Paracoccidioidomycosis (PCM) is the cause of many deaths from systemic mycoses. The etiological agents of PCM belong to the Paracoccidioides genus, which is restricted to Latin America.
KEYWORDS: Paracoccidioides, antifungal, targets, argentilactone, chemoproteomics, drug discovery
ABSTRACT
Paracoccidioidomycosis (PCM) is the cause of many deaths from systemic mycoses. The etiological agents of PCM belong to the Paracoccidioides genus, which is restricted to Latin America. The infection is acquired through the inhalation of conidia that primarily lodge in the lungs and may disseminate to other organs and tissues. The treatment for PCM is commonly performed via the administration of antifungals such as amphotericin B, co-trimoxazole, and itraconazole. The antifungal toxicity and side effects, in addition to their long treatment times, have stimulated research for new bioactive compounds. Argentilactone is a compound that was isolated from the Brazilian savanna plant Hyptis ovalifolia, and it has been suggested to be a potent antifungal, inhibiting the dimorphism of P. brasiliensis and the enzymatic activity of isocitrate lyase, a key enzyme of the glyoxylate cycle. This work was developed due to the importance of elucidating the putative mode of action of argentilactone. The chemoproteomics approach via affinity chromatography was the methodology used to explore the interactions between P. brasiliensis proteins and argentilactone. A total of 109 proteins were identified and classified functionally. The most representative functional categories were related to amino acid metabolism, energy, and detoxification. Argentilactone inhibited the enzymatic activity of malate dehydrogenase, citrate synthase, and pyruvate dehydrogenase. Furthermore, argentilactone induces the production of reactive oxygen species and inhibits the biosynthesis of cell wall polymers.
INTRODUCTION
The Paracoccidioides genus includes pathogenic fungi that cause paracoccidioidomycosis (PCM), which is commonly found in rural workers between 30 and 50 years of age who reside in areas where the disease is endemic. PCM manifests as a pneumopathology associated with mucosal and skin lesions, and it can spread via the blood or lymphatic system, affecting other organs and systems, such as the liver, spleen, bone, and central nervous system (1, 2). Control measures are not currently available for PCM. Thus, early diagnosis and correct treatment to prevent disease progression and complications remain the best strategies. The choice of an effective therapy for PCM is made by evaluating the degree of the lesion, disease severity, the patient's immunological capacity, hypersensitivity reaction, and contraindication of antifungal agents for pregnancy, due to the teratogenic effect, and for patients with hepatic and renal problems, due to its hepatotoxic and nephrotoxic effects (3). Currently, PCM therapy is a slow process involving months or years of administration using antifungal agents such as amphotericin B, co-trimoxazole, and itraconazole (2, 4). Those antifungals have limitations with regard to toxicity, leading to the frequent abandonment of treatment, which makes the search for new potential antifungals highly relevant to public health.
The Brazilian savanna is the second largest biome in Brazil, comprising more than 204 million hectares, and it has been explored extensively to identify new bioactive compounds (5, 6). The genus Hyptis, belonging to the family Lamiaceae, is composed of approximately 583 species that are exclusively neotropical, and they are distributed from the southern United States to Argentina. However, in the central-west region of Brazil, the genus is more frequent and they are important sources of bioactive constituents, with antimicrobial, inflammatory, cytotoxic, and insecticidal properties (7–9). The essential oil argentilactone is the major compound produced by Hyptis ovalifolia, showing antiproliferative activity against cancer cells (10), Leishmania amazonensis (11), and Trypanosoma cruzi (12) and antifungal activity against the dermatophytes Microsporum canis, Microsporum gypseum, Trichophyton mentagrophytes, and Trichophyton rubrum (9).
The inhibitory activity of argentilactone isolated from H. ovalifolia, as well as analog derivatives synthesized from argentilactone, were elucidated in relation to their actions against Paracoccidioides species and its native and recombinant isocitrate lyase (PbICL), an important enzyme for this fungus. Argentilactone and derivatives inhibited PbICL activity, yeast cell growth, and differentiation from mycelium to yeast (13). In addition, aiming to evaluate if argentilactone induces DNA damage in human cells, the comet assay was performed for MRC5 cells treated with different concentrations of argentilactone. In the MRC5 cells, argentilactone did not induce DNA damage, suggesting that this compound is safe to humans (14).
Target deconvolution is an important step in drug research and development, allowing the definition of selectivity and the early detection of potential side effects (15). Thus, considering the antiproliferative potential of argentilactone and the need to elucidate its mode of action in Paracoccidioides spp., Prado et al. (14) evaluated the proteomic response of Paracoccidioides lutzii after exposure to argentilactone. The study revealed that argentilactone downregulated the enzymes in important pathways, such as glycolysis, the tricarboxylic acid (TCA) cycle, and the glyoxylate cycle, leading the fungus to adopt alternative metabolic pathways to survival. In addition, P. lutzii upregulated the proteins involved in cell rescue, defense, and stress response. Araújo et al. used a transcriptome analysis as an experimental strategy to expand information about argentilactone's mode of action (16). That study suggested that argentilactone promoted dysfunction in mitochondrial membrane potential, leading to probable oxidative stress, and it may act on the cell wall. In addition, the ethanol level was reduced in the presence of argentilactone, suggesting that fermentation would also be suppressed.
In view of all the findings presented by previous studies, we aimed to identify the molecular targets of argentilactone in P. brasiliensis using chemoproteomics by affinity chromatography. Additionally, assays were performed to confirm the action of argentilactone on the activity of citrate synthase, malate dehydrogenase, and pyruvate dehydrogenase enzymes in the production of reactive oxygen species and in the biosynthesis of cell wall polymers. In addition, we propose an argentilactone model of action against Paracoccidioides spp. based on our chemoproteomics data and previously obtained proteomics and transcriptional data.
RESULTS AND DISCUSSION
Argentilactone, the major component of H. ovalifolia essential oil, was able to inhibit the growth of P. lutzii without promoting cytotoxic and genotoxic effects on MRC5 cells (14). Here, we expanded the evaluation of MIC and minimum fungicidal concentration (MFC) for other species, such as P. brasiliensis, P. americana, and P. restrepiensis. In addition, we evaluated the cytotoxic concentration (CC) to A549 (epithelial lung), BALB/c 3T3 clone A31 (fibroblast), and AMJ2-C11 (alveolar macrophage) cells, and the selectivity index (SI) of argentilactone was determined (Table 1). The results showed that argentilactone inhibited Paracoccidioides species growth at concentrations ranging from 4.5 μg/ml (P. restrepiensis strains PbEPM83 and Pb339) to 36 μg/ml (P. brasiliensis Pb18). Argentilactone was not cytotoxic to AMJ2-C11, BALB/c 3T3, and A549 cells at the concentrations used to inhibit Paracoccidioides species growth. In addition, argentilactone showed a higher selectivity index for these cells, confirming it to be a promising antifungal.
TABLE 1.
Biological activity of argentilactonea
| Species/isolate | MIC (μg/ml) | MFC (μg/ml) | CC (μg/ml)/SI for: |
||
|---|---|---|---|---|---|
| AMJ2-C11 | BALB/c 3T3 | A549 | |||
| P. lutzii | |||||
| Pb01 | 18b | 18 | 1,000/55.5 | 1,000/55.5 | 500/27.7 |
| P. brasiliensis | |||||
| Pb18 | 36 | 36 | 1,000/27.7 | 1,000/27.7 | 500/16.6 |
| P. americana | |||||
| Pb02 | 9 | 9 | 1,000/111.1 | 1,000/111.1 | 500/55.5 |
| Pb03 | 18 | 18 | 1,000/55.5 | 1,000/55.5 | 500/27.7 |
| P. restrepiensis | |||||
| PbEPM83 | 4.5 | 4.5 | 1,000/222.2 | 1,000/222.2 | 500/111.1 |
| Pb339 | 4.5 | 4.5 | 1,000/222.2 | 1,000/222.2 | 500/111.1 |
| Pb60855 | 18 | 18 | 1,000/27.7 | 1,000/27.7 | 500/16.6 |
AMJ2-C11, macrophage alveolar cells; BALB/c 3T3, fibroblast cells; A549, epithelial lung cells; CC, cytotoxic concentration.
Data are from Prado et al. (14).
The elucidation of the molecular target is an essential step for developing molecules with bioactive potentials (17). The proteomic and transcriptional responses of P. lutzii to argentilactone were previously evaluated by Prado et al. (14) and Araújo et al. (16). Although these analyses provided important information, the interactional level of argentilactone and proteins in P. brasiliensis is unknown. Methodologies based on chemoproteomics have been important tools for proposing drug targets (18). Thus, a chemoproteomics approach based on affinity chromatography was used in this work to identify the molecular targets of argentilactone in P. brasiliensis Pb18. To propose a robust model of action for argentilactone, we correlated our findings (argentilactone-interacting proteins) with the proteomic and transcriptomic data.
We considered argentilactone-interacting proteins, specifically those proteins that were identified exclusively in the column immobilized with argentilactone and absent from the control column. These proteins may have interacted directly or indirectly, resulting in a total of 109 proteins identified. Among the argentilactone-interacting proteins, 12 of them were downregulated and 6 were upregulated in response to argentilactone, according to the proteomics analysis of P. lutzii (14). Concerning the transcript levels, 8 genes were upregulated in the presence of argentilactone (16) (see Table S1 in the supplemental material).
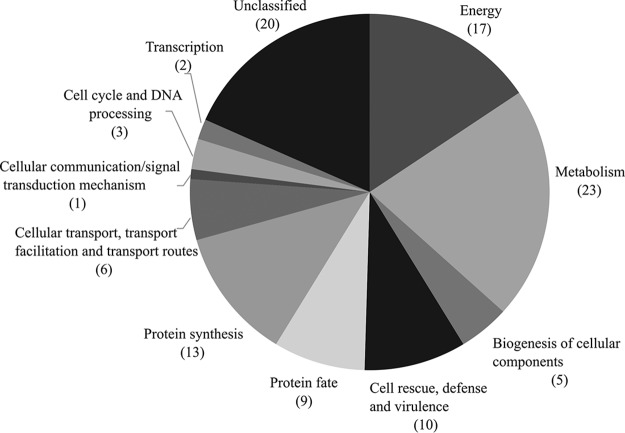
The argentilactone-interacting proteins were categorized according to the Munich Information Center for Protein Sequences (MIPS) functional category database, and their biological functions were proposed by PEDANT. Among the functional categories, the proteins from the most representative classes belong to metabolism (23 proteins); energy (17 proteins); synthesis protein (13 proteins); cell rescue, defense, and virulence (10 proteins); protein fate (9 proteins); and unclassified (20 proteins). The functional categories of amino acid metabolism (12 proteins) and lipid metabolism (6 proteins) were more representative in the category of metabolism (Fig. 1). The energy functional categories include proteins belonging to electron transport and membrane-associated energy conservation (6 proteins), glycolysis and gluconeogenesis (4 proteins), the pentose-phosphate pathway (4 proteins), the tricarboxylic acid pathway (2 proteins), and the methyl citrate cycle (1 protein). The functional categories of synthesis protein include proteins belonging to ribosome biogenesis (6 proteins), aminoacyl-tRNA-synthetases (4 proteins), and translation (3 proteins) (Table S1).
FIG 1.
Functional enrichment of proteins identified by chemoproteomics. The argentilactone-interacting proteins were categorized according to the Munich Information Center for Protein Sequences (MIPS) functional category database, available at geneontology.org.
Interaction of argentilactone with proteins from energy-related pathways.
Argentilactone interacted with glycolytic enzymes such as 6-phosphofructokinase (PFK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). PFK is an important regulatory enzyme of glycolysis, catalyzing the phosphorylation of fructose-6-phosphate to form expensive fructose-1,6-bisphosphate. In the presence of argentilactone, Paracoccidioides spp. were shown to downregulated PFK (14). Although its classical localization is cytoplasmic, GAPDH was identified in the vesicle proteome and cell wall of Paracoccidioides lutzii (19, 20). In addition to its role in metabolic reactions, GAPDH is considered a moonlighting protein of Paracoccidioides spp. with an important role as an adhesin, an immunoreactive molecule (21), and in response to internalization by macrophages (22). In the presence of argentilactone, GAPDH was upregulated by Paracoccidioides spp., suggesting that the interactions of argentilactone-GAPDH and argentilactone-PFK can interfere in both the primary way of obtaining energy and in processes of invasion and adhesion during infection. In fact, Prado et al. showed that glucose consumption was decreased during exposure to argentilactone (14), indicating that the glycolysis was partially blocked.
Linking the glycolytic pathway to the oxidative pathway of TCA cycle.
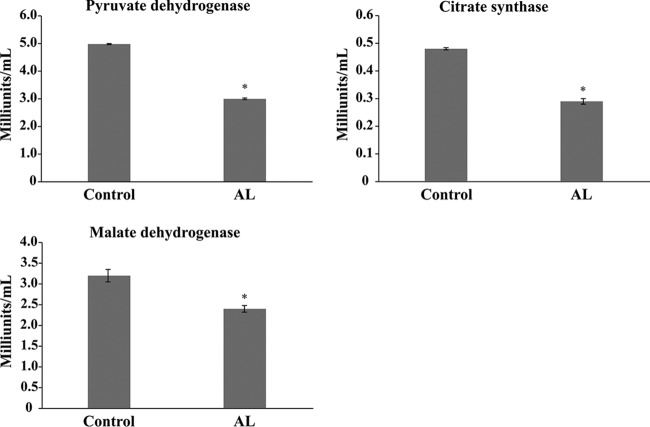
Tricarboxylic acid (TCA), the pyruvate dehydrogenase complex, which is composed of three enzymes (pyruvate dehydrogenase, dihydrolipoamide acetyltransferase, and dihydrolipoamide dehydrogenase), converts pyruvate into acetyl-coenzyme A (CoA) (23). The chemoproteomic analysis identified an argentilactone-interacting, pyruvate dehydrogenase complex component, Pdx1, a protein that is required for anchoring dihydrolipoamide dehydrogenase to the dihydrolipoamide transacetylase core of the pyruvate dehydrogenase complex activity. Depending on the cellular metabolic circumstance, pyruvate can have three fates, namely, acetyl-CoA that is oxidized by TCA, reduced to lactate, or converted to ethanol. The reaction mixtures of the TCA cycle are composed of eight enzymes, some of which are associated with the enzymatic complexes of the mitochondrial matrix. In relation to the proteins of TCA, argentilactone interacted with fumarate hydratase and isocitrate dehydrogenase, all of which were then downregulated by P. lutzii in the presence of argentilactone (14, 16). In addition, others proteins of the TCA cycle, such as citrate synthase, succinate dehydrogenase, and malate dehydrogenase, were also downregulated in the presence of argentilactone. Faced with these findings, we hypothesized that argentilactone could interfere in activity of the TCA modulating the enzymatic activity of important proteins. We investigated the effect of argentilactone on the enzymatic activity of citrate synthase, malate dehydrogenase, and pyruvate dehydrogenase (Fig. 2). In fact, it inhibits the activity of these proteins, which suggests that the pyruvate dehydrogenase complex and TCA are not fully functional.
FIG 2.
Effect of argentilactone on the enzymatic activity of citrate synthase, malate dehydrogenase, and pyruvate dehydrogenase. Enzymatic activity was determined using a protein extract of P. brasiliensis. A statistical analysis was performed using Student's t test, and samples with P values of ≤0.05 were considered statistically significant (*). The tests were performed in triplicate and are arranged on the graph as arithmetic means. The standard deviations between the triplicates of the samples are represented by a bar. AL, argentilactone.
An alternative for Paracoccidioides spp. to obtain acetyl-CoA and, thus, feed the TCA cycle to obtain energy could occur via β-oxidation. In fact, enzymes related to the degradation of lipids, such as 3-ketoacyl-CoA thiolase, interacted with argentilactone and were upregulated by P. lutzii in the presence of argentilactone. In addition, the lipid content was decreased during exposure to argentilactone (14), suggesting that, in response to the inhibition of the glycolytic pathway by argentilactone, Paracoccidioides spp. use β-oxidation to obtain acetyl-CoA.
Importantly, the β-oxidation of short-chain fatty acids exclusively generates acetyl-CoA units, which are intermediates in the TCA cycle, or, in the case of anaplerosis, in the glyoxylate cycle. During the β-oxidation of long-chain fatty acids besides acetyl-CoA, propionyl-CoA is generated and then is converted to pyruvate by the methylcitrate cycle. Argentilactone interacted with 2-methylcitrate synthase. 2-Methylcitrate synthase is the key enzyme of the methylcitrate cycle, and it is responsible for the condensation of propionyl-CoA with oxaloacetate to produce methylcitrate, which undergoes dehydration by 2-methylcitrate dehydratase, generating methyl cis-aconitate that is subsequently rehydrated to methylisocitrate. The cleavage of methylisocitrate occurs through the action of 2-methylcitrate lyase, generating pyruvate and succinate products, intermediates of the TCA cycle. Importantly, in addition to the production of TCA metabolic intermediates, the degradation of propionyl-CoA is essential because of its toxicity, as has been shown in Aspergillus spp. (24). In addition to the interactions of argentilactone-2-methylcitrate synthase identified by chemoproteomic analysis, the proteomic analyses of P. lutzii in the presence of argentilactone showed that 2-methylcitrate dehydratase was upregulated. These data suggest that, in the presence of argentilactone, the methylcitrate cycle was induced, probably to metabolize propionyl-CoA or to produce precursors for the TCA cycle.
Interaction of argentilactone with proteins related to amino acid metabolism.
Amino acids play important roles in various cellular processes, such as protein synthesis, nitrogen and carbon metabolism, and cell signaling. Thus, several pathways concerning amino acids essential to fungal metabolism, such as valine, threonine, tryptophan, and methionine, and nonessential amino acids, such as glutamine, glutamic acid, cysteine, and proline, have been proposed as targets for several compounds with antifungal activity, because some of those reactions are catalyzed by enzymes absent from humans (25). Proteins that catalyze the degradation reactions of valine and leucine, such as 3-hydroxyisobutyryl-CoA hydrolase, 3-hydroxybutyryl-CoA dehydrogenase, and 2-oxoisovalerate dehydrogenase, interacted with argentilactone. Those proteins participate in the pathway that converges in the production of malonyl-CoA, a metabolite that plays an important role in the synthesis and elongation of fatty acids (26).
The amino acid arginine is used as a precursor for the synthesis of proteins, besides compounds such as glutamate, creatine, polyamines, ornithine, and urea (27). Argininosuccinate synthase, an enzyme that catalyzes the condensation of citrulline and aspartame, which form argininosuccinate, the immediate precursor of arginine (28), was upregulated in the presence of argentilactone (14). Arginine is the precursor of 1-pyrroline-5-carboxylate, a substrate of 1-pyrroline-5-carboxylate dehydrogenase, an enzyme that catalyzes the irreversible conversion of 1-pyrroline-5-carboxylate to glutamate. This enzyme interacted with argentilactone, and its expression also was upregulated in the presence of argentilactone (14).
Cysteine dioxygenase is an important metalloenzyme in the regulation of cysteine levels (29), and it was identified by chemoproteomics as an argentilactone-interacting protein. In addition, in the presence of argentilactone, cystathionine synthase and cysteine synthase transcripts, which are important in cysteine biosynthesis and glutathione S-transferase, were downregulated (16), suggesting that argentilactone influences the detoxification system. In fact, transcripts involved in drug detoxification were downregulated in the presence of argentilactone, such as the MFS multidrug transporter, ABC multidrug transporter, trichothecene efflux pump, and benzoate 4-monooxygenase cytochrome P450 (16).
Interaction of argentilactone with stress response proteins.
Stress responses, such as nutrient depletion and osmotic and heat shock, as well as salt and oxidative stress, are essential requirements for fungal survival in the natural environment and during interactions with a host (30). Studies have shown the adaptation of Paracoccidioides spp. to stresses such as osmotic stress (31), oxidative stress (32), nutrient deprivation (33, 34), cell wall stressor agents (35), and drugs (36–39), suggesting prospects for the research and development of new antifungals.
Heat shock proteins (HSPs) are a large family of molecular chaperones that participate in the folding, stabilization, activation, and assembly of several proteins. Although the name HSP has been employed because of their high expression in cells after changes in temperature, their function is not only restricted to thermal responses; some HSPs are constitutively produced and have been associated with several environmental stresses (40). It was also demonstrated that HSPs play an important role during the infection process, because they are present on the cell surface. HSP70 and HSP90 are involved in the HSP90-calcineurin pathway, and they orchestrate the compensatory repair mechanisms of the cell wall in response to stress induced by caspofungin on A. fumigatus, thus being associated with resistance to this antifungal agent. In addition, the inhibition of HSP90, HSP70, and calcineurin may potentiate the effects of caspofungin, thus representing a novel and promising antifungal approach (41).
Argentilactone interacted with several HSPs, which were also upregulated in the presence of argentilactone (16), among them HSP70 and HSP90. In P. brasiliensis, the HSP70 and HSP90 family proteins were upregulated in yeast (pathogenic phase), suggesting its important role in the survival of the fungus for the establishment of infection (42). The reduced expression of HSP90 in P. brasiliensis by antisense RNA technology, the decreased yeast cell viability in culture, and interactions with macrophages, in addition to increased susceptibility to acid pH environments and oxidative stress, suggests the protective role of HSP90 during adaptation to hostile environments (43). In addition, HSP90 is involved in the cell differentiation of P. brasiliensis because it binds and stabilizes calcineurin, a protein associated with the dimorphism process (44).
Our results also showed that several enzymes involved in antioxidant defense and detoxification, such as peroxiredoxin and superoxide dismutase copper (SOD), interacted with argentilactone. In addition, Araújo et al. demonstrated that the enzymatic activity of SOD is increased in the presence of argentilactone and that the SOD mutants were extremely sensitive to this compound, suggesting that argentilactone can induce the production of reactive oxygen species (16). The increase in time-dependent ROS in the presence of argentilactone was investigated and confirmed by flow cytometry (Fig. 3).
FIG 3.
Evaluation of reactive oxygen species production. Flow cytometry analysis was used to measure the reactive oxygen species production. Numbered peaks indicate the autofluorescence of P. brasiliensis (1) and control cells grown in the absence (2) and presence (3) of argentilactone for 3, 6, and 12 h.
The mitochondria, the primary energetic machinery of cells, produce ATP via oxidative phosphorylation through the coordinated action of four enzymatic complexes and ATP synthase. In addition, its contribution to the maintenance of the redox potential of the cell and the generation of ROS is evident (45). Under various conditions, inhibition of electron transfer contributes to increased ROS production. Electron transfer can be inhibited if damage occurs in the electron transport chain or with a low ATP demand, which may result in an increase in NADH/NAD and the consequent generation of superoxide anions from the direct transfer of electrons from NADH via FMN to oxygen (46). Several subunits of ATP synthase interacted with argentilactone, and they also showed downregulation by P. lutzii in the proteome (14). In addition, Araújo et al. observed damage in the mitochondrial membrane potential of P. lutzii that was caused by argentilactone, similar to the effect of antimycin A, the compound inhibitor of the electron chain (16). These results lead us to believe that the possible cause of the increase in ROS production is the collapse in the chain of transport of electrons and the decrease in ATP synthesis.
Interaction of argentilactone with proteins related to cell wall polymer biosynthesis.
The cell wall is a structure that confers rigidity and protection to the cell against osmotic changes and environmental stresses, and it is the first site of interaction with the host cells, thus being considered an important factor of virulence in P. brasiliensis and a potential target for antifungals (47). The cell wall of Paracoccidioides spp. is composed of three types of polymers, glucans, mannans, and chitin, which vary according to their morphological structure (48).
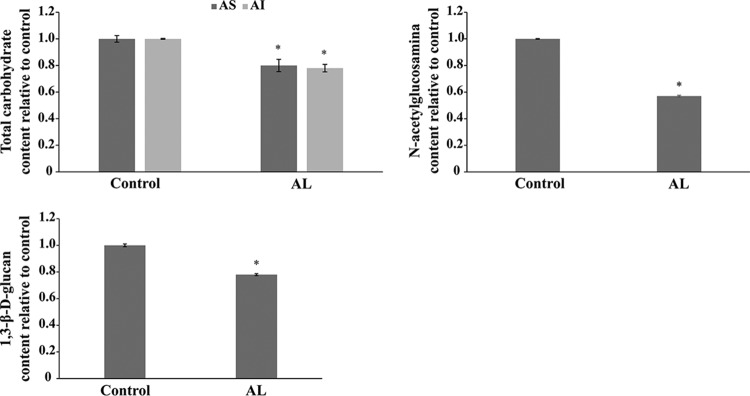
Argentilactone interacted with UDP-N-acetylglucosamine pyrophosphorylase, an enzyme that catalyzes the synthesis of UDP-acetylglucosamine from glycosamine-1-phosphate and UTP, which is directly related to the synthesis of chitin from N-glucosamine (49). This protein was downregulated in the presence of argentilactone by P. lutzii (14). Moreover, 21 transcripts related to the biogenesis of the P. lutzii cell wall were downregulated in the presence of argentilactone (16). Taken together, those data suggest that argentilactone interferes with cell wall metabolism. In fact, this hypothesis was corroborated in this work, because argentilactone reduced the total carbohydrate level of the cell wall and the N-acetylglucosamine and β-1,3-glycan polymers levels (Fig. 4). A similar result was observed for thioridazine (50) and oenotein B (38), compounds that inhibit the growth of P. brasiliensis by acting on cell wall biosynthesis.
FIG 4.
Quantification of cell wall polymers from P. brasiliensis. The levels of total carbohydrates, N-acetylglucosamine, and β-1,3-glycan were measured in the presence of argentilactone. The data were normalized using a control in the absence of argentilactone. AS, soluble alkali; AI, insoluble alkali; AL, argentilactone. A statistical analysis was performed using Student's t test, and samples with P values of ≤0.05 were considered statistically significant (*). The tests were performed in triplicate and are arranged on the graph as arithmetic means. The standard deviations between the triplicates of the samples are represented by a bar.
Conclusions.
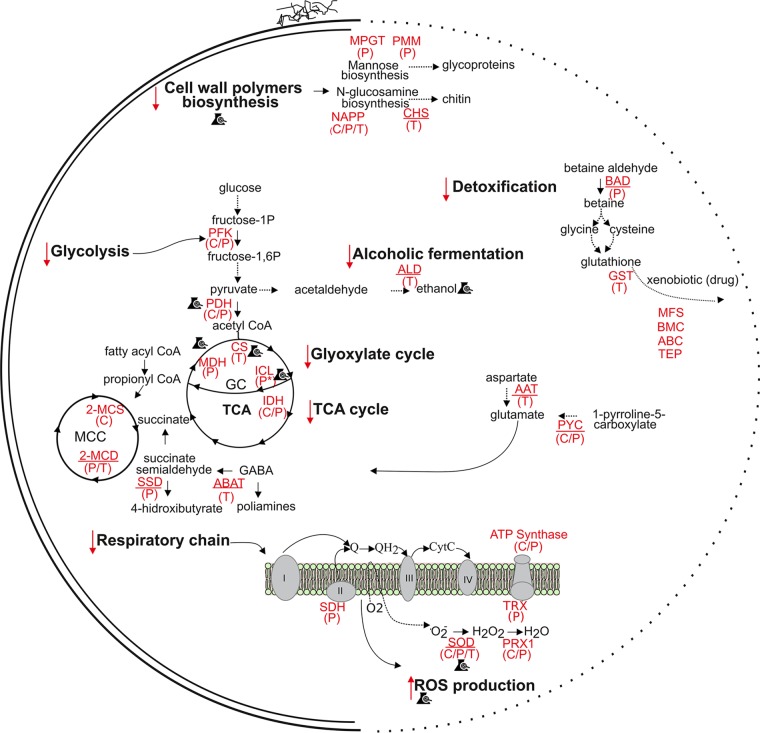
An overview of the metabolic changes in Paracoccidioides spp. in the presence of argentilactone is shown in Fig. 5. Altogether, the results of the transcriptional, proteomics, and chemoproteomics analyses suggest that argentilactone inhibits metabolic pathways such as those of glycolysis, the TCA cycle, and the glyoxylate cycle. In addition, it promotes damage in the electron transport chain, causing energetic depletion. Argentilactone leads to inhibition of polysaccharides synthesis, which is important for cell wall biosynthesis in P. brasiliensis. Additionally, argentilactone induces reactive oxygen species, leading to inhibition of the growth of P. brasiliensis.
FIG 5.
Overview of the metabolic changes in P. brasiliensis in the presence of argentilactone. Proteins interacting with argentilactone and modulated expression in the presence of argentilactone include the following: phosphofructokinase (PFK), pyruvate dehydrogenase (PDH), citrate synthase (CS), isocitrate dehydrogenase (IDH), malate dehydrogenase (MDH), succinate dehydrogenase (SDH), superoxide dismutase (SOD), peroxiredoxin (PRX1), thioredoxin (TRX), ATP synthase, mannose-1-phosphate guanyltransferase (MPGT), phosphomannomutase (PMM), UDP-N-acetylglucosamine pyrophosphorylase (NAPP), betaine aldehyde dehydrogenase (BAD), glutathione S-transferase (GST), alcohol dehydrogenase 1 (ALD), isocitrate lyase (ICL), malate synthase (MLS), aspartate aminotransferase (AAT), 4-aminobutyrate aminotransferase (ABAT), dehydrogenase (SSD), 1-pyrroline-5-carboxylate dehydrogenase (PYC), 2-methylcitrate dehydratase (2-MCD), 2-methylcitrate synthase (2-MCS), acyl-CoA dehydrogenase (ACD), MFS multidrug transporter (MFS), benzoate 4-monooxygenase cytochrome P450 (BMC), ABC multidrug transporter (ABC), and trichothecene efflux pump (TEP). The letters in parentheses indicate the following: C, argentilactone-interacting proteins revealed by chemoproteomics analysis; P, differentially expressed proteins in the presence of argentilactone (14); and T, differentially expressed transcripts in the presence of argentilactone (16). The underlined proteins are upregulated, and proteins that are not underlined are downregulated during proteomics or transcriptomics analysis. The symbol of an Erlenmeyer flask with a checkmark on it refers to the processes that were validated experimentally to confirm the influence of argentilactone (proteomic, transcriptomic, and chemoproteomics analysis). The red down arrows indicate processes downregulated/inhibited by argentilactone. Red up arrows indicate processes upregulated/induced by argentilactone. Asterisks refer to experimental validation of the inhibition of isocitrate lyase by the natural compound argentilactone (15). Dotted arrows summarize processes that have many steps.
MATERIALS AND METHODS
Argentilactone extraction.
The leaves of H. ovalifolia were harvested during the summer of 2016 in relevant areas of the savanna from the Goiânia municipality of Goiás, Brazil. Their essential oil was obtained by hydrodistillation, and argentilactone was isolated and identified as previously described by de Oliveira et al. (9).
Microorganisms and culture conditions.
P. lutzii Pb01, P. brasiliensis Pb18, P. americana Pb02 and Pb03, and P. restrepiensis (PbEPM83, Pb339, and Pb60855 were incubated in liquid Fava-Netto medium (51) (0.3% protease peptone, 1% peptone, 0.5% [wt/vol] meat extract, 0.5% [wt/vol] yeast extract, 1% brain heart infusion [BHI], 4% glucose, 0.5% NaCl, 5 μg/ml gentamicin), pH 7.2, for 48 h at 37°C under shaking. The cells then were centrifuged and washed with phosphate-buffered saline (0.09% Na2HPO4, 0.02% KH2PO4, 0.8% NaCl, 0.02% KCl, pH 7.2), transferred to chemically defined medium RPMI 1640 (Sigma-Aldrich), and incubated for 16 h at 37°C under shaking for the adaptation of the fungal cells.
Determination of the MIC.
The determination of the MIC was performed by following recommendations from the Clinical and Laboratory Standards Institute and adapted according to Paracoccidioides spp., as described by De Paula e Silva et al. (52). In each well of the microplate, dilutions of argentilactone were added, with addition of the fungal suspension to a final concentration of 1 × 105 cells/ml. To determine the maximum fungal growth (positive control), cells were placed in the presence of only RPMI 1640 medium. The plates were maintained at 36°C under agitation for 48 h. Subsequently, 20 μl of the 0.02% resazurin solution was added and incubated for 24 h under the same assay conditions. The MIC was visually determined at the point at which there was no change in the original blue color of the reagent.
Determination of the MFC.
The minimum fungicidal concentration was determined by following Takahagi-Nakaira et al. (53). The Paracoccidioides species cells were incubated with argentilactone, which was serially diluted while maintaining the same concentrations and culture conditions used in the MIC test. For each well, a subculture was performed by transferring 50 μl of the corresponding MIC material to a petri dish containing solid Fava-Netto medium. As a control for fungal growth, an aliquot of the positive control was also plated. The plates were incubated at 37°C for 7 days, with subsequent absorbance readout. The MFC was defined as the lowest concentration at which fungal growth was not visualized.
Cytotoxicity against mammalian cells.
The A549 (ATCC CCL-185), BALB/c 3T3 clone A31 (ATCC CCL-163), and AMJ2-C11 (ATCC CRL-2456) cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich) supplemented with 10% fetal bovine serum (Nutricell, São Paulo, Brazil). A total of 1 × 105 cells/ml were incubated with different concentrations of argentilactone for 48 h at 37°C and 5% CO2. A volume of 20 μl of 0.02% resazurin was added to each microwell and incubated for 24 h. The cytotoxic concentration (CC) was visually determined at the point at which there was no change in the original blue color of the reagent. The following expression determined the selectivity index of the argentilactone: SI = CC/MIC.
Cytoplasmic protein extraction.
P. brasiliensis cells of strain Pb18 (ATCC 32069) were grown in liquid Fava-Netto's medium (0.3% protease peptone, 1% peptone, 0.5% meat extract, 0.5% yeast extract, 1% BHI, 4% glucose, 0.5% NaCl, and 5 μg/ml gentamicin, pH 7.2) for 3 days at 37°C with shaking. The cells were subsequently centrifuged and resuspended in ammonium bicarbonate buffer (57 mM, pH 8.8). Protein extraction was performed by mechanical cell lysis using glass beads. The cell lysate was centrifuged at 5,000 × g for 15 min at 4°C to obtain a supernatant containing protein extract. The integrity of the protein extract was verified by SDS-PAGE and quantified with Bradford reagent (54).
Affinity chromatography.
Initially, a wavelength scanning of the argentilactone solution (1 mg/ml) was performed using wavelengths of 205 to 565 nm to verify the absorption range of argentilactone. For the assay, the wavelength of 265 nm was selected to monitor the absorbance of argentilactone. Argentilactone was immobilized on a chromatographic column containing an Octyl-Sepharose resin (GE Healthcare Life Sciences, Chicago, IL). Two milliliters of resin was added to a chromatographic column and then washed with 10 ml of ultrapure water. The resin was subsequently incubated with 1.5 ml of argentilactone (1 mg/ml) for 30 min, performed 5 times for complete immobilization. After each incubation, the absorbance of the solution exiting the system was measured. The saturation of the resin with argentilactone was verified by comparing the absorbance of argentilactone (wavelength, 265 nm) in the initial solution (1 mg/ml) and that after incubation with the resin.
The Octyl-Sepharose column containing immobilized argentilactone and the column without immobilized argentilactone (the control chromatographic column) were incubated with 6 mg of protein from Pb18 for 1 h on ice. After incubation, the column was washed with 10 ml of 50 mM ammonium bicarbonate buffer solution (pH 8.5). The presence of proteins was monitored during the washing process using the Bradford quantification method and polyacrylamide gel electrophoresis to ensure that all noninteracting proteins were removed. The argentilactone-interacting proteins and control column-interacting proteins were eluted by adding 4 ml of 75% acetonitrile. The samples were lyophilized and conditioned at −20°C until their identification. The sample was acquired from three biological replicates.
Tryptic digestion and data acquisition by nanoUPLC-MSE.
The proteins (argentilactone-interacting proteins and column-interacting proteins) were enzymatically digested and processed as described previously (55). To 50 μg of protein extract, 25 μl of RapiGEST 0.2% (vol/vol) (Waters Corporation, Milford, MA) was added, and the sample was vortexed and incubated at 80°C for 15 min. The reduction of disulfide bonds was subsequently performed by adding 2.5 μl of 100 mM dithiothreitol (DTT) (GE Healthcare), and the sample was incubated at 60°C for 30 min. Thereafter, 2.5 μl of a 300 mM iodoacetamide (GE Healthcare) solution was added for cysteine alkylation, and the sample was incubated in the dark at room temperature for 30 min. Ten μl of trypsin (Promega, Madison, WI, USA), prepared in 50 mM ammonium bicarbonate, next was added at 50 ng/μl. The sample was then digested at 37°C for 16 h. A RapiGEST precipitation was performed by adding 10 μl of 5% (vol/vol) trifluoroacetic acid (TFA) (Sigma-Aldrich, St. Louis, MO), followed by incubation at 37°C for 90 min. The sample was centrifuged at 13,000 × g at 6°C for 30 min. The supernatant was dried in a vacuum concentrator. All of the resulting peptides were suspended in 45 μl of a solution containing 20 mM ammonium formate and 150 fmol/μl PHB (rabbit phosphorylase B) (Waters Corporation). The MassPREP protein was used as an internal standard.
The nanoscale liquid chromatography separation of tryptic peptides was performed using a nanoACQUITY system (Waters Corporation) equipped with a nanoEase 5-μm Bridge BEH130 C18 300-μm by 50-mm precolumn, trap column (5 μm, 180 μm by 20 mm), and BEH130 C18 1.7-μm, 100-μm by 100-mm analytical reversed-phase column (Waters Corporation). The peptides were separated into five fractions, 10.8%, 14%, 16.7%, 20.4%, and 65%, in acetonitrile–0.1% (vol/vol) formic acid, at a flow rate of 2,000 μl/min. The source was operated in the positive ionization mode of nano-ESI(+). To perform external calibration, the masses were corrected based on Glu-fibrinopeptide B (GFP; Sigma-Aldrich) with a molecular mass of 785.8486. A GFP solution in 50% (vol/vol) methanol and 0.1% (vol/vol) formic acid was used at a final concentration of 200 fmol/μl, as delivered by the reference sprayer of the NanoLockSpray source for mass spectrometer. Mass spectrometry analysis was performed using a Synapt G1 mass spectrometer (Waters) equipped with a NanoElectronSpray source and two mass analyzers, a quadrupole, and a time-of-flight (TOF) sensor operating in the V-mode. Data were obtained using the instrument in MSE mode. The sample was analyzed from three technical replicates.
Data processing and protein identification.
MS raw data were processed using ProteinLynx Global Server, version 2.4 (Waters Corporation). The data were subjected to automatic background subtraction, deisotoping, and charge state deconvolution. After processing, each ion comprised an exact mass-retention time that contained the retention time, intensity-weighted average charge, inferred molecular weight based on charge, and m/z. The processed next spectra were searched against Pb18 protein sequences (Broad Institute; http://www.broadinstitute.org/annotation/genome/paracoccidioides_brasiliensis/Multiome.html) together with the reverse sequences.
The mass error tolerance for peptide identification was under 50 ppm. The parameters for protein identification included the following: (i) the detection of at least 2 fragment ions per peptide, (ii) 5 fragments per protein, (iii) the determination of at least 1 peptide per protein, (iv) carbamidomethylation of cysteine as a fixed modification, (v) the phosphorylation of serine, threonine, and tyrosine and the oxidation of methionine (considered variable modifications), (vi) maximum protein mass of 600 kDa, (vii) one missed cleavage site allowed for trypsin, and (viii) maximum false discovery rate (FDR) of 1%. The minimum repeat rate for each protein in all replicates was 2. The tables of peptides and proteins generated by the PLGS were merged, and the data on the dynamic range, peptide detection type, and mass accuracy were calculated for each sample, as previously described using the software MassPivot v1.0.1 (56), FBAT (57), Spotfire v8.0 (TIBCO software), and Microsoft Excel (Microsoft). The argentilactone-interacting proteins were categorized according to the functional category database Munich Information Center for Protein Sequences (MIPS), which is available at geneontology.org. Their biological functions were proposed by the PEDANT genome database (http://pedant.gsf.de/). The NCBI database was employed to annotate the uncharacterized proteins (https://www.ncbi.nlm.nih.gov/).
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (58) with the data set identifier PXD007680 for the sample argentilactone-interacting proteins and PXD008858 for the sample column-interacting proteins.
Enzymatic activity.
The enzymatic activities were explored using a crude extract of P. brasiliensis Pb18 cells.
Malate dehydrogenase.
The malate dehydrogenase activity was measured using a final concentration of 20 mM oxalacetic acid (Sigma-Aldrich), 2 mM NADH (Sigma-Aldrich), 50 mM Tris HCl, pH 7.5, and 36 μg/ml argentilactone. The control sample was prepared without argentilactone. The reaction was initiated by adding 10 μg of protein extract. The decrease in absorbance due to the oxidation of NADH was monitored at 340 nm for 30 min. A unit of malate dehydrogenase was defined as the oxidation of 1 μmol of NADH per minute at 37°C, based on the standard curve of NADH.
Pyruvate dehydrogenase.
The pyruvate dehydrogenase activity was measured by following the manufacturer's instructions (Sigma-Aldrich) at 37°C. Argentilactone at a final concentration of 36 μg/ml and 10 μg of protein extract were added to the assay. The NADH formation was monitored at 450 nm and was quantified using the standard curve. One unit of pyruvate dehydrogenase is the amount of enzyme that generated 1 μmol of NADH per min.
Citrate synthase.
The enzymatic activity of the citrate synthase was determined as previously described by Brock et al. (59). The reaction of 5,5′-dithiobis-2-nitrobenzoate (DTNB) with CoA, forming 5-thio-2-nitrobenzoic acid, gives a yellowish coloration to the reaction mixture detected at 412 nm. Final concentrations of 50 mM Tris-HCl, pH 8.0, 0.2 mM acetyl-CoA (Sigma-Aldrich), 1 mM oxaloacetate (Sigma-Aldrich), 1 mM DTNB (Sigma-Aldrich), and 36 μg/ml argentilactone were used, with the final volume being 150 μl. The sample control was set up and performed without argentilactone. The reaction was started by adding 10 μg of protein extract from Pb18, and the reading was completed after 35 min. One unit was defined as the release of 1 μmol CoA per min per ml at 37°C, based on the standard DTT curve, when varying the concentration from 0.5 mM to 25 mM.
Quantification of cell wall polymers.
The cell wall polymers were quantified as described by Tomazett et al. (35). A total of 1 × 105 cells were inoculated in the presence of 36 μg/ml argentilactone. Cells grown in the absence of the compound were used as the sample control. After 48 h of incubation, the cells were centrifuged and washed twice with 50 mM Tris-HCl buffer, pH 7.5, and then were resuspended in the same buffer with added glass beads for mechanical cell lysis. The cell wall was separated from the cytoplasmic fraction after centrifugation at 10,000 × g for 10 min. The soluble fraction was separated from the insoluble fraction after adding 1 M NaOH and 500 mM sodium borohydrate.
To determine the total carbohydrates, 1 mg of the dry weight of the cell walls was dissolved in 1 ml of 75% sulfuric acid and the mixture was stirred for 30 min. Subsequently, 30 μl of 5% phenol was added and the sample was incubated at 90°C for 5 min. The reading was performed using a wavelength of 490 nm. Quantification was performed based on the standard glucose curve.
To quantify N-acetylglucosamine, 2 mg of the cell wall sample was dissolved in 1 ml of 8 M HCl, followed by incubation at 100°C for 4 h. Thereafter, the solution was neutralized with 900 μl of 8 M NaOH. To the neutralized extract, 100 μl of 3 M Na2CO3 solution was added, followed by incubation for 20 min at 100°C. Subsequently, 700 μl of ethanol and 100 μl of Ehrlich's reagent were added. The reaction was quantified at 520 nm. The quantification of N-acetylglucosamine was performed in comparison with the standard curve of glycosamine.
To quantify the 1,3-β-d-glycan, 1 mg of the insoluble fraction was added to 200 μl of Tris DTT buffer (50 mM Tris HCl, pH 7.5, and 10 mM DTT) and 2 μl of glucanase, followed by incubation for 12 h at 37°C. The mixture was centrifuged, and the supernatant was collected for quantification. In one tube, 50 μl of the digestion was mixed with 1.5 ml of the 4-hydroxybenzoic acid hydrazide solution, and then the sample was incubated for 10 min at 100°C. Two hundred μl of the mixture was dispensed into the microplate, and the reading was performed at a wavelength of 410 nm. The quantification of 1,3-β-d-glycan was based on the results of the standard glucose curve. Laminarin was used as a negative control.
Evaluation of reactive oxygen species production.
A total of 1 × 105 cells were treated with 36 μg/ml of argentilactone for 3, 6, and 12 h at 37°C and then centrifuged at 5,000 × g and incubated with 20 μM 2′,7′-dichlorofluorescein diacetate for 30 min at room temperature under shelter from light. The cells next were washed and resuspended in 1 ml of phosphate-buffered saline. The fluorescence intensity was determined by flow cytometry using an Accuri C6, FL1-H channel (BD Biosciences, Ann Arbor, MI). A total of 10,000 events were considered for analysis.
Statistical analysis.
The experiments were performed in triplicate and were arranged on the graph as arithmetic means. Statistical analysis was performed using Student's t test, and samples with a P value of ≤0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work, performed at Universidade Federal de Goiás, was supported by MCTI/CNPq (Ministério da Ciência e Tecnologia/Conselho Nacional de Desenvolvimento Científico e Tecnológico), FNDCT (Fundo Nacional de Desenvolvimento Científico e Tecnológico), FAPEG (Fundação de Amparo à Pesquisa do Estado de Goiás), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), FINEP (Financiadora de Estudos e Projetos), PRONEX (Programa de Apoio a Núcleos de Excelência), and INCT-IF (Instituto Nacional de Ciência e Tecnologia para Inovação Farmacêutica).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00737-18.
REFERENCES
- 1.Silva-Vergara ML, Rocha IH, Vasconcelos RR, Maltos AL, de Freitas Neves F, de Almeida Silva Teixeira L, Mora DJ. 2014. Central nervous system paracoccidioidomycosis in an AIDS patient: case report. Mycopathologia 177:137–141. doi: 10.1007/s11046-014-9729-5. [DOI] [PubMed] [Google Scholar]
- 2.Shikanai-Yasuda MA, Mendes RP, Colombo AL, de Queiroz-Telles F, Kono ASG, Paniago AMM, Nathan A, do Valle ACF, Bagagli E, Benard G, Ferreira MS, de Teixeira MM, Silva-Vergara ML, Pereira RM, de Cavalcante RS, Hahn R, Durlacher RR, Khoury Z, de Camargo ZP, Moretti ML, Martinez R. 2017. Brazilian guidelines for the clinical management of paracoccidioidomycosis. Rev Soc Brasil Med Trop 50:715–740. doi: 10.1590/0037-8682-0230-2017. [DOI] [PubMed] [Google Scholar]
- 3.Shikanai-Yasuda MA. 2015. Paracoccidioidomycosis treatment. Rev Inst Med Trop São Paulo 57:31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Travassos LR, Taborda CP, Colombo AL. 2008. Treatment options for paracoccidioidomycosis and new strategies investigated. Expert Rev Anti Infect Ther 6:251–262. doi: 10.1586/14787210.6.2.251. [DOI] [PubMed] [Google Scholar]
- 5.Sano EE, Rosa R, Brito JLS, Ferreira LG. 2010. Land cover mapping of the tropical savanna region in Brazil. Environ Monit Assess 166:113–124. doi: 10.1007/s10661-009-0988-4. [DOI] [PubMed] [Google Scholar]
- 6.Violante IMP, Hamerski L, Garcez WS, Batista AL, Chang MR, Pott VJ, Garcez FR. 2012. Antimicrobial activity of some medicinal plants from the cerrado of the central-western region of Brazil. Braz J Microbiol 43:1302–1308. doi: 10.1590/S1517-83822012000400009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fragoso-Serrano M, Gibbons S, Pereda-Miranda R. 2005. Anti-staphylococcal and cytotoxic compounds from Hyptis pectinata. Planta Medica 71:278–280. doi: 10.1055/s-2005-837831. [DOI] [PubMed] [Google Scholar]
- 8.Bispo MD, Mourão RHV, Franzotti EM, Bomfim KBR, Arrigoni-Blank MF, Moreno MPN, Marchioro M, Antoniolli AR. 2001. Antinociceptive and antiedematogenic effects of the aqueous extract of Hyptis pectinata leaves in experimental animals. J Ethnopharmacol 76:81–86. doi: 10.1016/S0378-8741(01)00172-6. [DOI] [PubMed] [Google Scholar]
- 9.de Oliveira CMA, do Silva M, Kato L, da Silva CC, Ferreira HD, Souza LKH. 2004. Chemical composition and antifungal activity of the essential oil of Hyptis ovalifolia Benth (Lamiaceae). J Braz Chem Soc 15:756–759. doi: 10.1590/S0103-50532004000500023. [DOI] [Google Scholar]
- 10.de Fatima Â, Kohn LK, Antônio MA, de Carvalho JE, Pilli RA. 2004. Enantioselective syntheses of (R)- and (S)-argentilactone and their cytotoxic activities against cancer cell lines. Bioorg Med Chem 12:5437–5442. doi: 10.1016/j.bmc.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 11.Waechter A, Ferreira M, Fournet A, de Arias A, Nakayama H, Torres S, Hocquemiller R, Cavé A. 1997. Experimental treatment of cutaneous leishmaniasis with argentilactone isolated from Annona haematantha. Planta Medica 63:433–435. doi: 10.1055/s-2006-957728. [DOI] [PubMed] [Google Scholar]
- 12.de Fatima A, Modolo L, Conegero L, Pilli R, Ferreira C, Kohn L, de Carvalho J. 2006. Styryl lactones and their derivatives: biological activities, mechanisms of action and potential leads for drug design. Curr Med Chem 13:3371–3384. doi: 10.2174/092986706779010298. [DOI] [PubMed] [Google Scholar]
- 13.do Prado RS, Alves RJ, de Oliveira CMA, Kato L, da Silva RA, Quintino GO, do Desterro Cunha S, de Almeida Soares CM, Pereira M. 2014. Inhibition of Paracoccidioides lutzii Pb01 isocitrate lyase by the natural compound argentilactone and its semi-synthetic derivatives. PLoS One 9:e94832. doi: 10.1371/journal.pone.0094832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prado RS, Bailão AM, Silva LC, de Oliveira CMA, Marques MF, Silva LP, Silveira-Lacerda EP, Lima AP, Soares CM, Pereira M. 2015. Proteomic profile response of Paracoccidioides lutzii to the antifungal argentilactone. Front Microbiol 6:616. doi: 10.3389/fmicb.2015.00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raida M. 2011. Drug target deconvolution by chemical proteomics. Curr Opin Chem Biol 15:570–575. doi: 10.1016/j.cbpa.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 16.Araújo FS, Coelho LM, do Silva LC, da Silva Neto BR, Parente-Rocha JA, Bailão AM, de Oliveira CMA, da Fernandes GR, Hernández O, Ochoa JGM, de Soares CMA, Pereira M. 2016. Effects of argentilactone on the transcriptional profile, cell wall and oxidative stress of Paracoccidioides spp. PLoS Negl Trop Dis 10:e0004309. doi: 10.1371/journal.pntd.0004309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes J, Rees S, Kalindjian S, Philpott K. 2011. Principles of early drug discovery: principles of early drug discovery. Br J Pharmacol 162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen C, West GM, Geoghegan KF. 2017. Emerging methods in chemoproteomics with relevance to drug discovery, p 11–22. In Kasid U, Clarke R (ed), Cancer gene networks. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 19.Vallejo MC, Nakayasu ES, Matsuo AL, Sobreira TJP, Longo LVG, Ganiko L, Almeida IC, Puccia R. 2012. Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: comparative analysis with other pathogenic fungi. J Proteome Res 11:1676–1685. doi: 10.1021/pr200872s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbosa MS, Báo SN, Andreotti PF, de Faria FP, Felipe MSS, dos Santos Feitosa L, Mendes-Giannini MJS, de Soares CMA. 2006. Glyceraldehyde-3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect Immun 74:382–389. doi: 10.1128/IAI.74.1.382-389.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcos CM, de Oliveira HC, da Silva JF, Assato PA, Fusco-Almeida AM, Mendes-Giannini MJS. 2014. The multifaceted roles of metabolic enzymes in the Paracoccidioides species complex. Front Microbiol 5:719. doi: 10.3389/fmicb.2014.00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavares AHFP, Silva SS, Dantas A, Campos ÉG, Andrade RV, Maranhão AQ, Brígido MM, Passos-Silva DG, Fachin AL, Teixeira SMR, Passos GAS, Soares CMA, Bocca AL, Carvalho MJA, Silva-Pereira I, Felipe MSS. 2007. Early transcriptional response of Paracoccidioides brasiliensis upon internalization by murine macrophages. Microbes Infect 9:583–590. doi: 10.1016/j.micinf.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 23.Patel MS, Nemeria NS, Furey W, Jordan F. 2014. The pyruvate dehydrogenase complexes: structure-based function and regulation. J Biol Chem 289:16615–16623. doi: 10.1074/jbc.R114.563148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubey MK, Broberg A, Jensen DF, Karlsson M. 2013. Role of the methylcitrate cycle in growth, antagonism and induction of systemic defence responses in the fungal biocontrol agent Trichoderma atroviride. Microbiology 159:2492–2500. doi: 10.1099/mic.0.070466-0. [DOI] [PubMed] [Google Scholar]
- 25.Jastrzębowska K, Gabriel I. 2015. Inhibitors of amino acids biosynthesis as antifungal agents. Amino Acids 47:227–249. doi: 10.1007/s00726-014-1873-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WN, Tan KY. 2013. Malonate uptake and metabolism in Saccharomyces cerevisiae. Appl Biochem Biotechnol 171:44–62. doi: 10.1007/s12010-013-0334-8. [DOI] [PubMed] [Google Scholar]
- 27.Wu G, Morris SM. 1998. Arginine metabolism: nitric oxide and beyond. Biochem J 336:1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haines RJ, Pendleton LC, Eichler DC. 2011. Argininosuccinate synthase: at the center of arginine metabolism. Int J Biochem Mol Biol 2:8–23. [PMC free article] [PubMed] [Google Scholar]
- 29.Stipanuk MH, Ueki I, Dominy JE, Simmons CR, Hirschberger LL. 2009. Cysteine dioxygenase: a robust system for regulation of cellular cysteine levels. Amino Acids 37:55–63. doi: 10.1007/s00726-008-0202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kroll K, Pähtz V, Kniemeyer O. 2014. Elucidating the fungal stress response by proteomics. J Proteomics 97:151–163. doi: 10.1016/j.jprot.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 31.da Rodrigues LN, de Brito WA, Parente AFA, Weber SS, Bailão AM, Casaletti L, Borges CL, de Soares CMA. 2016. Osmotic stress adaptation of Paracoccidioides lutzii, Pb01, monitored by proteomics. Fungal Genet Biol 95:13–23. doi: 10.1016/j.fgb.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 32.de Arruda Grossklaus D, Bailão AM, Vieira Rezende TC, Borges CL, de Oliveira MAP, Parente JA, de Almeida Soares CM. 2013. Response to oxidative stress in Paracoccidioides yeast cells as determined by proteomic analysis. Microbes Infect 15:347–364. doi: 10.1016/j.micinf.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 33.de Lima PS, Casaletti L, Bailão AM, de Vasconcelos ATR, da Fernandes GR, de Soares CMA. 2014. Transcriptional and proteomic responses to carbon starvation in paracoccidioides. PLoS Negl Trop Dis 8:e2855. doi: 10.1371/journal.pntd.0002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parente AFA, de Rezende TCV, de Castro KP, Bailão AM, Parente JA, Borges CL, Silva LP, de Soares CMA. 2013. A proteomic view of the response of Paracoccidioides yeast cells to zinc deprivation. Fungal Biol 117:399–410. doi: 10.1016/j.funbio.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Tomazett PK, da Castro N, Lenzi SHL, de Almeida Soares CM, Pereira M. 2011. Response of Paracoccidioides brasiliensis Pb01 to stressor agents and cell wall osmoregulators. Fungal Biol 115:62–69. doi: 10.1016/j.funbio.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 36.do Carmo Silva L, Tamayo Ossa DP, da Castro SVC, Bringel Pires L, Alves de Oliveira CM, Conceição da Silva C, Coelho NP, Bailão AM, Parente-Rocha JA, de Soares CM, Ruiz OH, Ochoa JGM, Pereira M. 2015. Transcriptome profile of the response of Paracoccidioides spp. to a camphene thiosemicarbazide derivative. PLoS One 10:e0130703. doi: 10.1371/journal.pone.0130703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.da Neto BR, Carvalho PF, Bailão A, Martins W, de Almeida Soares C, Pereira M. 2014. Transcriptional profile of Paracoccidioides spp. in response to itraconazole. BMC Genomics 15:254. doi: 10.1186/1471-2164-15-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zambuzzi-Carvalho P, Tomazett P, Santos S, Ferri P, Borges C, Martins W, de Almeida Soares C, Pereira M. 2013. Transcriptional profile of Paracoccidioides induced by oenothein B, a potential antifungal agent from the Brazilian Cerrado plant Eugenia uniflora. BMC Microbiol 13:227. doi: 10.1186/1471-2180-13-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zambuzzi-Carvalho PF, Fernandes AG, Valadares MC, de Tavares PM, Nosanchuk JD, de Almeida Soares CM, Pereira M. 2015. Transcriptional profile of the human pathogenic fungus Paracoccidioides lutzii in response to sulfamethoxazole. Med Mycol 53:477–492. doi: 10.1093/mmy/myv011. [DOI] [PubMed] [Google Scholar]
- 40.Whitley D, Goldberg SP, Jordan WD. 1999. Heat shock proteins: a review of the molecular chaperones. J Vasc Surg 29:748–751. doi: 10.1016/S0741-5214(99)70329-0. [DOI] [PubMed] [Google Scholar]
- 41.Lamoth F, Juvvadi PR, Steinbach WJ. 2014. Heat shock protein 90 (Hsp90): a novel antifungal target against Aspergillus fumigatus. Crit Rev Microbiol 42:310–321. [DOI] [PubMed] [Google Scholar]
- 42.Nicola AM, Andrade RV, Dantas AS, Andrade PA, Arraes FB, Fernandes L, Silva-Pereira I, Felipe M. 2008. The stress responsive and morphologically regulated hsp90 gene from Paracoccidioides brasiliensis is essential to cell viability. BMC Microbiol 8:158. doi: 10.1186/1471-2180-8-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamayo D, Muñoz JF, Torres I, Almeida AJ, Restrepo A, McEwen JG, Hernández O. 2013. Involvement of the 90kDa heat shock protein during adaptation of Paracoccidioides brasiliensis to different environmental conditions. Fungal Genet Biol 51:34–41. doi: 10.1016/j.fgb.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Matos TGF, Morais FV, Campos CBL. 2013. Hsp90 regulates Paracoccidioides brasiliensis proliferation and ROS levels under thermal stress and cooperates with calcineurin to control yeast to mycelium dimorphism. Med Mycol 51:413–421. doi: 10.3109/13693786.2012.725481. [DOI] [PubMed] [Google Scholar]
- 45.Adam-Vizi V, Chinopoulos C. 2006. Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends Pharmacol Sci 27:639–645. doi: 10.1016/j.tips.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 46.Murphy MP. 2009. How mitochondria produce reactive oxygen species. Biochem J 417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amaral AC, Fernandes L, Galdino AS, Felipe MSS, de Soares CMA, Pereira M. 2005. Therapeutic targets in Paracoccidioides brasiliensis: post-transcriptome perspectives. Genet Mol Res 4:430–449. [PubMed] [Google Scholar]
- 48.Puccia R, Vallejo MC, Matsuo AL, Longo LVG. 2011. The Paracoccidioides cell wall: past and present layers toward understanding interaction with the host. Front Microbiol 2:257. doi: 10.3389/fmicb.2011.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bulik DA, Olczak M, Lucero HA, Osmond BC, Robbins PW, Specht CA. 2003. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot Cell 2:886–900. doi: 10.1128/EC.2.5.886-900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jabes DL, de Freitas Oliveira AC, Alencar VC, Menegidio FB, Reno DLS, Santos DS, Barbosa DA, Vilas Boas RO, de Oliveira Rodrigues Cunha RL, Rodrigues T, Costa de Oliveira R, Nunes LR. 2016. Thioridazine inhibits gene expression control of the cell wall signaling pathway (CWI) in the human pathogenic fungus Paracoccidioides brasiliensis. Mol Genet Genomics 291:1347–1362. doi: 10.1007/s00438-016-1184-1. [DOI] [PubMed] [Google Scholar]
- 51.Fava-Netto C. 1955. Estudos quantitativos sobre a fixação de complemento na blastomicose sul-americana, com antigeno polissacarídico. Arq Cir Clin Exp 18:197–254. [PubMed] [Google Scholar]
- 52.de Paula e Silva ACA, Oliveira HC, Silva JF, Sangalli-Leite F, Scorzoni L, Fusco-Almeida AM, Mendes-Giannini MJS. 2013. Microplate alamarBlue assay for paracoccidioides susceptibility testing. J Clin Microbiol 51:1250–1252. doi: 10.1128/JCM.02914-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahagi-Nakaira E, Sugizaki M, Peraçoli M. 2009. Microdilution procedure for antifungal susceptibility testing of Paracoccidioides brasiliensis to amphotericin B and itraconazole. J Venom Anim Toxins Incl Trop Dis 15:718–731. doi: 10.1590/S1678-91992009000400010. [DOI] [Google Scholar]
- 54.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 55.Murad AM, Souza GHMF, Garcia JS, Rech EL. 2011. Detection and expression analysis of recombinant proteins in plant-derived complex mixtures using nanoUPLC-MSE. J Sep Sci 34:2618–2630. doi: 10.1002/jssc.201100238. [DOI] [PubMed] [Google Scholar]
- 56.Murad AM, Rech EL. 2012. NanoUPLC-MSE proteomic data assessment of soybean seeds using the UniProt database. BMC Biotechnol 12:82. doi: 10.1186/1472-6750-12-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. 2004. PBAT: tools for family-based association studies. Am J Hum Genet 74:367–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu Q-W, Wang R, Hermjakob H. 2016. 2016 Update of the PRIDE database and its related tools. Nucleic Acids Res 44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brock M, Buckel W. 2004. On the mechanism of action of the antifungal agent propionate. Eur J Biochem 271:3227–3241. doi: 10.1111/j.1432-1033.2004.04255.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.