Abstract
Purpose
Targeted therapy is the cornerstone of treatment of advanced EGFR-mutant non–small-cell lung cancer (NSCLC). Next-generation sequencing (NGS), the preferred method for genotyping, typically requires several weeks. Here, we assessed workflows designed to rapidly identify patients with actionable EGFR mutations and reduce time to initiation (TTI) of epidermal growth factor receptor (EGFR)–directed therapy.
Patients and Methods
We performed rapid testing for EGFR L858R mutations and exon 19 deletions on paraffin-embedded or frozen section biopsy specimens from newly diagnosed patients with metastatic NSCLC by using an EGFR-specific assay (rapid test). To determine clinical utility, we assessed concordance with NGS results, turnaround time, and TTI of EGFR therapy, and we evaluated reimbursement data.
Results
Between January 2015 and September 2017, we performed 243 rapid EGFR tests and identified EGFR mutations in 43 patients (18%). With NGS results as a reference, sensitivity and specificity of the rapid EGFR polymerase chain reaction assay were 98% and 100%, respectively. The median turnaround time for NGS was 14 days, compared with 7 days for rapid testing (P < .001). In the rapid group, 95% of patients received an EGFR inhibitor in the first-line setting. The median TTI of EGFR therapy was significantly shorter in the rapid cohort when compared with 121 historical cases (22 v 37 days; P = .01). Escalation of the initiative into an interdisciplinary ultra-rapid next-day frozen-section workflow for highly symptomatic patients (n = 8) resulted in a reduction in the median (± standard deviation) turnaround time to 1 ± 0.4 days and allowed several patients to initiate therapy within 1 week of biopsy. An extended 9-month clinical evaluation phase confirmed operational sustainability (turnaround times: ultra-rapid, 0.81 ± 0.4 days; rapid, 3 ± 1.5 days), and a 63% reimbursement rate indicated financial sustainability.
Conclusion
Rapid genotyping facilitates earlier initiation of EGFR-directed therapies without compromising NGS workflows.
INTRODUCTION
Non–small-cell lung cancer (NSCLC) is a heterogeneous disease composed of unique molecular subsets with distinct clinical outcomes.1,2 Multiple randomized studies have established the superiority of molecularly targeted therapies versus chemotherapy for the treatment of EGFR-mutant and ALK-positive NSCLC.3-6 In other molecular subsets, single-arm studies confirm that treatment with targeted therapies can induce durable responses.7,8 As drugs that target these molecular drivers are approved for first-line treatment, genotyping in newly diagnosed NSCLC is considered the standard of care.9
Because of the need to interrogate a growing number of genes, next-generation sequencing (NGS) has largely replaced traditional single-gene assays.10 Guidelines endorsed by oncology and pathology societies recommend that molecular testing turnaround times not exceed 10 working days.9 Genotyping by NGS requires complex bioinformatics that can create treatment delays. Some patients with NSCLC present with symptomatic disease that requires initiation of treatment before molecular testing results are available.11 To our knowledge, the impact of molecular testing turnaround time on clinical decision making has not been formally assessed in NSCLC.
Here, we evaluated whether the addition of an epidermal growth factor receptor (EGFR)–specific assay to NGS at diagnosis produced accurate results and reduced time to initiation (TTI) of EGFR-directed therapy. We selected EGFR-mutant NSCLC on the basis of its relatively high prevalence (10% to 15%) among patients with NSCLC,12 the lack of overlap between EGFR mutations and other clinically relevant molecular alterations,13 and the fact that EGFR inhibitors were readily available for hospitalized patients. We hypothesized that concurrent rapid genotyping would improve clinical care without compromising comprehensive NGS-based genotyping efforts.
MATERIALS AND METHODS
Study Design
We conducted a multiphase, multidisciplinary quality improvement initiative from January 2015 to March 2018. In the first phase (January 2015 to May 2017), rapid EGFR testing was performed on tissue specimens from consecutive patients diagnosed with metastatic NSCLC who were never smokers/minimal smokers (≤ 10 pack years) or who were hospitalized at the time of diagnosis (Data Supplement). The second phase (May 2017 to September 2017) explored the impact of a multidisciplinary same-day fresh-tissue protocol (ultra-rapid testing). Specimens from both phases were submitted for NGS-based genotyping (Fig 1). We also identified a comparator population composed of 121 patients with EGFR-mutant NSCLC who were diagnosed before implementation of the rapid testing (ie, historical cohort; Data Supplement). We extracted clinicopathologic features, molecular testing results, and treatment histories from the medical record. Follow-up data were obtained through November 2017. To assess operational sustainability, we analyzed testing patterns during a subsequent standard of care testing period (July 2017 to March 2018). The Massachusetts General Hospital institutional review board approved this study; all participants provided written informed consent.
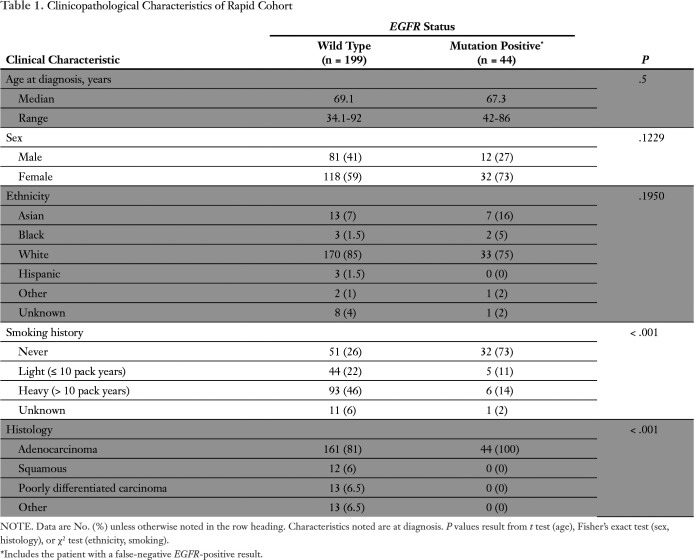
Fig 1.
Rapid EGFR testing approach. We implemented rapid EGFR testing in parallel to genotyping using next-generation sequencing (NGS; compare pathway A v B). As a result of differences in reporting times, detection of an actionable EGFR mutation with rapid testing might lead to a treatment decision before NGS results are obtained. Note that there is a (variable) delay from reporting to treatment decision and initiation of therapy because of cost-coverage determination, preauthorization requirements, etc.The ultra-rapid EGFR testing pathway (pathway C) is a multidisciplinary workflow designed to improve turnaround time using fresh tissue (frozen sections) to extract nucleic acids. Note that ultra-rapid testing combines preanalytical improvements with the optimized rapid workflow and allows coupling with NGS (Data Supplement). Dx, diagnosis; FNA, fine-needle aspiration; Neg, negative; PCR, polymerase chain reaction; Pos, positive; QC, quality control; TKI, tyrosine kinase inhibitor.
Molecular Analysis
Rapid assay. Tumor DNA was extracted from formalin-fixed paraffin-embedded samples (rapid) or fresh-frozen sections (ultra-rapid) in a Clinical Laboratory Improvement Amendments–certified laboratory and was analyzed using a multiplexed polymerase chain reaction (PCR) assay.14 Multiplex PCR single-base extension sequencing technology (Snapshot; Applied Biosystems, Foster City, CA) was used to analyze the EGFR c.2573T (p.L858) and c.2369C (p.T790) residues. A separate PCR reaction with primers that flanked EGFR exon 19 was used to detect in-frame activating insertions or deletions in EGFR.14 The analytical sensitivity for this assay was approximately 5%.
NGS. Isolated nucleic acids from tumor specimens were analyzed with our NGS assay that uses anchored multiplex PCR to detect single-nucleotide variants and insertions/deletions in a target set of cancer-related genes.15 In addition, we examined fusion transcripts and MET exon 14 skipping by using an RNA-based NGS solid-fusion assay.15
Statistical and Economic Analysis
For contingency analyses, we used t tests, Fisher’s exact tests, and χ2 statistics. We defined turnaround-time from the date of accessioning to the date of reporting, and we defined TTI from the date of the diagnostic biopsy to the date of ingestion of an EGFR tyrosine kinase inhibitor (TKI). We compared the TTI of EGFR-directed therapy in the rapid and historical groups by using event plots and log-rank statistics. For economic assessment, we reviewed line items of reimbursement data (January 3, 2017, to February 8, 2018) and extracted (by payor) the number of encounters and claim adjustment codes, and defined reimbursed as a payment greater than 0. For statistical computing and graphics, we used Prism 5 (GraphPad software, La Jolla, CA) and/or R (version 3.3.3; https://www.r-project.org/).
RESULTS
Patient Characteristics
Between January 2015 and May 2017, we performed rapid EGFR testing on 243 consecutive newly diagnosed patients with metastatic NSCLC (approximately two patients/week; Table 1). The median age at diagnosis was 69.1 years (range, 34 to 92 years). Most cancers were adenocarcinomas (n = 205; 84%). Never- or light-smokers comprised 54% (n = 132 of 243) of the rapid cohort. Forty-three (18%) of 243 tumors harbored EGFR mutations. One patient had an uncommon EGFR exon 19 deletion that did not involve the ELREA sequence (p.E746-A750).16 Figure 2A shows representative images of positive rapid test results. Although it was our intention that all specimens would also be submitted for NGS, specimens from nine patients were not referred for NGS. Among the remaining 234 patients, 23 (10%) did not have NGS results because of insufficient tissue or technical failure of NGS (Data Supplement).
Table 1.
Clinicopathological Characteristics of Rapid Cohort
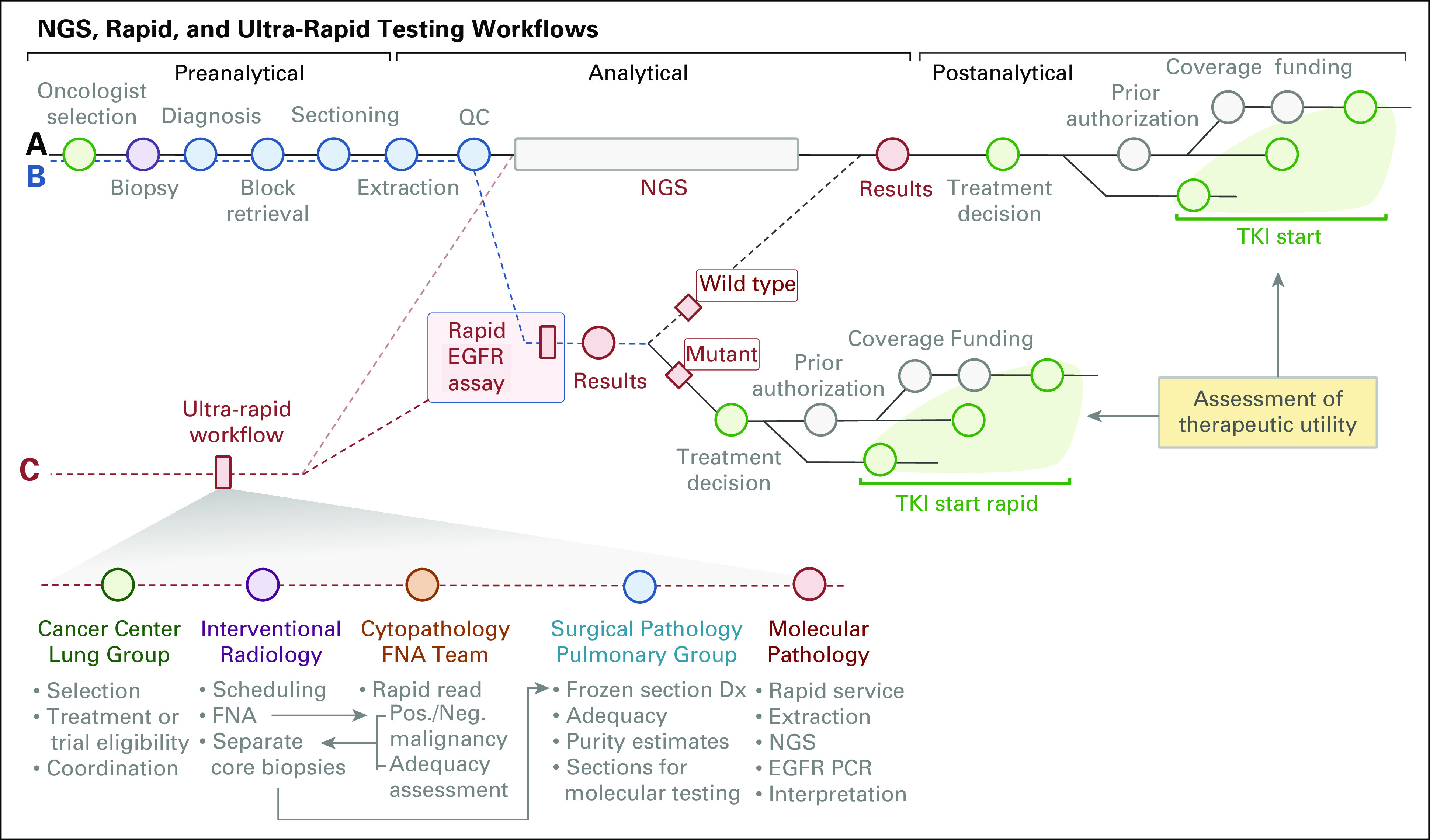
Fig 2.
Rapid EGFR assay and turnaround times compared with next-generation sequencing (NGS)–based genotyping. (A) The rapid EGFR assay consists of three separate reactions: a sizing assay to identify exon 19 (ELREA sequence) deletions and two single-nucleotide extension reactions to identify p.T790M and p.L858R missense mutations. (B) After validation (last quarter of 2014), we implemented rapid EGFR genotyping in January 2015. Scatter plots portray turnaround times of all 243 rapid EGFR samples (Jan 2015 to May 2017; black, EGFR wild type [WT]; red, EGFR mutation [mut] detected) and all specimens that underwent NGS (gray dots) during this period. Note that process improvements have led to a reduction in average turnaround times for both assays (lines).
Diagnostic Utility of the Rapid EGFR Assay
Improvement in turnaround time. First, we compared the turnaround times of the rapid and NGS assays by reviewing all specimens submitted for genotyping between July 2014 and January 2017 (Fig 2B). The median turnaround time in workdays for NGS genotyping was 14 days compared with 7 days for rapid testing (P < .001, t test). Forty-one of the 43 patients (95%) who tested positive for EGFR mutations with the rapid assay were also tested for mutations using NGS. In this group of patients, the median turnaround time for rapid EGFR and NGS was also 7 days and 14 days, respectively. Overall, rapid EGFR testing significantly reduced the time from biopsy to availability of EGFR results.
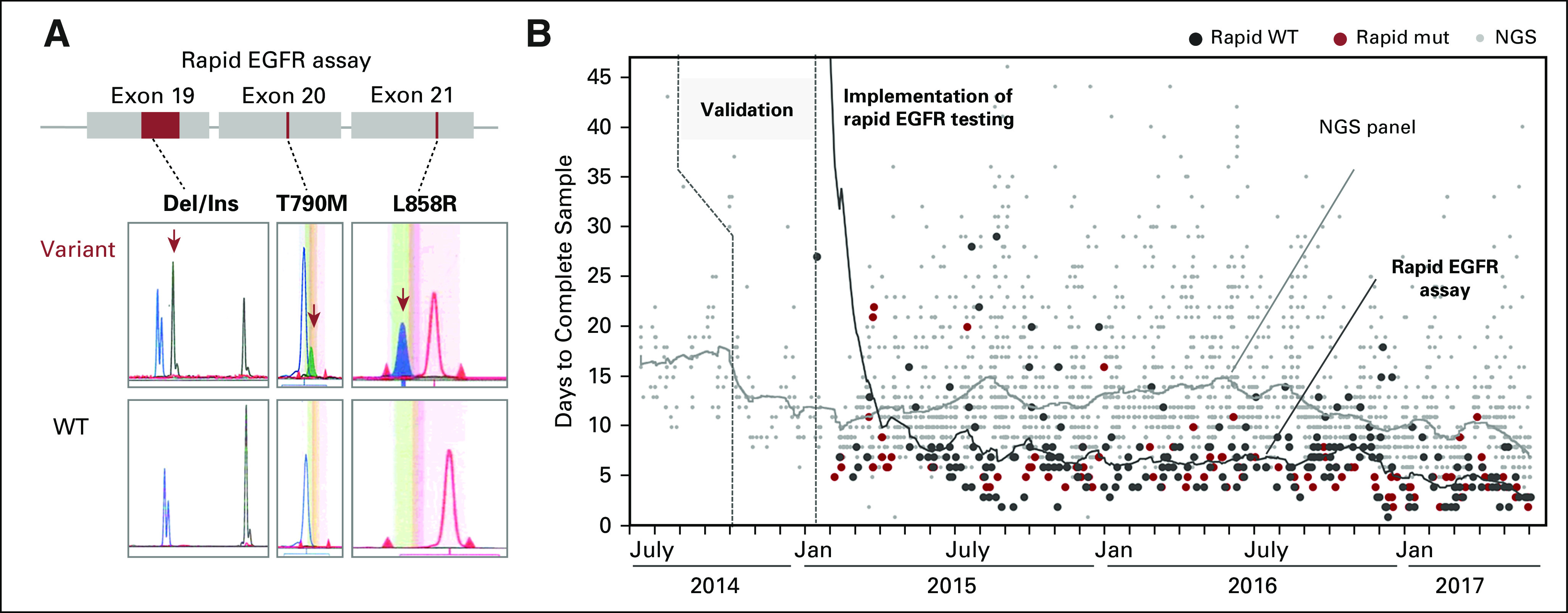
Concordance between rapid assay and NGS genotyping. Next, we assessed the technical performance of the rapid EGFR test by comparing rapid with NGS results. Through NGS, we identified one additional low-level (allelic fraction, approximately 4%) p.L858R case, which was missed by rapid EGFR testing (Fig 3). Compared with NGS, we did not detect any false-positive results with the rapid EGFR test. Overall, the sensitivity and specificity of the rapid EGFR test to detect the EGFR mutations of interest were 98% and 100%, respectively. The mutation-spectrum of the tumors that underwent rapid testing is depicted in Figure 3, and the Data Supplement shows probabilities of therapeutically actionable variants.
Fig 3.
Integration of molecular-genetic testing in 243 patients with non–small-cell lung cancer who underwent rapid EGFR genotyping. The heatmap portrays clinicopathologic features (top three rows), rapid EGFR results, and key molecular drivers along with the results of next-generation sequencing (NGS)–based fusion detection, fluorescence in-situ hybridization (FISH), and NGS panel results. Key findings include (1) an isolated false-negative rapid EGFR result (arrow), (2) the inability of the rapid EGFR test to detect EGFR mutations at other residues, (3) identification of at least one underlying driver mutation in more than 50% of all tested cases by using the integrated molecular diagnostic approach, and (4) the association between clinicopathologic features and certain key drivers (eg, never-smoking women with adenocarcinoma and EGFR v > 10 pack-year smoking history and KRAS). Adeno, adenocarcinoma; AdSQ, adenosquamous; Delins, insertion/deletion; Ex19del, exon 19 deletion; Ex20ins, exon 20 insertion; GCCa, giant cell carcinoma; LCNEC, large-cell neuroendocrine carcinoma; N/A, not applicable; Non-syn, nonsynonymous; PM, point mutation; py, pack year; SarcCa, sarcomatoid carcinoma; SQ, squamous.
Clinical Utility of Rapid EGFR Testing
TTI of TKI therapy. In 2009, a pivotal study demonstrated that empiric initiation of an EGFR TKI on the basis of clinical characteristics alone could be detrimental.17 As a result, demonstration of an EGFR mutation is a prerequisite for TKI initiation. Therefore, we selected time to EGFR TKI as the primary measure of clinical utility of the rapid EGFR test. To determine whether expedited reporting of EGFR results decreased the TTI of TKI therapy, we compared the median time to initiation of TKI therapy in our rapid cohort to that of a historical cohort of 121 patients diagnosed with EGFR-mutant NSCLC between 2011 and 2014 (before our rapid testing initiative); during this time, first-line treatment of EGFR-mutant NSCLC with an EGFR TKI was the standard of care at our institution.
Forty-one (95%) of 43 patients in the rapid cohort had sufficient follow-up after diagnosis to confirm the date of initiation of therapy. Of these patients, 39 (95%) received an EGFR TKI as first-line therapy, compared with 98 patients (81%) in the historical cohort (P = .04, Fisher’s exact). The median TTI of EGFR TKI therapy was 3.1 weeks for the rapid cohort compared with 5.3 weeks for the historical group (P < .001, log-rank; hazard ratio, 3.4; 95% CI, 2.1 to 5.6; Fig 4A). We reviewed the medical records of the 39 patients with EGFR-mutant disease who underwent both rapid testing and NGS to determine whether TKI therapy was initiated before NGS results. We were not able to assess this end point for four patients because of the following: (1) timing of TKI initiation could not be confirmed (n = 1); (2) an EGFR TKI was not initiated during the follow-up period (n = 1), or (3) initiation of TKI therapy was prompted by prior outside testing that predated results at our institution (n = 2). Among the remaining 35 patients, 17 (49%) started TKI therapy before NGS results (Fig 4B). Of note, apart from one patient who had de novo high-level MET amplification and a single case in which an EGFR T790M mutation was present at diagnosis,18 none of the other tumors harbored baseline co-alterations that would be expected to compromise the efficacy of EGFR TKI monotherapy (Fig 3).
Fig 4.
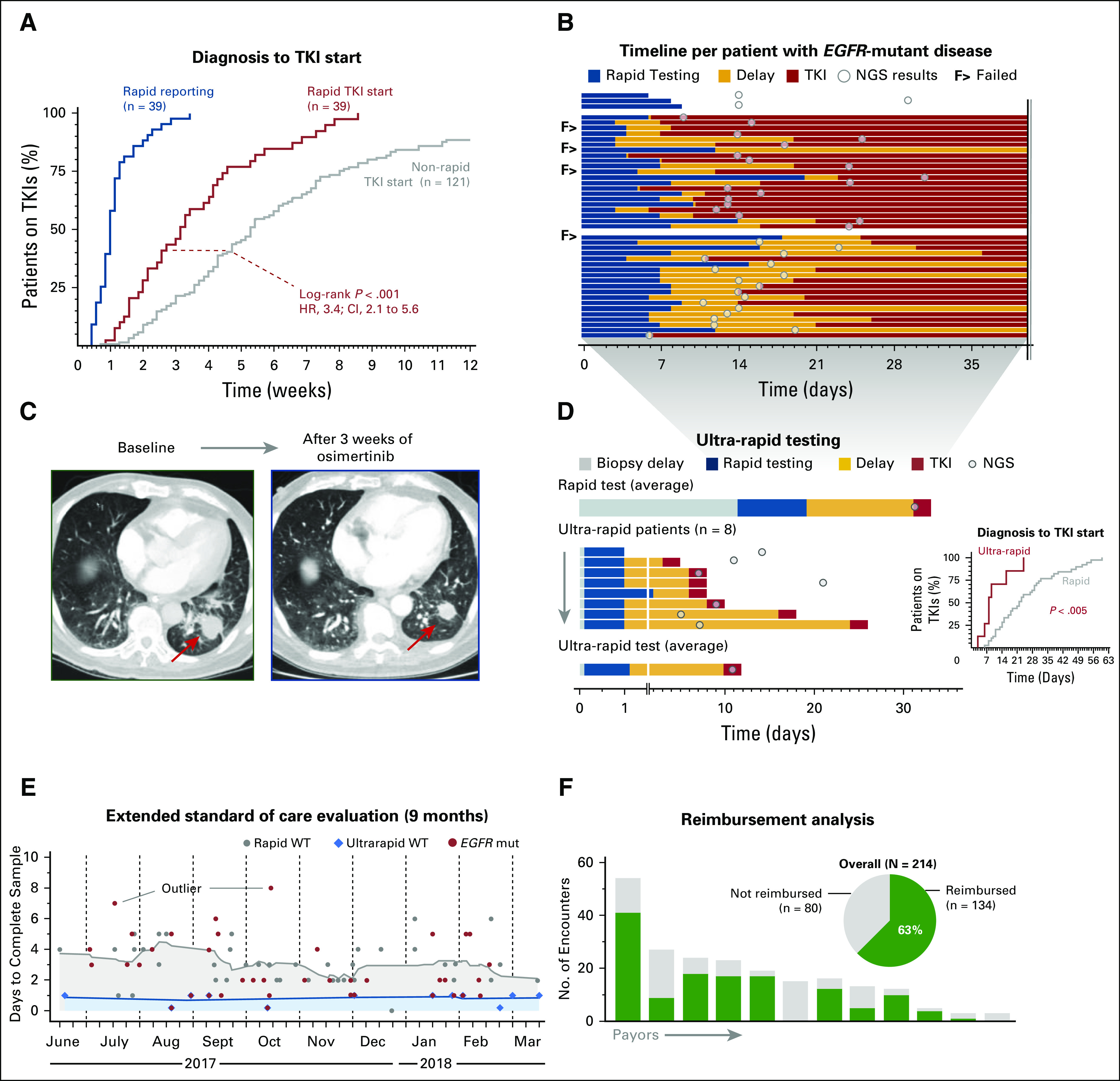
Therapeutic and clinical utility of rapid EGFR genotyping. (A) Event curve that shows rapid EGFR test reporting times and a comparison of the tyrosine kinase inhibitor (TKI) initiation times (relative to date of diagnosis) for patients in the rapid and historical cohorts. (B) Timeline of 43 patients with EGFR-mutant lung cancer. The top three patients did not receive epidermal growth factor receptor (EGFR)–directed therapy during the follow-up period. The second block shows the 49% of patients (n = 17 of 35 patients) with EGFR-mutant disease who started a TKI before next-generation sequencing (NGS) results were available. The third block shows patients who initiated EGFR-directed therapy after NGS results were available. (C) Response to the EGFR inhibitor osimertinib in a patient with non–small-cell lung cancer who underwent ultra-rapid EGFR testing: (left) pretreatment image and (right) response after 3 weeks; arrow indicates primary tumor. (D) Comparison of rapid test times (average) and the eight patients tested with the ultra-rapid protocol (Fig 1C); inset shows event curve comparison of time to initiation of TKI between the rapid and ultra-rapid subsets. (E) Turnaround times for rapid (gray) and ultra-rapid (blue) workflows in a 9-month extended standard-of-care evaluation phase; red, cases with an EGFR mutation. Outliers in reporting times are due to delays in block retrieval or repeated testing. (F) Reimbursement analysis: pie chart depicts the overall frequency of reimbursement; columns illustrate the payor-based number of reimbursed encounters. mut, mutated; WT, wild type.
Implementation of an Ultra-Rapid EGFR Testing Workflow
Certain patients with aggressive disease may benefit the most from rapid initiation of effective therapies with fast onset. For example, several of our patients presented with compromised neurologic function (eg, vision loss or ataxia) or severe symptoms that required hospitalization. Rapid EGFR testing can be rushed for these patients; however, our protocols have already been optimized and are not readily amenable for a reduction in turnaround time. To address this need, we launched a multidisciplinary ultra-rapid workflow to improve preanalytical processes (Fig 1C). The ultra-rapid EGFR testing pathway uses fresh tissue for confirmation of cancer diagnosis and same-day nucleic acid extraction. In cases where ultra-rapid testing fails, the formalin-fixed tumor sample serves as a safeguard and allows for traditional molecular testing. A case report of a patient who benefitted from this effort and a summary of the experience to date are described in the next section.
Case report.
Therapeutic utility of ultra-rapid testing.
A 79-year-old male never-smoker presented with 1 month of persistent cough and 1 day of inability to walk or swallow. Imaging studies revealed a 4-cm left lung mass (Fig 4C), lung nodules, and lesions throughout the brain and leptomeninges. The lung mass was sampled via percutaneous needle biopsy 4 days into admission, and cytopathology and frozen sections confirmed adenocarcinoma. Same-day extraction and next-day testing revealed an EGFR exon 19 deletion. He initiated treatment with osimertinib on day 7 of hospitalization. Osimertinib was selected on the basis of its activity against leptomeningeal disease.19 Within days, the cough resolved, and the patient experienced marked improvement in his swallowing and regained the ability to walk. Scans obtained 3 weeks into therapy demonstrated response in the chest (Fig 4C) and CNS, including leptomeningeal sites.
Ultra-rapid testing turnaround time and results.
Between May 2017 and September 2017, a total of eight patients participated in the ultra-rapid testing program (Fig 4D). Seven tumors harbored EGFR mutations, whereas one tumor contained a ROS1 rearrangement. Figure 4D illustrates the difference in turnaround time for the rapid and ultra-rapid tests and the time to initiation of TKI therapy for patients tested through each workflow (Fig 4D inset). The median turnaround time for EGFR results and the median TTI of an EGFR TKI were 1.5 days and 9 days, respectively. The sites of disease and presenting symptoms of these patients are detailed in the Data Supplement, which also summarizes our ultra-rapid workflow.
Rapid EGFR testing is operationally and financially sustainable.
After review of the data presented here, we offered the rapid (and ultra-rapid) workflows as standard of care tests beginning in July 2017. Between July 2017 and March 2018 (Fig 4E), the average (mean ± standard deviation) turnaround time was 3 ± 1.5 days for rapid (ie, formalin-fixed paraffin-embedded–based) testing and was 0.81 ± 0.4 days for the ultra-rapid (ie, frozen) workflow, respectively. Review of 214 clinical encounters with 1,475 line items of reimbursement data showed large variability between payors (range, 0% to 100%) and that, overall, 63% of encounters received reimbursement (Fig 4F). These data indicate that rapid EGFR testing in our practice is operationally and financially sustainable.
DISCUSSION
Here, we report the results of a quality improvement initiative to explore parallel testing with a rapid EGFR assay and NGS in newly diagnosed NSCLC. To our knowledge, this is the first report of systematic prospective implementation of co-testing. Our findings emphasize the diagnostic, clinical, and therapeutic utility of rapid EGFR testing.
Metastatic NSCLC is a devastating and incurable disease for which the prognosis is highly dependent on identification of actionable molecular drivers.2 The established efficacy of US Food and Drug Administration–approved targeted therapies in specific NSCLC subsets and the increasing number of investigational compounds underscore the necessity of identification of molecular alterations in a timely fashion. NGS panels deliver comprehensive genotypes, but reporting delays can be substantial. Several studies suggest that genotyping with multiple single-gene assays may reduce reporting time20,21; however,selective interrogation of the ever-expanding set of targets is impractical and may be limited by tissue availability.22
At the time our initiative began, consensus guidelines supported testing newly diagnosed patients with NSCLC for EGFR mutations and rearrangements in ALK and ROS1.9 These mutually exclusive genetic alterations are predominantly found in never- or light-smokers.13,23 Testing these alterations in two stages (ie, restriction of testing for rearrangements to patients whose disease is wild type for EGFR mutations) may lead to unnecessary delays between diagnosis and initiation of treatment for a significant proportion of patients and potentially steer symptomatic patients away from molecularly targeted treatments. Through co-testing, we detected ALK and ROS1 rearrangements in 11 and six patients, respectively, and identified molecular alterations with promising investigational therapies in more than 40 additional patients (Fig 3). The comprehensiveness of NGS results comes at a price of longer turnaround time. For example, in a recent retrospective analysis that involved 15 community oncology centers, the median turnaround time for EGFR testing and NGS was 23 days and 30 days, respectively.24 Although this practice is likely more common in the community setting, where in-house testing is infrequent, one study demonstrated that 19% of patients with EGFR mutations or ALK rearrangements at an academic medical center in Canada initiated first-line chemotherapy before testing results became available.11 The sequence of therapies may be particularly important for those patients initially treated with immunotherapy, because recent studies demonstrate that immunotherapy is seldom effective for EGFR-mutant NSCLC and that treatment with immunotherapy before targeted therapy increases the likelihood of the development of significant toxicities.25-27 Given these data, we believe that rapid testing should be performed in parallel with full NGS genotyping.
With a parallel-testing approach, we successfully performed rapid testing and NGS in 90% of patients in our initiative with material from a single biopsy. Because half of the patients with tissue/DNA that was insufficient for the full NGS were effectively tested for both ALK and ROS1 rearrangements using either fluorescence in-situ hybridization or the fusion assay, 95% of the patients in the rapid cohort met the minimum molecular testing requirement proposed by current ASCO guidelines. Because our success rate compares favorably with other studies that evaluated biopsy specimen adequacy,28 concerns about tissue availability should not deter implementation of similar testing strategies. However, we acknowledge that the tissue failure rate may be higher if express testing for other alterations is performed in conjunction with rapid EGFR testing and NGS. To offset this risk, the ultra-rapid testing protocol specifies that additional cores should be obtained if safe and feasible (Data Supplement). Nonetheless, limitations of tissue-based genotyping have kindled interest in plasma genotyping. Although PCR-based plasma genotyping is an expedient alternative to NGS-based tissue genotyping, the sensitivity of liquid biopsy for detection of EGFR mutations is lower than what we report here with the rapid assay.29,30
Despite the high sensitivity and specificity of the rapid assay, half of the patients in our study did not start TKI therapy before NGS results were available (Fig 4B). The inability to translate faster turnaround times into intervention could be the result of timing of drug shipment or socioeconomic factors. Participants in the rapid genotyping group started TKI therapy approximately 2 weeks before patients in the historical cohort. Because TTI was assessed retrospectively for the historical cohort, the median of 37 days between diagnosis and TKI therapy might be an overestimate; however, a similar delay was observed in another study.31 It also is conceivable that the difference in TTI might be explained by process improvements over time that have shortened the interval between requests for and receipt of a drug from specialty pharmacies. Because TKI use predated NGS results in approximately 50% of patients, the reduction in time to TKI initiation in the rapid group relative to the historical cohort might also reflect faster NGS reporting times. Indeed, our average turnaround time for NGS is approximately 10 days currently versus approximately 2 weeks in the past (Fig 2B). The scale of NGS panels currently precludes significant reduction, and the median and mean turnaround times for NGS results in the ultra-rapid cohort were 9 and 10 days, respectively. In contrast, we showed that PCR-based approaches can consistently produce rapid results within 24 hours of biopsy (Fig 4E)—a benchmark that is currently impossible to achieve with NGS.
It remains to be established whether initiation of an EGFR TKI earlier improves overall survival.32,33 The impact of the timing of EGFR therapy on overall survival has become even more difficult to assess because of widespread use of next-generation EGFR TKIs capable of overcoming TKI resistance in a subset of cases. As a result of the short duration of follow-up of our rapid cohort and imbalances between the rapid and historical groups (including enrollment of patients in the rapid cohort into a consolidation radiation protocol), we were unable to evaluate differences in progression-free survival on EGFR-directed therapy. Despite these limitations, there are notable advantages to implementation of an assay that increases first-line use of EGFR TKIs, including cost savings relative to up-front treatment with chemotherapy and the potential to induce faster and more robust responses in symptomatic patients.4,34-38 Although EGFR-mutant NSCLC is classically regarded as an indolent disease, many patients present with widely metastatic disease (Data Supplement).39,40 The superior efficacy of EGFR TKIs relative to chemotherapy against brain metastases and the potential to defer brain radiation for these patients underscore the importance of rapid identification of EGFR mutations.41,42 The small number of patients with EGFR-mutant disease in our study also limits the potential impact of our findings. Finally, we recognize that successful execution of this initiative was dependent on establishment of the infrastructure to support rapid testing (Data Supplement). Thus, our findings may not be generalizable to all practice settings. However, many laboratories are proficient in performing targeted EGFR genotyping,43 and our extended standard of care evaluation (Fig 4E) and reimbursement analysis (Fig 4F) indicate operational and financial sustainability.
In summary, we demonstrate that expedited EGFR genotyping enables early intervention with targeted therapies and allows symptomatic patients to access effective treatments. The growing number of actionable targets substantiates the need for diagnostic strategies that expedite molecular analysis.
ACKNOWLEDGMENT
We thank the entire clinical team of the Center for Integrated Diagnostics, in particular A. Baig, Z. Georgantis, B. Lemos, Y. Cao, C.E. Finn, and A.N. Raymond for expert technical assistance and A. Smith, K. Burke, and N. Jessop for expert administrative assistance. This work was funded in part by NIH Grant No. R01 CA225655 (J.K.L.), and the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank the staff of the interventional thoracic radiology division, in particular A. Sharma, M.D. Gilman, S. McDermott, M. Price, M. Petranovic, L.A., Howell, and V. Noah. We thank the members of the thoracic oncology department—C. Rizzo, M. Banwait, J. Gurski, J. James, and L. Ferris—for scheduling visits and coordinating biopsies. In addition, we thank the entire cytopathology division, in particular R. Arpin and B.J. Sweeney. We also thank the staff of the frozen section and surgical pathology division, in particular J. Botelho Houston and J. Patel.
AUTHOR CONTRIBUTIONS
Conception and design: Ibiayi Dagogo-Jack, Christopher G. Azzoli, Martha Pitman, Jo-Anne O. Shepard, Alice T. Shaw, A. John Iafrate, Jochen K. Lennerz
Collection and assembly of data: Ibiayi Dagogo-Jack, Christopher G. Azzoli, Florian Fintelman, Mari Mino-Kenudson, Anna F. Farago, Justin F. Gainor, Ginger Jiang, Zofia Piotrowska, Rebecca S. Heist, Inga T. Lennes, Jennifer S. Temel, Jessica J. Lin, Julie M. Batten, Hayley Robinson, Long P. Le, Lecia V. Sequist, Alice T. Shaw, A. John Iafrate, Jochen K. Lennerz
Provision of study material or patients: Ibiayi Dagogo-Jack, Christopher G. Azzoli, Anna F. Farago, Zofia Piotrowska, Inga T. Lennes, Jo-Anne O. Shepard, Lecia V. Sequist, Alice T. Shaw, A. John Iafrate, Jochen K. Lennerz
Data analysis and interpretation: Ibiayi Dagogo-Jack, Christopher G. Azzoli, Mari Mino-Kenudson, Justin F. Gainor, Meghan Mooradian, Subba R. Digumarthy, Vania Nose, Miguel Rivera, Valentina Nardi, Dora Dias-Santagata, Lecia V. Sequist, Alice T. Shaw, Jochen K. Lennerz
Administrative support: Christopher G. Azzoli, Vania Nose, A. John Iafrate, Jochen K. Lennerz
Financial support: A. John Iafrate, Jochen K. Lennerz
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Clinical Utility of Rapid EGFR Genotyping in Advanced Lung Cancer
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Ibiayi Dagogo-Jack
Honoraria: Foundation Medicine
Consulting or Advisory Role: Boehringer Ingelheim
Christopher G. Azzoli
Consulting or Advisory Role: Merck
Consulting or Advisory Role: ARIAD/Takeda
Florian Fintelmann
Consulting or Advisory Role: McKesson
Patents, Royalties, Other Intellectual Property: Royalties from writing a book with Elsevier (publisher)
Mari Mino-Kenudson
Consulting or Advisory Role: Merrimack, H3 Biomedicine
Anna F. Farago
Honoraria: Foundation Medicine
Consulting or Advisory Role: PharmaMar, Takeda, Abbvie, Loxo, Stemcentrx
Research Funding: PharmaMar (Inst), Abbvie (Inst), AstraZeneca (Inst), Bristol-Myers Squibb (Inst), Merck (Inst), Loxo (Inst), Ignyta (Inst)
Travel, Accommodations, Expenses: PharmaMar, Abbvie, Stemcentrx
Justin F. Gainor
Honoraria: Merck, Incyte, ARIAD, Novartis, Pfizer
Consulting or Advisory Role: Genentech, Bristol-Myers Squibb, Theravance, Loxo, Takeda, Array BioPharma, Amgen
Research Funding: Merck (Inst), Novartis (Inst), Genentech, Bristol-Myers Squibb (Inst), Adaptimmune (Inst), AstraZeneca (Inst), ARIAD, Jounce Therapeutics (Inst), Blueprint Medicines (Inst), Moderna Therapeutics (Inst), Tesaro (Inst)
Travel, Accommodations, Expenses: Affymetrix
Ginger Jiang
Employment: Novartis (I)
Zofia Piotrowska
Consulting or Advisory Role: Boehringer Ingelheim, AstraZeneca, ARIAD, Takeda, Superdimension, Guardant Health, Novartis, Abbvie
Research Funding: Novartis (Inst), ARIAD (Inst), Takeda (Inst), Guardant Health (Inst)
Rebecca S. Heist
Consulting or Advisory Role: Boehringer Ingelheim
Research Funding: Abbvie (Inst), Novartis (Inst), Roche (Inst), Incyte (Inst), Celgene (Inst), Mirati Therapeutics (Inst), Peregrine Pharmaceuticals (Inst), Exelixis (Inst), Millenium (Inst), Debiopharm Group (Inst), Corvus Pharmaceuticals (Inst)
Inga T. Lennes
Honoraria: Blue Cross and Blue Shield of Massachusetts
Consulting or Advisory Role: Kyruus
Jennifer S. Temel
Research Funding: Pfizer (Inst)
Meghan J. Mooradian
No relationship to disclose
Jessica J. Lin
Honoraria: Chugai Pharma
Consulting or Advisory Role: Boehringer Ingelheim
Subba R. Digumarthy
No relationship to disclose
Julie M. Batten
No relationship to disclose
Hayley Robinson
No relationship to disclose
Vania Nose
No relationship to disclose
Miguel Rivera
Consulting or Advisory Role: Loxom Asubio (I)
Speakers' Bureau: Pfizer (I)
Research Funding: Advanced Cell Diagnostics, Affymetrix
Patents, Royalties, Other Intellectual Property: Patents with Affymetrix
Valentina Nardi
Stock and Other Ownership Interests: KSQ Therapeutics (I), Navicor (I)
Consulting or Advisory Role: Thermo Fisher Scientific (I), Cell Signaling Technology (I), RAZE (I), Caloric Tests (I)
Dora Dias-Santagata
No relationship to disclose
Long P. Le
Stock and Other Ownership Interests: Archer Biosciences
Consulting or Advisory Role: Archer Biosciences
Patents, Royalties, Other Intellectual Property: I am a co-inventor of the Anchored Multiplex PCR technology which is licensed to ArcherDx. I receive royalty payments for this patent.
Travel, Accommodations, Expenses: Archer Biosciences
Lecia V. Sequist
Honoraria: AstraZeneca
Consulting or Advisory Role: AstraZeneca, Genentech, Roche, Bristol-Myers Squibb, Pfizer
Research Funding: Boehringer Ingelheim (Inst), Clovis Oncology (Inst), Genentech (Inst), Merrimack (Inst), Novartis (Inst), AstraZeneca (Inst), Johnson & Johnson (Inst), Merck (Inst), Pfizer (Inst), Guardant Health (Inst), Incyte (Inst)
Martha Pitman
Consulting or Advisory Role: Medtronic
Jo-Anne O. Shepard
No relationship to disclose
Alice T. Shaw
Honoraria: Pfizer, Novartis, Roche, Genentech, Foundation Medicine
Consulting or Advisory Role: Pfizer, Novartis, Genentech, Roche, ARIAD, Ignyta, Blueprint Medicines, Daiichi Sankyo, EMD Serono, Taiho Pharmaceutical, KSO Therapeutics, Natera, Loxo, Takeda
Research Funding: Pfizer, Novartis, Roche, Genentech,
A. John Iafrate
Stock and Other Ownership Interests: Archer Biosciences
Consulting or Advisory Role: Debiopharm Group, Chugai Pharma, Roche, Blueprint Medicines
Patents, Royalties, Other Intellectual Property: ArcherDx exclusive license to AMP technology
Jochen K. Lennerz
No relationship to disclose
REFERENCES
- 1.Jordan EJ, Kim HR, Arcila ME, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7:596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation–positive non–small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 4.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 5.Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371:2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 6.Soria JC, Tan DSW, Chiari R, et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non–small-cell lung cancer (ASCEND-4): A randomised, open-label, phase 3 study. Lancet. 2017;389:917–929. doi: 10.1016/S0140-6736(17)30123-X. [DOI] [PubMed] [Google Scholar]
- 7.Planchard D, Besse B, Groen HJM, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non–small-cell lung cancer: An open-label, multicentre phase 2 trial. Lancet Oncol. 2016;17:984–993. doi: 10.1016/S1470-2045(16)30146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non–small-cell lung cancer. N Engl J Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leighl NB, Rekhtman N, Biermann WA, et al. Molecular testing for selection of patients with lung cancer for epidermal growth factor receptor and anaplastic lymphoma kinase tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Guideline. J Clin Oncol. 2014;32:3673–3679. doi: 10.1200/JCO.2014.57.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pao W, Kris MG, Iafrate AJ, et al. Integration of molecular profiling into the lung cancer clinic. Clin Cancer Res. 2009;15:5317–5322. doi: 10.1158/1078-0432.CCR-09-0913. [DOI] [PubMed] [Google Scholar]
- 11.Lim C, Tsao MS, Le LW, et al. Biomarker testing and time to treatment decision in patients with advanced non–small-cell lung cancer. Ann Oncol. 2015;26:1415–1421. doi: 10.1093/annonc/mdv208. [DOI] [PubMed] [Google Scholar]
- 12.Piotrowska Z, Sequist LV. Epidermal growth factor receptor–mutant lung cancer: New drugs, new resistance mechanisms, and future treatment options. Cancer J. 2015;21:371–377. doi: 10.1097/PPO.0000000000000147. [DOI] [PubMed] [Google Scholar]
- 13.Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: An analysis of 1,683 patients with non–small-cell lung cancer. Clin Cancer Res. 2013;19:4273–4281. doi: 10.1158/1078-0432.CCR-13-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: A clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 16.Chung KP, Wu SG, Wu JY, et al. Clinical outcomes in non–small-cell lung cancers harboring different exon 19 deletions in EGFR. Clin Cancer Res. 2012;18:3470–3477. doi: 10.1158/1078-0432.CCR-11-2353. [DOI] [PubMed] [Google Scholar]
- 17.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 18.Gainor JF, Niederst MJ, Lennerz JK, et al. Dramatic response to combination erlotinib and crizotinib in a patient with advanced, EGFR-mutant lung cancer harboring de novo MET amplification. J Thorac Oncol. 2016;11:e83–e85. doi: 10.1016/j.jtho.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 19.Yang JC-H, Cho BC, Kim D-W, et al. Osimertinib for patients (pts) with leptomeningeal metastases (LM) from EGFR-mutant non–small-cell lung cancer (NSCLC): Updated results from the BLOOM study. J Clin Oncol. 2017;35 suppl; abstr 2020. [Google Scholar]
- 20.Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non–small-cell lung cancer: Results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT) Lancet. 2016;387:1415–1426. doi: 10.1016/S0140-6736(16)00004-0. [DOI] [PubMed] [Google Scholar]
- 21.DiStasio M, Chen Y, Rangachari D, et al. Molecular testing turnaround time for non–small-cell lung cancer in routine clinical practice confirms feasibility of CAP/IASLC/AMP guideline recommendations: A single-center analysis. Clin Lung Cancer. 2017;18:e349–e356. doi: 10.1016/j.cllc.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Drilon A, Wang L, Arcila ME, et al. Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin Cancer Res. 2015;21:3631–3639. doi: 10.1158/1078-0432.CCR-14-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutierrez ME, Choi K, Lanman RB, et al. Genomic profiling of advanced non–small-cell lung cancer in community settings: Gaps and opportunities. Clin Lung Cancer. 2017;18:651–659. doi: 10.1016/j.cllc.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Garassino MC, Cho BC, Kim JH, et al. Durvalumab as third-line or later treatment for advanced non–small-cell lung cancer (ATLANTIC): An open-label, single-arm, phase 2 study. Lancet Oncol. 2018;19:521–536. doi: 10.1016/S1470-2045(18)30144-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CK, Man J, Lord S, et al. Clinical and molecular characteristics associated with survival among patients treated with checkpoint inhibitors for advanced non–small-cell lung carcinoma: A systematic review and meta-analysis. JAMA Oncol. 2018;4:210–216. doi: 10.1001/jamaoncol.2017.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshima Y, Tanimoto T, Yuji K, et al. EGFR-TKI–Associated interstitial pneumonitis in nivolumab-treated patients with non–small-cell lung cancer. JAMA Oncol. doi: 10.1001/jamaoncol.2017.4526. [epub ahead of print on January 11, 2018] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sholl LM, Aisner DL, Varella-Garcia M, et al. Multi-institutional oncogenic driver mutation analysis in lung adenocarcinoma: The Lung Cancer Mutation Consortium experience. J Thorac Oncol. 2015;10:768–777. doi: 10.1097/JTO.0000000000000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oxnard GR, Paweletz CP, Sholl LM. Genomic analysis of plasma cell-free DNA in patients with cancer. JAMA Oncol. 2017;3:740–741. doi: 10.1001/jamaoncol.2016.2835. [DOI] [PubMed] [Google Scholar]
- 30.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016;2:1014–1022. doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacLean E, Louder A, Saverno K, et al. Molecular testing patterns in metastatic non–small-cell lung cancer. Am J Manag Care. 2016;22:e60–e67. [PubMed] [Google Scholar]
- 32.Lee CK, Davies L, Wu YL, et al. Gefitinib or erlotinib vs chemotherapy for EGFR mutation–positive lung cancer: Individual patient data meta-analysis of overall survival. J Natl Cancer Inst. 2017;109:109. doi: 10.1093/jnci/djw279. [DOI] [PubMed] [Google Scholar]
- 33.Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation–positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): Analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015;16:141–151. doi: 10.1016/S1470-2045(14)71173-8. [DOI] [PubMed] [Google Scholar]
- 34.Vergnenegre A, Massuti B, de Marinis F, et al. Economic analysis of first-line treatment with erlotinib in an EGFR-mutated population with advanced NSCLC. J Thorac Oncol. 2016;11:801–807. doi: 10.1016/j.jtho.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Ting J, Tien Ho P, Xiang P, et al. Cost-effectiveness and value of information of erlotinib, afatinib, and cisplatin-pemetrexed for first-line treatment of advanced EGFR mutation–positive non–small-cell lung cancer in the United States. Value Health. 2015;18:774–782. doi: 10.1016/j.jval.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 36.Lu S, Ye M, Ding L, et al. Cost-effectiveness of gefitinib, icotinib, and pemetrexed-based chemotherapy as first-line treatments for advanced non–small-cell lung cancer in China. Oncotarget. 2017;8:9996–10006. doi: 10.18632/oncotarget.14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Handorf EA, McElligott S, Vachani A, et al. Cost effectiveness of personalized therapy for first-line treatment of stage IV and recurrent incurable adenocarcinoma of the lung. J Oncol Pract. 2012;8:267–274. doi: 10.1200/JOP.2011.000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isla D, De Castro J, Juan O, et al. Costs of adverse events associated with erlotinib or afatinib in first-line treatment of advanced EGFR-positive non–small-cell lung cancer. Clinicoecon Outcomes Res. 2016;9:31–38. doi: 10.2147/CEOR.S121093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park K, Tan EH, O’Byrne K, et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation–positive non–small-cell lung cancer (LUX-Lung 7): A phase 2B, open-label, randomised controlled trial. Lancet Oncol. 2016;17:577–589. doi: 10.1016/S1470-2045(16)30033-X. [DOI] [PubMed] [Google Scholar]
- 40.Doebele RC, Lu X, Sumey C, et al. Oncogene status predicts patterns of metastatic spread in treatment-naive non–small-cell lung cancer. Cancer. 2012;118:4502–4511. doi: 10.1002/cncr.27409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuler M, Wu YL, Hirsh V, et al. First-line afatinib versus chemotherapy in patients with non–small-cell lung cancer and common epidermal growth factor receptor gene mutations and brain metastases. J Thorac Oncol. 2016;11:380–390. doi: 10.1016/j.jtho.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 42.Goss G, Tsai CM, Shepherd FA, et al. CNS response to osimertinib in patients with T790M-positive advanced NSCLC: Pooled data from two phase II trials. Ann Oncol. 2017 doi: 10.1093/annonc/mdx820. [DOI] [PubMed] [Google Scholar]
- 43.Kim AS, Bartley AN, Bridge JA, et al. Comparison of laboratory-developed tests and FDA-approved assays for BRAF, EGFR, and KRAS testing. JAMA Oncol. doi: 10.1001/jamaoncol.2017.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]