Abstract
Microorganisms growing at atmospheric pressures of 0.7 kPa may have a significant impact on the search for life on Mars. Data on their nutrient requirements in a simulated Martian environment are required to ascertain both the potential risk of forward contamination and the potential of past or present habitability of Mars. Serratia liquefaciens can grow at concomitant conditions of low pressure, low temperature, and anoxic atmosphere. Changes in the metabolic fingerprint of S. liquefaciens grown under varying physical conditions including diverse atmospheric pressures (0.7 kPa to 101.3 kPa), temperatures (30 °C or 0 °C), and atmospheric gas compositions (Earth or CO2) were investigated using Biolog GN2 assays. Distinct patterns for each condition were observed. Above 10 kPa S. liquefaciens performed similar to Earth-normal pressure conditions (101.3 kPa) whereas below 10 kPa shifts in metabolic patterns were observed. The differences indicated a physiological alteration in which S. liquefaciens lost its ability to metabolize the majority of the provided carbon sources at 0.7 kPa with a significant decrease in the oxidation of amino acids. By measuring the physiological responses to different carbon sources we were able to identify nutritional constraints that support cellular replication under simulated shallow Mars subsurface conditions.
Introduction
Identifying and protecting habitable zones (HZ) on Mars requires knowledge on the geochemical, environmental, and biological factors that may contribute to the colonization of HZ sites by hitchhiking spacecraft microorganisms1,2. In addition, characterizing the limits of terrestrial microbial life under realistic simulations of the Martian surface will inform the search for extant Martian life (if present) in plausible HZ terrains. Both efforts require data on the complex interactions of microbial metabolisms and the Martian environments being explored.
One constraint for robotic or crewed missions to Mars will be the question of whether Earth microorganisms have inherent abilities to survive, grow, and adapt to surface conditions on current-day Mars. One of the most challenging factors on Mars is low atmospheric pressure that ranges from 0.2 to 1 kPa, and equals about 1% of Earth sea-level pressure. Apart from the hypobaric conditions, microbial survival and growth also must cope with low water activities (aw) in soils, low temperatures, the presence of high salt concentrations and oxidizing compounds, low osmotic potentials in the regolith, high pCO2 and low pO2 atmospheres, as well as high doses of UV and ionizing radiation3–5. Recent studies have made progress in demonstrating microbial replication under a subset of the harsh conditions found on Mars by investigating whether bacteria can grow under hypobaric (close to 0.7 kPa), low temperature (near 0 °C for stable liquid water), and CO2-enriched anoxic conditions (henceforth called low-PTA conditions) similar to the Martian surface. Growth of 29 bacterial species under low-PTA conditions5–7 reveals active metabolism and growth at pressures encountered on the Martian surface, and consequently demonstrates the potential that some spacecraft microorganisms may be capable of colonizing hydrated terrains on Mars.
Serratia liquefaciens was one of the first bacteria shown capable of growth under low-PTA conditions5. Serratia liquefaciens, in the family Enterobacteriaceae, is a Gram-negative, motile, facultative anaerobic bacterium, and an ecological generalist. Its habitats include a variety of niches such as surface waters, soils, and the digestive tracts of rodents, plants, insects, fish, and humans8. Furthermore, Serratia spp. have been recovered from spacecraft assembly facilities9,10, Russian spacecraft11 and the International Space Station12,13 (ISS) which makes them plausible candidates to hitchhike on spacecraft to Mars. Experiments testing eight Serratia type strains confirmed the growth of S. ficaria, S. fonticola, S. grimesii, S. liquefaciens, S. plymuthica, and S. quinivorans, whereas S. marcescens and S. rubidaea failed to grow under low-PTA conditions14.
Low-PTA conditions represent a set of selective stressors which may introduce so far unknown alterations to cells rendering them unable to carry out normal metabolism and growth, or that may be lethal. Very little data exists on how microbial metabolism is affected by exposure to low-PTA conditions found on Mars. For example, it is unknown whether the same metabolic pathways (e.g., utilization of amino acids versus sugars) are active under low-PTA compared to Earth sea-level conditions.
Obtaining data on microbial metabolism and growth under simulated Martin conditions near 0.7 kPa is critical for modelling the potential risks of forward contamination in HZ regions on Mars2,15,16. In order to fill this knowledge gap we investigated carbon source utilization by S. liquefaciens using the Biolog GN2 assay system (Biolog, inc. Hayward, CA). The Biolog system was developed for bacterial identification based on species-specific metabolic fingerprints using the differential metabolism of 95 carbon sources17. Once a carbon substrate is metabolized (i.e., a substrate is oxidized) the colourless tetrazolium redox dye is reduced and forms a purple precipitate. The use of the tetrazolium reduction method allows for the quantification of differential substrate consumption providing estimates on the rates of metabolism on numerous organics under diverse conditions.
The objectives of the current study were to characterize carbon source utilization by S. liquefaciens under low-PTA conditions in order to (1) identify the effects of low pressure on metabolism, and (2) define unique organic substrates that are preferred by S. liquefaciens for metabolic activity and growth under simulated Mars surface conditions. Positive results from the latter objective were required to design ongoing complex simulations of microbial growth and adaptation to analogue soils and diverse environmental conditions on Mars (e.g., divergent aw, redox couples, and in situ organics).
Results
Metabolic activity and substrate richness (number of positive wells) of Serratia liquefaciens cells were evaluated under different pressures, temperatures, and gas compositions in which one set of conditions simulated a Martian surface environment (i.e., low-PTA condition). Biolog GN2 microarray plates were inoculated to simultaneously test a wide variety of 95 carbon substrates. The assays were conducted in hypobaric systems capable of maintaining pressures between 0.7 kPa and 101.3 kPa (+/−0.1 kPa). The Biolog GN2 plate includes two alcohols, six amines, 20 amino acids, four aromatic substances, one brominated substrate, 28 carbohydrates, 24 carboxylic acids, two esters, three phosphorylated substrates, five polymers and one control well containing only water17,18. Initially, a series of preliminary tests confirmed that the GN2 plates with 10 mM PO4 buffer alone as the carrier fluid (i.e., without bacteria) remained free of colour-shifts for up to 60 days at 0.7 kPa, 0 °C, and CO2-enriched atmosphere. Consequently, any colour changes measured were interpreted to be the result of microbial metabolism and not the spontaneous reduction of the tetrazolium dye.
Exp-1: Responses to various carbon sources in different atmospheric pressure conditions
In Exp-1, the temperature was held constant at 30 °C in an Earth-normal atmosphere of 78% pN2 and 21% pO2 present in the laboratory, and only pressure was modified to 2.5, 5.0, 10.0, 20.0, or 101.3 kPa (Earth-normal pressure). The Earth-normal atmosphere was created by allowing lab air to diffuse into the vacuum desiccators during the experiments. Due to the effects of low pressure on the evaporation rates of water, assays could not be conducted at pressures below 2.5 kPa, when incubated at 30 °C. Even at 2.5 kPa, the GN2 plates had to be partially rehydrated after 24 h of incubation during the 48-h assays. Rehydration was required to keep the nutrient concentrations and osmotic effects stable in each well throughout the experiment.
A principal component analysis (PCA) using the normalized absorbance values of the carbon sources as variables was performed to compare the carbon utilization profiles for the five tested pressures (Fig. 1). Principal component axis 1 accounted for 64% and PC axis 2 accounted for 9% of the total variance. The first PC separated the plates maintained at low pressures (2.5 and 5.0 kPa) from the plates incubated at higher pressures pointing towards different carbon utilization patterns amongst samples. Samples between 10.0 and 101.3 kPa grouped together indicating that pressures in this range only had a minor effect on carbon source utilization. There was no trend observed along the second principal component (PC2).
Figure 1.
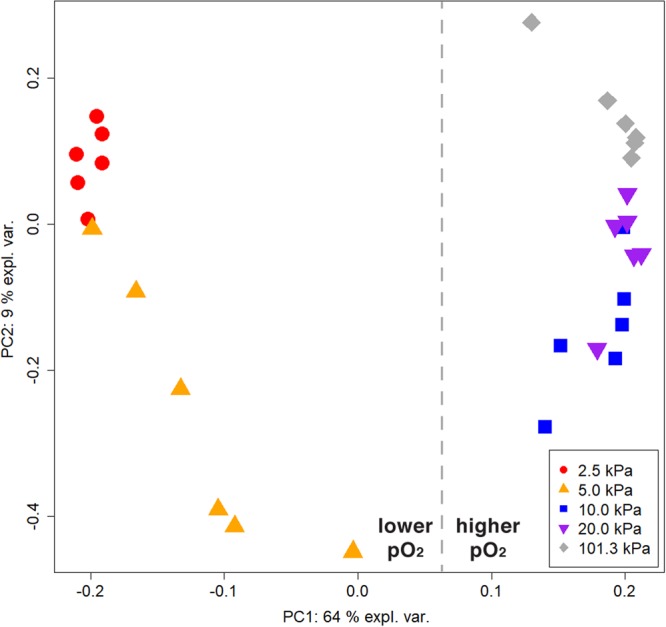
Substrate utilization patterns by Serratia liquefaciens under different pressures using Biolog GN2 microarrays. Ordination plot of metabolic fingerprints for cells grown at 30 °C; Earth-normal atmosphere; and pressures maintained at 2.5, 5.0, 10.0, 20.0, or 101.3 kPa (n = 6).
A summary of the overall carbon utilization patterns under different pressures is shown in Fig. 2a. A full list of substrates and their utilization can be found in Supplementary Table S1. Biolog GN2 plates kept at 2.5 and 5.0 kPa showed the smallest metabolic potential whereas samples incubated at Earth-normal pressure revealed the largest metabolic potential. While S. liquefaciens was able to utilize a total of 74 carbon sources at 101.3 kPa, the number of metabolized substrates decreased with decreasing atmospheric pressure to 41 at 2.5 kPa (Table 1). A similar trend was seen for the total well colour development (TWCD) with values ranging from 95 to 25 (i.e., for 101.3 to 2.5 kPa, respectively), and revealing only slight differences between 10.0 and 20.0 kPa (69 vs. 73). Thus, most changes occurred at pressures below 10.0 kPa. In addition, more than 50% of the positive reactions at 2.5 kPa showed very low OD values (i.e., <0.32) indicating low metabolic activity. Approximately half of the carbon sources used by S. liquefaciens at Earth-normal conditions supported metabolic activity at 2.5 kPa.
Figure 2.
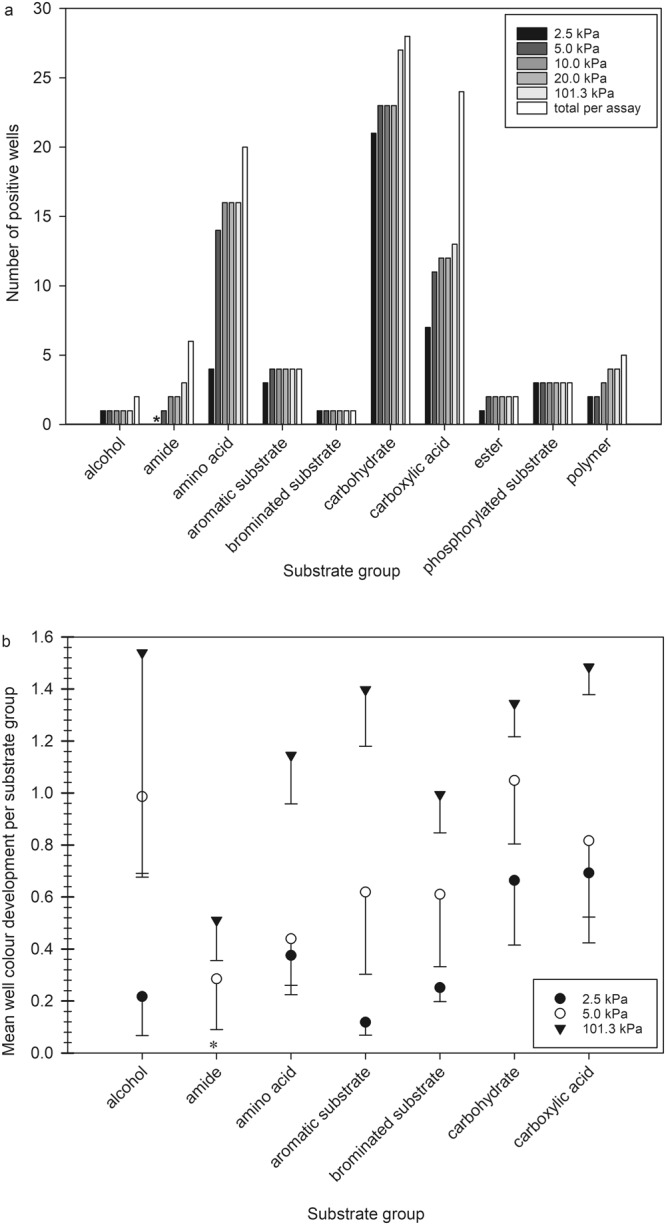
Overview of the metabolic fingerprints for Serratia liquefaciens cells grown under different atmospheric pressures using Biolog GN2 phenotypic microarrays. (a) The specific numbers of positive wells per substrate group are given for the following categories: alcohols, amides, amino acids, aromatic substrates, brominated substrates, carbohydrates, carboxylic acids, ester, phosphorylated substrates and polymers under the different pressure conditions (2.5, 5.0, 10.0, 20.0, 101.3 kPa) tested. (b) Comparison of OD readings between assays incubated at 2.5, 5.0, or 101.3 kPa. Error bars reflect standard deviations of the means between six independent replicates of the GN2 microarrays.
Table 1.
Total well colour development (TWCD) values and number of carbon sources utilized by Serratia liquefaciens under different pressures.
| Pressure (kPa) | TWCD | Carbon sources used |
|---|---|---|
| 2.5 | 25.32 ± 9.27 | 41 |
| 5.0 | 50.44 ± 15.32 | 62 |
| 10.0 | 69.02 ± 13.47 | 69 |
| 20.0 | 73.70 ± 14.40 | 68 |
| 101.3 | 94.90 ± 12.01 | 74 |
The Biolog GN2 plates were inoculated with S. liquefaciens and incubated at 30 °C at the pressures indicated. Data are expressed as the sums for TWCD including standard deviations (n = 6).
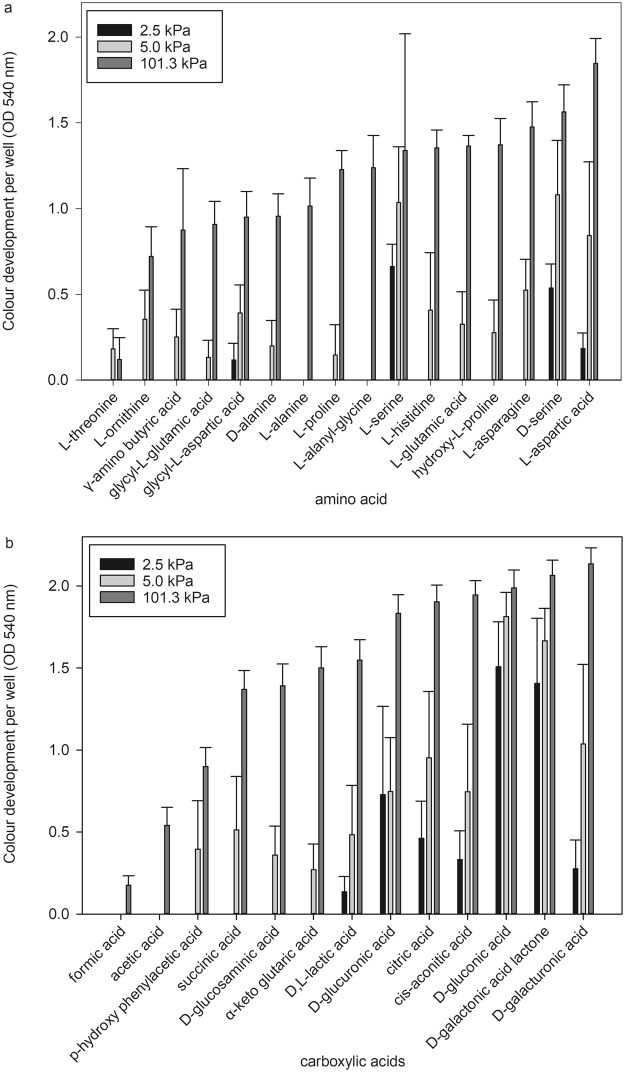
Under Earth-normal conditions, S. liquefaciens was able to utilize almost all available carbohydrate (96%) sources, 80% of the amino acids, but only 58% of the carboxylic acids. The strongest reactions (individual well values above 1) were observed for the nine metabolites: β-methyl-D-glucoside, D-galactose, D-raffinose, sucrose, N-acetyl-D-glucosamine, D-trehalose, D-galactonic acid lactone, D-gluconic acid, and glucose-6-phosphate. The substrates exclusively used at 101.3 kPa were L-alaninamide, L-rhamnose, adonitol, xylitol, D-arabitol, and formic acid. In contrast, when S. liquefaciens was grown at 2.5 kPa, a major reduction in the ability to metabolize amino acids was observed reducing the overall utilization to 20%, although for most of the other tested chemical groups (e.g., carbohydrates, alcohols, carboxylic acids) only small or no changes were observed with regard to carbon utilization (Fig. 2a,b). The greatest pressure-dependent changes in substrate richness were observed for amino acids and carboxylic acids. At 2.5 kPa, S. liquefaciens was not able to metabolize the following 12 amino acids: L-threonine, L-ornithine, γ-amino butyric acid, glycyl-L-glutamic acid, D- and L-alanine, L-proline, L-alanyl-glycine, L-histidine, L-glutamic acid, hydroxyl-L-proline, and L-asparagine (Fig. 3a), and the six carboxylic acids: formic acid, acetic acid, p-hydroxy phenylacetic acid, succinic acid, D-glucosaminic acid and α-keto glutaric acid (Fig. 3b). The highest OD, i.e., greatest response, was observed for the amino acids L- and D-serine, and the carboxylic acids D-gluconic acid and D-galactonic acid lactone. At 5.0 kPa, S. liquefaciens was not able to utilize two amino acids (L-alanine and L-alanyl-glycine) and two carboxylic acids (formic acid and acetic acid).
Figure 3.
Detailed results of the organic compounds utilized by Serratia liquefaciens cells for amino acids and carboxylic acids tested at different pressures. (a) The OD colour development per well for each amino acid at 2.5, 5.0, or 101.3 kPa. (b) The OD colour development per well for each carboxylic acid at 2.5, 5, or 101.3 kPa. Error bars are standard deviations of the means between six independent replicates of the Biolog GN2 microarray assays.
Exp-2: Metabolic fingerprints and the effect of incubation time for low-PTA conditions and controls
In a second series of experiments, the metabolic fingerprint for S. liquefaciens was investigated after incubation for 35, 42 and 49 days at: (1) low-PTA conditions, 0.7 kPa, 0 °C, CO2-enriched anoxic atmosphere, (2) Earth-control 1 (EC-1, see Methods) with Earth atmospheric pressure, 0 °C, CO2, (3) EC-2 with Earth atmospheric pressure, 0 °C, O2, (4) EC-3 with Earth atmospheric pressure, 30 °C, CO2-enriched anoxic atmosphere, and (5) EC-4 with Earth atmospheric pressure, 30 °C, O2. The specific carbon sources that are, or are not, utilized may indicate which metabolic pathways are able to function in hypobaric atmospheres like the Martian surface. After 35, 42, and 49 days of incubation OD readings were taken to determine whether incubation time had an effect on the metabolic fingerprint. After the completion of the three different 49-d assays at 0 °C, the plates were further incubated for 48 h at 101.3 kPa, 30 °C under an Earth-normal pO2 atmosphere. The extra 48 h incubations permitted us to evaluate if the cells in negative wells would return to normal metabolic functions as observed in optimal growth conditions.
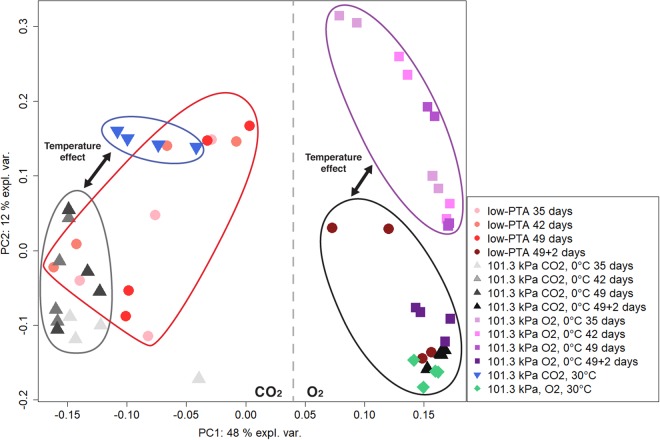
To identify the effects of the treatments (14 data sets, n = 4) on the metabolic fingerprints and whether there were distinct changes in substrate richness, PCA was performed. In the ordination of Fig. 4, the first two principal components accounted for 60% of the overall variance. The multivariate analysis revealed four clusters containing only data points from the conditions (EC-2) Earth atmospheric pressure, 0 °C, CO2, (EC-3) Earth atmospheric pressure, 0 °C, O2, (EC-4) Earth atmospheric pressure, 30 °C, CO2-enriched anoxic atmosphere, and (EC-5) Earth atmospheric pressure, 30 °C, O2, respectively. The assays from the low-PTA condition, which are spread among the samples from conditions EC-1 and EC-3, were the only exceptions. As expected all 49 + 2 d samples grouped with condition (EC-4) with some outliers from the low-PTA conditions.
Figure 4.
Overview of substrate utilization patterns in Serratia liquefaciens under low-PTA conditions and Earth controls at different incubation times. Ordination plot generated by principal component analysis of metabolic fingerprints at (1) low-PTA conditions of 0.7 kPa, 0 °C, and CO2-enriched anoxic atmosphere; (2) Earth atmospheric pressure, 0 °C, and CO2; (3) Earth atmospheric pressure, 0 °C, and lab-normal pO2; (4) Earth atmospheric pressure, 30 °C, and CO2-enriched anoxic atmosphere; and (5) Earth atmospheric pressure, 30 °C, lab-normal pO2. Data were taken on 35, 42, or 49 days. The 49 + 2 days treatments refer to Biolog GN2 plates incubated at 30 °C, Earth-normal pressures, and lab-normal atmospheres for an additional 2 d after termination of incubation conditions in (1) through (3) above.
Upon return of the plates from low-PTA conditions to 101.3 kPa at 30 °C, the cells in most substrate wells (i.e., 64 out of 74) resumed growth and the Earth-normal metabolic fingerprint pattern was observed with slight variations (see Table 2). Colour development in 10 wells failed when the Biolog plates were returned to optimal growth conditions. The reduced number of metabolized carbon sources demonstrated that cells of S. liquefaciens incubated for long periods at 0.7 kPa were not able to fully return to normal metabolic activity under optimal growth conditions. The results might indicate that some cellular machinery for carbon utilization was inhibited or turned off after long exposures to low pressures. In addition, a separation of the data set along PC1 accounting for 48% of the variance was identified that differentiated samples grown under O2 from those grown under CO2-enriched anoxic conditions (Fig. 4). Furthermore, a distinct effect of temperature on the metabolic fingerprint was observed when comparing samples grown in an aerobic environment at 30 °C (Fig. 4, green diamonds) compared to samples grown at 0 °C (Fig. 4, purple squares) but at the same atmospheric pressure. A similar effect was observed for samples grown in the CO2-enriched environment and Earth-normal pressure at 30 °C (Fig. 4, blue triangles) versus 0 °C (Fig. 4, grey triangles).
Table 2.
Total well colour development (TWCD) values and numbers of carbon sources utilized by Serratia liquefaciens under different pressures between 35 and 49 d.
| Condition | Days of incubation | TWCD | Carbon sources used |
|---|---|---|---|
| Low-PTA | 35 | 4.24 ± 1.35 | 16 |
| 42 | 4.67 ± 2.47 | 21 | |
| 49 | 7.49 ± 2.10 | 24 | |
| 49 + 2 | 48.66 ± 20.99 | 64 | |
| 101.3 kPa, CO2, 0 °C | 35 | 1.80 ± 0.22 | 5 |
| 42 | 2.92 ± 0.40 | 8 | |
| 49 | 4.32 ± 1.14 | 13 | |
| 49 + 2 | 74.94 ± 8.67 | 72 | |
| 101.3 kPa, O2, 0 °C | 35 | 24.39 ± 6.53 | 53 |
| 42 | 28.53 ± 4.90 | 54 | |
| 49 | 32.59 ± 4.25 | 55 | |
| 49 + 2 | 79.09 ± 17.62 | 66 | |
| 101.3 kPa, CO2, 30 °C | 2 | 32.95 ± 2.63 | 46 |
| 101.3 kPa, O2, 30 °C | 2 | 90.2 ± 8.18 | 74 |
After the experiments were terminated, the GN2 plates were incubated for an additional 48 h at 30 °C (i.e., the 49 + 2 treatments). The GN2 plates were inoculated with S. liquefaciens. Data are expressed as the sums (for TWCD) including standard deviations (n = 4).
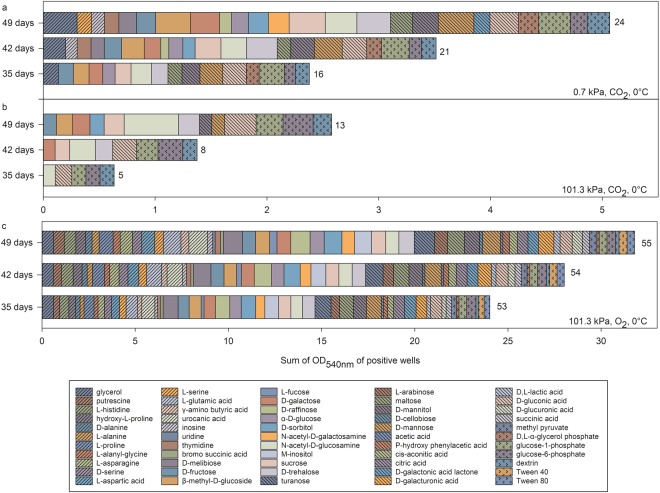
At 0 °C, colour development rates in many wells were delayed, and thus, presumably the growth or metabolic rates were retarded (Fig. 5 and TWCD values in Table 2). We observed a lag period of 28 d, after which, the colour development increased gradually until the final measurements were collected at 49 d. The lag period was likely due to slower growth rates at 0 °C compared to 30 °C, and that specific cell densities (i.e., ~108 cells/ml) had to be achieved before colour development was measureable17. The time course for colour development of the positive substrates under the various tested conditions is shown in Fig. 5. The results indicated that the rates of substrate oxidation were not linear throughout the incubation period. Consequently, the numbers of positive wells increased with incubation time which means that readings at earlier days led to an underestimation of the overall potential substrate richness for S. liquefaciens. For example, under low-PTA conditions at 0.7 kPa, substrate richness increased from 16 to 24 between days 35 and 49. In addition, metabolic activity was suppressed at all three sample dates for cells grown under CO2 at 0 °C atmospheres compared to low-PTA conditions, suggesting that low-PTA conditions may have “relaxed” a metabolic bottle-neck for many organics present in the Earth-normal conditions of 101.3 kPa, CO2, and 0 °C.
Figure 5.
Comparisons of substrate utilization patterns for Serratia liquefaciens cells grown at 0 °C under 0.7 or 101.3 kPa and under pO2 or pCO2. The OD readings were taken after 35, 42, and 49 d of incubation. The number at the end of each horizontal bar indicates substrate richness. The assay plates were incubated at (a) 0.7 kPa, CO2 atmosphere, and 0 °C; (b) 101.3 kPa, CO2 atmosphere, and 0 °C; or (c) 101.3 kPa, CO2 atmosphere, and 0 °C. The OD data represent average values of n = 3 replicates.
The highest OD values, hence the highest metabolic activities, were observed for glycerol, β-methyl-D-glucose, sucrose, D-mannose, n-acetyl-glucosamine (OD ~ 0.3) when cells were grown at low-PTA conditions; n-acetyl-glucosamine, D-gluconic acid, glucose-6-phosphate (OD = 0.48–0.26) when cells were grown at Earth atmospheric pressure, 0 °C, CO2; and D-melibiose, urocanic acid, D-raffinose, and turanose (OD > 1) when cells were grown in Earth atmospheric pressure, 0 °C, and O2 (Fig. 5). The observed differences suggest that different metabolic pathways were active under the diverse incubation conditions tested (Fig. 5, Supplementary Table S2).
After 49 d, the TWCD values were 7.49 (+/−2.10) at 0.7 kPa compared to 4.32 (+/−1.14) at 101.3 kPa, CO2, 0 °C (Table 2); a second example that low-PTA conditions seemed to relax a metabolic bottle-neck for cells grown under low temperature and CO2-enriched anoxic conditions. Accordingly, S. liquefaciens was able to use a total of 24 carbon sources at 0.7 kPa compared to only 13 at 101.3 kPa, CO2, 0 °C. In the latter case, nine carbohydrates, two phosphorylated substrates, one polymer, and one carboxylic acid were utilized. At 0.7 kPa S. liquefaciens used an additional six carbohydrates, two amino acids, one carbohydrate, one ester, one alcohol, one carboxylic acid, and two aromatic compounds. A similar trend was observed when comparing the two Earth controls (EC-3 and EC-4) incubated at 30 °C. A higher TWCD and a higher number of carbon sources were used in the aerobic environment (Table 2).
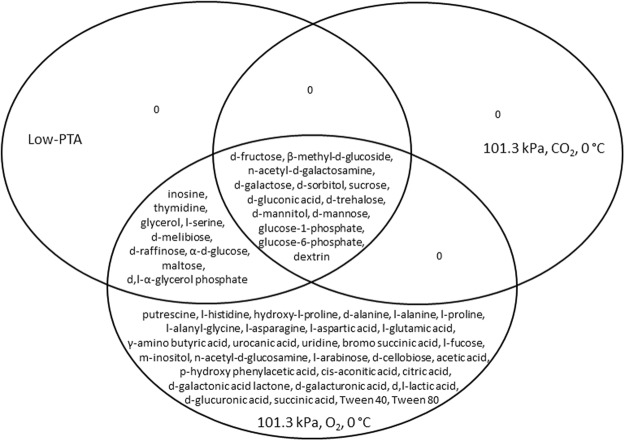
Comparison of metabolic fingerprints between O2 and CO2 atmospheres incubated at Earth-normal pressures and 30 °C demonstrated that the gas composition had an effect (Fig. 4). S. liquefaciens likewise used 46 substrates and an additional 28 substrates were metabolized when grown in an aerobic environment (Table 2). Comparison of carbon utilization of plates incubated at 0 °C and 30 °C, Earth normal pressure and O2 atmosphere highlighted the effects of temperature among the two temperatures tested (Fig. 4). S. liquefaciens likewise used 55 substrates at both temperatures tested, despite the fact that the metabolic rate was slower at 0 °C compared to 30 °C. Furthermore, S. liquefaciens was able to utilize an additional 19 carbon sources when grown at 30 °C (Table 2). Despite the observed differences, we were able to identify a core metabolic fingerprint for S. liquefaciens cells grown under all conditions (Fig. 6). The catabolism of 17% of the 95 organics, consisting mainly of carbohydrates, was not affected by either temperature, pressure, or gas composition. None of the tested substrates were exclusively utilized at either low-PTA, or under 101.3 kPa, CO2 and 0 °C conditions (Fig. 6).
Figure 6.
Venn diagram of the carbon sources utilized by Serratia liquefaciens after 49 d of incubation under three different growth conditions. Comparison of data from GN2 assay plates (n = 3) that were incubated at (1) low-PTA conditions at 0.7 kPa, CO2 atmosphere, and 0 °C; (2) 101.3 kPa, CO2 atmosphere, and 0 °C; or (3)101.3 kPa, CO2 atmosphere, and 0 °C.
Under optimal growth conditions (i.e., EC-4 at 101.3 kPa, O2, and 30 °C), S. liquefaciens metabolized the majority (n = 74) of the carbon sources. In contrast and under all of the tested conditions, S. liquefaciens was unable to utilize the following 18 substrates: 2,3-butanediol (alcohol), i-erythritol (carbohydrate); the amides succinamic acid, 2-aminoethanol, phenylethylamine; the amino acids L-leucine, L-phenylalanine, L-pyroglutamic acid; the carboxylic acids α-hydroxy butyric acid, α-keto butyric acid, β-hydroxy butyric acid, γ-hydroxy butyric acid, itaconic acid, malonic acid, propionic acid, quinic acid, sebacic acid; and the polymer α-cyclodextrin.
Discussion
If Earth bacteria can be shown to possess the metabolic range to grow in 0.7 kPa, 0 °C, and CO2 anoxic atmosphere this would increase the possibility that the Martian environment is habitable to Earth-like life, and may indicate that specific niches might be candidate locations to search for an extant Mars microbiota. While low pressure environments exist in high-altitude environments on Earth, or in extraterrestrial settings like the surface of Mars, hypobaric conditions cannot be found on Earth’s surface. Despite these ecological constraints, Earth bacteria have been identified with the ability to grow in low-pressure conditions5–7. One model organism for hypobaric studies is S. liquefaciens exhibiting a wide temperature growth range and versatility to tolerate various extremes, such as growth without oxygen or at hypobaric conditions. Thus, by fully characterizing the limits of growth under simulated Martian conditions, we can more effectively identify and protect niches on Mars that might harbour an extant microbial community. While many studies have been conducted to test microbial survivability under Martian conditions, with a focus on radiation and/or desiccation effects under Martian atmospheric gas composition19–24, only a few studies have looked into the ability of microorganisms to grow in low-pressure environmental settings5–7,25,26. These hypobaric studies revealed that the growth of the majority of microorganisms was inhibited indicating that low pressure may be a selective stress which constitutes a barrier for potentially growing on Mars. Consequently, assessing the metabolic responses to simulated Martian conditions and characterizing their carbon source utilization patterns are of critical importance when evaluating the risks for the forward contamination of HZ terrains.
To investigate the effects of low pressure under simulated Martian conditions on the metabolic fingerprint of S. liquefaciens, Biolog GN2 microarrays were used which have been successfully tested under low pO2 and low temperature conditions27. The microarrays are based on the measurements of redox potential changes in accordance with purple-colour development of a redox sensitive dye which has been proven very useful for qualitative and quantitative determination of microbial metabolism under aerobic and most anaerobic conditions28–30. However, the Biolog GN2 microarrays have not yet been tested under hypobaric conditions. Our experiments confirmed that the Biolog GN2 microarray system can be used to identify sole carbon sources for S. liquefaciens metabolism under simulated Martian conditions at 0.7 kPa.
Under optimal growth conditions (EC-4; 30 °C, pO2 (21%), Earth-normal pressure) S. liquefaciens displayed a high versatility in carbon utilization with the ability to metabolize 74 out of 95 substrates including alcohols, amines, amino acids, aromatic compounds, carboxylic acids, carbohydrates, esters, polymers, phosphorylated, and brominated substances. Serratia liquefaciens is a facultatively anaerobic heterotroph capable of growth under low pO2 and able to utilize a wide diversity of organics for metabolism. Isolates have been recovered from numerous ecological niches including the drinking water onboard the ISS12; plant rhizospheres, fruits, grasses, and vegetables31–33; processed foods34,35; meats34,36; or as opportunistic pathogens in fish37 and the human bloodstream38.
Apart from determining the substrate richness at optimal growth conditions, one of the main objectives investigated in Exp-1 was whether a single stressor such as pressure would alter the metabolic fingerprint of S. liquefaciens. Pressures from 2.5–101.3 kPa (at 30 °C) were tested to determine the physical limits in which low atmospheric pressures would begin to alter cellular metabolism. Compared to optimal growth conditions, the TWCD decreased at low pressures. The Biolog GN2 protocol allows the investigation of both metabolism alone, when starting cell densities are ≥108 cells/well, or cell growth plus metabolism, when starting cell densities are ~105 cells/well17,39. Because the GN2 assays described here were started with low cell densities at ~2 × 105 cells per well, results indicate lower metabolic activities and slower growth rates for S. liquefaciens cells incubated at low pressures.
The hypobaric effect of suppressing metabolism was confirmed by PCA (Fig. 1) separating the data into two groups with one cluster representing samples incubated at higher pO2 and the other representing samples incubated at lower pO2. Above 10.0 kPa, S. liquefaciens performed similarly to Earth-normal conditions whereas below 10.0 kPa a shift in the metabolic fingerprint and a reduction in growth rates were observed. The hypobaric effects of suppressing growth described here were similar to those observed for seven Bacillus spp. in which lowering atmospheric pressure led to reduction and then cessation of growth below 5 kPa7. Similarly, Deinococcus radiodurans R1 was found to grow at low pressures down to only 10.0 kPa4,5. The differences between the low pressures at 5 kPa or below and all higher pressures also may have been due to the effects of low pO2 (at 2.5 and 5 kPa) versus higher pO2 (10, 20, or 101.3 kPa), but the role of pO2 on carbon utilization was not independently studied here. These data indicate that pressures below 10.0 kPa induce a selective stress that can be described as a physical barrier for limiting growth in the majority of bacteria, while pressures above 10.0 kPa altered microbial activity to only a minor extent.
In Exp-2, a long lag phase was discovered when cultures were grown at 0 °C, and thus, quantitative microarray plate readings were started at 35 d. Based on early work in which colonies of S. liquefaciens on TSA agar surfaces occurred only after 10–14 days of incubation under low-PTA conditions5,14, the observed delay was anticipated. Haack et al.40 showed that colour development rates in wells are not always linear, and that substrate oxidation profiles follow similar sigmoidal functions as observed for bacterial growth curves. In addition, substrate oxidation profiles are not always directly linked with bacterial growth40.
When comparing all samples from Exp-2, we found that low-PTA conditions had a cumulative impact, as shown by PCA (Fig. 4), indicating that the metabolic fingerprints were affected by pO2, temperature, and pressure. The lowest rates of metabolism and carbon source utilization occurred when cells were incubated under 101.3 kPa, 0 °C, and CO2-enriched anoxic atmosphere. These results are consistent with previous observations that demonstrated S. liquefaciens5 and Carnobacterium sp.6 grew better under low-PTA conditions compared to 101.3 kPa at 0 °C and CO2-enriched anoxic atmospheres. The mechanism that increases growth rates of S. liquefaciens cells under low-PTA conditions compared to growth at 101.3 kPa, 0 °C, and CO2-enriched anoxic atmospheres is unknown.
Under low-PTA conditions S. liquefaciens favoured carbohydrates for metabolism over most other organic classes. In contrast, most of the amino acids used by S. liquefaciens under Earth-lab conditions of 101.3 kPa, 30 °C, and O2 were not metabolized by cells incubated at, or below, 2.5 kPa, regardless of the other conditions tested. For example, L-aspartic acid, glycyl-aspartic acids, D-serine, and L-serine were utilized at 2.5 kPa, but only L-serine was utilized under low-PTA conditions. The ability to metabolize carbohydrates and the inability to metabolize most amino acids under low-PTA conditions at 0.7 kPa are consistent with a transcriptomics study of S. liquefaciens conducted at the same low-PTA conditions as tested here41 in which the up-regulation of numerous genes in carbohydrate metabolic pathways and the down-regulation of various genes in amino acid pathways were demonstrated.
In the following sections, several hypotheses are discussed that may explain how the different metabolic rates of carbohydrates and amino acids are altered at low-PTA conditions; leading to the conclusion that the different metabolic rates are caused by the interplay of several factors rather than the effect of just one factor. At lower pressures and in CO2 atmosphere, the pO2 is significantly lower compared to the conditions under Earth-normal atmosphere. This leads to the induction of an anaerobic metabolism using carbohydrates for fermentation and anaerobic respiration processes. Results are consistent with the conclusion that S. liquefaciens favours carbohydrates over most other chemical classes under low-PTA conditions.
Incubation at temperatures below the optimum growth conditions has impacts not only on cell integrity and membrane fluidity but also on the metabolic rates due to altered enzyme kinetics. Thus, lower TWCD values for plates grown at 0 °C were expected, which is consistent with low temperatures having strong effects on amino acid and carbohydrate metabolisms42,43. However, in our study when comparing the metabolic fingerprints at 101.3 kPa, O2, 0 °C, with those at 30 °C, only small changes in the substrate utilization of amino acids were detected. Therefore, changes in the amino acid utilization patterns at low-PTA conditions observed here can be attributed to lower pressures.
Due to the paucity of data on bacterial metabolism under hypobaric conditions, a definitive explanation for why S. liquefaciens prefers carbohydrates over amino acids under low-PTA conditions is lacking. However, another approach to develop potential insights on microbial metabolism at low pressures is to compare the metabolisms of high-pressure tolerant bacteria (i.e., piezophiles) grown under their normal high-pressure environments (MPa range) with Earth-sea level pressures at 101.3 kPa (i.e., effectively a low-pressure environment for the piezophiles). For example, transcriptomes of Photobacterium profundum revealed an up regulation of genes involved in amino acid membrane transport and metabolism at 101.3 kPa (i.e., low pressure for P. profundum) compared to the optimal piezophilic environment at 28 MPa44. This upregulation was proposed as a strategy to compensate for the decreased activity of proteins and membrane functionality45.
Furthermore, membrane transporter systems are pressure sensitive with the majority of studies investigating the effects of high pressure. At 50 MPa, Paul and Morita46 reported up to a 90% inhibition for the transport of amino acids across membranes by Vibrio sp., which was suggested as the result of a conformational change of the membrane rather than the loss of function of the respiratory enzymes. In another example, Fajardo-Cavazos et al.4 found that low pressure led to changes in membrane composition when Bacillus subtilis was grown as 5.0 kPa compared to 101.3 kPa. In particular, while the ratio of unsaturated to saturated fatty acids decreased, the ratio of anteiso- to iso-fatty acids increased. The latter also has been implicated in microbial adaption to high pressure environments47. And finally, it is known that low temperature has a negative effect on membrane fluidity48. These observations suggest a mechanism in which low pressures alter membrane functionality by changing the permeability, fluidity, or protein movement. Furthermore, loss of membrane functionality might lead to interruptions in permeability, and therefore interact adversely with the uptake of certain classes of carbon sources. The exact role of low pressure and its potential effect on the membrane remains to be elucidated.
Apart from the influence that membrane permeability might have on amino acid uptake, the decreased utilization of amino acids at low pressures might be the result of physiological changes in cells. For example, S. liquefaciens cells possess flagella and are able to swarm49. Amino acids improve swarming capabilities in bacteria which require high demands for both organics and energy to synthesize and operate the flagella produced during swarming differentiation50. Under stressed conditions, like low pressures, S. liquefaciens cells may lose their motility and hence the demand for amino acids, and thus, prioritize vital functions that allow survival. A similar observation (e.g., a gradual decrease in cell motility) has been made for Shewanella oneidensis when grown at high pressure51.
Conclusions
The search for life on Mars remains a key objective of the Mars Exploration Program52, and numerous studies have proposed methods for characterizing HZ’s in the shallow subsurface2,3,53. Furthermore, it is imperative to avoid forward contamination of Special Regions on Mars with surface vehicles in order to protect the pristine terrains being explored2. Results from the current project are thus useful for both mitigating the forward contamination of exploration sites and the search for life on Mars.
Many of the carbon sources utilized by S. liquefaciens under low-PTA conditions (e.g., aliphatic compounds, PAHs, sugars, aldehydes, carboxylic acids) have been reported in interplanetary dust particles (IDPs) or carbonaceous chondrites54–56. And recently, indigenous aromatic and aliphatic organics were detected in lacustrine mudstones in Gale Crater57. Results here suggest that S. liquefaciens may be able to grow on Mars if transferred to stable hydrated niches that contain indigenous or accreted IDP/chondritic organics similar to the organics used in the Biolog GN2 microarrays. Furthermore, previous work showed that B. subtilis and Enterococcus faecalis58,59 and S. liquefaciens26,60 can survive contact with Martian regolith indicating that Martian regolith may not be overtly biotoxic to many of the bacteria typically recovered from spacecraft surfaces.
The Biolog GN2 microarray protocols developed here could be used to quickly screen spacecraft microorganisms for growth under simulated Martian conditions while also focusing on the utilization of the organics likely present on Mars. If specific organics (e.g., glycerol, β-methyl-D-glucose, sucrose, D-mannose, N-acetyl-glucosamine used by S. liquefaciens at 0.7 kPa) are found to be all utilized by a wider range of hypobarophiles, then Martian terrains that are shown to possess such organics (i.e., during in situ robotic landers or rovers experiments) could be identified as potential habitats for terrestrial life. Consequently, these sites might pose potential risks for the forward contamination of specific terrains. For example, if individual organics are shown to be present on Mars, pre-launch planetary protection protocols for follow-on missions could be enhanced for surface vehicles to prevent the forward contamination of the revisited sites.
And lastly, the search for life on Mars requires us to not only follow the water but also to follow the organics. Future rover/lander payloads must be developed that consider the ecological conditions for microbial growth in Special Regions being explored in order to prioritize niches that have a multiplicity of conducive conditions in which a putative extant Mars microbiota might be present. Organics that are part of the follow the water/organics strategy, and can be used by a diversity of Earth microorganisms under simulated low-PTA conditions, will inform the search for life on Mars and avoid microbial conflicts between the Earth (on spacecraft) and Martian (if present) microbiota.
Material and Methods
Microbial Procedures
Serratia liquefaciens (ATCC 27592) cells were grown on full-strength trypticase soy agar (TSA; 40 g L−1) (Difco media, Fisher Scientific, Pittsburgh, PA, USA) for 24 h at 30 °C. Cells were harvested in 10 mM sodium-phosphate buffer (PO4 buffer; pH 7.2), diluted to yield approx. 2.0 × 106 cells per mL (OD = 0.005–0.012 at 400 nm; average OD = 0.008), and stored at 24 °C for no more than 1 h before dispensing the cells into Biolog GN2 plates, as described below. Numbers of viable cells were estimated for each replicate by serially diluting inoculum through six, 10-fold dilutions and dispensing aliquots of each dilution into wells of a Most Probable Number (MPN) assay as described by Schuerger et al.23.
Biolog inoculation and incubation protocols
Biolog GN2 microarrays (Biolog, Inc., Haywood, CA, USA) were used for the current study, but a new low-pressure protocol had to be developed. The standard Biolog inoculation protocol uses a semi-liquid inoculating fluid obtained from Biolog, Inc. that contains a gelling agent (phytagel) to stabilize the inoculating fluid in the wells. During preliminary experiments with the Biolog GN2 system that included the gelling agent, significant bubbling activity was observed in most wells at 0.7 kPa that likely was caused by the retention of dissolved gases at higher pressures in the inoculating fluid. The gelling agent appeared to retard the outgassing of the inoculating fluid, thereby causing the formation of unwanted bubbles overflowing individual wells. The bubbling rendered the Biolog GN2 assay plates ineffective at low pressures.
The first step in the low-pressure Biolog GN2 assay protocol was to replace the inoculating fluid with a non-gelling alternative. We tested 10 mM PO4 buffer, sterile deionized water (SDIW), and 0.5% NaCl (all at pH 7.2) as carrier fluids. The 10 mM PO4 buffer was chosen because it produced slightly higher colour development by the tetrazolium dye for several organics (Schuerger, data not shown).
Biolog GN2 plates were prepared by dispensing 100 µL of cell suspensions in 10 mM PO4 buffer into each well (approx. 2 × 105 cells per well), adding 25 µL of SDIW to each well, and dispensing 150 µL of SDIW into the void spaces between wells in the Biolog GN2 plates. The addition of SDIW into void spaces between wells reduced the evaporation of water from the organics-containing wells at pressures between 0.7 and 5.0 kPa.
Replicates of GN2 plates were inserted into hypobaric desiccators, sealed, and maintained at 0.7 kPa for up to 49 d, as described below. At 0.7 kPa, the PO4 buffer in many wells would slowly evaporate, requiring the addition of 25–50 µL of SDIW (to maintain the osmotic strength and buffering capacity of the fluids) to each well as needed to return the volume of the incubating fluid to approx. 125 µL. The hypobaric desiccator at 0.7 kPa was vented twice weekly to examine and adjust the fluid levels in the wells.
And finally, negative controls were created without viable cells and co-incubated with inoculated GN2 plates for the duration of the experiments. When low pressure was not required, GN2-experimental and PO4-control plates were incubated in 2.5-L polycarbonate containers with gasket-sealed lids (AnaeroPak® System containers, Mitsubishi Gas Chemical, Co; obtained from Fisher Scientific).
Prior to quantitatively measuring the Biolog GN2 plate with a Genios Pro plate reader (Tecan USA, Research Triangle Park, NC, USA; Magellan v. 5.03 software), each well in the plate was stirred with sterile 10 µL pipette tips to resuspend the cells. The Genios system was set to measure the OD of wells in 10 spots within each well at 540 nm and 25 °C. The average of all 10 scan spots per well was automatically recorded and files converted to Excel CSV formatting for subsequent analysis.
Hypobaric desiccators
The design and operation of the hypobaric chambers were described previously5,14. In brief, 4-L polycarbonate desiccators (model 08-642-7, Fisher Scientific) were connected individually to a low pressure controller (model PU-842, KNF Neuberger, Trenton, NJ, USA) in which the controller and pump were placed outside a -20 °C microbial incubator. The Biolog GN2 plates were inserted into the desiccators and their lids raised and rotated approx. 20° such that the lids were not seated as per normal operations, but instead were slightly raised above the tops of the wells eliminating cross-contamination of wells caused by the condensation of water vapour around the well lips. Four anaerobic pouches61 (model 23-246-378 AnaeroPak® System sachets, Fisher Scientific) were added around the stacked GN2 plates, followed by the addition of 150 mL of SDIW in a 200 mL beaker to increase the vapour pressure of water at 0.7 kPa. The desiccator tops were then placed upon silicone O-rings coated with vacuum grease (Dow CorningTM, obtained from Fisher Scientific) to enhance the sealing of the desiccators at 0 °C.
The desiccators were fitted with 0.22 µm air filters on both the vacuum lines and the vent lines (see Fig. 1)5 to reduce microbial contamination during pump operations. Prior to inserting the desiccators into the -20 °C microbial incubator, the desiccators were flushed for 3–4 min with ultrahigh purity (UHP) CO2 gas passed through the filtered vent lines.
Experiment 1: Pressure-only effects between 2.5 and 101.3 kPa on carbon source utilization
To measure the effects of low pressure alone on the metabolism of individual carbon sources by S. liquefaciens, an experiment was designed to vary pressure between 2.5 and 101.3 kPa while holding gas composition at an Earth-normal 78% pN2 and 21% pO2 composition (lab atmosphere) and temperature (30 °C) constant. Due to high evaporation rates from the GN2 wells at 30 °C and 2.5 or 5.0 kPa, it was not possible to conduct assays at pressures below 2.5 kPa at 30 °C.
Biolog GN2 plates were prepared as described above, and inserted into separate hypobaric desiccators set at 2.5, 5.0, 10.0, 20.0, or 101.3 kPa. The GN2 plates were incubated for 48 h at 30 °C, the desiccators vented, the wells stirred, and the plates read with the Genios plate reader.
At 30 °C, the wells would lose approx. 1/3 of the starting volumes in each well. Thus, 40–50 µL of SDIW were added to each well, on an as needed basis, prior to reading the plates. Only two desiccators were available, and thus, hypobaric treatments were randomly assigned to each desiccator, and assays repeated until all five pressure treatments were conducted three times each with two GN2 replicates per assay (overall n = 6).
Experiment 2: Effects of Mars versus Earth pressure, gas composition, and temperature on carbon source utilization
In order to probe the effects of low pressure at 0.7 kPa on carbon source utilization, both temperature and pressure had to be regulated simultaneously. Thus, the experimental design was altered and the following five treatments created:
Mars simulations in which GN2 plates were incubated at low pressure (0.7 kPa), low temperature (0 °C), and CO2-enriched anoxic atmosphere61 (i.e., low pO2; <1% pO2). These conditions are referred to as low-PTA conditions that simulate a portion of the Mars surface environment. All low-PTA assays were conducted in the 4-L desiccator systems described above. All other assays with GN2 plates were maintained in 2.5-L polycarbonate, gasket-sealed, containers from Fisher Scientific.
Earth Control-1 (EC-1) assays were incubated at 101.3 kPa, 0 °C, and a CO2-enriched anoxic atmosphere (see above).
Earth Control-2 (EC-2) assays were incubated at 101.3 kPa, 0 °C, and an Earth-normal atmosphere of 78:21 pN2 to pO2 conditions.
Earth Control-3 (EC-3) assays were incubated at 101.3 kPa, 30 °C, and a CO2-enriched anoxic atmosphere (see above).
Earth Control-4 (EC-4) assays were incubated at 101.3 kPa, 30 °C, and an Earth-normal atmosphere of 78:21 pN2 to pO2 conditions.
The low-PTA assays were vented twice weekly, SDIW added to wells as required, new anaerobic pouches placed into the desiccators, the desiccators flushed with UHP CO2 gas, and the conditions equilibrated to 0.7 kPa at 0 °C. The tetrazolium dye colour shifts expected from the GN2 wells following metabolic activity were very slow to develop at low pressure and low temperature, so the GN2 plates were only measured with the Genios plate reader after 35, 42, and 49 d from the start of the experiment. The EC-1 and EC-2 assays were read at the same time-steps as GN2 plates incubated at low-PTA conditions. In contrast, colour development in the EC-3 and EC-4 assays progressed quickly because they were incubated at 30 °C, and thus, the plates were read at 48 h.
At the end of the 49 d (i.e., low-PTA, EC-1, and EC-2 conditions) or 48 h (i.e., EC-3 and EC-4) assays, the GN2 plates were placed under lab-normal conditions at 101.3 kPa, 30 °C, and 78:21 pN2 and pO2 atmospheres to determine if the incubation conditions killed cells in negative wells. The assays were randomly assigned to the available desiccators and polycarbonate containers and the experiment conducted twice with two replicates per treatment per experimental run (overall n = 4).
Data analysis
Absorbance values for each well containing a different carbon source were blanked against the values of the control well (i.e., the A1 well in the Biolog GN2 plates). All negative absorbance values were set to zero. A positive substrate response was taken to be an OD (at 540 nm) greater than 0.1 after the control well OD reading was subtracted. Total well colour development (TWCD) was calculated as the sum of the control well subtracted absorbance values of all 95 wells.
For principal component analysis (PCA) plots the data were normalized by dividing each absorbance value of a well by its average well colour development to minimize the influence of inoculum density difference as well as performance differences among plates62. The plots were created using the statistical environment RStudio63 and the default R package stats v3.4.4 applying the function princomp ().
All raw data for both experiments are provided in Supplementary Tables S1 (for Exp-1) and S2 (for Exp-2). The data are organized so that each replicate GN2 plate data forms a single column in each table.
Electronic supplementary material
Acknowledgements
The research was supported by two NASA grants (NNX12AJ84G and 80NSSC17K0263) from the Planetary Protection Research (PPR) program. We would also like to thank Michael Morrison and Joany Babilonia for their assistance in creating the PCA plots in R.
Author Contributions
P.S. analysed the data and wrote the manuscript. A.C.S. conceptualized the study, performed the experiments and helped write the manuscript. Both authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33856-3.
References
- 1.Kminek G, et al. Report of the COSPAR Mars Special Regions Colloquium. Adv. Space Res. 2010;46:811–829. doi: 10.1016/j.asr.2010.04.039. [DOI] [Google Scholar]
- 2.Rummel JD, et al. A new analysis of Mars “Special Region”: Findings of the Second MEPAG Special Regions Science Analysis Group SR-SAG2) Astrobiology. 2014;14:887–968. doi: 10.1089/ast.2014.1227. [DOI] [PubMed] [Google Scholar]
- 3.Cockell CS, et al. Habitability: A review. Astrobiology. 2016;16:89–117. doi: 10.1089/ast.2015.1295. [DOI] [PubMed] [Google Scholar]
- 4.Fajardo-Cavazos P, et al. Evolution of Bacillus subtilis to enhanced growth at low pressure: up-regulated transcription of des-desKR, encoding the fatty acid desaturase system. Astrobiology. 2012;12:258–270. doi: 10.1089/ast.2011.0728. [DOI] [PubMed] [Google Scholar]
- 5.Schuerger AC, Ulrich R, Berry BJ, Nicholson WL. Growth of Serratia liquefaciens under 7 mbar, 0 °C and CO2-enriched anoxic atmospheres. Astrobiology. 2013;13:115–131. doi: 10.1089/ast.2011.0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicholson WL, Krivushin K, Gilichinsky D, Schuerger AC. Growth of Carnobacterium spp. isolated from Siberian permafrost under simulated Mars conditions of pressure, temperature and atmosphere. PNAS. 2013;110:666–671. doi: 10.1073/pnas.1209793110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuerger AC, Nicholson WL. Interactive effects of hypobaria, low temperature, and CO2 atmospheres inhibit the growth of mesophilic Bacillus spp. under simulated martian conditions. Icarus. 2006;185:143–152. doi: 10.1016/j.icarus.2006.06.014. [DOI] [Google Scholar]
- 8.Grimont PAD, Grimont F. The genus Serratia. Annu. Rev. Microbiol. 1978;32:221–248. doi: 10.1146/annurev.mi.32.100178.001253. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh S, Osman S, Vaishampayan P, Venkateswaran K. Recurrent isolation of extremotolerant bacteria from the clean room where the Phoenix spacecraft components were assembled. Astrobiology. 2010;10:325–335. doi: 10.1089/ast.2009.0396. [DOI] [PubMed] [Google Scholar]
- 10.Moissl, C. et al. Molecular bacterial community analysis of clean rooms where spacecraft are assembled. FEMS Microbiol. Ecol., 10.1111/j.1574-69.2007.00360 (2007). [DOI] [PubMed]
- 11.Novikova ND. Review of the knowledge of microbial contamination of the Russian manned spacecraft. Microb. Ecol. 2004;47:127–132. doi: 10.1007/s00248-003-1055-2. [DOI] [PubMed] [Google Scholar]
- 12.La Duc MT, Sumner R, Pierson DL, Venkat P, Venkateswaran K. Evidence of pathogenic microbes in the International Space Station drinking water: reasons for concern? Habitation. 2004;10:39–48. doi: 10.3727/154296604774808883. [DOI] [PubMed] [Google Scholar]
- 13.Pierson, D. L. et al. Preflight and postflight microbiological results from 25 Space Shuttle crews. SAE Technical Paper Series Paper # 932139, pp1–5 (1993).
- 14.Schuerger AC, Nicholson WL. Twenty species of hypobarophilic bacteria recovered from diverse soils exhibit growth under simulated martian conditions at 0.7 kPa. Astrobiology. 2016;16:964–976. doi: 10.1089/ast.2016.1587. [DOI] [PubMed] [Google Scholar]
- 15.Costard F, Forget F, Mangold N, Peulvast JP. Formation of recent Martian debris flows by melting of near-surface ground ice at high obliquity. Science. 2002;295:110–113. doi: 10.1126/science.1066698. [DOI] [PubMed] [Google Scholar]
- 16.McEwen AS, et al. Seasonal flows on warm Martian slopes. Science. 2011;333:740–743. doi: 10.1126/science.1204816. [DOI] [PubMed] [Google Scholar]
- 17.Bochner BR. Sleuthing out bacterial identities. Nature. 1989;339:157–158. doi: 10.1038/339157a0. [DOI] [PubMed] [Google Scholar]
- 18.Garland JL, Mills AL. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 1991;57:2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rettberg P, Rabbow E, Panitz C, Horneck G. Biological space experiments for the simulation of Martian conditions: UV radiation and Martian soil analogues. Adv. Space Res. 2004;33:1294–1301. doi: 10.1016/j.asr.2003.09.050. [DOI] [PubMed] [Google Scholar]
- 20.Osman S, et al. Effect of Shadowing on survival of bacteria under conditions simulating the martian atmosphere and UV radiation. Appl. Environ. Microbiol. 2007;74:959–970. doi: 10.1128/AEM.01973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith DJ, et al. Survivability of Psychrobacter cryohalolentis K5 under simulated martian surface conditions. Astrobiology. 2009;9:221–228. doi: 10.1089/ast.2007.0231. [DOI] [PubMed] [Google Scholar]
- 22.Bak, E. N. et al. Silicates eroded under simulated Martian conditions effectively kill bacteria – a challenge for life on Mars. Front. Microbiol., 10.3389/fmicb.2017.01709 (2017). [DOI] [PMC free article] [PubMed]
- 23.Schuerger AC, Mancinelli RL, Kern RG, Rothschild LJ, McKay CP. Survival of endospores of Bacillus subtilis on spacecraft surfaces under simulated Martian environments: Implications for the forward contamination of Mars. Icarus. 2003;165:253–276. doi: 10.1016/S0019-1035(03)00200-8. [DOI] [PubMed] [Google Scholar]
- 24.Sinha N, Kral TA. Effect of UVC radiation on hydrated and desiccated cultures of slightly halophilic and non-halophilic methanogenic archaea: Implications for life on Mars. Microorganisms. 2018;6:43. doi: 10.3390/microorganisms6020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanervo E, Lehto K, Stahle K, Lehto J, Mäenpää P. Characterization of growth and photosynthesis of Synwxhoxstis sp. PCC6803 cultures under reduced atmospheric pressures and enhanced CO2 levels. Int. J. Astrobiol. 2005;4:979–1000. doi: 10.1017/S1473550405002466. [DOI] [Google Scholar]
- 26.Berry BJ, Jenkins DG, Schuerger AC. Effects of simulated Mars conditions on the survival and growth of Escherichia coli and Serratia liquefaciens. Appl. Environ. Microbiol. 2010;76:2377–2386. doi: 10.1128/AEM.02147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson SA, Sissons CH, Colemand MH, Wong L. Application of carbon source utilization patterns to measure the metabolic similarity of complex dental plaque biofilm microcosms. Appl. Environ. Microbiol. 2002;68:5779–5783. doi: 10.1128/AEM.68.11.5779-5783.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reichart O, Szakmár A, Jozwiak Á, Felföldi J, Baranyai L. Redox potential measurement as a rapid method for microbiological testing and its validation for coliform determination. Int. J. Food Microbiol. 2007;114:143–148. doi: 10.1016/j.ijfoodmicro.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 29.Bhupathiraju VK, Hernandez M, Landfear D, Alvarez-Cohen L. Application of a tetrazolium dye as an indicator of viability in anaerobic bacteria. J. Microbiol. Methods. 1999;37:231–243. doi: 10.1016/S0167-7012(99)00069-X. [DOI] [PubMed] [Google Scholar]
- 30.Konopa A, Oliver L, Turco RF., Jr. The use of carbon substrate utilization patterns in environmental and ecological microbiology. Microb. Ecol. 1997;35:103–115. doi: 10.1007/s002489900065. [DOI] [PubMed] [Google Scholar]
- 31.Grimont PAD, Grimont F, Starr MP. Serratia species isolated from plants. Curr. Microbiol. 1981;5:317–322. doi: 10.1007/BF01567926. [DOI] [Google Scholar]
- 32.Lalande R, Bissonnette N, Coutlee D, Antoun H. Identification of rhizobacteria from maize and determination of their plant-growth promoting potential. Plant Soil. 1989;115:7–11. doi: 10.1007/BF02220688. [DOI] [Google Scholar]
- 33.Ashelford KE, Fry JC, Bailey MJ, Jeffries AR, Day MJ. Characterization of six bacteriophages of Serratia liquefaciens CP6 isolated from the sugar beet phytosphere. Appl. Surf. Sci. 1999;65:1959–1965. doi: 10.1128/aem.65.5.1959-1965.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindberg A-M, Ljungh A, Löfdahl S, Molin G. Enterobacteriaceae found in high numbers in fish, minced meat, and pasteurized milk or cream and the presence of toxin encoding genes. Int. J. Food Microbiol. 1998;39:11–17. doi: 10.1016/S0168-1605(97)00104-9. [DOI] [PubMed] [Google Scholar]
- 35.Oh HI, Kim YJ, Chang EJ, Kim JY. Antimicrobial characteristics of chitosans against food spoilage microorganisms in liquid media and mayonnaise. Biosci. Biotechnol. Biochem. 2001;65:2378–2383. doi: 10.1271/bbb.65.2378. [DOI] [PubMed] [Google Scholar]
- 36.Stiles ME, Ng L-K. Biochemical characteristics and identification of Enterobacteriaceae isolated from meats. Appl. Environ. Microbiol. 1981;41:639–645. doi: 10.1128/aem.41.3.639-645.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aydin S, Erman Z, Bilgin OC. Investigation of Serratia liquefaciens infection in rainbow trout (Oncorhynchus mykiss Walbaum) Turk. J. Vet. Anim. Sci. 2001;25:643–650. [Google Scholar]
- 38.Grohskopf LA, et al. Serratia liquefaciens bloodstream infection from contamination of epoetin alfa at a hemodialysis center. N. Engl. J. Med. 2001;344:1491–1497. doi: 10.1056/NEJM200105173442001. [DOI] [PubMed] [Google Scholar]
- 39.Bochner BR, Gadzindky O, Panomitros E. Phenotype microarrays for high throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haak SK, Garchow H, Klug MJ, Forney LJ. Analysis of factors affecting the accuracy, reproducibility and interpretation of microbial community carbon source utilization patterns. Appl. Environ. Microbiol. 1995;61:1458–1468. doi: 10.1128/aem.61.4.1458-1468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fajardo-Cavazos, P., Morrison M. D., Miller, K. M., Schuerger, A. C. & Nicholson, W. L. Transcriptomic responses of Serratia liquefaciens cells grown under simulated Martian conditions of low temperature, low pressure, and CO2-enriched anoxic atmosphere. Sci. Rep. 8, 14938. [DOI] [PMC free article] [PubMed]
- 42.Campanaro S, et al. Temperature-dependent global gene expression in the Antarctic archaeon Methanococcoides burtonii. Environ. Microbiol. 2011;13:2018–2038. doi: 10.1111/j.1462-2920.2010.02367.x. [DOI] [PubMed] [Google Scholar]
- 43.De Maayer P, Anderson D, Cary C, Cowan DA. Some like it cold: understanding the survival strategies of psychrophiles. EMBO Rep. 2014;15:508–517. doi: 10.1002/embr.201338170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vezzi A, et al. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science. 2005;307:1459–1461. doi: 10.1126/science.1103341. [DOI] [PubMed] [Google Scholar]
- 45.Oger PM, Jebbar M. The many ways of coping with pressure. Res. Microbiol. 2010;161:799–809. doi: 10.1016/j.resmic.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 46.Paul KL, Morita RY. Effects of hydrostatic pressure and temperature on uptake and respiration of amino acids by a facultatively psychrophilic marine bacterium. J. Bacteriol. 1993;108:835–842. doi: 10.1128/jb.108.2.835-843.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang, J. & Bazylinski D. A. Deep sea geomicrobiology in High-Pressure Microbiology (eds Michiels, C., Bartlett, D. H. & Aertsen, A.) 237–264 (ASM Press, Washington DC, 2008).
- 48.Aguilar PS, Cronan JE, Jr., de Mendoza DA. Bacillus subtilis gene induced by cold shock encodes a membrane phospholipid desaturase. J. Bacteriol. 1998;180:2194–2200. doi: 10.1128/jb.180.8.2194-2200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eberl L, Molin S, Givskov M. Surface motility of Serratia liquefaciens MG1. J. Bacteriol. 1999;181:1703–1712. doi: 10.1128/jb.181.6.1703-1712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Givskov M, Eberl L, Molin S. Control of exoenzyme production, motility and cell differentiation in Serratia liquefaciens. FEMS Microbiol. Lett. 1997;148:118–122. doi: 10.1111/j.1574-6968.1997.tb10276.x. [DOI] [Google Scholar]
- 51.Sharma A, et al. Microbial Activity at Gigapascal Pressures. Science. 2002;295:1514–1516. doi: 10.1126/science.1068018. [DOI] [PubMed] [Google Scholar]
- 52.Des Marais DJ, et al. The NASA Astrobiology Roadmap. Astrobiology. 2008;8:715–730. doi: 10.1089/ast.2008.0819. [DOI] [PubMed] [Google Scholar]
- 53. Stoker, C. R. et al. Habitability of the Phoenix landing site. J. Geophys. Res. 115, 10.1029/2009JE003421 (2010).
- 54.Botta O, Glavin DP, Kminek G, Bada JL. Relative amino acid concentrations as a signature for parent body processing of carbonaceous chondrites. Orig. Life Evol. Biosph. 2002;32:143–163. doi: 10.1023/A:1016019425995. [DOI] [PubMed] [Google Scholar]
- 55.Ehrenfreund P, Glavin DP, Botta O, Cooper G, Bada JL. Extraterrestrial amino acids in Orgueil and Ivuna: Tracing the parent body of CI type carbonaceous chondrites. Proc. Natl. Acad. Sci. 2001;98:2138–2141. doi: 10.1073/pnas.051502898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ehrenfreund P, et al. Astrophysical and astrochemical insights into the origin of life. Rep. Prog. Phys. 2002;65:1427–1487. doi: 10.1088/0034-4885/65/10/202. [DOI] [Google Scholar]
- 57.Eigenbrode JL, et al. Organic matter preserved in 3-billion-year-old mudstones at Gale Crater, Mars. Science. 2018;360:1096–1101. doi: 10.1126/science.aas9185. [DOI] [PubMed] [Google Scholar]
- 58.Schuerger AC, Golden DC, Ming DW. Biotoxicity of Mars soils: 1. Dry deposition of analog soils on microbial colonies and survival under Martian conditions. Planet. Space Sci. 2012;72:91–101. doi: 10.1016/j.pss.2012.07.026. [DOI] [Google Scholar]
- 59.Schuerger AC, Ming DW, Golden DC. Biotoxicity of Mars soils: 2. Survival of Bacillus subtilis and Enterococcus faecalis in acqueous extracts derived from six Mars analog soils. Icarus. 2017;290:215–223. doi: 10.1016/j.icarus.2017.02.023. [DOI] [Google Scholar]
- 60.Mickol RL, Page JL, Schuerger AC. Magnesium sulfate salt solutions and ices fail to protect Serratia liquefaciens from the biocidal effects of UV irradiation under Martian conditions. Astrobiology. 2017;17:401–412. doi: 10.1089/ast.2015.1448. [DOI] [PubMed] [Google Scholar]
- 61.Van Horn KG, Warren K, Baccaglini EJ. Evaluation of the AnaeroPack System for growth of anaerobic bacteria. J. Clin. Microbiol. 1997;35:2170–2173. doi: 10.1128/jcm.35.8.2170-2173.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garland JL. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol. Biochem. 1996;28:231–221. doi: 10.1016/0038-0717(95)00131-X. [DOI] [Google Scholar]
- 63.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, http://www.R-project.org/ (2013).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.