Abstract
Cutaneous diffuse large B cell lymphomas (DLBCL) are aggressive lymphomas with a poor prognosis. To elucidate their genetic bases, we analyzed exome sequencing of 37 cutaneous DLBCLs including 31 DLBCL-leg type (DLBCL-LT) and 6 cutaneous DLBCL-not otherwise specified (DLBCL-NOS). As reported previously, 77% of DLBCL-LTs harbor NF-κB-activating MYD88 mutations. In nearly all MYD88-wild type DLBCL-LTs, we found cancer-promoting mutations which either activate the NF-κB pathway through alternative genes (NFKBIE or REL) or activate other canonical cancer pathways (BRAF, MED12, PIK3R1, and STAT3). After NF-κB, the second most commonly mutated pathway putatively enables immune evasion via mutations predicted to downregulate antigen processing (B2M, CIITA, HLA) or T cell co-stimulation (CD58). DLBCL-LTs have little genetic overlap with the genetically heterogeneous DLBCL-NOSs. Instead, they resemble primary CNS and testicular large B-cell lymphomas (PCNSLs and PTLs). Like PCNSLs/PTLs, 40% of DLBCL-LTs (vs. 0% of DLBCL-NOSs) harbored PDL1/PDL2 translocations, which lead to overexpression of PD-L1 or PD-L2 in 50% of the cases. Collectively, these data broaden our understanding of cutaneous DLBCLs and suggest novel therapeutic approaches (e.g. BRAF or PI3K inhibitors). Additionally, they suggest novel treatment paradigms, wherein DLBCL-LTs can be targeted with strategies (e.g. immune checkpoint blockers) currently being developed for genomically similar PCNSLs/PTLs.
INTRODUCTION
Diffuse large B cell lymphomas (DLBCLs) involving the skin include primary cutaneous diffuse large B cell lymphoma, leg type (DLBCL-LT) and DLBCL with secondary skin involvement (cutaneous DLBCL, not otherwise specified [DLBCL-NOS]). DLBCL-LT is the most aggressive primary cutaneous B cell lymphoma subtype and is associated with increased risk of extracutaneous spread and poor prognosis (overall 5-year survival of ~50%) (Kodama et al., 2005). Patients with DLBCL-LT usually present with rapidly growing red to violaceous tumors characteristically (but not always) on one or both lower legs of elderly patients (median age ~75 years) (Grange et al., 2014). In comparison, DLBCL-NOS tends to affect younger patients, with no anatomic preference, are more likely to present with advanced stage disease, and are associated with lower median survival than DLBCL-LTs (Lee et al., 2016).
DLBCL is a heterogeneous category of B-cell lymphomas that share large-cell morphology, and are comprised of distinct subtypes, defined by predilections for specific anatomic sites and extranodal tissues (e.g. the mediastinum, central nervous system, skin, etc.) and/or distinct molecular/genetic features (Swerdlow et al., 2016). Two groups of DLBCLs have been identified on the basis of gene expression profiles corresponding to cell of origin: germinal center B cell (GCB) type and activated B cell (ABC) types. Until recently, therapeutic strategies for all subtypes of DLBCLs were adapted from therapies originally validated in clinical trials for nodal DLBCL in part because of the relative rarity of and limited understanding of the differences between DLBCL subtypes (Senff et al., 2008). For cutaneous DLBCLs, use of therapies designed for nodal DLBCLs has important limitations for patients including a high risk for relapse (>58%) and dose-limiting toxicities (Nabhan et al., 2012, Suarez et al., 2013).
Cancer genomics have revolutionized our understanding of the pathophysiology and targetability of many cancers (Vogelstein et al., 2013). In part because of its relative rarity, our understanding of the genomics of DLBCL-LT and secondary cutaneous involvement by DLBCL-NOS remains incomplete. Recent efforts in DLBCL-LT have led to the discovery of MYD88, CD79B, and MYC as disease-relevant putative driver genes (Mareschal et al., 2017, Pham-Ledard et al., 2014a, Pham-Ledard et al., 2014b). However, the full landscape of targetable mutations remains unclear. To identify targetable mutations that can inform future therapeutic strategies, we analyzed exome sequencing data on 37 cutaneous DLBCLs and report the results of our efforts herein.
RESULTS
Clinical characteristics of cutaneous DLBCLs
We collected tissue from 25 patients with cutaneous DLBCLs. Diagnoses were confirmed by expert pathologists (LC, JG, AL). Nineteen patients (12 men, 7 women) had disease consistent with DLBCL-LT, with no evidence of systemic involvement at time of diagnosis. All DLBCL-LT cases were MUM1+ (100%) and CD10- (100%). The majority were positive for BCL2 (85%), BCL6 (92%), and FOXP1 (92%) (Tables 1, S1). This immunohistochemical profile was consistent with non-GCB (ABC-type) DLBCL. As expected, patients with DLBCL-LT were older (median age 80 years), presented with leg involvement (15 of 19, with 4 others appearing on the scalp, arm and abdomen) and had a poor prognosis (median survival of 41 months) (Tables 1, S1).
Table 1 -.
Clinical/immunophenotypical characteristics of individual patients with DLBCL-LT (n=31) versus DLBCL-NOS (n=6)
| SAMPLE | SEX | AGE | SITE | SKIN STAGE* | STATUS | F/U (MO) | INITIAL TX | BCL2 | MUM1 | BCL6 | CD10 | FOXP1 | CD20 | CD79A | CD5 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DLBCL-LT | Internal Samples (n=19) | AU02 | M | 76 | Leg | T2 | D+ | 12 | XRT | − | + | + | ND | − | + | ND | − |
| AU06 | M | 80 | Leg | T1 | D+ | 47 | XRT | + | + | + | ND | + | + | + | ND | ||
| AU10 | F | 78 | Leg | T2 | D+ | 13 | XRT | − | + | + | ND | ND | + | + | − | ||
| AU11 | F | 89 | Scalp | T1 | D+ | 8 | XRT | + | + | + | ND | + | + | + | − | ||
| AU14 | F | 81 | Leg | T2 | NA | NA | NA | + | + | + | ND | + | + | + | ND | ||
| AU18 | M | 79 | Trunk | T2 | NA | NA | NA | + | + | + | ND | + | + | + | − | ||
| AU20 | F | 89 | Leg | T2 | A+ | 1 | Supportive | + | + | + | ND | + | + | + | − | ||
| AU21 | F | 86 | Leg | T2 | A+ | 12 | XRT | + | + | + | ND | + | + | + | − | ||
| AU22 | M | 77 | Leg | T2 | A− | 37 | R-Chemo | + | + | + | ND | + | + | + | − | ||
| AU24 | M | 85 | Leg | T2 | D+ | 69 | R-Chemo | + | + | + | ND | + | + | + | − | ||
| AU25 | M | 88 | Leg | T2 | D+ | 10 | XRT | + | + | + | ND | + | + | + | ND | ||
| NU03 | M | 53 | Leg, Trunk, Arm | T3 | D+ | 208 | R-Chemo | + | + | − | ND | + | + | ND | − | ||
| NU07 | M | 89 | Leg | T2 | D+ | 41 | R-Chemo | + | + | + | ND | + | − | ND | − | ||
| MG73 | F | 90 | Leg | T2 | D− | 20.4 | Chemo | + | + | + | − | ND | + | ND | ND | ||
| MG76 | M | 76 | Leg, Arm | T3 | A− | 88.8 | R-Chemo | + | + | + | − | ND | + | + | − | ||
| MG77 | M | 50 | Scalp | T1 | A− | 54 | R-Chemo | + | + | + | − | ND | + | ND | ND | ||
| MG78 | M | 51 | Leg | T2 | A− | 33.6 | R-Chemo | + | + | + | − | ND | + | ND | ND | ||
| MG82 | M | 75 | Arm | T1 | D+ | 7.2 | R-Chemo | + | + | + | − | ND | + | ND | − | ||
| MG88 | F | 83 | Leg | T2 | NA | NA | NA | + | + | + | − | ND | + | ND | − | ||
| External Samples (n=12) | FR91 | F | 77 | Leg | T2 | D+ | 24 | R-Chemo | + | ND | − | − | ND | + | ND | ND | |
| FR08 | F | 70 | Leg | T2 | D+ | 101 | R-Chemo | + | ND | − | − | ND | + | ND | ND | ||
| FR74 | M | 88 | Leg | T2 | D+ | 79 | Supportive | + | + | + | − | ND | + | ND | ND | ||
| FR68 | F | 85 | Trunk | T1 | D+ | 78 | XRT | + | + | + | − | ND | + | ND | ND | ||
| FR24 | F | 88 | Leg | T2 | D+ | 61 | XRT | + | + | + | − | ND | + | ND | ND | ||
| FR01 | F | 92 | Leg | T2 | D+ | 9 | XRT | + | ND | − | − | ND | + | ND | ND | ||
| FR97 | F | 79 | Leg | T3 | A− | 6 | R-Chemo | + | + | + | − | ND | + | ND | ND | ||
| FR89 | F | 94 | Leg | T2 | D+ | 5 | Supportive | + | + | + | − | ND | + | ND | ND | ||
| FR81 | F | 77 | Leg | T2 | D+ | 84 | R-Chemo | + | − | − | − | ND | + | ND | ND | ||
| FR42 | M | 78 | Leg | T1 | A− | 64 | XRT | + | + | + | − | ND | + | ND | ND | ||
| FR82 | M | 78 | Leg, Arm | T3 | D+ | 40 | R-Chemo | + | + | + | − | ND | + | ND | ND | ||
| FR87 | F | 82 | Arm | T2 | D+ | 14 | XRT | + | + | + | − | ND | + | ND | ND | ||
| DLBCL-NOS | Internal Samples (n=6) | NU01 | M | 35 | Cheek | NA | NA | 0 | XRT | + | − | + | ND | − | + | ND | − |
| NU02 | M | 80 | Trunk | NA | D+ | 8 | R-Chemo | + | − | + | ND | ND | + | ND | − | ||
| NU04 | F | 36 | Scalp | NA | A− | 6 | R-Chemo | + | − | + | ND | ND | + | ND | ND | ||
| NU05 | M | 50 | Trunk | NA | A− | 8 | R-Chemo | − | − | + | − | + | + | ND | ND | ||
| NU06 | F | 58 | Leg | NA | D+ | 5 | R-Chemo | + | + | − | − | + | + | ND | + | ||
| MG31 | M | 82 | Trunk | NA | A− | 9.6 | R-Chemo | + | − | + | − | ND | + | ND | ND |
DLBCL-LT = diffuse large B cell lymphoma, leg type; DLBCL-NOS = secondary cutaneous diffuse large B cell lymphoma, not otherwise specified
Internal DLBCL-LT samples = samples sequenced in this study (n=19); External DLBCL-LT samples = samples from previously published DLBCL-LT exome datasets (n=12) (Mareschal et al 2017).
ND, not done; NA, not available; F/U = follow-up; TX = treatment; MO = months
D+, died of disease; D−, died of other causes; A+, alive with disease; A−, alive without disease
XRT, radiation therapy; R-Chemo, rituximab + chemotherapy
Skin staging based on TNM classification for primary cutaneous lymphomas other than mycosis fungoides/Sezary Syndrome (Kim et al., Blood 2008).
Six patients (4 men, 2 women) were found to have cutaneous disease with concurrent systemic involvement at the time of diagnosis, consistent with secondary cutaneous involvement by DLBCL-NOS (Table 1). DLBCL-NOS patients were generally younger than DLBCL-LT patients, and lesions were found in diverse anatomical locations (Table 1, S1), with only one occurring on the leg. Two of three DLBCL-NOS samples with available tissue were GCB type on the basis of immunohistochemical stains. Two of six patients died of progressive disease. Median survival could not be calculated as >50% of cases were censored. Patients in both groups were predominantly treated with radiation therapy or rituximab-based multiagent chemotherapy as first-line (Table 1).
Assessment of Copy Number Alterations in DLBCL-LT
Molecular inversion probe array analysis on tumor DNA was performed on six of 19 DLBCL-LT samples. Genomic alterations including copy-number gains, copy-number losses, and loss-of-heterozygosity were observed in all six cases. Clonal burden ranged from 30–90% (mean 62%) across cases; one case showed clonal diversity. The average number of alterations per sample was 26 (range: 16–42) and average proportion of the genome altered was 27% (range: 11–70%). Losses accounted for 63% of all calls (n=155), whereas loss-of-heterozygosity and copy number gains accounted for 14% and 23% of all calls, respectively. Homozygous losses accounted for 12% of all copy number calls, whereas high copy number gain, defined as >2 copies and consistent with amplification, was observed in only one case (2p16.1p15 region). Recurrent large genomic alterations observed in at least two cases included gains involving of chromosomes 1q, 7q, 10p, 10q, 11q, 12, 17q, 18, and X, losses of 1p, 6q, 8p and loss-of-heterozygosity of 9p and 18. Recurrent focal copy number alterations included gains of 2p16p15.1 (REL) and Xq28, and deletions of 2p11.2 (IGKV), 2q22.3 (ZEB2), 6p21 (HLA, NOTCH4), 8q12, 9p21.3 (CDKN2A), 14q32.33 (IGH locus), 15q15.1, 17q21.2 (RARA), and 22q11.23 (Figure S1, Table S2). Copy losses of HLA genes involved in MHC I/II antigen presentation affected 3 of 6 samples (50%).
Whole exome sequencing of DLBCL-LT and DLBCL-NOS tissue specimens
We performed whole exome sequencing on 19 DLBCL-LTs (6 with matched tumor-normal pairs, 13 without matched normal samples) and 6 DLBCL-NOS (1 matched, 5 unmatched). Consistent with previous reports on DLBCLs, the median numbers of mutations in DLBCL-LT and DLBCL-NOS were 175 mutations per case, with ranges of 46–380 and 90–308, respectively (Lohr et al., 2012, Mareschal et al., 2017) (Figure 1a).
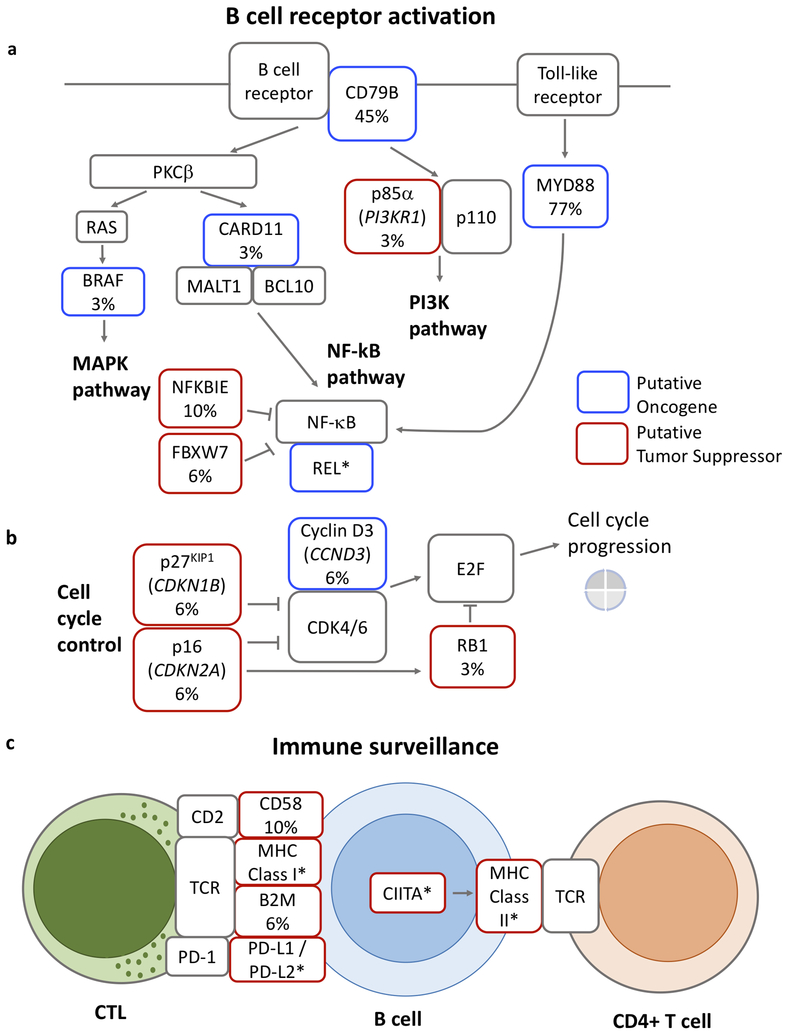
Figure 1. Landscape of somatic alterations in diffuse large B cell lymphoma, leg type (DLBCL-LT) and cutaneous diffuse large B cell lymphoma, not otherwise specified (DLBCL-NOS).
The number and type of single somatic nucleotide variants (SSNVs) in each sample of DLBCL-LT (a) and DLBCL-NOS (d). The tumor mutation burden as determined by the Foundation Medicine algorithm (see Methods) in DLBCL-LT (b) and DLBCL-NOS (e). Tumor mutation load is divided into three tiers (low, intermediate, and high). Recurrent somatic mutations identified by whole-exome sequencing in DLBCL-LT (c) and DLBCL-NOS (f). Blue denotes recurrent mutations in putative oncogenes; red denotes damaging mutations in putative tumor suppressor genes; grey denotes missense mutations that have not been functionally validated.
The tumor mutation burden (TMB), which correlates with tumor neoantigen burden and stratifies cancers based on their likelihood of responding to checkpoint blockade, was assessed using an established algorithm (Rizvi et al., 2015) (Johnson et al., 2016). Both DLBCL-LT and DLBCL-NOS cases were found to harbor an “intermediate” mutational load. suggesting potential therapeutic targetability with immunotherapies (Figure 1b and 1e, Table S3-S4).
Identification of putative driver genes in DLBCL-LT
To maximize our ability to detect cancer-promoting mutations, we employed an analytical pipeline that identifies putative driver mutations on the basis of the distribution of mutations and recurrences, both within our cohort and other published cohorts in the literature (Park et al., 2017, Vogelstein et al., 2013) (Tables S5-S13). In total, we analyzed 31 cases (19 cases of DLBCL-LT sequenced by us and 12 from publically available data) (Mareschal et al., 2017) (Table 1).
Using our analytical pipeline, we identified 30 somatic mutations in 21 putative driver genes across 8 biologically relevant pathways (Figure 1c). Ten of the mutations were validated gain of function mutations in 7 oncogenes (MYD88, CD79B, CARD11, BRAF, STAT3, MED12 and CCND3), some of which have been described in nodal DLBCL, but not DLBCL-LT (MYD88 p.S243N, CD79B p.L199P, STAT3 p.E616K and CARD11 p.R113Q (Figure 2; Table S6)). BRAF, STAT3, MED12, and CCND3 are genes newly implicated in DLBCL-LT. The remaining 20 mutations were loss of function or damaging mutations in 10 known tumor suppressor genes (PIK3R1, FBXW7, CREBBP, KMT2D, NFKBIE, BMF, PRDM1, CDKN1B, CDKN2A, CD58, ASXL1, RB1, B2M, and CIITA). Six of these genes are newly implicated in DLBCL-LT, but described in nodal DLBCL (PIK3R1, FBXW7, NFKBIE, CDKN1B), and other cancer types (BMF, ASXL1) (Bai et al., 2001, Boultwood et al., 2010, Hornsveld et al., 2016, Morin et al., 2016, Yao et al., 2017, Zhang et al., 2013) (Table 2).
Figure 2. Schematics of genes annotated with oncogenic missense mutations found in DLBCL-LT and DLBCL-NOS.
(a) Mutations marked with blue arrowheads denote validated gain of function missense mutations in putative oncogenes. (b) Mutations marked with red arrowheads denote loss of function missense mutations in putative tumor suppressor genes. Mutations marked with grey arrowheads denote missense mutations that have not been functionally validated.
Table 2.
Mutation prevalence and functional significance of DLBCL-LT putative driver genes
| Gene | #
Internal DLBCL-LT (n=19) |
%
Internal DLBCL-LT (n=19) |
#
External DLBCL-LT (n=12) |
%
External DLBCL-LT (n=12) |
# Total (n=31) |
% Total (n=31) |
ONC / TSG |
Validated
or damaging mutations |
Functional Domains Affected |
Previously described tumors |
Pathway | Function |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MYD88 | 14 | 74% | 10 | 83% | 24 | 77% | ONC | p.L265P, p.S243N | Toll/interleukin-1 (TIR) domain | DLBCL-LT, Nodal DLBCL (ABC>GCB) | NF-kB | Adapter protein involved in Toll-like receptor and IL-1 receptor signaling pathways. |
| CD79B | 8 | 42% | 6 | 50% | 14 | 45% | ONC | p.Y196D, p.Y196H, p.Y196N, p.Y196S, p.Y196C, p.L199P | Immunoreceptor tyrosine-based activation (ITAM) motif | DLBCL-LT, Nodal DLBCL (ABC>GCB) | NF-kB | Component of B cell receptor signaling complex |
| CREBBP | 4 | 21% | 1 | 8% | 5 | 16% | TSG | p.R1446C | Histone acetylation domain | DLBCL-LT, Nodal DLBCL (GCB>ABC) | Chromatin modification | Histone acetyltransferase and transcription co-activator |
| CD58 | 3 | 16% | 0 | 0% | 3 |
10% | TSG | p.V36fs, Splice | DLBCL-LT, Nodal DLBCL | Immune surveillance | Ligand for CD2 on T cells that aids in T cell activation | |
| KMT2D | 3 | 16% | 0 | 0% | 3 | 10% | TSG | p.K4778fs, p.R5448*, p.Q3333* | DLBCL-LT, Nodal DLBCL | Chromatin modification | H3K4 methyltransferase | |
| STAT3 | 3 | 16% | 0 | 0% | 3 | 10% | ONC | p.E616K | All-alpha domain | Nodal DLBCL | JAK-STAT | Transcription factor |
| NFKBIE | 2 | 11% | 1 | 8% | 3 | 10% | TSG | p.T253fs | Nodal DLBCL | NF-kB | IkB protein that inhibits NFkB | |
| PRDM1 | 2 | 11% | 1 | 8% | 3 | 10% | TSG | p.A391fs, Splice | DLBCL-LT, nodal DLBCL | B cell development | Transcriptional repressor of beta-interferon gene expression | |
| BMF | 2 | 11% | 0 | 0% | 2 | 6% | TSG | p.Q124* | Breast cancer | Apoptosis | Bcl-2 family protein that regulates apoptosis | |
| CCND3 | 2 | 11% | 0 | 0% | 2 | 6% | ONC | p.T283P, p.P284S | Conserved phosphorylation site used by proteosome | Nodal DLBCL | Cell cycle control | Component of cyclin D3-CDK4 complex that phosphorylates and inhibits RB family and regulates G1/S cell cycle transition |
| CDKN1B | 2 | 11% | 0 | 0% | 2 | 6% | TSG | p.W76*, p.R90fs | Nodal DLBCL | Cell cycle control | Encodes p27Kip1 inhibitor that prevents cyclin E-CDK2 or cyclin D-CDK4 complexes and controls cell cycle at G1. | |
| FBXW7 | 2 | 11% | 0 | 0% | 2 | 6% | TSG | p.R505C | WD40 repeated domain | Nodal DLBCL | Ubiquitination | Subunit of ubiquitin ligase complex involved in ubiquitin mediated degradation |
| B2M | 1 | 5% | 1 | 8% | 2 | 6% | TSG | p.R23fs | DLBCL-LT, Nodal DLBCL (GCB>ABC) | Immune surveillance | Subunit of MHC class I | |
| CDKN2A | 1 | 5% | 1 | 8% | 2 | 6% | TSG | p.R80* | DLBCL-LT, Nodal DLBCL (ABC>GCB) | Cell cycle control | Encodes p16 and p14ARF tumor suppressors; p16 inhibits CDK4/CDK6 and activates Rb; p14ARF activates p53 | |
| CIITA | 1 | 5% | 1 | 8% | 2 | 6% | TSG | p.Q476* | DLBCL-LT, Nodal DLBCL | Immune surveillance | Master transcription factor regulating MHC Class II expression. | |
| BRAF | 1 | 5% | 0 | 0% | 1 | 3% | ONC | p.V600E | Kinase domain | Melanoma | MAP kinase | Raf family kinase involved in MAP kinase pathway. |
| CARD11 | 1 | 5% | 0 | 0% | 1 | 3% | ONC | p.R113Q | LATCH inhibitory domain | DLBCL-LT, Nodal DLBCL (ABC>GCB) | NF-kB | Scaffold protein that associates with BCL10 and MALT1 to drive NF-kB activation. |
| MED12 | 1 | 5% | 0 | 0% | 1 | 3% | ONC | p.V41E | Undefined recurrently mutated domain on exon 2 | Leiomyoma | Transcription | Subunit of the Mediator transcription regulatory complex. |
| PIK3R1 | 1 | 5% | 0 | 0% | 1 | 3% | TSG | p.N564D | iSH2 domain | Nodal DLBCL | PI-3 kinase | Encodes regulatory subunit p85α of phosphoinositide 3-kinase (PI3K) |
| RB1 | 0 | 0% | 1 | 8% | 1 | 3% | TSG | p.Y173fs | DLBCL-LT, Nodal DLBCL | Cell cycle control | Repressor of E2F transcription factors | |
| ASXL1 | 0 | 0% | 1 | 8% | 1 | 3% | TSG | p.R417* | Nodal DLBCL, AML | Chromatin modification | Regulator of Polycomb-mediated gene silencing including H3K27me3 |
Internal DLBCL-LT samples (n=19) are those that were sequenced for this study. External DLBCL-LT samples (n=12) refer to previously published DLBCL-LT exome datasets (Mareschal et al 2017).
Nodal DLBCL ABC vs GCB based on data from 1001 nodal DLBCL whole exome samples (Reddy et al 2017)
Highlighting the clinical relevance of our findings, at least five signaling pathways with mutations in DLBCL-LTs are currently or potentially targetable (Figure 3). These include highly prevalent mutations in the NF-κB pathway (MYD88 (77%), CD79B (45%), CARD11 (3%), and NFKBIE (10%) (Figure 3). 29% have point or copy number mutations in genes predicted to affect tumor immune evasion including genes important for antigen presentation (HLA genes (50%), B2M (6%), CIITA (6%)) and genes important for T cell co-stimulation (CD58 (10%)).
Figure 3. Oncogenic pathways mutated in DLBCL-LT.
DLBCL-LT harbors mutations that are predicted to affect pathways downstream of B cell activation (a), including MAPK, NF-κB, and PI3K pathways, mutations that affect cell cycle control (b), and mutations that affect immune surveillance (c). Blue boxes denote putative oncogenes and red boxes denote putative tumors suppressor genes. Frequency (%) of somatic mutations in our expanded cohort of 31 DLBCL-LT is noted when relevant. (*) Putative oncogenes or tumor suppressors based on copy number data.
We also found less common mutations in the MAPK pathway (BRAF), the JAK-STAT pathway (STAT3), the PI-3K pathway (PIK3R1), cell cycle control pathways (CDKN2A, CDKN1B, CCND3, RB1), and chromatin modification pathways (KMT2D, CREBBP, ASXL1) (Figure 3; Tables S6-S12) (Vaque et al., 2014).
MYD88 wild-type tumors harbor targetable mutations
Seven of 31 DLBCL-LT (5 of 19 from our dataset, and 2 of 12 from a previous published dataset) did not have gain of function mutations in MYD88. Six of these harbored mutations functionally validated in other cancer types. Despite the smaller sample size, we were able to recognize distinct patterns. Three samples had mutations in other NF-κB signaling genes. One harbored a damaging mutation in tumor suppressor NFKBIE, one harbored copy number gains of REL, and one harbored both. Four samples (including 1 with REL amplification) had functionally validated mutations in other canonical oncogenic pathways, MAPK (BRAF) (Davies et al., 2002), PI3K (PIK3R1) (Jaiswal et al., 2009), JAK/STAT (STAT3) (Vallois et al., 2016), or transcriptional control (MED12) (Mittal et al., 2015) (Figure 1, Table 2). These non-NF-κB pathway mutations were all mutually exclusive of each other.
DLBCL-NOS
Among the DLBCL-NOS cohort, we identified mutations in 11 putative driver genes across 5 of 6 samples. None of these mutations are recurrent. These encompassed a range of signaling pathways including NF-κB (CARD11), JAK/STAT (STAT6), MAPK (KRAS), cell cycle control (RB1), immune surveillance (CIITA), DNA damage response (TP53, MSH6) and chromatin modification (CREBBP, KMT2D) (Figure 1f).
The CARD11 and STAT6 mutations were previously described in nodal ABC and GCB type DLBCLs respectively but not in cutaneous DLBCLs (Reddy et al., 2017). Finally, we found seven damaging mutations in tumor suppressor genes previously implicated in nodal DLBCL (KMT2D, TNFRSF14, CIITA, TP53, RB1, CREBBP and MSH6) (Cycon et al., 2009, de Miranda et al., 2013, Reddy et al., 2017) and a damaging mutation in ETNK1, previously implicated in atypical chronic myeloid leukemia (Gambacorti-Passerini et al., 2015) (Table S11, S13) (Figure 1f).
DLBCL-LT versus other nodal and extranodal DLBCL subtypes
In a pan-DLBCL analysis of nodal and extranodal DLBCL subtypes, we assessed and compared the distribution of mutations across these DLBCL subtypes. We measured the relative prevalence of mutations in DLBCL-LT putative driver genes in other DLBCL subtypes (defined as the similarity index; 1= exact correlation and 0 = no correlation; Materials and Methods). Our analysis confirmed that the mutational profile of DLBCL-LT overlapped with the ABC-subtype of nodal DLBCLs. However, it was most similar to two extranodal DLBCLs, primary central nervous system large B-cell lymphoma (PCNSL) (similarity index (SI) = 0.73) and primary testicular DLBCL (PTL) (SI=0.71) (Figure S2, Table S14). In addition, like PCNSL and PTL, DLBCL-LTs harbored a similarly high incidence of concurrent CD79B and MYD88 mutations (Braggio et al., 2015, Chapuy et al., 2016).
DLBCL-NOS had minimal similarity to DLBCL-LT. In fact, it was more similar to the GCB-subtype than to the ABC-subtype of nodal DLBCL. However, in general, it had little similarity (SI ≤ 0.42) compared to any other DLBCL subtype (Figure S3, Table S15).
PD-L1 and PD-L2 translocations
Given the genetic similarities with PCNSL and PTL, and the high frequency of PDL1/PDL2 alterations in these entities (Chapuy et al., 2016), we assessed the prevalence in DLBCL-LT of structural variants involving the PDL1/PDL2 locus. We performed fluorescent in situ hybridization (FISH) targeting the PD-L1/PD-L2 gene locus on 10 DLBCL-LTs and 5 DLBCL-NOSs with available tissue. Of the 10 DLBCL-LTs, 4 samples (40%) demonstrated break-apart translocations of PD-L1 and PD-L2 (Figure 4a). There were no structural variants in 5 of 5 DLBCL-NOS tested.
Figure 4. Chromosomal rearrangement of the PD-L1 and PD-L2 in DLBCL-LT.
(a) Break-apart fluorescent in situ hybridization (FISH) assay of DLBCL-LT with PD-L1 in red, PD-L2 in green, and chromosome 9 control in yellow. Samples with (+) and without (−) translocation are annotated. (b) Dual stain PD-L1 (brown, membranous) and PAX5 (red, nuclear) immunohistochemistry (IHC) of normal tonsil control (inset shows light PD-L1 staining of germinal center antigen-presenting cells), PD-L1 wild-type sample NU03, and PD-L1/PD-L2 mutant samples (NU07, AU25). Red PAX5 stains B-cells. (c) Dual stain PD-L2 (brown, membranous) and PAX5 (red, nuclear) IHC of normal tonsil, PD-L1 wild-type NU03, and PD-L1/PD-L2 mutant samples (NU07, AU25). The scale bar represents 25 μm. IHC images for AU10 and AU18 are unavailable due to inadequate tissue.
We performed dual immunohistochemistry staining (PDL1/PAX5 and PDL2/PAX5) on 9 DLBCL-LT samples to determine relative PD-L1 and PDL-2 expression in the tumor and in the microenvironment. There was an imperfect correlation between PD-L1/PD-L2 structural variants and tumor expression of PD-L1/PD-L2. Among the 4 samples with translocations, 2 (50%) showed upregulation of PD-L1 or PD-L2 (1 each) in both tumor and microenvironment (Figure 4, Table S16). These numbers are consistent with those reported in a recent study (Menguy et al., 2017). Among the 6 FISH-negative DLBCL-LT samples, there was no tumor expression of PD-L1 or PD-L2. The majority, 7 of 9 examined, had PD-L1 (7 cases) or PD-L2 expression (1 case) in the microenvironment (Figure 4b-4c). None of the DLBCL-NOS samples had evidence of tumor PD-L1 expression, but 4 of 5 had strong PD-L1 expression in the microenvironment (Table S16).
DISCUSSION
We performed exome sequencing of the largest cohort of cutaneous DLBCLs to date. We identified putative cancer promoting mutations in 21 genes, including 10 whose association with DLBCL-LT are, to our knowledge, previously unreported. More than 50% of mutations identified have not been previously described in DLBCL-LT.
Highlighting the clinical importance of our findings, our analyses of DLBCL-LT confirm the high prevalence of mutations in the potentially targetable NF-κB signaling pathway, with validated mutations in MYD88, CD79B, CARD11, or NFKBIE in 79% of tumors. We have identified NFKBIE mutations and REL copy gains as potential drivers that reactivate the NF-κB pathway in MYD88 wild-type tumors and occur exclusive of other NF-κB pathway mutations. Among the other MYD88-wild-type tumors without NF-κB pathway alterations, we identified mutations in canonical cancer pathway genes (e.g. BRAF (MAPK pathway), PIK3R1 (PI3K), STAT3 (JAK-STAT)) or pathways affecting the transcriptional machinery (MED12) that may serve as bypass pathways. These mutations have important clinical implications as they suggest that most of these tumors are targetable via inhibitors of these pathways and that non-NF-κB pathways can be primary drivers in a subset of DLBCL-LTs.
In addition, we found that DLBCL-LT bore hallmarks of ABC-type DLBCLs whereas DLBCL-NOS, if anything, bore hallmarks of GCB-type DLBCLs. These data suggest that 1) genetic tests at the patient bedside may distinguish these entities and 2) cutaneous DLBCL subtypes are different diseases with distinct pathophysiological mechanisms that may require distinct therapeutic strategies.
Our data highlight the importance of immune evasion in DLBCL-LT pathogenesis. DLBCL-LTs have sufficient neoantigens to be immunogenic. However, they harbor mutations in multiple pathways predicted to enable evasion from immune surveillance. First, PDL1/PDL2 locus alterations occurred in 40% of DLBCL-LTs. Secondly, we also found that 50% of samples harbored copy number deletions in either the MHC Class I or II loci, which have been shown to reduce the number of neoantigens that can be presented on the tumor cell surface (McGranahan et al., 2017). Lastly, we confirmed previous studies that found rare point mutations in genes associated with antigen processing [B2M (6% of samples), CIITA (6%)] or T cell co-stimulation [CD58 (10%)]. None of the samples were assessed by all three genetic tools (FISH, copy number analysis and exome sequencing). Nonetheless, to the extent we can, we infer that these mutations are mutually exclusive, and 50% or more DLBCL-LTs harbored mutations in one of these immune evasion pathways.
Lastly, we found broad similarities between DLBCL-LTs with two extranodal DLBCLs (PTL and PCNSL). In addition to frequent NF-κB activating mutations, all three DLBCL subtypes appear to employ multiple mechanisms in tumor cells and in the tumor microenvironment to evade immune surveillance. These data are particularly interesting in light of recent data in mice, which suggest that the lower legs, at times, can be an immunoprivileged site like the CNS and the testes. In particular, the lower legs support local upregulation of Treg cells that inhibit T cell responses in the setting of lymphedema (Garcia Nores et al., 2017).
Collectively, our findings suggest a novel therapeutic paradigm wherein patients with DLBCL-LT subtype can be included in clinical trials with genomically similar PCNSL and PTLs. As an example, our analyses led us to discover that 40% of DLBCL-LT had recurrent genetic alterations in PD-L1/PD-L2. This suggests that at least in a subset of these tumors, there is therapeutic value in utilizing checkpoint inhibitors and including these patients in currently active trials of similar DLBCL, such as nivolumab in relapsed/refractory PCNSL and PTL (NCT02857426).
MATERIALS AND METHODS
Sample preparation and sequencing analysis
All studies were approved by the Institutional Review Board (IRB) of Northwestern University. The DLBCL-LT samples were de-identified formaldehyde fixed paraffin embedded (FFPE) archival specimens from Medical University of Graz (Graz, Austria), Northwestern University (Chicago, USA) and Massachusetts General Hospital (Boston, MA) and were reviewed by expert dermatopathologists (JG, LC) or hematopathologists (AL, AB). Subject consent for this study was not required as all tissue was obtained from archival tissue that is IRB/Ethics Committee-approved for research purposes. 2mm cores (with >80% tumor cells) were obtained as described (Goh et al., 2016), matched normal skin was used as controls in 7 cases. Genomic DNA was prepared/sequenced/analyzed as previously described (Park et al., 2017). Genomic sequencing data were deposited in dbGaP (Accession number: phs001645).
Copy Number Aberrations
Genomic DNA was processed for molecular inversion probe array analysis using the OncoScan FFPE Assay kit (ThermoFisher, Santa Clara, CA, USA), as described (Paxton et al., 2015). Data analysis was performed using Chromosome Analysis software (ChAS) version 3.1 (ThermoFisher) and Nexus Express Software for OncoScan version 3.1 (Biodiscovery, Hawthorne, CA, USA) with reference to assembly GRCh37/hg19, as described (Andersen et al., 2017). All cases were processed using the TuScan segmentation algorithm except for case MG88, which was re-centered and processed using SNP-FASST2. Recurrent genomic alterations were calculated using the aggregate analysis in Nexus Express.
Comparison of DLBCL-LT and DLBCL-NOS with other DLBCL subtypes
We compared the frequency of mutations in the genes most commonly mutated in each DLBCL subtype with that of the same genes in DLBCL-LT or DLBCL-NOS. Vice-versa, we calculated the relative mutation prevalence in other DLBCL subtypes of 17 putative DLBCL-LT and 11 putative DLBCL-NOS driver genes. We normalized these values to a 0 to 1 similarity index scale where 1 would be exact correlation and 0 no correlation.
Tumor Mutation Burden Estimation
To assess relative tumor mutation burden, we quantified all somatic single nucleotide variants within 315 genes that are part of the Foundation Medicine panel (Campesato et al., 2015, Rosenberg et al., 2016). Relative tumor mutation burden cutoffs were ≥20 mutations per megabase (high), 6–19 (intermediate), or ≤5 (low).
Fluorescence in situ hybridization (FISH) of PD-1 ligands
Fluorescence in situ hybridization (FISH) was performed in collaboration with Empire Genomics (Buffalo, NY) on paraffin-embedded formalin-fixed tissue using probes developed to target PD-L1 (CD274, green) and PD-L2 (PDCD1LG2, red) within the chromosome 9p24.1 locus, and a control probe targeting the 9q arm (Con 9, yellow). The PDL1 probe does not overlap the PDL2 gene and the PDL2 probe does not overlap the PDL1 gene. The two probes consistently co-localized in normal metaphase spreads from all control tissues (human tonsils and peripheral blood mononuclear cells) tested. Over 90% of cells in these cases expressed 2 copies of PD-L1 adjacent to PD-L2.
Slides were hybridized according to established protocols (Empire Genomics) and reviewed by a pathologist. 200 cells were counted for all except one sample (100 cells) due to sample quality. Break-apart red/green signal pattern >10% of counted cells were considered above the threshold for translocation. Counts for amplification, relative copy gain, polysomy and normal copy numbers were also noted using criteria previously described (Roemer et al., 2016).
Double Immunohistochemistry Staining
Double staining of PD-L1 (1:50; clone E1L3N; Cell Signaling, Danvers, MA) or PD-L2 (1:50; clone D7U8C; Cell Signaling) and PAX5 (1:100; clone 24; BD Biosciences, San Jose, CA) was performed with an automated staining system (Bond III; Leica Biosystems, Buffalo Grove, IL) as previously described (Ansell et al., 2015). Stained slides were scored by an expert dermatopathologist (JG) and expert hematopathologist (AB), and percentages of both tumor PDL-1 or PDL-2 and microenvironment PDL-1 or PDL-2 were calculated by scoring 100–200 cells in each category. The threshold for PD-L1 and PD-L2 expression was defined at 30% for PAX5-positive tumor cells and at 20% for PAX5-negative immune cell microenvironment, as reported by others (Kiyasu et al., 2015).
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the patients who have contributed to this study, the Northwestern Skin Disease Research Center (DNA/RNA Delivery Core), Robert H. Lurie Comprehensive Cancer Center (Pathology Core), Center for Cancer Genome Discovery, Yale Center for Genome Analysis, Admera Health for their sequencing support, and Foglia Family Foundation for their support and the Northwestern University Research Computing Services for their invaluable contribution for providing effective solutions for the storage and analysis of data.
J.C. is the Ruth K. Freinkel Research Professor. He is supported in part by the National Institutes of Health (NIH), National Cancer Institute (NCI) (Grant #K08-CA191019–01A1), the NIH’s National Center for Advancing Translational Sciences (Grant #UL1TR001422). J.C. is a Doris Duke–Damon Runyon Clinical Investigator supported in part by the Damon Runyon Cancer Research Foundation (DRCRF #CI-84–16) and by the Doris Duke Charitable Foundation (DDCF #2016095). X.Z. is supported in part by grant #T32-AR060710 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the NIH and a Foglia Family Foundation grant. A.L. is supported in part by the NIH, NCI (Grant #K23CA184279). A.L also receives research support from American Society of Hematology/Amos Medical Faculty Development Program. A.L. gratefully acknowledges funding support from an anonymous foundation. A.W. is supported by grant #T15-LM011271 from the National Library of Medicine of the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations:
- DLBCL-LT
diffuse large B cell lymphoma, leg type
- DLBCL-NOS
secondary cutaneous diffuse large B cell lymphoma
- WES
whole exome sequencing
- PD-L1 and PD-L2
programmed death ligands 1 and 2
- SSNV
single somatic nucleotide variant
- PCNSL
primary central nervous system lymphoma
- PTL
primary testicular lymphoma
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
Co-first authors
REFERENCES
- Andersen EF, Paxton CN, O’Malley DP, Louissaint A Jr., Hornick JL, Griffin GK, et al. Genomic analysis of follicular dendritic cell sarcoma by molecular inversion probe array reveals tumor suppressor-driven biology. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2017;30(9):1321–34. [DOI] [PubMed] [Google Scholar]
- Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med 2015;372(4):311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Vlachonikolis J, Agnantis NJ, Tsanou E, Dimou S, Nicolaides C, et al. Low expression of p27 protein combined with altered p53 and Rb/p16 expression status is associated with increased expression of cyclin A and cyclin B1 in diffuse large B-cell lymphomas. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2001;14(11):1105–13. [DOI] [PubMed] [Google Scholar]
- Boultwood J, Perry J, Pellagatti A, Fernandez-Mercado M, Fernandez-Santamaria C, Calasanz MJ, et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia 2010;24(5):1062–5. [DOI] [PubMed] [Google Scholar]
- Braggio E, Van Wier S, Ojha J, McPhail E, Asmann YW, Egan J, et al. Genome-Wide Analysis Uncovers Novel Recurrent Alterations in Primary Central Nervous System Lymphomas. Clinical cancer research : an official journal of the American Association for Cancer Research 2015;21(17):3986–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campesato LF, Barroso-Sousa R, Jimenez L, Correa BR, Sabbaga J, Hoff PM, et al. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to PD-1 blockade in clinical practice. Oncotarget 2015;6(33):34221–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, Roemer MG, Stewart C, Tan Y, Abo RP, Zhang L, et al. Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood 2016;127(7):869–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cycon KA, Rimsza LM, Murphy SP. Alterations in CIITA constitute a common mechanism accounting for downregulation of MHC class II expression in diffuse large B-cell lymphoma (DLBCL). Exp Hematol 2009;37(2):184–94. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417(6892):949–54. [DOI] [PubMed] [Google Scholar]
- de Miranda NF, Peng R, Georgiou K, Wu C, Falk Sorqvist E, Berglund M, et al. DNA repair genes are selectively mutated in diffuse large B cell lymphomas. The Journal of experimental medicine 2013;210(9):1729–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambacorti-Passerini CB, Donadoni C, Parmiani A, Pirola A, Redaelli S, Signore G, et al. Recurrent ETNK1 mutations in atypical chronic myeloid leukemia. Blood 2015;125(3):499–503. [DOI] [PubMed] [Google Scholar]
- Garcia Nores GD, Ly CL, Savetsky IL, Kataru RP, Ghanta S, Hespe GE, et al. Regulatory T Cells Mediate Local Immunosuppression in Lymphedema. J Invest Dermatol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G, Walradt T, Markarov V, Blom A, Riaz N, Doumani R, et al. Mutational landscape of MCPyV-positive and MCPyV-negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget 2016;7(3):3403–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange F, Joly P, Barbe C, Bagot M, Dalle S, Ingen-Housz-Oro S, et al. Improvement of survival in patients with primary cutaneous diffuse large B-cell lymphoma, leg type, in France. JAMA dermatology 2014;150(5):535–41. [DOI] [PubMed] [Google Scholar]
- Hornsveld M, Tenhagen M, van de Ven RA, Smits AM, van Triest MH, van Amersfoort M, et al. Restraining FOXO3-dependent transcriptional BMF activation underpins tumour growth and metastasis of E-cadherin-negative breast cancer. Cell Death Differ 2016;23(9):1483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, Wang W, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer cell 2009;16(6):463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DB, Frampton GM, Rioth MJ, Yusko E, Xu Y, Guo X, et al. Targeted Next Generation Sequencing Identifies Markers of Response to PD-1 Blockade. Cancer Immunol Res 2016;4(11):959–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyasu J, Miyoshi H, Hirata A, Arakawa F, Ichikawa A, Niino D, et al. Expression of programmed cell death ligand 1 is associated with poor overall survival in patients with diffuse large B-cell lymphoma. Blood 2015;126(19):2193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama K, Massone C, Chott A, Metze D, Kerl H, Cerroni L. Primary cutaneous large B-cell lymphomas: clinicopathologic features, classification, and prognostic factors in a large series of patients. Blood 2005;106(7):2491–7. [DOI] [PubMed] [Google Scholar]
- Lee WJ, Won KH, Won CH, Chang SE, Choi JH, Moon KC, et al. Secondary Cutaneous Diffuse Large B-cell Lymphoma has a Higher International Prognostic Index Score and Worse Prognosis Than Diffuse Large B-cell Lymphoma, Leg Type. Acta Derm Venereol 2016;96(2):245–50. [DOI] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proceedings of the National Academy of Sciences of the United States of America 2012;109(10):3879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mareschal S, Pham-Ledard A, Viailly PJ, Dubois S, Bertrand P, Maingonnat C, et al. Identification of Somatic Mutations in Primary Cutaneous Diffuse Large B-Cell Lymphoma, Leg Type by Massive Parallel Sequencing. J Invest Dermatol 2017;137(9):1984–94. [DOI] [PubMed] [Google Scholar]
- McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell 2017;171(6):1259–71 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menguy S, Prochazkova-Carlotti M, Beylot-Barry M, Saltel F, Vergier B, Merlio JP, et al. PD-L1 and PD-L2 are Differentially Expressed by Macrophages or Tumor Cells in Primary Cutaneous Diffuse Large B-Cell Lymphoma, Leg Type. The American journal of surgical pathology 2017. [DOI] [PubMed] [Google Scholar]
- Mittal P, Shin YH, Yatsenko SA, Castro CA, Surti U, Rajkovic A. Med12 gain-of-function mutation causes leiomyomas and genomic instability. J Clin Invest 2015;125(8):3280–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Assouline S, Alcaide M, Mohajeri A, Johnston RL, Chong L, et al. Genetic Landscapes of Relapsed and Refractory Diffuse Large B-Cell Lymphomas. Clinical cancer research : an official journal of the American Association for Cancer Research 2016;22(9):2290–300. [DOI] [PubMed] [Google Scholar]
- Nabhan C, Smith SM, Helenowski I, Ramsdale E, Parsons B, Karmali R, et al. Analysis of very elderly (>/=80 years) non-hodgkin lymphoma: impact of functional status and co-morbidities on outcome. Br J Haematol 2012;156(2):196–204. [DOI] [PubMed] [Google Scholar]
- Park J, Yang J, Wenzel AT, Ramachandran A, Lee WJ, Daniels JC, et al. Genomic analysis of 220 CTCLs identifies a novel recurrent gain-of-function alteration in RLTPR (p.Q575E). Blood 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxton CN, Rowe LR, South ST. Validation of a Modified OncoScan Protocol for Use in a Clinical Laboratory. Cancer Genetics 2015;208(6):361. [Google Scholar]
- Pham-Ledard A, Beylot-Barry M, Barbe C, Leduc M, Petrella T, Vergier B, et al. High frequency and clinical prognostic value of MYD88 L265P mutation in primary cutaneous diffuse large B-cell lymphoma, leg-type. JAMA dermatology 2014a;150(11):1173–9. [DOI] [PubMed] [Google Scholar]
- Pham-Ledard A, Prochazkova-Carlotti M, Andrique L, Cappellen D, Vergier B, Martinez F, et al. Multiple genetic alterations in primary cutaneous large B-cell lymphoma, leg type support a common lymphomagenesis with activated B-cell-like diffuse large B-cell lymphoma. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2014b;27(3):402–11. [DOI] [PubMed] [Google Scholar]
- Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 2017;171(2):481–94 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348(6230):124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer MG, Advani RH, Ligon AH, Natkunam Y, Redd RA, Homer H, et al. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2016;34(23):2690–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 2016;387(10031):1909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senff NJ, Noordijk EM, Kim YH, Bagot M, Berti E, Cerroni L, et al. European Organization for Research and Treatment of Cancer and International Society for Cutaneous Lymphoma consensus recommendations for the management of cutaneous B-cell lymphomas. Blood 2008;112(5):1600–9. [DOI] [PubMed] [Google Scholar]
- Suarez AL, Querfeld C, Horwitz S, Pulitzer M, Moskowitz A, Myskowski PL. Primary cutaneous B-cell lymphomas: part II. Therapy and future directions. J Am Acad Dermatol 2013;69(3):343 e1–11; quiz 55–6. [DOI] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016;127(20):2375–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallois D, Dobay MP, Morin RD, Lemonnier F, Missiaglia E, Juilland M, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood 2016;128(11):1490–502. [DOI] [PubMed] [Google Scholar]
- Vaque JP, Martinez N, Batlle-Lopez A, Perez C, Montes-Moreno S, Sanchez-Beato M, et al. B-cell lymphoma mutations: improving diagnostics and enabling targeted therapies. Haematologica 2014;99(2):222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science 2013;339(6127):1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Xu F, Chen Y, Ge Y, Zhang F, Huang H, et al. Fbw7 regulates apoptosis in activated B-cell like diffuse large B-cell lymphoma by targeting Stat3 for ubiquitylation and degradation. J Exp Clin Cancer Res 2017;36(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America 2013;110(4):1398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.