Abstract
Long noncoding RNAs (LncRNAs) are involved in the pathogenesis of intervertebral disc degeneration (IDD). However, the biological function and expression of RMRP were still unclear. In our study, we showed that RMRP expression was up‐regulated in degenerated NP tissues compared to normal NP samples, and higher RMRP expression was associated with the disc degeneration grade. Further studies indicated that ectopic expression of RMRP enhanced NP cell growth and also enhanced the expression of ki‐67, PCNA and cyclin D1 in the NP cell. Moreover, overexpression of RMRP promoted the expression of Type II collagen and aggrecan and suppressed the expression of MMP13 and ADAMTS4. In addition, we found that the expression of miR‐206 was down‐regulated in degenerated NP tissues compared to normal NP samples, and lower miR‐206 expression was correlated with the disc degeneration grade. Interestingly, we indicated that miR‐206 expression in NP tissues was negatively correlated with the expression of RMRP. Ectopic expression of miR‐206 suppressed NP cell proliferation and suppressed the expression of Type II collagen and aggrecan and enhanced the expression of MMP13 and ADAMTS4. Furthermore, we demonstrated that overexpression of RMRP increased NP cell growth and regulated ECM expression through targeting miR‐206. These results suggested that lncRNA‐RMRP promoted the progression of IDD through targeting miR‐206, providing an attractive new therapeutic approach for the treatment of IDD disease.
Keywords: intervertebral disc degeneration, lncRNAs, miR‐206, RMRP
1. INTRODUCTION
Chronic low back pain (LBP) influenced more than 70% people at some time in their lives, with about 10% people becoming chronic disabled.1, 2, 3 The aetiology of LBP is considered to be multifactorial, and intervertebral disc degenerative (IDD) contributes to the development of LBP.3, 4, 5 The cause of IDD is thought to associate with many aetiological factors such as ageing, lifestyle and genetic predisposition.6, 7, 8, 9 Recent research in cell molecular biology may contemplate treating intervertebral disc itself at the molecular biology level to delay or prevent the development of IDD.10, 11, 12, 13, 14, 15 However, the underlying molecular and cellular mechanism of IDD progression is still unknown. Thus, it is important for us to study the mechanism of IDD and to find new treatment strategies for IDD.
Long noncoding RNAs (LncRNAs) are longer than 200 nucleotides (nts) in length with no or limited protein‐coding capacity and were found to play important roles in diverse human diseases and biological processes.16, 17, 18, 19, 20, 21 Mounting evidence has suggested that many lncRNAs are beregulated in diverse diseases such as tumours, diabetes, coronary heart disease, osteoarthritis and also IDD.22, 23, 24, 25, 26, 27, 28, 29 For example, Lan and colleagues30 profiled lncRNA expression in degenerate disc tissues and normal adults' lumbar discs by microarray and found a total of 3081 differentially expressed lncRNAs. Wang et al31 showed that knockdown of RP11‐296A18.3 suppressed human NP cell proliferation and decreased the expression of collagen I and matrix metalloproteinase (MMP)‐13. Tan et al32 studied the role of SNHG1 in the pathogenesis of IDD. They indicated that the expression of SNHG1 was up‐regulated in the IDD samples compared to the control samples. Recently, a novel lncRNA‐RMRP was found to be deregulated in several diseases.33, 34, 35 For example, Meng et al35 demonstrated that the expression of RMRP was up‐regulated in lung cancer tissues and overexpression of RMRP enhanced lung cancer cell growth, invasion and colony formation through regulating miR‐206 expression. Moreover, Shao et al36 found that the RMRP expression was deregulated in gastric tumour. In addition, Feng et al33 showed that RMRP expression was up‐regulated in glioma samples compared to normal brain samples. Down‐regulation of RMRP inhibited glioma cell growth, invasion and migration. However, the biological function and expression of RMRP are still unknown.
In this study, we first measured the expression of RMRP in the intervertebral disc tissues and we found that RMRP expression was up‐regulated in degenerated NP tissues compared to normal NP samples, and higher RMRP expression was associated with the disc degeneration grade.
2. MATERIALS AND METHODS
2.1. Tissues
Degenerated IVD tissues were collected from IDD patients undergoing discectomy surgery. Nondegenerated IVD tissues were obtained from cases with scoliosis undergoing surgery. Routine MRI of spine was performed for all these cases before operation and the modified Pfirrmann classification was used to determine the degree of disc degeneration according to T2‐weighted images. Sample was washed three times in the physiological saline and IVD was separated using stereotaxic microscope and then the tissue was immediately frozen in liquid nitrogen. Informed consent was obtained from each patient and this study was approved by the ethics committee of QingDao Central Hospital.
2.2. RNA isolation and qRT‐PCR
Total RNA from samples specimens or cells was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Reverse‐transcribed was performed with cDNA Synthesis Kit (GeneCopoeia Inc., Rockville, MD, USA) following the manufacturer's information. Quantitative real‐time reverse transcription‐PCR (qRT‐PCR) was carried out to determine the expression level of miRNA and LncRNA expression using SYBR‐green PCR kit (GenePharma, Shanghai, China) on PRISM 7900HT system. GAPDH or U6 snRNA was performed to as normalization control and the expression was determined with 2‐DDCT method. qRT‐PCR primers were listed as following: RMRP Forward: 5′‐ACTCCAAAGTCCGCCAAGA‐3′′ and 5′‐GTAACTAGA−GGGAGCTGAC‐3′; GADPH: Forward: 5′‐GTCAA‐CGGATTTGGTCTGTATT‐3′′ and 5′‐AGTCTTCTGGGTGGCAGTGAT‐3′.
2.3. Cell cultures and cell transfection
Human NP cell was isolated from human NP tissues following to previous standardized method. NP samples were washed in PBS for three times and NP was separated from IVD samples using stereotaxic microscope and then cut to pieces. Then, these tissues were incubated in Type II collagenase in DMEM medium. After isolation, cells were cultured in the DMEM medium with penicillin, streptomycin and 10% FBS. pcDNA‐RMRP and pcDNA‐control vector were purchased from the Biosune biotechnology (Shanghai, China) and then transfected to the cell using DharmaFECT1 kit (Dharmacon, TX, USA) following to instruction.
2.4. Cell proliferation and cell cycle assays
Cell growth of NP cell was determined using the 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) method. Cells were cultured in 96‐well plate and MTT medium was added to each well. The absorbance was detected at the 490 nm using enzyme‐labelled analyser. For the cell cycle, cells were washed twice in PBS and fixed with ethanol overnight at 4°C. DNA content staining was performed with propidium iodide and ribonuclease A for a half hour. Population in G2–M, S and G0‐G1 phase was determined by flow cytometry (Beckman Coulter, Indianapolis, USA).
2.5. Western blot assay
Total protein from corresponding cell or samples was extracted by RIPA buffer (Pierce) with Protease Inhibitor (Pierce, Waltham, MA, USA). The concentration of protein was determined with BCA Protein Kit. Equivalent protein was electrophoresed by 12% SDS‐PAGE and transferred to the PVDF membranes. The membrane was blocked with nonfat milk and then was immunoblotted with primary antibodies (anti‐HDAC6 and anti‐GAPDH, Abcam, Cambridge, UK). After washed for three times in PBS, the membrane was immunoblotted with HRP‐linked secondary antibodies (Cell Signaling, Boston, MA, USA). The signal was determined by ECL detection system (Millipore).
2.6. Statistical analysis
Data were measured by SPSS Data (version 18, Inc., Chicago, IL, USA) and were represented as mean ± SD. The comparison among multiple groups was determined with one‐way ANOVA and the difference between two groups was measured with Student's t test. P < 0.05 was considered to be statistically significant.
3. RESULTS
3.1. The expression of RMRP was up‐regulated in degenerated NP tissues
We first determined the expression level of RMRP in NP tissues. RMRP expression in the 5 normal NP tissues and 30 degenerated NP tissues was shown in Figure 1A,B. Furthermore, we indicated that the expression of RMRP was up‐regulated in degenerated NP tissues compared to normal NP samples (Figure 1C). Moreover, we showed that RMRP expression was higher in advanced the disc degeneration grade (Figure 1D).
Figure 1.
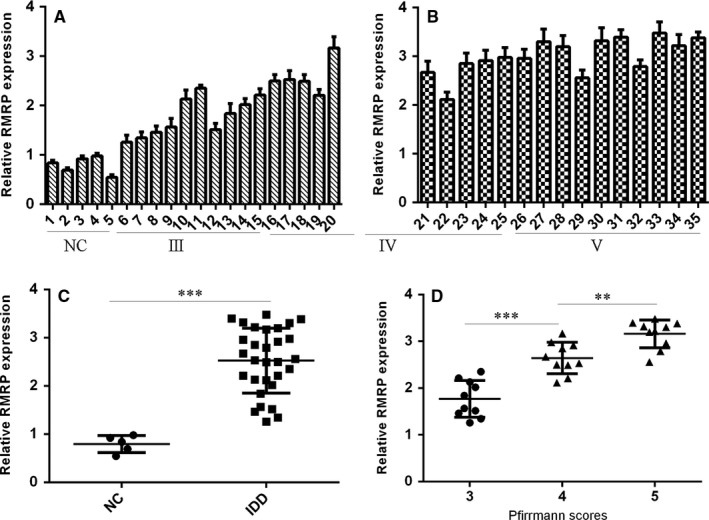
The expression of RMRP was up‐regulated in the degenerated NP tissues. A,B, The RMRP expression in the 5 normal NP tissues and 30 degenerated NP tissues was measured by qRT‐PCR. U6 was used as the internal control. C, The expression of RMRP was up‐regulated in degenerated NP tissues compared to normal NP samples. D, RMRP expression was higher with the disc degeneration grade. **P < 0.01 and ***P < 0.001
3.2. The expression of miR‐206 was down‐regulated in degenerated NP tissues
Then, we determined the expression level of miR‐206 in NP tissues. miR‐206 expression in 5 normal NP tissues and 30 degenerated NP tissues was shown in Figure 2A,B. Furthermore, we indicated that the expression of miR‐206 was down‐regulated in degenerated NP tissues compared to normal NP samples (Figure 2C). Moreover, we showed that miR‐206 expression was lower in early disc degeneration grade (Figure 2D). In addition, we indicated that miR‐206 expression in NP tissues was negatively correlated with the expression of RMRP (Figure 2E).
Figure 2.
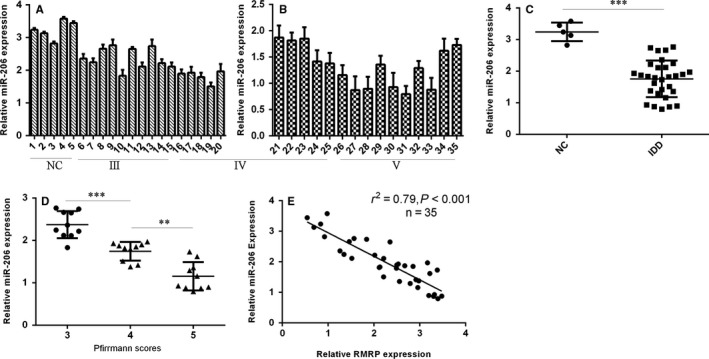
The expression of miR‐206 was down‐regulated in the degenerated NP tissues. A,B, The miR‐206 expression in the 5 normal NP tissues and 30 degenerated NP tissues was determined by qRT‐PCR assay. C, The expression of miR‐206 was down‐regulated in degenerated NP tissues compared to normal NP samples. D, miR‐206 expression was lower with the disc degeneration grade. E, miR‐206 expression in the NP tissues was negatively correlated with the expression of RMRP. **P < 0.01 and ***P < 0.001
3.3. Overexpression of RMRP suppressed the miR‐206 expression in the NP cell
We confirmed that the expression of RMRP was significantly up‐regulated in NP cell after transfected with pcDNA‐RMRP (Figure 3A). Overexpression of RMRP suppressed miR‐206 expression in NP cell (Figure 3B). In addition, we showed ectopic expression of RMRP promoted HDAC6 expression in NP cell (Figure 3C). We also indicated that RMRP overexpression enhanced HDAC6 protein expression in NP cell (Figure 3D).
Figure 3.

Overexpression of RMRP suppressed the miR‐206 expression in the NP cell. A, We confirmed that the expression of RMRP was significantly up‐regulated in the NP cell after transfected with pcDNA‐RMRP. B, Overexpression of RMRP suppressed the miR‐206 expression in the NP cell. C, Ectopic expression of RMRP promoted the HDAC6 expression in the NP cell. D, The protein expression of HDAC6 in the NP cell was detected by Western blot. GAPDH was used as the control
3.4. Ectopic expression of RMRP enhanced NP cell growth
We next showed that ectopic expression RMRP promoted NP cell proliferation (Figure 4A). The expression of ki‐67 was up‐regulated in NP cell after transfected with pcDNA‐RMRP (Figure 4B). Overexpression of RMRP enhanced PCNA expression in NP cell (Figure 4C). Moreover, ectopic expression of RMRP promoted the expression of cyclin D1 in NP cell (Figure 4D).
Figure 4.
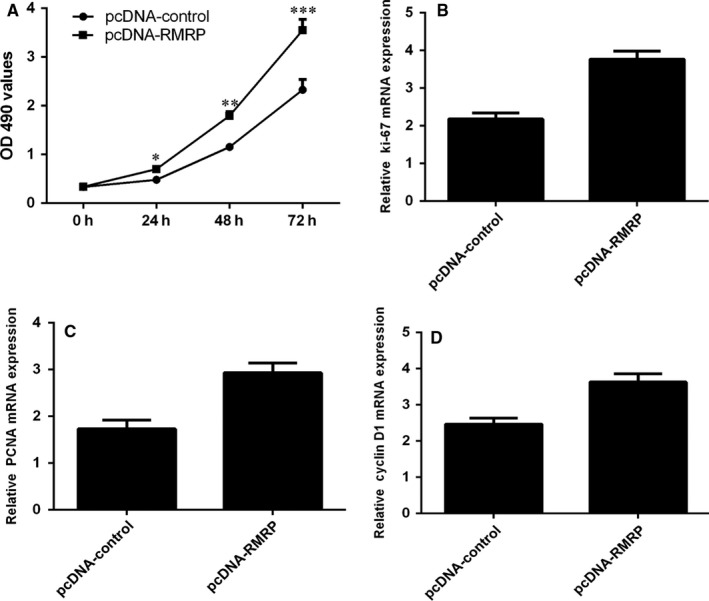
Ectopic expression of RMRP enhanced the NP cell growth. A, Ectopic expression RMRP promoted the NP cell proliferation. B, The expression of ki‐67 was measured by qRT‐PCR analysis. C, Overexpression of RMRP enhanced the PCNA expression in the NP cell. (D) Ectopic expression of RMRP promoted the expression of cyclin D1 in the NP cell. *P < 0.05, **P < 0.01 and ***P < 0.001
3.5. RMRP overexpression increased Type II collagen and aggrecan expression and suppressed MMP13 and ADAMTS4 expression
We next indicated that overexpression of RMRP increased Type II collagen and aggrecan expression in the NP cell (Figure 5A). In addition, we showed that ectopic expression of RMRP decreased the expression of MMP13 and ADAMTS4 (Figure 5B).
Figure 5.

RMRP overexpression increased Type II collagen and aggrecan expression and suppressed MMP13 and ADAMTS4 expression. A, The Type II collagen and aggrecan expression was determined by qRT‐PCR assay. GAPDH was used as the control. B, Ectopic expression of RMRP decreased the expression of MMP13 and ADAMTS4 expression in the NP cell
3.6. Ectopic expression of miR‐206 suppressed NP cell proliferation and regulated the extracellular matrix (ECM) expression
The expression of miR‐206 was significantly up‐regulated in NP cell after transfected with miR‐206 mimic (Figure 6A). We found that overexpression of miR‐206 suppressed NP cell proliferation (Figure 6B). The expression of ki‐67 was down‐regulated in NP cell after transfected with miR‐206 mimic (Figure 6C). Overexpression of miR‐206 suppressed PCNA expression in NP cell (Figure 6D). Moreover, ectopic expression of miR‐206 decreased the expression of cyclin D1 in NP cell (Figure 6E). Furthermore, we showed that overexpression of miR‐206 suppressed Type II collagen and aggrecan expression in NP cell (Figure 6F). In addition, we showed that ectopic expression of miR‐206 promoted the expression of MMP13 and ADAMTS4 in NP cell (Figure 6G).
Figure 6.
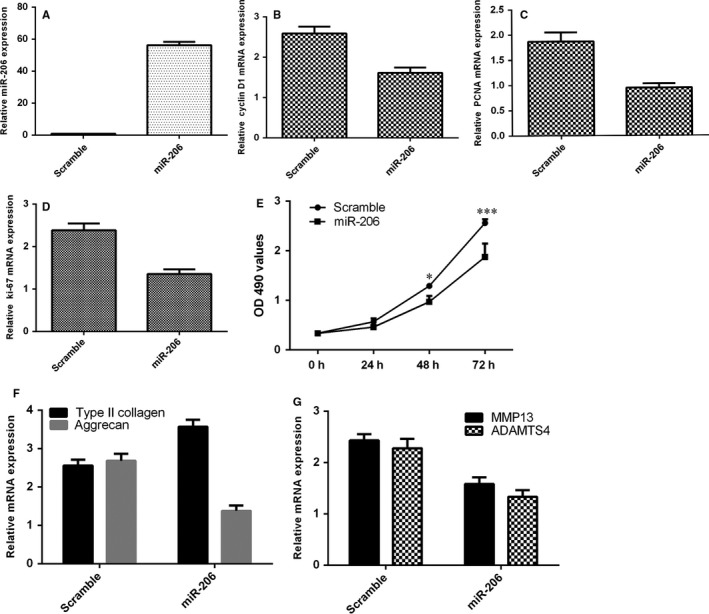
Ectopic expression of miR‐206 suppressed the NP cell proliferation and regulated the extracellular matrix (ECM) expression. A, The expression of miR‐206 was significantly up‐regulated in the NP cell after transfected with miR‐206 mimic. B, Overexpression of miR‐206 suppressed the NP cell proliferation. C, The ki‐67 expression was determined by qRT‐PCR assay. GAPDH was used as the control. D, The PCNA expression was measured by qRT‐PCR analysis. E, Ectopic expression of miR‐206 decreased the expression of cyclin D1 in the NP cell. F, Overexpression of miR‐206 suppressed the Type II collagen and aggrecan expression in the NP cell. G, Ectopic expression of miR‐206 promoted the expression of MMP13 and ADAMTS4 expression in the NP cell. *P < 0.05 and ***P < 0.001
3.7. Ectopic expression of RMRP increased NP cell growth and regulated ECM expression through targeting miR‐206
To study the biological function association between RMRP and miR‐206, we employed loss and gain‐of‐function approaches. We showed that the cell proliferation was down‐regulated in the group of pcDNA‐RMRP and scramble compared to pcDNA‐RMRP and miR‐206 mimic group (Figure 7A). In addition, we showed that ectopic expression of miR‐206 suppressed the expression of cyclin D1 (Figure 7B), ki‐67(Figure 7C) and PCNA (Figure 7D) in the pcDNA‐RMRP‐overexpressing NP cell. Moreover, we indicated that ectopic expression of RMRP increased Type II collagen and aggrecan expression (Figure 7E) and suppressed MMP13 and ADAMTS4 (Figure 7F) expression through inhibiting miR‐206 expression.
Figure 7.
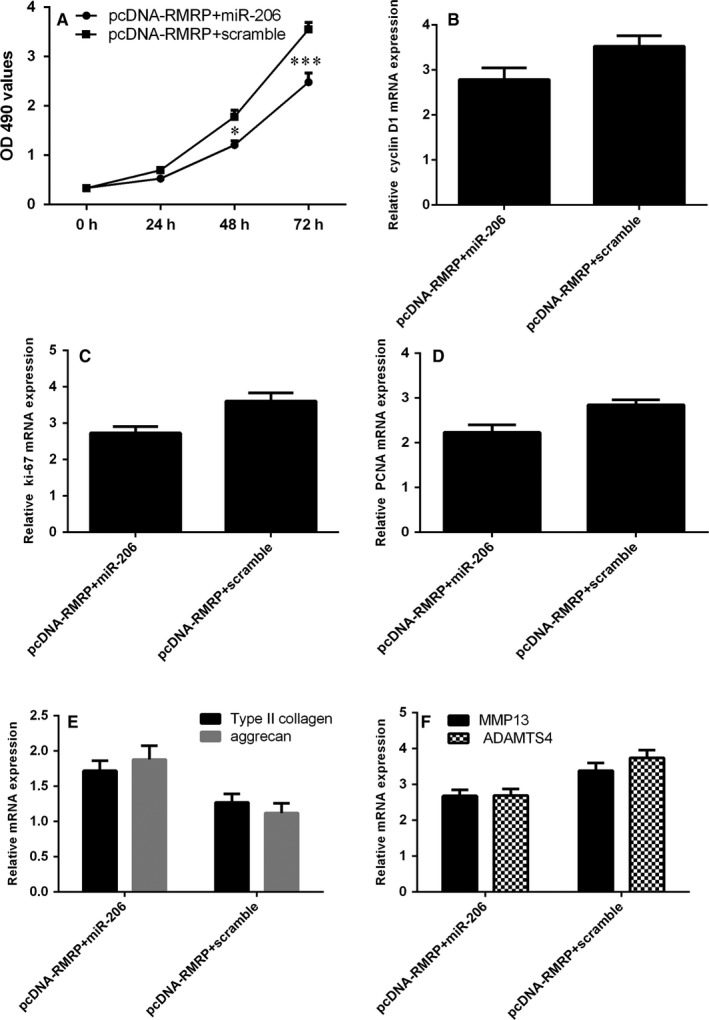
Ectopic expression of RMRP increased NP cell growth and regulated ECM expression through targeting miR‐206 expression. A, Cell proliferation was determined by MTT method. B, The cyclin D1 expression in the NP cell was detected by qRT‐PCR. C, The ki‐67 expression in the NP cell was detected by qRT‐PCR. D, The PCNA expression in the NP cell was detected by qRT‐PCR. E, Ectopic expression of RMRP increased Type II collagen and aggrecan expression in the pcDNA‐RMRP‐overexpressing NP cell. F, The expression of MMP13 and ADAMTS4 was measured by qRT‐PCR. *P < 0.05 and ***P < 0.001
4. DISCUSSION
In our study, we studied the expression and biological function of lncRNA‐RMRP in the degenerated intervertebral disc. We showed that RMRP expression was up‐regulated in degenerated NP tissues compared to normal NP samples and RMRP expression was associated with the disc degeneration grade. Further studies indicated that ectopic expression of RMRP enhanced NP cell growth and also enhanced the expression of ki‐67, PCNA and cyclin D1 in the NP cell. Moreover, overexpression of RMRP promoted Type II collagen and aggrecan expression and suppressed the expression of MMP13 and ADAMTS4. In addition, we found that the expression of miR‐206 was down‐regulated in degenerated NP tissues compared to normal NP samples and lower miR‐206 expression was correlated with early disc degeneration grade. Interestingly, we indicated that miR‐206 expression in NP tissues was negatively correlated with the expression of RMRP. Ectopic expression of miR‐206 suppressed NP cell proliferation and suppressed Type II collagen and aggrecan expression and enhanced the expression of MMP13 and ADAMTS4. Furthermore, we demonstrated that overexpression of RMRP increased NP cell growth and regulated ECM expression through targeting miR‐206.
A growing number of studies indicated that lncRNAs were correlated with the development of several diseases.23, 37, 38, 39 The role of lncRNAs in the development of IDD has also been explored.29, 31, 40 For example, Wang et al41 found that lncRNA linc‐ADAMTS5 cooperates with RREB1 to suppress ADAMTS5 expression, then affecting ECM degeneration of the IVD. Chen et al42 indicated that TUG1 expression was up‐regulated in degenerated IVD tissues and positively associated with β‐catenin and Wnt expression. Inhibited expression of TUG1 inhibited the expression of β‐catenin, Bax, ADAMTS5, MMP3, Caspase‐3 and Wnt1 and promoted the COL2A1, Aggrecan and Bcl‐2 expression. Xi et al43 demonstrated that the expression of HCG18 was up‐regulated in IDD patients and higher expression of HCG18 was correlated with the grade of disc degeneration. In addition, they showed that HCG18 inhibited NP cell growth and enhanced the development of IDD through the NFκB/TRAF6/miR‐146a‐5p axis. LncRNA‐RMRP is one new LncRNA that was deregulated in several diseases. For example, Shao et al36 found that RMRP expression was deregulated in gastric tumour. Moreover, Meng et al35 demonstrated that the expression of RMRP was up‐regulated in lung cancer tissues and overexpression of RMRP enhanced lung cancer cell growth, invasion and colony formation through regulating miR‐206 expression. In addition, Feng et al33 showed that RMRP expression was up‐regulated in glioma samples compared to normal brain samples. Down‐regulation of RMRP inhibited glioma cell growth, invasion and migration. However, the biological function and expression of RMRP were still unknown. In our study, we indicated that the expression of RMRP was up‐regulated in degenerated NP tissues compared to normal NP samples and higher RMRP expression was correlated with the disc degeneration grade. Further studies showed that overexpression of RMRP promoted NP cell proliferation and also increased the expression of ki‐67, PCNA and cyclin D1 in the NP cell. Moreover, ectopic expression of RMRP increased the Type II collagen and aggrecan expression and decreased the expression of MMP13 and ADAMTS4.
Recently, increasing evidence has suggested that lncRNAs function as competing endogenous RNAs (ceRNAs) to target miRNA expression.44, 45, 46 For examples, Shao et al36 showed that RMRP regulated gastric cell cycle and cyclin D2 expression through regulating miR‐206 expression. In line with this, we also showed that overexpression of RMRP suppressed miR‐206 expression in NP cell. Liu et al47 showed that miR‐206 suppressed head and neck squamous cell carcinoma cell growth, invasion and migration through targeting HDAC6 via mTOR/PTEN/AKT pathway. Pang et al48 indicated that miR‐206 overexpression suppressed hepatocellular carcinoma cell growth by regulating CDK9 expression. In our study, we indicated that the expression of miR‐206 was down‐regulated in degenerated NP tissues compared to normal NP samples, and lower miR‐206 expression was correlated with the disc degeneration grade. Interestingly, we indicated that miR‐206 expression in NP tissues was negatively correlated with the expression of RMRP. Ectopic expression of miR‐206 suppressed NP cell proliferation and suppressed the Type II collagen and aggrecan expression and enhanced the expression of MMP13 and ADAMTS4. Furthermore, we demonstrated that overexpression of RMRP increased NP cell growth and regulated ECM expression through targeting miR‐206.
Taken together, our study showed that RMRP expression was up‐regulated in degenerated NP tissues compared to normal NP samples, and higher RMRP expression was associated with the disc degeneration grade. Ectopic expression of RMRP increased NP cell growth and regulated ECM expression through targeting miR‐206. These results suggested that lncRNA‐RMRP promoted the progression of IDD through targeting miR‐206, providing an attractive new therapeutic approach for the treatment of IDD disease.
CONFLICT OF INTEREST
There is no conflict of interest statement.
Wang X, Peng L, Gong X, Zhang X, Sun R, Du J. LncRNA‐RMRP promotes nucleus pulposus cell proliferation through regulating miR‐206 expression. J Cell Mol Med. 2018;22:5468–5476. 10.1111/jcmm.13817
REFERENCES
- 1. Arana E, Kovacs FM, Royuela A, et al. Modic changes and associated features in Southern European chronic low back pain patients. Spine J. 2011;11:402‐411. [DOI] [PubMed] [Google Scholar]
- 2. Tegeder I, Lotsch J. Current evidence for a modulation of low back pain by human genetic variants. J Cell Mol Med. 2009;13:1605‐1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Millecamps M, Tajerian M, Naso L, Sage EH, Stone LS. Lumbar intervertebral disc degeneration associated with axial and radiating low back pain in ageing SPARC‐null mice. Pain. 2012;153:1167‐1179. [DOI] [PubMed] [Google Scholar]
- 4. Samartzis D, Karppinen J, Mok F, Fong DY, Luk KD, Cheung KM. A population‐based study of juvenile disc degeneration and its association with overweight and obesity, low back pain, and diminished functional status. J Bone Joint Surg Am. 2011;93:662‐670. [DOI] [PubMed] [Google Scholar]
- 5. Tajerian M, Alvarado S, Millecamps M, et al. DNA methylation of SPARC and chronic low back pain. Mol Pain. 2011;7:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Inoue N, Espinoza Orias AA. Biomechanics of intervertebral disk degeneration. Orthop Clin North Am. 2011;42:487‐499. vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clouet J, Pot‐Vaucel M, Grimandi G, et al. Characterization of the age‐dependent intervertebral disc changes in rabbit by correlation between MRI, histology and gene expression. BMC Musculoskelet Disord. 2011;12:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Song YQ, Karasugi T, Cheung KM, et al. Lumbar disc degeneration is linked to a carbohydrate sulfotransferase 3 variant. J Clin Investig. 2013;123(11):4909‐4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sobajima S, Shimer AL, Chadderdon RC, et al. Quantitative analysis of gene expression in a rabbit model of intervertebral disc degeneration by real‐time polymerase chain reaction. Spine J. 2005;5:14‐23. [DOI] [PubMed] [Google Scholar]
- 10. Rutges JP, Nikkels PG, Oner FC, et al. The presence of extracellular matrix degrading metalloproteinases during fetal development of the intervertebral disc. Eur Spine J. 2010;19:1340‐1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang M, Wang HQ, Zhang Q, Yan XD, Hao M, Luo ZJ. Alterations of ADAMTSs and TIMP‐3 in human nucleus pulposus cells subjected to compressive load: implications in the pathogenesis of human intervertebral disc degeneration. J Orthop Res. 2012;30:267‐273. [DOI] [PubMed] [Google Scholar]
- 12. Dahia CL, Mahoney E, Wylie C. Shh signaling from the nucleus pulposus is required for the postnatal growth and differentiation of the mouse intervertebral disc. PLoS ONE. 2012;7:e35944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu X, Li Z, Shen J, et al. MicroRNA‐10b promotes nucleus pulposus cell proliferation through RhoC‐Akt pathway by targeting HOXD10 in intervertebral disc degeneration. PLoS ONE. 2013;8:e83080. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Li Z, Yu X, Shen J, Chan MT, Wu WK. MicroRNA in intervertebral disc degeneration. Cell Prolif. 2015;48:278‐283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Z, Li X, Chen C, Chan M, Wu W, Shen J. Melatonin inhibits nucleus pulposus (NP) cell proliferation and extracellular matrix (ECM) remodeling via the melatonin membrane receptors mediated PI3K‐Akt pathway. J Pineal Res. 2017;63:e12435 10.1111/jpi.12435 [DOI] [PubMed] [Google Scholar]
- 16. Li Z, Yu X, Shen JX. Long non‐coding RNAs: emerging players in osteosarcoma. Tumor Biol. 2016;37:2811‐2816. [DOI] [PubMed] [Google Scholar]
- 17. Yu X, Li Z. Long non‐coding RNA HOTAIR: a novel oncogene (Review). Mol Med Rep. 2015;12:5611‐5618. [DOI] [PubMed] [Google Scholar]
- 18. Yu X, Li Z. Long non‐coding RNA growth arrest‐specific transcript 5 in tumor biology. Oncol Lett. 2015;10:1953‐1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dou J, Ni YY, He XF, et al. Decreasing lncRNA HOTAIR expression inhibits human colorectal cancer stem cells. Am J Transl Res. 2016;8:98‐108. [PMC free article] [PubMed] [Google Scholar]
- 20. Wan L, Kong J, Tang J, et al. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med. 2016;20:2036‐2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang S, Zhang W, Wu X, et al. The lncRNA MALAT1 functions as a competing endogenous RNA to regulate MCL‐1 expression by sponging miR‐363‐3p in gallbladder cancer. J Cell Mol Med. 2016;20:2299‐2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xin Y, Li Z, Shen J, Chan M, Wu W. CCAT1: a pivotal oncogenic long non‐coding RNA in human cancers. Cell Prolif. 2016;49:255‐260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao W, Dong S, Duan BS, et al. HOTAIR is a predictive and prognostic biomarker for patients with advanced gastric adenocarcinoma receiving fluorouracil and platinum combination chemotherapy. Am J Transl Res. 2015;7:1295‐1302. [PMC free article] [PubMed] [Google Scholar]
- 24. Huang CS, Liu SG, Wang HJ, Zhang ZC, Yang Q, Gao F. LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am J Transl Res. 2016;8:5025‐5034. [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang YH, Fu J, Zhang ZJ, Ge CC, Yi Y. LncRNA‐LINC00152 down‐regulated by miR‐376c‐3p restricts viability and promotes apoptosis of colorectal cancer cells. Am J Transl Res. 2016;8:5286‐5297. [PMC free article] [PubMed] [Google Scholar]
- 26. Qiu GZ, Tian W, Fu HT, Li CP, Liu B. Long noncoding RNA‐MEG3 is involved in diabetes mellitus‐related microvascular dysfunction. Biochem Biophys Res Comm. 2016;471:135‐141. [DOI] [PubMed] [Google Scholar]
- 27. He S, Gu WL, Li YZ, Zhu H. ANRIL/CDKN2B‐AS shows two‐stage clade‐specific evolution and becomes conserved after transposon insertions in simians. BMC Evol Biol. 2013;13:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Su W, Xie W, Shang Q, Su B. The long noncoding RNA MEG3 is downregulated and inversely associated with VEGF levels in osteoarthritis. Biomed Res Int. 2015;2015:356893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen WK, Yu XH, Yang W, et al. IncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50:e12313 10.1111/cpr.12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lan PH, Liu ZH, Pei YJ, et al. Landscape of RNAs in human lumbar disc degeneration. Oncotarget. 2016;7:63166‐63176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang XB, Lv GH, Li J, Wang B, Zhang QS, Lu C. LncRNA‐RP11‐296A18.3/miR‐138/HIF1A pathway regulates the proliferation ECM synthesis of human nucleus pulposus cells (HNPCs). J Cell Biochem. 2017;118:4862‐4871. [DOI] [PubMed] [Google Scholar]
- 32. Tan H, Zhao L, Song R, Liu Y, Wang L. The long noncoding RNA SNHG1 promotes nucleus pulposus cell proliferation through regulating miR‐326/CCND1. Am J Physiol Cell Physiol. 2018;315(1):C21‐C27. [DOI] [PubMed] [Google Scholar]
- 33. Feng W, Li L, Xu X, Jiao Y, Du W. Up‐regulation of the long non‐coding RNA RMRP contributes to glioma progression and promotes glioma cell proliferation and invasion. Arch Med Sci. 2017;13:1315‐1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Steinbusch M, Caron M, Surtel D, et al. Expression of RMRP RNA is regulated in chondrocyte hypertrophy and determines chondrogenic differentiation. Sci Rep. 2017;7:6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meng Q, Ren M, Li Y, Song X. LncRNA‐RMRP acts as an oncogene in lung cancer. PLoS ONE. 2016;11:e0164845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shao Y, Ye M, Li Q, et al. LncRNA‐RMRP promotes carcinogenesis by acting as a miR‐206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7:37812‐37824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li C, Lei BX, Huang SB, et al. H19 derived microRNA‐675 regulates cell proliferation and migration through CDK6 in glioma. Am J Transl Res. 2015;7:1747‐1764. [PMC free article] [PubMed] [Google Scholar]
- 38. Wang ZW, Jin YY, Ren HT, Ma XL, Wang BF, Wang YL. Downregulation of the long non‐coding RNA TUSC7 promotes NSCLC cell proliferation and correlates with poor prognosis. Am J Transl Res. 2016;8:680‐687. [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang XY, Zhang LX, Tian CJ, et al. LncRNAs BCYRN1 promoted the proliferation and migration of rat airway smooth muscle cells in asthma via upregulating the expression of transient receptor potential 1. Am J Transl Res. 2016;8:3409‐3418. [PMC free article] [PubMed] [Google Scholar]
- 40. Wan Z, Song F, Sun Z, et al. Aberrantly expressed long noncoding RNAs in human intervertebral disc degeneration: a microarray related study. Arthritis Res Ther. 2014;16:465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang K, Song Y, Liu W, et al. The noncoding RNA linc‐ADAMTS5 cooperates with RREB1 to protect from intervertebral disc degeneration through inhibiting ADAMTS5 expression. Clin Sci. 2017;131:965‐979. [DOI] [PubMed] [Google Scholar]
- 42. Chen J, Jia Y, Liu G, et al. Role of LncRNA TUG1 in intervertebral disc degeneration and nucleus pulposus cells via regulating Wnt/β‐catenin signaling pathway. Biochem Biophys Res Commun. 2017;491:668‐674. [DOI] [PubMed] [Google Scholar]
- 43. Xi Y, Jiang T, Wang W, et al. Long non‐coding HCG18 promotes intervertebral disc degeneration by sponging miR‐146a‐5p and regulating TRAF6 expression. Sci Rep. 2017;7:13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu XG, Jiang HJ, Jiang XJ. Downregulation of lncRNA ANRIL inhibits proliferation, induces apoptosis, and enhances radiosensitivity in nasopharyngeal carcinoma cells through regulating miR‐125a. Cancer Biol Ther. 2017;18:331‐338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li JQ, Hu SY, Wang ZY, et al. Long non‐coding RNA MEG3 inhibits microRNA‐125a‐5p expression and induces immune imbalance of Treg/Th17 in immune thrombocytopenic purpura. Biomed Pharmacother. 2016;83:905‐911. [DOI] [PubMed] [Google Scholar]
- 46. Zhang J, Yao T, Wang Y, Yu J, Liu Y, Lin Z. Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR‐21. Cancer Biol Ther. 2016;17:104‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu F, Zhao X, Qian Y, Zhang J, Zhang Y, Yin R. MiR‐206 inhibits Head and neck squamous cell carcinoma cell progression by targeting HDAC6 via PTEN/AKT/mTOR pathway. Biomed Pharmacother. 2017;96:229‐237. [DOI] [PubMed] [Google Scholar]
- 48. Pang C, Huang G, Luo K, et al. miR‐206 inhibits the growth of hepatocellular carcinoma cells via targeting CDK9. Cancer Med. 2017;6:2398‐2409. [DOI] [PMC free article] [PubMed] [Google Scholar]


