Abstract
Autophagy is an important process in endogenous substrate degradation by lysosomes within cells, with a degree of evolutionary conservation. Like apoptosis and cell senescence, cell autophagy is a very important biological phenomenon involving the development and growth of biological processes. Abnormal autophagy may lead to tumorigenesis. In recent years, increasing studies have demonstrated that long non-coding RNAs (lncRNAs) and miRNAs can regulate cell autophagy by modulating targetting gene expression. In this review, we will provide an overview of lncRNAs and miRNAs in autophagy modulation and new insights into the underlying mechanisms, as well as their potential utilization in disease diagnosis, prognosis, and therapy.
Keywords: autophagy, large intervening non-coding RNA, microRNA, oncogenesis, gene therapy
Introduction
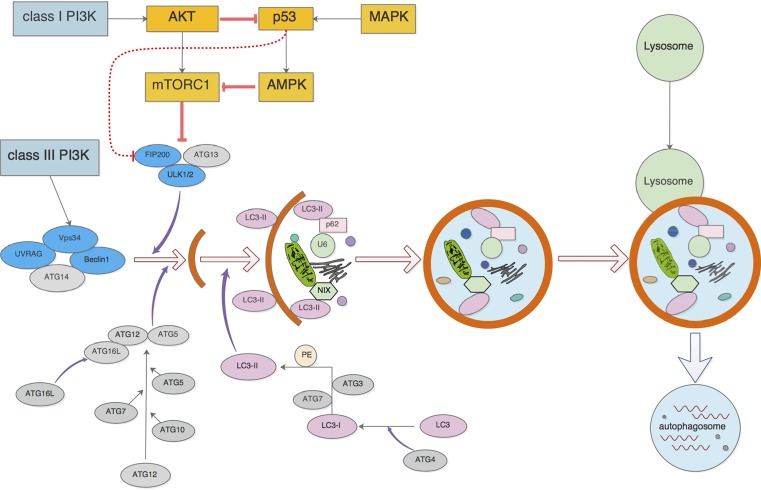
Autophagy is a process of sustaining metabolism and homeostasis by capturing and degrading intracellular components such as proteins and organelles. Autophagy plays an important role in protein and organelle quality control, knowing that a low level of basal autophagy can prevent gradual accumulation of damaged proteins and organelles in tissues, which is known to be toxic over time [1]. Three forms of autophagy are commonly described: macro-autophagy, micro-autophagy, and chaperone-mediated autophagy (CMA). Autophagy is an important cellular response to stress or starvation. The formation and development of autophagy involve many signaling pathways and related proteins (Figure 1), and the regulation of these signaling pathways and proteins can affect the process of autophagy [2]. In the initiation step of autophagy, the widely accepted sensor is the mechanistic target of rapamycin (mTOR) complex I (mTORCI) and many autophagy inducers trigger autophagy by initiating signal transduction cascades to tactfully inhibit mTORCI [3]. mTORCI plays a central role in the regulation of ULK1-ATG13-FIP200 (FAK family-interacting protein of 200 kDa) complex where inhibition of mTORCI upon rapamycin treatment enhances the kinase activity of ULK1 [4]. Phosphoinositide 3-kinases (PI3Ks) are divided into three isoforms (class I, class II, and class III). The class I PI3K triggers mTOR signaling pathway and inhibits autophagy, while the class III isoform activates the autophagy by corresponding to Vps34. The contribution of class II PI3K activity on autophagy is unclear [5]. Autophagy has been reported to either inhibit or promote cancer cell proliferation or tumorigenesis in model systems, suggesting that the role of autophagy in cancer is context dependent [6,7]. Moderate and effective autophagy can remove tumor cells and maintain homeostasis. However, impaired autophagy may reduce cell viability, and delayed elimination of apoptotic cells from the body may induce the development of cancer [8].
Figure 1. The process of autophagy and related regulatory proteins and signaling pathways.
Oncogenic and tumor suppressive signaling pathways are closely related to autophagy initiation. MAPK signaling can activate autophagy through AMPK activation and promotion of autophagy-related gene translation. PI3K-AKT signaling inhibits autophagy through mTOR activation and p53 inhibition. The Vps34 complex combines Beclin-1 promoting autophagic cell death which threatens the survival of cells. The linkage of ATG5 to ATG12 and ATG16L, and phosphatidyl ethanolamine (PE) to proteins of the microtubule-associated protein 1 light chain 3 (LC3) are vital for the initiation and development of autophagy.
Advances in human genome sequencing have shown that more than 90% of the human genomes are extensively transcribed but only approximately 2% of them serve as protein-coding genes [9]. The majority of the remaining transcripts known as non-coding RNAs (ncRNAs) have no protein-coding capacity, including small ncRNAs, especially miRNAs, and long ncRNAs (lncRNAs) based on their transcript size. LncRNAs are RNA transcripts that can be transcribed by polymerase II or III and are usually longer than 200 nts in length. Accumulating evidence shows that lncRNAs participate in the regulation of cell proliferation and apoptosis, and play crucial roles in tumorigenesis, progression, and drug resistance [10]. MiRNAs are endogenous RNAs, 19–25 nts in length, playing important gene-regulatory roles by pairing with the mRNAs of protein-coding genes to direct their post-transcriptional repression [11]. MiRNAs are reported to be dysregulated in the progression and invasion of various human cancers and play key roles in cell differentiation, proliferation, and cell death [12].
In recent years, increasing studies have demonstrated that lncRNAs and miRNAs can regulate cell autophagy in vitro and in vivo by modulating targetting gene expression at the level of chromatin organization, transcription, and post-transcription [13]. This review will provide an overview of lncRNAs and miRNAs in autophagy modulation and new insights into the underlying mechanisms of the lncRNA-autophagy axis and miRNA-autophagy axis and lncRNA-miRNA-autophagy axis in tumorigenesis.
Roles of miRNAs in regulating autophagy
The effect of miRNA on target gene mRNA depends mainly on the degree of complementarity between miRNA and target gene transcriptional sequence, mainly through three ways. The first is to cut-off the mRNA molecule of the target gene, knowing that miRNA is completely complementary to the target gene, which is similar to the siRNA, and finally cut the target mRNA. The second is to inhibit translation of the target gene, which is not completely complementary to the target gene, and suppress the translation without affecting the stability of mRNA. The third is binding inhibition, which has the above two modes: when combined with the target gene, it directly and targettedly cuts mRNA; when the target gene is not fully integrated, it plays a role in regulating gene expression [14].
miRNAs target autophagy-related proteins to regulate the autophagy
Beclin1 (ATG6) is a well-known key regulator of autophagy. Beclin1 can restore starvation-induced autophagy in ATG6-disrupted yeast strains and human breast carcinoma cells lacking detectable Beclin1 levels, whereas Beclin1 overexpression activates autophagy. Sufficient levels of Beclin1 are necessary for its autophagic function [15]. miR-30a, a member of the miR-30 family, is known to target Beclin1, and its expression was found to down-regulate Beclin1 expression and inhibit autophagy in medulloblastoma cell lines [16]. MiR-21 was up-regulated in chronic myeloid leukemia (CML), and treatment with antimiR-21 increased autophagy-related proteins Beclin-1, Vps34, and light chain 3 (LC3) II (LC3-II), eventually leading to an increase in autophagy flux [17].
Altered expression of miRNAs under hypoxia may induce a pathological process via modulating the expression of their downstream genes [18]. Hypoxia-induced up-regulation of miR-301a/b increased cell autophagy and viability of prostate cancer (PCa) cells by targetting N-myc downstream regulated gene 2 (NDRG2) which could lead to the increasing LC3-II [19]. In addition, Yu et al. [20] demonstrated that miR-24-3p accelerated cell proliferation, migration, invasion, and autophagy and inhibited apoptosis by inhibiting death effector domain containing (DEDD) and activating p62 (a crucial autophagy factor, play irreplaceable roles in eliminating the ubiquitinated damaged mitochondria by mitophagy) in bladder cancer. Myotubularin related protein 3 (MTMR3) is a member of myotubularin-related protein family, and has been demonstrated that MTMR3 decreased pattern recognition receptor (PRR)-induced PI3P and autophagy levels by targetting ATG5 and LC3-II in monocyte-derived macrophages [21]. miR-181a was found to be significantly up-regulated, while MTMR3 was found to be down-regulated in human gastric cancer (GC) tissues and cell lines [22]. Further investigations indicated that overexpression of miR-181a or depletion of MTMR3 attenuated starvation-induced autophagy in adenogastric sarcoma (AGS) cells, resulting in an increase in cell proliferation, colony formation, migration, invasion, as well as suppression of apoptosis [23].
Sirtuin 1 (Sirt1) is a histone deacetylase and mediates autophagy via LC3. Further research demonstrated that Sirt1-induced autophagy could deacetylate endogenous LC3 [24]. Deacetylation of LC3 by Sirt1 allows LC3 to interact with the nuclear protein DOR and return to the cytoplasm with DOR, where it is able to bind ATG7 and other autophagy factors and undergo phosphatidyl ethanolamine conjugation to preautophagic membranes. The association of deacetylated LC3 with autophagic factors shifts LC3’s distribution from the nucleus toward the cytoplasm [25]. The mimics of miR-152 and miR-24 induced autophagy by increasing the level of Sirt1 and deacetylated LC3, which may be important in preventing the development of uterine sarcoma [26]. In A549 and H460 cells of lung cancer, by increasing the levels of both LC3-II and Beclin1, miR-144 targetted the p53-induced glycolysis and apoptosis regulator (TIGAR), inhibited proliferation, enhanced apoptosis, and increased autophagy [27].
Smad2, a key downstream component of the TGF-β signaling pathway, was found to contribute to cancer initiation, invasion, metastasis, and self-renewal of colorectal cancer (CRC) stem cells [28]. MiR-140-5p could directly target Smad2 and its overexpression was found to decrease the Smad2 expression level, thus attenuating cell invasion and proliferation. Ectopic expression of miR-140-5p in cancer stem cells (CSCs) inhibited CSC growth and sphere formation in vitro by disrupting autophagy through suppressing ATG12 [29]. Recently, autophagy has been suggested to inhibit HPV infection, which is known to be closely related to cervical cancer. Further analysis revealed that increased level of miR-224-3p expression inhibited autophagy through targetting FIP200 in cervical cancer [30].
miRNAs target signaling pathways to regulate the autophagy
X-linked inhibitor of apoptosis (XIAP) was found to promote cell survival and suppress autophagy through XIAP-Mdm2-p53 pathway [31]. Forced expression of miR-23a significantly decreased the expression of XIAP and promoted autophagy, while down-regulation of miR-23a increased XIAP expression and suppressed autophagy in breast cancer cells [32]. But overexpressed miR-23a in melanoma suppressed the invasive and migratory properties of melanoma cells by abrogating autophagy through directly targetting ATG12 via autophagy-mediated AMPK-RhoA pathway [33].
As a protein kinase, mTOR is a confluence of upstream pathways regulating cell growth, proliferation, survival, and autophagy. It was reported [34] that activation of the mTOR pathway inhibited autophagy, indicating that some miRNAs can affect the process of cancers by regulating autophagy via the mTOR pathways. It was reported that the AMPK-mTOR signaling pathway regulated cell autophagy in non-small-cell lung cancer (NSCLC). Some studies [35] found that overexpression of miR-138 could inactivate lung cancer cell autophagy probably through the AMPK-mTOR signaling pathway, thus suppressing cell proliferation, invasion, and migration. In paclitaxel-resistant triple-negative breast cancer (TNBC) cells, miR-18a up-regulation enhanced autophagy via inhibiting the mTOR signaling pathway [36].
Paradoxically, either up-regulation or down-regulation of miR-96 suppressed PCa cell proliferation in vitro and tumor growth in vivo under hypoxia. Hypoxia increased the expression of miR-96 in PCa cells, and miR-96 stimulated autophagy by suppressing mTOR. But when ectopic overexpression of miR-96 reached a certain threshold, it abolished the hypoxia-induced autophagy through suppressing ATG7, a key autophagy-associated gene [37]. This observation might reveal a novel regulatory mode of autophagy by miRNAs. However, further studies are required to understand the complex role of miRNAs in autophagy regulation.
Some miRNAs regulate drug-resistance related autophagy
Yu et al. [38] found that activation of the CXCL12/CXCR4 axis promoted epithelial–mesenchymal transition (EMT) and concurrent up-regulation of miR-125b in human CRC. Further experiments indicated a role of miR-125b in conferring 5-fluorouracil (5-FU) resistance in CRC, probably through increasing autophagy by augmenting the cleavage of L3-I into LC3-II both in vitro and in vivo [38]. Beclin1, a key autophagy-promoting gene, could be inhibited by miR-30d, which sensitized anaplastic thyroid carcinoma (ATC) cells to cisplatin [39] and promoted cell apoptosis of human colon cancer cells [40]. Colon cancer cells often become resistant during chemotherapy. Some recent studies [41] provided evidence that miR-409-3p targetted and inhibited Beclin1, which further inhibited chemotherapy-induced autophagy and enhanced the chemosensitivity of colon cancer cells.
In terms of tumor therapy, low frequency magnetic fields (LF-MFs), which refer to magnetic fields with 3–30 Hz, have been shown to inhibit cancer cell proliferation in several studies [42]. LF-MFs could up-regulate the expression level of miR-486 by targetting B-cell adaptor for phosphatidylinositol 3-kinase (BCAP) and further inhibiting lung cancer through miR-486-induced autophagic cell death by inhibiting AKT/mTOR signaling pathway [43]. Activation of the PI3K-Akt-mTOR pathway may trigger endocrine resistance to tamoxifen and fluvestrant in breast cancer, while miR-214 could increase the sensitivity of breast cancer cells to 4-OHT/FUL through inhibition of autophagy via the PI3K-Akt-mTOR pathway [44]. In addition, miR-142-3p could inhibit starvation-induced autophagy and increase chemosensitivity of NSCLC in vitro and in vivo through the PI3K-Akt-mTOR pathway [45]. An HIF-1α-miR-21 positive feedback loop was observed through the PTEN/Akt/HIF-1α pathway whereby miR-21 decreased autophagy, resulting in increased radio-resistance in cervical cancer cells [46]. Similarly, miR-140-5p expression was highly induced during chemotherapy of osteosarcoma cells, and this was accompanied by autophagy up-regulation. Importantly, miR-140-5p regulated this context-specific autophagy through its target inositol 1,4,5-trisphosphate kinase 2 (IP3k2) and inhibited the IP3K2-mediated autophagy [47].
In spite of that, there are still many questions about the regulatory roles of miRNAs on autophagy that require further investigation. The same miRNA may have various influences on the different cancers through the dual function of autophagy, indicating that the exact role remains uncertain. Additionally, not all miRNAs concentrate on the levels of gene product, many of them only confirming to affect the entire network or signaling pathways. Therefore, future research should focus on a comprehensive understanding of the overall role of an miRNA in autophagy, thereby clarifying its role in tumorigenesis and development.
Roles of lncRNAs in regulating autophagy
LncRNAs execute their functions by interactions with other components such as proteins, RNAs, and DNAs. RNA–protein, RNA–RNA, and RNA–DNA interactions could be combined by a single lncRNA to build distinct functionality complexes [48]. Guide-, decoy-, and scaffold functions of lncRNAs have been identified. The guide function of lncRNAs mediates recruitment of chromatin-modifying enzymes to target genes, thereby affecting gene transcription and expression. The decoy function involves binding of miRNAs through lncRNAs, combining with transcription factors to interfere with the binding of gene promoter region. As a scaffold or bridge for protein interaction, lncRNAs affected protein polymer formation and regulated protein activity [49]. In addition to their well-established influence as regulators of transcription, lncRNAs were also effective modulators of pre-mRNA splicing, mRNA decay, and translation [50].
lncRNAs target autophagy-related proteins to regulate the autophagy
It is widely believed that HOX transcript antisense RNA (HOTAIR) mediates chromosomal remodeling and co-ordinates with polycomb repressive complex 2 (PRC2) to regulate gene expression in a variety of biological processes [51]. In hepatocellular carcinoma (HCC), HOTAIR was overexpressed in HCC tissues as compared with adjacent non-tumor tissues. HOTAIR overexpression could activate autophagy by increasing ATG3 and ATG7 expression, thus promoting HCC cell proliferation [52]. In addition, the expression level of HOTAIR was up-regulated in NSCLC. Further studies indicated that silencing of HOTAIR decreased drug resistance of NSCLC cells to crizotinib through inhibition of autophagy via suppressing phosphorylation of ULK1 which was an important component of ULK1–ATG13–FIP200 complex [53].
In papillary thyroid carcinoma (PTC), BRAF-activated lncRNA (BANCR) levels were significantly higher in those in PTC tissues, which contributed to PTC cell proliferation and activated autophagy [54]. This phenomenon was evaluated by observing the ratio of LC3-II/LC3-I, but we still donot know the specific regulation mechanism. In PTC, GAS8 antisense RNA 1 (GAS8-AS1) was down-regulated, and overexpression of GAS8-AS1 inhibited proliferation and activated autophagy by targetting ATG5 [55]. LncRNA highly up-regulated in liver cancer (HULC) could promote different pro-tumorigenic phenotypes, such as cell survival, proliferation, and invasion in vitro, as well as tumor growth and angiogenesis in vivo [56]. In epithelial ovarian carcinoma (EOC) tissues, the HULC expression level was higher than that in normal samples, which promoted tumorigenesis of ovarian carcinoma by inhibiting ATG7 to inhibit autophagy and induce progression by regulating ITGB1 [57]. HULC was also up-regulated in GC tissues and cell lines. However, up-regulation of HULC inhibited cell apoptosis by activating autophagy, and this overexpression was correlated with lymph node metastasis, distant metastasis, and advanced tumor node metastasis [58]. But, how HULC regulates autophagy and through which signaling pathway remains to be studied.
LncRNAs target signaling pathways to regulate the autophagy
p53 is one of the most famous tumor suppressors and it has an outstanding role in promoting autophagic cell death. In PI3K-AKT signaling pathway, the activation of PI3K/AKT could inhibit p53 and inhibited autophagic cell death through mTORC1 activation [2]. In the cytoplasm, p53 exerted direct autophagy-inhibitory functions through a direct molecular interaction with the human ortholog of yeast ATG17, namely RB1-inducible coiled-coil protein 1 (RB1CC1), also called FIP200, a protein that is essential for the very apical step of autophagy initiation [59]. Further investigations are required to better understand this dual aspect of p53 biology. Maternally expressed gene 3 (MEG3) is an imprinted gene located at 14q32 that encodes an ncRNA [60], which is expressed in normal tissues but is either lost or decreased in many human tumors and tumor derived cell lines. Studies have demonstrated that MEG3 is associated with cancer initiation, progression, metastasis, and chemoresistance [61,62]. In bladder cancer tissues, MEG3 levels were significantly reduced, which inhibited cell apoptosis and increased cell proliferation by activating autophagy because MEG3 regulates cancer cell proliferation by the p53 pathway and p53 negatively regulates autophagy [63].
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) plays an important role in cancer and acts as a transcriptional regulator for various genes, including those involved in cell proliferation, migration, and metastasis [64]. In pancreatic ductal adenocarcinoma (PDAC), increased expression of MALAT1 was identified as a diagnostic biomarker. Silencing of MALAT1 inhibited autophagy via HuR-TIA-1-mediated autophagic activation, thus inhibiting tumor proliferation and metastasis in vivo [65]. TIA-1 functioned as ancient DNA/RNA transacting regulator to broaden the transcriptome and proteome diversity, which triggered a series of biological processes, including cell invasion, migration, apoptosis, and autophagy through regulating several p53 signaling pathway-related genes [66].
Although numerous studies have demonstrated the importance of PI3K in the activation of AKT, there have been reports suggesting that AKT activation can proceed in a manner that is independent of PI3K. Liu et al. [67] provided the evidence that LINC00470 was required for AKT cytoplasm activation and the interaction of LINC00470 and fused in sarcoma (FUS) was critical for AKT activation.High pAKT activated by LINC00470 inhibited cell autophagy, which associated with GBM tumorigenesis and poor patient prognosis [67]. In hypoxic tumor cells, lincRNA-p21 knockdown induced G2/M phase arrest, promoted apoptosis, decreased cell proliferation and motility, and reduced autophagy through HIF-1/Akt/mTOR/P70S6K pathway [68].
Some lncRNAs regulate drug resistance related autophagy
In diffuse large B-cell lymphoma (DLBCL), MALAT1 was highly expressed. However, via elevating LC3-II/LC3-I protein expression and decreasing p62 expression, MALAT1 silencing could activate autophagy to inhibit adriamycin-induced EMT, thus suppressing tumor growth and reducing chemotherapy resistance [69]. In cisplatin-treated glioma cell line, MEG3 expression levels were increased. Elevated MEG3 enhanced the chemosensitivity of glioma cells to cisplatin through eliminating autophagy induced by cisplatin [70]. However, the molecular mechanisms underlying these effects remain unclear, and future studies will investigate this further.
Co-ordinative roles of lncRNAs and miRNAs in regulating autophagy
LncRNA and miRNA can induce the occurrence of disease by multisite and multitarget interactions, and regulate these interactions by adjusting their relative abundance. Most importantly, lncRNA can act as a competing endogenous RNA (ceRNA) by combining the same miRNA with other RNA transcripts to degrade the target gene mRNA. This lncRNA–miRNA combination could regulate the gene expression pattern to realize the communication and regulation between them, and promote the physiological and disease processes, such as cell differentiation, proliferation, apoptosis, and the development and progression of diseases [71,72].
LncRNA–miRNA axis targets autophagy-related proteins to regulate the autophagy
PVT1 was found to be highly expressed in glioma vascular endothelial cells. PVT1 overexpression increased the expression of ATG7 and Beclin1 by targetting miR-186, which induced protective autophagy, thus promoting glioma vascular endothelial cell proliferation, migration, and angiogenesis [73]. Additionally, MALAT1 expression was increased in glioma tissues compared with that in adjacent normal tissues. In addition, MALAT1 activated autophagy and promoted cell proliferation by sponging miR-101 and up-regulating STMN1, RAB5A and ATG4D expression in glioma cells [74].
HOTAIR expression was up-regulated in chondrosarcoma tissues and cell lines, which induced DNA methylation of miR-454-3p by recruiting EZH2 and DNMT1 to the miR-454-3p promoter regions to silence miR-454-3p expression. Through the HOTAIR-miR-454-3p-STAT3/ATG12 axis, HOTAIR knockdown-induced inhibition of autophagy indirectly promoted chondrosarcoma cell apoptosis [75]. Small nucleolar RNA host gene 15 (SNHG15) contributed to invasion, proliferation, migration, and autophagy by negatively regulating miR-141 in osteosarcoma through elevating the levels of ATG5 and LC3-II. This finding may provide a new potential target and prognostic biomarker for the treatment of OS [76]. In HCC, HNF1A-AS1 functioned as an oncogene in tumor growth and apoptosis through sponging tumor-suppressive miR-30b-5p and de-repressing Bcl-2 and HNF1A-AS1-miR-30b axis significantly promoted autophagy under starvation [77]. ATG5, as a target of miR-30b-5p, was regulated by HNF1A-AS1-miR-30b axis, resulting in positively regulating the autophagy.
LncRNA–miRNA axis targets signaling pathways to regulate the autophagy
In GC, lncRNA HAGLROS was a direct target of transcriptional factor STAT3 and could regulate autophagy-related gene expression of ATG9A and ATG9B by mTOR signals in two manners. On the one hand, HAGLROS acted as ceRNA of miR-100-5p to up-regulate mTOR expression by antagonizing miR-100-5p-mediated mTOR mRNA inhibition. On the other hand, HAGLROS could interact with mTORC1 components to activate mTORC1 signaling pathway. In these two manners, autophagy was inhibited, contributing to GC progression and poor prognosis [78]. LncRNA PTENP1 is a pseudogene of the tumor suppressor gene PTEN and its expression was down-regulated in several HCC cell lines. PTENP1 overexpression decoyed oncomirs miR-17, miR-19b, and miR-20a, which would otherwise target PTEN, suppress the oncogenic PI3K/AKT pathway, and induce autophagy and apoptosis [79].
LncRNA–miRNA axis regulates drug resistance related autophagy
More recent evidence has indicated that protective autophagy is an important reason for chemoresistance of cancer cells [80]. Chemoresistance has long been recognized as a major obstacle in cancer therapy. In GC cells, MALAT1 acted as a ceRNA for miR-23b-3p and attenuated the inhibitory effect of miR-23b-3p on ATG12, leading to chemoinduced autophagy and chemoresistance in GC cells [81]. Treatment with antitumor reagents such as oxaliplatin, 5-FU, and pirarubicin (THP) dramatically induced HULC expression and protective autophagy in HCC cells. MiR-6825-5p, miR-6845-5p, and miR-6886-3p were down-regulated by HULC, resulting in the elevation of Sirt1, USP22, and protective autophagy, thus attenuating the sensitivity of HCC cells to chemotherapeutic agents [82]. In pancreatic cancer, blockade of autophagy could reduce pancreatic CSCs activity and sensitize cancer cells to gemcitabine [83]. In pancreatic cancer, linc-ROR confers gemcitabine resistance at least partly via inducing autophagy, and further research reported a linc-ROR/miR-124/PTBP1/PKM2 axis that involved in the regulation of gemcitabine resistance in pancreatic cancer cells [84].
Myeloid cell leukemia sequence 1 (Mcl-1), an anti-apoptotic Bcl-2 family protein, has been reported to play a key role in autophagy. The anti-apoptotic Bcl-2 proteins including Bcl-2, Bcl-XL, Bcl-W, and Mcl-1 have been proposed to inhibit autophagy owing to their interaction with the autophagy regulator Beclin1 [85]. LncRNA-AC023115.3 was increased in cisplatin-resistant glioma cells and acted as a ceRNA for miR-26a which attenuated the inhibitory effect of miR-26a on GSK3β, thus increasing GSK3β, a proline-directed serine-threonine kinase that promotes the degradation of Mcl-1, leading to an increase in GSK3β and a decrease in autophagy [86]. LncRNA, cancer susceptibility candidate 2 (CASC2), was found as a tumor suppressor and down-regulated in various cancers [87]. CASC2 was down-regulated in glioma, and overexpression of CASC2 reduced TMZ-induced autophagy via mTOR up-regulation by targetting miR-193a-5p, thus enhancing the sensitivity of glioma cells to TMZ cytotoxicity [88]. Therefore, a better understanding about the molecular mechanisms of the interaction between lncRNA and miRNA would help enhance the protective autophagy and thus improve the outcome of chemotherapy for the treatment of cancers.
Applications of autophagy in tumor therapy
Autophagy inhibition is receiving more and more attention as a potentially new therapeutic approach in cancer. This field is looking forward to the clinical development of novel agents that target the upstream components of autophagy or lysosomes. In the past few years, significant progress was made with the discovery of inhibitors of the two main kinases involved in the autophagy process, Vps34 and ULK1, and key regulator of the autophagy initiation, PI3K and mTOR. In addition, with the in-depth study of the regulation of autophagy by ncRNAs, making it possible to utilize of the ncRNA system in cancer biology and pathology by targetting autophagic pathways.
Classical autophagy regulator
The seemingly paradoxical tumor-suppression and -promotion roles of autophagy provide more therapeutic opportunities for anticancer treatment. 3-methyladenine (3-MA), one of PI3K inhibitors, has been widely used as an autophagy inhibitor based on their inhibitory effect on class III PI3K activity [89]. But surprisingly, 3-MA was found to promote autophagy flux when treated under nutrient-rich conditions with a prolonged period of treatment due to its persistent inhibition on class I PI3K [90]. Understanding the dual role of 3-MA in autophagy may have important implications in autophagy study. In addition, hydroxychloroquine (HCQ) is the clinically available drug that could function as an autophagy inhibitor. HCQ inhibited autophagy by acting as a weak base and increased the pH of those compartments when trapped inside acidic cellular compartments (such as lysosomes) [91]. Rapamycin, an allosteric inhibitor of mTORC1, can selectively inhibit mTORC1 by binding to the 12-kDa immunophilin FK506-binding protein (FKBP12) to stimulate autophagy [92]. In addition to stimulating autophagic cancer cell death, the induction of autophagy by mTOR inhibitors could also sensitize cancer cells to other cancer therapeutics. But as we know, autophagy is important for some normal tissues, a critical question is whether systemic autophagy inactivation will be sufficiently selective to impair cancer growth while sparing normal tissues from the deleterious consequences.
LncRNA and miRNA modulation in autophagy-targetted anticancer therapeutics
Recently, accumulating evidence has suggested that lncRNAs and miRNAs play critical roles in cell survival/death and therefore show great potential in regulating autophagy at different stages. As mentioned above, in different periods of autophagy, including vesicle nucleation, elongation, and completion phase, lncRNAs and miRNAs all promoted or inhibited the process of autophagy to a certain extent. Therefore, these results of the research enhanced the possibilities of the utilization of the lncRNA and miRNA system in cancer therapy by targetting autophagic pathways. For example, during the early stage of autophagy, miR-376b can decrease the activity of Beclin1, thereby blocking vesicle nucleation. In addition, miR-376b was able to attenuate mTOR inhibitor rapamycin-induced autophagy [93]. Thus, targetting miR-376b, we can develop relevant targetted therapies.
Summary and future prospective
Autophagy is a classic mechanism of energy metabolism and self-renewal in cells, and plays an important role in biological development and homeostasis. Autophagy has a dual role in tumor therapy. As a tumor suppressor mechanism, autophagy can lead to cell death, limit the number of cells or reduce the mutation probability of DNA to prevent tumor formation. As a tumor protection mechanism, autophagy can protect cancer cells against chemotherapy drugs and delay apoptosis of tumor cells. However, there are still many problems to be solved in current research, such as the origin of autophagy, how signal transduction pathways co-ordinate and interact with each other, and their effects on cell survival.
It is well known that most cancer types can be cured, if diagnosed at an early stage. In recent years, many studies have shown that lncRNAs and miRNAs can be used as potential molecular diagnostic markers and therapeutic targets for disease. Their specific expression and regulation are closely related to many diseases, especially malignancies. The characteristics of lncRNA and miRNA, including disease specificity, cell type specificity and relative ease of detection, make them suitable for cancer patients. However, most ncRNAs are not tissue and cancer specific. For instance, HULC was dysregulated not only in EOC but also in GC. Furthermore, a group of patients with the same cancer type showed significant heterogeneity in ncRNA expression. Further studies should pay more attention to find more tissue and cancer-specific ncRNAs as potential molecular diagnostic markers and therapeutic targets. In addition, linking basic medical research with clinical treatment, namely translational medicine, is also the focus and difficulty in current scientific research. With the development of gene editing technology, especially the emergence of clustered regularly interspaced short palindromic repeats (CRISPR)/ RISPR associated (Cas), it is expected that more ncRNAs will be used as targets for clinical treatment.
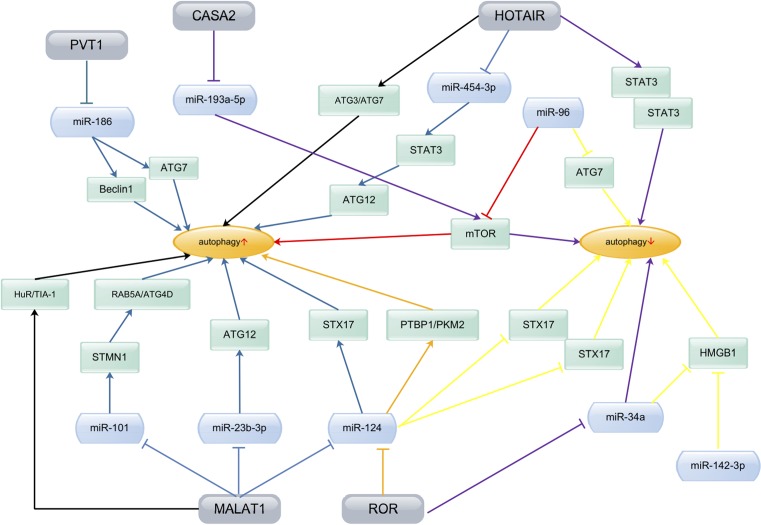
In this review, many studies have demonstrated that lncRNAs and miRNAs play important roles in autophagy (Table 1), and the regulation of autophagy forms a complex network (Figure 2). At present, the research on the involvement of lncRNAs and miRNAs in autophagy is still in its initial stage. Although more autophagy-related lncRNAs and miRNAs have been discovered, most of these studies mainly focussed on their expression and function. Therefore, more studies are required to address the interaction mechanism between lncRNAs and miRNAs, and the regulatory complexity of autophagy, including specific target genes, target proteins, and signaling pathways, for the sake of helping discover new targets, new drugs or new strategies for the diagnosis, treatment, and prognostic prediction of cancers.
Table 1. A list of lncRNAs and miRNAs associated with autophagy regulation.
| LncRNA/miRNA | Expression level | Target | Promote/suppress autophagy | Cancer type | Reference |
|---|---|---|---|---|---|
| miR-18a | ↑ | mTOR | Promote | TNBC | [36] |
| miR-24-3p | ↑ | DEDD/p62 | Promote | Bladder cancer | [20] |
| miR-152/miR-24 | ↑ | Sirt1 | Promote | Uterine sarcoma | [26] |
| miR-21 | ↑ | Beclin1/Vps34/LC3II | Suppress | CML | [17] |
| miR-138 | ↓ | AMPK/ mTOR | Suppress | Lung cancer | [35] |
| miR-30d | ↓ | ATG5/PI3K/Beclin1 | Suppress | Colon cancer | [40] |
| HOTAIR | ↑ | ATG3/ATG7 | Promote | HCC | [52] |
| HOTAIR | ↑ | ULK1 | Promote | NSCLC | [53] |
| MEG3 | ↓ | p53 | Suppress | Bladder cancer | [63] |
| LINC00470 | ↑ | AKT | Suppress | Glioblastoma | [67] |
Abbreviations: ↑, up-regulation; ↓, down-regulation.
Figure 2. The complex network of lncRNAs and miRNAs in regulating autophagy.
Different ncRNAs regulate autophagy through diverse signaling pathway by targetting relevant proteins which ultimately result in different outcomes. ↑, increase; ↓, decrease.
Abbreviations
- CASC2
cancer susceptibility candidate 2
- ceRNA
competing endogenous RNA
- CRC
colorectal cancer
- CRISPR
clustered regularly interspaced short palindromic repeat
- CSC
cancer stem cell
- EMT
epithelial–mesenchymal transition
- EOC
epithelial ovarian carcinoma
- GAS8-AS1
GAS8 antisense RNA 1
- GC
gastric cancer
- HCC
hepatocellular carcinoma
- HCQ
hydroxychloroquine
- HOTAIR
HOX transcript antisense RNA
- HULC
highly up-regulated in liver cancer
- LC3
light chain 3
- LF-MF
low frequency magnetic field
- lncRNA
long non-coding RNA
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- Mcl-1
myeloid cell leukemia sequence 1
- MEG3
maternally expressed gene 3
- MTMR3
myotubularin related protein 3
- mTOR
mechanistic target of rapamycin
- mTORC
mTOR complex
- ncRNA
non-coding RNA
- NSCLC
non-small-cell lung cancer
- PCa
prostate cancer
- PDAC
pancreatic ductal adenocarcinoma
- PI3K
phosphoinositide 3-kinase
- PTC
papillary thyroid carcinoma
- Sirt1
Sirtuin 1
- XIAP
X-linked inhibitor of apoptosis
- 3-MA
3-methyladenine
- 5-FU
5-fluorouracil
Author contribution
S.J. and Q.Y. designed and wrote the article. W.F. and X.S. revised the manuscript. The newly added author W.F. made suggestions in the first revision and participated in the revision of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Nature Science Foundation of China [grant number 81672099]; and the Key Project of Jiangsu Province [grant number ZDXKB2016011].
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Mizushima N. and Komatsu M. (2011) Autophagy: renovation of cells and tissues. Cell 147, 728–741 10.1016/j.cell.2011.10.026 [DOI] [PubMed] [Google Scholar]
- 2.Zhang J., et al. (2018) Mechanisms of autophagy and relevant small-molecule compounds for targeted cancer therapy. Cell. Mol. Life Sci., 75, 1803–1826, , 10.1007/s00018-018-2759-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laplante M. and Sabatini D.M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunlop E.A. and Tee A.R. (2014) mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Semin. Cell Dev. Biol. 36, 121–129 10.1016/j.semcdb.2014.08.006 [DOI] [PubMed] [Google Scholar]
- 5.Vanhaesebroeck B., et al. (2010) The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 11, 329–341 10.1038/nrm2882 [DOI] [PubMed] [Google Scholar]
- 6.White E. (2012) Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer 12, 401–410 10.1038/nrc3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White E., Mehnert J.M. and Chan C.S. (2015) Autophagy, metabolism, and cancer. Clin. Cancer Res. 21, 5037–5046 10.1158/1078-0432.CCR-15-0490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White E. (2015) The role for autophagy in cancer. J. Clin. Invest. 125, 42–46 10.1172/JCI73941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djebali S., et al. (2012) Landscape of transcription in human cells. Nature 489, 101–108 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu X., Sood A.K., Dang C.V. and Zhang L. (2017) The role of long noncoding RNAs in cancer: the dark matter matters. Curr. Opin. Genet. Dev. 48, 8–15 10.1016/j.gde.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel D.P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin S. and Gregory R.I. (2015) MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 15, 321–333 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z.H., et al. (2017) The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 24, 212–224 10.1038/cdd.2016.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 15.Wirawan E., et al. (2012) Beclin1: a role in membrane dynamics and beyond. Autophagy 8, 6–17 10.4161/auto.8.1.16645 [DOI] [PubMed] [Google Scholar]
- 16.Singh S.V., et al. (2017) Restoration of miR-30a expression inhibits growth, tumorigenicity of medulloblastoma cells accompanied by autophagy inhibition. Biochem. Biophys. Res. Commun. 491, 946–952 10.1016/j.bbrc.2017.07.140 [DOI] [PubMed] [Google Scholar]
- 17.Seca H., et al. (2013) Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Curr. Drug Targets 14, 1135–1143 10.2174/13894501113149990185 [DOI] [PubMed] [Google Scholar]
- 18.Bai R., Zhao A.Q., Zhao Z.Q., Liu W.L. and Jian D.M. (2015) MicroRNA-195 induced apoptosis in hypoxic chondrocytes by targeting hypoxia-inducible factor 1 alpha. Eur. Rev. Med. Pharmacol. Sci. 19, 545–551 [PubMed] [Google Scholar]
- 19.Guo Y.J., Liu J.X. and Guan YW. (2016) Hypoxia induced upregulation of miR-301a/b contributes to increased cell autophagy and viability of prostate cancer cells by targeting NDRG2. Eur. Rev. Med. Pharmacol. Sci. 20, 101–108 [PubMed] [Google Scholar]
- 20.Yu G., Jia Z. and Dou Z. (2017) miR-24-3p regulates bladder cancer cell proliferation, migration, invasion and autophagy by targeting DEDD. Oncol. Rep. 37, 1123–1131 10.3892/or.2016.5326 [DOI] [PubMed] [Google Scholar]
- 21.Taguchi-Atarashi N., et al. (2010) Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic 11, 468–478 10.1111/j.1600-0854.2010.01034.x [DOI] [PubMed] [Google Scholar]
- 22.Lin Y., et al. (2012) Genetic polymorphism at miR-181a binding site contributes to gastric cancer susceptibility. Carcinogenesis 33, 2377–2383 10.1093/carcin/bgs292 [DOI] [PubMed] [Google Scholar]
- 23.Lin Y., Zhao J., Wang H., Cao J. and Nie Y. (2017) miR-181a modulates proliferation, migration and autophagy in AGS gastric cancer cells and downregulates MTMR3. Mol. Med. Rep. 15, 2451–2456 10.3892/mmr.2017.6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Wang Y., Xiong Y., Wu J., Ding H., Chen X., Lan L. and Zhang H. (2016) Galangin induces autophagy via deacetylation of LC3 by SIRT1 in HepG2 cells. Sci. Rep. 6, 30496 10.1038/srep30496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang R., et al. (2015) Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 57, 456–466 10.1016/j.molcel.2014.12.013 [DOI] [PubMed] [Google Scholar]
- 26.Tong X., et al. (2018) Elevated levels of serum MiR-152 and miR-24 in uterine sarcoma: potential for inducing autophagy via SIRT1 and deacetylated LC3. Br. J. Biomed. Sci. 75, 7–12 10.1080/09674845.2017.1340225 [DOI] [PubMed] [Google Scholar]
- 27.Chen S., Li P., Li J., Wang Y., Du Y., Chen X. et al. (2015) MiR-144 inhibits proliferation and induces apoptosis and autophagy in lung cancer cells by targeting TIGAR. Cell. Physiol. Biochem. 35, 997–1007 10.1159/000369755 [DOI] [PubMed] [Google Scholar]
- 28.Gong Y., et al. (2014) Nodal promotes the self-renewal of human colon cancer stem cells via an autocrine manner through Smad2/3 signaling pathway. Biomed. Res. Int. 2014, 364134 10.1155/2014/364134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhai H., et al. (2015) Inhibition of colorectal cancer stem cell survival and invasive potential by hsa-miR-140-5p mediated suppression of Smad2 and autophagy. Oncotarget 6, 19735–19746 10.18632/oncotarget.3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang W., et al. (2016) miR-224-3p inhibits autophagy in cervical cancer cells by targeting FIP200. Sci. Rep. 6, 33229 10.1038/srep33229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang X., et al. (2013) XIAP inhibits autophagy via XIAP-Mdm2-p53 signalling. EMBO J. 32, 2204–2216 10.1038/emboj.2013.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen P., et al. (2017) MiR-23a modulates X-linked inhibitor of apoptosis-mediated autophagy in human luminal breast cancer cell lines. Oncotarget 8, 80709–80721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo W., et al. (2017) Down-regulated miR-23a contributes to the metastasis of cutaneous melanoma by promoting autophagy. Theranostics 7, 2231–2249 10.7150/thno.18835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallagher L.E., Williamson L.E. and Chan E.Y. (2016) Advances in autophagy regulatory mechanisms. Cells 5, 1135–1143 10.3390/cells5020024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye Z., et al. (2017) miR-138 suppresses the proliferation, metastasis and autophagy of non-small cell lung cancer by targeting Sirt1. Oncol. Rep. 37, 3244–3252 10.3892/or.2017.5619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan Y.X., Dai Y.Z., Wang X.L., Ren Y.Q., Han J.J. and Zhang H. (2016) MiR-18a upregulation enhances autophagy in triple negative cancer cells via inhibiting mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 20, 2194–2200 [PubMed] [Google Scholar]
- 37.Ma Y., et al. (2014) Biphasic regulation of autophagy by miR-96 in prostate cancer cells under hypoxia. Oncotarget 5, 9169–9182 10.18632/oncotarget.2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu X., et al. (2017) CXCL12/CXCR4 axis induced miR-125b promotes invasion and confers 5-fluorouracil resistance through enhancing autophagy in colorectal cancer. Sci. Rep. 7, 42226 10.1038/srep42226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., et al. (2014) Regulation of autophagy by miR-30d impacts sensitivity of anaplastic thyroid carcinoma to cisplatin. Biochem. Pharmacol. 87, 562–570 10.1016/j.bcp.2013.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang R., et al. (2017) Mir-30d suppresses cell proliferation of colon cancer cells by inhibiting cell autophagy and promoting cell apoptosis. Tumour Biol. 39, 10.1177/1010428317703984 [DOI] [PubMed] [Google Scholar]
- 41.Tan S., et al. (2016) miR-409-3p sensitizes colon cancer cells to oxaliplatin by inhibiting Beclin-1-mediated autophagy. Int. J. Mol. Med. 37, 1030–1038 10.3892/ijmm.2016.2492 [DOI] [PubMed] [Google Scholar]
- 42.Zimmerman J.W., et al. (2012) Cancer cell proliferation is inhibited by specific modulation frequencies. Br. J. Cancer 106, 307–313 10.1038/bjc.2011.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y., et al. (2017) Low frequency magnetic fields induce autophagy-associated cell death in lung cancer through miR-486-mediated inhibition of Akt/mTOR signaling pathway. Sci. Rep. 7, 11776 10.1038/s41598-017-10407-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X., et al. (2015) MiR-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Mol. Cancer 14, 208 10.1186/s12943-015-0480-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y., et al. (2017) MiR-142-3p overexpression increases chemo-sensitivity of NSCLC by inhibiting HMGB1-mediated autophagy. Cell. Physiol. Biochem. 41, 1370–1382 10.1159/000467896 [DOI] [PubMed] [Google Scholar]
- 46.Song L., et al. (2016) MiR-21 modulates radiosensitivity of cervical cancer through inhibiting autophagy via the PTEN/Akt/HIF-1alpha feedback loop and the Akt-mTOR signaling pathway. Tumour Biol. 37, 12161–12168 10.1007/s13277-016-5073-3 [DOI] [PubMed] [Google Scholar]
- 47.Wei R., et al. (2016) miR-140-5p attenuates chemotherapeutic drug-induced cell death by regulating autophagy through inositol 1,4,5-trisphosphate kinase 2 (IP3k2) in human osteosarcoma cells. Biosci. Rep. 36, e00392, , 10.1042/BSR20160238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guttman M. and Rinn J.L. (2012) Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 10.1038/nature10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weidle U.H., et al. (2017) Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics 14, 143–160 10.21873/cgp.20027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon J.H., Abdelmohsen K. and Gorospe M. (2013) Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 425, 3723–3730 10.1016/j.jmb.2012.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao Y., Li J. and Wang L. (2014) Large intervening non-coding RNA HOTAIR is an indicator of poor prognosis and a therapeutic target in human cancers. Int. J. Mol. Sci. 15, 18985–18999 10.3390/ijms151018985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L., et al. (2016) The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol. Biosyst. 12, 2605–2612 10.1039/C6MB00114A [DOI] [PubMed] [Google Scholar]
- 53.Yang Y., et al. (2018) Silencing of LncRNA-HOTAIR decreases drug resistance of Non-Small Cell Lung Cancer cells by inactivating autophagy via suppressing the phosphorylation of ULK1. Biochem. Biophys. Res. Commun., 497, 1003–1010, , 10.1016/j.bbrc.2018.02.141 [DOI] [PubMed] [Google Scholar]
- 54.Wang Y., et al. (2014) BRAF-activated long non-coding RNA contributes to cell proliferation and activates autophagy in papillary thyroid carcinoma. Oncol. Lett. 8, 1947–1952 10.3892/ol.2014.2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin Y., et al. (2018) LncRNA GAS8-AS1 inhibits cell proliferation through ATG5-mediated autophagy in papillary thyroid cancer. Endocrine 59, 555–564 10.1007/s12020-017-1520-1 [DOI] [PubMed] [Google Scholar]
- 56.Yu X., et al. (2017) HULC: an oncogenic long non-coding RNA in human cancer. J. Cell. Mol. Med. 21, 410–417 10.1111/jcmm.12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen S., et al. (2017) The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis. 8, e3118 10.1038/cddis.2017.486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Y., et al. (2014) Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol. Rep. 31, 358–364 10.3892/or.2013.2850 [DOI] [PubMed] [Google Scholar]
- 59.Morselli E., et al. (2014) p53 inhibits autophagy by interacting with the human ortholog of yeast Atg17, RB1CC1/FIP200. Cell Cycle 10, 2763–2769 10.4161/cc.10.16.16868 [DOI] [PubMed] [Google Scholar]
- 60.Balik V., et al. (2013) MEG3: a novel long noncoding potentially tumour-suppressing RNA in meningiomas. J. Neurooncol. 112, 1–8 10.1007/s11060-012-1038-6 [DOI] [PubMed] [Google Scholar]
- 61.Zheng Q., et al. (2018) Long noncoding RNA MEG3 suppresses liver cancer cells growth through inhibiting beta-catenin by activating PKM2 and inactivating PTEN. Cell Death Dis. 9, 253 10.1038/s41419-018-0305-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi Y., et al. (2018) MEG3 inhibits proliferation and invasion and promotes apoptosis of human osteosarcoma cells. Oncol. Lett. 15, 1917–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ying L., et al. (2013) Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol. Biosyst. 9, 407–411 10.1039/c2mb25386k [DOI] [PubMed] [Google Scholar]
- 64.Tian X. and Xu G. (2015) Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis. BMJ Open 5, e008653 10.1136/bmjopen-2015-008653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li L., et al. (2016) Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol. Cancer Ther. 15, 2232–2243 10.1158/1535-7163.MCT-16-0008 [DOI] [PubMed] [Google Scholar]
- 66.Sanchez-Jimenez C., Ludena M.D. and Izquierdo J.M. (2015) T-cell intracellular antigens function as tumor suppressor genes. Cell Death Dis. 6, e1669 10.1038/cddis.2015.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu C., et al. (2018) A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J. Hematol. Oncol. 11, 77 10.1186/s13045-018-0619-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen Y., et al. (2017) LincRNA-p21 knockdown enhances radiosensitivity of hypoxic tumor cells by reducing autophagy through HIF-1/Akt/mTOR/P70S6K pathway. Exp. Cell Res. 358, 188–198 10.1016/j.yexcr.2017.06.016 [DOI] [PubMed] [Google Scholar]
- 69.Li L.J., et al. (2017) The effects of the long non-coding RNA MALAT-1 regulated autophagy-related signaling pathway on chemotherapy resistance in diffuse large B-cell lymphoma. Biomed. Pharmacother. 89, 939–948 10.1016/j.biopha.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 70.Ma B., et al. (2017) Long noncoding RNA MEG3 contributes to cisplatininduced apoptosis via inhibition of autophagy in human glioma cells. Mol. Med. Rep. 16, 2946–2952 10.3892/mmr.2017.6897 [DOI] [PubMed] [Google Scholar]
- 71.Salmena L., et al. (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tay Y., Rinn J. and Pandolfi P.P. (2014) The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma Y., et al. (2017) PVT1 affects growth of glioma microvascular endothelial cells by negatively regulating miR-186. Tumour Biol. 39, 1010428317694326, , 10.1177/1010428317694326 [DOI] [PubMed] [Google Scholar]
- 74.Fu Z., et al. (2017) Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem. Biophys. Res. Commun. 492, 480–486 10.1016/j.bbrc.2017.08.070 [DOI] [PubMed] [Google Scholar]
- 75.Bao X., et al. (2017) Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth. Cell Death Dis. 8, e2605 10.1038/cddis.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu K., et al. (2017) LncRNA SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR-141. J. Biomed. Sci. 24, 46 10.1186/s12929-017-0353-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Z., et al. (2016) Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem. Biophys. Res. Commun. 473, 1268–1275 10.1016/j.bbrc.2016.04.054 [DOI] [PubMed] [Google Scholar]
- 78.Chen J.F., et al. (2018) STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol. Cancer 17, 6 10.1186/s12943-017-0756-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen C.L., et al. (2015) Suppression of hepatocellular carcinoma by baculovirus-mediated expression of long non-coding RNA PTENP1 and MicroRNA regulation. Biomaterials 44, 71–81 10.1016/j.biomaterials.2014.12.023 [DOI] [PubMed] [Google Scholar]
- 80.Bhat P., et al. (2018) Modulating autophagy in cancer therapy: advancements and challenges for cancer cell death sensitization. Biochem. Pharmacol. 147, 170–182 10.1016/j.bcp.2017.11.021 [DOI] [PubMed] [Google Scholar]
- 81.YiRen H., et al. (2017) Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol. Cancer 16, 174 10.1186/s12943-017-0743-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiong H., et al. (2017) LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 36, 3528–3540 10.1038/onc.2016.521 [DOI] [PubMed] [Google Scholar]
- 83.Yang M.C., et al. (2015) Blockade of autophagy reduces pancreatic cancer stem cell activity and potentiates the tumoricidal effect of gemcitabine. Mol. Cancer 14, 179 10.1186/s12943-015-0449-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li C., et al. (2016) Linc-ROR confers gemcitabine resistance to pancreatic cancer cells via inducing autophagy and modulating the miR-124/PTBP1/PKM2 axis. Cancer Chemother. Pharmacol. 78, 1199–1207 10.1007/s00280-016-3178-4 [DOI] [PubMed] [Google Scholar]
- 85.Hatok J. and Racay P. (2016) Bcl-2 family proteins: master regulators of cell survival. Biomol. Concepts 7, 10.1515/bmc-2016-0015 [DOI] [PubMed] [Google Scholar]
- 86.Ma B., et al. (2017) Long non-coding RNA AC0231153 suppresses chemoresistance of glioblastoma by reducing autophagy. Biochim. Biophys. Acta 1864, 1393–1404 10.1016/j.bbamcr.2017.05.008 [DOI] [PubMed] [Google Scholar]
- 87.Palmieri G., et al. (2017) Long non-coding RNA CASC2 in human cancer. Crit. Rev. Oncol. Hematol. 111, 31–38 10.1016/j.critrevonc.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 88.Jiang C., et al. (2018) Upregulation of CASC2 sensitized glioma to temozolomide cytotoxicity through autophagy inhibition by sponging miR-193a-5p and regulating mTOR expression. Biomed. Pharmacother. 97, 844–850 10.1016/j.biopha.2017.10.146 [DOI] [PubMed] [Google Scholar]
- 89.Pasquier B. (2015) Autophagy inhibitors. Cell. Mol. Life Sci. 73, 985–1001 10.1007/s00018-015-2104-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu Y.T., et al. (2010) Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J. Biol. Chem. 285, 10850–10861 10.1074/jbc.M109.080796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Onorati A.V., et al. (2018) Targeting autophagy in cancer. Cancer, 124, 3307–3318 10.1002/cncr.31335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li X., et al. (2013) Autophagy modulation as a target for anticancer drug discovery. Acta Pharmacol. Sin. 34, 612–624 10.1038/aps.2013.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Korkmaz G., et al. (2012) miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy 8, 165–176 10.4161/auto.8.2.18351 [DOI] [PubMed] [Google Scholar]