of Fetal Skin Reveals Key Genes Related to Hair Follicle Morphogenesis in Cashmere Goats. Plos One. 2016;11(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Botchkarev VA, Kishimoto J. Botchkarev VA, Kishimoto J. Molecular control of epithelial-mesenchymal interactions during hair follicle cycling. J Investig Dermatol Symp Proc 8:46–55. Journal of Investigative Dermatology Symposium Proceedings. 2003;8(1):46–55. [DOI] [PubMed] [Google Scholar]
- 3.Chen CC, Plikus MV, Tang PC, Widelitz RB, Chuong CM. The modulatable stem cell niche: Tissue interactions during hair and feather follicle regeneration. Journal of Molecular Biology. 2015;428(7):1423–40. 10.1016/j.jmb.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miao X, Luo Q, Qin X. Genome-wide transcriptome analysis in the ovaries of two goats identifies differentially expressed genes related to fecundity. Gene. 2016;582(1):69–76. 10.1016/j.gene.2016.01.047 [DOI] [PubMed] [Google Scholar]
- 5.Li YR, Fan WB, Li CQ, Yin J, Zhang YJ, Li JQ. Histomorphology research of the secondary follicle cycling of Inner Mongolia cashmere goat. Scientia Agricultura Sinica. 2008;41(11):3920–6. [Google Scholar]
- 6.Qi YX, Liu YB, Rong WH. RNA-Seq and its applications: a new technology for transcriptomics. Hereditas. 2011;33(11):1191–202. [DOI] [PubMed] [Google Scholar]
- 7.Marguerat S, Bähler J. RNA-seq: from technology to biology. Cellular & Molecular Life Sciences. 2010;67(4):569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geng R, Yuan C, Chen Y. Exploring differentially expressed genes by RNA-Seq in cashmere goat (Capra hircus) skin during hair follicle development and cycling. Plos One. 2013;8(4):e62704 10.1371/journal.pone.0062704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Cai B, Zhou J, Zhu H, Niu Y, Ma B, et al. Disruption of FGF5 in Cashmere Goats Using CRISPR/Cas9 Results in More Secondary Hair Follicles and Longer Fibers. Plos One. 2016;11(10):e0164640 10.1371/journal.pone.0164640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin M, Wang J, Chu MX, Piao J, Piao JA, Zhao FQ. The Study on Biological Function of Keratin 26, a Novel Member of Liaoning Cashmere Goat Keratin Gene Family. Plos One. 2016;11(12):e0168015 10.1371/journal.pone.0168015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004;116(6):769–78. [DOI] [PubMed] [Google Scholar]
- 12.Plikus MV, Mayer J, Cruz DDL, Baker RE, Maini PK, Maxson R, et al. Cyclic dermal BMP signaling regulates stem cell activation during hair regeneration. Nature. 2008;451(7176):340–4. 10.1038/nature06457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong Y, Xie M, Jiang Y, Xiao N, Du X, Zhang W, et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nature Biotechnology. 2013;31(2):135–41. 10.1038/nbt.2478 [DOI] [PubMed] [Google Scholar]
- 14.Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5(7):621 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 15.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29(16):1035–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Research. 2004;32(suppl_1):D258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogata H, Goto S, Fujibuchi W, Kanehisa M. Computation with the KEGG pathway database. Biosystems. 1998;47(1–2):119–28. [DOI] [PubMed] [Google Scholar]
- 18.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Research. 2004;32(suppl_1):277–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aoki-Kinoshita KF, Kanehisa M. Gene Annotation and Pathway Mapping in KEGG. Methods in Molecular Biology. 2007;396:71–91. 10.1007/978-1-59745-515-2_6 [DOI] [PubMed] [Google Scholar]
- 20.Torkamani N, Rufaut NW, Jones L, Sinclair RD. Beyond Goosebumps: Does the Arrector Pili Muscle Have a Role in Hair Loss? International journal of trichology. 2014;6(3):88 10.4103/0974-7753.139077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bie Q, Jin C, Zhang B, Dong H. IL-17B: A new area of study in the IL-17 family. Molecular Immunology, 2017, 90:50–56. 10.1016/j.molimm.2017.07.004 [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Fan YQ, Lv Z, Yao XJ, Huang KW, Meng Q, et al. Interleukin-25 promotes basic fibroblast growth factor expression by human endothelial cells through interaction with IL-17RB, but not IL-17RA. Clinical & Experimental Allergy. 2012;42(11):1604–14. [DOI] [PubMed] [Google Scholar]
- 23.Adachi T, Kobayashi T, Sugihara E, Yamada T, Ikuta K, Pittaluga S, et al. Hair follicle–derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nature Medicine. 2015;21(11):1272–9. 10.1038/nm.3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alinejad V, Dolati S, Motallebnezhad M, Yousefi M. The role of IL17B-IL17RB signaling pathway in breast cancer. Biomedicine & Pharmacotherapy. 2017;88:795–803. 10.1016/j.biopha.2017.01.120 [DOI] [PubMed] [Google Scholar]
- 25.Fabre J, Giustiniani J, Garbar C, Antonicelli F, Merrouche Y, Bensussan A, et al. Targeting the Tumor Microenvironment: The Protumor Effects of IL-17 Related to Cancer Type. International Journal of Molecular Sciences. 2016;17(9):1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds Joseph, nbsp Lee, YoungHee Shi, et al. Interleukin-17B Antagonizes Interleukin-25-Mediated Mucosal Inflammation. Immunity. 2015;42(4):692–703. 10.1016/j.immuni.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frankel AD, Berg JM, Pabo CO. Metal-dependent folding of a single zinc finger from transcription factor IIIA. Proceedings of the National Academy of Sciences. 1987;84(14):4841–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fantauzzo KA, Bazzi H, Jahoda CAB, Christiano AM. Dynamic expression of the zinc-finger transcription factor Trps1 during hair follicle morphogenesis and cycling. Gene Expression Patterns. 2008;8(2):51–7. 10.1016/j.modgep.2007.10.006 [DOI] [PubMed] [Google Scholar]
- 29.Oliver CH, Nichols J, Watson CJ. The KRAB domain zinc finger protein, Zfp157, is expressed in multiple tissues during mouse embryogenesis and in specific cells in adult mammary gland and skin. Genesis. 2013;51(3):179–86. 10.1002/dvg.22367 [DOI] [PubMed] [Google Scholar]
- 30.Jiang L, Wang H, Shi C, Liu K, Liu M, Wang N, et al. ZNF667/Mipu1 is a novel anti-apoptotic factor that directly regulates the expression of the rat Bax gene in H9c2 cells. Plos One. 2014;9(11):e111653–e. 10.1371/journal.pone.0111653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan YX, Wu RB, Qiao X, Zhang YJ, Wang RJ, Su R, et al. Hair follicle transcriptome profiles during the transition from anagen to catagen in Cashmere goat (Capra hircus). Genetics and Molecular Research. 2015;14(4):17904–15. 10.4238/2015.December.22.15 [DOI] [PubMed] [Google Scholar]
- 32.Xu T, Guo X, Wang H, Hao F, Du X, Gao X, et al. Differential gene expression analysis between anagen and telogen of Capra hircus skin based on the de novo assembled transcriptome sequence. Gene. 2013;520(1):30–8. 10.1016/j.gene.2013.01.068 [DOI] [PubMed] [Google Scholar]
- 33.Ji X-y, Wang J-x, Liu B, Zheng Z-q, Fu S-y, Tarekegn GM, et al. Comparative Transcriptome Analysis Reveals that a Ubiquitin-Mediated Proteolysis Pathway Is Important for Primary and Secondary Hair Follicle Development in Cashmere Goats. Plos One. 2016;11(10):e0156124 10.1371/journal.pone.0156124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Qu H, Jiang H, Zhao Z, Zhang Q. Transcriptome-Wide Comparative Analysis of microRNA Profiles in the Telogen Skins of Liaoning Cashmere Goats (Capra hircus) and Fine-Wool Sheep (Ovis aries) by Solexa Deep Sequencing. DNA and Cell Biology. 2016. 10.1089/dna.2015.3161 [DOI] [PubMed] [Google Scholar]
- 35.Bai WL, Dang YL, Wang JJ, Yin RH, Wang ZY, Zhu YB, et al. Molecular characterization, expression and methylation status analysis of BMP4 gene in skin tissue of Liaoning cashmere goat during hair follicle cycle. Genetica. 2016:1–11. [DOI] [PubMed] [Google Scholar]
- 36.Bai WL, Dang YL, Yin RH, Jiang WQ, Wang ZY, Zhu YB, et al. Differential Expression of microRNAs and their Regulatory Networks in Skin Tissue of Liaoning Cashmere Goat during Hair Follicle Cycles. Animal biotechnology. 2016;27(2):104–12. 10.1080/10495398.2015.1105240 [DOI] [PubMed] [Google Scholar]
- 37.Castela M, Linay F, Roy E, Moguelet P, Xu J, Holzenberger M, et al. IGF1R signaling acts on the anagen to catagen transition in the hair cycle. Experimental Dermatology. 2017. 10.1111/exd.13287 [DOI] [PubMed] [Google Scholar]
- 38.Ma X, Tian Y, Song Y, Shi J, Xu J, Xiong K, et al. Msi2 maintains quiescent state of hair follicle stem cells by directly repressing the Hh signaling pathway. Journal of investigative dermatology. 2017. 10.1016/j.jid.2017.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou G, Kang D, Ma S, Wang X, Gao Y, Yang Y, et al. Integrative analysis reveals ncRNA-mediated molecular regulatory network driving secondary hair follicle regression in cashmere goats. Bmc Genomics. 2018;19(1):222 10.1186/s12864-018-4603-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
The sequencing reads of each sequencing libraries have been deposited under NCBI with Project ID SUB3555047.