Abstract
Type 2 diabetes is typified by insulin-resistance in adipose tissue, skeletal muscle, and liver, leading to chronic hyperglycemia. Additionally, obesity and type 2 diabetes are characterized by chronic low-grade inflammation. Membrane-associated RING-CH-1 (MARCH1) is an E3 ubiquitin ligase best known for suppression of antigen presentation by dendritic and B cells. MARCH1 was recently found to negatively regulate the cell surface levels of the insulin receptor via ubiquitination. This, in turn, impaired insulin sensitivity in mouse models. Here, we report that MARCH1-deficient (knockout; KO) female mice exhibit excessive weight gain and excessive visceral adiposity when reared on standard chow diet, without increased inflammatory cell infiltration of adipose tissue. By contrast, male MARCH1 KO mice had similar weight gain and visceral adiposity to wildtype (WT) male mice. MARCH1 KO mice of both sexes were more glucose tolerant than WT mice. The levels of insulin receptor were generally higher in insulin-responsive tissues (especially the liver) from female MARCH1 KO mice compared to males, with the potential to account in part for the differences between male and female MARCH1 KO mice. We also explored a potential role for MARCH1 in human type 2 diabetes risk through genetic association testing in publicly-available datasets, and found evidence suggestive of association. Collectively, our data indicate an additional link between immune function and diabetes, specifically implicating MARCH1 as a regulator of lipid metabolism and glucose tolerance, whose function is modified by sex-specific factors.
Introduction
Metabolic syndrome represents one of the most pressing public health concerns worldwide. A general feature of metabolic syndrome is insulin resistance, hyperinsulinemia, and hyperglycemia [1, 2]. Insulin normally enhances glucose uptake by white adipose tissue and muscle [3], while suppressing hepatic gluconeogenesis [4] and encouraging whole body glucose disposal [5, 6]. Insulin-resistance, in turn, leads to a dysregulation of glucose metabolism and chronically results in overt type 2 diabetes mellitus [1].
Obesity increases the incidence of insulin resistance and type 2 diabetes [7]. Systemic and local (visceral adipose) inflammation has emerged as a key feature of obesity and the progression of type 2 diabetes [8, 9]. Adipocytes produce immune-active molecules, collectively termed adipokines, and adipose tissue contains a remarkable array of resident and infiltrating immune cells that can impact metabolism [10, 11]. Cellular stresses, such as ectopic lipid accumulation and elevated glucose exposure, instigate metabolic pathogenesis [12]. Notably, inflammation exacerbates both hyperglycemia and ectopic lipid accumulation [8, 13, 14]. Obesity increases macrophage and adaptive immune cell (B cell, CD8 T cell, and CD4 TH1 cell) infiltration into visceral adipose tissue, enhancing pro-inflammatory function and a pro-inflammatory cytokine expression profile [15–19]. This inflammatory state depresses insulin sensitivity, whereas anti-inflammatory T regulatory cells (Tregs) support insulin sensitivity [20].
The E3 ubiquitin ligase MARCH1 (Membrane-Associated RING-CH1; [21]) intersects with inflammation and insulin responsiveness. Its expression is strongest in lymphoid tissues and it functions in antigen-presenting cells (APCs) to negatively regulate, via ubiquitin-dependent effects on trafficking, the cell surface levels of Major Histocompatibility Complex class II (MHC-II) molecules and CD86 (a co-stimulatory protein) [21–23]. MARCH1 expression is decreased by APC activation/maturation signals [24, 25] and increased by anti-inflammatory stimuli, such as interleukin-10 [26]. Nagarajan, et al. recently showed that MARCH1 can target the insulin receptor for ubiquitin-dependent downregulation, negatively regulating insulin receptor signaling [27]. Accordingly, in vivo loss of MARCH1 in liver and adipose tissue correlated with improved glucose clearance, and MARCH1 over-expression impaired clearance [27].
Consistent with this new role for MARCH1, genome-wide association studies (GWAS) have provided modest evidence for an association between MARCH1 and type 2 diabetes. In a GWAS for type 2 diabetes in different Asian populations, MARCH1 was associated with type 2 diabetes in one ethnic group, but not two others [28]. Another GWAS in a Korean cohort detected an association between MARCH1 and body-mass index (BMI) [29].
During our experiments to characterize immunity in MARCH1 KO mice, we noted changes in adiposity that prompted us to investigate metabolic parameters. Herein, we report studies conducted to further assess the metabolic phenotype in male and female MARCH1 KO and wildtype mice. We also tested for a genetic association of variants in MARCH1 with type 2 diabetes in human case-control studies.
Materials and methods
Mice
Assessment of body weight, adiposity, glucose tolerance, and immune cell infiltration of adipose tissue
Sibling WT and MARCH1 KO experimental mice on the C57BL/6 background were derived through het-het breeding. Mice were reared ad libitum on NIH-31 chow (Harlan Laboratories; 7013). Body and food weights were taken weekly to assess body weight gain and food consumption. All mice were housed in the animal facility at the University of Arizona, and experiments were conducted under the approval and supervision of the Institutional Animal Care and Use Committee (IACUC). Mouse experiments were performed in a blinded fashion with respect to genotype.
Every effort was made to minimize animal suffering, in accordance with the University of Arizona IACUC guidelines. Mice were monitored regularly for any signs of disease and, if noted, veterinary staff were altered and mice promptly euthanized if they displayed signs of disease. Euthanasia was performed in accordance with the methods approved by the American Veterinary Medical Association panel on euthanasia, in which mice were exposed to CO2 from a compressed gas cylinder in a closed chamber in the University of Arizona animal facility. All participating personnel received training in the proper use of the CO2 chamber. Topical administration of ice-cold ethanol was used for anesthesia prior to blood collection from the tail vein, per IACUC guidelines.
Assessment of insulin receptor levels in liver, muscle, and adipose tissue
Littermate male and female C57BL/6 mice or littermate male and female MARCH1 KO mice (approximately 18 weeks old) were used in all tissue isolation experiments. Mice were bred and maintained in-house at the NCI-Frederick animal facility. All mice were cared for in accordance with National Institutes of Health guidelines with the approval of the National Cancer Institute Animal Care and Use Committee. Mice were euthanized by exposure to CO2 from a compressed gas cylinder in a closed chamber. All participating personnel received training in the proper use of the CO2 chamber.
Glucose measurements and glucose tolerance tests
Fasting blood glucose and intraperitoneal glucose tolerance tests (IPGTT; 1.5 mg/g body weight) were performed following an 8-hour fast, that was initiated at lights-on. Glucose was measured in blood collected from the tail vein using the FreeStyle Lite glucose monitoring system.
Collection of stromal-vascular fraction (SVF) of adipose and flow cytometry
Visceral adipose tissue (VAT) was dissociated with collagenase I in KRHB buffer for ~30–45 minutes at 37°C as described [30]. Digested tissue was filtered through a 100 μm cell strainer. Filtrate was centrifuged at 500 x g for 5 min, the pellet resuspended in KRHB, and the process repeated 4 times. The final pellet was collected as the SVF. Cell labeling for flow cytometry was performed as described [31]. Cells were analyzed on a FACSCalibur cytometer or an LSRII cytometer (BD Biosciences), and data were analyzed using FlowJo (Treestar). The gating schemes are given in S1 Fig and represent the gates used for quantitation of population percentages and cell numbers (obtained using CountBright Absolute Counting Beads according the manufacturer’s instructions; Life Technologies). The following antibodies were used: CD45.2 (clone 104), CD3 (17A2), CD11b (clone M1/70), MHC-II (clone M5/114.15.2), F4/80 (clone BM8), CD4 (clone L3T4), and CD8 (clone 53–6.7), all from eBioscience, and anti-mouse CD25 (clone PC61) from BioLegend.
Tissue isolation and immunoblotting
Mouse adipose tissue, skeletal muscle, and liver were isolated and immediately homogenized using a Precellys homogenizer (Bertin Corp.). Cells were solubilized in Triton X-100-containing lysis buffer (10 mM Tris-HCl,pH 7.4, 150 mM NaCl, 1% Triton X-100, 50 mM phenylmethylsulfonyl fluoride, 0.1 mM Nα-tosyl-L-lysine chloromethyl ketone hydrochloride, and 25 mM N-ethylmaleimide) for 1 h at 4°C. The protein content of each sample was determined using a Pierce BCA Protein Assay kit (Thermo Fisher Scientific). Proteins were separated by SDS-PAGE and transferred to PVDF membranes (Bio-Rad) as described previously [32]. Non-specific antibody binding sites on immunoblots were blocked by incubation overnight using 5% nonfat dry milk in PBS containing 0.1% Tween-20 (blotting buffer), washed, and incubated overnight with primary antibodies against insulin receptor β-subunit (Cell Signaling Technology) and appropriate secondary antibodies in blotting buffer. Immunoblots were re-probed using HRP-conjugated anti-GAPDH monoclonal antibody (Cell Signaling Technology) as a loading control. Immunoblots were quantitated and analyzed using a GS-900 calibrated densitometer and the software Image Lab (Bio-Rad) following the manufacturer's instructions. Quantitation of blot intensities was carried out by analysis of data obtained in the linear range of exposure. In each sample the ratio of insulin receptor β-subunit band intensity was expressed relative to the amount of GAPDH present.
MARCH1 single-nucleotide polymorphism (SNP) analyses
We performed a candidate gene investigation of MARCH1 SNPs in the following type 2 diabetes case-control datasets: the Northwestern NUgene (Vanderbilt University Medical Center, n = 3,357; Northwestern University, n = 5,812) and the Gene Environment Association Studies (GENEVA) in type 2 diabetes, which comprised participants from the Nurses’ Health Study (NHS, n = 3,314) and the Health Professional Follow-up Study (HPFS, n = 2,497). White participants in the NUgene study were genotyped on the Illumina 660W-Quadv1_A BeadChip platform, and African-American participants were genotyped on the Illumina 1M-Duo Beadchip platform [33]. All participants in the GENEVA study were genotyped on the Affymetrix 6.0 platform. The datasets for the analyses described in this manuscript were obtained through dbGaP (accession numbers phs000091.v2.p1 and phs000237.v1.p1). Details regarding the workflow and quality control decisions are described on the respective dbGaP webpages for each study.
Our objective was to identify SNPs associated with type 2 diabetes in two or more independent samples across platforms, as independent replication provides the strongest protection against false discovery. The four samples were first stratified by race/ethnicity to avoid confounding by population stratification, and 6 homogenous samples with a minimum sample size of 100 were identified for analysis (see Results and S1 Table). SNPs within the MARCH1 region of chromosome 4 (from position 165,300,000 to 164,445,000 of the GRCh37.p13 gene assembly) were considered. The number of SNPs in common across the groups was enumerated, given differing platforms, differing QC decisions across studies, and exclusion of SNPs with extreme allele frequency, to account for the potential for replication across studies/platforms.
Statistical analyses
All mouse data were analyzed in SAS Enterprise Guide 7.1 (SAS Institute, Cary, NC). Body weight and IPGTT were analyzed using a repeated-measures mixed-model ANOVA including genotype, time, and their interaction. Visceral adiposity as a proportion of body weight was analyzed separately for each sex using a one-way ANOVA with genotype as the main effect and age as a covariate. The results of the flow cytometric studies of the adipose SVF were analyzed by mixed ANOVA model including genotype, age, and genotype*age interactions. When appropriate, Tukey’s test was used to adjust for multiple comparisons.
The association between SNPs and type 2 diabetes was assessed using a log-additive logistic regression model, within each group. Figures were generated using the R package snp.plotter. Additional details are provided in the Results section. Overall, 497 SNPs were considered across the MARCH1 locus; only those SNPs shared between at least two platforms were considered.
The data used to produce the graphs in Figs 1–4 are housed in a public repository. The data are deposited within DRYAD (datadryad.org) with the accession number: doi:10.5061/dryad.r34k1.
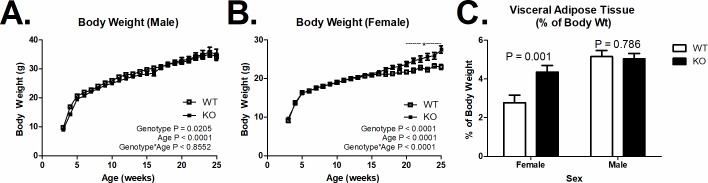
Fig 1. Body weight and visceral adiposity.
(A) Increased body weight in female mice MARCH1-deficient mice (KO) compared to wildtype mice (WT) on a chow diet after 20 weeks of age (WT n = 10–33; KO n = 12–41). (B) MARCH1-deficiency did not affect the age-related increase in body weight in male mice (WT n = 8–19; KO n = 7–28). (C) Increased visceral adiposity in female, but not male mice KO mice. Number (n) is indicated within bars). *indicates significant difference P < 0.05. Data are plotted at the mean +/- SEM.
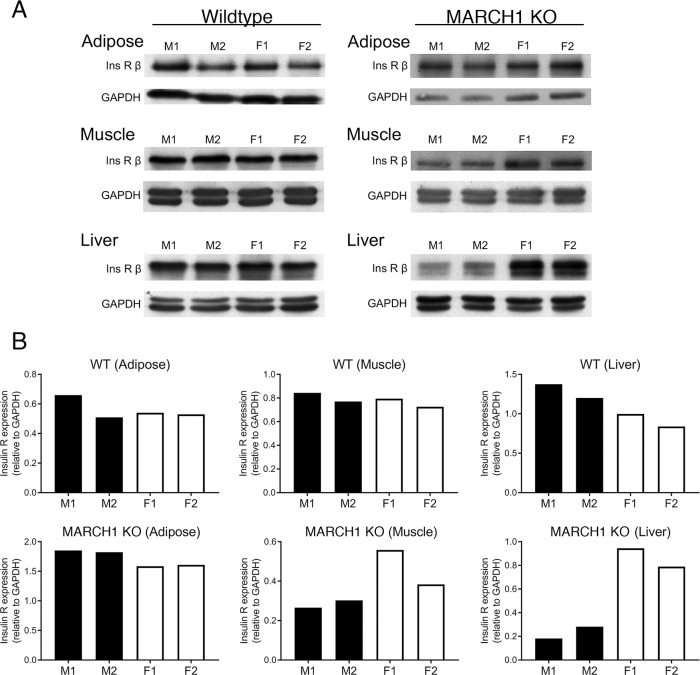
Fig 4. Comparison of insulin receptor levels in female versus male mice.
Proteins from adipose tissue, skeletal muscle, and liver from either WT or MARCH1 KO mice were isolated, separated by SDS-PAGE, and immunoblotted using antibodies recognizing insulin receptor β-chain and GAPDH (as a loading control). Each group consisted of two male and two female littermates. (A) Immunoblots showing the expression levels of insulin receptor β-chain (Ins R β) and GAPDH present in each sample. (B) Immunoblots were scanned by laser densitometry and band intensity was quantified. The expression level of insulin receptor β-chain was plotted relative to the amount of GAPDH present in each sample.
Results
MARCH1 deficiency results in sex-specific changes in body weight and adiposity
Body weight was similar throughout time in male KO versus WT mice (Fig 1A; P > 0.05). However, MARCH1 deficiency increased body weight in female mice from 20–25 weeks of age compared to WT female mice (Fig 1B; P < 0.05). This increase in body weight may be associated with increased adiposity, as the ratio of visceral adipose tissue to body weight was higher in female MARCH1 KO mice than in MARCH1 WT mice (Fig 1C; P = 0.001). In line with the lack of body weight change, MARCH1 deficiency did not affect visceral adiposity in male mice (P = 0.79).
MARCH1 deficiency increases glucose clearance
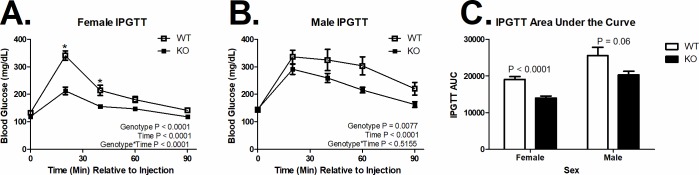
MARCH1 deficiency did not alter basal blood glucose (P > 0.5). However, MARCH1 deficiency improved glucose clearance assessed by IPGTT in both female (Fig 2A; P < 0.0001) and male (Fig 2B; P = 0.0077) mice. In fact, MARCH1 deficiency decreased the area under the curve (AUC) during the IPGTT by 26.7% (Fig 2C; P < 0.0001) in female mice and 20.5% (P = 0.06) in male mice.
Fig 2. Glucose tolerance.
(A) MARCH1 deletion (KO) decreased glucose clearance in female (WT n = 10, KO n = 12) and (B) male (WT n = 8, KO n = 7) mice. (C) Area under the glucose tolerance curve by sex and genotype (n is indicated within bars). *Indicates significant difference at that time point (P < 0.05). Data are plotted at the mean +/- SEM.
Leukocyte numbers in VAT are unaffected by MARCH1
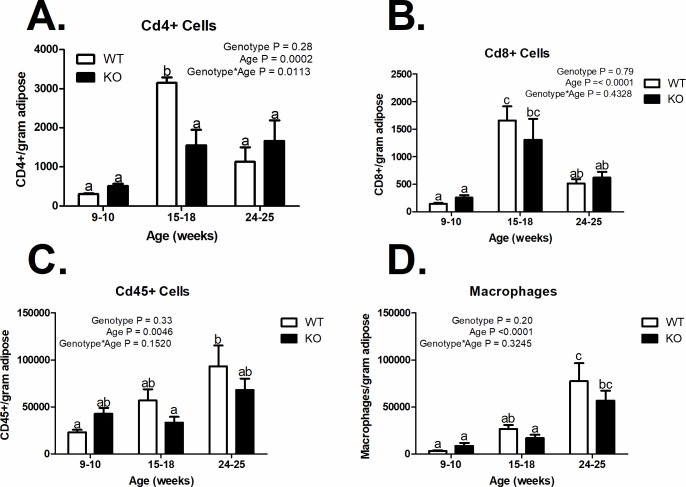
The increased visceral adipose tissue accumulation in MARCH1 KO females prompted us to study adipose tissue inflammation. We performed flow cytometric analysis of the stromal-vascular fraction (SVF) purified from VAT of MARCH1 KO and WT female mice to determine the extent of leukocyte infiltration. S1 Fig depicts a typical gating scheme for analysis of hematopoietic lineage cells in SVF. Total SVF leukocytes, CD4+ cells, CD8+ cells, and macrophages did not differ between KO and WT mice, irrespective of age (Fig 3). As expected, the MARCH1 KO macrophages expressed higher levels of MHC-II (S1 Fig). Data are plotted at the mean +/- SEM.
Fig 3. Immune cell infiltration of visceral adipose tissue.
(A) CD4+ cells (n = 3), (B) CD8+ cells (n = 3), (C) CDd45+ cells (n = 3–6), and (D) Macrophages/gram of visceral adipose tissue (n = 3–6). a, b, c = bars that share a common superscript do not differ significantly (P > 0.05). The numbers are based on flow cytometric analyses using the gating and scheme shown in S1 Fig. Data are plotted at the mean +/- SEM.
MARCH1 deficiency produces greater rescue of insulin receptor levels in females
To test whether the differences observed in female versus male mice could be explained, at least in part, by differential impact of MARCH1 deficiency on insulin receptor levels, immunoblots were performed with protein lysates from adipose, skeletal muscle, and liver (Fig 4). Receptor levels in tissues from female and male littermates were compared for wildtype or MARCH1 KO mice. In wildtype mice, the receptor levels were comparable in both sexes. In the absence of MARCH1, however, female mice had higher levels of the insulin receptor than males in the liver and to a lesser extent, in muscle; no difference was observed in adipose tissue.
MARCH1 and type 2 diabetes in human populations
Our findings in mice, along with published genetic associations studies [28, 29], implicating MARCH1 in metabolic disease, prompted us to test for an association between MARCH1 and type 2 diabetes in humans using GWAS datasets. Within the NUgene set, there were 139 non-Hispanic whites and 1,354 non-Hispanic blacks available for analysis. The NWU sample included 1,134 non-Hispanic whites and 242 non-Hispanic blacks. The HPFS included 2,397 non-Hispanic whites, and the NHS included 3,222 non-Hispanic whites (S1 Table). Other racial/ethnic groups were not sufficiently represented in these studies to allow for meaningful analyses. These six groups were then described in terms of case-control composition, sex, and family history of type 2 diabetes (S2 Table).
Logistic regression results are displayed graphically by sub-study (S2–S4 Figs). Complete results are also shown in tabular format, including minor allele frequency, odds-ratio for type 2 diabetes case status, raw p-value, Benjamini and Hochberg corrected p-value, and Bonferroni corrected p-value (S1 File). A total of 497 SNPs were available in at least two samples. Briefly, the number of SNPs with association (p<0.05, uncorrected for multiple comparisons) in each sample was as follows: HPFS, 15; NHS, 9; VU 660W platform, 5; NWU 660W platform, 6; VU 1M platform, 13; and NWU 1M platform, 26, though none were significant after correction for multiple comparisons (S1 File). Seven SNPs had significant associations (uncorrected) with type 2 diabetes in more than one sample, though not all of these associations were consistent across samples with respect to the direction of the association (Table 1). Two SNPs were found to be associated (p<0.05) with increased odds of type 2 diabetes in two analysis groups; rs12500778 in NHS and NWU, and rs17578337 in VU Whites and VU Blacks (Table 1). Both SNPs are intronic and lie at distance of ≈172 kb from each other.
Table 1. MARCH1 SNPs that are significantly associated with type 2 diabetes in more than one study.
| Study and Platforma | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPFS_Affy | NHS_Affy | VU_660W | NWU_660W | VU_1M | NWU_1M | |||||||||||||||||
| SNP | Locationb | MAF | OR | Raw p | MAF | OR | Raw p | MAF | OR | Raw p | MAF | OR | Raw p | MAF | OR | Raw p | MAF | OR | Raw p | |||
| rs13105536 | 164790122 | 28 | 0.98 | 0.8113 | 27.1 | 0.98 | 0.68329 | 31.3 | 0.93 | 0.78779 | 27.6 | 1.21 | 0.04539 | 6 | 0.64 | 0.00787 | 4.8 | 0.44 | 0.056 | |||
| rs17044105 | 164887559 | 2.3 | 1.49 | 0.03967 | 2.1 | 0.7 | 0.04321 | . | . | . | . | . | . | . | . | . | . | . | . | |||
| rs12500778 | 164939833 | 3.7 | 0.95 | 0.71279 | 4.2 | 1.32 | 0.02804 | . | . | . | . | . | . | 35.3 | 1.03 | 0.73125 | 35.1 | 1.98 | 0.0014 | |||
| rs10517789 | 164946712 | 2.6 | 1.57 | 0.01338 | 2.6 | 0.62 | 0.00408 | . | . | . | . | . | . | . | . | . | . | . | . | |||
| rs1494284 | 165016836 | . | . | . | . | . | 45.3 | 1.86 | 0.02901 | 46.1 | 1.17 | 0.06607 | 22.6 | 1.05 | 0.57169 | 23.1 | 0.64 | 0.0482 | ||||
| rs6821574 | 165021304 | . | . | . | . | . | . | 44.9 | 1.94 | 0.0193 | 46.5 | 1.16 | 0.07498 | 37.2 | 0.95 | 0.51311 | 38.4 | 0.66 | 0.0287 | |||
| rs17578337 | 165111724 | 28.5 | 0.91 | 0.13834 | 30.1 | 1.05 | 0.39701 | 30.2 | 1.94 | 0.03907 | 29.6 | 1.16 | 0.09526 | 7.7 | 1.35 | 0.03963 | 8.7 | 0.76 | 0.3962 | |||
OR greater than 1 (p<0.05) in orange; OR less than 1 (p<0.05) in blue; Raw p<0.05 in green
HPFS, Health Professional Follow-up Study; NHS, Nurses’ Health Study; VU, Vanderbilt University; NWU, Northwestern University; SNP, Single-nucleotide polymorphism; MAF, minor allele frequence; OR, odds ratio
aDesciption of the studies and platforms is provided in the Materials and Methods
bSNP location on chromosome 4
Given the sex differences we observed in our mice with respect to VAT accumulation, we stratified our data based on sex and BMI for the top 7 hits. Using a log-additive logistic regression model, none of the SNPs were significantly associated with sex or BMI, nor did age modify the association in these studies (P > 0.05). Further, genotype did not modify the association between BMI and type 2 diabetes (P > 0.05).
Discussion
Until recently, our knowledge of MARCH1 function came from the study of its role in antigen presentation to T cells [23, 34]. However, at least in overexpression studies, MARCH1 could regulate other molecules, as well [21]. Nagarajan, et al. recently found that MARCH1 causes degradation of the insulin receptor. MARCH1 loss improved insulin sensitivity in male and female mice on a normal chow diet, and MARCH1 did not alter weight gain or adiposity in male mice on chow diet [27]. We made similar findings in male mice, but noted that female chow-fed MARCH1 KO mice gained more weight and had more VAT than WT controls. The enhanced glucose uptake and resulting lipogenesis may explain the increased adiposity in female mice, but this same mechanism would be expected to induce similar effects in male mice. The increased adipose tissue lipid storage in females would reduce ectopic fat deposits and improve insulin action [35].
Sex differences in adipose tissue accumulation and function are multifactorial and many potential mechanisms may explain how MARCH1 exerts its sex-specific effect(s) [36, 37]. Both sexes of MARCH1 KO mice show improved glucose handling with loss of MARCH1, indicating that MARCH1 targets the insulin receptor in either case. However, it is possible that the magnitude of the MARCH1 effect on insulin receptor levels is more pronounced in females and could vary in different tissues. Indeed, when we compared insulin receptor levels between sexes in the absence of MARCH1, there were markedly higher levels of insulin receptor expression in the liver of female MARCH1 KO mice. This trend was less pronounced in skeletal muscle and not apparent in adipose tissue. Furthermore, it is known that visceral (perigonadal) adipocytes from female mice are more sensitive to insulin, in terms of activation of signaling cascades downstream for the insulin receptor and induction of lipogenesis [38]. Notably, this increased sensitivity in female adipocytes is not correlated with changes in insulin receptor expression levels [38]. Therefore, MARCH1-mediated downregulation of the insulin receptor may be greater in some tissues in female mice (and therefore MARCH1 loss leads to a greater restoration of insulin receptor levels), and/or an equivalent degree of insulin receptor upregulation with MARCH1 loss between sexes may have greater metabolic consequences in females; these possibilities remain to be fully tested, as does the manner in which sex differences impact the effect of MARCH1 on the insulin receptor. Regardless, the MARCH1 knockout mouse may be useful to identify sex-specific metabolic regulation or test for mechanisms regulating metabolic homeostasis in the absence of dietary stress.
In addition to the discovery that MARCH1 regulates the insulin receptor, it is possible that MARCH1 also impacts metabolism through its regulation of antigen presentation. Macrophages express relatively high levels of MARCH1 [22] and have come to occupy, along with adipocytes, a central position in immune homeostasis in adipose tissue [16, 39]. However, we did not detect obvious changes in leukocyte accumulation in adipose +/- MARCH1. Nonetheless, our findings warrant further analysis of the role for MARCH1 in metabolic substrate use and demands of different tissues.
The importance of MARCH1 in adipose tissue and liver was likely obscured by its relatively low expression in non-lymphoid tissues [21]. MARCH1 expression in APCs is regulated by multiple mechanisms; its levels generally decrease in pro-inflammatory settings and increase under anti-inflammatory conditions [24–26, 31, 40, 41]. Further, MARCH1 is an inherently unstable protein that is difficult to detect even in APCs [24, 31]. Though MARCH1 expression is relatively low in typical insulin-responsive tissues, this low level is clearly important to regulate the insulin receptor [27]. It is notable that MARCH1 mRNA in the liver is decreased by insulin treatment and that its levels increase in obese adipose tissue [27]. Thus, MARCH1 is broadly expressed, regulated by key metabolic hormones, and is a central regulator of insulin receptor homeostasis.
Two previous studies reported an association of MARCH1 with metabolic phenotypes in humans (type 2 diabetes and BMI; [28, 29]). Our human candidate gene investigation was not strongly supportive after correction for multiple testing, though two SNPs were nominally associated (p<0.05) with increased odds of type 2 diabetes in independent samples/platforms (Table 1). Whether these findings are indicative of causal variants or false discovery is unknown, since the associations were not statistically significant according to our significance threshold adjusted for multiple testing. We have made our results available so that other research groups may investigate further. The fact that the genetic associations reported thus far have all been in Asian populations, which were not part of our study, may indicate that MARCH1’s impact on metabolism varies by race/ethnicity.
The discovery that MARCH1 regulates glucose handling provides a new avenue to explore metabolic regulation and disease. The fact that, in mice, MARCH1 has sex-specific effects on lipid storage further indicates that the study of MARCH1 will yield insights into the differences between males and females with respect to metabolic diseases. Lastly, it will be important to leverage the MARCH1 KO mouse system to explore MARCH1’s dual roles in metabolism and immune regulation, and determine potential links between these processes.
Supporting information
Excel file containing the association data for all SNPs tested for association with type 2 diabetes. Abbreviations and legend are provided at the bottom of the table.
(XLSX)
(A) Light-scatter gating of the SVF stained with an isotype control (negative control) antibody. (B) SVF stained with anti-CD45 antibody. (C) CD11b and F4/80 staining of CD45+ cells. (D) MHC-II expression on the CD11b, F4/80 double-positive cells from wildtype (blue) and MARCH1-deficient mice. (E-G) Gating scheme for CD4+ and CD8+ T cells. (E) SVF stained with isotype control antibody. (F) CD45+ cell gate shown with a threshold applied so that the CD45-negative cells are not displayed. (G) CD4 and CD8 staining of the CD45+ cells.
(PDF)
SNP location is plotted on the x-axis, and the -ln(p-value) is on the y-axis. Up-pointing triangles indicate increased odds of type 2 diabetes, and down-pointing triangles indicate decreased odds of type 2 diabetes. Red triangles represent data from Northwestern University, and black triangles represent data from Vanderbilt University.
(PDF)
SNP location is plotted on the x-axis, and the -ln(p-value) is on the y-axis. Up-pointing triangles indicate increased odds of type 2 diabetes, and down-pointing triangles indicate decreased odds of type 2 diabetes. Red triangles represent data from Northwestern University, and black triangles represent data from Vanderbilt University.
(PDF)
SNP location is plotted on the x-axis, and the -ln(p-value) is on the y-axis. Up-pointing triangles indicate increased odds of type 2 diabetes, and down-pointing triangles indicate decreased odds of type 2 diabetes. Blue triangles represent data from the Health Professionals Follow-up Study, and red triangles represent data from the Nurse’s Health Study.
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors wish to thank all the organizers and participants associated with the GENEVA and NuGENE studies, and for making their data available through the dbGaP resource.
Funding support for the GWAS of Gene and Environment Initiatives in Type 2 Diabetes was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01HG004399).
The human subjects participating in the GWAS derive from The Nurses’ Health Study and Health Professionals’ Follow-up Study and these studies are supported by National Institutes of Health grants CA87969, CA55075, and DK58845. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the Gene Environment Association Studies, GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Funding support for genotyping, which was performed at the Broad Institute of MIT and Harvard, was provided by the NIH GEI (U01HG004424). The datasets used for the analyses described in this manuscript were obtained from dbGaP at [http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gap] through dbGaP accession number [phs000091].
Samples and data used in this study were provided by the NUgene Project (www.nugene.org). Funding support for the NUgene Project was provided by the Northwestern University’s Center for Genetic Medicine, Northwestern University, and Northwestern Memorial Hospital. Assistance with phenotype harmonization was provided by the eMERGE Coordinating Center (Grant number U01HG04603). This study was funded through the NIH, NHGRI eMERGE Network (U01HG004609). Funding support for genotyping, which was performed at The Broad Institute, was provided by the NIH (U01HG004424). Assistance with phenotype harmonization and genotype data cleaning was provided by the eMERGE Administrative Coordinating Center (U01HG004603) and the National Center for Biotechnology Information (NCBI). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/gap through dbGaP accession number phs000237.
Funding support for the Vanderbilt Genome-Electronic Records (VGER) project was provided through a cooperative agreement (U01HG004603) with the National Human Genome Research Institute (NHGRI) with additional funding from the National Institute of General Medical Sciences (NIGMS). The dataset and samples used for the VGER analyses were obtained from Vanderbilt University Medical Center's BioVU, which is supported by institutional funding and by the Vanderbilt CTSA grant UL1RR024975 from NCRR/NIH. Funding support for genotyping, which was performed at The Broad Institute, was provided by the NIH (U01HG004424). Assistance with phenotype harmonization and genotype data cleaning was provided by the eMERGE Administrative Coordinating Center (U01HG004603) and the National Center for Biotechnology Information (NCBI).
Data Availability
All relevant data to the mouse studies are within the manuscript, its Supporting Information files, or deposited within DRYAD (datadryad.org) with the accession number: doi:10.5061/dryad.r34k1. The genetic association study utilized data made available to approved users through The Database of Genotypes and Phenotypes (dbGaP) hosted by the The National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/gap). Approval may be requested from the following site: https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login, and requires that requesters satisfy the policies of dpGAP. The datasets utilized for this study had the following accession numbers: phs000091.v2.p1 and phs000237.v1.p1. The authors of this study did not have any special access privileges that other approved users would not have. Approved users are not permitted to distribute data to third parties.
Funding Statement
This project was supported by National Institutes of Health grant 1R56AI080756-01A2 (to L. Lybarger), start-up funds from the University of Arizona (to L. Lybarger), and the Intramural Research Program of the National Institutes of Health (to P. Roche and J. Bandola-Simon). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Johnson Andrew MF, Olefsky Jerrold M. The Origins and Drivers of Insulin Resistance. Cell. 2013;152(4):673–84. 10.1016/j.cell.2013.01.041 [DOI] [PubMed] [Google Scholar]
- 2.Muoio DM, Newgard CB. Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9(3):193–205. Epub 2008/01/18. 10.1038/nrm2327 . [DOI] [PubMed] [Google Scholar]
- 3.Basu A, Dalla Man C, Basu R, Toffolo G, Cobelli C, Rizza RA. Effects of type 2 diabetes on insulin secretion, insulin action, glucose effectiveness, and postprandial glucose metabolism. Diabetes care. 2009;32(5):866–72. Epub 2009/02/07. 10.2337/dc08-1826 ; PubMed Central PMCID: PMCPmc2671126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L, Sabet A, Djedjos S, Miller R, Sun X, Hussain MA, et al. Metformin and insulin suppress hepatic gluconeogenesis through phosphorylation of CREB binding protein. Cell. 2009;137(4):635–46. Epub 2009/05/20. 10.1016/j.cell.2009.03.016 ; PubMed Central PMCID: PMCPmc2775562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashcroft FM, Rorsman P. Diabetes mellitus and the beta cell: the last ten years. Cell. 2012;148(6):1160–71. Epub 2012/03/20. 10.1016/j.cell.2012.02.010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414(6865):799–806. Epub 2001/12/14. 10.1038/414799a . [DOI] [PubMed] [Google Scholar]
- 7.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444(7121):840–6. Epub 2006/12/15. 10.1038/nature05482 . [DOI] [PubMed] [Google Scholar]
- 8.Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol. 2011;11(2):98–107. 10.1038/nri2925 [DOI] [PubMed] [Google Scholar]
- 9.Arkan MC, Hevener AL, Greten FR, Maeda S, Li ZW, Long JM, et al. IKK-beta links inflammation to obesity-induced insulin resistance. Nat Med. 2005;11(2):191–8. Epub 2005/02/03. 10.1038/nm1185 . [DOI] [PubMed] [Google Scholar]
- 10.Mathis D. Immunological Goings-on in Visceral Adipose Tissue. Cell Metabolism. 2013;17(6):851–9. 10.1016/j.cmet.2013.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–45. Epub 2011/01/12. 10.1146/annurev-immunol-031210-101322 . [DOI] [PubMed] [Google Scholar]
- 12.Robertson RP, Harmon J, Tran PO, Poitout V. β-cell glucose toxicity, lipotoxicity, and chronic oxidative stress in type 2 diabetes. Diabetes. 2004;53:S119–S24. [DOI] [PubMed] [Google Scholar]
- 13.Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nature Rev Immunol. 2008;8:923–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donath MY, Storling J, Maedler K, Mandrup-Poulsen T. Inflammatory mediators and islet β-cell failure: a link between type 1 and type 2 diabetes. J Mol Med. 2003;81:455–70. 10.1007/s00109-003-0450-y [DOI] [PubMed] [Google Scholar]
- 15.Bhargava P, Lee CH. Role and function of macrophages in the metabolic syndrome. Biochemical Journal. 2012;442(2):253–62. 10.1042/BJ20111708 [DOI] [PubMed] [Google Scholar]
- 16.Dalmas E, Clément K, Guerre-Millo M. Defining macrophage phenotype and function in adipose tissue. Trends in Immunology. 2011;32(7):307–14. 10.1016/j.it.2011.04.008 [DOI] [PubMed] [Google Scholar]
- 17.Winer DA, Winer S, Shen L, Wadia PP, Yantha J, Paltser G, et al. B cells promote insulin resistance through modulation of T cells and production of pathogenic IgG antibodies. Nat Med. 2011;17(5):610–7. http://www.nature.com/nm/journal/v17/n5/abs/nm.2353.html#supplementary-information. 10.1038/nm.2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimura S, Manabe I, Nagasaki M, Eto K, Yamashita H, Ohsugi M, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15(8):914–20. http://www.nature.com/nm/journal/v15/n8/suppinfo/nm.1964_S1.html. 10.1038/nm.1964 [DOI] [PubMed] [Google Scholar]
- 19.Winer S, Chan Y, Paltser G, Truong D, Tsui H, Bahrami J, et al. Normalization of obesity-associated insulin resistance through immunotherapy. Nat Med. 2009;15(8):921–9. http://www.nature.com/nm/journal/v15/n8/suppinfo/nm.2001_S1.html. 10.1038/nm.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nature medicine. 2009;15(8):930–9. Epub 2009/07/28. 10.1038/nm.2002 ; PubMed Central PMCID: PMC3115752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartee E, Mansouri M, Hovey Nerenberg BT, Gouveia K, Fruh K. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. Journal of virology. 2004;78(3):1109–20. 10.1128/JVI.78.3.1109-1120.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuki Y, Ohmura-Hoshino M, Goto E, Aoki M, Mito-Yoshida M, Uematsu M, et al. Novel regulation of MHC class II function in B cells. The EMBO journal. 2007;26(3):846–54. 10.1038/sj.emboj.7601556 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishido S, Matsuki Y, Goto E, Kajikawa M, Ohmura-Hoshino M. MARCH-I: A new regulator of dendritic cell function. Molecules and Cells. 2010;29(3):229–32. 10.1007/s10059-010-0051-x [DOI] [PubMed] [Google Scholar]
- 24.De Gassart A, Camosseto V, Thibodeau J, Ceppi M, Catalan N, Pierre P, et al. MHC class II stabilization at the surface of human dendritic cells is the result of maturation-dependent MARCH I down-regulation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(9):3491–6. 10.1073/pnas.0708874105 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walseng E, Furuta K, Goldszmid RS, Weih KA, Sher A, Roche PA. Dendritic Cell Activation Prevents MHC Class II Ubiquitination and Promotes MHC Class II Survival Regardless of the Activation Stimulus. Journal of Biological Chemistry. 2010;285(53):41749–54. 10.1074/jbc.M110.157586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thibodeau J, Bourgeois-Daigneault MC, Huppe G, Tremblay J, Aumont A, Houde M, et al. Interleukin-10-induced MARCH1 mediates intracellular sequestration of MHC class II in monocytes. Eur J Immunol. 2008;38(5):1225–30. 10.1002/eji.200737902 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagarajan A, Petersen MC, Nasiri AR, Butrico G, Fung A, Ruan HB, et al. MARCH1 regulates insulin sensitivity by controlling cell surface insulin receptor levels. Nature communications. 2016;7:12639 10.1038/ncomms12639 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sim X, Ong RT-H, Suo C, Tay W-T, Liu J, Ng DP-K, et al. Transferability of Type 2 Diabetes Implicated Loci in Multi-Ethnic Cohorts from Southeast Asia. PLoS Genet. 2011;7(4):e1001363 10.1371/journal.pgen.1001363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M, Kwon DY, Kim M-S, Choi CR, Park M-Y, Kim A-j. Genome-wide association study for the interaction between BMR and BMI in obese Korean women including overweight. Nutr Res Pract. 2016;10(1):115–24. 10.4162/nrp.2016.10.1.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson ACS, Nunez M, Davidson R, Horm T, Schnittker K, Hart MV, et al. Mitigation of isolation-associated adipocyte interleukin-6 secretion following rapid dissociation of adipose tissue. Journal of Lipid Research. 2012. 10.1194/jlr.D031286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jabbour M, Campbell EM, Fares H, Lybarger L. Discrete Domains of MARCH1 Mediate Its Localization, Functional Interactions, and Posttranscriptional Control of Expression. J Immunol. 2009;183(10):6500–12. 10.4049/jimmunol.0901521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poloso NJ, Muntasell A, Roche PA. MHC Class II Molecules Traffic into Lipid Rafts during Intracellular Transport. The Journal of Immunology. 2004;173(7):4539–46. 10.4049/jimmunol.173.7.4539 [DOI] [PubMed] [Google Scholar]
- 33.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC medical genomics. 2011;4:13 10.1186/1755-8794-4-13 ; PubMed Central PMCID: PMC3038887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh J, Shin J-S. Molecular mechanism and cellular function of MHCII ubiquitination. Immunological Reviews. 2015;266(1):134–44. 10.1111/imr.12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9(5):367–77. 10.1038/nrm2391 ; PubMed Central PMCID: PMC2886982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White UA, Tchoukalova YD. Sex dimorphism and depot differences in adipose tissue function. Biochimica et Biophysica Acta (BBA)—Molecular Basis of Disease. 2014;1842(3):377–92. 10.1016/j.bbadis.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fried SK, Lee MJ, Karastergiou K. Shaping fat distribution: New insights into the molecular determinants of depot- and sex-dependent adipose biology. Obesity (Silver Spring). 2015;23(7):1345–52. 10.1002/oby.21133 ; PubMed Central PMCID: PMCPMC4687449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macotela Y, Boucher J, Tran TT, Kahn CR. Sex and Depot Differences in Adipocyte Insulin Sensitivity and Glucose Metabolism. Diabetes. 2009;58(4):803–12. 10.2337/db08-1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraakman MJ, Murphy AJ, Jandeleit-Dahm K, Kammoun HL. Macrophage Polarization in Obesity and Type 2 Diabetes: Weighing Down our Understanding of Macrophage Function? Frontiers in Immunology. 2014;5 10.3389/fimmu.2014.00470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tze LE, Horikawa K, Domaschenz H, Howard DR, Roots CM, Rigby RJ, et al. CD83 increases MHC II and CD86 on dendritic cells by opposing IL-10–driven MARCH1-mediated ubiquitination and degradation. The Journal of experimental medicine. 2011;208(1):149–65. 10.1084/jem.20092203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galbas T, Steimle V, Lapointe R, Ishido S, Thibodeau J. MARCH1 down-regulation in IL-10-activated B cells increases MHC class II expression. Cytokine. 2012;59(1):27–30. 10.1016/j.cyto.2012.03.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel file containing the association data for all SNPs tested for association with type 2 diabetes. Abbreviations and legend are provided at the bottom of the table.
(XLSX)
(A) Light-scatter gating of the SVF stained with an isotype control (negative control) antibody. (B) SVF stained with anti-CD45 antibody. (C) CD11b and F4/80 staining of CD45+ cells. (D) MHC-II expression on the CD11b, F4/80 double-positive cells from wildtype (blue) and MARCH1-deficient mice. (E-G) Gating scheme for CD4+ and CD8+ T cells. (E) SVF stained with isotype control antibody. (F) CD45+ cell gate shown with a threshold applied so that the CD45-negative cells are not displayed. (G) CD4 and CD8 staining of the CD45+ cells.
(PDF)
SNP location is plotted on the x-axis, and the -ln(p-value) is on the y-axis. Up-pointing triangles indicate increased odds of type 2 diabetes, and down-pointing triangles indicate decreased odds of type 2 diabetes. Red triangles represent data from Northwestern University, and black triangles represent data from Vanderbilt University.
(PDF)
SNP location is plotted on the x-axis, and the -ln(p-value) is on the y-axis. Up-pointing triangles indicate increased odds of type 2 diabetes, and down-pointing triangles indicate decreased odds of type 2 diabetes. Red triangles represent data from Northwestern University, and black triangles represent data from Vanderbilt University.
(PDF)
SNP location is plotted on the x-axis, and the -ln(p-value) is on the y-axis. Up-pointing triangles indicate increased odds of type 2 diabetes, and down-pointing triangles indicate decreased odds of type 2 diabetes. Blue triangles represent data from the Health Professionals Follow-up Study, and red triangles represent data from the Nurse’s Health Study.
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data to the mouse studies are within the manuscript, its Supporting Information files, or deposited within DRYAD (datadryad.org) with the accession number: doi:10.5061/dryad.r34k1. The genetic association study utilized data made available to approved users through The Database of Genotypes and Phenotypes (dbGaP) hosted by the The National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/gap). Approval may be requested from the following site: https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login, and requires that requesters satisfy the policies of dpGAP. The datasets utilized for this study had the following accession numbers: phs000091.v2.p1 and phs000237.v1.p1. The authors of this study did not have any special access privileges that other approved users would not have. Approved users are not permitted to distribute data to third parties.