Supplemental Digital Content is available in the text.
Keywords: biomarkers, gene expression, intensive care units, interleukin-7 receptor, septic shock
Abstract
Objectives:
Septic shock is the primary cause of death in ICUs. A better comprehension of its pathophysiology, in particular, the immune alteration mechanisms, opened new therapeutic perspectives such as the recombinant interleukin-7. The use of biomarkers could improve the identification of eligible patients for this therapy. The soluble form of the interleukin-7 appears as a promising candidate in this regard since an association between its high plasmatic level and mortality in critically ill patients has been demonstrated. Because there are no data available on the transcriptional regulation of the interleukin-7 receptor in such patients, this study aimed to explore the expression level of different interleukin-7 receptor transcripts after septic shock and evaluate their association with mortality.
Design:
Retrospective discovery cohort (30 patients) and validation cohort (177 patients).
Setting:
Two French ICUs (discovery study) and six French ICUs (validation study).
Patients:
Adult septic shock patients.
Interventions:
None.
Measurements and Main Results:
The quantification of several interleukin-7 receptor transcripts using specific reverse transcription quantitative polymerase chain reaction designs allowed for global evaluation of interleukin-7 receptor gene expression in whole blood. In the discovery cohort, all interleukin-7 receptor transcripts studied were expressed at lower levels in septic shock patients than in healthy volunteers. Interleukin-7 receptor gene expression at day 3 after septic shock diagnosis was associated with day 28 mortality. Patients at a lower risk of death showed higher expression levels. These results were confirmed in the independent validation cohort. Interestingly, using a threshold obtained on the discovery cohort, we observed in the validation cohort a high negative predictive value for day 28 mortality for the transcript encoding the membrane form of interleukin-7 receptor (0.86; 95% CI, 0.79–0.93).
Conclusions:
Interleukin-7 receptor transcripts appear as biomarkers of impaired adaptive immune response in septic shock patients and as a promising tool for patient stratification in clinical trials evaluating immunoadjuvant therapies.
Septic shock remains the primary cause of death in ICUs despite better understanding of the underlying pathophysiologic mechanisms and improved patient management. A recent study (1) using the new Sepsis-3 definitions (2) presented an ICU mortality rate for septic shock patients of 44%.
A disproportionate inflammatory response exceeding the concomitant anti-inflammatory response is observed in the early phase of septic shock, which is usually resolved within 72 hours. This phase may be followed by prolonged immunosuppression that accounts for over two thirds of deaths (3). Several studies reported impaired innate and adaptive immune responses following septic shock (4). Massive apoptosis of T cells leads to important lymphopenia (5). Also, the functionality of T lymphocytes (i.e., their proliferation and cytokine production capacity) is decreased (6).
Immunomodulating therapies have emerged as innovative and effective approaches to control dysregulated immune response following septic shock in view of either blocking an exaggerated response or boosting an altered one. It therefore appears crucial to efficiently and accurately determine the immune status of patients to provide the most effective treatment. There is, however, no specific clinical sign of impaired immune response. Ex vivo functional assays such as cytokine production or cell proliferation, although gold standards in the field, are not easily adaptable to clinical routine (7). There is now a great interest in easily measurable biomarkers to guide patient treatment.
Among the proposed immune-adjuvant treatments, the recombinant human interleukin (rhIL)-7 evaluated in HIV (8) or cancer (9) has already shown promising preclinical results in sepsis. The ex vivo use of rhIL-7 improved lymphocyte function following sepsis (6), and an in vivo treatment increased survival in a peritonitis mouse model (10).
The interleukin (IL)-7 signal is transduced via the IL-7 receptor (IL-7R), composed of the common γc and the IL-7–specific α (CD127) chains (11). As shown for other cytokine receptors, IL-7R can be found as a soluble protein (sCD127) (12). Studies have reported an association between high sCD127 plasma levels and mortality in septic shock (13) and critically ill patients (14). The mechanisms leading to the production of sCD127 are not fully understood as yet and may involve alternative splicing (12, 15) and posttranscriptional modifications (16). sCD127 is able to bind IL-7 (17), but its effect on IL-7 activity is still controversial; some studies reporting a decrease of IL-7 activity (18, 19), whereas others show a potentialization of its effects (17, 20). Because there is currently no available global transcriptional study of IL7R in septic shock patients, describing its expression modulation and evaluating its potential as a surrogate marker of septic shock-induced immune alterations appeared relevant.
The objectives of our study were to describe the transcriptional expression of IL7R during septic shock and assess the association between IL7R transcripts and day 28 (D28) mortality. This was explored in a discovery cohort and validated in a larger and independent cohort of septic shock patients.
MATERIALS AND METHODS
Patients and Samples
Septic Shock Patient Discovery and Validation Cohorts.
Septic shock patients were included from two independent existing cohorts, based on the following inclusion criteria: 1) predicted ICU length of stay of at least 2 days and 2) presence of Systemic Inflammatory Response Syndrome due to an ongoing infection. Septic shock was defined as a sepsis-induced hypotension persisting despite adequate fluid resuscitation requiring vasopressors (21). The exclusion criteria were aplasia for the discovery cohort, completed, for the validation cohort, with recent chemotherapy or immunosuppressive treatment, high-dose or prolonged corticosteroid treatment, primary immune deficiency, and extracorporeal circulation in the month preceding ICU admission. Protocols of the discovery (institutional review board [IRB] no. 11236) and validation (IRB no. 5044) cohorts were approved by local ethics committees. Nonopposition to inclusion in the protocols was systematically recorded from patients or next of kin (see details in supplemental patients description, Supplemental Digital Content 1, http://links.lww.com/CCM/D724).
Control Cohort.
Whole blood samples from 19 healthy volunteers (median age, 41 [35–50]; male, 42%) were obtained from the Etablissement Français du Sang in Lyon and used as controls. In accordance with standardized procedures for blood donation, informed consent was obtained and personal data were anonymized at the time of blood donation.
IL7R Transcripts Expression Level Measurement
Peripheral whole blood was collected in PAXgene tubes (PreAnalytix, Hilden, Germany). Briefly, after extraction, total RNA was reverse transcribed in complementary DNA. Three IL7R-specific polymerase chain reaction (PCR) assays were designed: the “IL7R-001” PCR is specific of the IL7R-001 transcript encoding the membrane form of CD127 and the “IL7R-003” PCR of the IL7R-003 transcript encoding a sCD127 form. The “IL7R-All” PCR quantifies all IL7R transcripts studied, including the two previously described (Supplemental Fig. 1, Supplemental Digital Content 1, http://links.lww.com/CCM/D724; and Supplemental Table 1, Supplemental Digital Content 1, http://links.lww.com/CCM/D724). Absolute concentrations (copies/µL) were used to compare the expression levels of the different transcripts. For analysis of the association with D28 mortality, gene expression was normalized using hypoxanthine phosphoribosyltransferase 1 as reference gene and results were expressed as Calibrated Normalized Relative Quantity (22). Experimental protocol is detailed in supplemental method (Supplemental Digital Content 1, http://links.lww.com/CCM/D724).
Statistics
Chi-square or Fisher exact tests were used for qualitative variables assessment. Quantitative variables were compared with a Student t test or Mann-Whitney U test. Paired Wilcoxon tests were used to compare the expression of IL7R transcripts between samples collected at two time-points. The association between D28 mortality and IL7R expression level was appreciated by univariate and multivariate Cox models. To allow comparison between models, hazard ratios were normalized to an increment from first to third quartile (interquartile range hazard ratio [IQR HR]). For multivariate analysis, confounding factors were selected using the following criteria: a correlation between the variable and IL7R expression level (Spearman r ≥ 0.15), a significant association with D28 mortality (p < 0.1, Cox model), and the absence of missing values. To avoid redundancy, variables used in Sequential Organ Failure Assessment (SOFA) or Simplified Acute Physiologic Score (SAPS) II calculation were not included in multivariate models including these scores. Area under receiver operating characteristics curves (AUROCs) were calculated to assess the predictive performance regarding D28 mortality. DeLong method was used for comparison of AUROCs. For IL7R transcripts expression, a threshold was determined in the discovery cohort in order to maximize: 1) the negative predictive value and 2) the specificity. In the validation cohort, survival curves with 95% CIs were drawn in patient groups defined according to IL7R expression threshold at D3 obtained in the discovery cohort. The log-rank test was used to compare the curves. A p value below 0.05 was considered significant. All statistics were performed with R software (version 3.2.4; R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
Discovery Cohort
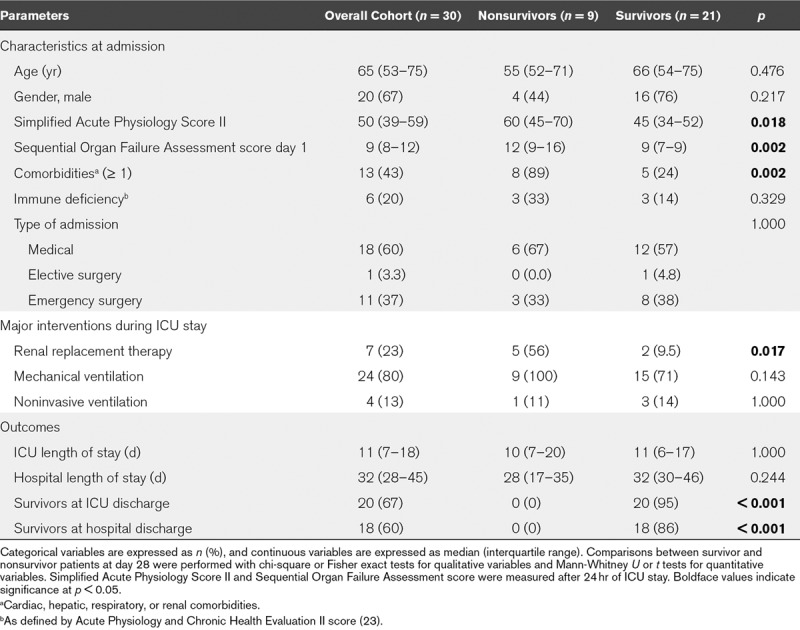
To study the modulation of IL7R transcripts after septic shock, we first retrospectively included 30 patients who were sampled both at day 1 (D1) and day 3 (D3) after septic shock diagnosis. This enabled the evaluation of the IL7R D3/D1 expression ratio for all selected patients. Nine patients (30%) died before D28 (Table 1). D28 nonsurvivors had more comorbidities and higher severity scores at admission than survivors.
TABLE 1.
Description of the Discovery Cohort According to Survival Status at Day 28 After Septic Shock Diagnosis
We measured IL7R expression levels using three PCR designs, of which one is specific of the IL7R-001 transcript that encodes the CD127 membrane and one of the sCD127-encoding IL7R-003 transcript. Expression levels of all IL7R transcripts studied were lower in septic shock patients than in healthy volunteers (p < 0.001) (Fig. 1A). In septic shock patients, we observed a moderate increase in the expression of all IL7R transcripts between D1 and D3 (p < 0.001). Overall, the expression level of the transcript encoding a sCD127 form was approximately 60-fold lower than the transcript encoding the CD127 membrane form.
Figure 1.
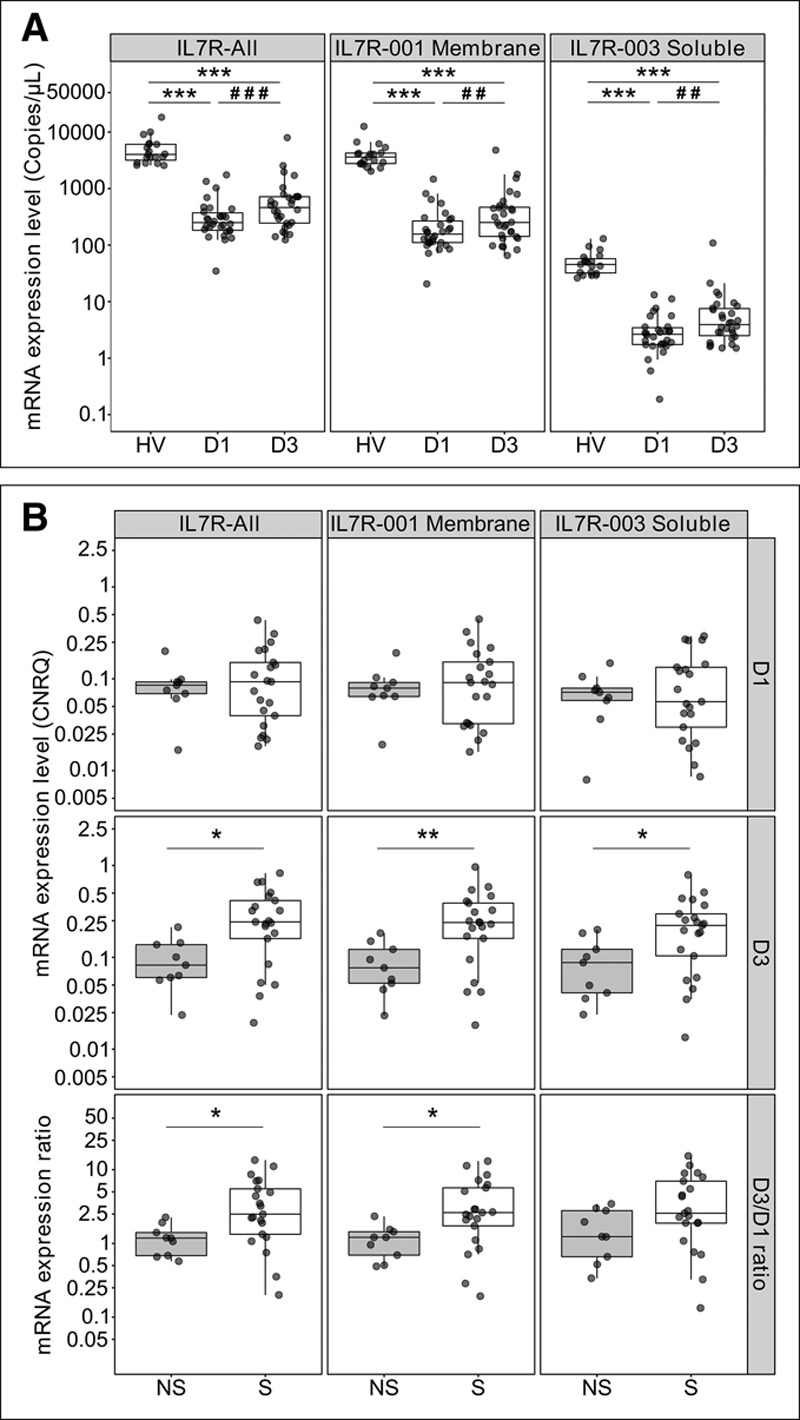
Interleukin-7 receptor (IL7R) transcripts expression levels in the discovery cohort. A, Comparison of messenger RNA (mRNA) expression between healthy volunteers (HV, n = 19) and septic shock patients at day 1 (D1) and day 3 (D3) after septic shock diagnosis (n = 30). mRNA levels are expressed as absolute concentration in copies/µL. B, Comparison of mRNA expression between day 28 nonsurvivor (NS, n = 9) and survivor (S, n = 21) patients, at D1 and D3 after septic shock diagnosis, and for the D3/D1 expression ratio. mRNA levels are expressed as calibrated normalized relative quantity (CNRQ) using hypoxanthine phosphoribosyltransferase 1 as reference gene. *p < 0.05, **p < 0.01, and ***p < 0.001 Mann-Whitney U test, ##p < 0.01 and ###p < 0.001 paired Wilcoxon test.
Although IL7R expression levels at D1 showed no difference between survivors and nonsurvivors, those of all transcripts at D3 were lower in nonsurvivors (p < 0.05) (Fig 1B). They also remained low in nonsurvivors but increased between D1 and D3 in survivors, as shown by higher D3/D1 ratios in survivors.
IL7R transcripts expression at D3 was significantly associated with D28 mortality, a higher expression level of IL7R transcripts indicating a lower risk of death (IQR HR [95% CI], 0.12 [0.02–0.81], 0.11 [0.02–0.74], and 0.20 [0.04–0.93] for IL7R-All, IL7R-001, and IL7R-003, respectively, p < 0.05) (Supplemental Table 2, Supplemental Digital Content 1, http://links.lww.com/CCM/D724).
To estimate the performance of IL7R as a prognostic biomarker, we calculated AUROC for each IL7R PCR assay at D3 and selected thresholds, in view of reaching a high negative predictive value and a high specificity (Supplemental Table 3, Supplemental Digital Content 1, http://links.lww.com/CCM/D724). All AUROCs were over 0.77, the highest one corresponding to the IL7R-001 membrane transcript.
As the IL7R-001 membrane transcript showed a high expression level and the most promising performances for D28 mortality prediction at D3, we specifically evaluated this transcript in the validation cohort.
Validation Cohort
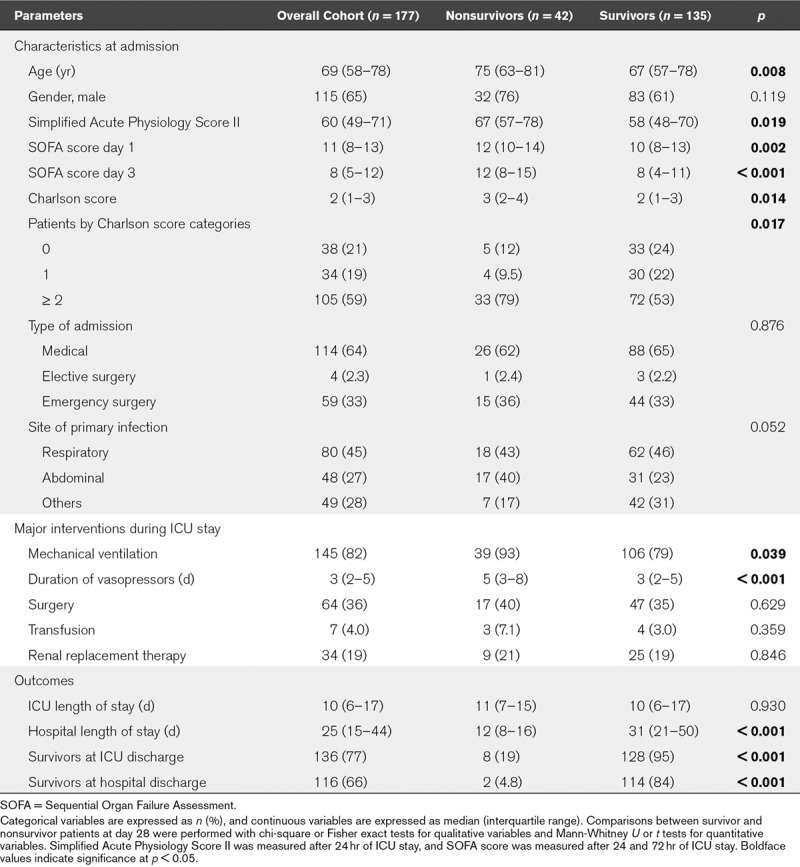
We included from a different cohort septic shock patients with available samples at D3 after septic shock diagnosis in order to confirm results from the discovery study (Supplemental Fig. 2, Supplemental Digital Content 1, http://links.lww.com/CCM/D724). Out of the 177 selected patients, 42 died within D28 (24%) (Table 2). Age, SAPS II, D1 and D3 SOFA, and Charlson scores were significantly higher in D28 nonsurvivors than in survivors. Lactate level, lymphocyte, and platelet counts at D3 were also significantly different between survivors and nonsurvivors (Supplemental Table 4, Supplemental Digital Content 1, http://links.lww.com/CCM/D724). The discovery and validation cohorts provided similar results in terms of comorbidities, biology, and severity (data not shown).
TABLE 2.
Clinical Characteristics of the Septic Shock Patients of the Validation Cohort According to Survival Status at Day 28
As observed in the discovery cohort, IL7R expression level at D3 was significantly lower in D28 nonsurvivors compared with survivors (p < 0.001) (Fig. 2A).
Figure 2.
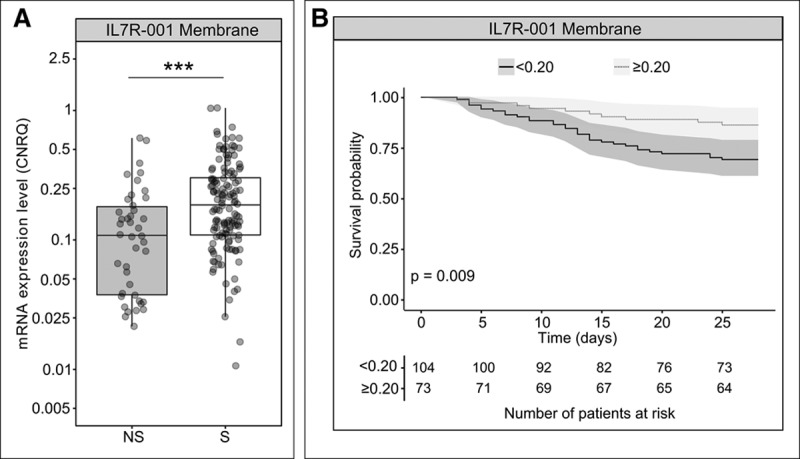
Interleukin-7 receptor (IL7R) membrane transcript (IL7R-001) expression levels in the validation cohort. A, Comparison of IL7R expression levels between day 28 nonsurvivor (NS, n = 42) and survivor (S, n = 135) patients. IL7R expression levels were measured at day 3 (D3) after septic shock diagnosis. Messenger RNA (mRNA) levels are expressed as calibrated normalized relative quantity (CNRQ) using hypoxanthine phosphoribosyltransferase 1 (HPRT1) as reference gene. ***p < 0.001 Mann-Whitney U test. B, Survival curves and 95% CIs in patient groups of the validation cohort defined according to IL7R membrane transcript (IL7R-001) expression levels at D3, using the threshold obtained on the discovery cohort. This threshold of 0.20 is expressed as a CNRQ using HPRT1 as reference gene. The log-rank test was used to compare survival between groups.
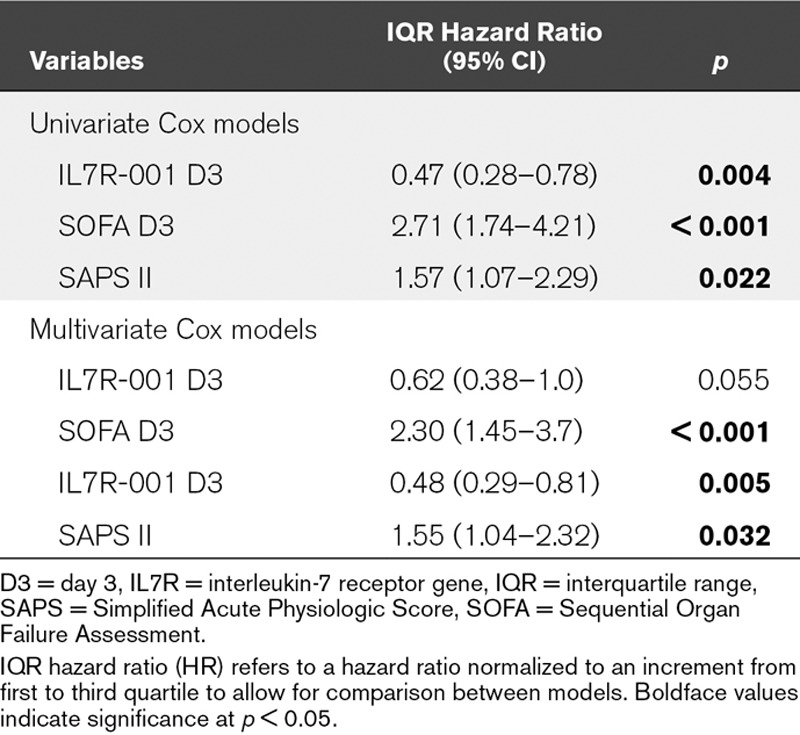
The validation cohort allowed confirming the significant association between D28 mortality and IL7R expression level at D3 (IQR HR, 0.47 [95% CI, 0.28–0.78]; p < 0.01) (Table 3). This association was still significant in a multivariate model adjusted for SAPS II (0.48 [0.29–0.81]; p < 0.01) (Table 3).
TABLE 3.
Association Between Interleukin-7 Receptor Membrane Transcript (IL7R-001) Expression Levels at Day 3 and Status at Day 28 After Septic Shock Diagnosis in the Validation Cohort
We assessed the performance of IL7R expression levels at D3 to identify D28 nonsurvivor patients and obtained an AUC of 0.68 (95% CI, 0.59–0.78). This AUC was not statistically different from that of SOFA (0.72 [0.63–0.80]) and SAPS II (0.63 [0.53–0.72]). Interestingly, using the threshold from the discovery study, a negative predictive value of 0.86 (0.79–0.93), a positive predictive value of 0.31 (CI, 0.26–0.36), a sensitivity of 0.76 (0.62–0.88), and a specificity of 0.46 (0.36–0.56) were obtained in the validation study.
Finally, survival probabilities in groups defined according to IL7R expression at D3 were significantly different (p < 0.01) (Fig. 2B).
DISCUSSION
Our study highlights for the first time the existing association between IL7R messenger RNA (mRNA) expression and D28 mortality in two independent cohorts of septic shock patients. Using a threshold obtained on the discovery cohort, IL7R expression level at D3 identified a group of septic shock patients from the validation cohort with a 2.2-fold lower risk of death. This suggests that IL7R mRNA quantification could help patient stratification in clinical trials evaluating immune-adjuvant therapies.
Given the important role of IL-7 in T cell development and homeostasis, rhIL-7 has been evaluated in clinical trials in HIV and cancer. The first clinical trial in septic shock patients has just been completed and demonstrated rhIL-7 efficacy in restoring lymphocyte count in the absence of any severe side effect (24). We previously suggested that IL7R could be a companion biomarker for such therapy (13). Previous studies have demonstrated that CD127 expression on T lymphocytes surface does not seem to undergo extensive regulation in septic shock (6, 25). In addition, CD127 is measured by flow cytometry, a technique that is not routinely implemented in clinical practice yet, consequently compromising its use as a biomarker in septic shock patients. Interestingly, we previously demonstrated a significant association between high plasma level of sCD127 and mortality (13, 14). Several IL7R transcripts have been identified and those lacking exon 6, the exon encoding transmembrane domain (12), may encode soluble forms of CD127.
To our knowledge, this study is the first to quantify different IL7R transcripts in septic shock patients. We set up specific PCR designs to measure 1) global IL7R expression; 2) the membrane CD127 encoding transcript; and 3) a soluble form of CD127 encoding transcript, whereas most of previous studies in autoimmune diseases (26, 27) or infectious diseases (28–30) evaluated only IL7R membrane transcript expression.
All transcripts studied, including that encoding a soluble form (IL7R-003), show decreased expression after septic shock. Its expression was interestingly lower in septic shock patients than in healthy volunteers, as also observed for the plasmatic level of sCD127. However, although the expression level of IL7R-003 was lower in nonsurvivors, the sCD127 plasma concentration was higher in nonsurvivors (13, 14). The expression of IL7R-007, another transcript lacking exon 6, was modulated similarly to IL7R-003 (data not shown).
Combining the results obtained on proteins (membrane and soluble CD127) and on IL7R transcripts underlines the complex mechanisms involved in the regulation of this receptor. The primary source of sCD127 has not yet been clearly determined but may be the result of concomitant shedding from the membrane and transcriptional regulation (15, 16). Furthermore, despite the absence of a comprehensive evaluation of IL7R forms, numerous molecules such as IL-7 itself, tumor necrosis factor, IL-6, or glucocorticoids, were shown to regulate its expression (31). This should be further considered when interpreting IL7R expression levels in pathologies leading to immune response modulation.
This study reveals the significant association of D3 IL7R expression with D28 mortality in two independent cohorts of septic shock patients and is in line with findings from Bauer et al (28), who reports the association of IL7R in a combination of three down-regulated genes in sepsis with long-term mortality.
The AUROCs obtained for all IL7R transcripts studied were similar to those obtained for SAPS II and SOFA scores. Specifically quantifying the transcript encoding an sCD127 form actually did not prove more informative toward the identification of patients at risk of death than the full-length transcript. An interesting finding in the validation study is the high negative predictive value for D28 mortality shown by the IL7R-001 membrane expression at D3. This highlights its possible use as an exclusion biomarker for rhIL-7 treatment: IL7R expression could indeed identify patients at a lower risk of death who may not benefit from immunotherapy in order to focus treatment on the high-risk population. Interestingly, we obtained similar results when considering nonseptic ICU patients (data not shown). This will also be evaluated in trauma, burn, and surgical patients in the REAnimation Low Immune Status Markers (REALISM) study (32).
Other markers of the adaptive immune response are associated with mortality in critically ill patients, the most studied being absolute lymphocyte count (33–35). Another indicator of the adaptive immune response is the lymphocyte proliferation assay that reflects T cell functionality and has been found associated with mortality in sepsis patients (36). Unfortunately, long time-to-results and standardization issues make its use for rapid immune status assessment difficult (7). In this specific context, the feasibility of measuring a biomarker such as IL7R in clinical routine using automated molecular biology platform (reverse transcription quantitative PCR) makes it a promising tool for immune status monitoring. The high multiplex capacity of such platforms offers the possibility to integrate IL7R in a panel of biomarkers from innate and adaptive immune response that have been shown to be associated with immune alterations in ICU patients, such as C-X3-C motif chemokine receptor 1 (37), IL10, and CD74 (38) or programmed cell death-1 (39). This could lead to the development of an integrated tool for immune characterization.
Our study, however, has some limitations. Healthy volunteers are younger than septic shock patients. Lymphocyte count and lactate level at D3, identified as potential confounding factors, might have been included in multivariate models in the validation study. In a model including IL7R-001 expression, SAPS II, lactate concentration, and lymphocyte count at D3, the only parameter significantly associated with D28 mortality was lactate concentration (data not shown). However, due to a high number of missing values (36%), interpretation of such model is limited. The lower levels of IL7R transcripts in patients compared with healthy volunteers may also reflect the lower number of T cells frequently observed after septic shock (40) and not only a down-regulation of the expression. Not surprisingly, IL7R expression in whole blood was correlated with lymphocyte count (Spearman r = 0.5). Besides the interest of IL7R expression by itself as a prognostic biomarker, its regulation in septic shock patients has to be specifically studied in isolated T lymphocytes.
CONCLUSIONS
Our results show that IL7R mRNA expression level is associated with D28 mortality in septic shock patients and presents a high negative predictive value. IL7R expression appears as a suitable marker of impaired adaptive immune response following septic shock. This marker could be evaluated in a panel of biomarkers combining innate and adaptive aspects of the immune response in order to precisely determine the immune status of patients and treat them more efficiently.
ACKNOWLEDGMENTS
We gratefully thank Sophie Blein for her precious advice on statistical analysis and Sophie Ablott for her careful review of the article. Marqueurs Inflammatoires Pronostiques en Réanimation (MIP Réa) study group members are as follows: Bernard Floccard, Guillaume Marcotte, Mathieu Page, and Christian Guillaume (Department of Anaesthesiology and Critical Care Medicine, Hospices Civils de Lyon, Groupement Hospitalier Edouard Herriot, University Claude Bernard Lyon 1, Lyon, France); Martin Cour, Laurent Argaud, Romain Hernu, Sylvie De La Salle, Marie Simon, and Thomas Baudry (Hospices Civils de Lyon, Medical Intensive Care Unit, Groupement Hospitalier Edouard Herriot, Lyon, France); Arnaud Friggeri, Hélène Vallin, Nathalie Panel, Marion Provent, Bernard Allaouchiche, Fabrice Thiolliere, Vincent Piriou, and Julien Bohé (Hospices Civils de Lyon, Intensive Care Unit, Centre Hospitalier Lyon Sud, Pierre Bénite, France); Frédéric Aubrun (Hospices Civils de Lyon, Intensive Care Unit, Hôpital de la Croix Rousse, Lyon, France); Emmanuelle Gallet-Gorius, Audrey Larue-Triolet, Christine Alberti-Segui, Marie-Angélique Cazalis, and Véronique Barbalat (Joint Research Unit HCL-bioMérieux, Groupement Hospitalier Edouard Herriot, Lyon, France); and Anne Portier (Hospices Civils de Lyon, Immunology Laboratory, Groupement Hospitalier Edouard Herriot, Lyon, France).
Supplementary Material
Footnotes
*See also p. 1867.
The experimental work was performed at the Hospices Civils de Lyon – bioMérieux - Université Claude Bernard Lyon 1, Joint Research Unit, Lyon, France.
Supported, in part, by grants from bioMérieux and Hospices Civils de Lyon, and was part of Advanced Diagnostic for New Therapeutic Approaches, a program dedicated to personalized medicine, coordinated by Institut Mérieux and supported by French public agency BPI France.
Dr. Delwarde and Peronnet contributed equally to this work.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccmjournal).
All authors work in a Joint Research Unit cofunded by the Hospices Civils de Lyon, bioMérieux, and Université Claude Bernard Lyon 1. Drs. Delwarde and Venet, Mr. Meunier, and Drs. Lepape, Rimmelé, and Monneret are employees of Hospices Civils de Lyon. Dr. Peronnet, Ms. Cerrato, and Drs. Mouillaux, Pachot, and Textoris are employees of bioMérieux. This study was part of Advanced Diagnostic for New Therapeutic Approaches (ADNA), a program dedicated to personalized medicine, coordinated by Institut Mérieux, and supported by the French public agency Banque Publique d'Investissement (BPI) France. Drs. Delwarde, Peronnet, and Venet, Ms. Cerrato, and Drs. Lepape, Monneret, and Textoris are coinventors on three patent families covering interleukin-7 receptor biomarkers. This does not alter the authors’ adherence to all Critical Care Medicine policies on sharing data and materials. Dr. Peronnet disclosed that she is a coinventor on the pending patent WO2017093672, and that this study was part of ADNA. Dr. Venet’s institution received funding from bioMerieux (he works in a joint research unit between bioMerieux and the Hospices Civils de Lyon, but does not receive any financial retribution from bioMerieux based on this collaboration). Dr. Lepape disclosed that he is a coinventors of patents WO2013140103 and WO2015040328. Drs. Pachot and Textoris’ institution received funding from ADNA. Dr. Pachot received funding from bioMérieux as an employee of bioMérieux; he received support for article research from ADNA, and he disclosed government work. Dr. Textoris disclosed that he is an employee of bioMeriaux and Hospices Civils de Lyon, and he is listed as an inventor of patent WO2017093672 (pending), for which he does not own rights. The remaining authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.SepNet Critical Care Trials Group: Incidence of severe sepsis and septic shock in German intensive care units: The prospective, multicentre INSEP study. Intensive Care Med 2016; 42:1980–1989. [DOI] [PubMed] [Google Scholar]
- 2.Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016; 315:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daviaud F, Grimaldi D, Dechartres A, et al. Timing and causes of death in septic shock. Ann Intensive Care 2015; 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hotchkiss RS, Monneret G, Payen D.Sepsis-induced immunosuppression: From cellular dysfunctions to immunotherapy. Nat Rev Immunol 2013; 13:862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Tulzo Y, Pangault C, Gacouin A, et al. Early circulating lymphocyte apoptosis in human septic shock is associated with poor outcome. Shock 2002; 18:487–494. [DOI] [PubMed] [Google Scholar]
- 6.Venet F, Foray AP, Villars-Méchin A, et al. IL-7 restores lymphocyte functions in septic patients. J Immunol 2012; 189:5073–5081. [DOI] [PubMed] [Google Scholar]
- 7.Duffy D, Rouilly V, Libri V, et al. Milieu Intérieur Consortium: Functional analysis via standardized whole-blood stimulation systems defines the boundaries of a healthy immune response to complex stimuli. Immunity 2014; 40:436–450. [DOI] [PubMed] [Google Scholar]
- 8.Sereti I, Dunham RM, Spritzler J, et al. ACTG 5214 Study Team: IL-7 administration drives T cell-cycle entry and expansion in HIV-1 infection. Blood 2009; 113:6304–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sportès C, Hakim FT, Memon SA, et al. Administration of rhIL-7 in humans increases in vivo TCR repertoire diversity by preferential expansion of naive T cell subsets. J Exp Med 2008; 205:1701–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unsinger J, Burnham CA, McDonough J, et al. Interleukin-7 ameliorates immune dysfunction and improves survival in a 2-hit model of fungal sepsis. J Infect Dis 2012; 206:606–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noguchi M, Nakamura Y, Russell SM, et al. Interleukin-2 receptor gamma chain: A functional component of the interleukin-7 receptor. Science 1993; 262:1877–1880. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin RG, Friend D, Ziegler SF, et al. Cloning of the human and murine interleukin-7 receptors: Demonstration of a soluble form and homology to a new receptor superfamily. Cell 1990; 60:941–951. [DOI] [PubMed] [Google Scholar]
- 13.Demaret J, Villars-Méchin A, Lepape A, et al. Elevated plasmatic level of soluble IL-7 receptor is associated with increased mortality in septic shock patients. Intensive Care Med 2014; 40:1089–1096. [DOI] [PubMed] [Google Scholar]
- 14.Peronnet E, Mouillaux J, Monneret G, et al. MIP Réa Study Group: Elevated soluble IL-7 receptor concentration in non-survivor ICU patients. Intensive Care Med 2016; 42:1639–1640. [DOI] [PubMed] [Google Scholar]
- 15.Rose T, Lambotte O, Pallier C, et al. Identification and biochemical characterization of human plasma soluble IL-7R: Lower concentrations in HIV-1-infected patients. J Immunol 2009; 182:7389–7397. [DOI] [PubMed] [Google Scholar]
- 16.Vranjkovic A, Crawley AM, Gee K, et al. IL-7 decreases IL-7 receptor alpha (CD127) expression and induces the shedding of CD127 by human CD8+ T cells. Int Immunol 2007; 19:1329–1339. [DOI] [PubMed] [Google Scholar]
- 17.Lundström W, Highfill S, Walsh ST, et al. Soluble IL7Rα potentiates IL-7 bioactivity and promotes autoimmunity. Proc Natl Acad Sci U S A 2013; 110:E1761–E1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawley AM, Faucher S, Angel JB.Soluble IL-7R alpha (sCD127) inhibits IL-7 activity and is increased in HIV infection. J Immunol 2010; 184:4679–4687. [DOI] [PubMed] [Google Scholar]
- 19.Monti P, Brigatti C, Krasmann M, et al. Concentration and activity of the soluble form of the interleukin-7 receptor α in type 1 diabetes identifies an interplay between hyperglycemia and immune function. Diabetes 2013; 62:2500–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Côté S, Matte J, Sad S, et al. Complexed soluble IL-7 receptor α and IL-7 increase IL-7-mediated proliferation and viability of CD8+ T-cells in vitro. Cell Immunol 2015; 293:122–125. [DOI] [PubMed] [Google Scholar]
- 21.Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest 1992; 101:1644–1655. [DOI] [PubMed] [Google Scholar]
- 22.Hellemans J, Mortier G, De Paepe A, et al. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 2007; 8:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knaus WA, Draper EA, Wagner DP, et al. APACHE II: A severity of disease classification system. Crit Care Med 1985; 13:818–829. [PubMed] [Google Scholar]
- 24.Francois B, Jeannet R, Daix T, et al. Interleukin-7 restores lymphocytes in septic shock: The IRIS-7 randomized clinical trial. JCI Insight 2018; 3:e98960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boomer JS, Shuherk-Shaffer J, Hotchkiss RS, et al. A prospective analysis of lymphocyte phenotype and function over the course of acute sepsis. Crit Care 2012; 16:R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rane L, Vudattu N, Bourcier K, et al. Alternative splicing of interleukin-7 (IL-7) and interleukin-7 receptor alpha (IL-7Ralpha) in peripheral blood from patients with multiple sclerosis (MS). J Neuroimmunol 2010; 222:82–86. [DOI] [PubMed] [Google Scholar]
- 27.Badot V, Luijten RK, van Roon JA, et al. Serum soluble interleukin 7 receptor is strongly associated with lupus nephritis in patients with systemic lupus erythematosus. Ann Rheum Dis 2013; 72:453–456. [DOI] [PubMed] [Google Scholar]
- 28.Bauer M, Giamarellos-Bourboulis EJ, Kortgen A, et al. A transcriptomic biomarker to quantify systemic inflammation in sepsis - A prospective multicenter phase II diagnostic study. EBioMedicine 2016; 6:114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sieling PA, Sakimura L, Uyemura K, et al. IL-7 in the cell-mediated immune response to a human pathogen. J Immunol 1995; 154:2775–2783. [PubMed] [Google Scholar]
- 30.Lill M, Kõks S, Soomets U, et al. Peripheral blood RNA gene expression profiling in patients with bacterial meningitis. Front Neurosci 2013; 7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazzucchelli R, Durum SK.Interleukin-7 receptor expression: Intelligent design. Nat Rev Immunol 2007; 7:144–154. [DOI] [PubMed] [Google Scholar]
- 32.Rol ML, Venet F, Rimmele T, et al. REALISM Study Group: The REAnimation Low Immune Status Markers (REALISM) project: A protocol for broad characterisation and follow-up of injury-induced immunosuppression in intensive care unit (ICU) critically ill patients. BMJ Open 2017; 7:e015734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heffernan DS, Monaghan SF, Thakkar RK, et al. Failure to normalize lymphopenia following trauma is associated with increased mortality, independent of the leukocytosis pattern. Crit Care 2012; 16:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drewry AM, Samra N, Skrupky LP, et al. Persistent lymphopenia after diagnosis of sepsis predicts mortality. Shock 2014; 42:383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung KP, Chang HT, Lo SC, et al. Severe lymphopenia is associated with elevated plasma interleukin-15 levels and increased mortality during severe sepsis. Shock 2015; 43:569–575. [DOI] [PubMed] [Google Scholar]
- 36.Heidecke CD, Hensler T, Weighardt H, et al. Selective defects of T lymphocyte function in patients with lethal intraabdominal infection. Am J Surg 1999; 178:288–292. [DOI] [PubMed] [Google Scholar]
- 37.Friggeri A, Cazalis MA, Pachot A, et al. MIP Rea Study Group: Decreased CX3CR1 messenger RNA expression is an independent molecular biomarker of early and late mortality in critically ill patients. Crit Care 2016; 20:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peronnet E, Venet F, Maucort-Boulch D, et al. MIP Rea Study Group: Association between mRNA expression of CD74 and IL10 and risk of ICU-acquired infections: A multicenter cohort study. Intensive Care Med 2017; 43:1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guignant C, Lepape A, Huang X, et al. Programmed death-1 levels correlate with increased mortality, nosocomial infection and immune dysfunctions in septic shock patients. Crit Care 2011; 15:R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venet F, Davin F, Guignant C, et al. Early assessment of leukocyte alterations at diagnosis of septic shock. Shock 2010; 34:358–363. [DOI] [PubMed] [Google Scholar]


