Abstract
Myeloid-derived suppressor cells (MDSC) are immature myeloid cells that accumulate in the circulation and the tumor microenvironment of most cancer patients. There, MDSC suppress both adaptive and innate immunity, hindering immunotherapies. The inflammatory milieu often present in cancers facilitates MDSC suppressive activity, causing aggressive tumor progression and metastasis. MDSC from tumor-bearing mice release exosomes, which carry biologically active proteins and mediate some of the immunosuppressive functions characteristic of MDSC. Studies on other cell types have shown that exosomes may also carry RNAs which can be transferred to local and distant cells, yet the mRNA and microRNA cargo of MDSC-derived exosomes has not been studied to date. Here, the cargo of MDSC and their exosomes was interrogated with the goal of identifying and characterizing molecules that may facilitate MDSC suppressive potency. Because inflammation is an established driving force for MDSC suppressive activity, we used the well-established 4T1 mouse mammary carcinoma system, which includes “conventional” as well as “inflammatory” MDSC. We provide evidence that MDSC-derived exosomes carry proteins, mRNAs, and microRNAs with different quantitative profiles than that of their parental cells. Several of these molecules have known or predicted functions consistent with MDSC suppressive activity, suggesting a potential mechanistic redundancy.
Keywords: myeloid-derived suppressor cells, exosomes, differential expression, mRNA, miRNA, next generation sequencing, shotgun proteomics
Graphical abstract
INTRODUCTION
Exosomes are membrane-bound extracellular vesicles of 30 to 100 nm in diameter that are secreted by all eukaryotic cells.1 Initially thought to remove unwanted cellular molecules, research in the past decade has demonstrated that exosomes can contain bioactive molecules such as proteins, lipids, mRNAs (mRNAs), and microRNAs (miRNAs).1,2 These nanosized vesicles can play a major role in intercellular communication because they mediate multiple biologic functions based on the cell type from which they originate.1
Exosomes can carry both miRNAs and mRNAs. miRNAs are noncoding RNAs composed of 18 to 25 nucleotides. They negatively regulate mRNA expression by repressing translation or by inducing mRNA degradation.3,4 These small RNAs exert their repressive function by binding to the untranslated region (UTR) or open reading frame of the target mRNAs.3,4 When transferred by exosomes into target cells, miRNAs can repress mRNA translation.5–8 In addition to their traditional repressive srole, miRNAs can also activate genes and thereby drive processes such as inflammation.9,10 Moreover, exosomes may carry and transfer protein-coding mRNAs to receiver cells.6,7,11–13 Transferred mRNAs can be translated in receiver cells if the translational machinery is accessible, and several studies have shown that those transferred mRNAs give rise to functional proteins.7,12,13
Myeloid-derived suppressor cells (MDSC) are immature myeloid cells that accumulate in the circulation and tumor microenvironment of most patients with cancer where they suppress both adaptive and innate immunity and support the growth and metastases of tumors.14,15 Inflammation is a major driving force for MDSC and enhances their abundance and their suppressive activity, thereby facilitating tumor progression.16–19 Because both inflammation and immune suppression facilitate cancer risk and tumor progression,20 we are studying the impact of inflammation on the induction and function of MDSC. To study the impact of inflammation, we have compared MDSC that develop in tumor-bearing mice under the standard inflammatory conditions associated with the presence of solid tumors (“conventional” MDSC) and MDSC that developed under heightened inflammatory conditions (“inflammatory” MDSC). The different inflammatory environments are generated in mice carrying the syngeneic 4T1 mammary carcinoma (conventional MDSC; 4T1 tumor) or 4T1 cells transfected with and expressing elevated levels of the proinflammatory mediator IL-1β (“inflammatory” MDSC; 4T1/IL-1β tumor).17,18,21,22 These previous studies established that inflammation increases both the number of MDSC as well as the suppressive potency of MDSC. We also know from our previous studies that the 25–30 nm diameter exosomes released by these MDSC contained proteins that chemo- attracted MDSC23–26 and that skewed antitumor immunity toward a tumor-promoting type 2 immune response.23
Because studies in other cellular systems have demonstrated that exosomes also carry miRNAs and mRNAs that impact receiver cells, we have interrogated MDSC and MDSC-derived exosomes for miRNA and mRNA cargo. Our goal is to characterize the mechanisms used by MDSC to mediate immune suppression and to determine the role of MDSC- derived exosomes in these mechanisms. To achieve this goal, the protein, mRNA and miRNA contents of conventional and inflammatory MDSC and MDSC-derived exosomes were interrogated. Two to five biological replicates of matched parental cells and released exosomes were analyzed by shotgun proteomics and next-generation sequencing to determine the protein and RNA cargos, respectively. Relative quantitation was performed in order to compare the RNA and protein content of exosomes with that of their parental cells and between MDSC-derived exosomes of conventional and inflammatory MDSC. The comprehensive qualitative and quantitative analysis of MDSC and MDSC-derived exosomal cargos identified multiple miRNAs and mRNAs whose known or predicted function is consistent with their involvement in MDSC-mediated immune suppression.
EXPERIMENTAL SECTION
Myeloid-Derived Suppressor Cells and Released Exosomes
BALB/c mice were injected with 7000 wild-type syngeneic 4T1 mammary carcinoma cells or 4T1 cells transduced to constitutively express the cytokine IL-1 (4T1/IL- 1β), as previously reported.17,18,21 Greater than 90% of the circulating leukocytes in the blood of BALB/c mice with large 4T1 or 4T1/IL-1β tumors are MDSC as assessed by plasma membrane markers (Gr1 and CD11b), content of arginase and reactive oxygen species, and immune suppressive activity.17,22 Each batch of MDSC consisted of 1 × 107 to 4 × 108 cells and was obtained from the blood of 1 to 3 mice with >1.5 cm diameter tumors. Isolated MDSC were stained with fluorescently tagged antibodies to the canonical MDSC markers Gr1 and CD11b. MDSC used in all experiments were >90% Gr1+ CD11b+ as assessed by flow cytometry. Each batch of MDSC was divided into two aliquots. One aliquot (1 × 106 to 4 × 107 MDSC) was used for shotgun proteomics and RNA analysis. For proteomics analysis, MDSC were frozen at −80 °C in 1 mL of (90:10) fetal calf serum-dimethyl sulfoxide and stored until used. In the case of RNA analysis, MDSC samples were processed immediately without being frozen or exposed to fetal calf serum. Exosomes were prepared as previously described23 from the second aliquot of MDSC (9 × 106-3.6 × 108 cells) that had not been frozen. Conventional and inflammatory MDSC-derived exo- somes prepared by our procedure banded on sucrose density gradients between 1.2 and 1.3 g/mL were 20–30 nm in diameter and had functional activity for some of the mechanisms mediated by their parental cells.23,26,27 RNAlater (Life Technologies, Carlsbad, CA) was added to the samples collected for RNA analyses. MDSC and MDSC-derived exosomes from 4T1 and 4T1/IL-1β tumor-bearing mice are termed “conventional” and “inflammatory”, respectively. All animal experiments were approved by University of Maryland Baltimore County and University of Maryland College Park Institutional Animal Care and Use Committees.
Experimental Design
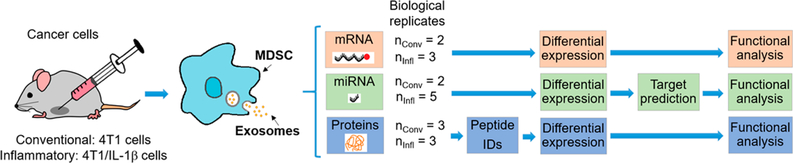
This study focused on interrogating the miRNA, mRNA, and protein cargo in MDSC and their released exosomes under conventional and heightened inflammation. Four sample types were analyzed: (1) conventional MDSC, (2) inflammatory MDSC, (3) conventional MDSC-derived exosomes, and (4) inflammatory MDSC-derived exosomes. To assess the effects of inflammation, conventional MDSC or their exosomes were compared to inflammatory MDSC or their exosomes. Additionally, MDSC-derived exosomes were compared to their corresponding parental cells. Figure 1 shows the experimental design. We analyzed mRNAs and miRNAs of 2 biological replicates from conventional MDSC and their corresponding exosomes. Three biological replicates of mRNAs and five biological replicates of miRNAs were analyzed from inflammatory MDSC and their released exosomes. Since our initial studies focused on the analysis of miRNAs under inflammatory conditions and later extended to include mRNAs and conventional conditions, miRNA samples derived from the same batch as the mRNA samples were included to reduce possible batch effects, resulting in an unbalanced number of biological replicates. In the case of proteins, 3 biological replicates were analyzed from conventional and inflammatory MDSC and their corresponding exosomes.
Figure 1.
Experimental design. MDSC and their released exosomes from tumor-bearing mice injected with 4T1 or 4T1/IL-1β mammary carcinoma cells were analyzed for protein, mRNA, and miRNA content. The number of biological replicates per condition is shown.
RNA Isolation and Library Creation
Total RNA was isolated using the mirVana miRNA Isolation Kit (Life Technologies, Carlsbad, CA) following manufacturer’s instructions with the addition of up to two additional phenol extraction steps, necessary to remove excess lipids. Large RNA species (>100nt) were isolated by precipitation in 33% ethanol, and small RNAs (>10nt) by precipitation in 80% ethanol. Complementary DNA libraries were created using the TruSeq Small RNA Library Preparation Kit and TruSeq RNA Library Preparation Kit version 2 (Illumina, San Diego, CA). Manufacturer’s instructions were followed except for the initial amounts of exosomal total RNA, which was adjusted to compensate for the paucity of rRNA and to ensure equivalent amounts across samples. The quality of MDSC large RNAs and concentrations of all RNA libraries was evaluated using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Libraries were sequenced using a HiSeq1500 sequencer at the University of Maryland sequencing core (Illumina). Since our sample preparation includes a polyadenylated (polyA) isolation step before sequencing, it is expected that a vast majority (>99%) of the sequenced mRNAs are capped and translationally competent. Additionally, we do not expect to detect nonexosome specific mRNA/miRNA reproducibly in our 2—5 biological replicates in order to pass the identification criteria set. Details on libraries created and sample replicates are summarized in Table S-1.
Next-Generation Sequencing Data Analysis
The quality of the reads obtained was evaluated using FastQC (version 0.11.2; http://www.bioinformatics.babraham.ac.uk) and biopieces (biopieces.org). RNA adapters were removed from polyA libraries using Trimmomatic (version 0.33)28 and small RNA adapters using Cutadapt (version 1.8.1).29 Libraries were separately mapped against the Ensembl Mus musculus precomputed transcriptome database (version GRCm38.79/ mm10, Dec 2015) using Kallisto revision 0.42.330 with 19 nt indexes for small RNAs and 31 nt indexes for large RNAs. The sorted pseudoalignments were counted against the Mus musculus immature miRNA database (http://www.mirbase.org, 28645 entries, version 21).31,32
RNA Data Visualization and Clustering
Possible sequencing depth biases due to the variability of biological replicates and sample batch effects were assessed. This process entailed the creation of density plots and boxplots, hierarchical clustering analysis based on Pearson’s correlation and Euclidian distance, and principal component analysis, before and after data normalization (Figure S-1—S4). Even though miRNA of the MDSC recovered after shedding exosomes was not evaluated, it was considered for hierarchical clustering as its replicates contributed to the measure of experimental variance for batch effect estimation. Several normalization approaches were evaluated including quantile,33 trimmed mean of M-values,34 relative log expression,35 upper quartile,36 and variance stabilized data.37 In the case of miRNA, quantile normalization was selected for data quality assessment. Normalization was not applied to the mRNA reads because the mRNA profiles between exosomes and MDSC were significantly different (see Figure S-3). In all cases, a log2 transformed counts per million (cpm) reads after low read count filtering was performed. A low read count was defined as any feature with counts less than twice the number of samples or cases where any single sample has less than 2 read counts. Moreover, bias due to batch effects was evaluated using several algorithms, including: surrogate variable analysis (sva), ComBat (combating batch effects when combining batches of gene expression microarray data),38 remove unwanted variation,38,39 and batch factor removal via residuals.
RNA Differential Expression Analysis
MicroRNAs and mRNAs differences in abundance were estimated for all possible pairwise comparisons using the Bioconductor packages linear model for microarray (limma),40 empirical analysis of digital gene expression data in R (edgeR),35 and differential gene expression analysis based on the negative binomial distribution (DESeq2).37 Our interpretation of results was primarily based on limma outputs. Biological replicates were performed with significant time between them. The data was initially treated as two separate experiments and later combined; therefore, a significant batch effect was introduced and needed to be addressed in the treatment of the data by the differential expression analyses. Initial attempts at including a batch factor in the statistical model failed because the experimental design was partially confounded, so the surrogate variable analysis (sva) package was used to adjust the statistical models and address batch effects. As a result, a good agreement was observed for limma, edgeR, and DESeq2 using the combined data. The resulting adjusted p-values were too conservative, so nonadjusted p-values were used to select genes for further analysis. Genes with differential abundances were defined as those with a fold- change ≥ 2 and (nonadjusted) p-value ≤ 0.05.
miRNA Target Gene Prediction
The miRNAtap package version 3.4 (https://bioconductor.org/packages/release/bioc/html/miRNAtap.html) was used to determine miRNA targets for those miRNA with statistically significant differences in abundance (fold-change ≥ 2, (nonadjusted) p-value ≤ 0.05). miRNAtap combines the output of 5 prediction tools: DIANA,41 PicTar,42 miRanda,43 TargetScan,44 and miRDB.45 Only those target genes that were predicted by at least 2 of the tools were considered for functional analysis.
Sample Preparation for Shotgun Proteomic analysis
Exosome lysates and tryptic digests were prepared following Burke et al.23 All digested lysates were lyophilized and reconstituted in solvent A (97.5:2.5 H2O-acetonitrile in 0.1% formic acid) prior to LC—MS/MS.
MDSC were thoroughly washed by centrifugation at 900g for 10 min at 4 °C using 10 mL of cold phosphate-buffered saline solution from Sigma-Aldrich (St. Louis, MO). MDSC pellets were lysed by incubation in 8 M urea in 50 mM ammonium bicarbonate buffer with protease inhibitor cocktail (Sigma- Aldrich) for 30 min at room temperature. In order to ensure that cells are completely lysed and the lysate is homogeneous, MDSC were further lysed mechanically using a set of syringes with needles of sequentially smaller gauge size (18, 20, and 21.5) obtained from Becton Dickinson & Co. (Franklin Lakes, NJ) and performing 5–10 strokes each time. Cell debris was removed by centrifugation at 14000g for 10 min. Supernatants were transferred to 3 kDa filters and buffer exchanged to reach an 8 mM urea concentration. The protein content of MDSC was measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL) and 25 μg aliquots were reduced, alkylated, and tryptic digested as reported in Burke et al.23
Protein Analysis by Liquid Chromatography Mass Spectrometry
Samples were analyzed using an Ultimate 3000 RSLCnano system (Dionex, Sunnyvale, CA) coupled to an orbitrap Fusion Lumos Tribrid (Thermo Fisher Scientific, Waltham, MA). Chromatographic separation was obtained using a C18 PepMap trap (0.3 × 5 mm, 5 μm particle size, 100 Å) and C18 Acclaim PepMap RSLC column (0.075 × 250 mm, 2 μm particle size, 100 Å), both obtained from Thermo Fisher Scientific. Samples (3 μg) were loaded into the trap, desalted, and concentrated using solvent A at a flow rate of 5 μL/min for 10 min and then eluted from the trap and further separated in the analytical column by linearly increasing solvent B (acetonitrile-H2O, (75:25) in 0.1% formic acid) from 5 to 55%, during the span of 150 min under a flow rate of 0.3 μL/min.
Precursor ion mass resolution of 120000 (at m/z 200) and product ion unit mass resolution were used. Ions were collected based on a target automatic gain control of 4 × 105 and 4 × 103 for precursor and product ions, respectively. The maximum injection times for precursor and product ions were set to 50 and 100 ms, respectively. Data dependent acquisition was carried out in a fixed 3 s duty cycle, in which the top n most abundant precursor ions (intensity >1 × 104) carrying charges from +2 to +7 were isolated by the quadrupole within a 1.6 m/z isolation window. Dynamic exclusion was set to exclude precursor ions for 60 s after being selected once in 30 s. The isolated precursor ions were fragmented using CID set to 35% normalized collision energy. Five technical replicates per biological replicate were injected in order to achieve in-depth protein identifications.
Protein Identification and Relative Quantitation
A total of 60 data files were converted into peak lists and formated as mzXML files using msconvert from ProteoWizard (version 3)46 and searched against the Uniprot Mus musculus reference protein sequence database including isoforms (release 01_2015, January 2015, 52654 entries) using three search engines (MSGF+ [version (v10072) (6/30/2014)], X!Tandem [version 2010.01.01.4], and OMSSA [version 2.1.1]) with PepArML (http://edwardslab.bmcb.georgetown.edu/PepArML).47 Search parameters allowed for up to 1 missed cleavage and a precursor (monoisotopic or first 13C peak) and product ion mass tolerance of 0.05 and 0.5 Da, respectively. The protease trypsin was selected. Carbamidomethylation was considered as a fixed modification, and oxidation of methionine, deamination of N-terminus glutamine, dehydration of N-terminus glutamic acid, and pyro-carbamidomethylation of N-terminus cysteine as variable modifications. Spectral FDR was estimated by PepArML following Elias and Gygi.48 Peptide spectrum matches (PSMs) were filtered at spectral FDR ≤ 0.31%, and global parsimony was applied, for which proteins were inferred on the basis of at least two unshared peptides in at least one of the four sample types. Protein FDR was estimated at ≤1% after applying the MAYU49 correction for large data sets.
Label-free relative quantitation was performed by spectral counting. In-house software was used to count the number of PSMs, also known as spectral counts, for each protein after filtering of PSMs by spectral FDR and protein inference by global parsimony. Differences in abundance between conditions were estimated following Old et al.50 Spectral counts ratios (Rsc) were estimated for each identified protein and statistically significant differences in abundance determined using Fisher’s Exact test with Benjamini-Hochberg adjustment for multiple testing.51 Three comparisons were carried out: (1) inflammatory exosomes versus conventional exosomes, (2) inflammatory MDSC versus conventional MDSC, and (3) exosomes versuss MDSC irrespective of inflammation condition. Rsc >1 means that a protein is present in greater abundance (fold-change ≥ 2) in “inflammatory exosomes” for comparison (1), “inflammatory MDSC” for comparison (2), and “exosomes” for comparison (3). Conversely, Rsc < −1 refers to an increase in abundance in “conventional exosomes” for comparison (1), “conventional MDSC” for comparison (2), and “MDSC” for comparison (3).
Gene Ontology and Pathway Analyses
mRNAs and putative mRNA targets of miRNAs with significant change in abundance [fold-change ≥2, (nonadjusted) p-value ≤0.05] for each pairwise comparison were annotated with respect to gene ontology (GO) categories and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database using the R package gProfiler,52 which uses the Mus musculus genome as background and the hypergeometric distribution with Benjamini-Hochberg adjustment for multiple testing to estimate the enrichment significance of differentially abundant gene-sets.
Identified proteins were annotated by their GO categories using the generic and PIR (Protein Information Resource, http://pir.georgetown.edu) GO slims and the KEGG pathway database.53 Enrichment of GO categories and KEGG pathways by differentially abundant proteins (fold-change ≥ 2, adjusted p-value ≤ 0.05) was evaluated by Fisher’s Exact test, using the entire set of identified proteins as background and the Benjamini-Hochberg adjustment for multiple testing. In all cases, GO gene-sets or KEGG pathways were considered statistically significantly enriched in differentially abundant RNAs or proteins for adjusted p-values ≤ 0.05.
Data Submission
The mRNA and miRNA sequences were submitted to the Sequence Real Archive NCBI with the following bioproject accession PRJNA363057. Mass spectrometric data was deposited into the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE54 partner repository with accession PXD006204 and doi: 10.6019/PXD006204.
RESULTS
MDSC-Derived Exosomes Carry RNAs
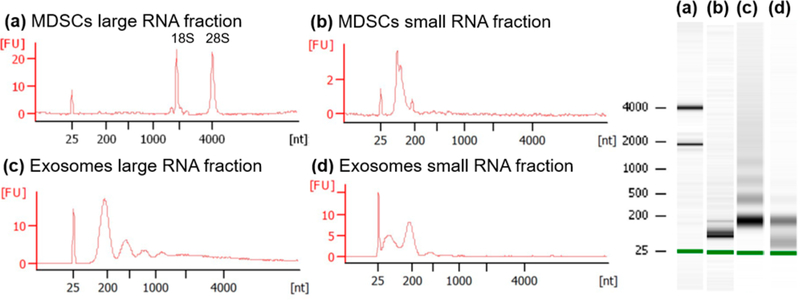
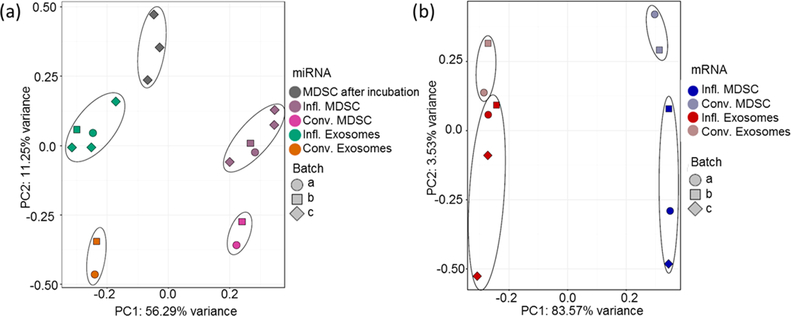
The RNA fractions isolated from MDSC and MDSC-derived exosomes were visualized by capillary electrophoresis using the Bioanalyzer. The characteristic 18S and 28S rRNA (rRNA) peaks were observed in the MDSC large RNA fraction as expected (Figure 2a), showing minimal RNA degradation during sample preparation. The observed distributions of isolated RNA in the MDSC small RNA fractions were unique to those samples and consistent, suggesting that the isolations were successful (Figure 2b). The large and small RNA fractions collected from MDSC-derived exosomes contain RNAs with nucleotide lengths that correspond to that of mRNA and miRNA, respectively (Figure 2, panels c and d). Ribosomal RNA was minimally detected in MDSC-derived exosomes, which is in agreement with previous small RNA studies on exosomes6,7,55,56 and supports the conclusion that our exosome samples were not contaminated with dead cells.57 The RNA libraries were sequenced and mapped against the Ensembl Mus musculus genome. Reads mapped on an average of 81130 and 69474 distinct annotated mRNA transcripts in exosomes and MDSC, respectively, and 2500 annotated miRNAs for both MDSC and their released exosomes. The pseudo counts of the RNA libraries were passed to SVA and PCA and plotted in order to visualize sample clustering (see Figure 3, panels a and b). The two first principal components accounted for 56 and 84% of the variance for miRNA and mRNA analyses, respectively. Clear clusters were observed by sample type (MDSC and exosomes), indicating a significant level of reproducibility between biological replicates and to a lesser degree by inflammation condition.
Figure 2.
RNA size distribution observed by capillary electrophoresis for MDSC (a) large RNA and (b) small RNA fractions; and exosomes (c) large RNA and (d) small RNA fractions. The y axis is labeled as [FU] for fluorescence units and the x axis as [nt] for nucleotide length.
Figure 3.
Principal component analyses (PCA) were performed using all (a) miRNAs and (b) mRNAs after filtering for low counts, normalization, and surrogate variable analysis to account for batch effects in the experimental model. In these PCA analyses, each point represents a sample color-coded by the experimental condition and shaped based on their experimental batch, as detailed in the figure.
Differential mRNA Profiles and Functional Analysis
Considering all analyzed samples, a total of 40433 mRNA transcript isoforms were confidently identified (Table S-2). The number of mRNA transcripts found to have differences in abundance for each pairwise comparison is shown in Table 1. On average, ~45% of the mRNA transcript isoforms identified showed statistically significant differences in abundance, when comparing exosomes to their parental cells irrespective of their inflammation conditions. Many of these mRNA transcript isoforms encoded for Uniprot TrEMBL predicted proteins. Differences between inflammation conditions are more subtle, with only ~3.5% of the total mRNA transcript isoforms showing differences in abundance. Enriched GO categories and KEGG pathways were identified for the differentially expressed mRNA species in each pairwise comparison (see Table S-3).
Table 1.
Number of miRNA, mRNA, and Proteins Found to Have Statistically Significant Differences in Abundance for Each Pairwise Comparisona
| pairwise comparison |
number of enriched |
|||
|---|---|---|---|---|
| mRNAs | miRNAs (predicted) |
proteins | ||
| MDSC | infl. vs conv. | 1847 | 53 (30) | 231 |
| conv. vs infl. | 2447 | 110 (18) | 123 | |
| exo. | infl. vs conv. | 339 | 41 (28) | 347 |
| conv. vs infl. | 1489 | 59 (9) | 69 | |
| conv. | exo. vs MDSC | 17783 | 199 (113) | - |
| MDSC vs exo. | 7262 | 106 (4) | ||
| infl. | exo. vs MDSC | 18858 | 499 (301) | - |
| MDSC vs exo. | 10457 | 84 (6) | ||
| combined infl. and conv. | exo. vs MDSC | - | - | 371 |
| MDSC vs exo. | - | - | 612 | |
Additionally, the numbers of enriched predicted miRNAs by Ensembl are specified in parentheses. Note that “Exo.” refers to exosomes, “Infl.” refers to inflammatory condition and “Conv.” refers to conventional condition. The total number of mRNA transcript isoforms and miRNAs identified across all samples were 40433 and 1435, respectively.
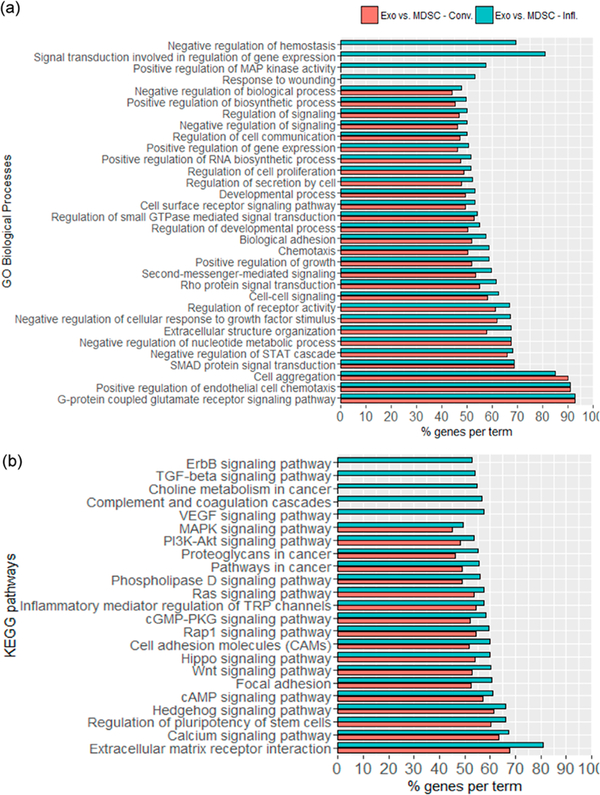
mRNA Transcripts differentially expressed in exosomes compared to their parental cells showed a similar enrichment in GO categories and KEGG pathways independently of the inflammation condition. Several of the enriched GO biological processes were related to intercellular communication, such as “chemotaxis”, “cell-cell signaling”, “cell surface signaling”, “cell aggregation”, and “biological adhesion” (see Figure 4a). Moreover, processes regulating growth, cell proliferation, cell migration, and cell survival were also significantly enriched. On the basis of the GO cellular compartment categories “cell surface” (53% of genes, adjusted p-value = 8.2 × 10−6), Cell periphery” (48% of genes, adjusted p-value = 3.6 × 10−7), and “plasma membrane” (47% of genes, adjusted p-value = 1.3 × 10−6), a total of 1804 mRNA transcripts identified encode for surface-associated proteins, including membrane receptors and ligands that could modulate various signaling pathways on the receiver cell. Additionally, a large number of KEGG signaling pathways were found to be enriched for the differentially expressed mRNA transcripts in exosomes when compared to their parental cells, including “proteoglycans in cancer”, “choline metabolism in cancer”, and “pathways in cancer” (Figure 4b). A few pathways were enriched in exosomes under inflammatory conditions only, including the “vascular endothelial growth factor (VEGF) signalling pathway”, “transforming growth factor-β (TGF-β) signalling pathway”, and “ErbB signaling pathway” associated with angiogenesis58 and chemotaxis.59
Figure 4.
Selected enriched (a) GO biological processes and (b) KEGG pathways for mRNA transcripts detected in greater abundance in exosomes, when compared against their parental cells, for conventional (red) and inflammatory (blue) conditions. Categories shown were statistically significant with adjusted p-value ≤ 0.05. The complete list of functional annotations of mRNA transcripts can be found in Table S-3.
Exosomal mRNA transcripts with statistically significant differences in abundance when inflammatory and conventional exosomes are compared and showed clear differences in their enriched GO biological processes. The enriched processes in inflammatory exosomes include “cell-cell signaling”, “cell surface receptor signaling pathway”, “macrophage differentiation”, “blood vessel endothelial cell differentiation”, “negative regulation of endothelial cell proliferation”, and several processes related to regulation of gene transcription. Moreover, KEGG “pathways in cancer” was also found to be enriched in inflammatory exosomes. No KEGG pathways were found to be significantly enriched in exosomes under conventional conditions (Table S-3).
Differential miRNA Profiles and Functional Analysis
A total of 1453 miRNAs were confidently identified and differential expression estimated (see Table S-4). The number of miRNAs found to be in greater and lower abundance for each of four pairwise comparisons are summarized in Table 1. Notably, 43% (~624) of the miRNAs were annotated as predicted by Ensembl-regions of the Mus musculus genome that can form a hairpin structure and are homologous to miRBase sequences. Hence, this study offers experimental evidence for the existence of these miRNAs. Additionally, approximately half of the predicted miRNAs were found to be in greater abundance in inflammatory exosomes.
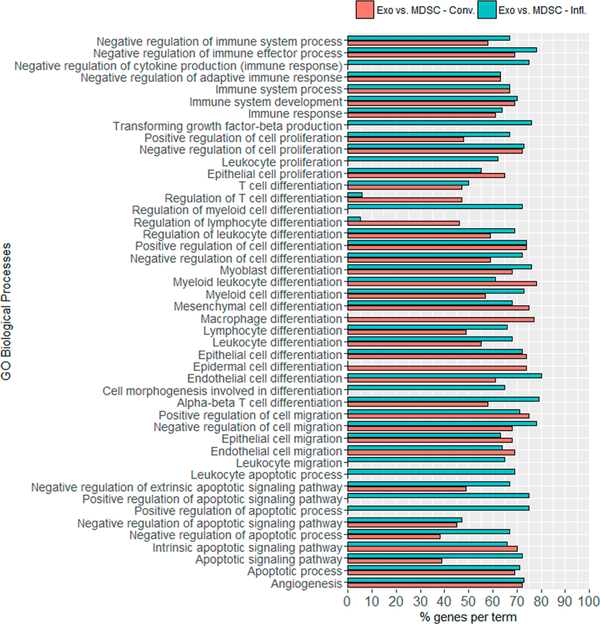
MicroRNA targets were predicted for those miRNAs with statistically significant differences in abundance for each comparison of interest. miRNA targets were assigned when at least 2 out of the 5 predictive tools agreed (see Table S-5). When immature miRNAs were identified, both the –3p and –5p strand were considered when performing target prediction, although just one may be present. Functional analyses were performed using all predicted targets for each selected miRNA, resulting in numerous KEGG pathways and GO biological processes that could be affected by the miRNAs’ repressive activity if transferred to a receiving cell (see Table S-6). Many miRNA targets that participate in biological processes associated with cell differentiation, cell proliferation, cell migration, apoptosis, immune system, and angiogenesis were associated with differentially abundant miRNAs found to be enriched in exosomes compared to their parental cells, as shown in Figure 5. These putative targets could negatively or positively regulate cellular processes in myeloid cells, macrophages, T cells, epithelial, endothelial, and mesenchymal cells; all of which play a role in tumor progression and metastasis. Moreover, considering those miRNAs enriched in exosomes when comparing their cargo based on inflammation condition, the KEGG pathway “apoptosis” (adjusted p-value = 0.021) was found to be enriched under heightened inflammation.
Figure 5.
Selected enriched GO biological processes of predicted miRNA targets by miRNAtap based on miRNAs detected in greater abundance in conventional (red) and inflammatory (blue) exosomes. Categories were statistically significant with adjusted p-value ≤ 0.05. The complete list of functional annotations of miRNA targets can be found in Table S-6.
Differential Protein Profiles and Functional Analysis
We have previously interrogated the protein content of conventional and inflammatory MDSC and MDSC-derived exosomes. These studies focused on determining the effect of inflammation on MDSC and exosomes separately.23,60 The present work provides the first protein relative quantitation between MDSC-derived exosomes and their matched parental cells independently of their inflammation condition, by searching and quantifying the combined data set of inflammatory and conventional samples. A total of 1726 proteins were identified with protein FDR of at most 1%, from which 1256 corresponded to exosomes and 1434 to MDSC, with a 58% overlap in proteins identified. As expected, many proteins considered exosome markers,1 such as CD9; heat shock proteins Hsp70 protein-4, Hsp cognate 71KDa, Hsp90α, and Hsp90β; MHC II molecules; and components of the ESCRT endosomal sorting machinery such as Alix (programmed cell death 6-interacting protein) and vacuolar protein sorting-associated proteins Vps25, Vps4B, and Vps37B were identified in exosomes. The proteins CD9, Vps4B, and Vps37B were found to be enriched in exosomes when compared to their parental cells, with CD9 presenting the largest difference in abundance of 89-fold (adjusted p-value = 1.6 × 10−32). Other proteins enriched in exosomes include annexins A4, A6, and A7 and the histones H2A.z, H2B, H3.1, H3.2, and H4.
Differences in protein abundance were estimated by spectral counting. All peptide-spectrum matches incident on a protein, including modified and shared peptides, were counted using inhouse software. A total of 261164 and 264500 PSMs were counted for MDSC and MDSC-derived exosomes, respectively. The median spectral count per identified protein was ~20, with ~260 proteins associated with more than 150 PSMs. Spectral count ratios (Rsc) were estimated as stated in the experimental procedures. A total of 371 proteins (21%) were found in greater abundance in exosomes, and 612 proteins (35%) were found in greater abundance in MDSC. Detailed lists of peptides identified, proteins inferred, and their relative abundances estimated by spectral counting are presented in Table S-7. GO and KEGG functional analyses were performed for those proteins found to be enriched in each pairwise comparison (Table S-8). Table 2 summarizes the GO categories and KEGG pathways enriched for proteins found in greater abundance in exosomes when compared to their parental cells. Notably, 64% of the proteins found in greater abundance in exosomes were localized to the “extracellular region”, “extracellular space”, and/or “cell surface”. We observed that many of the proteins corresponding to the “extracellular region” term included cytoplasmic and plasma membrane proteins. This can be explained by the fact that the GO term “extracellular region” includes “extracellular exosome”, which means that we observed an enrichment in proteins that have been previously reported to be present in exosomes. Additionally, many of the enriched surface proteins have been previously reported to be present in exosomes shed by MDSC,23,26,61 including 5 glycoproteins rigorously identified by Chauhan et al. to be on the surface of MDSC-derived exosomes.26 Significantly enriched GO biological processes included “immune system process”, “cell adhesion”, and “coagulation”.
Table 2.
GO Categories and KEGG Pathways Enriched with Proteins in Greater Abundance in Exosomes Compared to Parental Cells
| database | GO term/KEGG pathway | proteins | p-value |
| GO cellular compartment | extracellular region | 216 | 2.9 × 10−8 |
| extracellular space | 64 | 3.5 × 10−5 | |
| cell surface | 35 | 1.7 × 10−2 | |
| GO molecular function | antigen binding | 11 | 3.7 × 10−2 |
| signal transducer activity | 30 | 3.7 × 10−2 | |
| peptidase activity | 55 | 6.3 × 10−3 | |
| transferase activity | 86 | 4.4 × 10−2 | |
| GO biological processes | immune system process | 90 | 3.1 × 10−3 |
| carbohydrate metabolic process | 33 | 1.9 × 10−2 | |
| organic acid metabolic process | 58 | 2.3 × 10−3 | |
| proteolysis | 74 | 1.7 × 10−2 | |
| peptide metabolic process | 14 | 1.3 × 10−2 | |
| nitrogen compound metabolic process | 30 | 3.0 × 10−2 | |
| cell adhesion | 51 | 1.3 × 10−2 | |
| metabolic process | 161 | 2.4 ×10−2 | |
| nucleoside metabolic process | 30 | 2.1 × 10−3 | |
| nucleotide metabolic process | 46 | 2.3 × 10−3 | |
| protein metabolic process | 45 | 1.4 × 10−4 | |
| coagulation | 22 | 1.3 ×10−2 | |
| regulation of body fluid levels | 27 | 1.3 × 10−2 | |
| cofactor metabolic process | 31 | 1.3 × 10−2 | |
| KEGG | amino sugar and nucleotide sugar metabolism | 11 | 1.8 × 10−2 |
| complement and coagulation cascades | 14 | 1.0 × 10−3 | |
| cysteine and methionine metabolism | 8 | 4.0 × 10−2 | |
| pentose phosphate pathway | 11 | 2.8 × 10−2 | |
| proteasome | 19 | 1.0 × 10−3 | |
| purine metabolism | 18 | 1.8 × 10−2 |
DISCUSSION AND CONCLUSIONS
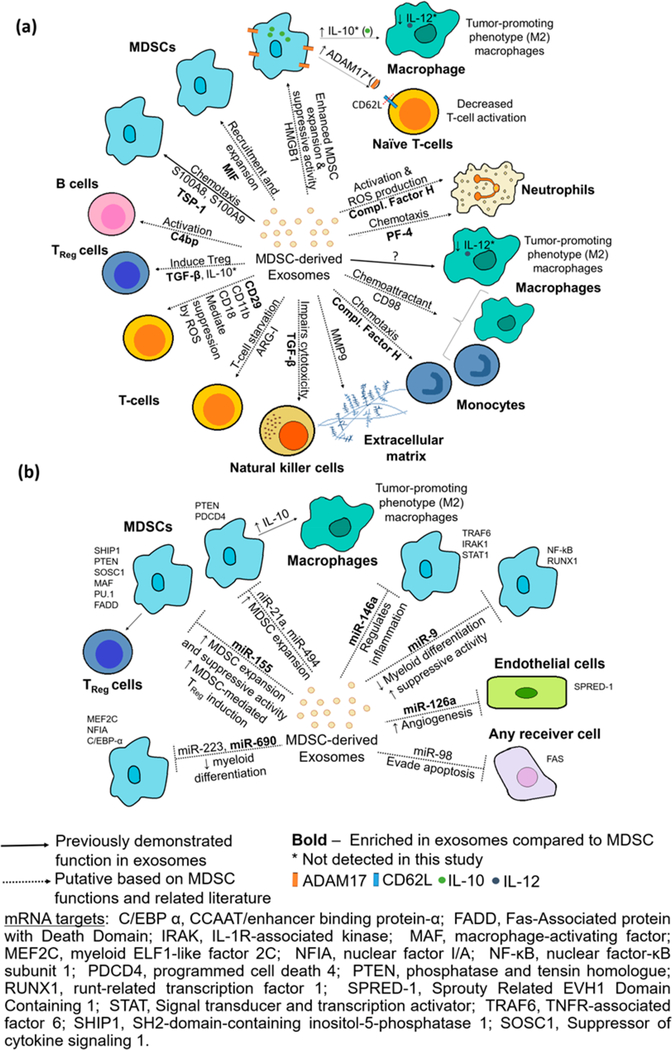
In this study, we provided evidence that MDSC-derived exosomes carry proteins, mRNAs, and miRNAs. We connected the predicted function of the RNAs and proteins in the exosome cargo with the known functions of MDSC, observing that the RNA cargo may in part mediate MDSC immunosuppressive activity and suggesting a potential mechanistic redundancy between functional proteins and RNAs. Through relative quantitation analyses, we demonstrated that there are significant qualitative and quantitative profile differences between MDSC and their released exosomes, with an average of 24%, 45%, and 30% of the identified miRNAs, mRNAs, and proteins estimated to be enriched in exosomes (irrespective of their inflammation condition). Moreover, exosomes were found to carry proteins reported to participate in (1) exosome biogenesis and protein loading,62 such as the enriched proteins CD9 and Vps4B, and (2) loading of particular miRNAs into exosomes,63–65 including the heterogeneous nuclear ribonucleoprotein A2/B1, argonaute-2, and the nuclease-sensitive element-binding protein-1 known to bind to miRNA-223, a miRNA identified in our samples. These observations support the hypothesis that certain proteins and miRNAs are preferentially loaded into exosomes through selective sorting mechanisms. The loading of selected proteins and RNAs into the exosomes suggests that MDSC and MDSC-derived exosomes may mediate some distinct immune suppressive functions.
Putative roles played by exosomes as intercellular communicators were proposed based on previously reported functions of proteins and miRNAs from MDSC, MDSC-derived exosomes and other cells, and based on the knowledge gathered in our study through GO functional annotations and KEGG pathway enrichment analyses (see Figure 6 and Table S-9, panels a and b for detailed references). Exosomes might deliver a wealth of mRNA transcripts that encode for proteins associated with biological processes such as “cell-cell signalling”, “chemotaxis”, “cell adhesion”, and “regulation of cell proliferation” to MDSC and other cells in the tumor microenvironment. Not surprisingly, some of the putative functions found for the mRNA transcripts overlap with that of the protein cargo, as most (91%) of the proteins carried in exosomes are also encoded by the mRNA transcripts present. Only 111 of the 1256 proteins identified in exosomes were not accompanied by their mRNA transcripts, from which 71 proteins belong to the GO cellular compartment categories “membrane”, “cell surface”, and “extracellular region”. Interestingly, only the biological process “complement activation, classical pathway” (adjusted p-value = 0.03) was found to be enriched in exosomes due to the presence of 5 immunoglobulins and C4B-binding protein (C4B-bp), all found in greater abundance in exosomes compared to their parental cells. Similarly, regarding mRNA transcript isoforms, the “complement and coagulation cascades” KEGG pathway was also found to be enriched in inflammatory exosomes when compared to their parental cells.
Figure 6.
Putative functions of (a) proteins and (b) miRNAs carried by MDSC-derived exosomes in the tumor microenvironment based on previously reported functions in exosomes and cells and GO functional annotation and KEGG pathway analyses. Demonstrated functions of MDSC-exosomes are shown with filled lines and putative functions with dotted lines. Proteins and miRNAs enriched in exosomes when compared to their parental cells are shown in bold. Additional information including references used to create this figure can be found in Table S-9, panels a and b.
Autocrine chemotaxis is very important for MDSC function. The proinflammatory proteins S100A8 and S100A9 present in exosomes mediate MDSC accumulation, enhance MDSC immunosuppressive function, and promote tumor growth and metastasis.23 These proteins are abundant in MDSC and MDSC-derived exosomes,23,25 and in our study they were found to be 4 to 6-fold (adjusted p-value ≈ 0) more abundant in MDSC. Additionally, our group has previously reported the chemotactic effect of leukocyte surface antigen CD47 and thrombospondin-1 (TSP-1) on the exosome surface, causing observable MDSC migration.28 In the current study, TSP-1 was found to be 12-fold (adjusted p-value ≈ 0) enriched in exosomes. The pro-inflammatory protein high mobility group box protein 1 (HMGB1), a highly abundant protein secreted by MDSC that also induces MDSC production and accumulation,66 showed no difference in abundance between MDSC and their released exosomes. Other proteins previously shown to have chemotactic activity for leukocytes when secreted are the cytokine macrophage migration inhibition factor (MIF) (2-fold, adjusted p-value = 9.2 × 10−3)67,68 and the chemokine platelet factor-4 (PF-4) (5-fold, adjusted p-value = 3.0 × 10−58) both found here to be enriched in exosomes.69
MDSC induce TReg or Th17 cells through the secretion of TGF-β1 together with IL-10 or IL-6, respectively.15 Moreover, membrane-bound TGF-ββ1 on MDSC can impair natural killer (NK) cell cytotoxicity.70 In this study, TGF-β1 was 4.3-fold (adjusted p-value = 8.4 × 10−14) enriched in exosomes when compared to their parental cells (irrespective of the inflammation conditions), allowing us to speculate that exosomal TGF-β1 may also play a role in these MDSC suppressive mechanisms.
The “complement and coagulation cascades” that are enriched in exosomes have been linked to inflammation and cancer, due to their roles in innate immunity and their ability to protect against injury and pathogens.7 Under chronic inflammatory conditions, these pathways also facilitate tumor progression and metastasis by supporting angiogenesis, cell proliferation, and apoptosis evasion.72 Markiewski et al. demonstrated that tumor progression in mice carrying TC-1 cervical carcinoma is reduced after knockout of complement proteins C3 or C4 and that complement component C5a drives the accumulation of MDSC.73 In our study, a notable enrichment of complement regulatory factor H (46-fold, adjusted p-value = 1.9 × 10−16) and C4B-bp (41-fold, adjusted p-value = 4.4 × 10−37) was observed in exosomes. This is interesting as several noncanonical functions have been reported for these proteins in human studies including: (1) activating B-cells through the binding of C4B-bp to CD40,74 (2) activating neutrophils and increasing their production of reactive oxygen species by binding CD11b/CD18,75 (3) promoting migration of monocytes by chemotaxis,75,76 and (4) escaping complement activation and immune surveillance in the tumor microenvironment.77
A large number of miRNAs were identified in our study, with almost half of the identifications corresponding to predicted miRNAs. Some of the miRNAs we identified in MDSC-derived exosomes have been previously detected in MDSC themselves. For example, miR-146a, which is 18-fold [(nonadjusted) p-value = 5.0 × 10−3] enriched in exosomes, binds to the 3ʹ-UTR of TNF receptor-associated factor 6 (TRAF6) and IL-1 receptor-associated kinase 1 (IRAK1) mRNAs, both relevant in the NF-κB pathway.78 The repression of these targets negatively regulates NF-κB activation, controlling inflammation78 and, in other cases, reducing myeloproliferation and suppressing the development of malignant tumors.79 Four miRNAs identified in MDSC-derived exosomes, miR-9, miR-494, miR-223, and miR-690, could play a role in regulating MDSC suppressive function, as they are capable of affecting the cell cycle, suppressing the differentiation of myeloid cells, and increasing MDSC proliferation.80,81 From these 4 miRNAs, only miR-690 was found in our study to be in greater abundance in exosomes when compared to their parental cells, irrespective of the inflammation condition. A key miRNA found to be enriched in exosomes is miRNA-155, which if delivered to MDSC could cause MDSC expansion and increase the production of IL-10.82 Exosomal delivery of miRNA-155 to surrounding MDSC therefore could contribute to the cross-talk between MDSC and macrophages through increased MDSC production of IL-10.83,84 Notably, MDSC-derived exosomes are known to increase MDSC production of IL-10, although the mechanism by which the exosomes exert this function was not determined. Additionally, IL-10 induces regulatory T cells (TReg), and miR-155 could increase their survival by suppressing the expression of cytokine signaling 1 (SOCS-1), which also contributes to suppression of antitumor immunity.15,85
RNAs responsible for several biological processes and pathways related to cancer were found to be enriched in inflammatory exosomes. Four differentially abundant miRNAs (miRNA-7022, miRNA-7062, miRNA-5134, and miRNA-704) had predicted mRNA targets that are part of the apoptotic pathway including Fas. Fas is also a validated target of miRNA- 98, a miRNA identified in exosomes and MDSC. miRNA-98 showed no difference in abundance between high and low inflammatory exosomes, no difference in abundance between exosomes and their parental cells, and a 3.7-fold enrichment in inflammatory MDSC when compared to conventional MDSC. Interestingly, it has been previously reported that MDSC are able to resist Fas-mediated apoptosis under heightened inflammation.60 Hence, if these exosomal miRNAs were delivered to MSDCs, they could contribute to the observed MDSC extended half-life by repressing Fas expression.
A significant overlap between the exosomal protein and mRNA cargo was observed with 91% of the proteins in exosomes being accompanied by their respective mRNA. From those mRNA-protein pairs, 54 corresponded to mRNAs and proteins that are both present in greater abundance (i.e., fold- change ≥2 and (nonadjusted) p-value ≤0.05) in exosomes when compared to their parental cells. As stated in the experimental section, the vast majority of the mRNA transcripts found are capped and translationally competent, hence, the transfer of mRNAs can be beneficial as many copies of the encoded protein can be produced from an mRNA transcript, offering a more lasting effect in the receiver cell. In the case of miRNAs, determining the degree of overlap between miRNAs and mRNA transcripts was made more challenging by the significant number of false positive predictions generated by currently available tools. Nevertheless, focusing on miRNAs previously studied in MDSC, we found that these miRNAs are not often accompanied by their enriched validated target mRNAs. This is important as it supports the idea that the miRNA observed is not bound to its mRNA and, hence, it is available to carry its repressive function. As discussed above, both proteins and RNAs carried in exosomes could mediate MDSC immune suppressive functions by promoting MDSC accumulation and expansion. Therefore, we hypothesize that the exosomal proteins and RNAs may have a synergistic effect and/or provide for mechanistic redundancy.
Supplementary Material
(Table S-2) Differential expression analysis of mRNA transcript isoforms
(Table S-5) miRNA targets predicted using miRNAtap
(Table S-1) Summary of samples collected for RNA 762 analysis and mapping statistics
(Table S-3) Functional analysis of enriched mRNA transcript isoforms
(Table S-4) Differential expression analysis of miRNAs
(Table S-6) Functional analysis of miRNA targets predicted
(Table S-7) PepArML peptide and protein identifications and label-free relative quantitation
(Table S-8) Functional analysis of enriched proteins
ACKNOWLEDGMENTS
L.G.-A., V.K.C, N.J.E., S.O.-R., and C.F. are supported by the National Institutes of Health Grants GM021248 and OD019938. L.G.-A. is additionally supported by the University of Maryland Ann G. Wylie Dissertation Fellowship. A.T.B. and N.M.E. are funded, in part, by NIH Grant AI094773. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ABBREVIATIONS
- ADAM
disintegrin and metalloproteinase domain-containing protein
- Alix (PDCD4)
programmed cell death 6-interacting protein
- ARG-I
arginase I
- C/EBPα
CCAAT/enhancer binding protein-α
- C4B-bp
C4B-binding proteins
- cpm
counts per million
- FADD
fas-associated protein with Death Domain
- Fas
tumor necrosis factor receptor superfamily member 6
- HMGB1
high mobility group box protein 1
- IRAK
IL-1R-associated kinase
- MAF
macrophage-activating factor
- MDSC
myeloid-derived suppressor cell(s)
- MEF2C
myeloid ELF1-like factor 2C
- MIF
macrophage migration inhibitor factor
- MMP9
matrix metallopeptidase 9
- NFIA
nuclear factor I/A
- NF-κB
nuclear factor-κB subunit 1
- PF-4
platelet factor-4
- PSM
peptide spectrum matches
- PTEN
phosphatase and tensin homologue
- RUNX1
runt-related transcription factor 1
- SPRED-1
sprouty Related EVH1 Domain Containing 1
- STAT
signal transducer and activator of transcription
- TGF-β
transforming growth factor-β
- TRAF6
tumor necrosis factor receptor-associated factor 6
- TSP-1,
thrombospondin-1
- SOCS- 1
suppressing the expression of cytokine signaling 1
- UTR
untranslated region
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Colombo M; Raposo G; Thery C Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol 2014, 30, 255–289. [DOI] [PubMed] [Google Scholar]
- (2).Gould SJ; Raposo G As we wait: coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles 2013, 2, 20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Bartel DP MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).He L; Hannon GJ MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet 2004, 5, 522–531. [DOI] [PubMed] [Google Scholar]
- (5).Montecalvo A; Larregina AT; Shufesky WJ; Beer Stolz D; Sullivan MLG; Karlsson JM; Baty CJ; Gibson GA; Erdos G; Wang Z; Milosevic J; Tkacheva OA; Divito SJ; Jordan R; Lyons-Weiler J; Watkins SC; Morelli AE Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Valadi H; Ekstrom K; Bossios A; Sjöstrand M; Lee JJ; Lötvall JO Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 2007, 9, 654–659. [DOI] [PubMed] [Google Scholar]
- (7).Ekstrom K; Valadi H; Sjöstrand M; Malmhäll C; Bossios A; Eldh M; Lötvall J Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J. Extracell. Vesicles 2012, 1, 18389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Pegtel DM; Cosmopoulos K; Thorley-Lawson DA; van Eijndhoven MAJ; Hopmans ES; Lindenberg JL; de Gruijl TD; Wurdinger T; Middeldorp JM Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci U. S. A 2010, 107, 63286333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Fabbri M; Paone A; Calore F; Galli R; Gaudio E; Santhanam R; Lovat F; Fadda P; Mao C; Nuovo GJ; Zanesi N; Crawford M; Ozer GH; Wernicke D; Alder H; Caligiuri MA; Nana-Sinkam P; Perrotti D; Croce CM MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U. S. A 2012, 109, E2110–E2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Fabbri M; Paone A; Calore F; Galli R; Croce CM A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol. 2013, 10, 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Chiba M; Kimura M; Asari S Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol. Rep 2012, 28, 1551–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kogure T; Lin W-L; Yan IK; Braconi C; Patel T Intercellular nanovesicle-mediated microRNA transfer: A mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology 2011, 54, 1237–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Tomasoni S; Longaretti L; Rota C; Morigi M; Conti S; Gotti E; Capelli C; Introna M; Remuzzi G; Benigni A Transfer of Growth Factor Receptor mRNA Via Exosomes Unravels the Regenerative Effect of Mesenchymal Stem Cells. Stem Cells Dev. 2013, 22, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ostrand-Rosenberg S Myeloid-derived suppressor cells: more mechanisms for inhibiting antitumor immunity. Cancer Immunol. Immunother 2010, 59, 1593–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Parker KH; Beury DW; Ostrand-Rosenberg S Myeloid- Derived Suppressor Cells: Critical Cells Driving Immune Suppression in the Tumor Microenvironment. Adv. Cancer Res. 2015, 128, 95–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ezernitchi AV; Vaknin I; Cohen-Daniel L; Levy O; Manaster E; Halabi A; Pikarsky E; Shapira L; Baniyash M TCR Down-Regulation under Chronic Inflammation Is Mediated by Myeloid Suppressor Cells Differentially Distributed between Various Lymphatic Organs. J. Immunol 2006, 177, 4763–4772. [DOI] [PubMed] [Google Scholar]
- (17).Bunt SK; Sinha P; Clements VK; Leips J; OstrandRosenberg S Inflammation Induces Myeloid-Derived Suppressor Cells that Facilitate Tumor Progression. J. Immunol 2006, 176, 284–290. [DOI] [PubMed] [Google Scholar]
- (18).Bunt SK; Yang L; Sinha P; Clements VK; Leips J; Ostrand-Rosenberg S Reduced Inflammation in the Tumor Microenvironment Delays the Accumulation of Myeloid-Derived Suppressor Cells and Limits Tumor Progression. Cancer Res. 2007, 67, 10019–10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Sinha P; Clements VK; Fulton AM; Ostrand-Rosenberg S Prostaglandin E2 Promotes Tumor Progression by Inducing Myeloid- Derived Suppressor Cells. Cancer Res. 2007, 67, 4507–4513. [DOI] [PubMed] [Google Scholar]
- (20).Hanahan D; Weinberg RA Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- (21).Pulaski BA; Ostrand-Rosenberg S Mouse 4T1 breast tumor model. Curr. Protoc. Immunol 2001, 39, 10.1002/0471142735.im2002s39. [DOI] [PubMed] [Google Scholar]
- (22).Elkabets NM; Ribeiro V; Dinarello C; Ostrand-Rosenberg S; Di Santo C; Apte R; Vosshenrich CAJ IL-1beta regulates a novel myeloid-derived suppressor cell subset that impairs NK cell development and function. Eur. J. Immunol 2010, 40, 3347–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Burke M; Choksawangkarn W; Edwards N; OstrandRosenberg S; Fenselau C Exosomes from Myeloid-Derived Suppressor Cells Carry Biologically Active Proteins. J. Proteome Res 2014, 13, 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Burke MC; Oei MS; Edwards NJ; Ostrand-Rosenberg S; Fenselau C Ubiquitinated Proteins in Exosomes Secreted by Myeloid-Derived Suppressor Cells. J. Proteome Res 2014, 13, 5965–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Geis-Asteggiante L; Dhabaria A; Edwards N; OstrandRosenberg S; Fenselau C Top-down analysis of low mass proteins in exosomes shed by murine myeloid-derived suppressor cells. Int. J. Mass Spectrom 2015, 378, 264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Chauhan S; Danielson S; Clements V; Edwards N; OstrandRosenberg S; Fenselau C Surface Glycoproteins of Exosomes Shed by Myeloid-Derived Suppressor Cells Contribute to Function. J. Proteome Res 2017, 16, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Sinha P; Clements V; Ostrand-Rosenberg S Reduction of myeloid-derived suppressor cells and induction of M1 macrophages facilitate the rejection of established metastastic disease. J. Immunol 2005, 174, 636–645. [DOI] [PubMed] [Google Scholar]
- (28).Bolger AM; Lohse M; Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Martin M Cutadapt removes adapter sequences from high- throughput sequencing reads. EMBnet.journal 2011, 17, 10. [Google Scholar]
- (30).Bray NL; Pimentel H; Melsted P; Pachter L Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 2016, 34, 525–527. [DOI] [PubMed] [Google Scholar]
- (31).Kozomara A; Griffiths-Jones S miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Griffiths-Jones S The microRNA Registry. Nucleic Acids Res. 2004, 32, 109D–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Dillies M-A; Rau A; Aubert J; Hennequet-Antier C; Jeanmougin M; Servant N; Keime C; Marot G; Castel D; Estelle J; Guernec G; Jagla B; Jouneau L; Laloe D; Le Gall C; Schaeffer B; Le Crom S; Guedj M; Jaffrezic F French StatOmique Consortium. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Briefings Bioinf. 2013, 14, 671–683. [DOI] [PubMed] [Google Scholar]
- (34).Robinson MD; Oshlack A A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Robinson MD; McCarthy DJ; Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Bullard JH; Purdom E; Hansen KD; Dudoit S Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinf. 2010, 11, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Anders S; Huber W Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Leek JT; Storey JD Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007, 3, 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Risso D; Ngai J; Speed TP; Dudoit S Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol 2014, 32, 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ritchie ME; Phipson B; Wu D; Hu Y; Law CW; Shi W; Smyth GK limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Paraskevopoulou MD; Georgakilas G; Kostoulas N; Vlachos IS; Vergoulis T; Reczko M; Filippidis C; Dalamagas T; Hatzigeorgiou AG DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Krek A; Grun D; Poy MN; Wolf R; Rosenberg L; Epstein EJ; MacMenamin P; da Piedade I; Gunsalus KC; Stoffel M; Rajewsky N Combinatorial microRNA target predictions. Nat. Genet 2005, 37, 495–500. [DOI] [PubMed] [Google Scholar]
- (43).Enright AJ; John B; Gaul U; Tuschl T; Sander C; Marks DS MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Friedman RC; Farh KK-H; Burge CB; Bartel DP Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008, 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Wong N; Wang X miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, D146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Chambers MC; Maclean B; Burke R; Amodei D; Ruderman DL; Neumann S; Gatto L; Fischer B; Pratt B; Egertson J; Hoff K; Kessner D; Tasman N; Shulman N; Frewen B; Baker TA; Brusniak M-Y; Paulse C; Creasy D; Flashner L; Kani K; Moulding C; Seymour SL; Nuwaysir LM; Lefebvre B; Kuhlmann F; Roark J; Rainer P; Detlev S; Hemenway T; Huhmer A; Langridge J; Connolly B; Chadick T; Holly K; Eckels J; Deutsch EW; Moritz RL; Katz JE; Agus DB; MacCoss M; Tabb DL; Mallick P A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol 2012, 30, 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Edwards NJ PepArML: A Meta-Search Peptide Identification Platform for Tandem Mass Spectra. Curr. Protoc. Bioinformatics 2013, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Elias JE; Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207–214. [DOI] [PubMed] [Google Scholar]
- (49).Reiter L; Claassen M; Schrimpf SP; Jovanovic M; Schmidt A; Buhmann JM; Hengartner MO; Aebersold R Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell. Proteomics 2009, 8, 2405–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Old WM; Meyer-Arendt K; Aveline-Wolf L; Pierce KG; Mendoza A; Sevinsky JR; Resing KA; Ahn NG Comparison of Label-free Methods for Quantifying Human Proteins by Shotgun Proteomics. Mol. Cell. Proteomics 2005, 4, 1487–1502. [DOI] [PubMed] [Google Scholar]
- (51).Benjamini Y; Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B 1995, 57, 289–300. [Google Scholar]
- (52).Reimand J; Arak T; Adler P; Kolberg L; Reisberg S; Peterson H; Vilo J g:Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016, 44, W83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Kanehisa M; Sato Y; Kawashima M; Furumichi M; Tanabe M KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Vizcaíno JA; Csordas A; del-Toro N; Dianes JA; Griss J; Lavidas I; Mayer G; Perez-Riverol Y; Reisinger F; Ternent T; Xu QW; Wang R; Hermjakob H 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016, 44 (D1), D447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Xiao D; Ohlendorf J; Chen Y; Taylor DD; Rai SN; Waigel S; Zacharias W; Hao H; McMasters KM Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS One 2012, 7, e46874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Ji H; Chen M; Greening DW; He W; Rai A; Zhang W; Simpson RJ Deep sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA-enrichment signatures. PLoS One 2014, 9, e110314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Crescitelli R; Lässer C; Szabó TG; Kittel A; Eldh M; Dianzani I; Buzás EI; Lötvall J Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2013, 2 (1), 20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).McMahon G VEGF Receptor Signaling in Tumor Angiogenesis. Oncologist 2000, 5, 3–10. [DOI] [PubMed] [Google Scholar]
- (59).Yarden Y; Sliwkowski MX Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol 2001, 2, 127–137. [DOI] [PubMed] [Google Scholar]
- (60).Chornoguz O; Grmai L; Sinha P; Artemenko KA; Zubarev RA; Ostrand-Rosenberg S Proteomic Pathway Analysis Reveals Inflammation Increases Myeloid-Derived Suppressor Cell Resistance to Apoptosis. Mol. Cell. Proteomics 2011, 10, M110.002980 10.1074/mcp.M110.002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Choksawangkarn W; Graham LM; Burke M; Lee SB; Ostrand-Rosenberg S; Fenselau C; Edwards NJ Peptide-based systems analysis of inflammation induced myeloid-derived suppressor cells reveals diverse signaling pathways. Proteomics 2016, 16, 1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Perez-Hernandez D; Gutierrez-Vazquez C; Jorge I; LopezMartin S; Ursa A; Sanchez-Madrid F; Vazquez J; Yanez-Mo M The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem 2013, 288, 11649–11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Zhang J; Li S; Li L; Li M; Guo C; Yao J; Mi S Exosome and exosomal microRNA: Trafficking, sorting, and function. Genomics, Proteomics Bioinf. 2015, 13, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Yang JM; Gould SJ The cis-acting signals that target proteins to exosomes and microvesicles. Biochem. Soc. Trans 2013, 41, 277–82. [DOI] [PubMed] [Google Scholar]
- (65).Villarroya-Beltri C; Gutierrez-Vazquez C; Sanchez-Cabo F; Perez-Hernandez D; Vazquez J; Martin-Cofreces N; Martinez-Herrera DJ; Pascual-Montano A; Mittelbrunn M; Sanchez- Madrid F Sumoylated HNRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun 2013, 4, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Parker KH; Sinha P; Horn LA; Clements VK; Yang H; Li J; Tracey KJ; Ostrand-Rosenberg S HMGB1 enhances immune suppression by facilitating the differentiation and suppressive activity of myeloid-derived suppressor cells. Cancer Res. 2014, 74, 5723–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Simpson KD; Cross JV MIF: metastasis/MDSC-inducing factor? Oncoimmunology 2013, 2, e23337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Simpson KD; Templeton DJ; Cross JV Macrophage Migration Inhibitory Factor Promotes Tumor Growth and Metastasis by Inducing Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. J. Immunol 2012, 189, 5533–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Deuel TF; Senior RM; Chang D; Griffin GL; Heinrikson RL; Kaiser ET Platelet factor 4 is chemotactic for neutrophils and monocytes. Proc. Natl. Acad. Sci. U. S. A 1981, 78, 4584–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Li H; Han Y; Guo Q; Zhang M; Cao X Cancer-expanded myeloid-derived suppressor cells induce anergy of NK cells through membrane-bound TGF-beta 1. J. Immunol 2009, 182, 240–249. [DOI] [PubMed] [Google Scholar]
- (71).Walport MJ Complement. First of two parts. N. Engl. J. Med 2001, 344, 1058–1066. [DOI] [PubMed] [Google Scholar]
- (72).Rutkowski MJ; Sughrue ME; Kane AJ; Mills SA; Parsa AT Cancer and the complement cascade. Mol. Cancer Res 2010, 8, 1453–1465. [DOI] [PubMed] [Google Scholar]
- (73).Markiewski M; DeAngelis R; Benencia F; Ricklin-Lichtsteiner S; Koutoulaki A; Coukos G; Lambris J; Gerard C Modulation of the anti-tumour immune response by complement. Nat. Immunol 2008, 9, 1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Brodeur SR; Angelini F; Bacharier LB; Blom AM; Mizoguchi E; Fujiwara H; Plebani A; Notarangelo LD; Dahlback B; Tsitsikov E; Geha RS C4b-binding protein (C4BP) activates B cells through the CD40 receptor. Immunity 2003, 18, 837–848. [DOI] [PubMed] [Google Scholar]
- (75).Kopp A; Hebecker M; Svobodová E; Józsi M Factor h: a complement regulator in health and disease, and a mediator of cellular interactions. Biomolecules 2012, 2, 46–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Nabil K; Rihn B; Jaurand MC; Vignaud JM; Ripoche J; Martinet Y; Martinet N Identification of human complement factor H as a chemotactic protein for monocytes. Biochem. J 1997, 326, 377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Ferreira VP; Pangburn MK; Cortes C Complement control protein factor H: the good, the bad, and the inadequate. Mol. Immunol 2010, 47, 2187–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Taganov KD; Boldin MP; Chang K-J; Baltimore D NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A 2006, 103, 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Zhao JL; Rao DS; Boldin MP; Taganov KD; O’Connell RM; Baltimore D NF-kB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 9184–9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Hegde VL; Tomar S; Jackson A; Rao R; Yang X; Singh UP; Singh NP; Nagarkatti PS; Nagarkatti M Distinct MicroRNA Expression Profile and Targeted Biological Pathways in Functional Myeloid-derived Suppressor Cells Induced by 9-Tetrahydrocannabinol in Vivo: regulation of CCAAT/enhancer-binding protein by microRNA-690. J. Biol. Chem 2013, 288, 36810–36826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Chen S; Zhang Y; Kuzel TM; Zhang B Regulating Tumor Myeloid-Derived Suppressor Cells by MicroRNAs. Cancer Cell Microenviron. 2015, 2, No. e637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Li L; Zhang J; Diao W; Wang D; Wei Y; Zhang C-Y; Zen K MicroRNA-155 and MicroRNA-21 Promote the Expansion of Functional Myeloid-Derived Suppressor Cells. J. Immunol 2014, 192, 1034–1043. [DOI] [PubMed] [Google Scholar]
- (83).Sinha P; Clements VK; Bunt SK; Albelda SM; OstrandRosenberg S Cross-talk between myeloid-derived suppressor cells and macrophages subverts tumor immunity toward a type 2 response. J. Immunol 2007, 179, 977–983. [DOI] [PubMed] [Google Scholar]
- (84).Bunt SK; Clements VK; Hanson EM; Sinha P; OstrandRosenberg S Inflammation enhances myeloid-derived suppressor cell cross-talk by signaling through Toll-like receptor 4. J. Leukocyte Biol 2009, 85, 996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Kroesen B-J; Teteloshvili N; Smigielska-Czepiel K; Brouwer E; Boots AMH; van den Berg A; Kluiver J Immuno-miRs: critical regulators of T-cell development, function and ageing. Immunology 2015, 144, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Table S-2) Differential expression analysis of mRNA transcript isoforms
(Table S-5) miRNA targets predicted using miRNAtap
(Table S-1) Summary of samples collected for RNA 762 analysis and mapping statistics
(Table S-3) Functional analysis of enriched mRNA transcript isoforms
(Table S-4) Differential expression analysis of miRNAs
(Table S-6) Functional analysis of miRNA targets predicted
(Table S-7) PepArML peptide and protein identifications and label-free relative quantitation
(Table S-8) Functional analysis of enriched proteins