Graphical Abstract

1. Introduction
The review focuses on recent aspects (last three years) of glycosylation analyses that provide relevant information about cancer. It includes recent development in glycan and protein enrichment methods for discovery of cancer markers. It will however focus on the recent technological developments in mass spectrometry (MS), bioinformatics and separation methods as they apply toward identifying cancer markers. More specifically, it will cover advances in matrix-assisted laser desorption/ionization (MALDI), electrospray ionization (ESI), capillary electrophoresis (CE), and liquid chromatography (LC) coupled to mass spectrometry. The discussions will include glycans, recently identified as potential markers for cancer that have been discovered using the highlighted technologies. We will also discuss emerging glycoproteomic techniques and site-specific methods, and how these methods are being utilized for cancer biomarker discovery. The large amount of data and the complexity of glycoproteomic analysis have been the impetus for developing bioinformatic methods for assigning glycosylation sites and characterizing the potentially very large site- or microheterogeneity.
This review will cover the most recent advancements in biomarker discovery of N- and O-glycosylation of proteins as well as the glycolipids. This group collectively constitutes glycosylation on the cell membrane or the glycocalyx. The review will also highlight methods that are highly reproducible, with low coefficient of variation (CV), and scalable for large sample sets. The reader is also referred to other notable earlier reviews on glycomic biomarkers for cancer. Mereiter et al. describe the recent glycomic effort in gastrointestinal cancer.1 A review focused on N-glycomic analysis of colorectal cancer has been published by Sethi and Fanayan.2 N-Glycan, O-glycan, and glycolipid characteristics of colorectal cancer were reviewed by Holst et al.3 Muchena et al. have provided a more general review of glycan biomarkers covering up to the current review period.4
The field of glycoscience also covers a broad area of structures and may include highly anionic (glycosaminoglycans) and monosaccharide (e.g. O-GlcNAc) modifications that require their specific and unique sets of analytical tools. The latter topics are not covered in this review.
1.1. Background of Glycosylation and Cancer
There is nearly 50 years of research illustrating that changes in glycosylation accompany cancer.5 Glycosylation is a dynamic process intimately involved in key processes in cells, including cell-cell and cell-extracellular communication as well as cell-cell adhesion, and cellular metabolism. Glycans expressed in several types of glycoconjugates are known to change during cancer genesis and progression.6 These changes increase the structural heterogeneity and alter the functions of cells.7 Glycosylation has been found to enable tumor-induced immunomodulation and metastasis.8–10 The cell-surface structures allow the immune cells to differentiate self/normal cells from non-self/abnormal cells.11 For example, terminal residues on N-glycans, such as sialic acids, are involved in immunity and cell-cell communication.12 Changes in glycosylation of adhesion proteins can largely influence their binding properties, leading to altered cell-cell or cell-matrix contacts.13 Other types of glycans are also involved in cancer. Gangliosides and sphingolipids are involved in transmembrane communication vital in tumor cell growth and invasion.14 Glycosaminoglycans are involved in tumor cell migration15 and motility.16–18
The search for effective markers is aided by the understanding of how glycans are synthesized. The glycan biosynthetic process is a non-template process involving multiple enzymes, some performing competing activities. It is estimated that more than 300 metabolic enzymes, composed of glycosyltransferases and glycosidases, are involved in the biosynthesis and processing of glycans.19–20 The best-known series of pathways belongs to the production of N-glycans (Figure 1). They illustrate the large degree of structural heterogeneity in glycosylation. N-Glycans are produced in a step-wise process beginning with the production of high mannose structures on a lipid, which are transferred to the nascent polypeptide chain to guide protein folding. Once folded, the glycans are then trimmed back and extended to form complex and hybrid structures. The folded protein can be secreted with glycans that range from early in the process to yield high mannose structures to later in the process corresponding to complex or hybrid structures. The number of structures for one glycosylation site can vary by a large degree, from a handful for transferrin21 to over 70 structures for IgG, the most abundant serum glycoprotein.22–23
Figure 1.
Representation of the glycosylation pathway of proteins. The pathway illustrates the complexity and heterogeneity of structures. The proteins may exit the pathway with various levels of glycosylation.
The glycan types and the extent of glycosylation differ between cells from the same tissue and between organs. Glycosylation may further depend on the physiological and/or pathological condition of the body.24–25 The aberrant changes of glycosylation may be due to over-expression, under-expression, or the localization of relevant glycosyltransferases, abnormal glycosidase activity, and the tertiary conformation of the protein or the peptide affecting the accessibility of the glycosylation site. Not surprisingly, most of the FDA approved markers for diseases are glycosylated proteins. Furthermore, antibody based therapies have strong glycomic component for treatment of cancers such as breast, lung, gastrointestinal system, and melanoma and lymphomas.26 While glycan-focused therapeutics has not been extensively pursued, the discovery of robust and verifiable markers could lead to more glycosylation-based cancer therapies.
1.2. Glycomic and glycoproteomic methods for cancer
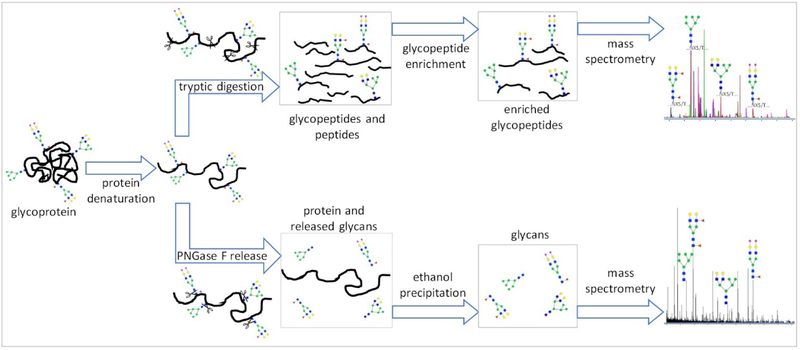
Rapid and reproducible glycomic analyses have been made possible by new technologies in separation, ionization, and mass analyzers. For glycosylation, two main strategies are commonly used (Figure 2). The glycans can be released from a protein mixture, globally from a fluid, or from a specific protein. N-Glycans are released from enriched or non-enriched biological samples like serum/plasma, urine, or saliva using PNGase F. O-Glycans are typically released chemically as there is no equivalent to PNGase F. Chemical approaches can also be used to globally release different types of glycans. For example, Song et. al. recently developed a method to simultaneously release free reducing N- and O-glycans from glycoproteins, and glycan nitriles from glycosphingolipids using household bleach.27 (Figure 3) The free glycans are generally analyzed using either MALDI-MS or LC-MS. Alternatively, glycoproteins can be analyzed as glycopeptides through protease digestion of the protein, thereby keeping a segment of the peptide backbone intact.
Figure 2.
Workflow scheme of glycan analysis in protein: Released glycan analysis by LC-MS and glycopeptide analysis by LC-MS/MS
Figure 3.
Strategies for glycan release from glycoproteins and glycosphingolipids: oxidative release of natural glycans (ORNG) vs. traditional glycomics methods. Reprinted with permission from Song, X.; Ju, H.; Lasanajak, Y.; Kudelka, M. R.; Smith, D. F.; Cummings, R. D., Oxidative release of natural glycans for functional glycomics. Nat Meth 2016, 13 (6), 528–534 ref(27). (Permission pending). Copyright 2016 Nature.
The first global glycomic analysis of serum was reported over 10 years ago for ovarian cancer.28 This glycomic profiling method involved MALDI-MS. There are intrinsic properties of glycans that need to be addressed in the global glycan analysis. The diversity of glycan species and similarities in structures pose unique challenges to mass spectrometry. The similarities in structures make it difficult to distinguish isomers, while the presence of both neutral residues such as galactose, mannose, fucose, N-acetylglucosamine, and N-acetylgalactosamine and anionic residues such as sialic acids and sulfated monosaccharides create ionization biases. Anionic oligosaccharides tend to give better signals in the negative mode due to deprotonation, while neutral oligosaccharides tend to be cationized with sodium in the positive mode. MALDI has two major limitations toward oligosaccharide analysis: it yields no isomer separation, and the energetic conditions of MALDI may cause the loss of labile groups such as sialic acids and fucoses. These issues have been largely resolved by stabilizing the glycans using derivatization29 or by decreasing the energy of the MALDI process with post-ionization cooling.30
Electrospray ionization (ESI or more specifically nanoESI) coupled with liquid chromatography overcomes many of the limitations in MALDI profiling. With ESI, singly and multiply protonated species are produced, in contrast to MALDI which produces sodium-coordinated species. Nanospray increases the sensitivity over MALDI and produces cooler ions, minimizing in-source fragmentation. When coupled with liquid chromatography (or nanoLC) it extends the number of identified glycans significantly. To further elucidate the exact glycan structures, however, other approaches such as exoglycosidase digestion31–32 and permethylation followed by acid hydrolysis and acetylation33–34 need to be used in combination with mass spectrometry techniques.
The next development of glycomic analysis will lie in the elucidation of glycopeptides and glycoproteins. In glycoproteomics, issues in proteomics are compounded with the difficulties in glycomics, and both will have to be addressed simultaneously. This approach requires new methods that can overcome the challenges associated with their specific analysis, including incomplete glycosite/glycopeptide identification, site occupancy, difficulties in elucidating all the structural details of glycan and its respective polypeptide, and limited glycan data interpretation algorithm/software. Despite these challenges, the results obtained from these analyses add in-depth knowledge regarding the identities of the glycans at their site of attachment within the proteins. This provides the glycan and the protein information that are essential in understanding the cancer biology. These types of analyses may provide more specific cancer markers and aid in the efforts to identify drug targets.
2. Methods for profiling glycans and glycoprotein cancer markers
Mass spectrometry has generally been at the core of the large glycomic effort. MS is ideal because of its sensitivity, and large peak capacity. It is also open to broad non-targeted and targeted approaches. However, the improvements in mass analyzers have not significantly advanced glycomic methods. There are several types of analyzers, but they all appear to be equally effective for glycomic analysis. Conversely, ionization and separation methods have significantly advanced glycomic and glycoproteomic analysis. The large number of structures in the glycome, as well as the large number of glycopeptides, require effective separation methods that increase the peak capacity and the information content of the analysis. The coupling of separation methods to mass spectrometry yielded an effective glycomic and glycoproteomic tool, which has also increased the information content even further.
2.1. Matrix-assisted laser desorption/ionization (MALDI – MS)
Several studies have already illustrated the use of MALDI as a tool to monitor alterations in glycosylation in cancer and diseases in various tissues including blood, serum, and muscle.4 In addition to discovering biomarkers for clinical diagnosis of cancer, characterization of protein glycosylation by MALDI-MS further helped elucidate the heterogeneity present in complicated biological samples.4, 35 MALDI-MS was the first method employed for global glycan profiling of serum. It is used primarily for N-glycans, and remains the most common method for glycomic profiling.36
MALDI-MS provides primarily a compositional profile yielding the combination of hexose (Hex, typically D-mannose and D-galactose), deoxyhexose (typically L-fucose, or Fuc), N-acetylhexosamine (HexNAc, typically N-acetyl-D-glucosamine and N-acetyl-D-galactosamine) and sialic acid (typically N-acetylneuraminic acid, or NeuAc).37 Because the biology of glycan formation is relatively well known, the glycan composition can be used to deduce the glycan type. For disease biomarker discovery, general changes in glycan types such as high mannose, complex, hybrid, bisecting GlcNAc, sialylation, and fucosylation are of direct biological significance.38 Coarse alterations in glycosylation have been shown to influence various cellular processes in many different cancers.39
MALDI imparts higher energy during ionization causing instability for analyte ions sometimes inducing unwanted fragmentation. This may further result in limited representation of anionic and sialylated glycan species.40 Permethylation is one of the most commonly used derivatization method for stabilizing glycans during ionization. Automated methods are emerging to make this process more suitable for glycan analysis. Shubhakar et al. developed a high-throughput format to monitor glycosylation in biologics, however the same method can be applied to cancer tissues.41 Derivatization of sialic acid to yield linkage information has been used by Holst et al. to examine formalin-fixed tissue for MALDI imaging.42 Specific stabilization and characterization of sialylated glycans was achieved with linkage-specific sialic acid derivatization employing ethyl esterification and dimethylamidation for differential analysis of α(2,3)- and α(2,6)-linked sialic acids in plasma and cancer tissues.42–43
A more recent innovation in MALDI-MS is the application of MALDI imaging on cellular glycosylation. Advancements in the characterization of N-linked glycans in cancer tissues using MALDI imaging was discussed in a recent review by Drake et al.44 Glycan profiles using tissue-based analysis have been used to determine expression in different cancers. Human formalin-fixed paraffin-embedded (FFPE) tissues have been used to characterize the spatial profile and localization of released N-glycans using MALDI-Imaging Mass Spectrometry (MALDI-IMS).45–46 FFPE tissues collected from human hepatocellular carcinomas, prostate, and pancreas have been found to have distinct glycan profiles when compared to adjacent non-tumor tissue slices.45, 47 For validation of alterations in glycan expression, tissue microarrays incorporating multiple FFPE tumor and normal tissue cores have been used in combination with MALDI-IMS.45 (Figure 4) A tetraantennary N-glycan Hex7HexNAc6 was found to be elevated, while a high mannose N-glycan Hex8HexNAc2 was decreased in hepatocellular carcinoma tissues. A similar approach using MALDI with ion mobility MS found high-mannose N-glycans expressed in FFPE ovarian cancer tissues.48 A new method for MALDI imaging combining IR MALDI with ESI (IR MALDESI) has been used by Nazari and Muddiman to image cancerous chicken ovarian tissues, although glycans were not directly examined.49
Figure 4.
MALDI-MS imaging of glycans on a liver tissue microarray with tumor (T) and normal (N) tissue cores from 16 hepatocellular carcinoma patients. The map corresponds to putative glycan structures (inset). Reprinted from Powers, T.; Holst, S.; Wuhrer, M.; Mehta, A.; Drake, R., Two-Dimensional N-Glycan Distribution Mapping of Hepatocellular Carcinoma Tissues by MALDI-Imaging Mass Spectrometry. Biomolecules 2015, 5 (4), 2554 (ref 42). Under Creative Commons license (CC BY 4.0) (http://creativecommons.org/licenses/by/4.0/).
2.2. Capillary Electrophoresis - MS
Capillary electrophoresis has long been used for separating oligonucleotides generally based on size with samples containing DNA.50 Minor modifications to instruments for genome sequencing have adapted these instruments to profiling oligosaccharides using photometric detectors.51 The high separation efficiency and resolution of CE opens the possibility of high throughput if the method can be robustly coupled to MS.
The coupling of CE to MS is difficult and complicated by several issues including the presence of buffers in CE that are not compatible with MS, and the low flow rates of CE. CE-MS for glycomic analysis has been previously reviewed by Mechref.52 CE performs generally better with anionic oligosaccharides. Sun et al. have shown that the coupling of CE to MS using an electrokinetic sheath pump offers better modes of analyses for heparin oligosaccharides and low molecular weight heparin.53 To couple the microfluidic CE device with MS, Kathri et al developed a removable microfluidic chip integrating the separation capillary and a nanoESI source as interface to analyze monosaccharides, glycans and glycopeptides.54 This method has been applied to standard glycoproteins, and the applicability and durability of the method for more complicated biological samples remain to be shown.
Capillary microchip electrophoresis was used along with MALDI-TOF-MS to profile serum glycans from patients with colorectal cancer.55 CE compliments the rapid compositional capabilities of MALDI-MS. The microchip electrophoresis can separate structural isomers, which could be more specific toward individual diseases. This study identified tri- and tetra-antennary fucosylated glycans that were significantly elevated in cancer samples when compared to the controls. 55
2.3. Liquid chromatography - MS
Liquid chromatography remains the ideal couple for mass spectrometry (LC-MS) in glycomic and glycoproteomic analysis. Liquid chromatography alone (with chromophoric labeling) has been used extensively to profile N-glycans for disease biomarkers and will not be discussed further here given the limited scope of this review.56 The coupling of LC with tandem MS (LC-MSn) allows confirmation of the monosaccharide composition and further yields structural information of both native and derivatized glycans.40 LC-MS further increases the depth of oligosaccharide profiling and is currently the most robust and reliable method for isomer separation and MS detection.57
There are several methods for separating oligosaccharides by LC depending on the types of stationary and mobile phases. Although C18 remains the most commonly used stationary phase for organic compounds, the hydrophilic nature of oligosaccharides prevents their retention on C18 column. Derivatization (typically permethylation) is required for retaining glycans on C18, however the separation of permethylated isomeric species is generally poor.58 A recent review by Vreeker and Wuhrer on the reverse phase chromatographic separation of oligosaccharides covers this topic.58
Hydrophilic interaction liquid chromatography, or HILIC, have been used to separate both native and labeled glycans. Ahn et al. employed 1.7-μm amide sorbent as the stationary phase for enhanced separation of 2-aminobenzamide labeled glycans including high mannose and multiply sialylated species.59 They have developed HILIC separation into a robust and repeatable platform for biomarker discovery with fluorescence detection. Although MS is not typically involved in these studies, the method is notable in the ability to perform rapid throughput analysis for large sample sizes.60–62 Native oligosaccharides can also be efficiently separated by HILIC. The hydrophilic nature of oligosaccharides dictate their elution properties allowing HILIC columns to separate certain oligosaccharide isomers.63
An alternative to HILIC for separating native oligosaccharides, is the porous graphitized carbon (PGC) stationary phase. Profiling of native oligosaccharides by LC-MS typically employs PGC as stationary phase. Shown in Figure 5 is an LC-MS profile of N-glycans released from cell membrane proteins combined from three cell lines.25 The profile shows more than 800 compounds over approximately four orders of magnitude in dynamic range.
Figure 5.
N-Glycan profile with LC-MS from a sample containing cell membranes combined from three cell lines including Caco-2, HT-29, HCC1954. Over 800 compounds are observed in a single the LC-MS run.
Isomeric separation of native (unlabeled, underivatized) compounds with PGC is often so effective that separation of anomers (at the reducing carbon) occurs and produces a splitting of the peaks. The splitting occurs greatest for smaller compounds and eventually vanishes for large multiantennary N-glycan structures. To address this issue, the glycans are often reduced to the alditol form. With the separation of the compounds, the retention time and accurate mass are unique for each structure and can be used to identify the compounds. In this way, glycans can be ordered according to their abundances based on peak areas (or volumes). PGC stationary phase in LC-MS was used by Sethi et al. to determine the reduced N-glycans on cell membrane from cell lines of colorectal cancer.64 In the study, increase of high mannose glycans and α(2,6)-sialylation was observed for the cancer. Song et al has used this method to determine the relative abundances of N-glycans released from serum (Figure 6) as well as biological variations between individuals.32
Figure 6.
N-Glycan profile of serum glycans with LC- QTOF MS. Peaks are numbered according to their abundances and correspond to individual annotated structures. Reprinted from Song, T.; Aldredge, D.; Lebrilla, C. B., A Method for In-Depth Structural Annotation of Human Serum Glycans That Yields Biological Variations. Anal Chem 2015, 87 (15), 7754–62 ref (32). Copyright 2015 American Chemical Society.
2.4. Quantitation of glycans for biomarkers
The ability to quantitate is key to the development of effective disease markers. Methods to quantitate glycans continue to be developed, however quantitation of glycans is generally easier than proteins but more difficult than metabolites. The major issue with quantitation of glycans is the lack of chemically pure and readily available standards.
There are several levels of quantitation that all have their utility in biomarker discovery. Relative quantitation is the simplest therefore the most commonly used. It essentially takes the absolute ion abundances and normalizes it to the total abundances. The most desirable method is for absolute quantitation where the absolute concentration is determined. This method has only been performed on oligosaccharides found in human milk.65 An attempt to obtain absolute quantitation of N-glycans has been performed by combining glycan types.66
Absent absolute quantitation methods, the measurement of absolute fold changes is the most accurate and convenient approach used in both MALDI-MS and LC-MS for biomarker discovery. Both methods can be used to measure fold changes of individual components with high repeatability.67–68 The limitations stem from the assumption that the same compound ionizes with the same efficiency in different samples, however this efficiency can vary depending on the ionization method. In MALDI where the glycans are all analyzed simultaneously, the presence of each glycan can affect the ionization of others. In LC-MS, ion suppression is greatly minimized as the compounds are separated, however it is not completely eliminated. Relative abundances are also distorted by the variation in ionization, although this has been shown to be minimal in the same group of glycans.69
An alternative approach to ion abundances has been to use isotopic labeling through derivatization. A recent review by Etxebarria and Reichardt70 on glycan quantitation addresses many of the issues in detail. Oligosaccharides in one sample is permethylated with 12CH3 while the other sample is derivatized with 13CH3 (see for example71). The approach is simple and robust, but it has limitations. Permethylation is never complete, and even 99% conversion can still produce partially methylated species that can severely decrease the dynamic range of the method. It also limits the comparison to two samples, unless a standard is added to each sample. It further adds another chemical reaction to a sample, which should be limited in biomarker discovery.
There are other issues that can complicate quantitation, but important in large sample set analysis. One is the enzyme PNGase F may have different reactivities depending on the supplier and the batch. In large sample sets, the same PNGase F batch has to be used to limit this variability. Similarly, other factors need to be concerned to maintain homogeneity. The other reagents and for native oligosaccharides, the solid phase extraction media such as porous graphitized carbon all need to come from the same manufacturing batch. These issues can affect quantitation in more profound ways than the quantitative method employed.
3. Sample preparation and methods to partition the glycoproteome
Sample preparation is an important consideration in marker discovery. The quality of the clinical sample and the precision of the work-up enhances the sensitivity and reproducibility of the overall method. Prolonged sample manipulation can alter the glycoform by the chemical release of labile groups such as sialic acids. Therefore, it is desirable to minimize sample preparation in steps and in time. Label-free in-situ analysis of N-glycans from hepatocellular carcinoma tissues was conducted using MALDI-IMS.47 N-glycans such as Hex8HexNAc2 and Hex7HexNAc6 were found to be differentially expressed in tumor and matched normal tissue samples. However, loss of sialic acid was observed when the results of native glycans were compared with the ethyl esterified glycans from the same samples.
A method that requires minimal sample preparation for targeted analysis is multiple reaction monitoring (MRM). Through the selection of targeted proteins, it is a method for partitioning the glycoproteome by focusing on relatively abundant species. The method is rapidly developing for label-free analysis with quantitation of glycans, peptides, and glycopeptides. It has already been applied to rapid-throughput analysis of serum glycopeptides from several cancers.22, 72–74
Nevertheless, enrichment methods are often required in cancer studies for extracting interesting glycoproteins or for untargeted biomarker discovery. Methods to enrich specific glycoproteins commonly involve the use of antibodies. Mass spectrometry readily couples with this method providing higher sensitivity for protein-specific glycosylation. Borchers and co-workers have recently reviewed the use of immunoaffinity in combination with mass spectrometry for quantitating proteins in biological fluids.75 A study by Gbormittah et al. using HPLC columns packed with anti-clusterin ligand immobilized agarose media for clusterin enrichment showed site-specific and glycan-specific changes in clear cell renal cell carcinoma.76 Immunoaffinity capture was performed on plasma samples from pre- and post-nephrectomy patients. The enriched clusterin was treated with trypsin to analyze the glycopeptides. Increased levels of biantennary, disialylated N-glycans with and without core fucose were observed in post-nephrectomy patients. The results were further confirmed using lectin blotting. Takahashi et al. isolated haptoglobin from the sera of a small number of cancer patients individually with esophageal, gastric, colon, gallbladder, pancreatic, and prostate cancer to determine the site-specific glycosylation associated with the cancer. In all cancers, monofucosylated N-glycans were increased significantly in all glycosylation sites.77 Immunoaffinity is also commonly used to purify proteins for intact protein analysis. Transferrin purified from plasma using anti-transferrin beads was employed to examine congenital disorders of glycosylation.78 Though not specifically cancer, the method shows the utility of native MS method to observe changes in protein glycosylation for diseases. Kim et al. performed intact haptoglobin analysis to monitor N-glycan alteration in gastric cancer patients and identified multiple markers with high diagnostic efficacy.79
Immunoaffinity enrichment is indeed an attractive method as it is highly specific for targeted enrichment and readily scalable and automated for reproducible rapid throughput analysis. It has been adapted to MALDI, MRM and other MS-based methods.80–81
A complimentary enrichment method employs lectins. Antibody enrichment typically targets protein, while lectins enrich through affinity for specific glycan structure. Lectins are non-antibody glycan binding proteins derived typically from microbes or plants. They generally have weaker affinity with only moderate specificity. They are used alone or with other lectins to capture groups of proteins with corresponding glycan motifs. Lectin microarrays and lectin magnetic bead arrays have also been used in combination with glycan release or glycopeptide production coupled with mass spectrometry.
A single lectin enrichment approach was applied to protease digest of serum to examine glycopeptides in pancreatic cancer. Maackia amerensis lectin II (MAL II) and Sambucus nigra lectin (SNA) were used to enrich for sialylated N-glycopeptides obtained from proteins of pancreatic cancer patients. Subsequent UPLC-MS analysis revealed 38 and 13 significantly changed glycopeptides from acute pancreatis and pancreatic cancer, respectively.82 There are limitations to this approach as it covers a subset of the glycome. Changes in non-sialylated glycopeptides of some proteins such as IgG have been shown to accompany cancers,72 which could be missed by this method.
Multi-lectin affinity chromatography (M-LAC) can be used to partition the glycoproteome while enriching for several glycan motifs simultaneously. Combined with depletion of abundant proteins, this method has been employed to simultaneously examine the depleted proteome, enriched glycoproteome, and N-glycome.83–84 A similar approach was employed for clear cell renal cell carcinoma, where a number of highly sialylated glycans, high mannose glycans, and low-level proteins were upregulated in the M-LAC fractions of plasma from cancer patients.85 Tandem affinity enrichment was used by Zhou et al. to identify glycoproteins through the MS/MS of glycopeptides.86 In the differences between prostate cancer cell types, they find increased fucosylated glycopeptides in the aggressive cell.
Lectin arrays have been previously used as a high-throughput stand-alone method for observing, albeit indirectly, changes in glycosylation in cancer.87 Coupled with mass spectrometry via in-situ proteolysis, it provides more specific determination of proteins.88 Lectin magnetic beads present an alternative form of the spatial arrays. Beads with lectins were used to pull out specific glycoproteins that can be probed using standard proteomic and glycoproteomic approaches.89 Glycoproteome of tissues from patients with colon cancer has been examined with this method.90 The method was also used to obtain markers in esaphagel adenocarcinoma patients.91 Using a semi-automated pipeline employing 20 unique lectins, Shah et al. obtained 246 lectin-protein candidates, with 45 showing significant differences between the groups. A panel consisting of eight glycoforms produced an area under receiver operating characteristic curve (AUROC) of 0.9425 corresponding to 95% specificity and 80% sensitivity.
The compliment method to lectins are glycan arrays.92 While these methods have not yet been used for cancer biomarkers, they may have future applications in detecting aberrant glycosylation in cancer.91
4. Glycoproteomic analysis for cancer
Glycoproteomic analysis of cancer combines both the glycan changes associated with cancer and the protein expression yielding potentially higher disease specificity. Glycoprotein cancer biomarkers are not new, with nearly all protein markers for cancer including CA-125 for ovarian, PSA for prostate, and HER2 for breast being just the most notable examples. While some of these markers are widely used, they may also be associated with inflammation and do not provide the necessary specificity for the cancer. However, glycosylation on proteins such as CA-12593 and PSA94 exhibit glycan differentiation in a cancer-specific manner for the respective diseases.
Glycoproteomic analysis of cancer is challenging in much of the same way as biomarker studies using only proteomic methods, but compounded further by the added complexity associated with glycosylation. The complexity of protein glycosylation due to the varying number of sites and the glycan heterogeneity associated with each site complicate the analytical determination but provide opportunity for observing small and unique biological changes. There are at least two potential methods for incorporating comprehensive glycan and protein information simultaneously into cancer biomarkers. A non-targeted approach employs proteomic methods adapted for glycosylation. Glycoproteomic methods generally provide broad coverage of the different sites but limited depth in the structural heterogeneity of the glycan. A more targeted approach involves a smaller number of proteins with more extensive site-specific glycan heterogeneity. Characterizing the protein and glycan heterogeneity simultaneously creates a considerable challenge that is slowly being addressed, however there have been notable advancements.
4.1. Non-targeted glycoproteomic method
Non-targeted glycoproteomic approach to cancer is relatively new but is recently enabled by the availability of bioinformatic tools that directly address glycosylation (See below). Biomarker discovery is limited to mainly cancer cell lines and small number of tissues. Shah et al compared two prostate cancer cells, an androgen-dependent strain LNCaP and an androgen-independent strain PC3.95 To quantitate the glycopeptides, isobaric tags for relative and absolute quantitation (iTRAQ) was used in conjunction with 2D-LC-MS to discover glycopeptides that differed for the two cell lines. Increased fucosylation was reported for PC3 relative to LNCaP. A subsequent cell line study comparing two ovarian cancer cell lines that are sensitive and resistant to doxorubicin treatment, OVCAR8 and NCI/ADR-RES, respectively yielded several N-glycosites that differed between the two cell lines.96 Glycosite occupancy was used to determine changes in glycosylation between three benign lesions and three ovarian cancer tissues97 and identified several glycoproteins that differed at specific glycosylation sites. The glycoproteomic analysis of urine was performed to detect aggressive prostate cancer.98 Unique glycosite containing peptides were found to correlate with Gleason scores of the cancer.
4.2. Targeted glycoproteomic method
Targeted glycoproteomic approaches typically involve the isolation of a single protein or a small group of proteins. These studies are more prevalent because they can be applied to large sample sets more readily. They typically involve a pre-enrichment step using either antibody or lectin affinity methods such as those described above.
An alternative approach is to monitor several proteins simultaneously without prior enrichment using multiple reaction monitoring (MRM) methods. MRM can target the glycopeptides of specific proteins after digestion of complicated protein mixtures such as serum. The ease of use and the high repeatability will increase the utility of this method in the future.
The key to effective MRM is knowing the glycosite and the glycans associated with it, or the resulting heterogeneous group of glycopeptides for the protein or group of proteins. A procedure for extensive glycosite mapping of serum proteins has been reported by Hong et al.99
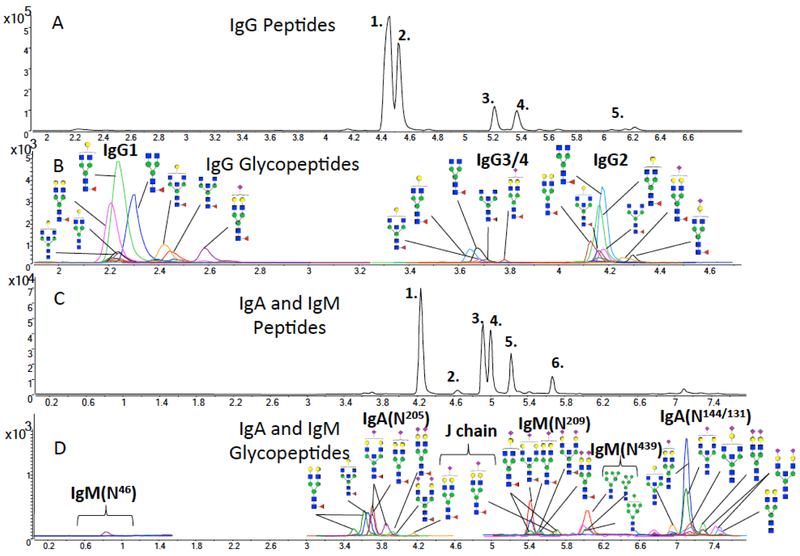
Glycopeptide MRM requires knowing the fragmentation behavior of glycopeptides. Glycopeptide fragmentation is complicated by the variation in the fragmentation aptitude of the glycan and the peptide backbone. Studies have shown that fragmentation of the glycopeptide under standard CID conditions (1–100 eV) generally produce glycan fragments as the major products.100 The most common fragments with high intensities during CID of multiply charged glycopeptides correspond to the glycan oxonium ions such as m/z 204.1 (HexNAc) and 366.1 (Hex1HexNAc1)99 (Figure 7). These fragments are present in nearly all glycopeptide CID spectra regardless of glycan type. To obtain greater specificity, a key feature of glycopeptides in reversed phase liquid chromatography is employed. The glycan part interacts poorly with the C18 stationary phase so that the hydrophobicity of the peptide backbone is the determining factor of the glycopeptide retention time. Shown in Figure 8 are the peptides and glycopeptides for IgG, IgA, and IgM from serum.99 Note that all glycoforms of a specific peptide elute at nearly the same retention times. This behavior means all the glycoforms of the glycopeptide can be searched at a specific retention time range using dynamic MRM (dMRM). This method increases the specificity of the glycopeptide assignments while significantly increasing the peak capacity of the method.
Figure 7.
Tandem MS of glycopeptides: (a) neutral glycopeptides, (b) sialylated glycopeptides, and (c) high mannose glycopeptides. Reprinted from Hong, Q.; Ruhaak, L. R.; Stroble, C.; Parker, E.; Huang, J.; Maverakis, E.; Lebrilla, C. B., A Method for Comprehensive Glycosite-Mapping and Direct Quantitation of Serum Glycoproteins. J Proteome Res 2015, 14 (12), 5179–92 ref (99). Copyright 2015 American Chemical Society.
Figure 8.
MRM of peptides and glycopeptides of immunoglobulins in serum. The spectra were extracted from a single LC-MS run with a total time of 10 minutes. Reprinted from Hong, Q.; Ruhaak, L. R.; Stroble, C.; Parker, E.; Huang, J.; Maverakis, E.; Lebrilla, C. B., A Method for Comprehensive Glycosite-Mapping and Direct Quantitation of Serum Glycoproteins. J Proteome Res 2015, 14 (12), 5179–92 ref (99). Copyright 2015 American Chemical Society.
Glycan abundances can be measured by directly monitoring the glycopeptides. However, the glycopeptide abundance alone provides no indication of whether the protein concentration or the abundance of a specific glycan at the site is changed. To elucidate the two possibilities, Hong et al. normalized the glycopeptide abundances to the protein abundances, thereby alleviating the lack of glycopeptide standards and removing the variation due to changes in protein expression.99 This method effectively decouples glycan changes from variation of protein abundances.
Protein glycosylation not only occurs with glycan site heterogeneity but with the degree of site occupancy. LC-MS based techniques have also improved methodologies for quantification of site occupancy, where both labeling and label-free methods can be used.101 MRM is also used for absolute quantification of site occupancy. In this method, the glycopeptide is deglycosylated with PNGase F converting the asparagine to aspartic acid. The intensities of standard peptides containing asparagine or aspartic acid are used for absolute quantification, while it needs to be kept in mind that aspartic acid can occur naturally in potential glycosylation sites.23
5. Bioinformatics in glycomic and glycoproteomic analysis
Glycans are synthesized through non-template bioprocesses making the structures of glycans and glycan-modified proteins both highly heterogeneous and diverse. Analyzing both glycans and proteins in biological samples with mass spectrometry generates even more data than standard proteomic methods. The combination of annotating glycan structures and assigning glycosylation sites on proteins is significantly more challenging than the analysis of unmodified peptides or peptides with only site-specific modifications such as oxidation and phosphorylation.
There have been significant attempts to develop software that can identify the proteins as well as the site heterogeneity and site occupancy. Bioinformatics software including a commercial product Byonic102, and those developed in various laboratories including GlycoPeptide Finder (GP Finder)103, pGlyco104–105, and Integrated GlycoProteomeAnalyzer (I-GPA)106 automate the annotation of glycopeptide structures from tandem MS data. Because glycosylation is known to be differentially expressed between normal and cancer disease cells, the proper software can provide comprehensive site-specific information for cancer biomarker studies.107 Glycan annotation tools have also been developed for assigning oligosaccharide structures from tandem MS data. They include GlycoMaid for automatic annotation of N-glycans,108 a de novo sequencing software for N- and O-glycans by Kumozaki et al.,109 and GAG-ID for glycosaminoglycans.110 These tools when automated and included in the workflow increase the sample flow and make it possible to study large sample sizes.
Glycan databases are needed to work with informatic software to assign glycan structures and site-specific heterogeneity. Databases for glycomic and glycoproteomic searches have become more available. For glycoproteomic analyses, databases such as Unipep and dbOGAP provide glycosylation sites with corresponding glycans.111 Available glycan databases include Carbohydrate Structure Database,112 UniCorn,113 and GlyTouCan.114 Most glycan databases however contain mainly putative glycan structures.
6. Glycosphingolipids as cancer markers
Glycosphingolipids (GSLs) are a class of glycolipid where the glycan moiety is bonded to a ceramide, a lipid composed of a sphingoid base and a fatty acid. GSLs are found in the cell membrane of all animals, and their compositions are known to be tissue- and species-specific. Aberrant glycosylation in these lipid-bound oligosaccharides has long been recognized as a hallmark of cancer. GSLs have shown changes in their composition in human and animal tumors. These changes arise from dysregulation of their synthetic and catabolic pathways, which are mediated by glycosyltransferases and glycosidases. They can exhibit either incomplete synthesis, which may lead to precursor accumulation, or neosynthesis of GSLs that are not expressed in healthy cells.115 The altered glycosphingolipidome can affect cellular processes such as signaling, growth, adhesion, and differentiation, among others.116 For a more comprehensive and recent perspective on the roles of glycosylation in cancer, the reader is referred to the review by Pinho and Reis.117 Figure 9 shows the schematic representations of root structures of GSLs found in mammalian systems, and the structures of some important GSLs that will be mentioned in the following paragraphs.
Figure 9.
Schematic representation of glycosphingolipid (GSL) root structures found in mammalian systems, and structures of some GSLs of interest discussed in the text.
GSL glycan epitopes already have clinical applications. A widely used marker is the sialyl-Lewis A antigen (also known as CA 19–9), which is present in O-glycans and glycolipds in the sera of gastrointestinal cancer patients.118 However, this field will benefit from further studies using the sensitivity of mass spectrometry-based methods. Candidates for biomarker studies include the GSL globotriaosylceramide (Gb3Cer), which has been correlated with increased cell invasiveness in colorectal cancer.119 Small cell lung cancer also showed increased levels of disialylated gangliosides GD2 and GD1b, and tri-sialylated GT1b.120 Breast cancer cells were found to have increased levels of gangliosides overall. They also expressed GSLs not usually found in healthy tissue, such as O-acetyl-GD3 and O-acetyl-GT3, as well as N-glycolyl-GM3, which is rarely found in human tissue.121 Increased levels of GM3 and GD3 were also found in brain tumors.122–123
Tumor onset and progression are also influenced by aberrant expression of gangliosides. Breast cancer stem cells have been found to express stage-specific embryonic antigen-3 (SSEA-3, also known as Gb5Cer), as well as two other closely related globo-series epitopes, SSEA-4 and globo-H.124 In melanoma, increased levels of disialylated gangliosides, GD3 and GD2, enhance the malignancy of cancer cells.125
The varied roles that GSLs play are reflected in the structural diversity of both their glycan headgroups and their ceramide tails. Of the glycan headgroups alone, about 450 different structures are possible based on what is currently known about GSL biosynthesis, and about half of these have already been reported.126 These numbers are compounded further by dozens of possible ceramide structures which can vary by lipid chain length, degree of unsaturation, and number of hydroxyl groups, leading to tens of thousands of possible GSL structures. As such, analysis and profiling of GSLs will often require multiple approaches for more complete structural elucidation and quantitation. Early methods employed thin layer chromatography (TLC) to separate the GSLs, followed by an immunoassay with lectins and antibodies that bind to specific glycan epitopes for detection and quantitation.127 However, this method is limited to identification of glycan structures for which there are known lectins. Limited sensitivity and specificity of immunoassay techniques can also lead to difficulty in detecting low-level compounds. Furthermore, the signal can be affected by accessibility of the glycan epitope to the antibody.
Mass spectrometry-based methods provide comprehensive detection of GSLs and significantly increased sensitivity, both of which are advantageous for discovering new biomarkers. Tandem MS methods can further provide structural information, such as the sequence and connectivity of the glycan and lipid characteristics of the ceramide. However, MS-based analysis can be complicated by the presence of isomeric and isobaric GSLs, and tandem MS data cannot reveal the monosaccharide epimer nor the type of linkages between them. Furthermore, one must be aware of possible glycan rearrangements when interpreting tandem MS data.128 Complete structural analysis of intact GSLs would require complementary methods such as enzymatic degradation.129 Often, prior studies on similar samples and knowledge on GSL biosynthetic pathways can supplement MS results and allow researchers to make reasonable inferences about GSL structures. Broader trends, such as changes in the level of certain GSLs or the degree of fucosylation and sialylation, can also be effectively monitored with MS-based methods without the need for more laborious structural elucidation.
GSLs are usually isolated from biological samples through Folch extraction.130 Efforts to avoid chloroform during the lipid extraction process have led to the development of less hazardous solvent systems such as the BuMe method.131 The extracted GSLs can then be subjected to further preparation steps depending on the information desired. When data on both the glycan and the lipid are required, intact GSLs can be analyzed through a variety of methods; the most common ones include reversed phase HPLC-ESI-MS and MALDI-MS. One quick and convenient method is to use TLC in conjunction with MALDI-MS.132 This has the advantage of requiring less sample preparation and cleanup, yet still having the high sensitivity of MALDI-MS. More recently, MALDI coupled with Orbitrap MS has opened new possibilities in localized profiling and imaging of normal and tumor tissues. With this method, Jirasko et al. have reported on novel sulfo-GSL structures in human renal cell cancer.133 In a study performed by Zamfir et al, a microfluidic device coupled to an ESI-Orbitrap MS was used to profile gangliosides in human astrocytoma from a single male patient.134 GSL structures were determined through multistage CID fragmentation; 37, 40, and 56 gangliosides were found in the astrocytes, its surrounding tissue, and normal brain tissue respectively.
An approach to studying the glycan heterogeneity is to cleave off the glycan from the ceramide with endoglycoceramidases (EGCase). This allows improved chromatographic separation of the glycans on a HILIC or PGC column with deeper structural analysis, albeit at the expense of information about the lipid. Holst et al. used this approach to analyze the GSL-derived glycans from colorectal cancer tissues of 13 patients, and reported on correlations between tumor progression and increased fucosylation, decreased sulfation and acetylation, and reduced expression of globo-GSLs and disialylated gangliosides.135 Albrecht et al. have recently outlined a method that uses the novel EGCase I from Rhodococcus triatomea.136 This broad-specificity EGCase can effectively cleave most GSLs, including globo-series GSLs and galactosylceramides that other known EGCases are unable to digest.
Researchers have been probing protein-carbohydrate interactions for biomarker discovery with novel MS methods. Complexes formed with gangliosides and the B subunit of cholera toxin (CTB) were studied with nano-ESI multistage MS by Capitan et al., and their specific noncovalent interactions were revealed by multistage CID and ETD fragmentation.137 An intriguing new technique for analyzing protein-GSL interactions is catch-and-release electrospray ionization-mass spectrometry (CaR-ESI-MS). The method was developed by Li et al. and involves the assembly of picodiscs of proteins, lipids, and GSLs in vitro. It further allows GSLs to be solubilized effectively in an aqueous environment.138 Proteins can then be introduced, and those that interact with the GSLs are captured by the picodiscs. The picodisc-bound protein complexes are then analyzed with a quadrupole-ion mobility separation-time-of-flight (Q-IMS-TOF) MS equipped with ESI. Mild CID conditions in the quadrupole can cause the detachment of the protein-GSL complex.
Advances in gene editing such as CRISPR-Cas9 have also provided new avenues for investigating the role of specific glycosyltransferases and glycosidases in tumor progression. A recent study by Alam et al. explored the effect of knocking out a key glycosyltransferase in the biosynthesis of lacto- and neolacto- series GSLs in several ovarian cancer cell lines, and found that it affected not only the biosynthesis of GSLs but also the α(2–6) sialylation on N-glycoproteins.139
An interesting avenue to explore is the role of exosomes in cancer progression. Exosomes are small vesicles released by cells to their extracellular milieu.140 Their composition is dependent on the cell type from which they are released, and they can contain proteins and other biomolecules, including GSLs. Exosomes can be released by most tissues, but some tumors are known to release more exosomes and may exhibit changes in their composition, and these changes can have immunomodulatory effects that prevent the destruction of the tumors by the immune system.141 In a study by Llorente and Sandvig, lipid structures in exosomes released by PC-3 prostate cancer cells were analyzed by hybrid triple quadrupole-linear ion trap MS.142 Exosomes were found to be enriched in certain classes of lipids, including GSLs, compared to their parent cells. Exosomal proteins and peptides have already been viewed as potential biomarkers for cancer and other diseases; more studies are currently underway to explore how exosomal lipid analysis can assist in forming a more conclusive set of data for detecting diseases.143
7. Emerging glycan markers
The methods and techniques for glycomic and glycoproteomic analyses of complicated biological samples continue to be improved, however the performance in at least glycomic profiling appears robust and sufficient for performing large samples sets. High-throughput methods for released N- or O-glycan analysis employing MALDI-MS or LC-MS techniques in combination with derivatization and enrichment protocols have been developed and applied to different types of biological samples. With the robustness of these methods, glycan markers have been discovered in various types of cancers, such as high mannose in breast cancer,144 truncated serum glycoforms in multiple cancer types.145
There are some general trends emerging from the current set of studies. Table 1 is a summary of cancer biomarker discovery studies involving glycans and glycoproteins in the review period. Increased levels of fucosylation, especially core fucosylation, appear to correlate with many types of cancers including esophageal, gastric, colon, gallbladder, pancreatic, liver, ovarian and breast cancer. Similar observations were reported for multi-fucosylated N-glycans with antenna fucosylation. Another type of glycan biomarker for cancer is the truncated N-glycans. Increased levels of truncated non-galactosylated N-glycans existing mainly on IgG in serum have been reported for lung, gastric, and ovarian cancer. Truncated bisecting glycans in tissues were also elevated for ovarian cancer along with amplified expression of the glycosyltransferase GnT-III for bisecting GlcNAc.
Table 1.
Summary of recent glycan biomarker studies for cancer
| Cancer | Specimen | No. of samples* |
Targets** | Enrichment method |
Analytical method |
Ref | Outcome |
|---|---|---|---|---|---|---|---|
| Kidney | Plasma | 20/20 pooled |
G, P, GP | 12P depletion, M-LAC | LC-MS | 85 | ↑ Highly sialylated and high mannose glycans |
| Ovarian | Serum | 100/147 | G | PGC SPE | LC-MS | 155 | ↓ High mannose, hybrid, galactosylated biantennary glycans |
| Pancreatic | Serum | 16/10 | GP (sialylated) | Albumin depletion, LAC |
LC-MS | 82 | ↑ Fucosylation of fetuin A, AGP, and transferrin ↓ Sialylation |
| Liver | Plasma | 40/41 | P | N/A | LC-MRM | 156 | ↓ Vitronectin and AGP |
| Pancreatic | Serum | 30/37 pooled |
De-GP (fucosylated) | 14P depletion LAC |
LC-MS | 157 | •19 proteins differentially expressed in cancer. |
| Liver | Tissue | 16/16 | G | N/A | MALDI-IMS | 45 | ↑Tetra-antennary ↓ Man8 |
| Ovarian | Tissue | 3/3 | G and P (bisecting) | Lectin | LC-MS | 145 | ↑ GnT-III and truncated bisecting glycans |
| Pancreatic | Serum | 4/6 | G (human AGP) |
Immunoaffinity | LC-MS | 158 | ↑ Fucosylated glycans |
| Liver | Tissue | 3 | G | N/A | MALDI-IMS | 47 | ↑ Core-fucosylation, Hex7HexNAc6 |
| Liver | Serum | 26/26 | De-GP | Lectin | LC-MS | 150 | ↑ Core-fucosylation of fibronectin |
| Ovarian | Plasma | 82/82 | G | PGC SPE | LC-MS | 159 | •Seven N-glycans correlated with ovarian cancer stage |
| Colorectal | Tissue | 5/18 | G | C18, PGC SPE | MALDI-MS | 146 | ↑ Monoantennary, sialylated&fucosylated and small high-mannose N-glycan |
| Pancreatic | Serum | 13/20 | G, P (sialyl-LeX) |
Immunoaffinity | LC-MS | 160 | ↑ Sialyl-Lewis X epitopes on ceruloplasmin |
| Pancreatic | Serum | 13/13 | De-GP | Lectin | LC-MS | 151 | •Eight core-fucosylated peptides exhibited significant difference |
| Colon | Tissue | 2 | G | N/A | MALDI-IMS | 42 | ↑ High mannose in various tumor regions, sialylated glycans in different regions |
| Ovarian | Cell | 1 | P | Biotin affinity | LC-MS | 161 | •589 differentially expressed glycoproteins upon GALNT3 knockdown |
| Colorectal | Serum | 20/42 | G | N/A | MALDI-MS | 55 | ↑ Fucosylated tri- and tetra-antennary glycans |
| Gastric Colon Prostate etc. |
Serum | 5/5–26 | G, de-GP (haptoglobin) |
Immunoaffinity Sepharose 4B | LC-MS | 149 | ↑ Monofucosylated N-glycans at all sites and difucosylated at sites N184, N207 and N241 |
| Prostate | Urine, serum | Urine: 40 Serum: 48/119 pooled |
De-GP | Hydrazide beads, HPLC fractionation | LC-MS | 98 | ↑ Aggressive prostate cancer associated glycoproteins in patients’ urine than serum samples |
| Liver | Serum | 27/42 | GP (fucosylated) |
LAC | LC-MS | 152 | ↑ Multifucosylated tetra-antennary glycans on AGP |
| Ovarian | Ascites fluid | 5 | G, P, GP | PGC SPE, lectins affinity | LC-MS | 162 | •Large, highly fucosylated and sialylated complex and hybrid glycans, and unusual glycopeptides identified |
| Breast | Serum | 43/91 | G | PGC SPE | LC-MS | 163 | ↑ Difucosylated N-glycans |
| Ovarian | Serum | 84/84 | GP (Igs) |
N/A | LC-MRM | 72 | ↑ Biantennary mono- or disialylated glycans on IgM site N209 ↓ Galactosylation on IgG, and biantennary glycans with bisecting GlcNAc on IgA2 site N205 |
| Gastric | Serum | 144/157 | O-glycome | PGC SPE | LC-MS | 147 | ↑ Large Core2 O-glycans |
| Pancreatic | Serum | 6/19 | G, protein (AGP) |
Immunoaffinity | LC-MS CZE-UV |
148 | ↑ α(1–3) fucosylation of AGP |
| Liver, colorectal | Serum | 10/10 | GP | Albumin depletion | LC-MS MRM | 164 | ↑ Fucosylated N- and disialylated O-glycopeptides of hemopexin in HCC patients previously diagnosed with HCV |
| Gastric | Serum | 44/44 | Protein (haptoglobin) | Immunoaffinity | LC-MS | 79 | •N-glycan variation of serum haptoglobin associated with patients with gastric cancer |
| Gastric | Serum | 40/163 | G | Sepharose beads | MALDI-MS | 165 | ↑ Hybrid and tri-, tetra-antennnary glycans; ↓ Monoantennary, bisecting type and core fucose |
| Liver | Saliva | 50/60 | G | Sepharose 4B | MALDI-MS | 166 | ↑ Fucosylated ↓ Sialylated |
| Prostate | Tissue | 8/8 | P (sialylated) |
Biotin affinity | LC-MS | 167 | •Detection of 21 unique proteins in cancer tissues |
| Liver | Serum | 40/40 pooled |
De-GP (β1,6-GlcNAc) |
LAC | LC-MS | 168 | •Altered N-glycosite occupancy for 11 proteins with β(1,6) GlcNAc branching N-glycans |
| Ovarian | Serum, ascetic fluid | 20/18 | G | C18, PGC SPE | MALDI-MS | 169 | ↑ Branching, sialylation and antennary fucosylation ↓ High mannose, bisecting GlcNAc in ascetic fluid than serum |
Other possible glycan markers include high-mannose, hybrid-type N-glycans, sialylated N- or O-glycans, and O-glycans with different core structures. High-mannose and hybrid-type N-glycans are less-processed compared to complex-type. Although high mannose glycans have long been recognized as potential markers for cancer, upregulation appears to be dependent on the cancer type. In recent studies, high mannose has been shown to significantly increase in colorectal cancer tissues and clear cell renal cell carcinoma plasma, while decrease in ovarian cancer serum in several independent studies. Alteration of sialylation is less consistent among studies because it is often structure- and protein-specific. For example, a study by Kaprio et al. showed that the total abundance of sialylated N-glycans was increased from rectal adenoma to carcinoma.146 But detailed glycan profiling showed that the increase was only significant for six glycans, while certain sialylated glycans were actually decreased. O-Glycan cancer markers are less studied compared to N-glycans. In a gastric cancer study, He et al. showed that the expression levels of several extended core2 O-glycans including Hex5HexNAc3 and Hex3HexNAc3 were significantly increased in the patients.147
For identifying protein specific glycan markers, the most widely used strategy is to isolate glycoprotein of interest using antibody and analyze the purified protein using glycomic or glycoproteomic approaches. This strategy has recently been applied to identify protein specific markers for several cancers, such as α(1–3) fucosylated glycans on alpha-1-acid glycoprotein in pancreatic cancer,148 monofucosylated N-glycans at all sites and difucosylated N-glycans at sites N184, N207 and N241 of haptoglobin in five types of operable gastroenterological cancer.149 Although immunoaffinity enrichment can efficiently isolate specific glycoproteins, the limited number of available antibodies has generally restricted the types of proteins that can be targeted. For more global analysis of protein specific markers, glycoproteomic approaches are needed. A common approach involving quantitative analysis of deglycosylated glycopeptides enriched by lectins have been used to study aberrant changes of core-fucosylation in hepatocellular carcinoma150 and pancreatic cancer.151 Upregulation of core-fucosylated glycopeptides from certain glycoproteins were observed, such as fibronectin for hepatocellular carcinoma and macrophage mannose receptor 1 for pancreatic cancer.
Site-specific glycan marker discovery for cancer is more challenging due to the lack of high-throughput sample preparation methods and data processing software for simultaneous identification and quantitation of intact glycopeptides in complicated biological samples. MRM has been regarded as the gold standard methodology for this purpose and applied to the quantitation of immunoglobulin glycopeptides in cancer with high speed, sensitivity and repeatability. Decrease in galactosylation on several subclasses of IgG, and biantennary glycans with bisecting GlcNAc on site N205 of IgA2, as well as increase in biantennary mono- or disialylated glycans on site N209 of IgM have been identified and validated for ovarian cancer. Untargeted approaches have also been used for glycopeptide biomarker studies, but either with very small number of samples or concentrating only on a few proteins. A study by Tanabe et al. profiled lectin-enriched fucosylated glycoproteins in hepatocellular carcinoma serum.152 They analyzed the data using an in-house software and found elevated levels of alpha-1-acid glycoprotein with multifucosylated tetraantennary N-glycans in patients.
Methods for the analysis of intact glycoproteins are also emerging for discovery of specific cancer biomarkers involving proteins with aberrant glycosylation. A recent example is a gastric cancer study on intact haptoglobin, where novel glycosylated intensity and frequency markers were discovered.79 The best predictive marker corresponding to Hex22HexNAc18Fuc2NeuAc10 exhibited AUC of 0.93.
8. Future outlook
Glycan biomarkers are emerging and will soon fulfill the initial promise for sensitive and specific diagnosis of cancer. The consistency of distinct compounds and classes of compounds between separate and independent studies point to the robustness of the methods, even when different methods are used. Research in this area will continue to grow toward higher throughput glycomic profiles that can be obtained consistently with high reproducibility. Future studies should further focus on large clinical studies to obtain precise markers with high accuracy. The glycomic studies using known chemical structures could further identify the enzymes that alter glycosylation providing potential targets for therapeutic intervention.
Glycoproteins and site-specific glycan markers will continue to develop. Glycoproteomic analysis will improve in accuracy as the software tools are further developed for simultaneous quantitation of protein and site-specific glycosylation. Further refinements in this area will include the utility of data independent acquisition (DIA) of glycopeptides, which is now just being explored to analyze glycoproteins.153–154 Included in this effort will be the area of native glycoprotein analysis.
ACKNOWLEDGMENTS
Funding provided by the National Institutes of Health (grant R01GM049077) is gratefully acknowledged.
REFERENCES
- 1.Mereiter S; Balmana M; Gomes J; Magalhaes A; Reis CA, Glycomic Approaches for the Discovery of Targets in Gastrointestinal Cancer. Front Oncol 2016, 6, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sethi MK; Kim H; Park CK; Baker MS; Paik YK; Packer NH; Hancock WS; Fanayan S; Thaysen-Andersen M, In-depth N-glycome profiling of paired colorectal cancer and non-tumorigenic tissues reveals cancer-, stage- and EGFR-specific protein N-glycosylation. Glycobiology 2015, 25 (10), 1064–78. [DOI] [PubMed] [Google Scholar]
- 3.Holst S; Wuhrer M; Rombouts Y, Glycosylation characteristics of colorectal cancer. Adv Cancer Res 2015, 126, 203–56. [DOI] [PubMed] [Google Scholar]
- 4.Kailemia MJ; Park D; Lebrilla CB, Glycans and glycoproteins as specific biomarkers for cancer. Analytical and Bioanalytical Chemistry 2017, 409 (2), 395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meezan E; Wu HC; Black PH; Robbins PW, Comparative studies on the carbohydrate-containing membrane components of normal and virus-transformed mouse fibroblasts. II. Separation of glycoproteins and glycopeptides by sephadex chromatography. Biochemistry 1969, 8 (6), 2518–2524. [DOI] [PubMed] [Google Scholar]
- 6.Vajaria BN; Patel PS, Glycosylation: a hallmark of cancer? Glycoconjugate Journal 2017, 34 (2), 147–156. [DOI] [PubMed] [Google Scholar]
- 7.Varki A, Biological roles of glycans. Glycobiology 2017, 27 (1), 3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stowell SR; Ju T; Cummings RD, Protein glycosylation in cancer. Annual Review of Pathology: Mechanisms of Disease 2015, 10, 473–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliveira-Ferrer L; Legler K; Milde-Langosch K In Role of protein glycosylation in cancer metastasis, Seminars in Cancer Biology, Elsevier: 2017. [DOI] [PubMed] [Google Scholar]
- 10.Häuselmann I; Borsig L, Altered tumor-cell glycosylation promotes metastasis. Frontiers in oncology 2014, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho W-L; Hsu W-M; Huang M-C; Kadomatsu K; Nakagawara A, Protein glycosylation in cancers and its potential therapeutic applications in neuroblastoma. Journal of Hematology & Oncology 2016, 9, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhide GP; Colley KJ, Sialylation of N-glycans: mechanism, cellular compartmentalization and function. Histochemistry and Cell Biology 2017, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassaganas S; Carvalho S; Dias AM; Pérez-Garay M; Ortiz MR; Figueras J; Reis CA; Pinho SS; Peracaula R, Pancreatic cancer cell glycosylation regulates cell adhesion and invasion through the modulation of α2β1 integrin and E-cadherin function. PloS one 2014, 9 (5), e98595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakomori S.-i., Tumor malignancy defined by aberrant glycosylation and sphingo (glyco) lipid metabolism. Cancer research 1996, 56 (23), 5309–5318. [PubMed] [Google Scholar]
- 15.Gao W; Kim H; Ho M, Human monoclonal antibody targeting the heparan sulfate chains of glypican-3 inhibits HGF-mediated migration and motility of hepatocellular carcinoma cells. PloS one 2015, 10 (9), e0137664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clausen TM; Pereira MA; Al Nakouzi N; Oo HZ; Agerbæk MØ; Lee S; Ørum-Madsen MS; Kristensen AR; El-Naggar A; Grandgenett PM, Oncofetal chondroitin sulfate glycosaminoglycans are key players in integrin signaling and tumor cell motility. Molecular Cancer Research 2016, 14 (12), 1288–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng X-B; Kohi S; Koga A; Hirata K; Sato N, Hyaluronan stimulates pancreatic cancer cell motility. Oncotarget 2016, 7 (4), 4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu M; Cao M; He Y; Liu Y; Yang C; Du Y; Wang W; Gao F, A novel role of low molecular weight hyaluronan in breast cancer metastasis. The FASEB Journal 2015, 29 (4), 1290–1298. [DOI] [PubMed] [Google Scholar]
- 19.Zoldoš V; Novokmet M; Bečeheli I; Lauc G, Genomics and epigenomics of the human glycome. Glycoconjugate journal 2013, 30 (1), 41–50. [DOI] [PubMed] [Google Scholar]
- 20.Wang X; Chen J; Li QK; Peskoe SB; Zhang B; Choi C; Platz EA; Zhang H, Overexpression of α (1, 6) fucosyltransferase associated with aggressive prostate cancer. Glycobiology 2014, 24 (10), 935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wada Y, Mass spectrometry of transferrin and apolipoprotein C-III for diagnosis and screening of congenital disorder of glycosylation. Glycoconj J 2016, 33 (3), 297–307. [DOI] [PubMed] [Google Scholar]
- 22.Ruhaak LR; Barkauskas DA; Torres J; Cooke CL; Wu LD; Stroble C; Ozcan S; Williams CC; Camorlinga M; Rocke DM; Lebrilla CB; Solnick JV, The Serum Immunoglobulin G Glycosylation Signature of Gastric Cancer. EuPA Open Proteom 2015, 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang N; Goonatilleke E; Park D; Song T; Fan G; Lebrilla CB, Quantitation of Site-Specific Glycosylation in Manufactured Recombinant Monoclonal Antibody Drugs. Anal Chem 2016, 88 (14), 7091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medzihradszky KF; Kaasik K; Chalkley RJ, Tissue-Specific Glycosylation at the Glycopeptide Level. Mol Cell Proteomics 2015, 14 (8), 2103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park D; Xu G; Barboza M; Shah IM; Wong M; Raybould H; Mills DA; Lebrilla CB, Enterocyte glycosylation is responsive to changes in extracellular conditions: implications for membrane functions. Glycobiology 2017, 27 (9), 847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuster MM; Esko JD, The sweet and sour of cancer: glycans as novel therapeutic targets. Nature Reviews Cancer 2005, 5 (7), 526–542. [DOI] [PubMed] [Google Scholar]
- 27.Song X; Ju H; Lasanajak Y; Kudelka MR; Smith DF; Cummings RD, Oxidative release of natural glycans for functional glycomics. Nat Meth 2016, 13 (6), 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An HJ; Miyamoto S; Lancaster KS; Kirmiz C; Li B; Lam KS; Leiserowitz GS; Lebrilla CB, Profiling of glycans in serum for the discovery of potential biomarkers for ovarian cancer. J Proteome Res 2006, 5 (7), 1626–35. [DOI] [PubMed] [Google Scholar]
- 29.Kailemia MJ; Ruhaak LR; Lebrilla CB; Amster IJ, Oligosaccharide analysis by mass spectrometry: a review of recent developments. Anal Chem 2014, 86 (1), 196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ninonuevo MR; Lebrilla CB, Mass spectrometric methods for analysis of oligosaccharides in human milk. Nutr Rev 2009, 67 Suppl 2, S216–26. [DOI] [PubMed] [Google Scholar]
- 31.Aldredge D; An HJ; Tang N; Waddell K; Lebrilla CB, Annotation of a serum N-glycan library for rapid identification of structures. J Proteome Res 2012, 11 (3), 1958–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song T; Aldredge D; Lebrilla CB, A Method for In-Depth Structural Annotation of Human Serum Glycans That Yields Biological Variations. Anal Chem 2015, 87 (15), 7754–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Y; Borges CR, A spin column-free approach to sodium hydroxide-based glycan permethylation. Analyst 2017, 142 (15), 2748–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaare S; Aguilar JS; Hu YM; Ferdosi S; Borges CR, Glycan Node Analysis: A Bottom-up Approach to Glycomics. Jove-J Vis Exp 2016, (111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruhaak LR; Miyamoto S; Lebrilla CB, Developments in the Identification of Glycan Biomarkers for the Detection of Cancer. Molecular & Cellular Proteomics 2013, 12 (4), 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harvey DJ, Matrix‐assisted laser desorption/ionization mass spectrometry of carbohydrates. Mass Spectrometry Reviews 1999, 18 (6), 349–450. [DOI] [PubMed] [Google Scholar]
- 37.An HJ; Peavy TR; Hedrick JL; Lebrilla CB, Determination of N-glycosylation sites and site heterogeneity in glycoproteins. Analytical chemistry 2003, 75 (20), 5628–5637. [DOI] [PubMed] [Google Scholar]
- 38.Svarovsky SA; Joshi L, Cancer glycan biomarkers and their detection–past, present and future. Analytical Methods 2014, 6 (12), 3918–3936. [Google Scholar]
- 39.Stowell SR; Ju T; Cummings RD, Protein Glycosylation in Cancer. Annual review of pathology 2015, 10, 473–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kailemia MJ; Ruhaak LR; Lebrilla CB; Amster IJ, Oligosaccharide analysis by mass spectrometry: a review of recent developments. Analytical chemistry 2013, 86 (1), 196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shubhakar A; Kozak RP; Reiding KR; Royle L; Spencer DI; Fernandes DL; Wuhrer M, Automated High-Throughput Permethylation for Glycosylation Analysis of Biologics Using MALDI-TOF-MS. Anal Chem 2016, 88 (17), 8562–9. [DOI] [PubMed] [Google Scholar]
- 42.Holst S; Heijs B; de Haan N; van Zeijl RJM; Briaire-de Bruijn IH; van Pelt GW; Mehta AS; Angel PM; Mesker WE; Tollenaar RA; Drake RR; Bovée JVMG; McDonnell LA; Wuhrer M, Linkage-Specific in Situ Sialic Acid Derivatization for N-Glycan Mass Spectrometry Imaging of Formalin-Fixed Paraffin-Embedded Tissues. Analytical Chemistry 2016, 88 (11), 5904–5913. [DOI] [PubMed] [Google Scholar]
- 43.Reiding KR; Blank D; Kuijper DM; Deelder AM; Wuhrer M, High-throughput profiling of protein N-glycosylation by MALDI-TOF-MS employing linkage-specific sialic acid esterification. Anal Chem 2014, 86 (12), 5784–93. [DOI] [PubMed] [Google Scholar]
- 44.Drake RR; Powers TW; Jones EE; Bruner E; Mehta AS; Angel PM, MALDI Mass Spectrometry Imaging of N-Linked Glycans in Cancer Tissues. Adv Cancer Res 2017, 134, 85–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Powers TW; Neely BA; Shao Y; Tang H; Troyer DA; Mehta AS; Haab BB; Drake RR, MALDI Imaging Mass Spectrometry Profiling of N-Glycans in Formalin-Fixed Paraffin Embedded Clinical Tissue Blocks and Tissue Microarrays. PLOS ONE 2014, 9 (9), e106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heijs B; Holst S; Briaire-de Bruijn IH; van Pelt GW; de Ru AH; van Veelen PA; Drake RR; Mehta AS; Mesker WE; Tollenaar RA; Bovée JVMG; Wuhrer M; McDonnell LA, Multimodal Mass Spectrometry Imaging of N-Glycans and Proteins from the Same Tissue Section. Analytical Chemistry 2016, 88 (15), 7745–7753. [DOI] [PubMed] [Google Scholar]
- 47.Powers T; Holst S; Wuhrer M; Mehta A; Drake R, Two-Dimensional N-Glycan Distribution Mapping of Hepatocellular Carcinoma Tissues by MALDI-Imaging Mass Spectrometry. Biomolecules 2015, 5 (4), 2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Everest-Dass AV; Briggs MT; Kaur G; Oehler MK; Hoffmann P; Packer NH, N-Glycan MALDI Imaging Mass Spectrometry on Formalin-Fixed Paraffin-Embedded Tissue Enables the Delineation of Ovarian Cancer Tissues. Molecular & Cellular Proteomics 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nazari M; Muddiman DC, Polarity switching mass spectrometry imaging of healthy and cancerous hen ovarian tissue sections by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Analyst 2016, 141 (2), 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durney BC; Crihfield CL; Holland LA, Capillary electrophoresis applied to DNA: determining and harnessing sequence and structure to advance bioanalyses (2009–2014). Anal Bioanal Chem 2015, 407 (23), 6923–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mantovani V; Galeotti F; Maccari F; Volpi N, Recent advances on separation and characterization of human milk oligosaccharides. Electrophoresis 2016, 37 (11), 1514–24. [DOI] [PubMed] [Google Scholar]
- 52.Mechref Y, Analysis of glycans derived from glycoconjugates by capillary electrophoresis‐mass spectrometry. Electrophoresis 2011, 32 (24), 3467–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun X; Lin L; Liu X; Zhang F; Chi L; Xia Q; Linhardt RJ, Capillary Electrophoresis–Mass Spectrometry for the Analysis of Heparin Oligosaccharides and Low Molecular Weight Heparin. Analytical Chemistry 2016, 88 (3), 1937–1943. [DOI] [PubMed] [Google Scholar]
- 54.Khatri K; Klein JA; Haserick JR; Leon DR; Costello CE; McComb ME; Zaia J, Microfluidic Capillary Electrophoresis-Mass Spectrometry for Analysis of Monosaccharides, Oligosaccharides, and Glycopeptides. Anal Chem 2017, 89 (12), 6645–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snyder CM; Alley WR Jr; Campos MI; Svoboda M; Goetz JA; Vasseur JA; Jacobson SC; Novotny MV, Complementary Glycomic Analyses of Sera Derived from Colorectal Cancer Patients by MALDI-TOF-MS and Microchip Electrophoresis. Analytical chemistry 2016, 88 (19), 9597–9605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guile GR; Rudd PM; Wing DR; Prime SB; Dwek RA, A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Analytical biochemistry 1996, 240 (2), 210–226. [DOI] [PubMed] [Google Scholar]
- 57.Stavenhagen K; Kolarich D; Wuhrer M, Clinical Glycomics Employing Graphitized Carbon Liquid Chromatography-Mass Spectrometry. Chromatographia 2015, 78 (5–6), 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vreeker GC; Wuhrer M, Reversed-phase separation methods for glycan analysis. Analytical and bioanalytical chemistry 2017, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahn J; Bones J; Yu YQ; Rudd PM; Gilar M, Separation of 2-aminobenzamide labeled glycans using hydrophilic interaction chromatography columns packed with 1.7μm sorbent. Journal of Chromatography B 2010, 878 (3), 403–408. [DOI] [PubMed] [Google Scholar]
- 60.Stöckmann H; Duke RM; Millán Martín S; Rudd PM, Ultrahigh Throughput, Ultrafiltration-Based N-Glycomics Platform for Ultraperformance Liquid Chromatography (ULTRA3). Analytical Chemistry 2015, 87 (16), 8316–8322. [DOI] [PubMed] [Google Scholar]
- 61.Adamczyk B; Tharmalingam-Jaikaran T; Schomberg M; Szekrényes Á; Kelly RM; Karlsson NG; Guttman A; Rudd PM, Comparison of separation techniques for the elucidation of IgG N-glycans pooled from healthy mammalian species. Carbohydrate Research 2014, 389, 174–185. [DOI] [PubMed] [Google Scholar]
- 62.Stockmann H; O’Flaherty R; Adamczyk B; Saldova R; Rudd PM, Automated, high-throughput serum glycoprofiling platform. Integrative Biology 2015, 7 (9), 1026–1032. [DOI] [PubMed] [Google Scholar]
- 63.Marino K; Bones J; Kattla JJ; Rudd PM, A systematic approach to protein glycosylation analysis: a path through the maze. Nat Chem Biol 2010, 6 (10), 713–723. [DOI] [PubMed] [Google Scholar]
- 64.Sethi MK; Hancock WS; Fanayan S, Identifying N-Glycan Biomarkers in Colorectal Cancer by Mass Spectrometry. Acc Chem Res 2016, 49 (10), 2099–2106. [DOI] [PubMed] [Google Scholar]
- 65.Xu G; Davis JC; Goonatilleke E; Smilowitz JT; German JB; Lebrilla CB, Absolute Quantitation of Human Milk Oligosaccharides Reveals Phenotypic Variations during Lactation. J Nutr 2017, 147 (1), 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nouso K; Amano M; Ito YM; Miyahara K; Morimoto Y; Kato H; Tsutsumi K; Tomoda T; Yamamoto N; Nakamura S; Kobayashi S; Kuwaki K; Hagihara H; Onishi H; Miyake Y; Ikeda F; Shiraha H; Takaki A; Nakahara T; Nishimura S; Yamamoto K, Clinical utility of high-throughput glycome analysis in patients with pancreatic cancer. J Gastroenterol 2013, 48 (10), 1171–9. [DOI] [PubMed] [Google Scholar]
- 67.Ruhaak LR; Taylor SL; Miyamoto S; Kelly K; Leiserowitz GS; Gandara D; Lebrilla CB; Kim K, Chip-based nLC-TOF-MS is a highly stable technology for large-scale high-throughput analyses. Anal Bioanal Chem 2013, 405 (14), 4953–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong Q; Lebrilla CB; Miyamoto S; Ruhaak LR, Absolute quantitation of immunoglobulin G and its glycoforms using multiple reaction monitoring. Anal Chem 2013, 85 (18), 8585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu S; Tao N; German JB; Grimm R; Lebrilla CB, Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res 2010, 9 (8), 4138–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Etxebarria J; Reichardt NC, Methods for the absolute quantification of N-glycan biomarkers. Biochimica Et Biophysica Acta-General Subjects 2016, 1860 (8), 1676–1687. [DOI] [PubMed] [Google Scholar]
- 71.Mechref Y; Hu Y; Desantos-Garcia JL; Hussein A; Tang H, Quantitative glycomics strategies. Mol Cell Proteomics 2013, 12 (4), 874–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruhaak LR; Kim K; Stroble C; Taylor SL; Hong Q; Miyamoto S; Lebrilla CB; Leiserowitz G, Protein-Specific Differential Glycosylation of Immunoglobulins in Serum of Ovarian Cancer Patients. J Proteome Res 2016, 15 (3), 1002–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruhaak LR; Stroble C; Dai J; Barnett M; Taguchi A; Goodman GE; Miyamoto S; Gandara D; Feng Z; Lebrilla CB; Hanash S, Serum Glycans as Risk Markers for Non-Small Cell Lung Cancer. Cancer Prev Res (Phila) 2016, 9 (4), 317–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ruhaak LR; Taylor SL; Stroble C; Nguyen UT; Parker EA; Song T; Lebrilla CB; Rom WN; Pass H; Kim K; Kelly K; Miyamoto S, Differential N-Glycosylation Patterns in Lung Adenocarcinoma Tissue. J Proteome Res 2015, 14 (11), 4538–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H; Popp R; Borchers CH, Affinity-mass spectrometric technologies for quantitative proteomics in biological fluids. TrAC Trends in Analytical Chemistry 2017, 90, 80–88. [Google Scholar]
- 76.Gbormittah FO; Bones J; Hincapie M; Tousi F; Hancock WS; Iliopoulos O, Clusterin Glycopeptide Variant Characterization Reveals Significant Site-Specific Glycan Changes in the Plasma of Clear Cell Renal Cell Carcinoma. Journal of Proteome Research 2015, 14 (6), 2425–2436. [DOI] [PubMed] [Google Scholar]
- 77.Takahashi S; Sugiyama T; Shimomura M; Kamada Y; Fujita K; Nonomura N; Miyoshi E; Nakano M, Site-specific and linkage analyses of fucosylated N-glycans on haptoglobin in sera of patients with various types of cancer: possible implication for the differential diagnosis of cancer. Glycoconjugate Journal 2016, 33 (3), 471–482. [DOI] [PubMed] [Google Scholar]
- 78.van Scherpenzeel M; Steenbergen G; Morava E; Wevers RA; Lefeber DJ, High-resolution mass spectrometry glycoprofiling of intact transferrin for diagnosis and subtype identification in the congenital disorders of glycosylation. Transl Res 2015, 166 (6), 639–649 e1. [DOI] [PubMed] [Google Scholar]
- 79.Kim JH; Lee SH; Choi S; Kim U; Yeo IS; Kim SH; Oh MJ; Moon H; Lee J; Jeong S; Choi MG; Lee JH; Sohn TS; Bae JM; Kim S; Min YW; Lee H; Lee JH; Rhee PL; Kim JJ; Lee SJ; Kim ST; Lee J; Park SH; Park JO; Park YS; Lim HY; Kang WK; An HJ; Kim JH, Direct analysis of aberrant glycosylation on haptoglobin in patients with gastric cancer. Oncotarget 2017, 8 (7), 11094–11104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Popp R; Malmstrom D; Chambers AG; Lin D; Camenzind AG; van der Gugten JG; Holmes DT; Pugia M; Jaremek M; Cornett S; Suckau D; Borchers CH, An automated assay for the clinical measurement of plasma renin activity by immuno-MALDI (iMALDI). Biochim Biophys Acta 2015, 1854 (6), 547–58. [DOI] [PubMed] [Google Scholar]
- 81.Chen YT; Chen HW; Wu CF; Chu LJ; Chiang WF; Wu CC; Yu JS; Tsai CH; Liang KH; Chang YS; Wu M; Ou Yang WT, Development of a Multiplexed Liquid Chromatography Multiple-Reaction-Monitoring Mass Spectrometry (LC-MRM/MS) Method for Evaluation of Salivary Proteins as Oral Cancer Biomarkers. Mol Cell Proteomics 2017, 16 (5), 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kontro H; Joenvaara S; Haglund C; Renkonen R, Comparison of sialylated N-glycopeptide levels in serum of pancreatic cancer patients, acute pancreatitis patients, and healthy controls. Proteomics 2014, 14 (15), 1713–1723. [DOI] [PubMed] [Google Scholar]
- 83.Zeng Z; Hincapie M; Pitteri SJ; Hanash S; Schakwijk J; Hogan JM; Wang H; Hancock WS, A Proteomics Platform Combining Depletion, Multi-lectin Affinity Chromatography (M-LAC), and Isoelectric Focusing to Study the Breast Cancer Proteome. Analytical Chemistry 2011, 83 (12), 4845–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Totten SM; Kullolli M; Pitteri SJ, Multi-Lectin Affinity Chromatography for Separation, Identification, and Quantitation of Intact Protein Glycoforms in Complex Biological Mixtures. Methods Mol Biol 2017, 1550, 99–113. [DOI] [PubMed] [Google Scholar]
- 85.Gbormittah FO; Lee LY; Taylor K; Hancock WS; Iliopoulos O, Comparative Studies of the Proteome, Glycoproteome, and N-Glycome of Clear Cell Renal Cell Carcinoma Plasma before and after Curative Nephrectomy. Journal of Proteome Research 2014, 13 (11), 4889–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou J; Yang W; Hu Y; Höti N; Liu Y; Shah P; Sun S; Clark D; Thomas S; Zhang H, Site-Specific Fucosylation Analysis Identifying Glycoproteins Associated with Aggressive Prostate Cancer Cell Lines Using Tandem Affinity Enrichments of Intact Glycopeptides Followed by Mass Spectrometry. Analytical Chemistry 2017, 89 (14), 7623–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Syed P; Gidwani K; Kekki H; Leivo J; Pettersson K; Lamminmaki U, Role of lectin microarrays in cancer diagnosis. Proteomics 2016, 16 (8), 1257–1265. [DOI] [PubMed] [Google Scholar]
- 88.Gray CJ; Sanchez-Ruiz A; Sardzikova I; Ahmed YA; Miller RL; Martinez JER; Pallister E; Huang K; Both P; Hartmann M; Roberts HN; Sardzik R; Mandal S; Turnbull JE; Eyers CE; Flitsch SL, Label-Free Discovery Array Platform for the Characterization of Glycan Binding Proteins and Glycoproteins. Analytical Chemistry 2017, 89 (8), 4444–4451. [DOI] [PubMed] [Google Scholar]
- 89.Dong L; Feng S; Li S; Song P; Wang J, Preparation of Concanavalin A-Chelating Magnetic Nanoparticles for Selective Enrichment of Glycoproteins. Anal Chem 2015, 87 (13), 6849–53. [DOI] [PubMed] [Google Scholar]
- 90.Li YG; Wen T; Zhu MZ; Li LX; Wei J; Wu XL; Guo MZ; Liu SP; Zhao HY; Xia SY; Huang WL; Wang PY; Wu ZZ; Zhao LQ; Shui WQ; Li Z; Yin ZN, Glycoproteomic analysis of tissues from patients with colon cancer using lectin microarrays and nanoLC-MS/MS. Molecular Biosystems 2013, 9 (7), 1877–1887. [DOI] [PubMed] [Google Scholar]
- 91.Shah AK; Le Cao KA; Choi E; Chen D; Gautier B; Nancarrow D; Whiteman DC; Saunders NA; Barbour AP; Joshi V; Hill MM, Serum Glycoprotein Biomarker Discovery and Qualification Pipeline Reveals Novel Diagnostic Biomarker Candidates for Esophageal Adenocarcinoma. Molecular & Cellular Proteomics 2015, 14 (11), 3023–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beloqui A; Calvo J; Serna S; Yan S; Wilson IBH; Martin-Lomas M; Reichardt NC, Analysis of Microarrays by MALDI-TOF MS. Angew Chem Int Edit 2013, 52 (29), 7477–7481. [DOI] [PubMed] [Google Scholar]
- 93.Saldova R; Struwe WB; Wynne K; Elia G; Duffy MJ; Rudd PM, Exploring the glycosylation of serum CA125. Int J Mol Sci 2013, 14 (8), 15636–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Llop E; Ferrer-Batalle M; Barrabes S; Guerrero PE; Ramirez M; Saldova R; Rudd PM; Aleixandre RN; Comet J; de Llorens R; Peracaula R, Improvement of Prostate Cancer Diagnosis by Detecting PSA Glycosylation-Specific Changes. Theranostics 2016, 6 (8), 1190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shah P; Wang X; Yang W; Toghi Eshghi S; Sun S; Hoti N; Chen L; Yang S; Pasay J; Rubin A; Zhang H, Integrated Proteomic and Glycoproteomic Analyses of Prostate Cancer Cells Reveal Glycoprotein Alteration in Protein Abundance and Glycosylation. Mol Cell Proteomics 2015, 14 (10), 2753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ji Y; Wei S; Hou J; Zhang C; Xue P; Wang J; Chen X; Guo X; Yang F, Integrated proteomic and N-glycoproteomic analyses of doxorubicin sensitive and resistant ovarian cancer cells reveal glycoprotein alteration in protein abundance and glycosylation. Oncotarget 2017, 8 (8), 13413–13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li QK; Shah P; Tian Y; Hu Y; Roden RBS; Zhang H; Chan DW, An integrated proteomic and glycoproteomic approach uncovers differences in glycosylation occupancy from benign and malignant epithelial ovarian tumors. Clin Proteomics 2017, 14, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jia X; Chen J; Sun S; Yang W; Yang S; Shah P; Hoti N; Veltri B; Zhang H, Detection of aggressive prostate cancer associated glycoproteins in urine using glycoproteomics and mass spectrometry. Proteomics 2016, 16 (23), 2989–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hong Q; Ruhaak LR; Stroble C; Parker E; Huang J; Maverakis E; Lebrilla CB, A Method for Comprehensive Glycosite-Mapping and Direct Quantitation of Serum Glycoproteins. J Proteome Res 2015, 14 (12), 5179–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mulagapati S; Koppolu V; Raju TS, Decoding of O-Linked Glycosylation by Mass Spectrometry. Biochemistry 2017, 56 (9), 1218–1226. [DOI] [PubMed] [Google Scholar]
- 101.Zhang S; Li W; Lu H; Liu Y, Quantification of N-glycosylation site occupancy status based on labeling/label-free strategies with LC-MS/MS. Talanta 2017, 170, 509–513. [DOI] [PubMed] [Google Scholar]
- 102.Bern M; Kil YJ; Becker C, Byonic: advanced peptide and protein identification software. Current protocols in bioinformatics 2012, 13.20. 1–13.20. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Strum JS; Nwosu CC; Hua S; Kronewitter SR; Seipert RR; Bachelor RJ; An HJ; Lebrilla CB, Automated Assignments of N- and O-Site Specific Glycosylation with Extensive Glycan Heterogeneity of Glycoprotein Mixtures. Analytical Chemistry 2013, 85 (12), 5666–5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zeng W-F; Liu M-Q; Zhang Y; Wu J-Q; Fang P; Peng C; Nie A; Yan G; Cao W; Liu C, pGlyco: a pipeline for the identification of intact N-glycopeptides by using HCD-and CID-MS/MS and MS3. Scientific reports 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu MQ; Zeng WF; Fang P; Cao WQ; Liu C; Yan GQ; Zhang Y; Peng C; Wu JQ; Zhang XJ; Tu HJ; Chi H; Sun RX; Cao Y; Dong MQ; Jiang BY; Huang JM; Shen HL; Wong CCL; He SM; Yang PY, pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat Commun 2017, 8 (1), 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park GW; Kim JY; Hwang H; Lee JY; Ahn YH; Lee HK; Ji ES; Kim KH; Jeong HK; Yun KN; Kim YS; Ko JH; An HJ; Kim JH; Paik YK; Yoo JS, Integrated GlycoProteome Analyzer (I-GPA) for Automated Identification and Quantitation of Site-Specific N-Glycosylation. Scientific Reports 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dutta SM; Yen T-Y; Semmes JO; Macher B, Abstract 3221: Bioinformatic approaches for large-scale identification and relative quantitation of N-glycosylation sites from breast cancer cell lines. Cancer Research 2013, 73 (8 Supplement), 3221–3221. [Google Scholar]
- 108.Xu G; Liu X; Liu QY; Li J; Zhou Y, Automatic annotation and visualization tool for mass spectrometry based glycomics. Rapid Communications in Mass Spectrometry 2016, 30 (23), 2471–2479. [DOI] [PubMed] [Google Scholar]
- 109.Kumozaki S; Sato K; Sakakibara Y, A Machine Learning Based Approach to de novo Sequencing of Glycans from Tandem Mass Spectrometry Spectrum. Ieee-Acm Transactions on Computational Biology and Bioinformatics 2015, 12 (6), 1267–1274. [DOI] [PubMed] [Google Scholar]
- 110.Chiu YL; Huang RR; Orlando R; Sharp JS, GAG-ID: Heparan Sulfate (HS) and Heparin Glycosaminoglycan High-Throughput Identification Software. Molecular & Cellular Proteomics 2015, 14 (6), 1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hizal DB; Wolozny D; Colao J; Jacobson E; Tian Y; Krag SS; Betenbaugh MJ; Zhang H, Glycoproteomic and glycomic databases. Clinical Proteomics 2014, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Egorova KS; Kalinchuk NA; Knirel YA; Toukach PV, Carbohydrate Structure Database (CSDB): new features. Russian Chemical Bulletin 2015, 64 (5), 1205–1210. [Google Scholar]
- 113.Akune Y; Lin CH; Abrahams JL; Zhang JY; Packer NH; Aoki-Kinoshita KF; Campbell MP, Comprehensive analysis of the N-glycan biosynthetic pathway using bioinformatics to generate UniCorn: A theoretical N-glycan structure database. Carbohydrate Research 2016, 431, 56–63. [DOI] [PubMed] [Google Scholar]
- 114.Aoki-Kinoshita K; Agravat S; Aoki NP; Arpinar S; Cummings RD; Fujita A; Fujita N; Hart GM; Haslam SM; Kawasaki T; Matsubara M; Moreman KW; Okuda S; Pierce M; Ranzinger R; Shikanai T; Shinmachi D; Solovieva E; Suzuki Y; Tsuchiya S; Yamada I; York WS; Zaia J; Narimatsu H, GlyTouCan 1.0-The international glycan structure repository. Nucleic Acids Research 2016, 44 (D1), D1237–D1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hakomori S.-i., Aberrant Glycosylation in Cancer Cell Membranes as Focused on Glycolipids: Overview and Perspectives. Cancer Research 1985, 45 (6), 2405–2414. [PubMed] [Google Scholar]
- 116.Hakomori S, Structure, organization, and function of glycosphingolipids in membrane. Current Opinion in Hematology 2003, 10 (1), 16–24. [DOI] [PubMed] [Google Scholar]
- 117.Pinho SS; Reis CA, Glycosylation in cancer: mechanisms and clinical implications. Nature reviews. Cancer 2015, 15 (9), 540. [DOI] [PubMed] [Google Scholar]
- 118.Safi F; Schlosser W; Kolb G; Beger HG, Diagnostic value of CA 19–9 in patients with pancreatic cancer and nonspecific gastrointestinal symptoms. Journal of Gastrointestinal Surgery 1997, 1 (2), 106–112. [DOI] [PubMed] [Google Scholar]
- 119.Kovbasnjuk O; Mourtazina R; Baibakov B; Wang T; Elowsky C; Choti MA; Kane A; Donowitz M, The glycosphingolipid globotriaosylceramide in the metastatic transformation of colon cancer. Proceedings of the National Academy of Sciences of the United States of America 2005, 102 (52), 19087–19092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yoshida S; Fukumoto S; Kawaguchi H; Sato S; Ueda R; Furukawa K, Ganglioside GD2 in Small Cell Lung Cancer Cell Lines. Enhancement of Cell Proliferation and Mediation of Apoptosis 2001, 61 (10), 4244–4252. [PubMed] [Google Scholar]
- 121.Marquina G; Waki H; Fernandez LE; Kon K; Carr A; Valiente O; Perez R; Ando S, Gangliosides Expressed in Human Breast Cancer. Cancer Research 1996, 56 (22), 5165–5171. [PubMed] [Google Scholar]
- 122.Hamasaki H; Aoyagi M; Kasama T; Handa S; Hirakawa K; Taki T, GT1b in human metastatic brain tumors: GT1b as a brain metastasis-associated ganglioside1. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1999, 1437 (1), 93–99. [DOI] [PubMed] [Google Scholar]
- 123.Shinoura N; Dohi T; Kondo T; Yoshioka M; Takakura K; Oshima M, Ganglioside Composition and Its Relation to Clinical Data in Brain Tumors. Neurosurgery 1992, 31 (3), 541–549. [DOI] [PubMed] [Google Scholar]
- 124.Cheung SKC; Chuang P-K; Huang H-W; Hwang-Verslues WW; Cho CH-H; Yang W-B; Shen C-N; Hsiao M; Hsu T-L; Chang C-F; Wong C-H, Stage-specific embryonic antigen-3 (SSEA-3) and β3GalT5 are cancer specific and significant markers for breast cancer stem cells. Proceedings of the National Academy of Sciences 2016, 113 (4), 960–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Furukawa K; Ohkawa Y; Matsumoto Y; Ohmi Y; Hashimoto N; Furukawa K, Regulatory Mechanisms for Malignant Properties of Cancer Cells with Disialyl and Monosialyl Gangliosides In Glycosignals in Cancer: Mechanisms of Malignant Phenotypes, Furukawa K; Fukuda M, Eds. Springer; Japan: Tokyo, 2016; pp 57–76. [Google Scholar]
- 126.Merrill AH, SphinGOMAP: Lipidomic analysis of [glyco]sphingolipid metabolism. The FASEB Journal 2006, 20 (5), A1472. [Google Scholar]
- 127.Bethke U; Müthing J; Schauder B; Conradt P; Mühlradt PF, An improved semi-quantitative enzyme immunostaining procedure for glycosphingolipid antigens on high performance thin layer chromatograms. Journal of Immunological Methods 1986, 89 (1), 111–116. [DOI] [PubMed] [Google Scholar]
- 128.Wuhrer M; Deelder AM; van der Burgt YEM, Mass spectrometric glycan rearrangements. Mass Spectrometry Reviews 2011, 30 (4), 664–680. [DOI] [PubMed] [Google Scholar]
- 129.Merrill AH, Sphingolipid and Glycosphingolipid Metabolic Pathways in the Era of Sphingolipidomics. Chemical Reviews 2011, 111 (10), 6387–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Folch J; Lees M; Sloane-Stanley G, A simple method for the isolation and purification of total lipids from animal tissues. The Journal of biological chemistry 1957, 226 (1), 497–509. [PubMed] [Google Scholar]
- 131.Löfgren L; Forsberg G-B; Ståhlman M, The BUME method: a new rapid and simple chloroform-free method for total lipid extraction of animal tissue. Scientific Reports 2016, 6, 27688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dreisewerd K; Müthing J; Rohlfing A; Meisen I; Vukelić Ž; Peter-Katalinić J; Hillenkamp F; Berkenkamp S, Analysis of Gangliosides Directly from Thin-Layer Chromatography Plates by Infrared Matrix-Assisted Laser Desorption/Ionization Orthogonal Time-of-Flight Mass Spectrometry with a Glycerol Matrix. Analytical Chemistry 2005, 77 (13), 4098–4107. [DOI] [PubMed] [Google Scholar]
- 133.Jirásko R; Holčapek M; Khalikova M; Vrána D; Študent V; Prouzová Z; Melichar B, MALDI Orbitrap Mass Spectrometry Profiling of Dysregulated Sulfoglycosphingolipids in Renal Cell Carcinoma Tissues. Journal of The American Society for Mass Spectrometry 2017, 28 (8), 1562–1574. [DOI] [PubMed] [Google Scholar]
- 134.Zamfir AD; Fabris D; Capitan F; Munteanu C; Vukelić Ž; Flangea C, Profiling and sequence analysis of gangliosides in human astrocytoma by high-resolution mass spectrometry. Analytical and Bioanalytical Chemistry 2013, 405 (23), 7321–7335. [DOI] [PubMed] [Google Scholar]
- 135.Holst S; Stavenhagen K; Balog CIA; Koeleman CAM; McDonnell LM; Mayboroda OA; Verhoeven A; Mesker WE; Tollenaar RAEM; Deelder AM; Wuhrer M, Investigations on Aberrant Glycosylation of Glycosphingolipids in Colorectal Cancer Tissues Using Liquid Chromatography and Matrix-Assisted Laser Desorption Time-of-Flight Mass Spectrometry (MALDI-TOF-MS). Molecular & Cellular Proteomics 2013, 12 (11), 3081–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Albrecht S; Vainauskas S; Stöckmann H; McManus C; Taron CH; Rudd PM, Comprehensive Profiling of Glycosphingolipid Glycans Using a Novel Broad Specificity Endoglycoceramidase in a High-Throughput Workflow. Analytical Chemistry 2016, 88 (9), 4795–4802. [DOI] [PubMed] [Google Scholar]
- 137.Capitan F; Robu AC; Popescu L; Flangea C; Vukelić Ž; Zamfir AD, B Subunit Monomers of Cholera Toxin Bind G1 Ganglioside Class as Revealed by Chip-Nanoelectrospray Multistage Mass Spectrometry. Journal of Carbohydrate Chemistry 2015, 34 (7), 388–408. [Google Scholar]
- 138.Li J; Fan X; Kitova EN; Zou C; Cairo CW; Eugenio L; Ng KKS; Xiong ZJ; Privé GG; Klassen JS, Screening Glycolipids Against Proteins in Vitro Using Picodiscs and Catch-and-Release Electrospray Ionization-Mass Spectrometry. Analytical Chemistry 2016, 88 (9), 4742–4750. [DOI] [PubMed] [Google Scholar]
- 139.Alam S; Anugraham M; Huang Y-L; Kohler RS; Hettich T; Winkelbach K; Grether Y; López MN; Khasbiullina N; Bovin NV; Schlotterbeck G; Jacob F, Altered (neo-) lacto series glycolipid biosynthesis impairs α2–6 sialylation on N-glycoproteins in ovarian cancer cells. Scientific Reports 2017, 7, 45367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Février B; Raposo G, Exosomes: endosomal-derived vesicles shipping extracellular messages. Current Opinion in Cell Biology 2004, 16 (4), 415–421. [DOI] [PubMed] [Google Scholar]
- 141.Vlassov AV; Magdaleno S; Setterquist R; Conrad R, Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochimica et Biophysica Acta (BBA) - General Subjects 2012, 1820 (7), 940–948. [DOI] [PubMed] [Google Scholar]
- 142.Llorente A; Skotland T; Sylvänne T; Kauhanen D; Róg T; Orłowski A; Vattulainen I; Ekroos K; Sandvig K, Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2013, 1831 (7), 1302–1309. [DOI] [PubMed] [Google Scholar]
- 143.Skotland T; Sandvig K; Llorente A, Lipids in exosomes: Current knowledge and the way forward. Progress in Lipid Research 2017, 66, 30–41. [DOI] [PubMed] [Google Scholar]
- 144.de Leoz ML; Young LJ; An HJ; Kronewitter SR; Kim J; Miyamoto S; Borowsky AD; Chew HK; Lebrilla CB, High-mannose glycans are elevated during breast cancer progression. Mol Cell Proteomics 2011, 10 (1), M110 002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Allam H; Aoki K; Benigno BB; McDonald JF; Mackintosh SG; Tiemeyer M; Abbott KL, Glycomic Analysis of Membrane Glycoproteins with Bisecting Glycosylation from Ovarian Cancer Tissues Reveals Novel Structures and Functions. Journal of Proteome Research 2015, 14 (1), 434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kaprio T; Satomaa T; Heiskanen A; Hokke CH; Deelder AM; Mustonen H; Hagstrom J; Carpen O; Saarinen J; Haglund C, N-glycomic profiling as a tool to separate rectal adenomas from carcinomas. Mol Cell Proteomics 2015, 14 (2), 277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.He Y; Xie Q; Wang Y; Liang Y; Xu X; Li Y; Miao J; Chen Z; Li Y, Liquid chromatography mass spectrometry-based O-glycomics to evaluate glycosylation alterations in gastric cancer. Proteomics Clin Appl 2016, 10 (2), 206–15. [DOI] [PubMed] [Google Scholar]
- 148.Balmana M; Gimenez E; Puerta A; Llop E; Figueras J; Fort E; Sanz-Nebot V; de Bolos C; Rizzi A; Barrabes S; de Frutos M; Peracaula R, Increased alpha1–3 fucosylation of alpha-1-acid glycoprotein (AGP) in pancreatic cancer. J Proteomics 2016, 132, 144–54. [DOI] [PubMed] [Google Scholar]
- 149.Takahashi S; Sugiyama T; Shimomura M; Kamada Y; Fujita K; Nonomura N; Miyoshi E; Nakano M, Site-specific and linkage analyses of fucosylated N-glycans on haptoglobin in sera of patients with various types of cancer: possible implication for the differential diagnosis of cancer. Glycoconj J 2016, 33 (3), 471–82. [DOI] [PubMed] [Google Scholar]
- 150.Yin H; Tan Z; Wu J; Zhu J; Shedden KA; Marrero J; Lubman DM, Mass-Selected Site-Specific Core-Fucosylation of Serum Proteins in Hepatocellular Carcinoma. J Proteome Res 2015, 14 (11), 4876–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tan Z; Yin H; Nie S; Lin Z; Zhu J; Ruffin MT; Anderson MA; Simeone DM; Lubman DM, Large-scale identification of core-fucosylated glycopeptide sites in pancreatic cancer serum using mass spectrometry. J Proteome Res 2015, 14 (4), 1968–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tanabe K; Kitagawa K; Kojima N; Iijima S, Multifucosylated Alpha-1-acid Glycoprotein as a Novel Marker for Hepatocellular Carcinoma. J Proteome Res 2016, 15 (9), 2935–44. [DOI] [PubMed] [Google Scholar]
- 153.Sanda M; Goldman R, Data Independent Analysis of IgG Glycoforms in Samples of Unfractionated Human Plasma. Anal Chem 2016, 88 (20), 10118–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pan KT; Chen CC; Urlaub H; Khoo KH, Adapting Data-Independent Acquisition for Mass Spectrometry-Based Protein Site-Specific N-Glycosylation Analysis. Anal Chem 2017, 89 (8), 4532–4539. [DOI] [PubMed] [Google Scholar]
- 155.Kim K; Ruhaak LR; Nguyen UT; Taylor SL; Dimapasoc L; Williams C; Stroble C; Ozcan S; Miyamoto S; Lebrilla CB; Leiserowitz GS, Evaluation of Glycomic Profiling as a Diagnostic Biomarker for Epithelial Ovarian Cancer. Cancer Epidemiology Biomarkers & Prevention 2014, 23 (4), 611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee JY; Kim JY; Cheona MH; Park GW; Ahn YH; Moon MH; Yoo JS, MRM validation of targeted nonglycosylated peptides from N-glycoprotein biomarkers using direct trypsin digestion of undepleted human plasma. Journal of Proteomics 2014, 98, 206–217. [DOI] [PubMed] [Google Scholar]
- 157.Nie S; Lo A; Wu J; Zhu JH; Tan ZJ; Simeone DM; Anderson MA; Shedden KA; Ruffin MT; Lubman DM, Glycoprotein Biomarker Panel for Pancreatic Cancer Discovered by Quantitative Proteomics Analysis. Journal of Proteome Research 2014, 13 (4), 1873–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gimenez E; Balmana M; Figueras J; Fort E; de Bolos C; Sanz-Nebot V; Peracaula R; Rizzi A, Quantitative analysis of N-glycans from human alfa-acid-glycoprotein using stable isotope labeling and zwitterionic hydrophilic interaction capillary liquid chromatography electrospray mass spectrometry as tool for pancreatic disease diagnosis. Analytica Chimica Acta 2015, 866, 59–68. [DOI] [PubMed] [Google Scholar]
- 159.Hecht ES; Scholl EH; Walker SH; Taylor AD; Cliby WA; Motsinger-Reif AA; Muddiman DC, Relative Quantification and Higher-Order Modeling of the Plasma Glycan Cancer Burden Ratio in Ovarian Cancer Case-Control Samples. J Proteome Res 2015, 14 (10), 4394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Balmana M; Sarrats A; Llop E; Barrabes S; Saldova R; Ferri MJ; Figueras J; Fort E; de Llorens R; Rudd PM; Peracaula R, Identification of potential pancreatic cancer serum markers: Increased sialyl-Lewis X on ceruloplasmin. Clin Chim Acta 2015, 442, 56–62. [DOI] [PubMed] [Google Scholar]
- 161.Sheta R; Woo CM; Roux-Dalvai F; Fournier F; Bourassa S; Droit A; Bertozzi CR; Bachvarov D, A metabolic labeling approach for glycoproteomic analysis reveals altered glycoprotein expression upon GALNT3 knockdown in ovarian cancer cells. Journal of Proteomics 2016, 145, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Miyamoto S; Ruhaak LR; Stroble C; Salemi MR; Phinney B; Lebrilla CB; Leiserowitz GS, Glycoproteomic Analysis of Malignant Ovarian Cancer Ascites Fluid Identifies Unusual Glycopeptides. J Proteome Res 2016, 15 (9), 3358–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ju L; Wang Y; Xie Q; Xu X; Li Y; Chen Z; Li Y, Elevated level of serum glycoprotein bifucosylation and prognostic value in Chinese breast cancer. Glycobiology 2016, 26 (5), 460–71. [DOI] [PubMed] [Google Scholar]
- 164.Darebna P; Novak P; Kucera R; Topolcan O; Sanda M; Goldman R; Pompach P, Changes in the expression of N- and O-glycopeptides in patients with colorectal cancer and hepatocellular carcinoma quantified by full-MS scan FT-ICR and multiple reaction monitoring. Journal of Proteomics 2017, 153, 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Qin R; Zhao J; Qin W; Zhang Z; Zhao R; Han J; Yang Y; Li L; Wang X; Ren S; Sun Y; Gu J, Discovery of Non-invasive Glycan Biomarkers for Detection and Surveillance of Gastric Cancer. J Cancer 2017, 8 (10), 1908–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Qin Y; Zhong Y; Ma T; Zhang J; Yang G; Guan F; Li Z; Li B, A pilot study of salivary N-glycome in HBV-induced chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Glycoconj J 2017, 34 (4), 523–535. [DOI] [PubMed] [Google Scholar]
- 167.Spiciarich DR; Nolley R; Maund SL; Purcell SC; Herschel J; Iavarone AT; Peehl DM; Bertozzi CR, Bioorthogonal Labeling of Human Prostate Cancer Tissue Slice Cultures for Glycoproteomics. Angew Chem Int Ed Engl 2017, 56 (31), 8992–8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu T; Shang S; Li W; Qin X; Sun L; Zhang S; Liu Y, Assessment of Hepatocellular Carcinoma Metastasis Glycobiomarkers Using Advanced Quantitative N-glycoproteome Analysis. Front Physiol 2017, 8, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Biskup K; Braicu EI; Sehouli J; Tauber R; Blanchard V, The ascites N-glycome of epithelial ovarian cancer patients. J Proteomics 2017, 157, 33–39. [DOI] [PubMed] [Google Scholar]