Abstract
To investigate the association between single nucleotide polymorphisms (SNPs) of A disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) 14 (ADAMTS14) gene and susceptibility to knee osteoarthritis (KOA) in Chinese Han population. Using a case–control design, we enrolled 346 KOA patients and 480 healthy controls. Peripheral blood samples were extracted from each subject. Genotype was determined by sequencing PCR products. The genotype frequencies between cases and controls were compared. The genotype distribution was in accordance with Hardy–Weinberg equilibrium. The minor G allele in case group was significantly higher than in the control group (21.4 compared with 8.8%, P=0.000, odds ratio (OR) = 1.71 (95% confidence interval (CI): 1.39–2.11). The GG genotype and the GG/AG combination were more common in the osteoarthritis (OA) group than in the control group. Compared with AA genotype, the GG (OR = 3.09, 95%CI: 2.01–4.75), AG (OR = 2.55, 95%CI: 1.64–3.96), and GG/AG (OR = 1.57, 95%CI: 1.19–2.07) increased the risk of OA. Multiple logistic confirmed the findings by adjusting some potential factors. Subgroup analysis indicated that the ras4747096 was still significantly associated with KOA. There were no significant differences in allele frequency or genotypes frequency for erythrocyte sedimentation rate and C-reaction protein in OA patients (P>0.05). ADAMTS14 gene polymorphism was associated with KOA, and the GG genotype increased the risk of KOA in Chinese Han population. The ADAMTS14 may be a diagnostic marker and therapeutic target for KOA treatment. The future study should explore the specific molecular mechanism.
Keywords: ADAMTS14, gene polymorphism, Hardy-Weinberg, knee osteoarthritis
Introduction
Knee osteoarthritis (KOA) is a serious rheumatic disease characterized by articular cartilage damage and joint space narrowing. In severe cases, when the knee completely loses cartilage, bone and soft tissue structure around the joint will start to change, which could lead to joint pain, swelling, and disability [1,2]. In the United States, symptomatic KOA occurs in approximately 10% of adults aged 60 or above. According to recent statistics, there are 9.3 million adults who are suffering from osteoarthritis (OA) of the knee in the United States. The overall number of KOA is as high as 350 million [3,4]. One in every six persons is suffering from the disease in Asia. With the development in ageing, KOA patients has increased every year, and estimated at approximately 200 million. The ratio of male to female is 1:2. As a result of the ageing process, the number of people with OA of the knee is expected to increase in the coming decades [5]. Pathogenesis of OA has not yet been fully elucidated up to now. It was widely acknowledged that that OA is related to gene and environment factors [6]. It was suggested that the gene accounted for more than 60% of diseases [7]. The human leukocyte antigen class II was the best-known gene in OA, accounting for approximately one-third of OA susceptibility [8]. More gene factors were waiting to be found.
The single nucleotide polymorphism (SNP) is more than 90% of human gene polymorphisms, which is the most common and stable gene variant in the human DNA strands [9]. The coding SNP consisted of synonymous coding SNP and non-synonymous coding SNP. The A disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) 14 (ADAMTS14) gene rs4747096 is a non-synonymous coding SNP, located on chromosome 10q22.1 [10]. This SNP can change the sequence of amino acid and affects the biological properties. It was known that the overloading, abnormal expression of protease and cytokines, and other factors could lead to imbalance of extracellular matrix decomposition and synthesis metabolism of cartilage, and finally lead to cartilage destruction [11]. ADAMTS play an important role in the degradation of cartilage extracellular matrix [12]. Colige et al. [13] reported that ADAMTS14 was involved in the formation process of collagen fiber II that was one of main components of joint cartilage extracellular matrix. The abnormal metabolism of collagen fiber II decreased the mechanical strength of joint cartilage while this process was one of the important factors affecting joint arthritis. Rodriguez-Lopez et al. [14] found that the G allele in rs4747096 was associated with symptomatic hand OA in Caucasian women requiring total knee replacement because of OA. Currently, there is no report about the relationship between ADAMTS14 gene rs4747096 and KOA. The present study investigates the association between SNPs of ADAMTS14 gene rs4747096 and KOA in Chinese Han population.
Materials and methods
Study population
We enrolled 346 KOA patients who were diagnosed from 2013 to 2017 in the People’s Hospital of Nanzhao County and the Center Hospital of Nanyang City. The diagnostic criteria of KOA were in accordance with the 2010 American College of Rheumatology/European League against Rheumatism criteria [15]. Patients with severe cardiovascular diseases, severe liver and kidney dysfunction, malignant tumor, and other autoimmune diseases were excluded. Grades 3–4 of radiographic signs of OA according to the Kellgren–Lawrence grading system [16]. The healthy control group was from outpatients of the two hospitals above without the history of OA and autoimmune diseases during the same period. The sample size was calculated using Quanto 1.2.4 and followed by the conditions: α = 0.05, β = 0.10, expected odds ratio (OR) = 1.8, the calculated sample size was 248 in the case group and control group, respectively. The present sample size was enough. We have added this description in the methods. The present study was approved by the institutional review board of Center Hospital of Nanyang City. Written informed consent was received from all study subjects.
Sample collection and DNA extraction
The general data of case group and two group were obtained via the questionnaire, which was conducted by the trained investigators. The general data included age, gender, smoking, body mass index (BMI), and drinking. Anti-cyclic citrullinated peptide antibodies (ACCP) > 5 RU/ml, antiperinuclear factor (APF) > 5 cells under the microscope, RF (rheumatoid factor) > 20 were positive, and erythrocyte sedimentation rate (ESR) ≥ 20 and C-reactive protein (CRP) > 8 were judged as abnormal [17].
The 5-ml peripheral vein blood was extracted from each study subject. The blood samples were put in EDTA anticoagulant tubes, stored in the refrigerator at −80°C until use. The genomic DNA extraction was completed via Blood DNA Master Kit (QIAamp DNA Blood Mini Kits, Qiagen, Germany). The extracted DNA was dissolved in TE buffer (10 mM Tris, 1 mM EDTA; pH = 7.8), quantitated by measuring the absorbance at 260 nm, and then stored at −20°C for genotyping.
SNP genotyping
The primer was designed by Generunner 6.2.07 Beta software and synthesized by Beijing Liuhe Huada Gene Company. The genotype was completed by the PCR-restriction fragment length polymorphism (PCR-RFLP). The forward primer sequence: 5′-TGTGCAGGACCAACGCCAACAG-3′ and the reverse primer sequence: 5′-GGAATTGCAGGTAACGGCTCATG-3′. The reaction system included 12 μl 2× Es Taq MasterMix, 2 μl forward and reverse and primer with 10 μl mol/l, 2 μl DNA templates. The PCR program consisted of the following steps: initial denaturation step at 94°C for 2 min, followed by 30 cycles: denaturation at 98°C for 10 s, annealing at 61°C for 30 s, extension at 72°C for 30 s, and extension again at 72°C for 2 min, and stored at 4°C. The PCR product was incubated overnight at 37°C with restriction enzyme. Then the mixtures were electrophoresed and visualized. The expected fragment was 196 bp in AA, 196, 158, and 38 bp in AG, and 158 and 38 bp in GG.
Statistical analysis
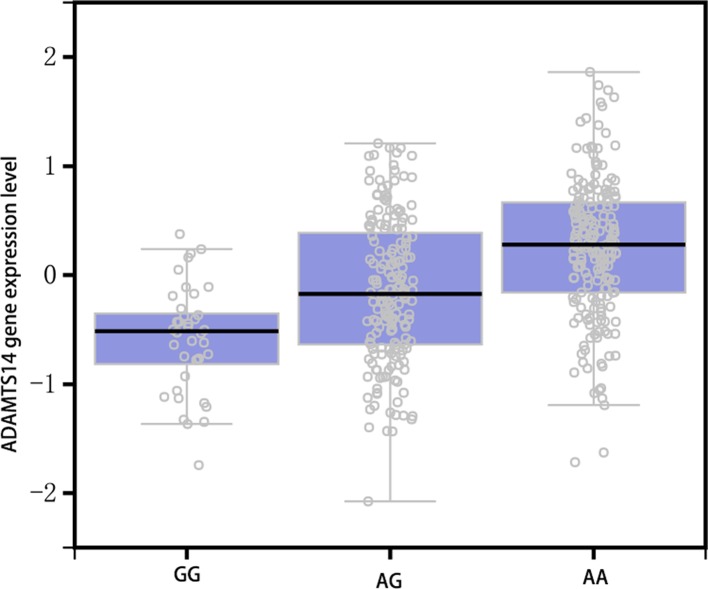
The continuous data were expressed using means ± S.D., and the comparison between case and control groups was performed by Student’s t test. The categorical data were expressed using count and percent, and Chi-square test was applied. The Hardy–Weinberg equilibrium test was conducted in the case and control groups. The risk of gene for OA was assessed by calculating OR with 95% confidence interval (95%CI). The stratification analysis was conducted in different ESR, age, gender, smoking, drinking, BMI, CRP. The adjusted ORs and their 95%CIs were also calculated, including age, gender, smoking, drinking, and BMI. We also used the Genotype Tissue Expression database to assess the correlation between SNP and ADAMTS14 (https://www.gtexportal.org/home/; Figure 1). All analyses were completed on SPSS 23.0, and P<0.05 was considered at significance level.
Figure 1. The GTEx dataset revealed a significant eQTL association between the rs4747096 genotype and expression of the ADAMTS14 in peripheral whole blood cells.
Results
The general characteristics were presented in Table 1; 49.1% of OA patients were males and the ratio of control was 51.7%. The mean age of case group and control group were 57.1 and 56.6 years, respectively. The rates of smoking and drinking were 3.29 and 28.9% in the case group and 39.2 and 31.7% in the control group. The mean courses of disease were 1.2 for case group and 1.1 for control group. There were no significant differences in gender ratio (P=0.472), age (P=0.278), BMI (P=0.534), course of disease (P=0.056), smoking (P=0.067), and drinking (P=0.394). The positive rates of ACCP, AKA, APF, and RF were 73.1, 36.7, 44.5, and 16.8% in the case group. The ratios of ESR > 20 and CRP > 8 were 52.0 and 66.2%, respectively.
Table 1. Comparison of general characteristics between case and control groups.
| Factors | Case group (n=346) | Control group (n=480) | χ2/t | P |
|---|---|---|---|---|
| Gender | 0.516 | 0.472 | ||
| Male | 170 (49.1%) | 248 (51.7) | ||
| Female | 176 (50.9%) | 232 (48.3) | ||
| Age | 57.1 ± 7.0 | 56.6 ± 7.0 | −1.086 | 0.278 |
| BMI | 23.6 ± 3.0 | 23.8 ± 3.1 | 0.622 | 0.534 |
| Course of disease | 1.2 ± 0.8 | 1.1 ± 0.7 | 1.907 | 0.056 |
| Smoking | 3.352 | 0.067 | ||
| Yes | 114 (32.9%) | 188 (39.2%) | ||
| No | 232 (67.1%) | 292 (60.8%) | ||
| Drinking | 0.725 | 0.394 | ||
| Yes | 100 (28.9%) | 152 (31.7%) | ||
| No | 246 (71.1%) | 328 (68.3%) | ||
| Kellgren–Lawrence grading | - | |||
| I/II | 206 (59.5%) | |||
| III/IV | 140 (40.5%) | |||
| ESR ≥ 20 | 180 (52.0%) | - | ||
| CRP > 8 mg/l | 229 (66.2%) | - | ||
| rs4747096 | 28.110 | 0.000 | ||
| AA | 146 (42.2%) | 256 (53.3%) | ||
| AG | 126 (36.4%) | 182 (37.9%) | ||
| GG | 74 (21.4%) | 42 (8.8%) |
Association of ADAMTS14 rs4747096 polymorphism with OA
Eight hundred and twenty-six samples were genotyped and the concordance rate of genotype was 100%. The distribution of genotype frequencies was 42.2% for AA, 36.4% for AG, and 21.4% for GG in the case group, and 53.3% for AA, 37.9% for AG, and 8.8% for GG. The Hardy–Weinberg equilibrium test indicated no significant differences in the case and in the control groups (P>0.05). Significant difference genotype differences were observed between case group and control group (χ2 = 28.110, P=0.000). The minor G allele in case group was significantly higher than in the control group (21.4 compared with 8.8%, P=0.000, OR = 1.71, 95%CI: 1.39–2.11). Significant differences in the distribution of genotypes were observed. The GG genotype and the GG/AG combination were more common in the OA group than in the control group. Compared with AA genotype, the GG (OR = 3.09, 95%CI: 2.01–4.75), AG (OR = 2.55, 95%CI: 1.64–3.96), and GG/AG (OR = 1.57, 95%CI: 1.19–2.07) increased the risk of OA. The codominant model also show the similar results (OR = 2.84, 95%CI: 1.89–4.27, P=0.000). We also conducted the multiple logistic to evaluate the relationship between rs4747096 and OA risk through adjusting some potential factors: age, gender, smoking, drinking, and BMI. The results were still significant in each model. The results were presented in Table 2.
Table 2. Logistic regression analysis of associations between ADAMTS14/rs4747096 gene polymorphism and KOA.
| Genotype | Cases | Control | OR (95% CI) | P | aOR (95%CI) | aP | ||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | |||||
| GG compared with AA | 74/146 | 21.4/42.2 | 42/256 | 8.8/53.3 | 3.09 (2.01–4.75) | 0.000 | 3.26 (2.11–5.04) | 0.000 |
| GG compared with AG | 74/126 | 21.4/36.4 | 42/182 | 8.8/37.9 | 2.55 (1.64–3.96) | 0.000 | 2.57 (1.65–4.02) | 0.000 |
| GG/AG compared with AA | 200/146 | 57.8/42.2 | 224/256 | 46.7/53.3 | 1.57 (1.19–2.07) | 0.002 | 1.60 (1.21–2.12) | 0.001 |
| GG compared with AG/AA | 74/272 | 21.4/78.6 | 42/438 | 8.8/91.2 | 2.84 (1.89–4.27) | 0.000 | 2.98 (1.98–4.51) | 0.000 |
| G compared with A | 274/418 | 39.6/60.4 | 266/694 | 27.7/72.3 | 1.71 (1.39–2.11) | 0.000 | - | - |
Abbreviation: aP, adjusted P value; aOR, adjust OR: adjust age, gender, smoking, drinking, and BMI.
Stratified analyses
We first conducted subgroup analyses in different age, gender, weight, smoking, and drinking. As shown in Table 3, significant differences were observed in AA compared with GG, AG compared with GG, AA/AG compared with GG, and A compared with G for male, and all models showed significant difference for female (P<0.05). For age, the models (AA compared with GG, AG compared with GG, A compared with G) showed significant difference (P<0.05). For smoking, there were significant differences between case group and control group. Patients with drinking and BMI ≥ 24 tend to show significant relationship between rs4747096 and OA. We further analyzed the relationship between these parameters and genotypes of rs4747096. There was no significant difference in allele frequency or genotypes frequency for these parameters in OA patients (P>0.05). Although the GG genotype was slightly higher in the CRP positive group than in CRP negative group, no significant difference was observed (P=0.376, Table 4).
Table 3. Subgroup analyses between ADAMTS14/rs4747096 gene polymorphism and the risk of KOA.
| Variables | rs4747096 (Case/control) | GG compared with AA | GG compared with AG | GG/AG compared with AA | GG compared with AG/AA | G compared with A | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | P | |
| Sex | |||||||||||||
| Male | 73/132 | 63/96 | 34/20 | 3.07 (1.65–5.73) | 0.020 | 2.59 (1.37–4.89) | 0.003 | 1.51 (1.02–2.24) | 0.039 | 2.85 (1.58–2.15) | 0.001 | 1.66 (1.24–2.23) | 0.001 |
| Female | 73/124 | 63/86 | 40/22 | 3.09 (1.70–5.60) | 0.000 | 2.48 (1.34–4.58) | 0.004 | 1.62 (1.09–2.41) | 0.017 | 2.80 (1.60–4.93) | 0.000 | 1.76 (1.31–2.36) | 0.000 |
| Age (years) | |||||||||||||
| <60 | 78/156 | 82/118 | 50/28 | 3.57 (2.09–6.11) | 0.000 | 2.57 (1.50–4.42) | 0.001 | 1.28 (0.82–2.01) | 0.277 | 3.05 (1.85–5.05) | 0.000 | 1.89 (1.46–2.45) | 0.000 |
| ≥60 | 68/100 | 40/64 | 24/14 | 2.52 (1.22–5.22) | 0.013 | 2.49 (1.16–5.35) | 0.019 | 1.81 (1.26–2.59) | 0.001 | 2.51 (1.25–5.06) | 0.010 | 1.43 (1.01–2.03) | 0.042 |
| Smoking | |||||||||||||
| Yes | 42/108 | 41/60 | 31/10 | 2.78 (1.57–4.93) | 0.000 | 2.81 (1.57–5.03) | 0.001 | 2.31 (1.43–3.73) | 0.001 | 2.79 (1.62–4.82) | 0.000 | 2.89 (2.01–4.15) | 0.000 |
| No | 104/148 | 85/122 | 43/22 | 3.99 (2.05–7.76) | 0.000 | 2.27 (1.14–4.51) | 0.020 | 1.26 (0.90–1.79) | 0.183 | 3.14 (1.69–5.84) | 0.000 | 1.47 (1.13–1.91) | 0.004 |
| Drinking | |||||||||||||
| Yes | 40/80 | 40/52 | 20/10 | 4.08 (2.35–7.07) | 0.000 | 3.71 (2.11–6.53) | 0.000 | 1.53 (1.10–2.13) | 0.012 | 3.91 (2.31–6.63) | 0.000 | 1.77 (1.38–2.28) | 0.000 |
| No | 106/176 | 86/130 | 54/22 | 2.00 (0.96–4.14) | 0.062 | 1.30 (0.62–2.73) | 0.490 | 1.67 (1.00–2.78) | 0.050 | 1.65 (0.84–3.25) | 0.148 | 1.47 (0.98–2.22) | 0.063 |
| BMI | |||||||||||||
| ≥24 | 63/98 | 55/74 | 27/18 | 3.72 (2.13–6.52) | 0.000 | 2.97 (1.68–5.30) | 0.000 | 1.70 (1.18–2.45) | 0.004 | 2.19 (1.15–4.15) | 0.017 | 0.93 (0.66–1.30) | 0.677 |
| <24 | 83/158 | 71/108 | 47/48 | 2.33 (1.19–4.58) | 0.014 | 2.02 (1.01–4.03) | 0.046 | 1.39 (0.90–2.14) | 0.140 | 3.38 (1.99–5.75) | 0.000 | 1.18 (0.90–1.55) | 0.219 |
Table 4. Comparison of genotypic and allelic frequencies of ADAMTS14/rs4747096 polymorphism amongst KOA subgroups stratified.
| Cases | GG | AG | AA | G | A |
|---|---|---|---|---|---|
| ESR ≥ 20 | 44 (24.4) | 58 (32.2) | 78 (43.3) | 146 (40.6) | 214 (59.4) |
| ESR < 20 | 30 (5.9) | 68 (41.0) | 68 (41.0) | 128 (38.6) | 204 (61.4) |
| P | 0.395 | 0.224 | 0.591 | ||
| OR (95%CI) | 1.28 (0.73–2.25) | 0.74 (0.46–1.20) | 1.00 | 1.09 (0.80–1.48) | 1.00 |
| CRP > 8 mg/l | 54 (23.6) | 77 (33.6) | 98 (42.8) | 185 (40.4) | 273 (59.6) |
| CRP ≤ 8 mg/l | 20 (17.1) | 49 (41.9) | 48 (41.0) | 89 (38.0) | 145 (62.0) |
| P | 0.376 | 0.302 | 0.548 | ||
| OR (95%CI) | 1.32 (0.71–2.45) | 0.77 (0.47–1.27) | 1.00 | 1.10 (0.80–1.53) | 1.00 |
| Grade I/II | 33 (21.4) | 52 (33.8) | 69 (44.8) | 118 (38.3) | 190 (61.7) |
| Grade III/IV | 41 (21.4) | 74 (38.5) | 66 (50.8) | 156 (43.1) | 206 (56.9) |
| P | 0.708 | 0.322 | 0.209 | ||
| OR (95%CI) | 0.90 (0.51–1.58) | 0.78 (0.49–1.27) | 1.00 | 0.82 (0.60–1.12) | 1.00 |
Discussion
It is well recognized that the susceptibility of KOA is influenced by genetic and environmental factors. In the past decades, a lot of studies had explored the associations between KOA and different candidate genes in different ethnic backgrounds [18,19]. However, results of most susceptibility genes identified have not been replicated in subpopulations different from population used in the previous study. Poonpet et al. [20] found the SNP rs4747096 in ADAMTS14 was associated with KOA in female Thai patients. After that, studies about association between ADAMTS14 and OA are few. To our best knowledge, this is the first study about association between ADAMTS14 and KOA in Chinese population, including males and females. Our study with more sample size indicated that the ADAMTS14 rs4747096 gene polymorphism was associated with KOA in Chinese Han population.
One of the primary pathology symptoms of OA is the degeneration of articular cartilage, characterized by degradation and synthesis imbalance of chondrocytes, extracellular matrix, and subchondral bone that was usually caused by mechanical and biological factors. The articular cartilage of the knee is fibrous cartilage, and collagen in the cartilage matrix accounts for 50–60% (mainly type II collagen) [21]. The type II collagen is essential to maintain the mechanical strength of cartilage because the collagen can hold on the stress of cartilage. The destruction of collagen fibers in superficial articular cartilage is an important histopathological signal of early stage OA. The experiments in vivo have suggested that the diameter of collagen fibers in early-stage OA articular cartilage increases, and the changes in the microstructure of usually occur earlier than subchondral bone destruction [22].
ADAMTS is a group of secretory proteases with the metalloproteinase domain, depolymerization domain, and platelet reactive protein domain, which is involved in the degradation of collagen polysaccharide in cartilage matrix. Studies on gene polymorphism of ADAMTS family member have become the focus of the pathogenesis of OA. Li et al. [23] found that the serum ADAMTSs level in the early KOA was significantly higher than that in the healthy people. However, the only serum ADAMTS5 level of advanced patients were significantly higher than that in the healthy people and speculated that the ADAMTS4 might be a serological marker of early KOA [23]. Kumar et al. [24] found that the OA severity mice with ADAMTS4/5 knockout was more significantly alleviated than that in the wild-type. Miller et al. [25] reported that the application of ADAMTS5 antibody could slow down the cartilage damage in mice with KOA. Gok et al. [26] reported that patients with >20 repeated sequences in ADAMTS9 gene had more serious KOA. The ADAMTS1/4/5/8/9/15 can promote the degradation of proteoglycan in articular cartilage while ADAMTS7/9/15 were involved in the degradation of oligomeric proteins in cartilage matrix [26]. These results indicated that ADAMTS gene was associated with OA.
ADAMTS14 gene is located on chromosome 10q222.1. At present, the function of ADAMTS14 is not fully clear. Since ADAMTS14 and ADAMTS3 genes (63%) are homologous, Fernandes et al. [27] speculated that ADAMTS14 and ADAMTS3 have similar functions, which can crack the N-terminal anterior peptide of collagen monomer, promote the aggregation of type II collagen monomer into type II collagen fiber, and participate in the repair process after cartilage damage. The protein encoded by ADAMTS14 gene consists of 1223 amino acids, which are composed of signal peptide, restructure domain, protease structural domain, poly structural domain, four thrombin-sensitive protein structural domains, and carboxyl terminal structural domain. The four thrombin-sensitive protein domains were separated by cysteine-rich amino acid fragments. The rs4747096 (A>G) G allele is the ancestral allele, but A allele accounts for larger frequency in European/Asian/African populations [28]. So G allele may be the dominant gene in the natural selection process. Changes in the sequence of bases result in the conversion of amino acids from glutamate (GAA) into GGA in the carboxyl terminal structure of the encoded protein molecule. We hypothesized that the polymorphism of ADAMTS14 gene rs747096 may cause the abnormal maturation process of type II collagen and lead to cartilage destruction, but this process still needs further research and verification. Results from GETs database indicated the GG genotype had lower level of ADAMTS gene expression, which suggested that the mutant gene cannot be expressed. The GG genotype made the ADAMTS14 expression level decreased. This results further indicated the close relationship existed between ADAMTS14 and KOA. We compared the genotype frequency of ADAMTS14 rs4747069 in Chinese Han population and found there was significant difference in genotype frequency between KOA patients and control group. The GG genotype frequency in the case group was significantly higher than that in the control group. Compared the control, the case have higher risk of KOA (OA = 3.09, 95%CI: 2.01–4.75). This result was different from previous two studies. Previous studies reported that the AA genotype was a risk gene. Poonpet et al. [20] reported a nearly significant association was found between the AG genotype and KOA in female patients (OR = 2.65; 95% CI = 0.99-7.19; P = 0.031). In contrast, the rs4747096 polymorphism was not associated with OA susceptibility in males in the present study. This result is doubtful. The upper CI is almost equal to 1. The significance was very weak. The sample size was so small in their study. We calculated the power of the present study and it is only 64.7% in the dominance model (significance model in female). The power of our study is more than 90% in the same model of inheritance [28]. Similarly, Wang et al. [29] investigate the association between SNPs of ADAMTS14 gene rs474096 and OA of the temporomandibular joint in Chinese Han females, and a quiet weak association was found (OR = 1.114, 95%CI:1.015-1.223) in the dominance model, and the power of the present study was 58.8%. More importantly, both of the studies did not adjust the confounding factors. Considering the sample size, power, and confounding factors, we believe previous studies may be false positive association. Our study results with adjusted several potential confounding factors give more strong evidence. A study with a variety of phenotypes totaling 3217 OA patients and 2214 healthy controls reported that the rare allele of the rs4747096 nsSNP in ADAMTS14 was overrepresented in women requiring joint replacement because of KOA (OR = 1.41, 95%CI = 1.1–1.8; P=0.002) and in patients with symptomatic hand OA (OR = 1.37, 95%CI = 1.0–1.9; P=0.047). The trend results of the present study was consistent with our reports [30]. Our results indicated that the rare allele gene increased the risk of KOA.
Our study has several limitations. A major limitation to the present study is that this is a case–control study design. This point could be found in all studies between gene polymorphism and diseases. This limitation might be overcome by the further exploring the mechanism. The other limitation is that some potential cofounding factors might be not included in the present study, which may overestimate or underestimate the effect of gene polymorphism. Further study is required.
In conclusion, our results indicated that ADAMTS14 gene polymorphism was associated with KOA, and the GG genotype of rs4747096 increased the risk of KOA in Chinese Han population. This is the first study to demonstrate that a correlation exists between ADAMTS14 gene polymorphisms and KOA. The ADAMTS14 may be a diagnostic marker and therapeutic target for KOA treatment. The future study should explore the specific molecular mechanism.
Acknowledgments
We thank the help from Department of Orthopedics People’s Hospital of Nanzhao County.
Abbreviations
- ACCP
anti-cyclic citrullinated peptide antibody
- ADAMTS
A disintegrin and metalloproteinase with thrombospondin motifs
- APF
antiperinuclear factor
- BMI
body mass index
- CI
confidence interval
- CRP
C-reactive protein
- KOA
knee osteoarthritis
- OA
osteoarthritis
- OR
odds ratio
- SNP
single nucleotide polymorphism
Author contribution
S.M. conceived and designed the research; C.O. and S.M. analyzed the data; C.O. created all tables and figures; S.M. drafted the manuscript; S.R. made critical revision of the manuscript; all authors read and approved the final manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Chen B., Deng Y., Tan Y., Qin J. and Chen L.B. (2014) Association between severity of knee osteoarthritis and serum and synovial fluid interleukin 17 concentrations. J. Int. Med. Res. 42, 138–144 10.1177/0300060513501751 [DOI] [PubMed] [Google Scholar]
- 2.Bajpayee A.G., Wong C.R., Bawendi M.G., Frank E.H. and Grodzinsky A.J. (2014) Avidin as a model for charge driven transport into cartilage and drug delivery for treating early stage post-traumatic osteoarthritis. Biomaterials 35, 538–549 10.1016/j.biomaterials.2013.09.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Losina E., Paltiel A.D., Weinstein A.M., Yelin E., Hunter D.J., Chen S.P. et al. (2015) Lifetime medical costs of knee osteoarthritis management in the United States: impact of extending indications for total knee arthroplasty. Arthritis Care Res. (Hoboken) 67, 203–215 10.1002/acr.22412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dillon C.F., Rasch E.K., Gu Q. and Hirsch R. (2006) Prevalence of knee osteoarthritis in the United States: arthritis data from the Third National Health and Nutrition Examination Survey 1991-94. J. Rheumatol. 33, 2271–2279 [PubMed] [Google Scholar]
- 5.Zhang Y., Xu L., Nevitt M.C., Aliabadi P., Yu W., Qin M. et al. (2001) Comparison of the prevalence of knee osteoarthritis between the elderly Chinese population in Beijing and whites in the United States: The Beijing Osteoarthritis Study. Arthritis Rheum. 44, 2065–2071 10.1002/1529-0131(200109)44:9%3c2065::AID-ART356%3e3.0.CO;2-Z [DOI] [PubMed] [Google Scholar]
- 6.Deane K.D., Demoruelle M.K., Kelmenson L.B., Kuhn K.A., Norris J.M. and Holers V.M. (2017) Genetic and environmental risk factors for rheumatoid arthritis. Best Pract. Res. Clin. Rheumatol. 31, 3–18 10.1016/j.berh.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacGregor A.J., Snieder H., Rigby A.S., Koskenvuo M., Kaprio J., Aho K. et al. (2000) Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 43, 30–37 10.1002/1529-0131(200001)43:1%3c30::AID-ANR5%3e3.0.CO;2-B [DOI] [PubMed] [Google Scholar]
- 8.Newton J.L., Harney S.M., Wordsworth B.P. and Brown M.A. (2004) A review of the MHC genetics of rheumatoid arthritis. Genes Immun. 5, 151–157 10.1038/sj.gene.6364045 [DOI] [PubMed] [Google Scholar]
- 9.Collins F.S., Brooks L.D. and Chakravarti A. (1998) A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 8, 1229–1231 10.1101/gr.8.12.1229 [DOI] [PubMed] [Google Scholar]
- 10.Verma P. and Dalal K. (2011) ADAMTS-4 and ADAMTS-5: key enzymes in osteoarthritis. J. Cell. Biochem. 112, 3507–3514 10.1002/jcb.23298 [DOI] [PubMed] [Google Scholar]
- 11.Martel-Pelletier J., Boileau C., Pelletier J.P. and Roughley P.J. (2008) Cartilage in normal and osteoarthritis conditions. Best Pract. Res. Clin. Rheumatol. 22, 351–384 10.1016/j.berh.2008.02.001 [DOI] [PubMed] [Google Scholar]
- 12.Apte S.S. (2009) A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J. Biol. Chem. 284, 31493–31497 10.1074/jbc.R109.052340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colige A., Vandenberghe I., Thiry M., Lambert C.A., Van Beeumen J., Li S.W. et al. (2002) Cloning and characterization of ADAMTS-14, a novel ADAMTS displaying high homology with ADAMTS-2 and ADAMTS-3. J. Biol. Chem. 277, 5756–5766 10.1074/jbc.M105601200 [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Lopez J., Pombo-Suarez M., Loughlin J., Tsezou A., Blanco F.J., Meulenbelt I. et al. (2009) Association of a nsSNP in ADAMTS14 to some osteoarthritis phenotypes. Osteoarthritis Cartilage 17, 321–327 10.1016/j.joca.2008.07.012 [DOI] [PubMed] [Google Scholar]
- 15.Cohen S. and Emery P. (2010) The American College of Rheumatology/European League against Rheumatism Criteria for the classification of rheumatoid arthritis: a game changer. Ann. Rheum. Dis. 69, 1575–1576 10.1136/ard.2010.138446 [DOI] [PubMed] [Google Scholar]
- 16.Kellgren J.H. and Lawrence J.S. (1957) Radiological assessment of osteo-arthrosis. Ann. Rheum. Dis. 16, 494–502 10.1136/ard.16.4.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez-Ramirez D.C., van der Leeden M., van der Esch M., Gerritsen M., Roorda L.D., Verschueren S. et al. (2013) Association of serum C-reactive protein and erythrocyte sedimentation rate with muscle strength in patients with knee osteoarthritis. Rheumatology (Oxford) 52, 727–732 10.1093/rheumatology/kes366 [DOI] [PubMed] [Google Scholar]
- 18.Soto-Hermida A., Fernandez-Moreno M., Oreiro N., Fernandez-Lopez C., Rego-Perez I. and Blanco F.J. (2014) mtDNA haplogroups and osteoarthritis in different geographic populations. Mitochondrion 15, 18–23 10.1016/j.mito.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 19.Takahashi H., Nakajima M., Ozaki K., Tanaka T., Kamatani N. and Ikegawa S. (2010) Prediction model for knee osteoarthritis based on genetic and clinical information. Arthritis Res. Ther. 12, R187 10.1186/ar3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poonpet T., Honsawek S., Tammachote N., Kanitnate S. and Tammachote R. (2013) ADAMTS14 gene polymorphism associated with knee osteoarthritis in Thai women. Genet. Mol. Res. 12, 5301–5309 10.4238/2013.November.7.5 [DOI] [PubMed] [Google Scholar]
- 21.Poole A.R., Kobayashi M., Yasuda T., Laverty S., Mwale F., Kojima T. et al. (2002) Type II collagen degradation and its regulation in articular cartilage in osteoarthritis. Ann. Rheum. Dis. 61, i78–i81 10.1136/ard.61.suppl_2.ii78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J., Gupta T., Barasz J.A., Kalajzic Z., Yeh W.C., Drissi H. et al. (2009) Analysis of microarchitectural changes in a mouse temporomandibular joint osteoarthritis model. Arch. Oral. Biol. 54, 1091–1098 10.1016/j.archoralbio.2009.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W., Du C., Wang H. and Zhang C. (2014) Increased serum ADAMTS-4 in knee osteoarthritis: a potential indicator for the diagnosis of osteoarthritis in early stages. Genet. Mol. Res. 13, 9642–9649 10.4238/2014.November.14.9 [DOI] [PubMed] [Google Scholar]
- 24.Kumar S., Chen M., Li Y., Wong F.H., Thiam C.W., Hossain M.Z. et al. (2016) Loss of ADAMTS4 reduces high fat diet-induced atherosclerosis and enhances plaque stability in ApoE(-/-) mice. Sci. Rep. 6, 31130 10.1038/srep31130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller R.E., Tran P.B., Ishihara S., Larkin J. and Malfait A.M. (2016) Therapeutic effects of an anti-ADAMTS-5 antibody on joint damage and mechanical allodynia in a murine model of osteoarthritis. Osteoarthritis Cartilage 24, 299–306 10.1016/j.joca.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gok K., Cemeroglu O., Cakirbay H., Gunduz E., Acar M., Cetin E.N. et al. (2018) Relationship between cytosine-adenine repeat polymorphism of ADAMTS9 gene and clinical and radiologic severity of knee osteoarthritis. Int. J. Rheum. Dis. 21, 821–827 10.1111/1756-185X.12849 [DOI] [PubMed] [Google Scholar]
- 27.Fernandes R.J., Hirohata S., Engle J.M., Colige A., Cohn D.H., Eyre D.R. et al. (2001) Procollagen II amino propeptide processing by ADAMTS-3. Insights on dermatosparaxis. J. Biol. Chem. 276, 31502–31509 10.1074/jbc.M103466200 [DOI] [PubMed] [Google Scholar]
- 28.Takeda S. (2016) ADAM and ADAMTS family proteins and snake venom metalloproteinases: a structural overview. Toxins (Basel) 8, 10.3390/toxins8050155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D.D., Gan Y.H., Ma X.C. and Meng J.H. (2018) Association between ADAMTS14 gene polymorphism and the temporomandibular joint osteoarthritis in Chinese Han females. Beijing Da Xue Xue Bao 50, 279–283 [PubMed] [Google Scholar]
- 30.Rodriguez-Lopez J., Pombo-Suarez M., Loughlin J., Tsezou A., Blanco F.J., Meulenbelt I. et al. (2009) Association of a nsSNP in ADAMTS14 to some osteoarthritis phenotypes. Osteoarthritis Cartilage 17, 321–327 10.1016/j.joca.2008.07.012 [DOI] [PubMed] [Google Scholar]