Abstract
Whole-genome duplication (WGD) has been a major evolutionary driver of increased genomic complexity in vertebrates. One such event occurred in the salmonid family ∼80 Ma (Ss4R) giving rise to a plethora of structural and regulatory duplicate-driven divergence, making salmonids an exemplary system to investigate the evolutionary consequences of WGD. Here, we present a draft genome assembly of European grayling (Thymallus thymallus) and use this in a comparative framework to study evolution of gene regulation following WGD. Among the Ss4R duplicates identified in European grayling and Atlantic salmon (Salmo salar), one-third reflect nonneutral tissue expression evolution, with strong purifying selection, maintained over ∼50 Myr. Of these, the majority reflect conserved tissue regulation under strong selective constraints related to brain and neural-related functions, as well as higher-order protein–protein interactions. A small subset of the duplicates have evolved tissue regulatory expression divergence in a common ancestor, which have been subsequently conserved in both lineages, suggestive of adaptive divergence following WGD. These candidates for adaptive tissue expression divergence have elevated rates of protein coding- and promoter-sequence evolution and are enriched for immune- and lipid metabolism ontology terms. Lastly, lineage-specific duplicate divergence points toward underlying differences in adaptive pressures on expression regulation in the nonanadromous grayling versus the anadromous Atlantic salmon. Our findings enhance our understanding of the role of WGD in genome evolution and highlight cases of regulatory divergence of Ss4R duplicates, possibly related to a niche shift in early salmonid evolution.
Keywords: Thymallus thymallus, genome assembly; salmonid; WGD; rediploidization; lineage-specific ohnolog resolution
Introduction
Whole-genome duplication (WGD) through spontaneous doubling of all chromosomes (autopolyploidization) has played a vital role in the evolution of vertebrate genome complexity (Van de Peer et al. 2009). However, the role of selection in shaping novel adaptations from the redundancy that arises from WGD is not well understood. The idea that functional redundancy arising from gene duplication sparks evolution of novel traits and adaptations was pioneered by Ohno (1970). Duplicate genes that escape loss or pseudogenization are known to acquire novel regulation and expression divergence (Lynch and Conery 2000; Zhang 2003; Conant and Wolfe 2008). Functional genomic studies over the past decade have demonstrated that large-scale duplications lead to the rewiring of regulatory networks through divergence of spatial and temporal expression patterns (Osborn et al. 2003; De Smet et al. 2017). As changes in gene regulation are known to be important in the evolution of phenotypic diversity and complex trait variation (Carroll 2000; Wray 2003), these post-WGD shifts in expression regulation may provide a substrate for adaptive evolution. Several studies have investigated the genome-wide consequences of WGD on gene expression evolution in vertebrates (Sémon and Wolfe 2008; Kassahn et al. 2009; Berthelot et al. 2014; Li et al. 2015; Acharya and Ghosh 2016; Lien et al. 2016; Robertson et al. 2017) and have revealed that a large proportion of gene duplicates have evolved substantial regulatory divergence of which, in most cases, one copy retains ancestral-like regulation (consistent with Ohno’s model of regulatory neofunctionalization). However, to what extent this divergence in expression is linked to adaptation remains to be understood. A major factor contributing to this knowledge gap is the lack of studies that integrate expression data from multiple species sharing the same WGD (Hermansen et al. 2016). Such studies would allow us to distinguish neutral evolutionary divergence in regulation from regulatory changes representing adaptive divergence and those maintained by purifying selection.
Salmonids have emerged as a model for studying consequences of autopolyploidization in vertebrates, owing to their relatively young WGD event (Ss4R, <100 Ma) (Ohno 1970; Alexandrou et al. 2013) and ongoing rediploidization (Macqueen and Johnston 2014; Lien et al. 2016; Limborg et al. 2016; Robertson et al. 2017). Directly following autopolyploidization, duplicated chromosomes pair randomly with any of their homologous counterparts resulting in an increased risk of formation of multivalents and consequently production of nonviable aneuploid gametes. Restoring bivalent chromosome pairing is therefore a critical step toward a functional genome post-WGD (Wolfe 2001). This can be achieved through, for example, structural rearrangements that suppress recombination, block multivalent formation, and drive the process of returning to a functional diploid state (i.e., rediploidization). As the mutational process is stochastic, rediploidization occurs independently for different chromosomes. As a result, the divergence of gene duplicates resulting from WGD (referred to as ohnologs) is also achieved independently for different chromosomes and hence occurs at different rates in various genomic regions. Recent studies on genome evolution subsequent to Ss4R have shown that the rediploidization process temporally overlaps with species radiation, resulting in lineage-specific ohnolog resolution (LORe) that may fuel differentiation of genome structure and function (Macqueen and Johnston 2014; Robertson et al. 2017). In fact, due to the delayed rediploidization, only 75% of the duplicated genome diverged before the basal split in the salmonid family ∼60 Ma (henceforth referred to as ancestral ohnolog resolution, AORe). Consequently, ∼25% of the Ss4R duplicates have experienced independent rediploidization histories after the basal salmonid divergence resulting in the Salmoninae and Thymallinae clades. Interestingly, the species within these two clades have also evolved widely different genome structures, ecology, physiology, and life history adaptations (Hendry and Stearns 2004). In contrast to the Thymallus lineage, the species in the subfamily Salmoninae have fewer and highly derived chromosomes resulting from large-scale chromosomal translocations and fusions (supplementary fig. S1, Supplementary Material online), display extreme phenotypic plasticity, and have evolved the capability of migrating between fresh and saltwater habitats (referred to as anadromy) (Nygren et al. 1971; Hartley 1987; Phillips and Ráb 2001; Alexandrou et al. 2013; Ocalewicz et al. 2013). This unique combination of both shared and lineage-specific rediploidization histories, and striking differences in genome structure and adaptations, provides an ideal study system for addressing key questions about the evolutionary consequences of WGD.
To gain deeper insights into how selection has shaped the evolution of gene duplicates post-WGD, we have sequenced, assembled, and annotated the genome of the European grayling (Thymallus thymallus Linnaeus, 1758), a species representative of an early diverging nonanadromous salmonid lineage, Thymallinae. We use this novel genomic resource in a comparative phylogenomic framework with the genome of Atlantic salmon (Salmo salar), of the Salmoninae lineage, to address the consequences of Ss4R WGD on lineage-specific rediploidization and selection on ohnolog gene expression regulation.
Our results reveal signatures of adaptive regulatory divergence of ohnologs, strong selective constraints on expression evolution in brain and neural-related genes, and lineage-specific ohnolog divergence. Moreover, diverse biological processes correspond to differences in evolutionary constraints during the 88–100 Myr of evolution post-WGD, pointing toward underlying differences in adaptive pressures in nonanadromous grayling and anadromous Atlantic salmon.
Materials and Methods
Sampling and Sequencing
A male grayling specimen was sampled outside of its spawning season (October 2012) from the River Glomma at Evenstad, Norway (61°25′0.1″N 11°9′49.7″E). The fish was humanely sacrificed, and various tissue samples were immediately extracted and conserved for later DNA and RNA analyses. Fin clips were stored on 96% ethanol for DNA sequencing. Tissues from the muscle, the gonad, the liver, the head kidney, the spleen, the brain, the eye, the gill, and the heart were stored in RNAlater for RNA extraction.
The DNA was extracted from fin clips using a standard high-salt DNA extraction protocol. A paired-end library with an insert size ∼180 bp (150 bp read length) and mate pair libraries of insert size ∼3 and 6 kb (100 bp read length) were sequenced using the Illumina HiSeq2000 platform (supplementary table S1, Supplementary Material online). Total RNA was extracted from the different tissue samples using the RNeasy mini kit (Qiagen) following the manufacturer’s instructions. The library construction and sequencing were carried out using Illumina TruSeq RNA Preparation kit on Illumina HiSeq2000 (supplementary table S2, Supplementary Material online). All the library preparation and sequencing were performed at the McGill University and the Génome Québec Innovation Centre.
Genome Assembly and Validation
The sequences were checked for their quality, and adapter trimming was performed using cutadapt (version 1.0) (Martin 2011). A de novo assembly was generated with Allpaths-LG (release R48777) (Gnerre et al. 2011) using the 180-bp paired-end library and the mate pair (3 and 6 kb) libraries. Assembly polishing was carried out using pilon (version 1.9) (Walker et al. 2014). The high copy number of mitochondrial DNA often leads to high read coverage and thus misassembly. The mitochondrial genome sequence in the assembly was thus reassembled by extracting the reads that mapped to the grayling (Thymallus thymallus) mtDNA sequence (GenBank accession number: NC_012928), followed by a variant calling step using Genome Analysis Toolkit (GATK) (version 3.4-46) (Van der Auwera et al. 2013). The consensus mtDNA sequence thus obtained was added back to the assembly.
To identify and correct possibly erroneous grayling scaffolds, we aligned the scaffolds against a repeat masked version of the Atlantic salmon genome (Lien et al. 2016) using megablast (e-value threshold 1e-250). Stringent filtering of the aligned scaffolds (representing 1.3 Gb of the 1.4-Gb assembly) identified 13 likely chimeric scaffolds mapping to two or more salmon chromosomes (supplementary file 1, Supplementary Material online), which were then selectively “broken” between, apparently, incorrectly linked contigs.
Transcriptome Assembly
The RNA-Seq data from all the tissue samples were quality checked using FastQC (version 0.9.2). The sequences were assembled using the following two methods. Firstly, a de novo assembly was performed using the Trinity (version 2.0.6) (Grabherr et al. 2011) pipeline with default parameters coupled with in silico normalization. This resulted in 730,471 assembled transcript sequences with a mean length of 713 bases. RSEM protocol-based abundance estimation within the Trinity package was performed where the RNA-Seq reads were first aligned back to the assembled transcripts using Bowtie2 (Faust and Hall 2012), followed by calculation of various estimates including normalized expression values such as FPKM (fragments per kilobase million). A script provided with Trinity was then used to filter transcripts based on FPKM, retaining only those transcripts with a FPKM of at least one.
Secondly, reference-guided RNA assembly was performed by aligning the RNA reads to the genome assembly using STAR (version 2.4.1b) (Dobin et al. 2013). Cufflinks (version 2.1.1) (Trapnell et al. 2010; Dobin et al. 2013) and TransDecoder (Haas et al. 2013) were used for transcript prediction and ORF (open reading frame) prediction, respectively. The resulting transcripts were filtered and retained based on homology against zebrafish and stickleback proteins, using BlastP and PFAM (1e-05). The de novo method resulted in 134,368 transcripts and the reference-based approach followed by filtering resulting in 55,346 transcripts.
Genome Annotation
A de novo repeat library was constructed using RepeatModeler with default parameters. Any sequence in the de novo library matching a known gene was removed using BlastX against the UniProt database. CENSOR and TEclass were used for classification of sequences that were not classified by RepeatModeler. Gene models were predicted using an automatic annotation pipeline involving MAKER (version 2.31.8), in a two-pass iterative approach (as described in https://github.com/sujaikumar/assemblage/blob/master/README-annotation.md). Firstly, ab initio gene predictions were generated using GeneMark ES (version 2.3e) (Lomsadze et al. 2005) and SNAP (version 20131129) (Korf 2004) trained on core eukaryotic gene data set (CEGMA). The first round of MAKER was then run using the following data as input: ab initio gene models, the UniProt database as protein evidence, the de novo identified repeat library and the de novo and reference guided transcriptome assemblies, as well as the transcript sequences from the recent Atlantic salmon annotation (Lien et al. 2016). The second pass involved additional data from training AUGUSTUS (Stanke et al. 2008) and SNAP models on the generated MAKER predictions.
Putative functions were added to the gene models using BlastP against the UniProt database (e-value 1e-5), and the domain annotations were added using InterProScan (version 5.4-47) (Quevillon et al. 2005). Using the MAKER standard filtering approach, the resulting set of genes was first filtered using the threshold of AED (Annotation Edit Distance), retaining gene models with AED score <1 and PFAM domain annotation. AED is a quality score given by MAKER that ranges from 0 to 1 and indicates the concordance between predicted gene model and the evidence provided, where an AED of 0 indicates that the gene models completely conforms to the evidence. Further, for the genes with AED score of 1 and no domain annotations, a more conservative BLAST search was performed against UniProt proteins and Atlantic salmon proteins with an e-value cut off of 1e-20. The genes with hits to either of these databases were also retained. The completeness of the annotations was again assessed using CEGMA (Parra et al. 2007) and benchmarking universal single-copy ortholog (BUSCO) (Simão et al. 2015).
Analysis of Orthologous Groups
We used orthofinder (version 0.2.8, e-value threshold at 1e-05) (Emms and Kelly 2015) to identify orthologous gene groups (i.e., orthogroup). As input to orthofinder, we used the MAKER-derived T. thymallus gene models as well as protein sequences from three additional salmonid species (Atlantic salmon, rainbow trout, and coho salmon), four nonsalmonid teleost species (Esox lucius, Danio rerio, Gasterosteus aculeatus, and Oryzias latipes), and two mammalian outgroups (Homo sapiens and Mus musculus). Rainbow trout protein annotations were taken from https://www.genoscope.cns.fr/trout/. Atlantic salmon (Annotation Release 100), Esox lucius (Annotation Release 101) data were downloaded from NCBI ftp server (ftp://ftp.ncbi.nlm.nih.gov/genomes/). The transcriptome data for Coho salmon were obtained from NCBI (GDQG00000000.1) and translated using TransDecoder. All other annotations were downloaded from ENSEMBL.
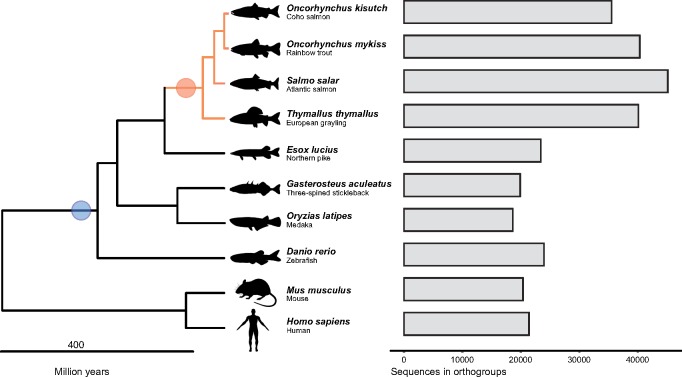
Each set of orthogroup proteins was then aligned using MAFFT(v7.130) (Katoh et al. 2002) using default settings, and the resulting alignments were then used to infer maximum-likelihood gene trees using FastTree (v2.1.8) (Price et al. 2010) (figs. 1a and b). As we were only interested in gene trees containing information on Ss4R duplicates, complex orthogroup gene trees (i.e., containing 2R or 3R duplicates of salmonid genes) were subdivided into the smallest possible subtrees. To this end, we developed an algorithm to extract all clans (defined as unrooted monophyletic clade) from each unrooted tree (Wilkinson et al. 2007) with two monophyletic salmonid tips as well as nonsalmonid outgroups resulting in a final set of 20,342 gene trees. In total, 31,291 grayling genes were assigned to a clan (fig. 1 and supplementary fig. S2, Supplementary Material online). We then identified homeology in the Atlantic salmon genome by integrating all-versus-all protein BLAST alignments with a priori information of Ss4R synteny as described by Lien et al. (2016). Using the homeology information, we inferred a set of high-confidence ohnologs originating from Ss4R. The scaffold length distribution and number of genes per scaffold containing the inferred Ss4R genes are plotted in supplementary figure S13, Supplementary Material online. The clans were grouped based on the gene tree topology into duplicates representing LORe and those with ancestrally diverged duplicates (AORe). The LORe regions were further categorized into two (duplicated or collapsed) based on the number of corresponding T. thymallus orthologs. These data were plotted on Atlantic salmon chromosomes using circos plot generated using OmicCircos (https://bioconductor.org/packages/release/bioc/html/OmicCircos.html). The LORe and AORe ohnologs with two copies in each species are hereafter referred to as ohnolog-tetrads (see supplementary fig. S14, Supplementary Material online, for the summary of the above steps).
Fig. 1.
—Species and genes in ortholog groups. Left: Phylogenetic relationship of species used for constructing ortholog groups and gene trees. The blue circle indicates the 3R-WGD event, while the Ss4R event is indicated with an orange circle. Right: Number of genes assigned to ortholog groups in each of the species used in the analysis.
Expression Divergence and Conservation
The grayling RNA-Seq reads from each of the eight tissues (liver, muscle, spleen, heart, head kidney, eye, brain, and gills) were mapped to the genome assembly using STAR (version 2.4.1b). The reads uniquely mapping to the gene features were quantified using htseq-count (Anders et al. 2015). The CPM (counts per million) value, here used as a proxy for expression, was then calculated using edgeR (Robinson et al. 2010). Similar CPM data sets were obtained from Atlantic salmon RNA-Seq data reported by Lien et al. (2016).
Filtering of ortholog groups (i.e., clans) was performed prior to analyses of expression evolution of Ss4R ohnologs: 1) We only considered Ss4R duplicates that were retained in both Atlantic salmon and grayling, and 2) the Ss4R duplicates were classified into AORe or LORe, based on topologies of the ortholog group gene trees, only gene pairs with non-zero CPM value were considered. This filtering resulted in a set of 5,070 duplicate pairs from both Atlantic salmon and grayling (ohnolog-tetrads) (summarized in supplementary fig. S14, Supplementary Material online). The gene expression values from the gene duplicates in the ohnolog-tetrads were clustered using hclust function in R, using Pearson correlation into eight tissue-dominated clusters. The expression pattern in the eight clusters of the genes in ohnolog-tetrads was used to further classify them into one of the ohnolog expression evolution categories (see table 2). The ohnolog-tetrads were further filtered based on expected topologies under LORe and AORe scenarios (see supplementary fig. S14, Supplementary Material online, for summary). Heatmaps of expression counts were plotted using pheatmap package in R (https://CRAN.R-project.org/package=pheatmap). To quantify the breadth of expression (i.e., the number of tissues a gene is expressed in), we calculated the tissue specificity index Tau (Yanai et al. 2005) for all the genes in ohnolog-tetrads, where a τ value approaching 1 indicates higher tissue specificity while 0 indicates ubiquitous expression.
Table 2.
Classification of Tissue Expression Divergence in the Ohnolog-Tetrads
| Evolutionary Scenario | AORe | LORe |
|---|---|---|
| I: Ancestral ohnolog divergence followed by purifying selection independently in both species | 199 (5.7%) | 24 (4.7%) |
| II: Conserved tissue regulation of all ohnologs | 869 (25%) | 131 (25.7%) |
| III: Atlantic salmon-specific divergence of one ohnolog | 375 (10.8%) | 70 (13.8%) |
| IV: Grayling-specific divergence of one ohnolog | 516 (14.8%) | 80 (15.7%) |
| V: Conserved tissue regulation among ohnologs within species but different between species | 195 (5.6%) | 51 (10.0%) |
| Unclassified (VI): Ohnolog-tetrads assigned to three or more tissue clusters | 1,326 (38.1%) | 153 (30.1%) |
| Total | 3,480 | 509 |
Note.—The number and percentages of genes in each category calculated based on the total number of topology-filtered ohnolog-tetrads.
Expression Comparison in Liver
Utilizing independent liver tissue samples, we compared differential expression in liver tissue gene expression among ohnologs of grayling and Atlantic salmon with their ohnolog-tetrad tissue expression evolution categories. The liver samples from four grayling individuals were sampled in the river Gudbrandsdalslågen (61°18′53.09″N 10°18′1.53″E). The samples were from two males (370,375 mm) and two females (330,360 mm). The fish was euthanized and dissected immediately after capture, and the liver was stored in RNAlater. Total RNA was extracted and 100-bp single-end read libraries were generated for two individuals and sequenced using the Illumina HiSeq4000 platform. For the other two individuals, 150-bp paired-end read libraries were generated and sequenced using the Illumina HiSeq2500 platform. RNA-Seq data for an additional four Atlantic salmon liver tissue samples were obtained from a feeding experiment (Gillard et al. 2018). Presmolt salmon were raised on fish oil-based diets under freshwater conditions.
The RNA-Seq read data were quality processed using CutAdapt (Martin 2011) before alignment we aligned the reads to grayling or Atlantic salmon (ICSASG_v2; Lien et al. 2016) genomes, respectively, using STAR (Dobin et al. 2013). RSEM (Li and Dewey 2011) expected counts were generated for gene features. EdgeR (Robinson et al. 2010) was used to generate normalized library sizes of samples (TMM normalization), followed by a differential expression analysis using the exact test method between the gene expression of both the grayling and Atlantic salmon ohnologs in each ohnolog-tetrad. The fold change (log2 scaled) and significance of differential expression (false discovery rate-corrected P-values) were produced for grayling and Atlantic salmon duplicates, as well as relative counts in the form of CPM.
Sequence Evolution
To estimate coding sequence evolution rates, we converted amino acid alignments to codon alignments using pal2nal (Suyama et al. 2006). The “seqinr” R package (http://seqinr.r-forge.r-project.org/) was used to calculate pairwise dN and dS values for all sequences in each alignment using the “kaks” function. For in-depth analyses of branch-specific sequence evolution of the cystic fibrosis transmembrane conductance regulator (CFTR) genes, we used the codeml in PAML (version 4.7a) (Yang 1997). To assess whether sequences in the CFTR gene tree evolved under similar selection pressure, we contrasted a fixed dN/dS ratio (1-ratio) model and a free-ratio model of codon evolution. A likelihood ratio test was conducted to assess whether a free-ratio model was a significantly better fit to the data. Branch-specific dN/dS values were extracted from the maximum likelihood results for the free ratios model.
The two Pacific salmon genes in the CFTR tree (fig. 5) correspond to a gene from rainbow trout and another from Coho salmon. A BLAT (Kent 2002) search of CFTR gene against the rainbow trout assembly (https://www.genoscope.cns.fr/trout/) resulted in hits on three different scaffolds, with one complete hit and two other partial hits on unplaced scaffolds. Additionally, Coho salmon data are based on a set of genes inferred from transcriptome data. Therefore, the presence of a single copy in the tree for the two species is likely an assembly artifact.
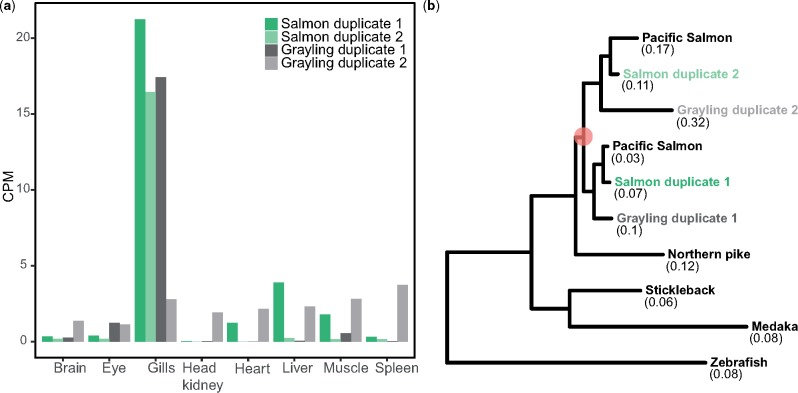
Fig. 5.
—Divergent selection on cystic fibrosis transmembrane conductance regulator. (a) Expression values, in terms of CPM, of the CFTR ohnologs in Atlantic salmon and grayling across eight tissues. (b) CFTR gene tree. The orange circle represents the Ss4R duplication. Branch-specific dN/dS values of the tip nodes are given in parentheses.
Genome-Wide Identification of Transcription Factor-Binding Sites
A total of 13,544 metazoan transcription factor protein sequences together with their binding site represented as position-specific scoring matrices (PSSMs referred to as motifs) were collected from transcription factor-binding profile databases such as CISBP, JASPAR, 3D-footprint, UniPROBE, HumanTF, HOCOMOCO, HumanTF2, and TRANSFAC.
DNA sequences from upstream promoter regions of Atlantic salmon (−1,000/+200 bp from TSS) were extracted. A first-order Markov model was created from the entire set of upstream promoter regions using the fasta-get-markov program in the MEME Suite (Bailey et al. 2009). This background model was used to convert frequency matrices into log-odds score matrices. We performed a genome-wide transcription factor-binding sites prediction in the Atlantic salmon genome using the PSSM collection and the Finding Individual Motif Occurrences (FIMO) (Grant et al. 2011) tool in the MEME Suite (P-value = 0.0001 and FDR = 0.2).
Motif similarity between Atlantic salmon ohnolog promoters was scored using the “Jaccard coefficient.” The promoter Jaccard coefficient is defined as
where A and B represents the type of motifs that were present in promoters of the A and B ohnolog copies. If A and B are empty, we set J(A, B) = 0 where 0 ≤ J(A, B) ≤ 1.
Gene Ontology Analysis
The gene ontology (GO) term enrichment analysis was performed using the “elim” algorithm implemented in the “topGO” R package (http://www.bioconductor.org/packages/2.12/bioc/html/topGO.html), with a significance threshold of 0.05 using the genes from all ohnolog-tetrad categories as the background. GO terms were assigned to salmon genes using Blast2GO (Conesa et al. 2005).
Results
Genome Assembly and Annotation
We sequenced the genome of a wild-caught male grayling individual, sampled from the Norwegian river Glomma (61°25′0.1″N 11°9′49.7″E), using the Illumina HiSeq 2000 platform (supplementary tables S1 and S2, Supplementary Material online). De novo assembly was performed using ALLPATHS-LG (Gnerre et al. 2011), followed by assembly correction using Pilon (Walker et al. 2014), resulting in 24,343 scaffolds with an N50 of 284 kb and a total size of 1.468 Gb (table 1). The scaffolds represent ∼85% of the k-mer-based genome size estimate of ∼1.8 Gb. The C-values estimated previously for European grayling are 2.1 pg (http://www.genomesize.com/) and 1.9 pg (Hartley 1987). To annotate gene structures, we used RNA-Seq data from nine tissues extracted from the sequenced individual. We constructed transcriptome assemblies using both de novo and reference-based methods. Repeat masking with a repeat library constructed using a combination of homology and de novo-based methods identified and masked ∼600 Mb (∼40%) of the assembly and was dominated by class1 DNA transposable elements (supplementary table S3 and a repeat landscape in supplementary fig. S2, Supplementary Material online). Finally, the transcriptome assemblies, the de novo-identified repeats along with the UniProt proteins (UniProt Consortium 2015), and Atlantic salmon coding sequences (Lien et al. 2016) were utilized in the MAKER annotation pipeline, predicting a total of 117,944 gene models, of which 48,753 protein-coding genes were retained based on AED score, homology with UniProt and Atlantic salmon proteins or presence of known domains. Assembly completeness was assessed at the gene level by looking for conserved genes using CEGMA and BUSCO. The assembly contains 236 (95.16%) out of 248 conserved eukaryotic genes (CEGs) with 200 (80.65%) complete CEGs. Of the 4,584 BUSCO (database: Actinopterygii, odb9), 4,102 complete (89.5%) and 179 (3.9%%) fragmented genes were found in the assembly (table 1).
Table 1.
Grayling Genome Assembly Statistics
| Assembly Statistics | Assembly Validation | ||
|---|---|---|---|
| Total size of scaffolds (bp) | 1,468,519,221 | Complete CEGMAa genes | 80.65% (200/248) |
| Number of scaffolds | 24,369 | Partial CEGMA genes | 95.16% (236/248) |
| Scaffold N50 (bp) | 283,328 | Complete BUSCOsb | 4,102 (89.5%) |
| Longest scaffold (bp) | 2,502,076 | Complete Duplicated BUSCOs | 1,724 (37.6%) |
| Total size of contigs (bp) | 1,278,330,545 | Fragmented BUSCOS | 179 (3.9%) |
| Number of contigs | 216,549 | Missing BUSCOS | 303 (6.6%) |
| Contig N50 (bp) | 11,206 | Total BUSCOS searched | 4,584 |
Based on 248 highly CEGS.
Based on a set of 4,584 Actinopterygii odb9 BUSCOs.
Divergent Rediploidization Rates among the Salmonid Lineages
Previous studies have suggested that up to 25% of the genome of the most recent common salmonid ancestor was still tetraploid when the grayling and Atlantic salmon lineages diverged (Lien et al. 2016; Robertson et al. 2017). To test this hypothesis, we used a phylogenomic approach to characterize rediploidization following Ss4R in grayling. We inferred 23,782 groups of orthologous genes (i.e., ortholog groups or orthogroups) using gene models from Homo sapiens (human), Mus musculus (mouse), Danio rerio (zebrafish), Gasterosteus aculeatus (three-spined stickleback), Oryzias latipes (medaka), Esox lucius (northern pike), Salmo salar (Atlantic salmon), Oncorhynchus mykiss (rainbow trout), and Oncorhynchus kisutch (coho salmon) (fig. 1). These orthogroups were used to infer gene trees. In total, 20,342 gene trees contained WGD events older than Ss4R (Ts3R or 2R) and were further subdivided into smaller subgroups (i.e., unrooted monophyletic clade termed as clans, see Materials and Methods for details and supplementary fig. S3, Supplementary Material online). To identify orthogroups with retained Ss4R duplicates, we relied on the high-quality reference genome of Atlantic salmon (Lien et al. 2016). A synteny-aware blast approach (Lien et al. 2016) was first used to identify Ss4R duplicate/ohnolog pairs in the Atlantic salmon genome, and this information was used to identify a total of 8,527 gene trees containing high-confidence ohnologs originating from Ss4R. Finally, gene trees were classified based on the tree topology into duplicates conforming to LORe and those with ancestrally diverged duplicates following the topology expected under AORe (fig. 2a). In total, 3,367 gene trees correspond to LORe regions (2,403 with a single copy in grayling) and 5,160 correspond to an AORe-like topology. These data were cross-checked with the LORe coordinates suggested by Robertson et al. (2017), and genes with LORe-type topologies from non-LORe regions of the genome were discarded. The final set (henceforth referred to as ohnolog-tetrads) consisted of 5,475 gene trees containing Ss4R duplicates from both species (4,735 AORe, 740 LORe). In addition, 482 ortholog sets contained Ss4R duplicates in Atlantic salmon but not in grayling.
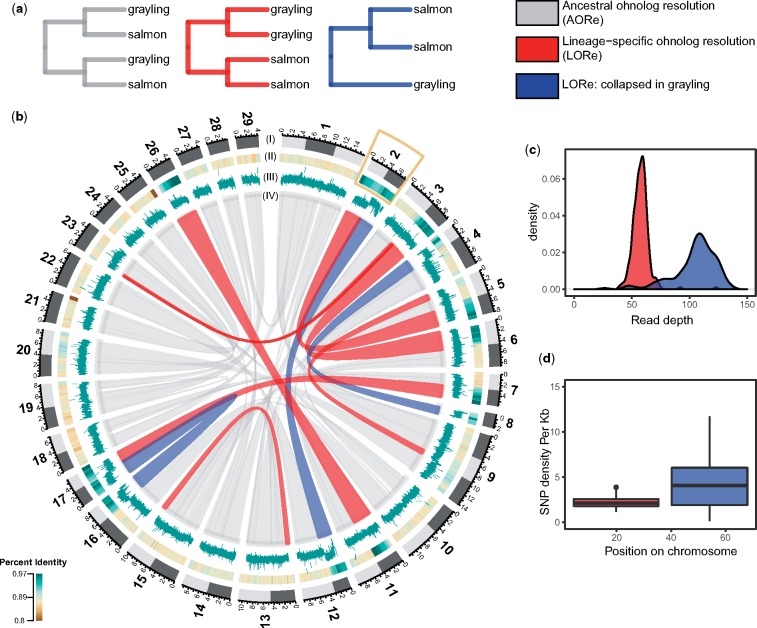
Fig. 2.
—Rediploidization in grayling genome. (a) Gene tree topologies corresponding to the different models of ohnolog resolution (ancestral divergence of ohnologs [AORe; gray] and lineage-specific divergence of ohnologs (LORe in red and LORe-like regions, with repeat collapse in grayling, in blue). (b) Circos plot: Outer track (I) represents the 29 chromosomes of Atlantic salmon with chromosome arms indicated using light and dark gray. (II) Percent identity between duplicated genomic regions in Atlantic salmon with darker green representing higher percent identity (see color scale). (III) Average number of reads mapped to grayling genes in the corresponding regions. (IV) The gray ribbons represent the ancestrally diverged gene duplicate pairs (AORe), while the red ribbons represent the LORe duplicate pairs and the blue ribbons correspond to LORe regions with a collapsed assembly in grayling. The inset plots show the distribution of average depth of reads mapped to the grayling genes (c) and SNP density per kb (d) across chromosome 2 (marked with a yellow box in b).
To identify regions of ancestral and lineage-specific rediploidization in the grayling genome, we assigned genes from gene trees that contained Ss4R duplicates to genomic positions on the Atlantic salmon chromosomes (fig. 2b). In Atlantic salmon, several homeologous chromosome arms (2p-5q, 2q-12qa, 3q-6p, 4p-8q, 7q-17qb, 11qa-26, and 16qb-17qa) have previously been described as homeologous regions under delayed rediploidization (Lien et al. 2016; Robertson et al. 2017) (indicated in fig. 2b as red and blue ribbons). Interestingly, for the homeologous LORe regions 2q-12qa, 4p-8q, and 16qb-17qa in Atlantic salmon, we identified only one orthologous region in grayling, suggesting either loss of large duplicated blocks or sequence assembly collapse in grayling. To test the “assembly collapse” hypothesis, we mapped the grayling Illumina paired-end reads that were used for the assembly back to the grayling genome sequence using BWA-MEM (Li 2013) and determined the mapped read depth for each of the grayling genes. Single-copy grayling genes in LORe regions consistently displayed read depths (∼100×) twice that of the LORe duplicates in grayling (fig. 2c and supplementary fig. S4a, Supplementary Material online), indicating assembly collapse rather than loss of large chromosomal regions. Additionally, the single nucleotide polymorphism (SNP) density of the scaffolds in these regions computed using FreeBayes (Garrison and Marth 2012) (quality filter of 30) displayed values that were on an average twice that of the background SNP density, albeit with a much wider distribution (fig. 2d and supplementary fig. S4b, Supplementary Material online).
Ohnolog Tissue Gene Expression Regulation
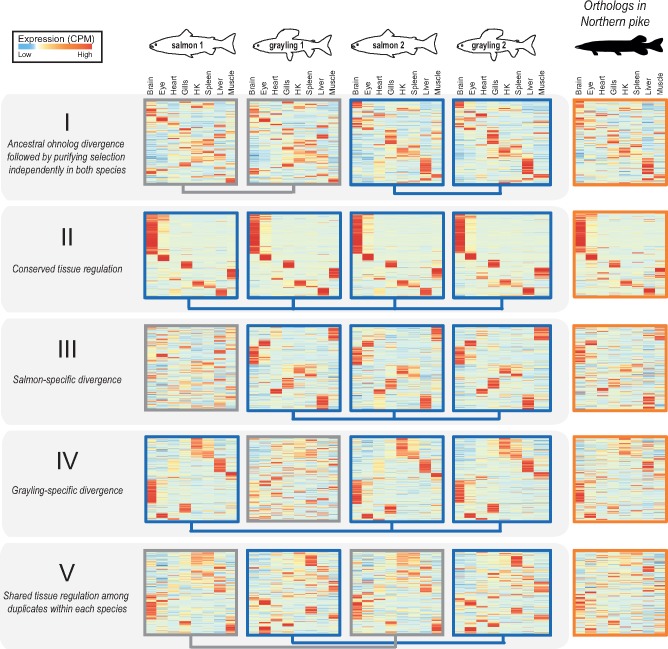
To investigate the regulatory divergence in tissue expression following Ss4R, we exploited tissue expression atlases of Atlantic salmon and grayling in a coexpression analysis. Individual genes from 5,070 “expressed” ohnolog-tetrads (20,280 genes in total) were assigned to eight “tissue-dominant” expression clusters (Materials and Methods, supplementary fig. S5, Supplementary Material online). These coexpression clusters were used to identify ohnolog-tetrads conforming to expectations of expression patterns under five evolutionary scenarios (see table 2, fig. 3 and supplementary fig. S6, Supplementary Material online): Ancestral ohnolog divergence followed by independent purifying selection in both species (I), conserved tissue regulation in both species (II), lineage-specific regulatory divergence of one duplicate (III and IV). In addition, a fifth (V) scenario where regulation among duplicates is shared within species but different between species is expected to be common in genomic regions with LORe. Further, the ohnolog-tetrads where three or all four of the duplicates were in different tissue-expression clusters were grouped into a sixth “unclassified” category.
Fig. 3.
—Selection on tissue expression regulation after whole-genome duplication. Heatmaps show clustering of tissue expression of the ohnolog-tetrads for each of the five evolutionary scenarios of tissue expression regulation following Ss4R WGD (see table 2). Within each category, the first four heatmaps represent one ohnolog-tetrad (i.e., four genes: salmon1, grayling1, salmon2, and grayling2) that were ordered based on similarity of expression profiles with the corresponding orthologs in northern pike (the fifth heatmap depicted on the right). Darker red corresponds to the highest expression level observed for one gene, and the darker blue to the lowest (scaled CPM). Connecting blue lines below the heatmaps indicate duplicates belonging to the same tissue clusters (conserved expression pattern). (An extended figure with ohnolog-tetrads subdivided into LORe and AORe in supplementary fig. S6, Supplementary Material online.)
After applying a gene tree topology-based filtering criterion to the 5,070 ohnolog-tetrads (see Materials and Methods), 509 conforming to the expectations of LORe and 3,480 conforming to AORe gene tree topologies were retained for further analyses. Of the five evolutionary scenarios, conserved tissue regulation was the most common (∼25%), followed by species-specific divergence of a single duplicate (∼11% in Atlantic salmon and ∼15% in grayling). Categories I and V were the least common categories (table 2), and as expected, category V was significantly enriched in LORe regions (Fisher’s exact test, two-sided, P-value < 0.0005).
To assess the directionality of the expression divergence relative to the presumed ancestral state, we compared tissue expression of the ohnolog-tetrads with that of the corresponding orthologs in northern pike (fig. 3). Previous studies have shown that genome-wide tissue-specific expression divergence among WGD ohnologs in teleosts mostly evolves through asymmetric divergence in tissue regulation (Lien et al. 2016; Sandve et al. 2018). The predominant expression pattern thus reflects one ohnolog copy retaining more regulatory similarity with unduplicated orthologs (regulatory neofunctionalization), with very small proportion (<1%) of ohnologs displaying characteristics of regulatory subfunctionalization (Lien et al. 2016). Under a model of subfunctionalization, the sum of expression levels of both ohnologs should correlate better to the assumed ancestral expression regulation than any of the individual ohnologs (Sandve et al. 2018). Therefore, we tested whether the divergence patterns leading to maintenance of the two ohnolog copies in the category I tetrads are associated with this atypical mode of expression divergence. Both the distribution of ohnolog tissue expression correlations (supplementary fig. S7, Supplementary Material online) and the patterns in the heatmaps (fig. 3) support the regulatory neofunctionalization pattern for all three evolutionary scenarios where we observe ohnolog divergence (categories I, III, and IV).
As different tissues are involved in different biological functions, we expect that the regulatory evolution is shaped by tissue-specific selective pressures (Gu and Su 2007). To test this, we evaluated the hypothesis that tissues are disproportionately contributing to ohnolog-tetrad divergence by comparing the “tissue-dominant” cluster distribution across all tetrads. For all evolutionary scenarios, between two and five tissue-expression clusters were significantly over- or underrepresented (Fisher’s tests, two sided, Bonferroni-corrected P-value ≤ 0.05), with the conserved category being the most skewed in tissue representation with a strong bias toward brain-specific expression (supplementary table S4, Supplementary Material online). The high tissue specificity (Tau score) of genes in ohnolog-tetrads associated with these genes (supplementary fig. S8, Supplementary Material online) corroborates the observed brain-specific expression bias.
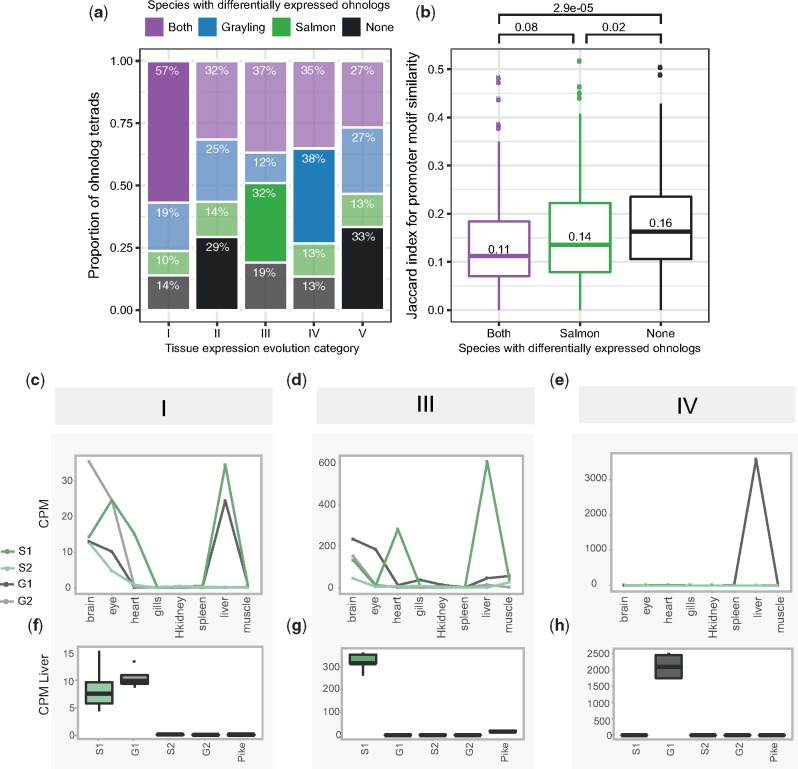
Further, we tested whether distinct ohnolog-tetrad divergence categories were coupled to patterns of protein-coding and promoter sequence evolution. Specifically, we tested the hypothesis that conserved regulation is associated with conserved protein-coding evolution. We estimated the dN/dS ratios for each duplicate pair within each species and compared the distribution of dN/dS statistics in each class with that of the “unclassified” category VI (supplementary fig. S9, Supplementary Material online). Low dN/dS (≪1) indicates strong purifying selection pressure. Categories I–V show variability in among-ohnolog dN/dS ratios, with category I having significantly higher dN/dS ratio compared with the “unclassified” category (Wilcoxon rank sum, P = 0.005) and categories II and V having significantly lower dN/dS ratios (Wilcoxon rank sum, P = 0.014 and P = 0.0017, respectively). The ohnolog pairs showing lineage-specific expression divergence (III and IV) did not have a significantly different dN/dS ratio compared with the unclassified category (Wilcoxon rank test, P = 0.36 and P = 0.26, respectively). Further, we used the Atlantic salmon genome to annotate and compare known transcription factor motifs divergence in the promoters (from 1,000 bp upstream to 200 bp downstream of the transcription start site) of ohnologs. Under the assumption that expression divergence is, at least partly, driven by changes in transcription factor-binding motifs located in proximal promoters, we tested whether ohnolog regulatory divergence in salmon (scenario I and III) was associated with divergence of promoter motifs. Indeed, the results add validation to the different expression evolution classifications (supplementary fig. S10, Supplementary Material online), with categories I and III having significantly less similar promoter motif content compared with ohnolog-tetrads with conserved tissue expression regulation in salmon (II, IV, and V) (Wilcoxon test all contrasts between I/III and II/IV/V, P < 0.04–0.002).
To evaluate whether the ohnologs in different classes were associated with distinct biological functions, we performed GO term enrichment tests. The ohnolog-tetrads of category II under strict selective constraints show highly brain-specific expression and are enriched for GO functions related to behavior and neural functions. In contrast, genes in category I, which represents ohnologs that underwent divergence in gene regulation following WGD, are associated with functions related to lipid metabolism, development, and immune system (supplementary file 2, Supplementary Material online).
Highly connected genes in protein–protein interaction (PPI) networks are often placed under strong constraints to maintain stoichiometry (Freeling and Thomas 2006; Sémon and Wolfe 2007). To test whether the strong constraints on the ohnolog-tetrads with conserved tissue expression (II) are associated with having higher PPIs, we extracted all the zebrafish genes from the gene trees corresponding to the ohnologs in the expression divergence categories and queried them against the STRING database (Szklarczyk et al. 2017) (version 10.5). Only associations with a score of above 7.0, suggesting high-confidence associations, were retained. As expected, we found that category II genes were indeed enriched for PPI (enrichment P-value = 1.05e-05) in comparison to the genes in the other classes (I, III, and IV) with diverged expression (enrichment P-value = 0.79).
Evolution of Gene Expression Levels Following WGD
The coexpression analyses leverage gene expression variation between tissues to classify ohnologs according to regulatory divergence. However, it is important to note that it does not explicitly test for significant changes in gene expression levels. To assess the divergence of ohnolog expression levels, we generated an RNA-Seq data set from additional liver samples from Atlantic salmon (n = 4) and grayling (n = 4). We tested for differences in liver expression levels between ohnolog pairs within both species and calculated absolute differences in fold change (FC) and the statistical significance (FDR-adjusted P-values) for these tests. Of the 2,467 ohnolog-tetrads in categories I–V (table 2), 54% (1,349) showed significant (FDR < 10−3) fold change differences (FC > 2) in liver expression in at least one species, with 19% (467) in both species, 18% (455) in grayling only, and 17% (427) in salmon only.
We then focused on the subset of ohnolog-tetrads where at least one ohnolog was assigned to an expression cluster displaying dominant expression in liver. From this subset of 552 ohnolog-tetrads, 80% (442) showed significant (FDR < 10−3) fold change differences (FC > 2) in liver expression in at least one species, 37% (204) both species, 25% (136) grayling only, and 18% (102) salmon only (supplementary table S5, Supplementary Material online). As tissue-dominance is the main factor in the analyses of tissue-regulatory evolution, we expected that the different evolutionary scenarios (fig. 3) should be associated with enrichments in certain patterns of expression level divergence, or alternatively, a lack thereof. We indeed found that the ohnolog-tetrads reflecting ancestral divergence followed by purifying selection in both species (scenario I) were significantly enriched in ohnologs being expressed at different levels in both species compared with other scenarios (fig. 4a, Fisher’s test P-value = 1.74 × 10−4). Conversely, those ohnolog pairs that show shared tissue regulatory patterns (scenarios II and V) were significantly enriched in ohnologs with no expression level divergence (P-values = 0.0117 and 6.79 × 10−3). Finally, ohnologs with tissue regulatory divergence in one species (scenario III and IV in fig. 3) also had the most pronounced enrichment of expression level divergence for that species (fig. 4a, P-values = 3.32 × 10−7 and 2.03 × 10−6). Three examples of putative liver-specific expression gains showing high correspondence between tissue regulation and expression level evolution are highlighted in figure 4c–h.
Fig. 4.
—Expression level evolution of the ohnolog-tetrads. (a, b) Ohnolog-tetrads were tested using liver expression data for cases of highly significant differential expression (FDR-adjusted P-value <10−3, absolute FC >2) between ohnologs of both species (purple), grayling ohnologs only (blue), Atlantic salmon ohnologs only (green), and no ohnologs of either species (black). Ohnolog tetrads shown in the figure had at least one ohnolog assigned to a tissue expression cluster displaying dominant expression in liver. The differential expression outcome expected to be the highest for each category is highlighted opaque, while the remaining outcomes are transparent. (a) The number and proportions of cases are given per tissue expression evolution category (see table 2). The differential expression outcome expected to be the highest for each category is highlighted opaque, while the rest are transparent. (b) Jaccard index score distributions for Atlantic salmon ohnolog promoter motif similarity, separated by differential expression outcome. P-values from pairwise comparisons testing for lower Jaccard index using the Wilcoxon test are indicated—as well as the median scores. The expression levels, in terms of CPM, from the tissue atlas data (c–e) and the corresponding data from the liver expression data are plotted (boxplots in f–h) for a selected example with a liver-specific gain of expression in each of the categories I, III, and IV. The examples indicated in (c–h) include ohnologs of ephrin type-B receptor 2-like (category I), contactin-1a-like gene (category III), and an E3 ubiquitin-protein ligase-like gene (category IV). The ohnologs in Atlantic salmon and grayling are represented as S1, S2 and G1 and G2, respectively.
Further, we assessed whether liver-specific expression level differences between ohnologs were associated with changes in transcription factor-binding motif presence in promoters. We partitioned the ohnologs into three categories; differentially expressed in both species (likely diverged in expression in an ancestor of all salmonids), species-specific expression level divergence (we only used salmon-specific cases as we had no promoter motif data for grayling), and no significant difference in expression level. The lowest promoter motif similarity was found among ohnologs where both species showed strong expression level divergence, followed by the salmon-specific and then no expression divergence (fig. 4b).
Selection on Chloride Ion Transporter Regulation
The most apparent difference in biology between grayling and Atlantic salmon is the ability of Atlantic salmon to migrate between freshwater and saltwater (anadromy), a trait that grayling has not evolved. A key feature in saltwater acclimation involves remodeling gill physiology to enable efficient ion secretion and maintenance of osmotic homeostasis (Evans et al. 2005). We therefore specifically investigated the ohnolog divergence in gill gene expression regulation for key ion transport-associated genes that perform critical function in the process of chloride ion secretion in sea water (Mackie et al. 2007; Nilsen et al. 2007). The Na+/K+/2Cl− cotransporter 1 (NKCC1a) gene showed an extreme gill-dominated expression of one of the ohnologs in salmon, while no such gill-specific expression was observed for the corresponding grayling ohnologs (supplementary fig. S11, Supplementary Material online). A particularly striking difference in expression pattern was observed for the cystic fibrosis transmembrane conductance regulator gene (CFTR; an ABC transporter-class conducting chloride ion transport), which exhibited grayling-specific regulatory divergence (Category IV). From the tissue expression profiles of this tetrad (fig. 5a), it was evident that the divergence of tissue regulation in grayling was associated with a loss of gill tissue expression specificity compared with Atlantic salmon. To determine whether the grayling CFTR duplicate with diverged expression also had signatures of coding sequence divergence, we computed branch-specific dN/dS. Notably, the grayling CFTR displaying diverged expression regulation also displays a 2-fold increase in dN/dS compared with its Ss4R duplicate copy with conserved expression regulation (fig. 5b). Na+/K+-ATPase subunit genes were not well represented in the annotation and orthogroups and hence not included in the analysis.
Discussion
A major limitation in previous studies of evolution of gene regulation following WGD in vertebrates has been the inability to distinguish between neutral and adaptive divergence (Hermansen et al. 2016). Here, we leverage gene expression data from two salmonid species and a close outgroup species (northern pike) in a comparative approach to identify shared expression evolution patterns following WGD in lineages evolving independently for ∼50 Myr. This allows us to identify evolutionarily long-term conservation of novel expression “phenotypes” arising after WGD—the hallmark of novel adaptive functions.
Although regulatory divergence of Ss4R ohnologs is widespread (Lien et al. 2016; Gillard et al. 2018), we show that ohnolog regulatory tissue divergence shared among species separated by ∼50 Myr of evolution is comparably rare (table 2).
Ohnolog-tetrads that include genes with liver- and gill-biased expression contribute disproportionately to signals that are consistent with adaptive expression evolution in a salmonid ancestor (supplementary table S4, Supplementary Material online). The genes predominantly expressed in liver have been shown to have a strong association with phyletic age (Kryuchkova-Mostacci and Robinson-Rechavi 2015), while also being associated with particularly fast expression evolution when compared with other tissues (Duret and Mouchiroud 2000; Khaitovich et al. 2006). However, in contrast to our results, this latter pattern is associated with signatures of neutral evolution rather than adaptive evolution of novel regulation in mammals (Kryuchkova-Mostacci and Robinson-Rechavi 2015). Future studies should therefore look into the forces that drive regulatory evolution in liver-centric genes, using broader comparative data sets, detailed characterization of the evolutionary turnover of liver-specific regulatory elements, and use of phylogenetic methods that are able to detect shifts in selection on gene regulation (Sandve et al. 2018)
Salmonids are suggested to have evolved from a pike-like ancestor, a relatively stationary ambush predator (Craig 2008). Under this assumption, early salmonid evolution must have involved adaptation to new pelagic and/or riverine habitats. Adaptations to new environments and evolution of different life history strategies are known to be associated with strong selective pressure on immune-related genes (Star et al. 2011; Haase et al. 2013; Solbakken et al. 2017). In line with this, we see an overrepresentation of immune-related genes among ohnologs that diverged in the common ancestor of salmon and grayling but have been under purifying selection in both species after speciation (category I, table 2 and supplementary file 2, Supplementary Material online). Furthermore, pikes are generally piscivorous throughout their life span, while salmonids depend more on aquatic and terrestrial invertebrate prey with significantly lower input of lipids, especially in early life (Carmona-Antoñanzas et al. 2013). Interestingly, duplicates with shared ancestral divergence (category I), which are candidates for adaptive divergence in regulation, are enriched for genes involved in lipid-homeostasis, metabolism, and energy storage (glycogen)-related functions (GO test results in supplementary file 2, Supplementary Material online).
The regulatory divergence of metabolism-related genes and its association with corresponding shifts in the prey nutrient profile have been previously described in other fish (McGirr and Martin 2017). In this study, we do find individual candidate genes for tissue remodeling of metabolism function, such as the ATP-binding cassette transporter gene linked to cholesterol metabolism (ABCA1, supplementary fig. S12, Supplementary Material online). However, in order to interpret these results from a perspective of gene regulation evolution related to novel lifestyle and diet adaptations in salmonid ancestor, a comprehensive analysis using, for example, liver coexpression network comparisons and controlled experiments with dietary modifications would be necessary.
Taken together, our results suggest a role of Ss4R ohnologs in adaptive evolution of novel gene expression regulation, possibly related to new pathogenic pressures in a new type of habitat, and optimization of lipid-homeostasis and glycogen metabolism-related functions in response to evolution of a more active pelagic/riverine life with limited lipid resources.
Purifying selection to maintain ancestral tissue regulation of ohnologs in both salmonid species was the most commonly observed fate of ohnolog expression evolution (category II, table 2 and fig. 3). These ohnologs were predominantly brain-specific and enriched for predicted PPIs. Several other studies in vertebrates have found similar results, with strong purifying selection on sequence and expression evolution in brain-dominant genes, as well as high retention probability following large-scale genome duplication (Khaitovich et al. 2005; Chan et al. 2009; Zheng-Bradley et al. 2010; Guschanski et al. 2017; Roux et al. 2017). As neuron function-related genes are involved in complex networks of signaling cascades and higher-order PPIs, this pattern is believed to be driven by either direct selection for maintaining novel gene copies due to dosage balance (relative or absolute) or indirectly through selection against toxic effects of misfolding or misinteracting proteins (Roux et al. 2017).
Recent analyses of salmonid genomes have revealed ∼25% LORe between Atlantic salmon and grayling (Lien et al. 2016; Robertson et al. 2017). Here, we find a set of LORe regions, corresponding to whole chromosome arms in Atlantic salmon, detected as single copy genes in grayling as a result of collapse during the assembly process. This strongly suggests that these sequences are in fact present as near-identical duplicated regions in the grayling genome. The larger chromosome arm-sized regions virtually indistinguishable at the sequence level (∼10% in total, i.e., blue ribbons in fig. 2b) are likely still recombining or have only ceased to do so in the recent evolutionary past. Large-scale chromosomal rearrangements often follow genome duplication to block or hinder recombination among duplicated regions (Comai 2005; Lien et al. 2016). The difference we observe in the rediploidization history between Atlantic salmon and grayling is thus likely linked to the distinctly different chromosome evolution in these species (supplementary fig. S1, Supplementary Material online) (Qumsiyeh 1994).
The genomic footprints of LORe also extend to ohnolog regulatory divergence. LORe regions showed a strong enrichment of species-specific tissue-specific expression pattern (category V, in table 2 and supplementary table S5, Supplementary Material online), as expected under lineage-specific rediploidization and subsequent regulatory divergence. However, we also find a small proportion (∼5%) of genes in AORe regions of the genome that reflect conserved tissue regulation among ohnologs within species but different regulation between species (category V). This observation is more difficult to explain, but it is likely real as the coding- and promoter sequence evolution analyses show that these ohnologs are biased toward high similarity in the coding and promoter sequences within each species (supplementary figs. S9 and S10, Supplementary Material online). Possible explanations for this observation could be a result of nonhomologous gene conversion (Hastings 2010) or evolution of species-specific tissue regulatory networks.
Finally, one fundamental difference between European grayling and Atlantic salmon is that Atlantic salmon has the ability to migrate between fresh- and seawater (anadromy). We demonstrate differences between European grayling and Atlantic salmon gill gene expression regulation for ohnologs of two key genes, NKCC1 and CFTR, involved in chloride ion homeostasis (Marshall and Singer 2002; Nilsen et al. 2007). NKCC1 is involved in entry of chloride ions into the basolateral membrane and is known to be regulated during migration to saltwater. Our finding that only Atlantic salmon displays a strong gill expression bias for one Ss4R copy of NKCC1a (supplementary fig. S11, Supplementary Material online) is congruent with adaptive evolution of novel gill expression regulation to ensure efficient ion transport in gills in anadromous salmonids.
As for the CFTR ohnologs, the results point toward an ancestral gill expression dominance in a salmonid ancestor, followed by a grayling lineage-specific loss of both tissue expression specificity and gill expression dominance, accompanied by increased accumulation of nonsynonymous substitutions (figs. 5a and b). Atlantic salmon on the other hand has retained both copies as “gill specific.” The diverged expression of the CFTR gene copy in European grayling could be related to evolution of novel function, with the elevated dN/dS reflecting a mixture of positive selection on some codons and relaxed purifying selection on others. However, a more parsimonious model of CFTR evolution in European grayling would be that there has been a relaxation of purifying selection pressure to maintain both CFTR copies in the nonanadromous species. We thus propose that maintaining two functional CFTR genes could be an adaptive trait in anadromous salmonids to improve their ability to remove excess chloride ions and maintain ion homeostasis in the sea.
Conclusions
We present the first genome assembly of European grayling and use it for comparative studies with the reference genome assembly of Atlantic salmon. We show that this draft genome assembly is a highly valuable resource for gene-based analyses of salmonids and their relatives. Our comparative genome and transcriptome analysis between Atlantic salmon and grayling provides novel insights into evolutionary fates of ohnologs subsequent to WGD and associations between signatures of selection pressures on gene duplicate regulation and the evolution of salmonid traits, including anadromy. The key candidate genes potentially involved in differences in lifestyle, dietary adaptations, and anadromy between salmon and grayling should be further followed up in future evolutionary and experimental studies. Hence, the genome resource of grayling opens up new exciting avenues for utilizing salmonids as a model system to understand the evolutionary consequences of WGD in vertebrates.
Availability of Data
The Illumina reads have been deposited at ENA under the project accession: PRJEB21333. The genome assembly and annotation data are available at https://doi.org/10.6084/m9.figshare.c.3808162. Atlantic salmon liver expression data are available at ENA or NCBI under sample accessions: SAMEA104483623, SAMEA104483624, SAMEA104483627, and SAMEA104483628.
Supplementary Material
Acknowledgments
This work was supported by funding from University of Oslo to the SAK project “Building a marine genome hub” and the Strategic Research Initiative, Center for Computational Inference in Evolutionary Life Science (CELS) to K.S.J. S.R.S. was funded partly by the Norwegian Research Council (project NFR 244164). We thank Kim M. Bærum for sampling of grayling and Marianne H.S. Hansen for excellent technical assistance. Sample preparation, library construction, and sequencing were carried out at the Norwegian Sequencing Centre (NSC), Norway and McGill University, and Génome Québec Innovation Centre, Canada. The computational work was performed on the Abel Supercomputing Cluster (Norwegian Metacenter for High-Performance Computing [NOTUR] and the University of Oslo), operated by the Research Computing Services group at USIT, the University of Oslo IT-department. We thank Geir O. Storvik and Kjetil L. Voje for helpful discussions. We greatly appreciate Daniel J. Macqueen, Marine S. Brieuc, and Monica H. Solbakken for critical reading of the manuscript.
Author Contributions
K.S.J., L.A.V., S.J., and S.L. conceived and planned the project and generation of the data. S.R.S. and S.V. performed all the analyses with help from A.J.N. and O.K.T. Differential expression analysis on the liver data set was performed by G.B.G. and T.R.H., and the promoter motif analysis was prepared by T.D.M. S.R.S., S.V., and A.J.N. drafted the manuscript. All authors read and commented on the manuscript.
Literature Cited
- Acharya D, Ghosh TC.. 2016. Global analysis of human duplicated genes reveals the relative importance of whole-genome duplicates originated in the early vertebrate evolution. BMC Genomics 17:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrou MA, Swartz BA, Matzke NJ, Oakley TH.. 2013. Genome duplication and multiple evolutionary origins of complex migratory behavior in Salmonidae. Mol Phylogenet Evol. 69(3):514–523. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, et al. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server issue):W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot C, et al. 2014. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 5:3657.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Antoñanzas G, Tocher DR, Taggart JB, Leaver MJ.. 2013. An evolutionary perspective on Elovl5 fatty acid elongase: comparison of Northern pike and duplicated paralogs from Atlantic salmon. BMC Evol Biol. 13:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2000. Endless forms: the evolution of gene regulation and morphological diversity. Cell 101(6):577–580. [DOI] [PubMed] [Google Scholar]
- Chan ET, et al. 2009. Conservation of core gene expression in vertebrate tissues. J Biol. 8(3):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. 2005. The advantages and disadvantages of being polyploid. Nat Rev Genet. 6(11):836–846. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH.. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 9(12):938–950. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. [DOI] [PubMed] [Google Scholar]
- Craig JF. 2008. A short review of pike ecology. Hydrobiologia 601(1):5–16. [Google Scholar]
- De Smet R, Sabaghian E, Li Z, Saeys Y, Van de Peer Y.. 2017. Coordinated functional divergence of genes after genome duplication in Arabidopsis thaliana. Plant Cell 29(11):2786–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D.. 2000. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 17(1):68–74. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DH, Piermarini PM, Choe KP.. 2005. The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev. 85(1):97–177. [DOI] [PubMed] [Google Scholar]
- Faust GG, Hall IM.. 2012. YAHA: fast and flexible long-read alignment with optimal breakpoint detection. Bioinformatics 28(19):2417–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Thomas BC.. 2006. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16(7):805–814. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. arXiv [q-Bio.GN]. Available from: http://arxiv.org/abs/1207.3907. [Google Scholar]
- Gillard G, et al. 2018. Life-stage-associated remodelling of lipid metabolism regulation in Atlantic salmon. Mol Ecol. 27(5):1200–1213. [DOI] [PubMed] [Google Scholar]
- Gnerre S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 108(4):1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS.. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27(7):1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Su Z.. 2007. Tissue-driven hypothesis of genomic evolution and sequence-expression correlations. Proc Natl Acad Sci U S A. 104(8):2779–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschanski K, Warnefors M, Kaessmann H.. 2017. The evolution of duplicate gene expression in mammalian organs. Genome Res. 27(9):1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase D, et al. 2013. Absence of major histocompatibility complex class II mediated immunity in pipefish, Syngnathus typhle: evidence from deep transcriptome sequencing. Biol Lett. 9(2):20130044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley SE. 1987. The chromosomes of salmonid fishes. Biol Rev Camb Philos Soc. 62(3):197–214. [Google Scholar]
- Hastings PJ. 2010. Mechanisms of ectopic gene conversion. Genes 1(3):427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry AP, Stearns SC.. 2004. Evolution illuminated: salmon and their relatives. Oxford University Press. [Google Scholar]
- Hermansen RA, Hvidsten TR, Sandve SR, Liberles DA.. 2016. Extracting functional trends from whole genome duplication events using comparative genomics. Biol Proced Online. 18:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassahn KS, Dang VT, Wilkins SJ, Perkins AC, Ragan MA.. 2009. Evolution of gene function and regulatory control after whole-genome duplication: comparative analyses in vertebrates. Genome Res. 19(8):1404–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-I, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12(4):656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Pääbo S.. 2006. Evolution of primate gene expression. Nat Rev Genet. 7(9):693–702. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, et al. 2005. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309(5742):1850–1854. [DOI] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryuchkova-Mostacci N, Robinson-Rechavi M.. 2015. Tissue-specific evolution of protein coding genes in human and mouse. PLoS One 10(6):e0131673.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [q-Bio.GN]. Available from: http://arxiv.org/abs/1303.3997. [Google Scholar]
- Li J-T, et al. 2015. The fate of recent duplicated genes following a fourth-round whole genome duplication in a tetraploid fish, common carp (Cyprinus carpio). Sci Rep. 5(1):8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S, et al. 2016. The Atlantic salmon genome provides insights into rediploidization. Nature 533(7602):200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limborg MT, Seeb LW, Seeb JE.. 2016. Sorting duplicated loci disentangles complexities of polyploid genomes masked by genotyping by sequencing. Mol Ecol. 25(10):2117–2129. [DOI] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M.. 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33(20):6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2000. The evolutionary fate and consequences of duplicate genes. Science 290(5494):1151–1155. [DOI] [PubMed] [Google Scholar]
- Mackie PM, Gharbi K, Ballantyne JS, McCormick SD, Wright PA.. 2007. Na+/K+/2Cl− cotransporter and CFTR gill expression after seawater transfer in smolts (0+) of different Atlantic salmon (Salmo salar) families. Aquaculture 272(1–4):625–635. [Google Scholar]
- Macqueen DJ, Johnston IA.. 2014. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc Biol Sci. 281(1778):20132881.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WS, Singer TD.. 2002. Cystic fibrosis transmembrane conductance regulator in teleost fish. Biochim Biophys Acta Biomembr. 1566(1–2):16–27. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10. [Google Scholar]
- McGirr JA, Martin CH.. 2017. Parallel evolution of gene expression between trophic specialists despite divergent genotypes and morphologies. Evol Lett. 2:62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TO, et al. 2007. Differential expression of gill Na+, K+ -ATPase α- and β-subunits, Na+,K+, 2Cl- cotransporter and CFTR anion channel in juvenile anadromous and landlocked Atlantic salmon Salmo salar. J Exp Biol. 210(16):2885–2896. [DOI] [PubMed] [Google Scholar]
- Nygren A, Nilsson B, Jahnke M.. 1971. Cytological studies in Thymallus thymallus and Coregonus albula. Hereditas 67(2):269–274. [DOI] [PubMed] [Google Scholar]
- Ocalewicz K, et al. 2013. Pericentromeric location of the telomeric DNA sequences on the European grayling chromosomes. Genetica 141(10–12):409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. New York: Springer-Verlag. [Google Scholar]
- Osborn TC, et al. 2003. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19(3):141–147. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I.. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23(9):1061–1067. [DOI] [PubMed] [Google Scholar]
- Phillips R, Ráb P.. 2001. Chromosome evolution in the Salmonidae (Pisces): an update. Biol Rev Camb Philos Soc. 76(1):1–25. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5(3):e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, et al. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33(Web Server issue):W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qumsiyeh MB. 1994. Evolution of number and morphology of mammalian chromosomes. J Hered. 85(6):455–465. [DOI] [PubMed] [Google Scholar]
- Robertson FM, et al. 2017. Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol. 18(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux J, Liu J, Robinson-Rechavi M.. 2017. Selective constraints on coding sequences of nervous system genes are a major determinant of duplicate gene retention in vertebrates. Mol Biol Evol. 34(11):2773–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandve SR, Rohlfs RV, Hvidsten TR.. 2018. Subfunctionalization versus neofunctionalization after whole-genome duplication. Nat Genet. 50(7):908–909. [DOI] [PubMed] [Google Scholar]
- Sémon M, Wolfe KH.. 2007. Consequences of genome duplication. Curr Opin Genet Dev. 17(6):505–512. [DOI] [PubMed] [Google Scholar]
- Sémon M, Wolfe KH.. 2008. Prefrential subfunctionalization of slow-evolving genes after allopolyploidization in Xenopus laevis. Proc Natl Acad Sci U S A. 105(24):8333–8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Solbakken MH, Voje KL, Jakobsen KS, Jentoft S.. 2017. Linking species habitat and past palaeoclimatic events to evolution of the teleost innate immune system. Proc Biol Sci. 284(1853):20162810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D.. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24(5):637–644. [DOI] [PubMed] [Google Scholar]
- Star B, et al. 2011. The genome sequence of Atlantic cod reveals a unique immune system. Nature 477(7363):207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server issue):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, et al. 2017. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45(D1):D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28(5):511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43(D1):D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A.. 2009. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 10(10):725–732. [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, et al. 2013. From FastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 43: 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M, McInerney JO, Hirt RP, Foster PG, Embley TM.. 2007. Of clades and clans: terms for phylogenetic relationships in unrooted trees. Trends Ecol Evol. 22(3):114–115. [DOI] [PubMed] [Google Scholar]
- Wolfe KH. 2001. Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet. 2(5):333–341. [DOI] [PubMed] [Google Scholar]
- Wray GA, et al. 2003. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 20(9):1377–1419. [DOI] [PubMed] [Google Scholar]
- Yanai I, et al. 2005. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21(5):650–659. [DOI] [PubMed] [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 13(5):555–556. [DOI] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol Evol. 18(6):292–298. [Google Scholar]
- Zheng-Bradley X, Rung J, Parkinson H, Brazma A.. 2010. Large scale comparison of global gene expression patterns in human and mouse. Genome Biol. 11(12):R124.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.