Abstract
The epithelial-mesenchymal transition (EMT) is a multistep dedifferentiation program important in tissue repair. Here, we examined the role of the transcriptional regulator NF-κB in EMT of primary human small airway epithelial cells (hSAECs). Surprisingly, transforming growth factor β (TGFβ) activated NF-κB/RELA proto-oncogene, NF-κB subunit (RELA) translocation within 1 day of stimulation, yet induction of its downstream gene regulatory network occurred only after 3 days. A time course of TGFβ-induced EMT transition was analyzed by RNA-Seq in the absence or presence of inducible shRNA-mediated silencing of RELA. In WT cells, TGFβ stimulation significantly affected the expression of 2,441 genes. Gene set enrichment analysis identified WNT, cadherin, and NF-κB signaling as the most prominent TGFβ-inducible pathways. By comparison, RELA controlled expression of 3,138 overlapping genes mapping to WNT, cadherin, and chemokine signaling pathways. Conducting upstream regulator analysis, we found that RELA controls six clusters of upstream transcription factors, many of which overlapped with a transcription factor topology map of EMT developed earlier. RELA triggered expression of three key EMT pathways: 1) the WNT/β-catenin morphogen pathway, 2) the JUN transcription factor, and 3) the Snail family transcriptional repressor 1 (SNAI1). RELA binding to target genes was confirmed by ChIP. Experiments independently validating WNT dependence on RELA were performed by silencing RELA via genome editing and indicated that TGFβ-induced WNT5B expression and downstream activation of the WNT target AXIN2 are RELA-dependent. We conclude that RELA is a master transcriptional regulator of EMT upstream of WNT morphogen, JUN, SNAI1–ZEB1, and interleukin-6 autocrine loops.
Keywords: NF-κB, epithelial-mesenchymal transition (EMT), gene regulation, transforming growth factor β (TGF-β), transcription regulation, lung disease, mucosal injury, RELA proto-oncogene, Wnt, ZEB
Introduction
The respiratory tract represents one of the largest surfaces of the human body exposed to the external environment (1, 2). Under normal conditions, this mucosal surface is a semi-impermeable barrier formed by highly differentiated epithelial cells that allow fluid conservation, permit gas exchange, and provide physical protection from pathogen invasion. Disruption of the epithelial barrier releases epithelial growth factors, including transforming growth factor (TGF),3 sequestered within the extracellular matrix that coordinate expression of gene regulatory networks (GRNs) to promote repair. Phenotypically, TGF orchestrates a dedifferentiation program known as the type II epithelial-mesenchymal transition (EMT). Type II EMT involves extensive cytosolic restructuring, resulting in the loss of apical-basal polarity, dissolution of adherens junctions, enhanced motility, and expression of fibrotic genes (for an in-depth review, see Ref. 3). As a consequence of this process, epithelial cells acquire stem cell–like properties, permitting the transitioned mesenchymal cell to repopulate regions of denuded epithelium, promoting tissue repair and extracellular matrix remodeling (3).
Transition to the mesenchymal state is not binary but rather the product of sequential cell-state changes beginning from the differentiated epithelial state transitioning into uncommitted “partial EMT (pEMT)” state(s) (4). Transition into pEMT is initiated by TGFβ binding to TGFβ receptor type II, signaling intracellularly via the SMAD-dependent, “canonical” and SMAD-independent, “noncanonical” pathways (5, 6). These two arms function coordinately to activate a cascade of transcription factors that result in ordered patterns of gene expression and inhibition, resulting in a metastable pEMT state (7). Gene expression patterns of SMAD are controlled in a cell type–specific manner by interaction with master transcription factors that function in coordinately regulated complexes to form acetylated histone-rich superenhancers on mesenchymal genes, promoting chromatin remodeling (8, 9). Persistent growth factor and microenvironmental stimulation is required to produce a stable mesenchymal transition. Time-resolved RNA profiling coupled with ChIP-sequencing studies have elucidated the role of the JUNB complex as an example of a “master” transcription factor of the mesenchymal state (9). Another master regulator is zinc finger E-box–binding homeobox 1 (ZEB1), a bifunctional transcriptional regulator activated by SNAI1 that down-regulates E-cadherin and the EMT-suppressing microRNA-200 family in a complex with the C-terminal binding protein repressor (10–12). Although the post-translational regulation of the SNAI1–ZEB1 autoamplification loop by microRNAs is well-described (10), the factor(s) controlling the initial transcriptional spike in SNAI1 expression required to trigger the amplification loop is not known in type II EMT.
In addition to master transcription factors, autocrine and paracrine growth factor signaling plays an important role in the maintenance of the mesenchymal state (9). For example, up-regulation of the WNT morphogen ligands that activate intracellular β-catenin (CTNNB)–transcription factor (TCF3) complex in neighboring cells to maintain SNAI1 and TGFβ expression is an important component of EMT (13, 14). Local up-regulation of IL6 promotes growth factor independence and expression of stem cell–like features in EMT via autocrine/paracrine signaling (15, 16). The identity of autocrine pathways, their timing of expression, and their relationship to the master transcription factors in type II EMT are not known.
Our previous work identified that persistent NF-κB action is central to the fully committed mesenchymal regulatory network in type II EMT (17, 18). However, the role of RELA in the initiation of type II EMT has not been determined. To address this gap, we conducted time course experiments during the initiation of TGFβ-induced mesenchymal transition in primary human small airway epithelial cells (hSAECs). Although nuclear translocation of RELA was detected within 1 day of TGFβ stimulation, the activating Ser-276 phosphorylation only significantly occurred after 3 days of stimulation, an event associated with activation of the NF-κB–dependent GRN. hSAECs expressing a doxycycline (Dox)-regulated shRNA were profiled by RNA-Seq in the absence or presence of RELA knockdown (KD). We observed that RELA controls the autocrine growth factors WNT and IL6 as well as the JUN and SNAI1 master transcription factors. These data implicate RELA as a master transcription regulator of type II EMT.
Results
We previously characterized a reproducible model of type II EMT using a continuously replicating line of hSAECs immortalized with human telomerase (6, 17). Unbiased genomic and proteomic profiling studies indicate that hSAEC expression patterns closely mimic those of primary small airway epithelial cells (19, 20). In the absence of stimulation, hSAECs exhibit cuboidal epithelial morphology, express cytokeratin 17, and form pseudostratified columnar epithelium in air–liquid interfaces (21).
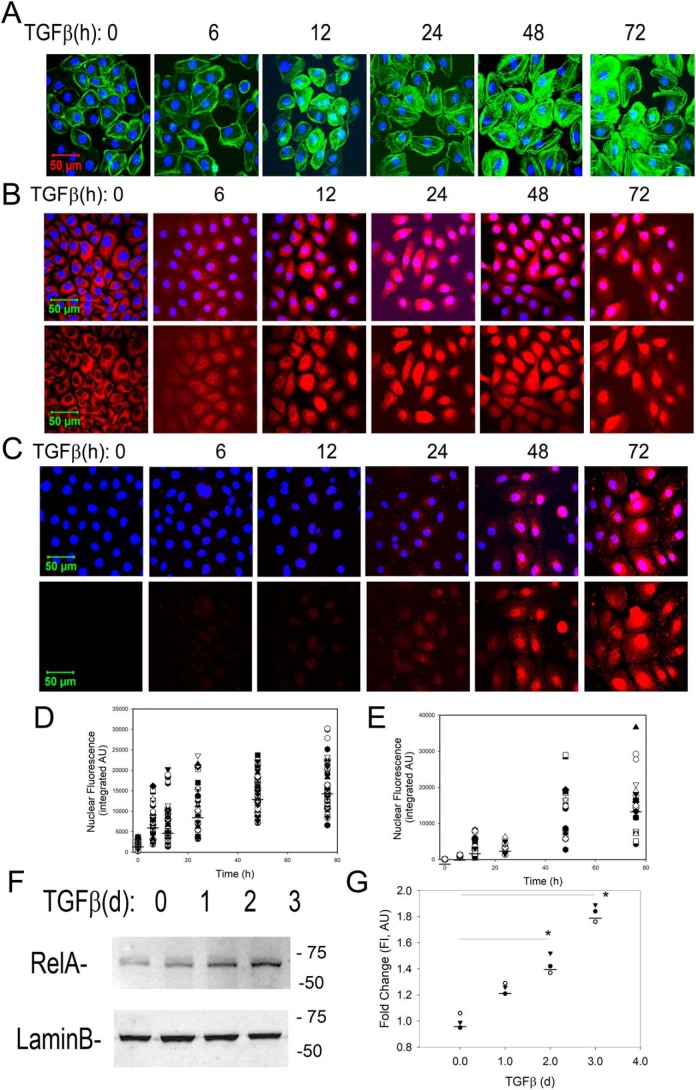
Upon TGFβ stimulation, hSAECs exhibit a time-dependent mesenchymal dedifferentiation program associated with formation of filamentous (F) actin stress fibers, detected by phalloidin staining. Significantly enhanced phalloidin staining was seen initially after 12 h and dramatically increased by 48 h (Fig. 1A).
Figure 1.
Kinetics of RELA translocation and activating phosphorylation. hSAECs were stimulated with TGFβ (10 ng/ml) for the indicated times (in hours). Untreated cells were negative controls (time 0). A, cells were fixed and stained with Alexa Fluor 488–phalloidin (green). Nuclei were counterstained with DAPI (blue) and imaged by CFM at 63× magnification. B, hSAECs were stained with rabbit anti-RELA Ab, and secondary detection was performed with Alexa Fluor 568–labeled goat anti-rabbit IgG (red). Nuclei were DAPI-counterstained and imaged by CFM. Top panel, Alexa Fluor 568 and DAPI merged image; bottom panel, Alexa Fluor 568 staining only. C, CFM for phospho-Ser-276 RELA. Top panel, Alexa Fluor 568 and DAPI merged image; bottom panel, Alexa Fluor 568 staining only. D and E, quantification of nuclear fluorescence intensity. Shown are scatter plots and median nuclear fluorescence intensities for RELA and phospho-Ser-276 RELA, respectively, for individual cell quantifications using ImageJ. AU, absorbance units. F, Western immunoblot for 65-kDa RELA (top). Lamin B was stained as a loading control (bottom). d, days. G, quantification of normalized fluorescence intensity of RELA signal. *, p < 0.05 (ANOVA, post hoc Tukey's test).
Kinetics of NF-κB/RELA activation
Our earlier RNA-Seq analysis of hSAECs in a fully committed EMT state after 15 days of TGFβ stimulation indicated the presence of activated RELA and a robust NF-κB GRN signature (17). To understand the early kinetics of NF-κB activation, we examined the effects of TGFβ on RELA nuclear translocation by confocal fluorescence microscopy (CFM) over a detailed time course from 0 to 72 h. In unstimulated control hSAECs, RELA was predominately cytoplasmic (Fig. 1B). A small fraction of nuclei were RELA-positive by 12 h, and RELA was predominately nuclear after 48 h (Fig. 1, B and D).
Earlier work has shown that activation of the RELA transcriptional activity requires phosphorylation of Ser-276 by ribosomal S6 kinases (22, 23). Ser-276 phosphorylation is a rate-limiting step required for p300-dependent acetylation and complex formation with bromodomain-containing protein 4 (BRD4) (18, 24, 25), a coactivator required for stable EMT (9, 18). In contrast to the kinetics of nuclear RELA translocation, the formation of activating phospho-Ser-276 RELA is initially detected after 2 days and dramatically induced by 3 days of TGFβ stimulation where a 15- and 42-fold increase is seen relative to unstimulated hSAECs, respectively (Fig. 1, C and E).
The changes in RELA nuclear translocation were validated by Western immunoblotting of nuclear extracts where TGFβ produced a time-dependent increase in nuclear RELA abundance after 2 and 3 days of TGFβ stimulation (Fig. 1, F and G). Collectively, these data indicate that TGFβ induces RELA translocation followed thereafter by RELA activating phosphorylation.
Validation of Dox-inducible RELA silencing
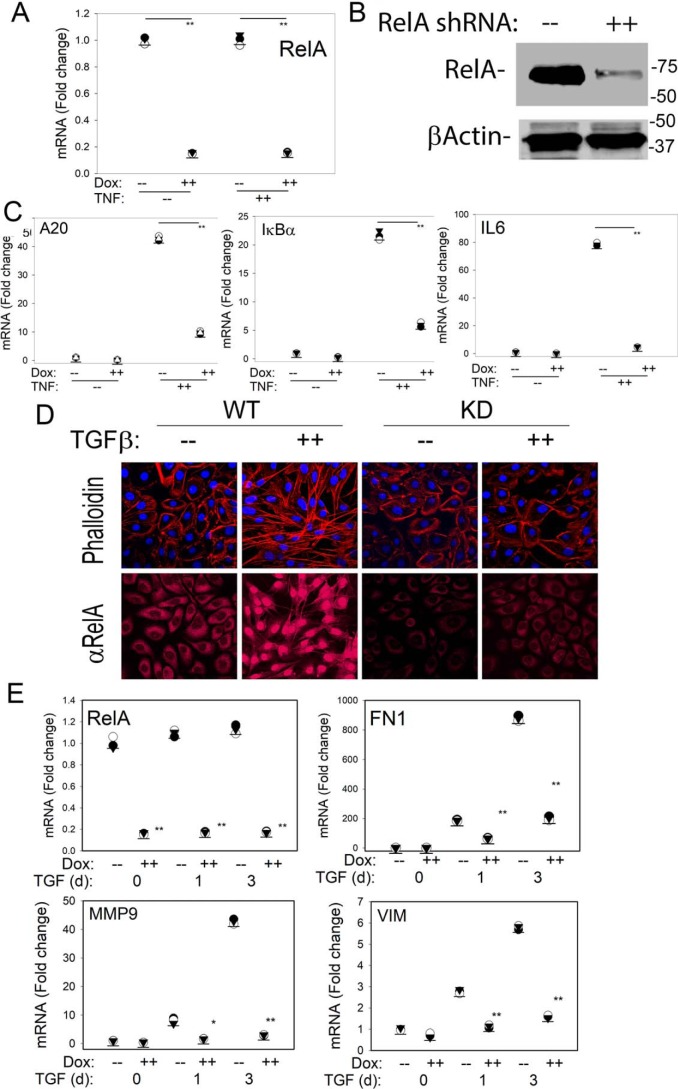
To systematically identify the direct genomic targets of RELA in the initiation of mesenchymal transition, we engineered hSAECs to stably express a Dox-inducible RELA shRNA. The rationale for selecting a Dox-inducible system was that this approach enables study of the effect of acute RELA depletion without the well-described compensation for NF-κB deficiency in knockout cell lines (26). We first confirmed the effect of Dox treatment on RELA abundance where 5 days of Dox treatment reduced endogenous RELA mRNA to 15% that of untreated hSAECs (Fig. 2A) and depleted total cellular protein to similar levels (Fig. 2B). To test whether this level of depletion functionally impaired RELA signaling, we examined the effect of TNF on the NF-κB–dependent GRN. For this purpose, we examined the response of TNFAIP3/A20, NFKBIA/IκBα, and IL6, representative genes directly regulated by NF-κB (27). We observed that an acute (1-h) TNFα stimulus induced TNFAIP3/A20 mRNA by 44-fold. By contrast, in RELA KD, TNFAIP3/A20 mRNA was only induced by 8-fold, representing a significant inhibition (p < 0.01, ANOVA, post hoc Tukey's t test). Similar findings were observed for the NF-κB/RELA–dependent NFKBIA/IκBα and IL6 genes where the 22-fold induction of NFKBIA/IκBα in WT cells was reduced to 5-fold after RELA silencing. The 80-fold induction of IL6 in WT cells was reduced to 5-fold in the RELA KD cells (p < 0.05, ANOVA, post hoc Tukey's test; Fig. 2A). These data indicate that the RELA KD functionally interfered with the NF-κB–dependent GRN and would be suitable for testing the role of NF-κB in the initiation of TGFβ-induced EMT.
Figure 2.
Characterization of RELA silencing by Dox-induced shRNA. A, effect on RELA expression. hSAECs stably expressing TRIPZ Tet-On inducible lentiviral RELA shRNA were isolated. Cells were untreated (−) or treated with Dox (2 μg/ml, 5 days) prior to TNF stimulation (20 ng/ml, 1 h). Shown are individual RELA mRNA abundance measurements in n = 3 samples expressed as -fold change relative to untreated cells. **, p < 0.01, ANOVA, post hoc Tukey's test. B, effect on RELA protein abundance. Shown is a Western immunoblot of untreated (−) or Dox-treated (2 μg/ml, 5 days) whole-cell lysates. Top panel, anti-RELA Ab; bottom panel, β-actin probed as a protein loading control. C, Q-RT-PCR for TNFAIP3/A20, NFKBIA/IκBα, and IL6 mRNA. Individual mRNA abundance in separate replicates normalized to POLB. *, p < 0.05; **, p < 0.01, ANOVA, post hoc Tukey's test. D, WT or RELA KD hSAECs were stimulated with TGFβ for 0, 1, or 3 days, respectively; cells were fixed, stained with Alexa Fluor 568–conjugated phalloidin (for distribution of F-actin, shown in red color, upper panel) or with primary rabbit anti-RELA antibody followed by staining with Alexa Fluor 568–conjugated anti-rabbit IgG (lower panel) as well as DAPI (in blue color); and examined by CFM. E, Q-RT-PCR was performed for RELA and mesenchymal signature genes FN1, MMP9, and VIM as indicated. Shown is individual replicate mRNA abundance normalized to β-polymerase relative to unstimulated cells. d, days. *, p < 0.05; **, p < 0.01, ANOVA, post hoc Tukey's test.
RELA KD blocks the initiation of TGFβ-induced mesenchymal transition
We next examined the response of the RELA KD hSAECs to TGFβ-induced mesenchymal transition. WT or RELA KD cells were subjected to a short time course of TGFβ induction (0, 1, and 3 days). Redistribution of F-actin was measured using phalloidin staining in CFM. In unstimulated controls, phalloidin staining is largely located in the cellular periphery (Fig. 2D, top). By contrast, TGFβ stimulation produced organized F-actin formation throughout the cytoplasm and nucleus after 3 days. RELA KD cells maintained a low level of phalloidin staining localized to the cellular periphery (Fig. 2D, top).
To confirm the persistence of RELA silencing over the 3-day time course, RELA abundance was measured by CFM and quantitative real-time RT-PCR (Q-RT-PCR) (primer sequences are given in Table S1). The cytoplasmic-to-nuclear redistribution of RELA and enhanced total RELA abundance were evident in the WT cells but significantly reduced in the RELA KD cells (Fig. 2D, bottom). Similarly, RELA mRNA was reduced to <15% that of WT cells over the 3 days of TGFβ stimulation (Fig. 2E). Compared with WT cells, RELA KD cells showed a significant reduction in TGFβ-induced fibronectin (FN1) expression from 900-fold in WT cells to 200-fold in RELA KD cells (Fig. 2E). Similarly, RELA silencing produced a significant inhibition of the 42-fold increase in matrix metalloproteinase 9 (MMP9) and the 5.5-fold increase in VIM expression to less than 2-fold (Fig. 2, E and F). We interpret these data to show that RELA plays an important role in initiating the early stages of type II EMT.
Systematic identification of NF-κB–dependent genes early in the mesenchymal transition
To systematically identify the RELA-induced GRN in the initial stages of the mesenchymal transition, WT or RELA KD cells were stimulated in the absence or presence of TGFβ for 1 and 3 days. Each experimental condition was reproduced in triplicate, representing 18 samples subjected to RNA-Seq analysis (Fig. S1). The quality metrics and numbers of mapped reads for the data set are given in Table S2.
Principal component analysis was used as an unsupervised approach to visualize sample reproducibility and gene expression similarity of biological treatments. The first principal component separated cells on the time of TGFβ treatment (Fig. 3B). By contrast, the second principal component separated cells on the basis of RELA activity. We observed that each biological replicate closely clustered with the other samples receiving the same treatment, indicating a high concordance of the biological replicates. Prior to TGFβ stimulation, RELA KD cells were widely separated from WT cells. After 1 day of treatment, both WT and RELA-silenced cells closely clustered; these populations then separated after 3 days of TGFβ treatment. We interpret these data to indicate that RELA controls global gene expression in the absence of stimulation, and its major effect is seen after 3 days of stimulation.
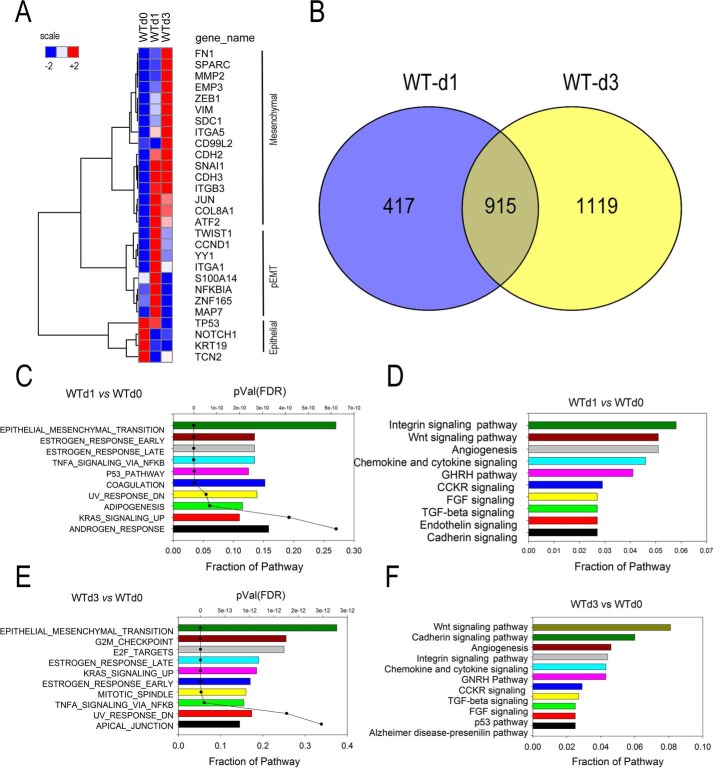
Figure 3.
Analysis of the TGFβ-induced gene regulatory network. A, behavior of EMT signature genes in WT cells. Expression patterns of 23 signature epithelial or mesenchymal genes in the WT cell time course were z-score–normalized and subjected to hierarchical clustering using Euclidian distance. Three major clusters, epithelial, pEMT, and mesenchymal genes, were identified. B, overlap of differentially expressed genes after 1 and 3 days of TGFβ stimulation. Genes with 4-fold change in transcripts per million and adjusted p value of <0.01 were compared. C and E, GSEA showing hallmark gene sets enriched in differentially expressed genes in WT cells for 1 and 3 days (d) after stimulation, respectively. For each gene set, the fraction of genes represented in the pathway and the significance are plotted (p value). FDR, false discovery rate. D and F, pathway enrichment analysis. Shown are the top 10 overrepresented pathways for the differentially expressed genes in WT cells for 1 and 3 days after stimulation as indicated. FGF, fibroblast growth factor; CCKR, cholecystokinin receptor; GHRH, growth hormone-releasing hormone.
Analysis of the TGFβ-induced transcriptional program
We first considered the analysis of the TGFβ-induced GRN in the absence of shRNA expression followed by identification of the network component that is RELA-dependent. TGFβ treatment changed the expression of 2,441 unique genes after filtering for a 4-fold change in gene expression and an adjusted p value of <0.01.
To ensure that we have captured the initial steps in mesenchymal transition, we examined the patterns of 28 well-established epithelial and mesenchymal “signature genes” (28) by hierarchical clustering. TGFβ induced at least two temporally coordinated waves of gene expression, identifying three major clusters: a group high in the untreated cells (Fig. 3A, WTd0), a transiently up-regulated group at 1 day of stimulation (Fig. 3A, WTd1), and a group later up-regulated after 3 days of stimulation (Fig. 3A, WTd3). The highly expressed genes in the WT untreated cells included characteristic epithelial genes, including keratin (KRT), NOTCH1, and TP53, whose expression fell after TGFβ stimulation. A smaller group of genes was transiently up-regulated at 1 day with expression falling by 3 days. These included integrin (ITGA)-1, the transcription factor YY1, and the S100 calcium-binding protein (S100A14). Also, a group of genes was up-regulated at 1 day and sustained for 3 days, included the master mesenchymal transcription factors SNAI1, TWIST1, and JUN.
Finally, the genes induced after 3 days of treatment included the characteristic mesenchymal genes FN1, VIM, secreted protein, acidic, cysteine-rich (SPARC), MMP, and others (Fig. 3). Collectively, these data indicate that we have captured components of the epithelial-pEMT-mesenchymal transition, consistent with the three-state transition observed in type III EMT in malignant cells (10). We also noted that the expression of the master transcription factors SNAI1, TWIST1, and JUN preceded that of ZEB1, consistent with the sequential expression of the SNAI1–ZEB1 regulatory network (10). We interpret these data to indicate that we have a robust data set that captures the early expression events in the initiation and pEMT-mesenchymal transition of primary cells.
To more systematically understand the temporal expression patterns, we compared the overlap of the differentially expressed genes on day 1 versus that of day 3. 417 genes were uniquely changed after 1 day of treatment, 915 genes were common, and 1,119 genes were regulated 3 days after treatment (Fig. 3B). To obtain information about the molecular processes underlying the TGFβ-induced gene regulatory network, we first performed gene set enrichment analysis (GSEA) on the highly differentially expressed genes in WT cells after 1 and 3 days. GSEA assumes that biological processes occur by coregulation of genes within common pathways and statistically identifies enriched pathways based on prior knowledge (29). The five most highly significant “hallmark” pathways included the EMT itself, “estrogen response,” “TNF signaling via the NF-κB pathway,” and the “p53” and “coagulation” pathways (Fig. 3C). Interestingly, 54 genes, over 25% of the hallmark EMT reference set, were identified in our RNA-Seq analysis; over half of these EMT genes contributed to the “estrogen response” and “TNFα signaling via NF-κB” gene sets. A separate functional classification analysis on the WT day 1 gene set was performed, examining for enrichment of signaling pathways, and resulted in the identification of “integrin,” “WNT signaling,” “angiogenesis,” and “chemokine/cytokine signaling” (Fig. 3D). We noted that the classifications by these two different analyses were highly concordant, with the genes contributing to the integrin and WNT signaling pathways also being contained within the GSEA/hallmark EMT gene set, and the chemokine/cytokine signaling within the hallmark GSEA “TNFα signaling” gene sets. Importantly, these data confirm our earlier observations that the NF-κB–dependent GRN is a core component of the epithelial-to-pEMT state transition (17, 30).
After 3 days of TGFβ stimulation in WT cells, GSEA again identified “EMT,” estrogen response, and TNFα signaling via NF-κB as dominant molecular processes; additional pathways that appeared included the cell cycle–regulatory pathways “G2M checkpoint” and “E2F targets” (Fig. 3E). Signaling pathway enrichment was predicted for WNT, “cadherin,” angiogenesis, and integrin pathways along with the chemokine/cytokine signaling pathway observed after 1 day of TGFβ treatment (Fig. 3F).
The NF-κB–dependent transcription program controls master EMT regulators
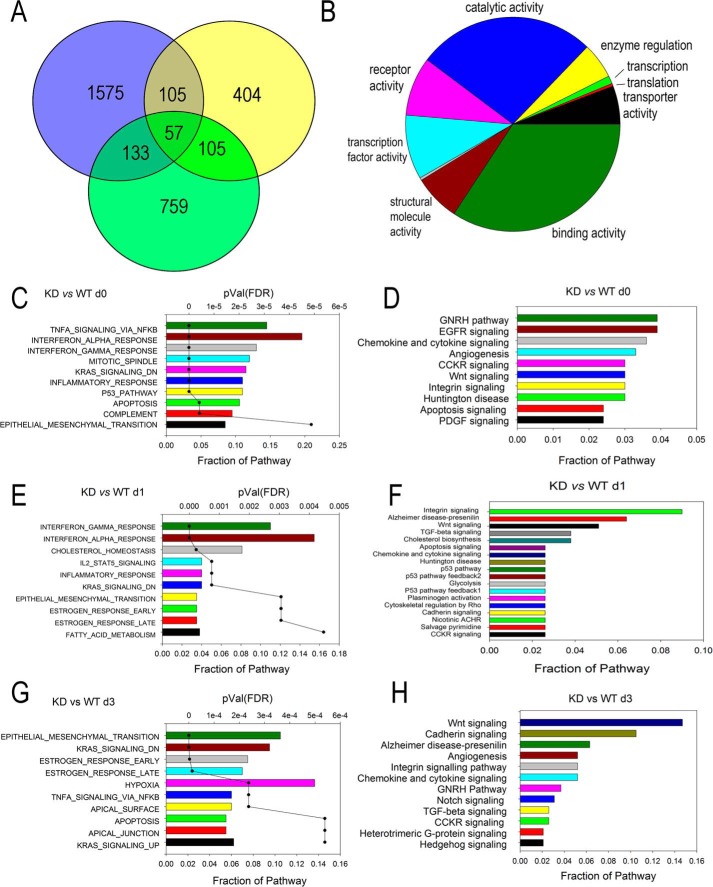
3,138 unique differentially expressed genes were identified as NF-κB–dependent by a 4-fold difference in comparing RELA KD cells versus WT for each sampling point in the experiment (Fig. 4A). The largest number of genes, 1,870, was identified in the RELA KD versus WT cells prior to TGFβ treatment, consistent with the distinct mapping of the control cells in the principal component analysis (cf. Figs. 4A and S1). After TGFβ stimulation, 671 genes were affected by RELA KD after 1 day of TGFβ treatment, and 1,055 genes were affected after 3 days of TGFβ treatment. Many are unique to each time point (Fig. 4A). Importantly, the number of NF-κB–dependent genes represented over half of those regulated by TGFβ at the same time point, indicating to us that NF-κB has a global effect on the EMT transcriptional program. The 3,138 unique NF-κB–dependent genes were subjected to analysis for molecular function by GO (Fig. 4B); the major functions identified included nucleic acid–binding transcriptional activity (10%) and binding (26%). GSEA and signaling pathway analyses were conducted for each KD versus WT comparison, day 0 (Fig. 4, C and D), day 1 (Fig. 4, E and F), and day 3 (Fig. 4, G and H). Here the rank-ordered hallmark gene sets underwent a time-dependent shift from TNFα signaling via NF-κB being the top ranked gene set in unstimulated cells (Fig. 4C) to EMT becoming the top ranked gene set in cells stimulated with TGFβ for 3 days (Fig. 4G). Similarly, the rank order of the signaling pathways changed where the most predominant signaling pathways in control (day 0),“gonadotropin-releasing hormone,” “epidermal growth factor receptor,” and “chemokine and cytokine signaling” (Fig. 4D), were replaced by the WNT and cadherin signaling pathways after 3 days of TGFβ treatment (Fig. 4H).
Figure 4.
Analysis of the RELA-dependent GRN. A, Venn diagram showing overlap of differentially expressed genes in RELA KD cells after 1 and 3 days of TGFβ stimulation. NF-κB–dependent genes are defined as those with 4-fold change between KD and WT cells, p < 0.01 compared for each treatment time. B, overrepresentation of the GO molecular functions for all unique genes in the RELA-dependent GRN. Note that the major GO term is transcription factor activity. C, E, and G, GSEA for RELA-dependent genes in control (day 0 (d0)) and 1 and 3 days after stimulation. D, F, and H, pathway enrichment analysis. Shown are the top 10 overrepresented pathways for the differentially expressed genes in RELA KD cells in control (day 0) and at 1 and 3 days as indicated. CCKR, cholecystokinin receptor; GHRH, growth hormone-releasing hormone; EGFR, epidermal growth factor receptor; PDGF, platelet-derived growth factor; ACHR, acetylcholine receptor; FDR, false discovery rate.
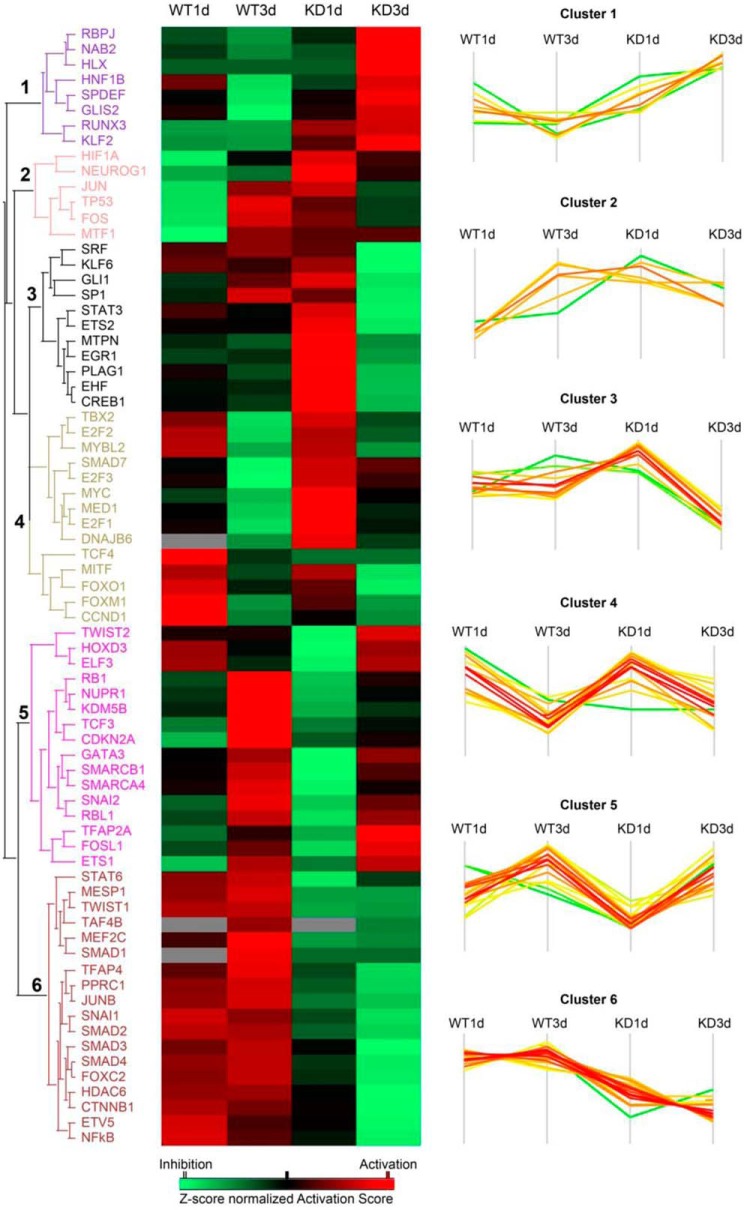
Inference of transcription factor networks under NF-κB/RELA control
To obtain more insight into the role of RELA in control of transcription factors in type II EMT, we conducted upstream regulator analysis in Ingenuity Pathway Analysis. Upstream regulator analysis uses the observed expression change in our data sets and prior knowledge of expected effects between transcriptional regulators and their target genes to identify the cascades of upstream regulators responsible (31). We predicted 73 upstream transcription regulators that show a significant activation or inhibition effect (activation z-score > or <2, p value of overlap <0.01) that clustered into six expression patterns (Fig. 5). Many master transcription factors of the EMT were identified in Clusters 2, 5, and 6. Of these, we noted that Cluster 2 contained the master transcription factor JUN/Fos (9). Similarly, Cluster 5 included TWIST2/SNAI2 and TCF3. Cluster 6 included JUNB, SNAI1, SMADs 2/3/4, CTNNB1, and NF-κB itself. From inspection of the effect of RELA KD, the actions of the transcription factors in Clusters 2 and 6 are the most significantly reduced. We also note that RELA KD results in the activation of mesenchymal-to-epithelial regulators SMAD7, MYC, and TBX2 (in Clusters 3 and 4).
Figure 5.
Upstream regulator analysis of NF-κB–dependent transcription factors in EMT. Upstream regulator analysis was performed on the differentially regulated genes identified in WT (1 or 3 days (d)) versus unstimulated cells and RELA KD (1 or 3 days) versus unstimulated RELA KD. Left, heat map of activation z-score of each transcription regulators. Red color, activation effect; green, inhibitory effect. Right, plot of z-score for individual regulators. Note the dramatic inhibitory effects of regulators in Cluster 6 in the RELA KD, including JUN, SNAI1, and CTNNB1.
Collectively, our data indicate that NF-κB signaling functions as a master regulator of type II EMT upstream of the mesenchymal state–promoting transcription factors and negatively regulates epithelial state–promoting factors. We next sought to validate the NF-κB dependence of the WNT pathway.
NF-κB/RELA regulates the WNT pathway in EMT
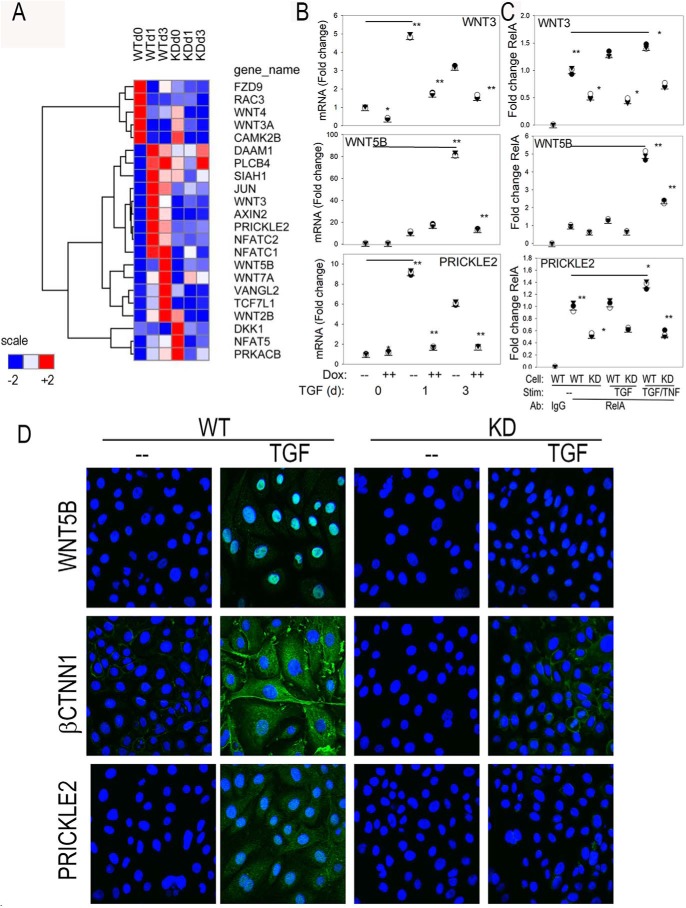
The WNT signaling pathway mediates cell–cell interactions controlling pattern formation in embryogenesis. The pathway consists of the WNT ligands encoded by 19 distinct human genes (14) whose products bind to a heterodimeric plasma membrane receptor consisting of Frizzled (FZ) and LRP5/6 proteins. WNT ligands activate two opposing pathways, depending on the ligand and context of stimulation. WNT3 signaling stabilizes CTNNB, functioning as a cofactor of T cell–regulatory factor (TCF), nuclear factor of activated T cells (NFAT) 5, and JUN. In contrast, WNT5B activates a noncanonical pathway through receptor tyrosine kinase–like orphan receptor 2 (ROR2) important in oncogenesis (13).
Experimentally observed changes in mRNA expression of the WNT pathway components are shown visually by hierarchical clustering of the RNA-Seq data. In WT cells, we observed that TGFβ stimulation down-regulated FZ component 9 receptor and WNT3a and -4 ligands and induced a strong up-regulation of WNT3A, PRICKLE2, and the NFATC isoforms within 1 day of stimulation (Fig. 6A). A later induction of WNT5B and -7A ligands was observed after 3 day of TGFβ stimulation. Interestingly, these major components of the WNT pathway required RELA; in the RELA KD cells, the TGFβ-induced up-regulation of WNT3, -5B, NFAT, and PRICKLE2 was blocked (Fig. 6A).
Figure 6.
Effect of RELA KD on the WNT signaling pathway. A, hierarchical clustering of temporal expression of WNT pathway members. Mean transcripts per million counts for each experimental condition were z-score–normalized by row and subjected to hierarchical clustering using Euclidean distance. Gene names are given by universal gene symbol. B, Q-RT-PCR assay was independently conducted to confirm RNA-Seq expression profiles for WNT3, WNT5B, and PRICKLE2 mRNA. Shown are individual mRNA abundance in replicates normalized to POLB. *, p < 0.05; **, p < 0.01, ANOVA, post hoc Tukey's test. C, XChIP assay was conducted to quantify RELA binding to the proximal promoter. Stimulation (Stim) conditions are control, TGFβ, or TGFβ + TNF. Antibodies used are normal anti-rabbit IgG or anti-RELA. Q-gPCR was conducted using gene-specific primer pairs (Table S3) corrected for input (70). Data are expressed as -fold change relative to control WT cells immunoprecipitated with IgG. Shown are individual replicates. D, expression of WNT5B, CTNNB1, and PRICKLE2 was determined by CFM. Ab detection was by secondary Alexa Fluor 488 (green); nuclei are stained with DAPI (blue). CFM was taken at 63×. d, day.
To confirm this observation, we conducted independent Q-RT-PCR assays of WNT3, -5B, and PRICKLE2 expression. Both WNT3 and PRICKLE2 were highly induced by TGFβ in WT cells, peaking after 1 day of stimulation at 4.5- and 9-fold over that of untreated cells, respectively (Fig. 6B). By contrast, in the RELA KD cells, the TGFβ induction of WNT3 was significantly reduced at all of time points (p < 0.01), whereas PRICKLE2 expression was reduced after 1 and 3 days of TGFβ stimulation. Similarly, WNT5B was highly induced in WT cells by 3 days of TGFβ treatment, increasing to 80-fold, and significantly inhibited after 1 and 3 days of stimulation in the RELA KD cells. These data confirm NF-κB/RELA dependence of these members of the WNT signaling pathway.
To provide a mechanism for their RELA dependence, we used two-step cross-linking ChIP (XChIP) assays to determine whether RELA directly interacted with these genes within their native chromatin environment. Q-gPCR assays were developed for high-affinity NF-κB–binding sites in the proximal regions of WNT3, WNT5B, and PRICKLE2 promoters (Table S3), corresponding to RELA peaks identified in ENCODE ChIP-Seq data (Fig. S2). We observed that RELA was strongly bound to these promoters in unstimulated cells by comparison of the signal from RELA immunoprecipitation with that of control IgG where a 10-fold increase in promoter signal was observed by Q-gPCR (p < 0.01; Fig. 6C). A 128-fold difference in signal intensity was observed when we examined the enrichment of an NF-κB–binding site relative to a distant region of the gene lacking an NF-κB–binding site (not shown). The effect of TGFβ stimulation did not significantly change the amount of RELA binding to the promoter over that of control cells, although an increase in RELA binding was observed by concomitant stimulation with TNF, a ligand that produces a much more robust RELA translocation than does TGFβ and potentiates TGFβ-induced EMT (17, 18, 32). For each treatment condition, less RELA binding was observed in the RELA KD versus WT cells, further indicating assay specificity (Fig. 6C). The inability to detect the 2-fold change in RELA abundance by 3 days of TGFβ stimulation reflects a limitation of the XChIP assay to small differences in RELA abundance/binding. These data indicate that the mechanisms of TGFβ-induced RELA activation early in the course of EMT are distinct from the robust nuclear translocation characteristic of the TNF-induced canonical pathway.
To further support the conclusion that RELA was required for WNT signaling in mesenchymal transition, we performed CFM experiments in control or TGFβ-stimulated WT or RELA KD cells. In the absence of stimulation, no WNT5B antigen was detected; however, TGFβ induced a dramatic up-regulation of WNT5B immunoreactivity within the nucleus (Fig. 6D, top row). This nuclear localization is consistent with WNT5B distribution within the kidney epithelium undergoing EMT (33). By contrast, in RELA KD cells, WNT5B immunoreactivity was undetectable in the absence or presence of TGFβ stimulation (Fig. 6D). Similarly, CTNNB1 immunoreactivity was strongly induced in the cytoplasm of TGFβ-stimulated WT cells, whereas its expression was also completely inhibited in RELA-silenced cells (Fig. 6D, middle row). Finally, PRICKLE2 immunoreactivity was TGFβ-inducible in WT cells in a cytoplasmic vesicle-bound distribution, consistent with its localization to Golgi bodies (Fig. 6D, bottom row). No PRICKLE2 immunostaining was detected in the RELA KD cells (Fig. 6D).
Independent validation of RELA dependence
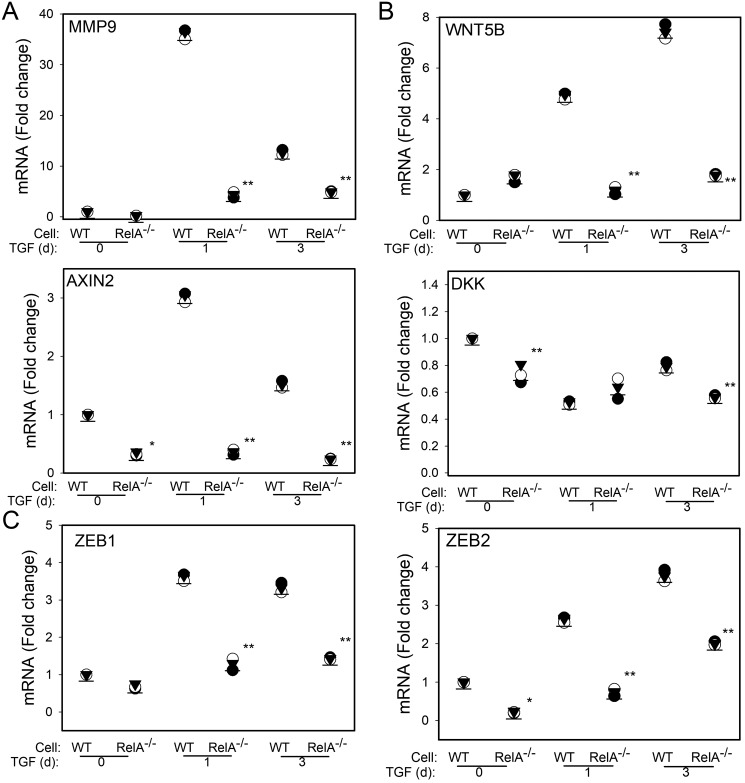
To independently confirm that the WNT pathway is RELA-dependent, we performed validation experiments using CRISPR/Cas9–mediated RELA silencing by genome editing. In these cells, RELA protein and mRNA are undetectable (34) and fail to induce MMP9 after TGFβ stimulation (see Figs. 2E and 7A). Because MMP9 is a hallmark gene of EMT, these data confirm the RELA dependence for the initiation of EMT. Next, WT or RELA−/− hSAECs were TGFβ-stimulated, and the expression of WNT ligand and downstream genes was measured by Q-RT-PCR. In a pattern similar to that observed in the RELA KD cells, the TGFβ-induced expression of WNT5B, AXIN2, and Dickkopf (DKK) was absent in RELA−/− hSAECs (Fig. 7B). Importantly, previous work has shown that AXIN2 expression is induced by WNT signaling, indicating that TGFβ functionally activates the WNT pathway (35). Finally, we confirmed that the up-regulation of the ZEB1- and -2 genes is RELA-dependent as basal and TGFβ-induced expression is reduced in RELA−/− hSAECs (Fig. 7C).
Figure 7.
Validation of RELA dependence of WNT signaling using RELA silencing by CRISPR/Cas9. CRISPR/Cas9–mediated genome editing was used to create a constitutive deletion of RELA in hSAECs. WT or RELA−/− cells were stimulated with TGFβ (10 ng/ml) for 0, 1, or 3 days (d) as indicated. Shown are -fold change mRNA in separate replicates normalized to POLB for MMP9, WNT5B, AXIN2, DKK, and ZEB1/2. *, p < 0.05; **, p < 0.01, ANOVA, post hoc Tukey's test.
NF-κB/RELA is upstream of the master EMT transcriptional regulators JUN and SNAI1
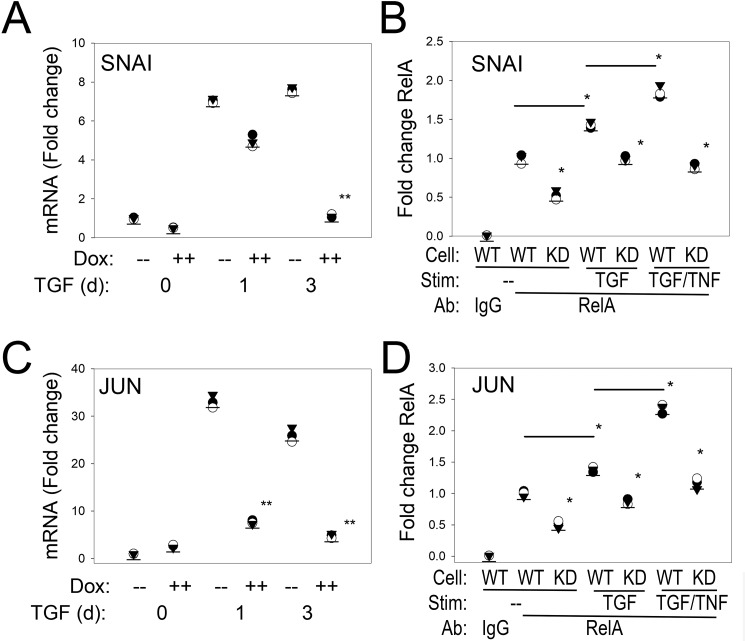
The pEMT-to-full mesenchymal transition depends on the activation of JUN and the SNAI1–ZEB1 master transcription factors (9, 11). In this regard, SNAI1 is one of the early expressed core EMT regulators important in triggering ZEB1 (10). SNAI1 mRNA expression was rapidly activated 7-fold within 1 day of TGFβ stimulation, peaking at 9-fold in WT cells, and was only transiently induced 5-fold in the RELA KD cells, falling to <2-fold after 3 days (Fig. 8A). A strong RELA signal was found associated with the endogenous SNAI1 promoter in control cells; TGFβ treatment slightly, but statistically significantly, increased this binding (Fig. 8B). In contrast, RELA binding was reduced for each treatment condition in the RELA KD cells (Fig. 8B).
Figure 8.
NF-κB/RELA is upstream of master transcription factors SNAI1 and JUN. A and C, Q-RT-PCR assay profile for SNAI1 and JUN, respectively. WT or RELA KD was cultured in the absence or presence of TGFβ stimulation (Stim) for 1 and 3 days (d). Shown is -fold change mRNA for individual replicates normalized to POLB. B and D, XChIP assay of RELA binding to the proximal promoter of SNAI1 and JUN, respectively. Q-gPCR was conducted using gene-specific primer pairs (Table S3) corrected for input (70). *, p < 0.05; **, p < 0.01, ANOVA, post hoc Tukey's test.
Because we noted that JUN was significantly affected by RELA silencing, we also sought to confirm its direct regulation by NF-κB. We confirmed a dramatic 30-fold up-regulation of JUN mRNA expression within 1 day of TGFβ stimulation (Fig. 8C). The TGFβ induction of JUN was significantly blunted in the RELA KD cells (Fig. 8C). We found a pattern of basal and TGFβ-increased RELA binding on the JUN promoter similar to that observed for SNAI1 in XChIP (Fig. 8D).
Discussion
Understanding the cellular pathways that trigger EMT is fundamental to the pathogenesis of asthma (6), chronic obstructive pulmonary diseases (36), and pulmonary fibrosis (37), diseases that affect over 300 million people worldwide (38). In the differentiated vertebrate airway, type II EMT plays a central role in the response to injury. Mucosal injury releases growth factors triggering the mesenchymal dedifferentiation program that produces extensive cytosolic restructuring resulting in the loss of apical-basal polarity, dissolution of adherens junctions, enhanced motility, and expression of mesenchymal fibrotic genes (3). These cells acquire stem cell–like properties, become resistant to reactive oxygen species stress, and become motile. This phenotypic change permits the transitioned mesenchymal cell to repopulate regions of denuded epithelium, promoting tissue repair and fibrosis (3). Currently, we understand that EMT is regulated by sequential activation of autocrine loops that are responsible for stepwise transition of epithelial to pEMT and subsequently to the full mesenchymal phenotype. However, the GRNs of the EMT program are highly cell type–dependent, initially determined through the interaction of TGFβ-induced SMAD3 with tissue-specific master transcription factors and modified by oncogenic GTPase signaling proteins (8, 17).
Our studies of type II EMT in primary human airway cells and those of type III EMT in transformed cells have demonstrated that NF-κB action is important for both types of mesenchymal transition (18, 39–41). NF-κB has multiple regulatory interfaces with EMT at the transcriptional and post-transcriptional levels (42). In this study, we used RNA-Seq to systematically identify the early transcriptional events under NF-κB control. We found that RELA controls expression of a significant fraction of those genes regulated by TGFβ. We have discovered and validated in two independent models of RELA silencing that RELA coordinately regulates ligands, receptors, and downstream targets of the WNT pathway as well as JUN and SNAI1–ZEB1 master transcription factors.
TGFβ induces a mode of NF-κB activation distinct from the canonical pathway
NF-κB is a pleiotropic, cytokine-inducible transcription factor regulated by post-translational modifications (43, 44). In resting cells, NF-κB/RELA is retained in the cytoplasm, but a small fraction undergoes continuous nuclear-cytoplasmic shuttling (45, 46). The well-understood canonical NF-κB pathway is rapidly (within minutes) triggered by cytokines to phosphorylate and ubiquitylate IκBα, resulting in RELA cytoplasmic release. RELA release is followed by a second activating step involving serine phosphorylation and acetylation, triggering complex formation with BRD4/CDK9 coactivator (47). Interestingly, our studies suggest that TGFβ activation of NF-κB is distinctly slower than the fast activation by cytokines where TGFβ-stimulated RELA translocation occurs much more slowly with activating Ser-276 phosphorylation even later.
In addition to the differences in kinetics, the 2-fold magnitude of RELA translocation in response to TGFβ is distinctly less than that produced by canonical NF-κB translocation (see Refs. 17 and 30). These data suggest that the major effect of TGFβ is post-translational regulation of the RELA phosphorylation state. Our previous work has suggested that RELA translocation is indirectly mediated by a paracrine factor secreted into the conditioned medium (18). The identity of this factor(s) is currently unknown and will require further study.
NF-κB mediates the WNT morphogen signaling
The mesenchymal transition is the product of sequential cascades involving the transition from epithelial to an uncommitted “partial” mesenchymal state followed by commitment to an irreversible mesenchymal state (11). These cell-state transitions are achieved by activating secondary (downstream) autocrine signaling loops, including IL6 and fibroblast growth factor (15, 16). In this study, we extend our understanding to the RELA dependence of the WNT pathway in TGFβ-induced type II mesenchymal transition.
The WNT pathway is important in the formation of cell fate determination, formation of cellular polarity in organ development, and control of cellular migration (14). The inducible WNT ligands are secreted, palmitoylated glycoproteins that function as morphogens by forming concentration gradients. All WNT ligands signal by binding heterodimeric FZ receptors, triggering intracellular signaling pathways, including the canonical and noncanonical pathways with the latter composed of both the WNT/Ca+ and planar cell polarity pathways (48). The canonical pathway stabilizes CTNNB1, which serves as a transcriptional coactivator inducing downstream homeodomain proteins important in pattern formation. By contrast, the noncanonical pathways are CTNNB1-independent, activating the tyrosine kinase ROR2 (49) and Rho GTPase, stimulating actin polymerization and cellular migration characteristic of the mesenchymal state (13, 14). The planar cell polarity pathway induces differential localization of PRICKLE2. Our RNA-Seq studies indicate that TGFβ suppresses expression of WNT3 and -4 but activates WNT5B and -7B ligands, suggesting that the mesenchymal state shifts the modes of WNT signaling. Consistent with this hypothesis, WNT5B partially inhibits canonical signaling (13), perhaps explaining the cytoplasmic CTNNB1 accumulation observed by CFM (Fig. 6D). A complete understanding of the functional effect of this differential ligand expression will require further study. Although RELA has been previously reported to activate WNT5A (50), our data are the first to show that NF-κB controls expression of WNT5B and -7A during mesenchymal transition, suggesting a much broader impact of RELA on WNT signaling.
Although our studies focus on RELA-dependent GRN in the initiation of EMT, our data permit us to infer the activity of the WNT pathway. The observations of selective up-regulation of AXIN2, a direct target of WNT signaling (35), and down-regulation of DKK, an inhibitor of the WNT signaling pathway (51), suggest that WNT signaling is activated early in the initiation of the mesenchymal transition by multiple mechanisms.
NF-κB/RELA is a master regulator of EMT upstream of SNAI1 and JUN
SNAI1 and ZEB1 are master transcriptional regulators of the mesenchymal transition whose expression are dramatically up-regulated by TGFβ (52). Currently, it is understood that SNAI1 and ZEB1 are silenced in epithelial cells by a double-negative feedback loop mediated by the microRNAs miR34 and miR200 (10, 53). Under basal conditions, miR34 post-transcriptionally inhibits SNAI1 expression, whereas miR200 blocks ZEB1 expression. Equilibrium of the double-negative feedback loops is perturbed by TGFβ-induced transcriptional up-regulation of SNAI1, which binds to the promoter and inhibits miR200 expression, releasing ZEB1. Our data here indicate that RELA plays a direct role in activating this SNAI1–ZEB1 negative feedback loop. Our data demonstrate that SNAI1 is directly activated by RELA binding to its proximal promoter. Initially, the TGFβ induction of SNAI1 after 1 day is not affected, but continued SNAI1 expression after 3 days requires RELA. This finding suggests a complex mode of SNAI1 regulation where SNAI1 is initially up-regulated directly by SMAD3 (7, 54) but later is functionally replaced by RELA for sustained expression. Our earlier studies have shown that RELA activation of SNAI1 is mediated by a RELA-dependent coactivator recruitment of the atypical histone acetyltransferase BRD4 (18). BRD4 activates SNAI1 by phosphorylation of the C-terminal repeat domain of RNA polymerase II, promoting its transcriptional elongation.
In addition to the SNAI1–ZEB1 double-feedback loop, a recent study has identified the essential role for a clique of JUNB–HNF4–ETS transcription factors in the pEMT-mesenchymal transition (9). JUNB–HNF4–ETS are induced by TGFβ stimulation, physically associate, and coordinately bind to the promoters of many mesenchymal genes (9). These factors are also associated with dense clusters of H3K27Ac loci, a.k.a. “superenhancers” in the A549 carcinoma cell line. Although JUNB is not induced by TGFβ in hSAECs, we provide evidence here that JUND, a homologous isoform with identical DNA binding specificity, is directly regulated by RELA. Collectively, these data show that RELA is upstream of these two core transcription factor loops necessary for mesenchymal transition, qualifying RELA to be considered as a “master transcriptional regulator” of EMT.
Master regulators are engaged in the coordinate regulation of cliques of downstream transcription factors by maintaining their expression by the formation of superenhancers (9, 55). Our data show that RELA directly binds to the WNT5B, JUND, SNAI, and ZEB1 promoters and is involved in their constitutive expression. Previous work has shown that NF-κB/RELA controls the activity of superenhancers important in innate inflammation (56). More work will be required to identify whether superenhancers mediate type II EMT and how the activated RELA complex maintains mesenchymal transition genes in open chromatin domains.
In summary, our study identifies the role of NF-κB/RELA as a master regulator of mesenchymal transition in primary human airway epithelial cells. Our studies demonstrate that RELA is required for the activation of the downstream WNT, JUN, and SNAI1–ZEB1 core regulatory factors required for transition from pEMT to the fully committed mesenchymal program.
Experimental procedures
hSAEC culture and induction of the mesenchymal transition
An immortalized hSAEC line established by infecting primary hSAECs with human telomerase and cyclin-dependent kinase-4 retrovirus constructs was described (20). The immortalized hSAECs were grown in small airway epithelial cell growth medium (Lonza, Walkersville, MD) in a humidified atmosphere of 5% CO2. For induction of EMT, TGFβ (10 ng/ml; PeproTech, Rocky Hill, NJ) was added to growth medium for the indicated times.
RELA silencing
hSAECs expressing Dox-regulated RELA shRNA were established using puromycin selection. Briefly, TRIPZ Tet-On inducible lentiviral RELA shRNA plasmid (Dharmacon, Thermo Fisher Scientific, Lafayette, CO) was reverse transfected with a packaging construct into BOS23 cell lines, and supernatants were harvested. Naïve hSAECs were infected and selected for puromycin resistance (4 μg/ml). Puromycin-resistant hSAECs were pooled and characterized. RELA depletion was produced by addition of Dox (2 μg/ml; Sigma-Aldrich) to the culture medium for 5 days.
RELA deletion was performed by CRISPR/Cas9 genome editing. A 20-nucleotide seed sequence in exon 4 of the RELA gene was used to introduce a stop codon that induces nonsense-mediated RNA decay. RELA−/− hSAECs lack detectable RELA mRNA and protein (34, 57).
Q-RT-PCR
Total RNA was extracted using acid guanidium phenol (TRI Reagent, Sigma-Aldrich). For gene expression analyses, 1 μg of RNA was reverse transcribed in a 20-μl reaction mixture using SuperScript III (58, 59). Relative changes in gene expression were quantified relative to control β-polymerase (POLB) transcripts using the ΔΔCt method (60). The forward and reverse gene-specific Q-RT-PCR primers are listed in Table S1.
Subcellular fractionation and Western immunoblot analyses
Nuclear and cytoplasmic proteins were fractionated as described previously (61). For Western blotting, equal amounts of nuclear protein were fractionated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. The membranes were incubated with affinity-purified rabbit polyclonal antibodies to RELA (Santa Cruz Biotechnology). Washed membranes were then incubated with IRDye 800–labeled anti-rabbit IgG antibodies (Rockland Immunochemicals, Gilbertsville, PA), and immune complexes were quantified using the Odyssey IR Imaging System (LI-COR Biosciences, Lincoln, NE).
Immunostaining and CFM
hSAECs incubated in the absence or presence of Dox (2 μg/ml, 5 days) were stimulated in the absence or presence of TGFβ (10 ng/ml, 3 days) and replated on rat-tail collagen-treated coverslips (Roche Applied Sciences). Cells were fixed with paraformaldehyde in PBS (4%, v/v; 30 min) and incubated in 0.1 m NH4Cl in PBS for 10 min. For visualization of cytoplasmic F-actin distribution, fixed cells were stained with Alexa Fluor 488– or Alexa Fluor 568–phalloidin (Life Technologies) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI) for visualizing the nucleus. The cells were then imaged with a Zeiss LSM510 fluorescence confocal microscope at a magnification of 63×.
For immunofluorescence staining, fixed cells were permeabilized with 0.5% Triton X-100 in PBS followed by incubation in blocking buffer (5% goat serum, 0.1% IGEPAL CA-630, 0.05% NaN3, and 1% BSA in PBS). Permeabilized cells were then incubated with 1:100 dilutions of the indicated antibodies in incubation buffer (0.1% IGEPAL CA-630, 0.05% NaN3, and 2% BSA in PBS) overnight at 4 °C. Antibodies used were: anti-RELA (Santa Cruz Biotechnology, sc-372), anti-TGFBR1 (Abcam, ab31013), anti-WNT5B (Abcam, ab150602), anti-SMAD7 (Sigma-Aldrich, HPA028897), anti-PRICKLE2 (Sigma-Aldrich, HPA035493), anti-CTNNB1 (Sigma-Aldrich, HPA029159), and phospho-SMAD2/3 (Cell Signaling Technology, 8828S). After washing, cells were stained with Alexa Fluor 488– or Alexa Fluor 568–conjugated goat anti-rabbit IgG (Life Technologies), respectively, in incubation buffer for 1 h. The cells were visualized as above.
RNA extraction and qualification
Total cellular RNA was extracted using TRI Reagent according to the manufacturer's recommendations. RNA was quantified spectrophotometrically using a NanoDrop ND-1000 (NanoDrop Technologies). Quality of the purified RNA was assessed by visualization of 18S and 28S RNA bands using an Agilent BioAnalyzer 2100 (Agilent Technologies, CA). The resulting electropherograms were used in the calculation of the 28S/18S ratio and the RNA integrity number.
Library construction and sequencing
A RiboZero Gold kit (Illumina, San Diego CA) was used to remove rRNA from 1 μg of total RNA. First and second strand synthesis, adapter ligation, and amplification of the library were performed using the Illumina TruSeq v2 Stranded RNA Sample Preparation kit under conditions prescribed by the manufacturer (Illumina). Samples were tracked using “index tags” incorporated into the adapters. Library quality was evaluated using an Agilent DNA-1000 chip on an Agilent 2100 Bioanalyzer. Quantification of library DNA templates was performed using Q-PCR and a known-size reference standard. Cluster formation of the library DNA templates was performed using the TruSeq PE Cluster kit v3 (Illumina) and the Illumina cBot work station using conditions recommended by the manufacturer. Template input was adjusted to obtain a cluster density of 700,000–900,000/mm2. Paired-end 50-base sequencing by synthesis was performed using TruSeq SBS kit v3 (Illumina) on three lanes of an Illumina HiSeq 1500 using the manufacturer's protocols.
RNA-Seq analysis
The raw next-generation sequencing analysis passed 11 separate quality analyses by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/,4 updated November 2013, accessed June 6, 2014). Reads were aligned to the human GRCh38 reference genome using STAR, an ultrafast splicing-aware alignment program, version 2.5.1b (63). Read counts per gene were obtained with the featureCounts function (v1.5.0-p1) of the subread software package (64) using the GENCODE version 24 basic annotation file. Counts were input into the DESeq2 differential gene expression analysis program (version 1.12.1) (65). Factors were RELA level (KD versus control) and TGFβ treatment (0, 1, and 3 days). Differentially expressed genes were called with a log2 -fold change greater than 2 or less than −2 and a Benjamini–Hochberg-adjusted p value (false discovery rate) of less than 0.01.
Data analysis
Genome ontology and signaling pathways were identified in Panther (62) (http://www.pantherdb.org/).4 Hierarchical clustering was performed on z-score–normalized data in GENE-E version 3.0.206 (66). Hallmark pathways were identified using gene set enrichment analysis (29). Protein–protein interaction network analysis was performed using NetworkAnalyst (67), mapping to 140,000 interactions from InnateDB (68). Networks were visualized and annotated in Cytoscape (69).
Two-step XChIP
XChIP was performed as described previously (70). hSAECs (4 × 106–6 × 106/100-mm dish) were washed twice with PBS. Protein–protein cross-linking was first performed with disuccinimidyl glutarate (2 mm, 45 min; Thermo Fisher Scientific) followed by protein–DNA cross-linking with formaldehyde (1%, 15 min; Thermo Fisher Scientific). Chromatin was fragmented by sonication, and equal amounts were immunoprecipitated overnight at 4 °C with 4 μg of the indicated Ab in ChIP dilution buffer. Immunoprecipitates were collected with 40 μl of protein A magnetic beads (Dynal Inc.), washed, and eluted in 250 μl of elution buffer for 15 min at room temperature. Samples were de-cross-linked in 0.2 m NaCl at 65 °C for 2 h. The precipitated DNA was phenol-chloroform–extracted, ethanol-precipitated, and dried.
Quantitation was performed by Q-gPCR using gene-specific primers (Table S3). These PCR primers were designed to amplify NF-κB–binding sites identified by TRANSFAC analysis and confirmed by ChIP-Seq tracks from ENCODE. Data are presented as -fold change relative to RELA IPs of untreated (control) cells. For this calculation, for each gene-specific primer set, the median Ct value of input was subtracted from the Ct value produced by RELA IP for each cellular treatment (ΔCt). For each cellular treatment, the -fold change relative to control cells was calculated by subtracting the ΔCt from the ΔCt of the RELA IP from control cells.
Statistical analysis
Differences among the means were tested for significance by ANOVA with Fisher's least significance difference test. Individual comparisons were made using post hoc Tukey's test. Significance was reached when the p value was <0.05. Values presented are means ± S.D.
Author contributions
B. T., S. G. W., J. Y., T. G. W., A. K., Y. Z., and A. R. B. conceptualization; B. T., S. G. W., J. Y., Y. Z., and A. R. B. data curation; B. T., S. G. W., J. Y., T. G. W., A. K., and A. R. B. formal analysis; B. T., S. G. W., and A. R. B. investigation; B. T., S. G. W., T. G. W., A. K., Y. Z., and A. R. B. writing-original draft; B. T., S. G. W., J. Y., T. G. W., Y. Z., and A. R. B. writing-review and editing; S. G. W., J. Y., T. G. W., and Y. Z. validation; S. G. W., J. Y., A. K., and A. R. B. methodology; A. K. and Y. Z. visualization; A. R. B. resources; A. R. B. funding acquisition; A. R. B. project administration.
Supplementary Material
Acknowledgment
Core support was provided from the University of Texas Medical Branch Next Generation Sequencing and Optical Imaging Core Laboratories.
This work was supported by NIAID, National Institutes of Health Grant AI062885; National Center for Advancing Translational Sciences, National Institutes of Health Grant ULTR00002373; and National Science Foundation Grant DMS-1361411. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Tables S1–S3.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE84135.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- TGF
- transforming growth factor
- EMT
- epithelial-mesenchymal transition
- hSAEC
- human small airway epithelial cell
- SNAI1
- Snail family transcriptional repressor 1
- GRN
- gene regulatory network
- pEMT
- partial EMT
- ZEB1
- zinc finger E-box–binding homeobox 1
- CTNNB
- β-catenin
- IL6
- interleukin-6
- Dox
- doxycycline
- KD
- knockdown
- CFM
- confocal fluorescence microscopy
- BRD4
- bromodomain-containing protein 4
- TNF
- tumor necrosis factor
- ANOVA
- analysis of variance
- Q-RT-PCR
- quantitative real-time RT-PCR
- GSEA
- gene set enrichment analysis
- GO
- gene ontology
- FZ
- Frizzled
- TCF
- T cell–regulatory factor
- NFAT
- nuclear factor of activated T cells
- ROR2
- receptor tyrosine kinase–like orphan receptor 2
- XChIP
- cross-linking ChIP
- Q-gPCR
- quantitative genomic PCR
- DKK
- Dickkopf
- miR
- microRNA
- DAPI
- 4′,6-diamidino-2-phenylindole
- IP
- immunoprecipitation
- POLB
- β-polymerase
- FN1
- fibronectin
- MMP9
- matrix metalloproteinase 9
- VIM
- vimentin
- Ab
- antibody.
References
- 1. Knight D. A., and Holgate S. T. (2003) The airway epithelium: structural and functional properties in health and disease. Respirology 8, 432–446 10.1046/j.1440-1843.2003.00493.x [DOI] [PubMed] [Google Scholar]
- 2. Lambrecht B. N., and Hammad H. (2012) The airway epithelium in asthma. Nat. Med. 18, 684–692 10.1038/nm.2737 [DOI] [PubMed] [Google Scholar]
- 3. Kalluri R., and Weinberg R. A. (2009) The basics of epithelial-mesenchymal transition. J. Clin. Investig. 119, 1420–1428 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jordan N. V., Johnson G. L., and Abell A. N. (2011) Tracking the intermediate stages of epithelial-mesenchymal transition in epithelial stem cells and cancer. Cell Cycle 10, 2865–2873 10.4161/cc.10.17.17188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Katsuno Y., Lamouille S., and Derynck R. (2013) TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Curr. Opin. Oncol. 25, 76–84 10.1097/CCO.0b013e32835b6371 [DOI] [PubMed] [Google Scholar]
- 6. Ijaz T., Pazdrak K., Kalita M., Konig R., Choudhary S., Tian B., Boldogh I., and Brasier A. R. (2014) Systems biology approaches to understanding epithelial mesenchymal transition (EMT) in mucosal remodeling and signaling in asthma. World Allergy Organ. J. 7, 13 10.1186/1939-4551-7-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang Y. C., Piek E., Zavadil J., Liang D., Xie D., Heyer J., Pavlidis P., Kucherlapati R., Roberts A. B., and Böttinger E. P. (2003) Hierarchical model of gene regulation by transforming growth factor β. Proc. Natl. Acad. Sci. U.S.A. 100, 10269–10274 10.1073/pnas.1834070100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mullen A. C., Orlando D. A., Newman J. J., Lovén J., Kumar R. M., Bilodeau S., Reddy J., Guenther M. G., DeKoter R. P., and Young R. A. (2011) Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147, 565–576 10.1016/j.cell.2011.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang H., Liu Y., Xue M., Liu H., Du S., Zhang L., and Wang P. (2016) Synergistic action of master transcription factors controls epithelial-to-mesenchymal transition. Nucleic Acids Res. 44, 2514–2527 10.1093/nar/gkw126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu M., Jolly M. K., Levine H., Onuchic J. N., and Ben-Jacob E. (2013) MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. Proc. Natl. Acad. Sci. U.S.A. 110, 18144–18149 10.1073/pnas.1318192110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang J., Tian X. J., Zhang H., Teng Y., Li R., Bai F., Elankumaran S., and Xing J. (2014). TGF-β-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci. Signal. 7, ra91 10.1126/scisignal.2005304 [DOI] [PubMed] [Google Scholar]
- 12. Postigo A. A., and Dean D. C. (1999) ZEB represses transcription through interaction with the corepressor CtBP. Proc. Natl. Acad. Sci. U.S.A. 96, 6683–6688 10.1073/pnas.96.12.6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kanzawa M., Semba S., Hara S., Itoh T., and Yokozaki H. (2013) WNT5A is a key regulator of the epithelial-mesenchymal transition and cancer stem cell properties in human gastric carcinoma cells. Pathobiology 80, 235–244 10.1159/000346843 [DOI] [PubMed] [Google Scholar]
- 14. Clevers H., and Nusse R. (2012) Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 15. Yao Z., Fenoglio S., Gao D. C., Camiolo M., Stiles B., Lindsted T., Schlederer M., Johns C., Altorki N., Mittal V., Kenner L., and Sordella R. (2010) TGF-β IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 107, 15535–15540 10.1073/pnas.1009472107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sullivan N. J., Sasser A. K., Axel A. E., Vesuna F., Raman V., Ramirez N., Oberyszyn T. M., and Hall B. M. (2009) Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 28, 2940–2947 10.1038/onc.2009.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tian B., Li X., Kalita M., Widen S. G., Yang J., Bhavnani S. K., Dang B., Kudlicki A., Sinha M., Kong F., Wood T. G., Luxon B. A., and Brasier A. R. (2015) Analysis of the TGFβ-induced program in primary airway epithelial cells shows essential role of NF-κB/RelA signaling network in type II epithelial mesenchymal transition. BMC Genomics 16, 529 10.1186/s12864-015-1707-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tian B., Zhao Y., Sun H., Zhang Y., Yang J., and Brasier A. R. (2016) BRD4 Mediates NFκB-dependent epithelial-mesenchymal transition and pulmonary fibrosis via transcriptional elongation. Am. J. Physiol. Lung Cell. Mol. Physiol. 311, L1183–L1201 10.1152/ajplung.00224.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao Y., Tian B., Sadygov R. G., Zhang Y., and Brasier A. R. (2016) Integrative proteomic analysis reveals reprograming tumor necrosis factor signaling in epithelial mesenchymal transition. J. Proteomics 148, 126–138 10.1016/j.jprot.2016.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramirez R. D., Sheridan S., Girard L., Sato M., Kim Y., Pollack J., Peyton M., Zou Y., Kurie J. M., Dimaio J. M., Milchgrub S., Smith A. L., Souza R. F., Gilbey L., Zhang X., et al. (2004) Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 64, 9027–9034 10.1158/0008-5472.CAN-04-3703 [DOI] [PubMed] [Google Scholar]
- 21. Vaughan M. B., Ramirez R. D., Wright W. E., Minna J. D., and Shay J. W. (2006) A three-dimensional model of differentiation of immortalized human bronchial epithelial cells. Differentiation 74, 141–148 10.1111/j.1432-0436.2006.00069.x [DOI] [PubMed] [Google Scholar]
- 22. Jamaluddin M., Tian B., Boldogh I., Garofalo R. P., and Brasier A. R. (2009) Respiratory syncytial virus infection induces a reactive oxygen species-MSK1-phospho-Ser-276 RelA pathway required for cytokine expression. J. Virol. 83, 10605–10615 10.1128/JVI.01090-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jamaluddin M., Wang S., Boldogh I., Tian B., and Brasier A. R. (2007) TNF-α-induced NF-κB/RelA Ser276 phosphorylation and enhanceosome formation is mediated by an ROS-dependent PKAc pathway. Cell. Signal. 19, 1419–1433 10.1016/j.cellsig.2007.01.020 [DOI] [PubMed] [Google Scholar]
- 24. Nowak D. E., Tian B., Jamaluddin M., Boldogh I., Vergara L. A., Choudhary S., and Brasier A. R. (2008) RelA Ser276 phosphorylation is required for activation of a subset of NF-κB-dependent genes by recruiting cyclin-dependent kinase 9/cyclin T1 complexes. Mol. Cell. Biol. 28, 3623–3638 10.1128/MCB.01152-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang B., Yang X. D., Zhou M. M., Ozato K., and Chen L. F. (2009) Brd4 coactivates transcriptional activation of NF-κB via specific binding to acetylated RelA. Mol. Cell. Biol. 29, 1375–1387 10.1128/MCB.01365-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoffmann A., Leung T. H., and Baltimore D. (2003) Genetic analysis of NF-κB/Rel transcription factors defines functional specificities. EMBO J. 22, 5530–5539 10.1093/emboj/cdg534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tian B., Nowak D. E., Jamaluddin M., Wang S., and Brasier A. R. (2005) Identification of direct genomic targets downstream of the nuclear factor-κB transcription factor mediating tumor necrosis factor signaling. J. Biol. Chem. 280, 17435–17448 10.1074/jbc.M500437200 [DOI] [PubMed] [Google Scholar]
- 28. Tan T. Z., Miow Q. H., Miki Y., Noda T., Mori S., Huang R. Y., and Thiery J. P. (2014) Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 6, 1279–1293 10.15252/emmm.201404208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kalita M., Tian B., Gao B., Choudhary S., Wood T. G., Carmical J. R., Boldogh I., Mitra S., Minna J. D., and Brasier A. R. (2013) Systems approaches to modeling chronic mucosal inflammation. Biomed. Res. Int. 2013, 505864 10.1155/2013/505864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Krämer A., Green J., Pollard J. Jr., and Tugendreich S. (2014) Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cieślik M., Hoang S. A., Baranova N., Chodaparambil S., Kumar M., Allison D. F., Xu X., Wamsley J. J., Gray L., Jones D. R., Mayo M. W., and Bekiranov S. (2013) Epigenetic coordination of signaling pathways during the epithelial-mesenchymal transition. Epigenetics Chromatin 6, 28 10.1186/1756-8935-6-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim M.-K., Maeng Y.-I., Sung W. J., Oh H.-K., Park J.-B., Yoon G. S., Cho C. H., and Park K. K. (2013) The differential expression of TGF-β1, ILK and wnt signaling inducing epithelial to mesenchymal transition in human renal fibrogenesis: an immunohistochemical study. Int. J. Clin. Exp. Pathol. 6, 1747–1758 [PMC free article] [PubMed] [Google Scholar]
- 34. Tian B., Yang J., Zhao Y., Ivanciuc T., Sun H., Wakamiya M., Garofalo R. P., and Brasier A. R. (2018) Central role of the NFκB pathway in the Scgb1a1-expressing epithelium in mediating respiratory syncytial virus-induced airway inflammation. J. Virol. 92, e00441–18 10.1128/JVI.00441-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jho E.-H., Zhang T., Domon C., Joo C.-K., Freund J.-N., and Costantini F. (2002) Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22, 1172–1183 10.1128/MCB.22.4.1172-1183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harris W. T., Kelly D. R., Zhou Y., Wang D., MacEwen M., Hagood J. S., Clancy J. P., Ambalavanan N., and Sorscher E. J. (2013) Myofibroblast differentiation and enhanced TGF-B signaling in cystic fibrosis lung disease. PLoS One 8, e70196 10.1371/journal.pone.0070196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim K. K., Kugler M. C., Wolters P. J., Robillard L., Galvez M. G., Brumwell A. N., Sheppard D., and Chapman H. A. (2006) Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc. Natl. Acad. Sci. U.S.A. 103, 13180–13185 10.1073/pnas.0605669103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vogelmeier C. F., Criner G. J., Martinez F. J., Anzueto A., Barnes P. J., Bourbeau J., Celli B. R., Chen R., Decramer M., Fabbri L. M., Frith P., Halpin D. M., López Varela M. V., Nishimura M., et al. (2017) Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. GOLD executive summary. Am. J. Respir. Crit. Care Med. 195, 557–582 10.1164/rccm.201701-0218PP [DOI] [PubMed] [Google Scholar]
- 39. Tian B., Patrikeev I., Ochoa L., Vargas G., Belanger K. K., Litvinov J., Boldogh I., Ameredes B. T., Motamedi M., and Brasier A. R. (2017) NF-κB mediates mesenchymal transition, remodeling and pulmonary fibrosis in response to chronic inflammation by viral RNA patterns. Am. J. Respir. Cell Mol. Biol. 56, 506–520 10.1165/rcmb.2016-0259OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huber M. A., Azoitei N., Baumann B., Grünert S., Sommer A., Pehamberger H., Kraut N., Beug H., and Wirth T. (2004) NF-κB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J. Clin. Investig. 114, 569–581 10.1172/JCI200421358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li C. W., Xia W., Huo L., Lim S. O., Wu Y., Hsu J. L., Chao C. H., Yamaguchi H., Yang N. K., Ding Q., Wang Y., Lai Y. J., LaBaff A. M., Wu T. J., and Lin B. R. (2012) Epithelial-mesenchymal transition induced by TNF-α requires NF-κB-mediated transcriptional upregulation of Twist1. Cancer Res. 72, 1290–1300 10.1158/1538-7445.AM2012-1290,10.1158/0008-5472.CAN-11-3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu Y., Deng J., Rychahou P. G., Qiu S., Evers B. M., and Zhou B. P. (2009) Stabilization of snail by NF-κB is required for inflammation-induced cell migration and invasion. Cancer Cell 15, 416–428 10.1016/j.ccr.2009.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brasier A. R. (2008) The nuclear factor-κB signaling network: insights from systems approaches, in Cellular Signaling and Innate Immune Responses to RNA Virus Infections (Brasier A. R., Lemon S. M., and Garcia-Sastre A., eds) pp. 119–135, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 44. Brasier A. R. (2006) The NF-κB regulatory network. Cardiovasc. Toxicol. 6, 111–130 10.1385/CT:6:2:111 [DOI] [PubMed] [Google Scholar]
- 45. Birbach A., Gold P., Binder B. R., Hofer E., de Martin R., and Schmid J. A. (2002) Signaling molecules of the NF-κB pathway shuttle constitutively between cytoplasm and nucleus. J. Biol. Chem. 277, 10842–10851 10.1074/jbc.M112475200 [DOI] [PubMed] [Google Scholar]
- 46. Kalita M. K., Sargsyan K., Tian B., Paulucci-Holthauzen A., Najm H. N., Debusschere B. J., and Brasier A. R. (2011) Sources of cell-to-cell variability in canonical nuclear factor-κB (NF-κB) signaling pathway inferred from single cell dynamic images. J. Biol. Chem. 286, 37741–37757 10.1074/jbc.M111.280925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brasier A. R., Tian B., Jamaluddin M., Kalita M. K., Garofalo R. P., and Lu M. (2011) RelA Ser276 phosphorylation-coupled Lys310 acetylation controls transcriptional elongation of inflammatory cytokines in respiratory syncytial virus infection. J. Virol. 85, 11752–11769 10.1128/JVI.05360-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Komiya Y., and Habas R. (2008) Wnt signal transduction pathways. Organogenesis 4, 68–75 10.4161/org.4.2.5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kanazawa A., Tsukada S., Kamiyama M., Yanagimoto T., Nakajima M., and Maeda S. (2005) Wnt5b partially inhibits canonical Wnt/β-catenin signaling pathway and promotes adipogenesis in 3T3-L1 preadipocytes. Biochem. Biophys. Res. Commun. 330, 505–510 10.1016/j.bbrc.2005.03.007 [DOI] [PubMed] [Google Scholar]
- 50. Katoh M., and Katoh M. (2009) Transcriptional mechanisms of WNT5A based on NF-κB, Hedgehog, TGFβ, and Notch signaling cascades. Int. J. Mol. Med. 23, 763–769 [DOI] [PubMed] [Google Scholar]
- 51. Bao J., Zheng J. J., and Wu D. (2012) The structural basis of DKK-mediated inhibition of Wnt/LRP signaling. Sci. Signal. 5, pe22 10.1126/scisignal.2003028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lamouille S., Xu J., and Derynck R. (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bracken C. P., Gregory P. A., Kolesnikoff N., Bert A. G., Wang J., Shannon M. F., and Goodall G. J. (2008) A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 68, 7846–7854 10.1158/0008-5472.CAN-08-1942 [DOI] [PubMed] [Google Scholar]
- 54. Hoot K. E., Oka M., Han G., Bottinger E., Zhang Q., and Wang X. J. (2010) HGF upregulation contributes to angiogenesis in mice with keratinocyte-specific Smad2 deletion. J. Clin. Investig. 120, 3606–3616 10.1172/JCI43304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Whyte W. A., Orlando D. A., Hnisz D., Abraham B. J., Lin C. Y., Kagey M. H., Rahl P. B., Lee T. I., and Young R. (2013) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brown J. D., Lin C. Y., Duan Q., Griffin G., Federation A., Paranal R. M., Bair S., Newton G., Lichtman A., Kung A., Yang T., Wang H., Luscinskas F. W., Croce K., Bradner J. E., et al. (2014) NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell 56, 219–231 10.1016/j.molcel.2014.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tian B., Liu Z., Yang J., Sun H., Zhao Y., Wakamiya M., Chen H., Rytting E., Zhou J., and Brasier A. R. (2018) Selective molecular antagonists of the bronchiolar epithelial NFκB-bromodomain-containing protein 4 (BRD4) pathway in viral-induced airway inflammation. Cell Rep. 23, 1138–1151 10.1016/j.celrep.2018.03.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tian B., Zhao Y., Kalita M., Edeh C. B., Paessler S., Casola A., Teng M. N., Garofalo R. P., and Brasier A. R. (2013) CDK9-dependent transcriptional elongation in the innate interferon-stimulated gene response to respiratory syncytial virus infection in airway epithelial cells. J. Virol. 87, 7075–7092 10.1128/JVI.03399-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu P., Lu M., Tian B., Li K., Garofalo R. P., Prusak D., Wood T. G., and Brasier A. R. (2009) Expression of an IKKγ splice variant determines IRF3 and canonical NF-κB pathway utilization in ssRNA virus infection. PLoS One 4, e8079 10.1371/journal.pone.0008079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 61. Forbus J., Spratt H., Wiktorowicz J., Wu Z., Boldogh I., Denner L., Kurosky A., Brasier R. C., Luxon B., and Brasier A. R. (2006) Functional analysis of the nuclear proteome of human A549 alveolar epithelial cells by HPLC-high resolution 2-D gel electrophoresis. Proteomics 6, 2656–2672 10.1002/pmic.200500652 [DOI] [PubMed] [Google Scholar]
- 62. Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., and Thomas P. D. (2017) PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liao Y., Smyth G. K., and Shi W. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 65. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gould J. Gene-e matrix visualization and analysis platform. http://www.broadinstitute.org/cancer/software/GENE-E/. Accessed July 12, 2016
- 67. Xia J., Gill E. E., and Hancock R. E. (2015) NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 10, 823–844 10.1038/nprot.2015.052 [DOI] [PubMed] [Google Scholar]
- 68. Breuer K., Foroushani A. K., Laird M. R., Chen C., Sribnaia A., Lo R., Winsor GL, Hancock R. E., Brinkman F. S., and Lynn D. J. (2013) InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res. 41, D1228–D1233 10.1093/nar/gks1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tian B., Yang J., and Brasier A. R. (2012) Two-step cross-linking for analysis of protein-chromatin interactions. Methods Mol. Biol. 809, 105–120 10.1007/978-1-61779-376-9_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.