Abstract
Aim:
The present study was conducted to detect and identify Helicobacter pylori within local cow breeds in the central region of Algeria.
Materials and Methods:
Two hundred (n=200) cows from three provinces of the central region of Algeria were studied, between January 2016 and September 2017. Each cow was subject to stool, milk, and blood sampling. Milk and fecal samples were used to detect and identify H. pylori using bacteriology culture method. Blood and milk samples were used to detect H. pylori immunoglobulin G (IgG) antibody using enzyme-linked immunosorbent assay. Polymerase chain reaction was used to confirm the abundance of H. pylori in milk by detecting glmM gene.
Results:
Out of 200 sera and 200 milk samples, 12% (24) and 4% (8/200) were positive for the H. pylori IgG antibody. glmM gene was detected in the milk of 13% of cows and was confirmed in all cows presenting IgG in milk.
Conclusion:
From the present study, we concluded that the glmM gene is an important marker for detecting H. pylori in milk. Moreover, Algerian local-breed cows are a source of H. pylori and could be responsible for serious zoonosis.
Keywords: Algeria, cows, feces, Helicobacter pylori, milk, serum
Introduction
Helicobacter pylori is a Gram-negative, microaerophilic bacterium. It has been detected in half of the human population [1-4]. Prevalence of H. pylori infection varies widely according to a geographic area, age, race, and ethnicity [5]. Developed countries are more exposed to H. pylori infection, as hygiene of animals, material, and personal is difficult to control in large herds, which would increase dramatically the transmission of the bacteria to humans [6]. Surprisingly, H. pylori infection seems to be in continuous increase in developing countries [7-9]. This increase might result from the efforts of increasing the production of animal originated food to respond to the increased population, without ensuring the quality of the production such as the hygiene if staff and herd.
H. pylori has a strong affinity to stomach mucosa and induce serious diseases in the gastrointestinal tract such as chronic gastritis, duodenal ulcer, and gastric cancer [10,11]. For that, the World Health Organization considers H. pylori as an important Class I carcinogen factor [11]. Even though humans are the principal reservoir of H. pylori [12,13], it could spread through food and water by fecal-oral and oral-oral routes and colonize the stomach and intestines of humans and several animal species [12,14].
Professionals dealing with animals and animal-originated food such as veterinarians, butchers, and slaughterhouse staff showed high levels of antibodies against H. pylori [15,16] suggesting that ruminants might be a source of contamination for humans. H. pylori was isolated from milk of different farming animals mainly cow, ewe, camel, and sow [17,18]. These findings confirm that farming animals are a potential source of H. pylori and represent a risk of contamination for humans handling them or consuming their originated products such as meat and milk.
As cow milk represents the most consumed milk in the world, we think that it may represent a potential source of human contamination by H. pylori. However, in Algeria milk is consumed mainly as raw milk or transferred to dairy products and its consumption is higher in the central part of Algeria which is known as the principal region of dairy farming. Therefore, we think that milk originated from cows in this region could be one of the main sources of transmission of H. pylori to humans. Our hypothesis is supported by the high prevalence of H. pylori infection in humans reported in different studies in the central region of Algeria [19,20].
H. pylori infection induces chronic inflammation leading to the development of immunoglobulin G (IgG) [21], which could be measured in blood easily and with high specificity of 90% [22]. Detecting pathogenic factors of H. pylori can be done by polymerase chain reaction (PCR) and glmM gene which encodes for a phosphoglucosamine mutase is often targeted [23,24].
According to our knowledge, there is no available data about studies that have been done in Algeria regarding the zoonotic aspect of H. pylori. Therefore, the main aim of the present study is to demonstrate whether H. pylori is abundant in local cow breeds and their milk.
Materials and Methods
Ethical approval
The experimental protocol was approved by the scientific council of the High National Veterinary School of Algiers, Algeria, 2012.
Sampling
The present study was conducted from January 2016 to September 2017 in the central region of Algeria. The existence of H. pylori was investigated in feces, serum, and fresh raw milk of 200 cows of the local Algerian breed. The selected cows come from six different regions located in three provinces of the central region of Algeria: Tizi-Ouzou, Bouira, and Boumerdes. Cows were selected according to lactation and age (at least 2 years old). All cows were under mixed indoor/outdoor housing condition and receiving the same diet composed of concentrate and forage.
Feces sampling
A small piece of stool (~5 mm in diameter; ~150 mg) was collected transrectally using a gloved hand and transferred into plastic tubes containing 1 ml of transport media (Stuart’s Transport Medium: Oxoid, France) and mixed thoroughly.
Blood sampling
5 ml of blood were collected from a coccygeal vein in Vacutainer tubes and let to clot. Serum was then transferred into sterile tubes after centrifugation. Serum samples were transported in the icebox and stored at 2-8°C for up to 3 days prior analysis. For H. pylori IgG antibodies detection, serum samples were stored at −80°C until further analysis.
Milk sampling
10 ml of fresh raw milk were collected into sterile sampling containers. Milk samples were filtered and stored at −20°C until further analysis, according to Bewley et al. [25].
Isolation of H. pylori
100 µl of filtered milk and feces transport media were cultured in Brucella agar plates containing 7% defibrinated horse blood (Pasteur Institute, Algiers) and Skirrow’s supplement (vancomycin, 0.01%, trimethoprim, 0.05%, and polymyxin B, 2.5 IU/ml) [23] (Cat. No. SR0069, Oxoid France). Plates were incubated at 37°C with 95-99% relative humidity under microaerophilic conditions (5% O2, 15% CO2, and 80% N2) (Cat. No. AGOO25A, bioMérieux, France). Humidity was obtained by placing a wet paper towel in the anaerobe jars. Plates were incubated, and the growth rate was observed daily for 5-7 days. Helicobacter spp. was identified either as small (1 mm or less in diameter), clear, dome-shaped colonies or as a fine, and translucent lawn. Suspected colonies were confirmed for the presence of H. pylori on the basis of colony morphology and positive biochemical reactions for catalase, urease, and oxidase tests.
Serological analysis
H. pylori IgG test was performed according to Safaei et al. [26]. Briefly, an enzyme-linked immunosorbent assay (ELISA) kit (H. pylori antibody [IgG] ELISA Kit; ABIN992624, GmbH Germany) was used and the results read by spectrophotometer at 450 nm. Reading <0.140 was considered negative; between 0.140 and 0.159 were considered equivocal, and ≥0.160 were considered positive. The equivocal results, according to the manufacturer’s instructions, must be repeated.
PCR analysis on milk
1 ml of each milk sample was used for extraction of cDNA by a DNA isolation kit (Cat. No. ABIN412492, Roche, Germany) according to the manufacturer’s instruction, with slight modification according to Quaglia et al. [18]. Extracted DNA was amplified according to Rahimi and Kheirabadi [20] for the glmM gene of 294 bp using 5-GAATAAGCTTTTAGGGGTGTTAGGGG-3 as primer forward and 5-GCTTACTTTCTAACACTAACGCGC-3 as primer reverse. PCR reactions were performed in a final volume of 50 μl containing 25 μl Green Master Mix (Sigma), 10 μl genomic DNA as a template, 13 μl free ionized water, and 1 μl of each primer. PCR was performed using a thermal cycler (Bio-Rad, France) under the following conditions: Initial denaturation for 10 min at 94°C, 35 cycles for 1 min at 94°C, 1 min at 55°C, 1 min at 72°C, and a final extension at 72°C for 10 min.
The PCR products were electrophoresed through a 1.5% agarose gels (Bio-Rad, France) containing ethidium bromide.
A DNA ladder (Bio-Rad, France) used to detect the molecular weight of observed bands under a UV lamp. Samples inoculated with H. pylori were used as positive controls, and sterile distilled water was used as negative control.
Statistical analysis
Comparisons of differences and similarities were conducted by the Chi-square test using SPSS software. Differences were considered significant where p<0.05.
Results
Using traditional bacteriology culture technique, no H. pylori strains were isolated from feces and milk. Based on the ELISA test, 12% (24/200) of cows were positive for the H. pylori IgG antibody (Table-1).
Table-1.
Occurrence of Helicobacter pylori among cows’ serum and milk samples using ELISA.
| Samples | Serum | Milk | ||
|---|---|---|---|---|
| Locality designation | Total number | Number of positive (%) | Total number | Number of positive (%) |
| Bouira1 | 24 | 8 (33.3) | 24 | 4 (16.7) |
| Bouira2 | 32 | 0 (0.0) | 32 | 0 (0.0) |
| Bouira3 | 44 | 4 (9.1) | 44 | 0 (0.0) |
| Tizi-Ouzou4 | 36 | 2 (5.5) | 36 | 0 (0.0) |
| Tizi-Ouzou5 | 32 | 6 (18.7) | 32 | 2 (6.25) |
| Boumerdes6 | 32 | 4 (12.5) | 32 | 2 (6.25) |
| Total | 200 | 24 (12.0) | 200 | 8 (4.0) |
ELISA=Enzyme-linked immunosorbent assay
The results of the detection of H. pylori IgG revealed that out of the 200 milk samples specimens, 4% (8/200) of cows were positive (Table-1). Among the 6 studied herds, 3 had 16.7% of H. pylori abundance each while the other 3 herds were healthy. No statistical difference was identified between the level of contamination of different regions (Figure-1) (p>0.05).
Figure-1.
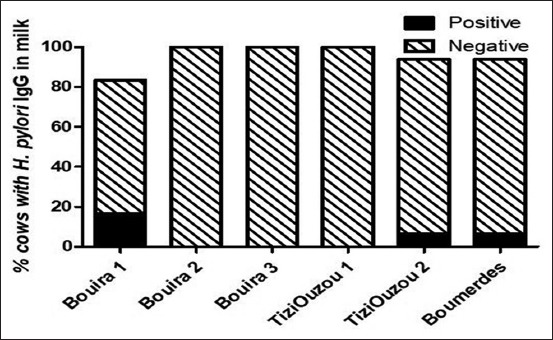
Occurrence of Helicobacter pylori among cows’ milk samples using enzyme-linked immunosorbent assay by region.
The results of the detection of H. pylori by PCR (Figure-2) revealed that 13% of the examined cow’s raw milk was positive for the presence of glmM gene (Figure-3). We found that all milk samples that were positive for H. pylori antibody were positive for the presence of the glmM gene.
Figure-2.
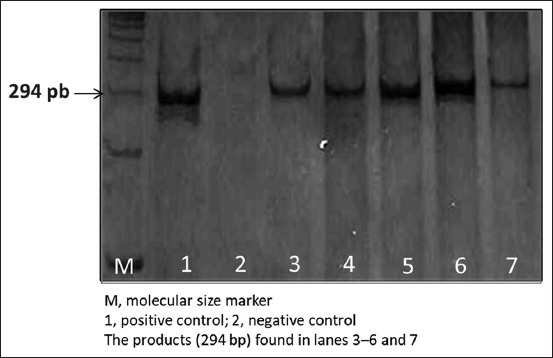
Results of the gel electrophoresis for polymerase chain reaction products of the glmM gene of the Helicobacter pylori isolates from cow milk.
Figure-3.
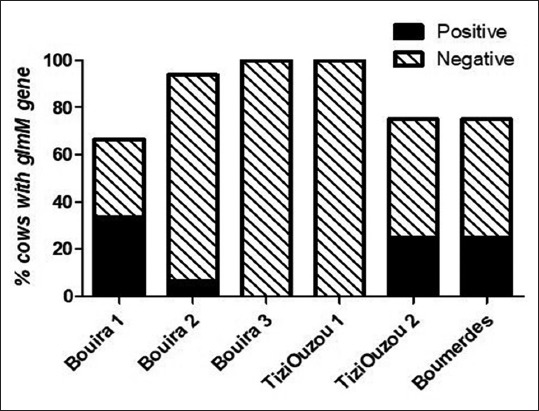
Occurrence of Helicobacter pylori in cows’ milk using polymerase chain reaction detection of the glmM gene.
Discussion
Helicobacter is considered emerging bacteria with major public implications [27,28]; however, its epidemiology is not fully understood [29]. In the present study, we evaluated whether cows of the local breed represent a reservoir for H. pylori. Different studies have been conducted in many countries, reporting the prevalence of H. pylori in animals with different results according to animal species and environmental factors [30,31]. This study is the first experimental work that confirms the presence of H. pylori in cows from different provinces of the central region of Algeria.
Determining H. pylori seroprevalence in cows is one of the best routine techniques to study the epidemiological aspect of the disease and the risk of human infection by these animals or their products [21,22]. As more sophisticated techniques, detecting glmM gene encoding for a phosphoglucosamine mutase using PCR is a potential tool for the direct detection of H. pylori or the confirmation of seropositive findings [24,32]. However, bacteriology is a widely used technique in the diagnosis of H. pylori as it gives additional information about antimicrobial susceptibility on the specific isolated strain [22].
In the present study, traditional bacteriology culture technique failed to identify H. pylori in raw milk and feces, leading to think that this method is not the best technique to detect H. pylori in animal originated products or as clinical diagnostic tool in live animals. Likewise, Tabatabaei [33] and Azevedo et al. [34] failed to isolate H. pylori from raw milk and feces of cows by classical bacteriology culture. Even in some studies where H. pylori were isolated, its bacteriological culture was very low [35] as only a few Helicobacter derived from feces of animals could be cultured according to the routine technique [36]. Furthermore, H. pylori can transform into coccoid form, a resistance form that makes it viable but uncultivable [11,34]. Therefore, using advanced technologies to detect the coccoid form is the best method for the study of H. pylori in animal-originated products.
ELISA assays revealed that 12% of the serum samples and 6% of milk samples showing the presence of IgG against H. pylori. The presence of IgG against H. pylori in bovine serum has been reported in several previous studies in different parts of the world [20,26]. In a study carried out in Iran, Safaei et al. [26] found that 27% of serum samples from cows were positive for H. pylori IgG. Likewise, Elhariri et al. [37] reported that 30% of cows were seropositive for H. pylori, using different ELISA kit.
To the best of our knowledge, there are only a few studies that have been done on animal originated food, particularly raw milk, as a source of contamination for humans by H. pylori [15,26]. Interestingly, we found that frequency of isolation of H. pylori from cows and their milk was different among the studied dairy herds and regions. Likewise, El-Gohary et al. [38] found a variation in the rate of contamination by H. pylori between different regions in Egypt. This statement could be explained by the different hygiene conditions that are mainly affected by the local culture and tradition.
In this work, we specifically targeted the glmM gene for molecular detection of H. pylori in raw milk. GlmM is implicated in the growth of H. pylori and is highly specific for the detection of H. pylori [39]. During this investigation, the glmM gene was detected in the raw milk of 13% cows. Our findings are similar to those recounted by Rahimi and Kheirabad [20], who reported that H. pylori glmM gene was detected in 14.1% of cow’s milk, in Iran. Interestingly, H. pylori was highly abundant in cow’s milk from countries with high milk production such as Japan, 72.2% [40] and Italy, 50% [18].
We think that the presence of H. pylori in cow’s milk is highly correlated with hygiene condition in dairy farms and the stress on the animals which is due to high milk production. Hygiene conditions are more difficult to be monitored and controlled in large herd size, as there are many materials and personally implicated in the work. Furthermore, cows with high milk yield are subject to strong stressful metabolism leading to decreasing the efficiency of the immune system [41] and systemic abundance and multiplication of H. pylori in cows and their milk.
Conclusion
This study shows that glmM can be successfully detected in raw milk using PCR, indicating that raw milk from local breed cows in the central region of Algeria could be a potential source of zoonosis by H. pylori. While IgG detection in blood and milk is effective techniques for the epidemiological study of H. pylori in large herds, bacteriology culture is not convenient for monitoring H. pylori abundance in dairy cows and their originated products.
Authors’ Contributions
MG conceived, designed the study and drafted the manuscript under the supervision of ZG. MG and MA designed the experiment protocol under the supervision of ZG. MG collected and analyzed samples. MG and MA revised the manuscript under the supervision of ZG. All authors read and approved the final manuscript.
Acknowledgments
The study was funded by the Ministries of Higher Education and Scientific Research of Algeria by a grant to MG for an internship at Central Laboratory of Clinical Biology, University Hospital Center of Nafisa Hamoud (Parnet), Hussein Dey, Algiers. The authors are thankful to the Microbiology team of Central Laboratory of Clinical Biology, University Hospital Center of Nafisa Hamoud (Parnet), Hussein Dey, Algiers.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Correa P, Piazuelo M.B. Helicobacter pylori infection and gastric adenocarcinoma. US Gastroenterol. Hepatol. Rev. 2001;7(1):59–64. [PMC free article] [PubMed] [Google Scholar]
- 2.Garza-González E, Perez-Perez G.I, Maldonado-Garza H.J, Bosques-Padilla F.J. A review of Helicobacter pylori diagnosis, treatment, and methods to detect eradication. World J. Gastroenterol. 2014;20(6):1438–1449. doi: 10.3748/wjg.v20.i6.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooi J.K.Y, Lai W.Y, Ng K.W, Suen M.M.Y, Underwood F.E, Tanyingoh D, Malfertheiner P, Graham D.Y, Wong V.W.S, Wu J.C.Y, Chan F.K.L, Sung J.J.Y, Kaplan G.G, Siew C.N.G. Global prevalence of Helicobacter pylori infection: Systematic review and meta-analysis. Gastroenterology. 2017;153(2):420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 4.Ghoshal U.C, Chaturvedi R, Correa P. The enigma of Helicobacter pylori infection and gastric cancer. Indian J. Gastroenterol. 2010;29(3):95–100. doi: 10.1007/s12664-010-0024-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gharavi M.J, Ebadi M, Fathi H, Yazdanyar Z, Valipor N.S, Afrogh P, Kalantar E. Trends in the prevalence of Helicobacter pylori infection in Fardis, Iran, 2011-2014. Int. J. Enteric. Pathog. 2016;4(1):11–32860. [Google Scholar]
- 6.Khalifa M.M, Sharaf R.R, Aziz R.K. Helicobacter pylori: A poor man's gut pathogen? Gut Pathog. 2010;2(1):2. doi: 10.1186/1757-4749-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drumm B, Rowland M. Por que o Helicobacter pylori estádesaparecendo? Mais perguntas do que respostas. J. Pediatr. 2011;87:369–370. doi: 10.2223/JPED.2130. [DOI] [PubMed] [Google Scholar]
- 8.Salih B.A. Helicobacter pylori infection in developing countries: The burden for how long? Saudi J. Gastroenterol. 2009;15(3):201–207. doi: 10.4103/1319-3767.54743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adlekha S, Chadha T, Krishnan P, Sumangala B. Prevalence of Helicobacter pylori infection among patients undergoing upper gastrointestinal endoscopy in a medical college hospital in Kerala, India. Ann. Med. Health Sci Res. 2013;3(4):559–563. doi: 10.4103/2141-9248.122109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham D.Y. History of Helicobacter pylori duodenal ulcer, gastric ulcer and gastric cancer. World J Gastroenterol. 2014;20(18):5191–5204. doi: 10.3748/wjg.v20.i18.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Testerman T.L, Morris J. Beyond the stomach: An updated view of Helicobacter pylori pathogenesis, diagnosis, and treatment. World J. Gastroenterol. 2014;20(36):12781–12808. doi: 10.3748/wjg.v20.i36.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payao S.L, Rasmussen L.T. Helicobacter pylori and its reservoirs: A correlation with the gastric infection. World J. Gastrointest. Pharmacol. Ther. 2016;7(1):126–132. doi: 10.4292/wjgpt.v7.i1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown L.M. Helicobacter pylori: Epidemiology and routes of transmission. Epidemiol. Rev. 2000;22(2):283–297. doi: 10.1093/oxfordjournals.epirev.a018040. [DOI] [PubMed] [Google Scholar]
- 14.Adler I, Muiño A, Aguas S, Harada L, Diaz M, Lence A, Labbrozzi M, Muiño J.M, Elsner B, Avagnina A, Denninghoff V. Helicobacter pylori and oral pathology: Relationship with the gastric infection. World J. Gastroenterol. 2014;20(29):9922–9935. doi: 10.3748/wjg.v20.i29.9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talaei R, Souod N, Momtaz H, Dabiri H. Milk of livestock as a possible transmission route of Helicobacter pylori infection. Gastroenterol. Hepatol. Bed. Bench. 2015;8(Suppl 1):S30–S36. [PMC free article] [PubMed] [Google Scholar]
- 16.Fox J.G, Wang T.C. Dietary factors modulate Helicobacter-associated gastric cancer in rodent models. Toxicol. Pathol. 2014;42(1):162–181. doi: 10.1177/0192623313512564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolodzieyski L, Kim B, Park H, Yoon HS, Lim CW. Prevalence of gastrospirillum-like organisms in pigs, cattle, and dogs: a comparison of diagnostic methods between species. Vet. Med. 2008;53:193–202. [Google Scholar]
- 18.Quaglia N.C, Dambrosio A, Normanno G, Parisi A, Patrono R, Ranieri G, Rella A, Celano G.V. High occurrence of Helicobacter pylori in raw goat, sheep and cow milk inferred by glmM gene: A risk of food-borne infection? Int. J. Food Microbiol. 2008;124(1):43–47. doi: 10.1016/j.ijfoodmicro.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Raaf N, Amhis W, Saoula H, Abid A, Nakmouche M, Balamane A, Arous N.A, Ouar-Korichi M, Vale F.F, Bénéjat L, Mégraud F. Prevalence, antibiotic resistance, and MLST typing of Helicobacter pylori in Algiers, Algeria. Helicobacter. 2017;22(6):e12446. doi: 10.1111/hel.12446. [DOI] [PubMed] [Google Scholar]
- 20.Rahimi E, Kheirabadi E.K. Detection of Helicobacter pylori in bovine, buffalo, camel, ovine, and caprine milk in Iran. Foodborne Pathog. Dis. 2012;9(5):453–456. doi: 10.1089/fpd.2011.1060. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell H.M, Bohane T.D, Berkowicz J, Hazell S.L, Lee A. Antibody to Campylobacter pylori in families of index children with gastrointestinal illness due to C pylori. Lancet. 1987;2(8560):681–682. doi: 10.1016/s0140-6736(87)92459-7. [DOI] [PubMed] [Google Scholar]
- 22.Megraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin. Microbiol. Rev. 2007;20(2):280–322. doi: 10.1128/CMR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dent J.C, McNulty C.A. Evaluation of a new selective medium for Campylobacter pylori. Eur. J. Clin. Microbiol. Infect. Dis. 1988;7(4):555–558. doi: 10.1007/BF01962615. [DOI] [PubMed] [Google Scholar]
- 24.Espinoza M.G, Vazquez R.G, Mendez I.M, Vargas C.R, Cerezo S.G. Detection of the glmM gene in Helicobacter pylori isolates with a novel primer by PCR. J. Clin. Microbiol. 2011;49(4):1650–1652. doi: 10.1128/JCM.00461-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bewley J.M, Harmon R.J, Locke S, Arnold M. Collection and preparation of milk samples for microbiological culturing. Agric. Natl. Resour. Publ. 2014:164. [Google Scholar]
- 26.Safaei H.G, Rahimi E, Zandi A, Rashidipour A. Helicobacter pylori as a zoonotic infection: The detection of H. pylori antigens in the milk and faeces of cows. J. Res. Med. Sci. 2011;16(2):184–187. [PMC free article] [PubMed] [Google Scholar]
- 27.Goh K.L, Wah-Kheong C, Shiota S, Yamaoka Y. Epidemiology of Helicobacter pylori infection and public health implications. Helicobacter. 2011;16(Suppl 1):1–9. doi: 10.1111/j.1523-5378.2011.00874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vouga M, Greub G. Emerging bacterial pathogens: The past and beyond. Clin. Microbiol. Infect. 2016;22(1):12–21. doi: 10.1016/j.cmi.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mladenova-Hristova I, Grekova O, Patel A. Zoonotic potential of Helicobacter spp. J. Microbiol. Immunol. Infect. 2017;50(3):265–269. doi: 10.1016/j.jmii.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Blaser M.J, Berg D.E. Helicobacter pylori genetic diversity and risk of human disease. J. Clin. Invest. 2001;107(7):767–773. doi: 10.1172/JCI12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanae S.Y.T. Helicobacter pylori in the Natural Environment. Scand. J. Infect. Dis. 2009;31(3):275–279. doi: 10.1080/00365549950163572. [DOI] [PubMed] [Google Scholar]
- 32.De Reuse H, Labigne A, Mengin-Lecreulx D. The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase. J. Bacteriol. 1997;179(11):3488–3493. doi: 10.1128/jb.179.11.3488-3493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabatabaei M. Application of molecular and cultural methods for identification of Helicobacter spp. In different animal sources. Glob. Vet. 2012;8(3):292–297. [Google Scholar]
- 34.Azevedo N.F, Almeida C, Cerqueira L, Dias S, Keevil C.W, Vieira M.J. Coccoid form of Helicobacter pylori as a morphological manifestation of cell adaptation to the environment. Appl. Environ. Microbiol. 2007;73(10):3423–3427. doi: 10.1128/AEM.00047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allahverdiyev A.M, Bagirova M, Caliskan R, Tokman H.B, Aliyeva H, Unal G, Oztel O.N, Abamor E.S, Toptas H, Yuksel P, Kalayci F, Aslan M, Erzin Y, Bal K, Kocazeybek B.S. Isolation and diagnosis of Helicobacter pylori by a new method: Microcapillary culture. World J. Gastroenterol. 2015;21(9):2622–2628. doi: 10.3748/wjg.v21.i9.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Momtaz H, Dabiri H, Souod N, Gholami M. Study of Helicobacter pylori genotype status in cows, sheep, goats and human beings. BMC Gastroenterol. 2014;14:61. doi: 10.1186/1471-230X-14-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elhariri M, Elhelw R, Hamza D, Sayed H. Serologic evidence and risk factors for Helicobacter pylori infection in animals and humans. J. Infect. Dev. Ctries. 2017;11(5):414–419. doi: 10.3855/jidc.9339. [DOI] [PubMed] [Google Scholar]
- 38.El-Gohary A.H, Yousef M.A, Mohamed A.A. Epidemiological study on H. pylori in cattle and its milk with special reference to its zoonotic importance. BLM. 2015;7(5):251. [Google Scholar]
- 39.Osman E.Y, El-Eragi A.M.S, Musa A.M, El-Magboul S.B, A/Rahman M.B, Abdo A.E. Detection of Helicobacter pylori glmM gene in bovine milk using nested polymerase chain reaction. Vet. World. 2015;8(7):913–917. doi: 10.14202/vetworld.2015.913-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujimura S, Kawamura T, Kato S, Tateno H, Watanabe A. Detection of Helicobacter pylori in cow's milk. Lett. Appl. Microbiol. 2002;35(6):504–507. doi: 10.1046/j.1472-765x.2002.01229.x. [DOI] [PubMed] [Google Scholar]
- 41.Bertoni G, Minuti A, Trevisi E. Immune system, inflammation and nutrition in dairy cattle. Anim. Prod. 2015;55(7):943. [Google Scholar]


