Abstract
Several inherited arrhythmias, including Brugada syndrome and arrhythmogenic cardiomyopathy, primarily affect the right ventricle and can lead to sudden cardiac death. Amongst many differences, right and left ventricular cardiomyocytes derive from distinct progenitors, prompting us to investigate how embryonic programming may contribute to chamber-specific conduction and arrhythmia susceptibility. Here, we show that developmental perturbation of Wnt signaling leads to chamber-specific transcriptional regulation of genes important in cardiac conduction that persists into adulthood. Transcriptional profiling of right versus left ventricles in mice deficient in Wnt transcriptional activity reveals global chamber differences, including genes regulating cardiac electrophysiology such as Gja1 and Scn5a. In addition, the transcriptional repressor Hey2, a gene associated with Brugada syndrome, is a direct target of Wnt signaling in the right ventricle only. These transcriptional changes lead to perturbed right ventricular cardiac conduction and cellular excitability. Ex vivo and in vivo stimulation of the right ventricle is sufficient to induce ventricular tachycardia in Wnt transcriptionally inactive hearts, while left ventricular stimulation has no effect. These data show that embryonic perturbation of Wnt signaling in cardiomyocytes leads to right ventricular arrhythmia susceptibility in the adult heart through chamber-specific regulation of genes regulating cellular electrophysiology.
Keywords: connexin43, transcriptional regulation, cardiac development, cardiac electrophysiology, Wnt signaling, ventricular tachycardia
1. Introduction
An emerging paradigm dictates that genetic pathways guiding morphologic patterning of the heart also program myocardial cellular electrical properties 1–5. Thus, it is not surprising that patients with structural congenital heart defects often suffer from arrhythmias 6. Wnt signaling regulates many aspects of heart development, including second heart field (SHF) progenitor proliferation and differentiation 7. Since the right ventricle (RV) and right ventricular outflow tract derive from SHF progenitors, this may at least in part underlie the observation of Wnt signaling perturbation in arrhythmogenic cardiomyopathy (ACM), which has an RV predilection 8. Genetic variants in several loci of the Wnt signaling pathway have been identified in genome-wide association studies of prolonged conduction intervals, a trait often associated with arrhythmias such as atrial fibrillation and sudden cardiac death 9–14. However, a precise understanding of how Wnt signaling regulates cardiac conduction in health and disease is not fully understood. Recent studies using gain-of-function (GOF) approaches demonstrated that canonical Wnt signaling regulates atrioventricular junction development and ion channel gene expression 15–17. Here, we seek to further define the role of Wnt signaling in regulation of cardiac conduction and arrhythmogenesis, with a particular focus on electrical programming of the right and left ventricles.
The Wnt signaling pathway involves 19 distinct Wnt ligands and 10 Frizzled receptors that signal through canonical or non-canonical Wnt pathways, the latter mediated through Ca2+ signaling 18. Canonical Wnt signaling is mediated by nuclear translocation of β-catenin, which binds to T-cell factor/lymphoid enhancer-binding factor (Tcf/Lef) cofactors bound to Wnt target gene regulatory elements, thereby de-repressing Wnt-sensitive transcription. Genetic manipulation of canonical Wnt signaling is often probed through tissue-specific knockout of the key transcriptional effector β-catenin. Since β-catenin also plays a key role in cell adhesion, here we utilize an allele of β-catenin that abrogates its transcriptional functions but preserves its cell adhesion functions in Wnt loss-of-function (LOF) studies, allowing us to specifically probe the transcriptional role of Wnt signaling in regulation of cardiac conduction genes in cardiomyocytes (CM) 19.
Utilizing both gain and loss-of-function approaches, we show that many genes, including those important for conduction, are regulated in a chamber-specific manner. NaV1.5, which mediates sodium currents, is down-regulated only in the RV in Wnt GOF mice, while connexin43 (Cx43), a component of gap junctions, is down-regulated in Wnt LOF mice only in the RV. In Wnt LOF mice, many transcripts are exclusively dysregulated in either the RV or LV, and very few transcripts are mutually altered in both the RV and LV. Both Wnt gain and loss-of-function models reveal dysregulation of Hairy/enhancer-of-split related with YRPW motif protein 2 (HEY2, also known as HRT2), a transcriptional repressor known to be a direct Notch target during cardiac development 20, in the RV only. Genome wide association studies link HEY2 variants with Brugada syndrome, a syndrome of ST elevation associated with right ventricular tachycardia 12. Wnt-mediated regulation of Hey2 expression is associated with β-catenin binding to enhancer elements in the RV but not LV. Differential β-catenin binding to the Hey2 enhancer in the RV can be seen as early as mid-gestation when Hey2 expression is high, and binding in the LV is not observed even with increased levels of active β-catenin. Wnt LOF chamber-specific transcriptional effects lead to conduction slowing only in the RV, associated with increased susceptibility to ventricular tachycardia (VT) with RV stimulation that persists into adulthood and may have implications for our understanding of right ventricular-predominant arrhythmias. Given that the Wnt pathway plays an important role in disease processes in the heart and more broadly, we expect these fundamental insights from signaling pathway to electrical phenotype may enable improvements in genetic testing and provide novel avenues for molecular-based therapeutics.
2. Methods
2.1. Mice
Mlc2vCre/+21, αMHC-Cre 22, αMHCrtTA 23, Ctnnb1dm19, Ctnnb1flox24, Ctnnb1fl(ex3)25, TetO-Cre 26 alleles were used to generate developmental Wnt LOF (Mlc2vCre/+; Ctnnb1dm/flox), developmental Wnt GOF (Mlc2vCre/+; Ctnnb1fl(ex3)/+ or αMHC-Cre; Ctnnb1fl(ex3)/+), and inducible Wnt GOF (αMHCrtTA; TetO-Cre; Ctnnb1fl(ex3)/+). To characterize Wnt signaling activity at different time points, Tcf/Lef:H2B-GFP embryos and mice were used 27. Animals were maintained on a mixed genetic background. Animal protocols were approved by the Animal Studies Committee at Washington University, and animals were handled in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Control data for molecular biology and electrophysiology were obtained from littermate animals with genotypes: Ctnnb1flox/+ for Wnt LOF, Ctnnb1fl(ex3)/+ for Wnt GOF, and TetO-Cre; Ctnnb1fl(ex3)/+ for inducible Wnt GOF (iWnt). Mice of both sexes were used in all studies, and though experiments were not powered to specifically detect sex differences, there were no obvious differences in any parameters between sexes.
| Mouse | Genotype | Phenotype | Figure Number(s) |
|---|---|---|---|
| Wnt GOF | Mlc2vCre/+;Ctnnb1fl(ex3)/+ | Survives to adulthood | 2, 3F, S7B, |
| Wnt GOF | αMHC-Cre; Ctnnb1fl(ex3)/+ | Embryonic lethal15 | 3A, 3C, S4 |
| Wnt GOF | αMHCrtTA; TetO-Cre; Ctnnb1fl(ex3)/+ | Adult Inducible, survives to adulthood | 3G |
| Wnt LOF | Mlc2vCre/+; Ctnnb1dm/flox | Survives to adulthood | 1B-F, 4, 5, 6, S2, S3, S8, S9, S10, S11 |
| Wnt Reporter | Tcf/Lef:H2B-GFP | Survives to adulthood (no overt phenotype) | 1A, S1 |
2.2. Surface EKG and Conscious Telemetry Recordings
Investigators were blinded to the sample group allocation during the experiment and analysis of the experimental outcome. Surface EKGs were performed on conscious mice using EKG Tunnel (EMKA Tech), and with isoflurane, as previously described 28. For pilot telemetry studies, developmental Wnt LOF (Mlc2vCre/+; Ctnnb1dm/flox, n=2 females; 1 male) mice and a littermate control (Ctnnb1flox/+, n=1 male) were between 3–5 months of age. ETA-F10 implantable radio frequency transmitters for EKG (Data Sciences International Inc.) were implanted subcutaneously in the posterior neck of adult mice. Leads were tunneled to the anterior chest in lead II position. After a post-implant recovery period of one week, ambulatory heart rhythm and HR were monitored in caged, moving mice. Once per night, mice were injected with isoproterenol intraperitoneally (0.2 mg/kg in weight). Recordings were obtained continuously at 1 kHz over a one-week period. HR and RR intervals were calculated at baseline and following isoproterenol injection using a custom Matlab program.
2.3. Histology and Immunohistochemistry
Immunohistochemistry was performed on paraffin sections with N-Cadherin antibody (1:100, Invitrogen 333900) and Alexa Fluor 488 secondary (1:200, Invitrogen 2–21202). Co-staining was performed with either an antibody recognizing Cx43 independent of phosphorylation status (1:50, Invitrogen 71–0700) or an antibody recognizing Cx43 phosphorylated at Ser368 (1:100, Cell Signaling Technology 3511) and then detected with signal amplification using anti-rabbit ImmPRESS (Vector Laboratories MP-7401) with TSA (Perkin Elmer SAT702001). For embryonic Wnt Reporter hearts, frozen sections were stained with α-actinin antibody (1:100, Sigma A2172), Alexa Fluor 568 secondary (1:200, Invitrogen A-11011), and DAPI (Sigma D9542).
To compare the ventricular CM size between genotypes, wheat germ agglutinin (WGA) staining was performed on frozen sections from Ctnnb1flox/+ controls (n=1 female; 2 males) and littermate Wnt LOF Mlc2vCre/+; Ctnnb1dm/flox mice (n=1 female; 2 males). The area of approximately 100 cells perpendicular to the plane of sectioning was outlined and quantified using Axiovision. The areas of only circular ventricular CMs were included in the analysis, while elongated cells were excluded, to minimize bias in the analysis from cells not perpendicular to the plane of sectioning. Quantification was performed in RV and LV from n=3 hearts of each genotype by an observer blinded to genotype. Statistical comparison was performed using unpaired t test with Welch’s correction.
2.4. RNA Sequencing
For adult RNA-sequencing, Wnt LOF (Mlc2vCre/+; Ctnnb1dm/flox) and littermate control (Ctnnb1flox/+) RV and LV were used (n=2 females; 1 male for each group). RV and LV were obtained from same animals. For embryonic RNA-sequencing, Wnt GOF (αMHC-Cre; Ctnnb1fl(ex3)/+) and littermate control (Ctnnb1fl(ex3)/+) E12.5 embryos were used, n=2 RV and LV were pooled for one sample, and 6 samples of each genotype were sequenced. Illumina libraries were constructed by the GTAC at Washington University School of Medicine. After ribosomal RNA depletion, reverse-transcription with random-priming, library construction and multiplexing, samples were sequenced on an Illumina HiSeq 2500 using indexed libraries multiplexed at 6 samples per lane. Single-end 50-base sequencing reads with a total target depth of >25 million reads were performed. Raw sequence reads were de-multiplexed, aligned to the reference sequence with Tophat v2.0.8 and Bowtie2 v2.1.0, and gene and exon-level abundances derived by HTSeq. GTAC analytical pipeline simultaneously performed a standard EdgeR and DEXSeq analysis of gene-level and exon-level features on the uniquely aligned, unambiguous count of reads per feature as determined by HTSeq using a negative-binomial test or generalized-linear negative-binomial model, respectively. Genes or exons were filtered to include only those that are expressed, and results were filtered to include only differentially expressed genes or exons. Lists of genes differentially regulated in the RV and LV of Wnt LOF compared to littermate control animals were used to generate a Venn diagram using an online resource: http://bioinformatics.psb.ugent.be/webtools/Venn/.
Accession number: Data have been deposited in the NCBI GEO database for release after publication. The following links have been created to allow review:
Wnt LOF in Figure 1, Supplemental Table 1: GSE106507 Token: qrihkmgonhyndef
Wnt GOF in Figure 3, Supplemental Table 2: GSE106506 Token: irmboueefjeblaf
2.5. Reverse Transcription-Quantitative Polymerase Chain Reaction
Total RNA was isolated from ventricles using Trizol (Invitrogen) and DNase treated using TURBO DNA-free DNase Treatment Kit (Ambion). First-strand cDNA was synthesized using a high Capacity cDNA Reverse Transcription kit (Applied Biosystems). Gene expression was assayed using the Power SYBR Green PCR Master Mix (Applied Biosystems) with primers listed below and quantified using the StepOne Plus Real-Time PCR system or ViiA™ 7 qRT-PCR system (Applied Biosystems). Relative fold changes were calculated using the comparative threshold cycle methods (2-ΔΔCt).
qRT-PCR oligonucleotide sequences
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Scn5a | 5’ GAAGAAGCTGGGCTCCAGA 3’ | 5’ CATCGAAGGCCTGCTTGGTC 3’ |
| Gja1 | 5’ ACAAGGTCCAAGCCTACTCCA 3’ | 5’ CCCCAGGAGCAGGATTCTGA 3’ |
| Axin2 | 5’ CAGCCCTTGTGGTTCAAGCT 3’ | 5’ GGTAGATTCCTGATGGCCGTAGT 3’ |
| Hey2 | 5’ CGACGTGGGGAGCGAGAACAAT 3’ | 5’ GGCAAGAGCATGGGCATCAAAGTA 3’ |
| Tbp | 5’ GGGATTCAGGAAGACCACATAG 3’ | 5’ CCTCACCAACTGTACCATCAG 3’ |
| Actc1 | 5’ GACCTCACTGACTACCTCATG 3’ | 5’ TCTCGTTCTCAAAATCCAGGG 3’ |
2.6. Chromatin Immunoprecipitation
Tissue samples were processed for ChIP as described previously with a few modifications 29. Briefly, for each biological replicate, 4 X 107 cells from single adult LV free wall, adult RV free wall, or embryonic ventricles were cross-linked with 1% formaldehyde for 30 mins at room temperature and flash frozen and stored at −80°C until further use. Chromatin was sheared to approximately 500 base pairs using a Bioruptor sonicator (Diagenode, Sparta, NJ). Chromatin was precleared using protein G, precipitated with anti-β-catenin antibody (Cell Signaling Technology 8814), or negative control anti-IgG antibody (Abcam ab46540). A motif search performed using Transfac Professional V10.2 identified a consensus Tcf/Lef binding motif with a helper site 30 within the Hey2 Enh3 genomic region (chr10:30,717,463–30,718,805), while the Hey2 Enh2 heart enhancer (chr10:30,948,697–30,949,670), Hey2 Enh1 heart enhancer (chr10:30,772,887–30,773,894), and Hey2 promoter region (640bp - 140bp upstream of TSS) did not contain a consensus Tcf/Lef binding motif. The Bhlh2 locus (108610745 in the 3’ UTR) was a positive control, and given that there are no consensus Tcf/Lef binding motifs in Scn10a proximal promoter (~500bp upstream of TSS), we chose this region as a negative control. For H3K27ac ChIP, the negative control was a gene desert region (chr15:70,649,224–70,649,887), and the positive control was a Gata4 peak (chr14:63,263,061–63,263,183). After reverse cross-linking, immunoprecipitated DNA was purified and analyzed using quantitative PCR (qPCR) and SYBR Green chemistry. Primer sequences for qPCR are indicated below. Fold enrichment was calculated as 2^(Ctinput − Cttest) and expressed relative to the IgG negative control.
ChIP-qPCR oligonucleotide sequences
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Hey2 TSS -140bp | 5’ CGTGGGAAAGAACCTCCAG 3’ | 5’ GCAGCTACTGTCGCCTA 3’ |
| Hey2 Enhancer 1 | 5’ AGTGAGATGGGAAGTCAGAGA 3’ | 5’ CATGGTGCCAGGGATAACTT 3’ |
| Hey2 Enhancer 2 | 5’ CTTAAGGCCACCTAGGATTCAG 3’ | 5’ GGAGTAGTGAGCGTAGATGTTTC 3’ |
| Hey2 Enhancer 3 | 5’ GACAGGCGAGTACATCCTATAAC 3’ | 5’ ATTTCCCTGGATTTCCGACTC 3’ |
| Bhlh2D | 5’ GCTAGACACGGATTTCAAGTGTATCC 3’ | 5’ CATCCACTGCTGGGCAAGAG 3’ |
| Scn10a | 5’ GCTGCCACTCAGACAAAGGAAT 3’ | 5’ GTAAAGCTATGCCCTGCACCCA 3’ |
| Gene desert region (Neg) | 5’ GGACTAGACTCTGAGATTTTGAGG 3’ | 5’ GGGCTACATCCTTAAACCTCAG3’ |
| Gata4 | 5’ AGGAAGGGAGGCACTCTATAG 3’ | 5’ GGTCAGACTGTACAACAGCTG 3’ |
2.7. Western Blotting
Protein lysates were prepared from RV and LV free wall and used for immunoblotting as previously described 29. Briefly, ventricles were harvested and homogenized (separately) in ice-cold 1X RIPA lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 1% Triton-X, 0.5% Sodium Deoxycholate, 0.1% SDS) with protease inhibitor cocktail tablet (1 tablet per 50 mL lysis buffer, Roche, Mannheim, Germany). Samples were centrifuged at 13200 g for 10 minutes at 4°C. The supernatants were used for electrophoresis following protein analysis via BCA assay (ThermoFisher). Immunoblotting was performed using anti-β-catenin (1:1000, Cell Signaling Technology 9562), anti-β-catenin (non-phosphorylated) (1:1000, Cell Signaling Technology 8814), anti-connexin43 (1:250, Invitrogen 71–0700), anti-phospho-connexin43Ser368 (1:1000, Cell Signaling Technology 3511), anti-NaV1.5 (1:1000, Alomone ASC-005), and anti-GAPDH (1:2000, Cell Signaling Technology 14C10) antibodies. Secondary detection was performed using HRP antibody (1:5000, abcam 6721) and Clarity Western ECL Substrate (Biorad).
2.8. Optical mapping with Programmed Electrical Stimulation
Changes in the membrane potential of murine cardiac tissue were recorded using optical mapping described previously 15. The cardiac tissue was perfused with oxygenated Tyrode’s solution at 37°C, pH 7.4. Blebbistatin, an excitation-contraction uncoupler, was used to eliminate motion artifacts in the recorded optical signal. Changes in the membrane potential were detected by Di-4-ANEPPS, a voltage-sensitive dye. A green LED light source with the wavelengths of 520 ± 5 nm was used to excite the voltage-sensitive dye. The emitted fluorescence was filtered by a long-pass filter at 650 nm and collected by the CMOS camera. The recorded data were analyzed using an open source custom MATLAB program. Using surface coordinates and time of the maximum derivatives (dVm/dtmax) of the optical signals, activation maps were generated and used to calculate the conduction velocity (CV). Longitudinal and transverse CV were calculated by distance traveled over the number of time isochrones along multiple lines originating from the site of stimulation. The fastest CV was defined as longitudinal, and the slowest as transverse (roughly 90 degrees apart).
The standard deviation from a previous optical mapping control cohort was used to perform a power calculation. It was estimated to detect a 20% difference in CV that six mice of each genotype (control and developmental Wnt LOF) would be required for 80% power. Investigators were blinded to the sample group allocation during the experiment and analysis of the experimental outcome. For optical mapping studies to measure CV, control Ctnnb1flox/+ mice (n=3 females; 4 males) and experimental Mlc2vCre/+; Ctnnb1dm/flox mice (n=4 females; 3 males) between 3 and 5 months of age were studied. The stimulus amplitude used is twice the diastolic capture threshold with a pulse duration of 2 ms. For determination of CV, the epicardial surface was stimulated with a 5 seconds long S1 drive train at 8 Hz, followed by a drive train at 10 Hz, and then a drive train at 12 Hz first on the RV followed by the LV. Episodes of ventricular tachycardia (VT) were defined as follows: rapid ventricular activity faster than atrial activity lasting longer than two seconds.
2.9. Microelectrode Recordings
Investigators were blinded to the sample group allocation during the experiment and analysis of the experimental outcome. Microelectrode recordings were obtained from control Ctnnb1flox/+ mice (n=3 females; 2 males) and experimental Mlc2vCre/+; Ctnnb1dm/flox mice (n=4 females; 1 male) between 3 and 5 months of age. Mouse hearts were Langendorff perfused and were recorded while in sinus rhythm. Using glass sharp microelectrodes, single RV or LV CMs were sampled near the epicardial surface. Thin glass pipettes with high resistance (10–20 MΩ) filled with 3M KCl were used to record action potentials from a minimum of 8 single cells from each region of the heart (technical replicates) in the intact perfused heart. To decrease noise from motion artifacts, blebbistatin was used to arrest motion and allow for stable microelectrode recording without requiring the use of floating electrodes. Electrical signals were digitized by a Power 1401 A/D converter. Experiments were performed and recordings were analyzed by an observer blinded to genotype. Sampled cells with a resting membrane potential (RMP) less than −70mV were averaged, and a minimum of 5 hearts were used for analysis for each group. The RMP, action potential amplitude (APA), maximum upstroke velocity (dVm/dtmax), and action potential duration (APD) at 90% repolarization are reported. Analysis of microelectrode recordings was performed using a custom MATLAB script. Cells with at least 20 action potentials and stable RMP were included in the data analysis. For each action potential in each cell, the peak potential value and time were calculated. The RMP was then calculated for each action potential using a histogram, where the maximum frequency value was defined as the RMP. The magnitude (amplitude) of each action potential was then defined as the difference between peak potential value and the RMP. The maximum change in voltage with respect to time (dVm/dtmax) was calculated using a first derivative. The point in time that corresponds to this value was then used as the start time for APD calculation. The end time was when the potential decreased to 90% of the amplitude value.
2.10. Intracardiac Electrophysiology with Programmed Stimulation
Protocols for in vivo mouse electrophysiology have been described in detail 28, 31. Investigators were blinded to the sample group allocation during the experiment and analysis of the experimental outcome. Briefly, control Ctnnb1flox/+ mice (n=6) and experimental Mlc2vCre/+; Ctnnb1flox/+ mice (n=6) between 2 and 4 months of age were studied. Mice were anesthetized with isoflurane, intubated, laid down on a heating pad to maintain core body temperature, and limb leads were placed subcutaneously to obtain multi-lead ECGs. An octapolar 1.7-French electrode catheter (CIBer mouse-EP, NuMED) was placed in the right atrium and ventricle under electrogram guidance through a jugular vein cutdown. A data acquisition unit (PowerLab, ADInstruments) was used to deliver electrical impulses at approximately twice the diastolic threshold, and to record surface ECGs and intracardiac electrograms. Electrograms were sampled at 2 kHz, high-pass filtered at 20 Hz and notch filtered at 60 Hz. To avoid aliasing, a 500 Hz low-pass filter was applied to the signal. Standard pacing protocols were used to assess right ventricular conduction, refractoriness and arrhythmia inducibility. For determination of arrhythmia susceptibility, a programmed pacing protocol that consists of single, double, and triple extrastimuli followed by burst pacing was performed on each heart. For single extrastimulus, a pacing drive train of 20 S1 at a cycle length of 100 ms was used, followed by an S2 with a 50 ms coupling interval, which was reduced in 5 ms intervals until loss of ventricular capture (ventricular effective refractory period, VERP). For double extrastimuli, a drive train of 20 S1 at a cycle length of 100 ms was followed by an S2 coupled at 50 ms and an S3 coupled at 50ms. S2 and S3 were brought in by 5 ms until reaching the VERP. For ventricular triple extrastimuli, a drive train of 20 S1 at a cycle length of 100 ms was followed by an S2, S2, S3, and S4 coupled at 50 ms. Similarly, S2, S3, and S4 were brought in by 5 ms until VERP. For burst pacing, a 2 second burst pacing at a cycle length of 50 ms was used for the first burst, followed by successive bursts with a 5 ms decrement of the cycle length until VERP. Studies were performed and arrhythmias were noted by two independent investigators who were blinded to the genotype of the mice. All mice were euthanized at end of study.
2.11. Statistical Analysis
All data are expressed as means ± standard error of the mean (SEM). For comparison of more than two experimental groups, the statistical significance of observed differences in mean was evaluated using a one-way or two-way analysis of variance (ANOVA), followed by post-hoc Tukey’s multiple comparison test. For comparison of two experimental groups, an equal variance Student’s t-test was used; a two-tailed P value of <0.05 was considered statistically significant. In analysis of contingency tables, a Fisher’s exact test was used.
2.12. Study Approval
Animal protocols were approved by the Animal Studies Committee at Washington University, and animals were handled in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
3. Results
3.1. Chamber-specific transcriptional regulation in Wnt LOF right versus left ventricles
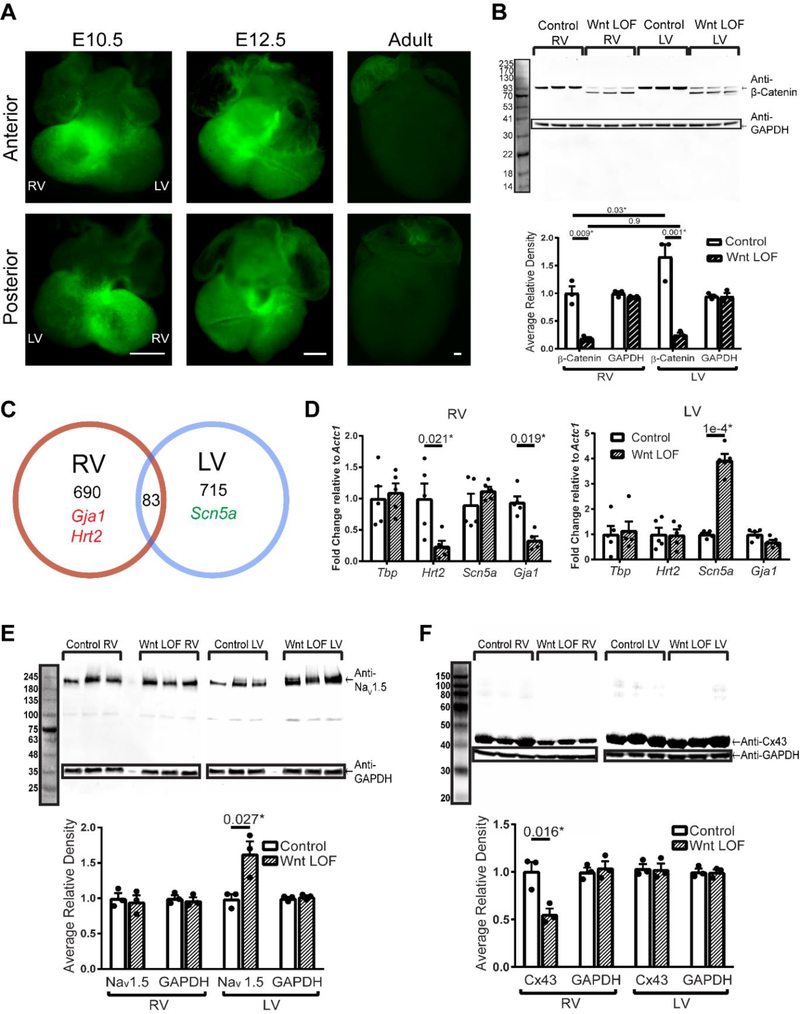
Binding of a Wnt ligand to its cell surface receptor(s) results in inactivation of a destruction complex that phosphorylates β-catenin and targets it for degradation 18. Inactivation of the destruction complex allows intracellular β-catenin to accumulate, eventually resulting in higher levels of β-catenin within the nucleus, where it is transcriptionally active. Several Wnt ligands are known to be expressed in the developing heart 7. To determine the regions within the developing heart where canonical Wnt signaling is most active, we assayed for Wnt activity using a transgenic mouse with reiterative Tcf/Lef binding sites located upstream of a nuclear-localized GFP (TCF/Lef:H2B-GFP 27). Early in development at embryonic day 10.5 (E10.5) and 12.5 (E12.5), Wnt activity is highest in the developing RV and atrioventricular regions, eventually becoming quiescent in the adult ventricles (Figure 1A). At E12.5, Wnt activity is found in a large number of myocytes in addition to non-myocytes, with higher density in the trabecular region when compared to the compact zone of the RV myocardium, as well as in CMs in the RV outflow tract (Supplemental Figure 1).
Figure 1 – Wnt signaling differentially regulates genes involved in cardiac conduction in right and left ventricles.
(A) Fluorescent images of TCF/Lef:H2B-GFP transgenic hearts where 6 Tcf/Lef responsive elements are upstream of an Hsp68 minimal promoter driving GFP. At E10.5 and E12.5, higher Wnt activity is noted in the primitive right ventricle (RV) and atrioventricular region when compared with the left ventricle (LV). Wnt activity is detectable in the right atrium but not in the ventricles of a 10-week old adult heart. Scale bar in each image = 500 μm. (B) Western blot for total β-catenin in adult control (Ctnnb1flox/+) and littermate Wnt LOF (Mlc2vCre/+; Ctnnb1dm/flox) (n=3 each) demonstrates higher levels of β-catenin protein in the LV at baseline when compared with RV. Wnt LOF mice express a truncated β-catenin protein at approximately 70kDa, in addition to a faint band representing wild type β-catenin at 93kDa, which likely represents residual β-catenin within non-myocytes. GAPDH is used for normalization and quantification of protein levels is based on wild type β-catenin band density. (C) Venn diagram of differentially altered transcripts in adult Wnt LOF (Mlc2vCre/+; Ctnnb1dm/flox) versus littermate control (Ctnnb1flox/+) RV and LV (n=3 each genotype). RNA sequencing revealed 690 transcripts dysregulated in Wnt LOF RV only, including Gja1 and Hey2, with 715 transcripts dysregulated in Wnt LOF LV only, including Scn5a. A small fraction of dysregulated transcripts (83 of 1488, <6%) are dysregulated in both ventricles. (D) qRT-PCR in adult Wnt LOF (Mlc2vCre/+; Ctnnb1dm/flox,) versus littermate control (Ctnnb1flox/+) mice for selected transcripts (n=5 each genotype). The TATA-box binding protein (Tbp, negative control) is unchanged in both ventricles. Hey2 expression is reduced 4-fold and Gja1 is reduced 3-fold in Wnt LOF RV only, while in contrast, Scn5a is upregulated 4-fold in Wnt LOF LV only. (E) Western blot of Nav1.5 (227 kDa) and superimposed GAPDH (37 kDa) with quantification based on band density normalized to GAPDH shows 60% increase in Nav1.5 in Wnt LOF LV when compared with littermate control LV, with no difference in the RV (n=3 each genotype). (F) Western blot of total Cx43 (43 kDa) normalized to GAPDH (n=3 each genotype) shows 50% reduction of Cx43 protein levels in Wnt LOF RV compared with littermate control RV, while Cx43 protein levels in the LV are unchanged, consistent with transcriptional changes seen in (C) and (D). One-way ANOVA and post hoc Tukey’s test was performed to determine statistical significance in (B). An equal variance Student’s t test was used for all other comparisons. Two-tailed P-values are indicated on the graphs, with *P<0.05 considered statistically significant.
We previously reported that developmental Wnt GOF mice have slowed CV predominantly affecting the RV 15. To further delineate the role of Wnt signaling in regulating cardiac conduction, we next assayed the effects in a LOF model. Since β-catenin is integral in transducing the Wnt signal into the nucleus but is also important for cell adhesion, we utilize an allele of β-catenin that contains a point mutation at the N-terminus and a truncation at the C-terminus rendering it transcriptionally inactive but preserving its role in cell adhesion (Ctnnb1dm19). When combined with a tissue-specific loss of function allele, the only remaining β-catenin expressed within ventricular CMs is transcriptionally inactive (Wnt LOF, Mlc2vCre/+; Ctnnb1dm/flox). Western blots for total β-catenin in wild type animals show higher amounts of β-catenin in the adult LV when compared with the RV (Figure 1B). As expected, Wnt LOF RV and LV reveal a significant decrease in β-catenin levels when compared with littermate controls, with a small amount of residual wild type β-catenin at 93 kDa either reflecting expression in non-myocytes or incomplete inactivation (Figure 1B). Wnt LOF mice also express an additional truncated band at 70 kDa from the dm allele (Figure 1B). Levels of non-phosphorylated, active β-catenin also parallel the decrease in total levels, as expected (Supplemental Figure 2).
To determine the global effects of transcriptionally inactive β-catenin within each ventricle, we performed RNA sequencing on tissue from adult Wnt LOF and littermate control RV and LV. Of the dysregulated transcripts in the RV, 690 of 773 are exclusively changed in the RV, while 715 of 798 transcripts are exclusively dysregulated in the LV (Figure 1C, Supplemental Figure 3, data accession tokens provided in Methods). Only 83 transcripts (<6% of the total) are dysregulated in both Wnt LOF RV and LV, and of these 83 transcripts, some change in discordant directions (Supplemental Table 1). Notably, RNA-sequencing demonstrates that Hey2 and Gja1 are down-regulated in the RV, while Scn5a is up-regulated in the LV. Validation via qRT-PCR demonstrates that Hey2 is downregulated more than 4-fold in the Wnt LOF RV only, while its expression in the LV remains unchanged (Figure 1D). Scn5a, which encodes the sodium channel NaV1.5, is upregulated more than 3-fold in Wnt LOF LV with no change in Wnt LOF RV (Figure 1D). Gja1, which encodes the predominant ventricular gap junction isoform connexin43 (Cx43), is reduced 4-fold within the RV while its expression is unchanged in the LV (Figure 1D). Consistent with the differential effect on transcript levels, Western blots show 60% increase in NaV1.5 protein in Wnt LOF LV compared with littermate controls and no change in the RV (Figure 1E). Similarly, there is a 50% reduction of total Cx43 in Wnt LOF RV compared with littermate controls, with no change in the LV (Figure 1F).
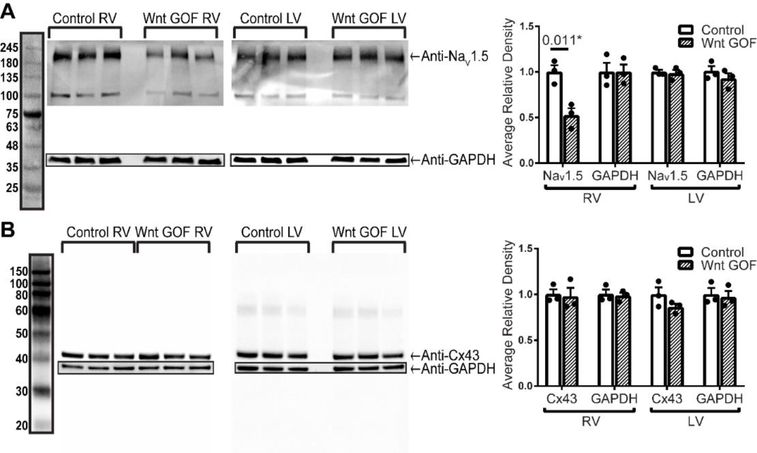
Given that we previously showed conduction slowing in Wnt GOF mice primarily affecting the RV 15, we asked whether these molecular determinants of conduction are similarly dysregulated with Wnt activation. Surprisingly, we found that NaV1.5 is reduced to 48% of littermate control levels in the Wnt GOF RV (Mlc2vCre; Ctnnb1flox(ex3)/+), while levels are unchanged in Wnt GOF LV (Figure 2A). Cx43 levels are unchanged in Wnt GOF mice in both the RV and LV (Figure 2B). This data reveals that the molecular determinants of cardiac conduction are dysregulated by Wnt signaling during development primarily within the RV, though GOF and LOF regulates distinct channels.
Figure 2 – NaV1.5 protein levels are decreased in Wnt gain-of-function mice only in the right ventricle, while Cx43 protein levels are unchanged.
(A) Western blot for NaV1.5 and GAPDH with quantification based on band density normalized to GAPDH shows a 48% decrease of NaV1.5 in Wnt GOF RV when compared with littermate control RV, with no differences in the LV (n=3 each genotype). (B) Western blot of Cx43 normalized to GAPDH (n=3 each genotype) shows no difference in Wnt GOF Cx43 protein levels compared to controls. An equal variance Student’s t test was used for comparisons. *P<0.05 was considered statistically significant.
3.2. Hey2 is a direct Wnt target in the right ventricle
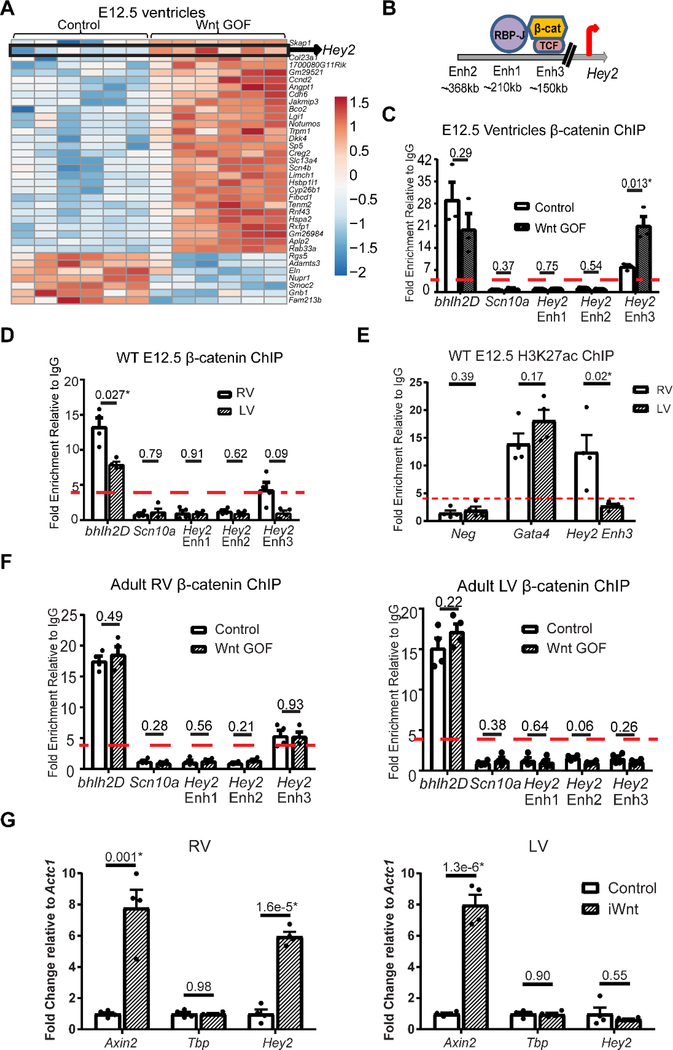
To preferentially identify upstream Wnt targets which may exert an effect during development, we performed RNA sequencing comparing global gene expression in developmental Wnt GOF (αMHC-Cre; Ctnnb1flox(ex3)/+) with littermate control E12.5 embryonic ventricles. Principal component analysis shows that all control samples cluster tightly together, while 5 of 6 Wnt GOF samples also cluster tightly together (Supplemental Figure 4). Differential expression analysis reveals 36 differentially regulated genes, a relatively short list likely due to the inclusion of all samples in the analysis (Figure 3A, Supplemental Table 2). Interestingly, Hey2 is up-regulated in Wnt GOF embryonic ventricles, and was also down-regulated in Wnt LOF RV (Figure 1C,D). Hey2 is a transcriptional repressor highly expressed in both ventricles during embryogenesis, with relatively lower expression in postnatal hearts 32. Hey2 is a known direct Notch target, however, it has long been postulated that regulation of Hey2 expression in the ventricles may not simply reflect a role of Notch signaling, and that Notch-independent mechanisms may regulate its expression.
Figure 3 – Hey2 is a direct target of Wnt signaling in the right ventricle with differential enhancer binding of β-catenin.
(A) Heat map representation of RNA sequencing of Wnt GOF (αMHC-Cre; Ctnnb1fl(ex3)/+) versus littermate control (Ctnnb1fl(ex3)/+) embryonic ventricles (n=6 each) reveals 36 differentially-regulated transcripts including Hey2. (B) Genomic regions of murine Hey2 delineating known upstream cardiac enhancers with location relative to the transcription start site, and putative Notch and Wnt transcriptional effector binding sites. (C) ChIP for β-catenin in embryonic day 12.5 (E12.5) Wnt GOF (αMHC-Cre; Ctnnb1fl(ex3)/+) and littermate control (Ctnnb1fl(ex3)/+) ventricles (RV and LV combined, n=3 each) shows basal binding to the positive control bhlh2D locus, without binding at the negative control Scn10a locus, Hey2 Enhancer (Enh) 1, nor Enh 2. 7-fold enrichment of β–catenin was detected in controls at the consensus Wnt-responsive element within Hey2 Enh 3, with further 19-fold enrichment in Wnt GOF ventricles (a threshold of 4-fold enrichment over IgG was considered binding). (D) ChIP for β-catenin in wild type (WT) E12.5 RV versus LV reveals 4-fold enrichment of β-catenin binding at Hey2 Enh3 in RV, without enrichment in the LV, at this early developmental stage (n=4). (E) ChIP for H3K27ac in WT E12.5 RV versus LV shows enrichment at the positive control Gata4 locus, but not at the negative control gene desert (neg) in both RV and LV. There is more than 12-fold enrichment of H3K27ac in the RV at Hey2 Enh3, however, in contrast there is less than 4-fold enrichment in the LV (n=4). (F) ChIP in adult RV shows β-catenin binding at the positive control bhlh2D locus, as well as Hey2 Enh3 in controls (Mlc2vCre/+), without further enrichment in littermate Wnt GOF mice (Mlc2vCre/+; Ctnnb1fl(ex3)/+). Binding is not detected at the negative control Scn10a promoter, nor at Hey2 Enh1,2 (n=4 each). In adult control and Wnt GOF LV, β-catenin binds at the positive control bhlh2D locus, without enrichment at the negative control Scn10a promoter, nor Hey2 Enh1–3. β-catenin binding at Hey2 Enh3 is not detected in Wnt GOF LV despite increased β-catenin levels, suggesting that levels alone may not account for the lack of Enh3 binding in the LV (n= 4 each). (G) qRT-PCR from inducible Wnt GOF mice (iWnt, αMHCrtTA; TetO-Cre; Ctnnb1fl(ex3)/+), where Wnt signaling was induced in adulthood, versus control (TetO-Cre; Ctnnb1fl(ex3)/+) mice, shows increased expression of the direct Wnt target Axin2 in both ventricles, no change in expression of Tbp (negative control), while Hey2 expression increases in the RV only (n=4 each). An equal variance Student’s t-test was used for comparisons, with all two-tailed P-values indicated. *P<0.05 considered statistically significant.
To determine whether Hey2 may be a direct Wnt target, we performed a bioinformatics analysis on Hey2 regulatory regions. Three Gata-responsive cardiac enhancers have previously been identified upstream of Hey2 with differential expression patterns in the embryonic heart (Supplemental Figure 5) 33. All three enhancers (Enh1–3) contain Gata4 binding sites, and we identified consensus RBP-J binding motifs within enhancer 1 (Enh1) and near the TSS, consistent with Hey2 as a known direct Notch target (Figure 3B). Members of the Tcf/Lef transcription factor family associate with DNA targets in repressive complexes in the absence of Wnt signaling. When canonical Wnt signaling is active, β-catenin complexes with Tcf/Lef to displace the repressive complex and activate transcription 18. We identified a putative Tcf/Lef Wnt response element30 within Hey2 enhancer 3 (Enh3) (Figure 3B), a 1343 base pair enhancer 82% conserved between mice and humans, with conserved Tcf/Lef and GATA binding sites (Supplemental Figure 6). To determine whether Enh3 may confer Wnt-responsiveness, we first determined whether β-catenin is associated with this region in embryonic ventricles by performing chromatin immunoprecipitation (ChIP) followed by qPCR. In control ventricles, we detected more than 20-fold enrichment of β-catenin at the positive control Bhlh2 locus relative to IgG, while we did not detect β-catenin binding to a negative control region within the Scn10a gene that functions as a Scn5a enhancer (Figure 3C) 34. Similarly, β-catenin does not bind to Enh1 nor Enh2, while there is 7-fold enrichment at Hey2 Enh3 (Figure 3C). We next asked whether Hey2 up-regulation (Figure 3A) correlates with increased β-catenin binding to the Hey2 enhancer in developmental Wnt GOF (αMHC-Cre; Ctnnb1flox(ex3)/+) embryonic ventricles when compared with littermate controls. While no further enrichment of basal β-catenin binding was seen in E12.5 Wnt GOF embryos at the Bhlh2D locus, 19-fold enrichment was observed at Hey2 Enh3, demonstrating dynamic binding (Figure 3C).
Given that Wnt LOF resulted in decreased Hey2 expression only in the RV (Figure 1C, 1D), we wondered whether chamber-specific patterns of Hey2 expression are present during embryonic stages. To address this, we dissected wild type E12.5 embryos and pooled 4 primitive RV (bulbus cordis region without the outflow tract region) and LV for gene expression studies, and 12 RV and LV for each ChIP technical replicate. We found approximately 1.8-fold higher Hey2 expression in the RV compared to the LV at E12.5 (p=0.034, Supplemental Figure 7A). ChIP for β-catenin shows binding at Hey2 Enh3 in the primitive RV but not in the LV (Figure 3D). We then performed ChIP for H3K27ac, a modification to Histone H3 that correlates with active enhancers, on wild type E12.5 RV and LV. We found over 12-fold enrichment in the RV at Hey2 Enh3, without significant fold enrichment in the LV, suggesting that even at this stage there is differential chromatin configuration in the RV and LV (Figure 3E).
Since Wnt activity is higher in early embryonic RV versus LV, we asked whether levels of active β-catenin can account for differential enhancer binding between ventricles. If levels alone are sufficient to dictate differential binding, we might expect to find enhanced β-catenin binding to the LV Hey2 Enh3 under conditions where active β-catenin levels are higher in the LV. In wild type adult mice, Western blots of total and non-phosphorylated (active) β-catenin demonstrate more than 1.5-fold higher levels of total β-catenin (Figure 1B) and active β-catenin (Supplemental Figure 2) in the adult LV compared with RV. Despite higher levels of active β-catenin in the adult LV, we observe over 5-fold β-catenin enrichment at the Hey2 Enh3 locus only in the RV, while binding to the LV enhancer is not observed (Figure 3F). To further test whether active β-catenin levels alone can overcome binding to Enh3 in the LV, we assayed for β-catenin binding in developmental Wnt GOF mice. While in the Wnt GOF RV we observe over 5-fold β-catenin enrichment at the Hey2 Enh3 locus (Figure 3F), despite increased levels of active β-catenin binding was not detected at Hey2 Enh3 in the LV (Figure 3F). Consistent with these findings, Hey2 gene expression is up-regulated in the RV only (Supplemental Figure 7B). Furthermore, in an adult “tet-on” inducible Wnt GOF model (iWnt, αMHCrtTA; TetO-Cre; Ctnnb1fl(ex3)/+), Hey2 gene expression is increased by 6-fold compared to control (TetO-Cre; Ctnnb1fl(ex3)/+) in the adult RV only, while the Wnt target Axin2 is up-regulated to a similar extent in both RV and LV (Figure 3G). Taken together, these results suggest that differential regulation of Hey2 by Wnt persists into adulthood and is not due to exclusion of active β-catenin from left ventricular nuclei, and is likely due to differential chromatin structure existing from early developmental stages.
3.3. Differential electrophysiologic effects in Wnt LOF right versus left ventricles
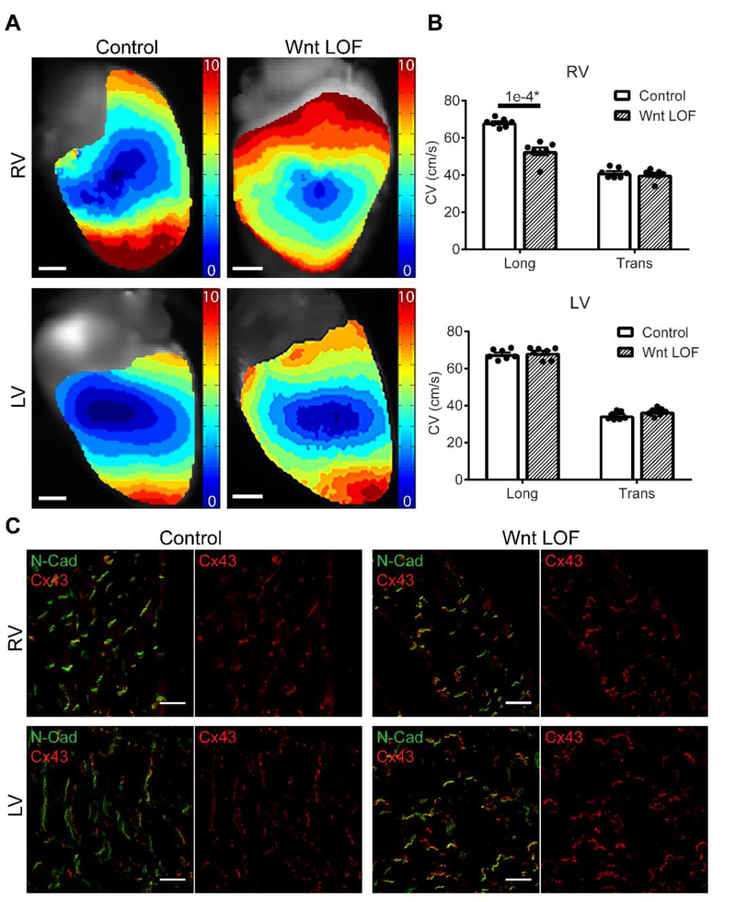
Given the observed differential expression of molecular determinants of conduction in Wnt LOF RV versus LV, we asked whether there may be chamber-specific electrophysiologic effects. Using optical mapping techniques together with programmed electrical stimulation, we measured the conduction velocity (CV) of Wnt LOF and littermate control mice (Figure 4A-B, n=7 each group). When stimulated within the physiological range (8 Hz Figure 4, 10 and 12 Hz Supplemental Figure 8), longitudinal CV in the RV is significantly slower in Wnt LOF mutants when compared with littermate controls (52.7±2.0 versus 68.2±0.9 cm/s, 8 Hz), while transverse CV is unchanged. In contrast, there is no change in either longitudinal or transverse CV in Wnt LOF LV.
Figure 4 – Conduction velocity is slower in Wnt loss-of-function right but not left ventricles.
(A) Representative reconstructed electrical activation maps from the RV (top) and LV (bottom) obtained during an optical mapping experiment with epicardial stimulation in Wnt LOF (Mlc2vCre/+; Ctnnb1dm/flox, right panel) and littermate control (Ctnnb1flox/+, left panel) hearts. Data were recorded during stimulation at 8Hz. More isochrones are visible on the Wnt LOF RV epicardial surface indicating slower conduction velocity (CV). Scale bars = 1 mm. (B) Calculated longitudinal and transverse CVs from control and Wnt LOF hearts (n=7 each genotype) show RV longitudinal CV is decreased by 23% in Wnt LOF RV compared with control (52.7 cm/s versus 68.2 cm/s) with decreased anisotropy (1.32 versus 1.66). In contrast, there is no difference in CV between Wnt LOF and control in the LV. An equal variance Student’s t test was used for comparisons, *P<0.05 were considered statistically significant. (C) Immunostaining of total Cx43 (red) and N-Cadherin (green) in control and littermate Wnt LOF shows co-localization without evidence of Cx43 lateralization in both RV and LV. Long = longitudinal, Trans = transverse, Scale bars = 20 μm.
Large changes in a single conduction parameter or modest changes in a combination of parameters are needed to exceed the limits of conduction reserve and impair conduction. To delineate whether factors in addition to reduced Cx43 levels may contribute to conduction slowing in Wnt LOF mice, we assessed cardiac morphology, fibrosis, and CM cell size (Supplemental Figure 9). While mice carrying the global Ctnnb1dm allele are uniformly smaller in size when compared with wild type littermates likely due to non-cardiac effects of this allele, the heart weight to tibia length ratio and CM cell size is unchanged in Wnt LOF mice (Supplemental Figure 9). In addition, no structural heart defects or increased fibrosis were observed in Mlc2vCre/+; Ctnnb1dm/flox Wnt LOF mice. Immunostaining shows normal localization of Cx43 at the intercalated disc, with strong co-localization together with N-Cadherin (Figure 4C). Previous studies have shown that phosphorylation of Cx43 at Serine 368 can lead to altered conductance and internalization of gap junctions 35, 36. Using an antibody specific for this phosphorylated Cx43 isoform, immunostaining demonstrates no significant change in Cx43 localization, while quantification by Western blot shows a similar level of protein reduction to that observed with total Cx43 in the RV (Supplemental Figure 10). Taken together, the absence of additional changes suggests that transcriptional down-regulation leading to reduced Cx43 protein is likely the primary reason for RV conduction slowing in Wnt LOF mice.
3.4. Increased excitability in Wnt LOF right ventricular cardiomyocytes
Conduction velocity is directly related to CM cell coupling through gap junctions and membrane sodium current. To gain further insight into the molecular basis for CV slowing, we performed intracellular sharp microelectrode recordings on Langendorff-perfused hearts (Figure 5A-G). Interestingly, in control hearts the maximum upstroke velocity (dVm/dtmax) is higher in RV subepicardial cells when compared with LV subepicardial cells (Figure 5D, Supplemental Table 3). Previous reports in dispersed cells have not reported this right versus left difference 37. We did not observe differences in the resting membrane potential (RMP) or action potential amplitude (APA) between control RV and LV (Figure 5F,G), while the action potential duration at 90% repolarization (APD90) is significantly longer in LV myocytes (Figure 5E), consistent with previous reports 38.
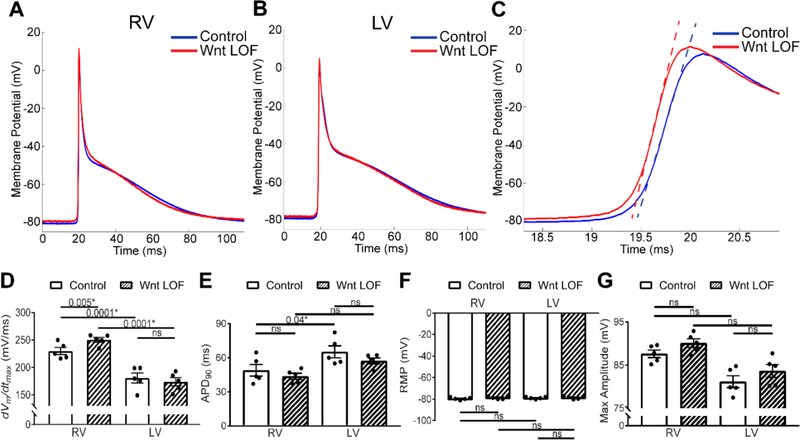
Figure 5 – Increased excitability in Wnt loss-of-function right ventricular cardiomyocytes.
Averaged action potential from control (blue) and Wnt LOF (red) RV (A) and LV (B) myocytes recorded with sharp microelectrodes reflecting a composite of all cells recorded. (C) Zoom of a portion of the action potential from (A) including Phase 0 with tangent lines drawn through the point of maximum upstroke velocity (dVm/dtmax) demonstrates a greater dVm/dtmax in Wnt LOF RV cells when compared with littermate controls (251 mV/ms vs. 229 mV/ms, p=0.005). (D-G) Graphical representation of RV and LV action potential parameters from control and littermate Wnt LOF hearts. dVm/dtmax and APD90 values differ between control RV and LV at baseline, consistent with previous reports. The dVm/dtmax is significantly higher in Wnt LOF RV compared to control RV cells, while maximal APA, RMP, and APD90 are unchanged in Wnt LOF RV cells. In contrast, no action potential parameters are different between control and Wnt LOF LV cells. At least ten cells per condition reflecting 20 to 100 APs per cell were averaged to yield one value per mouse. The average of n=5 mice per genotype is represented with mean±SEM. Control mice are Ctnnb1flox/+ and Wnt LOF are Mlc2vCre/+; Ctnnb1dm/flox. Statistics were performed using a two-way ANOVA followed by a post-hoc Tukey’s multiple comparisons test. RMP = resting membrane potential; APA = action potential amplitude; dVm/dtmax = maximal upstroke velocity; APD90 = duration of the action potential at 90% repolarization. *P<0.05 considered statistically significant, ns = not significant.
When compared with littermate controls, Wnt LOF RV CMs exhibit increased maximum upstroke velocity (dVm/dtmax 229±6.4 versus 251±4.3 V/sec, p=0.005, Figure 5A,C,D, Supplemental Table 3), while the LV maximum upstroke velocity remains unchanged (dVm/dtmax 180.6±9.2 versus 173.7±7.5 V/sec, Figure 5B,D). The APA, RMP, and APD90 are similar between control and Wnt LOF in both RV and LV (Figure 5E-G). In whole organ physiology, increased maximum upstroke velocity can be due to either enhanced phase 0 depolarization of the fast sodium current (which would result in increased conduction velocity), or from reduced intercellular gap junction coupling (which would result in decreased conduction velocity). While in an isolated cell INa,max is the sole determinant of dVm/dtmax, reduced coupling in tissue changes the source-sink relationship. Increased upstroke velocity in the setting of reduced coupling results from greater confinement of depolarizing charge to individual cells (decreased electrical load), thereby increasing the available charge for local depolarization and accelerating the rate of depolarization to cause a steeper AP upstroke. Therefore, taken together, increased maximum upstroke velocity in the absence of structural changes in Wnt LOF RV is consistent with decreased gap junction coupling as the leading contributor to conduction slowing.
3.5. Wnt LOF mice are predisposed to ventricular tachycardia originating from the right ventricle
To determine whether Wnt LOF mice have perturbation of baseline conduction intervals, EKGs were performed in mice while conscious or under light isoflurane sedation. Consistent with our previous report demonstrating that Wnt signaling programs the electrical phenotype of the atrioventricular junction 15, the PR interval is shorter in Wnt LOF mice when compared with littermate control (34.5±0.7 versus 37.7±0.8 ms) while the heart rate, P wave duration, QRS and QT intervals are unchanged (Table 1). It is often difficult to induce and sustain VT in mice in part due to the small ventricular surface area. To determine whether Wnt LOF mice exhibit spontaneous ventricular arrhythmias, we performed conscious telemetric monitoring on caged, moving mice. We did not detect spontaneous arrhythmias during one week of continuous telemetric monitoring. Given that increased sympathetic tone can often trigger arrhythmias in inherited and acquired cardiac disease, we injected Wnt LOF mice daily with isoproterenol. Though isoproterenol increased the heart rate as expected (Supplemental Figure 11), Wnt LOF mice did not develop spontaneous VT.
Table 1.
EKG intervals, Ex Vivo and In Vivo Programmed Electrical Stimulation EKG Intervals
| EKG Intervals | ||
| Control | Wnt LOF | |
| Conscious HR (bpm) | 692 ± 16 | 718 ± 12 |
| Isoflurane HR (bpm) | 488 ± 15 | 497 ± 23 |
| P wave duration (msec) | 12.5 ± 1.3 | 10.5 ± 0.7 |
| PR (msec) | 37.7 ± 0.8 | 34.5 ± 0.7* |
| QRS (msec) | 10.3 ± 0.7 | 10.1 ± 0.4 |
| QT (msec) | 17.5 ± 0.9 | 17.8 ± 8 |
| Ex Vivo Programmed Electrical Stimulation | ||
| Control | Wnt LOF | |
| Mice with VT following RV stim | 0/7 | 5/7* |
| Mice with VT following LV stim | 0/7 | 0/7 |
| Duration VT (s) | N/A | 94 ± 50 |
| VT cycle length (msec) | N/A | 48 ± 3 |
| Mice with spontaneous NSVT/VT | 0/7 | 2/7 |
| Total episodes spontaneous NSVT/VT | 0 | 734 |
| In Vivo Programmed Electrical Stimulation | ||
| Control | Wnt LOF | |
| Heart Rate (bpm) | 521 ± 21 | 572 ± 45 |
| V to A 1:1 (msec) | 90 ± 6 | 96 ± 6 |
| V to A Wenckebach (msec) | 83 ± 5 | 88 ± 5 |
| RVERP120 (msec) | 38 ± 3 | 28 ± 3* |
| Episodes NSVT/VT (S1S2) | 0 | 10* |
| Total episodes NSVT/VT | 0 | 31* |
EKG Intervals.
EKGs were performed on conscious Wnt LOF (Mlc2vCre/+; Ctnnb1dm/flox, n=6) and flox/+ littermate control (Ctnnb1, n=6) mice to determine HR, as well as during isoflurane sedation to determine HR and additional conduction parameters. Statistics were performed using unpaired t tests with Welch’s correction. *P<0.05, data are expressed as mean ± SEM.
Ex Vivo Programmed Electrical Stimulation.
Langendorff-perfused hearts from Wnt LOF (Mlc2vCre/+; Ctnnb1dm/flox, n=7) and littermate control (Ctnnb1flox/+, n=7) mice were stimulated with 40 beats of an S1 drive train at 8 Hz, 50 beats at 10 Hz and 60 beats at 12 Hz first on the RV followed by the LV. Episodes of VT were defined as ventricular activation faster than atrial activation with AV dissociation persisting longer than 2 s. Episodes of both monomorphic VT and polymorphic VT were observed, and representative activation maps and movies from the same hearts during sinus rhythm and ventricular tachycardia are shown in Figure 6 and Supplemental Videos 1 and 2. The average VT cycle length is shown for monomorphic VT episodes. 2 of 7 mice also developed frequent episodes of spontaneous NSVT/VT during the protocol. A Fisher’s exact test was used to determine significance between number of mice with and without VT. *P<0.05, data are expressed as mean ± SEM.
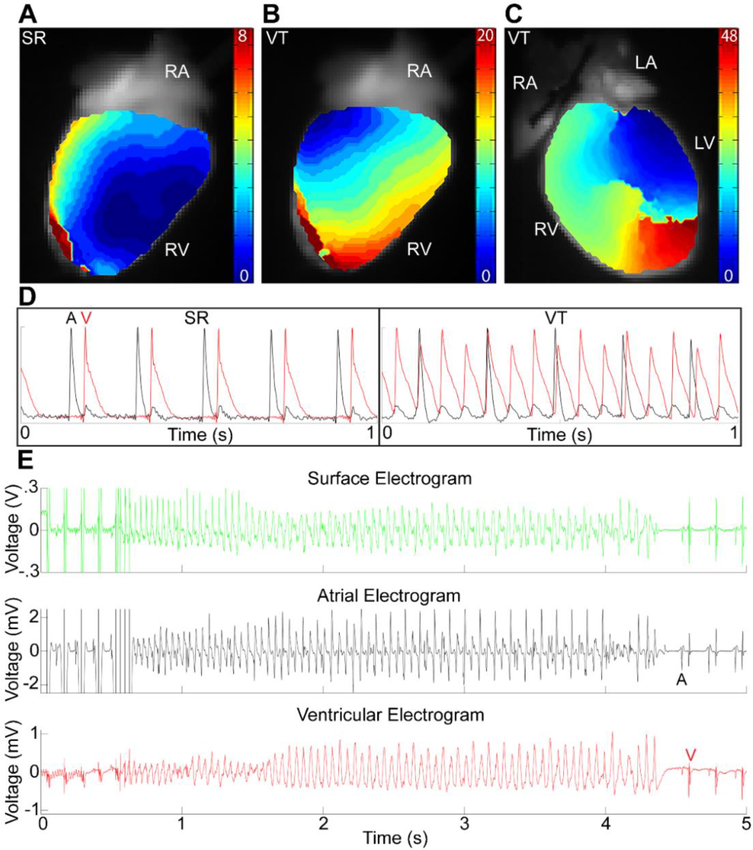
Figure 6 – Wnt loss-of-function mice are predisposed to ventricular tachycardia following right ventricular stimulation.
Reconstructed electrical activation maps showing the RV surface during sinus rhythm (SR, A), RV surface during ventricular tachycardia (VT, B), and anterior view of RV and LV during VT (C) from the same Wnt LOF mouse. VT was initiated following physiological drive train pacing on the RV epicardium (B) and was persistent. During SR (A) the earliest RV breakthrough site is near the apex. VT was initiated by stimulation of the RV epicardium with an S1 drive train at 8 Hz. During VT, electrical activation of the ventricles was altered both spatially and temporally. Note the different timescales for each activation map, with change in activation pattern from base to apex (B) and focal conduction block with reentry visible on the anterior surface during VT (C). (D) Optical action potentials during SR (left) show 1:1 activation of atria (black, A) followed by ventricle (red, V), whereas there is AV dissociation during VT (right) with more ventricular action potentials than atrial. 5 of 7 Wnt LOF (Mlc2vCre/+; Ctnnb1dm/flox) and 0 littermate control (Ctnnb1flox/+) hearts exhibited VT following RV stimulation while none of the hearts (both Wnt LOF and control) had VT following LV stimulation (Table 1) (E) Representative EKG (top, green), intracardiac atrial (middle, black) and intracardiac ventricular (bottom, red) traces during invasive electrophysiology studies showing an S1 drive train at 10 Hz followed by a triple extrastimulus initiating 4 s of polymorphic VT. 4 of 6 Wnt LOF and 0 of 6 littermate control mice had VT following RV stimulation (Table 1).
In Vivo Programmed Electrical Stimulation.
Isoflurane-sedated mice (same cohort used for EKGs, n=6 mice) were subjected to an invasive EP study via cannulation of the right internal jugular vein. The same stimulation protocol was performed on each mouse. The His electrogram was not visualized in the majority of mice, therefore AH and HV intervals are not shown. NSVT is defined as between 3 beats to 2 seconds of VT, while episodes of VT are defined as those lasting longer than 2 sec. Episodes of NSVT/VT elicited from 1 extrastimulus (S1S2) are delineated from total number of episodes, which includes those elicited from 2 or 3 extrastimuli and burst pacing. Statistics on heart rate, V to A conduction, and RVERP were performed using unpaired t tests with Welch’s correction. A Fisher’s exact test was used to determine significance between number of episodes of NSVT/VT and number of mice with NSVT/VT. *P<0.05, data are expressed as mean ± SEM.
It has previously been shown that intrinsic asymmetry in electric properties of tissue, for example excitability, refractory periods, or gap junction coupling, may widen the vulnerable window and increase the probability of unidirectional block after a premature stimulus 39. Once unidirectional block occurs, reentrant tachycardias can occur if the wavelength of excitation, defined as the product of CV and effective refractory period (ERP), is shorter than the dimensions of the circuit 40. Given that slowed conduction velocity in the setting of reduced gap junction coupling can predispose to reentrant VT, we performed programmed electrical stimulation on RV and LV of Langendorff-perfused hearts during optical mapping studies. A striking result is that five of seven Wnt LOF mice developed VT during RV stimulation at physiologic rates with just a single drive train, even without an extrastimulus, while none of the seven littermate controls developed VT in the setting of RV pacing (Figure 6A-E, Table 1). In some instances, a line of functional block with reentry on alternating beats occurred on the anterior surface during VT induced by RV stimulation (Supplemental Video 1, Supplemental Video 2). In contrast, the same pacing protocol applied to the LV did not lead to arrhythmias in Wnt LOF mice (Table 1). Predisposition to arrhythmias can be influenced by factors extrinsic to the heart including autonomic tone. To further probe whether Wnt LOF predisposes to an arrhythmic substrate in vivo, we performed invasive electrophysiology studies. Wnt LOF mice have a shorter RVERP when compared with littermate controls (28±3 vs 38±3 ms, Table 1) and increased susceptibility to VT following RV programmed electrical stimulation, similar to the results observed in ex vivo Langendorff-perfused hearts (Figure 5E, Table 1). These results demonstrate that Wnt transcriptional activity is required for electrical programming of the RV during development, and loss of function results in high predisposition to arrhythmias that persists in adulthood.
Discussion
In this study, we show that embryonic programming can differentially regulate adult electrophysiology in the right versus left ventricles. We specifically probe a Wnt LOF model where β-catenin is transcriptionally inactive but its cell adhesion properties are preserved. Loss of Wnt transcriptional activity during development results in a large, yet mostly mutually exclusive, number of genes dysregulated in the adult RV and LV. In this Wnt LOF model, CV is preferentially slowed in the RV, at least In part due to RV-specific loss of Cx43 (Figure 1, 4). Similarly, we previously reported that CV is slowed predominantly in the RV in Wnt GOF mice, and we now show that the mechanism is through reduced NaV1.5, while Cx43 levels are unchanged (Figure 2). Previous studies have shown using neonatal rat ventricular myocytes that Wnt activators can downregulate both Cx43 and NaV1.5 16, 17. Discrepancies with our Wnt GOF data could perhaps be explained by earlier developmental activation in our in vivo model, or modulation of β-catenin versus use of a Wnt agonist.
Interestingly, gap junction remodeling, evidenced by reduced immunostaining for Cx43 at cell-cell junctions appears to be a consistent feature of arrhythmogenic cardiomyopathy (ACM) 41. The classic right-dominant form of ACM, also known as arrhythmogenic right ventricular cardiomyopathy, is a leading cause of sudden cardiac death in the young 42. ACM is characterized by subepicardial fibro-fatty deposition, and is often associated with mutations in desmosomal proteins. ACM patients across a wide etiologic spectrum exhibit altered GSK-3β distribution, and pharmacologic inhibition of GSK-3β can rescue both the histologic and electrical phenotypes in animal models of the disease 43, 44. Among its diverse functions, GSK-3β regulates the Wnt signaling pathway and altered canonical Wnt signaling has been described in animal models and in patients with ACM 8, 45–47. ACM patients tend to have prolonged APDs 48, 49, while the RVERP is shorter in Wnt LOF mice (Table 1). Therefore, precisely how GSK-3β redistribution may impact Wnt signaling, and how Wnt signaling may relate to RV-predominant arrhythmias will likely remain an area of active interest.
Unbiased RNA-sequencing of RV and LV in Wnt LOF mice demonstrates distinct chamber-specific gene dysregulation, with relatively little overlap (Figure 1C). One interesting gene regulated by Wnt specifically within the RV is the previously well-studied Notch target Hey2. It has long been postulated that regulation of Hey2 expression in the ventricles may not reflect a role of Notch signaling, and that Notch-independent mechanisms may also regulate its expression. To our knowledge, this is the first study demonstrating Hey2 is a direct Wnt target in any tissue. We delineate RV-specific regulation of this gene that has been associated in GWAS studies with Brugada syndrome, characterized by sudden death from RV tachycardias 12, 50. Given that recent thoughts based on ultrastructural and electrophysiological changes have suggested that perhaps Brugada syndrome and ACM represent different spectrums of the same disease 51, it is intriguing that Hey2 is transcriptionally regulated by Wnt.
Differential chamber-specific β-catenin enhancer binding is present by E12.5 (Figure 3D), and even at this early developmental stage, there is already differential Hey2 enhancer activity (Figure 3E) that coincides with differential β-catenin binding. Increasing β-catenin protein levels from early developmental stages when myocytes turn on expression of Myosin light chain 2v onward could not overcome differential binding to the Hey2 enhancer (Mlc2vCre; Ctnnb1flox(ex3)/+, Figure 3G). Furthermore, Wnt induction in the adult activates Hey2 transcription only in the RV, while transcription of another Wnt target gene, Axin2, is similarly upregulated in both ventricles (Figure 3G). Since Axin2 is upregulated equally in both ventricles, this strongly suggests that the mechanism for differential Hey2 levels is not related to the amount of β-catenin in the cell or in the nucleus, but rather points to chamber-specific regulation at the level of active chromatin and enhancer binding at specific gene regulatory elements. Furthermore, even though β-catenin protein levels in the cell are similar in adulthood, transcriptional differences seen in the adult may result from the embryonic programming events that persist. While Hey2 is associated with Brugada syndrome and regulates important chamber-specific electrophysiological properties including dVm/dtmax 12, previous work has shown that Hey2 does not regulate the maximum upstroke velocity through Scn5a transcript levels 52. Therefore, dysregulation of Scn5a and Gja1 by Wnt could also occur through direct β-catenin binding to regulatory elements, or through other indirect mechanisms. The finding that early embryonic events pattern enhancer binding and regulate electrophysiology into adulthood may have implications for protocols involving sequential Wnt-pathway modulation to derive CMs from induced-pluripotent stem cells for downstream applications involving cellular electrophysiology 53. Specifically, a better understanding for the differential manner in which Wnt affects many genes relevant to electrophysiology in early right versus left ventricular progenitors may provide an opportunity to enhance differentiation and maturation of iPS-derived CMs.
Multiple previous studies have focused on the relationship between gap junction expression and arrhythmia susceptibility. Studies on Cx43+/− mice have yielded mixed results 54, 55, and cardiomyocyte-specific loss of function studies have shed further light on this relationship 56. While reduction of Cx43 protein levels to just under half normal levels did not significantly affect LV conduction velocity nor lead to ventricular tachyarrhythmias, further reduction to 18% of control levels was associated with decreased LV CV and inducible polymorphic ventricular tachycardia 57. Interestingly, though effects on RV CV were not reported in these studies, loss of Cx43 had a differential effect on K+ currents in the RV which predisposed to reentrant arrhythmias 37. The current manuscript examines the transcriptional role of Wnt signaling on cardiac conduction, noting a more robust reduction of Cx43 protein in the RV (approximately 50%, Figure 1F) with a significant 23% reduction in longitudinal CV (Figure 4A, 4B). Since the degree of RV conduction slowing was higher than we expected, we ruled out additional effects on excitability (Figure 5) or Cx43 protein mis-localization (Figure 4C, Supplemental Figure 10). One potential explanation for why this degree of Cx43 reduction could lead to marked RV conduction slowing may involve distinct safety factors for conduction between ventricles 58. In addition, the RVERP is significantly shorter in the Wnt LOF RV (Table 1), which would be expected to predispose to reentrant tachycardias given a shorter wavelength of excitation. Several genes encoding ion channel subunits important in cardiac repolarization are dysregulated in the Wnt LOF RV only (NCBI GEO submission GSE106507), including downregulation of genes encoding subunits of potassium channels (Kcnn2, Kcnmb1, Kcna4, and Kcnq1). Therefore, additional direct or indirect disturbances in ionic currents likely contribute to the marked differential arrhythmogenesis observed between the Wnt LOF RV and LV. Although we saw arrhythmia susceptibility in Wnt LOF RV in both in vivo and ex vivo experiments (Figure 6, Table 1), we did not observe spontaneous arrhythmias in telemetry-monitored mice injected with isoproterenol (Supplemental Figure 11). This suggests that, although Wnt LOF mice have a pro-arrhythmic substrate, they may lack calcium-mediated triggers. Consistent with this hypothesis, of the roughly 700 genes and transcripts dysregulated in the RV, only one calcium channel gene (Cacna2d3) was dysregulated.
In the healthy adult heart Wnt signaling is at low levels, but in response to pathologic stimuli such as myocardial infarction or pressure overload, Wnt signaling becomes activated 18. Though quite a bit is known regarding the effects of Wnt signaling on CM hypertrophy and fibrosis, relatively less is known about the effects of Wnt signaling on cardiac conduction and arrhythmogenesis 59. Indeed, therapeutic alteration of Wnt signaling in the post-myocardial infarction setting has produced conflicting results 18. Given that aberrant Wnt signaling is associated with diverse disorders giving rise to arrhythmias, including acquired disorders such as cardiac hypertrophy and myocardial infarction, as well as familial arrhythmias such as ACM, a better understanding of the role of Wnt signaling in cardiac conduction could enhance our approach to these diseases 60.
Supplementary Material
Highlights.
Embryonic programming via canonical Wnt signaling regulates electrophysiology
There is differential enhancer binding and activity in developing ventricles
Hey2 is a direct Wnt target in the right ventricle only
Wnt loss of function affects connexin43 and conduction in the right ventricle
Wnt loss of function mice have susceptibility to ventricular tachycardia
Acknowledgements:
This work was supported by the National Institutes of Health R01 HL130212 (SR). Dr. Rentschler holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. The funding sources had no involvement in the study design, collection, analysis and interpretation of data, in the writing of the report, nor in the decision to submit the article for publication. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for sequencing and genomic analysis.
Non-standard Abbreviations and Acronyms:
- SHF
Second heart field
- RV
Right ventricle
- LV
Left ventricle
- ACM
Arrhythmogenic cardiomyopathy
- GOF
Gain-of-function
- LOF
Loss-of-function
- Tcf/Lef
T-cell factor/lymphoid enhancer-binding factor
- TSS
Transcription start site
- CM
Cardiomyocyte
- Cx43
connexin43
- Hey2
Hairy/enhancer-of-split related with YRPW motif protein 2
- VT
Ventricular tachycardia
- NSVT
Non-sustained ventricular tachycardia
- HR
Heart Rate
- ChIP
Chromatin immunoprecipitation
- APD
Action potential duration
- APA
Action potential amplitude
- RMP
Resting membrane potential
- dVm/dtmax
Maximum upstroke velocity
- CV
Conduction velocity
- ERP
Effective refractory period
Footnotes
Disclosures:
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bakker ML, Boink GJ, Boukens BJ, Verkerk AO, van den Boogaard M, den Haan AD, Hoogaars WM, Buermans HP, de Bakker JM, Seppen J, Tan HL, Moorman AF, t Hoen PA and Christoffels VM. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res. 2012;94:439–49. [DOI] [PubMed] [Google Scholar]
- 2.Arnolds DE, Liu F, Fahrenbach JP, Kim GH, Schillinger KJ, Smemo S, McNally EM, Nobrega MA, Patel VV and Moskowitz IP. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J Clin Invest. 2012;122:2509–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rentschler S, Yen AH, Lu J, Petrenko NB, Lu MM, Manderfield LJ, Patel VV, Fishman GI and Epstein JA. Myocardial Notch signaling reprograms cardiomyocytes to a conduction-like phenotype. Circulation. 2012;126:1058–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bressan M, Liu G and Mikawa T. Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science. 2013;340:744–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs LE, Takeda M, Cuadra AE, Wakimoto H, Marks MH, Walker AJ, Seki T, Oh SP, Lu JT, Sumners C, Raizada MK, Horikoshi N, Weinberg EO, Yasui K, Ikeda Y, Chien KR and Kasahara H. Perinatal loss of Nkx2–5 results in rapid conduction and contraction defects. Circ Res. 2008;103:580–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh EP and Cecchin F. Arrhythmias in adult patients with congenital heart disease. Circulation. 2007;115:534–545. [DOI] [PubMed] [Google Scholar]
- 7.Tian Y, Cohen ED and Morrisey EE. The importance of Wnt signaling in cardiovascular development. Pediatr Cardiol. 2010;31:342–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Gras E, Lombardi R, Giocondo MJ, Willerson JT, Schneider MD, Khoury DS and Marian AJ. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest. 2006;116:2012–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Boogaard M, Wong LY, Tessadori F, Bakker ML, Dreizehnter LK, Wakker V, Bezzina CR, t Hoen PA, Bakkers J, Barnett P and Christoffels VM. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest. 2012;122:251930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, Tarasov KV, Muller M, Sotoodehnia N, Sinner MF, Verwoert GC, Li M, Kao WH, Kottgen A, Coresh J, Bis JC, Psaty BM, Rice K, Rotter JI, Rivadeneira F, Hofman A, Kors JA, Stricker BH, Uitterlinden AG, van Duijn CM, Beckmann BM, Sauter W, Gieger C, Lubitz SA, Newton-Cheh C, Wang TJ, Magnani JW, Schnabel RB, Chung MK, Barnard J, Smith JD, Van Wagoner DR, Vasan RS, Aspelund T, Eiriksdottir G, Harris TB, Launer LJ, Najjar SS, Lakatta E, Schlessinger D, Uda M, Abecasis GR, Muller-Myhsok B, Ehret GB, Boerwinkle E, Chakravarti A, Soliman EZ, Lunetta KL, Perz S, Wichmann HE, Meitinger T, Levy D, Gudnason V, Ellinor PT, Sanna S, Kaab S, Witteman JC, Alonso A, Benjamin EJ and Heckbert SR. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, Gudjonsson SA, Jonasdottir A, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Lochen ML, Kong A, Thorsteinsdottir U and Stefansson K. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–22. [DOI] [PubMed] [Google Scholar]
- 12.Bezzina CR, Barc J, Mizusawa Y, Remme CA, Gourraud JB, Simonet F, Verkerk AO, Schwartz PJ, Crotti L, Dagradi F, Guicheney P, Fressart V, Leenhardt A, Antzelevitch C, Bartkowiak S, Schulze-Bahr E, Zumhagen S, Behr ER, Bastiaenen R, Tfelt-Hansen J, Olesen MS, Kaab S, Beckmann BM, Weeke P, Watanabe H, Endo N, Minamino T, Horie M, Ohno S, Hasegawa K, Makita N, Nogami A, Shimizu W, Aiba T, Froguel P, Balkau B, Lantieri O, Torchio M, Wiese C, Weber D, Wolswinkel R, Coronel R, Boukens BJ, Bezieau S, Charpentier E, Chatel S, Despres A, Gros F, Kyndt F, Lecointe S, Lindenbaum P, Portero V, Violleau J, Gessler M, Tan HL, Roden DM, Christoffels VM, Le Marec H, Wilde AA, Probst V, Schott JJ, Dina C and Redon R. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet. 2013;45:1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Muller-Nurasyid M, Krijthe BP, Lubitz SA, Bis JC, Chung MK, Dorr M, Ozaki K, Roberts JD, Smith JG, Pfeufer A, Sinner MF, Lohman K, Ding J, Smith NL, Smith JD, Rienstra M, Rice KM, Van Wagoner DR, Magnani JW, Wakili R, Clauss S, Rotter JI, Steinbeck G, Launer LJ, Davies RW, Borkovich M, Harris TB, Lin H, Volker U, Volzke H, Milan DJ, Hofman A, Boerwinkle E, Chen LY, Soliman EZ, Voight BF, Li G, Chakravarti A, Kubo M, Tedrow UB, Rose LM, Ridker PM, Conen D, Tsunoda T, Furukawa T, Sotoodehnia N, Xu S, Kamatani N, Levy D, Nakamura Y, Parvez B, Mahida S, Furie KL, Rosand J, Muhammad R, Psaty BM, Meitinger T, Perz S, Wichmann HE, Witteman JC, Kao WH, Kathiresan S, Roden DM, Uitterlinden AG, Rivadeneira F, McKnight B, Sjogren M, Newman AB, Liu Y, Gollob MH, Melander O, Tanaka T, Stricker BH, Felix SB, Alonso A, Darbar D, Barnard J, Chasman DI, Heckbert SR, Benjamin EJ, Gudnason V and Kaab S. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnolds DE, Chu A, McNally EM, Nobrega MA and Moskowitz IP. The emerging genetic landscape underlying cardiac conduction system function. Birth Defects Res A Clin Mol Teratol. 2011;91:578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillers BS, Chiplunkar A, Aly H, Valenta T, Basler K, Christoffels VM, Efimov IR, Boukens BJ and Rentschler S. Canonical wnt signaling regulates atrioventricular junction programming and electrophysiological properties. Circ Res. 2015;116:398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang WB, Cho HC and Marban E. Wnt signalling suppresses voltage-dependent Na+ channel expression in postnatal rat cardiomyocytes. Journal of Physiology-London. 2015;593:1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ai ZW, Fischer A, Spray DC, Brown AMC and Fishman GI. Wnt-1 regulation of connexin43 in cardiac myocytes. Journal of Clinical Investigation. 2000;105:161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dawson K, Aflaki M and Nattel S. Role of the Wnt-Frizzled system in cardiac pathophysiology: a rapidly developing, poorly understood area with enormous potential. J Physiol. 2013;591:1409–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valenta T, Gay M, Steiner S, Draganova K, Zemke M, Hoffmans R, Cinelli P, Aguet M, Sommer L and Basler K. Probing transcription-specific outputs of beta-catenin in vivo. Genes Dev. 2011;25:2631–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin M, Small EM, van Rooij E, Qi X, Richardson JA, Srivastava D, Nakagawa O and Olson EN. Essential roles of the bHLH transcription factor Hrt2 in repression of atrial gene expression and maintenance of postnatal cardiac function. Proc Natl Acad Sci U S A. 2007;104:7975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Kubalak SW and Chien KR. Ventricular muscle-restricted targeting of the RXRalpha gene reveals a non-cell-autonomous requirement in cardiac chamber morphogenesis. Development. 1998;125:1943–9. [DOI] [PubMed] [Google Scholar]
- 22.Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA and Schneider MD. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100:169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valencik ML and McDonald JA. Codon optimization markedly improves doxycycline regulated gene expression in the mouse heart. Transgenic Res. 2001;10:269–75. [DOI] [PubMed] [Google Scholar]
- 24.Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O and Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–64. [DOI] [PubMed] [Google Scholar]
- 25.Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M and Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 1999;18:5931–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perl AKT, Wert SE, Nagy A, Lobe CG and Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10482–10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrer-Vaquer A, Piliszek A, Tian GN, Aho RJ, Dufort D and Hadjantonakis AK. A sensitive and bright single-cell resolution live imaging reporter of Wnt/beta-catenin signaling in the mouse. Bmc Dev Biol. 2010;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rentschler S, Harris BS, Kuznekoff L, Jain R, Manderfield L, Lu MM, Morley GE, Patel VV and Epstein JA. Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J Clin Invest. 2011;121:525–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khandekar A, Springer S, Wang W, Hicks S, Weinheimer C, Diaz-Trelles R, Nerbonne JM and Rentschler S. Notch-Mediated Epigenetic Regulation of Voltage-Gated Potassium Currents. Circulation Research. 2016;119:1324–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cadigan KM and Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol. 2012;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berul CI, Aronovitz MJ, Wang PJ and Mendelsohn ME. In vivo cardiac electrophysiology studies in the mouse. Circulation. 1996;94:2641–8. [DOI] [PubMed] [Google Scholar]
- 32.Koibuchi N and Chin MT. CHF1/Hey2 plays a pivotal role in left ventricular maturation through suppression of ectopic atrial gene expression. Circulation Research. 2007;100:850–855. [DOI] [PubMed] [Google Scholar]
- 33.He A, Gu F, Hu Y, Ma Q, Ye LY, Akiyama JA, Visel A, Pennacchio LA and Pu WT. Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease. Nat Commun. 2014;5:4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Boogaard M, Smemo S, Burnicka-Turek O, Arnolds DE, van de Werken HJG, Klous P, McKean D, Muehlschlegel JD, Moosmann J, Toka O, Yang XNH, Koopmann TT, Adriaens ME, Bezzina CR, de Laat W, Seidman C, Seidman JG, Christoffels VM, Nobrega MA, Barnett P and Moskowitz IR. A common genetic variant within SCN10A modulates cardiac SCN5A expression. Journal of Clinical Investigation. 2014;124:1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marquez-Rosado L, Solan JL, Dunn CA, Norris RP and Lampe PD. Connexin43 phosphorylation in brain, cardiac, endothelial and epithelial tissues. Biochim Biophys Acta. 2012;1818:1985–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth JW, Zhang SS, Sanchez JM, Lamouille S, Vogan JM, Hesketh GG, Hong T, Tomaselli GF and Shaw RM. A 14–3-3 mode-1 binding motif initiates gap junction internalization during acute cardiac ischemia. Traffic. 2014;15:684–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danik SB, Rosner G, Lader J, Gutstein DE, Fishman GI and Morley GE. Electrical remodeling contributes to complex tachyarrhythmias in connexin43-deficient mouse hearts. Faseb Journal. 2008;22:1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molina CE, Heijman J and Dobrev D. Differences in Left Versus Right Ventricular Electrophysiological Properties in Cardiac Dysfunction and Arrhythmogenesis. Arrhythm Electrophysiol Rev. 2016;5:14–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quan WL and Rudy Y. Unidirectional Block and Reentry of Cardiac Excitation - a Model Study. Circulation Research. 1990;66:367–382. [DOI] [PubMed] [Google Scholar]
- 40.Mines GR. On dynamic equilibrium in the heart. Journal of Physiology-London. 1913;46:349–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asimaki A, Kleber AG, MacRae CA and Saffitz JE. Arrhythmogenic Cardiomyopathy - New Insights into Disease Mechanisms and Drug Discovery. Prog Pediatr Cardiol. 2014;37:3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basso C, Corrado D, Bauce B and Thiene G. Arrhythmogenic right ventricular cardiomyopathy. Circ Arrhythm Electrophysiol. 2012;5:1233–46. [DOI] [PubMed] [Google Scholar]
- 43.Asimaki A, Kapoor S, Plovie E, Karin Arndt A, Adams E, Liu Z, James CA, Judge DP, Calkins H, Churko J, Wu JC, MacRae CA, Kleber AG and Saffitz JE. Identification of a new modulator of the intercalated disc in a zebrafish model of arrhythmogenic cardiomyopathy. Sci Transl Med. 2014;6:240ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chelko SP, Asimaki A, Andersen P, Bedja D, Amat-Alarcon N, DeMazumder D, Jasti R, MacRae CA, Leber R, Kleber AG, Saffitz JE and Judge DP. Central role for GSK3 beta in the pathogenesis of arrhythmogenic cardiomyopathy. Jci Insight. 2016;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lombardi R, Dong J, Rodriguez G, Bell A, Leung TK, Schwartz RJ, Willerson JT, Brugada R and Marian AJ. Genetic fate mapping identifies second heart field progenitor cells as a source of adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circ Res. 2009;104:1076–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lombardi R, da Graca Cabreira-Hansen M, Bell A, Fromm RR, Willerson JT and Marian AJ. Nuclear plakoglobin is essential for differentiation of cardiac progenitor cells to adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circ Res. 2011;109:1342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen SN, Gurha P, Lombardi R, Ruggiero A, Willerson JT and Marian AJ. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ Res. 2014;114:454–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finlay MC, Ahmed AK, Sugrue A, Bhar-Amato J and Quarta G. Dynamic Conduction and Repolarisation Changes in Early Arrhythmogenic Right Ventricular Cardiomyopathy versus Benign Outflow Tract Ectopy Demonstrated by High Density Mapping & Paced Surface ECG Analysis (vol 9, e99125, 2014). Plos One. 2014;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrews CM, Srinivasan NT, Rosmini S, Bulluck H, Orini M, Jenkins S, Pantazis A, McKenna WJ, Moon JC, Lambiase PD and Rudy Y. Electrical and Structural Substrate of Arrhythmogenic Right Ventricular Cardiomyopathy Determined Using Noninvasive Electrocardiographic Imaging and Late Gadolinium Magnetic Resonance Imaging. Circulation-Arrhythmia and Electrophysiology. 2017;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brugada P and Brugada J. Right Bundle-Branch Block, Persistent St Segment Elevation and Sudden Cardiac Death - a Distinct Clinical and Electrocardiographic Syndrome - a Multicenter Report. Journal of the American College of Cardiology. 1992;20:1391–1396. [DOI] [PubMed] [Google Scholar]
- 51.Corrado D, Zorzi A, Cerrone M, Rigato I, Mongillo M, Bauce B and Delmar M. Relationship Between Arrhythmogenic Right Ventricular Cardiomyopathy and Brugada Syndrome: New Insights From Molecular Biology and Clinical Implications. Circulation-Arrhythmia and Electrophysiology. 2016;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veerman CC, Podliesna S, Tadros R, Lodder EM, Mengarelli I, de Jonge B, Beekman L, Barc J, Wilders R, Wilde AAM, Boukens BJ, Coronel R, Verkerk AO, Remme CA and Bezzina CR. The Brugada Syndrome Susceptibility Gene HEY2 Modulates Cardiac Transmural Ion Channel Patterning and Electrical Heterogeneity. Circulation Research. 2017;121:537-+.. [DOI] [PubMed] [Google Scholar]
- 53.Lian XJ, Zhang JH, Azarin SM, Zhu KX, Hazeltine LB, Bao XP, Hsiao C, Kamp TJ and Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nature Protocols. 2013;8:162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guerrero PA, Schuessler RB, Davis LM, Beyer EC, Johnson CM, Yamada KA and Saffitz JE. Slow ventricular conduction in mice heterozygous for a connexin43 null mutation. Journal of Clinical Investigation. 1997;99:1991–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morley GE, Vaidya D, Samie FH, Lo C, Delmar M and Jalife J. Characterization of conduction in the ventricles of normal and heterozygous Cx43 knockout mice using optical mapping. Journal of Cardiovascular Electrophysiology. 1999;10:1361–1375. [DOI] [PubMed] [Google Scholar]
- 56.Gutstein DE, Morley GE, Tamaddon H, Vaidya D, Schneider MD, Chen J, Chien KR, Stuhlmann H and Fishman GI. Conduction slowing and sudden arrhythmic death in mice with cardiac-restricted inactivation of connexin43. Circulation Research. 2001;88:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danik SB, Liu FY, Zhang J, Suk HJ, Morley GE, Fishman GI and Gutstein DE. Modulation of cardiac gap junction expression and arrhythmic susceptibility. Circulation Research. 2004;95:1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw RM and Rudy Y. Ionic mechanisms of propagation in cardiac tissue - Roles of the sodium and L-type calcium currents during reduced excitability and decreased gap junction coupling. Circulation Research. 1997;81:727–741. [DOI] [PubMed] [Google Scholar]
- 59.Panakova D, Werdich AA and Macrae CA. Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca(2+) channel. Nature. 2010;466:874–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwon C and Tomaselli GF. Coins of the Realm in Atrioventricular Junction Development. Circulation Research. 2015;116:386–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.