Abstract
Studies of heterochronic parabiosis, where two animals of different ages are joined surgically, provided proof-of-principle results that systemic proteins have broad age-specific effects on tissue health and repair. In an effort to identify these systemic proteins, we previously developed a method to selectively label the proteome of only one animal joined in parabiosis utilizing bio-orthogonal non-canonical amino acid tagging (BONCAT), which can metabolically label proteins during their de novo synthesis by incorporating a methionine substitute, azido-nor-leucine (ANL), in cells expressing a mutant methionyl-tRNA synthetase (MetRSL274G). Once labeled, we can selectively identify the proteins produced by the MetRSL274G transgenic mouse in the setting of heterochronic parabiosis. This approach enabled the detection of several rejuvenating protein candidates from the young parabiont, which were transferred to the old mammalian tissue through their shared circulation. Although BONCAT is a very powerful technology, the challenges associated with its complexity including large starting material requirements and cost of ANL-labeled protein detection, such as modified Antibody Arrays and Mass Spectrometry, limit its application. Herein, we propose a lab-on-a-chip technology, termed Click-A+Chip for facile and rapid digital detection of ANL-labeled proteomes present in minute amount of sample, to replace conventional assays. Click-A+Chip is a graphene-based field effect biosensor (gFEB) which utilizes novel on-chip click-chemistry to specifically bind to ANL-labeled biomolecules. In this study, Click-A+Chip is utilized for the capture of ANL-labeled proteins transferred from young to old parabiotic mouse partners. Moreover, we were able to identify the young-derived ANL-labeled Lif-1 and Leptin in parabiotic systemic milieu, confirming previous data as well as providing novel findings on the relative levels of these factors in young versus old parabionts. Summarily, our results demonstrate that Click-A+Chip can be used for rapid detection and identification of ANL-labeled proteins, significantly reducing the sample size, complexity, cost and time associated with BONCAT analysis.
A. Introduction
Advances in modern medicine have increased the average lifespan worldwide which has highlighted the need to address the diseases of old age. These geriatric conditions have become endemic, posing unsustainable economic and social burdens1. Heterochronic parabiosis studies have provided proof-of-principle results that aging is both reversible and inducible through the specific composition of the circulatory milieu2,3. Specifically, a number of old tissues including muscle, heart, liver, brain, bones, etc. were rejuvenated by heterochronic parabiosis, and at the same time, young mice displayed a broad decline in tissue health and maintenance2–4. Recent studies utilizing heterochronic blood exchange in mice further confirmed that age-related changes in the blood play a major role in attenuating tissue health and function, and have uncoupled the rejuvenating properties of the endocrine–systemic milieu from the changes or adaptations caused by the sharing of organ systems and/or being joined for several weeks5. Combination of blood heterochronicity and BONCAT has the promise to directly and comprehensively identify the regulators of tissue aging and rejuvenation, aiding in the development of better diagnostics and treatments for a broad class of degenerative and metabolic diseases.
However, BONCAT demands downstream analysis such as Mass Spectrometry for specific protein identification. Mass Spectrometry requires milligrams of starting material, which is challenging to obtain in studies of small animals and stem cell research. Additionally, with Mass Spectrometry based profiling, the proteins6, which contain ANL, are conjugated to biotin through click-chemistry and can be enriched on affinity columns before their identification7. However, because ANL-labeling is not to saturation, the unlabeled proteins, which can bind non-specifically to the enrichment column, can introduce false positives. Furthermore, false positives can occur when a mixture of ANL-labeled and unlabeled proteins are processed for Mass Spectrometry directly, due to salt-adducts; the additional mass of ANL (172.19 g/mol) over methionine (149.21 g/mol) is ~22.98 g/mol, which is very close to the molecular weight of a sodium ion (22.99 g/mol) (See Table S1 in the Supporting Information).
An approach that utilizes ANL-adapted Antibody Array proteomics, takes advantage of biotin clicked to ANL-labeled proteins, which are detected by streptavidin-Cy3 fluorescence8. This method minimized false positives, however, it is limited by cost, the bias of antibody choice, and by the relatively low sensitivity i.e., intensity of fluorescence signal. Furthermore, in Mass Spectrometry and modified Antibody Arrays, some ANL-labeled proteins might be missed through their dilution by the unlabeled protein pool, conceivably generating false negative results. Finally, all of the above assays involve many steps and take many days to complete. Therefore, there is a need for a facile, accurate, digital proteomics technique, which can discriminate ANL-labeled proteins from unlabeled proteins and free ANL, enabling unambiguous direct detection of ANL-labeled proteins present in small starting samples of high biological complexity.
In this study, we have developed a digital biosensor, termed Click-A+Chip, for the rapid and unambiguous profiling of ANL-labeled proteins, which significantly minimizes cost, detection time, starting material, and false positives/negatives, as compared to Mass Spectrometry or modified Antibody Arrays. Click-A+Chip is a graphene-based field effect transistor (gFET) capable of specifically capturing ANL-labeled molecules (Figure 1). The gFET within the Click-A+Chip construct is composed of source and drain electrodes connected via a single graphene layer (graphene channel), where the current is modulated via a third electrode (gate). In order to detect the ANL-labeled proteomes, the surface of the graphene channel is functionalized with a novel click-based capture molecule, dibenzocyclooctyne-pyrene (DBOP). DBOP is capable of binding to ANL-labeled proteins from one end and associates with the surface of the graphene through π-π stacking to the other end9. The binding of the charged ANL-labeled protein to the surface of the graphene, via DBOP, changes the graphene channel conductivity, which is transduced into a signal in the form of a change in electrical parameter of the gFET such as the current flowing through the graphene channel from the source to the drain electrodes (IDS) (Figure 1). The captured ANL-labeled proteins on the surface of the graphene can be identified by subsequent introduction of antibodies specific to proteins of interest, which will further change the IDS in Click-A+Chip. Lif-1 and Leptin were chosen for detection, as they are promising protein candidates implicated in tissue rejuvenation8. Click-A+Chip facilitates the goal of identifying young rejuvenating factors from minute blood or cell samples, obtainable during small animal research, with the precision, simplicity, and speed required for timely and accessible monitoring.
Figure 1.
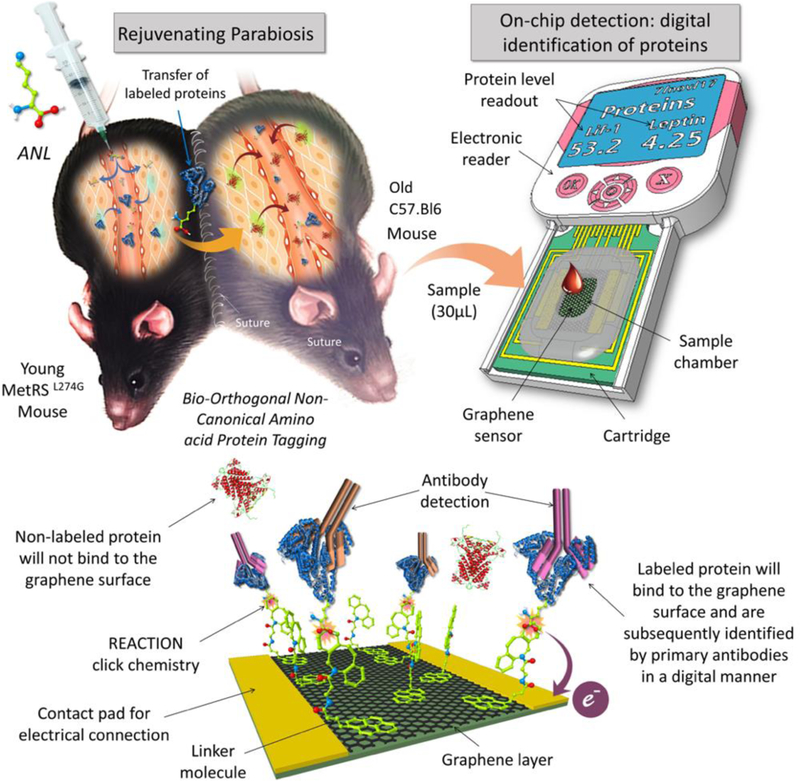
Click-A+Chip is a graphene-based biosensor for the digital identification of proteins after bio-orthogonal non-canonical amino acid tagging of the young proteome in the setting of heterochronic parabiosis. Click-A+Chip is composed of a field effect biosensor with a graphene channel which is functionalized with a novel synthetic molecule capable of selectively binding to ANL-labeled proteins via click-chemistry. The binding events between the linker molecule and the ANL-labeled protein result in the alteration of graphene conductivity and biosensor electrical parameters which can be detected using a handheld analyzer. Click-A+Chip is also capable of protein identification by further utilizing a range of primary antibodies to identify the labeled proteins within a few minutes and without the need for any secondary antibodies or any complex instrumentation.
B. Experimental
Chemicals and reagents
All commercially available compounds were purchased from Sigma Aldrich or Alfa Aesar and all solvents were purchased dry from Sigma Aldrich with a Sure/Seal system. NMR spectra were recorded on Bruker Avance 400 (400 MHz for 1H; 400 MHz for 13C) spectrometer. The chemical shifts (δ) are given in parts per million relative to CDCl3 (7.26 ppm for 1H) CDCl3 (77.16 ppm for 13C). Flash column chromatography was performed on silica gel (particle size 200–300 mesh, purchased from Sigma Aldrich) or using a Biotage Isolera Specktra system.
Synthesis of graphene click-linker (DBOP)
The click-linker is designed to bind to the surface of graphene on one end and react with the azide containing molecule on the other end via click-chemistry. The click-linker was synthesized by reacting N-hydroxysuccinimide (1) with 1-pyrenebutyric acid (2) to yield 2,5-dioxopyrrolidin- 1-yl 4-(pyren-1-yl)butanoate (3). Compound 3 was reacted with dibenzocyclooctyne-amine (4) to form a dibenzocyclooctyne-pyrene linker (DBOP) (5) as shown in Figure 2. The pyrene moiety of DBOP was chosen as the anchoring group for this application due to its high non-covalent affinity for the graphene surface, namely π-π stacking. The linker length was based on previous reports in the literature that utilized 1-pyrenebutyric acid in order to enable sufficient interaction with the azido containing molecules while maintaining high graphene sensitivity10.
Figure 2.
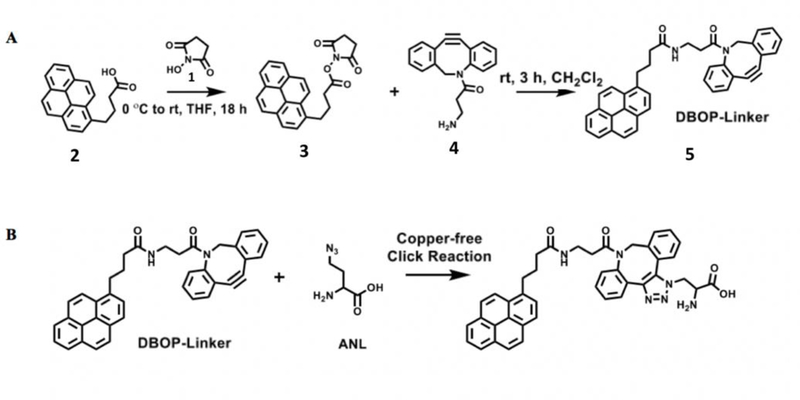
(A) The DBOP linker is the key component of Click-A+Chip and was synthesized in two steps. N-hydroxysuccinimide (1) was reacted with 1-pyrenebutyric acid (2) to generate 2,5-dioxopyrrolidin-1-yl 4-(pyren-1-yl) butanoate (3). Compound 3 was then reacted with commercially available dibenzocyclooctyne-amine (4) to form the dibenzocyclooctyne-pyrene linker (DBOP) (5). (B) The copper-free reaction between the DBOP linker and the ANL molecules.
Synthesis of compound 3: 2,5-dioxopyrrolidin-1-yl 4-(pyren-1-yl)butanoate
Compound 2, 1-pyrenebutyric acid (0.288 g, 1 mmol, 1 equiv), was added to a stirring solution of N-hydroxysuccinimide (1) (0.115 g, 1 mmol, 1 equiv) in THF (25 mL). The solution was cooled to 0 °C and added to N,N’-dicyclohexylcarbodiimide (0.206 g, 1 mmol, 1 equiv) in 5 mL of THF dropwise. After the addition, the reaction was allowed to warm to RT for 18 hr, filtered through celite, and the yellow filtrate was concentrated under reduced pressure. The crude material was recrystallized in ethanol to provide 0.099 g (0.26 mmol, 26%) of an orange solid. 1H NMR (400 MHz, CDCl3, ppm) δ 8.30 (d, J = 9.22 Hz, 1H), 8.17 (dd, J = 3.06, 7.57, 2H), 8.14 (d, J = 2.54 Hz, 1H), 8.12 (s, 1H), 3.48 (t, J = 7.59, 2H), 2.80–2.94 (m, 4H), 2.74 (t, J = 7.15 Hz, 2H), 2.31 (p, J = 7.15 Hz, 2H). 13C NMR (400 MHz, CDCl3) δ 169.3, 168.7, 131.0, 127.8, 127.6, 127.0, 126.0, 125.1, 125.0, 123.3, 32.4, 30.7, 26.6, 25.8. HRMS (ESI) m/z 385.1318 (calculated: 385.4190 [C24H19NO4] +).
Synthesis of compound 5: Dibenzocyclooctyne-pyrene
Compound 4, dibenzocyclooctyne-amine, (0.011 g, 0.040 mmol, 1 equiv) was dissolved in CH2Cl2 (5 mL) and added dropwise to a stirring solution of Compound 3 (0.020 g, 0.052 mmol, 1.3 equiv). After stirring at RT under N2 for 3 hr, the reaction solution was concentrated and purified by column chromatography (gradient elution using 1:1 hexanes/ethyl acetate then 100% ethyl acetate and finally 10% MeOH in CH2Cl2, Rf = 0.55). The compound was dried in vacuo to yield 0.014 g of white solid (64%). 1H NMR (400 MHz, CDCl3, ppm) δ 8.25 (d, J = 9.47 Hz, 1H), 8.05–8.19 (m, 4H), 7.91–8.05 (m, 3H), 7.82 (d, J = 7.82 Hz, 1H), 7.62 (d, J = 7.28 Hz, 1H), 7.20–7.38 (m, 5H), 7.15 (t, J = 7.37 Hz, 1H), 7.06 (d, J = 7.62 Hz, 1H), 5.91–6.07 (m, 1H), 5.06 (d, J = 14.19 Hz, 1H), 3.63 (d, J = 13.62 Hz, 1 H), 3.11–3.45 (m, 4H), 2.44 (ddd, J = 16.75, 7.40, 3.93 Hz, 1H), 2.02–2.23 (m, 4H), 1.97 (ddd, J = 17.12, 8.11, 3.93 Hz, 1H), 1.24 (t, ethyl acetate). 13C NMR (400 MHz, CDCl3, ppm) δ 172.4, 172.3, 151.0, 148.0, 136.0, 132.0, 132.0, 131.5, 131.0, 129.9, 129.0, 128.8, 128.6, 128.4, 128.3, 127.8, 127.5, 127.4, 127.4, 127.3, 126.7, 125.8, 125.6, 125.1, 125.0, 124.9, 124.8, 124.8, 123.5, 122.9, 122.6, 114.8, 107.7, 55.5, 36.1, 35.2, 34.9, 32.9, 27.4. HRMS (ESI) m/z 569.214 (calculated: 569.2191 [C38H30N2O2] +).
Click-A+Chip fabrication
Click-A+Chip is fabricated using conventional microfabrication technologies11 (Nanomedical Diagnostics, California, US). Briefly, the graphene chips of Click-A+Chip were fabricated using chemical vapor deposition (CVD) onto the surface of a SiO2 layer. Conventional photolithography techniques were used to create the source and drain consisting of gold (50 nm) with a layer of chrome (5 nm). To ensure the consistency of each batch of chips, the electrical resistance between the source and the drain was characterized using a conventional multimeter. Each chip was then attached to a custom printed circuit board (PCB) package using epoxy and wire bonds. The wire bonds were then encapsulated in epoxy, leaving an open cavity over the exposed graphene channels for biological sample placement. Finally, the entire device was annealed at 200°C to remove any potential residues from the process which completes the fabrication of the 3 terminal graphene field effect transistor (gFET).
Click-A+Chip functionalization
The graphene channel of the Click-A+Chip was functionalized by placing 20 μl of a 5 mM solution of DBOP in DMF directly atop the graphene surface (2 hr, at RT or overnight, 4 °C). They were subsequently washed with DMF to remove any unbound linker. DBOP was irreversibly adsorbed to the hydrophobic graphene surface in the presence of organic solvent due to the highly aromatic pyrene, which is stable against desorption in aqueous solutions12. We incorporated a strained alkyne into the linker because it is a well-established molecular structure that enables click-chemistry without the need for a copper catalyst which can be cytotoxic therefore interfering with the detection of certain sample types13,14 .
Measurement method
The mechanism of the Click-A+Chip sensing relies on field effect biosensing (FEB) technology. Any small electrostatic interaction or absorption of charged molecules such as proteins to the DBOP linker at the graphene surface will result in a shift in graphene conductivity and gFET electrical parameters. In this paper, the performance of the sensor was evaluated by continuous monitoring of I-Response, a measurement of the current flowing between the source and the drain within the graphene channel (IDS). The I-Response was measured utilizing a commercial electronic reader (Agile R100 system, Nanomedical Diagnostics)11. The Click-A+Chip response in the presence of biological samples and upon binding to ANL-labeled protein was characterized using the relative variation of the IDS, according to equation (1).
| 1 |
In this equation, IDS is the reduction peak current obtained in the presence of ANL-labeled molecule, and IDS0 is the current prior to exposure to the target molecule after sensor calibration.
Free ANL profiling
Click-A+Chip performance was first evaluated using commercial ANL and a synthesized PEG azide (see Supporting Information for synthetic details) to ensure adequate reactivity between the immobilized linker and the azido moiety. Briefly, functionalized Click-A+Chips were calibrated by incubating the chips with 30µl distilled water for 5 min. After calibration, a solution of varied concentrations of free ANL (50 μM, 100 µM, 200 µM, 400 µM, and 1 mM in 30 μl of distilled water) was placed atop the graphene surface and allowed to incubate (30 min, at RT) throughout which the current through the graphene channel was continuously monitored. In these experiments, the 30-minute incubation period was chosen to ensure the stabilization of the sensor response. However, the sensor response stabilized within 5 minutes (data not shown). Similar experiments were carried out where the performance of Click-A+Chip was evaluated in the presence of a synthesized PEG azide (Supporting Information).
ANL-labeled Leptin dose-curve on Click-A+Chip
Animal Procedures and Immunoprecipitation of ANL-labeled Leptin
For the purification of ANL-labeled Leptin, male MetRSL274G and male and female wild type C57.Bl6 mice (negative control) were injected with ANL subcutaneously at a dose of 0.2 mmol/kg, daily for 5 days. Mice were anaesthetized with isoflurane drop and euthanized by cervical dislocation. Blood samples were collected by heart puncture. Serum from these animals was collected and Leptin was purified using the ProteoExtract® Albumin/IgG Removal Kit (EMD Millipore) followed by the Pierce™ Crosslink IP Kit (Thermo Scientific) according to manufacturers’ instructions.
Detection of purified specific ANL-labeled protein using Click-A+Chip
ANL-labeled Leptin was purified by Immuno-precipitation from the blood serum of MetRSL274G mice that were treated for 6 days with ANL in vivo. Control unlabeled Leptin was purified by Immuno-precipitation from the blood serum of C57.Bl6 mice that were treated for 6 days with ANL in vivo. For evaluation with Click-A+Chip, the sensor was calibrated with 30µl of distilled water for 5 min. The sensor was subsequently incubated (15 minutes, at RT) with varied concentrations of the ANL-labeled Leptin (100 nM, 300 nM, 500 nM, 1 µM, 2.5 µM and 5 µM in ANL-free serum diluted by a factor of 5). Click-A+Chip was then rinsed three times with distilled water in order to remove unbound proteins. Click-A+Chip was then treated with 30µl of Leptin specific antibody (Abcam ab16227, 1/100 dilution). The I-Response was continuously monitored as described in the measurement method.
ANL-labeled protein profiling within parabiotic partners
Parabiosis animal procedure for the transfer of ANL-labeled proteins between old and young mice
Young male MetRSL274G mice (3–4 months) were joined in heterochronic parabiosis to old wild type male (22–24 months) C57.Bl6 mice. After 4 weeks in parabiosis, ANL was injected subcutaneously at a dose of 0.2 mmol/kg, daily for 6 days, after which cell-free serum was collected from the parabionts. For control studies, despite being identically treated with ANL, the parabiotic mouse partners (C57.Bl6) were not capable of incorporating ANL in their de novo synthesized proteomes as they lacked the mutant methionine tRNA synthase8,15.
Detection of total parabiotically transferred ANL-labeled proteomes using Click-A+Chip
To explore the capability of Click-A+Chip to capture ANL-labeled proteins, we first calibrated the graphene chip with 30µl distilled water (5 minutes, at RT). Then, 30µl of serum from old C57.Bl6 parabiotic mouse partners, which were joined with young MetRSL274G mice, was placed atop the graphene channel and analyzed. For control studies, 30µl of serum from old C57.Bl6 parabiotic mouse partners, which were joined to young C57.Bl6, was placed atop the graphene surface and analyzed.
Identification of specific ANL-labeled proteins in C57.Bl6 partners of MetRSL274G parabionts, using Click-A+Chip
To explore the capability of Click-A+Chip to identify ANL-labeled proteins, we first exposed the functionalized graphene chip to 30µl ANL-labeled serum obtained from young MetRSL274G parabionts and their old C57.Bl6 partners (30 minutes, at RT). The sensor was then washed and exposed to 30µl of Lif-1 (Abcam ab138002, 1/100 dilution) and Leptin (Abcam ab16227, 1/100 dilution) specific antibodies. We utilized Lif-1 and Leptin as model proteins, which were previously identified by ANL-adapted Antibody Arrays after heterochronic parabiosis8.
C. Results
Click-A+Chip response to free ANL
One of the key properties of Click-A+Chip is that it can discriminate between free ANL and proteins labeled with ANL metabolically (Figure 3). Namely, the sensor was able to detect the binding event between the linker and commercial ANL (Figure 3A), however, this signal was significantly lower than that produced by the binding event between ANL-labeled Leptin and the click-linker, which generated a strong I-Response at concentrations that were much lower than those tested of free-ANL (Figure 3A and 3B).
Figure 3. Free ANL calibration.
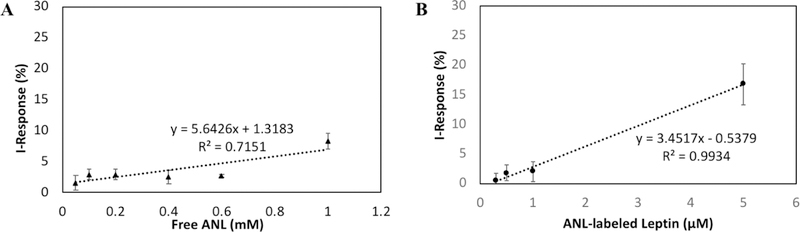
(A) The I-Response (%) + SD after exposure to varied concentrations of free ANL (50 μM, 100 µM, 200 µM, 400 µM, 600 µM and 1 mM in 30μl distilled water) (n=3 all points). (B) The I-Response (%) + SD after exposure to varied concentrations of purified ANL-labeled leptin (0.5 µM, 1 µM, 2.5 µM and 5 µM in ANL-free serum) (n=3 all points).
Click-A+Chip dose-curve to ANL-labeled purified protein
Demonstrating good sensitivity and excellent ANL-selectivity, Click-A+Chip was capable of detecting a broad range of concentrations (100 nM, 300 nM, 500 nM, 1 µM, 2.5 µM and 5 µM) of purified protein (Leptin) that had been metabolically labeled with ANL (Figure 4B). It was also clearly capable of discriminating between ANL-labeled and unlabeled protein. Namely, we have determined the I-Response after exposure of Click-A+Chip to purified Leptin, which was derived from the serum of MetRSL274G versus C57.Bl6 control mice (both administered with ANL in vivo) (as illustrated schematically in Figure 4A).
Figure 4. Click-A+Chip is capable of detecting low concentrations of ANL-labeled Leptin.
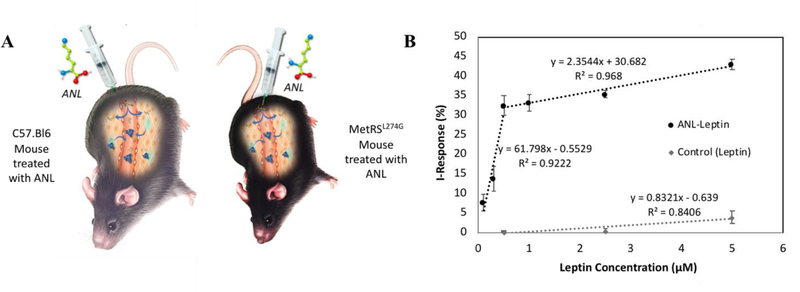
(A) MetRSL274G mice and control C57.Bl6 mice were identically treated with ANL in vivo. Serum was extracted from both transgenic and control mice and Leptin was purified via immunopercipitation. In the presence of ANL-free serum, purified Leptin was analyzed using Click-A+Chip. (B) I-Response + SD was detected by Click-A+Chip after exposure to varied concentrations of purified ANL-labeled and unlabeled Leptin (n=3 all points).
The sensor was able to detect nanomolar concentrations of ANL-labeled Leptin comparable to that of Antibody Arrays, but in a much faster, more sensitive, digitally quantifiable platform. This is an improvement upon the semi-quantification obtained from optical correlations of Antibody Arrays8. Importantly, the signal obtained from the control C57.Bl6-derived Leptin, where ANL could not be incorporated into the proteome, was ~5–20-fold lower than that of the experimental MetRSL274G derived Leptin (Figure 4B).
Click-A+Chip based detection and molecular identification of proteins in mixed heterochronic parabiotic blood serum
To test the ability of Click-A+Chip to detect ANL-labeled proteomes in mixed biological environments, we performed studies on both young and old mouse serum that was derived from heterochronic parabiosis between MetRSL247G and C57.Bl6 mice, where ANL was administered to the joined animals in vivo (as illustrated schematically in Figure 5A).
Figure 5.
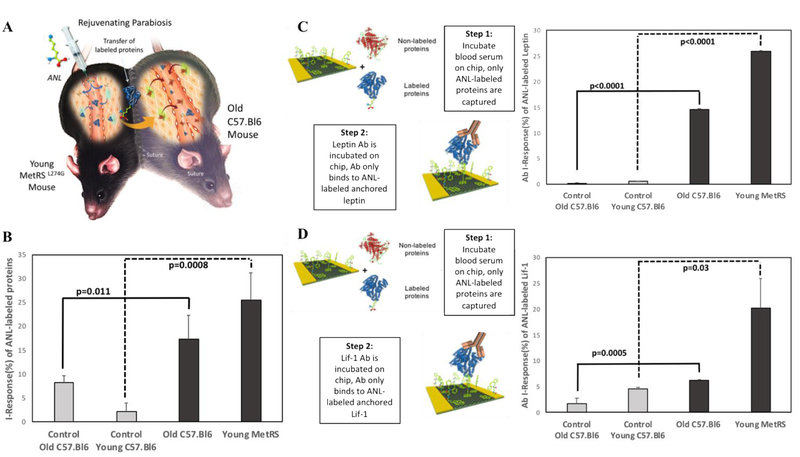
(A) Experimental setup of parabiosis between the young MetRS mouse and old C57.Bl6 mouse. (B) Click-A+Chip I-Response + SD in the presence of ANL-labeled proteins in both MetRSL274G / C57.Bl6 and control C57.Bl6 parabiotic mouse partners (old: p=0.01, n=3 control, experimental) (young: p=0.0008, n=2 control, 6 experimental). (C) The Click-A+Chip I-Response + SD after exposure to blood serum from young MetRSL274G and its old C57.Bl6 partner and subsequent exposure to Leptin specific antibodies indicated a 25% and 15% I-Response, respectively. The amount of labeled Leptin detected in serum from MetRSL274G and their C57.Bl6 parabiotic partners was significantly higher when compared with that of non-labeled young C57.Bl6 and old C57.Bl6 pair (old: p<0.0001, n=5 control, 3 experimental) (young: p<0.0001, n=2 control, 3 experimental). (D) Similarly, Click-A+Chip detected the presence of Lif-1 protein present in young MetRSL274G serum and old C57.Bl6 parabiotic partner serum. After exposure to Lif-1 antibodies, the sensor response indicated a 20% and 7% I-Response, respectively. The amount of labeled Lif-1 detected in serum from MetRSL274G and their C57.Bl6 parabiotic partners was significantly higher when compared with that of non-labeled young C57.Bl6 and old C57.Bl6 pair (old: 0.0005, n=5 control, 3, experimental) (young: p=0.03, n2 control, 3 experimental).
These results confirmed the specificity of the Click-A+Chip sensor to ANL-labeled proteomes and extrapolated it to the detection of metabolically-labeled proteins in complex mixtures of parabiotic blood, A 25% I-Response was observed after exposure to the young MetRSL274G parabiotic partner serum and an 18% I-Response was observed when exposed to the old C57.Bl6 parabiotic partner serum; Both of which were significantly higher (young: p=0.0008, old: p=0.01) than those of the control animals identically treated with ANL (Figure 5B).
To confirm these results and to extrapolate them to protein identification in mixed heterochronic blood serum, we examined the I-Response of the sensor after exposure to Leptin and Lif-1 specific antibodies, post incubation with in vivo ANL-labeled parabiosis-derived serum (Figure 5C and 5D). The results demonstrate that Click-A+Chip was able to molecularly identify ANL-labeled Lif-1 and Leptin in the shared circulation of old and young mouse parabionts.
There was only marginal change in Click-A+Chip I-Response after exposure to Leptin and Lif-1 specific antibodies on sensors used to analyze control serum obtained from young and old control C57.Bl6 mouse pairs. These data confirm the high signal to noise resolution of Click-A+Chip and further demonstrate that free ANL does not contaminate the digital proteomics as both MetRSL274G expressing and non-expressing animal pairs were treated with this metabolic label, and the minimal I-Response to the control C57.Bl6-derived parabiosis samples was similar to that of the free ANL (Figure 3 and 5B).
Moreover, these results not only identified these specific proteins in the MetRSL274G mice and their old C57.Bl6 parabiotic partners, in agreement with the Antibody Arrays8, but also yielded novel quantitative data on the relative amounts of Lif-1 and Leptin present in the old and young serum. Interestingly, even when circulation was shared and young ANL-labeled Leptin and Lif-1, were clearly detected in the old circulatory milieu, the young parabiotic serum had more Lif-1 and Leptin than old parabiotic serum, indicating the higher use, recycling and/or sequestering of these young-derived molecules by the old tissue. A similar albeit non-statistically significant trend was also observed when the total ANL-labeled proteomes of the young MetRSL274G and the old C57.Bl6 parabiotic partner (Figure 4B) were compared. As such, these results indicate that heterochronic parabiosis may not produce a 50/50 mixture of young and old proteins in old animals.
D. Discussion
Click-A+Chip is the first example of a graphene-based biosensor capable of detecting bio-orthogonally labeled proteins. It is also capable of directly and digitally quantifying the levels of azido-containing biomolecules such as ANL-labeled proteomes. In addition, the sensor is capable of molecular identification of any desired protein(s) through interactions with protein-specific antibodies post ANL-labeled protein capture. In this regard, Click-A+Chip has the potential to be used as a multiplex protein biosensor, with multiple graphene chips in one device, to enable multiple readouts for fast and comprehensive proteome profiling. This makes Click-A+Chip a promising new platform for digital proteomics, which is compatible with sub-microgram starting samples. With high resolution, Click-A+Chip quantifies the changes in de novo synthesized metabolically labeled proteomes, whether in response to environmental or experimental perturbations.
Click-A+Chip is a novel digital platform for the click-based detection and profiling of ANL-labeled proteins, which was developed in order to improve upon conventional methods such as Mass Spectrometry and modified Antibody Arrays. Specifically, we have greatly diminished the complexity, time involved, and the amount of starting material required by these experimental assays while increasing their accuracy through direct digital quantification and improving their signal to noise resolution. As Click-A+Chip can only recognize ANL-labeled proteins, our method is devoid of false positives, in contrast to Mass Spectrometry which reports all proteins that were eluted after biotin-affinity enrichment. Click-A+Chip also minimizes false negatives in contrast to modified Antibody Arrays, because of the greater sensitivity of digital detection as compared to optical correlations.
This work on digital detection of mammalian proteins that have been metabolically labeled in vivo and are derived from minute samples of complex biological mixtures, has the potential to innovate screening of treatments for age-related diseases and enable accurate longitudinal assessment of therapeutic interventions for a number of genetic and acquired pathologies16.
The specific detection and quantification of the relative levels of Leptin and Lif-1 in serum that was derived from the heart-bleads of heterochronic parabionts by Click-A+Chip demonstrated its capacity to profile and moreover, to quantitatively understand the specifics of blood heterochronicity.
Lif-1 and Leptin are implicated in health and regeneration of multiple organs including skeletal muscles, the thymus, and immune system organs, as well as in healthy metabolic homeostasis17. Interestingly, we observed that there was a net decrease in the young-derived Leptin and Lif-1 levels in the old heart-bleeds’ serum, as compared to the serum of the heart-bleeds of young parabiotic partners. These data suggest that ANL-labeled Lif-1 and Leptin, which likely contributed to the rejuvenation of old tissues, were sequestered or used more rapidly by the old parabionts, potentially deregulating Lif-1 and Leptin homeostasis in the young mouse through the blood sharing of parabiosis5,8.
Click-A+Chip has numerous potential applications due to the broad use of click-chemistry throughout the scientific community. Compared to conventional reactions, click-chemistry has been utilized in bio-conjugation chemistry for fragment-based drug discovery, high-throughput screening, receptor-ligand binding studies, activity-based protein profiling, DNA labeling, in vivo imaging, and more13,18–20. Thus, in addition to proteins, Click-A+Chip has the potential to be used as a multiplex biosensor, enabling a platform for the simultaneous detection of numerous metabolically or otherwise, azido-labeled analytes21. Since Click-A+Chip requires only a small sample volume with minimal sample preparation and can be arrayed in parallel, Click-A+Chip has the potential to reduce costs and assay time, making it a practical tool for a multitude of application, from biomarker detection and diagnosis to vaccine development22.
E. Conclusions
This communication presents a new portable digital device for biosensing based on functionalized graphene that can be employed for any click-able application. Here, we have synthesized a linker containing a strained alkyne that was successfully anchored to the surface of a graphene chip to generate a novel graphene-alkyne composite dubbed Click-A+Chip. We have demonstrated its ability to undergo a click-reaction with ANL-labeled proteins that is detectable through the change in electrical properties of the graphene surface. We have also demonstrated its ability to reliably discriminate between the free azide label, labeled proteins and unlabeled proteins with high resolution. Finally, we have demonstrated its ability to identify specific proteins through antibody binding without a need for secondary antibodies. Click-A+Chip has the potential to be utilized for monitoring and identifying biomarkers associated with tissue health and youth as well as those associated with pathology and aging. The click-able graphene surface utilized by Click-A+Chip has the potential to replace commercial labor-intensive optical assays and Mass Spectrometry with miniaturized samples and a more accurate digital format. The extensively utilized click-chemistry enables Click-A+Chip to be implemented across a broad spectrum of biosensing applications and we envision this technology being used for the digital measurement of diverse molecular targets.
Supplementary Material
Acknowledgements
We would like to acknowledge Nanomedical Diagnostics for our use of their Agile R100 technology. NIH RO1 HL139605–01, Roger’s family, and Open Philanthropy funds to IC supported this work.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Vaiserman A, Ed., R. Soc. Chem [Google Scholar]
- 2.Conboy MJ, Conboy IM and Rando TA, Aging Cell, 2013, 12, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weismann IL and Rando TA, Nature, 2005, 433, 760–764. [DOI] [PubMed] [Google Scholar]
- 4.Loffredo FS, Steinhauser ML, Jay SM, Gannon J, Pancoast JR, Yalamanchi P, Sinha M, Dall’Osso C, Khong D, Shadrach JL, Miller CM, Singer BS, Stewart A, Psychogios N, Gerszten RE, Hartigan AJ, Kim M-J, Serwold T, Wagers AJ and Lee RT, Cell, 2013, 153, 828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rebo J, Mehdipour M, Gathwala R, Causey K, Liu Y, Conboy MJ and Conboy IM, Nat. Commun, 2016, 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agard NJ, Prescher JA and Bertozzi CR, J. Am. Chem. Soc, 2004, 126, 15046–15047. [DOI] [PubMed] [Google Scholar]
- 7.Vellucci D, Kao A, Kaake RM, Rychnovsky SD and Huang L, J. Am. Soc. Mass Spectrom, 2010, 21, 1432–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Conboy MJ, Mehdipour M, Liu Y, Tran TP, Blotnick A, Rajan P, Santos TC and Conboy IM, Nat. Commun, , DOI: 10.1038/s41467-017-00698-y. [DOI]
- 9.Su Q, Pang S, Alijani V, Li C, Feng X and M??llen K, Adv. Mater, 2009, 21, 3191–3195. [Google Scholar]
- 10.Huang Y, Dong X, Shi Y, Li CM, Li L-J and Chen P, Nanoscale, 2010, 2, 1485. [DOI] [PubMed] [Google Scholar]
- 11.Lerner MB, Pan D, Gao Y, Locascio LE, Lee K-Y, Nokes J, Afsahi S, Lerner JD, Walker A, Collins PG, Oegema K, Barron F and Goldsmith BR, Sens. Actuators B Chem, 2017, 239, 1261–1267. [Google Scholar]
- 12.Li M, Xu P, Yang J, Ying H, Haubner K, Dunsch L and Yang S, J. Phys. Chem. C, 2011, 115, 4584–4593. [Google Scholar]
- 13.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA and Bertozzi CR, Proc. Natl. Acad. Sci, 2007, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prescher JA and Bertozzi CR, Nat. Chem. Biol, 2005, 1, 13–21. [DOI] [PubMed] [Google Scholar]
- 15.Mahdavi A, Hamblin GD, Jindal GA, Bagert JD, Dong C, Sweredoski MJ, Hess S, Schuman EM and Tirrell DA, J. Am. Chem. Soc, 2016, 138, 4278–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conboy IM, Conboy MJ and Rebo J, Aging, 2015, 7, 754–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruver AL and Sempowski GD, J. Leukoc. Biol, 2008, 84, 915–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horisawa K, Front. Physiol, 2014, 5, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma N, Wang Y, Zhao BX, Ye WC and Jiang S, Drug Des. Devel. Ther, 2015, 9, 1585–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martell J and Weerapana E, Molecules, 2014, 19, 1378–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richens JL, Lunt EAM and O’Shea P, Int. Immunopharmacol, 2015, 24, 166–168. [DOI] [PubMed] [Google Scholar]
- 22.Natesan M and Ulrich RG, Int. J. Mol. Sci, 2010, 11, 5165–5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


