Resistance to piperaquine has been associated with the amplification of the plasmepsin II gene in Cambodia. None of the 175 African isolates that we analyzed had plasmepsin II gene amplification (piperaquine 50% inhibitory concentration ranged from 0.94 to 137.5 nM), suggesting there is a low prevalence of piperaquine reduced susceptibility in Africa.
KEYWORDS: malaria, Plasmodium falciparum, antimalarial drug, resistance, in vitro, molecular marker, plasmepsin II gene, piperaquine
ABSTRACT
Resistance to piperaquine has been associated with the amplification of the plasmepsin II gene in Cambodia. None of the 175 African isolates that we analyzed had plasmepsin II gene amplification (piperaquine 50% inhibitory concentration ranged from 0.94 to 137.5 nM), suggesting there is a low prevalence of piperaquine reduced susceptibility in Africa. Additionally, the few isolates with reduced susceptibility to piperaquine did not harbor amplification of the plasmepsin II gene.
TEXT
Plasmodium falciparum resistance to most antimalarial drugs has emerged in Southeast Asia and spread to Africa (1, 2). Since 2005, the World Health Organization (WHO) recommended artemisinin-based combination therapies (ACT) as the first-line treatment for uncomplicated malaria, followed by artesunate for the treatment of severe malaria. However, artemisinin derivative-resistant P. falciparum strains emerged in western Cambodia, Myanmar, and Thailand, and eventually in all of Southeast Asia (3, 4). As soon as the last marketed ACT, dihydroartemisinin-piperaquine, was used, resistance emerged in Cambodia and later in Vietnam (5–8). In this context, it is essential to have markers of resistance to monitor the emergence and spread of resistance to dihydroartemisinin-piperaquine. Mutations (Y493H, F446I, and C580Y) in the propeller domain of the kelch 13 (K13) gene (PF3D71343700) were associated with in vivo and in vitro resistance to artemisinin in Southeast Asia (9, 10). Two recent studies showed that in vitro and in vivo resistance to piperaquine were associated with the amplification of the copy number of the plasmepsin II gene (PF3D7_1408000) (11, 12). However, these data were validated only on Cambodian isolates. The objective of the present study was to evaluate the copy numbers of the gene in African P. falciparum isolates and the gene's association with in vitro susceptibility to piperaquine.
A total of 175 P. falciparum isolates were successfully evaluated for the copy number of the plasmepsin II gene and assessed for ex vivo susceptibility to piperaquine. The isolates were collected from patients hospitalized in France from January 2015 to April 2017 with imported malaria from a country where malaria is endemic and, more particularly, from African French-speaking countries, such as Côte d'Ivoire, Cameroon, the Central African Republic, the Republic of the Congo, Guinea, Burkina Faso, Togo, Gabon, and Senegal (Table 1). The samples were sent from different civilian or military hospitals of the French National Reference Center for Imported Malaria Network (Aix en Provence, Bordeaux, Marseille, Montpellier, Nice, Toulon, and Toulouse) to the French National Reference Center for Malaria (Institut de Recherche Biomédicale des Armées [IRBA], Institut hospitalo-universitaire [IHU] Méditerranée Infection Marseille).
TABLE 1.
Geographical repartition of the 175 Plasmodium falciparum isolates
| Country | Isolate no. | Percentage |
|---|---|---|
| Cameroon | 47 | 26.9 |
| Côte d'Ivoire | 37 | 21.1 |
| Central African Republic | 18 | 10.3 |
| Guinea | 15 | 8.6 |
| Republic of the Congo | 14 | 8.0 |
| Togo | 8 | 4.6 |
| Burkina Faso | 7 | 4.0 |
| Gabon | 7 | 4.0 |
| Benin | 5 | 2.9 |
| Chad | 2 | 1.1 |
| Comores | 2 | 1.1 |
| Ghana | 1 | 0.6 |
| Madagascar | 2 | 1.1 |
| Mali | 2 | 1.1 |
| Nigeria | 2 | 1.1 |
| Angola | 1 | 0.6 |
| Democratic Republic of the Congo | 1 | 0.6 |
| Djibouti | 1 | 0.6 |
| Senegal | 1 | 0.6 |
Biobanking of human clinical samples used for malaria diagnostics and their secondary uses for scientific purposes are possible as long as the corresponding patients are informed and have not indicated any objections. This requirement was fulfilled here by giving verbal information to the patients, and no immediate or delayed patient opposition was reported to the hospital clinicians. Informed consent was not required for this study because the sampling procedures and testing are part of the French national recommendations for the care and surveillance of malaria.
The parasitemia, which ranged from 0.005% to 9.5%, was estimated on thin blood smears that were stained by eosin and methylene blue using a RAL kit (Réactifs RAL, Paris, France). The diagnosis of P. falciparum monoinfection was confirmed by real-time PCR (LightCycler 2.0; Roche Group, Basel, Switzerland), as previously described (13).
Piperaquine (PPQ) for the ex vivo drug susceptibility assay was obtained from Shin Poong Pharm Co. (Seoul, South Korea). PPQ was first dissolved in methanol and later diluted in water to final concentrations that ranged from 1.9 to 998 nM. The isolates were incubated for 72 h in a controlled atmosphere set at 85% N2, 10% O2, 5% CO2, and 37°C (maximum final parasitemia at 0.5% and final hematocrit of 1.5%). The isolates with parasitemia above 0.5% were diluted to parasitemia of 0.5% with fresh, uncontaminated erythrocytes. The drug susceptibility assay was revealed by the HRP2 enzyme-linked immunosorbent assay (ELISA)-based assay implemented in the Malaria Ag complement-enzyme linked immuno sorbent assay (CELISA) kit (reference KM2159; Cellabs Pty. Ltd., Brookvale, Australia) as previously described (14). The 50% inhibitory concentrations (IC50s) were validated only if the optical density (OD) ratio (OD at zero concentration/OD at maximum concentration of drug) was above 1.6 and the 95% confidence interval of the IC50 estimation was below 2.0. Each batch of plates was validated on the chloroquine (CQ)-resistant W2 clone of the Indochina strain (obtained from the Malaria Research and Reference Reagent Resource Center [MR4], VA, USA) in four independent experiments, using the same conditions as described below. The mean PPQ 50% inhibitory concentration values for the chloroquine-resistant W2 clone for the different batches used during the study was 54.1 ± 5.4 nM. There were no significant differences in the responses of the strains to PPQ between the different batches (P = 0.770).
The plasmepsin II gene copy number was estimated by TaqMan real-time PCR (LightCycler 2.0, Roche) using the single-copy β-tubulin gene (PF10_0084) as the control housekeeping gene. The following primers and probes were used: 5′-GGA GAT AAC CAA CAA CCA TTT AC-3′, 5′-GTT GTA CAT TTA ACA CTT GGG A-3′ and 5′-FAM-CCC ATA AAT TAG CAG ATC CTG TAT C-TAMRA-3′ for the plasmepsin II gene, and 5′-TGA TGT GCG CAA GTG ATC C-3′, 5′-TCC TTT GTG GAC ATT CTT CCT C-3′ and 5′-FAM-TAG CAC ATG CCG TTA AAT ATC TTC CAT GTC T-TAMRA-3′ for the β-tubulin gene (Eurogentec, Angers, France). PCRs were carried out using a 1× LightCycler TaqMan master mix (Roche, Germany), 900 nM forward primer, 900 nM reverse primer, 250 nM TaqMan probe, and 3 μl of template DNA. The thermal cycling conditions were 45 cycles of 95°C for 15 s and 60°C for 1 min. Each sample was assayed in duplicate. The 2−ΔΔCT method of relative quantification (where CT indicates cycle threshold) was used and adapted to estimate the copy number of the plasmepsin II gene, using the formula ΔΔCT = (CTplasmepsin II − CTβ-tubulin)sample − (CTplasmepsin II − CTβ-tubulin)calibrator. Genomic DNA extracted from the P. falciparum 3D7 strain, which has a single copy of each gene, was used as the calibrator, and the β-tubulin gene served as the control housekeeping gene in all experiments. We previously verified that the PCR efficiency was identical for the two genes. Samples were evaluated twice if the CT standard deviation was above 0.5 and if the copy number was above 1.5. Isolates with a copy number of ≥1.6 were classified as isolates with 2 copies (11, 15). A control parasite isolate harboring at least two copies of the plasmepsin II gene and sampled from a patient with malaria after returning from Cambodia was used to validate the quantitative PCR assay (sample kindly provided by Sandrine Houzé).
The ex vivo IC50 curves of PPQ were satisfactorily fitted to a sigmoidal function. Unlike the cultured-adapted Cambodian parasites (16), none of our isolates exhibited a bimodal dose-response curve when exposed to PPQ. The PPQ IC50 of the 175 isolates ranged from 0.94 to 137.5 nM (Fig. 1). A wide range of the PPQ responses was already observed in several studies, regardless of the methodology used. PPQ IC50s, assessed by a 72 h incubation with atmospheric generators for capnophilic bacteria and HRP2 ELISA in isolates from Dakar in 2013 to 2014 and 2013 to 2015, ranged from 2.5 to 168 nM and 3.9 to 241.9 nM, respectively (17, 18). The distribution of PPQ IC50 from 313 isolates obtained between 2008 and 2012 from patients hospitalized in France for imported malaria and assessed by a 42-h isotopic test ranged from 9.8 to 217.3 nM (19). A wide range of PPQ responses (3.1 to 188.9 nM) was also observed in isolates collected in 2010 to 2013 in Uganda and assessed using HRP2 ELISA detection (20). Only the isolates collected in 2016 in Uganda and assessed using a 72 h fluorescence assay with SYBR green I detection presented a narrow range for PPQ IC50, from 1.8 to 26.6 nM (21). In the absence of standardized ex vivo and in vitro tests, it is difficult to compare data from different laboratories. IC50 and cutoff values for in vitro resistance are specific to the methodology. The in vitro effects and the IC50s for antimalarial drugs depend on incubation and gas conditions and methodology (22–25). The isolates collected in Uganda in 2016 and assessed using a 72-h fluorescence assay with SYBR green I detection showed a narrow range for PPQ IC50, unlike those collected in 2010 to 2013 and assessed using HRP2 ELISA, which presented a wide range of PPQ responses (20, 21). Only the isolates that were assessed with SYBR green I detection showed a narrow range for PPQ IC50. Another hypothesis for a source of variation in drug responses may be the storage and the time between sample collection from patients and the completion of the ex vivo test. Blood was collected before therapy in an EDTA tube. Drug susceptibilities were assessed immediately or from samples stored at 4°C, even during transport, for a maximum of 48 h and without short-term culture. After collection from patients, the storage temperature of the samples was controlled by sensor. Additionally, Senegalese isolates, which were assessed immediately, presented the same wide range of in vitro responses (17, 18). One isolate had a reduced susceptibility to PPQ (IC50, >135 nM) (19). Another possible source of variation in the PPQ drug responses could result from the range of parasitemias used in the assay. This is because samples below 0.5% parasitemia were included in the assays and thus could not be adjusted to the standardized assay parasitemia.
FIG 1.
Distribution of ex vivo responses (IC50) of 175 African Plasmodium falciparum isolates to piperaquine.
Significant cross-susceptibilities were found between PPQ and mefloquine (coefficient of correlation [r] = 0.453; P < 0.0001), pyronaridine (r = 0.406; P < 0.0001), quinine (r = 0.247; P = 0.0011), and monodesethylamodiaquine (r = 0.189; P = 0.0138). Associations between PPQ and chloroquine or lumefantrine were not significant (r = 0.137; P = 0.0755; and and r = 0.114; P = 0.1413, respectively). The values of the coefficient of determination (r2) of the different associations were too low to explain the wide range of PPQ responses.
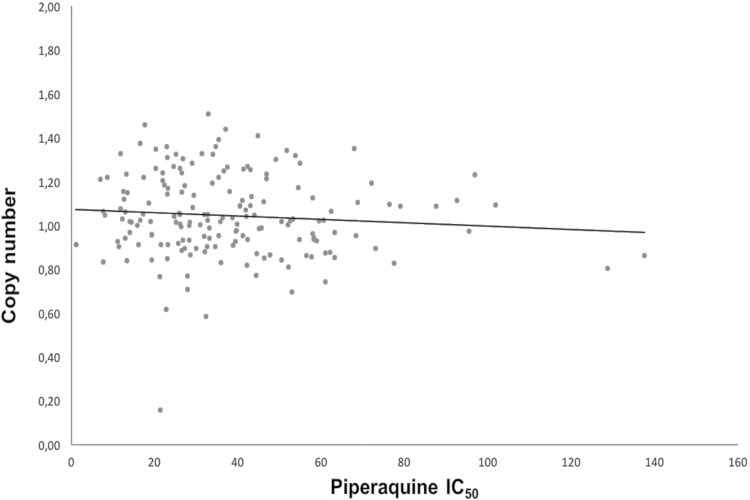
The copy number values ranged from 0.16 to 1.51 with a mean of 1.04 (Fig. 2). None of the isolates had more than one copy of the plasmepsin II gene.
FIG 2.
Copy number of the plasmepsin II gene as a function of piperaquine IC50 of 175 African Plasmodium falciparum isolates.
We did not observe any plasmepsin II gene amplification in the African isolates analyzed in this study. There was no association between the copy number of the plasmepsin II gene and susceptibility to PPQ. The most representative countries in terms of numbers were Cameroon and Côte d'Ivoire (47 and 37 isolates, respectively). Seven isolates (IC50, >90 nM) were outside the main scatter points representing the distribution of the in vitro responses to PPQ (Fig. 1) and could be considered parasites with reduced susceptibility to PPQ, although below 135 nM for six of them. The Cambodian P. falciparum strains, which were resistant ex vivo to PPQ and showed amplification of the plasmepsin II gene, presented IC50 values ranging from 89.3 to 159.6 nM (12). Among these seven isolates, four came from Cameroon and two from Côte d'Ivoire, suggesting the presence of few P. falciparum strains with reduced susceptibility to PPQ. The primary limitation of the present study is the low number of parasite isolates with reduced susceptibility to PPQ, probably due to the low use of dihydroartemisinin-piperaquine in Africa compared to that in Southeast Asia. Additionally, none of the seven isolates with reduced susceptibility to PPQ (IC50, >90 nM) harbored more than one copy of the plasmepsin II gene. This phenomenon was also shown in Cambodian isolates harboring a single copy of the plasmepsin II gene, for which a large range of PPQ IC50 was observed, as well as in some parasites with high IC50 values (12). The use of the standard ex vivo assay and not the PPQ survival assay (PSA) can be questionable (11, 26). However, Amato et al. identified the same association between ex vivo PPQ resistance and the amplification of the plasmepsin II gene by using a standard susceptibility drug assay (12). The resistant parasites showed ex vivo IC50s between 89.3 and 159.6 nM and in vitro IC50s between 55.9 and 79.7 nM after cultures had adapted, reflecting a high PSA survival % that ranged from 52.2 to 74.9% (12). Additionally, a novel mutation (F145I) on the P. falciparum chloroquine-resistance transporter gene (pfcrt) was recently identified and described as having an association with the PPQ IC90 values in Cambodian isolates by the standard in vitro assay (27). The influence of this mutation will have to be evaluated in African P. falciparum isolates.
The amplification of the plasmepsin II gene does not fully explain the decreased in vitro susceptibility to PPQ of some P. falciparum parasites in Africa. Indeed, no correlation was observed between the copy number of the plasmepsin II gene and ex vivo susceptibility to piperaquine in Ugandan P. falciparum isolates (21). The use of dihydroartemisinin-piperaquine as intermittent preventive treatment during pregnancy (IPTp) did not select for genotypes associated with amplification of the plasmepsin II gene in Uganda (28). Additionally, no amplification of the plasmepsin II gene was found in Cameroonian recrudescent Plasmodium falciparum parasites 2 years after treatment by dihydroartemisinin-piperaquine (29). Gupta et al. reported that 1.1% of the Plasmodium falciparum isolates circulating in Mozambique in 2015 harbored multiple copies of the plasmepsin II gene (30). However, none of these isolates were compared with in vitro data or clinical responses. These data do not allow assessment of the possible association between amplification of the plasmepsin II gene and reduced susceptibility to PPQ. Additionally, Amato et al. showed that more than half of isolates harboring multiple copies of the plasmepsin II gene presented low susceptibility to PPQ, suggesting that amplification of the plasmepsin II gene did not alone explain reduced susceptibility to PPQ (12).
These finding suggest that copy number variation of the plasmepsin II gene may not alone predict PPQ resistance in Africa. It seems that other resistance mechanisms and therefore other molecular markers may exist in Africa compared to those in Asia. This phenomenon has also been observed with resistance to artemisinin, which is associated with pfk13 polymorphisms in Asia, while no polymorphism is observed in most cases of clinical failure of ACT in Africa (10, 31–35). To overcome the limitation of this study, i.e., the low number of samples with reduced susceptibility, it is imperative to further assess more isolates from different geographical areas of Africa, and especially more P. falciparum strains resistant to PPQ from Africa.
ACKNOWLEDGMENTS
We thank the patients and the staff of the hospitals of the French National Reference Center for Imported Malaria Network. We thank Sandrine Houzé (Centre national de reference du Paludisme, Paris, France) for providing the clinical sample with two copies of the plasmepsin II gene.
This study was funded by the French Institute for Public Health Surveillance (Santé Publique France, grant CNR Paludisme). F.F.T. was supported by the Foundation Méditerranée Infection.
We declare that we have no competing interests.
The members of the French National Reference Centre for Imported Malaria Study Group are as follows: D. Basset (Centre Hospitalier Universitaire de Montpellier, Montpellier), P. Bastien (Centre Hospitalier Universitaire de Montpellier, Montpellier), F. Benoit-Vical (Centre Hospitalier Universitaire de Rangueil, Toulouse), A. Berry (Centre Hospitalier Universitaire de Rangueil, Toulouse), P. Brouqui (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), M. Cividin (Centre Hospitalier du Pays d'Aix, Aix en Provence), P. Delaunay (Centre Hospitalier Universitaire de l'Archet, Nice), L. Delhaes (Hôpital Pellegrin, Bordeaux), M. Drancourt (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), T. Gaillard (Hôpital d'Instruction des Armées Saint-Anne, Toulon), A. Genin (Centre Hospitalier du Pays d'Aix, Aix en Provence), E. Garnotel (Hôpital d'Instruction des Armées Laveran, Marseille), E. Javelle (Hôpital d'Instruction des Armées Laveran, Marseille), C. L'Ollivier (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), M. Leveque (Centre Hospitalier Universitaire de Montpellier, Montpellier), D. Malvy (Hôpital Pellegrin, Bordeaux), P. Marty (Centre Hospitalier Universitaire de l'Archet, Nice), G. Ménard (Hôpital d'Instruction des Armées Saint-Anne, Toulon), P. Millet (Hôpital Pellegrin, Bordeaux), P. Minodier (Hôpital Nord, Marseille), A. Mottard (Hôpital de Fréjus-Saint Raphael, Fréjus), P. Parola (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), R. Piarroux (Hôpital la Timone, Marseille), C. Pomares-Estran (Centre Hospitalier Universitaire de l'Archet, Nice), M.-C. Receveur (Hôpital Pellegrin, Bordeaux), A. Robin (Centre Hospitalier du Pays d'Aix, Aix en Provence), E. Sappa (Centre Hospitalier du Pays d'Aix, Aix en Provence), H. Savini (Hôpital d'Instruction des Armées Laveran, Marseille), F. Simon (Hôpital d'Instruction des Armées Laveran, Marseille), Y. Sterkers (Centre Hospitalier Universitaire de Montpellier, Montpellier), C. Surcouf (Hôpital d'Instruction des Armées Laveran, Marseille), E. Varlet (Centre Hospitalier Universitaire de Montpellier, Montpellier), and A. Wolff (Hôpital d'Instruction des Armées Laveran, Marseille).
Contributor Information
the French National Reference Centre for Imported Malaria Study Group:
D. Basset, P. Bastien, F. Benoit-Vical, A. Berry, P. Brouqui, M. Cividin, P. Delaunay, L. Delhaes, M. Drancourt, T. Gaillard, A. Genin, E. Garnotel, E. Javelle, C. L'Ollivier, M. Leveque, D. Malvy, P. Marty, G. Ménard, P. Millet, P. Minodier, A. Mottard, P. Parola, R. Piarroux, C. Pomares-Estran, M.-C. Receveur, A. Robin, E. Sappa, H. Savini, F. Simon, Y. Sterkers, C. Surcouf, E. Varlet, and A. Wolff
REFERENCES
- 1.Mita T, Venkatesan M, Ohashi J, Culleton R, Takahashi N, Tsukahara T, Ndounga M, Dysoley L, Endo H, Hombhanje F, Ferreira MU, Plowe CV, Tanabe K. 2011. Limited geographical origin and global spread of sulfadoxine-resistant dhps alleles in Plasmodium falciparum populations. J Infect Dis 204:1980–1988. doi: 10.1093/infdis/jir664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 3.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Seng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jttamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski D, Bozdech Z, Jeeyapant A, Cheah PY, Sakulhaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Des D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ. 2015. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spring MD, Lin JT, Manning JE, Vanachayangkul P, Somethy S, Bun R, Se Y, Chann S, Ittiverakul M, Sia-ngam P, Kuntawunginn W, Arsanok M, Buathong N, Chaorattanakawee S, Gosi P, Ta-aksorn W, Chanarat N, Sundrakes S, Kong N, Heng TK, Nou S, Teja-isavadharm P, Pichyangkul S, Phann ST, Balasubramanian S, Juliano JJ, Meshnick SR, Chour CM, Prom S, Lanteri CA, Lon C, Saunders DL. 2015. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect Dis 15:683–691. doi: 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- 6.Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, Sam B, Dek D, Try V, Amato R, Blessborn D, Song L, Tullo GS, Fay MP, Anderson JM, Tarning J, Fairhurst RM. 2016. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis 16:357–365. doi: 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phuc BQ, Rasmussen C, Duong TT, Dong LT, Loi MA, Ménard D, Traning J, Bustos D, Ringwald P, Galappaththy GL, Thieu NQ. 2017. Treatment failure of dihydroartemisinin/piperaquine for Plasmodium falciparum malaria, Vietnam. Emerg Infect Dis 23:715–717. doi: 10.3201/eid2304.161872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thanh NV, Thuy-Nhien N, Tuyen NT, Tong NT, Nha-Ca NT, Dong LT, Quang HH, Farrar J, Thwaites G, White NJ, Wolbers M, Hien TT. 2017. Rapid decline in the susceptibility of Plasmodium falciparum to dihydroartemisinin-piperaquine in the south of Vietnam. Malar J 16:27. doi: 10.1186/s12936-017-1680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fiarhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ménard D, Khim N, Beghain J, Adegnika AA, Alam MS, Amodu O, Awab Gr Barnadas C, Berry A, Boum Y, Bustos MD, Cao J, Chen JH, Collet L, Cui L, Das Thakur G, Dieye A, Djalle D, Dorkenoo MA, Eboumbou Moukoko CE, Espino E, Fandeur T, Ferreira-Da-Cruz Mf Fola AA, Fuehrer HP, Hassan Am Herrera S, Hongvanthong B, Houze S, Karim MJ, Jiang L, Kano S, Khan WA, Khanthavong M, Kremsner PG, Lacerda M, Leang R, Leelawong M, Li M, Lin K, Laminou IM, Mazarati JB, Menard S, Morlais I, Muhindo Mavoko H, Musset L, Na-Bangchang K, Nambozi M, Niare K, Noedl H, Ouedraogo JB, Pillai DR, Pradines B, Quang Phuc B, Ramharter M, Randrianarivelojosia M, Sattabongkot J, Sheikh Omar R, Silue KD, Sirima SB, Sutherland C, Syafruddin D, Tahar R, Tang LH, Toure OA, Tshibangu P, Vigan-Womas I, Warsame M, Wini L, Zakeri S, Kim S, Eam R, Berne L, Khean C, Chy S, Ken M, Loch K, Canier L, Duru V, Legrand E, Barale JC, Stokes B, Straimer J, Witkowski B, Fidock DA, Rogier C, Ringwald P, Ariey F, Mercereau-Puijalon O; for the K13 Artemisinin Resistance Multicenter Assessment consortium (KARMA). 2016. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med 374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, Chy S, Kim S, Ke S, Kloeung N, Eam R, Khean C, Ken M, Loch K, Bouillon A, Domergue A, Ma L, Bouchier C, Leang R, Huy R, Nuel G, Barale JC, Legrand E, Ringwald P, Fidock DA, Mercereau-Puijalon O, Ariey F, Ménard D. 2017. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis 17:174–183. doi: 10.1016/S1473-3099(16)30415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, Almagro-Garcia J, Neal AT, Sreng S, Suon S, Drury E, Jyothi D, Stalker J, Kwiatkowski DP, Fairhurst RM. 2017. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis 17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wurtz N, Fall B, Bui K, Pascual A, Fall M, Camara C, Diatta B, Ba Fall K, Saliou Mbaye P, Diémé Y, Bercion R, Wade B, Briolant S, Pradines B. 2013. Pfhrp2 and pfhrp3 polymorphisms in Plasmodium falciparum isolates from Dakar, Senegal: impact on rapid malaria diagnostic tests. Malar J 12:34. doi: 10.1186/1475-2875-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fall B, Madamet M, Camara C, Amalvict R, Fall M, Nakoulima A, Diatta B, Diémé Y, Wade B, Pradines B. 2016. Plasmodium falciparum in vitro resistance to monodesethylamodiaquine, Dakar, Senegal, 2014. Emerg Infect Dis 22:841–845. doi: 10.3201/eid2205.151321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price RN, Uhlemann AC, Brockman A, McReady R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bopp S, Magistrado P, Wong W, Schaffner SF, Mukherjee A, Lim P, Dhorda M, Amaratunga C, Woodrow CJ, Ashley EA, White NJ, Dondorp AM, Fairhurst RM, Ariey F, Menard D, Wirth DF, Volkman SK. 2018. Plasmepsin II–III copy number accounts for bimodal piperaquine resistance among Cambodian Plasmodium falciparum. Nat Commun 9:1769. doi: 10.1038/s41467-018-04104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fall B, Camara C, Fall M, Nakoulima A, Dionne P, Diatta B, Diémé Y, Wade B, Pradines B. 2015. Plasmodium falciparum susceptibility to standard and potential anti-malarial drugs in Dakar, Senegal, during the 2013–2014 malaria season. Malar J 14:60. doi: 10.1186/s12936-015-0589-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gendrot M, Diawara S, Madamet M, Kouta MB, Briolant S, Wade KA, Fall M, Benoit N, Nakoulima A, Amalvict R, Diémé Y, Fall B, Wade B, Diatta B, Pradines B. 2017. Association between polymorphisms in the Pfmdr6 gene and ex vivo susceptibility to quinine in Plasmodium falciparum parasites from Dakar, Senegal. Antimicrob Agents Chemother 61:e01183-16. doi: 10.1128/AAC.01183-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pascual A, Madamet M, Briolant S, Gaillard T, Amalvict R, Benoit N, Travers D, Pradines B. 2015. Multinormal in vitro distribution of Plasmodium falciparum susceptibility to piperaquine and pyronaridine. Malar J 14:49. doi: 10.1186/s12936-015-0586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tumwebase P, Conrad MD, Walakira A, LeClair N, Byaruhanga O, Nakazibwe C, Kozak B, Bloome J, Okiring J, Kakuru A, Bigira V, Kapisi J, Legac J, Gut J, Cooper RA, Kamya MR, Havlir DV, Dorsey G, Greehouse B, Nsobya SL, Rosenthal PJ. 2015. Impact of antimalarial treatment and chemoprevention on the drug sensitivity of malaria parasites isolated from Ugandan children. Antimicrob Agents Chemother 59:3018–3030. doi: 10.1128/AAC.05141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen SA, Ceja FG, Conrad MD, Tumwebaze PK, Byaruhanga O, Katairo T, Nsobya SL, Rosenthal PJ, Cooper RA. 2017. Changing antimalarial drug sensitivities in Uganda. Antimicrob Agents Chemother 61:e01516-17. doi: 10.1128/AAC.01516-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pradines B, Rogier C, Fusai T, Mosnier J, Daries W, Baret E, Parzy D. 2001. In vitro activities of antibiotics against Plasmodium falciparum are inhibited by iron. Antimicrob Agents Chemother 45:1746–1750. doi: 10.1128/AAC.45.6.1746-1750.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briolant S, Parola P, Fusai T, Madamet-Torrentino M, Baret E, Mosnier J, Delmont JP, Parzy D, Minodier P, Rogier C, Pradines B. 2007. Influence of oxygen on asexual blood cycle and susceptibility of Plasmodium falciparum to chloroquine: requirement of a standardized in vitro assay. Malar J 6:44. doi: 10.1186/1475-2875-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pascual A, Basco LK, Baret E, Amalvict R, Travers D, Rogier C, Pradines B. 2011. Use of the atmospheric generators for capnophilic bacteria Genbag-CO2 for the evaluation of in vitro Plasmodium falciparum susceptibility to standard anti-malarial drugs. Malar J 10:8. doi: 10.1186/1475-2875-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wein S, Maynadier M, Tran Van Ba C, Cerdan R, Peyrottes S, Fraisse L, Vial H. 2010. Reliability of antimalarial sensitivity tests depends on drug mechanisms of action. J Clin Microbiol 48:1651–1660. doi: 10.1128/JCM.02250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duru V, Khim N, Leang R, Kim S, Domergue A, Kloeung N, Ke S, Chy S, Eam R, Khean C, Loch K, Ken M, Lek D, Beghain J, Ariey F, Guerin PJ, Huy R, Mercereau-Puijalon O, Witkowski B, Ménard D. 2015. Plasmodium falciparum dihydroartemisinin-piperaquine failures in Cambodia are associated with mutant K13 parasites presenting high survival rates in novel piperaquine in vitro assays: retrospective and prospective investigations. BMC Med 13:305. doi: 10.1186/s12916-015-0539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agrawal S, Moser KA, Morton L, Cummings MP, Parihar A, Dwivedi A, Shetty AC, Drabek EF, Jacob CG, Henrich PP, Parobek CM, Jongsakul K, Huy R, Spring MD, Lanteri CA, Chaorattanakawee S, Lon C, Fukuda MM, Saunders DL, Fidock DA, Lin JT, Juliano JJ, Plowe CV, Silva JC, Takala-Harrison S. 2017. Association of a novel mutation in the Plasmodium falciparum chloroquine transporter with decreased piperaquine sensitivity. J Infect Dis 216:468–476. doi: 10.1093/infdis/jix334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conrad MD, Mota D, Foster M, Tukwasibwe S, Legac J, Tumwebaze P, Whalen M, Kakuru A, Nayebare P, Wallender E, Havlir DV, Jagannathan P, Huang L, Aweeka F, Kamya MR, Dorsey G, Rosenthal PJ. 2017. Impact of intermittent preventive treatment during pregnancy in Plasmodium falciparum drug resistance-mediating polymorphisms in Uganda. J Infect Dis 216:1008–1017. doi: 10.1093/infdis/jix421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malvy D, Torrentino-Madamet M, L'Ollivier C, Receveur MC, Jeddi F, Delhaes L, Piarroux R, Millet P, Pradines B. 2018. Plasmodium falciparum recrudescence two years after treatment of an uncomplicated infection without return to an area where malaria is endemic. Antimicrob Agents Chemother 62:e01892-17. doi: 10.1128/AAC.01892-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta H, Macete E, Bulo H, Salvador C, Warsame M, Carvalho E, Ménard D, Ringwald P, Bassat Q, Enosse S, Mayor A. 2018. Drug-resistant polymorphisms and copy numbers of Plasmodium falciparum, Mozambique, 2015. Emerg Infect Dis 24:40–48. doi: 10.3201/eid2401.170864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madamet M, Kouta MB, Wade KA, Lo G, Diawara S, Fall M, Bercion R, Nakoulima A, Fall KB, Benoit N, Gueye MW, Fall B, Diatta B, Pradines B. 2017. Absence of association between polymorphisms in the K13 gene and the presence of Plasmodium falciparum parasites at day 3 after treatment with artemisinin derivatives in Senegal. Int J Antimicrob Agents 49:754–756. doi: 10.1016/j.ijantimicag.2017.01.032. [DOI] [PubMed] [Google Scholar]
- 32.Dieye B, Affara M, Sangare L, Joof F, Ndiaye YD, Gomis JF, Ndiaye M, Mbaye A, Diakite M, Sy N, Mbengue B, Deme AB, Daniels R, Ahouidi AD, Dieye T, Abdullahi A, Doumbia S, Ndiaye JL, Diarra A, Ismaela A, Coulibaly M, Welty C, Ngwa AA, Shaffer J, D'Alessandro U, Volkman SK, Wirth DF, Krogstad DJ, Koita O, Nwakanma D, Ndiaye D. 2016. West Africa international centers of excellence for malaria research: drug resistance patterns to artemether-lumefantrine in Senegal, Mali, and The Gambia. Am J Trop Med Hyg 95:1054–1060. doi: 10.4269/ajtmh.16-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plucinski MM, Dimbu PR, Macaia AP, Ferreira CM, Samutondo C, Quivinja J, Afonso M, Kiniffo R, Mbounga E, Kelley JS, Patel DS, He Y, Talundzic E, Garrett DO, Halsey ES, Udhayakumar V, Ringwald P, Fortes F. 2017. Efficacy of artemether-lumefantrine, artesunate-amodiaquine, and dihydroartemisinin-piperaquine for treatment of uncomplicated Plasmodium falciparum malaria in Angola, 2015. Malar J 16:62. doi: 10.1186/s12936-017-1712-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plucinski MM, Talundzic E, Morton L, Dimbu PR, Macaia AP, Fortes F, Goldman I, Lucchi N, Stennies G, MacArthur JR, Udhayakumar V. 2015. Efficacy of artemether-lumefantrine and dihydroartemisinin-piperaquine for treatment of uncomplicated malaria in children in Zaire and Uíge Provinces, Angola. Antimicrob Agents Chemother 59:437–443. doi: 10.1128/AAC.04181-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutherland CJ, Lansdell P, Sanders M, Muwanguzi J, van Schalkwyk DA, Kaur H, Nolder D, Tucker J, Bennett HM, Otto TD, Berriman M, Patel TA, Lynn R, Gkrania-Klotsas E, Chiodina PL. 2017. pfk13-independent treatment failure in four imported cases of Plasmodium falciparum malaria treated with artemether-lumefantrine in the United Kingdom. Antimicrob Agents Chemother 61:e02382-16. doi: 10.1128/AAC.02382-16. [DOI] [PMC free article] [PubMed] [Google Scholar]