Abstract
Intra-pancreatic activation of the digestive proteases trypsin and chymotrypsin is an early event in the development of pancreatitis. Human genetic studies indicate that chymotrypsin controls trypsin activity via degradation, but there is no evidence of this from animal models. We used CRISPR-Cas9 to disrupt the chymotrypsinogen B1 gene (Ctrb1) in C57BL/6N mice and induced pancreatitis in CTRB1-deficient and C57BL/6N (control) mice by administration of cerulein. CTRB1-deficient mice given cerulein had significant increases in intra-pancreatic trypsin activity and developed more severe pancreatitis compared with control mice. CTRB1 therefore protects against secretagogue-induced pancreatitis by reducing trypsin activity. Protease inhibitors developed for treatment of pancreatitis should be designed to target trypsin but not chymotrypsin.
Keywords: mouse model, pancreas, inflammation, proteolysis
Activation of digestive proteases inside pancreatic acinar cells is an early event in experimental models of acute pancreatitis in rodents [1, 2]. The best-characterized example is the activation of trypsin and chymotrypsin in response to supramaximal stimulation with secretagogues such as cerulein, an analog of cholecystokinin. In the cerulein-induced pancreatitis model, the lysosomal cysteine protease cathepsin B appears to be responsible for activation of trypsinogen to trypsin [1–3]. In turn, trypsin can activate chymotrypsinogen to chymotrypsin, although an ATP-dependent activation mechanism unrelated to trypsin has been also proposed [4]. Trypsin activity has been implicated as a driver of the pathogenic process by inducing acinar cell apoptosis or necrosis either directly [5] or through the cytoplasmic release of cathepsin B [6]. Genetic deletion of cathepsin B or the mouse cationic trypsinogen (T7) moderately protected against pancreatitis, suggesting that trypsin-independent mechanisms also contribute to disease severity [3, 7]. The pathogenic role of chymotrypsin in the rodent pancreatitis models has been less well characterized; although the powerful digestive activity of this protease suggests significant harmful potential.
Human genetic studies together with biochemical investigations confirmed the pivotal role of trypsin and chymotrypsin in pancreatitis and revealed that chymotrypsin regulates trypsin activity through trypsinogen degradation and thereby exerts a protective function [8]. The clinically most common mutations in PRSS1 encoding human cationic trypsinogen cause hereditary pancreatitis by rendering trypsinogen resistant to protective degradation by chymotrypsin C (CTRC) and thereby increasing intra-pancreatic trypsin activity. Loss-of-function mutations in CTRC are independent risk factors for chronic pancreatitis, even in the absence of trypsinogen mutations, demonstrating that chymotrypsin-dependent trypsin control is essential for pancreas health. A regulatory role for chymotrypsin was further corroborated by the recent observations that a common inversion at the CTRB1-CTRB2 locus (encoding chymotrypsin B1 and B2) has a small but significant impact on pancreatitis risk [9].
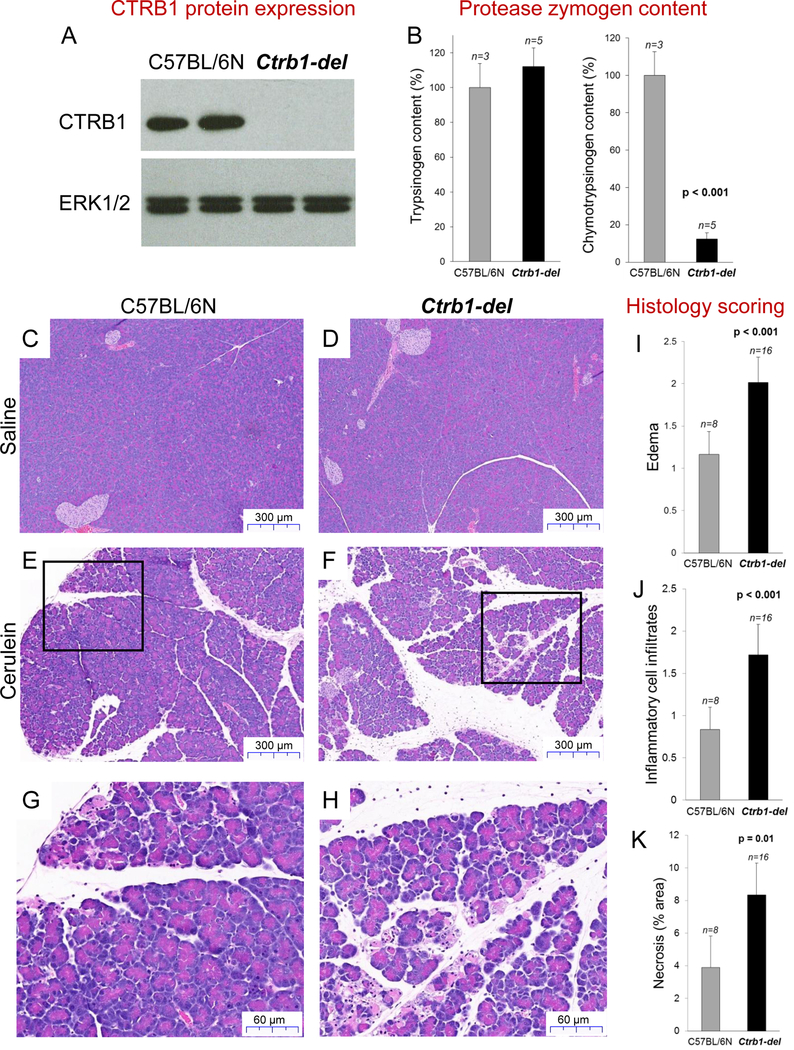
To confirm the presumed protective role of chymotrypsin in a mouse model in vivo, we used CRISPR-Cas9 based genome editing in C57BL/6N mice to disrupt the Ctrb1 gene. The mouse genome contains a single Ctrb1 gene, which encodes the functional ortholog of human CTRB2 [10, 11]. Genomic manipulation resulted in the surgical deletion of the 79 nucleotides long exon 4 from Ctrb1 (Supplementary Figure S1). Reverse-transcription PCR of pancreatic mRNA and Western blots of pancreas homogenates from homozygous Ctrb1-del mice confirmed successful elimination of CTRB1 expression (Figure 1A, Supplementary Figure S2). Total chymotrypsinogen content of pancreas homogenates from Ctrb1-del mice was reduced by almost 90%, relative to C57BL/6N controls, while trypsinogen content was unchanged (Figure 1B). Because the C57BL/6N strain is naturally deficient in CTRC, the remaining chymotrypsinogen is likely chymotrypsin-like protease (CTRL) [12, 13] or a non-acinar protease with chymotrypsin-like activity such as mast cell chymase. Mice deficient in CTRB1 showed no obvious phenotypic changes relative to their C57BL/6N parent strain; they exhibited normal weight gain and typical breeding behavior. Macroscopic pancreas morphology and pancreas histology, as judged on hematoxylin-eosin (H&E) stained sections, was also unchanged. No spontaneous pancreatitis or other pancreatic pathology was apparent up to 1 year of age.
Figure 1.
Chymotrypsin expression and cerulein-induced pancreatitis in Ctrb1-del and C57BL/6N control mice. A, Western blot analysis of CTRB1 protein in pancreas homogenates. ERK1/2 was measured as loading control. B, Trypsinogen and chymotrypsinogen content of pancreas homogenates. C-F, Representative hematoxylin-eosin (H&E) stained histological sections of the pancreas from mice treated with 10 hourly injections of saline (C, D) or cerulein (E, F). G, H, Higher magnification of indicated areas. I-K, Histology scoring of H&E stained sections for edema (I), inflammatory cell infiltration (J) and acinar cell necrosis (K). Mean values with S.D. are shown.
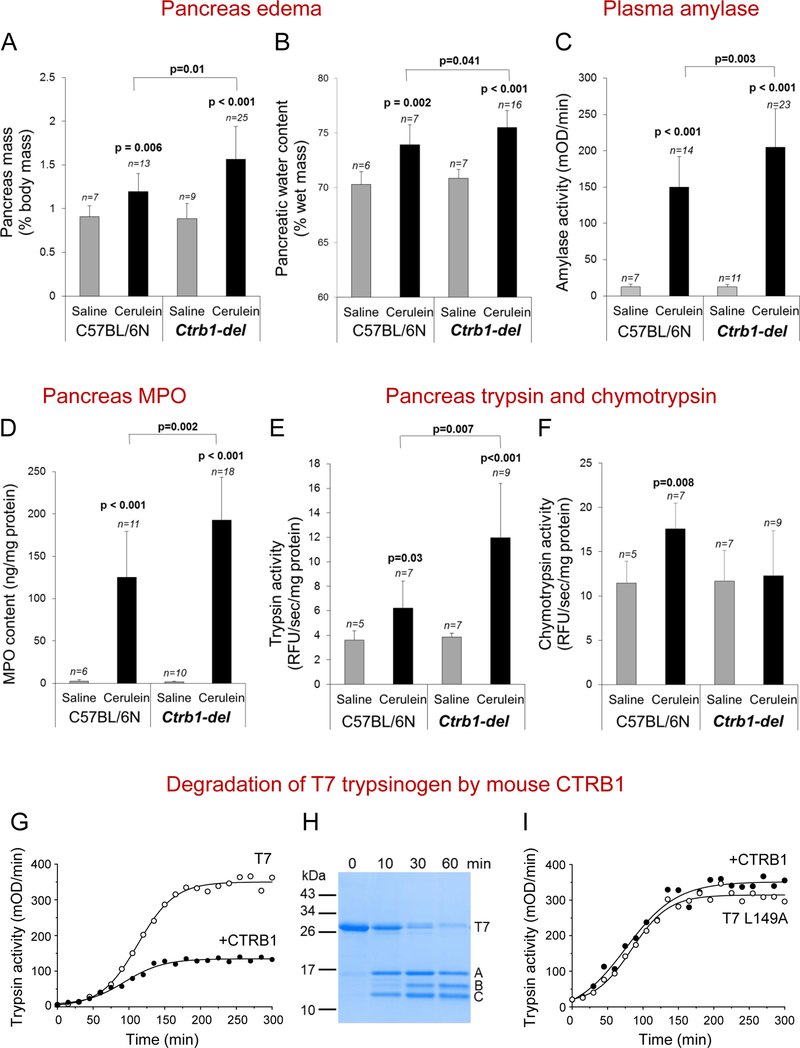
To examine whether the absence of CTRB1 alters pathological responses in experimentally induced pancreatitis, we challenged the mice with 10 hourly injections of cerulein and sacrificed them 1h after the last injection. Histological analysis of the pancreas indicated increased edema, stronger infiltration of inflammatory cells and more pronounced acinar cell necrosis in the Ctrb1-del mice compared to C57BL/6N controls (Figure 1C–K). These findings were confirmed and extended by direct assessment of edema and biochemical assays of pancreatitis severity. Thus, relative to controls, the pancreas of Ctrb1-del mice was visibly larger and weighed significantly more (Figure 2A). Measurement of pancreatic water content provided direct evidence for the more pronounced edema in Ctrb1-del mice (Figure 2B). Plasma amylase activity (Figure 2C) and pancreas tissue myeloperoxidase (MPO) content (Figure 2D) were significantly elevated in Ctrb1-del mice in comparison to controls, indicating more extensive acinar cell injury and inflammation. Systemic inflammation was also higher in Ctrb1-del mice, as judged by the lung MPO content (Supplementary Figure S3). In conclusion, all parameters of cerulein-induced pancreatitis were significantly worse in the absence of CTRB1.
Figure 2.
Cerulein-induced pancreatitis in Ctrb1-del and C57BL/6N control mice and degradation of T7 trypsinogen by CTRB1. Mean values with S.D. are shown. See Methods for experimental details. A, Pancreas mass. B, Pancreatic water content. C, Plasma amylase activity. D, Pancreas MPO content. E, F, Intra-pancreatic trypsin (E) and chymotrypsin (F) activation 30 min after a single saline or cerulein injection. G, Autoactivation of recombinant T7 trypsinogen in the absence and presence of mouse CTRB1. H, Digestion of T7 trypsinogen by CTRB1. Bands A and C are generated by cleavage of T7 at Leu149. Band B is the result of secondary CTRB1 cleavages at Tyr29 and Leu159. I, Autoactivation of T7 trypsinogen mutant L149A in the absence and presence of CTRB1.
To demonstrate that increased acinar cell damage and inflammation are due to higher trypsin activity, we measured cerulein-induced intra-pancreatic trypsin and chymotrypsin activities in Ctrb1-del and C57BL/6N control mice. Protease activities were determined at 30 min after a single cerulein injection when trypsin activity peaks [3, 7] and inflammatory cells do not influence trypsin activation yet. We found significantly higher trypsin activity in Ctrb1-del mice relative to controls (Figure 2E). As expected, chymotrypsin activity increased in C57BL/6N controls in response to cerulein but not in Ctrb1-del mice (Figure 2F). Interestingly, baseline chymotrypsin activity was similar in Ctrb1-del and C57BL/6N control mice, indicating that this is unrelated to CTRB1.
In contrast to the cerulein model, we observed no effect of Ctrb1 gene disruption in L-arginine-induced pancreatitis (Supplementary Figure S4). This result is consistent with recent studies showing that arginine acts through mitochondrial injury rather than early trypsin activation [14].
Finally, we performed biochemical experiments using purified proteins to demonstrate that CTRB1 degrades mouse cationic trypsinogen (T7) and thereby limits its activation to trypsin. T7 trypsinogen is the most abundant isoform comprising 40–50% of total mouse trypsinogen, and it autoactivates almost as readily as its human counterpart [15]. When autoactivation of T7 was measured in the presence of CTRB1, final trypsin levels were significantly reduced (Figure 2G). Digestion experiments revealed that CTRB1 cleaved T7 primarily at the Leu149-Ser150 peptide bond (Figure 2H). Mutation of Leu149 protected T7 against chymotryptic degradation and the L149A mutant autoactivated similarly in the absence and presence of CTRB1 (Figure 2I).
In summary, the present study demonstrates that the major mouse chymotrypsin CTRB1 protects against secretagogue-induced pancreatitis by degrading trypsinogen and reducing pathological trypsin levels. The findings should guide the design of anti-protease therapy in pancreatitis, which needs to spare protective chymotrypsin activity while targeting harmful trypsin.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to George Perides for advice and technical assistance with some of the pancreatitis models. Dóra Mosztbacher is acknowledged for sharing unpublished data on mouse chymotrypsin-like protease expression. The authors are thankful to Andrea Geisz for helpful discussions and critical reading of the manuscript.
This work was supported by the National Institutes of Health (NIH) grants R01 DK082412 and R01 DK058088 (to MST) and a grant from the National Pancreas Foundation (to EH).
Abbreviations:
- CTRB1
chymotrypsin B1
- CTRB2
chymotrypsin B2
- CTRC
chymotrypsin C
- CTRL
chymotrypsin-like protease
- H&E
hematoxylin-eosin
- MPO
myeloperoxidase
- PRSS1
serine protease 1, human cationic trypsinogen
- T7
mouse cationic trypsinogen
Footnotes
Conflict of interest: MST is a consultant for Takeda Pharmaceuticals, Inc. The other authors have no conflicts.
REFERENCES
- 1.Steer ML. Pancreas 1998; 17:31–37 [DOI] [PubMed] [Google Scholar]
- 2.Saluja AK et al. Annu Rev Physiol 2007; 69:249–269 [DOI] [PubMed] [Google Scholar]
- 3.Halangk W et al. J Clin Invest 2000; 106:773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thrower EC et al. Am J Physiol Gastrointest Liver Physiol 2006; 290:G894–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sendler M et al. J Biol Chem 2016; 291:14717–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talukdar R et al. Gastroenterology 2016; 151:747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawra R et al. Gastroenterology 2011; 141:2210–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hegyi E, Sahin-Tóth M. Dig Dis Sci 2017; 62:1692–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosendahl J. et al. Gut. 2017 Jul 28; [Epub ahead of print] [Google Scholar]
- 10.Hudáky P et al. Eur J Biochem 1999; 259:528–533 [DOI] [PubMed] [Google Scholar]
- 11.Szabó A, Sahin-Tóth M. FEBS J 2012; 279:4283–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reseland JE et al. J Biol Chem 1997; 272:8099–8104 [DOI] [PubMed] [Google Scholar]
- 13.Sogame Y et al. Pancreas 2002; 25:378–386 [DOI] [PubMed] [Google Scholar]
- 14.Biczo G et al. Gastroenterology 2018; 154:689–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Németh BC et al. J Biol Chem 2013; 288:24049–24062 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.