Abstract
The tuberous sclerosis complex (TSC) is a negative regulator of mTOR complex 1, a signaling node promoting cellular growth in response to various nutrients and growth factors. However, several regulators in TSC signaling still await discovery and characterization. Using pulldown and MS approaches, here we identified the TSC complex member, TBC1 domain family member 7 (TBC1D7), as a binding partner for PH domain and leucine-rich repeat protein phosphatase 1 (PHLPP1), a negative regulator of Akt kinase signaling. Most TBC domain–containing proteins function as Rab GTPase–activating proteins (RabGAPs), but the crystal structure of TBC1D7 revealed that it lacks residues critical for RabGAP activity. Sequence analysis identified a putative site for both Akt-mediated phosphorylation and 14-3-3 binding at Ser-124, and we found that Akt phosphorylates TBC1D7 at Ser-124. However, this phosphorylation had no effect on the binding of TBC1D7 to TSC1, but stabilized TBC1D7. Moreover, 14-3-3 protein both bound and stabilized TBC1D7 in a growth factor–dependent manner, and a phospho-deficient substitution, S124A, prevented this interaction. The crystal structure of 14-3-3ζ in complex with a phospho-Ser-124 TBC1D7 peptide confirmed the direct interaction between 14-3-3 and TBC1D7. The sequence immediately upstream of Ser-124 aligned with a canonical β-TrCP degron, and we found that the E3 ubiquitin ligase β-TrCP2 ubiquitinates TBC1D7 and decreases its stability. Our findings reveal that Akt activity determines the phosphorylation status of TBC1D7 at the phospho-switch Ser-124, which governs binding to either 14-3-3 or β-TrCP2, resulting in increased or decreased stability of TBC1D7, respectively.
Keywords: phosphorylation, Akt PKB, 14-3-3 protein, E3 ubiquitin ligase, protein stability, ubiquitylation (ubiquitination), tuberous sclerosis complex (TSC), mTORC1, phospho-switch
Introduction
Several nutrient-sensing mechanisms have evolved in organisms' adaptation to continuously changing environments. In mammals, the mTORC13 complex integrates environmental signals to positively control anabolic metabolism and cellular growth. mTORC1 is a multisubunit kinase that promotes protein synthesis through phosphorylation of ribosomal S6 kinase (S6K) and eIF4E-binding proteins (4E-BP) (1). Amino acids and growth factors are both necessary, but not sufficient, for the activation of mTORC1. For full activation of mTORC1, lysosomal localization is also needed. Once at the lysosome, mTORC1 can interact with its activator, the small GTPase Rheb, when found in its GTP-bound, active state. The tuberous sclerosis complex (TSC), composed of three proteins, TSC1, TSC2, and TBC1D7, has GTPase-activating protein (GAP) activity for Rheb, turning Rheb into a GDP-bound, nonactive state (2), thereby damping mTORC1 signaling. Growth factors activate mTORC1 through the PI3K-Akt pathway, and Akt directly phosphorylates TSC2, causing the TSC complex to dissociate from the lysosome (3). With loss of the TSC complex from the lysosome, Rheb exists in the GTP-bound form and is capable of activating mTORC1. Mutations of either TSC1 or TSC2 result in a tumor predisposition syndrome termed tuberous sclerosis complex (4).
Central to mTORC1 signaling and other important cellular pathways is the serine/threonine kinase, Akt (5). The Akt family of kinases is composed of three highly similar isoforms (Akt1, -2, and -3), which are thought to phosphorylate both redundant and unique substrates (6). Signaling through the PI3K-Akt pathway is often aberrantly regulated in cancer (7). Akt proteins are activated by growth factor-mediated increases in PI3K activity, leading to increased levels of PIP3 at cellular membranes, most notably the plasma membrane. Increased PIP3 levels drive localization of Akt to the plasma membrane through interaction with its pleckstrin homology domain (8). This membrane localization facilitates the interaction of Akt with two separate kinases, PDK1 and mTORC2, which phosphorylate the key residues Thr-308 and Ser-473 of Akt1, respectively (9), as well as the corresponding residues of Akt2 and Akt3. Phosphorylation of both Thr-308 and Ser-473 is needed for full activation of Akt activity (10). Deactivation of Akt signaling is facilitated by phosphatase-mediated dephosphorylation of these two important residues (11). The serine/threonine phosphatase, PP2A, dephosphorylates both Thr-308 and Ser-473 and is thought to represent the main phosphatase counteracting Akt activation (12–14). In addition to PP2A, PHLPP proteins (PHLPP1 and PHLPP2), which encode a PP2C-like phosphatase domain, have previously been shown to negatively regulate Akt and other members of the AGC family of kinases (15). PHLPP proteins have been suggested to preferentially dephosphorylate the Ser-473 site of Akt (16, 17).
Ubiquitin-proteasome system–mediated degradation is the major mechanism of regulating protein stability in the cell. Proteins targeted for destruction through the ubiquitin-proteasome system are tagged with polyubiquitin chains by a three-enzyme cascade composed of an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ubiquitin ligase (18). The substrate specificity of ubiquitination is usually determined by the E3 ligases. β-TrCP1 and the closely related β-TrCP2 are F-box proteins that act as substrate-binding subunits of the SCF (SKP1–cullin 1–F-box) RING E3 ligase complex, a family of ubiquitin ligases involved in phosphorylation-dependent ubiquitination of target proteins (19). β-TrCP1/2 are considered oncogenes and are important players in cancer progression (20). For β-TrCP1/2 to bind to substrates, the substrate must first be phosphorylated in a binding-specific degron motif. Various cellular stimuli cause activation of kinases that phosphorylate target proteins, leading to binding of F-box proteins and degradation of substrates (21, 22).
To identify novel substrate/binding partners of PHLPP1 and further clarify the role of PHLPP proteins in Akt signaling, we performed a pulldown/MS experiment. We identified the third member of the TSC complex, TBC1D7, as a novel binding partner of PHLPP1. In addition to being a structural component of the TSC complex, TBC1D7 has previously been suggested to be a Rab17-specific GAP (23). Structural comparison of TBC1D7 with typical TBC domain RabGAP proteins revealed that TBC1D7 is missing a key helix in the Rab GTPase binding groove and does not have the arginine/glutamine dual-finger residues that are critical for RabGAP activity. In addition, we found that Ser-124 of TBC1D7 is a substrate of Akt and that this phosphorylation positively regulates the stability of TBC1D7 protein via binding to 14-3-3 proteins. TBC1D7 also contains a degron N-terminal to Ser-124 that is recognized by the β-TrCP2 E3 ubiquitin ligase. We found that phosphorylation of Ser-124 is a molecular switch regulating the stability of TBC1D7.
Results
PHLPP proteins and TBC1D7 interact
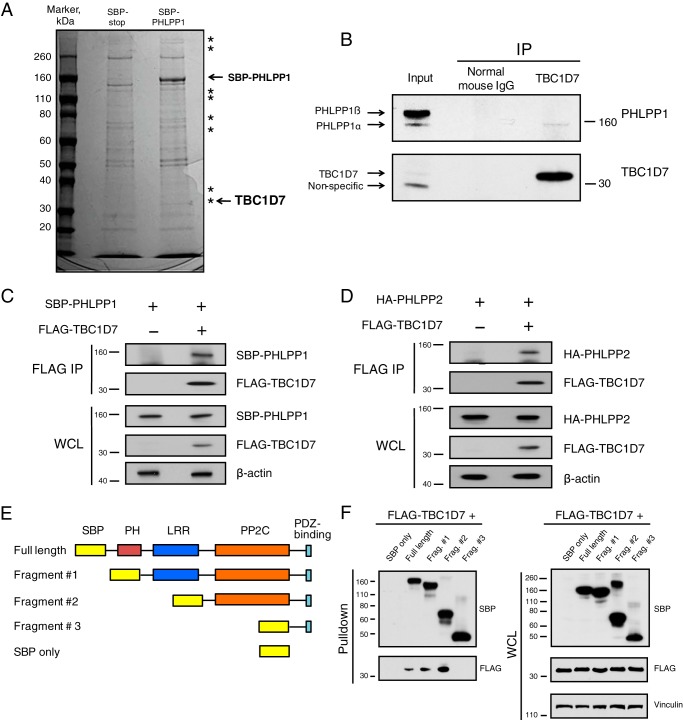
To expand understanding of the cellular roles of PHLPP proteins, we sought to identify and characterize novel proteins interacting with PHLPP1. Streptavidin-binding peptide (SBP)-tagged PHLPP was transfected into 293T cells, along with an SBP tag–expressing plasmid as a control for nonspecific binding to SBP. SBP-PHLLP1 and potential interacting proteins from cellular lysates were affinity-purified using streptavidin-agarose beads. As shown in Fig. 1A, colloidal blue staining of representative streptavidin pulldown samples revealed several bands present in the SBP-PHLPP1 lane and absent from the control pulldown lane. These unique bands were processed for identification by MS analysis. One such protein identified by peptide analysis was TBC1D7, a recently characterized third member of the TSC complex (24). Interaction between endogenous TBC1D7 and PHLPP1 proteins was confirmed by co-immunoprecipitation of PHLPP1 with TBC1D7 using a TBC1D7-specific antibody (Fig. 1B). TBC1D7 selectively co-immunoprecipitated the shorter α form of PHLPP1. The shorter form of PHLPP1 is transcribed from a downstream start codon and is missing the N-terminal intrinsically disordered region and a putative Ras association domain that exist in the longer β form of PHLPP1. The cause of the preferential binding of TBC1D7 with the α form over the β form is not known. Perhaps the β form of PHLPP1 is localized to a specific subcellular localization that is not in contact with TBC1D7, or the longer isoform forms an auto-inhibited conformation that prevents TBC1D7 binding. Furthermore, FLAG-TBC1D7 co-immunoprecipitated SBP-PHLPP1 when expressed together in 293T cells (Fig. 1C). As PHLPP1 and PHLPP2 are closely related, we show that in addition to PHLPP1, co-transfected FLAG-TBC1D7 can also co-immunoprecipitate HA-tagged PHLPP2 (Fig. 1D). To map the domain(s) of PHLPP1 essential for binding to TBC1D7, we constructed various N-terminal truncation mutants of SBP-tagged PHLPP1 (Fig. 1E). Streptavidin pulldown analysis, from lysates co-expressing SBP-tagged PHLPP1 truncation mutants and FLAG-TBC1D7, revealed that the PP2C-like phosphatase domain of PHLPP1 is necessary for binding to TBC1D7 (Fig. 1F). Collectively, these interaction studies characterize a novel protein–protein interaction between PHLPP1 and TBC1D7.
Figure 1.
PHLPP proteins and TBC1D7 are binding partners. A, 293T cells were transfected with either SBP or SBP-PHLPP1 expression plasmids. Lysates were subject to pulldown analysis using streptavidin beads. Affinity-purified complexes were resolved on SDS-PAGE, and the gel was stained with colloidal blue. Asterisks denote bands found exclusively in the SBP-PHLPP1 lane. Bands representing SBP-PHLPP1 and TBC1D7 proteins are highlighted. B, 293T cell lysates were immunoprecipitated with either a TBC1D7 mouse mAb or a control mouse IgG. Immunoprecipitates and input whole-cell lysates were resolved on SDS-PAGE and blotted with PHLPP1 and TBC1D7 antibodies. C, 293T cells were co-transfected with either vector and SBP-PHLPP1 or FLAG-TBC1D7 and SBP-PHLPP1 expression plasmids. Lysates were immunoprecipitated with FLAG antibody–conjugated beads. Immunoprecipitates and input whole-cell lysates were resolved on SDS-PAGE and blotted with SBP, FLAG, and β-actin antibodies. D, 293T cells were co-transfected with either vector and HA-PHLPP2 or FLAG-TBC1D7 and HA-PHLPP2 expression plasmids. Lysates were immunoprecipitated with FLAG antibody–conjugated beads. Immunoprecipitates and input whole-cell lysates were resolved on SDS-PAGE and blotted with HA, FLAG, and β-actin antibodies. E, diagram representing PHLPP1 domain deletions used in Fig. 1F. F, FLAG-TBC1D7 and various SBP-PHLPP1 expression constructs were co-transfected into 293T cells. Lysates were subject to pulldown analysis using streptavidin-agarose beads. Affinity-purified complexes and input whole-cell lysates were resolved on SDS-PAGE and blotted with SBP, FLAG, and vinculin antibodies.
TBC1D7 is an atypical TBC domain protein
The Tre-2/Bub2/Cdc16 (TBC) domain is widely observed in eukaryotic genomes and most proteins with TBC domains are believed to function as RabGAPs. Evolutionary analysis suggests that TBC1D7 belongs to a unique subclass of TBC proteins that exists only in metazoan genomes (25) but does not reveal how TBC1D7 is structurally different from typical TBC proteins. We purified recombinant full-length TBC1D7 from Escherichia coli, crystallized the protein using in situ proteolysis in the presence of dispase (26), and solved the crystal structure by single-wavelength anomalous diffraction of the selenomethionyl derivative. Statistics for the refined model are listed in Table S1.
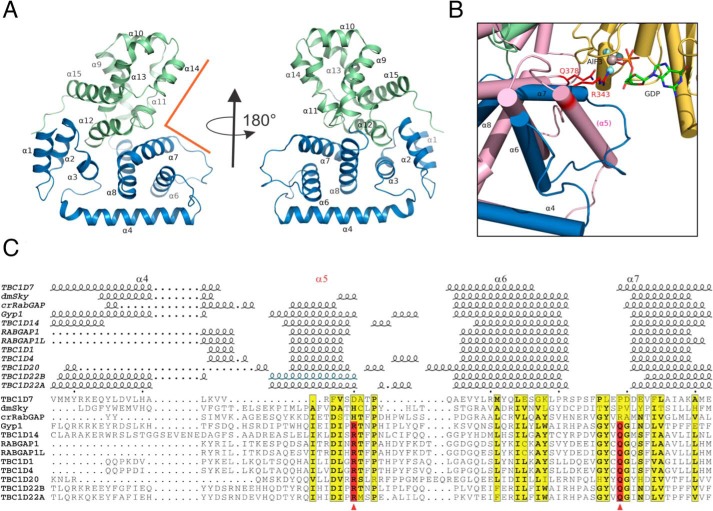
Overall, TBC1D7 is a globular protein that adopts an all-α fold (Fig. 2A). It contains 14 α-helices and several short 310 helices. A Dali search (27) revealed that the closest structural homologs are human TBC1D22A (28) (PDB code 2QFZ) and yeast Gyp1p (29) (PDB code 2G77) with high Z scores of 22.1 and 21.6, respectively. For the purpose of structure comparison, we numbered the α-helices according to the yeast Gyp1p structure as α1–α15 (Fig. 2A). From structure-based alignment (30) of all 12 known TBC domain structures, it is clear that helix α5 is missing in TBC1D7 (Fig. 2 (B and C) and Fig. S1). The linker between α4 and α6 is short, and the TBC1D7 polypeptide backbone takes a sharp turn after a long α-helix (α4) to form α6. The linker between α6 and α7 is, however, long and forms a relatively flexible protruding loop to partially fill the space occupied by α5 in other RabGAPs (Fig. 2B). Similar to Gyp1p, the helices are distributed between N-terminal and C-terminal subdomains spanning α1–α8 and α9–α15, respectively. Domain movement analysis using the Rapido (rapid alignment of protein structures in the presence of domain movements) server (31) to compare the structure of TBC1D7 with that of TBC1D22A identified two rigid body entities within the structure, roughly corresponding to the partitioning of the two subdomains. The two structures have a root mean square deviation of 3.0 Å over 235 residues, whereas a flexible alignment gives a root mean square deviation of 1.05 Å over the two rigid body entities.
Figure 2.
Structure of TBC1D7 and comparison with other TBC domains. A, overview of TBC1D7 structure. 15 α-helices were labeled based on the structure of yeast Gyp1 (89). The N-subdomain (α1–α8) is colored in blue, and the C-subdomain (α9–α15) is colored in light green. The rectangular groove that binds to Rab GTPase in Gyp1 is indicated using orange lines. B, an enlarged view of the region between α4 and α7 of TBC1D7 and compared with Gyp1 (salmon color) and Rab33 (yellow). Helix α5 is missing in TBC1D7. The dual-finger Arg-343/Gln-378 critical for Gyp1 RabGAP activity and the GDP molecule are shown as sticks. The AlF3 molecule is shown as a sphere. C, structure-based alignment of 12 known TBC domain structures of the region between α4 and α7. The active sites of the dual finger are indicated using red arrowheads. The structures are D. melanogaster Skywalker (dmSky, PDB code 5HJN), yeast Gyp1 (2G77), C. reinhardtii RabGAP (crRabGAP, 4P17), RABGAP1 (4NC6), RABGAP1L (3HZJ), TBC1D1 (3QYE), TBC1D4 (3QYB), TBC1D7 (3QWL), TBC1D14 (2QQ8), TBC1D20 (4HL4), TBC1D22A (2QFZ), TBC1D22B (3DZX). The α5 helix of the TBC1D22B crystal structure is presumed disordered or missing due to the use of protease and is indicated based on the high sequence identity to that of TBC1D22A.
Many TBC domains are known to possess RabGAP activity. A rectangular groove between the two subdomains is known to interact with Rab GTPases from known complex structures of the TBC domain with Rab GTPases (29, 32)(Fig. 2, A and B). To determine whether TBC1D7 is structurally primed for activating Rab GTPases, we overlaid the structure of TBC1D7 with that of the Gyp1p/Rab33 complex (Fig. 2B). Gyp1p uses a dual-finger Arg/Gln pair to promote the hydrolysis of GTP by Rab33, and the two key residues are located at the C terminus of α5 and N terminus of α7 (Fig. 2, B and C). It is clear from structure-based sequence alignment (Fig. 2C) that there are no equivalent residues for the Gyp1p Arg-343/Gln-378 dual finger in the vicinity. This suggests that TBC1D7 is an atypical TBC domain-containing protein that is unlikely to have RabGAP activity.
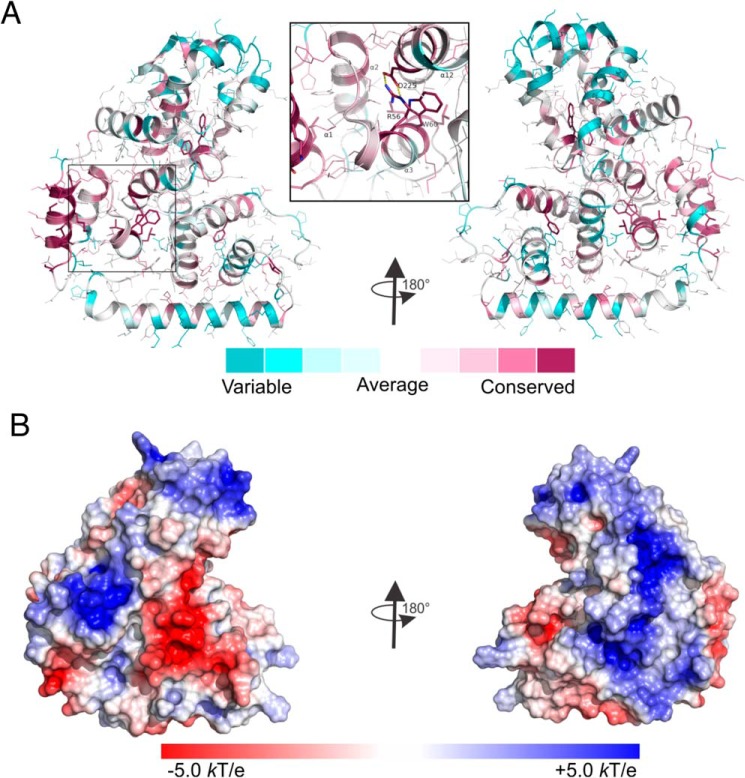
Analysis of the conservation of the residues of the TBC1D7 structure during evolution using the ConSurf server (33), based on the 250 closest homologs, revealed that the first helix α1 and residues involved in the packing of the structure are more conserved than the residues on the surface (Fig. 3A). Several hydrophobic and aromatic residues that are essential for forming hydrophobic cores are among the most conserved residues. A pair of Arg-Asp charged residues from N-subdomain α3 and C-subdomain α12 that form a salt bridge are also conserved during evolution, consistent with structural based alignment (Fig. S1), suggesting the importance of the bridge in maintaining the structure. The electrostatic potential surface of TBC1D7 (Fig. 3B) reveals positively and negatively charged patches on one side of the molecule and an extended positively charged patch on the other side of the molecule.
Figure 3.
Analysis of the TBC1D7 structure. A, conservation of TBC1D7 structure calculated using the ConSurf web server. Side chains of the most conserved residues are shown as sticks, whereas the rest are shown in thin lines. The enlarged insert shows the conserved salt bridge formed between residues Arg-56 and Asp-225 from helices α3 and α12 from the N- and C-subdomains, respectively. B, surface electrostatic potential of TBC1D7. Blue, positively charged residues; red, negatively charged residues.
Akt phosphorylates TBC1D7
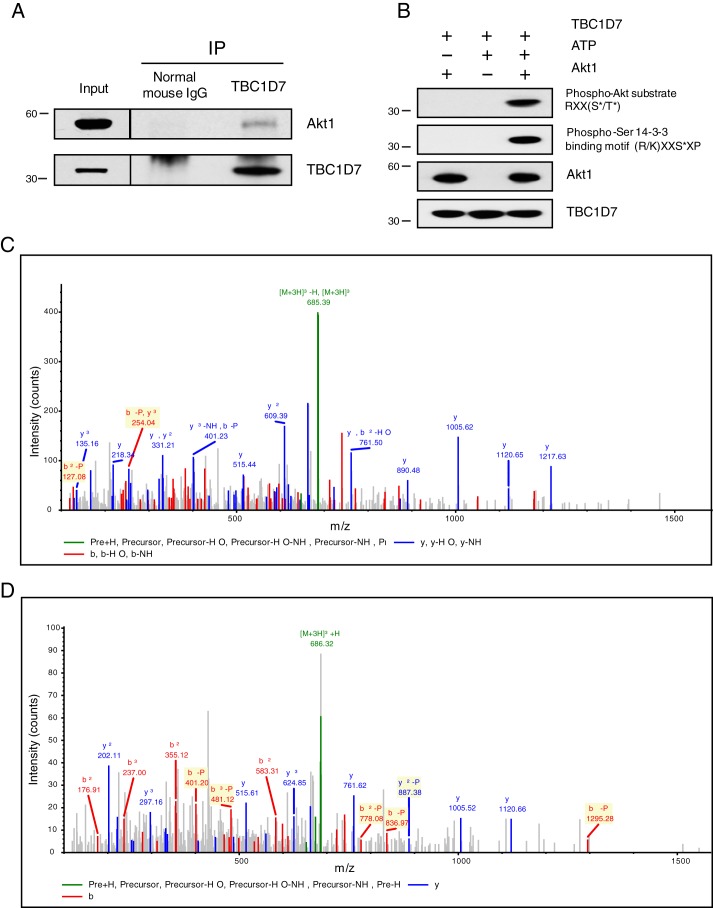
Both TSC1 and TSC2 are phosphoproteins, and TSC2 is a direct substrate of Akt-mediated inhibitory phosphorylation (34). Because TBC1D7 was pulled down using PHLPP1, a putative serine/threonine phosphatase, we considered the possibility that TBC1D7 protein might also be phosphorylated. Motif analysis of the TBC1D7 primary sequence, using the Scansite tool (35), revealed a high-probability site for Akt phosphorylation at Ser-124. The PhosphoSitePlus database reports that Ser-124 of TBC1D7 was identified as being phosphorylated via MS from lysates of Jurkat cells treated with the serine/threonine phosphatase inhibitor, calyculin A, and immunoprecipitated using a 14-3-3 binding motif antibody (36). Scansite analysis further predicted a high-probability 14-3-3-binding site centered upon Ser-124. A consensus Akt phosphorylation site is characterized by arginines at −3 and −5 from the target serine/threonine residue (RXRXX(pS/pT)) (37). Because TBC1D7 lacks the −5 arginine, it does not completely conform to a consensus Akt site. There are several examples in the literature of bona fide Akt substrates that contain the minimal Akt phospho-substrate sequence (RXX(pS/pT)), such as β-catenin, YAP, and p47PHOX (38–40). To determine whether TBC1D7 might be a target of Akt, we immunoprecipitated cell lysates using a TBC1D7-specific antibody and found that Akt was co-precipitated, confirming that TBC1D7 and Akt do interact in cells (Fig. 4A). Probing in vitro Akt kinase reactions with a minimal phospho-Akt substrate antibody (RXX(pS/pT)) demonstrated that TBC1D7 is a direct substrate for Akt (Fig. 4B). Mass spectrometry analysis of parallel kinase assay reactions confirmed that Ser-124 of TBC1D7 was phosphorylated directly by Akt (Fig. 4C). Furthermore, immunoprecipitated TBC1D7 from HeLa cells stimulated with insulin revealed that Ser-124 was phosphorylated in vivo (Fig. 4D). These data confirm that TBC1D7 is a direct target of Akt phosphorylation and that TBC1D7 is phosphorylated at Ser-124 in a growth factor–dependent manner.
Figure 4.
Akt interacts with and phosphorylates TBC1D7 at Ser-124. A, 293T cell lysates were immunoprecipitated with either a TBC1D7 mouse mAb or a control mouse IgG. Immunoprecipitates (IP) and input whole-cell lysates were resolved on SDS-PAGE and blotted with Akt1 and TBC1D7 antibodies. B, in vitro kinase assays were performed using active Akt1 and bacterially expressed recombinant TBC1D7 protein. As negative controls, either Akt1 or ATP was absent from the reaction mixtures. Assays were resolved on SDS-PAGE and probed with either a phospho-Akt substrate, phospho-Ser 14-3-3 binding motif, or Akt1 antibodies. The phospho-Ser 14-3-3 binding motif blot was stripped and reblotted with a TBC1D7-specific antibody. C and D, tandem mass spectrum showing phosphorylation of Ser-124 of TBC1D7 (C, in vitro; D, in vivo). Shown is the MS/MS spectrum of the peptide SPpSFPLEPDDEVFLAIAK, corresponding to residues 122–139. The identified b (red) and y (blue) ions are denoted in the spectrum; fragment ions important for localization of the site of phosphorylation are highlighted in yellow.
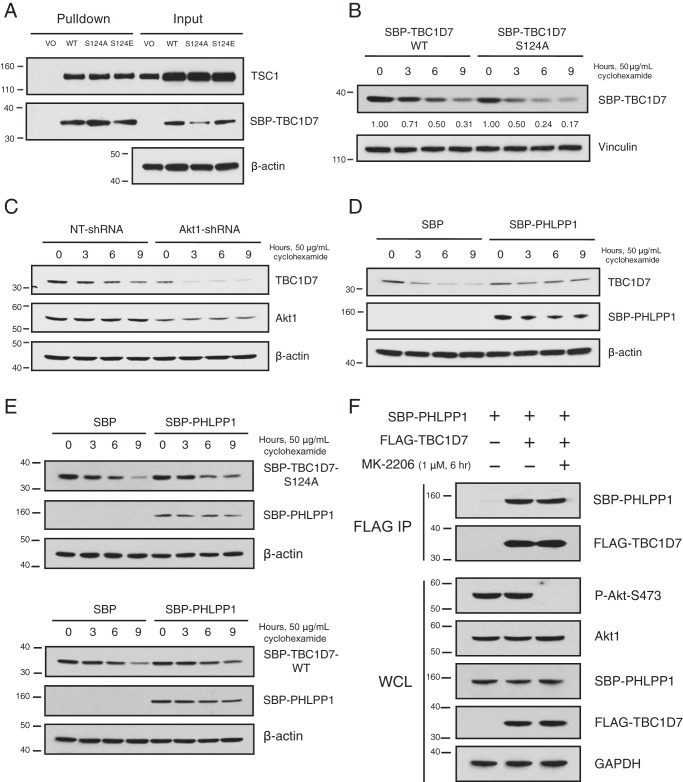
Phosphorylation at Ser-124 positively regulates TBC1D7 protein stability
Having confirmed that TBC1D7 is phosphorylated at Ser-124, we sought to determine the possible role(s) played by this site in TBC1D7 biology. Fluorescence microscopic analysis of GFP-tagged TBC1D7-WT, -S124A, and -S124E phospho-deficient and phosphomimetic mutants, respectively, revealed that phosphorylation at this site did not alter the subcellular localization of TBC1D7 (data not shown). Pulldown analysis from lysates expressing SBP-tagged TBC1D7-WT, -S124A, or -S124E mutants showed no difference in binding to TSC1 (Fig. 5A). These data complement work from a previous study showing that TBC1D7 binding to TSC1 was not affected by either phospho-deficient or phosphomimetic mutations of Ser-124 (41). Whereas no effect on binding to TSC1 was seen, steady-state levels of the S124A mutant TBC1D7 protein were reduced relative to the WT and S124E proteins (Fig. 5A). To determine a possible role of Ser-124 phosphorylation in the stability of TBC1D7 protein, a cyclohexamide treatment time course was performed, comparing the levels of WT and S124A proteins. As seen in Fig. 5B, S124A mutant TBC1D7 protein was less stable over time, compared with TBC1D7-WT protein. The TBC1D7-WT protein was twice as stable as the S124A mutant protein, with half-lives of ∼6 and 3 h, respectively. As we demonstrated that Akt1 could directly phosphorylate TBC1D7 at Ser-124 and that Ser-124 phosphorylation appears to stabilize TBC1D7, we explored the effect of stable Akt1 knockdown on TBC1D7 stability. Cyclohexamide chase analysis confirmed that stable Akt1 knockdown had a negative impact on TBC1D7 stability (Fig. 5C). Paradoxically, PHLPP1 had a similar effect on TBC1D7 as Akt1, with overexpression of PHLPP1 in cells positively regulating TBC1D7 protein stability (Fig. 5D). As Ser-124 phosphorylation of TBC1D7 and PHLPP1 overexpression both positively impact the stability of TBC1D7 protein and we have shown that the PP2C-like domain of PHLPP1 is needed for binding to TBC1D7 (Fig. 1F), we investigated whether the phospho-deficient S124A mutant might abrogate the positive effects of PHLPP1 overexpression on TBC1D7 stability. As demonstrated in Fig. 5E, increased stability of TBC1D7-S124A mutant protein was seen when PHLPP1 was overexpressed, similar to WT TBC1D7. These results suggest that the stabilizing effect of PHLPP1 overexpression on TBC1D7 protein stability is independent of the phosphorylation of TBC1D7 Ser-124. Furthermore, we demonstrate in Fig. 5F that binding between PHLPP1 and TBC1D7 is also independent of Ser-124 phosphorylation, as the interaction between PHLPP1 and TBC1D7 is not affected by Akt inhibition. Therefore, Ser-124 phosphorylation, mediated by Akt, leads to stabilization of TBC1D7 protein. However, both PHLPP1 interaction and the PHLPP1 overexpression–stabilizing effect are not dependent on Ser-124 phosphorylation.
Figure 5.
Ser-124 phosphorylation stabilizes TBC1D7. A, 293T cells were transfected with either SBP vector or SBP-TBC1D7 WT, S124A, S124E expression plasmids. Lysates were subject to pulldown analysis with streptavidin beads. Affinity-purified complexes and input whole-cell lysates were resolved on SDS-PAGE and blotted with TSC1, SBP, and β-actin antibodies. B, 293T cells were transfected with either SBP-TBC1D7-WT or -S124A expression plasmids. The following day, each dish was split into four separate dishes, and the day after, they were treated with 50 μg/ml cyclohexamide for various periods of time. Whole-cell lysates were separated on SDS-PAGE and blotted with SBP and vinculin antibodies. Zero-time points for both WT and S124A SBP-TBC1D7 were set to 1; relative expression of SBP-TBC1D7 proteins is indicated, as determined by densitometric analysis. Values were normalized to vinculin loading control. C, 293T cells stably expressing either NT- or Akt1-shRNA were treated with 50 μg/ml cyclohexamide for various periods of time. Whole-cell lysates were separated on SDS-PAGE and blotted with TBC1D7, Akt1, and β-actin antibodies. D, 293T cells were transfected with either SBP vector or SBP-PHLPP1 expression plasmids. The following day, each dish was split into four separate dishes, and the day after, they were treated with 50 μg/ml cyclohexamide for various periods of time. Whole-cell lysates were separated on SDS-PAGE and blotted with TBC1D7, SBP, and β-actin antibodies. E, 293T cells were co-transfected with SBP vector or SBP-PHLPP1 and SBP-TBC1D7-S124A (top) or SBP-TBC1D7-WT (bottom). The following day, each dish was split into four separate dishes, and the day after, they were treated with 50 μg/ml cyclohexamide for various periods of time. Whole-cell lysates were separated on SDS-PAGE and blotted with SBP and β-actin antibodies. F, 293T cells were co-transfected with either vector or FLAG-TBC1D7 and SBP-PHLPP1 expression plasmids. 42 h after transfection, cells were treated with either DMSO vehicle of MK-2206 (1 μm) for 6 h. Lysates were immunoprecipitated with FLAG antibody–conjugated beads. Immunoprecipitates (IP) and input whole-cell lysates (WCL) were resolved on SDS-PAGE and blotted with SBP, FLAG, P-Akt-Ser-473, and GAPDH antibodies. The P-Akt-Ser-473 blot was stripped and reprobed with an Akt1 antibody.
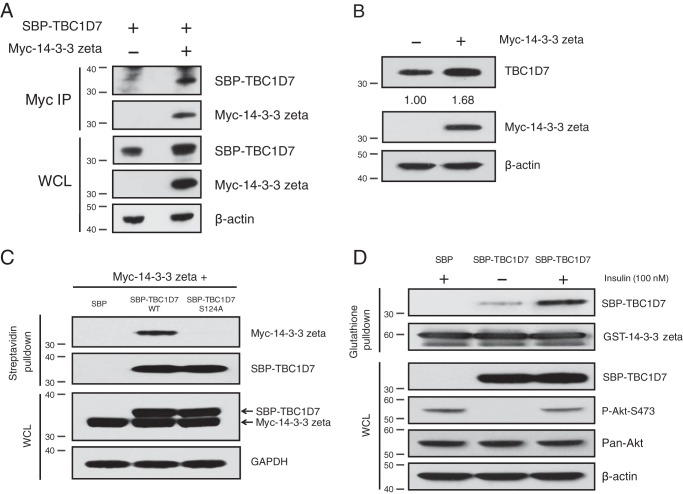
14-3-3ζ binds and stabilizes TBC1D7
14-3-3 proteins are an important family of phosphoserine/phosphothreonine-binding proteins that regulate numerous signaling pathways and control a myriad of cellular processes (42). In addition to predicting Akt phosphorylation at Ser-124, primary sequence analysis of TBC1D7 also predicted a high probability of binding to 14-3-3 proteins upon phosphorylation of Ser-124. Furthermore, Akt is a key kinase that generates 14-3-3 binding sites on a number of diverse proteins (43). Additional probing of in vitro kinase assay samples revealed that recombinant TBC1D7 protein was detected by a 14-3-3 phospho-substrate antibody recognizing a phosphorylated serine within the (R/K)XXpSXP epitope, which perfectly matches the sequence surrounding Ser-124 (RSPpSFP) (Fig. 4B). Transfection studies showed that Myc-14-3-3ζ could co-immunoprecipitate TBC1D7 (Fig. 6A). Interestingly, levels of tagged TBC1D7 were increased in lysate from cells that co-expressed Myc-14-3-3ζ, compared with those that were co-transfected with vector alone (Fig. 6A). Delving further into this observation, overexpression of Myc-tagged 14-3-3ζ resulted in increased steady-state levels of endogenous TBC1D7 protein, compared with mock vector–transfected cells (Fig. 6B). When co-expressed with Myc-14-3-3ζ, only SBP-TBC1D7-WT could pull down Myc-14-3-3ζ, but not SBP-TBC1D7-S124A (Fig. 6C). As TBC1D7 could be phosphorylated in cells in an insulin-dependent manner (Fig. 4D), we tested whether the binding of 14-3-3 to TBC1D7 could be modulated by the presence of growth factors. Compared with starvation alone, insulin stimulation of starved HeLa cells greatly increased the ability of SBP-TBC1D7 to be pulled down by GST-tagged 14-3-3ζ (Fig. 6D). Therefore, phosphorylation of TBC1D7 appears to be a requisite for 14-3-3 binding. These data suggest that growth factor–dependent Akt activity increases TBC1D7 stability through increased interaction with 14-3-3 proteins.
Figure 6.
14-3-3ζ interacts with and stabilizes TBC1D7. A, 293T cells were co-transfected with either vector and SBP-TBC1D7 or Myc-14-3-3ζ and SBP-TBC1D7 expression plasmids. Lysates were immunoprecipitated with Myc antibody. Immunoprecipitates (IP) and input lysates (WCL) were resolved on SDS-PAGE and blotted with SBP, Myc, and β-actin antibodies. B, 293T cells were transfected with either vector or Myc-14-3-3ζ. Whole-cell lysates were resolved on SDS-PAGE and blotted with TBC1D7, Myc, and β-actin antibodies. Relative expression of endogenous TBC1D7 is indicated, as determined by densitometric analysis. Values were normalized to β-actin loading control. The expression level of endogenous TBC1D7 from vector-transfected control cells was set to 1. C, 293T cells were co-transfected with Myc-14-3-3ζ and SBP vector, SBP-TBC1D7-WT, or SBP-TBC1D7-S124A. Lysates were subject to pulldown analysis with streptavidin beads. Affinity-purified complexes and input whole-cell lysates were resolved on SDS-PAGE and probed with Myc, SBP, and GAPDH antibodies. D, HeLa cells were transfected with either SBP vector or SBP-TBC1D7 expression plasmids. Cells were serum-starved (0% serum) overnight and treated for 15 min with 100 nm insulin or not. 10 μg of GSH Sepharose-conjugated recombinant GST-14-3-3ζ was incubated with cell lysates overnight. Co-purified complexes and input whole-cell lysates were resolved on SDS-PAGE and blotted with SBP, GST, P-Akt-Ser-473, and β-actin antibodies. The P-Akt-Ser-473 blot was stripped and reprobed with a pan-Akt antibody.
14-3-3ζ interacts with a TBC1D7 phosphopeptide directly
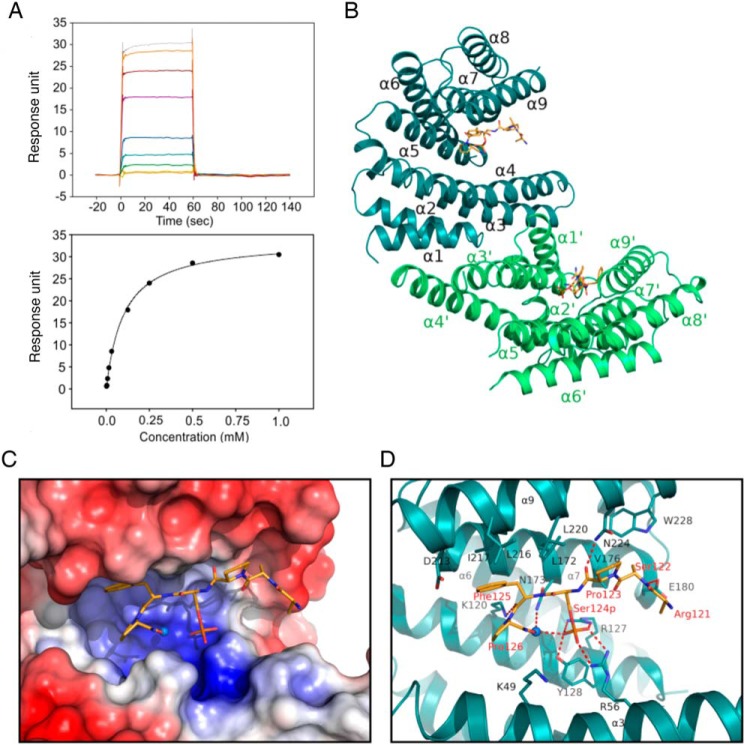
14-3-3 was the first protein known to specifically bind phosphoserine- or phosphothreonine-containing peptide motifs (44). To confirm that the TBC1D7–14-3-3 interaction is direct, we used surface plasmon resonance to measure the binding affinity between 14-3-3ζ and a TBC1D7-derived synthetic 12-mer phosphopeptide containing pSer-124, ESGKLPRSPpSFP, which corresponds to aa 115–126, the linker between α6 and α7. The phosphopeptide binds weakly but directly to 14-3-3 with a dissociation constant (KD) of 102 μm (Fig. 7A). This binding was further confirmed using isothermal titration calorimetry (Fig. S2). We co-crystallized the complex of 14-3-3ζ with the peptide. The structure was solved at 2.2 Å resolution using molecular replacement (Table S1).
Figure 7.
Interaction of TBC1D7 phosphopeptide with 14-3-3ζ. A, surface plasmon resonance measurement of a TBC1D7 phosphopeptide with 14-3-3. The sensorgram (top) shows the response unit when flowing peptide of increasing concentrations over a CM5 chip immobilized with 14-3-3 protein. The response was fitted to a one-site binding model (bottom) and shows a binding affinity of 102 μm. B, structure of 14-3-3 dimer with two TBC1D7 peptides. C, surface electrostatic potential map of the peptide binding groove of 14-3-3 at −5.0 kT/e to +5.0 kT/e scale. Blue, positively charged residues; red, negatively charged residues. D, stick model of the residues involved in the interaction between TBC1D7 peptide and 14-3-3 protein. The water molecule involved in binding is shown as a sphere. Salt bridges and hydrogen bonds are indicated with red dashed lines.
The overall structure is very similar to other reported 14-3-3/peptide structures. 14-3-3 protein forms a homodimer, and each monomer consists of nine α-helices (Fig. 7B). The first four helices (α1–α4) are essential for the dimerization. Side chains from helices α3, α5, α7, and α9 form an amphipathic binding groove that binds to the TBC1D7 peptide. Only the six C-terminal residues (121RSPpSFP126) of the TBC1D7 peptide are visible in the electron density map (Fig. S3), suggesting that the six N-terminal residues of the peptide remain flexible and may explain the low affinity between TBC1D7 and 14-3-3ζ. Detailed analysis of the complex structure revealed that residues within helices α3 and α5 of 14-3-3ζ formed a basic pocket to coordinate the phosphoserine side chain (Fig. 7C), which includes Arg-56 from α3 and Arg-127 and Tyr-128 from α5 (Fig. 7D). A water molecule coordinates hydrogen bonds with the side chain of Asn-173 and the phosphate group of the peptide. In addition, residues within α7 and α9 contribute to the binding through the side chains of aliphatic residues Leu-172, Leu-216, Ile-217, and Leu-220 with the side chain of Phe-125 of the peptide and a hydrogen bond between the side chain of Asn-224 with the carboxyl group of Pro-123 of the peptide (Fig. 7D).
The TBC1D7 peptide matches a mode I 14-3-3 binding motif (RSXpSXP) (45). A comparison of the 14-3-3/TBC1D7 complex structure with a reported crystal structure of 14-3-3ζ in complex with a mode I phosphopeptide (PDB code 1QJB) from polyoma middle T antigen indicates that the phosphoserine phosphate groups of both peptides are identically located and the peptide backbone follows a similar trace. The 14-3-3ζ and TBC1D7 peptide interface buries a 420-Å2 solvent-accessible area in the amphipathic groove of 14-3-3ζ, smaller than the 588-Å2 solvent-accessible area of 14-3-3/mT complex. This is consistent with the lower binding affinity of 14-3-3/TBC1D7 compared with that of 14-3-3/pS-mT complex.
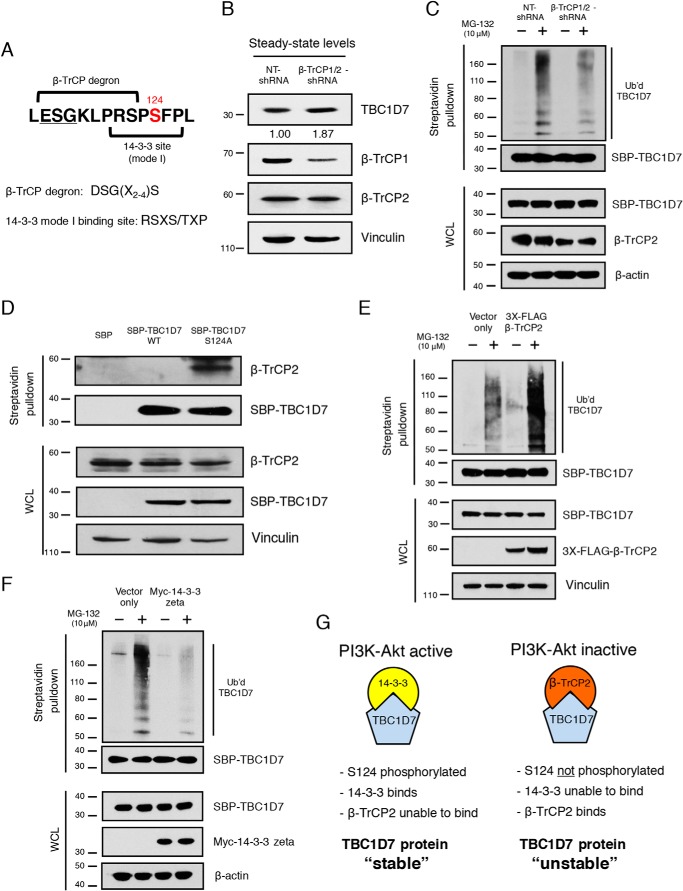
The E3 ubiquitin ligase, β-TrCP2, negatively regulates TBC1D7 stability
The sequence upstream of Ser-124 largely aligns with the canonical β-TrCP degron sequence, DSGX2–4S. The only difference is that the aspartic acid is replaced by a similarly charged glutamic acid residue (46) (Fig. 8A). To determine whether β-TrCP E3 ligases play a role in TBC1D7 stability, a specific shRNA that targets both β-TrCP1 and β-TrCP2 was employed. Stable knockdown of β-TrCP1/2 resulted in increased steady-state levels of TBC1D7, compared with nontargeting control shRNA stable cells (Fig. 8B). We also inquired whether the levels of ubiquitinated TBC1D7 were affected by stable β-TrCP1/2 knockdown. As seen in Fig. 8C, the extent of ubiquitination of affinity-purified TBC1D7 was decreased in cells stably expressing β-TrCP1/2 shRNA, compared with TBC1D7 purified from nontargeting control shRNA-expressing cells. Specific knockdown of β-TrCP1 alone had no effect on either steady-state TBC1D7 protein levels or TBC1D7 ubiquitination (data not shown), suggesting that β-TrCP2 is the main β-TrCP E3 ligase responsible for TBC1D7 ubiquitination.
Figure 8.
β-TrCP2 regulates TBC1D7 ubiquitination and stability. A, diagram of TBC1D7 primary sequence indicating overlapping β-TrCP degron and 14-3-3 mode I binding site. B, whole-cell lysates from 293T cells stably expressing either nontargeting (NT) or β-TrCP1/2-shRNA were resolved on SDS-PAGE and blotted with TBC1D7, β-TrCP1, β-TrCP2, and vinculin antibodies. Relative expression of endogenous TBC1D7 proteins is indicated, as determined by densitometric analysis. Values were normalized to vinculin loading control. The expression level of endogenous TBC1D7 from nontargeting shRNA-expressing control cells was set to 1. C, 293T cells stably expressing either NT- or β-TrCP1/2-shRNA were co-transfected with HA-ubiquitin and SBP-TBC1D7. Cells were treated or not with 10 μg/ml MG-132 for 4 h. Lysates were subject to pulldown analysis using streptavidin beads. Affinity-purified complexes and input whole-cell lysates (WCL) were resolved on SDS-PAGE and blotted with HA, SBP, β-TrCP2, and β-actin antibodies. D, 293T cells were transfected with either SBP, SBP-TBC1D7-WT, or SBP-TBC1D7-S124A expression plasmids. Lysates were subject to pulldown analysis using streptavidin-agarose beads. Affinity-purified complexes were resolved on SDS-PAGE and blotted with β-TrCP2, SBP, and vinculin antibodies. E, 293T cells were co-transfected with either HA-ubiquitin, SBP-TBC1D7, and empty vector or HA-ubiquitin, SBP-TBC1D7, and 3XFLAG-β-TrCP2 expression plasmids. Cells were treated, or not, with 10 μg/ml MG-132 for 4 h. Lysates were subject to pulldown analysis using streptavidin beads. Affinity-purified complexes and input whole-cell lysates were resolved on SDS-PAGE and blotted with HA, SBP, FLAG, and vinculin antibodies. F, 293T cells were co-transfected with either HA-ubiquitin, SBP-TBC1D7, and empty vector or HA-ubiquitin, SBP-TBC1D7, and Myc-14-3-3ζ expression plasmids. Cells were treated or not with 10 μg/ml MG-132 for 4 h. Lysates were subject to pulldown analysis using streptavidin beads. Affinity-purified complexes and input whole-cell lysates were resolved on SDS-PAGE and blotted with HA, SBP, Myc, and β-actin antibodies. G, diagram representing the role of TBC1D7 Ser-124 as a “phospho-switch” and controlling the stability of TBC1D7 protein. Phosphorylation of Ser-124, via PI3K-Akt signaling, determines binding of 14-3-3 and, thus, binding of β-TrCP2. Ser-124 phosphorylation-dependent binding of TBC1D7 controls the stability of TBC1D7 outside of the TSC complex.
Because the phosphorylation status of TBC1D7 on Ser-124 regulates its stability, we hypothesized that phosphorylation of Ser-124 restricts the ability of β-TrCP2 to bind to TBC1D7 within the β-TrCP degron adjacent to Ser-124. As seen in Fig. 8D, endogenous β-TrCP2 was selectively pulled down by phospho-deficient SBP-TBC1D7-S124A protein, but not SBP-TBC1D7-WT protein. Furthermore, ubiquitination levels of affinity-purified TBC1D7 were drastically increased in cells overexpressing β-TrCP2, compared with TBC1D7 purified from vector only–expressing control cells (Fig. 8E).
As shown in Fig. 6, 14-3-3ζ binding to TBC1D7 enhanced the stability of TBC1D7, suggesting that increased expression of 14-3-3 protein might result in decreased levels of ubiquitinated TBC1D7 protein. Co-transfection of Myc-14-3-3ζ indeed resulted in decreased levels of ubiquitinated SBP-TBC1D7, compared with SBP-TBC1D7 co-expressed with a vector-only control (Fig. 8F). These data demonstrate that the phosphorylation status of TBC1D7 at Ser-124 determines the ability of β-TrCP2 to bind, ubiquitinate, and, thus, control the relative stability of TBC1D7 protein. In this scenario, as outlined in Fig. 8G, binding of 14-3-3 to Ser-124–phosphorylated TBC1D7 would mask the β-TrCP degron and occlude binding of β-TrCP2, preventing ubiquitination of TBC1D7 and stabilizing cellular TBC1D7 protein levels. Conversely, when TBC1D7 is not phosphorylated on Ser-124, which occurs when signaling through the PI3K-Akt pathway is dampened, 14-3-3 is unable to bind and mask the β-TrCP degron motif. In this situation, β-TrCP2 would bind to TBC1D7, resulting in TBC1D7 ubiquitination and decreased protein stability.
Discussion
In this study, we identified a novel interaction between PHLPP proteins and the pseudo-RabGAP, TBC1D7. Because PHLPP proteins contain a PP2C-like phosphatase domain and have been reported to control the phosphorylation status of several proteins (15), we considered the possibility that TBC1D7 might be subject to phosphorylation. We demonstrate here that TBC1D7 is indeed phosphorylated at Ser-124 by Akt kinase. Data shown in Fig. 5 suggest that both PHLPP1 and Akt positively regulate the stability of TBC1D7. These results are surprising, as other studies concluded that PHLPP proteins antagonize Akt signaling by negatively regulating Akt itself as well as other downstream substrates (16, 17, 47).
During our studies, and despite much effort, we were unable to demonstrate any identifiable phosphatase activity associated with a recombinant PHLPP1 phosphatase domain, as was shown previously (16, 17). PHLPP proteins are missing several key residues necessary for catalysis (48) and therefore might be considered pseudophosphatases (49). Another member of the PP2C family of phosphatases, TAB1, which also is missing similar key catalytic residues, has been experimentally identified as a pseudophosphatase and has no catalytic activity (50). Comparable findings related to PHLPP2 protein and the strong likelihood of a lack of significant phosphatase activity have been noted (51). Data found in Fig. 1F revealed that the PP2C-like phosphatase domain of PHLPP1 is necessary for binding to TBC1D7. Our current hypothesis is that PHLPP proteins act as “pseudophosphatase adaptor” proteins, binding to phosphorylated serine/threonine residues of target proteins but not catalyzing dephosphorylation (52). We have demonstrated that both the positive effect of PHLPP1 overexpression on TBC1D7 protein stability and the interaction between PHLPP1 and TBC1D7 are not dependent upon phosphorylation of Ser-124 (Fig. 5, E and F). The possibility that PHLPP proteins bind to TBC1D7 and occlude binding of β-TrCP2 and/or prevent ubiquitin conjugation might help explain the positive effect seen on TBC1D7 protein stability when PHLPP1 was overexpressed. Although outside the realm of our current work, future studies involving precise determination of the regions and residues of TBC1D7 needed for PHLPP1 interaction may uncover phosphorylated Ser/Thr residues necessary for binding. Additionally, determining the precise lysine residue(s) that is being ubiquitinated may shed further light on PHLPP1's role in stabilizing TBC1D7 protein. With their unique protein architecture consisting of several protein–protein interaction modules, along with their PP2C-like phosphatase domain, PHLPP proteins may provide more specificity to numerous signaling pathways. Loss of PHLPP protein expression in several cancer types suggests that they are acting as tumor suppressor proteins (15). Current work is aimed at uncovering the precise mechanism(s) employed by PHLPP proteins to counteract Akt signaling and other important signaling pathways.
In search of functional relevance for phosphorylation of TBC1D7 at Ser-124, we found that a S124A phospho-deficient mutant of TBC1D7 was less stable, compared with WT TBC1D7 protein. We hypothesize that when cells preferentially signal through the PI3K-Akt pathway, as often occurs in numerous cancer types (53), there is an environment conducive to Akt-mediated phosphorylation of TBC1D7, allowing for 14-3-3 protein binding. When Akt activity is limited, Ser-124 is no longer phosphorylated or bound by 14-3-3 and is therefore subject to increased degradation. There are several reports in the literature documenting phosphorylation-dependent 14-3-3 binding resulting in stabilization of target proteins. For example, β-catenin, RALT, p27, HDAC7, Tiam1, YAP, and MDMX proteins are all stabilized via phosphorylation-mediated 14-3-3 binding (38, 54–59). In the case of Tiam1, YAP, and MDMX proteins, the kinase involved was determined to be Akt. Immediately N-terminal to Ser-124 is a putative β-TrCP degron motif. We found that TBC1D7 protein stability is regulated, in part, through β-TrCP–mediated ubiquitination. In particular, we found that β-TrCP2, but not β-TrCP1, bound to TBC1D7, and modulation of cellular β-TrCP2 levels could directly regulate TBC1D7 protein stability. β-TrCP degrons contain the canonical DSGX2–4S sequence motif and binding of β-TrCP1/2 to target substrates depends upon phosphorylation of these serines (20) (Fig. 8A).
Kinases likely involved in phosphorylating the β-TrCP degron adjacent to Ser-124 could be GSK3-β and the proline-directed kinase ERK1/2. Ser-122 of TBC1D7 fits the canonical ERK phosphorylation motif (PX(S/T)P) (60). Our MS analysis of immunoprecipitated TBC1D7 from insulin-stimulated HeLa cells also identified Ser-122 as being phosphorylated. Ser-116, if Ser-122 is already phosphorylated, may fit the consensus GSK3-β phosphorylation motif (SXXXpS) (61). GSK3-β needs prior priming of a site C-terminal to its target residue and is often found to be one of several kinases implicated in β-TrCP binding to target proteins and controlling their stability (62). Akt is a negative regulator of GSK3-β, and if PI3K-Akt signaling is dampened, then the activity of GSK3-β would be increased (63). Although outside the scope of this study, identifying the kinases necessary for β-TrCP2 binding may shed further light on the context of TBC1D7 cellular function.
Dibble et al. (24) identified TBC1D7 as a stable, third member of the TSC complex. They discovered that TBC1D7 occurred in two separate pools in the cell; one pool was bound to the TSC complex, and the other pool was a “free” pool, unbound to the TSC complex. They showed that the TBC1D7 protein in this TSC-free pool was less stable, compared with TBC1D7 in the TSC-bound pool, and demonstrated that TSC1 binding stabilized TBC1D7. Two previous reports had also shown that TBC1D7 was a binding partner of TSC1 and that TSC1 binding stabilized TBC1D7 (64, 65). In recent studies, structure–function assays have been performed to determine the sequences necessary for binding between the members of the TSC complex, particularly TBC1D7 and TSC1 (PDB codes 4Z6Y and 5EJC), the structures of which were solved using our structure model (PDB code 3QWL) (41, 66, 67). These studies further clarify how TBC1D7 stabilizes the TSC complex and demonstrate that TBC1D7 helices α4 and α6 are involved in binding to two TSC1 coiled-coil regions, allowing stable homodimerization of TSC1. The Ser-124 residue is located adjacent to several of the residues necessary to interface with TSC1. If Ser-124 is occluded by binding to TSC1, then TBC1D7 would be shielded from β-TrCP2 binding, and this would explain the stability of the TSC-bound pool of TBC1D7. When TBC1D7 is not bound to the TSC complex, it is no longer afforded the protection provided from this binding. Therefore, the phosphorylation status at Ser-124, which would determine 14-3-3 binding, may control the relative stability of the pool of TBC1D7 that is not bound to the TSC complex.
As mentioned previously, little is known about TBC1D7 function outside of the TSC complex. Although TBC1D7 was reported to inactivate Rab17 GTPase and be involved in primary cilia formation (23), our structural analysis reveals that TBC1D7 does not contain the catalytic residues of the dual finger seen in other active RabGAPs. Two other TBC domain proteins missing these residues, Drosophila melanogaster Skywalker (68) and Chlamydomonas reinhardtii RabGAP (69), were also shown to be incapable of acting as RabGAPs. Without solid enzymologic data, we hypothesize that TBC1D7 is a pseudo-RabGAP incapable of inactivating Rab GTPases. Our structure analysis indicated the residues involved in the packing of the core TBC1D7 structure are highly conserved, suggesting that the TBC domain may serve as a scaffold for protein–protein interactions, as has been described for the TSC complex (24, 41, 67). Therefore, the role(s) that TBC1D7 plays when unbound to the TSC complex are largely unknown. A recent report studying the Drosophila TBC1D7 homolog found that dTBC1D7 was involved in TSC-independent control of insulin signaling regulation (70). Current work is aimed at uncovering both the novel cellular roles played by TBC1D7 and the TBC1D7 binding partners that facilitate its biology outside of the TSC complex.
In summary, structural analysis suggests that TBC1D7 is a pseudo-RabGAP. Additionally, we identified TBC1D7 as being phosphorylated at Ser-124 by Akt. We demonstrate that Ser-124 phosphorylation has no effect on binding of TBC1D7 to TSC1. Rather, our data reveal that Ser-124 phosphorylation acts as a phospho-switch that regulates the stability of TBC1D7 when unbound to the TSC complex. Depending on the signaling output of the PI3K-Akt pathway, Ser-124 controls binding to 14-3-3 proteins and the ability to prevent binding of β-TrCP2, which would stabilize TBC1D7.
Experimental procedures
Antibodies and reagents
Primary antibodies used were as follows. TBC1D7, GAPDH, β-actin, phospho-Akt substrate, phospho-Ser 14-3-3 binding motif, pan-Akt, P-Akt-Ser-473, TSC1, and β-TrCP1 antibodies were from Cell Signaling Technology (Danvers, MA). Vinculin, TBC1D7, FLAG (M2) antibodies and FLAG (M2) beads were from Sigma-Aldrich. SBP and GST antibodies were from Santa Cruz Biotechnology, Inc. PHLPP2 antibody was from Bethyl Laboratories (Montgomery, TX). HA and Myc antibodies were from Covance/Biolegend (San Diego, CA). PHLPP1 antibody was from Cosmo Bio USA (Carlsbad, CA). β-TrCP2 antibody was from GeneTex (Irvine, CA). Protein A/G beads were from Santa Cruz Biotechnology. Streptavidin-agarose beads, isopropyl 1-thio-β-d-galactopyranoside, Polybrene, and common laboratory chemicals were from Sigma-Aldrich. ATP and kinase buffer were from Cell Signaling Technology. GSH-agarose beads were from GE Healthcare. Puromycin was from Gibco/Invitrogen (Grand Island, NY). DNA primers were from IDT (Coralville, IA). Restriction enzymes were from New England Biolabs (Ipswich, MA).
Cell culture
293T and HeLa cells (American Type Culture Collection, Manassas, VA) were maintained in Dulbecco's modified Eagle's medium (Gibco/Invitrogen) supplemented with 10% fetal bovine serum (Gibco/Invitrogen) and penicillin–streptomycin (Gibco/Invitrogen). Cells were maintained at 37 °C in a humidified atmosphere of 5% CO2.
Plasmid production and transfections
The cDNAs encoding full-length TBC1D7 (BC007054) and bovine 14-3-3ζ (Addgene catalog no. 13278) were subcloned into pET28-MHL vector (EF456735), which encodes an N-terminal His6 tag followed by a tobacco etch virus protease cleavage site. A serendipitous mutation in the original 14-3-3ζ cDNA template was fixed by QuikChange mutagenesis. Mouse 14-3-3ζ cDNA (Addgene, catalog no. 1944) was PCR-amplified and engineered with a Myc tag on the N terminus and inserted into the BamHI and EcoRI sites of the pcDNA3.1(+) expression vector (Myc-14-3-3ζ). HA-tagged PHLPP2 was obtained from Addgene (plasmid 22403) (HA-PHLPP2). HA-tagged ubiquitin was obtained from Addgene (plasmid 18712) (HA-ubiquitin). The pSBP vector has the SBP sequence inserted into the BamHI and EcoRI sites of pcDNA3.1(+), without a stop codon (71). As a control for transfection and nonspecific binding to the SBP tag, the pSBP-stop vector was constructed similarly to pSBP, but with a stop codon at the end of the SBP sequence. PHLPP1 α cDNA (Addgene, catalog no. 37100) was PCR-amplified and inserted into the EcoRI and XhoI sites of the pSBP expression vector (SBP-PHLPP1). Human TBC1D7 cDNA (Addgene catalog no. 32047) was PCR-amplified and engineered with a FLAG tag on the N terminus and inserted into the EcoRI and XhoI sites of pcDNA3.1(+) (FLAG-TBC1D7). Additionally, human TBC1D7 cDNA was PCR-amplified and inserted into the EcoRI and XhoI sites of pSBP (SBP-TBC1D7). Human β-TrCP2 cDNA (FBXW11 gene, clone accession number BC026213) (Transomics Technologies, Huntsville, AL) was PCR-amplified and inserted into the BamHI and XhoI sites of the pCMV-3Tag-6 expression vector (Agilent Technologies, Santa Clara, CA) (3XFLAG-β-TrCP2). Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit according to the manufacturer's instructions (Agilent). Cells were transiently transfected with expression plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Unless indicated, cells were routinely cultured for 48 h after transfections, before treatments and harvest.
shRNA-mediated gene knockdown
To stably knock down expression of Akt1, a pLKO.1-puro lentivirus plasmid-based shRNA targeting the sequence GGACTACCTGCACTCGGAGAA (clone ID TRCN0000010174, Sigma-Aldrich) was employed (Akt1-shRNA). To stably knock down expression of β-TrCP1, a pLKO.1-puro plasmid-based shRNA targeting the sequence GCGTTGTATTCGATTTGATAA (clone ID TRCN0000006543, Sigma-Aldrich) was employed (β-TrCP1-shRNA). To stably knock down expression of both β-TrCP1 and β-TrCP2, a primer containing the target sequence (AAGTGGAATTTGTGGAACATC), which targets both β-TrCP1 and β-TrCP2, along with a stem loop followed by the reverse target sequence, was annealed to a complementary primer and inserted into the EcoRI and AgeI sites of the pLKO.1-puro plasmid (Addgene number 10878) (β-TrCP1/2-shRNA). Additionally, a nontargeting shRNA plasmid (NT-shRNA) that targets no known human sequence was utilized as a control. A primer containing the target sequence (CTGGTTACGAAGCGAATCCTT) along with a stem loop followed by the reverse target sequence was annealed to a complementary primer and inserted into the EcoRI and AgeI sites of pLKO.1-puro. Lentiviral particles were produced via Lipofectamine 2000 (Invitrogen)-mediated triple transfection of 293T cells with the respective pLKO.1-puro shRNA plasmids along with the lentiviral envelope plasmid (pMD2.G, Addgene number 12259) and the lentiviral packaging plasmid (psPAX2, Addgene number 12260). Target cells were transduced with shRNA containing lentiviral particles in the presence of 8 μg/ml Polybrene, and stable cells were selected using 2 μg/ml puromycin.
Western blot analysis
After treatments, cells were washed with PBS and lysed in Nonidet P-40 lysis buffer: 50 mm Tris/HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40 supplemented with Complete Protease Inhibitor Mixture tablets and PhoStop phosphatase inhibitor tablets (Roche Applied Science) and centrifuged to remove insoluble material. 2× Laemmli sample buffer was added to equivalent amounts of cellular lysates, which were then heated at 95 °C for 5 min and resolved by SDS-PAGE on Novex 4–12% BisTris gels (Invitrogen) and transferred onto Immobilon polyvinylidene difluoride membrane (Millipore). Membranes were blocked in 5% (w/v) nonfat dried skimmed milk powder in TBS-Tween 20 (blocking buffer) (20 mm Tris/HCl, pH 7.6, 137 mm NaCl, and 0.2% Tween 20) and probed with appropriate primary antibodies followed by anti-mouse or anti-rabbit IgG-horseradish peroxidase–conjugated secondary antibody (Cell Signaling Technology). Membranes were washed in TBS-Tween 20 and incubated in Immobilon Western Chemiluminescent horseradish peroxidase substrate (Millipore), and the signal was developed on Biomax XAR film (Eastman Kodak Co.). When necessary, blots were stripped (62.5 mm Tris/HCl, pH 6.8, 2% SDS, and 100 mm β-mercaptoethanol), reblocked with blocking buffer, and reprobed to analyze total protein amounts. Band intensities were quantified using Image J software (National Institutes of Health) and normalized to loading control protein expression.
In vitro kinase assay
Purified TBC1D7 protein (2 μg) was incubated with or without 200 μm ATP (Cell Signaling Technology) or recombinant, active Akt1 protein (400 ng) (Millipore) in Kinase Assay Buffer (Cell Signaling Technology) at 37 °C for 30 min. Half of the kinase reactions were processed for MS, and half were processed for SDS-PAGE/Western blot analysis.
Protein purification and peptide synthesis
TBC1D7 or 14-3-3ζ in pET28-MHL plasmids were transformed in E. coli strain BL21(DE3) or the same strain harboring a pRARE2 plasmid. Protein overexpression was induced using 1 mm isopropyl 1-thio-β-d-galactopyranoside. Selenomethionine derivative of TBC1D7 used for X-ray crystallography phase determination was prepared using the M9 SeMEt growth medium kit from Medicilon following the manufacturer's instructions. Peptides corresponding to the linker loop between helices α6 and α7 of human TBC1D7 containing phosphorylated Ser-124 were synthesized by GenScript (>95% purity) with N-terminal acetylation and C-terminal amidation.
Crystallization
In situ proteolysis (26) was used for the crystallization of TBC1D7. Briefly, one volume of Dispase I from Bacillus polymyxa (Sigma-Aldrich, D4818) dissolved in a solution of 10 mm Tris-HCl at pH 7.5 with 100 mm NaCl at a final concentration of 1 mg/ml was mixed with 10 volumes of protein stock solution (15 mg/ml) immediately before setting up crystallization. The TBC1D7 crystal used for phasing was selenomethionine-labeled and grown in 30% PEG 1500, 0.2 m NaCl, and 0.1 m HEPES, pH 7.5, in a sitting-drop setup, using 0.5 μl of protein (protein/dispase mix) + 0.5 μl of well solution against 100 μl of reservoir buffer at room temperature. The native TBC1D7 crystal used for structure refinement was grown in 20% PEG 1500, 0.2 m MgCl2, and 0.1 m Tris, pH 8.5, in a sitting-drop setup, using 0.5 μl of protein (protein/dispase mix) + 0.5 μl of well solution. Paratone-N (Hampton Research) was used to cryoprotect the TBC1D7 crystals. For co-crystallization, a phosphopeptide corresponding to residues 115–126 of human TBC1D7 was dissolved at 10 mg/ml in gel filtration buffer (25 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm EDTA, and 1 mm DTT) and mixed at a 4-fold molar excess with 14-3-3ζ. The concentration of the 14-3-3ζ/TBC1D7 peptide complex was adjusted to a final concentration of 18 mg/ml. The complex was crystalized by sitting-drop vapor diffusion (well solution: 25% PEG 3350, 0.2 m NaCl, 0.1 m HEPES buffer, pH 7.5, and 5% ethylene glycol). Crystallization drops were set up using 1.5 μl of complex with 1.5 μl of well solution over 500 μl of well solution. Harvested crystal was flash-frozen in liquid nitrogen, with cryoprotection provided by the mother liquor.
Diffraction data collection and structure determination
X-ray diffraction data for 14-3-3ζ/TBC1D7 peptide complex were collected at 100 K at the Canadian Macromolecular Crystallography Facility beamline 08ID-1 of the Canadian Light Source (72). Data for TBC1D7 were collected at beamline 19-ID of the Advanced Photon Source (Argonne, IL). All data sets were processed with the HKL-3000 suite (73). The TBC1D7 structure was solved by the single-wavelength anomalous diffraction method (74) with AUTOSHARP (75)/SHELX (76)/SOLOMON (77)/DM (78). Phases were modified with RESOLVE (79), and a model was automatically built with BUCCANEER (80). The 14-3-3/peptide structure was solved by molecular replacement using PHASER (81) with PDB entry 1A38 as a search template. The graphics program COOT (82) was used for manual model building and visualization, and REFMAC (83) version 5.5 or BUSTER-TNT (84) version 2.9 was used for restrained refinement. The final models were validated by MOLPROBITY (85). The coordinates of TBC1D7 and 14-3-3ζ/TBC1D7 were deposited in the Protein Data Bank under codes 3QWL and 5ULO, respectively. All structure figures were prepared using PyMOL (Schrödinger, LLC, New York).
Bioinformatics analysis
The evolutionary conservation of TBC1D7 residues was analyzed on the ConSurf server (33) using default settings. Surface electrostatic potential of TBC1D7 was calculated using the PDB2PQR server followed by adaptive Poisson–Boltzmann solver (86). The display of the surface was rendered in PyMOL (Schrödinger). Structured-based sequence alignment was carried out using the Promals3D server (30) and rendered with ESPript (87).
Isothermal titration calorimetry (ITC)
Proteins were diluted with a buffer containing 25 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 1 mm tris(2-carboxyethyl)phosphine. All ITC measurements were performed at 25 °C either on a VP-ITC Micro Calorimeter (Malvern) or a Nano ITC (TA instruments). A total of 26 injections of 10 μl of protein, except the first injection of 5 μl, were delivered into a 1.4-ml sample cell containing the other protein. For Nano ITC measurement, a total of 25 injections, each of 2 μl, were delivered into a 0.167-ml sample cell at 180-s intervals. The data were analyzed using OriginTM for ITC on the instrument and fitted to a one-site binding model.
Surface plasmon resonance
Biacore T200 (GE Healthcare) was used to determine the binding affinities of TBC1D7 peptides with 14-3-3ζ. Purified 14-3-3ζ was immobilized on a CM5 chip at 7,000 RU using a standard NHS/EDC amine coupling protocol (GE Healthcare, catalog no. BR-1000-50). A concentration series of TBC1D7 peptides was injected over both captured 14-3-3ζ and reference channels. All of the experiments were carried out at 25 °C with a running buffer containing 25 mm NaH2PO4, pH 7.5, 500 mm NaCl, and 5% glycerol. Peptide samples were injected at a flow rate of 25 μl/min for 30 s, followed by a regeneration step with the flowing of the same buffer for 300 s. Sensorgrams were analyzed after reference extraction, and affinity curves were fitted by steady-state affinity analysis. Results were plotted using SigmaPlot version 11.0 (Systat Software).
Mass spectrometry
Proteins were separated by gel electrophoresis, and gel bands were cut and in-gel digested with trypsin (Thermo Scientific) at 37 °C for 16 h, as described previously (88). The dried peptides were separated on a 75 μm × 15 cm, 2-μm Acclaim PepMap reverse phase column (Thermo Scientific) at 300 nl/min using an UltiMate 3000 RSLCnano HPLC (Thermo Scientific). Peptides were eluted into a Thermo Orbitrap Fusion mass spectrometer using a linear gradient from 96% mobile phase A (0.1% formic acid in water) to 55% mobile phase B (20% water, 80% acetonitrile, 0.08% formic acid) over 30 min. Parent full-scan mass spectra were collected in the Orbitrap mass analyzer set to acquire data at 120,000 FWHM resolution; ions were isolated in the quadrupole mass filter and fragmented within the HCD cell (HCD normalized energy 32%, stepped ±3%), and the product ions were analyzed in the ion trap. Proteome Discoverer version 1.4 (Thermo) was used to search the data found in the Uniprot human database using SequestHT. The search was limited to tryptic peptides, with no more than two missed cleavages allowed. Cysteine carbamidomethylation was set as a fixed modification, with methionine oxidation and serine/threonine phosphorylation set as variable modifications. The precursor mass tolerance was 10 ppm, and the fragment mass tolerance was 0.6 Da. The Percolator node was used to score and rank peptide matches using a 1% false discovery rate. All modified spectra were manually validated.
Author contributions
J. P. M. and Y. T. conceptualization; J. P. M. and Y. T. data curation; J. P. M., M. M. G., and Y. T. supervision; J. P. M., F. H., L. Y., J. H., A. D., W. T., and L. M. M. J. investigation; J. P. M. and Y. T. writing-original draft; J. P. M., W. T., M. E. Y., P. A. R., L. M. M. J., M. M. G., and Y. T. writing-review and editing; A. D. and W. T. formal analysis; M. E. Y., P. A. R., and L. M. M. J. methodology; Y. T. funding acquisition.
Supplementary Material
Acknowledgments
We thank Amy K. Wernimont and John R. Walker for critical review of the TBC1D7 and 14-3-3ζ crystal structures, respectively. Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, is operated by UChicago Argonne, LLC, for the United States Department of Energy Office of Biological and Environmental Research under contract DE-AC02-06CH11357. We also thank George Leiman (Laboratory of Cell Biology) for editorial assistance.
This research was supported by the Intramural Research Program of NCI, National Institutes of Health, and by the Structural Genomics Consortium (SGC), a charity registered in the U.K. (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through the Ontario Genomics Institute, Innovative Medicines Initiative (EU/EFPIA), Janssen, Merck & Co., Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and the Wellcome Trust. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1–S3.
The atomic coordinates and structure factors (codes 3QWL and 5ULO) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- mTORC1
- mechanistic target of rapamycin complex 1
- TSC
- tuberous sclerosis complex
- GAP
- GTPase-activating protein
- SBP
- streptavidin-binding peptide
- ITC
- isothermal titration calorimetry
- FWHM
- full width at half-maximum
- PI3K
- phosphatidylinositol 3-kinase
- P-
- phosphorylated
- NT-shRNA
- nontargeting shRNA plasmid
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- PIP3
- inositol 1,4,5-trisphosphate
- ERK
- extracellular signal-regulated kinase
- HCD
- higher-energy C-trap dissociation.
References
- 1. Hara K., Yonezawa K., Weng Q. P., Kozlowski M. T., Belham C., and Avruch J. (1998) Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273, 14484–14494 10.1074/jbc.273.23.14484 [DOI] [PubMed] [Google Scholar]
- 2. Manning B. D., and Cantley L. C. (2003) Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 28, 573–576 10.1016/j.tibs.2003.09.003 [DOI] [PubMed] [Google Scholar]
- 3. Menon S., Dibble C. C., Talbott G., Hoxhaj G., Valvezan A. J., Takahashi H., Cantley L. C., and Manning B. D. (2014) Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785 10.1016/j.cell.2013.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Curatolo P., Bombardieri R., and Jozwiak S. (2008) Tuberous sclerosis. Lancet 372, 657–668 10.1016/S0140-6736(08)61279-9 [DOI] [PubMed] [Google Scholar]
- 5. Manning B. D., and Cantley L. C. (2003) United at last: the tuberous sclerosis complex gene products connect the phosphoinositide 3-kinase/Akt pathway to mammalian target of rapamycin (mTOR) signalling. Biochem. Soc. Trans 31, 573–578 10.1042/bst0310573 [DOI] [PubMed] [Google Scholar]
- 6. Toker A., and Marmiroli S. (2014) Signaling specificity in the Akt pathway in biology and disease. Adv. Biol. Regul. 55, 28–38 10.1016/j.jbior.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hers I., Vincent E. E., and Tavaré J. M. (2011) Akt signalling in health and disease. Cell. Signal. 23, 1515–1527 10.1016/j.cellsig.2011.05.004 [DOI] [PubMed] [Google Scholar]
- 8. Bellacosa A., Chan T. O., Ahmed N. N., Datta K., Malstrom S., Stokoe D., McCormick F., Feng J., and Tsichlis P. (1998) Akt activation by growth factors is a multiple-step process: the role of the PH domain. Oncogene 17, 313–325 10.1038/sj.onc.1201947 [DOI] [PubMed] [Google Scholar]
- 9. Manning B. D., and Toker A. (2017) AKT/PKB signaling: navigating the network. Cell 169, 381–405 10.1016/j.cell.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., and Hemmings B. A. (1996) Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15, 6541–6551 10.1002/j.1460-2075.1996.tb01045.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liao Y., and Hung M. C. (2010) Physiological regulation of Akt activity and stability. Am. J. Transl. Res. 2, 19–42 [PMC free article] [PubMed] [Google Scholar]
- 12. Rocher G., Letourneux C., Lenormand P., and Porteu F. (2007) Inhibition of B56-containing protein phosphatase 2As by the early response gene IEX-1 leads to control of Akt activity. J. Biol. Chem. 282, 5468–5477 10.1074/jbc.M609712200 [DOI] [PubMed] [Google Scholar]
- 13. Rodgers J. T., Vogel R. O., and Puigserver P. (2011) Clk2 and B56β mediate insulin-regulated assembly of the PP2A phosphatase holoenzyme complex on Akt. Mol. Cell 41, 471–479 10.1016/j.molcel.2011.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ugi S., Imamura T., Maegawa H., Egawa K., Yoshizaki T., Shi K., Obata T., Ebina Y., Kashiwagi A., and Olefsky J. M. (2004) Protein phosphatase 2A negatively regulates insulin's metabolic signaling pathway by inhibiting Akt (protein kinase B) activity in 3T3-L1 adipocytes. Mol. Cell Biol. 24, 8778–8789 10.1128/MCB.24.19.8778-8789.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Newton A. C., and Trotman L. C. (2014) Turning off AKT: PHLPP as a drug target. Annu. Rev. Pharmacol. Toxicol. 54, 537–558 10.1146/annurev-pharmtox-011112-140338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brognard J., Sierecki E., Gao T., and Newton A. C. (2007) PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol. Cell 25, 917–931 10.1016/j.molcel.2007.02.017 [DOI] [PubMed] [Google Scholar]
- 17. Gao T., Furnari F., and Newton A. C. (2005) PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol. Cell 18, 13–24 10.1016/j.molcel.2005.03.008 [DOI] [PubMed] [Google Scholar]
- 18. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 10.1146/annurev.biochem.78.081507.101607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee E. K., and Diehl J. A. (2014) SCFs in the new millennium. Oncogene 33, 2011–2018 10.1038/onc.2013.144 [DOI] [PubMed] [Google Scholar]
- 20. Fuchs S. Y., Spiegelman V. S., and Kumar K. G. (2004) The many faces of β-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene 23, 2028–2036 10.1038/sj.onc.1207389 [DOI] [PubMed] [Google Scholar]
- 21. Deshaies R. J., and Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- 22. Willems A. R., Schwab M., and Tyers M. (2004) A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695, 133–170 10.1016/j.bbamcr.2004.09.027 [DOI] [PubMed] [Google Scholar]
- 23. Yoshimura S., Egerer J., Fuchs E., Haas A. K., and Barr F. A. (2007) Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 178, 363–369 10.1083/jcb.200703047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dibble C. C., Elis W., Menon S., Qin W., Klekota J., Asara J. M., Finan P. M., Kwiatkowski D. J., Murphy L. O., and Manning B. D. (2012) TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell 47, 535–546 10.1016/j.molcel.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gabernet-Castello C., O'Reilly A. J., Dacks J. B., and Field M. C. (2013) Evolution of Tre-2/Bub2/Cdc16 (TBC) Rab GTPase-activating proteins. Mol. Biol. Cell 24, 1574–1583 10.1091/mbc.e12-07-0557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tong Y., Dong A., Xu X., and Wernimont A. (2014) Salvage or recovery of failed targets by in situ proteolysis. Methods Mol. Biol. 1140, 179–188 10.1007/978-1-4939-0354-2_14 [DOI] [PubMed] [Google Scholar]
- 27. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tempel W., Tong Y., Dimov S., Bochkarev A., and Park H. (2008) First crystallographic models of human TBC domains in the context of a family-wide structural analysis. Proteins 71, 497–502 10.1002/prot.21885 [DOI] [PubMed] [Google Scholar]
- 29. Pan X., Eathiraj S., Munson M., and Lambright D. G. (2006) TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442, 303–306 10.1038/nature04847 [DOI] [PubMed] [Google Scholar]
- 30. Pei J., Kim B. H., and Grishin N. V. (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 10.1093/nar/gkn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mosca R., and Schneider T. R. (2008) RAPIDO: a web server for the alignment of protein structures in the presence of conformational changes. Nucleic Acids Res. 36, W42–W46 10.1093/nar/gkn197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gavriljuk K., Gazdag E. M., Itzen A., Kötting C., Goody R. S., and Gerwert K. (2012) Catalytic mechanism of a mammalian Rab·RabGAP complex in atomic detail. Proc. Natl. Acad. Sci. U.S.A. 109, 21348–21353 10.1073/pnas.1214431110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., and Ben-Tal N. (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 10.1093/nar/gkw408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Manning B. D., Tee A. R., Logsdon M. N., Blenis J., and Cantley L. C. (2002) Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10, 151–162 10.1016/S1097-2765(02)00568-3 [DOI] [PubMed] [Google Scholar]
- 35. Obenauer J. C., Cantley L. C., and Yaffe M. B. (2003) Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31, 3635–3641 10.1093/nar/gkg584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., and Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alessi D. R., Caudwell F. B., Andjelkovic M., Hemmings B. A., and Cohen P. (1996) Molecular basis for the substrate specificity of protein kinase B: comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 399, 333–338 10.1016/S0014-5793(96)01370-1 [DOI] [PubMed] [Google Scholar]
- 38. Basu S., Totty N. F., Irwin M. S., Sudol M., and Downward J. (2003) Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell 11, 11–23 10.1016/S1097-2765(02)00776-1 [DOI] [PubMed] [Google Scholar]
- 39. Fang D., Hawke D., Zheng Y., Xia Y., Meisenhelder J., Nika H., Mills G. B., Kobayashi R., Hunter T., and Lu Z. (2007) Phosphorylation of β-catenin by AKT promotes β-catenin transcriptional activity. J. Biol. Chem. 282, 11221–11229 10.1074/jbc.M611871200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hoyal C. R., Gutierrez A., Young B. M., Catz S. D., Lin J. H., Tsichlis P. N., and Babior B. M. (2003) Modulation of p47PHOX activity by site-specific phosphorylation: Akt-dependent activation of the NADPH oxidase. Proc. Natl. Acad. Sci. U.S.A. 100, 5130–5135 10.1073/pnas.1031526100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Santiago Lima A. J., Hoogeveen-Westerveld M., Nakashima A., Maat-Kievit A., van den Ouweland A., Halley D., Kikkawa U., and Nellist M. (2014) Identification of regions critical for the integrity of the TSC1-TSC2-TBC1D7 complex. PLoS One 9, e93940 10.1371/journal.pone.0093940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Freeman A. K., and Morrison D. K. (2011) 14-3-3 proteins: diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 22, 681–687 10.1016/j.semcdb.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Manning B. D., and Cantley L. C. (2007) AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 10.1016/j.cell.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aitken A. (2006) 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 16, 162–172 10.1016/j.semcancer.2006.03.005 [DOI] [PubMed] [Google Scholar]
- 45. Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., and Cantley L. C. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91, 961–971 10.1016/S0092-8674(00)80487-0 [DOI] [PubMed] [Google Scholar]
- 46. Frescas D., and Pagano M. (2008) Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8, 438–449 10.1038/nrc2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qiao M., Wang Y., Xu X., Lu J., Dong Y., Tao W., Stein J., Stein G. S., Iglehart J. D., Shi Q., and Pardee A. B. (2010) Mst1 is an interacting protein that mediates PHLPPs' induced apoptosis. Mol. Cell 38, 512–523 10.1016/j.molcel.2010.03.017 [DOI] [PubMed] [Google Scholar]
- 48. Sierecki E., and Newton A. C. (2014) Biochemical characterization of the phosphatase domain of the tumor suppressor PH domain leucine-rich repeat protein phosphatase. Biochemistry 53, 3971–3981 10.1021/bi500428j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reiterer V., Pawłowski K., and Farhan H. (2017) STYX: a versatile pseudophosphatase. Biochem. Soc. Trans. 45, 449–456 10.1042/BST20160279 [DOI] [PubMed] [Google Scholar]
- 50. Conner S. H., Kular G., Peggie M., Shepherd S., Schüttelkopf A. W., Cohen P., and Van Aalten D. M. (2006) TAK1-binding protein 1 is a pseudophosphatase. Biochem. J. 399, 427–434 10.1042/BJ20061077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Agarwal N. K., Zhu X., Gagea M., White C. L. 3rd, Cote G., and Georgescu M. M. (2014) PHLPP2 suppresses the NF-κB pathway by inactivating IKKβ kinase. Oncotarget 5, 815–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reiterer V., Eyers P. A., and Farhan H. (2014) Day of the dead: pseudokinases and pseudophosphatases in physiology and disease. Trends Cell Biol. 24, 489–505 10.1016/j.tcb.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 53. Fruman D. A., and Rommel C. (2014) PI3K and cancer: lessons, challenges and opportunities. Nat. Rev. Drug Discov. 13, 140–156 10.1038/nrd4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen C. H., Chuang S. M., Yang M. F., Liao J. W., Yu S. L., and Chen J. J. (2012) A novel function of YWHAZ/β-catenin axis in promoting epithelial-mesenchymal transition and lung cancer metastasis. Mol. Cancer Res. 10, 1319–1331 10.1158/1541-7786.MCR-12-0189 [DOI] [PubMed] [Google Scholar]
- 55. Li X., Song S., Liu Y., Ko S. H., and Kao H. Y. (2004) Phosphorylation of the histone deacetylase 7 modulates its stability and association with 14-3-3 proteins. J. Biol. Chem. 279, 34201–34208 10.1074/jbc.M405179200 [DOI] [PubMed] [Google Scholar]
- 56. Lopez-Pajares V., Kim M. M., and Yuan Z. M. (2008) Phosphorylation of MDMX mediated by Akt leads to stabilization and induces 14-3-3 binding. J. Biol. Chem. 283, 13707–13713 10.1074/jbc.M710030200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Short J. D., Dere R., Houston K. D., Cai S. L., Kim J., Bergeron J. M., Shen J., Liang J., Bedford M. T., Mills G. B., and Walker C. L. (2010) AMPK-mediated phosphorylation of murine p27 at T197 promotes binding of 14-3-3 proteins and increases p27 stability. Mol. Carcinog. 49, 429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Takeda K., Takata T., Kawai Y., Ishigaki Y., and Kajinami K. (2013) Chk1-mediated phosphorylation of receptor-associated late transducer at serine 250 increases its stability by stimulating its interaction with 14-3-3. Genes. Cells 18, 369–386 10.1111/gtc.12043 [DOI] [PubMed] [Google Scholar]
- 59. Zhu G., Fan Z., Ding M., Zhang H., Mu L., Ding Y., Zhang Y., Jia B., Chen L., Chang Z., and Wu W. (2015) An EGFR/PI3K/AKT axis promotes accumulation of the Rac1-GEF Tiam1 that is critical in EGFR-driven tumorigenesis. Oncogene 34, 5971–5982 10.1038/onc.2015.45 [DOI] [PubMed] [Google Scholar]
- 60. Gonzalez F. A., Raden D. L., and Davis R. J. (1991) Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J. Biol. Chem. 266, 22159–22163 [PubMed] [Google Scholar]
- 61. Sutherland C. (2011) What are the bona fide GSK3 substrates? Int. J. Alzheimers Dis. 2011, 505607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xu C., Kim N. G., and Gumbiner B. M. (2009) Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle 8, 4032–4039 10.4161/cc.8.24.10111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., and Hemmings B. A. (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789 10.1038/378785a0 [DOI] [PubMed] [Google Scholar]
- 64. Nakashima A., Yoshino K., Miyamoto T., Eguchi S., Oshiro N., Kikkawa U., and Yonezawa K. (2007) Identification of TBC7 having TBC domain as a novel binding protein to TSC1-TSC2 complex. Biochem. Biophys. Res. Commun. 361, 218–223 10.1016/j.bbrc.2007.07.011 [DOI] [PubMed] [Google Scholar]
- 65. Sato N., Koinuma J., Ito T., Tsuchiya E., Kondo S., Nakamura Y., and Daigo Y. (2010) Activation of an oncogenic TBC1D7 (TBC1 domain family, member 7) protein in pulmonary carcinogenesis. Genes Chromosomes Cancer 49, 353–367 [DOI] [PubMed] [Google Scholar]
- 66. Gai Z., Chu W., Deng W., Li W., Li H., He A., Nellist M., and Wu G. (2016) Structure of the TBC1D7-TSC1 complex reveals that TBC1D7 stabilizes dimerization of the TSC1 C-terminal coiled coil region. J. Mol. Cell Biol. 8, 411–425 10.1093/jmcb/mjw001 [DOI] [PubMed] [Google Scholar]
- 67. Qin J., Wang Z., Hoogeveen-Westerveld M., Shen G., Gong W., Nellist M., and Xu W. (2016) Structural basis of the interaction between tuberous sclerosis complex 1 (TSC1) and Tre2-Bub2-Cdc16 domain family member 7 (TBC1D7). J. Biol. Chem. 291, 8591–8601 10.1074/jbc.M115.701870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fischer B., Lüthy K., Paesmans J., De Koninck C., Maes I., Swerts J., Kuenen S., Uytterhoeven V., Verstreken P., and Versées W. (2016) Skywalker-TBC1D24 has a lipid-binding pocket mutated in epilepsy and required for synaptic function. Nat. Struct. Mol. Biol. 23, 965–973 10.1038/nsmb.3297 [DOI] [PubMed] [Google Scholar]
- 69. Bhogaraju S., and Lorentzen E. (2014) Crystal structure of a Chlamydomonas reinhardtii flagellar RabGAP TBC-domain at 1.8 Å resolution. Proteins 82, 2282–2287 10.1002/prot.24597 [DOI] [PubMed] [Google Scholar]
- 70. Ren S., Huang Z., Jiang Y., and Wang T. (2018) dTBC1D7 regulates systemic growth independently of TSC through insulin signaling. J. Cell Biol. 217, 517–526 10.1083/jcb.201706027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fiordalisi J. J., Dewar B. J., Graves L. M., Madigan J. P., and Cox A. D. (2013) Src-mediated phosphorylation of the tyrosine phosphatase PRL-3 is required for PRL-3 promotion of Rho activation, motility and invasion. PLoS One 8, e64309 10.1371/journal.pone.0064309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Grochulski P., Fodje M. N., Gorin J., Labiuk S. L., and Berg R. (2011) Beamline 08ID-1, the prime beamline of the Canadian Macromolecular Crystallography Facility. J. Synchrotron Radiat. 18, 681–684 10.1107/S0909049511019431 [DOI] [PubMed] [Google Scholar]
- 73. Minor W., Cymborowski M., Otwinowski Z., and Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 10.1107/S0907444906019949 [DOI] [PubMed] [Google Scholar]
- 74. Rice L. M., Earnest T. N., and Brunger A. T. (2000) Single-wavelength anomalous diffraction phasing revisited. Acta Crystallogr. D Biol. Crystallogr. 56, 1413–1420 10.1107/S0907444900010039 [DOI] [PubMed] [Google Scholar]
- 75. Vonrhein C., Blanc E., Roversi P., and Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 76. Schneider T. R., and Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 10.1107/S0907444902011678 [DOI] [PubMed] [Google Scholar]
- 77. Abrahams J. P., and Leslie A. G. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52, 30–42 10.1107/S0907444995008754 [DOI] [PubMed] [Google Scholar]
- 78. Cowtan K., Zhang K., and Main P. (2006) 25.2.2 DM/DMMULTI software for phase improvement by density modification. in International Tables for Crystallography (Rossmann M., and Arnold E., eds) pp. 705–710, International Union of Crystallography, Chester, UK [Google Scholar]
- 79. Terwilliger T. (2004) SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 11, 49–52 10.1107/S0909049503023938 [DOI] [PubMed] [Google Scholar]
- 80. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 10.1107/S0907444906022116 [DOI] [PubMed] [Google Scholar]
- 81. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 10.1107/S0907444906045975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 83. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 84. Smart O. S., Womack T. O., Flensburg C., Keller P., Paciorek W., Sharff A., Vonrhein C., and Bricogne G. (2012) Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D Biol. Crystallogr. 68, 368–380 10.1107/S0907444911056058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., and Baker N. A. (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 10.1093/nar/gkm276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shevchenko A., Tomas H., Havlis J., Olsen J. V., and Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 89. Alipanahi B., Delong A., Weirauch M. T., and Frey B. J. (2015) Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33, 831–838 10.1038/nbt.3300 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.