Abstract
Advancements in microfluidic technologies have led to the development of many new tools for both the characterization and sorting of single cells without the need for exogenous labels. Label-free microfluidics reduce the preparation time, reagents needed, and cost of conventional methods based on fluorescent or magnetic labels. Furthermore, these devices enable analysis of cell properties such as mechanical phenotype and dielectric parameters that cannot be characterized with traditional labels. Some of the most promising technologies for current and future development towards label-free, single-cell analysis and sorting include electronic sensors such as Coulter counters and electrical impedance cytometry; deformation analysis using optical traps and deformation cytometry; hydrodynamic sorting such as deterministic lateral displacement, inertial focusing, and microvortex trapping; and acoustic sorting using traveling or standing surface acoustic waves. These label-free microfluidic methods have been used to screen, sort, and analyze cells for a wide range of biomedical and clinical applications, including cell cycle monitoring, rapid complete blood counts, cancer diagnosis, metastatic progression monitoring, HIV and parasite detection, circulating tumor cell isolation, and point-of-care diagnostics. Because of the versatility of label-free methods for characterization and sorting, the low-cost nature of microfluidics, and the rapid prototyping capabilities of modern microfabrication, we expect this class of technology to continue to be an area of high research interest going forward. New developments in this field will contribute to the ongoing paradigm shift in cell analysis and sorting technologies toward label-free microfluidic devices, enabling new capabilities in biomedical research tools as well as clinical diagnostics.
Graphical/Visual Abstract
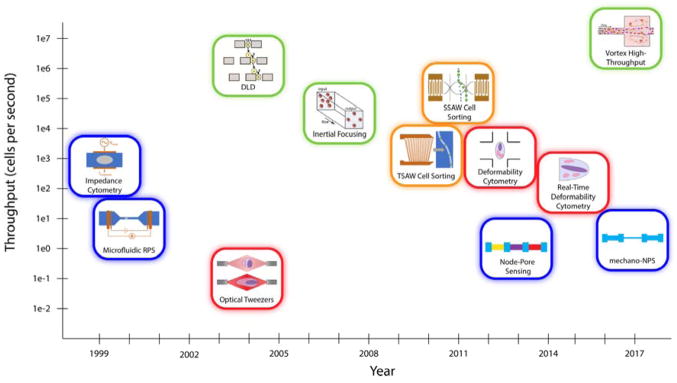
Many new tools that utilize microfluidic technologies for the label-free characterization and sorting of single cells have been developed in the last two decades. These methods can be broadly categorized as electrical (blue), optical (red), hydrodynamic (green), and acoustic (orange).
Introduction
Tools for cell sorting and subsequent characterization are indispensable in the life sciences and in medicine, as they enable rapid isolation of desired subpopulations and critical identification and monitoring for clinical diagnostics. Recently, single-cell isolation and analysis has gained much attention, as such analysis could potentially transform personalized medicine. Knowledge of the heterogeneity of a patient’s solid tumor at the single cell level could, for instance, enable therapies that target multiple cell subtypes (Kim et al., 2016), thereby improving survival rates. Identifying rare circulating tumor cells in patient blood could determine prognosis and efficacy of treatment (Miller, Doyle, & Terstappen, 2010). Current methods for single-cell analysis include flow cytometry and magnetic-activated cell sorting. However, both require 1) lengthy, resource-intensive sample preparation, leading to the potential loss of key cells; 2) cell labeling, for which multiplexing is limited by spectral emission overlap of fluorescent labels; and 3) a large population of cells. In the case of flow cytometry, dedicated technical support is often needed for instrument operation, and the instrument itself is sufficiently expensive to limit usage to core laboratories.
Beyond the difficulties discussed above, label-based methods for cell analysis and sorting may be hindered by even more fundamental issues. The use of labels inherently requires knowledge of the property or population that is being measured. It is impossible to search for new, undefined cell populations using only labels for known biomarkers. Perhaps an even more important consideration is that the biochemical process of a label binding a surface marker may alter the state of the cell, activating specific pathways. As discussed by Xi et al. (Xi, Yu, Wang, Xu, & Abassi, 2008), label-based tests in early drug development may be a contributing factor to the high rates of failure in later stages.
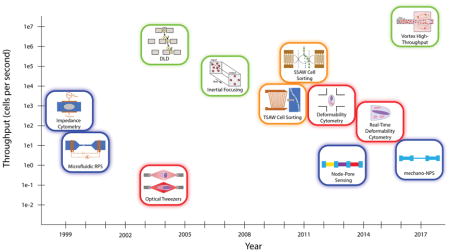
Label-free microfluidic techniques, which do not require exogenous or endogenous labels, offer an alternative approach to single-cell analysis. These techniques—highlighted in Fig. 1 and Table 1—can be classified under four broad areas: electrical, optical, hydrodynamic, and acoustic. While the throughput of many microfluidic screening and sorting technologies is not yet competitive with that of flow cytometry, their promise in identifying specific cells or small subpopulations of cells (e.g. circulating tumor cells or stem cells) make them highly attractive to the biomedical research and clinical diagnostics communities. Below, we highlight just a few exciting label-free techniques and their biomedical and clinical applications.
Figure 1.
Electrical (blue), optical (red), hydrodynamic (green), and acoustic (orange) methods of sorting cells. While hydrodynamic methods tend to offer higher throughputs, other methods typically provide more granular information about cells. It should be noted that the throughput values depicted are approximate and correspond to the first demonstration of that technology. Thus, current implementations of older technologies usually have higher throughput values than those shown here.
Table 1.
Microfluidic options for label-free cell analysis and sorting based on a variety of characteristics
| Criterion | Technology | Type | Description | References |
|---|---|---|---|---|
| Size | Inertial focusing | Sort | Inertial forces cause cells of a predetermined size to migrate to specific positions within a channel | (Di Carlo et al., 2007; Ozkumur et al., 2013) |
| Size | Vortex high throughput | Sort | Larger cells are trapped in microvortices that form in periodic wide sections of a microfluidic channel | (Che et al., 2016; Renier et al., 2017; Vortex Biosciences, 2018) |
| Size | Deterministic lateral displacement | Sort | Slightly offset rows of pillars deflect cells variably based on size | (Beech et al., 2012; Huang et al., 2004; Karabacak et al., 2014; Loutherback et al., 2012) |
| Size | Weir-type filter | Sort | Larger cells are captured in segments of a channel with a shallower depth | (Tu et al., 2016; Yeo et al., 2016; L. Zhu et al., 2004) |
| Size | Pillar-type filter | Sort | Larger cells or cell clusters are captured by narrowly-spaced pillars | (Au et al., 2017; Sarioglu et al., 2015) |
| Size | Resistive-pulse sensing | Measure | As cells pass through a channel, they cause a current drop that indicates cell size | (Becker et al., 1995; Satake et al., 2002) |
| Size | Electrical impedance cytometry / spectroscopy (EIC/EIS) | Measure | At low frequencies, measured impedance is dominated by cell size | (Cheung & Berardino, 2010; S Gawad et al., 2001) |
| Size | Standing surface acoustic waves | Sort | Cell velocity toward pressure node is proportional to the square of the cell radius | (Nam et al., 2011; Shi et al., 2009) |
| Size | Deformability cytometry | Measure | Size of deformed cells is measured with high-speed brightfield microscopy (100,000 FPS) | (Cytovale, 2018; Gossett et al., 2012) |
| Size | Real-time deformability cytometry | Measure | Size of deformed cells is measured with high-speed brightfield microscopy (1,000 FPS) | (Mietke et al., 2015; Otto et al., 2015; Xavier et al., 2016) |
| Size | Optofluidic rotation | Measure | Cells are continuously imaged with brightfield microscopy during trapping and rotation | (Kolb et al., 2015) |
| Size | Optical stretcher | Measure | Cells are imaged with brightfield microscopy before and after stretching | (Guck et al., 2005; Lincoln et al., 2004) |
| Size | Optical chromatography | Measure | Cells are imaged with brightfield microscopy before and after stretching | (Hebert et al., 2017; Imasaka et al., 1995; Kaneta et al., 2001; Lumacyte, 2018) |
| Deformability | Deterministic Lateral Displacement | Sort | Slightly offset rows of pillars in conjunction with a constrictive channel height sorts cells based on deformability | (Beech et al., 2012) |
| Deformability | Optical stretcher | Measure | Cells are stretched into an ellipsoid by optical traps; deformability is measured by the aspect ratio of this ellipsoid, imaged in a 2D plane | (Guck et al., 2005; Lincoln et al., 2004) |
| Deformability | Optical chromatography | Measure | Cells are stretched into an ellipsoid by opposing optical trap and flow force; deformability is measured by the aspect ratio of this ellipsoid, imaged in a 2D plane | Hebert et al., 2017; Imasaka et al., 1995; Kaneta et al., 2001; Lumacyte, 2018) |
| Deformability | Deformability cytometry | Measure | Two high-velocity fluid streams collide and deform a cell into an ellipse between them; deformability is measured by the aspect ratio of this ellipsoid, imaged in a 2D plane | (Cytovale, 2018; Gossett et al., 2012) |
| Deformability | Real-time deformability cytometry | Measure | Cells transiting a narrow channel are deformed into a bullet-like shape due to high shear; deformability is measured by the aspect ratio of this shape, imaged in a 2D plane | (Mietke et al., 2015; Otto et al., 2015; Xavier et al., 2016) |
| Deformability | Mechano-node-pore sensing | Measure | Cell volume and transit time through a constriction channel are measured using node-pore sensing to characterize deformability, resistance to deformation, and recovery | (J. Kim et al., 2018) |
| Compressibility | Standing surface acoustic waves | Sort | Lower cell compressibility results in faster cell velocity toward pressure node | (Ding et al., 2014) |
| Electrical properties | EIC/EIS (impedance, permittivity, membrane capacitance, cytoplasm conductivity) | Measure | AC signal is applied across the cell; the measured current response yields information about impedance, permittivity, membrane capacitance, and cytoplasm conductivity | (Cheung & Berardino, 2010; S. Gawad et al., 2004; Holmes et al., 2009; Sohn et al., 2000) |
| Surface markers | Node-pore sensing | Measure | Channel segments are coated in antibodies which react with cell surface markers. Cells expressing the corresponding marker traverse the channel segment more slowly | (Balakrishnan et al., 2015) |
| 3D tomography | Optofluidic rotation | Measure | Cells are immobilized with an optical trap, rotated with fluidic flow, and imaged with brightfield microscopy | (Kolb et al., 2015) |
ELECTRICAL ANALYSIS
Electronic sensors are an attractive choice for cell screening given the wide array of inexpensive, robust off-the-shelf electronic parts that are available. The cell’s dielectric (DE) properties reflect the biophysical parameters of the membrane and cytoplasm, and electronic sensors are easily integrated into microfluidic systems to measure these DE properties. These types of devices have been used to assess membrane morphology, ion channel status, intracellular ion flow, and nuclear size, and in turn, to identify physiological differences, track pathological changes, and discriminate cell subpopulations (Cheung & Berardino, 2010; Morgan, Sun, Holmes, Gawad, & Green, 2007; Valero, Braschler, & Renaud, 2010). For example, devices that measure cellular DE properties have been used to detect parasite-infected red blood cells (Valero et al., 2010), distinguish tumor cells from mononuclear blood cells (Becker et al., 1995; Gascoyne, Shim, Noshari, Becker, & Stemke-Hale, 2013; Han, Han, & Frazier, 2006; Kang, Yoo, Kim, & Lee, 2012); and track the metastatic progression of tumor cells (Zhao et al., 2014). In this section, we focus on a few general types of resistance- and impedance-based electronic sensors in development.
Resistive-pulse sensing
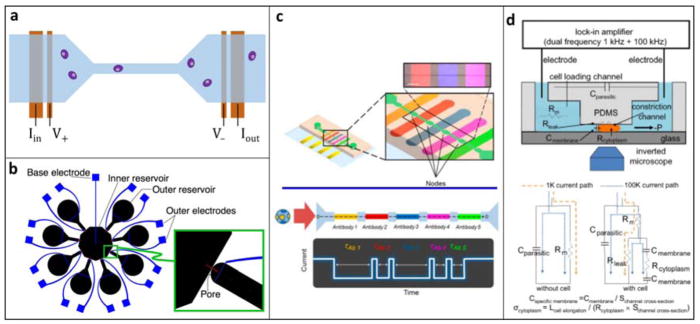
Resistive-pulse sensing (RPS), or the Coulter principle (Coulter, 1953), is one of the oldest methods for automated cell counting and analysis. RPS is still the most widely used method for particle counting and remains the industry standard for complete blood counts (CBCs). In traditional Coulter counters, particles pass through an aperture in the presence of a constant applied voltage. As a particle transits the aperture, it partially blocks the current flow. This causes a transient current drop whose magnitude corresponds to the size of the particle and whose duration indicates the transit time across the aperture. Microfluidic implementations of RPS, in which particles transit a microchannel instead of an aperture (Fig. 2a), were first successfully demonstrated in the early 2000s using standard micromachining as well as soft lithography (Omar Saleh, 2003). These devices have been shown to be a robust platform for counting specific subpopulations in blood—from circulating tumor cells to HIV particles (Becker et al., 1995; Watkins, Hassan, & Damhorst, 2013).
Figure 2.
A. Typical implementation of microfluidic RPS. A fluidic channel (blue), typically a PDMS mold, is bonded to a substrate containing microfabricated electrodes (orange). A voltage is applied across the channel while the current is monitored. A cell’s presence in the channel causes a current drop. B. Example of a multichannel RPS design using eight detection channels to improve throughput by multiplexing. Reprinted with permission from (Saleh, 2003) C. NPS, a variation of RPS, is used to measure five surface markers in one channel. Each section is functionalized with an antibody, and cells expressing the corresponding surface marker traverses that section more slowly. Adapted with permission from (Balakrishnan et al., 2015). Further permissions related to the material excerpted should be directed to the ACS. D. Schematic and electrical model of a constriction channel design for microfluidic EIC. Cells flow through the constriction channel while impedance and elongation are measured continuously. Two-frequency data at 1 kHz and 100 kHz allow calculation of specific membrane capacitance and cytoplasm conductivity. Adapted with permission from (Y. Zhao et al., 2014)
As recent advances have demonstrated, RPS can also be used to probe other cellular properties beyond size. Balakrishnan et al. used a novel method, node-pore sensing (NPS), to screen for multiple cell surface markers (Fig. 2c) (Balakrishnan et al., 2015). The sensing channel was coated with antibodies, and cells expressing surface markers that could specifically interact with these antibodies traversed the channel more slowly. To allow screening for multiple surface markers, the main microfluidic channel was divided into segments separated by nodes. Since the current density was greater in the segments than in the nodes, the current pulse was modulated, facilitating measurement of the cell’s transit times through each individual segment. Each segment was coated with a different antibody, and at least one segment was coated with an isotype control. This NPS device demonstrated the ability to screen for five surface markers simultaneously, and it was successfully used to identify leukemic blast subpopulations found in the bone marrow of acute myeloid leukemia patients.
More recently, a related technique called mechano-NPS was developed to measure the mechanical properties of cells. Mechano-NPS introduces a constriction segment whose width is much smaller than the diameter of a cell (K.-T. T. Kim et al., 2016). By analyzing the cell volume and transit time through the constriction segment, mechano-NPS can characterize cellular deformability, resistance to deformation, and recovery. Kim et al. have shown that mechano-NPS can discriminate between malignant and non-malignant breast and lung epithelial cells, as well as identify differences among pre- and post-menopausal primary human mammary epithelial cells.
Although microfluidic RPS devices characterize important cellular information, they do face tradeoffs among throughput, signal–noise ratio (SNR), and dynamic range. A common strategy to increase throughput without sacrificing SNR is to design a multiplexed RPS device with multiple measurement channels, as first described by Saleh in 2003 (Fig. 2b) (Omar Saleh, 2003). A variety of devices based on this concept have been developed (Jagtiani, Carletta, & Zhe, 2011; Jagtiani, Zhe, Hu, & Carletta, 2006; Zhe, Jagtiani, Dutta, Hu, & Carletta, 2007), but they require additional fabricated electrodes for each channel and are thus limited in scalability. Alternatively, phase-shift keying, or encoding data using changes in channel width, may be employed to improve performance. For example, specifically encoded channel geometries, coupled with signal processing strategies in post-processing, can dramatically improve SNR and dynamic range while allowing higher throughput by resolving coincidence events (Balakrishnan et al., 2013; Kellman, Rivest, Pechacek, Sohn, & Lustig, 2017; Rivest et al., 2015). Optimal performance may be achieved by combining multiple strategies, such as utilizing Barker-coded NPS to enable coincidence detection while incorporating code division multiplexing for multichannel design (Javanmard & Davis, 2013; Rivest et al., 2015).
Cellular impedance analysis
A natural extension of the Coulter principle is to apply an AC signal in order to extract cellular capacitance. This concept was first demonstrated at the single-cell level by Sohn et al., who designed a PDMS microfluidic device that could measure the DNA content of single cells based on total capacitance (Sohn et al., 2000). Many specific cellular properties can be extracted by varying the frequency of the excitation signal: Applying low-frequency signals (<1 MHz) yields information about cell size, while intermediate frequencies (~1–20 MHz) enable characterization of membrane properties such as capacitance, polarization, and ion channel activity. In the high frequency regime (~GHz), subcellular structures such as cytoplasm and vacuoles may be characterized (S Gawad, Schild, & Renaud, 2001; S. Gawad, Cheung, Seger, Bertsch, & Renaud, 2004). Impedance analysis can be implemented with the same standard microfabrication techniques as RPS, and its ability to characterize a wide range of cell properties enables its use for a variety of biological applications.
Techniques that measure the impedance response of cells at two or more frequencies may be called electrical impedance cytometry (EIC) or electrical impedance spectroscopy (EIS). Since papers in the literature do not use consistent terminology in the distinction between EIC and EIS, we will treat these terms as interchangeable. EIC/EIS can be used in conjunction with cell trapping and immobilization techniques to monitor cellular changes over time. This type of device has been used to enable single-cell monitoring of cell viability, size, cell cycle state, membrane properties, nuclear division, and cytokinesis (Becker et al., 1995; Gascoyne, Shim, Noshari, Becker, & Stemke-Hale, 2013; K.-H. Han, Han, & Frazier, 2006; Kang, Yoo, Kim, & Lee, 2012). Another class of EIC/EIS techniques aim to characterize many cells as they flow through a microchannel. These devices have been applied to a variety of applications, including cancer detection and monitoring (Kang et al., 2012; Y. Zhao et al., 2014), blood cell classification (X. Han, Berkel, Gwyer, Capretto, & Morgan, 2012; Holmes, Pettigrew, Reccius, & Gwyer, 2009; van Berkel, Gwyer, Deane, & Green, 2011; Watkins et al., 2013), stem-cell differentiation (Song, Wang, Rosano, & Prabhakarpandian, 2013), and parasite diagnosis (Du, Ha, Diez-Silva, Dao, & Suresh, 2013; Küttel, Nascimento, Demierre, & Silva, 2007).
Technological development continues to improve the capabilities of microfluidic EIC/EIS. For example, the constriction channel design for in-flow impedance analysis improves the sensitivity by ensuring a tight contact between the cells and electrodes (Fig. 2d) (Kang et al., 2012; Y. Zhao et al., 2014). However, the throughput of this technique is still limited to ~1 cell/sec ( Y Zhao, Chen, Li, et al., 2013). Haandbaek et al. developed a resonator-enhanced EIC/EIS microchip that characterize cells in flow at higher throughput ~100 cells/sec while maintaining high sensitivity (Haandbæk, Bürgel, Heer, & Hierlemann, 2014). The same group demonstrated another device that was able to analyze cells at very high frequencies up to 500 MHz, which enabled the characterization of small subcellular structures such as organelles (Haandbæk, Bürgel, Heer, & Hierlemann, 2014b). Sun et al. developed the maximum length sequence (MLS) approach to EIC/EIS to achieve quasi-real-time impedance characterization across a spectrum of frequencies simultaneously (Sun, Gawad, Bernabini, Green, & Morgan, 2007). In this technique, cells are exposed to a pseudorandom white noise signal, which is composed of many frequencies mixed together. The MLS device could thus simultaneously probe the impedance response single cells at 512 distinct frequencies in a window of ~1 ms.
Outlook: Electrical analysis
Electrical interfaces continue to be popular in microfluidic devices due to their robustness and ease of construction. Electrical properties of cells are tied to many important biophysical characteristics and are easily interrogated with electronic sensors that are compatible with microfluidic platforms. Many commercial technologies utilizing the techniques discussed in this section are already on the market, demonstrating that electrical devices for cellular analysis, in general, have great potential for commercialization. For example, the Millipore Scepter is a handheld tool that employs the Coulter principle for automated cell counting (Millipore Sigma, 2018), and Amphasys and xCELLigence both produce systems that employ impedance analysis for cell characterization and monitoring, respectively (ACEA Biosciences, 2018; Amphasys, 2018).
Coulter and RPS devices still dominate the field, and many recent studies have demonstrated novel methods for improving the throughput and performance of these devices. In future work, RPS technology could be applied to measure a wider array of cellular characteristics by designing novel devices which couple cell properties with channel transit time or cell size. Researchers also are rapidly expanding the capabilities of microfluidic EIC and EIS, both in terms of throughput and applications. It is possible that the signal-processing strategies that have been employed to create multichannel RPS devices could be utilized in EIC devices as well, which would aid in the goal of achieving true high-throughput impedance analyzers.
One application area of particular interest for these and other electronic devices is point-of-care (POC) diagnostics (Chin, Linder, & Sia, 2012; Kumar et al., 2013). For example, a number of groups have developed integrated POC systems using RPS or EIC for applications such as low-cost CBC and rapid T cell monitoring for HIV diagnosis (van Berkel et al., 2011; Watkins et al., 2013). In the interest of reducing production costs, some groups are working on new fabrication strategies that eliminate the need for microfabricated electrodes in these POC systems by utilizing printed circuit boards or off-the-shelf microneedles (S Emaminejad et al., 2016; S. Emaminejad et al., 2012; Mansor et al., 2017). With continuing improvements in hardware designs and manufacturing techniques, we expect electronics-based microfluidic devices to have great impact in clinical and POC diagnostics.
OPTICAL ANALYSIS OF CELL DEFORMATION
The visual observation of cells with a microscope has been advancing cell biology since van Leeuwenhoek’s work in the 17th century. Today, imaging of single cells typically requires the use of a fluorescent label to highlight certain features, both intracellularly and extracellularly. Here, we discuss the use of label-free imaging for mechanical phenotyping, or measuring a cell’s response to an applied force. Mechanical phenotyping can be a powerful parameter for diagnosing cancers where traditional biomarkers fall short. For example, in triple-negative breast cancers, the usual prognostic biomarkers (HER2, ER, PR), are not present, making it difficult to choose an effective treatment option without further, extensive testing (Foulkes, Smith, & Reis-Filho, 2010; Hudis & Gianni, 2011). However, malignant cells from breast as well as bladder tissue have been shown to be less stiff than their healthy counterparts; these discoveries suggest that mechanical phenotyping might be a critical consideration in diagnosing subtypes of cancers and guiding clinical decision-making (Lekka et al., 1999; Lincoln et al., 2004).
Until the 2000’s, the mechanical properties of cells were difficult or extremely time-consuming to measure. Techniques such as micropipette aspiration and atomic force microscopy were groundbreaking when they were first introduced, and though still used as gold standards, they are rapidly losing ground to higher-throughput assays (Binnig, Quate, & Gerber, 1986; Hochmuth, 2000). Today, single-cell mechanical testing takes place in microfluidic devices that apply stress to individual cells and measure their response. In the devices discussed here, cells are imaged before and after the application of a controlled force to estimate the cell’s propensity to deform.
Deformation using optical traps
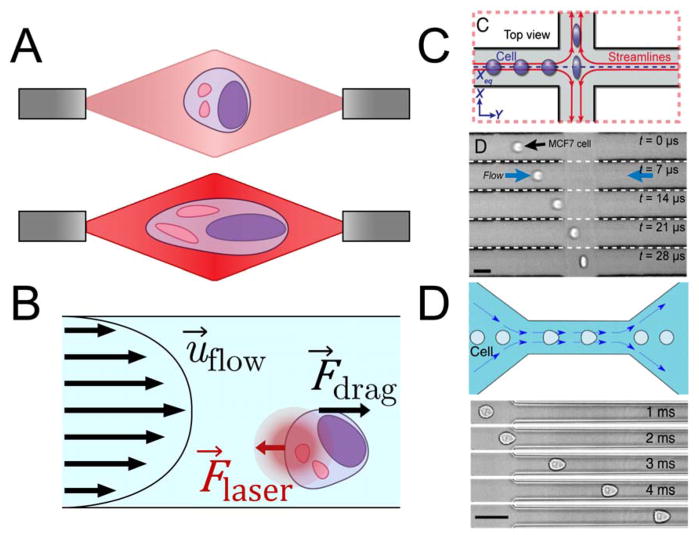
The optical stretcher was one of the first devices to use optical traps for mechanical testing. Optical stretchers use two divergent laser beams pointed towards each other such that they intersect in the middle of a microchannel (Fig. 3A) (Guck et al., 2005; Lincoln et al., 2004). Cells flow through the channel, perpendicular to the two beams, and are stably trapped at the midpoint between the two laser sources, which are tuned to a wavelength of 1064 nm to minimize cell damage. The photons colliding with the cell impart momentum to it, generating an appreciable net force. In the 1–10 mW range, this force anchors cells against the flow (Lincoln et al., 2004). As the power increases up to 1 W, the two opposite ends of the cell are pushed away from the cell’s center with a force around 200–500 pN, stretching the cell laterally (Fig. 3A) (Lincoln et al., 2004). The stress applied by the laser beams deforms the cell into an ellipsoid, and the strain is measured by analyzing images of the cell captured with a brightfield microscope. Optical trapping was used in early studies to show that malignant breast cancer cells were more deformable than non-malignant ones—an important finding that demonstrated the diagnostic potential of mechanical phenotyping (Guck et al., 2005; Lincoln et al., 2004).
Figure 3.
A. Schematic of a cell deformed in a dual-beam optical trap. At low laser power (top), the cell is simply trapped. At higher laser power (bottom), photons colliding with the cell provide enough momentum to physically stretch the cell. B. Free-body diagram describing optofluidic rotation. A dual-beam optical trap immobilizes a cell at a position offset from the center of the microchannel. The velocity profile applies a shear stress to one side of the cell, causing the cell to rotate around the axis of the laser beams. C. Schematic of a deformability cytometry stretching chamber (top) and time lapse of a cell in such a chamber (bottom) (Darling & Carlo, 2015). Cells enter an intersection of two high-speed flows from the left and right, and are deformed and imaged before exiting through the outlets at the top and bottom. Adapted with permission from (Darling & Carlo, 2015) D. Schematic of a real-time deformability cytometry constriction channel (top) and time lapse of a cell in such a channel (bottom) (Otto et al., 2015). Cells enter the narrow channel at high speed, where the shear rate is high enough to deform the cell into a bullet-like shape. Adapted with permission from (Otto et al., 2015)
While optical stretchers trap cells in the middle of a microchannel, Kolb et al. aligned lasers to trap cells slightly off-center a technique they called optofluidic rotation. Because cells are trapped off-center, the flow (1 nL/s) continuously applies a torque to one end of the cell, causing it to rotate at up to 15 RPM (Fig. 3B) (Kolb, Albert, Haug, & Whyte, 2015). By imaging cells with a brightfield or phase contrast microscope throughout a full rotation, one can visualize the cells’ tomography and the position of intracellular structures in 3D. Since optical traps can perform both optical stretching and optofluidic rotation, they have great potential for studying the tomography and 3D structures of cells undergoing deformation in just a few seconds. Because a cell is not a homogeneous material, and the applied forces are not isotropic, 3D spatial information might improve the measurement of anisotropic mechanical properties of cells. As many cells have a degree of spatial polarity, efforts to understand cell anisotropy might help refine mechanical phenotyping by including or controlling for differences in spatial directions.
A similar, but notable, class of microfluidic devices relies on optical chromatography (Imasaka, Kawabata, Kaneta, & Ishidzu, 1995). This method exploits opposing hydrodynamic and optical forces to deform cells in “optical channels” and has been used to study erythrocyte elasticity (Kaneta, Makihara, & Imasaka, 2001), among other cellular processes and properties (Hebert, Hart, Leski, Terray, & Lu, 2017).
Hydrodynamic deformation
Not all microfluidic mechanical phenotyping is done with optical traps. In other devices, the flow itself can apply stresses to cells. In a technique known as deformability cytometry (DC), cells are accelerated into a stretching chamber, where the high-velocity flows (around 10–20 μL/s) from two different microchannels meet (Fig. 3C) (Gossett et al., 2012). The opposing fluidic flow deforms a cell into an ellipsoid before pushing it toward an outlet. Gossett et al. demonstrated the ability of DC to differentiate among populations of normal and stimulated peripheral blood mononuclear cells and granulocytes (Gossett et al., 2012). Traditionally, discriminating between these phenotypes required immunostaining for clusters of differentiation or ELISAs to assay cytokine secretion. DC is able to quickly interrogate thousands of cells and measure distinguishable differences in population deformability.
While DC can analyze thousands of cells per second, it is difficult to capture images of cells moving through the device, as they persist in the stretching chamber for only a few microseconds (Gossett et al., 2012). As a result, a camera used for DC must be capable of acquiring images at over 100,000 frames per second (FPS); such cameras are often extremely expensive and require high-speed and high-density storage solutions to store and analyze data.
This drawback inspired Otto et al. to improve on the designs of Gosset et al. and develop real-time deformability cytometry (RT-DC) (Otto et al., 2015). In RT-DC, cells are deformed while in transit through a microchannel only slightly larger than the cell. The flow velocity is high enough such that the shear stress near the channel walls bends the cells into the shape of the flow velocity profile, which resembles a bullet (Fig. 3D) (Mietke et al., 2015). As in DC, cells in RT-DC are imaged with a high-speed camera, but because the flow velocity is considerably slower than in DC, a framerate of around 1,000 FPS is adequate. Cameras capable of imaging at this speed are more readily available than those needed for DC. The lower framerate also means RT-DC generates much smaller data sets for comparable experiments. As a result, deformation data can be analyzed in real time.
Like its predecessor, RT-DC has also been used to discriminate between leukocyte populations: a critical aspect of hematology (Otto et al., 2015). Another application of interest is the isolation of skeletal stem cells (SSCs) from bone marrow, where the SSC marker Stro-1 lacks the specificity needed to isolate SSCs with high purity (Xavier et al., 2016). Xavier et al. used RT-DC to discriminate between myeloid leukocytes and SSCs by demonstrating that SSCs were dramatically larger and stiffer (Otto et al., 2015; Xavier et al., 2016). Thus, RT-DC can be used to identify and isolate different cell populations in a single primary tissue sample based on deformability. This is particularly attractive if information from traditional biomarkers is difficult to acquire or inadequate for precise discrimination.
Outlook: Optical analysis of cell deformation
Methods for performing mechanical testing on single cells have existed for years; atomic force microscopy, for example, was introduced more than 30 years ago (Binnig et al., 1986). While this technique can acquire a single force curve in a few seconds, it can take an hour to generate a force map of an entire cell. By contrast, the microfluidics-based techniques discussed here enable much higher throughput. Optical stretchers, the first of these devices, are capable of performing a whole-cell mechanical test in just a few seconds (Ekpenyong et al., 2012). Although Lincoln et al. suggest that optical stretchers could be incorporated into flow cytometers to allow for deformability-based flow cytometry, modern flow cytometers operate at much faster rates, often analyzing thousands of cells per second (Lincoln et al., 2004). Hydrodynamic methods such as DC or RT-DC would be more appropriate for incorporating deformability measurements into flow cytometry.
Though the flow velocity of RT-DC is considerably slower than that of DC, it can still analyze hundreds of cells per second (Otto et al., 2015). This rate overlaps with the lower end of modern flow cytometers’ range of operating speeds. The large volume of high-speed video generated by DC necessitates offline processing and would thus be difficult to integrate with flow cytometry. RT-DC, however, can acquire and display deformability data on a single consumer-grade computer in real time. For this reason in particular, RT-DC would appear to be the best candidate for deformability-based flow cytometry and is primed for commercialization.
Nevertheless, older technologies have been commercialized already. LumaCyte’s Radiance takes advantage of optical chromatography to analyze and sort single cells based on their response to a laser force (Lumacyte, 2018). Cytovale seeks to use DC to diagnose sepsis at an early stage by analyzing activated neutrophils (Cytovale, 2018). Even so, we can expect additional commercial platforms in the future as RT-DC matures and new techniques are invented.
CELL SORTING
Beyond analyzing cells, it is often useful to sort cells into discrete populations based on a property of interest. Fluorescence-activated cell sorting (FACS), for example, is a widely used method to obtain relatively pure populations of cells that express a particular biomarker labeled with a fluorescent tag. In contrast, label-free sorting enables the downstream analysis and characterization of unmodified, viable cells. Without the use of labels, cells can be sorted based on size, shape, deformability, viscoelasticity, and compressibility. Label-free sorting of cells based on physical properties enables the study of associations such as that between the stiffness of tumor cells and biomarkers known to correlate with the aggressiveness of the tumor. We expect that label-free cell sorting will facilitate exciting discoveries that precede a new generation of therapies and diagnostics based on newfound associations.
Hydrodynamic sorting
Mixed populations of cells can be sorted based on their differing responses to hydrodynamic forces. Cells can be sorted based on a wide range of mechanical properties, including size, shape, deformability, and viscoelasticity (Beech, Holm, Adolfsson, & Tegenfeldt, 2012; Loutherback et al., 2012; Masaeli et al., 2016; Otto et al., 2015; Renier et al., 2017; Sarioglu et al., 2015). These properties are affected by cell characteristics such as cytoskeleton arrangement, chromatin configuration, and nuclear volume, among other factors. Differences in these cell characteristics arise in the course of several diseases (Lee & Lim, 2007; Li, Lee, Ong, & Lim, 2009) , and the label-free isolation of cells of a specific phenotype will allow novel treatments to be developed by enabling the detailed analysis of purified populations. Past discoveries that could have been expedited by label-free hydrodynamic sorting include the association of certain cell morphologies with expression levels of tumor suppression genes and the correlation of the mechanical stiffness of tumor cells with metastatic potential (Swaminathan et al., 2011; Yin et al., 2013). Other applications of hydrodynamic cell sorting include blood fractionation, isolation of CTCs, and separation of a single cell line into subpopulations (Delobel et al., 2010; Ozkumur et al., 2013; Tu et al., 2016).
In certain cases, the threshold for the property being measured is designed into the device itself, with multiple outlets used to collect populations above or below this threshold (continuous flow separation) (Beech et al., 2012; Loutherback et al., 2012). In other cases, a time differential is used wherein cells are trapped on the device indefinitely until a stimulus, such as a change in flow rate, is applied (trapping-based separation) (Sarioglu et al., 2015; Masaeli et al., 2016; Otto et al., 2015). We discuss in detail below examples of these different strategies.
Continuous-flow separation
Several groups have developed innovative continuous-flow techniques to sort cells into multiple outlets based on their physical properties. Continuous-flow techniques separate cells using a single flow rate through a channel. Here, we focus on two technologies that are promising for clinical translation: deterministic lateral displacement (DLD) and inertial focusing.
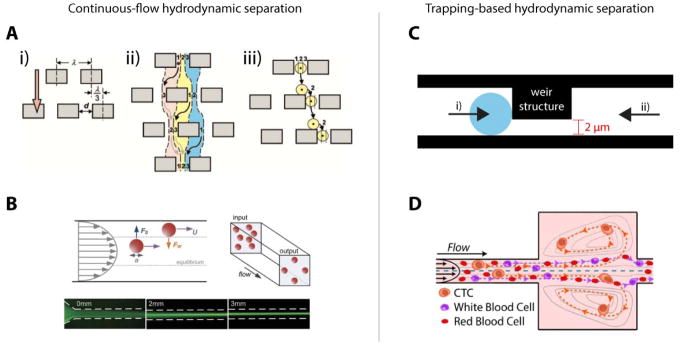
Deterministic lateral displacement (DLD) was first demonstrated for the separation of bacterial chromosomes of lengths 61 kb and 158 kb (Huang et al., 2004), but it has proven to be effective in fractionating whole blood into its components (Karabacak et al., 2014; Loutherback et al., 2012) and removing parasites from blood (Holm, Beech, Barrett, & Tegenfeldt, 2011). DLD relies on the asymmetric bifurcation of laminar flow around obstacles. By slightly offsetting repeating rows of obstacles or pillars in a microfluidic channel, particles can be deflected variably based on size (Fig. 4a). Because high cell concentrations at the input of a DLD array can cause cells to stack at the outlets leading to low purity in the sorted cell populations, a sheath flow is often necessary to achieve optimal cell concentrations. Up to 128-plex DLD arrays have been demonstrated; with this degree of multiplexing, a throughput of 15–20 million cells/second is possible. Despite flow rates of up to 80μL/min through each array, cells remain viable after processing (Loutherback et al., 2012). By reducing the height of the microfluidic channel to less than the height of cells being interrogated and adjusting the shear rate (from 40 to 16,400 Hz), DLD can also sort cells by deformability (Beech et al., 2012). The most promising application of DLD, however, is in rapid fractionation of blood, which would enable blood banking in areas where a centrifuge is not accessible. Such areas include low-resource settings or at the bedside; fractionation immediately following a blood draw could reduce blood degradation (Delobel et al., 2010).
Figure 4.
A. Mechanism of deterministic lateral displacement. Streamlines 1, 2, and 3 do not mix. If a cell or particle is large enough to be located primarily in streamline 3, as shown in the diagram, it will flow to the right of the pillars. Reprinted with permission from (Huang, Cox, Austin, & Sturm, 2004) . B. Free-body diagram of forces experienced by cells or particles during inertial focusing that pushes them toward equilibrium positions in a channel based on size. Reprinted with permission from (Zhou & Papautsky, 2013). C. Weir-type filter used to capture cells below a size/deformability threshold; once cells are trapped by centrifugal forces and negative pressure (i), back-flow is used to recover the cells from the device (ii). Adapted with permission from (Yeo et al., 2016). D. Trace of path of cells in Vortex device. At a constant, high flow rate, larger cells are trapped in large sections of the channel in microvortices, while smaller cells pass through to the outlet. When the flow rate is reduced, larger cells exit the vortices and are recovered at the outlet. Reprinted with permission from (Che et al., 2016).
Di Carlo et al. were the first to describe using inertial focusing for separation. Inertial focusing is a phenomenon that arises due to inertial effects of a fluid around a particle and the interaction of the flow field with the walls of a channel; the equilibrium positions within the channel exist where forces from the channel wall and a shear gradient lift force are equal (Fig. 4b) (Di Carlo, Irimia, Tompkins, & Toner, 2007). Whether or not inertial focusing occurs in a channel is primarily dependent on the channel’s Reynolds number, ReC, and the particle confinement ratio, λ, which is a function of the particle size and hydrodynamic diameter of the channel (Dh) (Martel & Toner, 2013). Inertial focusing occurs when λ > 0.07 and the particle Reynolds number ReP = (ReC)(λ2) ≥ 1 (Carlo, 2009). The number and location of the equilibrium positions within the channel can be controlled by creating a secondary Dean flow using curved channels. The strength of the Dean flow is characterized by the Dean number, De = ReC(Dh/2r)1/2, where r is the radius of curvature of the channel. Because inertial focusing is dependent on both particle size and the channel geometry, the range of particle sizes that are focused can be controlled, enabling separation of cells by size (Di Carlo, Irimia, Tompkins, & Toner, 2007). While there are many potential applications of inertial focusing for cell separation, the most recent and exciting one has been the isolation of rare CTCs from whole blood with high throughput (Fachin et al., 2017; Ozkumur et al., 2013). In this latest example, inertial focusing is used in conjunction with DLD and magnetic-activated cell sorting to isolate CTCs, independent of size, without labeling the CTCs. This technology was groundbreaking in its ability to reveal the heterogeneity of CTCs from a single patient. The device was able to recover 99.5% of input CTCs while sorting cells at up to 20 million cells/second (Fachin et al., 2017).
Trapping-based separation
While continuous-flow separation is convenient, the necessity of a sheath flow for technologies such as DLD can result in a dilute sample that necessitates additional concentration steps prior to molecular analysis (A. Han, Yang, & Frazier, 2007; James et al., 2008; Jang & Wang, 2007; Malleo, Nevill, Lee, & Morgan, 2010; Z. Zhu et al., 2015). In contrast, technologies that employ trapping strategies, such as filter and vortex technologies, inherently concentrate the target cells on the device. These technologies are typically used to capture and concentrate the largest cells in a mixture. We briefly discuss two types of traditional filters, weirs and pillars, have been used to separate cells based on size.
Weir-type filters consist of a channel which has two depths, one significantly deeper than the other (Fig. 4c). Cells that are too large to pass through the shallow section of the channel are trapped in the deeper section; smaller cells simply flow through to the outlet. Zhu et al. describe a basic weir-type filter for the detection of microbial cells between 2μm and 10μm in diameter in water (L. Zhu et al., 2004). Concentration of the cells of interest on the device allowed rapid immunofluorescence staining and verification of cell types within five minutes of capture. Tu et al. described developing a next-generation weir device in which interdigitated square-wave-shaped weirs are used to sort spiked leukemia cells from whole blood (Tu et al., 2016). Interdigitated weirs have a higher trapping capacity than straight weirs for a given channel width and are thus less prone to clogging. Yeo et al. used hydrodynamic focusing to drive cells toward weirs oriented perpendicularly to the main flow channel to capture single CTCs in individual chambers for subsequent single-cell analysis (Yeo et al., 2016). This device represents a significant improvement in throughput over cell picking using a micromanipulator, which is the current standard for isolating single CTCs from a sample that has undergone primary enrichment; cell picking can take up to 15 minutes per cell, while this device can trap CTCs as fast as 800 cells per minute.
Pillar-based filters consist of one or more arrays of closely-spaced structures that prohibit the passage of cells that are both too large and too stiff to pass between the structures. Flow can be reversed through these filters to recover cells or cell clusters trapped between the pillars. Mohamed et al. describe an early filter with four arrays of PDMS pillars with decreasing spacing from 20μm to 5μm; cells larger than 20μm in diameter were captured in the first array, while cells smaller than 5μm passed through all arrays to the outlet. This filter device proved capable of isolating neuroblastoma cells spiked into whole blood. Pillar-based filters have also been used to capture and study CTC clusters, which are thought to have greater metastatic potential than single CTCs (Au et al., 2017; Sarioglu et al., 2015). Triangle-shaped PDMS pillars caused bifurcation of flow at the tip; single cells flowed to either side through 12μm x 100μm gaps, while the strength of intracellular junctions resulted in cell clusters being captured on the tip of the triangle. Clusters of CTCs were discovered in 30–40% of patients with metastatic breast or prostate cancer.
Recently, hydrodynamic principles have been applied to selectively capture cells above a size threshold in microvortices. Once cells are trapped, a buffer flow is introduced at a higher flow rate, causing those cells to be released. Renier et al. successfully used the Vortex High Throughput chip to isolate and concentrate prostate CTCs from the diluted whole blood of prostate cancer patients (Fig. 4d) (Renier et al., 2017).
Outlook: Hydrodynamic sorting
Hydrodynamic sorting techniques are generally high-throughput, scalable, and cost-effective methods to separate cells based on physical properties. However, these techniques require precisely-controlled flow rates to perform according to their design specifications. This requirement can make integration into larger lab-on-a-chip systems challenging. For the techniques discussed, accurate cell sorting depends on the target cells having a distinct phenotype from other populations. Furthermore, an adequately large difference in phenotype distributions must exist to prevent reduced capture efficiency or purity. Despite these restrictions, hydrodynamic sorting techniques are likely to find widespread use in not only cancer biology studies but also clinical diagnostics (e.g. liquid biopsies). Commercially, hydrodynamic sorting has begun to enter the clinical market. For example, the Vortex Bioscience VTX-1 Liquid Biopsy System utilizes the Vortex High Throughput chip to isolate CTCs from cancer patient blood (Vortex Biosciences, 2018). Undoubtedly, many other platforms will similarly be available to clinicians and biomedical researchers in the near future.
Acoustic sorting
Utilizing acoustic waves is a versatile method to actuate the fluid and/or particles within a wide variety of microsystems. Applying an acoustic wave in a microfluidic system directly generates forces on both the particles and their surrounding medium, thereby enabling contact-free manipulation that does not introduce additional sources of contamination. As such, these techniques are ideal candidates for sorting cells, whether as an actuation mechanism for active-switching systems in which the property of interest is measured upstream, or for direct sorting based on a cell’s acoustic properties. Although researchers continue to develop microfluidic devices that employ bulk acoustic waves (BAWs) (Hammarström et al., 2010; Leibacher, Reichert, & Dual, 2015) , most recent work in acoustofluidics has focused on surface acoustic waves (SAWs). BAW technology is capable of higher throughput than SAW, but crucially, it is not compatible with PDMS-based microfluidics because it requires an acoustically reflective material for the fluidic channel wall. We thus focus on SAW devices.
SAW devices are typically fabricated by patterning one or more interdigital transducer (IDT) electrodes on a piezoelectric substrate, which is then bonded to a PDMS-molded microfluidic channel. When an alternating current (AC) signal is applied across an IDT at its resonant frequency (typically ~MHz), SAWs propagate along the substrate surface, traveling away from the IDT (Fig. 5a). Here we address devices that utilize two broad categories of SAWs: traveling surface acoustic waves and standing surface acoustic waves.
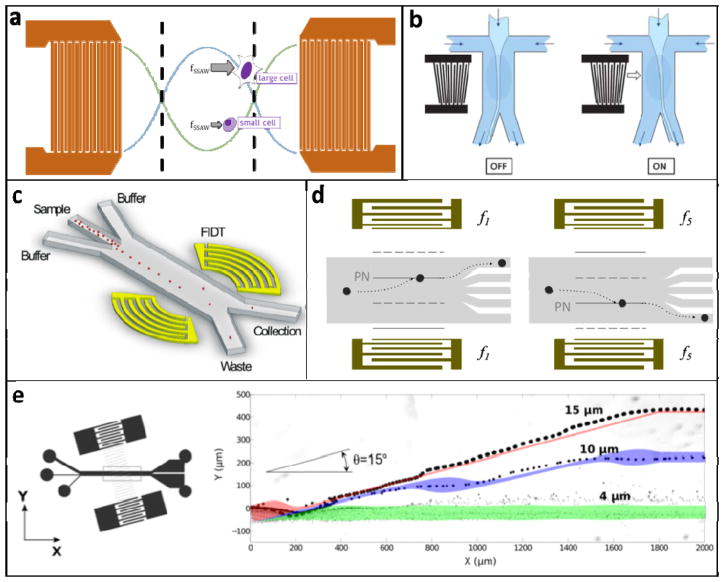
Figure 5.
A. Schematic of SSAW forces exerted on cells in the region between IDTs. All cells move toward the pressure nodes, but large cells experience a larger force and move with a faster velocity. B. Switchable TSAW system used as an actuator for cell sorting. The light blue stream, containing cells, flows into the left outlet channel unless the TSAW is turned on by applying an AC signal to the IDT electrodes. Reprinted with permission from (Franke, Braunmüller, Schmid, Wixforth, & Weitz, 2010) C. FIDT system for high-throughput cell sorting. The concentric design of the IDTs focuses the SSAW to a small region, allowing it to specifically actuate individual cells. This device sorted HeLa cells at ~7,000 Hz. Reprinted with permission from (Ren et al., 2015) D. Tunable SSAW device that employs chirped IDTs. Cells can be directed to one of five different outlets depending on the frequency of the signal applied to the IDTs. Adapted with permission from (Ding et al., 2012) E. The taSSAW device positions the fluidic channel at a small angle relative to the IDT fingers, thus positioning the pressure nodes at an angle relative to fluid flow. Whereas a traditional SSAW sorter can only achieve separation distances up to 1/4 of the acoustic wavelength, the taSSAW design can achieve greater separation and accordingly shows improved throughput and performance. The particle trajectories demonstrate that the 15μm beads were separated from the 4μm beads by >300μm (the acoustic wavelength was 300μm). Adapted with permission from (Ding et al., 2014)
Traveling surface acoustic waves & acoustic streaming
When an AC signal is applied across an IDT, the resulting traveling surface acoustic waves (TSAWs) move away from the IDTs in a direction transverse to the IDT fingers. When the TSAWs encounter a liquid interface, they exert a force on the fluid and induce flow (i.e. acoustic streaming) (Wixforth et al., 2004). While TSAWs also exert a primary acoustic radiation force on particles, acoustic streaming tends to dominate on the size scale of cells (Barnkob, Augustsson, Laurell, & Bruus, 2012). Thus, cell sorting and manipulation using TSAWs is limited to techniques that manipulate the surrounding medium rather than the cells themselves. For example, acoustic streaming has been utilized for droplet handling and manipulation in digital microfluidics and for switching the direction of a stream in continuous-flow devices (Fig. 5b) (Collignon, Friend, & Yeo, 2015; Franke et al., 2010). Because TSAW devices manipulate the fluid flow rather than the cells themselves, cells are not exposed to potentially harmful high shear stress. Furthermore, displacement is not affected by cell size. However, this also limits TSAW devices to active-switching configurations. TSAW-based active switching is very versatile and can achieve throughputs up to ~10 kHz, but it requires another method to measure the property of interest upstream before physically sorting cells.
Standing surface acoustic waves
In 2009, Shi et al. introduced the first device that utilized SSAWs to manipulate and sort particles directly in a continuous-flow configuration (Shi, Huang, Stratton, Huang, & Huang, 2009). This technology has since generated much interest, and the capabilities of SSAW devices have expanded tremendously because they are easy to fabricate and use. When a resonant AC signal is applied across two appropriately spaced sets of parallel IDTs, the resulting TSAWs interfere to generate a standing wave, which can be refracted into a microfluidic channel. In this case, the primary acoustic radiation force dominates over the viscous drag force, which causes cells to move toward the nearest pressure node with a velocity dependent on their physical properties (Fig. 5a). Specifically, cellular velocity Ur is proportional to the square of the cell radius Rp; increased density ρp and/or decreased compressibility βp will also result in a faster velocity (Equation 1). Thus, spatial separation based on these properties can be achieved by exposing cells to the SSAW-induced pressure field for an optimal length of time.
| Equation 1 |
Many groups have worked toward innovations in SSAW-based cell sorting technologies. Guldiken et al. developed a two-stage SSAW device that first focused particles into a stream of single cells before separating them by size, eliminating the need for precisely controlled hydrodynamic sheath flows (Guldiken, Jo, Gallant, Demirci, & Zhe, 2012). Ding et al. developed the first tunable SSAW device, which employed chirped IDTs that could produce variable-wavelength SSAWs depending on the applied AC frequency (Fig. 5d). This device was used as the actuator in a single-cell active-switching system that was precise enough to direct cells to one of five different outlets (Ding et al., 2012). In 2014, a new technique called tilted-angle SSAW (taSSAW) was developed in which the PDMS channel is positioned at an optimized angle (on the order of 10°) relative to the IDT fingers (Fig. 5e). This design enabled higher throughput and sensitivity due to its ability to separate cells by distances greater than the acoustic wavelength (Ding et al., 2014). Ren et al. achieved size-based sorting of polystyrene particles with throughput greater than 10 kHz using an active-switching device with a focused IDT design (Fig. 5c) (Ren et al., 2015).
The first successful application of SSAW for cell sorting was the separation of platelets from red and white blood cells based on size (Nam, Lim, Kim, & Shin, 2011). New technological developments have enabled new applications; for example, Ding et al. used taSSAW to separate cells based only on compressibility, which is thought to be an important feature of cancer phenotypes (as discussed throughout this review). The group was able to use this device to separate cancer cells from non-malignant cell lines (Ding et al., 2014). Using an improved taSSAW device, Li et al. successfully recovered circulating tumor cells (CTCs) from primary whole blood samples (Lee & Lim, 2007; Q. Li, Lee, Ong, & Lim, 2009). Although SSAWs have been used to separate synthetic microspheres of varying density (Jo & Guldiken, 2012), acoustic cell sorting based on density alone has not yet been demonstrated. This is likely because the variation in density between most cultured cell lines is not large enough relative to variations in size and compressibility. Thus, an inherent limitation in SSAW-based sorting is the difficulty in deconvolving these variables. One possible solution could be a multistage device that first sorts cells based on size, then by compressibility, and finally by density.
Outlook: Acoustic sorting
The advantages of using acoustofluidics to manipulate and sort cells are clear in that it is an inherently contact-free strategy and may be less harmful to cells than other methods. Although it is a relatively new field compared to other classes of technologies, there is clearly potential for commercial applications of acoustofluidics. For example, the Attune Flow Cytometer combines acoustic and hydrodynamic focusing to achieve higher throughput with less system clogging compared to traditional flow cytometers (ThermoFisher invitrogen, 2018).
Recent advancements in the field of SAWs have led to a wide range of new microfluidic devices with the capability to sort cells with greater throughput, specificity, and efficiency than ever before. SSAW-based devices hold particular promise for cell sorting applications due to their superior stability and robustness compared to TSAW technologies. Furthermore, their ability to exert force directly on cells makes them useful for both active-switching and direct sorting applications. However, since a cell’s SSAW-induced velocity depends on multiple properties, it may be difficult or even impossible to sort cells based on a single property without controlling for the others. Nevertheless, we believe that SSAW acoustofluidics is an area of particular promise in the development of label-free cell sorting methods.
Conclusion
For analyzing single cells, microfluidic devices offer numerous advantages over traditional benchtop assays. The most obvious benefits, such as rapid prototyping and high assay throughput, allow users to do more work in less time. An equally powerful advantage is the ease of integrating microfluidic devices with other transducers and sensors, such as metal electrodes, piezoelectric materials, and microscopes. Creative researchers have leveraged this compatibility to develop many diverse microfluidic platforms for analyzing and sorting cells.
Single-cell, label-free microfluidics may enable promising new disease diagnosis and prognosis technologies. Some cancers can be difficult to characterize based solely on protein expression due to high phenotypic heterogeneity or a complete lack of expression of traditional biomarkers (Foulkes et al., 2010; Hourigan, Gale, Gormley, Ossenkoppele, & Walter, 2017). Label-free methods for analyzing single cells offer clinicians new information which, when combined with existing tests, will enable more precise diagnoses. Label-free sorting methods can also be useful in clinical settings; they can be used to enrich cells of interest from blood or peripheral fluids like saliva, thus reducing sample volumes and streamlining diagnostic tests.
A valid criticism of the label-free, microfluidic single-cell analysis methods discussed here is that they are largely limited to measuring physical properties such as cell size, density, or deformability. However, each of these technologies has demonstrated some ability to correlate readouts of physical properties to a pathologically-relevant phenotype. Furthermore, technological advances are enabling new applications of these techniques to evaluate non-physical properties such as surface markers. Because microfluidic technologies are increasing in popularity, we are optimistic that label-free methods like those discussed here might soon be as commonplace in clinical pathology labs as they are today in research labs.
Acknowledgments
This work was partially funded by NIH 1R01CA190843-01, 1R01 EB024989-01, and 1R21CA182375-01A1. L.L.S. is supported by a Bakar Fellowship, K.L.C. and B.L. by NIH T32 GM00815532A1 and a National Science Foundation Graduate Research Fellowship, and T.C. by a Kang Family Graduate Award for Biotechnology and a Brodie Scholar Award.
Contributor Information
Thomas R. Carey, UC Berkeley–UC San Francisco Graduate Program in Bioengineering.
Kristen L. Cotner, UC Berkeley–UC San Francisco Graduate Program in Bioengineering.
Brian Li, UC Berkeley–UC San Francisco Graduate Program in Bioengineering.
Lydia L. Sohn, Dept. of Mechanical Engineering, University of California, Berkeley and UC Berkeley–UC San Francisco Graduate Program in Bioengineering
References
- ACEA Biosciences. xCELLigence RTCA systems. 2018. [Google Scholar]
- Amphasys. Ampha Z32 Impedance Flow Cytometry. 2018. [Google Scholar]
- Au S, Edd J, Stoddard A, Wong K, Fachin F, Maheswaran S, … Toner M. Microfluidic Isolation of Circulating Tumor Cell Clusters by Size and Asymmetry. Scientific Reports. 2017;7(1):2433. doi: 10.1038/s41598-017-01150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan KR, Anwar G, Chapman MR, Nguyen T, Kesavaraju A, Sohn LL. Node-pore sensing: a robust, high-dynamic range method for detecting biological species. Lab on a chip. 2013;13(7):1302–7. doi: 10.1039/C3LC41286E. [DOI] [PubMed] [Google Scholar]
- Balakrishnan KR, Whang JC, Hwang R, Hack JH, Godley LA, Sohn LL. Node-pore sensing enables label-free surface-marker profiling of single cells. Analytical chemistry. 2015;87(5):2988–95. doi: 10.1021/ac504613b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnkob R, Augustsson P, Laurell T, Bruus H. Acoustic radiation- and streaming-induced microparticle velocities determined by microparticle image velocimetry in an ultrasound symmetry plane. Physical Review E. 2012;86(5):056307. doi: 10.1103/PhysRevE.86.056307. [DOI] [PubMed] [Google Scholar]
- Becker F, Wang X, Huang Y, Pethig R, Vykoukal J, Gascoyne P. Separation of human breast cancer cells from blood by differential dielectric affinity. Proceedings of the National Academy of Sciences. 1995;92(3):860–864. doi: 10.1073/pnas.92.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech J, Holm S, Adolfsson K, Tegenfeldt J. Sorting cells by size, shape and deformability. Lab on a Chip. 2012;12(6):1048–1051. doi: 10.1039/C2LC21083E. [DOI] [PubMed] [Google Scholar]
- Binnig G, Quate C, Gerber C. Atomic Force Microscope. Physical Review Letters. 1986;56(9):930. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Carlo D. Inertial microfluidics. Lab on a Chip. 2009;9(21):3038–3046. doi: 10.1039/B912547G. [DOI] [PubMed] [Google Scholar]
- Che J, Yu V, Dhar M, Renier C, Matsumoto M, Heirich K, … Carlo D. Classification of large circulating tumor cells isolated with ultra-high throughput microfluidic Vortex technology. Oncotarget. 2016;7(11):12748–12760. doi: 10.18632/oncotarget.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KC, Berardino DM. Microfluidic impedance-based flow cytometry. Cytometry Part …. 2010 doi: 10.1002/cyto.a.20910. [DOI] [PubMed] [Google Scholar]
- Chin CD, Linder V, Sia SK. Commercialization of microfluidic point-of-care diagnostic devices. Lab on a Chip. 2012 doi: 10.1039/C2LC21204H. [DOI] [PubMed] [Google Scholar]
- Collignon S, Friend J, Yeo L. Planar microfluidic drop splitting and merging. Lab on a Chip. 2015;15(8):1942–1951. doi: 10.1039/C4LC01453G. [DOI] [PubMed] [Google Scholar]
- Coulter WH. Means for Counting Particles Suspended in a Fluid 1953 [Google Scholar]
- Cytovale. Cytovale. 2018. [Google Scholar]
- Darling EM, Carlo DD. High-throughput assessment of cellular mechanical properties. Annual review of biomedical …. 2015 doi: 10.1146/annurev-bioeng-071114-040545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delobel J, Rubin O, Prudent M, Crettaz D, Tissot JD, Lion N. Biomarker Analysis of Stored Blood Products: Emphasis on Pre-Analytical Issues. International Journal of Molecular Sciences. 2010;11(11):4601–4617. doi: 10.3390/ijms11114601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo D, Irimia D, Tompkins RG, Toner M. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proceedings of the National Academy of Sciences. 2007;104(48):18892–18897. doi: 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Lin SC, Lapsley M, Li S, Guo X, Chan C, … Huang T. Standing surface acoustic wave (SSAW) based multichannel cell sorting. Lab on a Chip. 2012;12(21):4228–4231. doi: 10.1039/C2LC40751E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Peng Z, Lin SC, Geri M, Li S, Li P, … Huang T. Cell separation using tilted-angle standing surface acoustic waves. Proceedings of the National Academy of Sciences. 2014;111(36):12992–12997. doi: 10.1073/pnas.1413325111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du E, Ha S, Diez-Silva M, Dao M, Suresh S. Electric impedance microflow cytometry for characterization of cell disease states. Lab on a Chip. 2013 doi: 10.1039/c3lc50540e. Retrieved from http://pubs.rsc.org/-/content/articlehtml/2013/lc/c3lc50540e. [DOI] [PMC free article] [PubMed]
- Ekpenyong A, Whyte G, Chalut K, Pagliara S, Lautenschläger F, Fiddler C, … Guck J. Viscoelastic Properties of Differentiating Blood Cells Are Fate- and Function-Dependent. PLoS ONE. 2012;7(9):e45237. doi: 10.1371/journal.pone.0045237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emaminejad S, Paik KH, Tabard-Cossa V. Portable cytometry using microscale electronic sensing. Sensors and Actuators B …. 2016 doi: 10.1016/j.snb.2015.08.118. Retrieved from http://www.sciencedirect.com/science/article/pii/S0925400515302951. [DOI] [PMC free article] [PubMed]
- Emaminejad S, Javanmard M, Dutton R, Davis R. Microfluidic diagnostic tool for the developing world: contactless impedance flow cytometry. Lab on a Chip. 2012;12(21):4499–4507. doi: 10.1039/C2LC40759K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachin F, Spuhler P, Martel-Foley J, Edd J, Barber T, Walsh J, … Toner M. Monolithic Chip for High-throughput Blood Cell Depletion to Sort Rare Circulating Tumor Cells. Scientific Reports. 2017;7(1):10936. doi: 10.1038/s41598-017-11119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. The New England journal of medicine. 2010;363(20):1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- Franke Braunmüller, Schmid Wixforth, Weitz Surface acoustic wave actuated cell sorting (SAWACS) Lab on a Chip. 2010;10(6):789–794. doi: 10.1039/B915522H. [DOI] [PubMed] [Google Scholar]
- Gascoyne P, Shim S, Noshari J, Becker F, Stemke-Hale K. Correlations between the dielectric properties and exterior morphology of cells revealed by dielectrophoretic field-flow fractionation. ELECTROPHORESIS. 2013;34(7):1042–1050. doi: 10.1002/elps.201200496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawad S, Schild L, Renaud P. Micromachined impedance spectroscopy flow cytometer for cell analysis and particle sizing. Lab on a Chip. 2001 doi: 10.1039/B103933B. [DOI] [PubMed] [Google Scholar]
- Gawad S, Cheung K, Seger U, Bertsch A, Renaud P. Dielectric spectroscopy in a micromachined flow cytometer: theoretical and practical considerations. Lab on a Chip. 2004;4(3):241–251. doi: 10.1039/B313761A. [DOI] [PubMed] [Google Scholar]
- Gossett D, Tse H, Lee S, Ying Y, Lindgren A, Yang O, … Carlo D. Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proceedings of the National Academy of Sciences. 2012;109(20):7630–7635. doi: 10.1073/pnas.1200107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guck J, Schinkinger S, Lincoln B, Wottawah F, Ebert S, Romeyke M, … Bilby C. Optical Deformability as an Inherent Cell Marker for Testing Malignant Transformation and Metastatic Competence. Biophysical Journal. 2005;88(5):3689–3698. doi: 10.1529/biophysj.104.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldiken R, Jo M, Gallant N, Demirci U, Zhe J. Sheathless Size-Based Acoustic Particle Separation. Sensors. 2012;12(1):905–922. doi: 10.3390/s120100905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haandbæk N, Bürgel SC, Heer F, Hierlemann A. Characterization of subcellular morphology of single yeast cells using high frequency microfluidic impedance cytometer. Lab on a Chip. 2014a doi: 10.1039/C3LC50866H. [DOI] [PubMed] [Google Scholar]
- Haandbæk N, Bürgel SC, Heer F, Hierlemann A. Resonance-enhanced microfluidic impedance cytometer for detection of single bacteria. Lab on a Chip. 2014b doi: 10.1039/C4LC00576G. [DOI] [PubMed] [Google Scholar]
- Hammarström B, Evander M, Barbeau H, Bruzelius M, Larsson J, Laurell T, Nilsson J. Non-contact acoustic cell trapping in disposable glass capillaries. Lab on a Chip. 2010;10(17):2251–2257. doi: 10.1039/C004504G. [DOI] [PubMed] [Google Scholar]
- Han A, Yang L, Frazier A. Quantification of the Heterogeneity in Breast Cancer Cell Lines Using Whole-Cell Impedance Spectroscopy. Clinical Cancer Research. 2007;13(1):139–143. doi: 10.1158/1078-0432.CCR-06-1346. [DOI] [PubMed] [Google Scholar]
- Han KH, Han A, Frazier A. Microsystems for isolation and electrophysiological analysis of breast cancer cells from blood. Biosensors and Bioelectronics. 2006;21(10):1907–1914. doi: 10.1016/j.bios.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Han X, Berkel C, Gwyer J, Capretto L, Morgan H. Microfluidic Lysis of Human Blood for Leukocyte Analysis Using Single Cell Impedance Cytometry. Analytical Chemistry. 2012;84(2):1070–1075. doi: 10.1021/ac202700x. [DOI] [PubMed] [Google Scholar]
- Hebert C, Hart S, Leski T, Terray A, Lu Q. Label-free Detection of Bacillus anthracis Spore Uptake in Macrophage Cells Using Analytical Optical Force Measurements. Analytical Chemistry. 2017 doi: 10.1021/acs.analchem.7b01983. [DOI] [PubMed] [Google Scholar]
- Hochmuth R. Micropipette aspiration of living cells. Journal of Biomechanics. 2000;33(1):15–22. doi: 10.1016/S0021-9290(99)00175-X. [DOI] [PubMed] [Google Scholar]
- Holm S, Beech J, Barrett M, Tegenfeldt J. Separation of parasites from human blood using deterministic lateral displacement. Lab on a Chip. 2011;11(7):1326–1332. doi: 10.1039/C0LC00560F. [DOI] [PubMed] [Google Scholar]
- Holmes D, Pettigrew D, Reccius CH, Gwyer JD. Leukocyte analysis and differentiation using high speed microfluidic single cell impedance cytometry. Lab on a Chip. 2009 doi: 10.1039/B910053A. [DOI] [PubMed] [Google Scholar]
- Hourigan C, Gale R, Gormley N, Ossenkoppele G, Walter R. Measurable residual disease testing in acute myeloid leukaemia. Leukemia. 2017;31(7):1482–1490. doi: 10.1038/leu.2017.113. [DOI] [PubMed] [Google Scholar]
- Huang L, Cox E, Austin R, Sturm J. Continuous Particle Separation Through Deterministic Lateral Displacement. Science. 2004;304(5673):987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- Hudis C, Gianni L. Triple-Negative Breast Cancer: An Unmet Medical Need. The Oncologist. 2011;16(Supplement 1):1–11. doi: 10.1634/theoncologist.2011-S1-01. [DOI] [PubMed] [Google Scholar]
- Imasaka T, Kawabata Y, Kaneta T, Ishidzu Y. Optical chromatography. Analytical Chemistry. 1995;67(11):1763–1765. doi: 10.1021/ac00107a003. [DOI] [Google Scholar]
- Jagtiani A, Carletta J, Zhe J. A microfluidic multichannel resistive pulse sensor using frequency division multiplexing for high throughput counting of micro particles. Journal of Micromechanics and Microengineering. 2011;21(6):065004. doi: 10.1088/0960-1317/21/6/065004. [DOI] [Google Scholar]
- Jagtiani A, Zhe J, Hu J, Carletta J. Detection and counting of micro-scale particles and pollen using a multi-aperture Coulter counter. Measurement Science and Technology. 2006;17(7):1706. doi: 10.1088/0957-0233/17/7/008. [DOI] [Google Scholar]
- James C, Reuel N, Lee E, Davalos R, Mani S, Carroll-Portillo A, … Apblett C. Impedimetric and optical interrogation of single cells in a microfluidic device for real-time viability and chemical response assessment. Biosensors and Bioelectronics. 2008;23(6):845–851. doi: 10.1016/j.bios.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Jang LS, Wang MH. Microfluidic device for cell capture and impedance measurement. Biomedical microdevices. 2007 doi: 10.1007/s10544-007-9084-0. Retrieved from http://link.springer.com/article/10.1007/s10544-007-9084-0. [DOI] [PubMed]
- Javanmard M, Davis RW. Coded corrugated microfluidic sidewalls for code division multiplexing. IEEE Sensors Journal. 2013 Retrieved from http://ieeexplore.ieee.org/abstract/document/6417941/
- Jo M, Guldiken R. Active density-based separation using standing surface acoustic waves. Sensors and Actuators A: Physical. 2012;187:22–28. doi: 10.1016/j.sna.2012.08.020. [DOI] [Google Scholar]
- Kaneta T, Makihara J, Imasaka T. An “Optical Channel”: A Technique for the Evaluation of Biological Cell Elasticity. Analytical Chemistry. 2001;73(24):5791–5795. doi: 10.1021/ac010441g. [DOI] [PubMed] [Google Scholar]
- Kang G, Yoo S, Kim HI, Lee JH. Differentiation between Normal and Cancerous Cells at the Single Cell Level Using 3-D Electrode Electrical Impedance Spectroscopy. IEEE Sensors Journal. 2012;12(5):1084–1089. doi: 10.1109/JSEN.2011.2167227. [DOI] [Google Scholar]
- Karabacak N, Spuhler P, Fachin F, Lim E, Pai V, Ozkumur E, … Toner M. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nature Protocols. 2014;9(3):694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellman M, Rivest F, Pechacek A, Sohn LL, Lustig M. Barker-coded node-pore resistive pulse sensing with built-in coincidence correction. 2017 doi: 10.1109/ICASSP.2017.7952317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Han S, Lei A, Miyano M, VMR, … Sohn LL. Characterizing cellular mechanical phenotypes with mechano-node-pore sensing. Nature Microsystems & Nanoengineering. 2018 doi: 10.1038/micronano.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KTT, Lee HW, Lee HOO, Song HJ, Jeong DEaE, Shin S, … Park W-YY. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome biology. 2016;17:80. doi: 10.1186/s13059-016-0945-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb T, Albert S, Haug M, Whyte G. Optofluidic rotation of living cells for single-cell tomography. Journal of Biophotonics. 2015;8(3):239–246. doi: 10.1002/jbio.201300196. [DOI] [PubMed] [Google Scholar]
- Kumar S, Kumar S, Ali M, Anand P, Agrawal V, John R, … Malhotra B. Microfluidic-integrated biosensors: Prospects for point-of-care diagnostics. Biotechnology Journal. 2013;8(11):1267–1279. doi: 10.1002/biot.201200386. [DOI] [PubMed] [Google Scholar]
- Küttel C, Nascimento E, Demierre N, Silva T. Label-free detection of Babesia bovis infected red blood cells using impedance spectroscopy on a microfabricated flow cytometer. Acta tropica. 2007 doi: 10.1016/j.actatropica.2007.03.002. Retrieved from http://www.sciencedirect.com/science/article/pii/S0001706X07000794. [DOI] [PubMed]
- Lee G, Lim C. Biomechanics approaches to studying human diseases. Trends in Biotechnology. 2007;25(3):111–118. doi: 10.1016/j.tibtech.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Leibacher I, Reichert P, Dual J. Microfluidic droplet handling by bulk acoustic wave (BAW) acoustophoresis. Lab on a Chip. 2015;15(13):2896–2905. doi: 10.1039/C5LC00083A. [DOI] [PubMed] [Google Scholar]
- Lekka M, Laidler P, Gil D, Lekki J, Stachura Z, Hrynkiewicz A. Elasticity of normal and cancerous human bladder cells studied by scanning force microscopy. European Biophysics Journal. 1999;28(4):312–316. doi: 10.1007/s002490050213. [DOI] [PubMed] [Google Scholar]
- Li P, Mao Z, Peng Z, Zhou L, Chen Y, Huang PH, … Huang T. Acoustic separation of circulating tumor cells. Proceedings of the National Academy of Sciences. 2015;112(16):4970–4975. doi: 10.1073/pnas.1504484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lee G, Ong C, Lim C. 13th International Conference on Biomedical Engineering; 2009. pp. 2122–2125. [DOI] [Google Scholar]
- Lincoln B, Erickson H, Schinkinger S, Wottawah F, Mitchell D, Ulvick S, … Guck J. Deformability-based flow cytometry. Cytometry Part A. 2004;59A(2):203–209. doi: 10.1002/cyto.a.20050. [DOI] [PubMed] [Google Scholar]
- Loutherback K, D’Silva J, Liu L, Wu A, Austin R, Sturm J. Deterministic separation of cancer cells from blood at 10 mL/min. AIP Advances. 2012;2(4):042107. doi: 10.1063/1.4758131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumacyte. LumaCyte Radiance 2018 [Google Scholar]
- Malleo D, Nevill JT, Lee LP, Morgan H. Continuous differential impedance spectroscopy of single cells. Microfluidics and nanofluidics. 2010 doi: 10.1007/s10404-009-0534-2. Retrieved from http://link.springer.com/article/10.1007/s10404-009-0534-2. [DOI] [PMC free article] [PubMed]
- Mansor M, Takeuchi M, Nakajima M, Hasegawa Y, Ahmad M. Electrical Impedance Spectroscopy for Detection of Cells in Suspensions Using Microfluidic Device with Integrated Microneedles. Applied Sciences. 2017;7(2):170. doi: 10.3390/app7020170. [DOI] [Google Scholar]
- Martel J, Toner M. Particle Focusing in Curved Microfluidic Channels. Scientific Reports. 2013;3(1):3340. doi: 10.1038/srep03340. [DOI] [Google Scholar]
- Masaeli M, Gupta D, O’Byrne S, Tse H, Gossett D, Tseng P, … Carlo D. Multiparameter mechanical and morphometric screening of cells. Scientific Reports. 2016;6(1):37863. doi: 10.1038/srep37863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietke A, Otto O, Girardo S, Rosendahl P, Taubenberger A, Golfier S, … Fischer-Friedrich E. Extracting Cell Stiffness from Real-Time Deformability Cytometry: Theory and Experiment. Biophysical Journal. 2015;109(10):2023–2036. doi: 10.1016/j.bpj.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Doyle G, Terstappen L. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. Journal of Oncology. 2010;2010:1–8. doi: 10.1155/2010/617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millipore Sigma. Scepter 2.0 Cell Counter. 2018. [Google Scholar]
- Morgan H, Sun T, Holmes D, Gawad S, Green N. Single cell dielectric spectroscopy. Journal of Physics D: Applied Physics. 2007;40(1):61. doi: 10.1088/0022-3727/40/1/S10. [DOI] [Google Scholar]
- Nam J, Lim H, Kim D, Shin S. Separation of platelets from whole blood using standing surface acoustic waves in a microchannel. Lab on a Chip. 2011;11(19):3361–3364. doi: 10.1039/C1LC20346K. [DOI] [PubMed] [Google Scholar]
- Otto O, Rosendahl P, Mietke A, Golfier S, Herold C, Klaue D, … Guck J. Real-time deformability cytometry: on-the-fly cell mechanical phenotyping. Nature Methods. 2015;12(3):199–202. doi: 10.1038/nmeth.3281. [DOI] [PubMed] [Google Scholar]
- Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, … Toner M. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Science translational medicine. 2013;5(179):179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Chen Y, Li P, Mao Z, Huang PH, Rufo J, … Huang T. A high-throughput acoustic cell sorter. Lab on a Chip. 2015;15(19):3870–3879. doi: 10.1039/C5LC00706B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier C, Pao E, Che J, Liu H, Lemaire C, Matsumoto M, … Sollier-Christen E. Label-free isolation of prostate circulating tumor cells using Vortex microfluidic technology. npj Precision Oncology. 2017;1(1):15. doi: 10.1038/s41698-017-0015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivest FR, Pechacek AP, Park R, Goodman K, Cho N, Lustig M, Sohn L. Toward real-time cell detection and characterization using Barker-coded Node-Pore Sensing. μTAS Conf Proc 2015 [Google Scholar]
- Saleh O. A Novel Resistive Pulse Sensor for Biological Measurements. Princeton University; 2003. [Google Scholar]
- Saleh O, Sohn L. Quantitative sensing of nanoscale colloids using a microchip Coulter counter. Review of Scientific Instruments. 2001;72(12):4449–4451. doi: 10.1063/1.1419224. [DOI] [Google Scholar]
- Saleh O, Sohn L. An Artificial Nanopore for Molecular Sensing. Nano Letters. 2003a;3(1):37–38. doi: 10.1021/nl0255202. [DOI] [Google Scholar]
- Saleh O, Sohn L. Direct detection of antibody–antigen binding using an on-chip artificial pore. Proceedings of the National Academy of Sciences. 2003b;100(3):820–824. doi: 10.1073/pnas.0337563100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarioglu A, Aceto N, Kojic N, Donaldson M, Zeinali M, Hamza B, … Toner M. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nature Methods. 2015;12(7):685–691. doi: 10.1038/nmeth.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake D, Ebi H, Oku N, Matsuda K, Takao H, Ashiki M, Ishida M. A sensor for blood cell counter using MEMS technology. Sensors and Actuators B: Chemical. 2002;83(1-3):77–81. doi: 10.1016/S0925-4005(01)01045-0. [DOI] [Google Scholar]
- Shi J, Huang H, Stratton Z, Huang Y, Huang T. Continuous particle separation in a microfluidic channel via standing surface acoustic waves (SSAW) Lab on a Chip. 2009;9(23):3354–3359. doi: 10.1039/B915113C. [DOI] [PubMed] [Google Scholar]
- Sohn L, Saleh O, Facer G, Beavis A, Allan R, Notterman D. Capacitance cytometry: Measuring biological cells one by one. Proceedings of the National Academy of Sciences. 2000;97(20):10687–10690. doi: 10.1073/pnas.200361297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Wang Y, Rosano JM, Prabhakarpandian B. A microfluidic impedance flow cytometer for identification of differentiation state of stem cells. Lab on a Chip. 2013 doi: 10.1039/C3LC41321G. [DOI] [PubMed] [Google Scholar]
- Sun T, Gawad S, Bernabini C, Green N, Morgan H. Broadband single cell impedance spectroscopy using maximum length sequences: theoretical analysis and practical considerations. Measurement Science and Technology. 2007;18(9):2859. doi: 10.1088/0957-0233/18/9/015. [DOI] [Google Scholar]
- Swaminathan V, Mythreye K, O’Brien T, Berchuck A, Blobe G, Superfine R. Mechanical Stiffness Grades Metastatic Potential in Patient Tumor Cells and in Cancer Cell Lines. Cancer Research. 2011;71(15):5075–5080. doi: 10.1158/0008-5472.CAN-11-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ThermoFisher invitrogen. Attune NxT Acoustic Focusing Flow Cytometry 2018 [Google Scholar]
- Tu J, Qiao Y, Xu M, Li J, Liang F, Duan M, … Lu Z. A cell sorting and trapping microfluidic device with an interdigital channel. AIP Advances. 2016;6(12):125042. doi: 10.1063/1.4972794. [DOI] [Google Scholar]
- Valero A, Braschler T, Renaud P. A unified approach to dielectric single cell analysis: Impedance and dielectrophoretic force spectroscopy. Lab on a Chip. 2010;10(17):2216–2225. doi: 10.1039/C003982A. [DOI] [PubMed] [Google Scholar]
- Van Berkel C, Gwyer JD, Deane S, Green N. Integrated systems for rapid point of care (PoC) blood cell analysis. Lab on a Chip. 2011 doi: 10.1039/C0LC00587H. [DOI] [PubMed] [Google Scholar]
- Vortex Biosciences. VTX-1 Liquid Biopsy System 2018 [Google Scholar]
- Watkins NN, Hassan U, Damhorst G. Microfluidic CD4+ and CD8+ T lymphocyte counters for point-of-care HIV diagnostics using whole blood. Science translational …. 2013 doi: 10.1126/scitranslmed.3006870. Retrieved from http://stm.sciencemag.org/content/5/214/214ra170.short. [DOI] [PubMed]
- Wixforth A, Strobl C, Gauer C, Toegl A, Scriba J, Guttenberg Z. Acoustic manipulation of small droplets. Analytical and Bioanalytical Chemistry. 2004;379(7–8):982–991. doi: 10.1007/s00216-004-2693-z. [DOI] [PubMed] [Google Scholar]
- Xavier M, Rosendahl P, Herbig M, Kräter M, Spencer D, Bornhäuser M, … Otto O. Mechanical phenotyping of primary human skeletal stem cells in heterogeneous populations by real-time deformability cytometry. Integrative Biology. 2016;8(5):616–623. doi: 10.1039/C5IB00304K. [DOI] [PubMed] [Google Scholar]
- Xi B, Yu N, Wang X, Xu X, Abassi Y. The application of cell-based label-free technology in drug discovery. Biotechnology Journal. 2008;3(4):484–495. doi: 10.1002/biot.200800020. [DOI] [PubMed] [Google Scholar]
- Yeo T, Tan S, Lim C, Lau D, Chua Y, Krisna S, … Lim C. Microfluidic enrichment for the single cell analysis of circulating tumor cells. Scientific Reports. 2016;6(1):srep22076. doi: 10.1038/srep22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Sadok A, Sailem H, McCarthy A, Xia X, Li F, … Bakal C. A screen for morphological complexity identifies regulators of switch-like transitions between discrete cell shapes. Nature Cell Biology. 2013;15(7):860–871. doi: 10.1038/ncb2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chen D, Li H, Luo Y, Deng B, Huang SB. A microfluidic system enabling continuous characterization of specific membrane capacitance and cytoplasm conductivity of single cells in suspension. Biosensors and …. 2013 doi: 10.1016/j.bios.2012.12.035. Retrieved from http://www.sciencedirect.com/science/article/pii/S0956566312008913. [DOI] [PubMed]
- Zhao Y, Chen D, Luo Y, Li H, Deng B, Huang SB. A microfluidic system for cell type classification based on cellular size-independent electrical properties. Lab on a Chip. 2013 doi: 10.1039/C3LC41361F. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhao XT, Chen DY, Luo YN, Jiang M, Wei C, … Chen J. Tumor cell characterization and classification based on cellular specific membrane capacitance and cytoplasm conductivity. Biosensors and Bioelectronics. 2014;57:245–253. doi: 10.1016/j.bios.2014.02.026. [DOI] [PubMed] [Google Scholar]
- Zhe J, Jagtiani A, Dutta P, Hu J, Carletta J. A micromachined high throughput Coulter counter for bioparticle detection and counting. Journal of Micromechanics and Microengineering. 2007;17(2):304. doi: 10.1088/0960-1317/17/2/017. [DOI] [Google Scholar]
- Zhou J, Papautsky I. Fundamentals of inertial focusing in microchannels. Lab on a Chip. 2013;13(6):1121–1132. doi: 10.1039/C2LC41248A. [DOI] [PubMed] [Google Scholar]
- Zhu L, Zhang Q, Feng H, Ang S, Chau F, Liu WT. Filter-based microfluidic device as a platform for immunofluorescent assay of microbial cells. Lab on a Chip. 2004;4(4):337–341. doi: 10.1039/B401834F. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Frey O, Haandbaek N, Franke F, Rudolf F, Hierlemann A. Time-lapse electrical impedance spectroscopy for monitoring the cell cycle of single immobilized S. pombe cells. Scientific Reports. 2015;5(1):17180. doi: 10.1038/srep17180. [DOI] [PMC free article] [PubMed] [Google Scholar]