Abstract
Early-onset coronary artery disease (CAD) has a strong genetic component. Although genome-wide association studies have identified various genes and loci significantly associated with CAD mainly in European populations, genetic variants that contribute toward susceptibility to this condition in Japanese patients remain to be definitively identified. In the present study, exome-wide association studies (EWASs) were performed to identify genetic variants that confer susceptibility to early-onset CAD in Japanese. A total of 7,256 individuals aged ≤65 years were enrolled in the present study. EWAS were conducted on 1,482 patients with CAD and 5,774 healthy controls. Genotyping of single nucleotide polymorphisms (SNPs) was performed using Illumina Human Exome-12 DNA Analysis BeadChip or Infinium Exome-24 BeadChip arrays. The association between allele frequencies for 31,465 SNPs that passed quality control and CAD was examined using Fisher's exact test. To compensate for multiple comparisons of allele frequencies with CAD, a false discovery rate (FDR) of <0.05 was applied for statistically significant associations. The association between allele frequencies for 31,465 SNPs and CAD, as determined by Fisher's exact test, demonstrated that 170 SNPs were significantly (FDR <0.05) associated with CAD. Multivariable logistic regression analysis with adjustment for age, sex, and the prevalence of hypertension, diabetes mellitus and dyslipidemia revealed that 162 SNPs were significantly (P<0.05) associated with CAD. A stepwise forward selection procedure was performed to examine the effects of genotypes for the 162 SNPs on CAD. The 54 SNPs were significant (P<0.05) and independent [coefficient of determination (R2), 0.0008 to 0.0297] determinants of CAD. These SNPs together accounted for 15.5% of the cause of CAD. Following examination of results from previous genome-wide association studies and linkage disequilibrium of the identified SNPs, 21 genes (RNF2, YEATS2, USP45, ITGB8, TNS3, FAM170B-AS1, PRKG1, BTRC, MKI67, STIM1, OR52E4, KIAA1551, MON2, PLUT, LINC00354, TRPM1, ADAT1, KRT27, LIPE, GFY and EIF3L) and five chromosomal regions (2p13, 4q31.2, 5q12, 13q34 and 20q13.2) that were significantly associated with CAD were newly identified in the present study. Gene ontology analysis demonstrated that various biological functions were predicted in the 18 genes identified in the present study. The network analysis revealed that the 18 genes had potential direct or indirect interactions with the 30 genes previously revealed to be associated with CAD or with the 228 genes identified in previous genome-wide association studies. The present study newly identified 26 loci that confer susceptibility to CAD. Determination of genotypes for the SNPs at these loci may prove informative for assessment of the genetic risk for CAD in Japanese patients.
Keywords: coronary artery disease, myocardial infarction, ischemic heart disease, genetics, exome-wide association study
Introduction
Coronary atherosclerosis is a chronic inflammatory vascular disease and is initiated as a result of endothelial damage and dysfunction, which lead to the accumulation and oxidation of low density lipoprotein (LDL)-cholesterol in the arterial wall (1,2) Monocytes migrate from the blood into the subendothelial intima and transform into macrophages, which then accumulate lipid particles (foam cells) to form the lipid core of atherosclerotic plaques (2,3). Inflammatory and thrombotic processes serve central roles in the formation of atherosclerotic lesions and subsequent plaque rupture, which lead toward acute coronary syndrome (2,3).
Coronary artery disease (CAD) and myocardial infarction (MI) are serious clinical conditions that remain the leading cause of mortality in the United States (4). Disease prevention is an important strategy for reducing the overall burden of CAD and MI, with the identification of biomarkers for disease risk being key for risk prediction and for potential intervention, in order to reduce the chance of future adverse coronary events. In addition to conventional risk factors for CAD, including hypertension, diabetes mellitus and dyslipidemia, the importance of genetic factors has been highlighted (5–7). Genes responsible for familial hypercholesterolemia and Tangier disease are prototypical examples of monogenic forms of CAD and MI with Mendelian inheritance (5,8). Familial hypercholesterolemia is an autosomal dominant disorder characterized by marked increases in the circulating concentrations of total cholesterol and LDL-cholesterol caused by mutations of the genes for LDL receptor (LDLR), apolipoprotein B (APOB), proprotein convertase subtilisin/kexin type 9 (PCSK9), cytochrome P450 family 7 subfamily A member 1 (CYP7A1) or LDL receptor adaptor protein 1 (LDLRAP1) (9,10). Tangier disease is an autosomal recessive disorder characterized by a decrease in the circulating concentration of high density lipoprotein (HDL)-cholesterol as a result of loss-of-function mutations in the ATP-binding cassette subfamily A member 1 gene (ABCA1) (11–13). The etiology of common forms of CAD is multifactorial and includes genetic components, as well as environmental and lifestyle factors (5–8). The heritability of common forms of CAD has been estimated to be 40–60% on the basis of family and twin studies (6,7,14).
Genome-wide association studies (GWASs) in European-ancestry (15–21), African American (22) or Han Chinese populations (23,24) have identified various genes and loci that confer susceptibility to CAD or MI. A meta-analysis of GWASs for CAD among European-ancestry populations, including low-frequency variants, identified 202 independent genetic variants at 129 loci with a false discovery rate (FDR) of <5% (25). These genetic variants together accounted for ~28% of the heritability of CAD, demonstrating that genetic susceptibility to this condition is largely determined by common variants with small effect sizes (6,25). A more recent meta-analysis for CAD in European-ancestry populations identified 304 independent genetic variants with an FDR of <5%, and these variants accounted for 21.2% of the heritability of CAD (26). In total, GWASs identified 163 loci associated with CAD at a genome-wide significance level and >300 possible loci for this condition with an FDR of <5% (7). Although several single nucleotide polymorphisms (SNPs) have been revealed to be significantly associated with MI in Japanese patients (27,28), genetic variants that contribute toward susceptibility to CAD and MI in Japanese patients remain to be definitively identified.
A study of monozygotic and dizygotic twins revealed that mortality from CAD at younger ages was significantly influenced by genetic factors in males and females, whereas the genetic effect was smaller at older ages (29,30). A family history of MI is also more apparent in individuals with early-onset MI than in those with late-onset MI, suggestive of a greater heritability in the former (31,32).
The present study included exome-wide association studies (EWASs) for CAD with the use of human exome array-based genotyping methods in order to identify genetic variants that confer susceptibility to this condition in Japanese patients. In order to increase the statistical power of the EWAS, patients with early-onset CAD were examined.
Materials and methods
Study subjects
In our previous EWAS, the median age of subjects with CAD was 69 years (33). Therefore, patients with an age of ≤65 years were defined as individuals with early-onset CAD in the present study. A total of 7,256 Japanese subjects aged ≤65 years [mean age, 51.7 years; age range, 18–65 years; males/females (%), 58.3/41.7; 1,482 with CAD, including 1,152 with MI, and 5,774 controls] were enrolled in the present study. The subjects were individuals who either visited outpatient clinics or were admitted to participating hospitals in Japan (Gifu Prefectural Tajimi Hospital, Tajimi; Gifu Prefectural General Medical Center, Gifu; Japanese Red Cross Nagoya First Hospital, Nagoya; Northern Mie Medical Center Inabe General Hospital, Inabe; and Hirosaki University Hospital and Hirosaki Stroke and Rehabilitation Center, Hirosaki, Japan) due to various symptoms or for an annual health check-up between October 2002 and March 2014, or who were community-dwelling individuals recruited to a population-based cohort study in Inabe between March 2010 and September 2014 (34).
The diagnosis of CAD was based on the detection of stenosis of >50% in any major coronary artery or in the left main trunk by coronary angiography. The diagnosis of MI was based on typical electrocardiographic changes and on increases in the serum activity of creatine kinase (MB isozyme) and in the serum concentration of troponin T. The diagnosis was confirmed by identification of the responsible stenosis in any of the major coronary arteries or in the left main trunk by coronary angiography. The control individuals had no history of MI, CAD, aortic aneurysm or peripheral artery disease; of ischemic or hemorrhagic stroke; or of other atherosclerotic, thrombotic, embolic or hemorrhagic disorders. Although certain control individuals had conventional risk factors for CAD, including hypertension, diabetes mellitus, dyslipidemia and CKD, they did not have any cardiovascular complications.
EWAS
Venous blood (5 or 7 ml) was collected into tubes containing 50 mmol/l ethylenediaminetetraacetic acid (disodium salt), peripheral blood leukocytes were isolated, and genomic DNA was extracted from these cells with the use of a DNA extraction kit (Genomix; Talent SRL, Trieste, Italy; or SMITEST EX-R&D; Medical & Biological Laboratories, Co., Ltd., Nagoya, Japan). The EWASs for CAD (1,482 cases and 5,774 controls) was performed with the use of a Human Exome-12 v1.2 DNA Analysis BeadChip or Infinium Exome-24 v1.0 BeadChip (Illumina, Inc., San Diego, CA, USA). These exome arrays include putative functional exonic variants selected from ~12,000 individual exome and whole-genome sequences. The exonic content consists of ~244,000 SNPs from European, African, Chinese and Hispanic individuals (35). SNPs contained in only one of the exome arrays (~2.6% of all SNPs) were excluded from analysis. Quality control was performed as follows (36): i) Genotyping data with a call rate of <97% were discarded, with the mean call rate for the remaining data being 99.9%; ii) gender specification was checked for each sample, and those for which gender phenotype in the clinical records was inconsistent with genetic sex were discarded; iii) duplicate samples and cryptic relatedness were checked by calculation of identity by descent, and all pairs of DNA samples exhibiting an identity by descent of >0.1875 were inspected and one sample from each pair was excluded; iv) the frequency of heterozygosity for SNPs was calculated for all samples, and those with extremely low or high heterozygosity (>3 standard deviations from the mean) were discarded; v) SNPs in sex chromosomes or mitochondrial DNA were excluded from the analysis, as were nonpolymorphic SNPs or SNPs with a minor allele frequency of <1.0%; vi) SNPs whose genotype distributions deviated significantly (P<0.01) from Hardy-Weinberg equilibrium in control individuals were discarded; and vii) genotype data were examined for population stratification by principal components analysis (37), and population outliers were excluded from the analysis. A total of 31,465 SNPs passed quality control for the EWASs of CAD and these SNPs were subjected to analyses.
Statistical analysis
For analysis of the characteristics of the study subjects, quantitative data were compared between subjects with CAD and controls using the unpaired Student's t-test. Categorical data were compared between the two groups using the Pearson's χ2 test. Allele frequencies were estimated by the gene counting method, and Fisher's exact test was applied to identify departure from the Hardy-Weinberg equilibrium. In the EWAS, the association between allele frequencies of each SNP and CAD was examined using the Fisher's exact test. The genomic inflation factor (λ) was 0.93. To compensate for multiple comparisons of genotypes with CAD, an FDR was applied for statistical significance of association (38). The significance level was set at an FDR of <0.05 for the EWAS. Multivariable logistic regression analysis was performed with CAD as a dependent variable and independent variables, including age, sex (0, female and 1, male), the prevalence of hypertension, diabetes mellitus, and dyslipidemia (0, no history of these conditions; 1, positive history), as well as the genotype of each SNP. Genotypes of the SNPs were assessed according to dominant [0, AA; 1, AB + BB (A, major allele; B, minor allele)] and recessive (0, AA + AB; 1, BB) genetic models, and the P-value, odds ratio and 95% confidence interval were calculated. A stepwise forward selection procedure was also performed to examine the effects of genotypes on CAD. The P-levels for inclusion in and exclusion from the model were 0.25 and 0.1, respectively. In the stepwise forward selection procedure, each genotype was examined according to a dominant or recessive model on the basis of statistical significance in the multivariable logistic regression analysis. The association between genotypes of SNPs and intermediate phenotypes of CAD was examined using the Pearson's χ2 test. With the exception of the initial EWAS by the Fisher's exact test (FDR <0.05), P<0.05 was considered to indicate a statistically significant difference. Statistical tests were performed using JMP Genomics version 9.0 software (SAS Institute, Inc., Cary, NC, USA).
Association between genes, chromosomal loci and SNPs identified in the present study and phenotypes previously reported by GWASs
The genes, chromosomal loci, and SNPs identified in the present study were compared with the cardiovascular disease-related phenotypes previously reported by GWASs available in the Genome-Wide Repository of Associations Between SNPs and Phenotypes (GRASP) Search database v. 2.0.0.0 (https://grasp.nhlbi.nih.gov/Search.aspx), developed by the Information Technology and Applications Center at the National Center for Biotechnology Information (National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, USA) (39,40).
Gene Ontology analysis
Biological functions of the genes were examined by the use of the Gene Ontology and GO Annotations databases (QuickGO version 2018; https://www.ebi.ac.uk/QuickGO/; European Bioinformatics Institute, European Molecular Biology Laboratory, Hinxton, Cambridgeshire, UK) (41,42).
Network analysis of gene-gene interactions
Network analyses were performed to predict functional gene-gene interactions by the use of GeneMANIA Cytoscape plugin (http://apps.cytoscape.org/apps/genemania; Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, Canada) (43–45) using Cytoscape v3.4.0 software (http://www.cytoscape.org/; The Cytoscape Consortium, San Diego, CA, USA) (46). To begin with, the 30 genes (ACE, NOS3, CCL2, PON1, CD40LG, LOX, APOB, CRP, APOA1, LPA, ESR1, LDLR, APOC3, VEGFA, LTA, HMOX1, MMP3, APOA5, PCSK9, CDKN2B, TLR4, GNB3, PTGS2, NPPB, ABCG8, ESR2, CXCL12, MIA3, IRS1 and ABO) were selected from the DisGeNET database (http://www.disgenet.org/web/DisGeNET; Integrative Biomedical Informatics Group, Research Programme on Biomedical Informatics, Barcelona Biomedical Research Park, Barcelona, Spain) (47,48), according to the rank order of high scores in association with CAD. Next, the 234 genes previously identified by GWASs (7) were selected, among which six genes were not included in GeneMANIA database and had no interaction with other genes. Therefore, the 228 genes (SKI, PRDM16, FHL3, PCSK9, PPAP2B, SORT1, NGF, CASQ2, TDRKH, IL6R, ATP1B1, NME7, DDX59, CAMSAP2, LMOD1, HHAT, SERTAD4, DIEXF, MIA3, AGT, APOB, ABCG5, ABCG8, PRKCE, VAMP5, VAMP8, GGCX, ZEB2, FIGN, CALCRL, TFPI, WDR12, NBEAL1, FN1, TNS1, IRS1, KCNJ13, COL6A3, FGD5, ALS2CL, RTP3, CDC25A, SPINK8, MAP4, ZNF589, RHOA, ITGB5, DNAJC13, STAG1, MSL2, NCK1, PPP2R3A, MRAS, ARHGEF26, TIPARP, FNDC3B, RGS12, REST, NOA1, STBD1, PRDM8, FGF5, HNRNPD, UNC5C, MAD2L1, PDE5A, ZNF827, EDNRA, PALLD, SEMA5A, MAP3K1, LOX, SLC22A4, IL5, RAD50, ARHGAP26, FOXC1, PHACTR1, EDN1, HDGFL1, C2, ANKS1A, PI16, KCNK5, VEGFA, RAB23, FAM46A, CENPW, TCF21, PLEKHG1, LPA, PLG, MAD1L1, DAGLB, RAC1, KDELR2, TMEM106B, HDAC9, CCM2, BCAP29, GPR22, CFTR, ZC3HC1, KLHDC10, PARP12, TBXAS1, NOS3, NAT2, LPL, BMP1, ZFPM2, TRIB1, KLF4, SVEP1, DAB2IP, ABO, CDC123, KIAA1462, CXCL12, TSPAN14, FAM213A, LIPA, CYP17A1, CNNM2, NT5C2, SH3PXD2A, HTRA1, TRIM5, TRIM22, TRIM6, SWAP70, CTR9, ARNTL, HSD17B12, SIPA1, SERPINH1, ARHGAP42, PDGFD, APOA1, APOC3, APOA4, APOA5, C1S, PRPF31, HOXC4, LRP1, FGD6, SH2B3, KSR2, HNF1A, CCDC92, SCARB1, FLT1, N4BP2L2, PDS5B, COL4A1, COL4A2, MCF2L, CUL4A, ARID4A, PSMA3, TMED10, SERPINA1, HHIPL1, YY1, TRIP4, SMAD3, ADAMTS7, MFGE8, FURIN, FES, CETP, HP, CFDP1, BCAR1, PLCG2, CDH13, SMG6, PEMT, CORO6, BLMH, ANKRD13B, GIT1, SSH2, EFCAB5, COPRS, RAB11FIP4, DHX58, KAT2A, RAB5, NKIRAS2, DNAJC7, KCNH4, HCRT, GHDC, GOSR2, UBE2Z, GIP, BCAS3, PECAM1, DDX5, TEX2, ACAA2, RPL17, PMAIP1, MC4R, LDLR, SMARCA4, FCHO1, COLGALT1, ZNF507, HNRNPUL1, TGFB1, APOE, APOC1, PVRL2, COTL1, SNRPD2, PROCR, EIF6, ZHX3, PLCG1, PLTP, MMP9, ZNF831, BACH1, KCNE2 and ADORA2A) were applied to analysis.
Results
Characteristics of subjects
The characteristics of the 7,256 subjects enrolled in the present study are presented in Table I. The age, the frequency of males, and the prevalence of obesity, hypertension, diabetes mellitus (DM), dyslipidemia, chronic kidney disease (CKD) and hyperuricemia, as well as body mass index, systolic and diastolic blood pressure, fasting plasma glucose level, blood glycosylated hemoglobin (hemoglobin A1c) content, and the serum concentrations of triglycerides, creatinine, and uric acid were greater, whereas the serum concentration of HDL-cholesterol and estimated glomerular filtration rate were lower, in patients with CAD than in controls.
Table I.
Characteristics of control subjects and patients with coronary artery disease.
| Characteristic | Control | Coronary artery disease | P-value |
|---|---|---|---|
| No. subjects | 5,774 | 1,482 | |
| Age, years | 50.6±10.2 | 55.9±7.4 | <0.0001 |
| Sex, males/females, % | 52.1/47.9 | 82.5/17.5 | <0.0001 |
| Smoking, % | 42.5 | 43.0 | 0.7719 |
| Obesity, % | 31.0 | 43.0 | <0.0001 |
| Body mass index, kg/m2 | 23.2±3.5 | 24.5±3.5 | <0.0001 |
| Hypertension, % | 31.7 | 70.0 | <0.0001 |
| Systolic BP, mmHg | 121±18 | 139±27 | <0.0001 |
| Diastolic BP, mmHg | 75±13 | 78±15 | <0.0001 |
| Diabetes mellitus, % | 12.7 | 58.7 | <0.0001 |
| Fasting plasma glucose, mmol/l | 5.66±1.78 | 7.55±3.39 | <0.0001 |
| Blood hemoglobin A1c, % | 5.72±0.96 | 6.89±1.75 | <0.0001 |
| Dyslipidemia, % | 56.9 | 84.1 | <0.0001 |
| Serum triglycerides, mmol/l | 1.32±0.98 | 1.84±1.34 | <0.0001 |
| Serum HDL-cholesterol, mmol/l | 1.65±0.45 | 1.20±0.36 | <0.0001 |
| Serum LDL-cholesterol, mmol/l | 3.18±0.83 | 3.18±0.98 | 0.9770 |
| Chronic kidney disease, % | 10.3 | 29.4 | <0.0001 |
| Serum creatinine, µmol/l | 69.8±61.0 | 95.5±119.3 | <0.0001 |
| eGFR, ml min−1 1.73 m−2 | 78.7±17.1 | 70.7±26.9 | <0.0001 |
| Hyperuricemia, % | 15.2 | 25.5 | <0.0001 |
| Serum uric acid, µmol/l | 321±89 | 353±102 | <0.0001 |
Quantitative data represent the mean ± standard deviation and were compared between subjects with coronary artery disease and controls with the unpaired Student's t-test. Categorical data were compared between the two groups using Pearson's χ2 test. P<0.05 was considered to indicate a statistically significant difference. Obesity was defined as a body mass index of ≥25 kg/m2; hypertension as a systolic BP of ≥140 mmHg, diastolic BP of ≥90 mmHg, or the taking of anti-hypertensive medication; diabetes mellitus as a fasting plasma glucose level of ≥6.93 mmol/l, blood hemoglobin A1c content of ≥6.5%, or the taking of anti-diabetes medication; dyslipidemia as a serum triglyceride concentration of ≥1.65 mmol/l, serum HDL-cholesterol concentration of <1.04 mmol/l, serum LDL-cholesterol concentration of ≥3.64 mmol/l or the taking of anti-dyslipidemic medication; chronic kidney disease as an estimated glomerular filtration rate (eGFR) of <60 ml min−1 1.73 m−2; and hyperuricemia as a serum uric acid concentration of >416 µmol/l or the taking of uric acid-lowering medication. BP, blood pressure; HDL, high-density lipoprotein; LDL, low-density lipoprotein; eGFR, estimated glomerular filtration rate.
EWAS for CAD
The association between allele frequencies for 31,465 SNPs that passed quality control and CAD was examined using the Fisher's exact test, and the 170 SNPs were significantly (FDR <0.05) associated with CAD (Table II).
Table II.
170 SNPs significantly (FDR <0.5) associated with coronary artery disease in the exome-wide association study.
| Gene | SNP | Nucleotide substitutiona | Amino acid substitution | Chromosome | Position | MAF, % | Allele OR | P-value, allele frequency | FDR, allele frequency |
|---|---|---|---|---|---|---|---|---|---|
| PLCB2 | rs200787930 | C/T | E1106K | 15 | 40289298 | 1.2 | 0.03 | 1.24×10−29 | 1.56×10−26 |
| MARCH1 | rs61734696 | G/T | Q137K | 4 | 164197303 | 1.2 | 0.03 | 2.09×10−29 | 2.54×10−26 |
| VPS33B | rs199921354 | C/T | R80Q | 15 | 91013841 | 1.2 | 0.03 | 2.76×10−29 | 3.30×10−26 |
| CXCL8 | rs188378669 | G/T | E31* | 4 | 73741568 | 1.2 | 0.03 | 3.15×10−29 | 3.70×10−26 |
| TMOD4 | rs115287176 | G/A | R277W | 1 | 151170961 | 1.2 | 0.03 | 1.21×10−28 | 1.39×10−25 |
| COL6A3 | rs146092501 | C/T | E1386K | 2 | 237371861 | 1.2 | 0.04 | 2.93×10−28 | 3.27×10−25 |
| ZNF77 | rs146879198 | G/A | R340* | 19 | 2934109 | 1.2 | 0.04 | 2.92×10−28 | 3.27×10−25 |
| ADGRL3 | rs192210727 | G/T | R580I | 4 | 61909615 | 1.3 | 0.10 | 2.92×10−23 | 3.06×10−20 |
| OR52E4 | rs11823828 | T/G | F227L | 11 | 5884973 | 36.6 | 1.54 | 3.40×10−21 | 3.35×10−18 |
| ALDH2 | rs671 | G/A | E504K | 12 | 111803962 | 27.6 | 1.41 | 4.12×10−15 | 3.78×10−12 |
| ACAD10 | rs11066015 | G/A | 12 | 111730205 | 27.5 | 1.41 | 4.92×10−15 | 4.45×10−12 | |
| BRAP | rs3782886 | A/G | 12 | 111672685 | 29.3 | 1.37 | 4.38×10−13 | 3.71×10−10 | |
| HECTD4 | rs11066280 | T/A | 12 | 112379979 | 29.0 | 1.37 | 6.94×10−13 | 5.73×10−10 | |
| HECTD4 | rs2074356 | C/T | 12 | 112207597 | 25.4 | 1.36 | 1.21×10−11 | 9.78×10−9 | |
| NAA25 | rs12231744 | C/T | R876K | 12 | 112039251 | 35.1 | 0.77 | 1.68×10−9 | 1.24×10−6 |
| GOSR2 | rs1052586 | T/C | 17 | 46941097 | 48.7 | 0.79 | 3.94×10−8 | 2.61×10−5 | |
| ATXN2 | rs7969300 | T/C | N248S | 12 | 111555908 | 38.8 | 0.79 | 4.41×10−8 | 2.87×10−5 |
| LILRB2 | rs73055442 | C/T | R103H | 19 | 54279838 | 1.6 | 44.10 | 2.00×10−7 | 1.20×10−4 |
| rs12229654 | T/G | 12 | 110976657 | 22.5 | 1.28 | 2.09×10−7 | 1.24×10−4 | ||
| LOC107987429 | rs2844533 | T/C | 6 | 31383025 | 15.3 | 1.32 | 3.49×10−7 | 1.95×10−4 | |
| MTFR2 | rs143974258 | G/A | R360* | 6 | 136231355 | 3.3 | 0.05 | 6.66×10−7 | 3.60×10−4 |
| PSORS1C1 | rs3130559 | C/T | 6 | 31129524 | 44.2 | 0.82 | 1.51×10−6 | 7.74×10−4 | |
| rs2596548 | G/T | 6 | 31362769 | 5.4 | 1.51 | 1.83×10−6 | 9.21×10−4 | ||
| EIF3L | rs9466 | T/C | 22 | 37877742 | 21.6 | 1.28 | 1.96×10−6 | 9.77×10−4 | |
| LPGAT1 | rs150552771 | T/C | K200E | 1 | 211783358 | 5.0 | 7.14 | 2.26×10−6 | 0.0011 |
| LAIR2 | rs34429135 | T/A | F115Y | 19 | 54508164 | 2.5 | ND | 2.70×10−6 | 0.0013 |
| rs2523644 | A/G | 6 | 31374707 | 8.1 | 1.40 | 2.75×10−6 | 0.0013 | ||
| rs10757278 | A/G | 9 | 22124478 | 49.5 | 0.83 | 2.92×10−6 | 0.0014 | ||
| CCHCR1 | rs130067 | T/G | E328D | 6 | 31150734 | 33.2 | 0.81 | 3.10×10−6 | 0.0015 |
| TCHP | rs74416240 | G/A | 12 | 109904793 | 13.3 | 1.30 | 3.25×10−6 | 0.0015 | |
| rs1333049 | G/C | 9 | 22125504 | 49.4 | 1.20 | 3.95×10−6 | 0.0018 | ||
| CDKN2B-AS1 | rs4977574 | A/G | 9 | 22098575 | 47.1 | 1.21 | 4.18×10−6 | 0.0019 | |
| CDKN2B-AS1 | rs2383207 | G/A | 9 | 22115960 | 33.7 | 0.81 | 4.86×10−6 | 0.0022 | |
| SLC16A1 | rs1049434 | T/A | D490E | 1 | 112913924 | 34.7 | 0.82 | 5.76×10−6 | 0.0025 |
| GIT2 | rs925368 | T/C | N389S | 12 | 109953174 | 12.5 | 1.30 | 6.02×10−6 | 0.0026 |
| rs1333048 | A/C | 9 | 22125348 | 49.6 | 1.20 | 6.46×10−6 | 0.0028 | ||
| rs2523578 | T/C | 6 | 31360765 | 8.1 | 1.39 | 6.54×10−6 | 0.0028 | ||
| rs404890 | G/T | 6 | 32231090 | 30.5 | 1.22 | 8.90×10−6 | 0.0037 | ||
| APOE | rs7412 | C/T | R176C | 19 | 44908822 | 4.3 | 0.60 | 1.06×10−5 | 0.0043 |
| CCHCR1 | rs130071 | G/A | 6 | 31148433 | 5.1 | 1.52 | 1.06×10−5 | 0.0043 | |
| rs602633 | C/A | 1 | 109278889 | 7.6 | 0.69 | 1.15×10−5 | 0.0046 | ||
| CELSR2 | rs12740374 | G/T | 1 | 109274968 | 7.7 | 0.69 | 1.15×10−5 | 0.0046 | |
| MKI67 | rs145121731 | G/A | S2722L | 10 | 128102595 | 1.5 | 2.04 | 1.20×10−5 | 0.0047 |
| CUBN | rs78201384 | C/T | E304K | 10 | 17111024 | 2.7 | 0.52 | 1.38×10−5 | 0.0054 |
| PSORS1C3 | rs887466 | T/C | 6 | 31175734 | 41.1 | 1.20 | 1.38×10−5 | 0.0054 | |
| PSORS1C1 | rs3094663 | G/A | 6 | 31139310 | 30.9 | 1.20 | 1.40×10−5 | 0.0054 | |
| rs10853110 | A/G | 17 | 49241052 | 39.2 | 1.20 | 1.49×10−5 | 0.0057 | ||
| WDR37 | rs10794720 | C/T | 10 | 1110225 | 8.5 | 0.71 | 1.52×10−5 | 0.0057 | |
| CELSR2 | rs629301 | A/C | 1 | 109275684 | 7.8 | 0.70 | 1.52×10−5 | 0.0057 | |
| SKIV2L | rs592229 | G/T | 6 | 31962664 | 42.4 | 1.20 | 1.57×10−5 | 0.0058 | |
| rs12182351 | T/C | 6 | 32233930 | 29.8 | 1.22 | 1.59×10−5 | 0.0059 | ||
| POU5F1 | rs3130503 | G/A | 6 | 31169388 | 29.5 | 1.20 | 1.64×10−5 | 0.0060 | |
| PSORS1C3 | rs1265155 | T/C | 6 | 31175917 | 41.1 | 1.19 | 1.68×10−5 | 0.0061 | |
| CELSR2 | rs646776 | A/G | 1 | 109275908 | 7.7 | 0.70 | 1.70×10−5 | 0.0062 | |
| rs2596503 | C/T | 6 | 31353033 | 19.3 | 1.24 | 1.75×10−5 | 0.0063 | ||
| TRPM1 | rs2241493 | T/C | N54S | 15 | 31070149 | 12.6 | 0.76 | 1.81×10−5 | 0.0065 |
| CCDC141 | rs13419085 | T/C | N1170S | 2 | 178837710 | 1.8 | 0.46 | 1.92×10−5 | 0.0068 |
| VARS2 | rs9394021 | A/G | Q777R | 6 | 30925350 | 44.9 | 0.84 | 1.98×10−5 | 0.0069 |
| SFTA2 | rs2286655 | T/C | 6 | 30931969 | 44.9 | 1.19 | 1.99×10−5 | 0.0069 | |
| rs3873334 | T/C | 6 | 30928370 | 44.9 | 1.19 | 1.98×10−5 | 0.0069 | ||
| rs9261800 | C/G | 6 | 30408822 | 2.8 | 7.21 | 2.02×10−5 | 0.0069 | ||
| TCF19 | rs3130453 | C/T | 6 | 31157072 | 34.4 | 0.83 | 2.10×10−5 | 0.0072 | |
| C21orf59 | rs76974938 | C/T | D67N | 21 | 32609946 | 2.4 | 0.00 | 2.14×10−5 | 0.0073 |
| DDR1 | rs2239518 | T/C | 6 | 30897948 | 44.9 | 1.19 | 2.19×10−5 | 0.0074 | |
| CDSN | rs3130984 | C/T | S143N | 6 | 31117187 | 13.4 | 1.29 | 2.20×10−5 | 0.0074 |
| rs197932 | T/C | 17 | 46896981 | 26.9 | 0.82 | 2.23×10−5 | 0.0075 | ||
| CDSN | rs3130981 | C/T | D527N | 6 | 31116036 | 13.6 | 1.29 | 2.30×10−5 | 0.0075 |
| MICB-DT | rs3132469 | C/T | 6 | 31488790 | 5.3 | 1.46 | 2.41×10−5 | 0.0078 | |
| HLA-DQB1 | rs1049056 | C/A | A6S | 6 | 32666592 | 11.9 | 1.30 | 2.51×10−5 | 0.0081 |
| DDR1 | rs2239517 | A/G | 6 | 30897338 | 44.6 | 1.19 | 2.59×10−5 | 0.0083 | |
| CCHCR1 | rs1265110 | G/A | 6 | 31151645 | 30.2 | 0.83 | 2.69×10−5 | 0.0085 | |
| CCDC63 | rs10774610 | T/C | 12 | 110902439 | 23.7 | 1.22 | 2.76×10−5 | 0.0087 | |
| GTF2H4 | rs2284176 | C/T | 6 | 30907845 | 44.6 | 1.19 | 2.80×10−5 | 0.0088 | |
| GTF2H4 | rs3909130 | G/A | 6 | 30906388 | 44.6 | 1.19 | 2.84×10−5 | 0.0089 | |
| GTF2H4 | rs916920 | G/A | 6 | 30909425 | 44.7 | 1.19 | 2.85×10−5 | 0.0089 | |
| rs1264569 | A/G | 6 | 30397543 | 4.6 | 1.49 | 2.98×10−5 | 0.0092 | ||
| CACNA1D | rs35874056 | G/A | G460S | 3 | 53702798 | 2.0 | 25.00 | 3.09×10−5 | 0.0094 |
| rs9468845 | A/G | 6 | 30901816 | 44.7 | 1.19 | 3.12×10−5 | 0.0094 | ||
| DDR1 | rs8408 | C/T | 6 | 30899889 | 44.7 | 1.19 | 3.11×10−5 | 0.0094 | |
| DDR1 | rs7756521 | C/T | 6 | 30880476 | 44.7 | 1.19 | 3.10×10−5 | 0.0094 | |
| CDKN2B-AS1 | rs1011970 | G/T | 9 | 22062135 | 5.6 | 1.41 | 3.15×10−5 | 0.0095 | |
| ADAT1 | rs145161932 | T/C | R57G | 16 | 75612670 | 1.4 | 0.39 | 3.29×10−5 | 0.0098 |
| POU5F1 | rs885950 | T/G | 6 | 31172375 | 34.0 | 0.83 | 3.28×10−5 | 0.0098 | |
| DDR1 | rs4618569 | A/G | 6 | 30887474 | 44.7 | 1.19 | 3.41×10−5 | 0.0101 | |
| KRT13 | rs146918776 | A/G | Y281H | 17 | 41502993 | 1.5 | 1.94 | 3.51×10−5 | 0.0103 |
| rs2523638 | G/A | 6 | 31376496 | 43.1 | 1.19 | 3.53×10−5 | 0.0103 | ||
| PSRC1 | rs599839 | A/G | 1 | 109279544 | 7.9 | 0.71 | 3.52×10−5 | 0.0103 | |
| rs9275141 | G/T | 6 | 32683340 | 26.4 | 1.21 | 3.62×10−5 | 0.0105 | ||
| CCDC63 | rs10849915 | T/C | 12 | 110895818 | 23.6 | 1.22 | 3.63×10−5 | 0.0105 | |
| HLA-DRA | rs3177928 | G/A | 6 | 32444658 | 5.9 | 1.41 | 3.81×10−5 | 0.0108 | |
| OAS3 | rs2072134 | C/T | 12 | 112971371 | 17.6 | 1.24 | 4.06×10−5 | 0.0114 | |
| USP45 | rs41288947 | C/G | T521R | 6 | 99446210 | 14.9 | 1.26 | 4.11×10−5 | 0.0115 |
| CCHCR1 | rs1265109 | A/C | 6 | 31151812 | 48.2 | 1.18 | 4.16×10−5 | 0.0116 | |
| LOC101929163 | rs6930777 | C/T | 6 | 32383789 | 5.5 | 1.43 | 4.45×10−5 | 0.0122 | |
| rs7333181 | G/A | 13 | 111568950 | 2.5 | 0.54 | 4.45×10−5 | 0.0122 | ||
| DDR1 | rs1264323 | T/C | 6 | 30888130 | 38.8 | 1.19 | 4.48×10−5 | 0.0122 | |
| LINC00243 | rs3094111 | G/A | 6 | 30820414 | 14.7 | 1.25 | 4.52×10−5 | 0.0123 | |
| rs10484561 | T/G | 6 | 32697643 | 5.9 | 1.41 | 4.55×10−5 | 0.0123 | ||
| PSORS1C1 | rs3130558 | G/C | 6 | 31129406 | 13.7 | 1.27 | 4.59×10−5 | 0.0124 | |
| HLA-DQB1 | rs1049060 | T/A | S27T | 6 | 32666529 | 28.8 | 1.20 | 4.92×10−5 | 0.0131 |
| rs2844650 | G/A | 6 | 30934756 | 4.7 | 1.47 | 4.99×10−5 | 0.0131 | ||
| DDR1 | rs3132572 | T/C | 6 | 30893952 | 4.7 | 1.47 | 4.99×10−5 | 0.0131 | |
| CCHCR1 | rs1265115 | T/G | 6 | 31149298 | 47.7 | 1.18 | 4.96×10−5 | 0.0131 | |
| CCHCR1 | rs3094225 | T/C | 6 | 31145275 | 48.4 | 1.18 | 4.93×10−5 | 0.0131 | |
| LOC107987453 | rs3129987 | C/T | 6 | 30798427 | 14.5 | 1.25 | 5.05×10−5 | 0.0132 | |
| DPCR1 | rs2517451 | A/G | 6 | 30946974 | 4.7 | 1.47 | 5.11×10−5 | 0.0133 | |
| KIAA1551 | rs10771894 | A/G | S352G | 12 | 31982009 | 32.4 | 1.19 | 5.18×10−5 | 0.0134 |
| rs13427905 | C/T | 2 | 71846585 | 18.5 | 0.80 | 5.22×10−5 | 0.0134 | ||
| ABCA1 | rs1883025 | G/A | 9 | 104902020 | 28.8 | 0.83 | 5.46×10−5 | 0.0139 | |
| SFTA2 | rs2253705 | G/A | 6 | 30932317 | 18.0 | 1.23 | 5.60×10−5 | 0.0141 | |
| PLUT | rs954750 | G/A | 13 | 27889801 | 48.3 | 1.18 | 5.86×10−5 | 0.0146 | |
| TCF19 | rs1419881 | T/C | 6 | 31162816 | 48.1 | 1.18 | 6.37×10−5 | 0.0156 | |
| rs13209234 | G/A | 6 | 32448198 | 5.9 | 1.41 | 6.47×10−5 | 0.0158 | ||
| PSORS1C1 | rs1265100 | T/C | 6 | 31137533 | 32.2 | 0.83 | 6.55×10−5 | 0.0159 | |
| YEATS2 | rs76174573 | G/T | C1232F | 3 | 183804099 | 3.7 | 0.61 | 6.74×10−5 | 0.0162 |
| ABO | rs1053878 | C/T | P156L | 9 | 133256264 | 22.8 | 1.20 | 6.78×10−5 | 0.0162 |
| rs4014195 | C/G | 11 | 65739351 | 16.6 | 1.24 | 6.78×10−5 | 0.0162 | ||
| SFTA2 | s2253588 | C/G | 6 | 30931600 | 23.6 | 1.21 | 6.93×10−5 | 0.0165 | |
| CYP4F8 | rs201166643 | C/A | R488S | 19 | 15629257 | 1.1 | ND | 7.00×10−5 | 0.0165 |
| NAXE | rs7516274 | C/G | L19V | 1 | 156591859 | 1.8 | 0.48 | 7.18×10−5 | 0.0169 |
| rs10757283 | T/C | 9 | 22134173 | 33.8 | 0.84 | 7.25×10−5 | 0.0170 | ||
| BTNL2 | rs28362680 | G/A | A202V | 6 | 32403039 | 39.7 | 0.85 | 7.40×10−5 | 0.0171 |
| BTNL2 | rs10947262 | C/T | 6 | 32405535 | 39.7 | 0.85 | 7.40×10−5 | 0.0171 | |
| KRT27 | rs17558532 | C/T | A284T | 17 | 40779624 | 3.6 | 0.62 | 7.71×10−5 | 0.0176 |
| GTF2H4 | rs3130780 | G/T | 6 | 30906531 | 18.0 | 1.23 | 7.71×10−5 | 0.0176 | |
| rs2532934 | T/C | 6 | 30926982 | 24.1 | 1.20 | 7.74×10−5 | 0.0176 | ||
| VARS2 | rs753725 | G/A | 6 | 30923094 | 24.1 | 1.20 | 7.68×10−5 | 0.0176 | |
| PLUT | rs11619319 | A/G | 13 | 27913462 | 48.1 | 1.18 | 7.64×10−5 | 0.0176 | |
| rs3095273 | C/T | 6 | 29598592 | 5.5 | 1.41 | 8.16×10−5 | 0.0184 | ||
| TNS1 | rs918949 | C/T | V1590I | 2 | 217809974 | 42.8 | 0.85 | 8.39×10−5 | 0.0188 |
| LINC00243 | rs3130785 | C/T | 6 | 30828961 | 14.6 | 1.24 | 8.37×10−5 | 0.0188 | |
| VARS2 | rs2249464 | C/T | R309W | 6 | 30920384 | 24.1 | 1.20 | 9.39×10−5 | 0.0207 |
| rs3095345 | A/G | 6 | 30854636 | 17.9 | 1.22 | 9.37×10−5 | 0.0207 | ||
| ITGB8 | rs80015015 | G/A | C481Y | 7 | 20401881 | 7.1 | 1.35 | 1.01×10−4 | 0.0220 |
| VARS2 | rs885905 | C/T | 6 | 30922654 | 23.4 | 1.20 | 1.07×10−4 | 0.0232 | |
| LIPE | rs34052647 | G/A | R611C | 19 | 42407617 | 5.5 | 1.39 | 1.16×10−4 | 0.0249 |
| PHACTR1 | rs9369640 | A/C | 6 | 12901209 | 9.1 | 0.74 | 1.30×10−4 | 0.0275 | |
| BTNL2 | rs41417449 | T/C | M295V | 6 | 32396234 | 23.0 | 0.83 | 1.35×10−4 | 0.0280 |
| BTNL2 | rs41441651 | C/T | D336N | 6 | 32396111 | 23.0 | 0.83 | 1.35×10−4 | 0.0280 |
| BTNL2 | rs28362675 | C/A | E454* | 6 | 32394744 | 23.0 | 0.83 | 1.35×10−4 | 0.0280 |
| BTNL2 | rs78587369 | G/A | T165I | 6 | 32403150 | 23.0 | 0.83 | 1.35×10−4 | 0.0280 |
| BTNL2 | rs3763315 | G/T | 6 | 32408877 | 23.0 | 0.83 | 1.35×10−4 | 0.0280 | |
| BTNL2 | rs2076528 | T/G | 6 | 32396417 | 23.0 | 0.83 | 1.35×10−4 | 0.0280 | |
| PRKG1 | rs9414827 | G/A | 10 | 51137314 | 10.1 | 0.76 | 1.37×10−4 | 0.0282 | |
| rs6537384 | T/G | 4 | 145949613 | 28.8 | 1.19 | 1.43×10−4 | 0.0294 | ||
| rs6067640 | G/A | 20 | 51092837 | 38.5 | 0.85 | 1.48×10−4 | 0.0302 | ||
| rs10514995 | A/G | 5 | 66443611 | 48.7 | 1.16 | 1.51×10−4 | 0.0306 | ||
| BTNL2 | rs34423804 | T/A | V283D | 6 | 32396269 | 23.0 | 0.83 | 1.63×10−4 | 0.0329 |
| PHACTR1 | rs9349379 | G/A | 6 | 12903725 | 34.2 | 0.85 | 1.69×10−4 | 0.0341 | |
| STIM1 | rs116855870 | A/G | 11 | 4055527 | 1.1 | 1.93 | 1.71×10−4 | 0.0343 | |
| ZNF142 | rs3821033 | C/T | A1313T | 2 | 218642579 | 11.2 | 1.26 | 1.78×10−4 | 0.0355 |
| LINC00354 | rs4907518 | G/A | 13 | 111898209 | 45.6 | 0.85 | 1.82×10−4 | 0.0362 | |
| TNS3 | rs11763932 | G/A | 7 | 47567880 | 42.0 | 0.85 | 1.91×10−4 | 0.0378 | |
| BTRC | rs2270439 | C/A | P566H | 10 | 101550817 | 3.5 | 0.63 | 1.94×10−4 | 0.0381 |
| MIA3 | rs2936051 | A/G | E881G | 1 | 222629862 | 40.1 | 0.85 | 1.96×10−4 | 0.0384 |
| rs6825911 | C/T | 4 | 110460482 | 45.9 | 0.86 | 2.01×10−4 | 0.0391 | ||
| VNN1 | rs2294757 | G/A | T26I | 6 | 132713959 | 37.4 | 0.85 | 2.02×10−4 | 0.0393 |
| ZNF860 | rs140232911 | C/T | S161L | 3 | 31989561 | 10.4 | 0.44 | 2.09×10−4 | 0.0406 |
| rs838880 | C/T | 12 | 124777047 | 47.5 | 1.16 | 2.23×10−4 | 0.0430 | ||
| MIA3 | rs2936052 | A/G | K605R | 1 | 222629034 | 34.4 | 0.85 | 2.26×10−4 | 0.0430 |
| DTNBP1 | rs2743868 | G/A | 6 | 15625577 | 31.6 | 1.18 | 2.26×10−4 | 0.0430 | |
| MON2 | rs11174549 | A/G | I1385V | 12 | 62565357 | 5.0 | 1.40 | 2.26×10−4 | 0.0430 |
| rs507666 | G/A | 9 | 136149399 | 27.8 | 1.18 | 2.26×10−4 | 0.0430 | ||
| FAM170B | rs73302786 | G/T | D252E | 10 | 49131709 | 3.5 | 1.47 | 2.36×10−4 | 0.0445 |
| PSORS1C3 | rs3131018 | G/T | 6 | 31175805 | 15.7 | 1.23 | 2.36×10−4 | 0.0445 | |
| PIEZO2 | rs35033671 | C/A | C1148F | 18 | 10759842 | 11.0 | 1.27 | 2.39×10−4 | 0.0448 |
| SLC22A3 | rs1810126 | C/T | 6 | 160451119 | 49.1 | 0.86 | 2.46×10−4 | 0.0460 | |
| PANK1 | rs11185790 | G/A | 10 | 89612776 | 46.9 | 1.16 | 2.57×10−4 | 0.0481 | |
| GFY | rs73053944 | C/G | T203S | 19 | 49427038 | 2.9 | 1.51 | 2.58×10−4 | 0.0481 |
| RNF2 | rs1046592 | A/G | 1 | 185100429 | 33.9 | 0.85 | 2.63×10−4 | 0.0488 |
Allele frequencies were analyzed using Fisher's exact test.
Major allele/minor allele. SNP, single nucleotide polymorphisms; MAF, minor allele frequency; OR, odds ratio; FDR, false discovery rate; ND, not determined.
Multivariable logistic regression analysis of the association between SNPs and CAD
The association between the 170 SNPs identified in the EWAS for CAD and this condition was examined by multivariable logistic regression analysis with adjustment for age, sex and the prevalence of hypertension, diabetes mellitus and dyslipidemia (Table III). The 162 SNPs were significantly (P<0.05 in a dominant or recessive model) associated with CAD.
Table III.
162 SNPs associated with coronary artery disease as determined by multivariable logistic regression analysis.
| Dominant model | Recessive model | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | SNP | P-value | OR | 95% CI | P-value | OR | 95% CI | |
| PLCB2 | rs200787930 | C/T | <0.0001 | 0.02 | 0.01–0.09 | |||
| MARCH1 | rs61734696 | G/T | <0.0001 | 0.02 | 0.01–0.10 | |||
| VPS33B | rs199921354 | C/T | <0.0001 | 0.02 | 0.01–0.09 | |||
| CXCL8 | rs188378669 | G/T | <0.0001 | 0.02 | 0.01–0.09 | |||
| TMOD4 | rs115287176 | G/A | <0.0001 | 0.02 | 0.01–0.10 | |||
| COL6A3 | rs146092501 | C/T | <0.0001 | 0.02 | 0.01–0.10 | |||
| ZNF77 | rs146879198 | G/A | <0.0001 | 0.02 | 0.01–0.10 | |||
| ADGRL3 | rs192210727 | G/T | <0.0001 | 0.07 | 0.03–0.16 | 0.9959 | ||
| OR52E4 | rs11823828 | T/G | <0.0001 | 1.66 | 1.41–1.97 | <0.0001 | 2.44 | 2.01–2.97 |
| ALDH2 | rs671 | G/A | <0.0001 | 1.73 | 1.50–2.01 | <0.0001 | 1.80 | 1.44–2.26 |
| ACAD10 | rs11066015 | G/A | <0.0001 | 1.73 | 1.49–2.01 | <0.0001 | 1.79 | 1.42–2.25 |
| BRAP | rs3782886 | A/G | <0.0001 | 1.71 | 1.48–1.99 | <0.0001 | 1.70 | 1.36–2.12 |
| HECTD4 | rs11066280 | T/A | <0.0001 | 1.73 | 1.49–2.01 | <0.0001 | 1.73 | 1.38–2.17 |
| HECTD4 | rs2074356 | C/T | <0.0001 | 1.61 | 1.39–1.87 | <0.0001 | 1.76 | 1.38–2.26 |
| NAA25 | rs12231744 | C/T | <0.0001 | 0.63 | 0.54–0.73 | <0.0001 | 0.55 | 0.43–0.70 |
| GOSR2 | rs1052586 | T/C | 0.0003 | 0.73 | 0.62–0.87 | <0.0001 | 0.64 | 0.53–0.77 |
| ATXN2 | rs7969300 | T/C | <0.0001 | 0.63 | 0.55–0.74 | <0.0001 | 0.57 | 0.45–0.71 |
| rs12229654 | T/G | <0.0001 | 1.46 | 1.26–1.69 | <0.0001 | 1.72 | 1.31–2.25 | |
| LOC107987429 | rs2844533 | T/C | <0.0001 | 1.36 | 1.17–1.59 | 0.8616 | ||
| MTFR2 | rs143974258 | G/A | 0.0014 | 0.04 | 0.01–0.28 | |||
| PSORS1C1 | rs3130559 | C/T | 0.0127 | 0.82 | 0.70–0.96 | 0.0629 | ||
| rs2596548 | G/T | <0.0001 | 1.76 | 1.41–2.20 | 0.2047 | |||
| EIF3L | rs9466 | T/C | 0.0053 | 1.24 | 1.07–1.44 | 0.0199 | 1.47 | 1.06–2.04 |
| LPGAT1 | rs150552771 | T/C | 0.9970 | <0.0001 | 2.20 | 1.83–2.64 | ||
| rs2523644 | A/G | <0.0001 | 1.59 | 1.31–1.92 | 0.9088 | |||
| rs10757278 | A/G | <0.0001 | 0.71 | 0.60–0.83 | 0.0023 | 0.77 | 0.65–0.91 | |
| CCHCR1 | rs130067 | T/G | 0.0010 | 0.78 | 0.68–0.91 | 0.0183 | 0.73 | 0.57–0.95 |
| TCHP | rs74416240 | G/A | 0.0002 | 1.35 | 1.15–1.58 | 0.1725 | ||
| rs1333049 | G/C | 0.0031 | 1.29 | 1.09–1.53 | <0.0001 | 1.41 | 1.20–1.66 | |
| CDKN2B-AS1 | rs4977574 | A/G | 0.0003 | 1.36 | 1.15–1.60 | <0.0001 | 1.43 | 1.21–1.69 |
| CDKN2B-AS1 | rs2383207 | G/A | <0.0001 | 0.75 | 0.65–0.87 | 0.0171 | 0.75 | 0.59–0.95 |
| SLC16A1 | rs1049434 | T/A | 0.0106 | 0.83 | 0.71–0.96 | <0.0001 | 0.57 | 0.45–0.73 |
| GIT2 | rs925368 | T/C | 0.0001 | 1.37 | 1.16–1.61 | 0.3189 | ||
| rs1333048 | A/C | 0.0036 | 1.29 | 1.09–1.53 | <0.0001 | 1.40 | 1.19–1.64 | |
| rs2523578 | T/C | <0.0001 | 1.55 | 1.27–1.88 | 0.8694 | |||
| rs404890 | G/T | 0.0005 | 1.29 | 1.12–1.50 | 0.0160 | 1.35 | 1.06–1.72 | |
| APOE | rs7412 | C/T | 0.0001 | 0.56 | 0.42–0.76 | 0.2259 | ||
| CCHCR1 | rs130071 | G/A | 0.0149 | 1.36 | 1.06–1.73 | 0.2668 | ||
| rs602633 | C/A | 0.0001 | 0.64 | 0.51–0.80 | 0.1782 | |||
| CELSR2 | rs12740374 | G/T | <0.0001 | 0.63 | 0.51–0.79 | 0.1708 | ||
| MKI67 | rs145121731 | G/A | 0.0014 | 1.94 | 1.29–2.91 | 0.9957 | ||
| CUBN | rs78201384 | C/T | 0.0003 | 0.50 | 0.34–0.73 | 0.9959 | ||
| PSORS1C3 | rs887466 | T/C | 0.0013 | 1.29 | 1.11–1.52 | 0.1666 | ||
| PSORS1C1 | rs3094663 | G/A | <0.0001 | 1.41 | 1.21–1.63 | 0.6404 | ||
| rs10853110 | A/G | 0.0026 | 1.26 | 1.09–1.47 | 0.0102 | 1.29 | 1.06–1.56 | |
| WDR37 | rs10794720 | C/T | 0.0003 | 0.68 | 0.55–0.84 | 0.0734 | ||
| CELSR2 | rs629301 | A/C | 0.0002 | 0.65 | 0.52–0.82 | 0.1708 | ||
| SKIV2L | rs592229 | G/T | 0.0154 | 1.22 | 1.04–1.42 | 0.0138 | 1.26 | 1.05–1.51 |
| rs12182351 | T/C | 0.0008 | 1.28 | 1.11–1.48 | 0.0138 | 1.37 | 1.07–1.75 | |
| POU5F1 | rs3130503 | G/A | <0.0001 | 1.41 | 1.22–1.63 | 0.6308 | ||
| PSORS1C3 | rs1265155 | T/C | 0.0017 | 1.29 | 1.10–1.50 | 0.1666 | ||
| CELSR2 | rs646776 | A/G | 0.0002 | 0.65 | 0.52–0.82 | 0.2660 | ||
| rs2596503 | C/T | 0.0204 | 1.19 | 1.03–1.38 | 0.1828 | |||
| TRPM1 | rs2241493 | T/C | 0.0002 | 0.71 | 0.60–0.85 | 0.0410 | 0.49 | 0.25–0.97 |
| CCDC141 | rs13419085 | T/C | 0.0005 | 0.43 | 0.27–0.69 | |||
| VARS2 | rs9394021 | A/G | 0.0261 | 0.82 | 0.69–0.98 | 0.0117 | 0.81 | 0.69–0.95 |
| SFTA2 | rs2286655 | T/C | 0.0097 | 1.24 | 1.05–1.45 | 0.0277 | 1.22 | 1.02–1.45 |
| rs3873334 | T/C | 0.0117 | 1.23 | 1.05–1.44 | 0.0261 | 1.22 | 1.02–1.45 | |
| TCF19 | rs3130453 | C/T | 0.0077 | 0.82 | 0.71–0.95 | 0.0024 | 0.68 | 0.53–0.87 |
| DDR1 | rs2239518 | T/C | 0.0103 | 1.23 | 1.05–1.45 | 0.0282 | 1.22 | 1.02–1.45 |
| CDSN | rs3130984 | C/T | <0.0001 | 1.39 | 1.18–1.64 | 0.1220 | ||
| rs197932 | T/C | 0.0042 | 0.81 | 0.70–0.93 | 0.0348 | 0.73 | 0.54–0.98 | |
| CDSN | rs3130981 | C/T | <0.0001 | 1.39 | 1.18–1.64 | 0.1226 | ||
| MICB-DT | rs3132469 | C/T | <0.0001 | 1.63 | 1.30–2.05 | 0.3960 | ||
| HLA-DQB1 | rs1049056 | C/A | 0.0050 | 1.28 | 1.08–1.52 | 0.1586 | ||
| DDR1 | rs2239517 | A/G | 0.0115 | 1.23 | 1.05–1.44 | 0.0326 | 1.21 | 1.02–1.44 |
| CCHCR1 | rs1265110 | G/A | 0.0096 | 0.83 | 0.71–0.95 | 0.0948 | ||
| CCDC63 | rs10774610 | T/C | 0.0006 | 1.29 | 1.12–1.49 | 0.0214 | 1.38 | 1.05–1.82 |
| GTF2H4 | rs2284176 | C/T | 0.0153 | 1.22 | 1.04–1.43 | 0.0306 | 1.21 | 1.02–1.45 |
| GTF2H4 | rs3909130 | G/A | 0.0138 | 1.22 | 1.04–1.43 | 0.0326 | 1.21 | 1.02–1.44 |
| GTF2H4 | rs916920 | G/A | 0.0139 | 1.22 | 1.04–1.43 | 0.0326 | 1.21 | 1.02–1.44 |
| rs1264569 | A/G | 0.0004 | 1.54 | 1.21–1.97 | 0.5339 | |||
| rs9468845 | A/G | 0.0141 | 1.22 | 1.04–1.43 | 0.0332 | 1.21 | 1.02–1.44 | |
| DDR1 | rs8408 | C/T | 0.0141 | 1.22 | 1.04–1.43 | 0.0326 | 1.21 | 1.02–1.44 |
| DDR1 | rs7756521 | C/T | 0.0155 | 1.22 | 1.04–1.43 | 0.0303 | 1.21 | 1.02–1.45 |
| CDKN2B-AS1 | rs1011970 | G/T | 0.0047 | 1.36 | 1.10–1.69 | 0.2199 | ||
| ADAT1 | rs145161932 | T/C | 0.0104 | 0.48 | 0.27–0.84 | 0.9960 | ||
| POU5F1 | rs885950 | T/G | 0.0049 | 0.81 | 0.70–0.94 | 0.0129 | 0.73 | 0.57–0.94 |
| DDR1 | rs4618569 | A/G | 0.0141 | 1.22 | 1.04–1.43 | 0.0333 | 1.21 | 1.02–1.44 |
| KRT13 | rs146918776 | A/G | <0.0001 | 2.21 | 1.50–3.26 | |||
| rs2523638 | G/A | 0.0197 | 1.20 | 1.03–1.41 | 0.1032 | |||
| PSRC1 | rs599839 | A/G | 0.0004 | 0.67 | 0.54–0.83 | 0.1023 | ||
| rs9275141 | G/T | 0.0134 | 1.20 | 1.04–1.39 | 0.0065 | 1.43 | 1.11–1.85 | |
| CCDC63 | rs10849915 | T/C | 0.0005 | 1.30 | 1.12–1.50 | 0.0458 | 1.33 | 1.01–1.76 |
| HLA-DRA | rs3177928 | G/A | <0.0001 | 1.58 | 1.27–1.96 | 0.7635 | ||
| OAS3 | rs2072134 | C/T | 0.0004 | 1.31 | 1.13–1.53 | 0.0268 | 1.51 | 1.05–2.16 |
| USP45 | rs41288947 | C/G | 0.0002 | 1.35 | 1.15–1.58 | 0.1360 | ||
| CCHCR1 | rs1265109 | A/C | 0.0007 | 1.35 | 1.14–1.61 | 0.0334 | 1.20 | 1.01–1.41 |
| LOC101929163 | rs6930777 | C/T | <0.0001 | 1.60 | 1.28–2.00 | 0.9815 | ||
| rs7333181 | G/A | 0.0219 | 0.64 | 0.44–0.94 | 0.9969 | |||
| DDR1 | rs1264323 | T/C | 0.0240 | 1.19 | 1.02–1.39 | 0.0423 | 1.22 | 1.01–1.48 |
| LINC00243 | rs3094111 | G/A | 0.0061 | 1.24 | 1.06–1.45 | 0.5293 | ||
| rs10484561 | T/G | <0.0001 | 1.59 | 1.28–1.98 | 0.7637 | |||
| PSORS1C1 | rs3130558 | G/C | <0.0001 | 1.39 | 1.18–1.64 | 0.2712 | ||
| HLA-DQB1 | rs1049060 | T/A | 0.0676 | 0.0077 | 1.38 | 1.09–1.75 | ||
| rs2844650 | G/A | <0.0001 | 1.66 | 1.30–2.11 | 0.4729 | |||
| DDR1 | rs3132572 | T/C | <0.0001 | 1.66 | 1.30–2.11 | 0.4729 | ||
| CCHCR1 | rs1265115 | T/G | 0.0004 | 1.36 | 1.15–1.62 | 0.0443 | 1.19 | 1.00–1.40 |
| CCHCR1 | rs3094225 | T/C | <0.0001 | 1.46 | 1.23–1.74 | 0.5822 | ||
| LOC107987453 | rs3129987 | C/T | 0.0037 | 1.26 | 1.08–1.47 | 0.5667 | ||
| DPCR1 | rs2517451 | A/G | <0.0001 | 1.65 | 1.30–2.11 | 0.4729 | ||
| CCHCR1 | rs1265115 | T/G | 0.0004 | 1.36 | 1.15–1.62 | 0.0443 | 1.19 | 1.00–1.40 |
| CCHCR1 | rs3094225 | T/C | <0.0001 | 1.46 | 1.23–1.74 | 0.5822 | ||
| LOC107987453 | rs3129987 | C/T | 0.0037 | 1.26 | 1.08–1.47 | 0.5667 | ||
| DPCR1 | rs2517451 | A/G | <0.0001 | 1.65 | 1.30–2.11 | 0.4729 | ||
| KIAA1551 | rs10771894 | A/G | 0.1122 | 0.0085 | 1.35 | 1.08–1.69 | ||
| rs13427905 | C/T | 0.0024 | 0.78 | 0.67–0.92 | 0.1397 | |||
| ABCA1 | rs1883025 | G/A | 0.0051 | 0.81 | 0.70–0.94 | 0.0080 | 0.68 | 0.51–0.90 |
| SFTA2 | rs2253705 | G/A | 0.0015 | 1.28 | 1.10–1.48 | 0.6682 | ||
| PLUT | rs954750 | G/A | 0.0409 | 1.19 | 1.01–1.41 | 0.0008 | 1.33 | 1.12–1.57 |
| TCF19 | rs1419881 | T/C | 0.0009 | 1.34 | 1.13–1.60 | 0.0487 | 1.18 | 1.00–1.39 |
| rs13209234 | G/A | <0.0001 | 1.55 | 1.25–1.93 | 0.7617 | |||
| PSORS1C1 | rs1265100 | T/C | 0.0055 | 0.81 | 0.70–0.94 | 0.4135 | ||
| YEATS2 | rs76174573 | G/T | 0.0031 | 0.61 | 0.44–0.85 | 0.1756 | ||
| ABO | rs1053878 | C/T | 0.0033 | 1.25 | 1.08–1.44 | 0.1723 | ||
| rs4014195 | C/G | 0.0083 | 1.23 | 1.05–1.43 | 0.2586 | |||
| SFTA2 | rs2253588 | C/G | 0.0021 | 1.26 | 1.09–1.45 | 0.7016 | ||
| rs10757283 | T/C | 0.0079 | 0.82 | 0.71–0.95 | 0.0181 | 0.74 | 0.58–0.95 | |
| BTNL2 | rs28362680 | G/A | 0.1143 | 0.0082 | 0.76 | 0.62–0.93 | ||
| BTNL2 | rs10947262 | C/T | 0.1143 | 0.0082 | 0.76 | 0.62–0.93 | ||
| KRT27 | rs17558532 | C/T | 0.0004 | 0.57 | 0.41–0.78 | 0.5002 | ||
| GTF2H4 | rs3130780 | G/T | 0.0022 | 1.26 | 1.09–1.47 | 0.6706 | ||
| rs2532934 | T/C | 0.0023 | 1.25 | 1.08–1.45 | 0.5320 | |||
| VARS2 | rs753725 | G/A | 0.0021 | 1.26 | 1.09–1.45 | 0.5274 | ||
| PLUT | rs11619319 | A/G | 0.0482 | 1.18 | 1.00–1.40 | 0.0007 | 1.33 | 1.13–1.58 |
| rs3095273 | C/T | 0.0045 | 1.38 | 1.11–1.73 | 0.1778 | |||
| TNS1 | rs918949 | C/T | 0.0028 | 0.79 | 0.68–0.92 | 0.1301 | ||
| LINC00243 | rs3130785 | C/T | 0.0060 | 1.24 | 1.06–1.45 | 0.5417 | ||
| VARS2 | rs2249464 | C/T | 0.0028 | 1.25 | 1.08–1.44 | 0.5320 | ||
| rs3095345 | A/G | 0.0021 | 1.27 | 1.09–1.47 | 0.7469 | |||
| ITGB8 | rs80015015 | G/A | <0.0001 | 1.56 | 1.28–1.91 | 0.2511 | ||
| VARS2 | rs885905 | C/T | 0.0028 | 1.25 | 1.08–1.44 | 0.7687 | ||
| LIPE | rs34052647 | G/A | 0.0001 | 1.53 | 1.23–1.90 | 0.0074 | 3.70 | 1.42–9.65 |
| PHACTR1 | rs9369640 | A/C | 0.0025 | 0.73 | 0.60–0.90 | 0.1527 | ||
| BTNL2 | rs41417449 | T/C | 0.1641 | 0.0017 | 0.55 | 0.38–0.80 | ||
| BTNL2 | rs41441651 | C/T | 0.1586 | 0.0017 | 0.55 | 0.38–0.80 | ||
| BTNL2 | rs28362675 | C/A | 0.1584 | 0.0017 | 0.55 | 0.38–0.80 | ||
| BTNL2 | rs78587369 | G/A | 0.1590 | 0.0018 | 0.55 | 0.38–0.80 | ||
| BTNL2 | rs3763315 | G/T | 0.1637 | 0.0017 | 0.55 | 0.38–0.80 | ||
| BTNL2 | rs2076528 | T/G | 0.1524 | 0.0017 | 0.55 | 0.38–0.80 | ||
| PRKG1 | rs9414827 | G/A | 0.0003 | 0.70 | 0.58–0.85 | 0.0395 | 0.46 | 0.22–0.96 |
| rs6537384 | T/G | 0.0191 | 1.19 | 1.03–1.38 | 0.1787 | |||
| rs6067640 | G/A | 0.0323 | 0.85 | 0.73–0.99 | 0.0054 | 0.74 | 0.60–0.91 | |
| rs10514995 | A/G | 0.0467 | 1.19 | 1.00–1.42 | 0.0825 | |||
| BTNL2 | rs34423804 | T/A | 0.1675 | 0.0018 | 0.55 | 0.38–0.80 | ||
| PHACTR1 | rs9349379 | G/A | 0.0029 | 0.80 | 0.69–0.93 | 0.0820 | ||
| STIM1 | rs116855870 | A/G | 0.0133 | 1.76 | 1.13–2.76 | 0.9967 | ||
| ZNF142 | rs3821033 | C/T | 0.0015 | 1.32 | 1.11–1.57 | 0.3469 | ||
| LINC00354 | rs4907518 | G/A | 0.0138 | 0.82 | 0.70–0.96 | 0.0050 | 0.77 | 0.64–0.92 |
| TNS3 | rs11763932 | G/A | 0.0054 | 0.81 | 0.69–0.94 | 0.0018 | 0.73 | 0.60–0.89 |
| BTRC | rs2270439 | C/A | 0.0063 | 0.64 | 0.47–0.88 | 0.6622 | ||
| MIA3 | rs2936051 | A/G | 0.1239 | 0.0002 | 0.67 | 0.55–0.83 | ||
| rs6825911 | C/T | 0.0020 | 0.78 | 0.67–0.91 | 0.0446 | 0.83 | 0.70–1.00 | |
| VNN1 | rs2294757 | G/A | 0.2269 | 0.0334 | 0.79 | 0.63–0.98 | ||
| ZNF860 | rs140232911 | C/T | 0.0013 | 0.14 | 0.04–0.46 | |||
| rs838880 | C/T | 0.0338 | 1.20 | 1.01–1.41 | 0.0308 | 1.20 | 1.02–1.43 | |
| MIA3 | rs2936052 | A/G | 0.0927 | 0.0044 | 0.71 | 0.56–0.90 | ||
| DTNBP1 | rs2743868 | G/A | 0.0208 | 1.19 | 1.03–1.37 | 0.0941 | ||
| MON2 | rs11174549 | A/G | 0.0427 | 1.28 | 1.01–1.61 | 0.1917 | ||
| rs507666 | G/A | 0.0098 | 1.21 | 1.05–1.40 | 0.1384 | |||
| FAM170B | rs73302786 | G/T | 0.0003 | 1.62 | 1.25–2.11 | 0.6351 | ||
| PSORS1C3 | rs3131018 | G/T | 0.0002 | 1.35 | 1.15–1.58 | 0.9209 | ||
| PIEZO2 | rs35033671 | C/A | 0.0035 | 1.30 | 1.09–1.54 | 0.1909 | ||
| PANK1 | rs11185790 | G/A | 0.0065 | 1.26 | 1.07–1.48 | 0.0457 | 1.19 | 1.00–1.41 |
| GFY | rs73053944 | C/G | 0.0011 | 1.60 | 1.21–2.13 | 0.0698 | ||
| RNF2 | rs1046592 | A/G | 0.0301 | 0.85 | 0.74–0.98 | 0.7186 | ||
Multivariable logistic regression analysis was performed with adjustment for age, sex, and the prevalence of hypertension, diabetes mellitus and dyslipidemia. P<0.05 was considered to indicate a statistically significant difference. SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
Stepwise forward selection procedure of the effects of SNPs on CAD
A stepwise forward selection procedure was performed to examine effects of genotypes for the 162 SNPs associated with CAD by multivariable logistic regression analysis on this condition (Table IV). The 54 SNPs were significant (P<0.05) and independent [coefficient of determination (R2), 0.0008 to 0.0297] determinants of CAD. These SNPs together accounted for 15.5% of the cause of CAD.
Table IV.
54 SNPs associated with coronary artery disease as determined by a stepwise forward selection procedure.
| Gene | SNP | P-value | R2 (individual) | R2 (accumulated) |
|---|---|---|---|---|
| PLCB2 | rs200787930 | <0.0001 | 0.0297 | 0.0297 |
| ALDH2 | rs671 | <0.0001 | 0.0061 | 0.0358 |
| GOSR2 | rs1052586 | <0.0001 | 0.0053 | 0.0411 |
| PSORS1C1 | rs3094663 | <0.0001 | 0.0052 | 0.0463 |
| CCHCR1 | rs130071 | <0.0001 | 0.0059 | 0.0522 |
| rs13427905 | <0.0001 | 0.0047 | 0.0569 | |
| OR52E4 | rs11823828 | <0.0001 | 0.0043 | 0.0612 |
| EIF3L | rs9466 | <0.0001 | 0.0042 | 0.0654 |
| KIAA1551 | rs10771894 | <0.0001 | 0.0039 | 0.0693 |
| CCDC141 | rs13419085 | <0.0001 | 0.0035 | 0.0728 |
| MIA3 | rs2936051 | 0.0001 | 0.0033 | 0.0761 |
| rs602633 | 0.0001 | 0.0033 | 0.0794 | |
| KRT27 | rs17558532 | 0.0001 | 0.0032 | 0.0826 |
| TRPM1 | rs2241493 | 0.0002 | 0.0030 | 0.0856 |
| rs7333181 | 0.0002 | 0.0030 | 0.0886 | |
| ADAT1 | rs145161932 | 0.0002 | 0.0029 | 0.0915 |
| APOE | rs7412 | 0.0003 | 0.0028 | 0.0943 |
| YEATS2 | rs76174573 | 0.0004 | 0.0026 | 0.0969 |
| SLC16A1 | rs1049434 | 0.0005 | 0.0025 | 0.0994 |
| RNF2 | rs1046592 | 0.0007 | 0.0025 | 0.1019 |
| rs6825911 | 0.0006 | 0.0024 | 0.1043 | |
| ITGB8 | rs80015015 | 0.0007 | 0.0024 | 0.1067 |
| USP45 | rs41288947 | 0.0007 | 0.0024 | 0.1091 |
| PHACTR1 | rs9369640 | 0.0007 | 0.0024 | 0.1115 |
| rs1333048 | 0.0008 | 0.0024 | 0.1139 | |
| rs838880 | 0.0011 | 0.0022 | 0.1161 | |
| STIM1 | rs116855870 | 0.0017 | 0.0021 | 0.1182 |
| rs2523644 | 0.0016 | 0.0021 | 0.1203 | |
| MKI67 | rs145121731 | 0.0020 | 0.0020 | 0.1223 |
| FAM170B-AS1 | rs73302786 | 0.0019 | 0.0020 | 0.1243 |
| rs6067640 | 0.0022 | 0.0020 | 0.1263 | |
| GFY | rs73053944 | 0.0024 | 0.0019 | 0.1282 |
| WDR37 | rs10794720 | 0.0033 | 0.0018 | 0.1300 |
| SKIV2L | rs592229 | 0.0037 | 0.0018 | 0.1318 |
| rs6537384 | 0.0041 | 0.0017 | 0.1335 | |
| rs10757283 | 0.0058 | 0.0016 | 0.1351 | |
| CDKN2B-AS1 | rs1011970 | 0.0110 | 0.0014 | 0.1365 |
| PRKG1 | rs9414827 | 0.0087 | 0.0014 | 0.1379 |
| rs197932 | 0.0127 | 0.0013 | 0.1392 | |
| LINC00354 | rs4907518 | 0.0125 | 0.0013 | 0.1405 |
| LIPE | rs34052647 | 0.0151 | 0.0013 | 0.1418 |
| BTRC | rs2270439 | 0.0143 | 0.0013 | 0.1431 |
| TNS3 | rs11763932 | 0.0163 | 0.0013 | 0.1444 |
| TNS1 | rs918949 | 0.0158 | 0.0012 | 0.1456 |
| rs12229654 | 0.0184 | 0.0011 | 0.1467 | |
| rs4014195 | 0.0213 | 0.0011 | 0.1478 | |
| PANK1 | rs11185790 | 0.0227 | 0.0011 | 0.1489 |
| rs507666 | 0.0279 | 0.0010 | 0.1499 | |
| MON2 | rs11174549 | 0.0359 | 0.0009 | 0.1508 |
| HECTD4 | rs2074356 | 0.0396 | 0.0009 | 0.1517 |
| CUBN | rs78201384 | 0.0484 | 0.0009 | 0.1526 |
| PLUT | rs954750 | 0.0497 | 0.0008 | 0.1534 |
| ABCA1 | rs1883025 | 0.0479 | 0.0008 | 0.1542 |
| rs10514995 | 0.0493 | 0.0008 | 0.1550 |
SNP, single nucleotide polymorphisms; R2, coefficient of determination.
Association between SNPs associated with CAD and intermediate phenotypes
The association between the 54 SNPs associated with CAD and intermediate phenotypes of this condition, including hypertension, DM, hypertriglyceridemia, hypo-HDL-cholesterolemia, hyper-low density lipoprotein (LDL)-cholesterolemia, CKD, obesity, and hyperuricemia, was examined using Pearson's χ2 test (Table V).
Table V.
Association between SNPs associated with coronary artery disease and intermediate phenotypes.
| Gene | SNP | Hypertension | DM | Hyper-TG | Hypo-HDL | Hyper-LDL | CKD | Obesity | Hyperuricemia |
|---|---|---|---|---|---|---|---|---|---|
| PLCB2 | rs200787930 | <0.0001a | 0.0004a | 0.3432 | <0.0001a | <0.0001a | <0.0001a | 0.0405a | 0.9639 |
| ALDH2 | rs671 | 0.0039a | 0.0074a | 0.0298a | <0.0001a | <0.0001a | 0.0273a | 0.0350a | <0.0001a |
| GOSR2 | rs1052586 | 0.3498 | 0.0167a | 0.4457 | 0.2898 | 0.2638 | 0.6185 | 0.3670 | 0.4679 |
| PSORS1C1 | rs3094663 | 0.0069a | 0.0670 | 0.0947 | 0.0020a | 0.3869 | 0.1080 | 0.7345 | 0.5091 |
| CCHCR1 | rs130071 | 0.0865 | 0.0008a | 0.2247 | 0.0143a | 0.0141a | 0.5894 | 0.8651 | 0.0423a |
| rs13427905 | 0.0149a | 0.0149a | 0.0524 | 0.0545 | 0.6318 | 0.9487 | 0.1197 | 0.0920 | |
| OR52E4 | rs11823828 | 0.0024a | <0.0001a | 0.0265a | 0.1186 | 0.4445 | 0.0027a | 0.1141 | 0.2815 |
| EIF3L | rs9466 | 0.0008a | 0.0204a | 0.0054a | 0.2905 | 0.0114a | 0.2368 | 0.2435 | 0.0312a |
| KIAA1551 | rs10771894 | 0.2439 | 0.0091a | 0.9562 | 0.0343a | 0.6934 | 0.3869 | 0.0974 | 0.5419 |
| CCDC141 | rs13419085 | 0.3387 | 0.1255 | 0.6537 | 0.1647 | 0.7447 | 0.2483 | 0.8101 | 0.7938 |
| MIA3 | rs2936051 | 0.4092 | 0.5246 | 0.9990 | 0.0475a | 0.1222 | 0.6614 | 0.8787 | 0.1949 |
| rs602633 | 0.4468 | 0.2375 | 0.7350 | 0.0005a | 0.0021a | 0.0842 | 0.6617 | 0.9338 | |
| KRT27 | rs17558532 | 0.1706 | 0.2358 | 0.3643 | 0.3607 | 0.7663 | 0.0133a | 0.2306 | 0.5325 |
| TRPM1 | rs2241493 | 0.3861 | 0.2332 | 0.7465 | 0.7106 | 0.0815 | 0.1387 | 0.5698 | 0.9502 |
| rs7333181 | 0.2308 | 0.0487a | 0.0379a | 0.1185 | 0.2010 | 0.0795 | 0.2182 | 0.6544 | |
| ADAT1 | rs145161932 | 0.4468 | 0.0160a | 0.3611 | 0.3357 | 0.5412 | 0.7534 | 0.5836 | 0.1202 |
| APOE | rs7412 | 0.3680 | 0.9184 | 0.6322 | 0.1157 | <0.0001a | 0.6367 | 0.5319 | 0.2528 |
| YEATS2 | rs76174573 | 0.1305 | 0.0687 | 0.0380a | 0.0606 | 0.8313 | 0.6458 | 0.6338 | 0.1706 |
| SLC16A1 | rs1049434 | 0.8319 | 0.0016a | 0.1897 | 0.0686 | 0.9212 | 0.0646 | 0.8850 | 0.2672 |
| RNF2 | rs1046592 | 0.0007a | 0.0140a | 0.5319 | 0.0544 | 0.4098 | 0.7276 | 0.4643 | 0.1040 |
| rs6825911 | 0.0317a | 0.4070 | 0.5755 | 0.1068 | 0.1423 | 0.4050 | 0.2325 | 0.6717 | |
| ITGB8 | rs80015015 | 0.4001 | 0.5178 | 0.4838 | 0.0075a | 0.5169 | 0.3341 | 0.1339 | 0.2408 |
| USP45 | rs41288947 | 0.4383 | 0.1373 | 0.2641 | 0.0162a | 0.6636 | 0.1341 | 0.0063a | 0.0682 |
| PHACTR1 | rs9369640 | 0.1667 | 0.4673 | 0.8831 | 0.5247 | 0.8191 | 0.7417 | 0.6674 | 0.4133 |
| rs1333048 | 0.2947 | 0.0251a | 0.5799 | 0.0156a | 0.3204 | 0.6650 | 0.5825 | 0.2450 | |
| rs838880 | 0.9565 | 0.0108a | 0.3044 | 0.0045a | 0.7818 | 0.7699 | 0.8126 | 0.2552 | |
| STIM1 | rs116855870 | 0.1425 | 0.2455 | 0.6418 | 0.8631 | 0.7116 | 0.5285 | 0.7357 | 0.3365 |
| rs2523644 | 0.4105 | 0.2101 | 0.6604 | 0.0773 | 0.0627 | 0.0449a | 0.2968 | 0.2580 | |
| MKI67 | rs145121731 | 0.0903 | 0.2528 | 0.2203 | 0.0138a | 0.4030 | 0.0048a | 0.2459 | 0.5059 |
| FAM170B-AS1 | rs73302786 | 0.2662 | 0.6366 | 0.2511 | 0.2687 | 0.8989 | 0.8203 | 0.2323 | 0.5404 |
| rs6067640 | 0.0380a | 0.2144 | 0.7990 | 0.0077a | 0.6995 | 0.1809 | 0.6901 | 0.9347 | |
| GFY | rs73053944 | 0.0145a | 0.5731 | 0.5880 | 0.2471 | 0.8788 | 0.6316 | 0.4534 | 0.9116 |
| WDR37 | rs10794720 | 0.6272 | 0.0103a | 0.9125 | 0.6954 | 0.6528 | 0.1352 | 0.7804 | 0.0458a |
| SKIV2L | rs592229 | 0.0014a | 0.0754 | 0.0752 | 0.0157a | 0.7557 | 0.1230 | 0.3617 | 0.1727 |
| rs6537384 | 0.3890 | 0.1107 | 0.2980 | 0.3818 | 0.0344a | 0.0808 | 0.0620 | 0.7745 | |
| rs10757283 | 0.8792 | 0.0082a | 0.9667 | 0.0420a | 0.4745 | 0.0342a | 0.8876 | 0.4636 | |
| CDKN2B-AS1 | rs1011970 | 0.0443a | 0.0628 | 0.6524 | 0.0834 | 0.5456 | 0.4293 | 0.6420 | 0.3237 |
| PRKG1 | rs9414827 | 0.6287 | 0.8365 | 0.6694 | 0.0424a | 0.1488 | 0.6060 | 0.0291a | 0.0549 |
| rs197932 | 0.1918 | 0.0272a | 0.5146 | 0.5673 | 0.2395 | 0.3102 | 0.7843 | 0.4625 | |
| LINC00354 | rs4907518 | 0.2933 | 0.8434 | 0.3915 | 0.1285 | 0.0028a | 0.6454 | 0.6846 | 0.7513 |
| LIPE | rs34052647 | 0.1525 | 0.7148 | 0.0040a | 0.4199 | 0.0801 | 0.0879 | 0.0940 | 0.0081a |
| BTRC | rs2270439 | 0.3191 | 0.9636 | 0.1684 | 0.1393 | 0.9515 | 0.2852 | 0.4689 | 0.5780 |
| TNS3 | rs11763932 | 0.4812 | 0.6129 | 0.9920 | 0.8857 | 0.1424 | 0.9598 | 0.9307 | 0.9591 |
| TNS1 | rs918949 | 0.1509 | 0.0510 | 0.3218 | 0.5993 | 0.7044 | 0.5484 | 0.6955 | 0.9461 |
| rs12229654 | 0.0203a | 0.3290 | 0.1080 | <0.0001a | 0.0171a | 0.1167 | 0.2297 | <0.0001a | |
| rs4014195 | 0.1622 | 0.0445a | 0.0270a | 0.1811 | 0.7090 | 0.3732 | 0.5607 | 0.2233 | |
| PANK1 | rs11185790 | 0.3638 | 0.4169 | 0.1750 | 0.2583 | 0.3889 | 0.0149a | 0.6355 | 0.3282 |
| rs507666 | 0.9872 | 0.0084a | 0.7080 | 0.0370a | 0.0129a | 0.4210 | 0.9126 | 0.6992 | |
| MON2 | rs11174549 | 0.0283a | 0.0133a | 0.8790 | 0.3587 | 0.0325a | 0.2617 | 0.6406 | 0.9153 |
| HECTD4 | rs2074356 | 0.0285a | 0.1092 | 0.0109a | <0.0001a | 0.0002a | 0.0174 | 0.1786 | <0.0001a |
| CUBN | rs78201384 | 0.1429 | 0.9473 | 0.7525 | 0.0027a | 0.0269a | 0.0327a | 0.9812 | 0.3888 |
| PLUT | rs954750 | 0.9214 | 0.0212a | 0.6905 | 0.8004 | 0.8585 | 0.0264a | 0.2382 | 0.8865 |
| ABCA1 | rs1883025 | 0.8085 | 0.2006 | 0.0134a | 0.3092 | 0.0667 | 0.8891 | 0.4019 | 0.3377 |
| rs10514995 | 0.0014a | 0.0353a | 0.2529 | 0.3708 | 0.7542 | 0.0855 | 0.5232 | 0.0365a |
Data are P-values. The association between genotypes of each SNP and intermediate phenotypes was examined using Pearson's χ2 test. SNP, single nucleotide polymorphism; DM, diabetes mellitus; hyper-TG, hypertriglyceridemia; hypo-HDL, hypo-HDL-cholesterolemia; hyper-LDL, hyper-LDL-cholesterolemia; CKD, chronic kidney disease.
P<0.05 was considered to indicate a statistically significant difference.
The SNP rs671 of ALDH2 was significantly (P<0.05) associated with all the intermediate phenotypes; rs200787930 of PLCB2 and rs2074356 of HECTD4 to six of the eight phenotypes; rs9466 of EIF3L to five of the eight phenotypes; rs130071 of CCHCR1, rs11823828 of OR52E4 and rs12229654 to four of the eight phenotypes; rs11174549 of MON2, rs10514995, rs507666, rs10757283 and rs78201384 of CUBN to three of the eight phenotypes; rs1046592 of RNF2, rs13427905, rs3094663 of PSORS1C1, rs6067640, rs592229 of SKIV2L, rs4014195, rs7333181, rs838880, rs1333048, rs10771894 of KIAA1551, rs954750 of PLUT, rs10794720 of WDR37, rs34052647 of LIPE, rs602633, rs145121731 of MKI67, rs41288947 of USP45, and rs9414827 of PRKG1 to two of the eight phenotypes; and rs73053944 of GFY, rs6825911, rs1011970 of CDKN2B-AS1, rs1049434 of SLC16A1, rs145161932 of ADAT1, rs1052586 of GOSR2, rs197932, rs1883025 of ABCA1, rs76174573 of YEATS2, rs80015015 of ITGB8, rs2936051 of MIA3, rs7412 of APOE, rs4907518 of LINC00354, rs6537384, rs17558532 of KRT27, rs11185790 of PANK1, and rs2523644 to one of the eight phenotypes.
Linkage disequilibrium analyses
Linkage disequilibrium (LD) was examined among SNPs associated with CAD. There was significant LD among rs12229654 at 12q24.1, rs671 of ALDH2, and rs2074356 of HECTD4 [square of the correlation coefficient (r2), 0.564 to 0.882)].
Association between genes, chromosomal loci and SNPs identified in the present study and phenotypes previously reported by GWASs
The association between genes, chromosomal loci and SNPs identified in the present study and cardiovascular disease-related phenotypes previously reported by GWASs available in the GRASP Search database (Table VI). Chromosomal region 1p13.3, MIA3, PHACTR1, SKIV2L, CDKN2B-AS1, 9p21, ALDH2 and HECTD4 were previously revealed to be associated with CAD or MI. SLC16A1, PSORS1C1, CCHCR1, 6p21.3, ABCA1, 9q34.2, CUBN, PANK1, 12q24.1, 12q24.31, PLCB2 and APOE were previously associated with circulating concentrations of LDL-cholesterol, HDL-cholesterol, triglycerides or insulin, or type 1 diabetes mellitus. Chromosome 4q24, 17q21.3 and GOSR2 were previously associated with systolic or diastolic blood pressure or pulse pressure. CCDC141, TNS1, WDR37 and 11q13.1 were previously associated with cardiac, pulmonary or renal function. The remaining 21 genes (RNF2, YEATS2, USP45, ITGB8, TNS3, FAM170B-AS1, PRKG1, BTRC, MKI67, STIM1, OR52E4, KIAA1551, MON2, PLUT, LINC00354, TRPM1, ADAT1, KRT27, LIPE, GFY and EIF3L) and five chromosomal regions (2p13, 4q31.2, 5q12, 13q34 and 20q13.2) identified in the present study have not been revealed to be associated with CAD or cardiovascular disease-related phenotypes in previous GWASs.
Table VI.
Association between genes, chromosomal loci and SNPs associated with coronary artery disease in the present study and previously examined cardiovascular disease-related phenotypes.
| Gene/chr. locus | SNP | Chr. | Position | Previously examined phenotypes |
|---|---|---|---|---|
| 1p13.3 | rs602633 | 1 | 109278889 | CAD (23202125, 20032323), LDL-cholesterol (20686565, 23063622, 19060906, 21943158, 18193043, 18262040, 19913121, 21977987, 20339536), HDL-cholesterol (23063622, 20686565), total cholesterol (20686565, 23063622) |
| SLC16A1 | rs1049434 | 1 | 112913924 | HDL-cholesterol (23063622) |
| RNF2 | rs1046592 | 1 | 185100429 | None |
| MIA3 | rs2936051 | 1 | 222629862 | CAD (19198612, 21347282, 23364394, 21378990, 17554300, 22319020, 21966275), MI (19198609) |
| 2p13 | rs13427905 | 2 | 71846585 | None |
| CCDC141 | rs13419085 | 2 | 178837710 | Heart rate (23583979, 20639392), left ventricular mass (19584346) |
| TNS1 | rs918949 | 2 | 217809974 | Lung function, forced expiratory volume in |
| 1 second (20010834, 21946350, 23284291) | ||||
| YEATS2 | rs76174573 | 3 | 183804099 | None |
| 4q24 | rs6825911 | 4 | 110460482 | Systolic BP (21572416), diastolic BP (21572416) |
| 4q31.2 | rs6537384 | 4 | 145949613 | None |
| 5q12 | rs10514995 | 5 | 66443611 | None |
| PHACTR1 | rs9369640 | 6 | 12901209 | CAD (21378988, 23202125, 22745674, 21347282, 23364394, 21378990, 22751097, 22745674), MI (19198609, 21378990), ischemic stroke (22306652) |
| PSORS1C1 | rs3094663 | 6 | 31139310 | Type 1 diabetes (17554300, 17632545), triglycerides (20686565), total cholesterol (20686565) |
| CCHCR1 | rs130071 | 6 | 31148433 | Triglycerides (20686565) |
| 6p21.3 | rs2523644 | 6 | 31374707 | Type 1 diabetes (17554300, 17632545), LDL-cholesterol (23063622, 20686565), triglycerides (23063622, 20686565), total cholesterol (23063622, 20686565) |
| SKIV2L | rs592229 | 6 | 31962664 | CAD (21971053), type 1 diabetes (17554300, 17632545), LDL-cholesterol (20686565), triglycerides (20686565), total cholesterol (20686565) |
| USP45 | rs41288947 | 6 | 99446210 | None |
| ITGB8 | rs80015015 | 7 | 20401881 | None |
| TNS3 | rs11763932 | 7 | 47567880 | None |
| CDKN2B-AS1 | rs1011970 | 9 | 22062135 | CAD (21347282), LDL-cholesterol (23063622), abdominal aortic aneurysm (20622881), type 2 diabetes (17463249) |
| 9p21 | rs1333048 | 9 | 22125348 | CAD (23202125, 21606135, 19198612, 17634449, 20032323, 23364394), MI (17478679), intracranial aneurysm (22961961) |
| 9p21 | rs10757283 | 9 | 22134173 | Type 2 diabetes (20581827) |
| ABCA1 | rs1883025 | 9 | 104902020 | HDL-cholesterol (20686565, 23505323, 23063622, 21909109, 19060911, 21347282, 19060906, 18193043, 18193044, 18193046, 22629316, 20864672, 21347282, 23726366), LDL-cholesterol (20686565), total cholesterol (20686565, 23063622, 20339536) |
| 9q34.2 | rs507666 | 9 | 136149399 | Venous thrombosis (22675575), VLDL-cholesterol |
| small lipoprotein fraction concentration (19936222), LDL-cholesterol lipoprotein fraction concentration (19936222) | ||||
| WDR37 | rs10794720 | 10 | 1110225 | Estimated glomerular filtration rate (20383146, 22479191), serum creatinine (20383146) |
| CUBN | rs78201384 | 10 | 17111024 | LDL-cholesterol (23063622), HDL-cholesterol (23063622), total cholesterol (23063622) |
| FAM170B-AS1 | rs73302786 | 10 | 49131709 | None |
| PRKG1 | rs9414827 | 10 | 51137314 | None |
| PANK1 | rs11185790 | 10 | 89612776 | Insulin concentration (19060910) |
| BTRC | rs2270439 | 10 | 101550817 | None |
| MKI67 | rs145121731 | 10 | 128102595 | None |
| STIM1 | rs116855870 | 11 | 4055527 | None |
| OR52E4 | rs11823828 | 11 | 5884973 | None |
| 11q13.1 | rs4014195 | 11 | 65739351 | Serum urate (23263486), serum creatinine (20383146), estimated glomerular filtration rate (20383146) |
| KIAA1551 | rs10771894 | 12 | 31982009 | None |
| MON2 | rs11174549 | 12 | 62565357 | None |
| 12q24.1 | rs12229654 | 12 | 110976657 | HDL-cholesterol (21909109) |
| ALDH2 | rs671 | 12 | 111803962 | CAD (21971053, 21572416, 23202125), MI (21971053), LDL-cholesterol (21572416, 20686565), HDL-cholesterol |
| (21572416, 21372407), total cholesterol (20686565), systolic BP | ||||
| (21572416), diastolic BP (21572416, 21909115), serum creatinine | ||||
| (22797727), estimated glomerular filtration rate (22797727), type 1 diabetes (17554300) | ||||
| HECTD4 | rs2074356 | 12 | 112207597 | CAD (21971053, 21572416, 22751097, 19820697, 23364394, 23202125), MI (19820697), LDL-cholesterol (21572416, 20686565), HDL-cholesterol (21572416, 21909109, 22751097), total cholesterol |
| (20686565), systolic BP (21572416, 21909115), diastolic BP | ||||
| (21572416, 21909115, 19862010, 19430479, 22751097), hypertension (21572416), serum creatinine (22797727), estimated | ||||
| glomerular filtration rate (22797727), type 1 diabetes (18978792) | ||||
| 12q24.31 | rs838880 | 12 | 124777047 | HDL-cholesterol (20686565) |
| PLUT | rs954750 | 13 | 27889801 | None |
| 13q34 | rs7333181 | 13 | 111568950 | None |
| LINC00354 | rs4907518 | 13 | 111898209 | None |
| TRPM1 | rs2241493 | 15 | 31070149 | None |
| PLCB2 | rs200787930 | 15 | 40289298 | Triglycerides (23063622) |
| ADAT1 | rs145161932 | 16 | 75612670 | None |
| KRT27 | rs17558532 | 17 | 40779624 | None |
| 17q21.3 | rs197932 | 17 | 46896981 | Pulse pressure (21909110), systolic BP (21909110, 21909115) |
| GOSR2 | rs1052586 | 17 | 46941097 | Pulse pressure (21909110), systolic BP (21909110, 21909115) |
| LIPE | rs34052647 | 19 | 42407617 | None |
| APOE | rs7412 | 19 | 44908822 | LDL-cholesterol (23100282, 23063622, 20686565, 22629316, 19060911, 23067351, 23696881, 20838585), HDL-cholesterol |
| (21386085), triglycerides (23063622, 20686565, 22629316, 19060911, 21386085), total cholesterol (23063622, 20686565) | ||||
| GFY | rs73053944 | 19 | 49427038 | None |
| 20q13.2 | rs6067640 | 20 | 51092837 | None |
| EIF3L | rs9466 | 22 | 37877742 | None |
Data were obtained from the GRASP Search database (https://grasp.nhlbi.nih.gov/Search.aspx) with a P-value of <1.0×10−6. Numbers in parentheses are PubMed IDs. SNP, single nucleotide polymorphisms; Chr., chromosome; HDL, high density lipoprotein; LDL, low density lipoprotein; CAD, coronary artery disease; MI, myocardial infarction; BP, blood pressure.
Gene Ontology analysis of genes identified in the present study
Biological functions of the 21 genes identified in the present study were estimated using the database of Gene Ontology and GO Annotations (QuickGO; Table VII). Given that FAM170B-AS1 is the gene for non-coding RNA, FAM170B was examined. Various biological functions were predicted in the 18 genes (RNF2, YEATS2, USP45, ITGB8, TNS3, FAM170B, PRKG1, BTRC, MKI67, STIM1, OR52E4, MON2, TRPM1, ADAT1, KRT27, LIPE, GFY and EIF3L), although those of KIAA1551, PLUT and LINC00354 were not. Gene ontology analysis revealed that ITGB8, PRKG1, STIM1 and LIPE may be involved in the development of CAD.
Table VII.
Gene ontology analysis of the 21 genes identified in the present study.
| Gene | Function | Biological process |
|---|---|---|
| RNF2 | Ubiquitin-protein transferase activity, chromatin binding, zinc ion binding, transferase activity, metal ion binding, ubiquitin protein ligase activity, RING-like zinc finger domain binding | Histone H2A-K119 monoubiquitination, negative regulation of transcription by RNA polymerase II, regulation of DNA-templated transcription, germ cell development, negative regulation of DNA binding transcription factor activity, negative regulation of G0 to G1 transition |
| YEATS2 | Modification-dependent protein binding, RNA polymerase II transcription factor activity, sequence-specific DNA binding | Negative regulation of transcription by RNA polymerase II, histone H3 acetylation, negative regulation of DNA-templated transcription |
| USP45 | Thiol-dependent ubiquitin-specific protease activity, cysteine-type peptidase activity, zinc ion binding, thiol-dependent ubiquitinyl hydrolase activity | Protein deubiquitination, ubiquitin-dependent protein catabolic process, DNA repair, global genome nucleotide-excision repair |
| ITGB8 | Extracellular matrix protein binding, signaling receptor binding | Ganglioside metabolic process, cell adhesion, integrin-mediated signaling pathway, regulation of gene expression, positive regulation of angiogenesis, cartilage development, extracellular matrix organization, cell-matrix adhesion |
| TNS3 | Protein binding, focal adhesion | Positive regulation of cell proliferation, cell migration, lung alveolus development |
| FAM170B | Protein binding, outer acrosomal membrane | Positive regulation of acrosome reaction, regulation of fertilization |
| PRKG1 | cGMP-dependent protein kinase activity, calcium channel regulator activity, nucleotide binding, ATP binding, transferase activity, cGMP binding, protein serine/threonine kinase activity, cGMP-dependent protein kinase activity | Negative regulation of vascular smooth muscle cell proliferation and migration, neuron migration, cGMP-mediated signaling, dendrite development, forebrain development, relaxation of vascular smooth muscle, regulation of GTPase activity, negative regulation of platelet aggregation, actin cytoskeleton organization |
| BTRC | Ubiquitin-protein transferase activity, ubiquitin protein ligase activity, β-catenin binding, protein phosphorylated amino acid binding, protein dimerization activity | Protein polyubiquitination, ubiquitin-dependent protein catabolic process, regulation of circadian rhythm, regulation of canonical Wnt signaling pathway, protein dephosphorylation, mammary gland epithelial cell proliferation, regulation of I-κB kinase/NF-κB signaling, positive regulation of DNA-templated transcription, G2/M transition of mitotic cell cycle, negative regulation of DNA binding transcription factor activity, stress-activated MAPK cascade, interleukin-1-mediated signaling pathway |
| MKI67 | RNA binding, DNA binding, ATP binding, protein binding | Regulation of mitotic nuclear division, regulation of chromosome segregation and organization, cell proliferation |
| STIM1 | Calcium channel regulator activity, calcium ion binding, microtubule plus-end binding, metal ion binding, protein binding | Cellular calcium ion homeostasis, activation of store-operated calcium channel activity, regulation of calcium ion transport, positive regulation of angiogenesis, regulation of cardiac conduction |
| OR52E4 | Olfactory receptor activity, G-protein coupled receptor activity | Signal transduction, G-protein coupled receptor signaling pathway, detection of chemical stimulus involved in sensory perception of smell |
| KIAA1551 | Uncharacterized | |
| MON2 | Protein binding, protein transport | Golgi to endosome transport |
| PLUT | Uncharacterized | |
| LINC00354 | Uncharacterized | |
| TRPM1 | Ion channel activity | G-protein coupled glutamate receptor signaling pathway, ion transmembrane transport, protein tetramerization, cellular response to light stimulus |
| ADAT1 | RNA binding, tRNA-specific adenosine deaminase activity, hydrolase activity, metal ion binding | tRNA processing |
| KRT27 | Structural molecule activity, intermediate filament | Hair follicle morphogenesis, keratinization, cornification |
| LIPE | Triglyceride lipase activity, serine hydrolase activity, protein kinase binding, hormone-sensitive lipase activity | Protein phosphorylation, lipid metabolic process, steroid metabolic process, cholesterol metabolic process, triglyceride catabolic process, long-chain fatty acid catabolic process, diacylglycerol catabolic process |
| GFY | Protein localization to non-motile cilium, non-motile cilium assembly | Sensory perception of smell, response to stimulus |
| EIF3L | Translation initiation factor activity, RNA binding, protein binding | Translational initiation, viral translational termination-reinitiation |
Data for predicted functions and biological processes of the genes were obtained from database of Gene Ontology and GO Annotations (QuickGO; http://www.ebi.ac.uk/QuickGO/).
Network analysis of newly identified genes
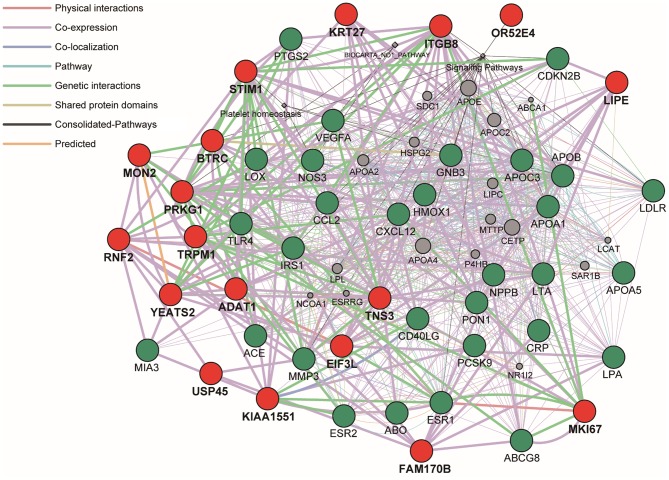
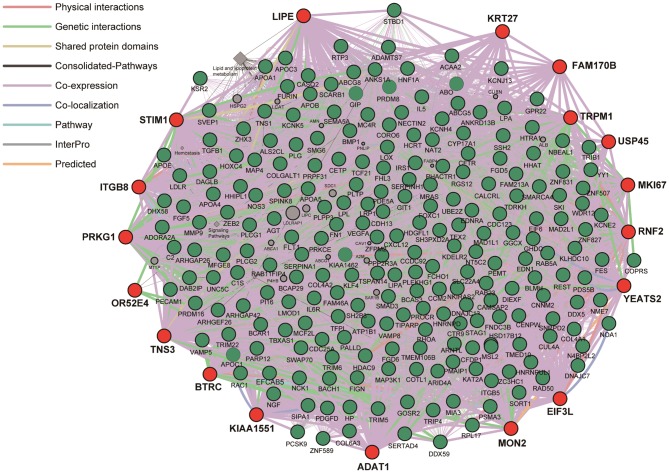
Network analysis of the 21 genes identified in the present study was performed using the GeneMANIA Cytoscape plugin with Cytoscape v3.4.0 software (Figs. 1 and 2). FAM170B was applied to the analysis instead of FAM170B-AS1. PLUT and LINC00354 were not included in the GeneMANIA database. GFY had no interaction with other genes. The network analysis revealed that the 18 genes identified in the present study had potential direct or indirect interactions with the 30 genes previously revealed to be associated with CAD (Fig. 1). Similar analysis revealed that complex networks were observed between the 18 genes identified in the present study and the 228 genes identified in previous GWASs (Fig. 2).
Figure 1.
Network analysis of the 18 genes identified in the present study (closed red circle) was performed to predict functional gene-gene interactions by the use of GeneMANIA Cytoscape plugin (http://apps.cytoscape.org/apps/genemania) using Cytoscape v3.4.0 software (http://www.cytoscape.org/). The 30 genes (closed green circle) were selected from the DisGeNET database (http://www.disgenet.org/web/DisGeNET) according to the rank order of high scores in association with CAD and applied to analysis.
Figure 2.
Network analysis of the 18 genes identified in the present study (closed red circle) was performed to predict functional gene-gene interactions by the use of GeneMANIA Cytoscape plugin (http://apps.cytoscape.org/apps/genemania) using Cytoscape v3.4.0 software (http://www.cytoscape.org/). The 228 genes previously identified by GWASs (closed green circle) were applied to analysis. Interactions between closed red circles or between closed red and green circles are shown with bold lines. Molecules shown in closed grey circles represent putative mediators of interactions between the genes.
Discussion
Despite recent advances in therapy for acute coronary syndrome, including coronary stent implantation (49), CAD remains the leading cause of mortality and is therefore a key public health problem (4). The identification of genetic variants that confer susceptibility to CAD is therefore clinically important for the prevention and management of this condition.
The EWAS was performed for patients with early-onset CAD, with genetic factors serving a greater role in such patients compared with those with late-onset CAD. The present study identified the 54 SNPs as significant and independent determinants of CAD. These SNPs together accounted for 15.5% of the cause of CAD. Among these loci, 21 genes (RNF2, YEATS2, USP45, ITGB8, TNS3, FAM170B-AS1, PRKG1, BTRC, MKI67, STIM1, OR52E4, KIAA1551, MON2, PLUT, LINC00354, TRPM1, ADAT1, KRT27, LIPE, GFY and EIF3L) and 5 chromosomal regions (2p13, 4q31.2, 5q12, 13q34 and 20q13.2) that confer susceptibility to CAD have been newly identified.
Among 26 SNPs identified, 14 SNPs were significantly associated with two to five of the eight intermediate phenotypes. The SNP rs9466 of EIF3L was associated with hypertension, DM, hypertriglyceridemia, hyper-LDL-cholesterolemia and hyperuricemia; rs11823828 of OR52E4 with hypertension, DM, hypertriglyceridemia, and CKD; rs11174549 of MON2 with hypertension, DM, hyper-LDL-cholesterolemia; rs10514995 at 5q12 with hypertension, DM and hyperuricemia; rs1046592 of RNF2 and rs13427905 at 2p13 with hypertension and DM; rs6067640 at 20q13.2 with hypertension and hypo-HDL-cholesterolemia; rs7333181 at 13q34 with DM and hypertriglyceridemia; rs10771894 of KIAA1551 with DM and hypo-HDL-cholesterolemia; rs954750 of PLUT with DM and CKD; rs34052647 of LIPE with hypertriglyceridemia and hyperuricemia; rs145121731 of MKI67 with hypo-HDL-cholesterolemia and CKD; rs41288947 of USP45 and rs9414827 of PRKG1 with hypo-HDL-cholesterolemia and obesity. The seven SNPs were significantly related to one of the eight intermediate phenotypes. The rs73053944 of GFY was associated with hypertension; rs145161932 of ADAT1 with DM; rs76174573 of YEATS2 with hypertriglyceridemia; rs80015015 of ITGB8 with hypo-HDL-cholesterolemia; rs4907518 of LINC00354 and rs6537384 at 4q31.2 with hyper-LDL-cholesterolemia; rs17558532 of KRT27 with CKD. Given that these intermediate phenotypes are risk factors for CAD (4), the association between these loci and CAD may be attributable, at least in part, to their effects on intermediate phenotypes. By contrast, five SNPs in TNS3, FAM170B-AS1, BTRC, STIM1 and TRPM1 were not associated with intermediate phenotypes. The underlying molecular mechanisms of the association between these loci and CAD remain to be elucidated.
Recent GWASs have identified potential biological pathways underlying the association between genetic loci and CAD, including metabolism of LDL-cholesterol, triglycerides and lipoprotein (a); insulin resistance; thrombosis; inflammation, cell adhesion and transendothelial migration; cellular proliferation, vascular remodeling and extracellular matrix metabolism; and vascular tone and nitric oxide signaling (50,51). Network analysis of functional gene-gene interactions may be informative to clarify biological process of CAD and to identify therapeutic targets for this condition (52). Therefore, the present study performed gene ontology and network analyses to predict biological processes of the identified genes and interactions between these genes and those previously revealed to be associated with CAD. Gene ontology analysis revealed that biological functions of ITGB8 (integrin-mediated signaling pathway), PRKG1 (relaxation of vascular smooth muscle), STIM1 (activation of store-operated calcium channel activity) and LIPE (cholesterol and triglyceride metabolism) may serve roles in the development of CAD. However, the roles of the remaining 17 genes in CAD remain unclear. The network analysis revealed that the 18 genes identified in the present study had direct or indirect interactions with the 30 genes selected from the DisGeNET database (47,48), as well as complex networks with 228 genes previously identified by the GWASs (7). However, the underlying molecular mechanisms of these interactions remain to be elucidated.
It was previously demonstrated that six SNPs were associated with CAD (P<0.01), as determined by multivariable logistic regression analysis with adjustment for covariates following an initial EWAS screening of allele frequencies among subjects with early-onset and late-onset forms of this condition (33). The associations between three of the six SNPs [rs202069030 (P=2.58×10−6), rs7188 (P=0.0098) and rs2271395 (P=0.0042)] and CAD were replicated (P<0.05) in the present study. These results suggested that genetic variants associated with CAD differ, in part, between early-onset and late-onset patients with this condition. We also examined nine SNPs associated with MI (P<0.01) in a previous study (33). Associations between five of the nine SNPs [rs202103723 (P=0.0033), rs188212047 (P=0.0034), rs1265110 (P=2.69×10−5), rs9258102 (P=0.0374) and rs439121 (P=0.0108)] and CAD (P<0.05) were identified in the present study.
There are several limitations to the present study: i) Given that the results were not replicated, their validation will be necessary in independent study populations or in other ethnic groups; ii) it is possible that SNPs identified in the present study are in LD with other genetic variants in the same gene or in other nearby genes that are actually responsible for the development of CAD; and iii) the functional relevance of identified SNPs to the pathogenesis of CAD remains to be elucidated.
In conclusion, the present study identified the 54 SNPs as significant and independent determinants of CAD. Among these loci, 21 genes (RNF2, YEATS2, USP45, ITGB8, TNS3, FAM170B-AS1, PRKG1, BTRC, MKI67, STIM1, OR52E4, KIAA1551, MON2, PLUT, LINC00354, TRPM1, ADAT1, KRT27, LIPE, GFY and EIF3L) and 5 chromosomal regions (2p13, 4q31.2, 5q12, 13q34 and 20q13.2) that confer susceptibility to CAD were newly identified in the present study. Determination of genotypes for the SNPs at these loci may prove informative for assessment of the genetic risk for CAD in Japanese patients.
Acknowledgements
Not applicable.
Funding
The present study was supported by CREST, Japan Science and Technology Agency, Kawaguchi, Japan (grant no. JPMJCR1302).
Availability of data and materials
All datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YY contributed to the conception and design of the study; to acquisition, analysis and interpretation of the data; and to drafting of the manuscript. KK, MO, HH and TF all contributed to the acquisition of the data and to the revision of the manuscript. YY, IT and JS contributed to the analysis and interpretation of the data, as well as to the revision of the manuscript.
Ethics approval and consent to participate
The study protocol complied with the Declaration of Helsinki and was approved by the Committees on the Ethics of Human Research of Mie University Graduate School of Medicine, Hirosaki University Graduate School of Medicine, and participating hospitals (Gifu Prefectural Tajimi Hospital, Gifu Prefectural General Medical Center, Japanese Red Cross Nagoya First Hospital, Northern Mie Medical Center Inabe General Hospital, and Hirosaki Stroke and Rehabilitation Center). Written informed consent was obtained from all subjects.
Patient consent for publication
All authors approved submission of the final version of the article for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 3.Libby P. Mechanisms of acute coronary syndromes and their implications for therapy. N Engl J Med. 2013;368:2004–2013. doi: 10.1056/NEJMra1216063. [DOI] [PubMed] [Google Scholar]
- 4.Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling FN, Deo R, et al. American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee: Heart disease and stroke statistics-2018 update: A report from the American Heart Association. Circulation. 2018;137:e67–e492. doi: 10.1161/CIR.0000000000000558. [DOI] [PubMed] [Google Scholar]
- 5.Kathiresan S, Srivastava D. Genetics of human cardiovascular disease. Cell. 2012;148:1242–1257. doi: 10.1016/j.cell.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McPherson R, Tybjaerg-Hansen A. Genetics of coronary artery disease. Circ Res. 2016;118:564–578. doi: 10.1161/CIRCRESAHA.115.306566. [DOI] [PubMed] [Google Scholar]
- 7.Erdmann J, Kessler T, Venegas Munoz L, Schunkert H. A decade of genome-wide association studies for coronary artery disease: The challenges ahead. Cardiovasc Res. 2018;114:1241–1257. doi: 10.1093/cvr/cvy084. [DOI] [PubMed] [Google Scholar]
- 8.Dai X, Wiernek S, Evans JP, Runge MS. Genetics of coronary artery disease and myocardial infarction. World J Cardiol. 2016;8:1–23. doi: 10.4330/wjc.v8.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soutar AK, Naoumova RP. Mechanisms of disease: Genetic causes of familial hypercholesterolemia. Nat Clin Pract Cardiovasc Med. 2007;4:214–225. doi: 10.1038/ncpcardio0836. [DOI] [PubMed] [Google Scholar]
- 10.Paththinige CS, Sirisena ND, Dissanayake V. Genetic determinants of inherited susceptibility to hypercholesterolemia - a comprehensive literature review. Lipids Health Dis. 2017;16:103. doi: 10.1186/s12944-017-0488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodzioch M, Orsó E, Klucken J, Langmann T, Böttcher A, Diederich W, Drobnik W, Barlage S, Büchler C, Porsch-Ozcürümez M, et al. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 12.Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO, et al. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 13.Rust S, Rosier M, Funke H, Real J, Amoura Z, Piette JC, Deleuze JF, Brewer HB, Duverger N, Denèfle P, et al. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat Genet. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- 14.Peden JF, Farrall M. Thirty-five common variants for coronary artery disease: The fruits of much collaborative labour. Hum Mol Genet. 2011;20(R2):R198–R205. doi: 10.1093/hmg/ddr384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 17.Wellcome Trust Case Control Consortium, corp-author. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, et al. WTCCC and the Cardiogenics Consortium: Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, et al. Cardiogenics; CARDIoGRAM Consortium: Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CARDIoGRAMplusC4D Consortium, corp-author. Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, Bennett F, Bowden DW, Chakravarti A, Dreisbach A, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: The NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, Xiong X, Liao YH, Zeng QT, Yang YZ, et al. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 24.Lu X, Wang L, Chen S, He L, Yang X, Shi Y, Cheng J, Zhang L, Gu CC, Huang J, et al. Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Consortium: Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, et al. EPIC-CVD Consortium; CARDIoGRAMplusC4D; UK Biobank CardioMetabolic Consortium CHD working group: Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi F, Yokota M, Yamamoto K, Nakashima E, Katsuya T, Asano H, Isono M, Nabika T, Sugiyama T, Fujioka A, et al. Genome-wide association study of coronary artery disease in the Japanese. Eur J Hum Genet. 2012;20:333–340. doi: 10.1038/ejhg.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirokawa M, Morita H, Tajima T, Takahashi A, Ashikawa K, Miya F, Shigemizu D, Ozaki K, Sakata Y, Nakatani D, et al. A genome-wide association study identifies PLCL2 and AP3D1-DOT1L-SF3A2 as new susceptibility loci for myocardial infarction in Japanese. Eur J Hum Genet. 2015;23:374–380. doi: 10.1038/ejhg.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 30.Zdravkovic S, Wienke A, Pedersen NL, Marenberg ME, Yashin AI, De Faire U. Heritability of death from coronary heart disease: A 36-year follow-up of 20 966 Swedish twins. J Intern Med. 2002;252:247–254. doi: 10.1046/j.1365-2796.2002.01029.x. [DOI] [PubMed] [Google Scholar]
- 31.Nora JJ, Lortscher RH, Spangler RD, Nora AH, Kimberling WJ. Genetic-epidemiologic study of early-onset ischemic heart disease. Circulation. 1980;61:503–508. doi: 10.1161/01.CIR.61.3.503. [DOI] [PubMed] [Google Scholar]
- 32.Roncaglioni MC, Santoro L, D'Avanzo B, Negri E, Nobili A, Ledda A, Pietropaolo F, Franzosi MG, La Vecchia C, Feruglio GA, GISSI-EFRIM Investigators Role of family history in patients with myocardial infarction. An Italian case-control study. Circulation. 1992;85:2065–2072. doi: 10.1161/01.CIR.85.6.2065. [DOI] [PubMed] [Google Scholar]
- 33.Yamada Y, Sakuma J, Takeuchi I, Yasukochi Y, Kato K, Oguri M, Fujimaki T, Horibe H, Muramatsu M, Sawabe M, et al. Identification of STXBP2 as a novel susceptibility locus for myocardial infarction in Japanese individuals by an exome-wide association study. Oncotarget. 2017;8:33527–33535. doi: 10.18632/oncotarget.16945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada Y, Matsui K, Takeuchi I, Oguri M, Fujimaki T. Association of genetic variants with hypertension in a longitudinal population-based genetic epidemiological study. Int J Mol Med. 2015;35:1189–1198. doi: 10.3892/ijmm.2015.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD, Hansen M, Borecki IB, Cupples LA, Fornage M, et al. Best practices and joint calling of the HumanExome BeadChip: The CHARGE Consortium. PLoS One. 2013;8:e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 39.Leslie R, O'Donnell CJ, Johnson AD. GRASP: Analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics. 2014;30:i185–i194. doi: 10.1093/bioinformatics/btu273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eicher JD, Landowski C, Stackhouse B, Sloan A, Chen W, Jensen N, Lien JP, Leslie R, Johnson AD. GRASP v2.0: An update on the Genome-Wide Repository of Associations between SNPs and phenotypes. Nucleic Acids Res. 2015;43(D1):D799–D804. doi: 10.1093/nar/gku1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Binns D, Dimmer E, Huntley R, Barrell D, O'Donovan C, Apweiler R. QuickGO: A web-based tool for Gene Ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huntley RP, Binns D, Dimmer E, Barrell D, O'Donovan C, Apweiler R. QuickGO: A user tutorial for the web-based Gene Ontology browser. Database (Oxford) 2009;2009:bap010. doi: 10.1093/database/bap010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(suppl_2):W214–20. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montojo J, Zuberi K, Rodriguez H, Bader GD, Morris Q. GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000Res. 2014;3:153. doi: 10.12688/f1000research.4572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piñero J, Queralt-Rosinach N, Bravo À, Deu-Pons J, Bauer-Mehren A, Baron M, Sanz F, Furlong LI. DisGeNET: A discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015;2015:bav028. doi: 10.1093/database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz F, Furlong LI. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bønaa KH, Mannsverk J, Wiseth R, Aaberge L, Myreng Y, Nygård O, Nilsen DW, Kløw NE, Uchto M, Trovik T, et al. NORSTENT Investigators: Drug-eluting or bare-metal stents for coronary artery disease. N Engl J Med. 2016;375:1242–1252. doi: 10.1056/NEJMoa1607991. [DOI] [PubMed] [Google Scholar]
- 50.Klarin D, Zhu QM, Emdin CA, Chaffin M, Horner S, McMillan BJ, Leed A, Weale ME, Spencer CCA, Aguet F, et al. CARDIoGRAMplusC4D Consortium: Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet. 2017;49:1392–1397. doi: 10.1038/ng.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howson JMM, Zhao W, Barnes DR, Ho WK, Young R, Paul DS, Waite LL, Freitag DF, Fauman EB, Salfati EL, et al. CARDIoGRAMplusC4D; EPIC-CVD: Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet. 2017;49:1113–1119. doi: 10.1038/ng.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lempiäinen H, Brænne I, Michoel T, Tragante V, Vilne B, Webb TR, Kyriakou T, Eichner J, Zeng L, Willenborg C, et al. CVgenes@target consortium: Network analysis of coronary artery disease risk genes elucidates disease mechanisms and druggable targets. Sci Rep. 2018;8:3434. doi: 10.1038/s41598-018-20721-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.