Abstract
Immune therapy of cancer is among the most promising recent advances in medicine. Whether the immune system can keep cancer in check depends on, among other factors, the efficiency of immune cells to recognize and eliminate cancer cells. We describe a time-resolved single-cell assay that reports the quality, quantity, and kinetics of target cell death induced by single primary human natural killer (NK) cells. The assay reveals that single NK cells induce cancer cell death by apoptosis and necrosis but also by mixed forms. Inhibition of either one of the two major cytotoxic pathways, perforin/granzyme release or FasL/FasR interaction, unmasked the parallel activity of the other one. Ca2+ influx through Orai channels is important for tuning killer cell function. We found that the apoptosis/necrosis ratio of cancer cell death by NK cells is controlled by the magnitude of Ca2+ entry and furthermore by the relative concentrations of perforin and granzyme B. The possibility to change the apoptosis/necrosis ratio employed by NK cells offers an intriguing possibility to modulate the immunogenicity of the tumor microenvironment.
Keywords: natural killer cells (NK cells), cancer, apoptosis, necrosis (necrotic death), cell death, calcium, caspase, cellular immune response, imaging, immunology
Introduction
State-of-the-art immunotherapy against cancer includes CAR T-cell therapy (1, 2), checkpoint inhibitor therapy (3, 4), or other antibody-based therapies (5, 6), such as rituximab against CD20+ lymphoma (7, 8). Cancer therapies commonly converge on the cytotoxic arm of the immune system (3, 9, 10). Both cytotoxic T lymphocytes (CTL)3 and natural killer (NK) cells are involved in the immune surveillance of cancer. Whereas CTL recognize specific peptides presented by MHC-I using the T-cell receptor, NK cells analyze both activating and inhibitory ligands on the target cell with their germ line–encoded receptor repertoire and integrate these signals. Upon activation, killer cells form a tight contact with the target called the immune synapse (IS) and polarize lytic granules (LG) as well as other organelles and molecules toward the contact site (11, 12). LG contain the pore-forming protein perforin, which, upon release, generates holes in the target cell membrane and allows serine proteases called granzymes to enter the cytosol of the target cell. If this local membrane disruption cannot be neutralized by the target cell, cell death by rapid swelling and lysis follows (necrosis). If membrane integrity can be restored, the injected granzymes induce cell death by caspase activation and subsequent caspase-dependent apoptosis (13). In addition to the granule-based killing pathway, another well-established killing mechanism involves the interaction of Fas ligand (FasL) on cytotoxic cells with Fas receptor (FasR)-positive target cells inducing target cell apoptosis (14). Whereas CAR T-cell therapy has been well-established in the clinic for several years, much work is currently focused on exploring the anti-tumor potential of primary human NK cells and also the cell line NK-92 in the clinic (9, 15).
Malfunctioning of NK cells' and CTL's perforin is, in its severest case, associated with life-threatening hyperinflammation as part of hemophagocytic lymphohistiocytosis (16). Whereas the complete absence of perforin function is a rare disease presenting with severe symptoms during infancy (familial hemophagocytic lymphohistiocytosis 2), hypomorphic mutations with reduced perforin concentration or function are more frequent in the population (16, 17). Although not immediately life-threatening, a reduced perforin function is linked with increased susceptibility to late-onset hemophagocytic lymphohistiocytosis (16, 18) and development of cancer (17–19). These findings highlight the importance of analyzing NK cell and CTL cytotoxicity in a detailed and quantitative manner.
Results
Casper-GR as a single-cell apoptosis marker
Jurkat E6-1 and K562 cells are well-characterized targets for NK cytotoxicity. The FRET-based apoptosis reporter Casper3-GR (20) was stably transfected into both cell lines to create stable, monoclonal target cell lines named Jurkat pCasper and K562 pCasper to analyze NK cell–mediated cytotoxicity on a single-cell level. Casper-GR consists of a GFP and a red fluorescent protein (RFP) fused by a linker containing the caspase recognition sequence DEVD. Because the DEVD sequence is not caspase-3–specific but cleaved by caspase-3 and -7 as well as other caspases (21, 22), we prefer to refer to the construct as Casper and not Casper3 to acknowledge this fact. Due to the close proximity of GFP and RFP in Casper-GR and their matched emission and excitation spectra, a considerable part of the GFP energy will be transferred to RFP in a radiationless manner by FRET, leading to red fluorescence upon excitation with blue light. Upon cleavage of the linker by caspase activity, this red fluorescence will decrease as a result of the increasing distance of the fluorophores, whereas an increase in green fluorescence will be detected. These two target cell lines were chosen because they greatly differ in their expression of the Fas receptor (FasR) CD95. Jurkat E6-1 cells express FasR and K562 cells do not express FasR (23, 24).
To test the viability of Jurkat pCasper and K562 pCasper cells, both cell lines were imaged for 6 h at 37 °C, 5% CO2, acquiring GFP, RFP, and FRET emission. Counting dead cells after 6 h revealed that under these conditions, 4.1% of Jurkat pCasper and 1.8% of K562 pCasper cells (n = 296 or n = 284, respectively) showed signs of apoptosis (Fig. 1, a and b), comparable with control cells. Thus, monoclonal viable Jurkat pCasper and K562 pCasper cells positive for both GFP and RFP expression (Fig. S1) were successfully generated.
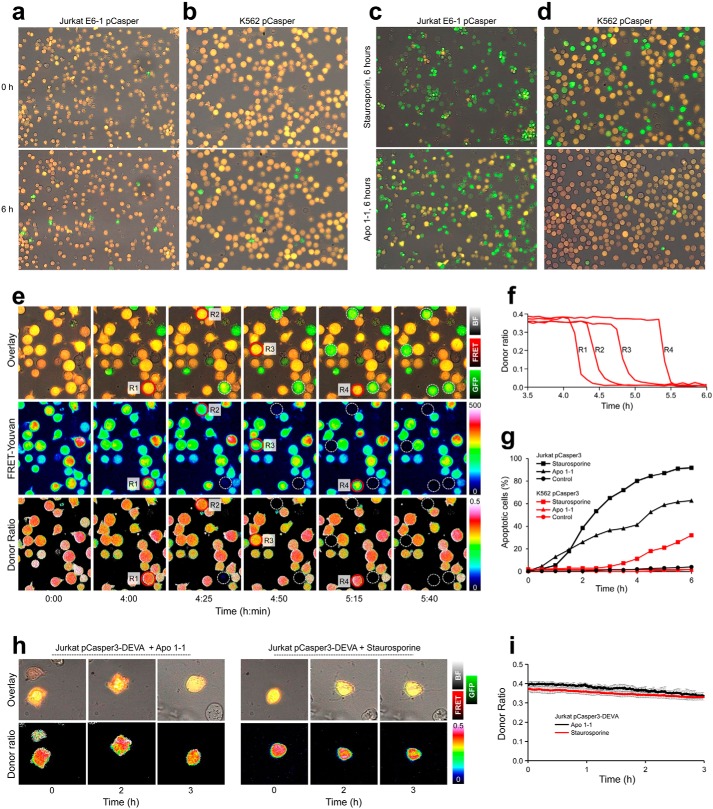
Figure 1.
Viability and apoptosis analysis of Jurkat pCasper (Jurkat pC) and K562 pCasper (K562 pC) cells. a and b, viability of Jurkat pC and K562 pC cells was analyzed in 96-well plates. Cells were imaged every 5 min in the brightfield, FRET fluorescence of pCasper, and GFP fluorescence of pCasper with a ×20 objective over 6 h with the Cell Observer microscope under incubator conditions (37 °C, 5% CO2). Spontaneous signs of apoptosis are indicated by the reduction of the FRET signal and corresponding gain of the GFP signal (green color in the overlay of the brightfield, GFP, and FRET channels). Counting dead cells after 6 h revealed that in these conditions, >95% were viable. c and d, the same experimental setup as in a or b, except that 2.5 μm staurosporine or 5 μg/ml anti-CD95 antibody Apo 1-1 was applied at time 0. e, quantification of FRET signals. As in c, 2.5 μm staurosporine was applied to K562 pC cells at time 0. In addition to the brightfield, GFP, and FRET overlay as shown in d, FRET-Youvan and FRET donor ratio are calculated (both color-coded). Low values are shown in black to dark blue, and high ones are shown in red to white. R1–R4 are marked and turn apoptotic during the experiment. f, FRET donor ratio kinetics of the cells R1–R4 from e over 6 h. g, quantification of all cells from a–d. The time point of apoptosis induction in each cell was quantified by the abrupt reduction of the FRET donor ratio signal as shown in f. h, Jurkat cells were transfected with the Casper-GR construct with a DEVA mutation in the caspase-binding site. Overlay of brightfield, GFP, and FRET channel and the FRET donor ratio is depicted for two cells stimulated with 2.5 μm staurosporine or 5 μg/ml anti-CD95 antibody Apo 1-1. Both cells are clearly apoptotic after 2 h, as indicated by morphological changes but show no change in fluorescence signals or the FRET donor ratio (h, quantification of n = 10 cells per condition in i).
Using flow cytometry analysis and quantitative RT-PCR, we confirmed the findings by others (23, 24) that Jurkat E6-1 but not K562 cells express FasR (Fig. S2 (a and b) and Table S1). Importantly for our study, this expression profile was not changed by stable pCasper-GR transfection (Fig. 2, c and d).
Figure 2.
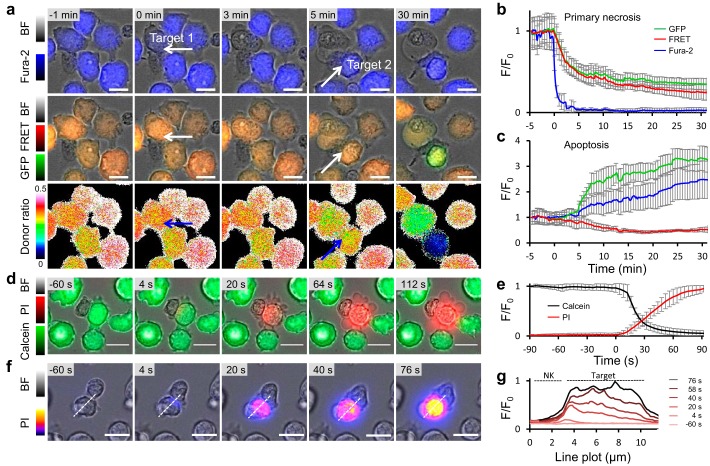
Quantification of target cell apoptosis and necrosis by pCasper induced by primary human NK cells. a, nonlabeled NK cells were applied to Jurkat pCasper target cells loaded in addition with Fura2-AM. Cells were imaged in AIMV medium with the Cell Observer every 10 s, and overlays of brightfield + Fura-2 (at 360 nm) or brightfield + GFP + FRET and the FRET donor ratio are shown over time. White arrows mark the two contact sites between one NK cell and two different target cells. b, average Fura-2, GFP, and FRET fluorescence of five target cells from three experiments with typical necrotic morphological changes (cell swelling, loss of cytosol and organelles), such as the one marked by the white arrow in a at time 0. c, average Fura-2, GFP, and FRET fluorescence of five target cells from three experiments with typical apoptotic morphological changes (cell shrinking, blebbing, stop of organelle movement), such as the one marked by the white arrow in a at time 5 min. d, calcein (500 nm)-loaded Jurkat E6-1 target cells were imaged with the Cell Observer in 0.5 mm Ca2+ Ringer's solution containing 100 μm propidium iodide. Time 0 was set to the start of fluorescence changes of 1%. One representative killing event is depicted. e, average fluorescence of seven cells from five experiments, such as the one shown in d. f, same as d except that calcein fluorescence is not shown, and propidium iodide is indicated by a color code. One representative killing event is depicted. g, relative fluorescence changes (F/F0) of the cell shown in f along the dotted line starting from the nonlabeled NK cell. Scale bars, 10 μm. Error bars, S.E.
To verify pCasper-GR functionality, Jurkat pCasper (Fig. 1c) or K562 pCasper (Fig. 1d) cells were incubated for 6 h with the apoptosis inducer staurosporine (2.5 μm) or the CD95-activating antibody Apo 1-1 (5 μg/ml). In both cell lines, staurosporine caused cell death (Fig. 1 (c and d), top panels), which was verified by cell blebbing and cell shrinking. Fluorescence intensity decreased in the FRET channel (red) and correspondingly increased in the GFP channel (green), indicating caspase dependent cleavage of the recognition sequence DEVD. The functionality of the CD95 antibody Apo 1-1, however, was restricted to Jurkat pCasper cells (Fig. 1c, bottom). K562 pCasper cells showed no sign of cell death following Apo 1-1 incubation, and cells remained orange (Fig. 1d, bottom). These results confirm functionality of Jurkat pCasper and K562 pCasper cell lines to specifically measure caspase-dependent apoptosis and also provide further functional evidence for the presence of FasR in Jurkat pCasper but not in K562 pCasper cells.
To define time points of apoptosis induction, we analyzed the FRET donor ratio according to Youvan et al. (25), which has the great advantage that it is intensity-independent. Fig. 1e shows the time course of an experiment similar to the one shown in Fig. 1d. Application of 2.5 μm staurosporine induces the change from orange to green fluorescence in K562 pCasper cells at different time points (four cells are marked R1–R4). The FRET signal and the FRET donor ratio after Youvan et al. (25) were calculated (both color-coded). Fig. 1f shows that in the donor ratio time series, the signals of cells R1–R4 disappear at different time points but remain constant before this time point. The change in slope can be easily quantified and is a very reliable measure for apoptosis induction. This method was used to quantify data as shown in Fig. 1 (a–d). The results are shown in Fig. 1g and represent the time course of apoptosis induction in Jurkat pCasper and K562 pCasper cells by staurosporine or Apo 1-1. In Jurkat pCasper cells, both staurosporine and Apo 1-1 induce apoptosis without much delay. In K562 pCasper cells, staurosporine induces apoptosis after a delay of 2–3 h. Apo 1-1 has no effect, which is expected, considering the absence of the FasR CD95.
To test the specificity of the Casper-GR construct, we mutated its DEVD binding site to DEVA, which cannot be cleaved by caspase. Jurkat E6-1 cells were transiently transfected and exposed to the same Apo 1-1 or staurosporine concentrations as in Fig. 1c. Two representative cells are shown in Fig. 1h. Despite clear morphological signs of apoptosis induction (blebbing and loss of organelle movement), there was no change in the overlay of FRET fluorescence and GFP signal of Casper-GR (top) or in the donor ratio (bottom), as also evident from the average of 10 cells for each condition (Fig. 1i). We did not observe a single transfected cell with any sign of fluorescence changes. In conclusion, Jurkat pCasper and K562 pCasper cells are reliable tools to report the exact time point of caspase-dependent apoptosis induction. The change of the caspase-sensitive recognition sequence DEVD to the insensitive DEVA sequence rendered the FRET sensor inactive during apoptosis.
Casper-GR reports apoptosis and necrosis in target cells following contact with NK cells
NK cells can induce apoptotic or necrotic cell death in their respective targets and potentially also induce “mixed” forms of target cell death. A main difference between apoptotic and necrotic cell death is the integrity of the cell membrane, which is at least initially kept intact during apoptosis but is immediately disrupted during necrosis (26).
Because Jurkat E6-1 and K562 cells differ in the expression of the FasR (23, 24), they can be used to analyze NK cell–mediated perforin/granzyme cytotoxicity either in combination with (Jurkat pCasper) or without (K562 pCasper) concomitant FasL/FasR-induced apoptosis.
To quantify target cell death by primary human NK cells, Jurkat pCasper target cells were additionally loaded with Fura-2, a small-molecule calcium indicator with excitation spectra well-separated from GFP and RFP. Fura-2 was excited close to its isosbestic point, where fluorescence does not depend on the calcium concentration. It was thus used here as a relatively small cytosolic dye to report target cell lysis by necrosis, because it will rapidly diffuse out of the cell once significant membrane damage has been inflicted. Fig. 2a shows three different fluorescence signals of the same Fura-2–loaded Jurkat pCasper target cells: overlay of bright field signal and Fura-2 fluorescence (top); overlay of bright field signal, FRET fluorescence of Casper-GR, and GFP signal of Casper-GR (middle); donor ratio (FRET/GFP) (bottom). In this example, one NK cell approaches two target cells (marked by white arrows). The Fura-2 signal sharply deceases at 0 min in target 1 once contacted by the NK cell (Fig. 2a), averaged in Fig. 2b for five targets. In parallel, the Casper-GR fluorescence signal starts to decline, albeit with slower kinetics due the large molecular weight of the sensor protein. These changes imply that target cell integrity is lost, indicative of necrosis. The donor ratio of target 1 (Fig. 2a, bottom) is maintained over time with no abrupt decrease, indicating that the target is not apoptotic.
The Fura-2 signal of target 2 does not decrease over time; it even slightly increases (because the target slightly shrinks). However, 5 min after contact with the same NK cell, the FRET signal of target 2 decreases, which is paralleled by an increase of the GFP signal, indicative of a caspase-dependent cleavage of the Casper-GR DEVD site, which diminishes the FRET efficiency and increases the GFP signal as averaged in five targets (Fig. 2c). Combining the drop of the FRET signal and the increase of the GFP signal, the donor ratio sharply drops (Fig. 2a, bottom). Target 2 thus goes into apoptosis.
Because the rapid, parallel loss of both GFP and FRET fluorescence proved sufficient to determine killing by necrosis and because of potential harm to cell viability caused by UV light excitation, we proceeded to perform all subsequent imaging without Fura-2.
To further test membrane disruption as observed by dye loss during NK cell–induced killing of target 1, two dyes were combined, as measured by Lopez et al. (27). Calcein was loaded in Jurkat E6-1, and propidium iodide was kept in the supernatant. Imaging revealed that after encountering a target cell, NK cells can induce the loss of the target cell's cytosolic dye (calcein) and the parallel intrusion of the supernatant dye (propidium iodide; Fig. 2d and quantified in Fig. 2e). To verify that disruption of the target cell's integrity is caused by the NK cell at the contact site, the fluorescence intensity of the intruding dye, propidium iodide, was analyzed. Using a line plot, which starts at the IS and dissects the target cell in half (Fig. 2f), propidium iodide fluorescence intensity was plotted over time. Results confirm that propidium iodide enters the target cell at the IS and then gradually distributes to the distant part of the target cell (Fig. 2g), as described previously by Lopez et al. (27). This shows that NK cells are able to induce target necrosis by a disruption in the target cell's membrane at the contact site, the IS.
In conclusion, Casper-GR can reliably be used as an apoptosis and necrosis sensor that reports single-target cell death induced by primary human NK cells. Interestingly, the same NK cell is able to kill two different targets within minutes by two different killing modes during serial killing (see also Video S1).
Distinguishing the target cell death spectrum by Casper-GR fluorescence pattern following NK cell contact
Analyzing cell morphology and Casper-GR fluorescence in parallel offers the possibility to categorize the target cell death spectrum following contact with primary human NK cells. We have analyzed, classified, and quantified different modes of cell death in Jurkat pCasper cells killed by human NK cells. Fig. 3a depicts a target cell that shows typical apoptosis signs after NK cell contact; it shrinks and blebs. This is paralleled by a prominent increase of GFP fluorescence and a decrease in the FRET signal. Quantification is shown in Fig. 3b. In particular, the GFP signal increase and the concomitant donor ratio decrease offer an exact temporal quantification of apoptosis initiation as shown before in Fig. 2.
Figure 3.
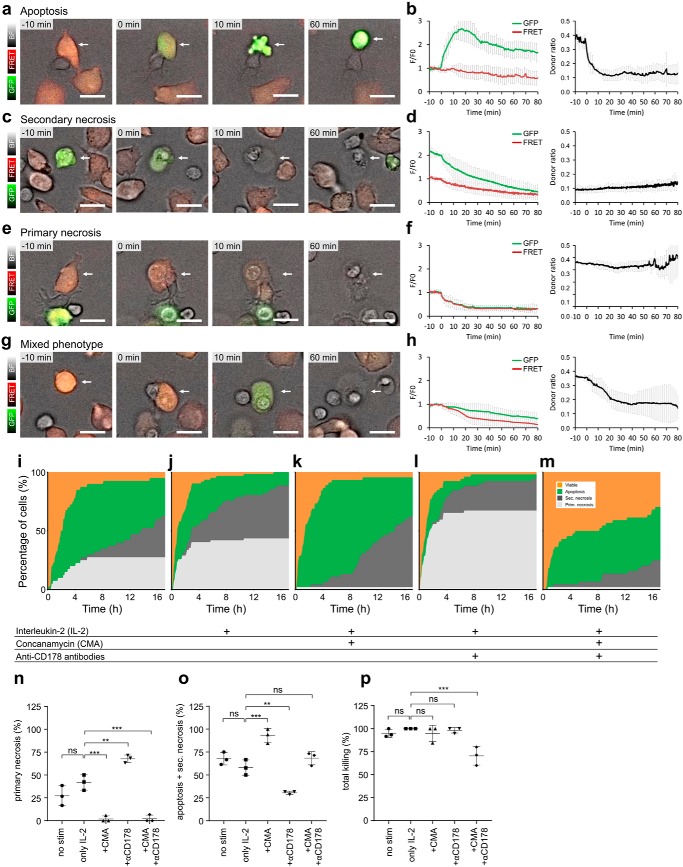
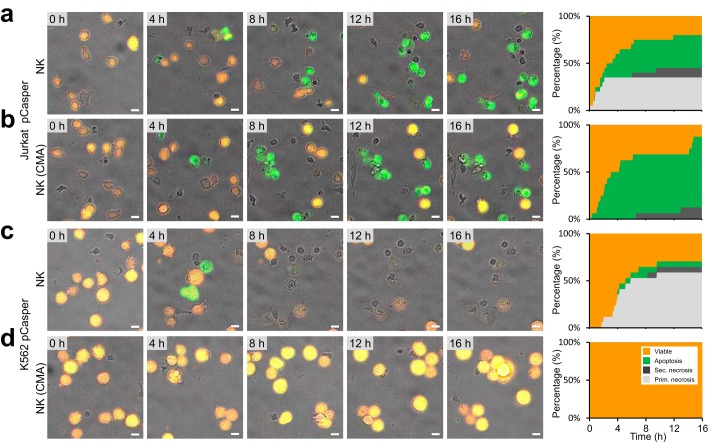
NK cell contact–dependent target cell death plot. Jurkat pCasper cells can be used to distinguish target cell apoptosis, primary necrosis, secondary necrosis (after initial apoptosis), and a mixed phenotype (parallel apoptosis and necrosis) induced by NK cells. Cells are from seven experiments for a–h. a, example of target cell apoptosis (white arrow), as evident by the morphological changes (blebbing, cell shrinking, stop of organelle and cell movement). b, time-resolved averaged normalized GFP and FRET fluorescence and the calculated FRET donor ratio for 8 cells. c, example of target cell secondary necrosis following an earlier apoptosis, as evident from the increased GFP signal (white arrow) evident by the morphological changes (cell swelling, loss of cytosol, and organelles). d, time-resolved averaged normalized GFP and FRET fluorescence and the calculated FRET donor ratio for five cells. e, example of target cell primary necrosis (with no evident apoptosis; white arrow), as evident by the morphological changes (cell swelling and loss of cytosol and organelles). f, time-resolved averaged normalized GFP and FRET fluorescence and the calculated FRET donor ratio for six cells. g, example of target cell parallel apoptosis and necrosis (mixed phenotype, necrosis with caspase activity; white arrow), as evident by the morphological changes (blebbing and cell swelling) and fluorescence changes. h, time-resolved averaged normalized GFP and FRET fluorescence and the calculated FRET donor ratio for 5 cells. i–m, primary NK cells were left untreated or stimulated with 100 units/ml IL-2 for 24 h. Cells were settled in a 96-well plate and observed for 17 h with the high-content imaging microscope under incubator conditions (37 °C, 5% CO2). For each condition, six experiments (two experiments from each of three blood donors to exclude individual donor effects) were analyzed. The color code for target cells is as follows: viable (orange), apoptosis (green), secondary necrosis after apoptosis (dark gray), and primary necrosis including the mixed phenotype (light gray). Time point 0 marks the onset of apoptosis or primary/secondary necrosis, respectively. i, analysis of 40 NK cells with target cell contacts. j, 62 NK cells stimulated with 100 units/ml IL-2. k, 45 NK cells treated with 100 nm concanamycin A to inhibit necrosis. l, 52 NK cells treated with 20 μg/ml CD178 antibodies (two different clones) to inhibit apoptosis. m, 44 NK cells treated with 100 nm concanamycin A and 20 μg/ml CD178 antibodies (two different clones) to inhibit necrosis and apoptosis. NK to target cell ratio was 1:1, 10,000 each), about 62,000 cells/cm2. Scale bars, 10 μm. n, percentage of Jurkat pCasper target cells killed by primary necrosis after 17 h. o, percentage of Jurkat pCasper target cells killed by apoptosis and secondary necrosis after 17 h. p, percentage of total killing of Jurkat pCasper target cells after 17 h. Data are shown as mean ± S.D. (error bars) (n = 3 donors). p values were calculated using ordinary one-way analysis of variance. **; p < 0.01; ***, p < 0.001; ns, no significant difference.
In some of the initially apoptotic target cells, a sudden drop of both the higher GFP signal (after apoptosis) and FRET fluorescence was observed (compare GFP fluorescence between 0 and 10 min in Fig. 3c). This was paralleled by a volume increase of the target cell, indicating that the cell was lysed. We refer to this type of cell death as secondary necrosis after the target cell was initially killed by apoptosis. Initiation of secondary necrosis can be exactly quantified by the abrupt decline in both GFP and FRET signals as indicated at time 0 in Fig. 3d, whereas the donor ratio remains unchanged.
In target cells that show an increase of volume after NK cell contact without any sign of apoptosis (Fig. 3e), a parallel and quantitatively identical decrease of GFP and FRET signals was observed (Fig. 3f), indicating that target cell lysis was initiated. The donor ratio remains constant with no signs of apoptosis. We refer to this type of cell death as primary necrosis because cells lose their integrity and burst open without prior caspase activity.
In some target cells, we observed a mix of morphological signs indicative of apoptosis and necrosis in the target cells (volume increase and cells turning from orange to green). In these target cells, both GFP and FRET signals decreased (Fig. 3, g and h); however, the GFP signal decreased to a lesser extent compared with the FRET signal, which results in a loss of RFP fluorescence and less orange cell appearance (and thus more green). This is also evident by the decrease in donor ratio, indicative of apoptosis. Target cell necrosis and apoptosis are initiated within 1 min (at approximately the same time point; Fig. 3h), and we thus refer to this mode of cell death as a mixed phenotype.
In summary, Casper-GR reliably reports a spectrum of target cell death modes following contact with primary human NK cells. Our data also reveal that NK cells kill target cells in multiple ways ranging from apoptosis with or without secondary necrosis via mixed phenotypes to primary necrosis. Interestingly, the very same NK cell can kill one target by primary necrosis and the next one within very few minutes by apoptosis. The target cell death spectrum is not “black and white” with only apoptosis and necrosis, but intermediates exist, and primary human NK cells make full use of the spectrum. By a combined quantitative analysis of GFP, FRET, and donor ratio, the exact starting point of the cell death mode can be determined.
We used Casper-GR fluorescence changes to create a target cell death plot after NK cell contact for more than 50 cells analyzed from three different blood donors (Fig. 3i). To create the target cell death plot, target cells were randomly chosen, with the only exclusion criterion that they did not escape from the field of observation during the entire length of the experiment. The total number of randomly chosen target cells was set to 100%, and they are presented as a stacked line plot. During long-term experiments of 17 h, NK cells initially kill their targets by either primary necrosis (light gray) or apoptosis (green), whereas at later times, apoptosis, partially followed by secondary necrosis (dark gray), is predominant. Mixed cell death forms are included in primary necrosis in this analysis. Total killing by either primary necrosis or apoptosis after 17 h is also quantified as mean ± S.D. of three donors (Fig. 3, n and o). 68% of all target cells were apoptotic or secondary necrotic after 17 h. 27% were killed via primary necrosis. Total killing was 95%; 5% of target cells survived. Without NK cells, almost all targets remain viable over the time of an experiment, as also indicated in Fig. 1a.
In cell culture, IL-2 is routinely added to primary NK cells to increase their perforin content and make them more killing-competent (28). After 24 h of IL-2 stimulation, there was a tendency of an increased share of primary necrosis by 15% to a total of 42% (Fig. 3, j and n), although this difference did not reach statistical significance (p = 0.077). There is, however, a clear increase in the speed of early necrosis induction within the first 2–3 h. The respective share of apoptotic and secondary necrotic cells was lowered to 58% (Fig. 3o). Total killing was 100% (Fig. 3, j and p). In conclusion, necrotic killing was enhanced by IL-2 stimulation at the expense of apoptotic killing, with the total killing remaining constant over time.
Quantification of perforin/granzyme versus FasL/FasR-induced target cell death by NK cells
NK cells can kill their targets by perforin/granzyme stored in lytic granules or by the FasL/FasR mechanism (14, 28). We tested the function of both pathways in four different ways using the single-cell death assay.
Pharmacological intervention
At low nanomolar concentrations, concanamycin A (CMA) can be used to interfere with the perforin/granzyme pathway without interfering with the FasL/FasR mechanism (14, 29). Application of CMA indeed inhibited primary necrosis in target cells induced by NK cells almost completely (down to 2%) (Fig. 3, k and n). Interestingly, the apoptotic portion including secondary necrosis increased to 93%, unmasking FasL/FasR activity following inhibition of the perforin/granzyme pathway (Fig. 3, k and o).
Whereas CMA proved to be a very potent inhibitor of perforin based killing, it is more difficult to pharmacologically interfere with FasL/FasR killing. Antibodies against FasL have been shown to partially inhibit FasL toxicity. Using a combination of two clones (NOK-1 and NOK-2) to bind and block two similar but distinct FasL epitopes (30), the apoptotic portion was lowered by 28% to a total of 30% apoptotic and secondary necrotic cells compared with the IL-2 control (Fig. 3, l and o). Thus, as expected, anti-CD178 antibodies could partially reduce apoptosis. Interestingly, the portion of primary necrosis killing was increased by 26–68%, unmasking the perforin/granzyme activity in the case of FasL/FasR inhibition (Fig. 3, l and n).
Finally, combining CD178 antibodies and CMA to inhibit both apoptosis and necrosis, primary necrosis was blocked almost completely to 2% as by CMA alone (Fig. 3, m and n), whereas apoptosis combined with secondary necrosis remained largely unchanged (Fig. 3, m and o). Only under these conditions was total killing significantly reduced; otherwise, total killing remained the same (Fig. 3p).
Induction of apoptosis and necrosis via purified perforin and granzyme B
To elucidate the potential and ratio of necrosis or apoptosis induction by perforin and granzyme B, we tested a concentration series of purified perforin and granzyme B on target cells in the absence of NK cells (Fig. 4).
Figure 4.
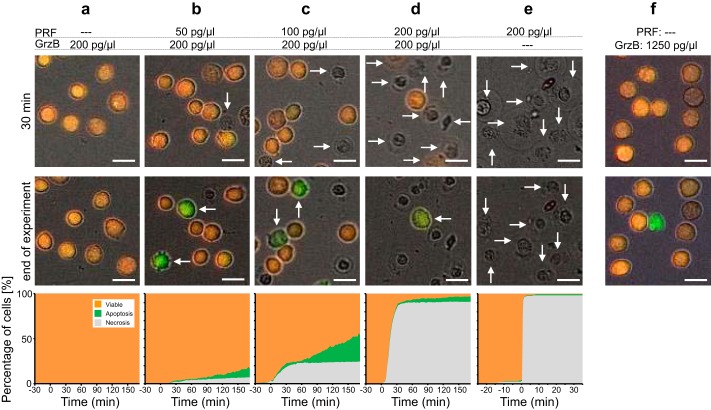
Induction of apoptosis and necrosis via purified perforin and granzyme B. Target cell death of Jurkat pCasper cells was analyzed following the addition of different combinations of recombinant perforin and granzyme B. a, 0 pg/μl perforin and 200 pg/μl granzyme B (63 cells); b, 50 pg/μl perforin and 200 pg/μl granzyme B (445 cells); c, 100 pg/μl perforin and 200 pg/μl granzyme B (516 cells); d, 200 pg/μl perforin and 200 pg/μl granzyme B (393 cells); e, 200 pg/μl perforin and 0 pg/μl granzyme B (167 cells). f, example with 0 pg/μl perforin and 1250 pg/μl granzyme B. Each experiment was carried out in a single well at incubator conditions (37 °C, 5% CO2) in Ca2+-free Ringer's. At −30 min, perforin and granzyme B were added, and at 0 min, 1.25 mm Ca2+ was added, which is required for perforin activity. Two time points and the target cell death plot of all cells are shown for each condition. Scale bars, 10 μm.
Without perforin, a concentration of 200 pg/μl granzyme B did not induce apoptosis, and all target cells remained viable (Fig. 4a). The addition of increasing perforin concentrations with granzyme B kept at 200 pg/μl led to a dose-dependent induction of necrosis (Fig. 4, b–d) up to 91% at 200 pg/μl perforin (Fig. 4d) with varying amounts of apoptosis induction, which was highest at 100 pg/μl perforin (Fig. 4c). Perforin alone only induced necrosis with no apoptosis present (Fig. 4e). We conclude that at high perforin concentrations, target cell necrosis is predominant, probably because target cells cannot repair membrane damage by perforin, whereas at intermediate concentrations, perforin and granzyme B also induce apoptosis probably by the perforin-dependent uptake of granzyme B.
To test whether granzyme B may enter the cell in the absence of perforin via endocytosis and induce apoptosis, granzyme B concentration was increased to 1250 pg/μl. At this concentration, granzyme B was able to induce apoptosis in a low number of cells (11%), as indicated in the example in Fig. 4f.
In conclusion, perforin and granzyme B, besides inducing necrosis in a perforin concentration–dependent way, are also able to induce a significant amount of target cell apoptosis and thus very likely contribute to target cell apoptosis induced by NK cells.
Lytic granules accumulate at the immune synapse before necrosis induction
The complete inhibition of primary necrosis by interfering with perforin release following CMA incubation, in combination with the finding that perforin acts exclusively at the immune synapse (Fig. 2f) (27, 31), predicts that lytic granules should always be present at the IS in case NK cells kill by necrosis but not necessarily if they kill by apoptosis. To correlate target cell death with NK lytic granule movement, we stained lytic granules with LysoTracker Red and acquired three-dimensional (3D) time-lapse data. LysoTracker Red not only marks lytic granules (and lysosomal compartments) in NK cells but (unfortunately) also in target cells. Therefore, fluorescently labeled structures in the NK cell are marked by a white arrow to distinguish them from the structures in the target cells, in which the plasma membrane is highlighted with a white line. Fig. S3a shows that the bulk of lytic granules in NK cells are translocated as a group. For every immune synapse analyzed, necrotic target cell death occurred only after the group of lytic granules arrived at the IS, evident by the LysoTracker fluorescence signal within the NK cell close to the plasma membrane of the target cell (Fig. S3a, six of six cells). To kill several target cells, the group of lytic granules moved sequentially from IS to IS (Fig. S3a) from the first IS formed with the right target to the second one formed with the left target. In case NK cells killed targets by apoptosis, lytic granules were not necessarily relocated to the IS. Fig. S3b shows that apoptosis of the target is initiated, as indicated by the increased GFP signal, without any LysoTracker signal being close to the IS. The statistics confirm that in 50% of apoptosis cases, lytic granules were not localized at the IS, whereas in the other 50% they were (n = 8 cells). Thus, there is no correlation between target cell apoptosis and lytic granule localization at the IS. This result is in very good agreement with the finding that perforin release from lytic granules is not required for apoptosis (Fig. 3k). However, perforin and granzyme are able to induce apoptosis without FasL/FasR activation (Fig. 4c).
Analysis of Fas-deficient target cells
To analyze the role of FasR, K562 pCasper cells that express FasR were compared with Jurkat pCasper cells. We used NK cells from the same blood donor to compare target cell death in both cell lines. Figs. 5 (a and b) confirms what was already shown for other blood donors in Fig. 3; NK cells kill Jurkat pCasper cells both ways, by apoptosis and primary necrosis, whereas in CMA-treated Jurkat pCasper cells, primary necrosis is completely inhibited.
Figure 5.
Ratio of apoptosis and necrosis in FasR-expressing compared with FasR-deficient target cells. 75.000 Jurkat pCasper (a and b) or K562 pCasper (c and d) target cells were settled in each well of a 96-well plate and observed for 17 h with the high-content imaging microscope under incubator conditions in AIMV medium (37 °C, 5% CO2) on a fibronectin-coated surface. NK cells (a and c) or NK cells incubated with 100 nm CMA (b and d) from the same blood donor were added at time 0. Target cell death following NK contact was monitored in the brightfield + GFP + FRET overlay and quantified as shown in Fig. 3. For quantification, 213 (a), 236 (b), 88 (c), and 143 (d) cells were analyzed from four experiments. Scale bars, 10 μm.
The same experiments were carried out with FasR-deficient K562 pCasper targets (Fig. 5, c and d). The probability of primary necrosis induced by NK cells was found to be very high, with only very little apoptosis (and secondary necrosis) present. Following incubation with CMA, neither apoptosis nor necrosis of target cells were observed. This confirms that the perforin/granzyme pathway is responsible for all primary necrosis but in addition also initiates apoptosis (and secondary necrosis) in some targets (Fig. 5c), as already suggested by the experiments with purified perforin and granzyme (Fig. 4c).
Calcium influx changes the apoptosis/necrosis ratio
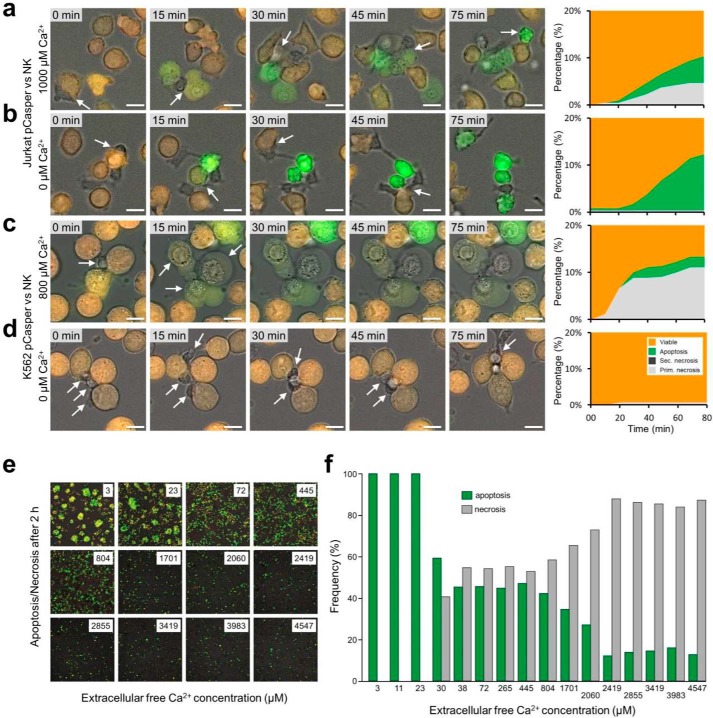
Ca2+ entry through Orai channels in NK cells and the concomitant Ca2+ signals are highly relevant for target cell death following immune synapse formation between NK cells and their targets (29, 32–34). Using the single-cell assay, we tested whether Ca2+ entry influences the apoptosis/necrosis ratio. With 1 mm extracellular Ca2+ present, Jurkat pCasper target cells were killed by NK cells similarly as shown before in AIMV medium, which contains 800 μm free Ca2+ (29) with a balanced ratio of apoptosis and primary necrosis, analyzed during 80 min (Fig. 6a). With no external Ca2+ added and the remaining Ca2+ buffered by EGTA, primary necrosis was completely inhibited (Fig. 6b) because release of lytic granules and the action of perforin both require Ca2+ (29, 33, 35, 36). Apoptosis was increased under these conditions, unmasking the activity of the FasL/FasR pathway.
Figure 6.
Calcium influx regulates the apoptosis to necrosis ratio. a–d, Jurkat pCasper and K562 pCasper were settled on a fibronectin-coated coverslip in the presence (a and c) or absence (b and d) of external calcium. NK cells were added at time 0, and killing events were observed for 75 min via brightfield + GFP + FRET overlay. a, in the presence of external calcium, the FasR-positive target cells Jurkat pCasper cells were killed in a mixture of necrosis, apoptosis, and mixed forms via the cumulative effect of perforin, granzymes, and FasL/FasR. b, in the absence of calcium, Jurkat pCasper cells were only killed via apoptosis due to the perforin's inability to adhere at the target cell's membrane. The residual FasL/FasR pathway is able to induce apoptosis in the absence of external calcium. c, in the presence of external calcium, the FasR-deficient K562 pCasper cells are killed via necrosis, accompanied by some mixed forms caused by perforin-mediated granzymes. d, in the absence of external calcium, K562 pC cells are not killed at all, because perforin cannot attach to the K562's membrane; nor can apoptosis be induced due to the lack of FasR. e, during cytotoxicity experiments as outlined in a, the external Ca2+ concentration was varied over a wide range by adding EGTA or CaCl2 to AIMV. Apoptotic cells are visible in a green color. f, frequency of apoptotic and necrotic Jurkat pCasper cell killing as a function of the external free Ca2+ concentration taken from pictures as shown in e 2 h after incubation with NK cells. Scale bars, 10 μm.
In cases where FasR-deficient K562 pCasper cells were used as targets, NK cells killed the target mostly by primary necrosis with very little apoptosis in the presence of 800 μm free Ca2+ in AIMV medium (Fig. 6c), induced by the perforin/granzyme mechanism. However, with drastically reduced external Ca2+ (buffered with 1 mm EGTA, which decreases free Ca2+ in AIMV below 3 μm (29)), both necrosis and apoptosis were basically absent (Fig. 6d), because 1) perforin release at the IS and perforin-mediated target cell lysis both require Ca2+, and 2) FasR is not expressed in K562 cells.
We tested the Ca2+ dependence of the apoptosis/necrosis ratio in more detail. External Ca2+ concentrations between 3 and 4547 μm were applied to the Jurkat pCaper3 target cells, and NK cell–dependent target cell apoptosis and necrosis were quantified after 2 h. Whereas apoptosis is more frequent at low Ca2+ (green cells and green bars in Fig. 6 (e and f)), necrosis frequency is significantly increased at higher Ca2+ (gray bars in Fig. 6f). At very low Ca2+, perforin action is impaired (36), and thus necrosis is completely absent below 30 μm. Ca2+ influx largely controls the apoptosis/necrosis ratio.
Manual versus automated apoptosis and necrosis analysis
As described under “Experimental procedures,” we analyzed each cell presented in this work individually and manually confirmed positioning of ROIs and fluorescence values. This analysis method is very reliable but, obviously, time-consuming; therefore, we developed a semi-automated analysis routine that can be performed with standard software packages. Target cells were detected using Imaris (Bitplane), and the required mathematical calculations were automated in Excel as described under “Experimental procedures.”
We compared the results of manual and automated analysis for three blood donors (Fig. S4) using the same experimental conditions as shown in Fig. 3 (i–m). Between blood donors there are slight differences regarding cytotoxicity of their respective NK cells. The automated analysis matches the manual one qualitatively very well and, in most cases, also quantitatively with two exceptions. First, the automated analysis used here does not distinguish between primary and secondary necrosis. Both are counted simply as necrosis but compare well with the sum of primary and secondary necrosis from manual analysis. The second difference is that the onset of necrotic events appears to be slightly earlier in the automated compared with the manual analysis, which is most likely attributable to threshold settings.
Single-target cell death analysis in a 3D collagen matrix
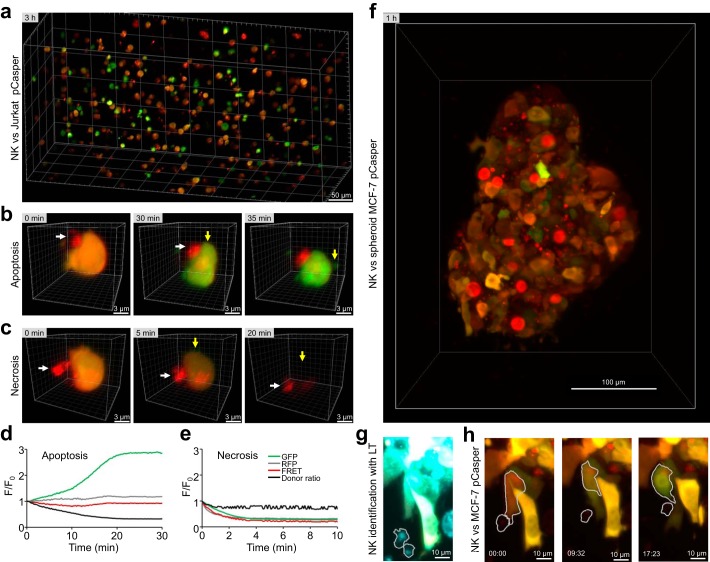
One disadvantage of working with human cells is that in vivo experiments are not feasible. To mimic physiologically relevant conditions more closely, we established the single-cell death assay in a 3D environment. A collagen matrix–based system was established to analyze interactions between single NK cells and Jurkat pCasper cells in a 3D environment (37). For these measurements, light-sheet microscopy was chosen because it offers very fast measurements of larger three-dimensional samples with minimal dye bleaching and phototoxicity (38). Fig. 7a depicts a reconstructed image of a three-dimensional matrix at one time point obtained by light-sheet microscopy. Target cells are orange (viable) or green (already apoptotic), and NK cells are stained by LysoTracker Red (red color). In the 3D matrix measurements, target cell apoptosis (Fig. 7b) and primary necrosis (Fig. 7c) following NK cell contact can be easily distinguished and quantified. We performed the same analysis as for 2D measurements. Consistent with caspase-dependent target cell apoptosis, the GFP signal increased, whereas the FRET slightly decreased and, most importantly, the donor ratio decreased (Fig. 7d). Consistent with target cell necrosis, fluorescence in the GFP and FRET decreased in parallel, whereas the donor ratio remained relatively unchanged (Fig. 7e). These examples show that the single-cell apoptosis/necrosis assay can be transferred into a 3D matrix–based environment. The activity of primary human cytotoxic immune cells can be analyzed at a single-cell level in a large 3D matrix under well-controlled temperature and pH conditions, which together better resemble the physiological in vivo environment than 2D-coated flat surfaces.
Figure 7.
Single target cell death analysis in a 3D collagen matrix. a, overview of NK cells 3 h after starting the experiment (stained by LysoTracker Red; red) and Jurkat pCasper target cells in a collagen matrix imaged by light-sheet microscopy. Some targets are still viable as indicated by the orange fluorescence, some are killed by apoptosis as indicated by green fluorescence (loss of the FRET signal, compare b), and others are killed by necrosis, which can be detected by loss of fluorescence (compare c). b, example of an individual target cell in the collagen matrix killed by apoptosis following NK cell contact. Contact time between the target cell and NK cell was defined as 0 min. The loss of FRET fluorescence after 30 min is evident by the change from orange to green color. c, example of an individual target cell in the collagen matrix killed by necrosis following NK cell contact. Contact time between the target cell and NK cell was defined as 0 min. The loss of fluorescence after 5 min is evident. d and e, kinetics of GFP fluorescence, RFP fluorescence, FRET signals, and donor ratios of the cells shown in b and c. f, MCF-7 cells transfected with pCasper, grown into spheroids, and embedded in a collagen matrix together with NK cells, 1 h after start of gelation. Shown is a maximum intensity projection of pCasper-FRET in red, pCasper-GFP in green, and LysoTracker Red also in red. Most target cells express pCasper. Most cells are still viable, but some apoptotic cells can be identified. Some MCF-7 cells show a strong LysoTracker signal. g, NK cells at the border of the spheroid are migrating actively and are identified and marked by their LysoTracker signal (depicted in cyan). h, a representative killing event of an MCF-7 cell is depicted. After contact with the NK cell, the target undergoes shrinking and a change from red to green fluorescence, indicative of apoptosis.
We further explored the 3D experimental assay by growing spheroids of MCF-7 cells, a cell line that does not contain caspase-3 (39) but can be killed by NK cell–mediated apoptosis (Fig. S5, a and b). These experiments also confirm that other caspases cleave the DEVD sequence (21, 22). Transfection of caspase-3 into MCF-7 cells, however, had a tendency to accelerate apoptosis (Fig. S5c), indicating that caspase-3 plays an important role following NK cell–mediated target cell death. NK cells embedded in a collagen matrix together with MCF-7 spheroids that we observed to migrate toward the edge of the spheroid and to induce target cell death, as shown in Fig. 7 (f–h). Thus, the assay can be used to monitor target cell death in 3D spheroids.
Discussion
The assay developed here allows quantification of target cell death induced through contact with primary human natural killer cells regarding kinetics, modes (apoptosis or necrosis or mixed forms), and mechanisms (perforin/granzyme versus FasL/FasR) on a single-cell level. We found that inhibition of either one of the two major cytotoxic pathways, perforin/granzyme release or FasL/FasR interaction, unmasked the parallel activity of the other one. This may explain why mild perforin malfunction apparently does not cause strong phenotypes but may correlate with long-term cancer development (17).
The prevailing mode of cancer cell death has a large impact on the local tumor environment by shaping the profile of released inflammatory versus tolerogenic substances. For instance, the release of inflammatory substances like ATP (40) is higher in the case of necrosis. The possibility of controlling the apoptosis/necrosis ratio offers an intriguing opportunity to influence the tumor microenvironment. Modification of Ca2+ influx is one possibility. Ca2+ influx through Orai1 channels is important for killer cell function (33–35), and there is an apparent Ca2+ optimum for cytotoxicity (29). We found that Ca2+ influx regulates the ratio between NK cell–induced cancer cell apoptosis and necrosis. An Orai1 channel blocker could thus be an interesting tool to guide the mode of killer cell cytotoxicity by changing the apoptosis/necrosis ratio, thereby regulating the immunogenicity of cancer cell death.
Screening cytotoxicity of CTL or NK cells is currently receiving attention, considering the many advances of immunotherapy against cancer (1–4). This is particularly important because human NK cells have recently become more relevant for adoptive cellular immune therapy against cancer (9). Considering the substantial interspecies differences between mice and humans regarding NK cell subsets and receptors, which cause major problems for clinical translation (9), in vitro assays for human NK cells or humanized mice gain relevance. The 2D and 3D assays reported here are ideally suited to analyze the efficiency of natural killer cells of cancer patients, including kinetics, mode, and mechanism of target cell killing to predict treatment efficiency.
Experimental procedures
Ethical approval
Research was approved by the local ethics committee (approval 84/15; Prof. Dr. Rettig-Stürmer). The local blood bank in the Institute of Clinical Hemostaseology and Transfusion Medicine at Saarland University Medical Center provided leukocyte reduction system chambers, a by-product of platelet collection from healthy blood donors. All blood donors provided written consent to use their blood for research purposes.
Reagents, buffers, and solutions
Calcein-AM (C3100), and Fura-2/AM (F1211) were purchased from Thermo Fisher Scientific. Anti-human CD178 was from BD Biosciences. Anti-human CD95 Apo 1-1 was from Enzo Life Sciences. Mouse IgG1 APC isotype control, mouse IgG1 FITC isotype control, and mouse IgG2a PE isotype control were from Biolegend. IL-2 was from Invitrogen. Nucleofector Kit V was from Lonza. Vectors for pCasper-GR, TagGFP-N, and TagRFP-N were from Evrogen. BSA was from Sigma-Aldrich. BSA buffer contained 2 ml of 1 m HEPES stock, 5 ml of 3 m NaCl stock, 1 g of BSA, pH 7.4, and 100 ml of double-distilled H2O. Dulbecco's PBS was from Thermo Fisher Scientific, the Dynabeads® UntouchedTM human NK cell isolation kit was from Thermo Fisher Scientific, fetal calf serum (FCS) (EU-approved) was from Thermo Fisher Scientific, fibronectin was from Sigma-Aldrich, human granzyme B (ALX-200-602-C010) and perforin (ALX-200-604-C001) were from Enzo, collagen was from Advanced BioMatrix, LysoTracker® Red DND-99 was from Thermo Fisher Scientific, PBS was from Thermo Fisher Scientific, penicillin/streptomycin was from Thermo Fisher Scientific, poly-l-ornithine was from Sigma-Aldrich, propidium iodide was from Thermo Fisher Scientific, and concanamycin A was from Santa Cruz Biotechnology, Inc. RPMI 1640 was from Thermo Fisher Scientific, G418 was from Thermo Fisher Scientific, staurosporine from Streptomyces sp. was from Sigma-Aldrich, and TetraSpeckTM microspheres were from Thermo Fisher Scientific.
0 mm Ca2+ Ringer's solution contained 155 mm NaCl, 4.5 mm KCl, 10 mm glucose, 5 mm Hepes, 3 mm MgCl2, 1 mm EGTA, pH 7.4 (1 n NaOH). 0.5 mm Ca2+ Ringer's solution contained 155 mm NaCl, 4.5 mm KCl, 10 mm glucose, 5 mm Hepes, 2.5 mm MgCl2, 0.5 mm CaCl2, pH 7.4 (1 n NaOH). 1 mm Ca2+ Ringer's solution contained 155 mm NaCl, 4.5 mm KCl, 10 mm glucose, 5 mm Hepes, 2 mm MgCl2, 1 mm CaCl2, pH 7.4 (1 n NaOH). All other chemicals and reagents not specifically mentioned were from Merck, J.T. Baker, or Sigma-Aldrich.
Primary human NK cells
Human peripheral blood mononuclear cells (PBMCs) of healthy donors were isolated from leukocyte reduction system chambers used for thrombocyte apheresis (Institute of Clinical Hematology and Transfusion Medicine, Homburg, Germany) by density gradient centrifugation using Lymphocyte Separation Medium 1077 (PromoCell) as described previously (41). Primary NK cells were isolated from PBMC using Dynabead® UntouchedTM NK cell isolation kits (Thermo Fisher Scientific) according to the manufacturer's specifications as described previously (41). NK cells were incubated at 37 °C, 5% CO2 in AIM V® medium (Thermo Fisher Scientific) substituted with 10% FCS at 2 × 106 cells/ml and, depending on the experiment, either used untreated or substituted with 100 units/ml IL-2 (Thermo Fisher Scientific). Purity of NK cells after isolation was checked via flow cytometry analysis. 93.3 ± 1.6% of the negatively isolated cells were NK cells, compared with 7.49 ± 1.68% within the PBMC (mean ± S.D. of three donors) (Fig. S6). Contamination of the purified NK population by monocytes and B and T cells was excluded via CD14, CD19, CD3, and CD4 flow cytometry analysis (Fig. S6). In many publications, NK cells are stimulated by IL-2 (28, 42, 43). To check the effect of IL-2 stimulation on freshly isolated primary human NK cells, cells were cultivated for 1–8 days in the presence of 100 units/ml IL-2 before use.
Generation of stable target cell lines with Casper-GR
3 × 106 Jurkat E6-1 (ATCC, TIB-15TM) or K562 (ATCC 1994) were transfected with 2 μg of pCasper3-GR vector (Evrogen) (20) using a Nucleofector® II (Lonza, program C-016). The transfected population was treated for 6 days with 2.5 mg/ml and then for 12 days with 1.25 mg/ml G418. Subsequently, the population was seeded in 96-well plates after dilution to 0.5 cells/well. Wells that contained only one cell were marked, and fluorescence signals of growing colonies were assessed. Fluorescent, monoclonal colonies were transferred to bigger flasks. Jurkat E6-1 and K562 cell clones were maintained in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% FCS, 1% penicillin/streptomycin (Thermo Fisher Scientific), and 1.25 mg/ml G418. To check the proportion of fluorescent cells, both target cell lines, referred to as Jurkat pCasper and K562 pCasper were analyzed by flow cytometry. Comparing them with the original cell lines, flow cytometry analysis confirmed that, in both cases, >93% of cells are expressing the fluorescent sensor protein at easily detectable, homogeneous levels (Fig. S1).
2D life cell imaging
2D life cell imaging was carried out with three different imaging systems: Cell Observer (Zeiss), DMI 6000 B (Leica), or ImageXpress Micro XLS (Molecular Devices). All systems are equipped for 37 °C, 5% CO2 incubation.
For Cell Observer, excitation was realized via Colibri® LEDs 470 nm and neutral white LED with excitation BP 560/40 nm. The GFP filter set was Zeiss 38 HE (excitation BP 470/40 nm, emission BP 525/50 nm). RFP filter set was 45 HQ (excitation BP 560/40 nm, emission BP 630/75 nm). The RFP excitation filter was removed from cube and mounted in front of the neutral white LED to enable GFP excitation to pass the 45 HQ filter cube during FRET experiments.
For the DMI 6000 B (Leica), excitation was realized via a mercury short-arc reflector lamp (EL6000) using Ex 490/20 nm and Em 525/36 nm filters for GFP and Ex 552/24 nm and Em 600/32 nm filters for RFP.
For the ImageXpress, excitation was realized via Spectra X LED illumination (Lumencor) using LEDs 470/24 and 542/27 nm for GFP and RFP excitation, respectively. The filter sets for FRET were Ex 472/30 nm and Em 520/35 nm for GFP and Em 641/75 nm for RFP. A 20 × S Fluor 0.75 numerical aperture objective (Nikon) was used.
For imaging, target cells were settled on 25-mm glass coverslips (Kindler) or in black 96-well plates with transparent bottom (Sigma-Aldrich). Both were coated with 50 μl of 0.1 mg/ml fibronectin (Sigma-Aldrich), which facilitates cell migration. Fibronectin was coated onto the coverslip or well bottom for 30 min at room temperature and then aspirated. The coated surface was dried via sterile bench airflow for 10 min, and then the target cells (1 × 105 up to 8 × 105 cells, depending on desired density) were settled onto the coated surface in cell medium for 30 min at 37 °C, 5% CO2. Cells were covered with cell medium or Ca2+ buffer solution, 950 μl in coverslip-based microscopy and 100 μl in 96-well plate–based microscopy. Either RPMI® or AIM V® was used; both were supplemented with 10% FCS, 10 mm HEPES, and 1% penicillin/streptomycin for long-term experiments. For coverslip-based microscopy experiments, NK cells were added after the acquisition start in a volume of 50 μl, adding up to a total medium volume of 1 ml. In 96-well (BD Biosciences) plate-based microscopy, NK cells were added before acquisition start in 100 μl of cell medium, adding up to a total volume of 200 μl/well. NK cells sank to the coated surface, adhered, started migration, and interacted with target cells.
For certain experiments, target cells were stained with calcein (Thermo Fisher Scientific), Fura-2 AM (Thermo Fisher Scientific), or LysoTracker® Red (Thermo Fisher Scientific). For calcein or Fura-2 AM staining, 5 × 105 target cells were suspended in 1 ml of RPMI® + 10 mm Hepes and incubated for 15 min at room temperature on a shaker with 500 nm calcein or 1 μm Fura-2. Cells were centrifuged for 8 min at 300 × g, resuspended in 50 μl of RPMI®, and settled on a coated coverslip or 96-well plate as described. Fura-2 AM was not imaged ratiometrically but, in this case, was used only as cytosolic label, excited at 360 nm. NK cells were stained with LysoTracker® Red by 30-min preincubation of 2 × 105 NK cells in 1 ml of AIM V® + FCS + 100 nm LysoTracker® Red at 37 °C, 5% CO2. NK cells were centrifuged for 8 min at 300 × g and then resuspended in 50 μl of AIM V® and added to the measurement. For long-term measurements, LysoTracker® Red was added to the supernatant. To visualize dye influx in target cells following membrane disruption, 100 μm propidium iodide (Thermo Fisher Scientific) was added to the supernatant.
MCF-7 cell culture and transfection
MCF-7 cells were cultured in Dulbecco's modified Eagle medium containing 10% FCS. They were transfected by nucleofection (Amaxa Nucleofector® II, Lonza Kit NHEM Neo, program A024) with 2 μg of pcDNA3-Casp3-myc (Addgene plasmid 11813) or 2 μg of empty pcDNA3 vector and 2 μg of pCasper3-GR vector. pcDNA3-Casp3-myc (44) was a gift from Guy Salvesen.
3D life cell imaging with light-sheet microscopy
For selective plane illumination microscopy with the Z.1 light-sheet microscope (Zeiss), excitation was realized by two lasers, 488 and 561 nm. Emission was filtered via Em 525/40 nm and Em 585 LP filters.
Nutragen® (Advanced Biomatrix) was used to create 3D collagen matrices according to the manufacturer's specifications. 0.25 × 106 NK cells were preincubated for 30 min with 500-nm LysoTracker® Red, mixed with 1.5 × 106 Jurkat pCasper cells in a 50-μl collagen matrix, and sucked into a glass capillary (Zeiss). The capillary was incubated vertically for 1 h at 37 °C, 5% CO2 for gelation. For spheroid experiments, MCF-7 cells were transfected as described above and cultured for 24 h in a low binding 24-well plate (VWR 734–1584) to form spheroids. NK cells and spheroids were mixed in the collagen matrix and incubated horizontally in a capillary for gelation. Imaging was performed in AIM V® substituted with 500 nm LysoTracker® red at 37 °C, 5% CO2 at 10- or 20-s intervals per stack, depending on the imaged volume.
Image analysis
Image analysis and post-processing was done with Axiovision version 4.8 (Zeiss), Zen version 11.0.0.190 (Zeiss), ImageJ version 1.5i (National Institutes of Health), Imaris version 8.1.2 (Bitplane), Igor Pro version 6.2.2.2 (Wavemetrics), Huygens version 4.5.1p3 (SVI), and Excel 2010 (Microsoft). Background subtraction, bleaching correction, F/F0 normalization, and donor ratio calculation were performed with ImageJ and Excel. Using ImageJ, background was subtracted via a rolling ball algorithm, and cells were tracked manually via the Speckle TrackerJ plugin (45). ROI intensities were assessed via the Speckle Intensity Trajectories plugin (45). ROI intensity data were processed and visualized via Excel 2010.
Long-term imaging assay (with concanamycin A and anti-CD178)
The bottom of a black, clear 96-well plate was coated with fibronectin at room temperature and then aspirated. The coated surface was dried via sterile bench airflow for 10 min. Per well, 1 × 105 Jurkat pCasper target cells in 100 μl of AIM V® + 10% FCS + 10 mm HEPES were settled for 30 min at 37 °C, 5% CO2. NK cells of different donors were used 1 day after isolation and either used unstimulated or stimulated with 100 units/ml IL-2 (Thermo Fisher Scientific). Stimulated NK cells (1 × 105 cells/condition) were preincubated for 4 h with 200 nm CMA (Santa Cruz Biotechnology, Inc.), 20 μg/ml CD178 antibody (BD Biosciences; clones NOK-1 and NOK-2 at 1:1), or both at 37 °C, 5% CO2 in a round-bottom 96-well plate (BD Biosciences). After preincubation, NK cells were added to the targets at 1:1 to a total volume of 200 μl/well, thereby lowering CMA concentration to 100 nm and antibody concentration to 10 μg/ml during the experiment. Total acquisition time was 17 h in 4-min intervals using the ImageXpress Micro XLS system (Molecular Devices) at 37 °C, 5% CO2. Acquired image data were loaded in ImageJ as time series, and background subtraction was performed via a rolling ball algorithm.
FRET evaluation, correction factors, and donor ratio calculation
To evaluate FRET correction factors, Jurkat E6-1 cells were transfected separately with TagGFP-N or TagRFP-N (Evrogen) as described (46). After cultivation, cells were seeded and imaged on coated coverslips as described using Cell Observer Z1 (Zeiss). Each transfection was checked in donor, acceptor, and FRET channel using equal excitation times (300 ms), LED intensities of 25%, and a 20×/0.75 Fluar objective (Zeiss) (Fig. S7). The percentages of donor bleed through and acceptor cross-talk of the previously described filter setting were determined at 3.6 and 22.6% using the FRET and colocalization analyzer plugin for ImageJ (National Institutes of Health). The corrected FRET signal Fc was calculated by linear unmixing, using the correction factors mentioned as Fc = (Chfret − BGfret) − (Chdon − BGdon) × cfdon − (Chacc − BGacc) × cfacc. Each fluorescent signal (Ch) was corrected by background signals (BG). cfdon and cfacc are the slopes of donor bleed-through and acceptor cross-talk (Fig. S7). Based on this, the intensity-independent donor ratio was calculated by dividing the FRET signal by the background-corrected fluorescence intensity of the donor fluorophore, GFP, according to Youvan et al. (25) as FDN = Fc/(Chdon − BGdon) using the AxioVision physiology module (Zeiss).
Manual target cell tracking and cell death analysis
Target cells were tracked manually using the Speckle TrackerJ plugin for ImageJ. Cells were chosen on a random basis but were discarded in cases where they left the field of view before the end of the experiment. xy positions for each observed cell were tracked for each time point. Fluorescence intensities were read out using the Speckle intensity plugin. Data were imported into Excel, fluorescence values were normalized to the start value of each track, and the donor ratio for each cell was calculated, as described above. Judging the fluorescence and donor ratio values as explained under “Results,” the cell status was determined for each time point, being viable, apoptotic, necrotic, or secondary necrotic. This information was pooled for all observed cells for each time point. The total number of all observed cells was set to 100%. For each time point, the proportion of apoptotic, necrotic, or secondary necrotic cells was calculated and displayed in a color-coded diagram over time (death plot).
Automated target cell death analysis
Target cells were detected using Imaris (Bitplane), but this could be done with most available software packages, including ImageJ. For each time point, cells were detected via spot detection in the GFP fluorescence channel. To avoid tracking errors due to cell collisions and to keep the automated analysis as simple as possible, spots were not connected over time. Therefore, secondary necrosis cannot be distinguished from primary necrosis. Fluorescence values for all channels were extracted, and data were imported into Excel. For each spot, the donor ratio was calculated as described above. Spots with fluorescence values near the background (mean background fluorescence + 10%) were defined as necrotic (lysed cell debris). Donor ratio and FRET data of all remaining spots (viable and apoptotic) were pooled for each time point. Histograms are shown in Fig. S8. Comparing the donor ratio over time, it is obvious that next to the distribution of living cells at time 0, another population of cells with a donor ratio < 0.6 develops, indicative of apoptosis. Similar as for manual analysis, cells with a clear drop of the FRET signal (in this case <600 units) are counted as being necrotic. During the very first pictures (before cytotoxicity by NK cells starts), the total number of cells is identical to the total number of detected spots. Unfortunately, necrotic cells often cannot be recognized by the spot detection algorithm because their fluorescence is indistinguishable from the background. Therefore, once the number of detected spots decreases due to cytotoxicity, the last detected maximum is kept as the total sum of cells (100%). This allows us to approximate the proportion of undetected necrotic cells as the number of cells missing to complete the total sum. For each time point, the number of viable, apoptotic, or necrotic cells is displayed in a color-coded, stacked diagram over time (death plot; Fig. S4).
Automated 3D cell tracking was performed with Imaris (Bitplane) using the GFP fluorescence of target cells with the autoregressive movement model. Maximum gap size was 3 time points, and maximum movement was 5 μm between time points. Based on this tracking analysis, spherical ROIs for all fluorescence channels were created. The ROI's diameter was intentionally set smaller than the real target cell's diameter to exclude RFP and FRET fluorescence interference from nearby LysoTracker® Red–labeled NK cells during synapse formation. For all tracked cells, fluorescence intensities were extracted, sorted with IgorPro (Wavemetrics), and processed via Excel.
Purified perforin and granzyme B
Black, clear-bottom 398-well plates (BD Biosciences) were coated with 0.1 mg/ml poly-l-ornithine (Sigma-Aldrich) analog to the fibronectin coating. Perforin and granzyme B, purified from YT-NK cells, were obtained from Enzo. Ca2+-HEPES buffer and BSA-HEPES buffer were prepared according to the manufacturer's specifications (Enzo). Per well, 2 × 105 Jurkat pCasper cells were settled in 20 μl of BSA-HEPES. Image acquisition was performed at 37 °C, 5% CO2. 15 min after acquisition start, 20 μl of BSA-HEPES buffer, containing the desired perforin and granzyme B concentration, were added to each well. 15 min after adding perforin and granzyme B, 40 μl of Ca2+-HEPES buffer were added to each well to induce perforin activation, whereas image acquisition was continued to monitor cell viability. Background correction was performed via the rolling ball algorithm in ImageJ (National Institutes of Health). Cells were tracked automatically using Imaris (Bitplane). The number and type of cell death were calculated from fluorescence values using Excel (Microsoft).
Variation of extracellular free Ca2+ concentration in AIMV medium
AIMV medium with different extracellular free Ca2+ concentration was prepared as described by Zhou et al. (29). The calculated extracellular free Ca2+ concentrations are presented, and they differ modestly from the ones measured by Zhou et al. (29).
Flow cytometry
For details, please see Figs. S1, S2, and S5. To check the purity of isolated NK cells, cells were stained with an antibody mixture containing anti-human CD3 (clone SK7) from Biolegend, CD4 (clone RPA-T4), CD14 (clone MϕP9), CD16 (clone B73.1), CD19 (clone HIB19), CD45 (clone HI30), and C56 (clone NCAM16.2) all from BD Biosciences, washed twice in PBS + 0.5% BSA. Jurkat pCasper and K562 pCasper cells were stained with an allophycocyanin-conjugated anti-human CD95 antibody (clone DX2; Biolegend) for 25 min at room temperature in the dark in PBS + 0.5% BSA. Data acquisition was performed on a FACSVerse flow cytometer (BD Biosciences) and analyzed using FlowJo version 10.
Quantitative PCR
Quantitative RT-PCR was carried out with the QuantiTect SYBR® Green PCR kit (Qiagen) using the CFX96TM Real-Time SystemC1000TM thermal cycler (Software Bio-Rad CFX Manager, version 3.0) as described previously (46). For the expression analysis of CD95 (FasR), the following primer pairs were used: CD95_1_forw (5′-CAAGGGATTGGAATTGAGGA-3′) and CD95_1_rev (5′-TGG AAG AAA AAT GGG CTT TG-3′); CD95_2_forw (5′-GTC CAA AAG TGT TAA TGC CCA AGT-3′) and CD95_2_rev (5′-ATG GGC TTT GTC TGT GTA CTC CT-3′). RNAPol and TBP were used as reference genes. Sequences are given by Wenning et al. (47).
To change Evrogen's pCasper-GR bp at position 1400 from A to C, QuikChange® PCR (Agilent) was used with the following primers: primer BAN1033 QC caspase cleavage site forward (pCasper: DEVA), 5′-CAG CGG TGA CGA GGT CGC CGG TAC CTC AGT CGC CAC-3′; primer BAN1034 QC caspase cleavage site reverse (pCasper: DEVA), 5′-GTG GCG ACT GAG GTA CCG GCG ACC TCG TCA CCG CTG-3′. Primers were synthesized by MWG-Biotech.
Statistical analysis
Data are presented as mean ± S.E. (n = number of experiments) if not stated otherwise. Data were tested for significance using one-way analysis of variance or Student's t test (the latter if Gaussian distribution was confirmed): *, p < 0.05; **; p < 0.01; ***, p < 0.001; ns, no significant difference. Statistics were calculated using Microsoft Excel 2010, GraphPad Prism version 7.01, and Igor Pro version 6 (Wavemetrics) software.
Author contributions
C. S. B., K. S. F., S. M., A. K., M. H., and C. K. conceptualization; C. S. B., K. S. F., S. M., A. K., M. H., and C. K. formal analysis; C. S. B. and A. K. validation; C. S. B., K. S. F., S. M., A. K., M. H., and C. K. investigation; C. S. B. and K. S. F. visualization; C. S. B., K. S. F., S. M., A. K., M. H., and C. K. methodology; C. S. B., M. H., and C. K. writing-original draft; C. S. B., K. S. F., A. K., M. H., and C. K. writing-review and editing; M. H. and C. K. supervision; M. H. funding acquisition; M. H. and C. K. project administration.
Supplementary Material
Acknowledgments
We very much appreciate the help of Prof. Hermann Eichler and the Institute of Clinical Hemostaseology and Transfusion Medicine at Saarland University Medical Center in obtaining human blood cells. We gratefully acknowledge help with flow cytometry from Cora Hoxha and help with quantitative PCR and generation of stable cell lines from Eva C. Schwarz, Dalia Alansary, and Barbara A. Niemeyer. We thank Gertrud Schwär, Cora Hoxha, and Carmen Hässig for cell preparation and excellent technical support. Renping Zhao was of great help with light-sheet microscopy. MCF-7 cells were a gift from Frank Neumann (Internal Medicine I, Saarland University). We thank all laboratory members for insightful discussions.
This project was supported by Deutsche Forschungsgemeinschaft (DFG) Grants SFB 894 (project A1 MH), SFB 1027 (project A2 MH), and IRTG 1830 (to M. H. and Barbara Niemeyer). This work was also supported by Bundesministerium für Bildung und Forschung (BMBF) Grant 031LO133. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Video S1, Table S1, and Figs. S1–S8.
- CTL
- cytotoxic T lymphocyte(s)
- NK
- natural killer
- IS
- immune synapse
- LG
- lytic granule(s)
- FasL
- Fas ligand
- FasR
- Fas receptor
- RFP
- red fluorescent protein
- CMA
- concanamycin A
- 2D and 3D
- two- and three-dimensional, respectively
- ROI
- region of interest
- FCS
- fetal calf serum
- PBMC
- peripheral blood mononuclear cell
- BP
- bandpass.
References
- 1. Kershaw M. H., Westwood J. A., and Darcy P. K. (2013) Gene-engineered T cells for cancer therapy. Nat. Rev. Cancer 13, 525–541 10.1038/nrc3565 [DOI] [PubMed] [Google Scholar]
- 2. Smaglo B. G., Aldeghaither D., and Weiner L. M. (2014) The development of immunoconjugates for targeted cancer therapy. Nat. Rev. Clin. Oncol. 11, 637–648 10.1038/nrclinonc.2014.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pardoll D. M. (2012) The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharma P., and Allison J. P. (2015) The future of immune checkpoint therapy. Science 348, 56–61 10.1126/science.aaa8172 [DOI] [PubMed] [Google Scholar]
- 5. Weiner L. M. (2007) Building better magic bullets–improving unconjugated monoclonal antibody therapy for cancer. Nat. Rev. Cancer 7, 701–706 10.1038/nrc2209 [DOI] [PubMed] [Google Scholar]
- 6. Thakur A., Huang M., and Lum L. G. (2018) Bispecific antibody based therapeutics: strengths and challenges. Blood Rev. 32, 339–347 10.1016/j.blre.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 7. Hernandez-Ilizaliturri F. J., Reddy N., Holkova B., Ottman E., and Czuczman M. S. (2005) Immunomodulatory drug CC-5013 or CC-4047 and rituximab enhance antitumor activity in a severe combined immunodeficient mouse lymphoma model. Clin. Cancer Res. 11, 5984–5992 10.1158/1078-0432.CCR-05-0577 [DOI] [PubMed] [Google Scholar]
- 8. Griffin M. M., and Morley N. (2013) Rituximab in the treatment of non-Hodgkin's lymphoma–a critical evaluation of randomized controlled trials. Expert Opin. Biol. Ther. 13, 803–811 10.1517/14712598.2013.786698 [DOI] [PubMed] [Google Scholar]
- 9. Guillerey C., Huntington N. D., and Smyth M. J. (2016) Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 17, 1025–1036 10.1038/ni.3518 [DOI] [PubMed] [Google Scholar]
- 10. Sharma P., and Allison J. P. (2015) Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 161, 205–214 10.1016/j.cell.2015.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stinchcombe J. C., Majorovits E., Bossi G., Fuller S., and Griffiths G. M. (2006) Centrosome polarization delivers secretory granules to the immunological synapse. Nature 443, 462–465 10.1038/nature05071 [DOI] [PubMed] [Google Scholar]
- 12. de Saint Basile G., Ménasché G., and Fischer A. (2010) Molecular mechanisms of biogenesis and exocytosis of cytotoxic granules. Nat. Rev. Immunol. 10, 568–579 10.1038/nri2803 [DOI] [PubMed] [Google Scholar]
- 13. Voskoboinik I., Smyth M. J., and Trapani J. A. (2006) Perforin-mediated target-cell death and immune homeostasis. Nat. Rev. Immunol. 6, 940–952 10.1038/nri1983 [DOI] [PubMed] [Google Scholar]
- 14. Kataoka T., Shinohara N., Takayama H., Takaku K., Kondo S., Yonehara S., and Nagai K. (1996) Concanamycin A, a powerful tool for characterization and estimation of contribution of perforin- and Fas-based lytic pathways in cell-mediated cytotoxicity. J. Immunol. 156, 3678–3686 [PubMed] [Google Scholar]
- 15. Boudreau J. E., and Hsu K. C. (2018) Natural killer cell education and the response to infection and cancer therapy: stay tuned. Trends Immunol. 39, 222–239 10.1016/j.it.2017.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Janka G. E. (2012) Familial and acquired hemophagocytic lymphohistiocytosis. Annu. Rev. Med. 63, 233–246 10.1146/annurev-med-041610-134208 [DOI] [PubMed] [Google Scholar]
- 17. Voskoboinik I., Whisstock J. C., and Trapani J. A. (2015) Perforin and granzymes: function, dysfunction and human pathology. Nat. Rev. Immunol. 15, 388–400 10.1038/nri3839 [DOI] [PubMed] [Google Scholar]
- 18. Chia J., Yeo K. P., Whisstock J. C., Dunstone M. A., Trapani J. A., and Voskoboinik I. (2009) Temperature sensitivity of human perforin mutants unmasks subtotal loss of cytotoxicity, delayed FHL, and a predisposition to cancer. Proc. Natl. Acad. Sci. U.S.A. 106, 9809–9814 10.1073/pnas.0903815106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Imai K., Matsuyama S., Miyake S., Suga K., and Nakachi K. (2000) Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: an 11-year follow-up study of a general population. Lancet 356, 1795–1799 10.1016/S0140-6736(00)03231-1 [DOI] [PubMed] [Google Scholar]
- 20. Shcherbo D., Souslova E. A., Goedhart J., Chepurnykh T. V., Gaintzeva A., Shemiakina I. I., Gadella T. W., Lukyanov S., and Chudakov D. M. (2009) Practical and reliable FRET/FLIM pair of fluorescent proteins. BMC Biotechnol. 9, 24 10.1186/1472-6750-9-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benkova B., Lozanov V., Ivanov I. P., and Mitev V. (2009) Evaluation of recombinant caspase specificity by competitive substrates. Anal. Biochem. 394, 68–74 10.1016/j.ab.2009.07.012 [DOI] [PubMed] [Google Scholar]
- 22. Talanian R. V., Quinlan C., Trautz S., Hackett M. C., Mankovich J. A., Banach D., Ghayur T., Brady K. D., and Wong W. W. (1997) Substrate specificities of caspase family proteases. J. Biol. Chem. 272, 9677–9682 10.1074/jbc.272.15.9677 [DOI] [PubMed] [Google Scholar]
- 23. Caricchio R., Reap E. A., and Cohen P. L. (1998) Fas/Fas ligand interactions are involved in ultraviolet-B-induced human lymphocyte apoptosis. J. Immunol. 161, 241–251 [PubMed] [Google Scholar]
- 24. Owen-Schaub L. B., Zhang W., Cusack J. C., Angelo L. S., Santee S. M., Fujiwara T., Roth J. A., Deisseroth A. B., Zhang W. W., and Kruzel E. (1995) Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol. Cell. Biol. 15, 3032–3040 10.1128/MCB.15.6.3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Youvan D. C., Silva C., and Bylina E. (1997) Calibration of fluorescence resonance energy transfer in microscopy using genetically engineered GFP derivatives on nickel chelating beads. Biotechnology 3, 1–18 [Google Scholar]
- 26. Kroemer G., Galluzzi L., Vandenabeele P., Abrams J., Alnemri E. S., Baehrecke E. H., Blagosklonny M. V., El-Deiry W. S., Golstein P., Green D. R., Hengartner M., Knight R. A., Kumar S., Lipton S. A., Malorni W., et al. (2009) Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 16, 3–11 10.1038/cdd.2008.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lopez J. A., Jenkins M. R., Rudd-Schmidt J. A., Brennan A. J., Danne J. C., Mannering S. I., Trapani J. A., and Voskoboinik I. (2013) Rapid and unidirectional perforin pore delivery at the cytotoxic immune synapse. J. Immunol. 191, 2328–2334 10.4049/jimmunol.1301205 [DOI] [PubMed] [Google Scholar]
- 28. Zhu Y., Huang B., and Shi J. (2016) Fas ligand and lytic granule differentially control cytotoxic dynamics of natural killer cell against cancer target. Oncotarget 7, 47163–47172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou X., Friedmann K. S., Lyrmann H., Zhou Y., Schoppmeyer R., Knörck A., Mang S., Hoxha C., Angenendt A., Backes C. S., Mangerich C., Zhao R., Cappello S., Schwär G., Hässig C., et al. (2018) A calcium optimum for cytotoxic T lymphocyte and natural killer cell cytotoxicity. J. Physiol. 596, 2681–2698 10.1113/JP274964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nisihara T., Ushio Y., Higuchi H., Kayagaki N., Yamaguchi N., Soejima K., Matsuo S., Maeda H., Eda Y., Okumura K., and Yagita H. (2001) Humanization and epitope mapping of neutralizing anti-human Fas ligand monoclonal antibodies: structural insights into Fas/Fas ligand interaction. J. Immunol. 167, 3266–3275 10.4049/jimmunol.167.6.3266 [DOI] [PubMed] [Google Scholar]
- 31. Gwalani L. A., and Orange J. S. (2018) Single degranulations in NK cells can mediate target cell killing. J. Immunol. 200, 3231–3243 10.4049/jimmunol.1701500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoth M. (2016) CRAC channels, calcium, and cancer in light of the driver and passenger concept. Biochim. Biophys. Acta 1863, 1408–1417 10.1016/j.bbamcr.2015.12.009 [DOI] [PubMed] [Google Scholar]
- 33. Maul-Pavicic A., Chiang S. C., Rensing-Ehl A., Jessen B., Fauriat C., Wood S. M., Sjoqvist S., Hufnagel M., Schulze I., Bass T., Schamel W. W., Fuchs S., Pircher H., McCarl C. A., Mikoshiba K., et al. (2011) ORAI1-mediated calcium influx is required for human cytotoxic lymphocyte degranulation and target cell lysis. Proc. Natl. Acad. Sci. U.S.A. 108, 3324–3329 10.1073/pnas.1013285108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schwarz E. C., Qu B., and Hoth M. (2013) Calcium, cancer and killing: the role of calcium in killing cancer cells by cytotoxic T lymphocytes and natural killer cells. Biochim. Biophys. Acta 1833, 1603–1611 10.1016/j.bbamcr.2012.11.016 [DOI] [PubMed] [Google Scholar]
- 35. Lyubchenko T. A., Wurth G. A., and Zweifach A. (2001) Role of calcium influx in cytotoxic T lymphocyte lytic granule exocytosis during target cell killing. Immunity 15, 847–859 10.1016/S1074-7613(01)00233-3 [DOI] [PubMed] [Google Scholar]
- 36. Voskoboinik I., Thia M. C., Fletcher J., Ciccone A., Browne K., Smyth M. J., and Trapani J. A. (2005) Calcium-dependent plasma membrane binding and cell lysis by perforin are mediated through its C2 domain: a critical role for aspartate residues 429, 435, 483, and 485 but not 491. J. Biol. Chem. 280, 8426–8434 10.1074/jbc.M413303200 [DOI] [PubMed] [Google Scholar]
- 37. Schoppmeyer R., Zhao R., Hoth M., and Qu B. (2018) Light-sheet microscopy for three-dimensional visualization of human immune cells. J. Vis. Exp. 10.3791/57651 10.3791/57651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Strobl F., Schmitz A., and Stelzer E. H. K. (2017) Improving your four-dimensional image: traveling through a decade of light-sheet-based fluorescence microscopy research. Nat. Protoc. 12, 1103–1109 10.1038/nprot.2017.028 [DOI] [PubMed] [Google Scholar]
- 39. Jänicke R. U., Sprengart M. L., Wati M. R., and Porter A. G. (1998) Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem. 273, 9357–9360 10.1074/jbc.273.16.9357 [DOI] [PubMed] [Google Scholar]
- 40. Junger W. G. (2011) Immune cell regulation by autocrine purinergic signalling. Nat. Rev. Immunol. 11, 201–212 10.1038/nri2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kummerow C., Schwarz E. C., Bufe B., Zufall F., Hoth M., and Qu B. (2014) A simple, economic, time-resolved killing assay. Eur. J. Immunol. 44, 1870–1872 10.1002/eji.201444518 [DOI] [PubMed] [Google Scholar]
- 42. Clausen J., Vergeiner B., Enk M., Petzer A. L., Gastl G., and Gunsilius E. (2003) Functional significance of the activation-associated receptors CD25 and CD69 on human NK-cells and NK-like T-cells. Immunobiology 207, 85–93 10.1078/0171-2985-00219 [DOI] [PubMed] [Google Scholar]
- 43. Frey M., Packianathan N. B., Fehniger T. A., Ross M. E., Wang W. C., Stewart C. C., Caligiuri M. A., and Evans S. S. (1998) Differential expression and function of L-selectin on CD56bright and CD56dim natural killer cell subsets. J. Immunol. 161, 400–408 [PubMed] [Google Scholar]
- 44. Stennicke H. R., and Salvesen G. S. (1997) Biochemical characteristics of caspases-3, -6, -7, and -8. J. Biol. Chem. 272, 25719–25723 10.1074/jbc.272.41.25719 [DOI] [PubMed] [Google Scholar]
- 45. Smith M. B., Karatekin E., Gohlke A., Mizuno H., Watanabe N., and Vavylonis D. (2011) Interactive, computer-assisted tracking of speckle trajectories in fluorescence microscopy: application to actin polymerization and membrane fusion. Biophys. J. 101, 1794–1804 10.1016/j.bpj.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bhat S. S., Friedmann K. S., Knörck A., Hoxha C., Leidinger P., Backes C., Meese E., Keller A., Rettig J., Hoth M., Qu B., and Schwarz E. C. (2016) Syntaxin 8 is required for efficient lytic granule trafficking in cytotoxic T lymphocytes. Biochim. Biophys. Acta 1863, 1653–1664 10.1016/j.bbamcr.2016.04.014 [DOI] [PubMed] [Google Scholar]
- 47. Wenning A. S., Neblung K., Strauss B., Wolfs M. J., Sappok A., Hoth M., and Schwarz E. C. (2011) TRP expression pattern and the functional importance of TRPC3 in primary human T-cells. Biochim. Biophys. Acta 1813, 412–423 10.1016/j.bbamcr.2010.12.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.