Abstract
Implementation of Biofire FilmArray Blood Culture Identification Multiplex PCR panel (BCID) at a cancer hospital was associated with reduced time to appropriate antimicrobial therapy. Additional reductions were not observed when BCID was coupled with antimicrobial stewardship intervention.
Keywords: antimicrobial stewardship, hematopoietic stem cell transplant, immunocompromised host, molecular-based rapid diagnostic testing, neutropenic fever
Molecular-based rapid diagnostic testing (mRDT) has the ability to decrease time between microbiologic sampling and organism identification to guide and direct antimicrobial therapy. Use of multiplex polymerase chain reaction (PCR) assays for bloodstream infections (BSIs) has been associated with shorter durations of empiric broad-spectrum antimicrobial therapy, reduced time to appropriate antimicrobial therapy, and shorter durations of inpatient stays for isolates considered blood culture contaminants [1, 2]. Utilization of mass spectroscopy for the identification of BSIs has demonstrated not only a benefit in mortality, but also a potential cost savings for the institution when results are sent to and acted upon by an Antimicrobial Stewardship Program (ASP) [3–5]. Recent Infectious Diseases Society of America (IDSA) guidelines for implementing an ASP recommend rapid diagnostic testing on blood cultures with ASP support; however, many ASPs may not have resources to employ all recommended interventions. Evidence supporting which clinical areas to target ASP involvement is needed [6].
The impact of mRDT in patients with cancer or other immunocompromising conditions is unknown. Many infections in immunocompromised hosts require longer durations of therapy, have guideline recommendations for extended durations of broad-spectrum empiric antimicrobials, and lack comprehensive evaluations of therapeutic selection and duration [7, 8]. These factors complicate antimicrobial de-escalation and may limit the benefit of mRDT utilization. Yet, with rising rates of Clostridium difficile infection, drug-resistant infections, and antimicrobial-related adverse drug events, antimicrobial stewardship (AS) has the potential to improve patient outcomes while preserving antimicrobials for this vulnerable population [9–13]. The primary objective of this study was to evaluate the impact of a multiplex PCR assay for BSIs on antimicrobial therapy in immunocompromised patients at a cancer hospital with and without AS intervention.
METHODS
Study Design
This 3-arm pre/post intervention study evaluating a multiplex PCR-based blood culture identification (BCID) panel with and without AS intervention was performed at the University of Utah Huntsman Cancer Institute between July 2014 and January 2017. Biofire FilmArray Multiplex PCR BCID was implemented in November 2014 without AS intervention, followed by AS notification of results and active guidance on therapy communicated to providers in October 2016. Patients were evaluated immediately before (PRE) and after (POST) BCID implementation in 2014 for 100 days in each arm as a convenience sample. The effect of AS intervention coupled with BCID (POST-AS) was evaluated for 100 days after a formal AS protocol for reviewing all new BCID results was implemented in 2016. During this time, there were no changes in institutional protocols for neutropenic fever management, changes to the formulary, or significant changes in rates of antimicrobial resistance that would have affected local prescribing practices. The University of Utah Institutional Review Board reviewed and approved the study protocol.
All inpatients with a first positive blood culture in an aerobic bottle were eligible for inclusion. If multiple positive blood cultures were identified during an admission, only the first positive blood culture was included for the assessment. Patients were excluded if they were younger than 18 years of age at the time of BSI, not admitted at the time of BSI identification, transferred from an outside hospital with an active BSI, discharged before the clearance of BSI, had Gram stain–positive but culture-negative infections, had BSI organisms identified only in anaerobic bottles, or if the BCID did not identify an organism or was not performed in BCID groups.
All microbiological analysis was completed at the Associated Regional University Pathologists (ARUP) Laboratory. Blood cultures were incubated using a BACTEC automated blood culture system for up to 96 hours. All positive growth underwent Gram staining, with critical results called to inpatient medical teams. In the POST and POST-AS arms, an aliquot from the aerobic bottles with positive growth was run on the FilmArray BCID for organism identification. Results from the FilmArray BCID were called to the inpatient medical teams in both the POST and POST-AS cohorts. An aliquot from all aerobic or anaerobic bottles with positive growth was streaked on solid media via a standard laboratory protocol. Organisms grown from media were identified via matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) mass spectrometry and underwent antimicrobial susceptibility testing using BD Phoenix.
The ASP in the POST-AS arm evaluated all positive BCID results. Theradoc alerts for all positive blood cultures were sent to AS team members via email. These alerts listed patient information and the organism identified. Alerts did not include antimicrobial recommendations, and each case required chart review by the ASP. Clinical pharmacists could sign up to receive these alerts, but the alerts did not contain antimicrobial recommendations, and no specific education or training was provided on interpretation or recommendations for each alert. The ASP consisted of an infectious diseases (ID) physician and an ID pharmacy resident who reviewed cases together. Antimicrobial therapy for all patients with a BCID result were evaluated by the ASP from 0800 to 1700 Monday through Friday, with recommendations for antimicrobial selection, duration of therapy, and ID consultation, when indicated, relayed directly to the primary medical team via telephone or in person. In addition, a progress note was left in the electronic medical record to document the BCID result and AS recommendations. Any BCID result posted between 1700 and 0759 was evaluated by ASP team members during their next scheduled shift.
Data Collection
All data were collected via retrospective chart review, including patient demographics, laboratory, microbiology, antimicrobial use, and outcomes.
Outcomes and Definitions
The primary outcome was time to appropriate antimicrobial therapy, defined as the narrowest-spectrum antimicrobial to treat an infection taking into consideration evidence-based treatment guidelines, patient allergies, and the need for polymicrobial coverage. Based on this definition, broad-spectrum therapy was deemed appropriate in neutropenic patients until neutrophil count recovery. Time to appropriate therapy was calculated from time of Gram stain result to order time of appropriate antimicrobial therapy based upon identified organism, source of infection, and the immunocompromised status of the patient. Appropriate antibiotics for microbiological contaminants included removal of antibiotic therapy or guideline-based treatment for neutropenic fever if appropriate. Immunocompromising conditions were defined as the following: (1) receiving corticosteroids for 3 months or more at a dose equivalent to prednisone 20 mg daily immediately before the BSI, (2) active hematological malignancy or solid tumor, (3) history of hematopoietic stem cell transplant or solid organ transplant, or (4) absolute neutrophil count (ANC) <500 cells/mm3 at any time 30 days before the BSI. Secondary outcomes included the time from Gram stain to organism identification, percentage of patients ordered appropriate antimicrobials at any time after blood cultures were collected, in-hospital mortality, 30-day all-cause mortality, and 30-day readmission rates.
Statistical Analysis
Descriptive statistics of patient demographics and microbiology, including mean and standard deviation for continuous variables and counts and percentages for categorical variables, were summarized. Univariate comparisons of categorical outcomes were assessed by chi-square test. Median values were reported for continuous outcomes, and nonparametric testing (Kruskal-Wallis) was performed due to the non-normal distribution of the data. All statistical tests were evaluated at an alpha level of .05.
A multivariate analysis was employed to analyze the effect of BCID with and without AS intervention on the time to appropriate antimicrobial therapy, controlling for a set of covariates. There was a left shift in data distribution, with 25% of patients receiving appropriate antibiotics before Gram stain result. To account for left censoring of the outcome variable, a Tobit regression model was employed. Demographic characteristics including age and gender and clinical characteristics including ANC, multiplex PCR BCID with and without AS, admitting medical service, Charlson Comorbidity Index, immunocompromised status, patients seen by an ID consult service, methicillin-resistant Staphylococcus aureus (MRSA), methicillin-susceptible Staphylococcus aureus (MSSA), or Pseudomonas aeruginosa infection, surgical encounters, and patients with any Charlson comorbidity or cancer were included as control variables in the model.
RESULTS
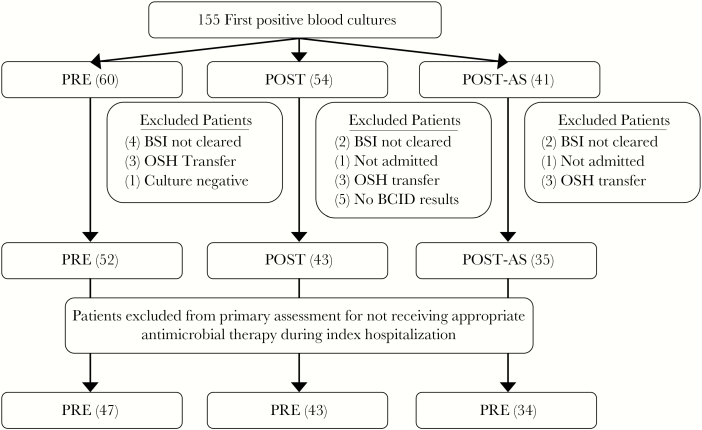
One hundred fifty-five patients were identified with unique first positive blood cultures during the study (Figure 1). Twenty-five patients were excluded (8 PRE, 11 POST, 6 POST-AS). Eight patients were discharged before documented microbiologic clearance (4 PRE, 2 POST, 2 POST-AS), 9 patients were transferred from an outside hospital with an active BSI (3 PRE, 3 POST, 3 POST-AS), 2 patients were not directly admitted after blood draw for culture (1 POST, 1 POST-AS), 1 patient in the PRE group had a Gram-positive stain but culture-negative infection, and 5 patients had infections in the POST cohort with either no organisms detected by PCR or no PCR performed.
Figure 1.
Flow diagram for participants included in the study. Unadjusted mean time to appropriate antimicrobial therapy using a 1-way analysis of variance test as a comparison: no statistical difference in uncontrolled means between PRE, POST, and POST-AS groups (44 hours, 25 hours, 26 hours; P = .069) at an alpha = .05 level. Abbreviations: BCID, multiplex polymerase chain reaction–based blood culture identification; BSI, bloodstream infection; PRE, before BCID implementation; OSH, outside hospital; POST, after BCID implementation; POST-AS, BCID coupled with antimicrobial stewardship.
Patient Demographics
The mean age and sex of patients within each cohort were similar (Table 1). There were differences between groups in the number of patients with leukemia (PRE 37%, POST 19%, POST-AS 14%; P = .03), breast cancer (0%, 12%, 5.7%; P = .04), unclear baseline infection source (14%, 5%, 29%; P = .01), and immunosuppression (65%, 72%, 91%; P = .02). The mean Charlson Comorbidity Index score, number of patients with BSI while neutropenic, number of patients managed by an ID consult service, and patients with an intensive care unit (ICU) encounter during the index hospital stay were similar between the 3 groups.
Table 1.
Patient Demographics
| PRE (n = 52), No. (%)a |
POST (n = 43), No. (%)a |
POST-AS (n = 35), No. (%)a |
P Valueb | |
|---|---|---|---|---|
| Age, mean (SD), y | 62 (13) | 57 (17) | 58 (16) | .439 |
| Female | 18 (35) | 21 (49) | 18 (51) | .217 |
| Any Charlson comorbidity | 51 (98) | 40 (93) | 34 (97) | .416 |
| Charlson Comorbidity Score, mean (SD) | 5.6 (2.5) | 5.5 (2.8) | 6.3 (2.7) | .453 |
| Malignancy characteristics | ||||
| Leukemia | 19 (37) | 8 (19) | 5 (14) | .033 |
| Lymphoma | 7 (14) | 5 (12) | 10 (29) | .096 |
| Breast cancer | 0 | 5 (12) | 2 (5.7) | .044 |
| Any cancer | 41 (79) | 34 (79) | 33 (94) | .118 |
| ANC <500 cells/mm3 | 14 (27) | 15 (35) | 10 (29) | .685 |
| Immunosuppressed | 34 (65) | 31 (72) | 32 (91) | .021 |
| Transplant type | ||||
| Allogeneic | 8 (15) | 2(4.7) | 3 (8.6) | .210 |
| Autologous | 2 (3.9) | 3 (7.0) | 4 (11) | .393 |
| Medical treatment service | ||||
| ICU admission | 9 (17) | 10 (23) | 7 (20) | .833 |
| Oncology admission | 15 (29) | 16 (37) | 13 (37) | |
| Hematology admission | 12 (23) | 6 (14) | 7 (20) | |
| BMT admission | 8 (15) | 5 (12) | 6 (17) | |
| ICU encounter | 16 (31) | 19 (44) | 11 (31) | .336 |
| Surgical encounter | 6 (12) | 1 (2.3) | 0 | .190 |
| Infection characteristics | ||||
| Line-related source | 18 (35) | 14 (33) | 6 (17) | .180 |
| Urinary source | 7 (14) | 11 (26) | 4 (11) | .175 |
| Respiratory source | 0 | 3 (7.0) | 1 (2.9) | .146 |
| Abdomen source | 18 (35) | 6 (14) | 9 (26) | .070 |
| Unclear source | 7 (14) | 2 (4.7) | 10 (29) | .012 |
| ID Ccnsult | 22 (42) | 18 (42) | 14 (40) | .976 |
Abbreviations: ANC, absolute neutrophil count; BCID, multiplex polymerase chain reaction–based blood culture identification; BMT, blood and marrow transplant; ICU, intensive care unit; ID, infectious diseases; PRE, before BCID implementation; POST, after BCID implementation; POST-AS, BCID coupled with antimicrobial stewardship.
aUnless otherwise specified.
bKruskal-Wallis test (continuous variables) or chi-square test (categorical variables).
Microbiology
The majority of infections based on final organism identification were with Gram-positive bacteria (53%), with an even distribution of organisms between study arms (Table 2). Forty-one of 72 (57%) Gram-positive infections were from Staphylococcus species, with 3 MRSA, 25 coagulase-negative Staphylococcus species, and 13 MSSA or Staphylococcus lugdunensis isolated. Three of 13 (23%) Enterococcus species infections were from Enterococcus faecium. Gram-negative bacteria caused fewer infections overall but had the highest single pathogen incidence, with Escherichia coli identified in 24% of blood cultures. Pseudomonas aeruginosa infections were more prevalent in the POST than PRE or POST-AS groups (0%, 6.7%, 0%; P = .05).
Table 2.
Microbiologya
| PRE (n = 54 organisms), No. (%) |
POST (n = 45 organisms), No. (%) |
POST-AS (n = 37 organisms), No. (%) |
P Valueb | |
|---|---|---|---|---|
| Gram positivec | 27 (50) | 24 (52) | 21 (57) | .816 |
| Coag.-negative Staphylococcus spp. | 6 (11) | 9 (20) | 10 (27) | .148 |
| Enterococcus faecalis | 3 (5.6) | 5 (11) | 0 | .103 |
| Enterococcus faecium | 3 (5.6) | 0 | 0 | .097 |
| MRSA | 2 (3.7) | 1 (2.2) | 0 | .498 |
| MSSA | 4 (7.4) | 5 (11) | 3 (8.1) | .798 |
| Streptococcus anginosus group | 1 (1.9) | 0 | 2 (5.4) | .246 |
| Streptococcus pneumoniae | 0 | 1 (2.2) | 2 (5.4) | .226 |
| Streptococcus viridans group | 2 (3.7) | 0 | 1 (2.7) | .445 |
| Gram negatived | 26 (48) | 21 (47) | 15 (41) | .762 |
| Enterobacter cloacae complex | 2 (3.7) | 1 (2.2) | 0 | .498 |
| Escherichia coli | 17 (31) | 8 (18) | 8 (22) | .259 |
| Klebsiella oxytoca | 1 (1.9) | 2 (4.4) | 3 (8.1) | .361 |
| Klebsiella pneumoniae | 4 (7.4) | 5 (11) | 4 (11) | .786 |
| Pseudomonas aeruginosa | 0 | 3 (6.7) | 0 | .045 |
| Yeast | 1 (1.9) | 0 | 1 (2.7) | .573 |
| Polymicrobial | 2 (3.7) | 2 (4.4) | 1 (2.7)e | .917 |
Abbreviations: BCID, multiplex polymerase chain reaction–based blood culture identification; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus; PRE, before BCID implementation; POST, after BCID implementation; POST-AS, BCID coupled with antimicrobial stewardship.
aFinal blood culture identification.
bKruskal-Wallis test (continuous variables) or chi-square test (categorical variables).
cGram-positive organisms not listed: Enterococcus gallinarum, Enterococcus casseliflavus, Granulicatella adiacens, Listeria monocytogenes, Staphylococcus lugdunensis, Streptococcus group C (2), Streptococcus group G (2), Streptococcus mitis group (2), Streptococcus pyogenes.
dGram-negative organisms not listed: Serratia marcescens, Citrobacter freundii, Pantoea agglomerans, Citrobacter Koseri.
eThree organisms identified from blood culture.
Primary and Secondary Outcomes
Patients received effective antibiotic therapy against the identified bloodstream pathogen with empiric antimicrobials in 45/52 (87%) PRE, 40/43 (93%) POST, and 32/35 (91%) POST-AS BSI encounters. All identified patients received effective antibiotic therapy during their encounter. Patients were ordered appropriate antimicrobial therapy in 47/52 (90%) PRE, 43/43 (100%) POST, and 34/35 (97%) POST-AS BSI encounters (P = .07) (Table 3). Eighty-five of the 124 patients in the primary analysis (69%) required antibiotic adjustment for appropriate antimicrobial therapy, whereas 39 patients (31%) had appropriate antibiotics ordered at the time of Gram stain. Sixty-nine patients underwent antibiotic de-escalation (21 PRE, 27 POST, 21 POST-AS), and 16 patients required dose escalation (9 PRE, 3 POST, 4 POST-AS). In patients who received appropriate antimicrobial therapy, there was no statistical difference in uncontrolled median time to appropriate antimicrobial therapy from Gram stain between the PRE, POST, and POST-AS groups (30 hours, 17 hours, 20 hours; P = .43).1 The multiplex PCR rapid diagnostic test improved median time to organism identification from positive Gram stain by >40 hours between the BCID and non-BCID arms (44 hours, 2.8 hours, 1.5 hours; P < .001). There was no statistically significant mortality or readmission difference between the 3 arms.
Table 3.
Primary and Secondary Outcomes
| PRE (n = 52), No. (%)a |
POST (n = 43), No. (%)a |
POST-AS (n = 35), No. (%)a |
P Valueb | |
|---|---|---|---|---|
| Appropriate antimicrobial therapy | 47 (90) | 43 (100) | 34 (97) | .071 |
| Time to appropriate antimicrobial therapy,c median (IQR), h | 30 (0–59) | 17 (0–41) | 20 (0–43) | .432 |
| Time to organism identification, median (IQR), h | 44 (30–57) | 2.8 (1.4–5.1) | 1.5 (1.3–2.0) | <.001 |
| In-hospital mortality | 2 (3.9) | 3 (7.0) | 2 (5.7) | .793 |
| 30-d mortality | 5 (9.6) | 6 (14) | 5 (14) | .747 |
| 30-d readmission | 15 (29) | 8 (19) | 11 (31) | .374 |
| 30-d readmission with bacteremia episode | 2 (3.9) | 1 (2.3) | 3 (8.6) | .401 |
Abbreviations: BCID, multiplex polymerase chain reaction–based blood culture identification; IQR, intequartile range; PRE, before BCID implementation; POST, after BCID implementation; POST-AS, BCID coupled with antimicrobial stewardship.
aUnless otherwise specified.
bKruskal-Wallis test (continuous variables) or chi-square test (categorical variables).
cMean: PRE (43.5 hours), POST (24.6 hours), POST-AS (25.9 hours) 1-way analysis of variance P = .069.
Multivariate Regression Model
A Tobit regression analysis evaluating the time to appropriate antimicrobial therapy was performed (Table 4). Using sample distribution of other covariate values, the predicted time to appropriate antimicrobial therapy from Gram stain for the BCID intervention cohorts (POST, 13.1 hours; P = .02; POST-AS, 8.3 hours; P = .02) was significantly less than for the PRE cohort (38.1 hours). As reported in Table 4, these values represent a decrease in time to appropriate antimicrobial therapy of 25.0 hours (POST) and 29.8 hours (POST-AS) when compared with PRE patients. Other variables associated with decreased time to appropriate antimicrobial therapy on adjusted analysis included neutropenia (58 hours earlier than patients without severe neutropenia; P < .001), surgery during the index encounter (83 hours earlier than patients who did not have surgery; P = .01), and patients admitted to an oncology service (31 hours earlier than patients admitted to an ICU; P = .02).
Table 4.
Time to Appropriate Antimicrobial Tobit Regression Model
| Variable Group | Variable | Reference Variable | Tobit Coefficient, h | Standard Error | P Value |
|---|---|---|---|---|---|
| BCIDa | POST | PRE | –25.0 | 10.7 | .021 |
| POST-AS | PRE | –29.8 | 11.6 | .012 | |
| Organism | MRSA | No | 44.2 | 28.6 | .125 |
| MSSA | No | 2.0 | 15.9 | .899 | |
| Pseudomonas aeruginosa | No | 15.8 | 29.6 | .594 | |
| Age, y | ≥65 | <65 | 10.5 | 9.8 | .284 |
| Gender | Female | Male | –0.7 | 9.1 | .944 |
| Charlson comorbidity | Any | No | 26.9 | 30.5 | .380 |
| Charlson Comorbidity Index | Score | 0.7 | 2.1 | .756 | |
| Cancer | Any | No | 36.3 | 18.6 | .053 |
| Immunocompromised | Yes | No | –18.6 | 14.9 | .214 |
| ANC | ≥500 | <500 | 57.6 | 12.9 | <.001 |
| Admission service | Oncology | ICU | –30.7 | 13.2 | .022 |
| Hematology | ICU | –7.2 | 15.3 | .637 | |
| BMT | ICU | –17.0 | 16.5 | .304 | |
| Surgical encounter | Yes | No | –83.2 | 29.4 | .005 |
| ID consult | Yes | No | 1.4 | 9.7 | .884 |
Abbreviations: ANC, absolute neutrophil count; BMT, blood and marrow transplant; BCID, multiplex polymerase chain reaction–based blood culture identification; ICU, intensive care unit; ID, infectious diseases; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus; PRE, before BCID implementation; POST, after BCID implementation; POST-AS, BCID coupled with antimicrobial stewardship.
aPredicted time to appropriate antimicrobial therapy for intervention cohorts with sample distribution of other covariate values: PRE (38.1 hours), POST (13.1 hours), POST-AS (8.3 hours).
DISCUSSION
Although a number of studies have shown benefit from combining mRDT with AS interpretation and intervention, to our knowledge this is the first evaluation of the impact of rapid diagnostics for BSIs in immunocompromised patients. We found that implementation of BCID for BSIs decreased the time to appropriate antimicrobial therapy (25 hours POST and 30 hours POST-AS) with and without AS intervention in a multivariable regression analysis, but without significance in the primary analysis (13 hours POST and 10 hours POST-AS) when compared with the PRE intervention cohort. The addition of AS in both analyses did not significantly affect the time to appropriate antimicrobial therapy. Importantly, our interventions were not associated with any increase in in-hospital or 30-day mortality. Additionally, there are likely clinically meaningful outcomes unmeasured in this study affected by shortening time to appropriate therapy, such as adverse drug events, secondary antimicrobial resistance, and Clostridium difficile infection.
Recent studies evaluating the impact of implementing mRDT technology have identified a benefit from AS intervention on blood culture results in a general population [1, 4]. Factors in this study that may have minimized the impact of BCID with AS on time to appropriate therapy other than patient population include notification of BCID results via email rather than a page or call to the ASP, lack of 24-hour ASP availability for consultation, and a third party notification system (Theradoc) that clinical pharmacists had the ability to subscribe to in order to receive real-time notification of all blood culture results. Given the urgent need to improve antimicrobial prescribing and the need to target ASP resources to clinical areas with the largest impact, our findings and clinical experience suggest that additional studies with BCID or other rapid diagnostic tests for BSIs are warranted in this patient population, with possible tailoring of additional AS intervention to non-neutropenic or solid tumor patients.
Interestingly, there was a notable difference in median time to appropriate therapy between the uncontrolled study population and the multivariate analysis (PRE 30 hours vs 38 hours; POST 17 hours vs 13 hours; POST-AS 20 hours vs 8 hours). All 3 arms had a substantial number of patients with neutropenic fever who were on appropriate antimicrobial therapy at the time of Gram stain. However, patients with surgical encounters (6 PRE, 1 POST, 0 POST-AS) were likely also a large driver of shorter time to appropriate therapy. Once all independent variables were controlled, a large difference was noted primarily in the POST-AS group for time to appropriate antimicrobial therapy.
To evaluate the results of this study, nonparametric statistical methods were employed due to non–normally distributed data for our primary outcome. The regression analysis required a Tobit assessment due to the left censoring of our data, with more than one-fourth of our patients receiving appropriate antibiotic administration before Gram stain results. This left shift in data was likely influenced by the high number of patients with neutropenic fever receiving appropriate empiric antibiotic administration.
There are several limitations that restrict the generalizability of this study. We evaluated a small number of patients and may not have been powered to detect additional benefit from AS intervention with BCID compared with BCID alone. The underlying disease, BSI source, and immunosuppression status were unbalanced between the 3 groups and could have contributed to differences in antimicrobial use that affected time to appropriate therapy. Patients in the POST-AS cohort had more BSIs with an unidentified source and higher immunosuppression status than the PRE or POST cohorts, which may have influenced antibiotic prescribers’ decision to de-escalate antimicrobials. Moreover, we recently developed an ASP with 1 of the first interventions including acting upon blood culture results. It is possible that providers were unfamiliar and unwilling to take recommendations from the ASP; however, review of acceptance of ASP recommendations 2 weeks into the intervention found an 80% acceptance rate. The primary outcome of appropriate antimicrobial therapy is a subjective outcome evaluated by reviewers who were not blinded to the patients, study, intervention, or outcome. Lastly, neutropenic fever patients showed overwhelmingly appropriate antimicrobial utilization based upon our internal and IDSA guidelines, which promote broad-spectrum antimicrobial use until neutrophil count recovery, thereby limiting the opportunity for ASP intervention. The continued use of broad-spectrum antimicrobial therapy in neutropenic fever patients with documented BSI with susceptible organisms warrants further evaluation to assess outcomes associated with antimicrobial de-escalation, thereby informing future ASP intervention in immunocompromised patients.
In conclusion, this single-site, retrospective study found reductions in time to organism identification and time to appropriate antimicrobial therapy with mRDT for BSI in immunocompromised patients. Studies with greater power are needed to assess the additional benefit of AS with BCID vs BCID alone in order to improve ASP resource allocation and prioritization.
Acknowledgments
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Prior presentations: Preliminary data from this research were presented at the ASM Microbe Conference (New Orleans, LA) and ID Week (San Diego, CA) in 2017.
References
- 1. Banerjee R, Teng CB, Cunningham SA, et al. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 2015; 61:1071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pardo J, Klinker KP, Borgert SJ, et al. Clinical and economic impact of antimicrobial stewardship interventions with the FilmArray blood culture identification panel. Diagn Microbiol Infect Dis 2016; 84:159–64. [DOI] [PubMed] [Google Scholar]
- 3. Huang AM, Newton D, Kunapuli A, et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis 2013; 57:1237–45. [DOI] [PubMed] [Google Scholar]
- 4. Patel TS, Kaakeh R, Nagel JL, et al. Cost analysis of implementing matrix-assisted laser desorption ionization-time of flight mass spectrometry plus real-time antimicrobial stewardship intervention for bloodstream infections. J Clin Microbiol 2017; 55:60–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Timbrook TT, Morton JB, McConeghy KW, et al. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 2017; 64:15–23. [DOI] [PubMed] [Google Scholar]
- 6. Barlam TF, Cosgrove SE, Abbo LM, et al. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis 2016; 62:e51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Freifeld AG, Bow EJ, Sepkowitz KA, et al. ; Infectious Diseases Society of America Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 2011; 52:e56–93. [DOI] [PubMed] [Google Scholar]
- 8. National Comprehensive Cancer Network. Prevention and treatment of cancer-related infections website (version 2.2017). https://www.nccn.org/professionals/physician_gls/pdf/infections.pdf. Accessed 7 August 2017. [Google Scholar]
- 9. Tamma PD, Avdic E, Li DX, et al. Association of adverse events with antibiotic use in hospitalized patients. JAMA Intern Med 2017; 177:1308–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davey P, Brown E, Charani E, et al. Interventions to improve antibiotic prescribing practices for hospital inpatients. Cochrane Database Syst Rev 2013; 4:CD003543. [DOI] [PubMed] [Google Scholar]
- 11. Baur D, Gladstone BP, Burkert F, et al. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis. Lancet Infect Dis 2017; 17:990–1001. [DOI] [PubMed] [Google Scholar]
- 12. Barlam TF, Cosgrove SE, Abbo LM, et al. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis 2016; 62:e51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akova M. Epidemiology of antimicrobial resistance in bloodstream infections. Virulence 2016; 7:252–66. [DOI] [PMC free article] [PubMed] [Google Scholar]