The impact of direct (physical) versus indirect (metabolic) interactions between different yeast species has attracted significant research interest in recent years. This is due to the growing interest in the use of multispecies consortia in bioprocesses of industrial relevance and the relevance of interspecies interactions in establishing stable synthetic ecosystems. Compartment bioreactors have traditionally been used in this regard but suffer from numerous limitations. Here, we provide independent evidence for the importance of physical contact by using a genetic system, based on the FLO gene family, to modify the degree of physical contact and, therefore, the degree of asexual intraspecies and interspecies adhesion in yeast. Our results show that interspecies contact significantly impacts population dynamics and the survival of individual species. Remarkably, different members of the FLO gene family often lead to very different population outcomes, further suggesting that FLO gene expression may be a major factor in such interactions.
KEYWORDS: adhesion, cell-cell interaction, interspecies, yeast
ABSTRACT
Physical contact between yeast species, in addition to better-understood and reported metabolic interactions, has recently been proposed to significantly impact the relative fitness of these species in cocultures. Such data have been generated by using membrane bioreactors, which physically separate two yeast species. However, doubts persist about the degree that the various membrane systems allow for continuous and complete metabolic contact, including the exchange of proteins. Here, we provide independent evidence for the importance of physical contact by using a genetic system to modify the degree of physical contact and, therefore, the degree of asexual intraspecies and interspecies adhesion in yeast. Such adhesion is controlled by a family of structurally related cell wall proteins encoded by the FLO gene family. As previously shown, the expression of specific members of the FLO gene family in Saccharomyces cerevisiae dramatically changes the coadhesion patterns between this yeast and other yeast species. Here, we use this differential aggregation mediated by FLO genes as a model to assess the impact of physical contact between different yeast species on the relative fitness of these species in simplified ecosystems. The identity of the FLO gene has a marked effect on the persistence of specific non-Saccharomyces yeasts over the course of extended growth periods in batch cultures. Remarkably, FLO1 and FLO5 expression often result in opposite outcomes. The data provide clear evidence for the role of physical contact in multispecies yeast ecosystems and suggest that FLO gene expression may be a major factor in such interactions.
IMPORTANCE The impact of direct (physical) versus indirect (metabolic) interactions between different yeast species has attracted significant research interest in recent years. This is due to the growing interest in the use of multispecies consortia in bioprocesses of industrial relevance and the relevance of interspecies interactions in establishing stable synthetic ecosystems. Compartment bioreactors have traditionally been used in this regard but suffer from numerous limitations. Here, we provide independent evidence for the importance of physical contact by using a genetic system, based on the FLO gene family, to modify the degree of physical contact and, therefore, the degree of asexual intraspecies and interspecies adhesion in yeast. Our results show that interspecies contact significantly impacts population dynamics and the survival of individual species. Remarkably, different members of the FLO gene family often lead to very different population outcomes, further suggesting that FLO gene expression may be a major factor in such interactions.
INTRODUCTION
Microbial cell walls are the cells’ primary interface with the environment and other organisms. Research into the interactions between different yeast species indicates that direct physical contact between cells contributes significantly to ecological interactions, such as inhibition or stimulation (1, 2). It has been suggested that the early death of two non-Saccharomyces yeasts, namely Kluyveromyces thermotolerans and Torulaspora delbrueckii, in mixed fermentations with S. cerevisiae was due to cell-to-cell contact (1). Lopez et al. (3) similarly assessed the direct and indirect interactions between two yeast species, namely S. cerevisiae and Kluyveromyces marxianus, and found that both were inhibited in terms of growth and cell numbers only when in direct contact with one another. However, the systems used to establish the importance of physical contact in interspecies interactions in yeast have been based on the use of so-called membrane bioreactors. In these systems, two species are inoculated in separate compartments separated by a membrane, designed in such ways as to allow for the exchange of metabolites but not for mixing of cells. Other systems use different membranes and manners to ensure metabolic homogeneity between the compartments, including using peristaltic pumps (2) and other tools. These systems do not allow for an immediate and complete transfer of all relevant metabolites and macromolecules, such as proteins, which poses limitations on the nature and extent of the interactions investigated. Furthermore, no data exist regarding the mechanisms or genes that are involved in supporting physical interaction-driven fitness. Investigating genes or gene families which could potentially modulate or regulate interspecies cell-cell contact would be ideal to fill this knowledge gap. This would allow for controlled, directed physical interactions between selected species, enabling the determination of viability impacts on the species involved, compared with noninteracting control scenarios.
FLO genes, which encode cell wall-anchored adhesion proteins are mostly, if not entirely, responsible for the modifications of sex-independent adhesion properties of yeast cell walls (4, 5). Flo proteins are lectin-like proteins which bind to cell wall mannans on adjacent cells (6–8). In this process, Ca2+ ions act as cofactors in maintaining the active conformation of surface proteins, thereby enhancing the capacity of lectins to interact with α-mannan carbohydrates (9).
In S. cerevisiae, FLO genes are represented by a family of subtelomeric genes (FLO1, FLO5, FLO9, and FLO10) as well as the nonsubtelomeric gene FLO11/MUC1. The different Flo proteins are structurally very similar, and all data thus far show a strong functional overlap in terms of broad phenotypic impacts (4, 10, 11). Recently, it has been suggested that the FLO gene family may be involved in building niche ecosystems or associations of different yeast species in natural ecosystems (12).
The exact role of single species floc or aggregate formation is unclear. However, data suggest that these multicellular aggregates may be a defense mechanism adopted by some yeast strains to generate nutritionally rich microenvironments by selective lysis in order to survive such adverse conditions (13). It has also been suggested that they provide the organism with a competitive advantage (14), as studies show that cell-cell adhesion plays a role in self-recognition and the social organization of S. cerevisiae. Thus, adhesion promotes recognition and the physical connection between cells to trigger survival responses, such as alterations to cell wall composition (10, 14).
While the genetic regulation of FLO genes in S. cerevisiae has been fairly well elucidated, questions remain regarding the origins and roles of this multigene family with seemingly overlapping, even redundant functions. Given the compact and efficient organization of the S. cerevisiae genome, it is unlikely that the different members of the FLO gene family would have been retained from an evolutionary perspective if some unique and critical functions were not imparted by these genes in conditions the yeast would encounter in its natural habitat, a habitat copopulated by numerous other genera and species of yeast.
Furthermore, although the cell-cell adhesion behavior of S. cerevisiae has been widely studied, very little is known regarding the adhesion behavior of other species of yeast. For example, nine genes (named KaFLO1 to KaFLO9) in the yeast Hanseniaspora uvarum were found to contain adhesion-related domains as well as repeated sequences in a study by Pu et al. (15). This suggests that H. uvarum and likely other species of yeast also have a large FLO gene family that controls cell-cell adhesion. Genome sequences from other yeast species show that these species also harbor PA14 lectin-binding domains (16).
Rossouw et al. (12) reported that adhesion can occur between different species of yeast and that these interactions show specificity for different combinations of yeast. Moreover, the different members of the FLO gene family differentially impact the aggregation outcomes for different pairings of yeast species. While Rossouw et al. (12) investigated the degree to which individual FLO genes influence coaggregation between species, here we seek to utilize and explore these attributes further. Indeed, since the differentially expressed FLO genes lead to selective aggregation and adhesion, they can be used to evaluate the consequences of physical associations on population dynamics and species fitness in model consortia.
Laboratory yeast strains (genetic background FY23) overexpressing individual FLO genes (FLO1, FLO5, and FLO11) under the control of the HSP30 promoter and three non-Saccharomyces yeast species were used to model the impact of selective aggregation on population outcomes (17). The FY23 strain is a flo8 deletion mutant that presents a null adhesion background with which to assess the impact of selective yeast-yeast adhesion in the model consortia. The term overexpression as applied to this study simply refers to the controlled expression of FLO genes at sufficient levels to induce a consistent adhesion reponse, driven by a single FLO gene only under standard laboratory and fermentation conditions. Expressing the FLO genes individually in a null background presents the most rational control system, since individually deleting FLO genes from an adhesion-competent background would mean that several different FLO genes would be expressed together. This expression would obscure any findings linked to specific members of the FLO gene family.
The three non-Saccharomyces yeasts, namely Lachancea thermotolerans, Wickerhamomyces anomalus, and Hanseniaspora opuntiae, were selected based on the results of a previous study which showed interesting trends with regard to their interspecies adhesion behavior (12). Importantly, these non-Saccharomyces species are part of the vineyard and wine fermentation ecosystem, and their presence, sometimes in dominant numbers, has been reported in numerous studies (18, 19).
The selective nature of the different members of the FLO gene family in terms of interspecies physical aggregation (12) is a useful property that allows for the control and manipulation of interspecies cell-cell contact. Our results show that interspecies contact significantly impacts population dynamics and the survival of individual species in simplified wine-like ecosystems. The data suggest that selective physical interactions between multiple species play a major role in multispecies yeast ecosystem outcomes. While the mechanistic basis for these outcomes is not clear, the impact of differential physical aggregation is clear and pronounced.
RESULTS
Pairwise strain interactions.
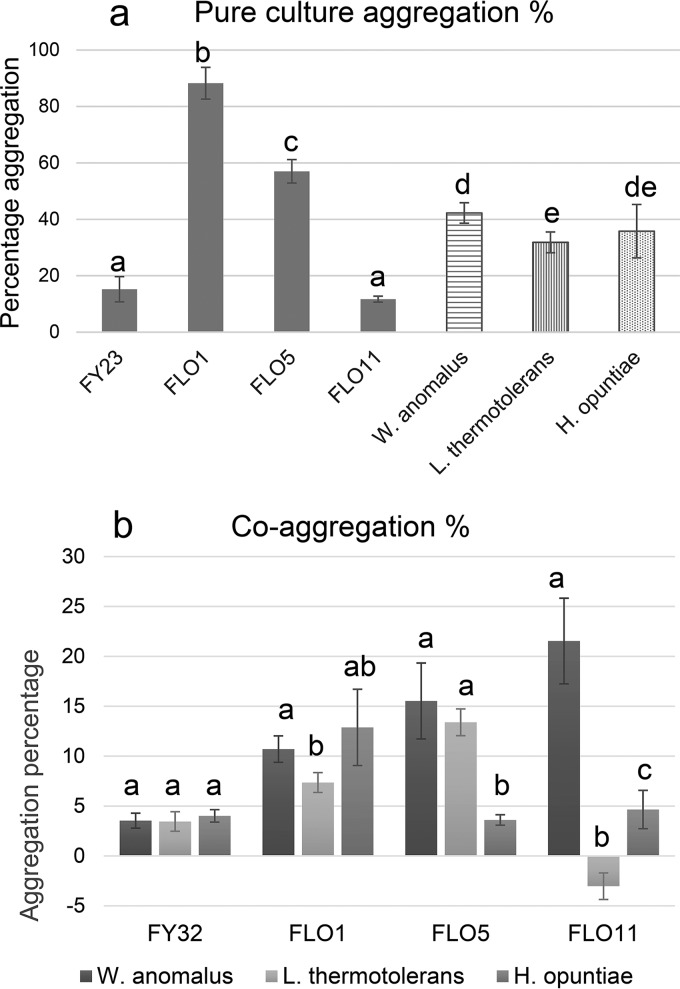
Individual or pure-culture adhesion of the selected non-Saccharomyces yeast strains as well as their coaggregation with the S. cerevisiae mutant strains (the percent increase or decrease in adhesion in coculture compared with the respective individual pure cultures) were assessed in yeast nitrogen base (YNB) cultures, and the results are shown in Fig. 1.
FIG 1.
(a) Pure culture aggregation of strains used in this study, (b) as well as coaggregation of the three non-Saccharomyces yeasts in combination with each of the FLOoverexpressing strains and control FY23. All values are the average of five repeats ± standard deviation. (a) Lowercase letters indicate significant differences (P < 0.05) between all strains in pure culture and (b) between the aggregation percentages of the non-Saccharomyces yeasts for each of the FLO treatments separately.
The pure-culture aggregation, percentages of these strains are aligned with the results of previous studies (12, 17), while all three non-Saccharomyces yeasts sediment at higher levels than the unmodified S. cerevisiae strain (Fig. 1a). W. anomalus yeast has the highest sedimentation percentage of 40% in pure culture. All three FLO-overexpressing strains coaggregate with W. anomalus (Fig. 1b), while only the FLO1-overexpressing strain coaggregates with H. opuntiae. Both the FLO1 and FLO5 strains coaggregate with L. thermotolerans, although FLO11 does not. The negative coaggregation percent shown for the FLO11-L. thermotolerans coculture means that the degree of adhesion and sedimentation of these species combined is less than would be expected based on the individual pure culture sedimentation percentages of the two strains.
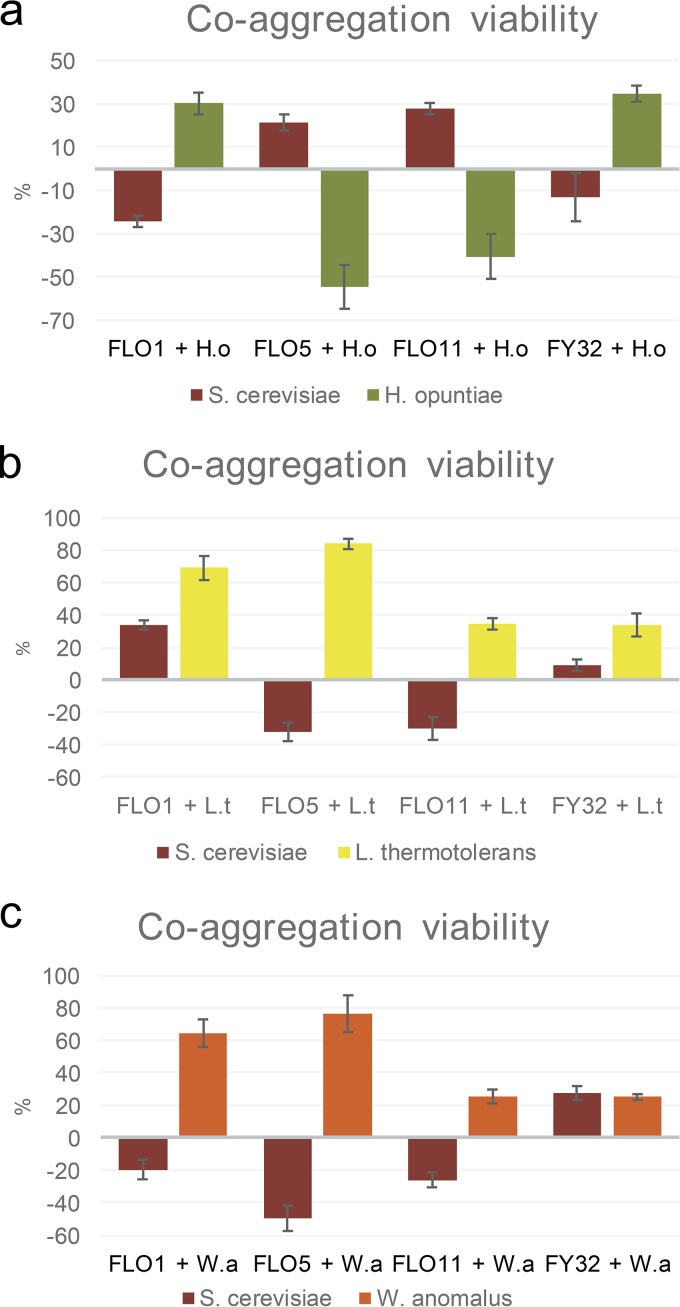
To investigate the impact of this selective adhesion in pairwise combinations on the survival of one or both partners, pairs of assays were set up to compare cell viability after 16 h in saline solution containing either no (nonadhesive conditions) or a small amount of CaCl2 to induce coaggregation. Figure 2 shows the impact (percent increase or decrease) on cell surivival for both species in the coaggregating cultures compared with that of the nonaggregating cell suspensions where no cell-cell adhesion occurs. The percent increase or decrease in the viability of the S. cerevisiae or non-Saccharomyces yeast strains is shown for the aggregating conditions (adhesion-inducing) relative to nonaggregating conditions. While this system is oversimplified and does not reflect the complexity of a natural system, it provides a means to assess the direct impact of interspecies adhesion without confounding factors, such as competition for nutrients influencing the outcomes, and is in scope equivalent to the previously published data based on membrane bioreactors.
FIG 2.
Percent increase (or decrease) in 24-h survival of individual species grown under aggregating conditions compared with nonaggregating conditions. Pairwise combinations were set up between the three overexpressing strains and control FY23 and each non-Saccharomyces yeast, namely (a) H. opuntiae (H.o), (b) L. thermotolerans (L.t), and (c) W. anomalus (W.a). Data for S. cerevisiae are indicated by red bars and for the non-Saccharomyces yeast by green, yellow, and orange bars for H. opuntiae, L. thermotolerans, and W. anomalus respectively. P values are shown in Table S1.
P values for pairwise S. cerevisiae and non-Saccharomyces yeast (L. thermotolerans, H. opuntiae, or W. anomalus) comparisons of viability percents after 24 h of two-species coculture (data shown in Fig. 2). Download Table S1, DOCX file, 0.02 MB (19.1KB, docx) .
Copyright © 2018 Rossouw et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Under aggregating conditions, the FLO5 and FLO11 strains (which do not coaggregate with H. opuntiae) showed improved survival (20% to 25% greater number of viable cells) compared with the S. cerevisiae in the parallel nonaggregating conditions. This suggests that S. cerevisiae benefits by self-adhesion under these circumstances (Fig. 2a). Keeping in mind that flo11p does not have a PA14 lectin domain, the cell-cell interactions between the FLO11-expressing S. cerevisiae and non-Saccharomyces yeasts may involve lectin-indepedent or mannan-independent aggregation. While the interactions in the case of the FLO11 treatments may be aspecific, different than FLO1 and FLO5 in this regard, the adhesion outcomes are nonetheless different for different species combinations, regardless of whether this is due to the direct action of the Flo11p.
On the other hand, coaggregation between the FLO1 and H. opuntiae strains is detrimental to S. cerevisiae, leading to a 20% lower viability of S. cerevisiae under these conditions than the same cultures in nonaggregating conditions. In contrast, direct physical contact between the FLO1 strain of S. cerevisiae and H. opuntiae, while detrimental to S. cerevisiae, provides an advantage of some sort to the H. opuntiae in terms of survival, as the H. opuntiae yeasts in the coaggregating mixed cultures show, on average, a 30% greater viability than the same treatment in nonaggregating conditions. This indicates that the effect of the interaction on cell viability is based on direct and sustained physical contact between the different species, contrasted to the nonaggreating conditions where the two cocultured species are only able to interact transiently with one another.
In contrast, a very different trend is observed in the presence of L. thermotolerans yeasts, where the S. cerevisiae FLO5 and FLO11 strains have decreased survival (greater than 30% decline) under aggregating conditions compared with the same mixed cultures grown under nonaggregating conditions. The L. thermotolerans strain, on the other hand, shows an increase in survival of up to 80% in the coaggregating cultures compared with nonaggregating conditions for the paired cultures (Fig. 2b). Interestingly, coaggregation between the FLO1-overexpressing S. cerevisiae and L. thermotolerans strains is beneficial for both parties. This contrasts strongly with the results of the W. anomalus strain pairings (Fig. 2c), where coaggregation between the FLO-overexpressing strains and W. anomalus strains is in all cases to the detriment of S. cerevisiae, and the benefit of the W. anomalus. These findings highlight the complexity of physical interactions between different species of yeast and their impact on the growth and viability of the species involved.
Population dynamics in multispecies consortia.
In order to assess the impact of the FLO gene-dependent physical interactions on population outcomes in a model system, a simplified design was implemented using a defined synthetic grape must, reflecting the composition of a grape must after pressing. Pressed grape must is an important environmental niche for industrially relevant fermentation microorganisms. The fermentation environment has arguably played an important role in the domestication and evolution of commercial yeasts (20, 21), an important consideration when evaluating the impact of gene families related to intraspecies and interspecies interactions. This sytem also allows for an extended (more than 2 weeks) period of batch culture and growth and provides the opportunity to observe population dynamics over a longer time course. Different combinations of the four S. cerevisiae strains and the three non-Saccharomyces yeasts (multifactorial three-way pairings, as well as all four species together) were inoculated into the synthetic must to create multispecies systems more representative of the complexity of a natural environment, yet simple enough to monitor and characterize adequately. In each treatment, all strains were inoculated at equal cell densities (CFU ml−1). Depending on the particular combination of species, the identity of the FLO gene overexpressed, as well as the stage of the fermentation process, significant differences were observed in the composition of the yeast population.
S. cerevisiae, L. thermotolerans, and H. opuntiae.
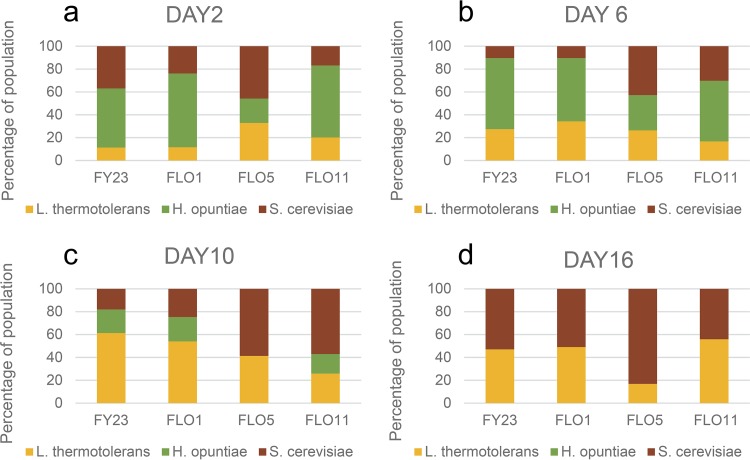
In the absence of W. anomalus, H. opuntiae is generally the dominant species (or codominant) in the early stages of fermentation (Fig. 3a and b). Under these circumstances, coaggregation with H. opuntiae (as the FLO1 strain and control FY23 do) is to the detriment of S. cerevisiae. However, the FLO11 and FLO5 strains do not coaggregate with H. opuntiae and constitute a large proportion (40% and 50%, respectively) of the yeast population by day 6 of growth in these consortia (Fig. 3c). H. opuntiae is present at the lowest levels of all three species in the FLO5 treatment (Fig. 3c). Coaggregation between the FLO5-overexpressing S. cerevisiae and L. thermotolerans strains (Fig. 1b) appears to be advantageous to both species under these conditions, enabling them to outcompete H. opuntiae, compared with FLO1, FLO11, and the control (Fig. 3c).
FIG 3.
Percent composition of S. cerevisiae, L. thermotolerans, and H. opuntiae in three-species cocultures by days 2 (a), 6 (b), 10 (c), and 16 (d) of fermentative growth. Four parallel sets of cultures were inoculated with either the control FY23 or one of the three FLO-overexpressing strains of S. cerevisiae. Values are the average of three repeats.
In the absence of W. anomalus, the dominant species is S. cerevisiae in the FLO5 treatment (90%). This is also the only one of the four FLO treatments where H. opuntiae is absent (or below detection levels) by day 10 (Fig. 3c). In FY23, FLO1, and FLO11, S. cerevisiae and L. thermotolerans are present at more or less equal levels by the end of fermentation (Fig. 3d), with H. opuntiae no longer present in any of the treatments.
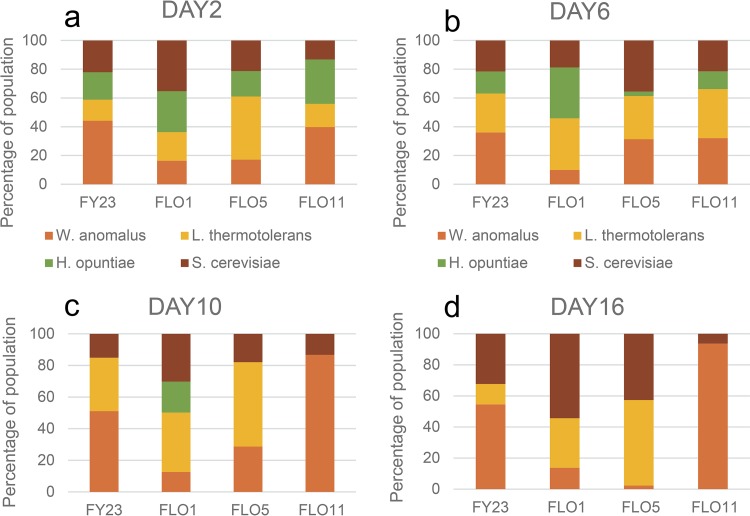
S. cerevisiae, L. thermotolerans, and W. anomalus.
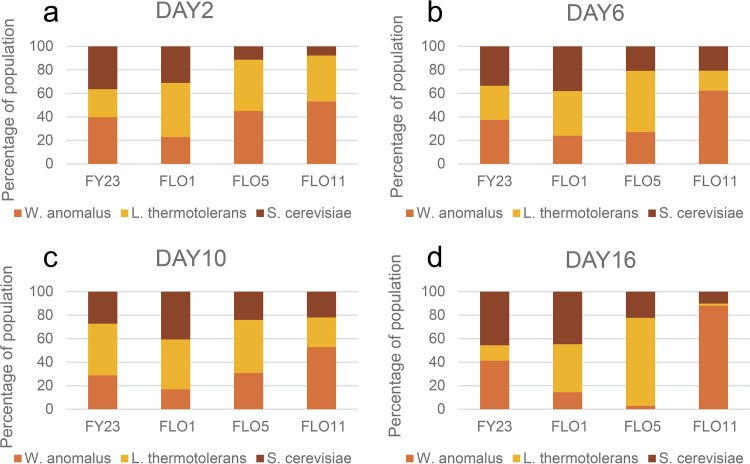
In strain combinations that include W. anomalus, different dynamics can be observed. In the absence of H. opuntiae (Fig. 4a to d), S. cerevisiae in the FLO5 and FLO11 treatments show the lowest survival (20% and 10% abundance, respectively, by day 16), while in the FLO1 treatment, S. cerevisiae consitutes 40% of the total population and W. anomalus constitutes less than 20%. In contrast, W. anomalus is the dominant species in the FLO11 treatment, both at day 2 and day 6 (Fig. 4a and b). Interestingly, while the levels of S. cerevisiae in the FLO5 and FLO11 treatments are similar at day 6 (Fig. 4b), the dominant species (at 50%) in the FLO5 treatment is L. thermotolerans, contrasted with the FLO11 treatment where W. anomalus is dominant (60%).
FIG 4.
Percent composition of S. cerevisiae, W. anomalus, and L. thermotolerans in three-species cocultures by days 2 (a), 6 (b), 10 (c), and 16 (d) of fermentative growth. Four parallel sets of cultures were inoculated with either the control FY23 or one of the three FLO-overexpressing strains of S. cerevisiae. Values are the average of three repeats.
In the absence of H. opuntiae, coaggregation between FLO1 and L. thermotolerans appears to benefit both parties, enabling them to outcompete W. anomalus, compared with the control FY32 and FLO11 treatment (Fig. 4a to d). Although FLO5 does coaggregate with L. thermotolerans (Fig. 1b), this appears to be to the detriment of the S. cerevisiae but to the benefit of L. thermotolerans, which is able to outcompete W. anomalus (compared with the control strain) in the early stages and outcompete S. cerevisiae in the later growth stages (Fig. 4c and d). This broadly aligns with the results of the pairwise inhibition/viability assays (Fig. 2b), which pointed toward a mutually beneficial association between the FLO1-overexpressing S. cerevisiae and L. thermotolerans.
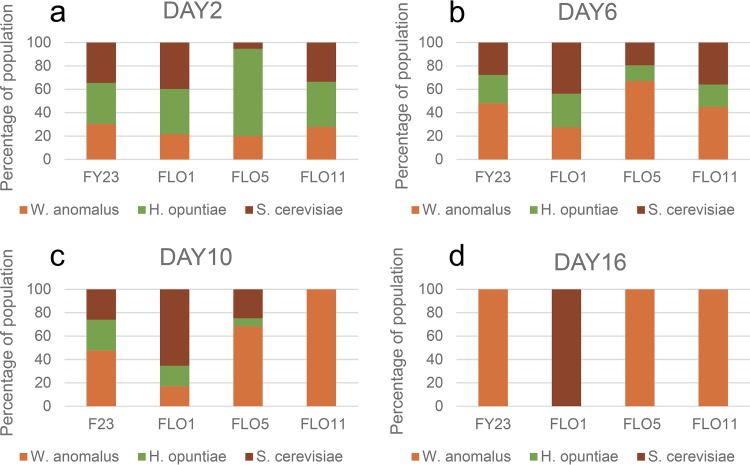
S. cerevisiae, H. opuntiae, and W. anomalus.
When W. anomalus and H. opuntiae are present in consortia lacking L. thermotolerans, the proportion of H. opuntiae and S. cerevisiae in the populations of these two treatments decline as W. anomalus increases by day 6 of growth (Fig. 5b). The exception is the FLO1 treatment, where S. cerevisiae and H. opuntiae are codominant at approximately 40% and 30% for days 2 and 6, respectively (Fig. 5a and b). The results for FLO1-overexpressing S. cerevisiae are the most starkly contrasted with the control and other treatments: When L. thermotolerans is not present in the ecosystem, W. anomalus dominates the fermentation in all treatments (at 100%), but for the FLO1-overexpressing treatment, it is S. cerevisiae which is dominant at 100% abundance by day 16 (Fig. 5c and d).
FIG 5.
Percent composition of S. cerevisiae, W. anomalus, and H. opuntiae in three-species cocultures by days 2 (a), 6 (b), 10 (c), and 16 (d) of fermentative growth. Four parallel sets of cultures were inoculated with either the control FY23 or one of the three FLO-overexpressing strains of S. cerevisiae. Values are the average of three repeats.
This strongly suggests that FLO1-mediated physical interaction provides a competitive advantage over W. anomalus in a scenario where other strong competitors (such as L. thermotolerans) are absent. Coaggregation between the FLO1 strain and H. opuntiae in the early stages of fermentation may have provided the S. cerevisiae in this treatment with an ally initially to restrict the growth of the W. anomalus. In support of this speculative federation, H. opuntiae is still present in the FLO1 treatment by day 10 of fermentation in significant amounts but has all but disappeared in the FLO5 and FLO11 treatments.
Four species consortia.
S. cerevisiae and L. thermotolerans are the dominant species in the FLO1 treatments by day 10 (Fig. 6c), with S. cerevisiae dominating at more than 50% of the total population by day 15 (Fig. 6d). At this point, very little (less than 15%) W. anomalus is present. However, W. anomalus is the dominant species toward and at the end of fermentative growth for the FY23 and FLO11 treatments, almost exclusively so in the case of FLO11 (Fig. 6d). The FLO5-overexpressing S. cerevisiae shows a greater percent contribution to the overall population (40%) than the control and FLO11 strain; however, in this case, L. thermotolerans is the dominant species by day 16 (Fig. 6d). In all FLO5 treatments containing W. anomalus, the dominant species is W. anomalus in the absence of L. thermotolerans, but L. thermotolerans dominates when both W. anomalus and L. thermotolerans are present in consortia (Fig. 3 to 6).
FIG 6.
Percent composition of S. cerevisiae, W. anomalus, L. thermotolerans, and H. opuntiae in four-species cocultures at days 2 (a), 6 (b), 10 (c), and 16 (d) of fermentative growth. Four parallel sets of cultures were inoculated with either the control FY23 or one of the three FLO-overexpressing strains of S. cerevisiae. Values are the average of three repeats.
In general, the FLO1 and FLO5 strains appear to follow different strategies for collegial associations with other strains at different stages of fermentation. Both strategies are successful (to a greater or lesser degree), as S. cerevisiae levels for FLO1 and FLO5 are higher at most stages of fermentative growth than the control for the 4-strain treatment. However, the relative levels of the other three species are strongly influenced by the identity of the overexpressed FLO gene.
DISCUSSION
Although the number of species and strains used in our simplified consortia does not represent the natural complexity of most yeast ecosystems, the results highlight the dramatic impact of differential physical interaction (as mediated by different members of the FLO adhesion protein family), compared with noninteracting yeasts, on population dynamics and survival. Importantly, while the data are based on a highly simplified ecosystem in well-controlled environments, the inoculation ratios and cell concentrations of our simplified consortia are within the range in which these species are sometimes encountered in spontaneously fermenting grape juice, for example.
While it has been reported that FLO genes cause differential adhesion between Saccharomyces cerevisiae and specific species of non-Saccharomyces yeast (12), here we show that these different adhesion relationships have very significant consequences for the balance of species in the ecosystem. The results show that, in this controlled model system, FLO5 overexpression aligns S. cerevisiae with L. thermotolerans, providing it with an advantage in competition with W. anomalus. This strain does not coaggregate with H. opuntiae and in the absence of L. thermotolerans is rapidly outcompeted by W. anomalus. Although the FLO5 strain does coaggregate with W. anomalus, this appears to be to its detriment.
The FLO1-overexpressing S. cerevisiae, in contrast, coaggregates with H. opuntiae, which appears to be a beneficial association in the early stages of fermentation, providing the FLO1 strain with an advantage compared with the other three S. cerevisiae strains (particularly in the presence of W. anomalus). In the H. opuntiae-containing treatments paired with the FLO1-overexpressing strain, the H. opuntiae also persists to later stages of fermentation compared with the other strain treatments.
W. anomalus rapidly outcompetes the FLO11-overexpressing S. cerevisiae strain in all treatments where it is present, as FLO11 does not coaggregate with either H. opuntiae or L. thermotolerans. Partnering with one of these strains (H. opuntiae in early fermentation or L. thermotolerans in later stages of fermentation) appears to be necessary to mount a defensive against the otherwise dominant W. anomalus (or simply to reduce physical interactions with W. anomalus, which appears inhibitory to S. cerevisiae).
Considered together, the results clearly show that different members of the FLO gene family exert a notable influence in terms of yeast species demographics at different stages of fermentation in a model system. While these genes only mediate adhesion, the resulting physical interaction leads to a species-specific growth response, mediated by mechanisms and means yet to be elucidated. Considering that intermicrobial interactions constitute one of the main selection pressures in natural ecosystems, it is reasonable to speculate that the FLO gene family (the only gene family responsible for asexual adhesion) may play a role in the evolution of cooperativity and antagonism between different species of yeast.
In support of this theory, The FLO1, FLO5, FLO9, and FLO10 genes are carried in subtelomeric loci (22), which holds important implications for the evolution of FLO genes, as subtelometric loci are subject to increased recombination frequencies (23). In addition, FLO genes contain up to 20 tandemly repeated sequences in their middle region which can lead to high mutation frequencies by recombination events (24, 25). The frequent recombination of FLO genes is thought to be an important mechanism for the rapid adaptation of adhesion properties of natural yeast in changing environments (26). Indeed, it has been shown that the FLO gene family has evolved and expanded extraordinarily fast (27). Both interchromosomal and intrachromosomal ectopic recombination are considered to occur for FLO gene paralogs (28, 29). More specifically, two types of recombination events occur between FLO genes. First, recombination events occur across small regions of homology in the N-terminal or C-terminal domain of FLO genes. Recombination events in the N-terminal can alter the strength and preference of substrate binding and hold implications for the function of modified FLO genes. Second, recombination across the central repeat domains of FLO genes leads to variation in the length and sequence of the repeat regions (28).
Understanding the molecular mechanisms and regulation of interspecies adhesion processes, as well as the impacts thereof on interspecies interaction dynamics, is important in terms of potential industrial application. Considering that cell-cell adhesion appears to play a pivotal role in the survival and social dynamics of yeast populations in natural environments, this information is also important to our understanding of possible evolutionary mechanisms linked to physical interactions between different microorganisms in shared ecological niches.
Evolutionary studies have clearly demonstarted that S. cerevisiae has undergone significant evolutionary changes, sometimes referred to as “domestication” due to the opportunities provided by human-made fermentation environments (20, 21) which would include adaptations that favor its relative fitness in these multispecies fermentation ecosystems.
To the best of our knowledge, no other gene family has yet shown such dramatic effects on population dynamics in multispecies systems. The data clearly demonstrate that the assortment of FLO genes at the disposition of S. cerevisiae would allow this yeast to selectively adapt to challenges presented by differing and rapidly changing yeast-rich environmental niches. Indeed, it can be argued that no other genetic system in S. cerevisiae would provide the same type of flexibility, responsiveness, and advantages for rapid adaptation. The large number of FLO genes, mostly located in subtelomeric recombination hotspots, combined with epigenetic regulation allowing for population-wide adjusted switches of FLO gene expression, would allow for rapid adjustment to the challenges of interspecies competition in changing yeast ecosystems. Considering that S. cerevisiae is present at less than 1% (sometimes even undetectable levels) of the yeast population at the start of fermentation, selective associations with dominant yeast species could provide S. cerevisiae with an advantage in the initial fermentation stages.
Future work should seek to investigate the genetics and expression of FLO-equivalent adhesion genes in species of non-Saccharomyces yeast, focusing in particular on the impacts which different species combinations have on the expression of these genes as population dynamics evolve over time. In addition, the role of the members of the FLO gene family in multispecies biofilms should be investigated, given the importance of biofilm formation in microbial persistence in humans and hospital equipment (30).
MATERIALS AND METHODS
Strains, media, and culture conditions.
The yeast strains used in this study were selected from the strain collection at the Institute for Wine Biotechnology (Table 1). The S. cerevisiae strains used are described by Govender et al. (17). They include the FY23 laboratory strain which is nonflocculent due to a mutation in the FLO8 gene, as well as three strains each overexpressing one FLO gene, namely FLO1, FLO5, and FLO11, under the control of the HSP30 promoter construct, which is induced at the onset of stationary phase as well as under certain stresses, such as heat shock (17). The non-Saccharomyces yeast strains used were Wickerhamomyces anomalus, Lachancea thermotolerans, and Hanseniaspora opuntiae, which were previously described in Rossouw et al. (12). Strains were maintained on YPD agar from pure frozen cultures. Liquid overnight cultures were grown in 5 ml YPD broth (BioLab, South Africa) to exponential phase at 30°C. Wallerstein nutrient (WLN) agar (BioLab) was used for culturing and enumerating yeast from fermentations and assays.
TABLE 1.
| Species | Strain or isolate | Genotype |
|---|---|---|
| Saccharomyces cerevisiae | FY23 | MATa leu2 trp1 ura3 flo8-1 |
| FY23-F1H | MATa leu2 trp1 ura3 flo8-1 FLO1p::SMR1-HSP30p | |
| FY23-F5H | MATa leu2 trp1 ura3 flo8-1 FLO5p::SMR1-HSP30p | |
| FY23-F11H | MATa leu2 trp1 ura3 flo8-1 FLO11p::SMR1-HSP30p | |
| Wickerhamomyces anomalus | IWBT-Y934 | |
| Lachancea thermotolerans | IWBT-Y983 | |
| Hanseniaspora opuntiae | IWBT-Y1055 |
Ca2+-dependent aggregation assays.
To quantify the degree to which individual strains aggregate, flocculation assays were carried out as described previously (6, 7, 17). Since FLO lectin-dependent aggregation only occurs in the presence of Ca2+, these assays are based on measuring the optical density of cell suspensions before and after the addition of Ca2+. Greater differences in the optical densities before and after Ca2+ addition reflect greater aggregation and sedimentation rates, and vice versa. Initially, yeast colonies for each isolate were inoculated (6 repeats) in test tubes containing 5 ml soyabean casein digest (SCD) medium and grown to stationary phase. An aqueous solution of EDTA (pH 8.0) was added to these cultures to a final concentration of 50 mM, and the cultures were agitated vigorously by vortexing at the maximum speed setting. The optical density at 600 nm (OD600) was determined immediately (reading A). Ca2+-dependent aggregation was subsequently induced by spinning down 1 ml of the liquid cultures in a microcentrifuge, followed by washing in 1 ml ddH2O and resuspension in 1 ml of 40 mM CaCl2. The samples were then vigorously agitated as before and left undisturbed for 60 s. A sample was taken from below the meniscus in the microcentrifuge tube of each sample and mixed thoroughly with 160 µl of a 40 mM CaCl2 solution. A second spectrophotometric measurement was then taken at a wavelength of 600 nm as before (reading B). For more information see Bester et al. (7). The extent of Ca2+-dependent aggregation was then calculated using the following formula:
To calculate the extent of coaggregation between different species of yeast in mixed cultures, S. cerevisiae strains and the non-Saccharomyces yeasts under investigation were combined in a 1:1 cell:cell ratio and the assay carried out using the mixed culture as described in the preceding section. The total cell concentrations in the coaggregation assays (i.e., S. cerevisiae plus non-Saccharomyces strain) were the same as for pure cultures. The aggregation percent was calculated as before, and the coaggregation percent was calculated by subtracting the expected aggregation rate (based on the combined average percentages of the pure cultures) from the experimentally determined aggregation percent obtained for the combined cultures.
Microscopy.
Alexa Fluor wheat germ agglutinin (WGA) conjugate (Invitrogen) staining of cells and fluorescence microscopy were carried out as described by Wright (31). Image acquisition was performed on an Olympus cell system attached to an IX 81 inverted fluorescence microscope equipped with an F-view II cooled CCD camera (Soft Imaging Systems). The excitation lasers used were the 495-nm wavelength for WGA 488 (green) and 679 nm for WGA 680 (red), and the emission filters used were 519 nm and 702 nm, respectively. Images were processed and background subtracted using the Cell software and presented in a maximum intensity projection. Cell cultures were combined in a 1:1 ratio of the non-Saccharomyces yeast under investigation in combination with each of the FLO gene-overexpressing strains and control FY32 separately (1 × 107 cells/ml of each). Species were individually prestained (S. cerevisiae in red, non-Saccharomyces yeast in red) and the species combined under aggregating (containing Ca2+) and nonaggregating (no Ca2+, containing EDTA) conditions. Samples of cell sediments were taken for microscopic evaluation as described.
Pairwise interaction assays.
The three non-Saccharomyces strains and overexpression strains (Table 1) were washed three times (after preculture in YPD) and coinoculated into buffered saline solution containing either 5 mM EDTA (nonaggregating conditions) or 10 mM CaCl2 (inducing Flo protein-driven aggregation). Culturing paired species with and without CaCl2 allows for the determination of cell survival of both species after 16 h when in direct physical contact in multispecies aggregates. After 16 h, serial dilutions were plated onto WLN agar to allow for differential identification and quantification (CFU ml−1) of the S. cerevisiae and W. anomalus, L. thermotolerans, and H.opuntiae in the various pairings under coaggregating versus nonaggregating conditions. Interaction assays were performed in quadruplicate. The percent increase/decrease of the yeast species in these assays was calculated under the aggregating conditions relative to the nonaggregating conditions.
Multispecies growth experiments.
Cells were inoculated and grown in a chemically defined synthetic must under fermentative conditions, mimicking a natural environment for multispecies yeast communities. These growth conditions allow for an extended growth period and observation window for the yeast-yeast interactions over time, compared with those of conventional rich medium and aerobic growth conditions. The medium used is based on the formulation of the Australian Wine Research Institute (32), with amino acid additions as described by Bely et al. (33). Sugar concentrations were 100 g/liter each of glucose and fructose, and the pH of the medium was adjusted to 3.3 with NaOH. Strains were precultured onto YPD and coinoculated in 80 ml fermentation flasks at an OD600 of 0.1 each. The following combinations were used:
L. thermotolerans, W. anomalus, S. cerevisiae (FY23/FLO1/FLO5/FLO11)
L. thermotolerans, H. opuntiae, S. cerevisiae (FY23/FLO1/FLO5/FLO11)
W. anomalus, H. opuntiae, S. cerevisiae (FY23/FLO1/FLO5/FLO11)
L. thermotolerans, W. anomalus, H. opuntiae, S. cerevisiae (FY23/FLO1/FLO5/FLO11)
All treatments were carried out in triplicate. Samples were taken at days 1, 2, 5, 10, and 16 (the end of alcoholic fermentation) for analysis of sugars and for DNA extraction.
Automated ribosomal intergenic spacer analysis.
DNA extraction was carried out on samples taken from the multispecies fermentations as described by Hoffman (34). Automated ribosomal intergenic spacer analysis (ARISA) was subsequently performed using 50 ng of DNA template and carboxy-fluorescein-labeled forward (ITS1-6FAM) and ITS4 primers (35, 36). The labeled PCR products were separated by capillary electrophoresis on an ABI 3,010 × I Genetic analyzer (Applied Biosystems) at the Central Analytical Facility, Stellenbosch University. The raw data were converted to electropherograms and further analyzed in Genemapper 4.1 (Applied Biosystems). Peak areas for each species in the consortium as well as S. cerevisiae were calculated to determine the relative species abundance in each fraction. The average abundance of each of the individual peaks was calculated and represented as a percentage of the total number of peak heights displayed in each sample. Statistical analyses were conducted using XLStat 2017.
Significance values for different FLO treatments in S. cerevisiae, L. thermotolerans, and H. opuntia cocultures at days 2, 6, 10, and 16 of growth. Download Table S2, DOCX file, 0.02 MB (18.9KB, docx) .
Copyright © 2018 Rossouw et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Significance values for different FLO treatments in S. cerevisiae, L. thermotolerans, and W. anomalus cocultures at days 2, 6, 10, and 16 of growth. Download Table S3, DOCX file, 0.02 MB (19.1KB, docx) .
Copyright © 2018 Rossouw et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Significance values for different FLO treatments in S. cerevisiae, H. opuntiae, and W. anomalus cocultures at days 2, 6, 10, and 16 of growth. Download Table S4, DOCX file, 0.02 MB (17.7KB, docx) .
Copyright © 2018 Rossouw et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Significance values for different FLO treatments in S. cerevisiae, H. opuntiae, L. thermotolerans, and W. anomalus cocultures at days 2, 6, 10, and 16 of growth. Download Table S5, DOCX file, 0.02 MB (21KB, docx) .
Copyright © 2018 Rossouw et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
ARISA analyses were carried out at the Central Analytical Facility (CAF) of Stellenbosch University.
This work was supported by the National Research Foundation (NRF) of South Africa through a SARCHI grant (UID 83471) to F.F.B. and an RCA grant (91448) to D.R.
We declare that we have no competing financial interests.
D.R. and F.F.B. conceptualized the project; D.R. and S.P.M. performed the experiments; D.R., S.P.M., and F.F.B. analyzed and interpreted the data; and D.R. and F.F.B. wrote the manuscript.
REFERENCES
- 1.Nissen P, Nielsen D, Arneborg N. 2003. Viable Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-Saccharomyces yeasts in mixed cultures by a cell-cell contact-mediated mechanism. Yeast 20:331–341. doi: 10.1002/yea.965. [DOI] [PubMed] [Google Scholar]
- 2.Renault PE, Albertin W, Bely M. 2013. An innovative tool reveals interaction mechanisms among yeast populations under oenological conditions. Appl Microbiol Biotechnol 97:4105–4119. doi: 10.1007/s00253-012-4660-5. [DOI] [PubMed] [Google Scholar]
- 3.Lopez CLF, Beaufort S, Brandam C, Taillandier P. 2014. Interactions between Kluyveromyces marxianus and Saccharomyces cerevisiae in tequila must type medium fermentation. World J Microbiol Biotechnol 30:2223–2229. doi: 10.1007/s11274-014-1643-y. [DOI] [PubMed] [Google Scholar]
- 4.Bester MC, Jacobson D, Bauer FF. 2012. Many Saccharomyces cerevisiae cell wall protein encoding genes are coregulated by Mss11, but cellular adhesion phenotypes appear only Flo protein dependent. G3 (Bethesda) 2:131–141. doi: 10.1534/g3.111.001644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sampermans S, Mortier J, Soares EV. 2005. Flocculation onset in Saccharomyces cerevisiae: the role of nutrients. J Appl Microbiol 98:525–531. doi: 10.1111/j.1365-2672.2004.02486.x. [DOI] [PubMed] [Google Scholar]
- 6.Bauer FF, Govender P, Bester MC. 2010. Yeast flocculation and its biotechnological relevance. Appl Microbiol Biotechnol 88:31–39. doi: 10.1007/s00253-010-2783-0. [DOI] [PubMed] [Google Scholar]
- 7.Bester MC, Pretorius IS, Bauer F. 2006. The regulation of Saccharomyces cerevisiae FLO gene expression and Ca2+-dependent flocculation by Flo8p and Mss11p. Curr Genet 49:375–383. doi: 10.1007/s00294-006-0068-z. [DOI] [PubMed] [Google Scholar]
- 8.Goossens K, Willaert R. 2010. Flocculation protein structure and cell-cell adhesion mechanism in Saccharomyces cerevisiae. Biotechnol Lett 32:1571–1585. doi: 10.1007/s10529-010-0352-3. [DOI] [PubMed] [Google Scholar]
- 9.Miki BL, Poon NH, James AP, Seligy VL. 1982. Possible mechanism for flocculation interactions governed by gene FLO1 in Saccharomyces cerevisiae. J Bacteriol 150:878–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goossens KVY, Lelasi FS, Nookaew I, Stals I, Alonso-Sarduy L, Daenen L, Van Mulders SE, Stassen C, Rge v. E, Siewers V, Delvaux FR, Kasas S, Nielsen J, Devreese B, Willaert RG. 2015. Molecular mechanism of flocculation self-recognition in yeast and its role in mating and survival. mBio 6:1–16. doi: 10.1128/mBio.00427-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soares EV. 2011. Flocculation in Saccharomyces cerevisiae: a review. J Appl Microbiol 110:1–18. doi: 10.1111/j.1365-2672.2010.04897.x. [DOI] [PubMed] [Google Scholar]
- 12.Rossouw D, Bagheri B, Setati ME, Bauer FF. 2015. Co-flocculation of yeast species, a new mechanism to govern population dynamics in microbial ecosystems. PLoS One 10:1–17. doi: 10.1371/journal.pone.0136249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose AH. 1984. Physiology of cell aggregation: flocculation by Saccharomyces cerevisiae as a model system, p 323–335. In Marshall KC. (ed), Microbial adhesion and aggregation. Springer, Berlin, Germany. [Google Scholar]
- 14.Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, Prevost MC, Latgé J-P, Fink GR, Foster KR, Verstrepen KJ. 2008. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pu L, Jingfan F, Kai C, Chao-An L, Yunjiang C. 2014. Phenylethanol promotes adhesion and biofilm formation of the antagonistic yeast Kloeckera apiculata for the control of blue mold on citrus. FEMS Yeast Res 14:536–546. doi: 10.1111/1567-1364.12139. [DOI] [PubMed] [Google Scholar]
- 16.Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. 2007. A biochemical guide to yeast adhesins: glycoproteins for social and anticosial occasions. Microbiol Mol Biol Rev 71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govender P, Domingo JL, Bester MC, Pretorius IS, Bauer FF. 2008. Controlled expression of the dominant flocculation genes FLO1, FLO5 and FLO11 in Saccharomyces cerevisiae. Appl Environ Microbiol 74:6041–6052. doi: 10.1128/AEM.00394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagheri B, Bauer FF, Setati EC. 2017. The impact of Saccharomyces cerevisiae on a wine yeast consortium in natural and inoculated fermentations. FEMS Yeast Res 8:01988. doi: 10.3389/fmicb.2017.01988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Setati ME, Jacobson D, Andong U-C, Bauer FF, Bauer F. 2012. The vineyard yeast microbiome, a mixed model microbial map. PLoS One 7:e52609. doi: 10.1371/journal.pone.0052609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camarasa C, Sanchez I, Brial P, Bigey F, Dequin S. 2011. Phenotypic landscape of Saccharomyces cerevisiae during wine fermentation: evidence for origin-dependent metabolic traits. PLoS One 6:e25147. doi: 10.1371/journal.pone.0025147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsit S, Dequin S. 2015. Diversity and adaptation of Saccharomyces wine yeast: a review. FEMS Yeast Res 15:fov067. doi: 10.1093/femsyr/fov067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teunissen AW, Steensma HY. 1995. Review: the dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 11:1001–1013. doi: 10.1002/yea.320111102. [DOI] [PubMed] [Google Scholar]
- 23.Verstrepen KJ, Fink GR. 2009. Genetic and epigenetic mechanisms underlying cell-surface variability in protozoa and fungi. Annu Rev Genet 43:1–24. doi: 10.1146/annurev-genet-102108-134156. [DOI] [PubMed] [Google Scholar]
- 24.Verstrepen KJ, Reynolds TB, Fink GR. 2004. Origins of variation in the fungal cell surface. Nat Rev Microbiol 2:533–540. doi: 10.1038/nrmicro927. [DOI] [PubMed] [Google Scholar]
- 25.Verstrepen KJ, Jansen A, Lewitter F, Fink GR. 2005. Intragenic tandem repeats generate functional variability. Nat Genet 37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rando OJ, Verstrepen K. 2007. Timescales of genetic and epigenetic inheritance. Cell 128:655–668. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 27.Hahn MW, De Bie T, Stajich JE, Nguyen C, Cristianini N. 2005. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res 15:1153–1160. doi: 10.1101/gr.3567505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christiaens JF, Van Mulders SE, Duitama J, Brown CA, Ghequire MG, De Meester L, Michiels J, Wenseleers T, Voordeckers K, Verstrepen KJ. 2012. Functional divergence of gene duplicates through ectopic recombination. EMBO Rep 13:1145–1151. doi: 10.1038/embor.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rolland T, Dujon B, Richard G-F. 2010. Dynamic evolution of megasatellites in yeasts. Nucleic Acids Res 38:4731–4739. doi: 10.1093/nar/gkq207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkel JS, Mitchell AP. 2011. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol 9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright CS. 1984. Structural comparison of the two distinct sugar binding sites in wheat germ agglutinin isolectin II. J Mol Biol 178:91–104. doi: 10.1016/0022-2836(84)90232-8. [DOI] [PubMed] [Google Scholar]
- 32.Jiranek V, Langridge P, Henschke PA. 1995. Amino acid and ammonium utilization by Saccharomyces cerevisiae wine yeasts from a chemically defined medium. Am J Enol Vitic 46:75–83. [Google Scholar]
- 33.Bely M, Sablayrolles JM, Barre P. 1990. Description of alcoholic fermentation kinetics: its variability and significance. Am J Enol Vitic 41:319–324. [Google Scholar]
- 34.Hoffman S. 2003. Rapid isolation of yeast chromosomal DNA, p 13.11.2–13.11.4. In Ausubel FM, Brent R, Kingstone RE, Moore DD, Seidman JG, Smith JA, Struhl K (ed), Current protocols in molecular biology. John Wiley and Sons, Somerset, NJ. [Google Scholar]
- 35.Chovanová K, Kraková L, Zenisová K, Turcovská V, Brežná B, Kuchta T, Pangallo D. 2011. Selection and identification of autochthonous yeasts in Slovakian wine samples using a rapid and reliable threestep approach. Lett Appl Microbiol 53:231–237. doi: 10.1111/j.1472-765X.2011.03097.x. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh S, Bagheri B, Morgan HH, Divol BD, Setati ME. 2015. Assessment of wine microbial diversity using ARISA and cultivation-based methods. Ann Microbiol 65:1833–1840. doi: 10.1007/s13213-014-1021-x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P values for pairwise S. cerevisiae and non-Saccharomyces yeast (L. thermotolerans, H. opuntiae, or W. anomalus) comparisons of viability percents after 24 h of two-species coculture (data shown in Fig. 2). Download Table S1, DOCX file, 0.02 MB (19.1KB, docx) .
Copyright © 2018 Rossouw et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Significance values for different FLO treatments in S. cerevisiae, L. thermotolerans, and H. opuntia cocultures at days 2, 6, 10, and 16 of growth. Download Table S2, DOCX file, 0.02 MB (18.9KB, docx) .
Copyright © 2018 Rossouw et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Significance values for different FLO treatments in S. cerevisiae, L. thermotolerans, and W. anomalus cocultures at days 2, 6, 10, and 16 of growth. Download Table S3, DOCX file, 0.02 MB (19.1KB, docx) .
Copyright © 2018 Rossouw et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Significance values for different FLO treatments in S. cerevisiae, H. opuntiae, and W. anomalus cocultures at days 2, 6, 10, and 16 of growth. Download Table S4, DOCX file, 0.02 MB (17.7KB, docx) .
Copyright © 2018 Rossouw et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Significance values for different FLO treatments in S. cerevisiae, H. opuntiae, L. thermotolerans, and W. anomalus cocultures at days 2, 6, 10, and 16 of growth. Download Table S5, DOCX file, 0.02 MB (21KB, docx) .
Copyright © 2018 Rossouw et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.