Abstract
Raman microspectroscopy provides chemo-selective image contrast, sub-micrometer resolution, and multiplexing capabilities. However, it suffers from weak signals resulting in image-acquisition times of up to several hours. Surface-enhanced Raman scattering (SERS) can dramatically enhance signals of molecules in close vicinity of metallic surfaces and overcome this limitation. Multimodal, SERS-active nanoparticles are usually labeled with Raman marker molecules, limiting SERS to the coating material. In order to realize multimodal imaging while acquiring the rich endogenous vibronic information of the specimen, a core–shell particle based on “Nanorice”, where a spindle-shaped iron oxide core is encapsulated by a closed gold shell, is developed. An ultrathin layer of silica prevents agglomeration and unwanted chemical interaction with the specimen. This approach provides Raman signal enhancement due to plasmon resonance effects of the shell while the optical absorption in the near-infrared spectral region provides contrast in photoacoustic tomography. Finally, T2-relaxation of a magnetic resonance imaging (MRI) experiment is altered by taking advantage of the iron oxide core. The feasibility for Raman imaging is evaluated by nearfield simulations and experimental studies on the primate cell line COS1. MRI and photo acoustics are demonstrated in agarose phantoms illustrating the promising translational nature of this strategy for clinical applications in radiology.
Keywords: molecular imaging, magnetic resonance imaging (MRI), photoacoustic imaging, radiology, surface-enhanced Raman scattering
1. Introduction
Today’s state-of-the-art imaging techniques for clinical application, such as magnetic resonance imaging (MRI), positron emission tomography (PET), X-ray computed tomography (CT), and different types of microscopy, are usually selected depending on the individual task, weighing the advantages and drawbacks of each method.[1] In case of complementary information, the combination of individual methods has been proven to be highly beneficial for the diagnostic outcome.[2]
It follows that the ability of multimodal image contrast is highly desired while developing new nanoparticle-based contrast agents,[3] allowing for their potential use in intraoperative, theranostic imaging. Raman spectroscopy is a nontoxic and label-free optical method probing the specimen with light in the visible spectral region that can highly benefit from recent nanoparticle-based imaging concepts. Compared to brightfield or even fluorescence microscopy, the so-called Raman micro-spectroscopy provides key advantages such as multiplexing capabilities and the ability to reveal vibrational spectra of the sample that result in images based on molecular information rather than the presence of marker mole cules (e.g., fluorescent dyes). In contrast to IR spectroscopy, Raman can probe outside the optical absorption of water, an ideal property for in vivo applications. Therefore, it has been widely used in material and life sciences[4] where it holds great potential for molecular imaging and brain surgery.[5] For the latter, the discrimination of tumor margins is essential for improving surgical outcomes and the Raman-based differentiation of malignant from normal tissues has already been achieved.[6] Fast chemo selective Raman imaging could complement methods such as MRI at a higher spatial resolution and at time-consuming ex vivo classification of resected tissue.
However, despite its enormous potential, Raman spectroscopy still suffers from weak signals due to the low efficiency of inelastic scattering,[7] and image acquisition times of up to several hours currently prevent imaging applications of clinical relevance.
Over the last decades, many efforts have been made to overcome the limitations summarized above; nonlinear Raman methods[8] such as stimulated Raman[9] and coherent anti-Stokes Raman scattering[10] increased the speed of data acquisition up to video rates.[11] Unfortunately, these methods require complex hardware and are difficult to implement in clinical settings. The so-called surface-enhanced Raman scattering (SERS)[12] can easily be combined with conventional hardware: It is based on surface plasmon resonance effects (SPRE) where electrons of a metallic surface oscillate in resonance with the electric field of the irradiated light. This phenomenon leads to local field enhancement that can reasonably alter the Raman cross section of neighboring molecules. The effect is accompanied by Raman signal enhancement of several orders of magnitude.[13] The increased signal is beneficial for increasing the speed of measurement by decreasing the acquisition time. Unfortunately, the range of nearfield enhancement is limited to a few nanometers which requires the molecules of interest to be located in close vicinity to active sites (the so-called hot spots) of the SERS substrate.
A promising strategy is to directly include Raman marker molecules into the substrate, resulting in perfect control of the metal–particle interaction[14] at the expense of collecting the rich intrinsic spectral information that arise from endogenous sample molecules such as lipids, proteins, or DNA.[15] A different strategy is to expose the sample to plain (nonlabeled) gold or silver nanoparticles which frequently results in strong SERS effects.[16] Those spectra can be different from the spontaneous Raman signatures, for example due to a change of selection rules for molecules positioned close to surfaces[17] or superimposed by surface-enhanced fluorescence. Besides that, it is rather difficult to evaluate the analytical value of SERS spectra in cases where the reference data (spontaneous Raman) of the specimen are not available. As a consequence, intrinsic SERS of biological samples is still not very common.
Last but not least, particle-based imaging agents for clinical applications have to be stable in aqueous solutions and free of cytotoxicity. Especially when MRI contrast is pursued (which might be frequently the case for clinical practice), the common strategy of functionalization by gadolinium compounds has to be reevaluated considering discussions regarding secondary toxicity.[18]
In this study, we address above-mentioned challenges by the development of a SERS imaging agent that can enhance Raman signals of arbitrary biological samples. We provide detailed simulations of nearfield scattering and control experiments that compare the SERS spectra to intrinsic Raman measurements of the same specimen. In addition, we explicitly show MRI contrast that is realized by incorporating iron oxide in the particle, neither requiring gadolinium nor additional steps of surface functionalization.
2. Results and Discussion
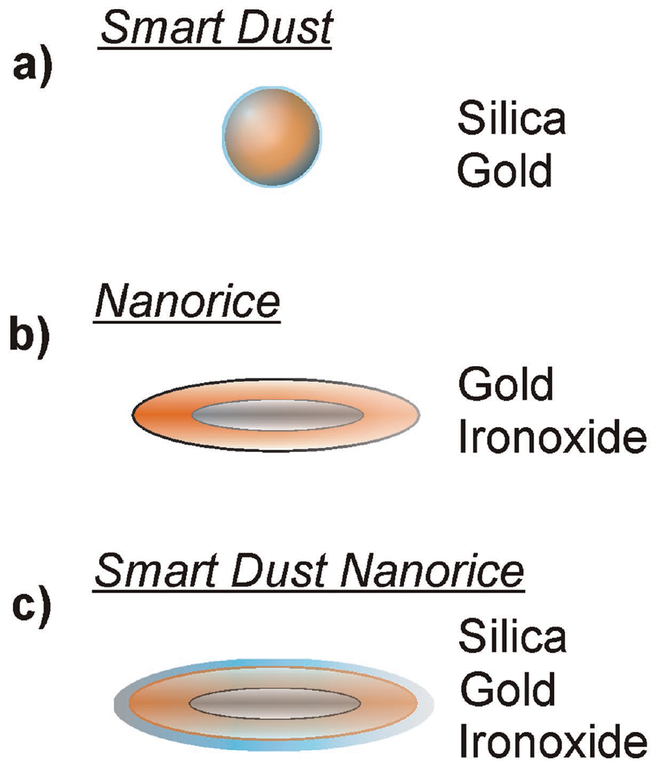
In order to realize the demands described above, we based our approach on two recent concepts of plasmonic nanoparticles: a “smart dust” concept[19] uses gold spheres that are stabilized by an ultrafine layer of silica and randomly distributed over the specimen, see also the illustration in Figure 1a. A concept called “Nanorice”[20] presents a core–shell nanoparticle of gold-coated iron(III)oxide (hematite, illustrated in Figure 1b), which is synthesized while plasmon hybridizations are calculated. The material has also been simulated in terms of considering its properties for plasmonic waveguides.[21] As we hypothesize that the iron oxide core can affect the T2-relaxation time of neighboring protons, Nanorice has been complemented by an ultrathin silica shell (Figure 1c) to prevent agglomeration and direct sample–gold interaction. In the following sections, we refer to this product as Smart-Dust-Nanorice or SDN. Please see Section S1 (Supporting Information) for a brief review of the historical aspects of gold nanoparticle solutions.
Figure 1.
The architecture of the particles: a) “Smart Dust,” a gold sphere of 50 nm diameter that is coated by an ultrafine layer of silica. b) “Nanorice,” a core–shell particle with a core of iron oxide and a shell of gold. c) A combination of both concepts is synthesized in this study and introduced as Smart Dust Nanorice or “SDN.”
2.1. Results of the Chemical Synthesis
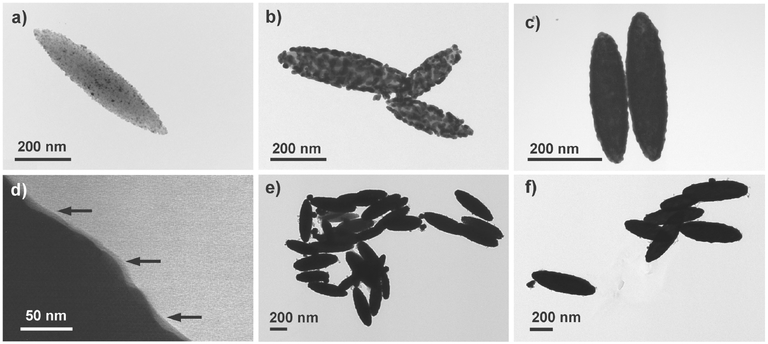
The chemical synthesis of the Nanorice particle was carried out according to literature. We illustrate the process by the following transmission electron microscopy (TEM) images where the gold–shell formation has been stopped at intermediate levels for a better visualization: Figure 2a shows an iron oxide core that has been functionalized by ultrafine gold spheres that were already grown further by electroless deposition. Further growth led to a state shown in Figure 2b, and Figure 2c finally shows how a closed gold shell of ≈30 nm thicknesses has been formed. The product of each stage of the synthesis was controlled before moving to the next level of synthesis. While the formation of the hematite spindles and the gold seed deposition on the surface were monitored via TEM, UV–vis spectroscopy was used to validate the size of the Duff gold-sol which is crucial for the success of the coating procedure of the next step. Please also see Section S2 (Supporting Information) for further details. While using formaldehyde and carbon monoxide as reducing agent, we also confirm that carbon monoxide resulted in a better batch-to-batch reproducibility, as previously recommended.[22] Finally, an ultrathin layer of silica was subsequently added on the product by modifying the “smart-dust-method,” please see the Experimental Section for details. The ultrathin silica coating of ≈5 nm was verified via TEM, Figure 2d, where black arrows point out the silica layer. Figure 2e,f shows groups of finished particles at lower magnification.
Figure 2.
a) TEM image of spindle-shaped hematite particles after seed deposition with ultrafine gold spheres. b) Seed particles grown by electroless deposition of gold. c) Proper growth conditions result in a closed shell of gold, encapsulating the iron oxide core. d) The ultrathin layer of silica (≈5 nm) appears as a gray structure on the gold surface (black arrows). e,f) Cluster of finished SDN particles.
2.2. Nearfield Simulations of Individual Particles
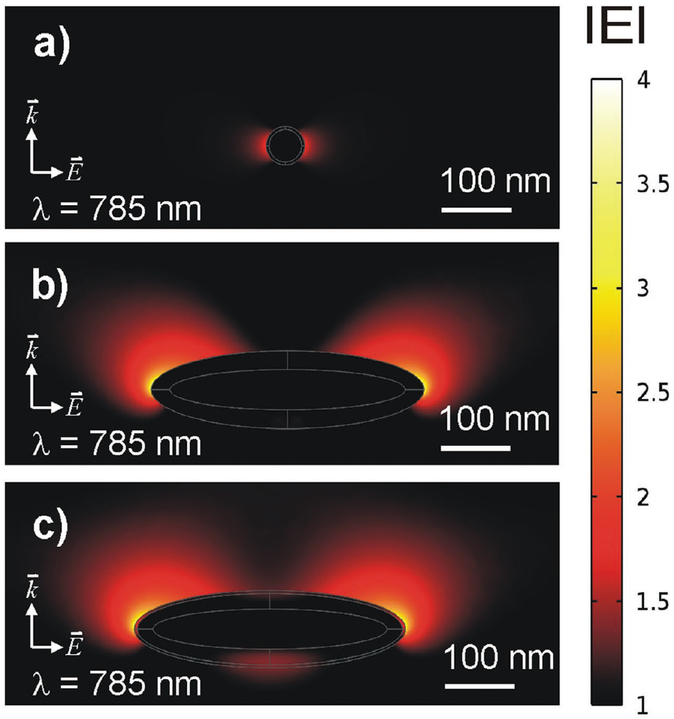
Nearfield simulations (finite element method) of a Smart Dust, the Nanorice, and the SDN particle are shown in Figure 3a–c, respectively. Further examples of nearfield calculation using open-source code are given in Section S1 (Supporting Information). The particle dimensions of 50 nm in diameter (sphere) and 350 nm (a “grain of rice” along its longitudinal axis) were taken from literature. The excitation wavelength is set to 785 nm, the silica layer is modeled to a thickness of 5 nm. The electrical wave propagates from the bottom to top (with the wavevector ) and with a polarization of the electrical field in parallel to the image plane. Phase shifts lead to interference and this is why plotting the modulus of results in nearfield patterns slightly “pushed” from bottom to top. Magnitudes greater than the ambient electrical field strength (set to 1, shown as black background color) were associated with a thermal color scheme at linear scale. As shown in Figure 3a, the gold sphere emits a dipole pattern.[23] Given the small size of the sphere, the area where sample molecules are exposed to enhanced field strength is small. Nevertheless, the electrical field right above the silica surface (≈10 nm) is 1.8 times stronger (bright red). This result shows that the silica coating still allows sample molecules to benefit from potential SERS effects as the introduced spacing to the particle is smaller than the range, to which the ambient electrical field is enhanced. The longitudinal plasmon resonance of Nanorice is simulated in Figure 3b. This particle dimension is seven times larger than the diameter of the sphere and a field enhancement covers longer distances as well. This effect might be highly beneficial for intrinsic SERS applications as it is different from a spherical approach, where increasing the diameter does not automatically result in an increased range of nearfield enhancement due to a shift from dipole to quadrupole resonances (please see Section S1, Supporting Information). In addition, the factor of local field enhancement measures 3.2 (readout 10 nm above the surface), which is almost twice as high as in case of the spherical particle. In Figure 3c, an ultrafine layer of silica has been added to the simulation; the nearfield enhancing properties are preserved and measure a factor of about 3. These simulations can serve as a model for visualizing qualitative nearfield effects of different particle geometries; however, they do not provide a qualitative prediction of SERS, as stated below.
Figure 3.
Nearfield simulations of the particles according to the sketches in Figure 1. The results are shown for an excitation wavelength of 785 nm with a polarization of the electrical field along the image plane and a wave propagating from the bottom to top along . a) Dipole pattern of a single gold sphere with a diameter of 50 nm and 4 nm silica coating. b) Longitudinal plasmon resonance of “Nanorice” with a size of 350 nm along the longitudinal axis, shown true to scale according to (a). c) Longitudinal plasmon resonance with additional silica coating (4 nm).
2.3. Nearfield Simulations of Particle Clusters
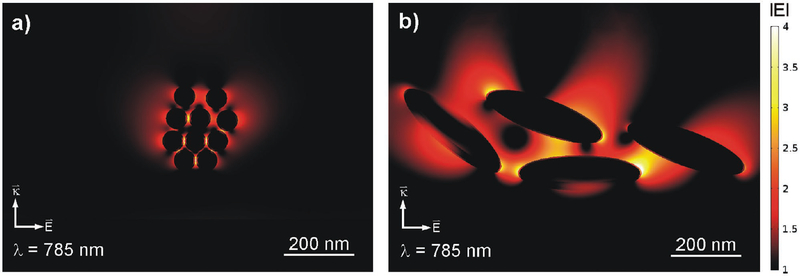
A random cluster of 50 nm gold spheres with an ultrafine layer of silica is modeled in Figure 4a. The nearfield simulation is performed for excitation at 785 nm. Several spots of enhanced field intensity of up to a factor of 4 (again modulus of ) light up at positions, where individual spheres come close to each other. Just as the particle size, these areas are small and the range of enhanced field strength is limited to a few nanometers. Figure 4b shows a completely different behavior for SDN: Although the distances between individual particles measure up to 100 nm, the simulation of four individual particles shows strong nearfield enhancement for large areas between the particles, covering thousands of nm2 and up to 300 nm in range.
Figure 4.
a) Simulated nearfield scattering of a cluster of silica-coated 50 nm gold spheres. Several “hot spots” appear in the gaps between the spheres. b) The same study for a cluster of four SDN particles. In contrast to spheres, local field enhancement occurs for large areas between the particles, covering thousands of nm2 and up to 300 nm in range.
As cited above and supported by these simulations, agglomeration of particles and the resulting “hot spots” play a key role for the signal enhancing properties of a SERS substrate. This raises the question whether or not individual particles can actually provide Raman enhancement at all. This, however, requires thorough control experiments and the ability to control and to measure a single-particle environment. The challenge has recently been mastered for individual spherical particles that were in fact proven to be inactive in terms of Raman enhancement.[24] Based on our simulations, we also consider the SERS effects of SDN as the result of agglomeration and do not assume that the signal arises from individual particles. We also hypothesize that the ability of Nanorice and SDN, namely, to transfer the large particle size into large areas of nearfield enhancement, is an advantage over a spherical approach in case of larger biological structures.
While quantifying the Raman enhancement that can be obtained by SERS effects, recent studies agree to a more moderate approach where Raman signal enhancement occurs up to the power of four of the local electrical field enhancement (modulus).[13] Given the result of Figure 4b, we assume that SDN can enhance Raman signal of sample molecules by a factor of about 250.
2.4. Raman Imaging
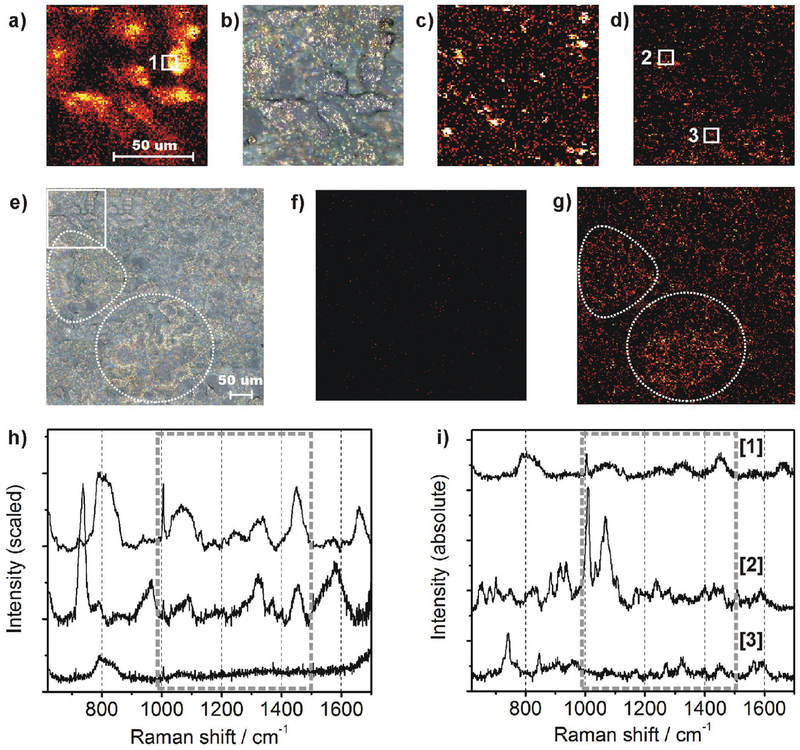
As an intrinsic reference experiment, plain COS cells were scanned with a step size of 1 μm, an incident laser power of 17 mW in focal volume, and an acquisition time of 5 s pixel−1. Accordingly, the overall scan time of 101 × 101 μm took about 14 h. The maximum intensity projection at 1450 wavenumbers (a typical Raman shift for C–H deformation vibrational modes where components such as lipids and proteins contribute to the signal) is shown in Figure 5a. Raman imaging of the same area was conducted again, after spray-coating with SDN particles. The brightfield image in Figure 5b shows the cellular structures that correspond to the Raman image in (a). It also shows evenly distributed SDN particles that exhibit intense backward scattering under white-light illumination. The second Raman scan was carried out at a reduced laser power (by a factor of 50 due to photodamage observed in the presence of SDN) and at a pixel dwell time of 0.5 s. Although these settings should result in 500 times less signal, the maximum intensity projection in Figure 5c shows a large number of bright pixels resulting from SERS effects of the SDN particles. Since the particle density is limited, not every Raman-probed pixel hosts a SERS-active site which explains the dark areas. Since we cannot assume that SERS events are purely selective to the molecules of interest (the cellular compounds), we defined a filter algorithm to select only for spectra that contain endogenous signal of the sample: For each spectrum collected in (c), Pearson’s linear correlation with an intrinsic reference spectrum of COS cells was calculated within the spectral region between 1000 and 1500 cm−1. The latter one was obtained by averaging 18 spectra from the cell membrane collected in Figure 5a (white square no. 1). Finally, pixels showing a correlation coefficient below 0.5 were not considered for image processing while higher values were assigned to a thermal color scheme. Figure 5d shows an image corrected for nonspecific SERS events. Subsequently, the increased speed of data acquisition enabled us to scan a larger area of the sample (402 × 402 μm, step size of 2 μm). A bright-field image (already SDN coated) is shown in Figure 5e. The former scan area is indicated by a dashed square in the upper left. Prominent morphological structures in the middle left and the lower sections of the image are indicated by dashed circles. Laser power and acquisition time were kept as in (c) and (d) for fast scanning. As a negative control, we demonstrate that the absence of SDN particles, Figure 5f, does not contain any comparable Raman results while a high density of SERS results arise after SDN application and rescanning the same area as shown in Figure 5g. The sample structures of Figure 5e are still present (see white circles). The scan was accomplished within 9 h while more than 72 h would have been necessary by using the scan parameters of Figure 3a.
Figure 5.
a) Intrinsic Raman image of COS cells, 101 × 101 μm, maximum intensity projection at 1450 cm−1. b) The same area as in panel (a) using brightfield microscopy (500× magnification) and after application of SDN particles (bright spots). c) Raman experiment after SDN coating, acquisition time and laser intensity were reduced by factor 10 and 50, respectively. d) Correlation analysis (Pearson) between SERS spectra and an intrinsic Raman reference spectrum of the cell membrane. Only correlation coefficients above 0.5 were considered and arranged in a thermal color scheme. e) Bright-field image of a large area (402 × 402 μm) after SDN application. Prominent sample structures are highlighted by dashed circles. f) Raman result of the same area without SDN particles, acquisition time as in (c). g) Scan after SDN application, the data were processed as in (c). Sample structures from (e) are highlighted by dashed circles again. h) Averaged spectra collected from the sample. Upper line: Average of 18 intrinsic spectra collected within the white square in (b). Middle line: Average of 40.400 SERS spectra collected after SDN application. Lower line: Control experiment, average of 40.400 spectra from the image shown in (f), collected before SDN application, scan parameters as in (c). i) Manual selection of three prominent spectra without averaging and scaling. Upper line: Intrinsic reference spectrum of the cell membrane without SDN. Middle and lower line: two single-shot SERS spectra, collected at the white squares in (d).
2.5. Raman Spectroscopy
The following section focuses on the spectral discussion of the SERS results in order to validate that intrinsic Raman information has been retrieved rather than random spectral signatures. The intrinsic reference spectrum of COS cells is shown in Figure 5h (upper line) and the multiple signals arising in the fingerprint region are typical for intrinsic Raman spectra of eukaryotic cells. The averaged SERS spectrum (10.000 spectra) from the spectra collected in Figure 5f is shown below (middle line). The region between 1000 and 1500 cm−1 (dashed frame) matches very well with the intrinsic reference. The SERS spectrum exhibits lower intensity around 800 cm−1, higher intensities around 950 and 1600 cm−1, and an additional sharp feature at 730 cm−1. The signal at 800 cm−1 can be associated to the O–P–O vibrational mode of the DNA backbone and it is assumed that due to spray-coating, the SDN particles mostly attached to the cellular surface and did not get in proximity to the nucleus and mitochondria, where the DNA is located. The strong signal around 730 cm−1 is frequently reported for SERS experiments and indicates successful interaction between SERS substrate and cellular compounds of the sample.[25] The averaged spectral result (40.400 spectra) of the negative control (imaging result shown in Figure 4f is shown in Figure 5h, lower line, and here most of the vibrational features are absent. Only a weak vibrational mode at 800 cm−1 and a few minor features in the spectral region between 1000 and 1500 cm−1 are still present. This result proves that the observed SDN signal enhancement is not just an effect of averaging a large number of spectra and indeed originates from intrinsic SERS effects. The experimental results clearly demonstrate the utility of the SDN particle for efficient and intrinsic Raman imaging of biological samples. Additional control experiments are provided in the Supporting Information where we show enhancement of different chemical compounds, provide a discussion of potential “background” SERS spectra, and evaluate intrinsic Raman enhancement based on the plain spherical “smart dust” particle (Sections S3–S5, Supporting Information). As shown in Figure 5i, individual (“single-shot”) spectra originating from the SERS active sites show well-resolved Raman signals. Different from Figure 5h, the spectral intensities were not scaled so that absolute intensities (counts CCD) can be compared. Their intensities are in the same order of magnitude than spectra obtained from the spontaneous Raman control experiment (which was used at ten times longer acquisition time and 50 times higher laser power). Starting with a single, non-SDN reference spectrum of the cell membrane (5 s acquisition time, upper line), two SDN spectra (collected within 0.5 s) are plotted as the middle and lower lines. They were collected from the positions marked by square nos. 2 and 3 in Figure 5d. They either show high-intensity features around 1000 and 1100 cm−1 or a comprehensive set of vibrational features that, within 1000 and 1500 cm−1, again correspond very well to the intrinsic reference.
2.6. MRI and Photoacoustic Imaging with SDN
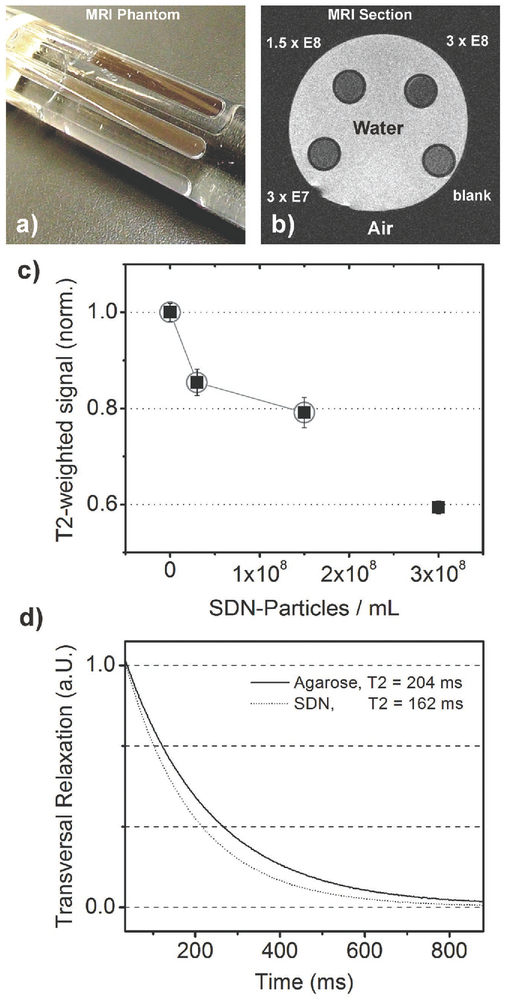
The SDN particle is composed of iron oxide (hematite) and a gold nanoshell that absorbs light in the near-infrared region. We therefore assumed that the substrate can also provide multi modal image contrast using both MRI and photoacoustic imaging. For nanoparticles of iron oxide (magnetite), it has already been shown that they can alter the transversal relaxation time (T2) within an MRI experiment[26] which has been used to increase MRI contrast without the need of gadolinium in recent studies.[27] In case of SDN, this would even allow MRI contrast without the need for additional surface functionalization. While the motivation for MRI contrast is obvious due to its superior role in functional imaging of living subjects,[28] photoacoustic tomography is rather inexpensive and overcomes the drawback of depth-limited imaging associated with purely optical methods.[29] For validating MRI effects of SDN, a test sample (“phantom”) was used as shown in Figure 6a. It contains water and four NMR test tubes filled with plain water and different concentrations of SDN in water, each of them stabilized by 2% agarose. The T2-weighted MRI image (cross section of the phantom) is shown in Figure 6b where the tubes appear as dark circles. The contrast increases with the SDN concentration. The results of averaging multiple slices are shown in the diagram of Figure 6c where contrast values are normalized to the reference tube. It follows that a concentration of 1.5 × 108 particles mL−1 decreases the MRI signal to 80% of its reference value. For this SDN concentration, the transversal relaxation has been measured accordingly, shown in Figure 6d. Time constants of 204 ms were found for plain agarose while 162 ms were measured in presence of SDN. This result shows that SDN indeed affects T2 relaxation and that the imaging results are not just based on secondary effects, for example, a lower proton density of the concentrated samples.
Figure 6.
a) Photograph of a phantom for MRI. Test tubes containing different concentrations of SDN in aqueous solution and plain agarose in water (as a reference) were arranged in parallel and placed in a larger plastic tube filled with water. b) Axial section of this phantom shows darkening of the test tubes while SDN concentrations increase. c) Plot of T2-contrast relative to the reference. d) T2-relaxation curves for plain agarose (solid line) and an SDN concentration of 3 × 107 particles mL−1.
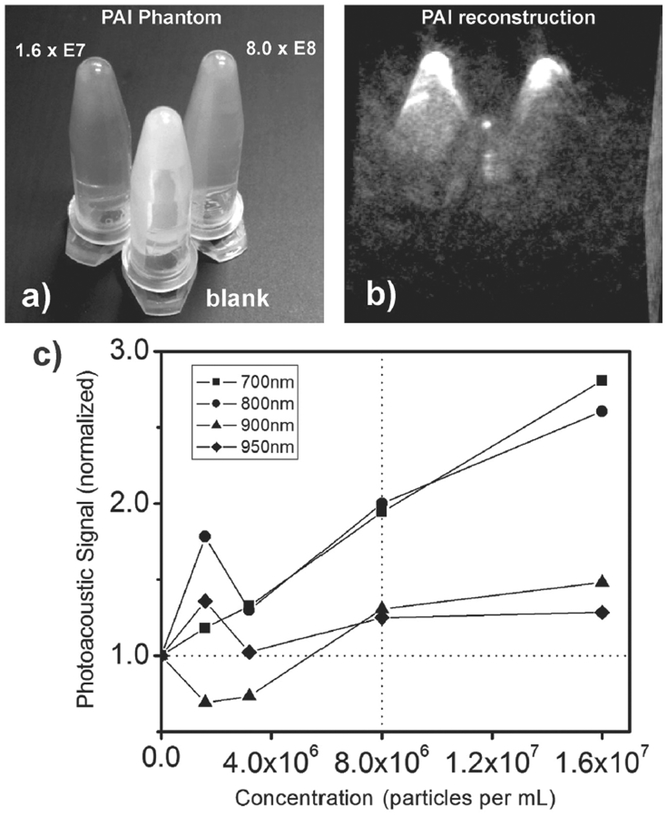
Photoacoustic tomography was performed by filling Eppendorf tubes with four different concentrations of SDN in aqueous suspension, stabilized by 2% agarose. The samples were scanned in two groups, each time in the presence of a reference tube filled with plain agarose. A photograph of the tubes is shown in Figure 7a. The same arrangement was kept during the measurement so that the main area of laser irradiation was located between the tips for homogeneous irradiation. Figure 7b shows the 3D reconstruction of the photoacoustic data, collected at a probe wavelength of 700 nm, and an acquisition time of 12 s. The strongest concentration of SDN (rear left, 1.6 × 107 particles mL−1) shows the strongest signal intensity. Half of the initial concentration still results in a prominent signal (rear right, 8 × 106 particles mL−1). The weak signal emerging from the reference tube (in front) confirms that in fact the SDN particles cause the enhanced signal. A quantitative examination is presented in Figure 7c for four different excitation wavelengths. For each scan, the signals emitted from the tips were plotted against the SDN concentration. The reference value (set to one) is shown as dashed horizontal line. An increasing SDN concentration also increased the photoacoustic signal and 8 × 106 particles mL−1 resulted in a strong signal intensity compared to plain agarose. This trend holds for all of the four different wavelengths, however, it is less pronounced for 900 and 950 nm due to the increasing optical absorption of water.
Figure 7.
a) Photograph of Eppendorf tubes filled with different SDN concentrations (rear left and rear right) and plain agarose (in front) for photoacoustic imaging. b) The tomographic reconstruction (3D volume rendering) of the photoacoustic data. c) Plot of the photoacoustic signal as a function of SDN concentrations at four different wavelengths of excitation. The results are shown as square (700 nm), circle (800 nm), triangle (900 nm), and diamond (950 nm).
3. Conclusion
We exploited the respective advantages of core–shell nanoparticles with ultrathin silica coating to create a plasmonic nano-tool for imaging application. The resulting particle enhanced the spectral signatures of COS cells by two orders of magnitude and Raman imaging was based on purely intrinsic Raman signals rather than by specifically incorporated Raman marker molecules. The SERS properties were combined with gadolinium-free MRI contrast; T2-shortening was demonstrated for magnetic resonance imaging in agarose phantoms, darkening the reference signal by 20%. Photoacoustic signal was enhanced by a factor of 2. This is the first experimental validation and characterization of the SERS effects of Nanorice and the first time that intrinsic Raman signal enhancement is combined with a gadolinium-free MRI contrast modality within a plasmonic nanoparticle.
Subsequent projects studies could use spectral information to discriminate between biological components such as gray and white matter classification of brain tissue based on the corresponding Raman spectra.[30] Moreover, we expect greatest potential for the combination of SERS with widefield Raman illumination, recently described as a highly promising concept[31] which could finally increase the speed of data acquisition to become sufficiently short for intrinsic in vivo Raman imaging at the simplicity of spontaneous Raman hardware. The transfer from hematite to magnetite should result in a better MRI performance and would allow additional functionality by manipulating particle properties and temperature by external magnetic fields.[32] Potential CT-contrast due to the gold shell is expected as well. We finally aim to decrease the particle size to enable tumor selective uptake due to EPR effects and/or by recent approaches of controlled cell particle interaction.[33]
For example, a synthesis of ultrafine magnetide nanorods by ultrasound irradiation as presented by Kumar et al.[34] could point out the way toward a promising strategy to address the remaining challenges above.
Future studies will have to put the focus on potential cytotoxicity. As mentioned above, the silica shell is intended as a protection in two different ways; on the one hand from particle agglomeration, on the other hand from unwanted interaction of the gold surface with the specimen. The concept of silica coating has shown in the recent past that Raman-labeled gold spheres have got a very low cytotoxicity.[35] A potential long-term decomposition of the ultrafine silica layer, however, will have to be carefully monitored.
In conclusion, this study emphasizes the significant multi-modal potential of advanced core–shell substrates as contrast agents for clinical molecular imaging.
4. Experimental Section
Chemical compounds were purchased from Sigma-Aldrich, carbon monoxide was generated in situ by injecting formic acid (53%) in concentrated sulfuric acid. Glassware was cleaned with aqua regia and flushed with deionized water (Siemens Millipore).
Transmission electron microscopy of particles was performed by using by a JEOL 1230 microscope (Jeol USA Inc.).
The chemical synthesis of the initial “Nanorice” particle was carried out according to literature.[20] In brief, iron chloride was hydrolyzed in the presence of phosphate ions resulting in spindle-shaped hematite particles with a length of ≈300–400 nm and a diameter of 120–160 nm. As a second step, an ultrafine gold sol was obtained using the Duff method.[36] After surface functionalization by aminopropyltrimethoxysilane (APTMS), Nanorice was obtained by coating of 0.9 mL of the seed-coated iron oxide spindle suspension under vigorous stirring and gentle warming. To proceed, 20 μL APTMS (97% stock solution) were injected in the reaction mixture immediately after Nanorice formation. While continuing stirring, 2 mL of a sodium silica solution of 0.54% weight were added after 1 min. The solution was kept under gentle stirring for 48 h and frequently sonicated. Afterward, the particles were washed in deionized water and stored at room temperature in aqueous suspension. SDN was observed to be stable in aqueous suspensions for a period of up to six weeks. Afterward, irreversible aggregation occurred within a few days. The same effect was observed for the original SHINERS particles (synthesized in parallel experiments, data not shown) and might be explained by the fact that the silica coating is an order of magnitude thinner than in current substrates for in vivo imaging[37] and can dissolve within the observed time frame.
Nanoparticle tracking analysis was carried out by using a Nanosight LM300 setup (Malvern Instruments Ltd.) and allowed to determine the initial particle concentration.
COS cells COS1, ATCC CRL-1650 were selected as representative of mammalian cells that can be grown directly on quartz slides without additional surface treatment that could superimpose the cellular Raman signatures. Cells were grown in Dulbecco’s modified Eagle medium (DMEM; Life Technologies) containing fetal bovine calf serum, glutamine, glucose, and penicillin/streptomycin and propagated at 37 °C in a humidified atmosphere with 5% CO2. Slides were flushed with phosphate buffered saline and air dried without further treatment. For a uniform deposition of SDN particles, an aqueous suspension of particles was sprayed on the sample by using an airbrush setup with 0.3 mm nozzle diameter.
Hyperspectral Raman data sets were collected between 620 and 1700 wavenumbers. A Raman microscope (ReniShaw inVia, UK) was used, equipped with a 785 nm CW laser, neutral density filters to reduce the laser power, a CCD detector, and dry objective (Leica NPlan 50×, NA = 0.75). The spectra were recorded in point-scanning mode and baseline corrected by fitting with polynomial basis functions (Matlab Software, MathWorks). Raman images were displayed in LabView software (National Instruments). The laser power was measured at the backside aperture of the microscope objective by using a power meter (Coherent Inc.). To estimate the power within focal volume, the initial value was scaled by a factor of 0.6 as a typical value for transmission efficiencies at 785 nm for standard microscope objectives. Spectra and graphs were displayed by using Origin Software (OriginLab Inc.).
The photoacoustic data sets were collected on a Nexus 128 photoacoustic scanner (Endra Life Sciences, USA) via agarose phantom imaging. Eppendorf tubes (0.6 mL) were filled with 500 μL of aqueous SDN suspensions at four different concentrations, namely, 1,6 × 107, 8 × 106, 3.2 × 106, and 1.6 × 106 particles mL−1 and 2% agarose to prevent from precipitation. A fifth tube served as reference containing 2% agarose in plain water. The results were visualized by using Amide software[38] (GNU license).
MRI data sets were collected on an Icon scanner (1T, Bruker Corp., USA). Scans of the sample were performed using a T2-RARE scheme[39] with a time to repeat of 3000 ms, an echo time of 200 ms, and averaging six scans. The DICOM images were further processed with ImageJ software (Wayne Rasband, public domain, National Institute of Health, USA). To validate T2 effects, glass tubes for NMR spectroscopy were filled with aqueous suspensions of SDN at concentrations of 3 × 108, 1.5 × 108, and 3 × 107 particles mL−1. The suspension was stabilized by 2% agarose to prevent from precipitation. A fourth tube contained plain agarose solution and was used as a reference. The tubes were arranged as an array within a plastic tube filled with water.
Nearfield simulations of the main particle were performed by defining the particles in a virtual environment and solving for the scattering problem by using Comsol Multiphysics Finite Element Method software (Comsol Inc.), Version 4.4. The open source code DDSCAT[40] was used for calculating the examples in the Experimental Section.
Supplementary Material
Acknowledgements
The authors acknowledge Dr. Laura J. Pisani and Dr. Ryan Spitler for assisting in collection of MRI data. The authors further acknowledge Dr. Jung Ho Yo for his useful comments of the manuscript. This work was funded by the NCI CCNE-T U54 U54CA151459 grant. The authors would also like to thank the Canary Foundation, Ben & Catherine Ivy Foundation, and Sir Peter Michael Foundation. J.V.J. acknowledges NIH HL117048. The authors acknowledge the Victorian Government of Australia for providing partial funding support for this work in the form of a Victorian postdoctoral research fellowship for J.L.C. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
References
- [1].Aisen AM, Martel W, Braunstein EM, McMillin KI, Phillips WA, Kling T, Am. J. Roentgenol 1986, 146, 749; Kinkel K, Lu Y, Both M, Warren RS, Thoeni RF, Radiology 2002, 224, 748,; Boss A, Bisdas S, Kolb A, Hofmann M, Ernemann U, Claussen CD, Pfannenberg C, Pichler BJ, Reimold M, Stegger L, J. Nucl. Med. 2010, 51, 1198,; Jermyn M, Mok K, Mercier J, Desroches J, Pichette J, Saint-Arnaud K, Bernstein L, Guiot M-C, Petrecca K, Leblond F, Sci. Transl. Med 2015, 7, 274ra19. [DOI] [PubMed] [Google Scholar]
- [2].Cherry SR, in Multimodality Imaging: Beyond PET/CT and SPECT/CT, Seminars in Nuclear Medicine, Elsevier 2009, 39, p. 348; Alaboudy A, Inoue T, Hatanaka K, Chung H, Hyodo T, Kumano S, Murakami T, Moustafa EFA, Kudo M, Oncology 2011, 81, 66. [Google Scholar]
- [3].Kircher MF, de la Zerda A, Jokerst JV, Zavaleta CL, Kempen PJ, Mittra E, Pitter K, Huang R, Campos C, Habte F, Nat. Med 2012, 18, 829; Liu H, Lu W, Wang H, Rao L, Yi Z, Zeng S, Hao J, Nanoscale 2013, 5, 6023,; Yi Z, Li X, Xue Z, Liang X, Lu W, Peng H, Liu H, Zeng S, Hao J, Adv. Funct. Mater. 2015, 25, 7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Petry R, Schmitt M, Popp J, ChemPhysChem 2003, 4, 14; [DOI] [PubMed] [Google Scholar]; b) Krafft C, Popp J, Anal. Bioanal. Chem. 2015, 407, 699. [DOI] [PubMed] [Google Scholar]
- [5].a) Jokerst JV, Pohling C, Gambhir SS, MRS Bull. 2013, 38, 625; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Koljenovic S, Bakker Schut T, Wolthuis R, Vincent A, Hendriks-Hagevi G, Santos L, Kros J, Puppels G, Anal. Chem. 2007, 79, 557. [DOI] [PubMed] [Google Scholar]
- [6].a) Bergner N, Romeike BF, Reichart R, Kalff R, Krafft C, Popp J, in European Conf. Biomedical Optics, OSA publishing, Washington D.C, USA: 2011, p. 80870X; [Google Scholar]; b) Gajjar K, Heppenstall LD, Pang W, Ashton KM, Trevisan J, Patel II, Llabjani V, Stringfellow HF, Martin-Hirsch PL, Dawson T, Anal. Methods 2013, 5, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Long DA, Quantum Mechanical Theory of Rayleigh and Raman Scattering, Wiley, West Sussex, England: 2002. [Google Scholar]
- [8].a) Chung C-Y, Boik J, Potma EO, Annu. Rev. Phys. Chem. 2013, 64, 77; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cheng J-X, Xie XS, Coherent Raman Scattering Microscopy, CRC Press, Boca Raton, Florida, USA: 2016; [Google Scholar]; c) Kiskis J, Fink H, Nyberg L, Thyr J, Li J-Y, Enejder A, Sci. Rep 2015, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Volkmer A, Emerging Raman Applications and Techniques in Biomedical and Pharmaceutical Fields, Springer International Publishing AG, Switzerland: 2010, p. 111. [Google Scholar]
- [10].a) Maker P, Terhune R, Phys. Rev. 1965, 137, A801; [Google Scholar]; b) Camp CH Jr., Cicerone MT, Nat. Photonics 2015, 9, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Evans CL, Potma EO, Puoris’ haag M, Côté D, Lin CP, Xie XS, Proc. Natl. Acad. Sci. USA 2005, 102, 16807; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Saar BG, Freudiger CW, Reichman J, Stanley CM, Holtom GR, Xie XS, Science 2010, 330, 1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Albrecht MG, Creighton JA, J. Am. Chem. Soc. 1977, 99, 5215; [Google Scholar]; b) Jeanmaire DL, Van Duyne RP, J. Electroanal. Chem. Interfacial Electrochem 1977, 84, 1; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Stiles PL, Dieringer JA, Shah NC, Van Duyne RP, Annu. Rev. Anal. Chem 2008, 1, 601; [DOI] [PubMed] [Google Scholar]; d) Fan M, Andrade GF, Brolo AG, Anal. Chim. Acta 2011, 693, 7; [DOI] [PubMed] [Google Scholar]; e) Lakowicz JR, Anal. Biochem. 2005, 337, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Le Ru EC, Etchegoin PG, MRS Bull. 2013, 38, 631. [Google Scholar]
- [14].Gandra N, Singamaneni S, Adv. Mater. 2013, 25, 1022. [DOI] [PubMed] [Google Scholar]
- [15].Matthews Q, Brolo A, Lum J, Duan X, Jirasek A, Phys. Med. Biol. 2011, 56, 19. [DOI] [PubMed] [Google Scholar]
- [16].Huang X, El-Sayed IH, Qian W, El-Sayed MA, J. Am. Chem. Soc. 2006, 128, 2115. [DOI] [PubMed] [Google Scholar]
- [17].Otto A, Mrozek I, Grabhorn H, Akemann W, Phys J.: Condens. Matter 1992, 4, 1143. [Google Scholar]
- [18].Rogosnitzky M, Branch S, BioMetals 2016, 29, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li JF, Huang YF, Ding Y, Yang ZL, Li SB, Zhou XS, Fan FR, Zhang W, Zhou ZY, Wu DY, Nature 2010, 464, 392. [DOI] [PubMed] [Google Scholar]
- [20].Wang H, Brandl DW, Le F, Nordlander P, Halas NJ, Nano Lett. 2006, 6, 827. [DOI] [PubMed] [Google Scholar]
- [21].Cui X, Erni D, in 14th European Conf. Integrated Optics (ECIO’08) Jakajima B.V, Eindhoven, The Netherlands: 2008, p. 11. [Google Scholar]
- [22].Brinson BE, Lassiter JB, Levin CS, Bardhan R, Mirin N, Halas NJ, Langmuir 2008, 24, 14166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kelly KL, Coronado E, Zhao LL, Schatz GC, J. Phys. Chem. B 2003, 107, 668. [Google Scholar]
- [24].Zhang Y, Walkenfort B, Yoon JH, Schlücker S, Xie W, Phys. Chem. Chem. Phys. 2015, 17, 21120. [DOI] [PubMed] [Google Scholar]
- [25].a) Papadopoulou E, Bell SE, Analyst 2010, 135, 3034; [DOI] [PubMed] [Google Scholar]; b) Li B, Sirimuthu N, Ray BH, Ryder AG, J. Raman Spectrosc. 2012, 43, 1074; [Google Scholar]; c) Cialla D, Pollok S, Steinbrücker C, Weber K, Popp J, Nanophotonics 2014, 3, 383. [Google Scholar]
- [26].a) Lee N, Yoo D, Ling D, Cho MH, Hyeon T, Cheon J, Chem. Rev. 2015, 115, 10637; [DOI] [PubMed] [Google Scholar]; b) Simon GH, Bauer J, Saborovski O, Fu Y, Corot C, Wendland MF, Daldrup-Link HE, Eur. Radiol. 2006, 16, 738. [DOI] [PubMed] [Google Scholar]
- [27].a) Buxton RB, Edelman RR, Rosen BR, Wismer GL, Brady TJ, J. Comput. Assisted Tomogr. 1987, 11, 7; [DOI] [PubMed] [Google Scholar]; b) Seyfer P, Pagenstecher A, Mandic R, Klose KJ, Heverhagen JT, J. Magn. Reson. Imaging 2013, 39, 665. [DOI] [PubMed] [Google Scholar]
- [28].a) Sinke MR, Dijkhuizen RM, Caimo A, Stam CJ, Otte WM, NeuroImage 2016, 135, 79; [DOI] [PubMed] [Google Scholar]; b) Bipat S, Glas AS, van der Velden J, Zwinderman AH, Bossuyt PM, Stoker J, Gynecol. Oncol. 2003, 91, 59; [DOI] [PubMed] [Google Scholar]; c) Sosnovik DE, Weissleder R, Curr. Opin. Biotechnol. 2007, 18, 4; [DOI] [PubMed] [Google Scholar]; d) Winkler SA, Schmitt F, Landes H, DeBever J, Wade T, Alejski A, Rutt BK, Neuro-image 2016, 10.1016/j.neuroimage.2016.11.033; [DOI] [PMC free article] [PubMed]; e) Kraff O, Fischer A, Nagel AM, Mönninghoff C, Ladd ME, J. Magn. Reson. Imaging 2015, 41, 13. [DOI] [PubMed] [Google Scholar]
- [29].a) Zackrisson S, van de Ven S, Gambhir S, Cancer Res. 2014, 74, 979; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Van de Sompel D, Sasportas LS, Jokerst JV, Gambhir SS, PLoS One 2016, 11, e0152597; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Bohndiek SE, Bodapati S, Van De Sompel D, Kothapalli S-R, Gambhir SS, PLoS One 2013, 8, e75533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].a) Ji M, Orringer DA, Freudiger CW, Ramkissoon S, Liu X, Lau D, Golby AJ, Norton I, Hayashi M, Agar NY, Sci. Transl. Med 2013, 5, 201ra119; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pohling C, Buckup T, Pagenstecher A, Motzkus M, Biomed. Opt. Express 2011, 2, 2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bohndiek SE, Wagadarikar A, Zavaleta CL, Van de Sompel D, Garai E, Jokerst JV, Yazdanfar S, Gambhir SS, Proc. Natl. Acad. Sci. USA 2013, 110, 12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Thakor AS, Jokerst JV, Ghanouni P, Campbell JL, Mittra E, Gambhir SS, J. Nucl. Med. 2016, 57, 1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].a) Kempen PJ, Kircher MF, de la Zerda A, Zavaleta CL, Jokerst JV, Mellinghoff IK, Gambhir SS, Sinclair R, Micron 2015, 68, 70; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Parak WJ, Science 2016, 351, 814. [DOI] [PubMed] [Google Scholar]
- [34].Kumar RV, Koltypin Y, Xu X, Yeshurun Y, Gedanken A, Felner I, J. Appl. Phys. 2001, 89, 6324. [Google Scholar]
- [35].a) Thakor AS, Gambhir SS, CA: Cancer J. Clin. 2013, 63, 395; [DOI] [PubMed] [Google Scholar]; b) Campbell JL, SoRelle ED, Ilovich O, Liba O, James ML, Qiu Z, Perez V, Chan CT, de la Zerda A, Zavaleta C, Biomaterials 2017, 135, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Duff DG, Baiker A, Edwards PP, Langmuir 1993, 9, 2301. [Google Scholar]
- [37].Zavaleta CL, Hartman KB, Miao Z, James ML, Kempen P, Thakor AS, Nielsen CH, Sinclair R, Cheng Z, Gambhir SS, Small 2011, 7, 2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Loening AM, Gambhir SS, Mol. Imaging 2003, 2, 131. [DOI] [PubMed] [Google Scholar]
- [39].Hennig J, Nauerth A, Friedburg H, Magn. Reson. Med. 1986, 3, 823. [DOI] [PubMed] [Google Scholar]
- [40].a) Draine BT, Flatau PJ, J. Opt. Soc. Am. A 2008, 25, 2693; [DOI] [PubMed] [Google Scholar]; b) Flatau P, Draine B, Opt. Express 2012, 20, 1247; [DOI] [PubMed] [Google Scholar]; c) Flatau PJ, Draine B, J. Opt. Soc. Am. A 1994, 11, 1491; [Google Scholar]; d) Goodman J, Draine BT, Flatau PJ, Opt. Lett. 1991, 16, 1198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.