Abstract
Nutrition is paramount in shaping all aspects of animal biology. In addition, the influence of the intestinal microbiota on physiology is now widely recognized. Given that diet also shapes the intestinal microbiota, this raises the question of how the nutritional environment and microbial assemblages together influence animal physiology. This research field constitutes a new frontier in the field of organismal biology that needs to be addressed. Here we review recent studies using animal models and humans and propose an integrative framework within which to define the study of the diet- physiology-microbiota systems and ultimately link it to human health. Nutritional Geometry sits centrally in the proposed framework and offers means to define diet compositions that are optimal for individuals and populations.
eTOC Blurb
How the nutritional environment and microbial assemblages together influence our physiology is a frontier in the field of organismal biology that needs to be addressed. Leulier et al. propose an integrative framework within which to define the study of the diet-physiology-microbiota systems and ultimately link it to human health.
Physiology is the scientific discipline that focuses on the body’s capacity to regulate its internal environment by studying individual organ functions, organ to organ communication and systemic regulation. 160 years ago, Claude Bernard proposed the concept of an “internal milieu” ensuring the stability of organ functions whereby external variations are compensated for and equilibrated (Gross, 1998). This notion led to the emergence of the concept “physiological homeostasis”, in which organic processes regulate the maintenance of steady states in the body (Cannon, 1932). It was later recognized that signals arising from the changing environments where animals live interact with the body’s various homeostatic systems, ultimately resulting in different physiological and biological outcomes (Bartholomew, 1986; Schmidt-Nielsen, 1997). Among such environmental factors, nutrition is paramount in shaping all aspects of biology, from cellular and physiological processes to behavioral and ecological interactions (Simpson and Raubenheimer, 2012). In addition, the influence of the intestinal microbial communities, or the microbiota, on animal physiology and behavior is now widely recognized (Fraune and Bosch, 2010; McFall-Ngai et al., 2013; Sommer and Backhed, 2013). Since diet and nutrition shape not only animal physiology but also the ecology of the gut microbiota, it raises the question of how the nutritional environment and microbial assemblages together affect animal physiology. These relationships are made more complex by the fact that the microbiota are themselves part of the animal’s nutritional environment, serving as both a supply and a drain on the host’s nutrition, thereby potentially altering the host’s feeding behavior and physiology directly and indirectly. This research field is still in its infancy and we believe it constitutes a new frontier in the field of organismal biology in need of a unifying vision and guidelines.
In this context, a conference was organized from April 25–29th 2016 hosted by the “Fondation des Treilles” at the “Domaine des Treilles” in Tourtour, France. We discussed the emerging links and interdependencies among the topics of animal nutrition, microbiota, growth, metabolism and health. At the meeting we explored a vision for a research program in integrative physiology that aims to define and delineate the complex interactions comprising the “host physiology - nutrition - microbiota” axis. The research field studying these interrelationships is relatively young, but some impressive progress has been made at the level of phenomenological studies. We recognize the need for sophisticated descriptions of such phenomena, and for an increasing emphasis on underlying mechanisms to propel the field from correlational observations to causal links.
We believe the use of model organisms has played, and will continue to play, a decisive role in this challenge. Animal models have long enabled us to identify the shared biological functions among living organisms, and facilitated the discovery of conserved molecular mechanisms governing the fundamental principles of biology. While discussing the relevant angles in integrative physiology, we reached the consensus that not all models are suitable for all biological questions, and therefore emphasized the importance of choosing the appropriate model organism to study a given biological question (Figure 1). We also noted the feasibility of applying research findings from one model organism to another, as part of a coherent translational pathway leading to human health applications. In this Perspective, we first review recent studies using established animal models and consider future prospects for the use of such model organisms; next we present new approaches in human studies to discover mechanisms underlying the relationships between nutrition, physiology and the microbiota (Figure 2). Finally, we propose an integrative framework derived from nutritional ecology within which to define the study of the diet-physiology-microbiota system and ultimately to link it to human health and precision nutrition (Figure 3).
Figure 1: Integrative physiology:
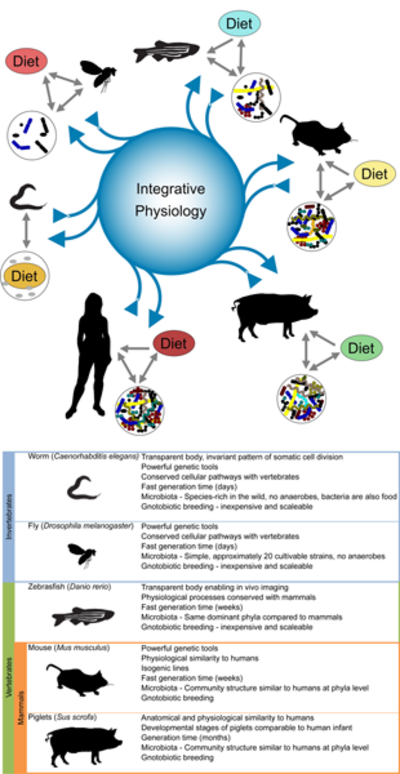
From model systems to human to models. The nutritional environment and microbial assemblages together influence animal physiology.
Figure 2: “Bedside to Bench to Bedside”-.

Strategy for dissecting the role of the gut microbiota in human health and disease.
Figure 3:
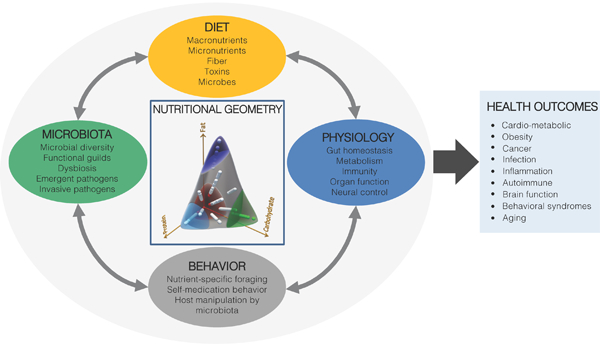
An integrative framework linking nutritional ecology to diet-physiology- behavior-microbiota interactions and health.
The bacterivorous worm
Caenorhabditis elegans has proven instrumental in delineating conserved genetic pathways, characterizing novel genes and identifying novel gene functions. The simple body plan, invariant pattern of somatic cell division and transparent body allows for the characterization of many phenotypes without the need for dissection or extensive sample processing. Although evolutionarily distant to humans, C. elegans and humans share many conserved cellular pathways. Notably, Insulin and TOR pathways function in C. elegans, as in other organisms, to coordinate nutrient and metabolic state with cellular processes (Long et al., 2002; Ogg et al., 1997).
For C. elegans, bacteria are both diet and microbiota (Figure 1). C. elegans are bacterivores that can survive on a variety of bacterial species. These bacteria influence metabolism, life history traits, and gene expression (Coolon et al., 2009; MacNeil et al., 2013; Samuel et al., 2016; Shtonda and Avery, 2006; Soukas et al., 2009). However, not all bacterial strains can colonize the animal. The microbiome of wild C. elegans is largely composed of Proteobacteria but includes a phylogenetically diverse set of microorganisms (Dirksen et al., 2016). Sampling of animals from different geographical locations revealed a common core microbiota that differs in bacterial representation from surrounding soils, and from the microbiota of related species of Caenorhabditis, C. remanei isolated from the same sites (Dirksen et al., 2016). Similarly, a core microbiota, and two identifiable enterotypes, were established in laboratory strains of C. elegans exposed to diverse laboratory-generated bacterial environments (Berg et al., 2016). Altogether, these studies demonstrate that colonization is not determined solely by the prevalence of bacterial species in the environment, but is a guided process that is likely driven by the interaction of host and bacterial factors.
As with many other organisms, co-evolution of C. elegans with its microbiota likely resulted in the generation of interspecies relationships that are advantageous for the host. For instance, C. elegans does not produce nitric oxide (NO), owing to the fact that NO synthase genes are absent from the C. elegans genome. However, NO produced by Bacillus subtilis functions in C. elegans to enhance lifespan and stress response (Gusarov et al., 2013). While NO is not required for the viability of C. elegans, colonization of the gut by B. subtilis provides a distinct advantage to the animal.
C. elegans offers many advantages in studying the integration of microbiota, diet and metabolism. One of the biggest challenges in this endeavour is complexity; necessitating simple models that can be used for high-throughput screening and that would enable genetic modification of host and microbe. C. elegans are propagated as hermaphrodites, have a large brood size and a short generation time (3 days). As a result, large numbers of isogenic animals can be generated. C. elegans offers a unique advantage in that the animals inhabit the bacterial lawn and thus are continuously exposed to bacterial products even without colonization of the gut. Lastly, C. elegans can be made germ-free using a hypochlorite treatment for egg collection, thus eliminating the complication of pre-existing microbiota that occurs in other organisms.
As with any system there are limitations. The C. elegans microbiome differs significantly from the human microbiome. Bacteria identified in the wild C. elegans microbiome are predominantly aerobes or facultative anaerobes, suggesting that the colonized C. elegans intestine is an aerobic environment, which may not support growth of anaerobes.However, the fundamental principles that can be derived from studying C. elegans microbiome will likely shed light on biological concepts conserved in others animal- microbiome interactions.
The commensal fly
Drosophila melanogaster has a long and illustrious career as a model organism to evaluate the host-pathogen interaction, and how such interactions affect host development and physiology (Buchon et al., 2014). For example, the past research in Drosophila led to the identification of a novel role of the Toll receptor in innate immunity, which rapidly catalyzed the discovery of the Toll-like receptor in humans (Lemaitre, 2004). Now, this simple and genetically tractable model animal is actively contributing to the understanding of ‘animal-gut microbiota-nutrition’ interactions (Erkosar et al., 2013; Lee and Brey, 2013). In addition to its powerful genetic toolkit, the fly model offers the decisive advantage of having a simple and culturable microbiota comprising approximately 20 strains mainly from the families Acetobacteraceae and Lactobacillaceae. Such low diversity makes it easier not only for analyzing the changes in commensal gut community membership, but also for identifying host and/or nutritional factors capable of influencing commensal community structure (Figure 1). Although most Drosophila gut commensal species are distinct from those of humans, members of the Lactobacillaceae family have conserved probiotic effects from flies to humans. Therefore, the observations from a Lactobacilli-Drosophila interaction model are directly relevant for understanding the underlying mechanistic events of probiotic effects in more complex vertebrate models, including humans. Indeed, it has been found that upon chronic undernutrition, specific strains of Lactobacilli sustain juvenile Drosophila growth by boosting insulin-like peptides (ILPs) production (Storelli et al.,2011), in part via enhancing the host’s capacity for dietary protein digestion and uptake of amino-acids (Erkosar et al., 2015). Moreover, the same Lactobacillus strain can also partly restore the growth dynamics of chronically undernourished mono-colonized mice by stimulating Insulin-like Growth Factor-1 (IGF-1) activity (Schwarzer et al., 2016). These observations suggest that some Lactobacilli exert their probiotic effects by modifying the host’s hormonal and nutritional status, and the mechanism behind such beneficial effect has an ancient evolutionary origin. With the demonstration that Lactobacilli-Drosophila study can be readily translated into a mammalian model, further investigations in flies will provide a unique opportunity to evaluate such evolutionarily conserved mechanism and extend such studies to discover how specific commensal strains influence diverse aspects of animal physiology. From a therapeutic point of view, evaluating the strain-specific beneficial effects of Lactobacilli also provides a possible explanation on why some bacterial strains confer more potent beneficial effects on health than others.
An added advantage of the fly is that most Drosophila commensal bacteria can be cultured and genetically manipulated in vitro (e.g., for generating random mutant library or performing targeted mutagenesis), which greatly facilitates the identification of the specific functionalities of the microbiome that can be attributed to a corresponding host physiological trait. For example, by combining the facile production of gnotobiotic flies with a mutant library of a specific gut microbiota member, large-scale genetic screens can uncover the specific microbiome requirement for an animal’s development and host physiology. Subsequent functional studies of these candidates will yield the molecular mechanisms by which the microbiota impacts host physiology in a microbiome-specific and possibly also in a metabolome-specific manner. Using this methodological concept, Shin et al. (2011) successfully discovered an Acetobacter pomorum gene product that is essential to sustain animal growth during severe undernutrition. By individually monoassociating 3000 individual clones of an Acetobacter pomorum mutant library to Drosophila germ-free larvae, the authors identified the periplasmic pyrroloquinoline quinone (PQQ)-alcohol dehydrogenase-dependent electron transport system that is required for A.pomorum to mediate systemic activation of insulin signaling leading to larval growth promotion. Using a similar approach, Chaston et al. monoassociated gnotobiotic flies with different commensal strains and conducted a metagenome-wide association study. The outcome linked specific microbiome functions to microbiota- modulated host traits, e.g. bacterial PQQ system for larval growth promotion, and the glucose oxidation pathway enzymes for reduction of lipid storage in the host (Chaston et al., 2014).
However, to effectively study the physiological consequences of host-microbiota interaction, it is critical to also account for the contributions from the host’s dietary and nutritional environment. In Drosophila, it is long established that nutrition profoundly alters different physiological parameters in the fly, such as metabolic homeostasis, longevity and immune competence (Baker and Thummel, 2007; Tatar et al., 2014). The presence or absence of the gut microbiota adds another layer of complexity to this line of investigation. For example, in the presence of varying glucose and yeast concentration in the diet, germ-free flies and their conventional or gnotobiotic siblings display drastically different larval growth rate, developmental timing and survival (Shin et al., 2011; Storelli et al., 2011; Wong et al., 2014). To pinpoint the molecular mechanism behind these observations, recent fly studies have revealed that the microbiota directly or indirectly (e.g., by modulating host digestive ability) alters the quality of nutrients (Erkosar et al., 2015; Wong et al., 2014), so that the host senses and responds differently to nutrition in the presence of microbial influences. Furthermore, a specific nutrient itself may promote or hamper the growth of certain microbiota members, thereby modify the community structure and function, and may in turn affect host physiology. These microbiome-modified or -derived nutrients are sensed in distinct organs (e.g., fat body, intestine and brain), which initiate complex organ-to-organ communication, resulting in a systemic regulation via hormones. However, deconstructing the tripartite interaction among the host, gut microbiota and nutrition, and studying the inter-organ signaling pathways in such context are a complex and fledgling field. It is still unclear how the host senses nutritional-microbial environments and integrates such environmental input into a genetic program to shape its physiology. In this regard, Drosophila gnotobiotic models can play a vital role in evaluating such complexity. First, Drosophila is rapidly recognized as a powerful model to study inter-organ communication, as it is equipped with most of the vital organs that work in concert to regulate metabolic homeostasis and systemic growth (Boulan et al., 2015; Droujinine and Perrimon, 2016). As an example, at this meeting, Pierre Leopold presented an elegant study showing that stunted, a larval fat-body derived ligand, circulates in the hemolymph, is uptaken up by the Insulin Producing Cells (IPCs) in the brain by methuselah, a G-protein coupled receptor (GPCR), and hence triggers Dilp2 release from the IPCs to control systemic growth during the well-fed state (Delanoue et al., 2016). Similar fly studies have already uncovered different inter-organ circuits regulating host physiological responses in different nutritional context; the next challenge is to integrate the microbial contribution to such interaction. Using gnotobiotic flies, large-scale genetic screens and specific functional analyses of single genes can be carried-out in a tissue- specific manner, while synchronized with precise timing and strict control over nutritional and microbial inputs. Such controlled experimental system will provide novel insights into complex ‘animal physiology-gut microbiota-nutrition’ interactions.
The pathophysiological zebrafish
The zebrafish (Danio rerio) is emerging as a powerful vertebrate model for integrative physiology. The same advantages of the zebrafish that led to its success as a model for developmental genetics and toxicology over the past four decades (Grunwald and Eisen, 2002) have recently been leveraged to explore physiological processes. These include its small size, high fecundity, rapid external development, and amenability to in vivo imaging of physiologic processes in a transparent vertebrate. Zebrafish physiology and metabolism display extensive conservation with humans and other mammals. This permits the zebrafish to effectively model key pathophysiological processes involved in human diseases such as inflammatory bowel disease, insulin resistance, diabetes, hepatic steatosis, dyslipidemia, atherosclerosis, and obesity (Hill et al., 2016; Marjoram and Bagnat, 2015; Renshaw and Trede, 2012; Schlegel and Gut, 2015). In addition to identification of novel disease genes and therapies, these models can be used to define the physiologic function of genes implicated by genome-wide association studies in humans (Minchin et al., 2015).
Within this evolving context of zebrafish as a model system for vertebrate physiology, researchers are beginning to explore the impact of nutrition. Studies in adult zebrafish have identified tissue-specific responses to starvation (Drew et al., 2008; Tian et al., 2015) and feeding (Seiliez et al., 2013), and the impact of diet composition on growth, metabolism, inflammation, and behavior (O’Brine et al., 2015; Powell et al., 2015; Smith et al., 2013a). Interestingly, adult phenotypes can also be influenced by nutritional exposures during earlier larval stages (Perera and Yufera, 2016; Rocha et al., 2015). The features of the zebrafish model are well suited to define mechanisms underlying short- and long-term physiologic impact of nutritional exposures. For example, over-nutrition using glucose or chicken egg yolk feeding was found to stimulate pancreatic beta-cell expansion through mTOR and insulin/IGF-1 pathways respectively (Maddison and Chen, 2012) . To define roles of nutrition on microbiota and host physiology, it will be important to continue improving our understanding of zebrafish nutritional requirements, to establish standardized open-formulation diets, and to improve reporting standards in the scientific literature for diet composition and post-embryonic staging (Watts et al., 2016).
Similar to other animal models, the study of the zebrafish microbiota has been empowered by advances in high-throughput sequencing and development of gnotobiotic methods. 16S rDNA sequence-based studies of bacterial communities in the zebrafish intestine have identified taxa that vary as a function of host development (Burns et al., 2016; Rawls et al., 2004), domestication status (Roeselers et al., 2011), starvation (Semova et al., 2012), and dietary fat level (Wong et al., 2015b). Compared to humans and other mammals, zebrafish gut bacterial communities reach similar levels of phylogenetic diversity with shared dominant phyla, but with little overlap at shallower taxonomic levels (Hacquard et al., 2015; Rawls et al., 2006) (Figure 1). An important frontier in the field is to define the factors determining gut microbiota composition, including the relative contributions of selective and neutral processes. Recent studies found that the influence of selective processes on zebrafish gut microbiota composition increases as hosts age (Burns et al., 2016), prompting studies into underlying physiologic mechanisms.
Establishment of gnotobiotic husbandry methods in zebrafish has empowered analysis of microbial ecology and the influence of microbiota on host physiology (Pham et al., 2008). High-resolution in vivo imaging of gnotobiotic zebrafish colonized with defined communities of fluorescently labeled bacteria has revealed complex behaviors and spatial organization within the intestinal lumen (Jemielita et al., 2014; Rawls et al., 2007; Stephens et al., 2015), and identified host intestinal motility as an important factor governing microbiota composition (Wiles et al., 2016). Initial comparisons of germ-free zebrafish to those colonized with conventional microbiota revealed diverse roles for microbiota on host physiology (Bates et al., 2006; Rawls et al., 2004). Many of the host responses to microbiota in zebrafish are shared with humans and other animals, including stimulation of intestinal epithelial renewal (Cheesman et al., 2011; Rawls et al., 2004), enhanced harvest and metabolism of dietary nutrients (Semova et al., 2012), behavior (Davis et al., 2016), and stimulation of innate immune effectors (Bates et al., 2007; Galindo-Villegas et al., 2012; Kanther et al., 2011; Kanther et al., 2014). Interestingly, microbiota from zebrafish and mouse can evoke many of the same responses in zebrafish hosts, suggesting that the respective microbial stimuli may be shared features of these distinct microbial communities (Rawls et al., 2006). Colonization of gnotobiotic zebrafish with simple defined bacterial communities revealed that specific host responses can be determined by community composition, but that the relative abundance of a community member does not necessarily predict its relative impact (Rolig et al., 2015). Gnotobiotic zebrafish can be used to discover novel microbial products that control important aspects of host physiology, as recently demonstrated in the identification of a conserved bacterial protein that promotes pancreatic β-cell number (Hill et al., 2016). Evidence for microbial control of zebrafish physiology is also derived from studies in which administration of probiotic bacteria to zebrafish altered behavior, growth bone and metabolic biomarkers (Borrelli et al., 2016; Falcinelli et al., 2015).
Physiology, nutrition, and microbiota are still relatively new fields of study for the zebrafish model, however the intersection of these fields is already ripe for exploration. For example, in vivo imaging of fluorescently labeled dietary fatty acids (FA) fed to gnotobiotic zebrafish revealed that microbiota stimulate absorption of dietary FA in a feeding-dependent manner. Feeding enriched the Firmicutes members of the microbiota as compared to starved animals, and a single Firmicutes member (Exiguobacterium acetylicum ZWU0009) was sufficient to stimulate increased FA absorption (Semova et al., 2012). A fundamental challenge in integrative physiology is to define microbial factors that shape microbial ecology and host physiology. For members of the microbiota that can be cultivated, this can be addressed through genetic manipulation. Transposon mutagenesis has been successfully deployed in γ-Proteobacteria members of the zebrafish microbiota, and used to identify genes required for colonization in vivo (Stephens et al., 2015). However, the majority of microbes that colonize the vertebrate intestine are not amenable to genetic manipulation due to a lack of methods for DNA transformation, insertional mutagenesis and transgenesis. To address this challenge, a recent study combined chemical mutagenesis, phenotypic selection and population- based genome sequencing in E. acetylicum to identify genes associated with motility (Bae et al., 2016). Further refinement and application of these genetic approaches can be expected to lead to new insights into bacterial mechanisms involved in integrative physiology in zebrafish as well as other animals.
The integrative mouse
Mice and humans shared a common ancestor approximately 65–110 million years ago (Emes et al., 2003). As a consequence up to 99% of the mouse genes have a counterpart in the human genome (Waterston et al., 2002). Thanks to its genetic and physiological similarities to humans and its rapid and prolific breeding, mouse has been a classic mammalian model of choice for the past hundred years. Since the establishment of the first inbred DBA line in 1909, over 450 isogenic mouse strains have been derived and characterized over the past century (Beck et al., 2000). The purity of the isogenic background, combined with the ever-advancing technologies such as the inducible tissue and cell specific gene deletions, enabled investigators to generate multitudes of transgenic and knock-out mouse models to study virtually every aspect of mammalian development, physiology and disease aetiology (Danielian et al., 1998; Krupke et al., 2008; Sellers et al., 2012). In this context the Knock-out Mouse Project (KOMP) and International Mouse Phenotyping Consortium (IMPC) propose to obtain and characterize a mutant of each of the 20,000 known and predicted mouse protein coding genes, in attempt to characterize each gene’s biological functions at large scale (Austin et al., 2004; Brown and Moore, 2012). Among the mouse models, gnotobiotic mice derived from isogenic strains deserve special attention for their instrumental contribution to the understanding of mammalian physiology. The first long term germ-free mouse colonies were established after the Second World War (Reyniers, 1959). Currently, genetically manipulating axenic animals and their gnotobiotic counterparts while varying environmental cues such as the diet, temperature and microbial input represents a powerful and indispensable approach to study integrative physiology. As an example, MyD88 is an adaptor protein required for TLRs signalling upon detection of microbial molecules. Tissue-specific deletion of MyD88 in the intestinal epithelial cells or in distant organs such as the liver revealed a new role for TLRs signalling in diet-induced obesity and metabolic disorders (Duparc et al., 2016; Everard et al., 2014).
Besides genetics, environment is a key factor affecting mammalian physiology. In the last decade the influence of the microbial environment, and in particular the intestinal microbiota on mammalian biology has become evident (Shanahan, 2012). Mice and humans both harbour complex gut microbiota with similar community structure at the phyla level (Nguyen et al., 2015); most metabolites produced by the mouse gut microbiota are also found in humans (Xiao et al., 2015). This level of conservation, together with the ability to generate gnotobiotic mouse models made it possible to dissect the specific interactions amongst the intestinal microbiota, environmental fluctuations and the host genetic makeup (Macpherson and McCoy, 2015; Tlaskalova- Hogenova et al., 2011). This line of investigation yielded novel discoveries of the microbiota-related pathophysiology in several human diseases such as Inflammatory Bowel Syndrome (Manichanh et al., 2012), arthritis or colitis (Stepankova et al., 2007; Wu et al., 2010), metabolic syndromes such as obesity and diabetes (Backhed et al., 2007), and more recently, childhood undernutrition (Blanton et al., 2016a). Specifically, Smith at al. used germ-free mice to show that susceptibility to acute undernutrition can be transmitted by microbiota isolated from human monozygotic twins discordant for kwashiorkor (Smith et al., 2013b). Further, recent papers have highlighted the importance of microbiota and specific bacterial strains for the mammalian juvenile growth (Blanton et al., 2016b; Schwarzer et al., 2016).
Along these lines, future research in integrative physiology would benefit from the proposed establishment of isobiotic mouse lines, namely mouse lines harbouring complex microbiota with defined composition resistant to immigrating strains, so that the physiological status would be the same as that of a mouse colonised with a diverse microbiota (Macpherson and McCoy, 2015). Efforts are already underway to establish a comprehensive collection of mouse gut bacteria with associated genome sequences. The preliminary data suggest that a minimal consortium of 18 strains can cover up to 75% of the known functional potential of the mouse microbial metagenome (Lagkouvardos et al., 2016).
While the presence of gut microbiota has been shown to be important for many aspects of host physiology, such presence is far from static. Indubitably, the composition and the metabolism of the intestinal microbiota are dependent on the diet (David et al., 2014). For example, high-fat diet alters the gut microbiota composition, which in turn induces low-grade inflammation via intestinal leakage of lipopolysaccharides (LPS), a phenomenon called metabolic endotoxemia (Cani et al., 2007). This is one of the first studies that mechanistically links diet-induced changes in gut microbiota with the onset of inflammation and diabetes. Later, these findings were further confirmed in extended human studies (Lassenius et al., 2011; Pussinen et al., 2011). Thus nutrition also influences host physiology indirectly, by inducing changes in the microbiota (For review see: (Tremaroli and Backhed, 2012)). However, we may not rule out that dietary fats influence the gut microbiota via indirect mechanisms, such as by changing the bile acid flux and release into the intestinal lumen and thereby affecting the gut microbiota composition (Duparc et al., 2016). Recent evidence also suggests that vitamin D directly impact on the production of antimicrobial peptides and immunity which in turn may affect the gut microbiota composition (Su et al., 2016). Evidence of the rapid changes that the microbiota experiences upon diet change also comes from gnotobiotic mice colonized with complete human microbiota. Shifts from a fibre rich diet to a high fat diet changes the relative abundance of different taxa as well as the microbiome transcriptional responses (Turnbaugh et al., 2009). Gnotobiotic mice can be further helpful in elucidating how different bacteria resembling simplified human microbiota respond to defined diet with variable ratios of protein:fat:sugars (Faith et al., 2011). As a result, the obtained data can be used to create models predicting the behaviour of more complex intestinal microbial systems, possibly leading to future nutritional therapies to revert a dysbiotic microbiota to healthy configuration in humans.
Taken together, the physiological similarity between mice and humans, the ability of genetic manipulation and the feasibility of gnotobiotic breeding enable integrative studies of genes-diet-microbiota interactions (Figure 1). These advantages make mice an attractive model in the field of integrative physiology for the understanding of human health and disease.
The human-microbiota-associated piglet
As described above, murine models have made significant contributions in the field of gut microbiota. These mammalian models are attractive because they are cost-effective and relatively easy to manage on a large scale, yet they are limited by several important physiological, immunological and metabolic differences from humans. Rodent and other animal models sometimes do not faithfully replicate the clinical manifestations of human diseases. As a model for understanding gut microbial ecology and host physiology, piglets have unique advantages: first, piglets undergo developmental stages comparable to that of human infants (i.e., the US Food Drug and Administration defined four stages in early human development which are telescoped into the first 6.5 months of postnatal swine life (Garthoff et al., 2002)); they have a human-sized omnivorous dietary regimen with comparable nutritional requirements; their digestive system is very similar to that of humans in terms of anatomy, histology and physiology; their immune system closely resemble that of humans; and finally, piglets manifest similar disease symptoms and progression as humans when suffering diabetes, atherosclerosis, hypertension and necrotising enterocolitis (NEC) (Heinritz et al., 2013). These traits place the piglets in an ideal position to study related human conditions influencing the interactions between nutrition, microbiota and host physiology (Figure 1).
In addition, the recent generation of human-microbiota associated (HMA) piglets makes this model particularly attractive. HMA piglets were established by inoculating fresh fecal sample preparations of a healthy ten-year-old boy to neonatal piglets delivered by Caesarean section and housed in a temperature and humidity-controlled, air-filtrated barrier-system facility. It was demonstrated that the bacterial composition of HMA piglets was highly homologous with that of their human donor, but significantly different from that of conventional piglets. Two predominant groups - Bacteroides and Bifidobacteria successfully colonize HMA piglets (Pang et al., 2007). The intestinal immunity of HMA piglets is well developed (Che et al., 2009) and the metabolomic features of their colonic content revealed by 1H NMR more closely resemble those of humans (Shen, 2008). This work established a novel HMA piglet model, which exhibited high similarity of physiology, metabolism and gut microbiota to humans. HMA piglets were used to study the effects of the prebiotic ingredient fructo-oligosaccharides (FOS) on the gut microbiota and host metabolism (Shen et al., 2010). In parallel, it was found that HMA piglets showed high sensitivity to opportunistic pathogens in donor’s gut (Wei et al., 2008), which suggests its potential application in the safety evaluation of fecal inoculum used in “fecal microbiota transplantation” for humans. Thus, the HMA piglets are now introduced to screen the efficacy of pre- and probiotic interventions (Wen et al., 2014). Furthermore, HMA piglets studies can confirm phenotypes caused by specific microbiota configurations observed in gnotobiotic mice. For example, special HMA piglet models associated with artificial bacterial consortium cultured from infant stools were introduced to study the impact of microbiota and sialylated milk oligosaccharides on infant growth patterns (Charbonneau et al., 2016). Thus, HMA piglets have the potential to be a robust model not only for investigating how the gut microbiota composition changes in response to environmental factors, such as age, diet, antibiotic use and infection, but also for elucidating the interactions between host, diet and gut bacteria in human health and disease (Wang and Donovan, 2015).
The multiorganismal human
Human physiology can be seen as the integration between functions encoded in human genome and the microbiome, particularly the gut microbiome (Qin et al., 2010; Zhao, 2013). Nutrients taken into the digestive system are partitioned between human cells and gut bacteria. The gut bacteria rely on non-digestible or un-digested nutrients from the diet together with mucin and sloughed cells locally produced in the gut to maintain their population levels (Flint et al., 2012; Marcobal et al., 2013). Metabolically active gut bacteria can impact host physiology and physiopathology by delivering various bioactive compounds into the systemic circulation (Nicholson et al., 2012; Wikoff et al., 2009).
To understand and dissect the contribution of gut microbiota to human health, a top- down, systems approach may be employed (Zhao and Shen, 2010) (Figure 2). In this strategy, blood, urine and fecal samples served as three windows for monitoring and measuring health dynamics of the superorganism human hosts at both the personalized level and cohort level. Non-targeted, multi-omics technologies can be used to profile molecular variations in these samples along time or upon interventions. Multivariate statistics can help correlate variations of gut microbiota with changes of host health phenotypes, such as inflammatory cytokines for immunity, urine metabolites for metabolism. In this way, key functional bacteria, whose population shifts are associated with changes of immunity or metabolism, can be isolated into pure culture or defined consortium. The isolates can be inoculated into various model organisms, from worms, to flies, to mice, to piglets for reproducing relevant disease/health phenotypes. Such established models, which recapitulate some of the relevant superorganism phenotypes, can facilitate molecular mechanistic studies, eventually leading to a thorough understanding of the molecular chain of causation from the active colonization of these key functional bacteria in the gut to the eventual development of disease/health phenotypes of the superorganism hosts. Nutritional factors, which may drive population changes of key functional gut bacteria and thus modulate molecular cross-talk between host and gut microbiota, can be evaluated and identified via such a strategy. This strategy follows the logic of Koch’s postulates for identifying members of the gut microbiota, which may make causative contributions to host physiology or physiopathology (Zhao, 2013). It can be used for individual case studies or for following a cohort of people throughout an intervention. For example, in one case study, a population of the opportunistic pathogen Enterobacter spp. was reduced from more than 30% of the total gut bacterial population to almost non-detectable soon after the human host followed a dietary intervention aiming to change the gut microbiota. The volunteer lost 51.4 kg from the 175kg initial bodyweight over 23 weeks on the intervention. One strain Enterobacter cloacae B29 was isolated from his gut and found to induce obesity when mono-associated with germfree mice, indicating that this strain may contribute causatively to the extreme obesity phenotype of the host (Fei and Zhao, 2013). (Koeth et al., 2013; Wang et al., 2011)(Zhang and Zhao, 2016)
In a clinical trial on nutritional intervention of genetically obese children with Prader- Willi syndrome, high complex carbohydrates in the diet induced significant changes of the gut microbiota with concomitant weight loss and alleviation of metabolic deteriorations (Zhang et al., 2015). NMR-based metabolomic analysis of the urine samples showed a significant decrease of TMAO concentration. TMAO is a putative risk factor for atherosclerosis, generated by oxidation of TMA a metabolite converted from dietary choline by gut bacteria (Koeth et al., 2013; Wang et al., 2011). 118 high quality draft genomes of bacteria shared by more than 20% of the samples were directly assembled from metagenomic datasets of 110 time-series fecal samples via the canopy- based algorithm. 31 of the 118 genomes were positively correlated with urine concentration of TMAO. 13 of the correlated genomes were found to encode the two genes needed for converting choline into TMA (Craciun and Balskus, 2012). These 13 genomes would represent candidate strains as the key functional bacteria possibly contributing to the risk of atherosclerosis in humans. The next step would be to isolate these strains based on their nutritional requirements for growth, which can be deduced from their genomes. The isolated candidate strains can be mono-associated in the gut of an appropriate animal model such as germfree ApoE knockout mice or germ-free piglets. Dietary availability of choline can be adjusted to demonstrate the mechanistic link between choline-TMA conversion and plaque formation in host’s blood vessels. Genetic manipulation of the strains for knocking-out their choline-TMA conversion genes can also be used to confirm their causative role in development in host’s atherosclerosis.
Thus, despite the sheer complexity of the integrated physiology between human host and the gut microbiota, we propose that the combined use of strain-level metagenomics analysis and metabolite-specific metabolomic profiling will pinpoint specific genomes representing gut bacterial strains, which may mechanistically contribute to a particular disease phenotype in the host. Reproduction of the disease phenotype(s) by colonizing germ-free animals with pure isolates of these genomes under appropriate nutritional conditions can validate their causative role in the nutrition-related pathology and provide an ideal model for elucidating the molecular mechanisms involved (Figure 2). Only after such rigorous causative studies, these bacterial genomes may become both biomarkers for prediction and diagnosis and targets for developing new therapies.
It should be noted that the gut microbiota consists of not just bacteria but also fungi, viruses and parasites. Metagenomic sequencing of total DNAs from fecal samples can capture the genomic level changes of all the members of the gut microbiota. Given appropriate bioinformatic and multivariate statistic tools, interactions among all these members and between their hosts can be evaluated to provide a whole picture of how the gut microbiota variations may impact host physiology or physiopathology. This top- down, systems strategy can significantly facilitate our understanding on how nutritional modulation of the gut microbiota can contribute to the health recovery and maintenance of human hosts.
From model organisms to an integrative framework for human health
The relationship between diet, animal physiology and gut microbiota is fundamental to health, and the organic processes of host physiology, behavior and microbiota are entwined to such an extent that they can effectively be considered parts of the same homeostatic system. Understanding the processes and mechanisms that underlie these relationships holds great promise to improve human health - and gaining such understanding requires extensive research in model animals.
We have briefly reviewed some of the major model organisms, considering their relative advantages and disadvantages as experimental systems and their strengths and limitations as models for human health (Figure 1). Clearly, no model is universal - in fact, they are not necessarily even typical representatives of their own taxa. For example, Drosophila is in many respects an atypical insect, and inbred laboratory mouse lines represent only part of the genetic diversity found in the wild. There are caveats to using model organisms, of course, including the danger of over-extending the model analogy by treating species as if they were more human than they are - a form of physiological anthropomorphism. Examples here include testing diet formulations that are beyond the realms of the natural environment for the model species, or imposing unrepresentative physiological tests. Another potential problem comes from presuming that phenomena and their underlying mechanisms are conserved (homologous), when in fact they are independently derived and analogous (Bolker, 2012). Gut bacteria and their hominid hosts have co-evolved over millions of years (Moeller et al., 2016). This co-evolution has likely driven the formation of interspecies relationships between humans and bacteria that do not exist in other animals. Similarly, non-human animals may have evolved interspecies relationships that do not exist in humans. These relationships may promote colonization, inhibit activation of immune response, or result in optimized nutrient sharing between organisms. In using non-human models to study human-associated bacteria, we must consider that some host-specific effects will be lost and that others, not reflecting the human response, may be gained.
No single model is likely to recapitulate every aspect of human-physiology-diet- microbiota interactions. However, experimental programs integrating several model species can forge a ‘consilience of inductions’ (Whewell, 1840; Wilson, 1998), strengthening the case for testing mechanistic hypotheses in humans. Extending this multi-species approach beyond the standard model systems may offer yet further opportunities, by exploring phylogenetic and ecological patterns that can yield unexpected breakthroughs in the study of human health. This principle is well illustrated by the field of nutritional ecology, where comparative analysis of a wide range of species has illuminated processes in human regulatory physiology and appetite control that predispose the subjects to obesity and metabolic disease in the modern world (Raubenheimer and Simpson, 2016). A corollary is that species-specific solutions to ecological challenges, even though not directly relevant to humans, may provide new understanding for humans by illustrating how such problems have been solved by evolution (Bolker, 2012).
Taming the complexity of nutrition
Central to any investigation of the diet-host-microbiota system is defining the nutritional environment. Host organisms must acquire multiple nutrients in appropriate amounts and proportions to perform optimally. Failure to attain dietary balance has profound consequences for health, growth and reproduction, with these effects reverberating across all levels in the hierarchy of biological organization, from molecular and cellular processes to physiological and behavioral responses - and beyond the organism to shape group dynamics, trophic interactions and ecosystem function (Simpson and Raubenheimer, 2012). The same nutritional imperatives face the microbiota, members of which gain their primary nutrition from food ingested by the host or from host-derived secretions, and may in turn provision the host with metabolites and nutrients. The interests of the host and microbiota need not align, however, nor need those of the different species comprising the heterogeneous microbial assemblage within the gut, setting up the potential for competition, evolutionary conflict, the emergence of pathogenicity among otherwise commensal bacteria, behavioral manipulation of the host, and self-medication by the host (Ponton et al., 2011; Wong et al., 2015a).
Despite the fundamental importance of nutrition to the animal-microbiota association, traditional approaches in nutrition science have usually been “one variable at a time”, focusing on the roles of single dietary components rather than the entire mixture, thereby not taking account of the inter-dependence of nutrients and other dietary constituents (fiber, toxins, microbes, etc.) within diets and their interactive effects on biological outcomes (Raubenheimer and Simpson, 2016). An integrative framework for considering such interactions has been provided by Nutritional Geometry (NG), which uses graphical models to: a) quantify changing requirements for multiple, potentially interacting macro- and micro-nutrients and other dietary constituents; b) demonstrate how animals select foods, control food intake and utilize ingested nutrients to attain those requirements, and c) map as response surfaces the consequences of different multi-dimensional nutrient intakes on multiple response measures, across scales from molecular to ecological (Simpson and Raubenheimer, 2012).
Defining the nutritional phenotype
Nutritional Geometry offers a means to define the nutritional phenotypes of both host and microbiota, and to integrate these within a single experimental and conceptual framework. For example, NG has been used in flies and mice to show that macronutrient intakes (amounts, quality and balance) profoundly impact appetite, growth, reproduction, aging, cardio-metabolic health, obesity and immune function (Solon-Biet et al., 2016; Solon-Biet et al., 2014; Solon-Biet et al., 2015). NG has also been used to map the microbial ecology of the gut in flies and mice as a function of macronutrient intakes (Holmes et al., 2016; Ponton et al., 2015). Such mapping allows comprehensive associations to be made between diet composition, microbial assemblage structure and multiple host health outcomes; and to help define community diversity, functional guilds and keystone species among bacterial taxa as related to ingested and host-derived sources of nutrients (Holmes et al., 2016). As a result, concepts such as microbial ‘dysbiosis’ and the ‘holobiont’ become more tractable to mechanistic exploration (Holmes et al., 2016; Wong et al., 2015a).
Using Nutritional Geometry in precision nutrition
It follows that Nutritional Geometry also offers a powerful tool for optimizing diet to support specified metabolic and health outcomes for individuals, particular age groups and populations. By mapping the association between intakes of multiple nutrients (macro- and micro-nutrients as well as other dietary constituents) and different measures of health, it becomes possible to identify combinations of intakes that yield a desired outcome, then to translate these intakes into combinations of foods, meals and dietary patterns to achieve the desired outcome. Factors such as individual food preferences, food culture, economic constraints and the environmental impacts of food production can be factored into this translation, given that there are many ways to attain a given combination of nutrient intakes (Raubenheimer and Simpson, 2016). Similarly, aspects such as the digestibility and bio-availability of nutrients can be factored into NG models, as can temporal patterns of feeding and interactions with variables such as ambient temperature and levels of physical activity (Simpson and Raubenheimer, 2012).
A systems approach
We envisage a program of research in which experiments coordinated across multiple model organisms systematically explore nutritional relationships to define the phenotype of the animal-microbiota system (Figure 3). Such descriptions will be used to uncover signature patterns linking diet to health and microbial ecology. These studies will provide new mechanistic hypotheses, which can then be tested using computational systems modeling, gnotobiotic animals, molecular genetics and pharmacological tools.Associational data from human cohorts can be used to seek evidence of the same phenomenological signatures, leading in turn to clinical trials and other experimental interventions. Such a translational pathway, united under an integrating theoretical framework and involving multiple model systems, promises a truly integrative physiology.
Acknowledgements
The authors would like to thank the Fondation des Treilles for hosting us during our seminar and all attendees for fruitful discussions and the pleasant atmosphere of the meeting. F.L. is particularly thankful to the Fondation Schlumberger pour l’Education et la Recherche for its generous support and Dali Ma for critical reading and editing of the manuscript. We apologize to our colleagues whose work cannot be cited due to the space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, et al. (2004). The knockout mouse project. Nature genetics 36, 921–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Manchester JK, Semenkovich CF, and Gordon JI (2007). Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proceedings of the National Academy of Sciences of the United States of America 104, 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Mueller O, Wong S, Rawls JF, and Valvidia RH (2016). Genomic sequencing- based mutational enrichment analysis identifies motility genes in a genetically intractable gut microbe. Proceedings of the National Academy of Sciences of the United States of America 113, 14127–14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KD, and Thummel CS (2007). Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab 6, 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew GA (1986). The Role of Natural-History in Contemporary Biology. Bioscience 36, 324–329. [Google Scholar]
- Bates JM, Akerlund J, Mittge E, and Guillemin K (2007). Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell host & microbe 2, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, and Guillemin K (2006). Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Developmental biology 297, 374–386. [DOI] [PubMed] [Google Scholar]
- Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, and Fisher EM (2000). Genealogies of mouse inbred strains. Nature genetics 24, 23–25. [DOI] [PubMed] [Google Scholar]
- Berg M, Stenuit B, Ho J, Wang A, Parke C, Knight M, Alvarez-Cohen L, and Shapira M (2016). Assembly of the Caenorhabditis elegans gut microbiota from diverse soil microbial environments. The ISME journal 10, 1998–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton LV, Barratt MJ, Charbonneau MR, Ahmed T, and Gordon JI (2016a). Childhood undernutrition, the gut microbiota, and microbiota-directed therapeutics. Science 352, 1533. [DOI] [PubMed] [Google Scholar]
- Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, et al. (2016b). Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolker J (2012). Model organisms: There’s more to life than rats and flies. Nature 491, 31–33. [DOI] [PubMed] [Google Scholar]
- Borrelli L, Aceto S, Agnisola C, De Paolo S, Dipineto L, Stilling RM, Dinan TG, Cryan JF, Menna LF, and Fioretti A (2016). Probiotic modulation of the microbiota-gut-brain axis and behaviour in zebrafish. Scientific reports 6, 30046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulan L, Milan M, and Leopold P (2015). The Systemic Control of Growth. Cold Spring Harbor perspectives in biology 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, and Moore MW (2012). The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mammalian genome : official journal of the International Mammalian Genome Society 23, 632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Silverman N, and Cherry S (2014). Immunity in Drosophila melanogaster-- from microbial recognition to whole-organism physiology. Nature reviews Immunology 14, 796–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, and Bohannan BJ (2016). Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. The ISME journal 10, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et al. (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772. [DOI] [PubMed] [Google Scholar]
- Cannon WB (1932). The wisdom of th ebody. (New York: W.W. Norton & Company, Inc.). [Google Scholar]
- Charbonneau MR, O’Donnell D, Blanton LV, Totten SM, Davis JC, Barratt MJ, Cheng J, Guruge J, Talcott M, Bain JR, et al. (2016). Sialylated Milk Oligosaccharides Promote Microbiota-Dependent Growth in Models of Infant Undernutrition. Cell 164, 859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston JM, Newell PD, and Douglas AE (2014). Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. mBio 5, e01631–01614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che C, Pang X, Hua X, Zhang B, Shen J, Zhu J, Wei H, Sun L, Chen P, Cui L, et al. (2009). Effects of human fecal flora on intestinal morphology and mucosal immunity in human flora-associated piglet. Scandinavian journal of immunology 69, 223–233. [DOI] [PubMed] [Google Scholar]
- Cheesman SE, Neal JT, Mittge E, Seredick BM, and Guillemin K (2011). Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proceedings of the National Academy of Sciences of the United States of America 108Suppl 1, 4570–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon JD, Jones KL, Todd TC, Carr BC, and Herman MA (2009). Caenorhabditis elegans genomic response to soil bacteria predicts environment-specific genetic effects on life history traits. PLoS genetics 5, e1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craciun S, and Balskus EP (2012). Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proceedings of the National Academy of Sciences of the United States of America 109, 21307–21312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, and McMahon AP (1998). Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Current biology : CB 8, 1323–1326. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DJ, Bryda EC, Gillespie CH, and Ericsson AC (2016). Microbial modulation of behavior and stress responses in zebrafish larvae. Behavioural brain research 311, 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoue R, Meschi E, Agrawal N, Mauri A, Tsatskis Y, McNeill H, and Leopold P (2016). Drosophila insulin release is triggered by adipose Stunted ligand to brain Methuselah receptor. Science 353, 1553–1556. [DOI] [PubMed] [Google Scholar]
- Dirksen P, Marsh SA, Braker I, Heitland N, Wagner S, Nakad R, Mader S, Petersen C, Kowallik V, Rosenstiel P, et al. (2016). The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC biology 14, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew RE, Rodnick KJ, Settles M, Wacyk J, Churchill E, Powell MS, Hardy RW, Murdoch GK, Hill RA, and Robison BD (2008). Effect of starvation on transcriptomes of brain and liver in adult female zebrafish (Danio rerio). Physiological genomics 35, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droujinine IA, and Perrimon N (2016). Interorgan Communication Pathways in Physiology: Focus on Drosophila. Annual review of genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duparc T, Plovier H, Marrachelli VG, Van Hul M, Essaghir A, Stahlman M, Matamoros S, Geurts L, Pardo-Tendero MM, Druart Cv et al. (2016). Hepatocyte MyD88 affects bile acids, gut microbiota and metabolome contributing to regulate glucose and lipid metabolism. Gut. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emes RD, Goodstadt L, Winter EE, and Ponting CP (2003). Comparison of the genomes of human and mouse lays the foundation of genome zoology. Human molecular genetics 12, 701–709. [DOI] [PubMed] [Google Scholar]
- Erkosar B, Storelli G, Defaye A, and Leulier F (2013). Host-intestinal microbiota mutualism: “learning on the fly”. Cell host & microbe 13, 8–14. [DOI] [PubMed] [Google Scholar]
- Erkosar B, Storelli G, Mitchell M, Bozonnet L, Bozonnet N, and Leulier F (2015). Pathogen Virulence Impedes Mutualist-Mediated Enhancement of Host Juvenile Growth via Inhibition of Protein Digestion. Cell host & microbe 18, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Geurts L, Caesar R, Van Hul M, Matamoros S, Duparc T, Denis RG, Cochez P, Pierard F, Castel J, et al. (2014). Intestinal epithelial MyD88 is a sensor switching host metabolism towards obesity according to nutritional status. Nature communications 5, 5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, McNulty NP, Rey FE, and Gordon JI (2011). Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 333, 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcinelli S, Picchietti S, Rodiles A, Cossignani L, Merrifield DL, Taddei AR, Maradonna F, Olivotto I, Gioacchini G, and Carnevali O (2015). Lactobacillus rhamnosus lowers zebrafish lipid content by changing gut microbiota and host transcription of genes involved in lipid metabolism. Scientific reports 5, 9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei N, and Zhao L (2013). An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. The ISME journal 7, 880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, and Forano E (2012). Microbial degradation of complex carbohydrates in the gut. Gut microbes 3, 289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune S, and Bosch TC (2010). Why bacteria matter in animal development and evolution. Bioessays 32, 571–580. [DOI] [PubMed] [Google Scholar]
- Galindo-Villegas J, Garcia-Moreno D, de Oliveira S, Meseguer J, and Mulero V (2012). Regulation of immunity and disease resistance by commensal microbes and chromatin modifications during zebrafish development. Proceedings of the National Academy of Sciences of the United States of America 109, E2605–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garthoff LH, Henderson GR, Sager AO, Sobotka TJ, O’Dell R, Thorpe CW, Trotter WJ, Bruce VR, Dallas HL, Poelma PL, et al. (2002). The Autosow raised miniature swine as a model for assessing the effects of dietary soy trypsin inhibitor. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association 40, 487–500. [DOI] [PubMed] [Google Scholar]
- Gross CG (1998). Claude Bernard and the constancy of the internal environment. Neuroscientist 4, 380–385. [Google Scholar]
- Grunwald DJ, and Eisen JS (2002). Headwaters of the zebrafish -- emergence of a new model vertebrate. Nature reviews Genetics 3, 717–724. [DOI] [PubMed] [Google Scholar]
- Gusarov I, Gautier L, Smolentseva O, Shamovsky I, Eremina S, Mironov A, and Nudler E (2013). Bacterial nitric oxide extends the lifespan of C. elegans. Cell 152, 818–830. [DOI] [PubMed] [Google Scholar]
- Hacquard S, Garrido-Oter R, Gonzalez A, Spaepen S, Ackermann G, Lebeis S, McHardy AC, Dangl JL, Knight R, Ley Rv et al. (2015). Microbiota and Host Nutrition across Plant and Animal Kingdoms. Cell host & microbe 17, 603–616. [DOI] [PubMed] [Google Scholar]
- Heinritz SN, Mosenthin R, and Weiss E (2013). Use of pigs as a potential model for research into dietary modulation of the human gut microbiota. Nutrition research reviews 26, 191–209. [DOI] [PubMed] [Google Scholar]
- Hill JH, Franzosa EA, Huttenhower C, and Guillemin K (2016). A conserved bacterial protein induces pancreatic beta cell expansion during zebrafish development. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AJ, Chew YV, Colakoglu F, Cliff JB, Klaassens E, Read MN, Solon-Biet SM, McMahon AC, Cogger VC, Ruohonen Kv et al. (2016). Diet-microbiome interactions in health are controlled via intestinal nitrogen source constraints. Cell Metabolism 25, 140–151. [DOI] [PubMed] [Google Scholar]
- Jemielita M, Taormina MJ, Burns AR, Hampton JS, Rolig AS, Guillemin K, and Parthasarathy R (2014). Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. mBio 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, Sun X, Muhlbauer M, Mackey LC, Flynn EJ 3rd, Bagnat M, Jobin C, and Rawls JF (2011). Microbial colonization induces dynamic temporal and spatial patterns of NF-kappaB activation in the zebrafish digestive tract. Gastroenterology 141, 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, Tomkovich S, Xiaolun S, Grosser MR, Koo J, Flynn EJ 3rd, Jobin C, and Rawls JF (2014). Commensal microbiota stimulate systemic neutrophil migration through induction of serum amyloid A. Cellular microbiology 16, 1053–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, et al. (2013). Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature medicine 19, 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupke DM, Begley DA, Sundberg JP, Bult CJ, and Eppig JT (2008). The Mouse Tumor Biology database. Nature reviews Cancer 8, 459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagkouvardos I, Pukall R, Abt B, Foesel BU, and Meier-Kolthoff JP (2016). The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. 1, 16131. [DOI] [PubMed] [Google Scholar]
- Lassenius MI, Pietilainen KH, Kaartinen K, Pussinen PJ, Syrjanen J, Forsblom C, Porsti I, Rissanen A, Kaprio J, Mustonen J, et al. (2011). Bacterial endotoxin activity in human serum is associated with dyslipidemia, insulin resistance, obesity, and chronic inflammation. Diabetes care 34, 1809–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WJ, and Brey PT (2013). How microbiomes influence metazoan development: insights from history and Drosophila modeling of gut-microbe interactions. Annual review of cell and developmental biology 29, 571–592. [DOI] [PubMed] [Google Scholar]
- Lemaitre B (2004). The road to Toll. Nature reviews Immunology 4, 521–527. [DOI] [PubMed] [Google Scholar]
- Long X, Spycher C, Han ZS, Rose AM, Muller F, and Avruch J (2002). TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Current biology : CB 12, 1448–1461. [DOI] [PubMed] [Google Scholar]
- MacNeil LT, Watson E, Arda HE, Zhu LJ, and Walhout AJ (2013). Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, and McCoy KD (2015). Standardised animal models of host microbial mutualism. Mucosal immunology 8, 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison LA, and Chen W (2012). Nutrient excess stimulates beta-cell neogenesis in zebrafish. Diabetes 61, 2517–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Borruel N, Casellas F, and Guarner F (2012). The gut microbiota in IBD. Nature reviews Gastroenterology & hepatology 9, 599–608. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Southwick AM, Earle KA, and Sonnenburg JL (2013). A refined palate: bacterial consumption of host glycans in the gut. Glycobiology 23, 1038–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjoram L, and Bagnat M (2015). Infection, Inflammation and Healing in Zebrafish: Intestinal Inflammation. Current pathobiology reports 3, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences of the United States of America 110, 3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin JE, Dahlman I, Harvey CJ, Mejhert N, Singh MK, Epstein JA, Arner P, Torres-Vazquez J, and Rawls JF (2015). Plexin D1 determines body fat distribution by regulating the type V collagen microenvironment in visceral adipose tissue. Proceedings of the National Academy of Sciences of the United States of America 112, 4363–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, Caro-Quintero A, Mjungu D, Georgiev AV, Lonsdorf EV, Muller MN, Pusey AE, Peeters M, Hahn BH, and Ochman H (2016). Cospeciation of gut microbiota with hominids. Science 353, 380–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TLA, Vieira-Silva S, Liston A, and Raes J (2015). How informative is the mouse for human gut microbiota research? Disease Models & Mechanisms 8 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, and Pettersson S (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. [DOI] [PubMed] [Google Scholar]
- O’Brine TM, Vrtelova J, Snellgrove DL, Davies SJ, and Sloman KA (2015). Growth, Oxygen Consumption, and Behavioral Responses of Danio rerio to Variation in Dietary Protein and Lipid Levels. Zebrafish 12, 296–304. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, and Ruvkun G (1997). The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994–999. [DOI] [PubMed] [Google Scholar]
- Pang X, Hua X, Yang Q, Ding D, Che C, Cui L, Jia W, Bucheli P, and Zhao L (2007). Inter-species transplantation of gut microbiota from human to pigs. The ISME journal 1, 156–162. [DOI] [PubMed] [Google Scholar]
- Perera E, and Yufera M (2016). Soybean Meal and Soy Protein Concentrate in Early Diet Elicit Different Nutritional Programming Effects on Juvenile Zebrafish. Zebrafish 13, 61–69. [DOI] [PubMed] [Google Scholar]
- Pham LN, Kanther M, Semova I, and Rawls JF (2008). Methods for generating and colonizing gnotobiotic zebrafish. Nature protocols 3, 1862–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponton F, Wilson K, Cotter SC, Raubenheimer D, and Simpson SJ (2011). Nutritional immunology: a multi-dimensional approach. PLoS pathogens 7, e1002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponton F, Wilson K, Holmes A, Raubenheimer D, Robinson KL, and Simpson SJ (2015). Macronutrients mediate the functional relationship between Drosophila and Wolbachia. Proceedings Biological sciences 282, 20142029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell ML, Pegues MA, Szalai AJ, Ghanta VK, D’Abramo LR, and Watts SA (2015). Effects of the Dietary omega3:omega6 Fatty Acid Ratio on Body Fat and Inflammation in Zebrafish (Danio rerio). Comparative medicine 65, 289–294. [PMC free article] [PubMed] [Google Scholar]
- Pussinen PJ, Havulinna AS, Lehto M, Sundvall J, and Salomaa V (2011). Endotoxemia is associated with an increased risk of incident diabetes. Diabetes care 34, 392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raubenheimer D, and Simpson SJ (2016). Nutritional Ecology and Human Health. Annual review of nutrition 36, 603–626. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Goodman AL, Trent CM, and Gordon JI (2007). In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proceedings of the National Academy of Sciences of the United States of America 104, 7622–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, and Gordon JI (2006). Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Samuel BS, and Gordon JI (2004). Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proceedings of the National Academy of Sciences of the United States of America 101, 4596–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw SA, and Trede NS (2012). A model 450 million years in the making: zebrafish and vertebrate immunity. Disease models & mechanisms 5, 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyniers JA (1959). The pure-culture concept and gnotobiotics. Annals of the New York Academy of Sciences 78, 3–16. [Google Scholar]
- Rocha F, Dias J, Engrola S, Gavaia P, Geurden I, Dinis MT, and Panserat S (2015). Glucose metabolism and gene expression in juvenile zebrafish (Danio rerio) challenged with a high carbohydrate diet: effects of an acute glucose stimulus during late embryonic life. The British journal of nutrition 113, 403–413. [DOI] [PubMed] [Google Scholar]
- Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, and Rawls JF (2011). Evidence for a core gut microbiota in the zebrafish. The ISME journal 5, 1595–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolig AS, Parthasarathy R, Burns AR, Bohannan BJ, and Guillemin K (2015). Individual Members of the Microbiota Disproportionately Modulate Host Innate Immune Responses. Cell host & microbe 18, 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Rowedder H, Braendle C, Felix MA, and Ruvkun G (2016). Caenorhabditis elegans responses to bacteria from its natural habitats. Proceedings of the National Academy of Sciences of the United States of America 113, E3941–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A, and Gut P (2015). Metabolic insights from zebrafish genetics, physiology, and chemical biology. Cellular and molecular life sciences : CMLS 72, 2249–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Nielsen K (1997). Animal physiology : adaptation and environment (Cambridge: Cambridge University Press; ). [Google Scholar]
- Schwarzer M, Makki K, Storelli G, Machuca-Gayet I, Srutkova D, Hermanova P, Martino ME, Balmand S, Hudcovic T, Heddi A, et al. (2016). Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 351, 854–857. [DOI] [PubMed] [Google Scholar]
- Seiliez I, Medale F, Aguirre P, Larquier M, Lanneretonne L, Alami-Durante H, Panserat S, and Skiba-Cassy S (2013). Postprandial regulation of growth- and metabolism-related factors in zebrafish. Zebrafish 10, 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers RS, Clifford CB, Treuting PM, and Brayton C (2012). Immunological variation between inbred laboratory mouse strains: points to consider in phenotyping genetically immunomodified mice. Veterinary pathology 49, 32–43. [DOI] [PubMed] [Google Scholar]
- Semova I, Carten JD, Stombaugh J, Mackey LC, Knight R, Farber SA, and Rawls JF (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell host & microbe 12, 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan F (2012). The gut microbiota-a clinical perspective on lessons learned. Nature reviews Gastroenterology & hepatology 9, 609–614. [DOI] [PubMed] [Google Scholar]
- Shen J (2008). Modulating effects of Fructo-oligosaccharides on Gut Microbiota and Host metabolism in Human Flora-Associated Piglet Mode (Shanghai Jiao Tong University; ). [Google Scholar]
- Shen J, Zhang B, Wei H, Che C, Ding D, Hua X, Bucheli P, Wang L, Li Y, Pang X, et al. (2010). Assessment of the modulating effects of fructo-oligosaccharides on fecal microbiota using human flora-associated piglets. Archives of microbiology 192, 959–968. [DOI] [PubMed] [Google Scholar]
- Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, and Lee WJ (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674. [DOI] [PubMed] [Google Scholar]
- Shtonda BB, and Avery L (2006). Dietary choice behavior in Caenorhabditis elegans. The Journal of experimental biology 209, 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson SJ, and Raubenheimer D (2012). The Nature of Nutrition: A Unifying Framework from Animal Adaptation to Human Obesity (Princeton and Oxford: Princeton University Press; ). [Google Scholar]
- Smith DL Jr., Barry RJ, Powell ML, Nagy TR, D’Abramo LR, and Watts SA (2013a). Dietary protein source influence on body size and composition in growing zebrafish. Zebrafish 10, 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. (2013b). Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, Cogger VC, Pulpitel T, Heblinski M, Wahl D, McMahon AC, Warren A, Durrant-Whyte J, Walters KA, Krycer JR, et al. (2016). Defining the Nutritional and Metabolic Context of FGF21 Using the Geometric Framework. Cell Metab 24, 555–565. [DOI] [PubMed] [Google Scholar]
- Solon-Biet SM, McMahon AC, Ballard JW, Ruohonen K, Wu LE, Cogger VC, Warren A, Huang X, Pichaud N, Melvin RG, et al. (2014). The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab 19, 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, Walters KA, Simanainen UK, McMahon AC, Ruohonen K, Ballard JW, Raubenheimer D, Handelsman DJ, Le Couteur DG, and Simpson SJ (2015). Macronutrient balance, reproductive function, and lifespan in aging mice. Proceedings of the National Academy of Sciences of the United States of America 112, 3481–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, and Backhed F (2013). The gut microbiota--masters of host development and physiology. Nature reviews Microbiology 11, 227–238. [DOI] [PubMed] [Google Scholar]
- Soukas AA, Kane EA, Carr CE, Melo JA, and Ruvkun G (2009). Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes & development 23, 496–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepankova R, Powrie F, Kofronova O, Kozakova H, Hudcovic T, Hrncir T, Uhlig H, Read S, Rehakova Z, Benada Ov et al. (2007). Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflammatory bowel diseases 13, 1202–1211. [DOI] [PubMed] [Google Scholar]
- Stephens WZ, Wiles TJ, Martinez ES, Jemielita M, Burns AR, Parthasarathy R, Bohannan BJ, and Guillemin K (2015). Identification of Population Bottlenecks and Colonization Factors during Assembly of Bacterial Communities within the Zebrafish Intestine. mBio 6, e01163–01115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storelli G, Defaye A, Erkosar B, Hols P, Royet J, and Leulier F (2011). Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR dependent nutrient sensing. . Cell Metabolism 14, 403–414. [DOI] [PubMed] [Google Scholar]
- Su D, Nie Y, Zhu A, Chen Z, Wu P, Zhang L, Luo M, Sun Q, Cai L, Lai Y , et al. (2016). Vitamin D Signaling through Induction of Paneth Cell Defensins Maintains Gut Microbiota and Improves Metabolic Disorders and Hepatic Steatosis in Animal Models. Frontiers in physiology 7, 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Post S, and Yu K (2014). Nutrient control of Drosophila longevity. Trends in endocrinology and metabolism: TEM 25, 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, He G, Mai K, and Liu C (2015). Effects of postprandial starvation on mRNA expression of endocrine-, amino acid and peptide transporter-, and metabolic enzyme-related genes in zebrafish (Danio rerio). Fish physiology and biochemistry 41, 773–787. [DOI] [PubMed] [Google Scholar]
- Tlaskalova-Hogenova H, Stepankova R, Kozakova H, Hudcovic T, Vannucci L, Tuckova L, Rossmann P, Hrncir T, Kverka M, Zakostelska Zv et al. (2011). The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cellular & molecular immunology 8, 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V, and Backhed F (2012). Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, and Gordon JI (2009). The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science translational medicine 1, 6ra–14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, and Donovan SM (2015). Human microbiota-associated swine: current progress and future opportunities. ILAR journal 56, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, et al. (2011). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- Watts SA, Lawrence C, Powell M, and D’Abramo LR (2016). The Vital Relationship Between Nutrition and Health in Zebrafish. Zebrafish 13 Suppl 1, S72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Shen J, Pang X, Ding D, Zhang Y, Zhang B, Lu H, Wang T, Zhang C, Hua X, et al. (2008). Fatal infection in human flora-associated piglets caused by the opportunistic pathogen Klebsiella pneumoniae from an apparently healthy human donor. The Journal of veterinary medical science 70, 715–717. [DOI] [PubMed] [Google Scholar]
- Wen K, Tin C, Wang H, Yang X, Li G, Giri-Rachman E, Kocher J, Bui T, Clark- Deener S, and Yuan L (2014). Probiotic Lactobacillus rhamnosus GG enhanced Th1 cellular immunity but did not affect antibody responses in a human gut microbiota transplanted neonatal gnotobiotic pig model. PloS one 9, e94504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whewell W (1840). The Philosophy of the Inductive Sciences, Founded Upon Their History. (London, J. W. Parker; ). [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, and Siuzdak G (2009). Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proceedings of the National Academy of Sciences of the United States of America 106, 3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles TJ, Jemielita M, Baker RP, Schlomann BH, Logan SL, Ganz J, Melancon E, Eisen JS, Guillemin K, and Parthasarathy R (2016). Host Gut Motility Promotes Competitive Exclusion within a Model Intestinal Microbiota. PLoS biology 14, e1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EO (1998). Consilience: the unity of knowledge. (New York: Knopf; ). [Google Scholar]
- Wong AC, Dobson AJ, and Douglas AE (2014). Gut microbiota dictates the metabolic response of Drosophila to diet. The Journal of experimental biology 217, 1894–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AC, Holmes A, Ponton F, Lihoreau M, Wilson K, Raubenheimer D, and Simpson SJ (2015a). Behavioral Microbiomics: A Multi-Dimensional Approach to Microbial Influence on Behavior. Frontiers in microbiology 6, 1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Stephens WZ, Burns AR, Stagaman K, David LA, Bohannan BJ, Guillemin K, and Rawls JF (2015b). Ontogenetic Differences in Dietary Fat Influence Microbiota Assembly in the Zebrafish Gut. mBio 6, e00687–00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-J, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, and Mathis D (2010). Gut-Residing Segmented Filamentous Bacteria Drive Autoimmune Arthritis via T Helper 17 Cells. Immunity 32, 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Feng Q, Liang S, Sonne SB, Xia Z, Qiu X, Li X, Long H, Zhang J, Zhang D et al. (2015). A catalog of the mouse gut metagenome. Nat Biotech 33, 1103–1108. [DOI] [PubMed] [Google Scholar]
- Zhang C, Yin A, Li H, Wang R, Wu G, Shen J, Zhang M, Wang L, Hou Y, Ouyang H, et al. (2015). Dietary Modulation of Gut Microbiota Contributes to Alleviation of Both Genetic and Simple Obesity in Children. EBioMedicine 2, 968–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, and Zhao L (2016). Strain-level dissection of the contribution of the gut microbiome to human metabolic disease. Genome medicine 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L (2013). The gut microbiota and obesity: from correlation to causality. Nature reviews Microbiology 11, 639–647. [DOI] [PubMed] [Google Scholar]
- Zhao L, and Shen J (2010). Whole-body systems approaches for gut microbiota- targeted, preventive healthcare. Journal of biotechnology 149, 183–190. [DOI] [PubMed] [Google Scholar]


