Abstract
ERK3 is an atypical mitogen-activated protein kinase (MAPK) that has recently gained interest for its role in promoting cancer cell migration and invasion. However, the molecular regulation of ERK3 functions in cancer cells is largely unknown. ERK3 has a single phospho-acceptor site (Ser189) in its activation motif rather than the TXY conserved in conventional MAPKs such as ERK1/2. Although dual phosphorylation of the TXY motif is known to be critical for the activation of conventional MAPKs, the role of Ser189 phosphorylation in ERK3 activity and its function in cancer cells remain elusive. In this study, we revealed that activation loop phosphorylation is important for ERK3 in promoting cancer cell invasiveness, as the S189A mutation greatly decreased the ability of ERK3 to promote migration and invasion of lung cancer cells. Interestingly, a catalytically inactive ERK3 mutant was still capable of increasing migration and invasion, although to a lesser extent compared with WT ERK3, suggesting that ERK3 promotes cancer cell invasiveness by both kinase-dependent and kinase-independent mechanisms. To elucidate how the S189A mutation reduces the invasiveness-promoting ability of ERK3, we tested its effect on the kinase activity of ERK3 toward steroid receptor coactivator 3 (SRC3), a recently identified substrate of ERK3 critical for cancer cell invasiveness. Compared with ERK3, ERK3-S189A exhibited a dramatic decrease in kinase activity toward SRC3 and a concomitantly reduced ability to stimulate matrix metalloproteinase expression. Taken together, our study unravels the importance of Ser189 phosphorylation for intramolecular regulation of ERK3 kinase activity and invasiveness-promoting ability in lung cancer cells.
Keywords: extracellular signal–regulated kinase (ERK), cell migration, cell invasion, lung cancer, mitogen-activated protein kinase (MAPK), activation loop phosphorylation
Introduction
Mitogen-activated protein kinases (MAPKs)2 are Ser/Thr kinases that play important roles in signal transduction. They are classified into conventional MAPKs, such as extracellular signal–regulated kinase (ERK) 1/2 and p38 isoforms (α, β, γ, and δ), and atypical MAPKs, such as ERK3 and ERK4. The conventional MAPKs have a highly conserved Thr-XAA-Tyr (TXY) motif in their activation loops, whose dual phosphorylation by an upstream kinase results in a dramatic increase in kinase activity and translocation into the nucleus (1, 2). Unlike the conventional MAPKs, ERK3 has a single phospho-acceptor site, Ser189, in its SEG activation motif. ERK3-Ser189 is constitutively phosphorylated in resting cells and is not affected by common cell stimuli (3). In addition, the subcellular distribution of ERK3 is not affected by phosphorylation of ERK3-Ser189, nor in response to common mitogenic or stress stimuli, including serum, epidermal growth factor, lysophosphatidic acid, arsenite, and hydrogen peroxide (4–6). ERK3-Ser189 phosphorylation can occur in cis as a result of autophosphorylation (4, 7) or in trans by the group I p21-activated protein kinases (PAKs) (8, 9). The phosphorylation of ERK3-Ser189 is reduced by dual-specificity phosphatase 2 (DUSP2) (10).
The atypical MAPK ERK3 has important physiological functions, including pulmonary differentiation, T cell activation, and angiogenesis (11–13). Recently, the role of ERK3 signaling in cancer has been a subject of significant interest. ERK3 increases the migration and invasion of lung cancer cells both in vitro and in vivo (14). The underlying molecular mechanism involves phosphorylation of steroid receptor coactivator 3 (SRC3) at Ser857 by ERK3 (14). SRC3 is overexpressed in several types of cancers, including breast cancer, lung cancer, and prostate cancer (15), and is considered a bona fide oncogene, as it promotes cell proliferation and transformation, cancer cell migration and invasion, as well as tumorigenesis and metastasis in mice (16–18). Phosphorylation of SRC3-Ser857 by ERK3 increases the interaction of SRC3 with the transcription factor PEA3 and subsequently leads to the up-regulation of matrix metalloproteinases (MMPs), enzymes critical for cell invasion by degrading the extracellular matrix (14). Similar to its role in lung cancer cells, ERK3 promotes the migration of breast cancer cells by regulating cell morphology and spreading (19). Furthermore, ERK3 increases the chemoresistance of lung cancer cells to topoisomerase II inhibitors (20). Several mutations (or variants) of ERK3 have been found in cancers, including ERK3-L290P and ERK3-L290V, which were found to confer ERK3 an increased ability to promote migration and invasion of cancer cells (21). In line with its role in promoting the migration and invasion of lung cancer and breast cancer cells, the expression level of ERK3 is up-regulated in multiple types of cancers, including lung cancer, gastric cancer, and oral squamous cell carcinoma (14, 22–24).
In contrast to conventional MAPKs, for which a large number of substrates have been identified, ERK3 has a very restricted substrate specificity, and it does not phosphorylate many generic MAPK substrates in vitro, including c-Jun, Tal1, MyoD, c-Elk, and papilloma virus protein E7 (4, 7). A well-characterized substrate of ERK3 is MAPK-activated protein kinase 5 (MK5) (6, 25). ERK3 phosphorylates MK5 at Thr182, leading to MK5 activation (6, 25). MK5 activation by ERK3 is regularly determined in a coupled kinase assay by measuring the phosphorylation of MK5 peptide substrate or its protein substrates Hsp27 or Hsp25 in the presence of both ERK3 and MK5 kinases (3, 6, 25). Importantly, substrate phosphorylation by MK5 was greatly reduced when incubated with the ERK3-S189A mutant compared with that in the presence of WT ERK3, suggesting that the Ser189 phospho-site is important for ERK3 to activate MK5 (3). It remains to be further elucidated, however, whether Ser189 phosphorylation has a direct regulation on ERK3 enzymatic activity per se, given that Ser189 is required for ERK3 to bind to the substrate MK5 (3, 8). With recent identification of additional ERK3 substrates, including SRC3, it would be interesting to determine whether the Ser189 phospho-site is only involved in substrate binding or also directly regulates the catalytic activity of ERK3. In addition, it is unclear whether ERK3-Ser189 phosphorylation is required for the migration/invasion-promoting function of ERK3 in cancer cells.
In this study, we investigated the importance of Ser189 phosphorylation for ERK3 in promoting cancer cell migration and invasion as well as its role in regulating the catalytic activity of ERK3 by directly measuring phosphorylation of the substrates in an in vitro kinase assay. We found that the S189A mutation significantly reduced the ability of ERK3 to promote lung cancer cell migration and invasion. Interestingly, the kinase-inactive ERK3 mutant was still capable of significantly promoting cell migration and invasion, although the promoting effect was significantly reduced compared with WT ERK3, suggesting that ERK3 promotes cell migration and invasion in both kinase-dependent and kinase-independent manners. The in vitro kinase activity of ERK3 and ERK3-S189A was compared using proteins purified from bacteria or mammalian cells. Interestingly, bacterially expressed recombinant ERK3 and ERK3-S189A proteins showed low but equivalent kinase activity. However, when ERK3 proteins were expressed and purified from 293T mammalian cells, S189A mutation led to a great decrease in the kinase activity of ERK3 toward substrates, including SRC3 and myelin basic protein (MBP). Intriguingly, the S189A mutation does not seem to affect the interaction between ERK3 and SRC3. Consistent with its effect in decreasing the ability of ERK3 to promote cell invasiveness, the S189A mutation significantly diminished the ability of ERK3 to up-regulate the levels of MMP9 and MMP10. Our study demonstrates the importance of activation loop phosphorylation in regulating the kinase activity and cellular functions of ERK3.
Results
Activation loop phosphorylation is important for the migration-promoting ability of ERK3 in lung cancer cells
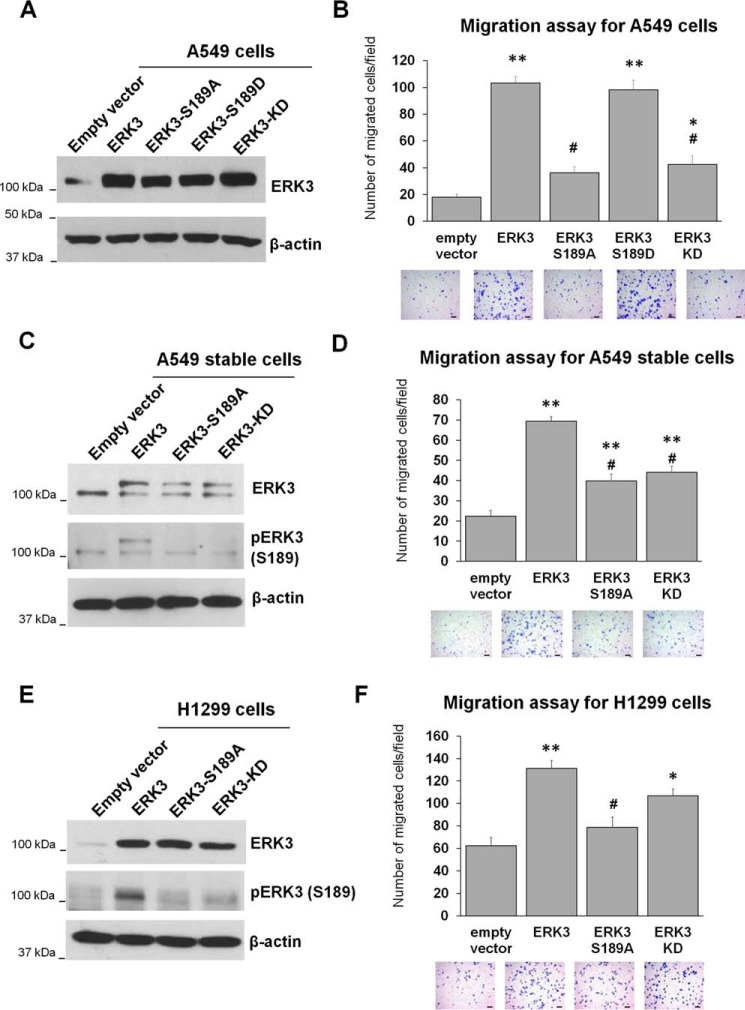
Recent studies have revealed that ERK3 increases the migration of lung cancer and breast cancer cells (14, 19). However, the role of ERK3 activation loop phosphorylation in this process is unclear. Hence, we investigated the effect of mutating the activation loop phosphorylation site on the ability of ERK3 to promote cancer cell migration. As expected, overexpression of ERK3 with an HA tag by transient lentiviral transduction significantly increased the migration of A549 lung cancer cells (Fig. 1, A and B). The kinase-dead (KD) mutant (ERK3-KD) defective in ATP binding showed a much reduced ability to promote the migration of A549 cells. Importantly, similar to the effect of ERK3-KD, the ERK3-S189A mutant had a dramatic decrease in the ability to promote migration of A549 cells compared with that of WT ERK3. However, overexpression of ERK3-S189D, a potential phosphomimic of ERK3 at Ser189, increased the migration of A549 cells at a similar level as that by ERK3 (Fig. 1, A and B). In line with these results obtained by transiently overexpressing HA-ERK3, stable overexpression of myc-tagged ERK3 by lentiviral transduction significantly increased the migration of A549 cells, whereas the myc-ERK3-S189A and myc-ERK3-KD mutants had a significantly reduced ability to promote migration compared with myc-ERK3 (Fig. 1, C and D). Similar results were obtained in another lung cancer cell line, H1299. Although transient overexpression of HA-ERK3 increased the migration of H1299 cells, both the S189A and KD mutants showed a decrease in the number of migrated cells compared with that of ERK3 (Fig. 1, E and F).
Figure 1.
Activation loop phosphorylation is important for the migration-promoting ability of ERK3 in lung cancer cells. A and B, A549 cells were transiently transduced with lentiviruses expressing an empty vector or HA-tagged WT or mutant ERK3 (S189A, S189D, or KD). The overexpression of ERK3 was verified by Western blotting using an anti-ERK3 antibody (A), and migration was assessed by Transwell cell migration assay (B). C and D, A549 cells were stably transduced with lentiviruses expressing an empty vector, myc-ERK3, myc-ERK3-S189A, or myc-ERK3-KD. Overexpression of myc-ERK3 proteins in stable cell lines was verified by Western blotting using anti-ERK3 antibody (C). Phospho-ERK3 (Ser189) was analyzed by immunoprecipitation of ERK3 using an ERK3 Ab, followed by Western blotting using anti-phospho-ERK3 (Ser189) antibody. In both the ERK3 and pERK3 (Ser189) immunoblots, the lower bands represent endogenous ERK3 (or endogenous phospho-ERK3-Ser189), whereas the higher bands represent the overexpressed myc6-tagged ERK3 (or phospho-myc6-ERK3-Ser189) proteins. Cell migration ability was assessed by Transwell migration assay (D). E and F, H1299 cells were transiently transfected with an empty vector or HA-tagged WT or mutant ERK3, as indicated. Two days post-transfection, the cells were lysed and analyzed by Western blotting using an anti-ERK3 antibody and an anti-phospho-ERK3 (Ser189) antibody (E). The migration ability of the cells was determined by Transwell migration assay (F). The quantitated migration ability is presented as the number of migrated cells per field. Values in the bar graphs represent mean ± S.E. (n ≥ 6 fields). **, p < 0.001 (significantly different compared with empty vector); *, p < 0.05 (significantly different compared with empty vector); #, p < 0.001 (significantly different compared with ERK3); one-way ANOVA. Representative images of migrated cells stained with crystal violet are shown below each bar graph. Scale bars, 100 μm.
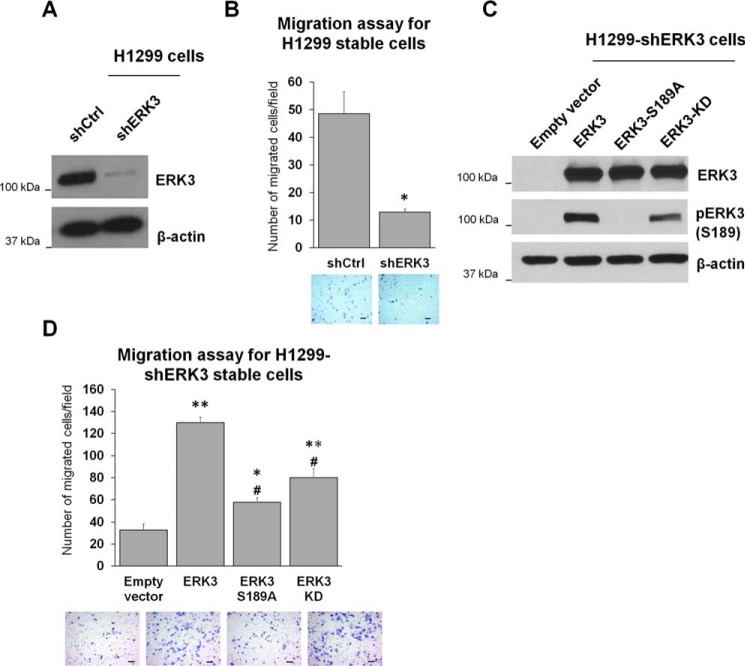
Next, to further confirm the importance of Ser189 phosphorylation and the kinase activity of ERK3 in promoting cancer cell migration, we generated H1299 cells with stable depletion of endogenous ERK3 using an shRNA that targets the 3′ UTR of ERK3 mRNA (Fig. 2A), followed by re-expression of WT ERK3, ERK3-S189A, or ERK3-KD (Fig. 2C). Consistent with our previous finding (14), stable knockdown of ERK3 greatly reduced the migration ability of H1299 cells (Fig. 2B). Importantly, re-expression of WT ERK3 rescued the migration ability of H1299-shERK3 cells (Fig. 2D, compare WT ERK3 with the empty vector control). Similar to their effects in parental H1299 cells, overexpression of ERK3-S189A or ERK3-KD had a significantly reduced ability to stimulate migration of H1299-shERK3 cells compared with that of WT ERK3 (Fig. 2D).
Figure 2.
Activation loop phosphorylation is important for the migration-promoting ability of ERK3 when re-expressed in H1299 cells with depletion of endogenous ERK3. A, generation of H1299 cells with stable expression of shCtrl) or shERK3. Knockdown of ERK3 was verified by Western blot analysis using an anti-ERK3 Ab. B, Transwell migration assay of stable H1299-shCtrl and H1299-shERK3 cells. Quantitated migration ability is presented as the number of migrated cells per field. Values in the bar graph represent mean ± S.E. (n = 6 fields). *, p < 0.001 (significantly different compared with shCtrl); Student's t test. C and D, H1299-shERK3 cells were transiently transfected with an empty vector or HA-tagged WT or mutant ERK3, as indicated. Two days post-transfection, cells were lysed and analyzed by Western blotting using an anti-ERK3 antibody and an anti-phospho-ERK3 (Ser189) antibody (C). Cell migration ability was determined by Transwell migration assay and is presented as the number of migrated cells per field (D). Values in the bar graph represent mean ± S.E. (n ≥ 6 fields). **, p < 0.0001 (significantly different compared with empty vector); *, p < 0.05 (significantly different compared with empty vector); #, p < 0.0001 (significantly different compared with ERK3); one-way ANOVA. For all migration assays, representative images of migrated cells stained with crystal violet are shown below the bar graphs. Scale bars, 100 μm.
Collectively, these results demonstrate that both activation loop phosphorylation and kinase activity are important for ERK3 to promote lung cancer cell migration. Notably, in the experiments described above, although migration induced by the ERK3-KD mutant was significantly less than that of WT ERK3, ERK3-KD still significantly increased cancer cell migration compared with the empty vector control, suggesting that ERK3 may have a kinase-independent role in promoting cancer cell migration.
Activation loop phosphorylation is important for the invasion-promoting ability of ERK3 in lung cancer cells
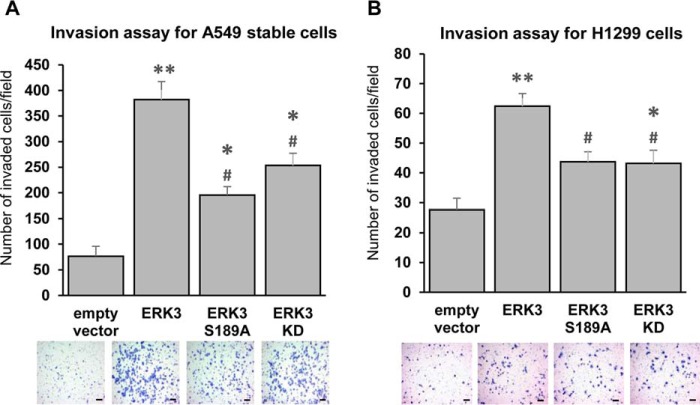
To metastasize, cancer cells leave their primary site and invade through the basement membrane, which comprises an extracellular matrix. Increased migration/motility of cancer cells does not always correspond to an increased invasion ability. Previous in vitro and in vivo studies showed that ERK3 increases the invading capability of lung cancer cells (14). However, the importance of activation loop phosphorylation and kinase activity in the invasion-promoting ability of ERK3 remains unclear. Hence, we compared the invasion ability of A549 lung cancer cells that have stable overexpression of either ERK3-S189A or ERK3-KD with that of A549 cells with stable overexpression of WT ERK3. As expected, A549 cells with stable overexpression of myc-ERK3 showed a significant increase in invasion compared with cells expressing an empty control vector. On the other hand, compared with cells with overexpression of ERK3, A549 cells with stable overexpression of ERK3-S189A or ERK3-KD showed a significant reduction in invasion ability (Fig. 3A). Similarly, overexpression of HA-ERK3 significantly increased the invasion of H1299 cells, whereas ERK3-S189A or ERK3-KD mutants had significantly decreased invasion-promoting ability compared with WT ERK3 (Fig. 3B). Interestingly, similar to its role in promoting cancer cell migration, the ERK3-KD mutant significantly increased the invasion of both A549 and H1299 cells compared with that of cells expressing the empty vector control, suggesting that ERK3 promotes cancer cell invasion in both kinase-dependent and kinase-independent manners. Taken together, these results demonstrate the importance of activation loop phosphorylation and catalytic activity in the ability of ERK3 to promote lung cancer cell invasion.
Figure 3.
Activation loop phosphorylation is important for the invasion-promoting ability of ERK3 in lung cancer cells. A, Transwell Matrigel invasion assay of A549 cells stably expressing an empty vector, WT, or mutant myc-ERK3 as described in Fig. 1C. B, Transwell Matrigel invasion assay of H1299 cells with transient overexpression of an empty vector or WT or mutant HA-ERK3 as described in Fig. 1E. Quantitated invasion ability is presented as the number of invaded cells per field. Values in the bar graphs represent mean ± S.E. (n ≥ 6 fields). **, p < 0.0001 (significantly different compared with empty vector); *, p < 0.05 (significantly different compared with empty vector); #, p < 0.05 (significantly different compared with ERK3); one-way ANOVA. Representative images of invaded cells stained with crystal violet are shown below the bar graphs. Scale bars, 100 μm.
Overexpression of WT or mutant ERK3 does not affect lung cancer cell proliferation
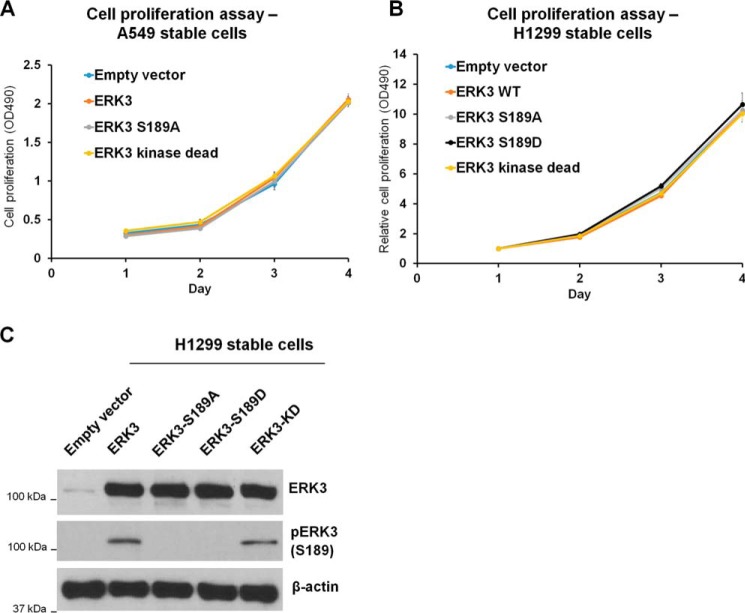
The role of ERK3 in cancer cell proliferation remains elusive. Although some studies showed that ERK3 negatively regulated cell cycle progression in fibroblasts, squamous cell carcinoma, and hepatocarcinoma lines (5, 26, 27), another study showed that knockdown of ERK3 has little effect on lung cancer cell proliferation (14). Hence, we wanted to clarify the role of ERK3 in lung cancer cell growth by determining the effects of overexpression of either WT ERK3 or each of the ERK3 mutants in lung cancer cell proliferation. In A549 cells, stable overexpression of myc-ERK3 showed no significant effect on cell proliferation compared with the empty vector control. In addition, cells with stable overexpression of ERK3-S189A or ERK3-KD mutants proliferated at a similar rate (Fig. 4A). Similarly, stable overexpression of HA-ERK3 did not significantly change H1299 cell proliferation compared with the empty vector control; neither did the ERK3-S189A, ERK3-S189D, or ERK3-KD mutants (Fig. 4, B and C). Therefore, we conclude that, although ERK3 has an important role in promoting lung cancer cell migration and invasion, it does not affect lung cancer cell proliferation.
Figure 4.
Overexpression of WT or mutant ERK3 does not affect lung cancer cell proliferation. A, an MTS cell proliferation assay was performed for A549 cells with stable overexpression of an empty vector or WT or mutant myc-ERK3 as described in Fig. 1C. Cell viability at different time points (days) was measured and expressed as A490. B, an MTS cell proliferation assay was performed for H1299 cells with stable overexpression of an empty vector, WT, or mutant HA-ERK3 as indicated. Cell viability is expressed as A490 normalized to values of day 1 for each condition. C,ERK3 overexpression and ERK3-Ser189 phosphorylation in the generated H1299 stable cells were verified by Western blotting. Values represent mean ± S.D. (n ≥ 3). Statistical analysis was conducted to compare A490 of the different cell lines at each time point by two-way ANOVA. No significant difference was detected between the cell lines at p < 0.05.
Mutation of the phosphorylation site in the activation motif does not affect the in vitro kinase activity of bacterially expressed ERK3
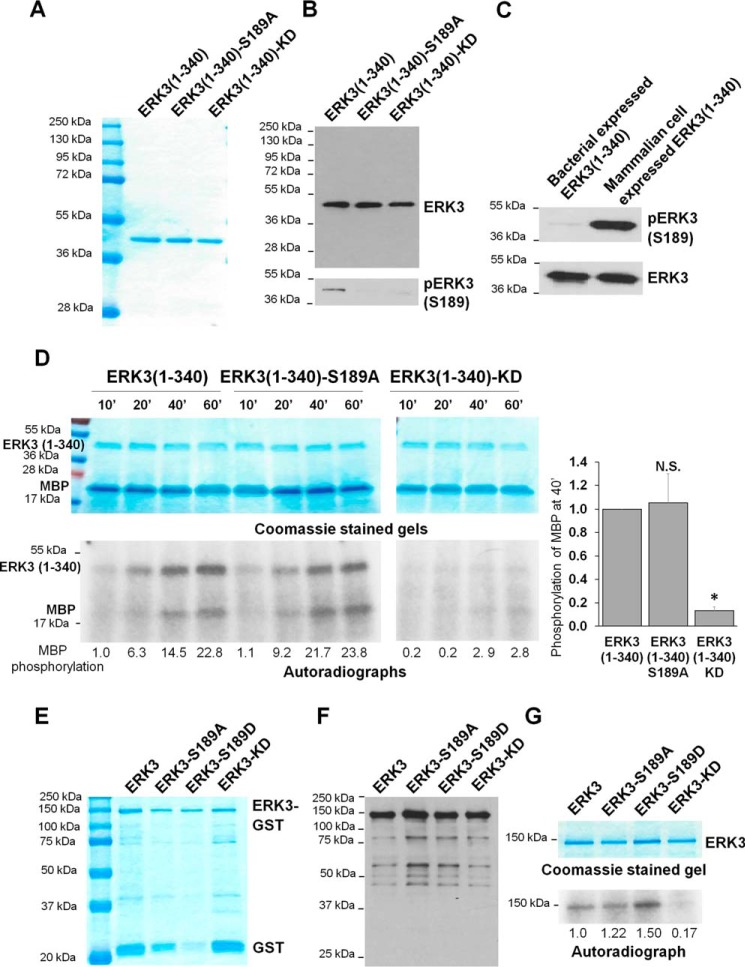
Ser189 phosphorylation has been shown to be important for ERK3 in activating MK5 (3). It remains to be further elucidated, however, whether Ser189 phosphorylation directly regulates ERK3 enzymatic activity per se, given that Ser189 is required for ERK3 to bind to the substrate MK5 (3, 8). To further elucidate this, we expressed and purified recombinant WT or mutant ERK3 proteins from bacteria and compared their kinase activities in vitro.
First, we expressed and purified the kinase domain of ERK3 alone, which lies within the first 340 amino acids of ERK3. His-tagged ERK3(1–340), ERK3(1–340)-S189A, and ERK3(1–340)-KD recombinant proteins were expressed and purified with a high purity and yield (Fig. 5A). Western blot analysis of the purified recombinant proteins confirmed the expression of the proteins and the phosphorylation of ERK3(1–340) at Ser189 (Fig. 5B). However, the level of Ser189 phosphorylation of ERK3(1–340) expressed in bacteria was much lower than that of ERK3(1–340) protein expressed and immunoprecipitated from 293T mammalian cells (Fig. 5C). Predictably, the bacterially expressed ERK3(1–340)-S189A mutant did not display phosphorylation at Ser189; neither did ERK3(1–340)-KD (Fig. 5B). The kinase activity of the purified recombinant ERK3(1–340) proteins was then analyzed by an in vitro kinase assay using MBP as the substrate in the presence of [γ-32P]ATP (Fig. 5D). ERK3(1–340) was catalytically active, as shown by increasing phosphorylation of the substrate MBP and increasing ERK3 autophosphorylation with increasing reaction time. As anticipated, the ERK3(1–340)-KD mutant, which served as a negative control for the kinase assay, neither phosphorylated MBP nor was autophosphorylated. Interestingly, bacterially expressed ERK3(1–340)-S189A was as active as its WT counterpart, as demonstrated by phosphorylation of the substrate MBP and its autophosphorylation. These data demonstrate that the activity of the bacterially expressed ERK3 kinase domain is not affected by activation loop phosphorylation.
Figure 5.
The S189A mutation does not affect the in vitro kinase activity of recombinant ERK3 protein expressed in bacteria. A, purification of recombinant His-ERK3(1–340) proteins expressed in bacteria. The proteins were purified using Ni-NTA resin and analyzed by SDS-PAGE and subsequent Coomassie Blue staining. The molecular mass of protein markers is indicated. B, His-ERK3(1–340) proteins expressed in bacteria were subjected to Western blot analysis using either an anti-ERK3 mAb that targets an N-terminal epitope in ERK3 (top panel) or an anti-phospho-ERK3 (S189) antibody (bottom panel). C, Western blot analysis of His-ERK3(1–340) protein expressed in bacteria and HA-ERK3(1–340) expressed in 293T cells using anti-ERK3 and anti-phospho-ERK3 (Ser189) antibodies. D, in vitro kinase assay of bacterially expressed ERK3(1–340) proteins. 1 μg of each purified protein was incubated with 2 μg of recombinant MBP and [γ-32P]ATP for different times (minutes). The samples were analyzed by SDS-PAGE, followed by Coomassie staining (above) and autoradiography (below). Quantification of MBP phosphorylation by WT or mutant ERK3(1–340) proteins is shown below the autoradiograph. For the purpose of comparison, the normalized phosphorylation level of MBP by WT ERK3(1–340) at 10 min was arbitrarily set as 1.0. Quantification of MBP phosphorylation after kinase reaction for 40 min (40′) is presented on the right. The MBP phosphorylation level by WT ERK3(1–340) was arbitrarily set as 1.0. The bar graph represents the mean ± S.E. of three independent experiments. *, p < 0.05; N.S., not significantly different by one-way ANOVA. E, purification of recombinant His-ERK3-GST proteins expressed in bacteria. The proteins were purified using GSH beads, followed by elution with reduced GSH. The eluted proteins were analyzed by SDS-PAGE, followed by Coomassie Blue staining. F, Western blot analysis of the purified His-ERK3-GST proteins using an anti-ERK3 antibody that targets an N-terminal epitope in ERK3. G, in vitro kinase assay of bacterially expressed recombinant His-ERK3-GST proteins. 350 ng of purified ERK3 protein (WT or mutant) was incubated with [γ-32P]ATP in kinase assay buffer for measuring autophosphorylation. Protein samples were analyzed by SDS-PAGE and Coomassie staining (top) and autoradiography (bottom). Quantification of ERK3 phosphorylation is shown below the autoradiograph; the normalized phosphorylation level of WT ERK3 was arbitrarily set as 1.0.
Next we attempted to express and purify full-length ERK3 from bacteria. Full-length WT or mutant ERK3 recombinant proteins with an N-terminal His6 tag and a C-terminal GST tag were purified, albeit at a lower yield because of marked degradation, which could be expected for larger proteins such as ERK3 (Fig. 5, E and F). The in vitro kinase activity of these bacterially expressed full-length ERK3 proteins toward known ERK3 substrates was barely detectable (data not shown). Because bacteria do not endogenously express ERK3 or its upstream kinase, the phosphorylation of bacterially expressed ERK3 in an in vitro kinase assay is deduced to be mainly catalyzed by ERK3 itself and can represent ERK3 activity. Hence, an in vitro kinase assay was performed to compare the autophosphorylation of WT ERK3 protein with that of Ser189 mutant proteins (Fig. 5G). Autophosphorylation was detected for ERK3 but not for the ERK3-KD mutant. Similar to the results of the assays of purified kinase domain of ERK3, mutation of Ser189 to alanine or aspartate did not change the autophosphorylation of full-length ERK3. This suggests that mutation of Ser189 in the activation loop does not affect the enzymatic activity of bacterially expressed ERK3 and that the Ser189 residue is not the main autophosphorylation site under these conditions.
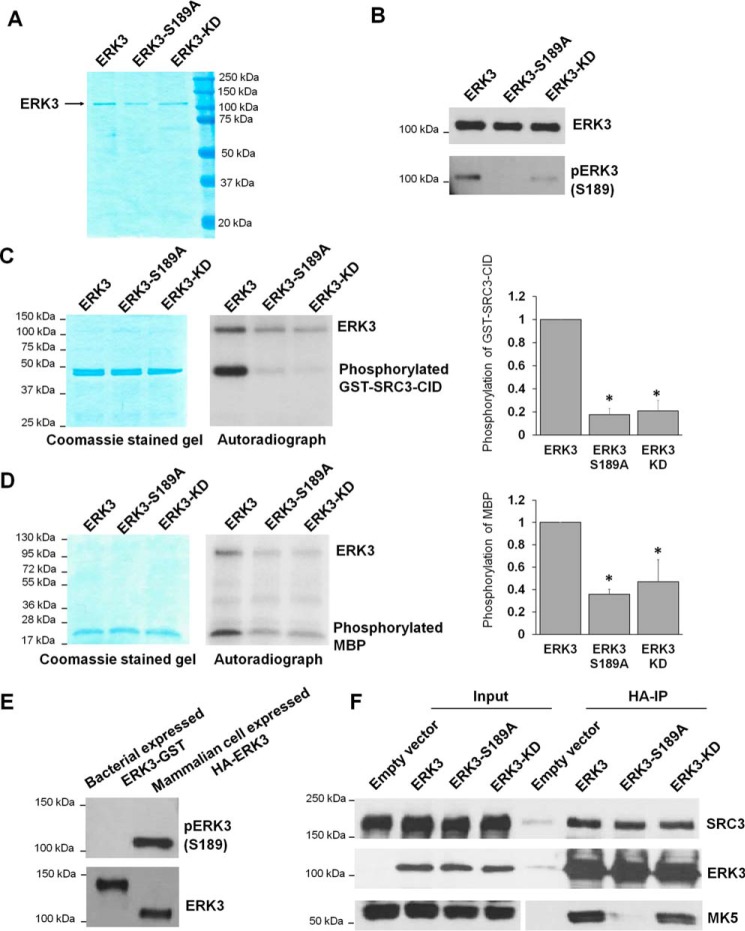
Activation loop phosphorylation is important for in vitro enzymatic activity of ERK3 immunoprecipitated from mammalian cells
Recombinant protein kinases expressed in bacteria display minimal posttranslational modifications, including phosphorylation (28). On the contrary, protein kinases expressed in mammalian cells are subjected to a greater extent of posttranslational modifications that likely regulate their kinase activity. Hence, we decided to examine the importance of Ser189 phosphorylation in the kinase activity of ERK3 by expressing and immunoprecipitating WT and mutant ERK3 proteins from mammalian cells. HA-tagged ERK3 proteins were expressed and immunoprecipitated from 293T cells using HA antibody (Ab)-conjugated agarose beads, followed by elution of the proteins off the beads using HA peptide. Analysis by SDS-PAGE showed that ERK3 proteins migrated on the gels at the expected molecular mass of nearly 100 kDa and were immunoreactive to anti-ERK3 antibody (Fig. 6, A and B). Immunoblotting with an antibody specific for pERK3 (Ser189) revealed that ERK3, but not the S189A mutant, is phosphorylated on the Ser189 residue in 293T cells. As Ser189 can be phosphorylated by other kinase(s), such as PAKs, in mammalian cells, ERK3-KD still had a detectable level of phosphorylation at Ser189, although it was much lower than that of WT ERK3 (Fig. 6B).
Figure 6.
The S189A mutation decreases the in vitro kinase activity of ERK3 protein expressed and immunoprecipitated from 293T cells. A, purification of WT or mutant ERK3 proteins by immunoprecipitation from 293T cells. 293T cells were transfected with the HA-ERK3, HA-ERK3-S189A, or HA-ERK3-KD plasmid, followed by immunoprecipitation of exogenously expressed ERK3 proteins using HA Ab–conjugated agarose beads and elution of the proteins with HA peptide. The purified proteins (300 ng each) were analyzed by SDS-PAGE, followed by Coomassie Blue staining. B, Western blot analysis of proteins purified from mammalian cells using anti-ERK3 and anti-phsopho-ERK3 (Ser189) antibodies. C and D, in vitro kinase assay for WT or mutant HA-ERK3 proteins immunoprecipitated from mammalian cells using GST-SRC3-CID (C) or MBP (D) as substrates. The assays were performed by incubating 100 ng of WT or mutant ERK3, as indicated, together with 1 μg of recombinant protein substrate in the presence of [γ-32P]ATP. Total protein levels of substrates in the reactions are shown by Coomassie staining (left panels). ERK3 proteins are barely seen in the Coomassie-stained gels because of their small amounts (100 ng each). Phosphorylation of the substrates was detected by autoradiography (center panels). Quantification of substrate phosphorylation by WT or mutant ERK3 proteins is shown in the right panels. For the purpose of comparison, the normalized phosphorylation level of substrates by WT ERK3 was arbitrarily set as 1.0. The bar graphs represent the mean ± S.E. of three independent experiments. *, p < 0.05 by one-way ANOVA. E, Western blot analysis of His-ERK3-GST recombinant protein expressed in bacteria and HA-ERK3 protein expressed in 293T cells using anti-phospho-ERK3 (Ser189) and anti-ERK3 antibodies. F, activation loop phosphorylation and kinase activity are not required for interaction of ERK3 with its substrate SRC3 in cells. 293T cells were co-transfected with SRC3 and an HA tag–expressing empty vector, WT HA-ERK3, or mutant HA-ERK3 plasmids, as indicated. Two days post-transfection, cells were lysed, and the interactions of ERK3 with SRC3 and MK5 were analyzed by co-immunoprecipitation using HA Ab–conjugated beads, followed by Western blotting using antibodies against ERK3, SRC3, and MK5.
Following the successful purification of WT and mutant ERK3 proteins from 293T cells, an in vitro kinase assay was performed to compare their kinase activities using MBP or a fragment of SRC3 as a substrate. Previous work showed that ERK3 phosphorylates SRC3 on the Ser189 residue, which locates within the CBP-interacting domain (CID) (14). Hence, we used GST-tagged SRC3-CID as a substrate. Consistent with previous studies, ERK3 efficiently phosphorylated both MBP and SRC3, whereas ERK3-KD showed much reduced activity of phosphorylating these substrates (Fig. 6, C and D). The weak but detectable phosphorylation of substrates by ERK3-KD could be attributed to its weak residual activity and/or the presence of minimal amounts of other kinases in the immunoprecipitates. Interestingly, although mutation of S189A did not affect the kinase activity of bacterially expressed ERK3 (Fig. 5, D and G), ERK3-S189A protein purified from 293T cells, compared with ERK3, had greatly decreased kinase activity toward the substrates MBP and SRC3 and also toward itself (autophosphorylation) (Fig. 6, C and D). Notably, ERK3 expressed in 293T cells was highly phosphorylated at Ser189, whereas Ser189 phosphorylation of ERK3 expressed in bacteria was undetectable (Fig. 6E). In line with the levels of Ser189 phosphorylation, ERK3 expressed in 293T cells (100 ng per reaction) readily phosphorylated MBP and SRC3-CID (Fig. 6, C and 6D), whereas ERK3 expressed in bacteria at an even higher amount (350 ng/reaction) did not exhibit detectable activity toward MBP or SRC-3-CID (data not shown). When expressed in bacteria, ERK3 has low or undetectable level of Ser189 phosphorylation and low kinase activity; hence, mutation of Ser189 to alanine had little effect on the kinase activity of ERK3. Taken together, these results suggest that Ser189 phosphorylation is critical for the kinase activity of ERK3.
Previous studies showed that Ser189 phosphorylation is necessary for the binding of ERK3 to MK5 (3, 8). To test whether Ser189 phosphorylation is similarly important for the binding of ERK3 to its substrate SRC3, we examined the interactions of ERK3 or ERK3-S189A mutant with SRC3 by performing a co-immunoprecipitation experiment (Fig. 6F). 293T cells were co-transfected with the FLAG-SRC3 plasmid in addition to an empty vector or WT or mutant HA-ERK3 plasmids, followed by pulldown of ERK3 proteins using HA Ab–conjugated agarose beads. The interaction of MK5 with ERK3 was also examined as an experimental control. Consistent with previous reports, both ERK3 and ERK3-KD interacted with MK5, whereas mutation of Ser189 to alanine almost abolished this interaction (3, 8). Also, as reported previously, SRC3 was co-immunoprecipitated with ERK3 (14). Although ERK3-S189A had greatly reduced activity in phosphorylating SRC3 (Fig. 6C), it did not show a clear difference in interaction with SRC3, suggesting that activation loop phosphorylation is not required for binding of ERK3 to SRC3 but is likely to have another role in the regulation of ERK3 enzymatic activity. ERK3-KD also showed interaction with SRC3, similar to that of ERK3.
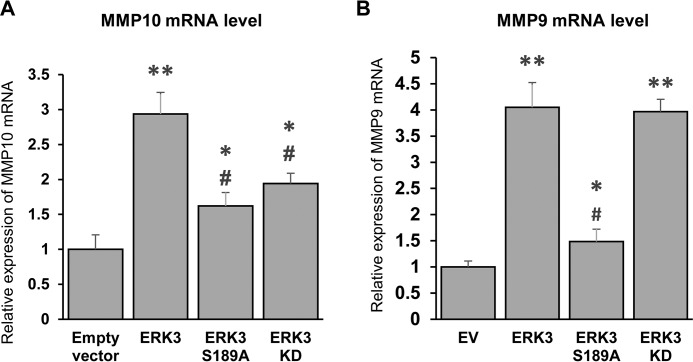
Activation loop phosphorylation is important for ERK3-induced expression of MMP9 and MMP10 genes in lung cancer cells
Previous work has shown that ERK3 increases lung cancer cell invasiveness through phosphorylating SRC3 and up-regulating MMP10 gene expression. The same study showed that ERK3 up-regulates MMP9 expression by an SRC3-independent mechanism (14). However, the importance of ERK3-Ser189 phosphorylation for MMP up-regulation has not been reported. Hence, we compared the effects of overexpression of ERK3 versus ERK3-S189A on MMP9 and MMP10 gene expression in lung cancer cells. In line with previous findings, overexpression of ERK3 increased MMP9 and MMP10 gene expression in A549 cells. Importantly, the S189A mutation significantly decreased the ability of ERK3 to up-regulate both MMP10 and MMP9 (compare ERK3-S189A versus ERK3 in Fig. 7, A and B, respectively). Although the MMP10 mRNA level was significantly lower in cells with ERK3-KD expression versus those with ERK3 expression, ERK3-KD and ERK3 showed a similar ability to up-regulate the MMP9 mRNA level (Fig. 7, A and B), suggesting that up-regulation of MMP9 by ERK3 is independent of its kinase activity.
Figure 7.
The S189 mutation decreases the ability of ERK3 to up-regulate the expression of MMP10 and MMP9 genes. A and B, quantitative RT-PCR analysis of MMP10 (A) and MMP9 (B) gene expression in A549 cells with stable expression of an empty vector, ERK3, ERK3-S189A, or ERK3-KD as described in Fig. 1C. Values in the bar graphs represent mean ± S.D. (n = 3). **, p < 0.0001 (significantly different compared with empty vector); *, p < 0.05 (significantly different compared with empty vector); #, p < 0.005 (significantly different compared with ERK3); one-way ANOVA.
Discussion
Protein kinases are involved in a myriad of cellular processes and signaling pathways; hence, they are typically under tight control. There are various mechanisms that regulate the activity of kinases, including posttranslational modifications, physical interactions with inhibitory or activating protein partners, and changes in cellular localization that alter the availability of substrates and activators (29). The activity of the conventional MAPKs such as ERK1/2 is tightly regulated by dual phosphorylation of the TXY motif in their activation loop. In response to extracellular signals, MAPK kinases phosphorylate conventional MAPKs on threonine and tyrosine residues in the activation loop, resulting in increased activity and nuclear translocation (2). On the other hand, the regulation of the enzymatic activity of the atypical MAPK ERK3 is currently poorly understood. ERK3 has a single phospho-acceptor site (Ser189) in its SEG motif in the activation loop that is constitutively phosphorylated in resting mammalian cells and not affected by many known cell stimuli (3). Also, Ser189 phosphorylation does not alter the subcellular localization of ERK3 in the nucleus (5). Although Ser189 is known to be important for the binding of ERK3 to MK5 and the subsequent activation of MK5 by ERK3 (3), it remains to be further elucidated whether Ser189 directly regulates the kinase activity of ERK3. Recent studies have demonstrated an important role for ERK3 in promoting cancer cell migration and invasion, but it is unclear whether Ser189 phosphorylation regulates the functions of ERK3 in cancer cells.
This work clearly demonstrates that activation loop phosphorylation is important for the ability of ERK3 to promote the migration and invasion of lung cancer cells. This can be attributed to multiple mechanisms. First, it is due to its importance for kinase activity, as demonstrated by the great reduction in kinase activity by S189A mutation in ERK3 expressed and purified from mammalian cells. SRC3 is a substrate of ERK3 and has been shown to be an important downstream mediator of ERK3 function in cancer cells. Phosphorylation of SRC3 by ERK3 leads to up-regulation of MMPs, increasing the invasion of cancer cells (14). Consistently, we found that, compared with ERK3, both ERK3-S189A and a catalytically inactive ERK3-KD mutant had significantly decreased abilities to up-regulate MMP10 gene expression and to promote cancer cell invasion. Also, Ser189 phosphorylation might be important for kinase-independent role of ERK3 in promoting cancer cell invasiveness, given that ERK3-S189A had decreased ability to up-regulate MMP9 gene expression, whereas ERK3-KD showed ability equivalent to that of ERK3. The kinase-independent mechanism by which ERK3 increases MMP9 expression and cell invasiveness remains to be elucidated. Interestingly, a recent study revealed the presence of ERK3, but not ERK3-S189A, in the cell membranes of breast cancer cells (19). Because the membrane localization of ERK3 is potentially important for its ability to induce morphological changes and enhance cell motility (19), this may be another mechanism underlying our finding that Ser189 phosphorylation is important for cancer cell migration. Further studies are needed to elucidate how ERK3 is localized to the cell membrane and regulates cell morphology and motility.
Importantly, our study also reveals that ERK3 possesses a kinase-independent ability to promote lung cancer cell migration and invasion. Although ERK3-KD, compared with ERK3, displayed a reduced ability to promote lung cancer cell migration and invasion, it significantly increased both migration and invasion compared with the empty vector control. Interestingly, the up-regulation of MMP9 expression by ERK3-KD is equivalent to that by WT ERK3, which can be one of the mechanisms underlying the kinase-independent role of ERK3 in promoting cancer cell invasion. In fact, a kinase-independent role for ERK3 in regulating the morphology of breast cancer cells was noted in another study; a catalytically inactive mutant of ERK3 decreased the breast cancer cell spread area comparably with the effect of WT ERK3 (19). In contrast, the catalytic activity of ERK3 is required for the survival and proper differentiation of T cells in the thymus, as knock-in of the ERK3-KD allele recapitulated the phenotype in T cell survival observed in mice with ERK3 deficiency (30). The elucidation of catalytic versus noncatalytic functions of ERK3 would entail greater knowledge about the substrates and the interacting partners of ERK3 in different cellular contexts.
Consistent with previously published work demonstrating that knockdown of ERK3 did not affect lung cancer cell proliferation, we found in this study that lung cancer cell proliferation was not affected by exogenous expression of WT ERK3, the catalytically inactive ERK3 mutant (ERK3-KD), or ERK3 with mutation of the activation loop phosphorylation site (ERK3-S189A and ERK3-S189D). However, some studies have shown an inhibitory role for ERK3 in the proliferation of fibroblasts, squamous carcinoma cells, and hepatocarcinoma cells (5, 26, 27, 31). On the other hand, some studies have demonstrated that ERK3 promotes proliferation of endothelial cells and T cells (12, 13). Taken together, these findings suggest that ERK3 has a differential role in cell proliferation depending on cell type.
In this study, we also attempted to address a gap in the current understanding of the regulation of the kinase activity of ERK3 by activation loop phosphorylation. Study of the mechanisms that regulate the activation of ERK3 has been hampered by insufficient knowledge about the structure and bona fide substrates of ERK3. Until recently, MK5 was the best-characterized downstream target of ERK3. However, the mechanism of MK5 activation by ERK3 is not fully understood, and debates remain whether ERK3 directly phosphorylates MK5 or only enhances its autophosphorylation (6, 25). Mutation of ERK3-S189 to alanine abolishes the interaction between ERK3 and MK5 and, hence, decreases ERK3-mediated MK5 activation (3). However, it is unclear whether Ser189 phosphorylation directly regulates the catalytic activity of ERK3 and whether Ser189 is important for the binding of ERK3 to other substrates. To address these questions, we first expressed and purified the full length or the kinase domain alone of ERK3 proteins with or without mutations of Ser189 from bacteria. Intriguingly, mutation of Ser189 to the nonphosphorylatable alanine in either the full-length ERK3 or the kinase domain alone produced in bacteria did not affect the in vitro kinase activity of ERK3. In contrast, when expressed and immunoprecipitated from mammalian cells, the kinase activity of ERK3 was almost abolished because of the S189A mutation. In addition, the S189A mutation did not affect the interaction of ERK3 with the substrate SRC3. Notably, the kinase activity of ERK3 purified from 293T mammalian cells was much higher than that of bacterially expressed ERK3 protein. One possible reason for this difference in activity is that the phosphorylation level of ERK3 on Ser189 is different in bacteria versus mammalian cells. Indeed, bacterially expressed ERK3 protein had a very low level of Ser189 phosphorylation compared with that of ERK3 expressed in mammalian cells. These results suggest that bacterially expressed ERK3 protein has low kinase activity, at least partly because of a low level of phosphorylation in the activation loop, whereas ERK3 protein expressed in mammalian cells has high kinase activity, as it is highly phosphorylated in the activation loop. Taken together, these findings suggest that Ser189 phosphorylation in the activation loop is critical for the kinase activity of ERK3. Another potential mechanism underlying the regulation of the kinase activity of ERK3 by Ser189 in mammalian cells is that this residue is important for recruiting activator(s) of ERK3. ERK3 protein expressed in bacteria, however, may lack such activator(s). Hence, this Ser189-dependent activation mechanism for ERK3 would not exist in bacteria.
To conclude, this study provides important insights into the regulation of the kinase activity and cell migration/invasion-promoting abilities of ERK3 by activation loop phosphorylation. In addition, we revealed a kinase-independent role for ERK3 in promoting cancer cell migration and invasion. These fundamental findings provide a better understanding of the molecular regulation of ERK3 activity and function in cancer cells, which is critical for endeavors to develop therapeutic agents that target ERK3 signaling in cancer. To better understand ERK3 signaling in cancer, future work is needed to further elucidate the interacting partners that regulate the activity and cellular functions of ERK3, including those that mediate its kinase-independent roles.
Experimental procedures
Cell culture
A549 and H1299 lung cancer cells were maintained in RPMI 1640 growth medium, and 293T human embryonic kidney cells were maintained in Dulbecco's modified Eagle's medium. The growth medium was supplemented with 10% fetal bovine serum and 1% antibiotics (penicillin–streptomycin). All culture media and supplements were purchased from Gibco/Thermo Fisher Scientific.
Expression plasmids
The mammalian expression constructs of WT ERK3, ERK3-S189A, and ERK3-KD with an HA tag at the N terminus (pSG5-HA-ERK3) were described previously (21). ERK3-KD has mutations of K49A/K50A in the ATP binding site of ERK3. pSG5-HA-ERK3 was used as a template for generating the pSG5-HA-ERK3-S189D mutant plasmid by site-directed mutagenesis using the QuikChange II site-directed mutagenesis kit (Agilent Technologies) and the following primer: 5′-ccatttagtaaccaatccttcatcaagatgacccttatgggaataatg.
The lentiviral expression construct of ERK3 with an HA tag at the N terminus (pCDH-HA-ERK3) was generated by PCR amplification of HA-ERK3 using pSG5-HA-ERK3 as a template and the following primers, which have NheI restriction sites: 5′-cggctagcgccaccatggcatatccatatgatg and 5′-cggctagcttagttcagatgtttcagaatgctgctg. The PCR product was inserted into pCR® 2.1-TOPO® TA vector (Invitrogen), followed by NheI digestion of the resultant plasmid and ligation of the HA-ERK3 released fragment into NheI-digested pCDH-CMV-MCS-EF1-Puro (System Biosciences). The lentiviral expression constructs ERK3-S189A, ERK3-S189D, and ERK3-KD were generated similarly by using the appropriate pSG5-HA-ERK3 template plasmids. The lentiviral expression construct ERK3 with six Myc tags at the N terminus (pCDH-Myc6-ERK3) was generated as described previously (14) and used as a template for generating S189A and KD mutant plasmids using the QuikChange II site-directed mutagenesis kit and the primers described previously (21). The nontargeting control shRNA pLKO.1-puro (catalog no. SHC016) and the shRNA that targets a sequence within the 3′ UTR of ERK3 mRNA (SHCLND, clone ID NM_002748.x-3734s1c1) were purchased from Sigma-Aldrich.
The bacterial expression construct ERK3(1–340) with an N-terminal His6 tag, pET-28b(+)-ERK3(1–340), was generated as follows. Complementary DNA encoding amino acids 1–340 of ERK3 was amplified by PCR using pSG5-HA-ERK3 as a template and the following primers with EcoRI sites: 5′-ggaattcggcagagaaatttgaaag and 5′-ggaattcttaatcatcaacttcatcttcaatatg. The PCR product was inserted into the pCR® 2.1-TOPO® TA vector and digested with EcoRI. The released fragment was subcloned into the EcoRI site of the pET-28b(+) plasmid (Novagen). S189A and KD mutants of this plasmid were generated similarly using the appropriate mutant pSG5-HA-ERK3 plasmids as templates. The bacterial expression construct ERK3 with an N-terminal His6 tag and a C-terminal GST tag, pHGST.1 ERK3, was a kind gift from Dr. Sylvain Meloche (Université de Montréal, Canada) (32). This plasmid was used as a template to generate the three mutant plasmids pHGST.1 ERK3-S189A, pHGST.1 ERK3-S189D, and pHGST.1 ERK3-KD by site-directed mutagenesis as described above. The pGEX-4T-1-SRC3-CID plasmid was used for bacterial expression of GST-SRC3-CID, which contains amino acids 841–1080 of SRC3 with a GST tag (14). The mammalian expression plasmid pSG5-FLAG-SRC3 was described previously (14). The sequences of all mutant plasmids generated by site-directed mutagenesis were verified by sequencing.
Generation of stable cell pools
Pseudotyped lentiviral particles were produced in 293T cells by cotransfecting a lentiviral expression construct with Trans-Lentiviral Packaging Plasmid Mix (Open Biosystems). The pseudoviral particles were harvested 48 h after transfection and concentrated using PEG-it virus precipitation solution (System Biosciences), following the manufacturer's instructions. H1299 cells with stable expression of nontargeting control shRNA (shCtrl) or shRNA that targets an UTR in ERK3 mRNA (shERK3) were generated by transducing cells with the appropriate lentivirus in the presence of 5 μg/ml Polybrene. Two days post-transduction, a pool of cells was selected by puromycin (1 μg/ml) for 10 days and used for subsequent experiments. A549 cells with stable overexpression of the control empty vector pCDH or WT or mutant myc6-ERK3 were generated following the same procedure. H1299 cells with stable overexpression of the empty vector, WT HA-ERK3, or the different mutant HA-ERK3 constructs were also generated similarly.
Transient lentiviral transductions and plasmid transfections
A549 cells were transduced with lentiviruses expressing an empty vector pCDH, WT pCDH-HA-ERK3, or mutant pCDH-HA-ERK3 in the presence of 5 μg/ml Polybrene for 2 days. Transient transfections with plasmids were performed using Lipofectamine 3000 reagent (Invitrogen/Thermo Fisher Scientific) or FuGENE HD reagent (Active Motif) following the manufacturers' instructions.
Western blotting
Cells were lysed with EBC lysis buffer (50 mm Tris (pH 7.5), 150 mm NaCl, 0.5% NP-40, 1 mm Complete protease inhibitors (Roche Diagnostics), and 1 mm phosphatase inhibitor mixture III (Sigma-Aldrich)). Western blotting was performed by SDS-PAGE, followed by transferring the proteins onto nitrocellulose membranes and blocking the membranes with 5% nonfat milk in PBS with Tween 20 (PBS-T) for 30 min. Afterward, the membranes were incubated overnight with the primary antibodies at 4 °C, followed by 1-h incubation with the appropriate secondary antibodies at room temperature. The following primary antibodies were used: anti-ERK3 (Abcam, catalog no. ab53277), anti-p-ERK3 (Ser189) generated by our laboratory as described previously (21), anti-MK5 (Cell Signaling Technology, catalog no. D70A10), anti-SRC3 (Cell Signaling Technology, catalog no. 5E11), and anti-β-actin (Sigma-Aldrich, catalog no. A5316). The following secondary antibodies were used: anti-mouse (Bio-Rad, catalog no. 170-6516) and anti-rabbit (Bio-Rad, catalog no. 170-6515). The Western blots were visualized by chemiluminescence (Thermo Fisher). β-Actin was used as a loading control.
Two-chamber Transwell cell migration and invasion assays
Cell migration was analyzed using a modified two chamber Transwell system (BD Biosciences) following the manufacturer's instructions. Cells were detached by trypsin/EDTA, washed once with serum-free medium, and then resuspended in serum-free medium. Complete culture medium with 10% fetal bovine serum was added to each bottom well. 40,000 cells were added in each Transwell insert and allowed to migrate for 16 h (for A549) or 18 h (for H1299 cells) in a 37 °C cell incubator, and then the cells on the upper surface of the insert were removed using cotton swabs. The migrated cells attached on the undersurface were fixed with 4% paraformaldehyde for 15 min and stained with crystal violet solution (0.5% in water) for 10 min. Migrated cells were then photographed and counted under a microscope at ×50 magnification. The cell invasion assay was performed following the same procedures as for the cell migration assay, except that the Transwell inserts were precoated with 1 mg/ml growth factor-reduced Matrigel (BD Biosciences). 60,000 cells were added to each insert and allowed to invade for 16 h (for A549) or 19 h (for H1299 cells).
Cell proliferation assay
The proliferation of stable A549 cells or H1299 cells was determined using the CellTiter 96® AQueous One Solution Cell Proliferation Assay Kit (Promega) following the manufacturer's instructions.
Recombinant protein expression in Escherichia coli
Recombinant WT and mutant His-ERK3(1–340) proteins with an N-terminal His6 tag were purified as follows. The appropriate pET-28b(+) expression plasmids were transformed into E. coli BL-21 (DE3) (Novagen). A single colony of transformed cells was inoculated in Luria broth medium and cultured at 37 °C overnight, followed by 100-fold dilution and further growth at 37 °C until an A600 of 0.6 was reached. Protein synthesis was induced by adding 1 mm isopropyl β-d-1-thiogalactopyranoside for 3 h at 30 °C. The cells were harvested and lysed using BugBuster® protein extraction reagent (Millipore). The clarified lysates were then incubated with Ni-NTA beads, followed by elution of the His-tagged proteins off the beads with imidazole following the manufacturer's conditions (Ni-NTA His ·Bind® resin, Millipore). The eluted proteins were concentrated and further purified using an Amicon Ultra-0.5 centrifugal 10-kDa cutoff filter (Millipore).
Recombinant WT and mutant His-ERK3-GST proteins and GST-SRC3-CID protein were purified as follows. E. coli BL-21 (DE3) were transformed with WT or mutant pHGST.1 ERK3 plasmids or the pGEX-4T-1-SRC3-CID plasmid. A single colony of transformed cells was inoculated in Luria broth medium and cultured at 37 °C overnight. Then the culture was diluted 50-fold and allowed to grow at 37 °C until an A600 of 0.6 was reached. Protein synthesis was induced by addition of 0.4 mm isopropyl β-d-1-thiogalactopyranoside for 6 h at room temperature, and then the cells were harvested and lysed using buffer A (20 mm HEPES (pH 7.6), 150 mm KCL, 10% glycerol, 1 mm DTT, 0.1 mm phenylmethanesulfonyl fluoride and Complete protease inhibitor mixture (Roche)), followed by sonication. The clarified lysates were then incubated with GSH-Sepharose 4B (GE Healthcare), and the proteins were eluted off the beads with reduced GSH. For the eluted ERK3 recombinant proteins, an Amicon Ultra centrifugal 30-kDa cutoff filter (Millipore) was used to concentrate and further purify the proteins. The protein purity was assessed by SDS-PAGE, followed by staining with Coomassie Blue solution (InstantBlue, Expedeon). Recombinant MBP was a kind gift from Dr. Yong-jie Xu (Wright State University).
Immunoprecipitation of ERK3 protein from mammalian cells
WT or mutant HA-tagged ERK3 proteins were purified from mammalian cells as described previously (14). The appropriate pSG5-HA-ERK3 plasmids were transfected in 293T cells. Two days post-transfection, cells were lysed, and the protein lysate supernatants were incubated with anti-HA affinity agarose beads (Sigma-Aldrich, catalog no. E6779) for 3 h. After washing the beads, the proteins were eluted using HA peptide (Sigma-Aldrich). The protein purity was assessed by SDS-PAGE, followed by staining with Coomassie Blue solution (InstantBlue, Expedeon).
In vitro kinase assay
Each in vitro kinase assay reaction contained ERK3 protein, a substrate, 5 μCi [γ-32P]ATP (PerkinElmer Life Sciences), and 25 μm cold ATP. The amounts of ERK3 and each substrate are indicated for each experiment. The reactions were carried out at 30 °C for 30 min (except for Fig. 5D) and then stopped by SDS sample buffer and boiling. Proteins were resolved by SDS-PAGE, stained with Coomassie Blue solution, and visualized by autoradiography. Quantification of substrate phosphorylation is determined by calculating the ratio of the band intensity of phosphorylated substrate in the autoradiograph over that of the corresponding total substrate protein in the Coomassie-stained gel.
Co-immunoprecipitation
293T cells were transfected with plasmids. Two days post-transfection, the cells were lysed with EBC lysis buffer as described under “Western blotting.” The protein lysate supernatant was precleared for 1 h using protein A affinity gel beads (Sigma-Aldrich, catalog no. P6486), and then HA-tagged ERK3 proteins were immunoprecipitated using anti-HA affinity agarose beads (Sigma-Aldrich, catalog no. E6779) for 3 h. Afterward, the beads were washed, and the proteins were boiled off the beads in SDS sample buffer. The proteins were resolved by SDS-PAGE and Western blotting as described before. 1% of the amount of protein supernatant for immunoprecipitation was loaded as the input control.
RNA extraction and quantitative RT-PCR
For gene expression analysis, total RNA was extracted from cells using TRIzol reagent (Ambion), and reverse transcription was carried out using SuperScript VILO Master Mix (Invitrogen) according to the manufacturer's protocol. Quantitative PCR was performed using the TaqMan probe system (Roche Diagnostics) on the Applied Biosystems 7500 with glyceraldehyde-3-phosphate dehydrogenase as the internal control. Relative expression to the normalizer sample was calculated using the ΔΔCT method.
Statistics
Data are expressed as mean ± S.D. or S.E., as specified in the figure legends. All experiments were repeated at least three times, and a representative figure is presented. Statistical significance was determined by two-sided Student's t test, one-way analysis of variance (ANOVA), or two-way ANOVA, as indicated in each figure legend, and a p value of less than 0.05 was considered statistically significant.
Author contributions
L. E. data curation; L. E., H. A., and M. M. investigation; L. E. methodology; L. E. and W. L. writing-original draft; L. E., H. A., M. M., and W. L. writing-review and editing; W. L. conceptualization; W. L. resources; W. L. supervision.
This work was supported by a start-up fund of Wright State University and NCI, National Institutes of Health grant 1R01CA193264-01 (to W. L.) and by the Biomedical Sciences Ph.D. Program of Wright State University (to L. E.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal–regulated kinase
- PAK
- p21-activated protein kinase
- MMP
- matrix metalloproteinase
- MBP
- myelin basic protein
- HA
- hemagglutinin
- KD
- kinase-dead
- shRNA
- short hairpin RNA
- GST
- glutathione S-transferase
- Ab
- antibody
- CID
- CBP-interacting domain
- Ni-NTA
- nickel-nitrilotriacetic acid
- ANOVA
- analysis of variance
- MTS
- [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt].
References
- 1. Cargnello M., and Roux P. P. (2011) Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol. Biol. Rev. 75, 50–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raman M., Chen W., and Cobb M. H. (2007) Differential regulation and properties of MAPKs. Oncogene 26, 3100–3112 10.1038/sj.onc.1210392 [DOI] [PubMed] [Google Scholar]
- 3. Déléris P., Rousseau J., Coulombe P., Rodier G., Tanguay P. L., and Meloche S. (2008) Activation loop phosphorylation of the atypical MAP kinases ERK3 and ERK4 is required for binding, activation and cytoplasmic relocalization of MK5. J. Cell Physiol. 217, 778–788 10.1002/jcp.21560 [DOI] [PubMed] [Google Scholar]
- 4. Cheng M., Boulton T. G., and Cobb M. H. (1996) ERK3 is a constitutively nuclear protein kinase. J. Biol. Chem. 271, 8951–8958 10.1074/jbc.271.15.8951 [DOI] [PubMed] [Google Scholar]
- 5. Julien C., Coulombe P., and Meloche S. (2003) Nuclear export of ERK3 by a CRM1-dependent mechanism regulates its inhibitory action on cell cycle progression. J. Biol. Chem. 278, 42615–42624 10.1074/jbc.M302724200 [DOI] [PubMed] [Google Scholar]
- 6. Seternes O. M., Mikalsen T., Johansen B., Michaelsen E., Armstrong C. G., Morrice N. A., Turgeon B., Meloche S., Moens U., and Keyse S. M. (2004) Activation of MK5/PRAK by the atypical MAP kinase ERK3 defines a novel signal transduction pathway. EMBO J. 23, 4780–4791 10.1038/sj.emboj.7600489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coulombe P., and Meloche S. (2007) Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim. Biophys. Acta 1773, 1376–1387 10.1016/j.bbamcr.2006.11.001 [DOI] [PubMed] [Google Scholar]
- 8. De la Mota-Peynado A., Chernoff J., and Beeser A. (2011) Identification of the atypical MAPK Erk3 as a novel substrate for p21-activated kinase (Pak) activity. J. Biol. Chem. 286, 13603–13611 10.1074/jbc.M110.181743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Déléris P., Trost M., Topisirovic I., Tanguay P. L., Borden K. L., Thibault P., and Meloche S. (2011) Activation loop phosphorylation of ERK3/ERK4 by group I p21-activated kinases (PAKs) defines a novel PAK-ERK3/4-MAPK-activated protein kinase 5 signaling pathway. J. Biol. Chem. 286, 6470–6478 10.1074/jbc.M110.181529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perander M., Al-Mahdi R., Jensen T. C., Nunn J. A., Kildalsen H., Johansen B., Gabrielsen M., Keyse S. M., and Seternes O. M. (2017) Regulation of atypical MAP kinases ERK3 and ERK4 by the phosphatase DUSP2. Sci. Rep. 7, 43471 10.1038/srep43471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Klinger S., Turgeon B., Lévesque K., Wood G. A., Aagaard-Tillery K. M., and Meloche S. (2009) Loss of Erk3 function in mice leads to intrauterine growth restriction, pulmonary immaturity, and neonatal lethality. Proc. Natl. Acad. Sci. U.S.A. 106, 16710–16715 10.1073/pnas.0900919106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marquis M., Boulet S., Mathien S., Rousseau J., Thébault P., Daudelin J. F., Rooney J., Turgeon B., Beauchamp C., Meloche S., and Labrecque N. (2014) The non-classical MAP kinase ERK3 controls T cell activation. PLoS ONE 9, e86681 10.1371/journal.pone.0086681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang W., Bian K., Vallabhaneni S., Zhang B., Wu R. C., O'Malley B. W., and Long W. (2014) ERK3 promotes endothelial cell functions by upregulating SRC-3/SP1-mediated VEGFR2 expression. J. Cell Physiol. 229, 1529–1537 10.1002/jcp.24596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Long W., Foulds C. E., Qin J., Liu J., Ding C., Lonard D. M., Solis L. M., Wistuba I. I., Qin J., Tsai S. Y., Tsai M. J., and O'Malley B. W. (2012) ERK3 signals through SRC-3 coactivator to promote human lung cancer cell invasion. J. Clin. Invest. 122, 1869–1880 10.1172/JCI61492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yan J., Tsai S. Y., and Tsai M. J. (2006) SRC-3/AIB1: transcriptional coactivator in oncogenesis. Acta Pharmacol. Sin. 27, 387–394 10.1111/j.1745-7254.2006.00315.x [DOI] [PubMed] [Google Scholar]
- 16. Kuang S. Q., Liao L., Zhang H., Lee A. V., O'Malley B. W., and Xu J. (2004) AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res. 64, 1875–1885 10.1158/0008-5472.CAN-03-3745 [DOI] [PubMed] [Google Scholar]
- 17. Torres-Arzayus M. I., Font de Mora J., Yuan J., Vazquez F., Bronson R., Rue M., Sellers W. R., and Brown M. (2004) High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell 6, 263–274 10.1016/j.ccr.2004.06.027 [DOI] [PubMed] [Google Scholar]
- 18. Yan J., Erdem H., Li R., Cai Y., Ayala G., Ittmann M., Yu-Lee L. Y., Tsai S. Y., and Tsai M. J. (2008) Steroid receptor coactivator-3/AIB1 promotes cell migration and invasiveness through focal adhesion turnover and matrix metalloproteinase expression. Cancer Res. 68, 5460–5468 10.1158/0008-5472.CAN-08-0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Al-Mahdi R., Babteen N., Thillai K., Holt M., Johansen B., Wetting H. L., Seternes O. M., and Wells C. M. (2015) A novel role for atypical MAPK kinase ERK3 in regulating breast cancer cell morphology and migration. Cell Adh. Migr. 9, 483–494 10.1080/19336918.2015.1112485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bian K., Muppani N. R., Elkhadragy L., Wang W., Zhang C., Chen T., Jung S., Seternes O. M., and Long W. (2016) ERK3 regulates TDP2-mediated DNA damage response and chemoresistance in lung cancer cells. Oncotarget 7, 6665–6675 10.18632/oncotarget.6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alsaran H., Elkhadragy L., Shakya A., and Long W. (2017) L290P/V mutations increase ERK3's cytoplasmic localization and migration/invasion-promoting capability in cancer cells. Sci. Rep. 7, 14979 10.1038/s41598-017-15135-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kostenko S., Dumitriu G., and Moens U. (2012) Tumour promoting and suppressing roles of the atypical MAP kinase signalling pathway ERK3/4-MK5. J. Mol. Signal 7, 9 10.1186/1750-2187-7-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rai R., Mahale A., and Saranath D. (2004) Molecular cloning, isolation and characterisation of ERK3 gene from chewing-tobacco induced oral squamous cell carcinoma. Oral Oncol. 40, 705–712 10.1016/j.oraloncology.2004.01.010 [DOI] [PubMed] [Google Scholar]
- 24. Liang B., Wang S., Zhu X. G., Yu Y. X., Cui Z. R., and Yu Y. Z. (2005) Increased expression of mitogen-activated protein kinase and its upstream regulating signal in human gastric cancer. World J. Gastroenterol. 11, 623–628 10.3748/wjg.v11.i5.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schumacher S., Laass K., Kant S., Shi Y., Visel A., Gruber A. D., Kotlyarov A., and Gaestel M. (2004) Scaffolding by ERK3 regulates MK5 in development. EMBO J. 23, 4770–4779 10.1038/sj.emboj.7600467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crowe D. L. (2004) Induction of p97MAPK expression regulates collagen mediated inhibition of proliferation and migration in human squamous cell carcinoma lines. Int. J. Oncol. 24, 1159–1163 [PubMed] [Google Scholar]
- 27. Xiang Z., Wang S., and Xiang Y. (2014) Up-regulated microRNA499a by hepatitis B virus induced hepatocellular carcinogenesis via targeting MAPK6. PLoS ONE 9:e111410, 10.1371/journal.pone.0111410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Terpe K. (2006) Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 72, 211–222 10.1007/s00253-006-0465-8 [DOI] [PubMed] [Google Scholar]
- 29. Endicott J. A., Noble M. E., and Johnson L. N. (2012) The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 81, 587–613 10.1146/annurev-biochem-052410-090317 [DOI] [PubMed] [Google Scholar]
- 30. Marquis M., Daudelin J. F., Boulet S., Sirois J., Crain K., Mathien S., Turgeon B., Rousseau J., Meloche S., and Labrecque N. (2014) The catalytic activity of the mitogen-activated protein kinase extracellular signal-regulated kinase 3 is required to sustain CD4+ CD8+ thymocyte survival. Mol. Cell Biol. 34, 3374–3387 10.1128/MCB.01701-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ling S., Xie H., Yang F., Shan Q., Dai H., Zhuo J., Wei X., Song P., Zhou L., Xu X., and Zheng S. (2017) Metformin potentiates the effect of arsenic trioxide suppressing intrahepatic cholangiocarcinoma: roles of p38 MAPK, ERK3, and mTORC1. J. Hematol. Oncol. 10, 59 10.1186/s13045-017-0424-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Coulombe P., and Meloche S. (2002) Dual-tag prokaryotic vectors for enhanced expression of full-length recombinant proteins. Anal. Biochem. 310, 219–222 10.1016/S0003-2697(02)00319-6 [DOI] [PubMed] [Google Scholar]