Abstract
The respiratory pathogen, Moraxella catarrhalis, is a human-specific commensal that frequently causes acute otitis media in children and stimulates acute exacerbations in chronic obstructive pulmonary disease (COPD) patients. The exact molecular mechanisms defining host-pathogen interactions promoting pathogenesis are not clearly understood. Limited knowledge hampers vaccine and immunotherapeutic development required to treat this emerging pathogen. Here we reveal in detail a novel antibacterial role displayed by short leucine–rich proteoglycans (SLRPs) in concert with complement. We show that fibromodulin (FMOD), osteoadherin (OSAD) and biglycan (BGN) but not decorin (DCN) enhance serum killing of M. catarrhalis. Our results suggests that M. catarrhalis binding to SLRPs is a conserved feature as the overwhelming majority of clinical and laboratory strains bound all four SLRPs. Furthermore, we resolve the binding mechanism responsible for this interaction and highlight the role of the ubiquitous surface protein (Usp), A2/A2H in mediating binding to host SLRPs. A conserved immune evasive strategy used by M. catarrhalis and other pathogens is the surface acquisition of host complement inhibitors such as C4b-binding protein (C4BP). We observed that FMOD, OSAD and BGN competitively inhibit binding of C4BP to the surface of M. catarrhalis, resulting in increased C3b/iC3b deposition, membrane attack complex (MAC) formation and subsequently decreased bacterial survival. Furthermore, both OSAD and BGN promote enhanced neutrophil killing in vitro, both in a complement dependent and independent fashion. In summary, our results illustrate that SLRPs, FMOD, OSAD and BGN, portray complement-modulating activity enhancing M. catarrhalis killing, defining a new antibacterial role supplied by SLRPs.
Introduction
Evolutionary pressure has dictated the development of several key features to protect the mammalian host from infection from the billions of endogenous and exogenous microflora. The innate immune system governs the first response to any potentially infectious agent. Physical barriers lined with intricate detection and signaling systems, ancient elaborate effector pathways and responder phagocytic and antigen presenting cells mediate overall protection. One critical element of innate immunity in mediating this detection, response and subsequent elimination of foreign species is complement.
The complement system is composed of a multitude of soluble or surface expressed proteins with defined activators and inhibitors embroiled in a constant flux to maintain homeostasis. Complement components circulate in the blood and extracellular fluids. Microbial activation of complement occurs through various means but converges at the level of C3 activated through the formation of C3 convertases. These complexes instigate the cleavage of C3 into the anaphylatoxin and antimicrobial C3a peptide and major opsonin C3b/iC3b, responsible for mediating phagocytosis of foreign bodies by professional phagocytes. The next major step in complement activation is the formation of C5 convertases via binding of C3b to C3 convertases resulting in a new enzymatic platform directing the cleavage of C5 into C5a and C5b. Whereas C5a is a potent anaphylatoxin, C5b deposits onto the bacterial membrane initiates the formation of the membrane attack complex, resulting in lysis of susceptible cells, such as Gram-negative bacteria (1). To prevent host cell attack, complement inhibitors regulate complement activation in a strict manner. Two soluble inhibitors, factor H (FH) and C4b-binding protein (C4BP) (2) prevent formation of C3 convertase through binding of C3b and C4b respectively and serving as cofactors for the serine protease, factor I.
Microbes, particularly bacteria, have evolved several mechanisms to inhibit complement activation and examples of bacteria targeting every feature of complement have been reported (3). The Gram-negative opportunistic respiratory pathogen, Moraxella catarrhalis, is no exception. M. catarrhalis is a human specific commensal and a recognized respiratory pathogen (4, 5). M. catarrhalis causes significant morbidity and economic burden as a common etiological agent of otitis media and exacerbations in patients with chronic obstructive pulmonary disease (COPD) (4, 5). One major immune evasion strategy employed by M. catarrhalis is the recruitment of the complement inhibitor C4BP (6). Inhibiting C4BP acquisition by M. catarrhalis may provide a novel therapeutic avenue to treat infections, which is urgently required given the increasing problem of failed therapy due to antibiotic resistance.
Short-leucine rich proteoglycans (SLRPs) such as fibromodulin (FMOD), osteoadherin (OSAD), biglycan (BGN) and decorin (DCN) are extracellular matrix (ECM) components containing a distinct central leucine – rich repeat region (LRR) flanked by disulphide bridges at the N- and C-termini (7). SLRPs are highly versatile molecules displaying differences in glycosylation of the core region and amino acid sequence and charge at the terminal ends. Classically, SLRPs function as important components in maintaining and regulating the ECM structure and cellular adhesion through interaction with integrins (7). More recently, the role of SLRPs, specifically BGN and DCN, as regulators of the innate immune system in response to tissue injury or cellular stress has been illustrated. Under normal physiological conditions matrix-bound SLRPs are not capable of immune activation, however in soluble form, following limited proteolysis of the ECM or secretion from macrophages, SLRPs act as endogenous ligands of toll-like receptors triggering a rapid sterile inflammatory response (8, 9).
SLRPs also function as complement modulators, both as activators and inhibitors (10). Both FMOD and OSAD interact with the globular head domain of C1q stimulating activation of the classical complement pathway (11). In contrast, both BGN and DCN bind primarily to the stalk region of C1q, inhibiting classical pathway activation, presumably through inhibition of C1s/C1r activity (11, 12). Additionally, both FMOD and OSAD capture C4BP and FH and therefore may limit complement activation at early stages of the classical pathway (11, 13). Whether these SLRPs interact with M. catarrhalis and alter complement activity and bacterial elimination is currently unknown and provided the motivation for the current study
Materials and Methods
Bacteria and culture conditions
A list of bacterial strains used in this study is shown in Table 1. Moraxella catarrhalis clinical and laboratory strains, Haemophilus influenzae type b (Hib) strain RM804 and non-typeable H. influenzae (NTHi) strain 3655 were grown on chocolate agar plates for 24 h at 37 °C with 5 % CO2. Bacteria were subsequently streaked onto new chocolate agar plates for 6 h, scraped from plates, resuspended in 25 % (v/v) brain-heart infusion (BHI) broth/glycerol and stored in aliquots at - 80 °C. Pseudomonas aeruginosa ATCC27853 and KR601were grown in LB broth for 24 h at 37 °C with shaking.
Table 1:
List of strains used in this study
| Clinical isolate / strain | Description | Reference |
|---|---|---|
| Moraxella catarrhalis | ||
| KR529 | Clinical isolate | (14) |
| KR485 | Clinical isolate | (14) |
| O35E | Clinical isolate | (14) |
| KR516 | Clinical isolate | (14) |
| KR531 | Clinical isolate | (14) |
| KR540 | Clinical isolate | (14) |
| KR503 | Clinical isolate | (14) |
| KR488 | Clinical isolate | (14) |
| KR509 | Clinical isolate | (14) |
| KR484 | Clinical isolate | (14) |
| KR480 | Clinical isolate | (14) |
| BBH18 | Clinical isolate | (14) |
| O46E | Clinical isolate | (14) |
| CCUG353 | Clinical isolate | (14) |
| KR483 | Clinical isolate | (14) |
| Bc5 | Laboratory strain | (34) |
| RH4 | Laboratory strain | (35) |
| RH4ΔuspA1 | RH4 devoid of ubiquitous surface protein A1 | (6) |
| RH4ΔuspA2 | RH4 devoid of ubiquitous surface protein A2 | (6) |
| RH4ΔuspA1ΔuspA2 | RH4 devoid of both ubiquitous surface protein A1 and A2 | (6) |
| RH4Δmid | RH4 devoid of immunoglobulin D (IgD)-binding protein (MID) | (36) |
| Pseudomonas aeruginosa | ||
| ATCC27853 | Laboratory strain | ATCC |
| KR601 | Clinical isolate | This study |
| Haemophilus influenzae | ||
| type b strain RM804 | Clinical isolate, capsule-deficient | (37) |
| non-typeable (NTHi) strain 3655 | Clinical isolate, encapsulated | CCUG |
Abbreviations: CCUG; Culture Collection University of Gothenburg, ATCC; American Type Culture Collection
Proteins, antibodies and sera
Human recombinant small leucine-rich proteoglycans (SLRPs) including fibromodulin (FMOD), osteoadherin (OSAD), biglycan (BGN), and decorin (DCN) were expressed with a hexa histidine tag from the pCEP4 vector in FreeStyle 293-F cells (Invitrogen) and purified using a similar protocol as described (14). The pCEP4 vector containing fibromodulin (FMOD) was a gift from Dr. Sebastian Kalamajski (Uppsala University, Sweden) (15). Briefly, FreeStyle 293 Expression Medium (Invitrogen) containing secreted SLRPs was collected and adjusted to 0.3 M NaCl and 50 mM Tris-HCl, pH 8.0. Medium was then filtered through a 0.45-μm membrane, and concentrated using a 10-kDa cellulose membrane in a stirred ultrafiltration system (Amicon). The concentrated medium was then applied to a Ni2+-NTA column equilibrated with 50 mM Tris-HCl, pH 8.0 with 0.3 M NaCl. After washing with 5 volumes of 50 mM Tris-HCl, pH 8.0, the protein in the column was eluted with a linear gradient of 0–500 mM imidazole in 50 mM Tris-HCl, pH 8.0. The eluted proteins were analyzed by SDS-PAGE, dialyzed against PBS, and stored at −80°C in aliquots. SLRPs were confirmed by Western blotting with polyclonal rabbit anti-bovine SLRPs Abs (homemade). The yield of protein from 1 liter of conditioned medium was 17 mg for FMOD, 10 mg for OSAD, 7 mg for BGN, and 14 mg for DCN. C4BP was purified from human plasma as described previously (16). Biotinylation of SLRPs was achieved using the EZ-Link™ Sulfo-NHS-LC-Biotinylation kit (ThermoFisher) as per manufacturers’ instructions. Bovine serum albumin (A8806; Sigma) was used as control protein.
The following primary antibodies (Abs) were used for flow cytometric analysis of complement deposition on the surface of M. catarrhalis: polyclonal rabbit anti-human C1q (A0136, Dako), monoclonal mouse anti-human C4BP MK104 (homemade, (17)), mouse anti-human MAC (aE11, Hycult Biotech), and polyclonal rabbit anti-human C3d (A0063, Dako). Primary Abs were detected using fluorescently labeled secondary F(ab’)2 goat anti-rabbit AF647 (A21246, Invitrogen) or goat anti-mouse AF647 (A21235, Invitrogen). For the detection of biotinylated proteins, streptavidin AF647 conjugate (S21374, ThermoFisher) was used.
Normal human serum (NHS) was prepared from freshly drawn blood obtained form at least 10 healthy volunteers. Blood was allowed to clot for 30 min at room temperature and then incubated on ice for 1 h. Following two rounds of centrifugations at 700 x g, at 4 °C for 8 min, serum fractions were collect, pooled and stored immediately at - 80 °C. All healthy volunteers provided written informed consent according to the recommendations of the local ethical committee in Lund (permit 2017/582) and the Declaration of Helsinki (18). To prepare C4BP-depleted human serum (C4BP-dpl), freshly pooled human serum from four donors was passed through HiTrap affinity column coupled with the monoclonal C4BP antibody MK104. Resulting serum samples were verified to be C4BP-depleted through ELISA analysis as described previously (19). Plasma-purified C1q was added (20 μg/mL) to restore C1q concentration to normal levels, as C1q is partially lost during C4BP depletion due to C1q binding to Ab-column. C4BP, purified from the serum from which it was depleted, was replenished at physiological concentrations (200 μg/mL).
Binding of SLRPs to bacteria
To screen binding of SLRPs to pathogenic bacteria, bacteria were grown on corresponding agar plates, washed, and suspended in PBS. After staining with 10 μM CFSE (Sigma-Aldrich), bacteria were resuspended into 1% (w/v) BSA/PBS. Bacterial suspension with 5 × 106 CFU in 50 μl was then mixed with an equal volume of 1% (w/v) BSA/PBS containing 100 μg/ml biotinylated FMOD (2.3 μM), OSAD (1.94 μM), BGN (2.35 μM), and 200 μg/ml biotinylated DCN (4.94 μM). After an incubation at 37°C for 1 h, bacteria were centrifuged at 5000 × g for 10 min, washed once with 1% (w/v) BSA/PBS, and incubated with streptavidin-AF647 at room temperature. After incubation for 1 h in the dark, bacteria were centrifuged, washed once with 1% (w/v) BSA/PBS, and bound SLRPs on bacteria were detected using a Cyflow space flow cytometer (Partec). To examine binding of SLRPs to clinical isolates of bacteria, binding assays were performed as described above, and bound SLRPs on bacteria were detected in a 96-well plate using a CytoFLEX flow cytometer (Beckman Coulter). Bacteria directly incubated with Streptavidin Alexa Fluor 647 conjugate were used as treatment control for background binding. CFSE+ bacteria were detected based on their fluorescence signal and a gating region was set to exclude debris. Geometric mean fluorescence intensity (gMFI) was used to determine amount of SLRPs binding to bacteria.
To assess bacterial cell surface proteins responsible for binding of SLRPs, wild-type (RH4) and isogenic mutants of uspA1, uspA2, mid and double mutant uspA1uspA2 and (Table 1) were grown, stained with CFSE and binding performed in identical fashion to the above conditions using a CytoFLEX flow cytometer.
To assess direct binding of SLRPs with UspA2, MaxiSorp microtiter plates (Nunc) were coated overnight at 4 °C with recombinant UspA2 (0.14 μM; 10 μg/mL), cloned from M. catarrhalis strain RH4 and expressed in E. coli as described previously (20). Plates were washed three times with 300 μL of wash buffer (50 mM Tris, 150 mM NaCl, 0.1 % Tween 20, pH 8). Plates were blocked to prevent non-specific binding by using 250 μL of quench (wash buffer containing 3 % fish gelatin) and incubated at room temperature for 2 h. Plates were further washed three times in wash buffer and biotinylated SLRPs were added at increasing amounts (0.012 – 1.98 μM; 0.6 – 80 μg/mL) in binding buffer (50 mM HEPES, 150 mM NaCl, 2 mM CaCl2, pH 7.4) for 30 min at room temperature. Following incubation, plates were washed three times in wash buffer and wells were incubated with 50 μL streptavidin – HRP (1:200) in quench for 1 h at room temperature. Plates were further washed three times in wash buffer, developed using TMB substrate solution (ThermoFisher) and reaction stopped using 0.5 M H2SO4. Binding of SLRPs was detected using a Cytation 5 Cell Imaging plate reader (BioTek) at 450nm.
Serum bactericidal assay
Serum bactericidal assay was performed as described previously (21). Briefly, M. catarrhalis (2.5 × 105 CFU) were incubated with either 5 or 50 μg/mL SLRPs or BSA at 37 °C for 30 min in GVB++ buffer (5 mM veronal buffer pH 7.3, 0.1% (w/v) gelatin, 140 mM NaCl, 1 mM MgCl2, and 0.15 mM CaCl2). Following incubation, SLRPs/bacteria solution was washed in PBS or not and further incubated with pooled NHS to a final concentration of 10% (strain RH4) or 20% (strain Bc5) in GVB++ buffer. For calculation of bacterial survival, aliquots of bacteria were removed at time 0 and following incubation at 37°C for 30 min, diluted in PBS and spread onto BHI agar for colony enumeration. Serum treated with complement C5 inhibitor OmCI (10 μg/ml; 0.625 μM) (Swedish Orphan Biovitrum) (22) on ice for 30 min or compstatin CP40 (20 μM) (23) were used as serum controls. BSA at 50 μg/mL (0.75 μM), which has no effect on complement activation, was used as negative protein control. Bacteria were incubated with SLRPs alone at 37°C for 30 min in GVB++ buffer to determine whether SLRPs have antimicrobial activity.
Complement deposition assay
CFSE-labeled M. catarrhalis was incubated with pooled NHS in a 96-well plate in the presence of SLRPs, as described in the serum bactericidal assay. After incubation, bacteria were washed once with 1%-BSA/PBS, and deposited complement components were detected with primary Abs incubated at room temperature for 30 min at a dilution of 1:1000 in 1%-BSA/PBS. Bacteria were centrifuged and washed once in 1%-BSA/PBS followed by fluorescently labeled secondary Abs staining for 30 min at room temperature in the dark using a dilution of 1:1000. Bacteria were again centrifuged and washed once in 1%-BSA/PBS and finally resuspended in 150 μL of 1%-BSA/PBS. Deposited complement components were assessed using a CytoFLEX flow cytometer (Beckman Coulter). Geometric mean fluorescence intensity (gMFI) was used to determine amount of complement deposition. Heat-inactivated serum, primary isotype antibody and secondary antibody only controls were used to assess specificity of antibodies used. Stained and unstained bacteria were used for gating bacteria and a minimum of 20,000 events was examined.
Neutrophil bactericidal assays
Human neutrophils were isolated using firstly Histopaque®−1119 (Sigma) separation of peripheral venous blood drawn from healthy volunteers and secondly a Percoll-based gradient method as previously described (24). Neutrophils were resuspended in RPMI 1640 plus 10 mM HEPES and viability was assessed by trypan blue staining typically yielding greater than 95%. For neutrophil bactericidal assays, neutrophils (5 × 105) were incubated with M. catarrhalis (5 × 106 CFU) (MOI 10) in the presence of 200 μg/mL SLRPs (4.5 μM (FMOD), 3.9 μM (OSAD), 4.7 μM (BGN), 4.9 μM (DCN)) or BSA with either 5% OmCI-treated or compstatin-treated serum in a final volume of 300 μL. Plates were incubated at 37°C, 5% CO2 and at time 30 and 60 min neutrophils were lysed using 1 % saponin/PBS for 15 min on ice. Bacteria were diluted in PBS and plated onto BHI agar plates and incubated for 24 h at 37°C with 5% CO2. Colonies were counted and intra- and extracellular bacterial survival was assessed by dividing CFU at time 30 or 60 min by CFU at time 0.
Statistical analysis
A one-way or two-way ANOVA was used to examine the difference between experimental results (GraphPad Prism v7.0) where a p value <0.05 was considered to be statistically significant. The p values reported in figure legends represent the post-hoc tests.
Results
SLRPs specifically bind M. catarrhalis
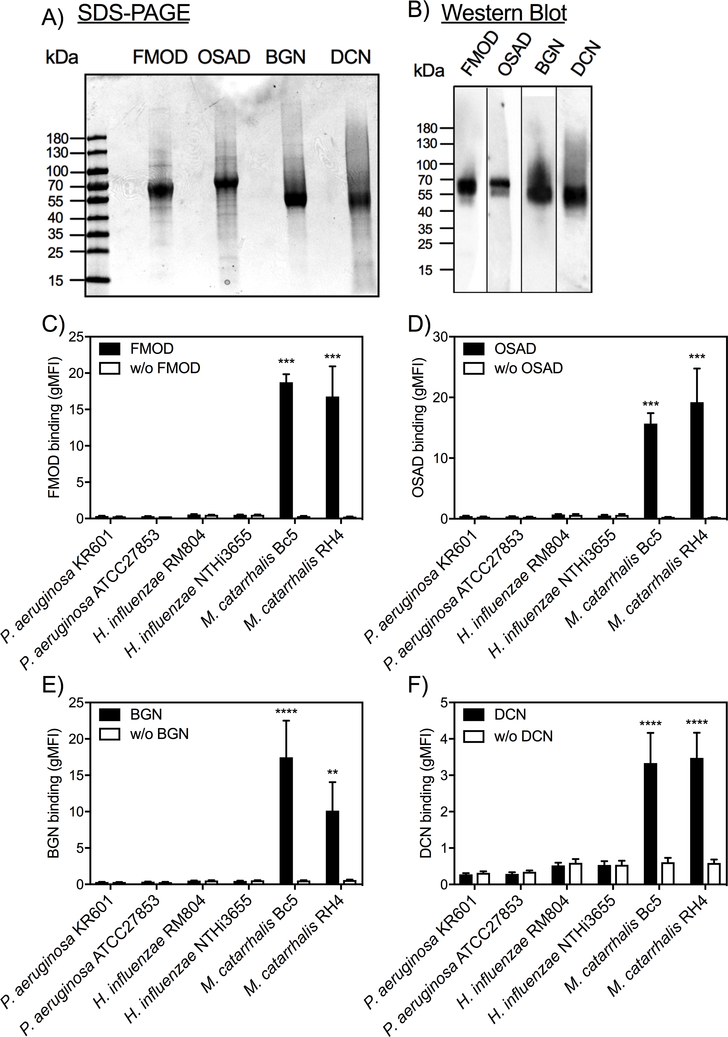
SLRPs have been shown to regulate various extracellular matrices and modulate cellular functions and innate immunity via interaction with cell surface receptors (7–9). We previously reported that SLRPs FMOD, OSAD, BGN and DCN could regulate complement activity through interaction with C1q, C4BP and FH (11, 13). However, whether these SLRPs play a role in modulating innate immune responses directed against pathogenic bacteria remains unclear. To understand the role of SLRPs in innate immunity, we expressed recombinant human SLRPs in eukaryotic cells and purified them using affinity chromatography. The purified SLRPs were estimated with a purity of ≥ 90% by SDS-PAGE under reducing conditions (Fig. 1A) and confirmed by Western blotting using our in-house rabbit anti-bovine SLPRs, which are highly similar to human SLRPs (Fig. 1B). Recombinant his-tagged FMOD, OSAD, BGN, and DCN are predicted be 44.0, 50.4, 40.6, and 38.7 kDa, respectively. However, all proteins are larger than the predicted molecular mass in SDS-PAGE gel due to glycosylation. Next, we determined the binding of biotinylated SLRPs to major Gram-negative bacterial species important in respiratory infections, namely Pseudomonas aeruginosa, Haemophilius influenzae and M. catarrhalis. We found that of these pathogens only M. catarrhalis (laboratory strains Bc5 and RH4) bound the four SLRPs (Fig. 1C-F).
Figure 1. Small leucine-rich proteoglycans (SLRPs) interact with M. catarrhalis.
Recombinant human SLRPs detected by A) reducing SDS-PAGE (5 μg of each protein) and B) Western blotting (0.5 μg). Biotinylated C) FMOD, D) OSAD, E) BGN and F) DCN were incubated with major respiratory pathogens P. aeruginosa, H. influenzae and M. catarrhalis and bound SLRPs were detected with fluorescently labeled streptavidin by measuring fluorescence intensity using a Cyflow space flow cytometer (Partec). Mean values and standard deviation (SD) of at least 3 individual experiments are shown. Statistical differences were calculated using a one-way ANOVA analysis with Bonferroni’s post-test in comparison to control without SLRPs. ** p < 0.01, *** p < 0.001, **** p < 0.0001.
FMOD, OSAD and BGN enhance complement-mediated killing of M. catarrhalis
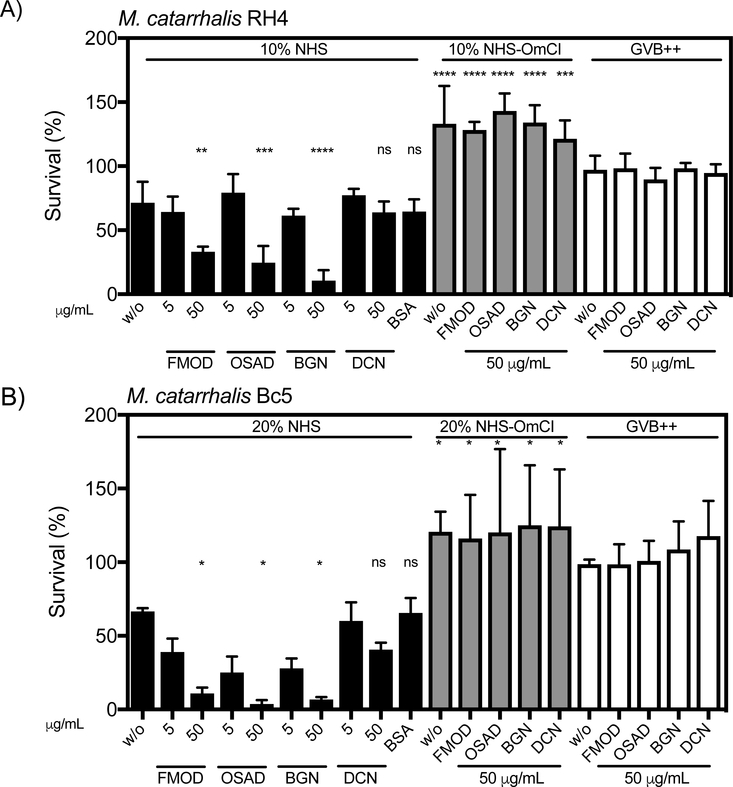
As SLRPs can both regulate complement activity and bind M. catarrhalis, we aimed to determine whether SLRPs affect survival of M. catarrhalis in pooled normal human serum (NHS). We found that SLRPs FMOD, OSAD and BGN when supplemented at 50μg/mL significantly decreased survival of both M. catarrhalis RH4 (Fig. 2A) and Bc5 (Fig. 2B) in NHS. Despite being not statistically significant, DCN led to a slight reduction in survival in the Bc5 strain compared to BSA, but no difference was observed in strain RH4, suggesting that DCN does not enhance complement-mediated killing of M. catarrhalis. Furthermore, inhibition of MAC formation by previous treatment of serum with the C5 inhibitor OmCI prevented killing of M. catarrhalis under any SLRP condition illustrating that SLRPs enhance killing through complement mediated lysis (Fig 2A-B). Lastly, no antimicrobial activity was observed when SLRPs were incubated with M. catarrhalis in GVB++ buffer in the absence of serum, confirming that the enhanced killing was mediated by complement. To verify that excess unbound SLRPs were not causing a by-stander complement activation effect and contributing to enhanced killing, we also measured the effect of washing bacteria following SLRP binding prior to incubation with serum (Fig. S1). As in the above results, a significant decrease in survival was observed for FMOD, OSAD and BGN but not DCN, indicating the SLRPs bound to the bacterial surface promoted enhanced bacterial killing in the presence of serum.
Figure 2. FMOD, OSAD and BGN enhance serum killing of M. catarrhalis.
Bacterial survival in human serum was defined as the ratio (%) of the colony-forming units (CFU) at 30 min to time 0. Error bars represent SD of three independent experiments. Serum treated by C5 inhibitor OmCI was used as serum control, and 50 μg/ml BSA was used as a negative protein control. Statistical differences were calculated using a one-way ANOVA with Dunnett’s post-test versus bacteria without SLRPs. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
SLRPs interact directly with UspA2/2H of M. catarrhalis
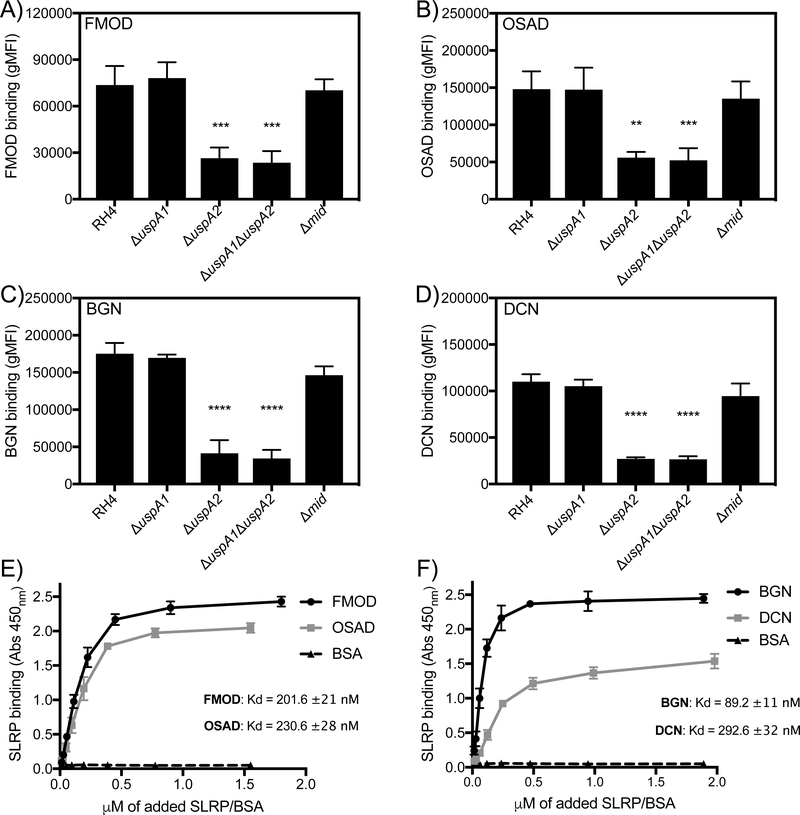
M. catarrhalis interacts with human proteins via major surface proteins such as UspA1/A2, MID, and outer membrane porins such as OmpCD and Mha (4). Given that previous work has shown that UspA1, UspA2 and MID can interact with soluble extracellular matrix proteins we investigated the interaction of wild type (RH4) and isogenic mutants lacking the above surface proteins with biotinylated SLRPs through flow cytometry (Fig. 3A-D). We found that deletion of the uspA2 gene resulted in a significant decrease in binding of all SLRPs in question highlighting the importance of UspA2 as a ligand for SLRP binding. No difference in binding was observed with neither the uspA1 nor mid mutants.
Figure 3. SLRPs interact directly with UspA2/2H of M. catarrhalis.
Biotinylated SLRPs were incubated with M. catarrhalis mutants devoid of selected surface proteins UspA1, UspA2/2H, and MID that interact with various host proteins. Bound SLRPs were detected with fluorescently labeled streptavidin by measuring fluorescence intensity using a CytoFLEX flow cytometer (Beckman Coulter). UspA2/2H mutant showed significantly reduced binding of all SLRPs (A-D). Biotinylated SLRPs E) (FMOD and OSAD) and F) (BGN and DCN) bind immobilized recombinant UspA2 with differing affinities. Error bars represent SD of three independent experiments. Statistical differences were calculated using a one-way ANOVA with Dunnett’s posttest versus wild type (RH4) bacteria. ** p < 0.01, *** p < 0.001, **** p < 0.0001.
To further elucidate the interaction between SLRPs and M. catarrhalis we employed a direct biochemical binding assay using immobilized recombinant UspA2, derived from strain RH4, and increasing concentrations of biotinylated SLRPs and BSA (Fig. 3E-F). In accordance with our binding results above employing wild-type and uspA2 mutant, all four SLRPs bound UspA2, with the highest affinity observed for BGN (Kd = 89 ±11 nM), similar affinities seen for FMOD (Kd = 202 ±21 nM) and OSAD (Kd = 231 ±28 nM) and the lowest affinity seen for DCN (Kd = 293 ±32 nM).
SLRPs bind to the majority of clinical isolates of M. catarrhalis
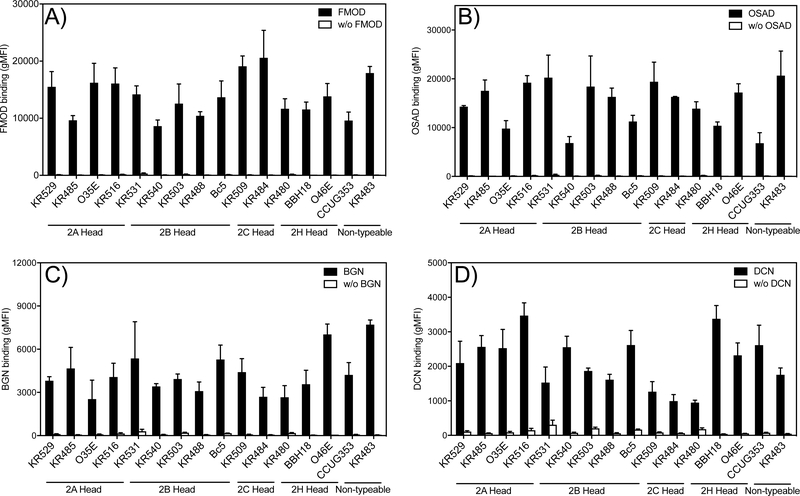
To determine the clinical relevance of M. catarrhalis interaction with SLRPs we evaluated the binding capacity of a panel of clinical isolates (n=16) to all four SLRPs (Fig. 4A-D). These clinical isolates were chosen based on their respective differences in the N- terminal sequence motif of the UspA2 protein, in order to capture a significant diversity of important clinical M. catarrhalis strains. This domain is classified into the different groups 2A, 2B, 2C and ‘nontypeable’ based on the domain distribution and sequence similarity (25). We found that the overwhelming majority of clinical isolates bound all four SLRPs whereby there was a general trend for increased binding in the order of FMOD ≥OSAD >BGN >DCN. However, isolates that express UspA2/2H with different N-terminal repeats of head domains showed no significant difference in binding of SLRPs (Fig. S2).
Figure 4. SLRPs bind to M. catarrhalis clinical isolates expressing UspA2/2H.
The highly diverse head domains of UspA2/2H are classified into N-terminal repeats (NTER) 2A, 2B, 2C, and 2H, and non-typeable. All tested clinical strains bound SLRPs at varying degrees. Negative control consisted of bacterial straining with Streptavidin Alexa Fluor 647 in the absence of biotinylated proteins. Error bars represent the SD of three individual experiments.
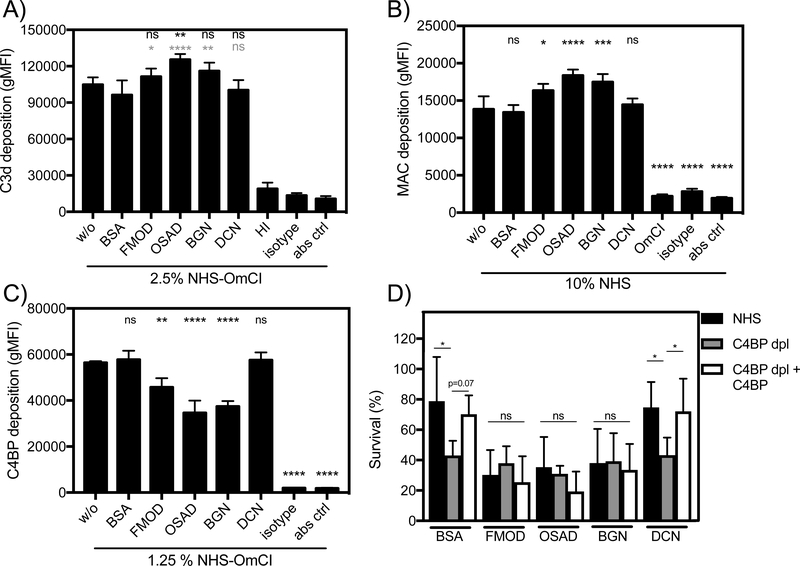
FMOD, OSAD and BGN increase C3b and MAC deposition by preventing C4BP binding to M. catarrhalis
To further understand how SLRPs regulate complement leading to the enhanced serum sensitivity of M. catarrhalis, we measured deposition of complement components on the bacterial surface in the presence of SLRPs, BSA or no added protein, using flow cytometry. In agreement with decreased survival of M. catarrhalis in serum, FMOD, OSAD and BGN significantly increased C3b deposition compared to BSA (gray stars) whereas only OSAD significantly increased C3b deposition compared to no protein control (Fig. 5A). Next, we looked at MAC deposition following incubation for 20 min in serum, a shortened time to prevent significant lysis. Complementing the serum killing and C3b deposition results, FMOD, OSAD and BGN had significantly more MAC deposited on the bacterial surface compared to BSA or DCN (Fig. 5B).
Figure 5. FMOD, OSAD and BGN increase complement deposition through inhibition of C4BP binding.
Deposition of complement components A) C3b B) MAC and C) C4BP on M. catarrhalis RH4 was analyzed using flow cytometry. D) Bacterial survival in 5 % C4BP depleted sera (C4BP dpl) with matched survival in 5 % NHS and C4BP dpl replenished with C4BP (C4BP dpl + C4BP) at physiological concentrations. Error bars represent SD of three (A-C) and 5 (D) independent experiments. Serum treated by C5 inhibitor OmCI or heat-inactivated was used as serum control, and BSA was used as a negative protein control. Grey stars (BSA) and black starts (without protein w/o) indicate statistical calculations using a one-way ANOVA with Dunnett’s post-test. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns, not significant.
As acquisition of C4BP by M. catarrhalis is an efficient strategy to prevent complement-mediated lysis and is facilitated through interaction with UspA1 and UspA2 (6), with hypothesized that SLRPs FMOD, OSAD and BGN may competitively inhibit binding of C4BP and thus render M. catarrhalis more susceptible to serum killing. We measured C4BP binding following incubation in OmCI-treated serum and showed a significant decrease when bacteria were previously incubated with FMOD, OSAD and BGN, with again, no difference observed with DCN or BSA (Fig. 5C).
To confirm our results that SLRPs inhibit binding of C4BP and thus disrupt a major immune evasive strategy of M. catarrhalis, we depleted C4BP from NHS using an anti-C4BP mAb MK104 coupled column, which interacts with high affinity to the α-chain complement control protein (CCP) domain 1 of C4BP (17). Depletion of C4BP from NHS (C4BP-dpl) resulted in increased killing of M. catarrhalis RH4 compared to NHS in the presence of both BSA and DCN (Fig. 5D). Increased survival comparable to NHS was observed following replenishment of purified C4BP to physiological levels (200 μg/mL) when BSA and DCN were present. In comparison, FMOD, OSAD and BGN enhanced serum bactericidal activity in NHS compared to both BSA and DCN as observed previously (Fig 2A-B). Importantly, no significant change in serum killing was observed between BSA/DCN and FMOD/OSAD/BGN in C4BP-dpl (Fig. 5D) confirming that prevention of C4BP binding by FMOD, OSAD and BGN to the bacterial surface is responsible for the increased complement-mediated killing of M. catarrhalis.
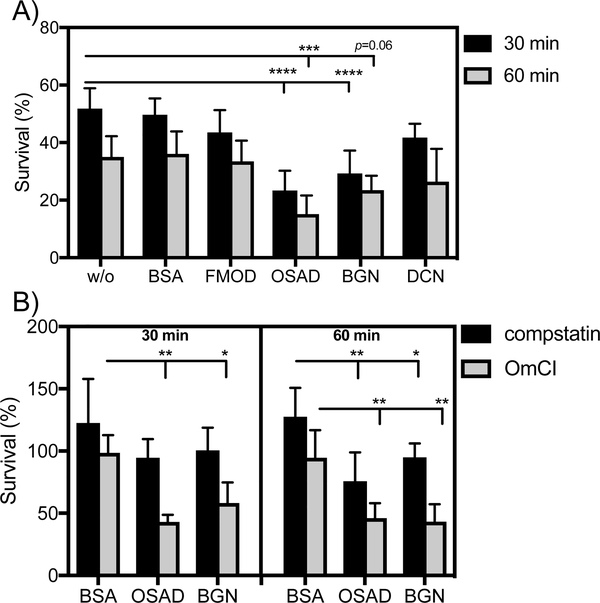
OSAD and BGN enhance neutrophil killing of M. catarrhalis in a complement dependent and independent manner
Neutrophils represent a critical phagocytic cell type in innate immunity, central to host defense against invading pathogens. Additionally, complement mediated opsonisation accelerates phagocytosis and removal of pathogenic bacteria. Considering that SLRP-bound bacteria had increased complement deposition in the presence of serum, we wanted to investigate whether this translated into increased killing in a neutrophil bactericidal assay. Interestingly, in the presence of human neutrophils and OmCI-treated serum, both OSAD and BGN significantly enhanced M. catarrhalis RH4 killing, observed at both 30 and 60 min incubation period (Fig. 6A). Incubation with FMOD or DCN did not significantly increase bacterial killing compared to BSA. Next we wanted to investigate whether this enhanced neutrophil killing was dependent on complement opsonisation or whether SLRPs themselves could serve as mediators of enhanced neutrophil killing. Using OSAD and BGN, we repeated the neutrophil bactericidal assays with either OmCI-treated serum (inhibiting complement at the C5 level) or compstatin-treated serum (inhibiting complement at the C3 level). At 30 min we observed only a decrease in bacterial survival in the OmCI-treated serum conditions and not in the presence of compstatin (Fig. 6B). Surprisingly, after 60 min we observed a statistically significant decrease in survival both with the OmCI- and compstatin-treated sera compared to BSA. This suggests that the main mechanism of SLRP-dependent enhanced neutrophil killing is via complement activation. After a prolonged incubation time, however, SLRPs promote a bactericidal killing effect in concert with neutrophils, which is independent of complement.
Figure 6. SLRPs OSAD and BGN enhance neutrophil killing of M. catarrhalis.
Human neutrophils were incubated with M. catarrhalis RH4 in the presence of A) OmCI-treated serum and SLRPs or BSA or B) OmCI or compstatin-reated serum and OSAD, BGN or BSA for 30 or 60 min at 37°C and 5%CO2. Following incubation total viable bacteria was enumerated following lysis of neutrophils and bacterial survival was calculated by diving CFU at time 30 or 60 with that of time 0. Graphs are presented as the mean and SD of 5 independent experiments and analyzed using a two-way ANOVA with Bonferroni post-hoc tests comparing SLRP/BSA condition to that of no protein control (without, w/o) A) or SLRPs to BSA control B). * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
As compstatin-treated serum still contains C1q, which can act as an opsonin and promote phagocytosis, and as previous work has shown that SLRPs can interact with C1q (11, 12), we investigated the binding of C1q from serum in the presence of SLRPs (Supp Fig. 3). We observed no difference in binding of C1q to the bacterial surface when bacteria were incubated with FMOD or DCN compared to BSA. In contrast, a significant reduction in C1q binding was shown when bacteria were bound with OSAD and BGN. Therefore, these results indicate that enhanced neutrophil killing under compstatin-treated serum conditions in the presence of OSAD and BGN was not due to increased C1q binding
Discussion
M. catarrhalis causes significant morbidity in children and COPD patients, and is responsible for a plethora of respiratory infections and occasionally, systemic diseases (26). The exact molecular mechanisms governing M. catarrhalis pathogenicity are not fully understood. However, mounting evidence suggests that immune evasion, directed primarily at circumventing the complement system, is an essential feature of pathogenic strains (4–6, 14, 21, 27). Therefore, future treatment intervention directed at hampering complement inhibitor recruitment is a promising avenue of research. In this study, we highlight a novel antimicrobial role displayed by specific SLRPs, namely FMOD, OSAD and BGN and in detail revealed the molecular mechanisms resulting in enhanced innate immunity against M. catarrhalis.
SLRPs such as BGN and DCN are considered bi-functional proteoglycans, acting both as central structural components of the ECM and danger-associated molecular patterns (DAMPs) stimulating immune reactions (28). SLRPs are abundantly present in the ECM and distributed in numerous tissues throughout the body (7). Previous immumohistochemical analysis has shown that BGN and DCN are expressed in the human lung and bronchial tissue (29–31). Furthermore, mining of the Human Protein Atlas (www.proteinatlas.org) (32), a genome-wide analysis of RNA and protein expression from samples representing major tissues and organs, confirmed the expression of all SLRPs used in this study in lung tissue. RNA expression data generated from 320 individual tissue samples showed that for this set of SLRPs, BGN and DCN had the highest expression, followed by FMOD with OSAD having the lowest expression (Suppl Fig 4). Combined, these expression data and previous immunohistochemical analysis indicate that these SLRPs are present in sites anatomically important for M. catarrhalis infection and therefore may play a role in host innate immune defense. The exact concentrations of SLRPs present in human tissues and/or plasma are difficult to estimate. One reason for this is that SLRPs are present in higher concentrations following trauma, proteolysis of the ECM and under sterile and non-sterile inflammatory conditions. Previous work in the field has shown that both BGN and DCN expression is enhanced during experimental sepsis in murine models following LPS challenge (8, 9). Macrophages were observed to be the main secretory cell responsible for enhanced expression. Following stimulation with IL-1β and Il-6, macrophage increased BGN secretion, which in turn induced expression of TNFα and MIP-2, contributing to the overall pro-inflammatory environment and increased SLRP expression (8). Furthermore, DCN expression was higher in cohorts of septic patients compared to healthy controls (9). In this study DCN concentration in plasma were estimated to be at 10 ng/mL in septic patients. Previous studies by this group also estimated another SLRP, PRELP, to be at a similar range present in bronchoalveolar lavage fluid (14). It is tempting to speculate that during infection and particularly sepsis, SLRP expression is increased as a result of secretion of pro-inflammatory cytokines stimulating macrophages and other immune and non-immune cells while at the same time the highly inflamed environment contributes to increased SLRP proteolysis from the ECM. Therefore during infection the local concentration of SLRPs may be higher than the surrounding environment, which could influence complement and innate immune activity and bacterial survival.
Of the three main Gram-negative respiratory pathogens screened in this study, only M. catarrhalis was bound by SLRPs. M. catarrhalis expresses numerous surface proteins, which bind an array of ECM proteins, plasma and complement components permitting colonization and evasion of the host innate and adaptive immunity (4, 26). The major surface proteins of M. catarrhalis are the UspA1 and UspA2/2H. Both UspA1 and A2/2H interact with C4BP, however UspA2/2H is more strongly expressed than UspA1 and therefore plays a more prominent role in C4BP binding and in conferring a complement resistant phenotype (6). Through mutational analysis we determined that all four SLRPs bound to M. catarrhalis predominantly through UspA2/2H. This suggested that SLRP binding to UspA2/2H could competitively inhibit C4BP binding resulting in reduced complement inhibition. Using flow cytometry we illustrated that prior binding of FMOD, OSAD, BGN but not DCN to M. catarrhalis effective reduced C4BP binding thus explaining the increased serum sensitivity.
Given the similarity between BGN and DCN it is surprising that BGN and not DCN competitively inhibits C4BP binding. Both BGN and DCN are members of the class I SLRP family, possessing significant homology at both the protein and genetic level. BGN contains two N-terminal tissue-specific chondroitin/dermatan sulfate side chains whereas DCN contains one, and both differ in the pattern and level of glycosylation (7). These differences permit both SLRPs to perform different tasks in terms of ECM maintenance and cell signaling and possibly binding to different regions of UspA2, resulting in differential inhibition of C4BP. Our results show that M. catarrhalis can bind both DCN and C4BP simultaneously. UspA2 is a trimeric autotransporter adhesin, which interacts with C4BP specifically at the CCP2, CCP5 and CCP7 domains (6). UspA2 is composed of a globular head and stalk domain and therefore it is feasible that DCN but not the other SLRPs bind to specific regions within UspA2 that are not required for C4BP binding. Future biochemical studies are required to fully confirm this hypothesis.
Incubation of bacteria with SLRPs in the presence of serum resulted in significant opsonisation with C3b/iC3b (Fig. 5A). As these opsonins are recognized by complement receptors present on neutrophils we wished to examine where this resulted in enhanced neutrophil bactericidal killing. Here we observed that only OSAD and BGN effectively increased neutrophil killing of M. catarrhalis but not FMOD or DCN. This was surprising considering that FMOD enhanced C3b opsonisation in the presence of serum. Therefore, we checked whether this enhanced neutrophil killing was independent of complement by using compstatin-treated serum, effectively blocking complement at the C3 level. We observed that the majority of the neutrophil killing was complement (opsonisation) mediated (Fig. 6A). However with prolonged incubation both OSAD and BGN enhanced killing in a complement (opsonisation) independent manner. It is known that certain ECM proteins such as the SLRP lumican can enhance phagocytosis by interacting with both bacteria and phagocytes via surface expressed integrins (33). Additionally, it has been shown that other SLRPs such as BGN and DCN can bind to toll-like receptors expressed on professional phagocytes and induce a pro-inflammatory response (8, 9). Two questions arise that require future molecular dissection: 1) Can FMOD and OSAD interact with professional phagocytes and induce an immune response analogous to BGN and DCN and 2) can the SLRPs in question mediate an interaction between bacteria and phagocytes which facilitates enhanced phagocytosis and subsequent killing. As such future molecular characterization is underway to elucidate fully the mechanisms of SLRPs mediated neutrophil bactericidal activity.
Recent work by our lab has shown that respiratory pathogens such as M. catarrhalis can interact with ECM components whereby two opposing scenarios may result, namely attenuated or enhanced complement activity. M. catarrhalis interacts with cartilage oligomeric matrix protein (COMP) preventing complement deposition and interfering with complement-independent phagocytosis, enhancing survival (21). Conversely, M. catarrhalis can be bound by the SLRP, Proline/arginine-rich end leucine-rich repeat protein (PRELP), which disrupts C4BP binding, significantly augmenting complement-mediated lysis and neutrophil killing (14). To this complex interaction between complement, M. catarrhalis and ECM components we introduce the newfound antibacterial role of FMOD, OSAD and BGN, which through interaction with the surface expressed UspA2/2H and in concert with complement, accelerate the eradication of an important respiratory pathogen. Finally, the elucidation of the molecular basis for SLRP-mediated enhanced killing may provide novel research avenues to devise therapies to treat infection.
Supplementary Material
Acknowledgements
We would like to thank Dr Sebastian Kalamajski (Uppsala University) for the kind gift of the pCEP4:FMOD plasmid and the Swedish Orphan Biovitrum for the OmCI vector. Dr Sara Nilsson and Dr Chrysostomi Gialeli (Lund University) are thanked for their helpful discussions on the manuscript.
Funding
This work was supported by Swedish Research Council Grant (2016–01142), King Gustav Vth 80-years anniversary foundation, Österlunds Foundation (A.B.), Lars Hierta Memorial Foundation, Tore Nilssons Foundation and the Royal Physiographic Society of Lund (M.L.) and NIH grant (AI 068730) (J.D.L).
Abbreviations
- BHI
brain-heart infusion
- BGN
biglycan
- C4BP
C4b-binding protein
- CCP
complement control protein
- DCN
decorin
- dpl
depleted serum
- FH
factor H
- FMOD
fibromodulin
- MAC
membrane attack complex
- NHS
normal human serum
- NTER
N-terminal repeat
- OSAD
osteoadherin
- SLRP
short leucine-rich proteoglycan
- Usp
ubiquitous surface protein
References
- 1.Merle NS, Church SE, Fremeaux-Bacchi V, and Roumenina LT 2015. Complement System Part I - Molecular Mechanisms of Activation and Regulation. Front Immunol 6: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blom AM, Kask L, and Dahlback B 2001. Structural requirements for the complement regulatory activities of C4BP. J Biol Chem 276: 27136–27144. [DOI] [PubMed] [Google Scholar]
- 3.Blom AM, Hallstrom T, and Riesbeck K 2009. Complement evasion strategies of pathogens-acquisition of inhibitors and beyond. Mol Immunol 46: 2808–2817. [DOI] [PubMed] [Google Scholar]
- 4.Su YC, Singh B, and Riesbeck K 2012. Moraxella catarrhalis: from interactions with the host immune system to vaccine development. Future Microbiol 7: 1073–1100. [DOI] [PubMed] [Google Scholar]
- 5.Murphy TF, and Parameswaran GI 2009. Moraxella catarrhalis, a human respiratory tract pathogen. Clin Infect Dis 49: 124–131. [DOI] [PubMed] [Google Scholar]
- 6.Nordstrom T, Blom AM, Forsgren A, and Riesbeck K 2004. The emerging pathogen Moraxella catarrhalis interacts with complement inhibitor C4b binding protein through ubiquitous surface proteins A1 and A2. J Immunol 173: 4598–4606. [DOI] [PubMed] [Google Scholar]
- 7.McEwan PA, Scott PG, Bishop PN, and Bella J 2006. Structural correlations in the family of small leucine-rich repeat proteins and proteoglycans. J Struct Biol 155: 294–305. [DOI] [PubMed] [Google Scholar]
- 8.Schaefer L, Babelova A, Kiss E, Hausser HJ, Baliova M, Krzyzankova M, Marsche G, Young MF, Mihalik D, Gotte M, Malle E, Schaefer RM, and Grone HJ 2005. The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. J Clin Invest 115: 2223–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merline R, Moreth K, Beckmann J, Nastase MV, Zeng-Brouwers J, Tralhao JG, Lemarchand P, Pfeilschifter J, Schaefer RM, Iozzo RV, and Schaefer L 2011. Signaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and MicroRNA-21. Sci Signal 4: ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Happonen KE, Heinegard D, Saxne T, and Blom AM 2012. Interactions of the complement system with molecules of extracellular matrix: relevance for joint diseases. Immunobiology 217: 1088–1096. [DOI] [PubMed] [Google Scholar]
- 11.Sjoberg AP, Manderson GA, Morgelin M, Day AJ, Heinegard D, and Blom AM 2009. Short leucine-rich glycoproteins of the extracellular matrix display diverse patterns of complement interaction and activation. Mol Immunol 46: 830–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groeneveld TW, Oroszlan M, Owens RT, Faber-Krol MC, Bakker AC, Arlaud GJ, McQuillan DJ, Kishore U, Daha MR, and Roos A 2005. Interactions of the extracellular matrix proteoglycans decorin and biglycan with C1q and collectins. J Immunol 175: 4715–4723. [DOI] [PubMed] [Google Scholar]
- 13.Happonen KE, Sjoberg AP, Morgelin M, Heinegard D, and Blom AM 2009. Complement inhibitor C4b-binding protein interacts directly with small glycoproteins of the extracellular matrix. J Immunol 182: 1518–1525. [DOI] [PubMed] [Google Scholar]
- 14.Liu G, Ermert D, Johansson ME, Singh B, Su YC, Paulsson M, Riesbeck K, and Blom AM 2017. PRELP Enhances Host Innate Immunity against the Respiratory Tract Pathogen Moraxella catarrhalis. J Immunol 198: 2330–2340. [DOI] [PubMed] [Google Scholar]
- 15.Kalamajski S, Bihan D, Bonna A, Rubin K, and Farndale RW 2016. Fibromodulin Interacts with Collagen Cross-linking Sites and Activates Lysyl Oxidase. J Biol Chem 291: 7951–7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahlback B 1983. Purification of human C4b-binding protein and formation of its complex with vitamin K-dependent protein S. Biochem J 209: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardig Y, Hillarp A, and Dahlback B 1997. The amino-terminal module of the C4b-binding protein alpha-chain is crucial for C4b binding and factor I-cofactor function. Biochem J 323 ( Pt 2): 469–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Medical A 2013. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 310: 2191–2194. [DOI] [PubMed] [Google Scholar]
- 19.Potempa M, Potempa J, Okroj M, Popadiak K, Eick S, Nguyen KA, Riesbeck K, and Blom AM 2008. Binding of complement inhibitor C4b-binding protein contributes to serum resistance of Porphyromonas gingivalis. J Immunol 181: 5537–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan TT, Nordstrom T, Forsgren A, and Riesbeck K 2005. The respiratory pathogen Moraxella catarrhalis adheres to epithelial cells by interacting with fibronectin through ubiquitous surface proteins A1 and A2. J Infect Dis 192: 1029–1038. [DOI] [PubMed] [Google Scholar]
- 21.Liu G, Gradstedt H, Ermert D, Englund E, Singh B, Su YC, Johansson ME, Aspberg A, Agarwal V, Riesbeck K, and Blom AM 2016. Moraxella catarrhalis Evades Host Innate Immunity via Targeting Cartilage Oligomeric Matrix Protein. J Immunol 196: 1249–1258. [DOI] [PubMed] [Google Scholar]
- 22.Nunn MA, Sharma A, Paesen GC, Adamson S, Lissina O, Willis AC, and Nuttall PA 2005. Complement inhibitor of C5 activation from the soft tick Ornithodoros moubata. J Immunol 174: 2084–2091. [DOI] [PubMed] [Google Scholar]
- 23.Mastellos DC, Yancopoulou D, Kokkinos P, Huber-Lang M, Hajishengallis G, Biglarnia AR, Lupu F, Nilsson B, Risitano AM, Ricklin D, and Lambris JD 2015. Compstatin: a C3-targeted complement inhibitor reaching its prime for bedside intervention. Eur J Clin Invest 45: 423–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leffler J, Martin M, Gullstrand B, Tyden H, Lood C, Truedsson L, Bengtsson AA, and Blom AM 2012. Neutrophil extracellular traps that are not degraded in systemic lupus erythematosus activate complement exacerbating the disease. J Immunol 188: 3522–3531. [DOI] [PubMed] [Google Scholar]
- 25.Su YC, Hallstrom BM, Bernhard S, Singh B, and Riesbeck K 2013. Impact of sequence diversity in the Moraxella catarrhalis UspA2/UspA2H head domain on vitronectin binding and antigenic variation. Microbes Infect 15: 375–387. [DOI] [PubMed] [Google Scholar]
- 26.Verduin CM, Hol C, Fleer A, van Dijk H, and van Belkum A 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin Microbiol Rev 15: 125–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hallstrom T, Nordstrom T, Tan TT, Manolov T, Lambris JD, Isenman DE, Zipfel PF, Blom AM, and Riesbeck K 2011. Immune evasion of Moraxella catarrhalis involves ubiquitous surface protein A-dependent C3d binding. J Immunol 186: 3120–3129. [DOI] [PubMed] [Google Scholar]
- 28.Frey H, Schroeder N, Manon-Jensen T, Iozzo RV, and Schaefer L 2013. Biological interplay between proteoglycans and their innate immune receptors in inflammation. FEBS J 280: 2165–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallgren O, Nihlberg K, Dahlback M, Bjermer L, Eriksson LT, Erjefalt JS, Lofdahl CG, and Westergren-Thorsson G 2010. Altered fibroblast proteoglycan production in COPD. Respir Res 11: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weitoft M, Andersson C, Andersson-Sjoland A, Tufvesson E, Bjermer L, Erjefalt J, and Westergren-Thorsson G 2014. Controlled and uncontrolled asthma display distinct alveolar tissue matrix compositions. Respir Res 15: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J, Olivenstein R, Taha R, Hamid Q, and Ludwig M 1999. Enhanced proteoglycan deposition in the airway wall of atopic asthmatics. Am J Respir Crit Care Med 160: 725–729. [DOI] [PubMed] [Google Scholar]
- 32.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, and Ponten F 2015. Proteomics. Tissue-based map of the human proteome. Science 347: 1260419. [DOI] [PubMed] [Google Scholar]
- 33.Shao H, Lee S, Gae-Scott S, Nakata C, Chen S, Hamad AR, and Chakravarti S 2012. Extracellular matrix lumican promotes bacterial phagocytosis, and Lum−/− mice show increased Pseudomonas aeruginosa lung infection severity. J Biol Chem 287: 35860–35872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forsgren A, Brant M, Mollenkvist A, Muyombwe A, Janson H, Woin N, and Riesbeck K 2001. Isolation and characterization of a novel IgD-binding protein from Moraxella catarrhalis. J Immunol 167: 2112–2120. [DOI] [PubMed] [Google Scholar]
- 35.Christensen JJ, Ursing J, and Bruun B 1994. Genotypic and phenotypic relatedness of 80 strains of Branhamella catarrhalis of worldwide origin. FEMS Microbiol Lett 119: 155–159. [DOI] [PubMed] [Google Scholar]
- 36.Nordstrom T, Jendholm J, Samuelsson M, Forsgren A, and Riesbeck K 2006. The IgD-binding domain of the Moraxella IgD-binding protein MID (MID962–1200) activates human B cells in the presence of T cell cytokines. J Leukoc Biol 79: 319–329. [DOI] [PubMed] [Google Scholar]
- 37.Kroll JS, and Moxon ER 1988. Capsulation and gene copy number at the cap locus of Haemophilus influenzae type b. J Bacteriol 170: 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.