Abstract
Introduction
There is increasing evidence that the microbiome contributes to esophageal disease. Diet, especially fiber and fat intake, is a known potent modifier of the colonic microbiome, but its impact on the esophageal microbiome is not well described. We hypothesized that dietary fiber and fat intake would be associated with a distinct esophageal microbiome.
Methods
We collected esophageal samples from 47 ambulatory patients scheduled to undergo endoscopy who completed a validated food frequency questionnaire quantifying dietary fiber and fat intake. Using 16S high-throughput sequencing, we determined composition of the esophageal microbiome and predicted functional capacity of microbiota based on fiber and fat intake.
Results
Among all samples, the most abundant phyla were Firmicutes (54.0%), Proteobacteria (19.0%), Bacteroidetes (17.0%), Actinobacteria (5.2%), and Fusobacteria (4.3%). Increasing fiber intake was significantly associated with increasing relative abundance of Firmicutes (p = 0.04) and decreasing relative abundance of Gram-negative bacteria overall (p = 0.03). Low fiber intake was associated with increased relative abundance of several Gram-negative bacteria, including Prevotella, Neisseria, and Eikenella. Several predicted metabolic pathways differed between highest and lowest quartile of fiber intake. Fat intake was associated with altered relative abundance of few taxa, with no alterations at the phylum level and no changes in microbiome functional composition.
Conclusions
Dietary fiber, but not fat, intake was associated with a distinct esophageal microbiome. Diet should be considered an important modifier of the esophageal microbiome in future studies. Studies are also needed to elucidate how the effects of dietary fiber on the esophageal microbiome may contribute to esophageal disease.
Introduction
An increasing body of literature links esophageal diseases to the esophageal microbiome. Eosinophilic esophagitis (EOE), gastroesophageal reflux disease (GERD) and Barrett’s esophagus (BE), esophageal adenocarcinoma (EAC), and esophageal squamous cell carcinoma (ESCC) and its precursor lesions have all been associated with altered composition of the esophageal microbiome1–5. Given the proposed role of the microbiome in esophageal pathology, it is important to characterize the host and environmental factors that may shape esophageal microbiota. One such potential factor is diet.
Diet is a potent modulator of the distal gut microbiome, and plays an important role in disease pathogenesis. In animal models of the colonic microbiome, diet—especially fat and fiber intake—causes profound alterations of microbial composition. High-fat diet causes significant disruption of colonic microbiota, including inverted proportions of the two dominant phyla, Bacteroidetes to Firmicutes6–8. Such changes are associated with obesity, increased intestinal energy harvest, and intestinal inflammation6,7. Conversely, dietary fiber contributes to different microbial alterations, and is associated with reduced weight gain and improved glycemic and lipid control9–11. The microbiome associated with a fiber-deprived diet may correlate with reduced intestinal mucosal barrier robustness and increased gut permeability12.
In humans, increased caloric intake and dietary fiber intake are both associated with altered colonic microbial composition. Increased fiber intake and associated changes to the microbiome are correlated with decreased colorectal cancer risk and increased insulin sensitivity13–15. High fiber consumption may confer a lower risk of Fusobacterium nucleatum-positive, but not F. nucleatum-negative, colon cancer, suggesting that the effect of diet on cancer risk is mediated in part by microbiome composition16.
The impact of diet on the esophageal microbiome, on the other hand, has not been extensively studied. Dietary intake is associated with risk or management of a variety of esophageal pathologies, including EOE and the reflux-associated esophageal diseases (GERD, BE, and EAC). The mechanism through which diet produces this impact on esophageal health is unknown, raising the possibility that its effects are mediated by the esophageal microbiome.
In this study, we tested the hypothesis that dietary fiber and fat are associated with alterations to the esophageal microbiome. We performed a cross-sectional study using squamous esophagus brushings from patients undergoing upper endoscopy who completed a food frequency questionnaire to quantify dietary fiber and fat intake. Microbiome composition of esophageal samples was determined using 16S rRNA sequencing. We characterized differences in the esophageal microbiome and in predicted functional capacity based on dietary fiber or fat intake.
Methods
Study population
Patient data and samples used for the current study had been previously collected for a case-control study of the esophageal microbiome in adult patients with and without BE, described elsewhere in detail17. Briefly, patients ≥18 years old who were scheduled to undergo upper endoscopy for clinical indications were included. Demographics and clinical data collected prior to endoscopy included height, weight, waist and hip circumference, smoking history, history of reflux symptoms (using a modified version of the Mayo Gastro-Esophageal Reflux Questionnaire18), and medications. Patients were excluded if they had a history of gastric or esophageal cancer, gastric or esophageal surgery, endoscopic therapy for BE or esophageal cancer, immunosuppressed state, or use of antibiotics, steroids, or other immunosuppressants within the previous 3 months. All participants provided written informed consent. This study was approved by the Columbia University Medical Center Institutional Review Board.
Dietary intake assessment
On the day of endoscopy, each patient completed a food frequency questionnaire (FFQ) comprised of 17 questions regarding dietary intake over the preceding 4 weeks. This FFQ was derived from the National Health Interview Survey and validated for assessment of dietary fiber and fat19,20. Results were used to calculate daily fiber intake (grams per day) and fat intake (percentage of calories per day) for each patient. While this FFQ is not specifically validated for fruit and vegetable intake, Pyramid servings per day of fruit/vegetable intake were also calculated21.
Sample collection
All subjects were fasting at the time of endoscopy. Saliva and oral swabs were collected prior to endoscopy. During the endoscopy and prior to tissue sampling, the endoscope channel was flushed with 20 mL sterile water. Each microbiome sample was collected by passing a brush 10 times back and forth in four quadrants (Endoscopy Cytology Brush, model G22174; Cook Medical, Bloomington, IN). Two brushings were taken from the following sites: squamous esophagus (3 cm proximal to the squamo-columnar junction), gastric cardia (within 1 cm of the top of the gastric folds), and mid-BE segment in patients with BE. Brush tips were cut using sterile wire cutters and samples were stored in sterile Eppendorf tubes at −80 °C. For purposes of this study, only esophageal squamous brushings were analyzed.
Bacterial DNA isolation and microbiome analyses
DNA was extracted from the study samples using the Mo Bio DNeasy PowerSoil kit (Qiagen, Valencia, CA). Bacterial DNA was isolated by PCR targeting the V4 hypervariable variable region of the 16S rRNA gene using 515F/806R primers, as described22. The V4 region for all samples was sequenced using the Illumina HiSeq 2500 in Rapid Mode with 2 × 250 bp read length (Illumina, San Diego, CA). The Greengenes reference database was used23. Taxonomic units were clustered at 97% sequence similarity using USEARCH and taxonomic assignments were made using mothur. A phylogenetic tree of the contigs was generated with FastTree version 2.1.724, and diversity indices and unweighted and weighted UniFrac distances were calculated25. Sequence data were uploaded to the NCBI Sequence Read Archive (SRP141237).
For analyses of relative abundance of Gram-negative bacteria, Gram-negative genera and species were identified using a reference list assembled by our group (Supplementary Table 1). The relative abundances of these taxa were summed for each sample.
For analyses of relative abundance of short-chain fatty acid-producing bacteria, relative abundance of genera from Clostridial clusters IV and XIVa were summed for each sample.
Statistical analysis
Continuous variables were summarized using means/medians, and categorical variables were summarized using proportions. Continuous variables were compared using t tests or rank-sum tests where appropriate, and categorical variables were compared using Fisher’s exact tests. Analyses were performed separately according to fiber or fat intake. Spearman correlation coefficients were calculated to assess for associations between dietary fiber or fat intake and relative abundance of individual phyla. Patients were also grouped into quartiles based on daily fiber or fat intake, with the first quartile (Q1) denoting the lowest daily intake and the fourth quartile (Q4) denoting the highest daily intake. Alpha diversity was assessed by calculating Shannon diversity indices. Average linkage (UPGMA) method was used to cluster patients based on unweighted and weighted UniFrac metrics. Rank-sum tests were used to compare pairwise distances within and between the lowest and highest quartiles of fiber or fat intake. Taxon-level differences between groups were assessed via linear discriminant analysis effect size (LEfSe)26,27. Functional composition of the esophageal microbiome was assessed using predicted metabolic pathways derived by phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) analysis28.
Multivariable linear regression was performed to assess effect of age, sex, smoking, PPI use, aspirin use, BE status, waist:hip ratio, or body-mass index (BMI) on the association between dietary fiber or fat intake and phylum relative abundance. A reduced model was generated by sequentially removing variables with the highest p-value that was also > 0.15. As acid suppression was felt to represent a potential effect modifier, a priori secondary analyses were performed stratified by PPI use. Statistical significance was defined as p < 0.05. Bonferroni correction was performed in instances of multiple comparisons, with adjusted p-values reported. Analyses were performed using Stata 14.1 (StataCorp, College Station, TX) and SPSS Statistics v23.0 (IBM, Armonk, NY).
Results
Clinical data
A total of 47 patients met inclusion criteria, completed the FFQ, and underwent endoscopy with squamous esophagus sample collection. Baseline characteristics of the study population are shown in Table 1. No subjects had visible esophagitis on endoscopy. Median dietary fat intake was 33.6% daily calories from fat (IQR 32.4–35.6), with mean 30.9% daily calories from fat in the lowest quartile of fat intake (Q1) and mean 37.7% daily calories from fat in the highest quartile of fat intake (Q4). Median dietary fiber intake was 16.0 g/day of fiber (IQR 13.1–18.4), with mean 11.2 g/day in the lowest quartile of fiber intake (Q1) and mean 24.3 daily calories from fat in the highest quartile of fiber intake (Q4). Within individuals, there was no significant correlation between dietary intakes of fat and fiber. There was a strong positive correlation between daily servings of fruit/vegetable intake and dietary fiber intake (rho = 0.76, p < 0.0001).
Table 1.
Characteristics of study population (n = 47 patients)
| Age (years), median (IQR) | 65.0 (54.0–71.5) |
| Male sex, no. (%) | 37 (78.7) |
| Body mass index, median (IQR)a | 28.4 (25.7–32.5) |
| Waist:hip ratio, median (IQR) | 0.96 (0.94–1.00) |
| Ever smoker, no. (%)b | 27 (57.4) |
| PPI use, no. (%) | 37 (78.7) |
| Daily aspirin use, no. (%)b | 14 (29.8) |
| History of reflux symptoms, no. (%)c | 39 (83.0) |
| Indication for endoscopy, no. (%) | |
| GERD | 5 (10.6) |
| Barrett’s esophagus | 31 (66.0) |
| Other | 11 (23.4) |
| Dietary intake, median (IQR) | |
| Grams per day of fiber | 16.0 (13.1–18.4) |
| Percent daily calories from fat | 33.6% (32.4–35.6%) |
IQR interquartile range, PPI proton pump inhibitor, GERD gastroesophageal reflux disease
aData missing for three patients
bData missing for one patient
cPer symptom questionnaire
Squamous esophageal microbiome
The most abundant bacterial phyla among all samples were Firmicutes (mean 54.0%, SD 21.1%), Proteobacteria (mean 19.0%, SD 15.5%), Bacteroidetes (mean 17.0%, SD 12.1%), Actinobacteria (mean 5.2%, SD 5.9%), and Fusobacteria (mean 4.3%, SD 4.7%). All other phyla had relative abundance <1%. At the genus level, Streptococcus was most abundant, with a mean relative abundance of 37.9% (SD 21.5%). Gram-negative bacteria comprised a mean of 49.0% (SD 22.6%) of bacteria in the esophagus.
There was a non-significant inverse association between fiber intake and alpha diversity by Shannon index (rho = −0.26, p = 0.07), and no association between fat intake and alpha diversity (rho = −0.15, p = 0.32). When stratified by PPI use, the trend toward inverse correlation between fiber intake and alpha diversity neared significance among PPI users (rho = −0.34, adjusted p = 0.08).
Beta diversity analyses were performed comparing the lowest (Q1) and highest (Q4) quartiles of dietary fat or fiber intake. In the unweighted analysis of dietary fiber, there were significantly lower UniFrac distances within Q1 than within Q4 (adjusted p = 0.005) or than within Q1 versus Q4 (adjusted p = 0.03; Supplementary Figure 1). There were no significant differences in the weighted analysis, or in comparisons of lowest and highest dietary fat quartiles.
Dietary fat intake and the esophageal microbiome
There were no significant correlations between fat intake and relative abundance of any phyla (Supplementary Figure 2A–E) or relative abundance of Gram-negative bacteria overall. When stratified by PPI use, there remained no association between fat intake and relative abundance of any phyla. To identify differentially abundant taxa, comparisons were made of lowest (Q1) versus highest (Q4) quartile of fat intake. There were few differences in relative abundance of specific taxa in the two groups. Patients in Q1 had increased relative abundance of Leptotrichia and of unclassified genera within families Streptococcaceae and Neisseriaceae (Supplementary Figure 3).
Dietary fiber intake and the esophageal microbiome
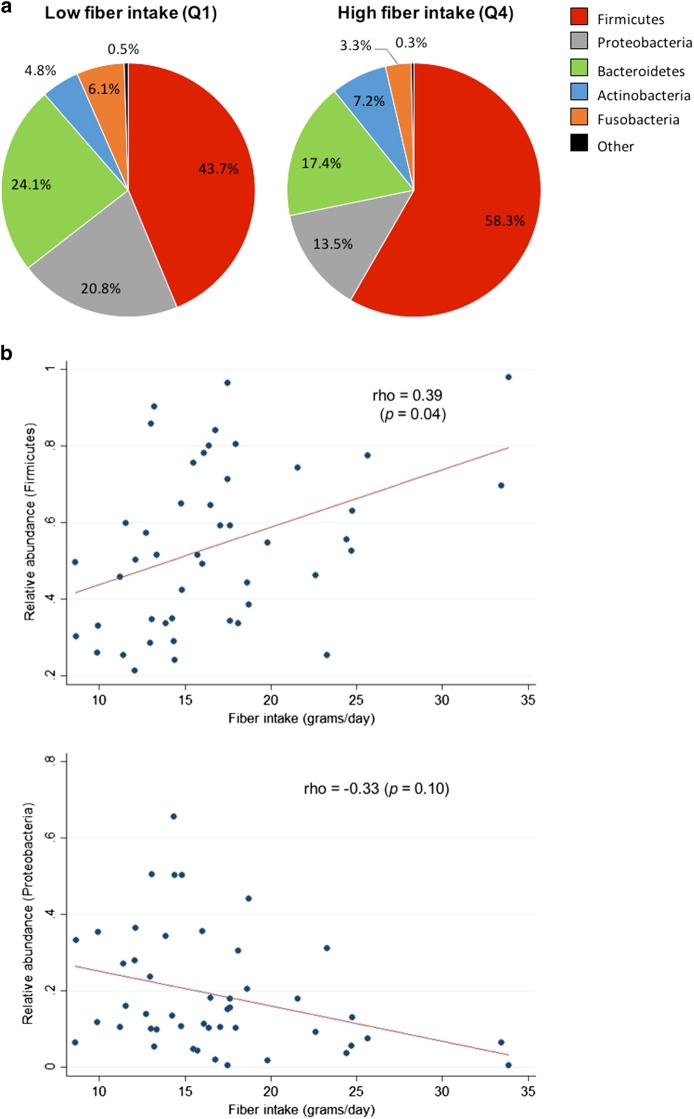
Shifts in mean relative abundance of major phyla were noted in response to dietary fiber intake (Fig. 1a). Increasing fiber intake was significantly correlated with increasing relative abundance of Firmicutes (rho = 0.39, adjusted p = 0.04), and trended toward an inverse correlation with relative abundance of Proteobacteria (rho = −0.33, adjusted p = 0.10) (Fig. 1b, c). Fiber intake was not associated with relative abundance of other phyla (Supplementary Figure 2F–H). In multivariate analyses, the associations between fiber intake and relative abundance of Firmicutes and Proteobacteria were not altered by adjustment for patient characteristics, including BMI or history of GERD symptoms, or medication use. Increasing BMI was independently associated with decreasing relative abundance of Firmicutes (p = 0.03) and increasing relative abundance of Proteobacteria (p = 0.05), the opposite pattern as observed for dietary fiber. Other patient characteristics, including age, sex, smoking status, history of BE, and aspirin use, were not associated with relative abundance of Firmicutes or Proteobacteria. Similar to fiber, dietary fruit/vegetable intake was positively correlated with relative abundance of Firmicutes (rho = 0.48, adjusted p = 0.004) and inversely correlated with relative abundance of Proteobacteria (rho = −0.35, adjusted p = 0.09) and Gram-negative bacteria (rho = −0.51, adjusted p = 0.002).
Fig. 1. Relative abundances of esophageal Firmicutes and Proteobacteria were associated with dietary fiber intake.
a Mean relative abundances of all phyla in samples from patients in the lowest (Q1) and highest (Q4) quartiles of dietary fiber intake. b Increasing dietary fiber intake was associated with increasing relative abundance of Firmicutes and decreasing relative abundance of Proteobacteria (all subjects). p-values are adjusted for multiple comparisons
Among PPI users, there was a trend toward a positive correlation between fiber intake and Firmicutes (rho = 0.35, adjusted p = 0.15), and there was a significant inverse correlation between fiber intake and Proteobacteria (rho = −0.40, adjusted p = 0.05). In PPI non-users, the correlations were attenuated and not significant (Firmicutes: rho = 0.22, adjusted p = 1.00; Proteobacteria: rho = −0.13, adjusted p = 1.00).
There was a significant inverse correlation between fiber intake and relative abundance of Gram-negative bacteria overall (rho = −0.39, adjusted p = 0.03). This association was not altered by adjustment for patient characteristics, including BMI or history of GERD symptoms, or medication use. Increased BMI was independently associated with increased relative abundance of Gram-negative bacteria (p = 0.007), again with an association opposite that of fiber. When stratified by PPI use, the inverse association between fiber intake and relative abundance of Gram-negative bacteria trended towards significance among PPI users (rho = −0.35, adjusted p = 0.08).
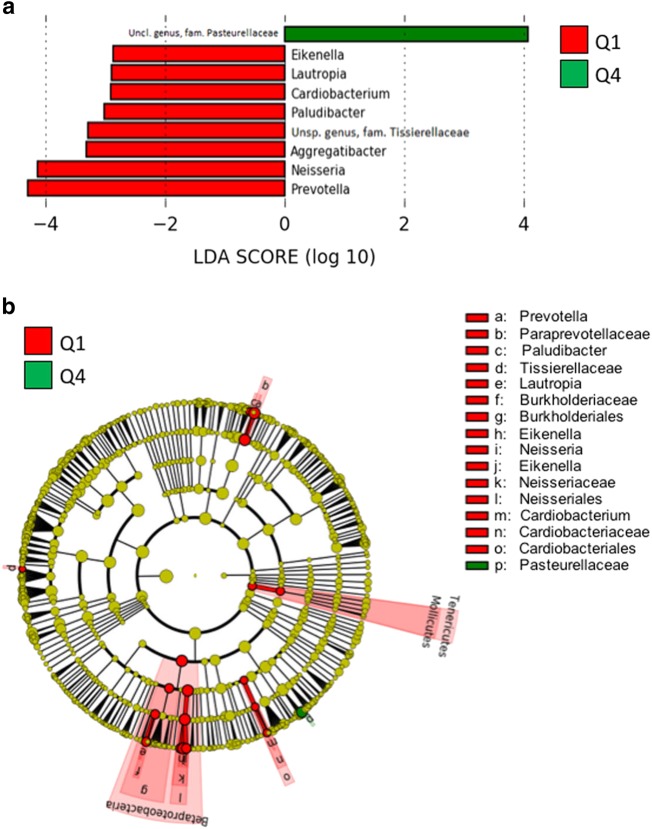
When the lowest quartile of fiber intake (Q1) was compared to the highest quartile of fiber intake (Q4), subjects in Q1 had significantly increased relative abundance of numerous taxa (Fig. 2). These increases occurred predominately within class Betaproteobacteria, including genera Neisseria, Lautropia, and Eikenella. Subjects in Q1 also had increased relative abundance of the genus Prevotella, among other taxa. Subjects in Q4 had increased relative abundance of an unspecific genus in family Pasteurellaceae.
Fig. 2. There were several differentially abundant taxa comparing subjects in the lowest and highest quartiles of fiber intake.
a Linear discriminant analysis effect size (LEfSe) representation of differentially abundant genera, and b cladogram representation of all differentially abundant taxa in esophageal samples between subjects in the lowest quartile (Q1) compared to the highest quartile (Q4) of fiber intake
Short-chain fatty acid-producing bacteria
We explored the possibility that diet may be associated with relative abundance of short-chain fatty acid (SCFA)-producing bacteria, as has been observed in the colon. SCFA producers were very rare in the esophagus; of the 47 subjects included in the study, only 10 had a specimen containing any SCFA-producing bacteria. The highest combined relative abundance in a single sample was 0.9%. Comparing subjects with and without any SCFA-producing bacteria, there was no difference in intake of fiber (p = 0.18) or fat (p = 0.96). There was also no association with PPI use (p = 0.64).
Functional composition of the esophageal microbiome
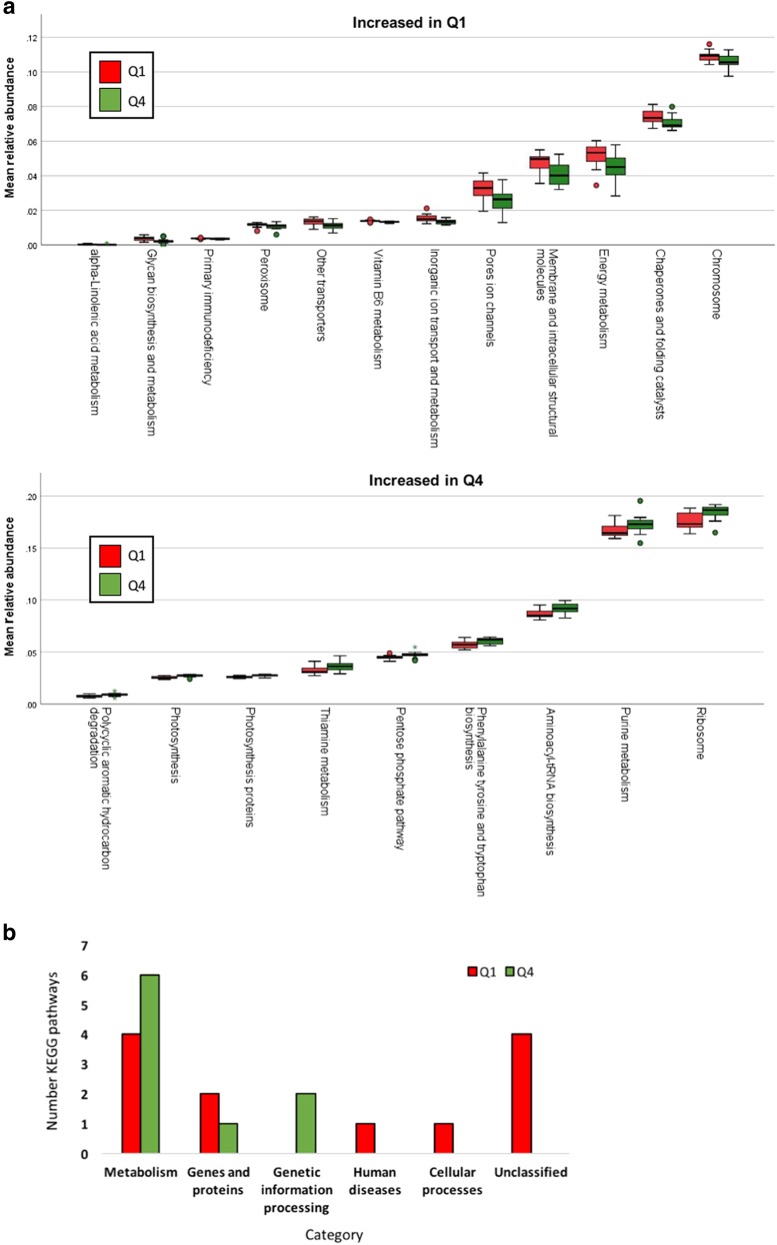
Relative abundances of predicted metabolic pathways were compared between patients in the lowest (Q1) versus highest (Q4) quartiles of fat or fiber intake. Fat intake was not associated with differential relative abundance of any metabolic pathways. Fiber intake was significantly associated with altered relative abundance of a number of predicted metabolic pathways (Fig. 3a). The majority of pathways affected were involved in cell metabolism, with more of these pathways increased in Q4 than in Q1 (Fig. 3b).
Fig. 3. Functional changes to the esophageal microbiome were associated with fiber intake.
a Predicted metabolic pathways increased in lowest quartile (Q1) or highest quartile (Q4) of fiber intake. b Number of pathways increased in a given KEGG category in Q1 compared to Q4
Discussion
In this prospective study of the association between dietary fat and fiber intake and esophageal microbial composition, fiber intake was associated with a distinct microbiome. Increased fiber intake was associated with increased relative abundance of Firmicutes and decreased relative abundance of Gram-negative bacteria overall. Many taxonomic alterations among patients with the lowest quartile of fiber intake were observed within class Betaproteobacteria. Relative abundance of numerous predicted metabolic pathways varied based on fiber intake. Fat intake was associated with only minor alterations to the esophageal microbiome and no predicted functional changes.
When patients were stratified according to PPI use, the significant associations between fiber intake and microbiome composition were restricted to patients taking PPIs. PPIs are known to impact the upper gastrointestinal microbiome29, so it is possible that this exposure either acts synergistically with dietary fiber to magnify the impact on the microbiome or makes the microbiome more susceptible to effects of fiber. Ultimately, the differential observations between PPI users and non-users must be interpreted with caution due to small sample size. Many of the trends observed among PPI non-users were similar to those of PPI users, and the former group may have been underpowered to detect significant alterations.
The mechanisms for the association between dietary intake and esophageal microbiome composition may be multifactorial. Given that food passes only very transiently through the esophagus, it is possible that dietary intake impacts the esophageal microbiome indirectly. One plausible mechanism is through effects on the microbiome of adjacent areas of the gastrointestinal tract. Diet—such as high sugar intake or intake of certain vitamins—is known to affect composition of the oral microbiome30,31. Oral and esophageal microbial compositions are closely related, possibly due to movement of oral microbes to more distal gastrointestinal sites;32 as such, it is possible that the associations between dietary fiber and esophageal microbiota may be mediated by effects of fiber or other related factors on the oral microbiome. Similarly, perhaps through association with obesity, fiber intake could be associated with frequency or severity of acid reflux and reflux-associated disease, another factor with known associations with altered upper gastrointestinal microbiome composition3,17,33. On the other hand, dietary fiber could impact the esophageal microbiome via its effects on host metabolism. In our study, increased BMI had significant independent associations with esophageal microbiota. The phyla affected by increased BMI were the same as those associated with fiber intake, but in the opposite direction; for example, increased relative abundance of Firmicutes was associated with increasing fiber intake but decreasing BMI. It is possible that the impact of fiber on the esophageal microbiome is mediated in part by its impact on the distal GI tract or on systemic metabolism, which in turn influences BMI. Low fiber intake can lead to weight gain and obesity34, and high fiber intake may contribute to increased SCFA production in the colon and improved systemic insulin sensitivity;35 these changes might in turn have effects on the esophageal microbiome.
Changes to the esophageal microbiome associated with dietary fiber may have important implications for esophageal disease. In this study, fiber intake was positively associated with relative abundance of Firmicutes and negatively associated with relative abundance of Gram-negative bacteria, most notably Betaproteobacteria. Some studies of the distal esophagus microbiome have posited the presence of two predominant “types”: one dominated by Streptococcus, a species within phylum Firmicutes, and associated with a healthy, phenotypically normal esophagus, and one with high abundance of Gram-negative anaerobes and microaerophiles and associated with abnormal esophagus, including reflux esophagitis and BE2,3. The inverse association between dietary fiber and prevalence of Gram-negative bacteria in this study provides one mechanism through which increased fiber consumption might protect against inflammation-mediated esophageal disease.
In studies of the distal GI microbiome, the beneficial effects of fiber to colonic health are driven in part by promotion of SCFA production by Clostridial clusters IV and XIVa36. These microbes are thought to contribute to colonic health via SCFA—specifically butyrate—production, which provides energy to colonocytes and contributes to mucosal barrier integrity, thus increasing resistance to opportunistic colonic infections37,38. We explored whether diet may impact esophageal health via effects on SCFA-producing bacteria. In our study, however, SCFA-producing bacteria were only detected in 10 of 47 patients, and the sum relative abundance of SCFA-producing bacteria for all individuals was very low. A significant impact of dietary fiber on esophageal SCFA production is therefore unlikely, or at least difficult to detect due to low abundance.
This study had several strengths. This is one of the first studies to investigate the impact of diet on the upper GI microbiome. Dietary intake in our study population closely mirrored that of US adults, among whom mean dietary fat intake is 33.6% daily calories from fat39 and mean dietary fiber intake is 17.3 g/day40. There was a broad spread of fiber intake in the study population. Data about patient medication use, smoking history, and anthropometrics were collected, which allowed for assessment of how these factors affected the association between fat or fiber intake and microbiome composition. Several significant associations between fiber intake and esophageal microbiota were identified.
There were certain limitations. Some analyses, such as stratification by PPI use, were limited by sample size. There was a relatively modest difference in daily dietary fat intake between the highest and lowest quartiles; the interquartile range of median daily percent calories from fat was only 3.1%. As such, it is possible that the effect of dietary fat on the esophageal microbiome was underestimated. While the median 16.0 g/day of dietary fiber intake in our study was representative of average intake in the United States, this amount is substantially lower than the FDA-recommended 25 g/day of fiber. Future studies of populations with more varied dietary intake may reveal a stronger association between fat and fiber and esophageal microbiome composition. As PPIs substantially impact the esophageal microbiome and most patients in our study with a history of reflux symptoms were taking PPIs, meaningful analyses assessing the impact of reflux symptoms in the absence of PPIs could not be performed. Finally, while the FFQ used for this study was validated for fat and fiber intake, a more extensive questionnaire in future studies may provide more granular detail with regard to the impact of specific nutrients or food products on the esophageal microbiome. The FFQ did allow for calculation of fruit and vegetable intake, which correlated closely with dietary fiber; it is possible that the associations between dietary fiber and esophageal microbiome composition in this study are attributable in part to other aspects of fruit and vegetable consumption.
Dietary fiber intake was associated with distinct esophageal microbiome composition, while fat intake appeared to have a much lesser impact. In the current study, increased fiber intake was associated with increased relative abundance of Firmicutes and decreased relative abundance of Gram-negative bacteria, as well as altered abundance of several specific taxa. These findings may indicate an association between low fiber intake and an esophageal microbiome that promotes chronic inflammation. Dietary fiber may play a powerful role in shaping the esophageal microbiome, and should be considered an important modifier in studies of the esophageal microbiome. Future studies are needed to elucidate how the effects of dietary fiber on the esophageal microbiome contribute to esophageal health and disease.
Study Highlights
What is current knowledge
Microbiome composition is associated with a variety of esophageal diseases.
Diet profoundly impacts the colonic microbiome, but its effect on the esophageal microbiome is unknown.
What is new here
Dietary fiber intake was associated with a distinct human esophageal microbiome.
Fiber intake was associated with increased Firmicutes relative abundance and decreased Gram-negative bacteria relative abundance.
Dietary fat intake was associated with only minimal changes to the esophageal microbiome.
Electronic supplementary material
Financial support
The authors were supported in part by a Columbia Physician’s and Surgeon’s Dean’s Research Fellowship (E.J.S.), a Career Development Award from NIDDK (K23 DK111847; D.E.F.), a U54 award from NCI (U54 CA163004; J.A.A.), and the Price Family Foundation (J.A.A.).
Conflict of interest
Guarantor of the article: Julian A. Abrams, MD, MS.
Specific author contributions: Y.R.N.—study concept and design, interpretation of data, manuscript preparation, critical revision of manuscript, approval of final draft submitted. E.J.S.—study conduct, interpretation of data, critical revision of manuscript, approval of final draft submitted. G.C.—study conduct, critical revision of manuscript, approval of final draft submitted. D.E.F.—analysis and interpretation of data, critical revision of manuscript, approval of final draft submitted. H.K.—analysis and interpretation of data, critical revision of manuscript, approval of final draft submitted. C.J.L.—study conduct, interpretation of data, critical revision of manuscript, approval of final draft submitted. N.C.T.—analysis and interpretation of data, critical revision of manuscript, approval of final draft submitted. J.A.A.—study concept and design, study conduct, analysis and interpretation of data, manuscript preparation, critical revision of manuscript, approval of final draft submitted.
Potential competing interests
None.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information: The online version of this article (10.1038/s41424-018-0067-7) contains supplementary material, which is available to authorized users.
References
- 1.Harris JK, et al. Esophageal microbiome in eosinophilic esophagitis. PLoS. One. 2015;10:e0128346. doi: 10.1371/journal.pone.0128346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang L, et al. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137:588–597. doi: 10.1053/j.gastro.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gall A, et al. Bacterial composition of the human upper gastrointestinal tract microbiome is dynamic and associated with genomic instability in a Barrett’s esophagus cohort. PLoS. One. 2015;10:e0129055. doi: 10.1371/journal.pone.0129055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackett KL, et al. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett’s and oesophageal carcinoma: association or causality? Aliment. Pharmacol. Ther. 2013;37:1084–1092. doi: 10.1111/apt.12317. [DOI] [PubMed] [Google Scholar]
- 5.Yu G, et al. Association between upper digestive tract microbiota and cancer-predisposing states in the esophagus and stomach. Cancer Epidemiol. Biomark. Prev. 2014;23:735–741. doi: 10.1158/1055-9965.EPI-13-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy EA, Velazquez KT, Herbert KM. Influence of high-fat diet on gut microbiota: a driving force for chronic disease risk. Curr. Opin. Clin. Nutr. Metab. Care. 2015;18:515–520. doi: 10.1097/MCO.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobel YR, et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat. Commun. 2015;6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox LM, et al. The nonfermentable dietary fiber hydroxypropyl methylcellulose modulates intestinal microbiota. FASEB J. 2013;27:692–702. doi: 10.1096/fj.12-219477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parnell JA, Reimer RA. Prebiotic fiber modulation of the gut microbiota improves risk factors for obesity and the metabolic syndrome. Gut Microbes. 2012;3:29–34. doi: 10.4161/gmic.19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, et al. Bamboo shoot fiber prevents obesity in mice by modulating the gut microbiota. Sci. Rep. 2016;6:32953. doi: 10.1038/srep32953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai MS, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339–1353. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen HM, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am. J. Clin. Nutr. 2013;97:1044–1052. doi: 10.3945/ajcn.112.046607. [DOI] [PubMed] [Google Scholar]
- 14.Zeng H, Lazarova DL, Bordonaro M. Mechanisms linking dietary fiber, gut microbiota and colon cancer prevention. World J. Gastrointest. Oncol. 2014;6:41–51. doi: 10.4251/wjgo.v6.i2.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brahe LK, et al. Dietary modulation of the gut microbiota--a randomised controlled trial in obese postmenopausal women. Br. J. Nutr. 2015;114:406–417. doi: 10.1017/S0007114515001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehta RS, et al. Association of dietary patterns with risk of colorectal cancer subtypes classified by Fusobacterium nucleatum in tumor tissue. JAMA Oncol. 2017;3:921–927. doi: 10.1001/jamaoncol.2016.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snider E, et al. Barrett’s esophagus is associated with a distinct oral microbiome. Clin. Transl. Gastroenterol. 2018;9:135. doi: 10.1038/s41424-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Locke GR, et al. A new questionnaire for gastroesophageal reflux disease. Mayo Clin. Proc. 1994;69:539–547. doi: 10.1016/S0025-6196(12)62245-9. [DOI] [PubMed] [Google Scholar]
- 19.Thompson FE, et al. Dietary intake estimates in the National Health Interview Survey, 2000: methodology, results, and interpretation. J. Am. Diet. Assoc. 2005;105:352–363. doi: 10.1016/j.jada.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 20.Williams GC, et al. Performance of a short percentage energy from fat tool in measuring change in dietary intervention studies. J. Nutr. 2008;138:212S–217S. doi: 10.1093/jn/138.1.212S. [DOI] [PubMed] [Google Scholar]
- 21.Thompson FE, et al. Performance of a short tool to assess dietary intakes of fruits and vegetables, percentage energy from fat and fibre. Public Health Nutr. 2004;7:1097–1106. doi: 10.1079/PHN2004642. [DOI] [PubMed] [Google Scholar]
- 22.Nossa CW, et al. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 2010;16:4135–4144. doi: 10.3748/wjg.v16.i33.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 24.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS. One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyob G, et al. Drug susceptibility of Mycobacterium tuberculosis in HIV-infected and -uninfected Ethiopians and its impact on outcome after 24 months of follow-up. Int. J. Tuberc. Lung. Dis. 2004;8:1388–1391. [PubMed] [Google Scholar]
- 27.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freedberg DE, Lebwohl B, Abrams JA. The impact of proton pump inhibitors on the human gastrointestinal microbiome. Clin. Lab. Med. 2014;34:771–785. doi: 10.1016/j.cll.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller MK, et al. Oral microbial profiles of individuals with different levels of sugar intake. J. Oral. Microbiol. 2017;9:1355207. doi: 10.1080/20002297.2017.1355207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato I, et al. Nutritional correlates of human oral microbiome. J. Am. Coll. Nutr. 2017;36:88–98. doi: 10.1080/07315724.2016.1185386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segata N, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amir I, et al. Gastric microbiota is altered in oesophagitis and Barrett’s oesophagus and further modified by proton pump inhibitors. Environ. Microbiol. 2014;16:2905–2914. doi: 10.1111/1462-2920.12285. [DOI] [PubMed] [Google Scholar]
- 34.Ludwig DS, et al. Dietary fiber, weight gain, and cardiovascular disease risk factors in young adults. JAMA. 1999;282:1539–1546. doi: 10.1001/jama.282.16.1539. [DOI] [PubMed] [Google Scholar]
- 35.Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 2015;11:577–591. doi: 10.1038/nrendo.2015.128. [DOI] [PubMed] [Google Scholar]
- 36.Lange K, et al. Comparison of the effects of five dietary fibers on mucosal transcriptional profiles, and luminal microbiota composition and SCFA concentrations in murine colon. Mol. Nutr. Food Res. 2015;59:1590–1602. doi: 10.1002/mnfr.201400597. [DOI] [PubMed] [Google Scholar]
- 37.Lopetuso LR, et al. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. doi: 10.1186/1757-4749-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antharam VC, et al. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 2013;51:2884–2892. doi: 10.1128/JCM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.National Center for Health Statistics. Health, United States, 2016: With Chartbook on Long-term Trends in Health. Centers for Disease Control and Prevention; https://www.cdc.gov/nchs/data/hus/hus16.pdf#056 (2017). [PubMed]
- 40.NHANES. What we eat in America, NHANES 2015–2016. United States Department of Agriculture Agricultural Research Service; https://www.ars.usda.gov/ARSUserFiles/80400530/pdf/1516/Table_1_NIN_GEN_15.pdf(2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.