Abstract
RNF144A is a single-pass transmembrane RBR E3 ligase that interacts with and degrades cytoplasmic DNA-PKcs, which is an epidermal growth factor receptor (EGFR)-interacting partner. Interestingly, RNF144A expression is positively correlated with EGFR mRNA and protein levels in several types of cancer. However, the relationship between RNF144A and EGFR is poorly understood. This study reports an unexpected role for RNF144A in the regulation of EGF/EGFR signaling and EGF-dependent cell proliferation. EGFR ligands, but not DNA-damaging agents, induce a DNA-PKcs-independent interaction between RNF144A and EGFR. RNF144A promotes EGFR ubiquitination, maintains EGFR protein, and prolongs EGF/EGFR signaling during EGF stimulation. Moreover, depletion of RNF144A by multiple independent approaches results in a decrease in EGFR expression and EGF/EGFR signaling. RNF144A knockout cells also fail to mount an immediate response to EGF for activation of G1/S progression genes. Consequently, depletion of RNF144A reduces EGF-dependent cell proliferation. These defects may be at least in part due to a role for RNF144A in regulating EGFR transport in the intracellular vesicles during EGF treatment.
Keywords: E3 ubiquitin ligase, ubiquitylation (ubiquitination), epidermal growth factor receptor (EGFR), vesicles, trafficking, DNA-dependent serine/threonine protein kinase (DNA-PK), RBR
Introduction
RNF144A is a less characterized member of the ring between ring fingers (RBR)3 family proteins that possess ubiquitin E3 ligase activities (1–5). Most, if not all, well-characterized RBR E3 ligases, such as Parkin and HOIP, play important roles in many cellular functions and pathogenesis of human diseases. It has been shown that several self-regulating or autoinhibitory mechanisms can precisely control their physiological functions (1, 6–9). Among 12 human RBR family members, RNF144A along with RNF144B, RNF19A/Dorfin, RNF19B, and RNF217, contain one or two transmembrane (TM) domains, and may have other unique physiological functions and regulation mechanisms (4). Indeed, RNF144A has a specific self-regulation and activation mechanism through its RBR-TM(GXXXG) superstructure (10). The single TM domain of RNF144A also makes it localized mainly in the membranes and transported between the cell surface and the membranes of intracellular vesicles including endosomes, lysosomes, endoplasmic reticulum, perinuclear vesicles, and others (11). The membrane localization and intracellular transport suggest that RNF144A may be involved in regulation of receptors and their signaling. Moreover, RNF144A is the first identified mammalian E3 ligase for DNA-dependent protein kinase, catalytic subunit (DNA-PKcs), which is an interacting partner of the epidermal growth factor receptor (EGFR) and also plays roles in cell metabolism and proliferation (11–14). However, the relationship between RNF144A and the EGF/EGFR signaling and the EGF-dependent cell proliferation remains unexplored.
EGFR belongs to the EGFR family of receptor tyrosine kinases (15). Containing one TM domain, EGFR is mainly localized on the cell membrane and transported between different subcellular organelles to carry out its physiological functions or undergo degradation. Binding of EGFR ligands such as EGF, transforming growth factor-α (TGF-α), and others will initiate EGFR dimerization, autophosphorylation, ubiquitination, activation, and endocytosis (16–18). Activated cell-surface EGFR is internalized and sorted in the early endosomes or macropinosomes (19, 20), where the receptor is either recycled back to the cell surface or degraded through the multivesicular bodies-lysosome axis pathway (19).
During EGF/EGFR signaling, ligand-activated EGFR recruits several initiators of major signal pathways including GRB2 for the mitogen-activated protein kinase pathway, GAB1 for the phosphoinositide 3-kinase pathway, and others at the cell surface. These major signaling pathways lead to many essential cellular functions including migration, proliferation, differentiation, and others (16–18). Moreover, EGF binding also activates the expression of the EGFR gene itself (21–23). Activated EGFR can also be transported from the cell surface into the nucleus and directly act as a transcription coregulator to activate CCND1 (24) and MYBL2 (25) and other genes for the G1/S phase progression and cell proliferation. Additionally, EGFR may have other yet to be determined roles in different subcellular organelles. For example, the mitochondrial EGFR can phosphorylate mitochondrial cytochrome c oxidase subunit II (CoxII) and regulate mitochondria function (26). Endosome-activated EGFR can promote cell proliferation and survival (27) or induce cell apoptosis (28). Thus, in addition to surface EGFR, the cytosolic active EGFR also contributes to EGFR cellular functions. However, more research is needed to fully understand EGFR regulation. Furthermore, EGFR can interact with DNA-PKcs and promote DNA-PK-dependent error-prone nonhomologous end-joining (NHEJ) DNA repair upon radiation treatment (29, 30). Hence, aberrant EGFR activity is usually involved in the pathogenesis of human cancer, and is a target for cancer therapy (15). Certainly, a better understanding of the complex regulation of EGFR activity and downstream signaling will benefit both basic scientific knowledge and therapy options for cancer patients.
In this study, the functional roles of RNF144A in the regulation of the EGF/EGFR signaling were investigated using various biochemical and cell biological approaches. Multiple lines of evidence support that RNF144A interacts with EGFR and promotes its signaling. First, EGFR ligands, but not DNA damaging agents, induce the cytosolic RNF144A-EGFR interaction. This interaction requires the RNF144A TM domain that is important for RNF144A activation (10). Furthermore, RNF144A promotes ubiquitination of EGFR and regulates its transport in the intracellular vesicles. This mechanism is important for sustaining EGFR protein level and EGF/EGFR signaling after EGF treatment. Consequently, RNF144A promotes EGF-dependent cell cycle gene activation and proliferation. Taken together, these findings reveal a novel role for RNF144A in the regulation of the EGF-dependent cell proliferation and sustaining EGF/EGFR signaling.
Results
EGF induces colocalization of RNF144A and EGFR in the intracellular vesicles
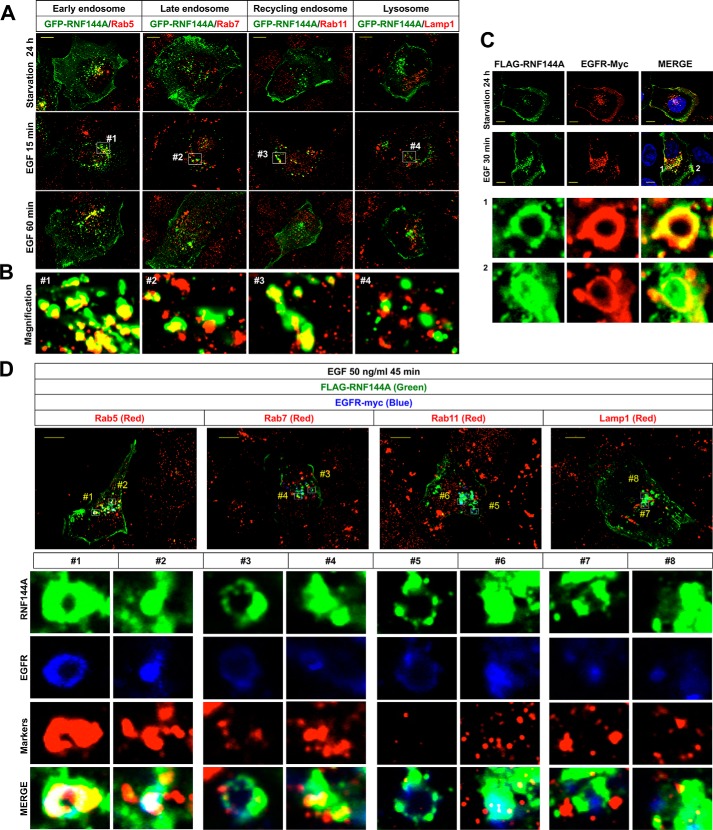
To investigate the relationship between RNF144A and EGF/EGFR signaling, we examined subcellular localization of RNF144A after EGF stimulation (Fig. 1A). Consistent with previous work on RNF144A subcellular localization (10, 11), RNF144A is localized to the plasma membrane and subcellular vesicles under serum starvation (Fig. 1A, top panel). Interestingly, EGF stimulation caused a rapid change in RNF144A localization between the plasma membrane and the cytoplasm. RNF144A moved into the cytoplasm (Fig. 1A, middle panel) at 15 min after EGF stimulation and then moved again to the plasma membrane at 60 min after EGF stimulation (Fig. 1A, bottom panel). We further showed that RNF144A is mainly colocalized with Rab5 (early endosomes), Rab7 (late endosomes), and Rab11 (recycling endosomes) under EGF stimulation (Fig. 1B). Thus, there is dynamic relocalization of RNF144A upon EGF stimulation.
Figure 1.
EGF stimulation induces colocalization of RNF144A and EGFR in the intracellular vesicles. A and B, immunofluorescence analysis (×100) shows that EGF stimulation resulted in the transport of RNF144A between the plasma membrane and the intracellular vesicles. U2OS cells were starved in serum-free, 0.1% BSA-containing medium for 24 h and then stimulated with EGF (50 ng/ml). Yellow bars, 10 μm (A). High magnification of RNF144A and EGFR colocalization in different intracellular vesicles (white blank boxes from EGF treated for 15 min) (B). C, immunofluorescence analysis (×100) shows that EGF dramatically induced colocalization of FLAG-tagged RNF144A (green) and EGFR (red) in the intracellular vesicles of U2OS cells. The bottom panel shows high magnification of the colocalization of RNF144A and EGFR from the white blank boxes 1 and 2 in 30 min EGF stimulation in the merged image. Yellow bars, 10 μm. D, immunofluorescence analysis (×100) shows that EGF significantly induced colocalization of FLAG-tagged RNF144A (green) and EGFR (blue) in intracellular Rab5-containing vesicles (red) in U2OS cells. Yellow bars, 10 μm. The bottom panel shows high magnification of the colocalization of RNF144A, EGFR, and different intracellular vesicle markers from the white open boxes 1–8.
We also demonstrated that RNF144A is colocalized with EGFR in the plasma membrane and intracellular vesicles (Fig. 1C and Fig. S1). Furthermore, EGF stimulation also resulted in colocalization of RNF144A and EGFR in the big ring-shaped vesicles (Fig. 1C, bottom panel, and Fig. 4F). We characterized these intracellular big ring-shaped vesicles as predominantly vesicles containing Rab5 protein (Fig. 1D, #1). Notably, RNF144A and EGFR are also colocalized to intracellular nonring shape dots (Fig. 1D, #2, #4, #6, and #8) and transported together in the cytoplasm (Movie S1). These results indicate that EGF stimulation induces colocalization of RNF144A and EGFR in different intracellular vesicles, and suggest a possible relationship between RNF144A and EGFR upon EGF stimulation.
Figure 4.
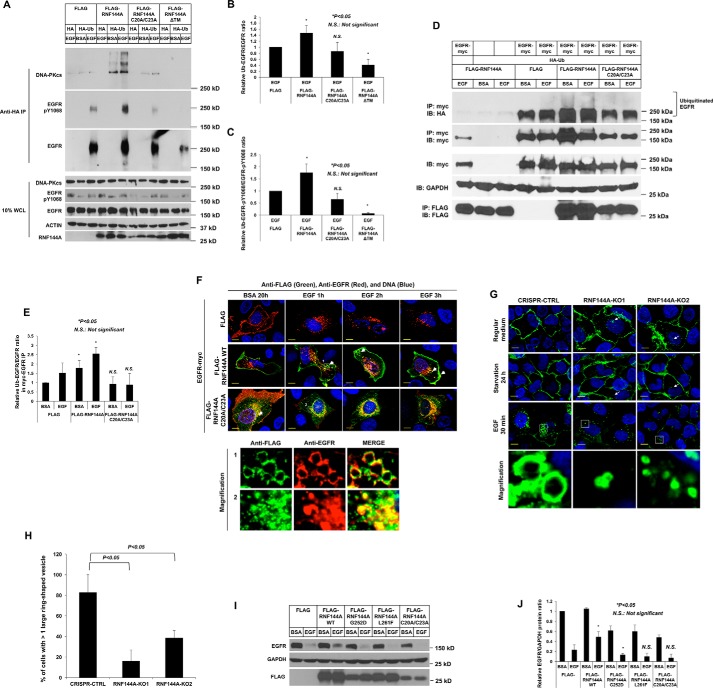
RNF144A promotes ubiquitination of EGFR. A–E, Western blot analysis shows that overexpression of WT, but not a ligase-dead mutant RNF144A(C20A/C23A) or ΔTM mutant, could promote mono- and poly-ubiquitination of EGFR after EGF stimulation. Transfected cells were pretreated with proteasome inhibitor, MG132, and lysosome inhibitor, CQ (A) or MG132 alone (D), for 30 min prior to EGF treatment. B and C are quantifications of the Ub-EGFR/EGFR ratio and Ub-EGFR-Tyr(P)1068/EGFR-Tyr(P)1068 ratio from three independent experiments. A representative set of data are shown in A. E is quantification of the Ub-EGFR/EGFR ratio from D. F, immunofluorescence analysis (×100) shows that overexpression of WT, but not ligase-dead mutant (C20A/23A), RNF144A induced colocalization of RNF144A-EGFR in both intracellular small and big ring-shaped vesicles (white arrows) upon EGF treatment in U2OS cells. The bottom panel shows a high magnification of colocalization of RNF144A and EGFR in these intracellualr ring-shaped vesicles (1) or corresponding controls (2) (white blank box from EGF treatment for 1 h). Yellow bars: 10 μm. G and H, knockout of RNF144A reduces EGF-induced formation of the intracellular big ring-shaped EGFR vesicles. G, fluorescence analysis (×100) shows EGF treatment significantly induced big ring-shaped EGFP-EGFR positive vesicles in the CRISPR-CTRL cells, but not in two RNF144A-depleted cells. Yellow bars: 10 μm. H, quantification of the number of the cells with more than one big ring-shaped EGFP-EGFR positive vesicles. 200 EGFP-EGFR positive cells from each group in G were scored. *, p value <0.05. I, Western blot analysis shows that overexpression of RNF144A mutants (but not WT) decreased EGF-stimulated EGFR protein levels. J, quantitation of the EGFR/GAPDH ratio from I; quantification data represent mean ± S.D. from three biological replicates. *, p < 0.05; N.S., not significant. HA, HA-tagged empty vector; HA-Ub, HA-tagged ubiquitin; FLAG, FLAG-tagged control empty vector in FLAG-tagged proteins overexpression experiments. BSA, 0.1% BSA in serum-free medium. EGF, EGF stimulation.
EGFR ligands induce a dynamic interaction between RNF144A and EGFR
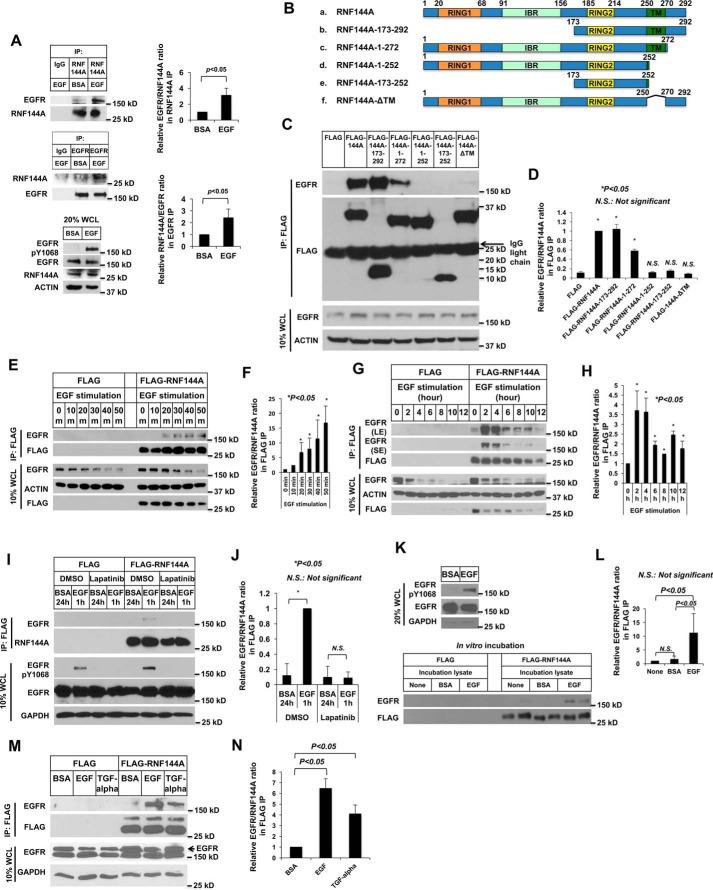
In view of the EGF-induced colocalization of RNF144A-EGFR in different intracellular vesicles (Fig. 1), we then investigated whether RNF144A interacted with EGFR, particularly after EGF stimulation (Fig. 2). Indeed, EGF stimulation significantly promoted endogenous RNF144A-EGFR interaction (Fig. 2A). In addition, RNF144A was able to interact with EGFR through its C terminus (amino acids 173–292), and its TM domain was required for this interaction (Fig. 2, B–D). We also examined the kinetic of RNF144A-EGFR association. The complex was induced after 20 min of EGF stimulation, sustained for up to 4 h, and then decreased (Fig. 2, E–H). Moreover, treatment with a dual EGFR/HER2 kinase inhibitor, Lapatinib, blocked the EGF-dependent RNF144A-EGFR interaction, supporting that EGFR kinase activity is required for RNF144A-EGFR interaction (Fig. 2, I–J). In addition, an in vitro association assay confirmed that RNF144A only associated with EGF-stimulated EGFR (Fig. 2, K–L). In this in vitro assay, affinity purified FLAG-RNF144A was incubated with EGFR prepared from separately cultured EGF-stimulated cells. Thus, the data demonstrate their interaction can happen in vitro independently of their membrane localization. Moreover, the interaction was not observed under starvation conditions when both proteins are localized in the plasma membrane (Fig. 1C). Together, these data argue that the RNF144A-EGFR interaction is not simply due to colocalization in the same intracellular compartment. We further demonstrated that both EGFR ligands, EGF and TGF-α, were able to greatly induce RNF144A-EGFR association (Fig. 2, M and N). Taken together, these results indicate that EGFR ligands can induce a dynamic interaction between RNF144A and EGFR.
Figure 2.
EGFR ligands induce a dynamic interaction between RNF144A and EGFR. A, Western blot analysis demonstrates association between RNF144A and EGFR in the endogenous level in U2OS cells. B–D, the TM domain of RNF144A is required for the RNF144A-EGFR interaction. B, a scheme of the FLAG-tagged human RNF144A and different deletion constructs used in mapping the RNF144A-EGFR interacting region. C, deletion of the TM domain, but not the RBR domain, of RNF144A blocked the RNF144A-EGFR interaction. D, quantification of the EGFR/RNF144A ratio in the FLAG IP from C. E–H, Western blotting shows that EGF induced RNF144A-EGFR association in a time-dependent manner (E and G). LE: long exposure; SE, short exposure. F and H, quantification of EGFR/RNF144A ratio in the FLAG IP from E and G. I and J, EGFR kinase inhibitor, lapatinib, blocked the EGF-induced RNF144A-EGFR association. K and L, in vitro binding experiment shows that RNF144A could only pulldown EGFR from the cell lysates in which the cells had been stimulated by EGF. M and N, Western blot analysis shows that EGFR ligands, EGF and TGF-α, induced RNF144A-EGFR association. FLAG, FLAG-tagged empty vector; BSA, 0.1% BSA in serum-free medium; EGF, EGF stimulation; DMSO is a control solution of lapatinib. All quantification data represent mean ± S.D. from three independent experiments. *, p < 0.05; N.S., not significant.
RNF144A interacts with endocytosed EGFR in early endosomes
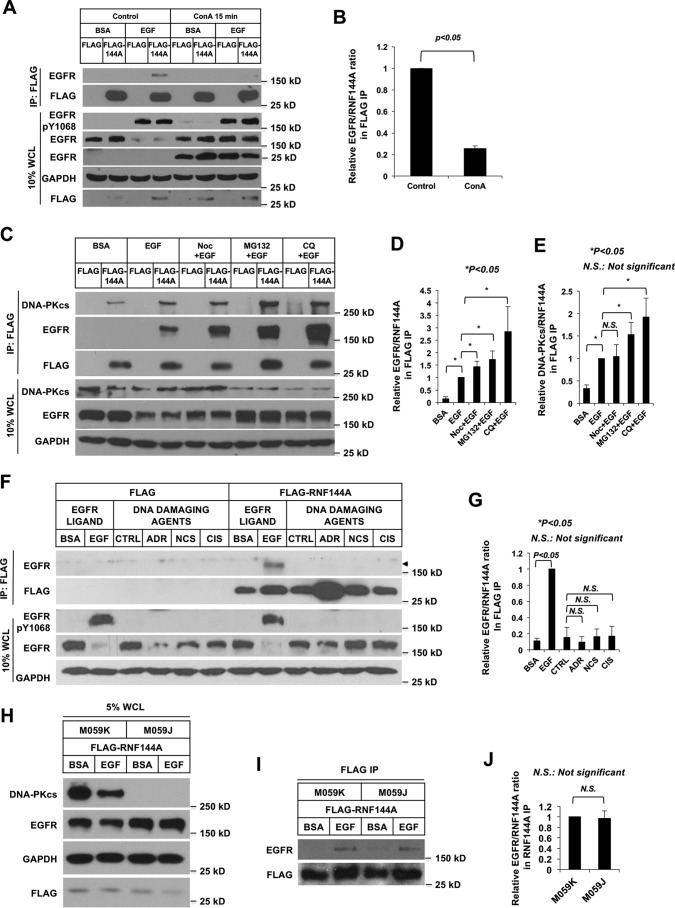
To further investigate the RNF144A-EGFR interaction in detail, we used various inhibitors to map the subcellular localization of the RNF144A-EGFR interaction. Interestingly, an endocytosis inhibitor, concanavalin A (ConA), reduced the EGF stimulation-dependent RNF144A-EGFR interaction, suggesting that RNF144A interacted with EGFR in the cytosolic vesicles rather than in the plasma membrane (Fig. 3, A and B). Consistent with previous studies, ConA treatment not only blocked endocytosis of the activated EGFR and prevented its subsequent degradation but also resulted in cleavage of a small fraction of EGFR in the plasma membrane to a 26-kDa fragment (31), which is shown here as a positive control for ConA treatment. Because the microtubule-depolymerizing agent is able to block transfer of endocytosed components from the early to late endosomes (32, 33), nocodazole (Noc) was used to further determine the effect of early-to-late endocytosis trafficking on the RNF144A-EGFR interaction. As expected, lapatinib, but not Noc, treatment blocked EGF-mediated endocytosis of EGFR (Fig. S2). Unlike ConA, which reduced RNF144A-EGFR interaction (Fig. 3, A and B), Noc treatment led to accumulation of the RNF144A–EGFR complex, indicating that this interaction may occur in early endosomes, rather than in late endosomes (Fig. 3, C and D). Moreover, both proteasome and lysosome inhibitors also greatly led to the accumulation of RNF144A–EGFR complex. These data, along with the results of Fig. 1D, indicate that upon EGF stimulation, RNF144A interacts with endocytosed EGFR in early endosomes and both are transported together to late endosomes and lysosomes.
Figure 3.
The interaction of RNF144A with EGFR requires endocytosis and is independent of its interaction with DNA-PKcs nor is RNF144A-EGFR association induced by DNA damage. A and B, endocytosis inhibitor, ConA, reduced the EGF-induced RNF144A-EGFR interaction. C–E, EGF induced both RNF144A-EGFR and RNF144A–DNA-PKcs interaction. Noc, MG132, and lysosome inhibitor, chloroquine (CQ), could further enhance the accumulation of the RNF144A–EGFR complex. Cells were pretreated with the indicated inhibitors for 30 min prior to EGF stimulation. F and G, DNA damaging agents, adriamycin (ADR), neocarzinostatin (NCS), and cisplatin (CIS) could not induce RNF144A-EGFR association. H–J, EGF-induced RNF144-EGFR interaction in both DNA-PK-proficient (M059K) and DNA-PK-deficient (M059J) cells. B, D–E, G, and J are quantifications of EGFR/RNF144A or DNA-PKcs/RNF144A ratio in the FLAG IP from A, C, F, and I, respectively. All quantification data represent mean ± S.D. from three biological replicates. *, p < 0.05; N.S., not significant. FLAG, FLAG-tagged empty vector; BSA, 0.1% BSA in serum-free medium; EGF, EGF stimulation; CTRL is a control solution of DNA damaging agents.
RNF144A-EGFR interaction is not induced upon DNA damage and does not require DNA-PKcs
A severe DNA damage may also trigger both EGFR endocytosis and downstream survival signaling (34), and RNF144A accumulation (11). We examined RNF144A-EGFR association after treatment with various DNA damaging agents to see whether RNF144A–EGFR complex formation is specific to EGF induction. DNA damaging agents only induced RNF144A accumulation but not RNF144A-EGFR association, supporting that ligand-dependent endocytosis is required for this interaction (Fig. 3F). This EGF stimulation-dependent RNF144A-EGFR interaction was also observed at the endogenous levels (Fig. 2A). Therefore, these results strongly suggest that RNF144A interacts with activated EGFR and regulates its functions.
DNA-PKcs can interact with both RNF144A (11, 35) and EGFR (36–38). EGFR kinase activity inhibitor, gefitinib, induces DNA-PKcs cytosolic redistribution, EGFR-DNA-PKcs association, and inhibits DNA-PKcs activity (38, 39). Hence, we determined the relationship between RNF144A, DNA-PKcs, and EGFR upon EGF treatment. Consistent with our previous study (11), RNF144A interacted with DNA-PKcs (Fig. 3, C and E). Interestingly, EGF treatment also promoted RNF144A–DNA-PKcs complex accumulation (Fig. 3, C and E, and Fig. S3A). Moreover, treatment of either a lysosome inhibitor or a proteasome inhibitor led to accumulation of the RNF144A–DNA-PKcs complex, suggesting a lysosome and/or proteasome-dependent degradative process of RNF144A–DNA-PKcs complex after EGF stimulation. RNF144A knockdown also increased the DNA-PKcs protein level in both cytosol and nucleus, and EGF stimulation dramatically increased DNA-PKcs in the RNF144A knockdown cell (Fig. S3B). Conversely, RNF144A overexpression promoted EGF-dependent degradation of DNA-PKcs, and the proteasome inhibitor was able to rescue this degradation (Fig. S3C). These data support that RNF144A promotes DNA-PKcs degradation upon EGF stimulation. To investigate whether DNA-PKcs is involved in RNF144A-EGFR interaction, we examined their interaction in DNA-PKc-deficient cells (M059J) and DNA-PKc-proficient cells (M059K). The results demonstrated that DNA-PKcs is not required for the RNF144A-EGFR interaction, because their interaction was detectable to a similar extent in both cell lines (Fig. 3, H–J, and Fig. S4). In the present study, we focused on the effect of RNF144A on EGFR protein and signaling. Future study will be required to address whether there is any relationship between RNF144A–EGFR and RNF144A–DNA-PKcs complexes.
RNF144A promotes ubiquitination of EGFR
Because RNF144A is an EGFR-associated E3 ligase in the intracellular vesicles, the ubiquitination level of EGFR upon overexpression of RNF144A was also examined. As shown in Fig. 4, A–C, overexpression of WT RNF144A promoted ubiquitination of EGFR and phosphorylated Tyr1068-EGFR after EGF stimulation. In addition, overexpression of WT RNF144A significantly enhanced ubiquitination of Tyr(P)1068-EGFR and resulted in a higher molecular weight smear pattern in total EGFR ubiquitination. Consistent with our previous study (10, 11), overexpression of WT RNF144A (but not ligase-dead C20A/C23A mutant or ΔTM mutant) can also promote ubiquitination of DNA-PKcs. Similar results were obtained in the overexpression of exogenous EGFR and WT RNF144A under starvation and EGF stimulation conditions (Fig. 4, D and E). However, overexpression of a ligase-dead mutant failed to promote EGFR ubiquitination (Fig. 4, A–E). Interestingly, this increased ubiquitination level of EGFR may be coupled with EGF-stimulated EGFR subcellular localization (Fig. 4, F–H) and protein levels (Fig. 4, I and J). Overexpression of WT RNF144A, but not a ligase-dead mutant, promoted the formation of intracellular ring-shaped vesicles in these EGFR-positive cells following EGF stimulation (Fig. 4F). Of note, RNF144A is colocalized with EGFR in these intracellular ring-shaped vesicles, which are also Rab5-containing vesicles (Fig. 1D). Both WT RNF144A and RNF144A(C20A/C23A) can co-localize with Rab5 (Fig. S6). Thus, the difference observed in Fig. 4F is not because the mutant localizes at different vesicles, but rather suggest that formation of EGFR(+) ring-shaped vesicles depends on the E3 ligase activity of RNF144A. We also made use of the RNF144A knockout U2OS cells (described in next section) to investigate a role for RNF144A in the formation of these EGFR-positive ring vesicles. The EGFP-tagged EGFR is mainly located on the cell surface of RNF144A-proficient and -deficient cells (Fig. 4G, upper two panels) under regular medium and serum starvation conditions. EGF stimulation induced big ring-shaped EGFR-positive vesicles in the control cells. However, depletion of RNF144A significantly reduced the number of these large EGFR-positive vesicles (Fig. 4, G and H, and Fig. S5). We also explored EGFR protein levels by overexpression of WT or mutant RNF144A. Overexpression of WT RNF144A increased EGFR protein, but overexpression of ligase-dead mutant RNF144A(C20A/C23A) decreased EGF-stimulated EGFR protein (Fig. 4, I and J). Our previous study has characterized that the TM domain of RNF144A is important for its E3 ligase activation and subcellular localization, we further determined the effect of two somatic TM domain mutations (RNF144A(G252D) and RNF144A(L261F)) on EGFR protein. RNF144A(G252D) originally came from gastric adenocarcinoma. It has been shown that RNF144A(G252D) lost its membrane localization and had a very short half-life (∼20 min) (10). RNF144A(L261F) is derived from cutaneous melanoma. We did not find significant changes in subcellular localization and half-life (10). Interestingly, these two somatic RNF144A TM mutations, G252D and L261F, either completely inhibited or reduced the interaction of RNF144A and EGFR (Fig. S7). Moreover, overexpression of these two tumor-derived RNF144A mutants also decreased the EGFR protein level (Fig. 4, I and J). Because RNF144A WT provides positive regulation of EGFR protein, the loss or decrease of RNF144A-EGFR interaction inhibits or mitigates the effect of RNF144A on the EGFR protein. These results indicate that RNF144A can regulate EGFR protein levels and localization through its E3 ligase activity.
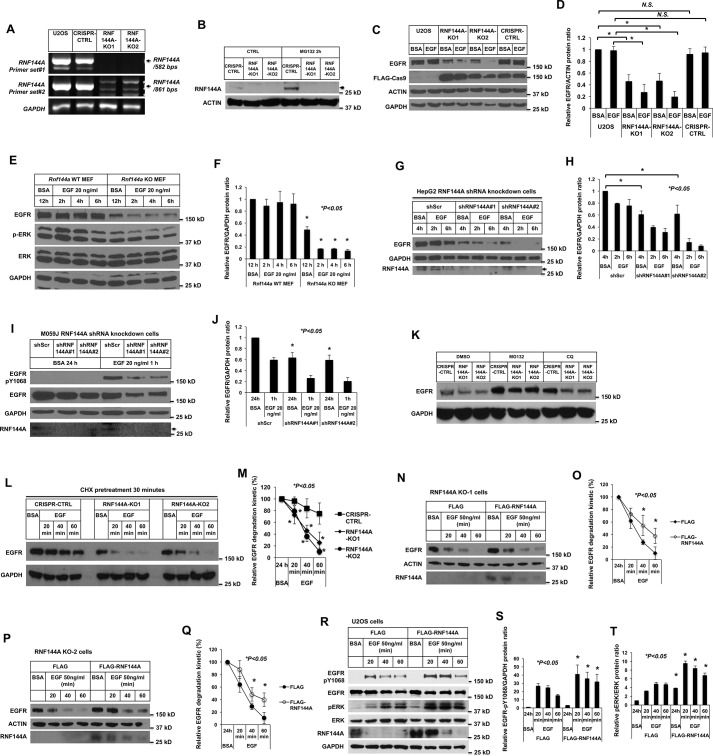
RNF144A plays a key role for maintaining EGFR protein levels and signaling
To further determine the potential role of RNF144A in EGF/EGFR-dependent functions, two independent RNF144A-depleted clones, RNF144A-KO1 and RNF144A-KO2, were generated in U2OS cells using the CRISPR/Cas9 system (Fig. 5, A and B) (40, 41). Consistent with the previous studies (10, 11), RNF144A was unstable and can be rescued in CRISPR-CTRL cells by the proteasome inhibitor MG132 (Fig. 5B). We first examined the EGFR protein levels in these knockout cells. As shown in Fig. 5, C and D, RNF144A knockout decreased the EGFR protein levels in U2OS cells, both in basal (prior to EGF stimulation) and EGF stimulation conditions. Similarly, a decrease of EGFR protein basal level was also observed in the transformed Rnf144a KO mouse embryonic fibroblasts (MEFs) (Fig. 5, E and F, and Fig. S8) and in RNF144A-depleted human HepG2 and M059J cells (Fig. 5, G–J, and Fig. S9). In addition, proteasome and lysosome inhibitors can partially rescue the EGFR protein levels (Fig. 5K). This rescue supports that EGFR protein stability can be modulated by RNF144A via a post-transcriptional mechanism. Indeed, depletion of RNF144A also promoted EGF-induced degradation of EGFR protein under inhibition of newly synthesized EGFR by cycloheximide treatment (Fig. 5, L and M). Furthermore, reintroduction of exogenous RNF144A attenuated EGF-stimulated EGFR degradation in both RNF144A KO cells (Fig. 5, N–Q). Overexpression of RNF144A further stabilized both EGFR-Tyr(P)1068 and total EGFR and promoted their downstream signaling of phosphorylation of ERK (Fig. 5, R–T, and Fig. S10). Overall, these results suggest that RNF144A plays a key role in stabilizing the EGFR protein during the EGF stimulation cycle.
Figure 5.
RNF144A is important for maintaining EGFR protein and the EGF/EGFR signaling. A and B, generation of two independent RNF144A stable knockout clones in U2OS cells using the CRISPR/Cas9 system. DNA-agarose gel (1%) shows that the RNF144A transcript was undetectable in two different primer sets even after 50 cycles of RT-PCR amplification. Primer set (# 1) RNF144A-407F/RNF144A-988R and (# 2) RNF144A-749F/RNF144A-1609R (A). Western blot analysis shows the expression of RNF144A in the CRISPR-CTRL system, but not in the two RNF144A knockout cell lines. The proteasome inhibitor MG132 could only further accumulate RNF144A in CRISPR-CTRL cells (B). The arrow indicates RNF144A bands. C–F, Western blot analysis shows that knockout of RNF144A reduced the EGFR protein level in U2OS cells (C and D) and in the transformed MEFs (E and F). Quantitation of Western blots by ImageJ software from three independent experiments of C or E is shown in D or F, respectively. *, p < 0.05; N.S., not significant. G–J, Western blot analysis shows that knockdown of RNF144A reduced EGFR protein level in HepG2 cells (G) and M059J cells (I). The arrows in G and I indicate RNF144A bands. Depletion of RNF144A in these two cell lines were also independently confirmed by qRT-PCR assays (Fig. S9). H and J, quantitation of Western blots by ImageJ software from three independent experiments of G or I is shown in H or J, respectively. K, Western blot analysis shows that proteasome inhibitor (MG132) and lysosome inhibitor (CQ) could partially rescue EGFR protein levels from the RNF144A knockout U2OS cells. L and M, depletion of RNF144A promoted EGF-induced degradation of EGFR protein under inhibition of newly synthesized EGFR by cycloheximide (CHX) treatment. The data shown represent mean ± S.D. (n = 3 biological replicates for each condition). *, p < 0.05; N.S., not significant. N–Q, reintroduction of exogenous RNF144A attenuated EGF-stimulated EGFR degradation in both RNF144A KO cells. O and Q, quantitation of Western blots by ImageJ software from three experiments of N or P is shown in O or Q, respectively. R–T, overexpression of WT RNF144A decreased EGFR degradation and induced the EGF-dependent phosphorylation of ERK Thr202/Tyr204 by EGF stimulation. S and T are quantitations of EGFR-Tyr(P)1068/GAPDH ratio and pERK/ERK ratio from R; all quantification data represent mean ± S.D. from three independent experiments. *, p < 0.05; N.S., not significant.
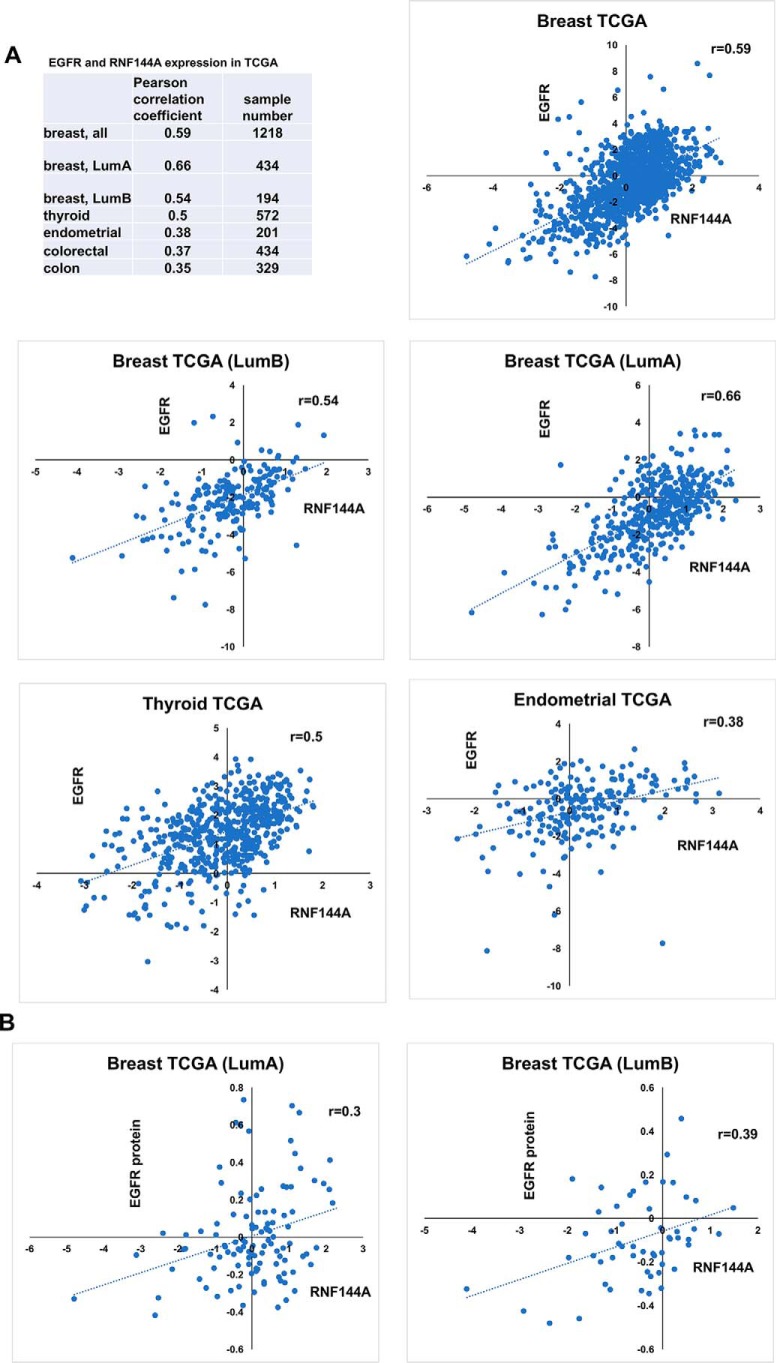
Positive correlation between the expression of EGFR and RNF144A in several types of cancer
Given the positive regulation of EGFR protein and signaling by RNF144A, we also examined the relationship between RNF144A and EGFR in several types of clinical cancer samples (Fig. 6). Because EGF binding to EGFR activates expression of the EGFR gene itself (21–23), EGFR mRNA levels may in part reflect EGF/EGFR signaling activity. Indeed, we found a positive correlation between RNF144A and EGFR mRNA expression in breast, thyroid, colon, endometrial, and colorectal cancers (Fig. 6A). In addition, RNF144A transcript levels were also positively correlated with EGFR protein levels in luminal A and luminal B breast cancers (Fig. 6B). The correlation in many cancer types in conjunction with the data presented in cultured cells strongly support that RNF144A up-regulates EGFR protein and its signaling function.
Figure 6.
Positive correlation between the expression of EGFR and RNF144A in several types of cancer. A and B, Pearson correlation coefficients (r) between the mRNA expression (A) or protein expression (B) of EGFR and that of RNF144A in TCGA database (data extracted from xena.ucsc.edu). LumA, luminal A subtype breast cancer; LumB, luminal B subtype breast cancer. The correlations in some cancer types are also shown as scatter plots. x axis, RNF144A-gene expression RNAseq-IlluminaHiSeq pancan normalized; Unit, pan-cancer normalized log2(norm_count+1); y axis, EGFR-gene expression RNAseq-IlluminaHiSeq pancan normalized; Unit, pan-cancer normalized log2(norm_count+1). The pancan-normalized dataset subtracts out the mean expression across all cancer types and shows which genes are up-regulated or down-regulated compared with all other tissues. B, scatter plots show the correlation between EGFR protein levels and RNF144A transcript levels in luminal A and luminal B breast cancers. x axis, RNF144A-gene expression RNAseq-IlluminaHiSeq pancan normalized; Unit, Pan-cancer normalized log2(norm_count+1); y axis, EGFR-protein expression RPPA-RPPA; unit, normalized RPPA value.
A role for RNF144A in EGF-dependent gene activation and cell proliferation
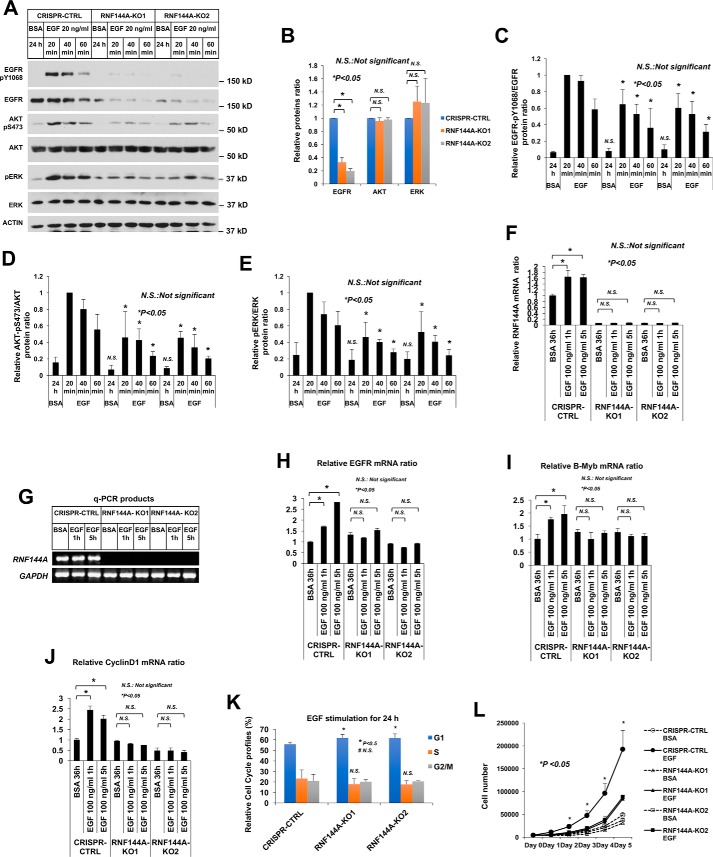
To further determine a physiological role for RNF144A in EGF/EGFR signaling, we examined EGF-dependent gene activation, cell proliferation, and other functions in two RNF144A knockout cell lines. Contrary to RNF144A overexpression, which promoted EGF/EGFR signaling (Fig. 5, R–T, and Fig. S10), RNF144A depletion reduced EGFR signaling responses, including phosphorylation of EGFR–Tyr1068, AKT–Ser473, and ERK–Thr202/Tyr204 (Fig. 7, A–E). Notably, depletion of RNF144A only resulted in a decrease in EGFR protein levels, but did not result in a decrease in AKT or ERK protein levels (Fig. 7B). We then examined RNF144A mRNA and the activation of EGF/EGFR-dependent G1/S progression genes after EGF stimulation. Interestingly, EGF stimulation induced RNF144A gene expression (Fig. 7F and Fig. S8), supporting the involvement of RNF144A in EGF/EGFR-dependent functions. Consistent with the well-established EGF-dependent functions, EGF stimulation activated EGFR (21–23), CCND1 (24), and MYBL2 (25) genes in the CRISPR-CTRL cells (Fig. 7, H–J). Importantly, the response of EGFR, B-Myb, and Cyclin D1 to EGF stimulation was blunted in RNF144A-depleted cells (Fig. 7, H–J). Knockout of Rnf144a in mouse embryonic fibroblasts also inhibited EGF-dependent up-regulation of Egfr expression (Fig. S8). Moreover, RNF144A depletion affected EGF-dependent cell cycle progression, caused an increase of G1 phase cell population (Fig. 7K), and reduced EGF-dependent cell growth (Fig. 7L). These results demonstrate a role for RNF144A in EGF-stimulated gene activation and cell proliferation.
Figure 7.
Knockout of RNF144A inhibits EGF-dependent gene activation and cell cycle progression. A–E, knockout of RNF144A reduced the EGF-dependent phosphorylation of EGFR-Tyr1068 (C), AKT-Ser473 (D), and ERK-Thr202/Tyr204 (E). B, relative proteins ratio shows knockout of RNF144A only reduced EGFR protein level, but not AKT or ERK protein levels. All quantification data represent mean ± S.D. from three biological replicates. *, p < 0.05; N.S., not significant. F–J, real-time quantitative RT-PCR analysis shows that EGF induced the expression of RNF144A (F), EGFR (H), B-Myb (I), and Cyclin D1 (J) in the RNF144A CRISPR-CTRL cells but not in the two RNF144A knockout cell lines. G, DNA-agarose gel shows the RNF144A transcript could not be detected even after 40 cycles of qPCR amplification from F. Results were normalized to GAPDH levels and the mean ± S.D. (n = 3) are expressed relative to the expression of genes in CRISPR control cells. K, cell cycle profile analysis shows that depletion of RNF144A slightly increased G1 phase population upon 24 h of EGF treatment. L, cell proliferation analysis shows that RNF144A was required for a full response of EGF-dependent cell proliferation. Data in K and L represent mean ± S.D. from three biological replicates.
Discussion
The current study uncovers unexpected functions for an RBR E3 ligase RNF144A. To our knowledge, this is the first study demonstrating that RNF144A physically interacts with EGFR and regulates its signaling and EGF-dependent cell proliferation. Several lines of evidence are presented to support this conclusion. First, by a combination of biochemical and cell biological studies we identify the formation of the RNF144A–EGFR complex upon EGFR ligand stimulation in endocytic vesicles and early endosomes where ubiquitination of activated EGFR by RNF144A maintains EGFR protein level and sustains EGF/EGFR signaling. Second, using murine genetic knockout, CRISPR knockout, and small hairpin RNA approaches, a physiological role for RNF144A in maintaining EGFR protein level and EGF/EGFR signaling is demonstrated in both human and mouse cells. Consistently, overexpression of RNF144A can promote EGF/EGFR signaling. Moreover, the expression levels of RNF144A and EGFR are positively correlated in several types of cancer. Third, depletion of RNF144A blocks EGF-dependent G1/S progression genes activation and reduces the EGF-dependent cell proliferation. Collectively, this study reveals a fundamental function of RNF144A in the regulation of growth factor signaling and cell proliferation.
RNF144A contains an RBR domain and a single TM domain. Our previous study has revealed that the single TM domain is responsible for RNF144A membrane localization and self-association, both required for its ligase function (10). This single TM domain not only restricts RNF144A subcellular localization and activation but also physiologically restricts its target proteins to the ones that are localized in the membrane or at least exist on the same subcellular compartment of RNF144A RBR domain for ubiquitination. The data that the TM domain, but not RBR domain, of RNF144A is required for the RNF144A-EGFR interaction further supports a role for this TM domain (Fig. 2, B–D). Interestingly, the correct TM domain and its juxtamembrane region of RNF144A are also important for RNF144A-EGFR interaction and/or RNF144A plasma membrane localization. RNF144A(1–272), RNF144A(G252D), and RNF144A(L261F) reduce their interaction with EGFR (Fig. 2, B–D, and Fig. S7), and both RNF144A(1–272) and RNF144A(G252D) alters their subcellular localization pattern (Fig. S11) (10). In addition, overexpression of the RNF144A-ΔTM mutant fails to promote ubiquitination of EGFR and DNA-PKcs (Fig. 4, A–C). Because the RNF144A-ΔTM mutant still contains a complete RBR domain, these data indicate that both RBR and TM domains of RNF144A are required for its E3 ligase activity in the cells. Additionally, we show that EGF can induce the expression of RNF144A, which in turn plays a role in maintaining the EGFR protein level and signaling. Effective EGF signaling in the presence of RNF144A further induces EGFR gene expression (Fig. 7H and Fig. S8). Therefore, through RNF144A, EGF and EGFR induce a positive feedback loop to achieve a maximal response during growth factors stimulation. Indeed, in several types of cancer, such as breast, thyroid, endometrial cancers, etc., RNF144A expression is highly correlated with the levels of EGFR transcript and protein (Fig. 6).
It is worth noting that a correlation between RNF144A and EGFR expression is seen in several breast cancer subtypes, (luminal A, r = 0.66; luminal B, r = 0.54; Her2, r = 0.32, normal-like; r = 0.41), but it is not seen in basal-like subtypes (r = −0.057). EGFR is overexpressed in most triple-negative or basal-like subtype breast cancer through other well-characterized mechanisms. Thus, it is possible that due to its overexpression via other mechanisms, EGFR levels no longer correlate with RNF144A expression. However, given the fact that a role for RNF144A in sustaining EGFR function is demonstrated in multiple cancer cell lines, we suspect that RNF144A knockout in basal-like breast cancer would still affect EGFR function. Additional research will be needed to address the RNF144A–EGFR complex in triple-negative breast cancer.
EGFR endocytosis and transport can precisely control its cellular functions in response to different conditions. Endocytosed EGFR-containing vesicles fuse with different cytosolic vesicles and wait for sorting in the multivesicular bodies that usually store and hold EGFR for a while. It has been shown that EGF induces the formation of enlarged EGFR-positive ring-shaped vesicles that colocalize with different endosome residents, including Rab5 and others (42, 43). These enlarged ring-shaped vesicles may sequester EGFR and determine its destination and functions (44). Interestingly, our data show that overexpression of WT RNF144A, but not the ligase-dead mutant (C20A/C23A), dramatically induces the formation of these large ring-shaped vesicles where RNF144A, EGFR, and Rab5 are colocalized during EGF treatment (Figs. 1, C and D, and 4, F–H, and Fig. S5). RNF144A interacts with EGFR in early endosomes and traffic together to late endosomes and lysosome (Fig. 3, A–D). RNF144A ubiquitinates EGFR (Fig. 4, A–E) and is required to maintain the EGFR protein level (Fig. 5). Given that both proteasome and lysosomal inhibitors can cause RNF144A–EGFR complex accumulation (Fig. 3, C and D), and also partially rescue the effect of RNF144A knockout on EGFR protein reduction (Fig. 5K), RNF144A might regulate EGFR proteins through both proteasome and lysosome degradation pathways. Interestingly, it has also been shown that both proteasome and lysosome inhibitors can block EGFR degradation (45–47). However, most data still suggest that EGFR is mainly degraded in lysosomes. The effect of proteasome inhibitors on the degradation of EGFR may be due to the indirect effect of regulatory proteins and/or free ubiquitin on the EGFR trafficking to the lysosome (48). Because RNF144A is unstable and can be accumulated by a proteasome inhibitor (Fig. 5B), the effect of the proteasome inhibitor on RNF144A-EGFR regulation may be also due to indirect regulation of EGFR transport to lysosomes. All together, these data suggest that during endocytosis of EGFR after EGF stimulation, RNF144A binds and ubiquitinates EGFR in early endosomes. This process may promote the transport or fusion of EGFR-containing vesicles and form the large ring-shaped EGFR(+) vesicles, which ultimately is important for maintaining the overall EGFR protein level and EGFR signaling function. We postulate that ubiquitination of EGFR by RNF144A may signal for EGFR trafficking and/or compete with degradative ubiquitination by other E3 ligases. Future work is warranted to elucidate the detailed mechanism. As an important signaling initiator of many essential cellular functions, EGFR and other receptor tyrosine kinases have been reported to be the targets of several E3 ligases during different transport stages (49). Our study also shows that RNF144A is required for a full response to EGF in cell proliferation (Fig. 7L). It will be very interesting to determine whether these E3 ligases can coordinate with each other to regulate the EGF/EGFR signaling.
DNA-PKcs is a key component for error-prone NHEJ DNA repair process for cell survival. The cytosolic DNA-PKcs is the first identified target of RNF144A (11). Although RNF144A can interact with both DNA-PKcs and EGFR, the interactions appear to be independent of each other (Fig. 3, H–J, and Fig. S4). Through these two independent interactions, RNF144A promotes different cell fates in response to different stimuli. During normal cell proliferation, EGF induces RNF144A for maximizing EGF/EGFR signaling. When cells suffer from persistent or severe DNA damage insults, RNF144A is induced by p53 and promotes apoptosis and possibly inhibits errors-prone NHEJ DNA repair by down-regulating DNA-PKcs (11). Interestingly, EGFR can interact with DNA-PKcs and promote NHEJ DNA repair upon radiation or cisplatin treatment (29, 30). This regulation may have clinical implications because blockage of EGFR signaling can sensitize cancer cells to radio- or chemotherapies by decreasing the DNA-PKcs DNA repair capacity (38, 50). EGFR is frequently overexpressed in human epithelial tumors and is associated with therapy resistance. Therefore, EGFR inhibitors in combination with other DNA damaging agents are tested in the cancer harboring activating mutant EGFR or overexpressing EGFR (51–55). Interestingly, transiently overexpression of RNF144A also lessened the down-regulating effect of the DNA damaging agents on the EGFR protein level (Fig. 3, F and G), suggesting a role of RNF144A for maintaining the EGFR level even under DNA damage. Although RNF144A expression is down-regulated in many types of cancer, it is up-regulated in certain cancers, such as gastric cancer and melanoma (Oncomine). Enhanced EGFR levels/activity in RNF144A-overexpressing tumors (Fig. 6) may contribute to resistance to chemotherapy. Thus, our new findings suggest a benefit of combining EGFR inhibitors with DNA damaging agents in the cancer with high RNF144A expression. In conclusion, the current study elucidates a novel physiological role of RNF144A in EGF/EGFR signaling and EGF-dependent cell proliferation.
Experimental procedures
Cell culture
All cells used in this study were maintained in media supplemented with (10% (v/v) fetal bovine serum, penicillin (50 IU/ml), and streptomycin (50 μg/ml)), and grown in a humidified incubator at 37 °C with 5% CO2 and 95% air as previously described (11). Specifically, HEK293T, HepG2, and transformed Rnf144A WT and knockout MEF cells were maintained in DMEM. M059K and M059J cells were maintained in DMEM:F-12 at 1:1 medium. U2OS cells were grown in McCoy's 5A medium.
Plasmids and transfection
Mammalian expressing FLAG-tagged RNF144A WT and FLAG-tagged RNF144A mutant plasmids were generated as described previously (10, 11). The validated RNF144A hairpin shRNA constructs (V2LHS_254611 and V2LHS_72643) were purchased from Open Biosystems (11). The lentiCRISPR v2 (number 52961) (41), EGFR-GFP (number 32751) (56), and pcDNA6A-EGFR WT (number 42665) (57) were purchased from Addgene. Standard Lipofectamine 2000 (Life Technologies) or polyethylenimine (Sigma) (58) methods were used for transfection of all cells. If desired, the cells were transfected with the indicated tagging construct plasmid or its original empty vector as a control.
Reagents and antibodies
All reagents were purchased from Sigma unless indicated otherwise. Recombinant human EGF and TGF-α were purchased from R&D Systems. Lapatinib ditosylate was purchased from OXCHEM Corporation. Adriamycin (doxorubicin hydrochloride) was purchased from Calbiochem. RNF144A antibody was purchased from Abcam (ab75054 and ab89260), Novus Biological (NBP1-01049), and LifeSpan BioSciences (LS-C162648). EGFR antibody (D38B1, number 4267), Phospho-EGF Receptor Pathway Antibody Sampler Kit (number 9789), and Endosomal Marker Antibody Sampler Kit (number 12666) were purchased from Cell Signaling. DNA-PKcs antibody was purchased from NeoMarkers. GAPDH (sc-47724), EGFR (sc-03), and HA (sc-805) antibodies were purchased from Santa Cruz Biotechnology. EGFR mAb (199.12) was purchased from Thermo Fisher Scientific. The p84 (5E10) antibody was purchased from GeneTex. Rabbit or mouse FLAG antibody (F7245 and F3165), actin antibody (A2066), mouse monoclonal anti-HA-agarose antibody (A2095), and anti-FLAG® M2 Affinity Gel (A2220) were purchased from Sigma. Myc-Trap® was purchased from ChromoTek. BrdU antibody (B44) was purchased from BD Biosciences.
Generation of RNF144A CRISPR knockout stable cells
The stable cell lines were generated using the standard Target Guide Sequence Cloning Protocol (40, 41). Briefly, two pairs of single guide oligos (sgRNF144A_4F, 5′-CAC CGC AGA GCT TGC AAG ACA CCA G-3′ and sgRNF144A_4R, 5′-AAA CCT GGT GTC TTG CAA GCT CTG C-3′; sgRNF144A_10F, 5′-CAC CGG CCG GGC ACC AAG TCC GAC A-3′ and sgRNF144A_10R, 5′-AAA CTG TCG GAC TTG GTG CCC GGC C-3′) were synthesized to target RNF144A RING1 and IBR domains, respectively. The pair of oligos were annealed and cloned into the lentiCRISPR v2 vector using the BsmBI restriction enzyme site. The cloned vectors or empty vector were then transfected into U2OS cells and selected by puromycin (1 μg/ml) for 3 weeks. The resistant clones were screened for RNF144A depletion using RT-PCR with two different oligo sets: RNF144A-407F/RNF144A-988R and RNF144A-749F/RNF144A-1609R. RNF144A-depleted cells were further confirmed with different qPCR sets of oligos: RNF144A-qPCR-1F/RNF144A-qPCR-1R and RNF144A-qPCR-2F/RNF144A-qPCR-2R.
Generation of transformed Rnf144a knockout MEF cells
Rnf144a heterozygote knockout mouse was generated in the Baylor College of Medicine mouse core facility. In brief, the Rnf144a knockout first construct (HTGR6005_A_5_G08) was purchased from KOMP Repository, and C57BL/6J ES-cell electroporation was performed at the Baylor College of Medicine mouse core facility. Mouse was genotyping used different oligos: LoxP-F, 5′-ACT GAT GGC GAG CTC AGA CC-3′; LoxP-R, 5′-GCC TTT GAA TCC TAC AAC AG-3′; and LoxP-F-genotyping, 5′-CTG CAA GGA GTA CCC AGC TT-3′. To make a pair of Rnf144a knockout and WT MEF cells, Rnf144a heterozygote knockout mice were mated and mouse embryonic fibroblasts were isolated with day 13.5 to day 14.5 mouse embryos. The genotype of MEF was confirmed by genotyping using genotyping oligo sets and real-time RT-PCR using the mouse Rnf144a PCR primer set: Rnf144a-qPCR-F/Rnf144a-qPCR-R. Primary Rnf144a WT and knockout MEF were transformed by transfecting with simian virus 40 (SV40) T antigen and maintained in DMEM.
Real-time RT-PCR and PCR
RNA was extracted using TRIzol reagent (Invitrogen) as described previously (11). Real-time RT-PCR was then performed in triplicate on an MX3005P Thermal Cycler (Stratagene) using SYBR Green dye to measure amplification and ROX as a reference dye. Transcript levels were normalized with GAPDH levels, which were analyzed in parallel with test genes. Results were analyzed with MxPro 4.1 Quantitative PCR software (Stratagene). The PCR primer sequences are listed in Table 1.
Table 1.
PCR primers used in this study
| PCR primer names | PCR primer sequences |
|---|---|
| Human EGFR-F | 5′-GGCACTTTTGAAGATCATTTTCTC-3′ |
| Human EGFR-R | 5′-CTGTGTTGAGGGCAATGAG-3′ |
| Human B-Myb-F | 5′-TGAAGGAGGTGCTCCGTTCTGA-3′ |
| Human B-Myb-R | 5′-CCAGAGACTTGCGGACCTTCTT-3′ |
| Human cyclin D1-F | 5′-GGAACACCAGCTCCTGTGCTGCGAAG-3′ |
| Human cyclin D1-R | 5′-CCACTTGAGCTTGTTCACCAG-3′ |
| Human RNF144A-qPCR-1F | 5′-CTGTTTGATCCCTGTCGGACT-3′ |
| Human RNF144A-qPCR-1R | 5′-GATGGGCGCGTCATCTTCTT-3′ |
| Human RNF144A-qPCR-2F | 5′-AAACCGCAATTAGCTGCCCA-3′ |
| Human RNF144A-qPCR-2R | 5′-CGACAGGGATCAAACAGCAC-3′ |
| Human RNF144A-407F | 5′-CTCCTCCCCTCTGGATTTCT-3′ |
| Human RNF144A-988R | 5′-GATGGGCGCGTCATCTTCTT-3′ |
| Human RNF144A-749F | 5′-CTGTTTGATCCCTGTCGGACT-3′ |
| Human RNF144A-1609R | 5′-AGAGAGAAAGCTGCCTGGTG-3′ |
| Mouse Rnf144a-qPCR-F | 5′-TTGAGTGTATGGTTGCCGCAGAA-3′ |
| Mouse Rnf144a-qPCR-R | 5′-CCTATGTCCTGGAGTTGGCACA-3′ |
Immunoprecipitation and Western blot analysis
For co-immunoprecipitation (co-IP), the 24–72-h transfected cells or un-transfected cells were harvested with TNN buffer (50 mm Tris, 0.25 m NaCl, 5 mm EDTA, and 0.5% Nonidet P-40) as described previously (11). An aliquot of the cell lysates was saved for protein input control, and immunoprecipitation was carried out using the indicated antibodies overnight and resolved by 10, 6, or 4–10% (v/v) gradient SDS-PAGE (Bio-Rad). Cells prepared for Western blot analysis were lysed in SDS lysis buffer (1% SDS, 60 mm Tris, pH 6.8), and equal amounts of proteins were resolved by SDS-PAGE. The specific signals of both co-IP and Western blotting were detected with indicated antibodies.
Immunofluorescence staining
Cells were seeded on cover glasses 1 day before experiments. Cells were treated with specific conditions as described in each experiment and then fixed with 4% paraformaldehyde for 10 min followed by permeabilization with 0.5% Triton X-100 in 1× PBS for 10 min. The fixed cells were then subjected to blocking with 2% BSA in 1× PBS for 2 h before overnight incubation with the indicated primary antibodies. The cells were then stained with a FITC- or Texas Red X-conjugated secondary antibody. The nuclei were stained with Hoechst 33258. Images were captured on a Zeiss fluorescence microscope (Axio Observer Inverted Microscope) equipped with ApoTome 2 (Zeiss). For ApoTome 2 acquisition settings, images were taken with “optical sectioning” in the ApoTome acquisition mode. Averaging number was set at “two,” and the ApoTome filter option was set at “strong.”
Cell cycle analysis and flow cytometry
Cells were fixed with 70% ethanol and then stained with propidium iodide followed by flow cytometry. At least 10,000 cells were analyzed for each sample using FlowJo software (Tree Star).
Cell proliferation assay
Cells were seeded into 60-mm plates at 2,000 cells/plate. After incubation for 24 h (day 0), the medium was aspirated and the plates were washed twice with 1× PBS. Plates were added with different condition mediums (serum-free, 0.1% BSA medium with/without 25 ng/ml of EGF) for the indicated days. Conditional mediums were replaced daily. For cell counting, the medium was aspirated and wells were rinsed once with PBS. Cells were then trypsinized and counted with a trypan blue method for live cells. All assays were performed in triplicate.
In vitro protein–protein association
For in vitro RNF144A-EGFR association experiments, the FLAG-tagged empty control vector and FLAG-tagged RNF144A were transfected into HEK 293T cells and cultured in regular media for 24 h. Transfected cells were lysed using RIPA lysis buffer and the lysates were used in FLAG-tagged protein immunoprecipitation overnight in Eppendorf tubes using an anti-FLAG M2 affinity-agarose beads (Sigma, A2220). Tubes were washed 5 times using RIPA lysis buffer and aliquoted into 6 tubes for each incubation of the indicated cell lysates or control incubation buffer. To prepare EGFR-containing cell lysates, a separate set of HEK 293T cells were starved for 24 h in 0.1% BSA serum-free media. Starved cells were then treated with or without EGF for 1 h and lysed with RIPA lysis buffer. 0.5 mg of 0.1% BSA or EGF-stimulated cell lysate was added to the affinity purified FLAG-RNF144A M2 beads and reconstituted by addition of 10-fold TNN lysis buffer for in vitro association assays. The tubes were incubated at 4 °C for 4 h, then were washed three times with TNN buffer and subjected to immunoblot analysis with 10% SDS-PAGE buffer.
Statistical analysis
All experiments were performed at least three times. Data are presented as mean ± S.D. and represent three independent experiments. Student's t test (two-tailed) was used for comparing group means, and p values < 0.05 were considered significant. Gene expression data in TCGA RNA-Seq database (IlluminaHiSeq pancan normalized) and EGFR protein levels in the TCGA RPPA database were extracted through https://xena.ucsc.edu/4 server and Pearson correlation coefficients were calculated to evaluate correlations. This study used public gene expression TCGA datasets including TCGA thyroid carcinoma (THCA), TCGA Colon Cancer (COAD), TCGA endometrioid carcinoma (UCEC), TCGA colon, and rectal cancer (COADREAD), and TCGA breast cancer (BRCA) and others. The information of breast cancer subtypes classified according to PAM50 RNA-Seq in TCGA breast cancer cohort was also obtained from xena.ucsc.edu server.4 For more detailed information on RNA-Seq and RPPA experiments, protocols and software, please visit the TCGA Data Portal at https://cancergenome.nih.gov/. In this study, the Pearson correlation coefficients >0.35 were considered to be a positive linear correlation.
Author contributions
S.-R. H. and W.-C. L. conceptualization; S.-R. H. and W.-C. L. data curation; S.-R. H. and W.-C. L. formal analysis; S.-R. H. and W.-C. L. validation; S.-R. H. and W.-C. L. investigation; S.-R. H. and W.-C. L. visualization; S.-R. H. and W.-C. L. methodology; S.-R. H. and W.-C. L. writing-original draft; S.-R. H. and W.-C. L. writing-review and editing; W.-C. L. resources; W.-C. L. software; W.-C. L. supervision; W.-C. L. funding acquisition; W.-C. L. project administration.
Supplementary Material
Acknowledgments
We thank the members of the W.-C. L. and Fang-Tsyr (Fannie) Lin laboratories for discussion. We also thank the BCM Genetically Engineered Mouse (GEM) core for the Rnf144a Knockout First mouse project.
This work was supported by National Institutes of Health Grants R01CA100857, R01CA203824, and R01CA138641 and Department of Defense Grants W81XWH-14-1-0339 and W81XWH-14-1-0306. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1-S11 and Movie S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- RBR
- ring between ring fingers
- EGFR
- epidermal growth factor receptor
- TM
- transmembrane
- DNA-PKcs
- DNA-dependent protein kinase catalytic subunit
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- aa
- amino acids
- Ub
- ubiquitin
- ADR
- adriamycin
- NCS
- neocarzinostatin
- ConA
- concanavalin A
- CQ
- chloroquine
- Noc
- nocodazole
- NHEJ
- nonhomologous end-joining
- MEF
- mouse embryonic fibroblast
- IP
- immunoprecipitation
- DMEM
- Dulbecco's modified Eagle's medium
- HA
- hemagglutinin
- qPCR
- quantitative PCR.
References
- 1. Marin I., Lucas J. I., Gradilla A. C., and Ferrús A. (2004) Parkin and relatives: the RBR family of ubiquitin ligases. Physiol. Genomics 17, 253–263 10.1152/physiolgenomics.00226.2003 [DOI] [PubMed] [Google Scholar]
- 2. Smit J. J., and Sixma T. K. (2014) RBR E3-ligases at work. EMBO Rep 15, 142–154 10.1002/embr.201338166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Spratt D. E., Walden H., and Shaw G. S. (2014) RBR E3 ubiquitin ligases: new structures, new insights, new questions. Biochem. J. 458, 421–437 10.1042/BJ20140006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakamura N. (2011) The role of the transmembrane RING finger proteins in cellular and organelle function. Membranes 1, 354–393 10.3390/membranes1040354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eisenhaber B., Chumak N., Eisenhaber F., and Hauser M. T. (2007) The ring between ring fingers (RBR) protein family. Genome Biol. 8, 209 10.1186/gb-2007-8-3-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boisson B., Laplantine E., Prando C., Giliani S., Israelsson E., Xu Z., Abhyankar A., Israël L., Trevejo-Nunez G., Bogunovic D., Cepika A. M., MacDuff D., Chrabieh M., Hubeau M., Bajolle F., et al. (2012) Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat. Immunol. 13, 1178–1186 10.1038/ni.2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duda D. M., Olszewski J. L., Schuermann J. P., Kurinov I., Miller D. J., Nourse A., Alpi A. F., and Schulman B. A. (2013) Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure 21, 1030–1041 10.1016/j.str.2013.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kelsall I. R., Duda D. M., Olszewski J. L., Hofmann K., Knebel A., Langevin F., Wood N., Wightman M., Schulman B. A., and Alpi A. F. (2013) TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes. EMBO J. 32, 2848–2860 10.1038/emboj.2013.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaugule V. K., Burchell L., Barber K. R., Sidhu A., Leslie S. J., Shaw G. S., and Walden H. (2011) Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 30, 2853–2867 10.1038/emboj.2011.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ho S. R., Lee Y. J., and Lin W. C. (2015) Regulation of RNF144A E3 ubiquitin ligase activity by self-association through its transmembrane domain. J. Biol. Chem. 290, 23026–23038 10.1074/jbc.M115.645499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ho S. R., Mahanic C. S., Lee Y. J., and Lin W. C. (2014) RNF144A, an E3 ubiquitin ligase for DNA-PKcs, promotes apoptosis during DNA damage. Proc. Natl. Acad. Sci. U.S.A. 111, E2646–2655 10.1073/pnas.1323107111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park S. J., Gavrilova O., Brown A. L., Soto J. E., Bremner S., Kim J., Xu X., Yang S., Um J. H., Koch L. G., Britton S. L., Lieber R. L., Philp A., Baar K., Kohama S. G., Abel E. D., Kim M. K., and Chung J. H. (2017) DNA-PK promotes the mitochondrial, metabolic, and physical decline that occurs during aging. Cell Metab. 25, 1135–1146e7 10.1016/j.cmet.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ray L. B. (2017) DNA damage linked to fitness loss in aging. Science 356, 1041 [DOI] [PubMed] [Google Scholar]
- 14. An J., Yang D. Y., Xu Q. Z., Zhang S. M., Huo Y. Y., Shang Z. F., Wang Y., Wu D. C., and Zhou P. K. (2008) DNA-dependent protein kinase catalytic subunit modulates the stability of c-Myc oncoprotein. Mol. Cancer 7, 32 10.1186/1476-4598-7-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takeuchi K., and Ito F. (2011) Receptor tyrosine kinases and targeted cancer therapeutics. Biol. Pharm. Bull. 34, 1774–1780 10.1248/bpb.34.1774 [DOI] [PubMed] [Google Scholar]
- 16. Brand T. M., Iida M., Li C., and Wheeler D. L. (2011) The nuclear epidermal growth factor receptor signaling network and its role in cancer. Discov. Med. 12, 419–432 [PMC free article] [PubMed] [Google Scholar]
- 17. Tomas A., Futter C. E., and Eden E. R. (2014) EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 24, 26–34 10.1016/j.tcb.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lemmon M. A., and Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henriksen L., Grandal M. V., Knudsen S. L., van Deurs B., and Grøvdal L. M. (2013) Internalization mechanisms of the epidermal growth factor receptor after activation with different ligands. PLoS ONE 8, e58148 10.1371/journal.pone.0058148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bryant D. M., Kerr M. C., Hammond L. A., Joseph S. R., Mostov K. E., Teasdale R. D., and Stow J. L. (2007) EGF induces macropinocytosis and SNX1-modulated recycling of E-cadherin. J. Cell Sci. 120, 1818–1828 10.1242/jcs.000653 [DOI] [PubMed] [Google Scholar]
- 21. Clark A. J., Ishii S., Richert N., Merlino G. T., and Pastan I. (1985) Epidermal growth factor regulates the expression of its own receptor. Proc. Natl. Acad. Sci. U.S.A. 82, 8374–8378 10.1073/pnas.82.24.8374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Earp H. S., Austin K. S., Blaisdell J., Rubin R. A., Nelson K. G., Lee L. W., and Grisham J. W. (1986) Epidermal growth factor (EGF) stimulates EGF receptor synthesis. J. Biol. Chem. 261, 4777–4780 [PubMed] [Google Scholar]
- 23. Kudlow J. E., Cheung C. Y., and Bjorge J. D. (1986) Epidermal growth factor stimulates the synthesis of its own receptor in a human breast cancer cell line. J. Biol. Chem. 261, 4134–4138 [PubMed] [Google Scholar]
- 24. Lin S. Y., Makino K., Xia W., Matin A., Wen Y., Kwong K. Y., Bourguignon L., and Hung M. C. (2001) Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3, 802–808 10.1038/ncb0901-802 [DOI] [PubMed] [Google Scholar]
- 25. Hanada N., Lo H. W., Day C. P., Pan Y., Nakajima Y., and Hung M. C. (2006) Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol. Carcinog. 45, 10–17 10.1002/mc.20147 [DOI] [PubMed] [Google Scholar]
- 26. Demory M. L., Boerner J. L., Davidson R., Faust W., Miyake T., Lee I., Hüttemann M., Douglas R., Haddad G., and Parsons S. J. (2009) Epidermal growth factor receptor translocation to the mitochondria: regulation and effect. J. Biol. Chem. 284, 36592–36604 10.1074/jbc.M109.000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Y., Pennock S., Chen X., and Wang Z. (2002) Endosomal signaling of epidermal growth factor receptor stimulates signal transduction pathways leading to cell survival. Mol. Cell Biol. 22, 7279–7290 10.1128/MCB.22.20.7279-7290.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rush J. S., Quinalty L. M., Engelman L., Sherry D. M., and Ceresa B. P. (2012) Endosomal accumulation of the activated epidermal growth factor receptor (EGFR) induces apoptosis. J. Biol. Chem. 287, 712–722 10.1074/jbc.M111.294470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Szumiel I. (2006) Epidermal growth factor receptor and DNA double strand break repair: the cell's self-defence. Cell Signal. 18, 1537–1548 10.1016/j.cellsig.2006.03.010 [DOI] [PubMed] [Google Scholar]
- 30. Liccardi G., Hartley J. A., and Hochhauser D. (2011) EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 71, 1103–1114 10.1158/0008-5472.CAN-10-2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang H., Nishishita T., Fitzgerald T., Landon E. J., and Inagami T. (2000) Inhibition of AT1 receptor internalization by concanavalin A blocks angiotensin II-induced ERK activation in vascular smooth muscle cells: involvement of epidermal growth factor receptor proteolysis but not AT1 receptor internalization. J. Biol. Chem. 275, 13420–13426 10.1074/jbc.275.18.13420 [DOI] [PubMed] [Google Scholar]
- 32. Bomsel M., Parton R., Kuznetsov S. A., Schroer T. A., and Gruenberg J. (1990) Microtubule- and motor-dependent fusion in vitro between apical and basolateral endocytic vesicles from MDCK cells. Cell 62, 719–731 10.1016/0092-8674(90)90117-W [DOI] [PubMed] [Google Scholar]
- 33. Gruenberg J., and Howell K. E. (1989) Membrane traffic in endocytosis: insights from cell-free assays. Annu. Rev. Cell Biol. 5, 453–481 10.1146/annurev.cb.05.110189.002321 [DOI] [PubMed] [Google Scholar]
- 34. Oksvold M. P., Huitfeldt H. S., Østvold A. C., and Skarpen E. (2002) UV induces tyrosine kinase-independent internalisation and endosome arrest of the EGF receptor. J. Cell Sci. 115, 793–803 [DOI] [PubMed] [Google Scholar]
- 35. Wu Y. H., Hong C. W., Wang Y. C., Huang W. J., Yeh Y. L., Wang B. J., Wang Y. J., and Chiu H. W. (2017) A novel histone deacetylase inhibitor TMU-35435 enhances etoposide cytotoxicity through the proteasomal degradation of DNA-PKcs in triple-negative breast cancer. Cancer Lett. 400, 79–88 10.1016/j.canlet.2017.04.023 [DOI] [PubMed] [Google Scholar]
- 36. Bandyopadhyay D., Mandal M., Adam L., Mendelsohn J., and Kumar R. (1998) Physical interaction between epidermal growth factor receptor and DNA-dependent protein kinase in mammalian cells. J. Biol. Chem. 273, 1568–1573 10.1074/jbc.273.3.1568 [DOI] [PubMed] [Google Scholar]
- 37. Dittmann K., Mayer C., Fehrenbacher B., Schaller M., Raju U., Milas L., Chen D. J., Kehlbach R., and Rodemann H. P. (2005) Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J. Biol. Chem. 280, 31182–31189 10.1074/jbc.M506591200 [DOI] [PubMed] [Google Scholar]
- 38. Friedmann B. J., Caplin M., Savic B., Shah T., Lord C. J., Ashworth A., Hartley J. A., and Hochhauser D. (2006) Interaction of the epidermal growth factor receptor and the DNA-dependent protein kinase pathway following gefitinib treatment. Mol. Cancer Ther. 5, 209–218 10.1158/1535-7163.MCT-05-0239 [DOI] [PubMed] [Google Scholar]
- 39. Friedmann B., Caplin M., Hartley J. A., and Hochhauser D. (2004) Modulation of DNA repair in vitro after treatment with chemotherapeutic agents by the epidermal growth factor receptor inhibitor gefitinib (ZD1839). Clin. Cancer Res. 10, 6476–6486 10.1158/1078-0432.CCR-04-0586 [DOI] [PubMed] [Google Scholar]
- 40. Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelsen T., Heckl D., Ebert B. L., Root D. E., Doench J. G., and Zhang F. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanjana N. E., Shalem O., and Zhang F. (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barbieri M. A., Roberts R. L., Gumusboga A., Highfield H., Alvarez-Dominguez C., Wells A., and Stahl P. D. (2000) Epidermal growth factor and membrane trafficking: EGF receptor activation of endocytosis requires Rab5a. J. Cell Biol. 151, 539–550 10.1083/jcb.151.3.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baumdick M., Brüggemann Y., Schmick M., Xouri G., Sabet O., Davis L., Chin J. W., and Bastiaens P. I. (2015) EGF-dependent re-routing of vesicular recycling switches spontaneous phosphorylation suppression to EGFR signaling. Elife 4, e12223 10.7554/eLife.12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vanlandingham P. A., and Ceresa B. P. (2009) Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J. Biol. Chem. 284, 12110–12124 10.1074/jbc.M809277200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ettenberg S. A., Magnifico A., Cuello M., Nau M. M., Rubinstein Y. R., Yarden Y., Weissman A. M., and Lipkowitz S. (2001) Cbl-b-dependent coordinated degradation of the epidermal growth factor receptor signaling complex. J. Biol. Chem. 276, 27677–27684 10.1074/jbc.M102641200 [DOI] [PubMed] [Google Scholar]
- 46. Levkowitz G., Waterman H., Zamir E., Kam Z., Oved S., Langdon W. Y., Beguinot L., Geiger B., and Yarden Y. (1998) c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12, 3663–3674 10.1101/gad.12.23.3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carpenter G., and Cohen S. (1976) 125I-labeled human epidermal growth factor: binding, internalization, and degradation in human fibroblasts. J. Cell Biol. 71, 159–171 10.1083/jcb.71.1.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lipkowitz S. (2003) The role of the ubiquitination-proteasome pathway in breast cancer: ubiquitin mediated degradation of growth factor receptors in the pathogenesis and treatment of cancer. Breast Cancer Res. 5, 8–15 10.1186/bcr667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haglund K., and Dikic I. (2012) The role of ubiquitylation in receptor endocytosis and endosomal sorting. J. Cell Sci. 125, 265–275 10.1242/jcs.091280 [DOI] [PubMed] [Google Scholar]
- 50. Dittmann K., Mayer C., and Rodemann H. P. (2005) Inhibition of radiation-induced EGFR nuclear import by C225 (Cetuximab) suppresses DNA-PK activity. Radiother. Oncol. 76, 157–161 10.1016/j.radonc.2005.06.022 [DOI] [PubMed] [Google Scholar]
- 51. Wong H., and Yau T. (2013) Molecular targeted therapies in advanced gastric cancer: does tumor histology matter? Therap. Adv. Gastroenterol. 6, 15–31 10.1177/1756283X12453636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leung L., Mok T. S., and Loong H. (2012) Combining chemotherapy with epidermal growth factor receptor inhibition in advanced non-small cell lung cancer. Ther. Adv. Med. Oncol. 4, 173–181 10.1177/1758834012440015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ayyappan S., Prabhakar D., and Sharma N. (2013) Epidermal growth factor receptor (EGFR)-targeted therapies in esophagogastric cancer. Anticancer Res. 33, 4139–4155 [PubMed] [Google Scholar]
- 54. Nakai K., Hung M. C., and Yamaguchi H. (2016) A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am. J. Cancer Res. 6, 1609–1623 [PMC free article] [PubMed] [Google Scholar]
- 55. Miyamoto Y., Suyama K., and Baba H. (2017) Recent advances in targeting the EGFR signaling pathway for the treatment of metastatic colorectal cancer. Int. J. Mol. Sci. 18, e752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Carter R. E., and Sorkin A. (1998) Endocytosis of functional epidermal growth factor receptor-green fluorescent protein chimera. J. Biol. Chem. 273, 35000–35007 10.1074/jbc.273.52.35000 [DOI] [PubMed] [Google Scholar]
- 57. Hsu S. C., and Hung M. C. (2007) Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J. Biol. Chem. 282, 10432–10440 10.1074/jbc.M610014200 [DOI] [PubMed] [Google Scholar]
- 58. Longo P. A., Kavran J. M., Kim M. S., and Leahy D. J. (2013) Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol. 529, 227–240 10.1016/B978-0-12-418687-3.00018-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.