Abstract
Antibacterial-guided fractionation of the Dictyoceratid sponges Lamellodysidea sp. and two samples of Dysidea granulosa yielded 14 polybrominated, diphenyl ethers including one new methoxy-containing compound (8). Their structures were elucidated by interpretation of spectroscopic data of the natural product and their methoxy derivatives. Most of the compounds showed strong antimicrobial activity with low- to sub-microgram mL−1 minimum inhibitory concentrations against drug-susceptible and drug-resistant strains of Staphylococcus aureus and Enterococcus faecium, and two compounds inhibited Escherichia coli in a structure-dependent manner.
Graphical Abstract
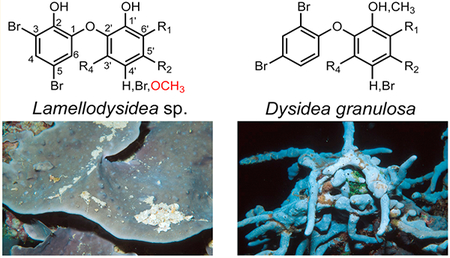
The occurrence of bacterial infections by antibiotic-resistant organisms in hospital as well as community settings continues to be a major burden in public health. Knowing that our current arsenal of antibiotics comes almost exclusively from bacterial- and fungal-derived natural products1 and that high-throughput screening of synthetic small-molecule libraries for the discovery of new antibiotics has met with limited success, some experts have proposed continued investigations into natural products libraries for discovery of new antibiotics.2–4 While screening organic extracts from the NCI Open Repository for antibiotic activity against the model Gram-negative bacterium Escherichia coli, we identified three antibacterial extracts that originated from related marine sponges of the family Dysideidae. These included two separate collections of Dysidea granulosa and one collection of Lamellodysidea sp.
Marine sponges of the Dysideidae family are known for containing a rich array of diverse natural products. Examples include unusual sterols,5–7 glycolipids,8 sesterterpenes,9 and sesquiterpene quinones;10 the peptidic natural products dysinosins,11,12 dysideaprolines, and barbaleucamides;13 and numerous polybrominated and hydroxylated diphenyl ethers (PBDEs).14–17 Halogenated diphenyl ethers in general have garnered attention from different fields. Revealed by natural products chemists, they represent some of the earliest discoveries of polyhalogenated compounds coming from nature.18 The distribution of synthetic polychlorinated and polybrominated biphenyls continues to be studied by environmental toxicologists.19,20 Most recently Agarwal et al. described a biosynthetic scheme that can lead to formation of PBDEs in a widespread marine bacterium.21 In addition to isolating known PBDEs 1–7 from D. granulosa and Lamellodysidea sp., we describe the structural assignment of a new PBDE, compound 8 (Figure 1), along with an approach to assign bromination and hydroxylation patterns in substituted diphenyl ethers. Unexpected trends in antibacterial activities were observed for these compounds.
Figure 1.
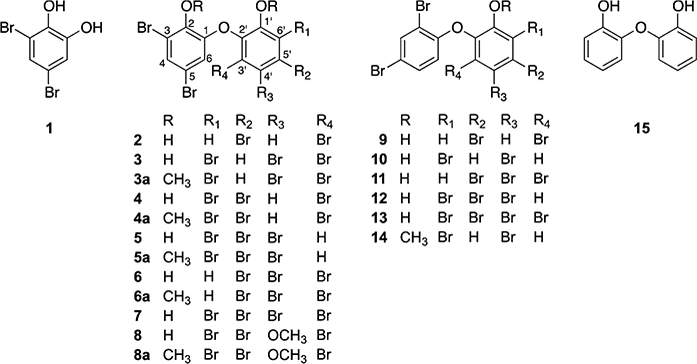
Compounds isolated from Lamellodysidea sp. (1–8), two collections of Dysidea granulosa (9–14), synthetic methylates 3a–6a and 8a, and a commercial 2,2′-dihydroxydiphenyl ether (15).
The extracts of Lamellodysidea sp. and two collections of D. granulosa inhibited the growth of both Gram-positive and Gram-negative bacteria. The Gram-positive bacteria included Bacillus subtilis and drug-susceptible and drug-resistant strains of Staphylococcus aureus and Enterococcus faecium, with E. coli representing Gram-negative bacteria. With an emphasis on identifying the compounds responsible for the Gram-negative inhibitory activity, we fractionated these extracts on HP20SS eluting with H2O to MeOH followed by acetone. LC-MS and NMR analyses of the E. coli-active fractions identified a number of known PBDEs, along with a new compound, 8. Compound 8 was isolated as a white solid, and the low-resolution ESIMS spectrum of 8 showed a distribution of negatively charged ions at m/z 620.6, 622.6, 624.6, 626.6, 628.6, and 630.6 having relative intensities of 1:4:6:6:4:1, which indicated the presence of five bromine atoms in the molecule. The molecular formula of C13H7Br5O4 was determined by HRESIMS, indicating eight degrees of unsaturation.
The 1H NMR spectroscopic data of compound 8 (Table 1) exhibited a pair of meta-coupled (2.2 Hz) signals at δH 6.54 and 7.37 and one methoxy group at δH 3.87 (s, 3H). Two broad singlets (δH 7.08 and 7.56) in the 1H NMR spectrum were tentatively assigned as hydroxy groups. The presence of one methoxy carbon, two sp2 methines, and 10 sp2 nonprotonated carbons was apparent from the 13C NMR and HSQC spectra, consistent with 8 being a polybrominated diphenyl ether.22 Methylation of 8 to give 8a confirmed the presence of two hydroxy groups (Table 1).
Table 1.
1H (500 MHz) and 13C NMR (126 MHz) Data for 8 and 8aa
| 8 | 8a | ||||
|---|---|---|---|---|---|
| position | δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | ΔδC(δ8a−δ8) |
| 1 | 144.9, C | 150.8, C | 5.9 | ||
| 2 | 142.7, C | 145.5, C | 2.8 | ||
| 3 | 111.3, C | 119.2, C | 7.9 | ||
| 4 | 116.3, CH | 7.37, d (2.2) | 116.9, CH | 7.40, d (2.2) | 0.6 |
| 5 | 111.8, C | 116.8, C | 5.0 | ||
| 6 | 129.5, CH | 6.54, d (2.2) | 129.4, CH | 6.49, d (2.2) | −0.1 |
| 1′ | 145.1, C | 148.5, C | 3.4 | ||
| 2′ | 138.8, C | 145.0, C | 6.2 | ||
| 3′ | 119.2, C | 119.4, C | 0.1 | ||
| 4′ | 149.0, C | 152.6, C | 3.6 | ||
| 5′ | 113.4, C | 113.5, C | 0.2 | ||
| 6′ | 114.2, C | 121.7, C | 7.5 | ||
| −OH | 7.08, br s | ||||
| −OH | 7.56, br s | ||||
| 1′-OCH3 | 61.6, CH3 | 3.79, s | |||
| 4′-OCH3 | 61.1, CH3 | 3.87, s | 61.0, CH3 | 3.91, s | |
| 2-OCH3 | 61.3, CH3 | 4.00, s | |||
Chemical shifts (δ) are referenced to residual CDCl3 (1H: 7.26/13C: 77.16 ppm).
The locations of the bromines and hydroxy groups in ring A of 8 and 8a were clear from the chemical shifts, meta-coupling, and HMBC correlations. The HMBC spectra (Figure 2) showed couplings from H-4 to C-2, C-3, C-5, and C-6 and from H-6 to C-1, C-2, C-4, and C-5 in 8 and 8a. Additionally, the coupling from the methoxy group at δH 4.00 to C-2 in derivative 8a established the positions of the ether and hydroxy groups in ring A and the chemical shifts of C-1 and C-2. These data indicated that the meta-coupled protons were para to the ether and to a brominated carbon, leaving only the H-4/H-6 arrangement possible.
Figure 2.
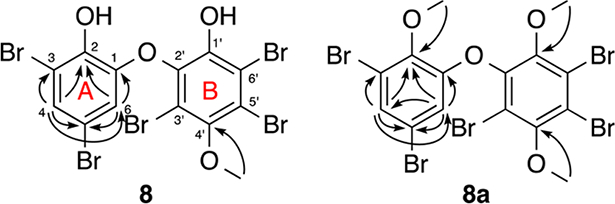
HMBC correlations used in structure determination of compounds 8 and 8a.
Assignment of the substitution pattern in ring B was more challenging due to the lack of HMBC correlations. In addition, the 13C chemical shifts did not match any known PBDEs. This was likely due to the presence of a methoxy group that affects 13C chemical shifts in PBDEs.23 We tentatively assigned the position of the naturally occurring −OCH3 group in ring B using a combination of NOEs and distance comparisons from molecular modeling. In selective 1D-NOE spectra of 8 an NOE between the OCH3 and H-6 was the only one observed (Figure S13, Supporting Information). As shown in Figure 3, this suggests that the −OCH3 group is located meta to the ether and attached at C-4′ or C-6′. In the NOE spectra of methylated derivative 8a, NOEs between the new methoxy group in ring B and H-6 and −OCH3 in ring A were present (Figure 3D); however no NOEs were observed between the two −OCH3 groups in ring B (Supporting Information). This suggested that the hydroxy group in 8 was para to the −OCH3 group and thus located at C-1′.
Figure 3.
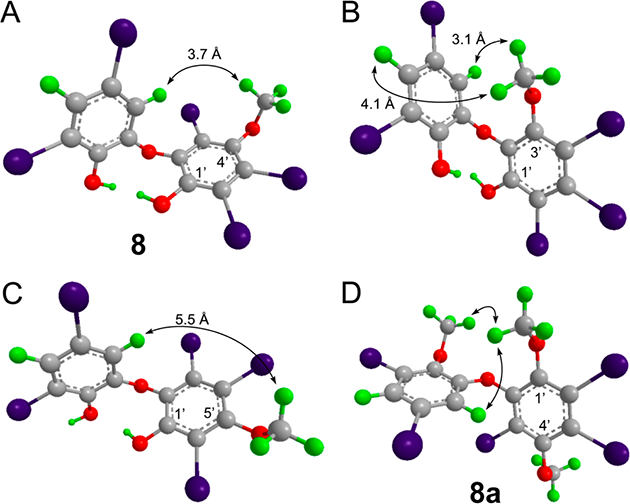
Interproton distances of three possible isomers of 8 and NOEs observed for 8a. Coordinates for the three possible structures of compound 8 were energy minimized using Chem3D, and interproton distances between H-4 and H-6 of ring A and the −OCH3 group in ring B were measured in each and compared to the experimental NOE data (A–C). The isomer shown in B (1′-hydroxy-3′-methyoxy) predicts NOEs that are not observed, and the isomer in C (1′-hydroxy-5′-methyoxy) shows a distance greater than 5.5 Å for an NOE that is present. Panel D shows the NOEs observed for methylated 8a, in agreement with experimental data. 1D NOE spectra are shown in Figure S13.
To provide additional support for the location of substituents in ring B of compound 8, we prepared the methoxy derivatives of known compounds 3–6 by treatment with iodomethane to give compounds 3a–6a, with 3a and 5a being new compounds (Supporting Information), and compared their 13C chemical shifts. Compared to the parent compounds 3–6, chemical shifts for C-1, C-3, and C-5 in ring A and C-2′, C-4′, and C-6′ in ring B were deshielded by an average of 6.4, 8.3, 4.6, 4.7, 3.2, and 4.0 ppm, respectively. Similarly, the corresponding carbons in 8 and 8a were deshielded by 5.9, 7.9, 5.0, 6.2, 3.6, and 7.5 ppm (Table 1). These trends in chemical shifts are consistent with the substitution pattern suggested above and confirm the locations of the bromine, hydroxy, and methoxy substituents. We note that these chemical shift changes at positions ortho and para to a methoxy group are seen at both protonated and halogenated carbons, making this a useful approach for assigning these complex substitution patterns. Interestingly, compounds originating from Lamellodysidea all contained a 2-hydroxy-3,5-dibromophenyl moiety for ring A, while compounds from D. granulosa uniformly contained a 2,4-dibromophenyl moiety, making this chemistry specific for taxonomically distinct marine sponge samples (Table 2).
Table 2.
Occurrence of Compounds in Three Dysideae Sponges
| sponge | NPID numbera |
collection site | compounds |
|---|---|---|---|
| Lamellodysidea sp. | C024121 | Papua New Guinea | 1–8 |
| Dysidea granulosa | C031381 | Palau | 9–13 |
| Dysidea granulosa | C024075 | Papua New Guinea | 9, 10, and 14 |
Corresponds to the NCI Open Repository Natural Products ID number.
We tested compounds 1–14 for antibacterial activities against the panel of drug-susceptible and drug-resistant bacteria and fungi shown in Table 3. Compounds 2–13 showed strong antibacterial effects toward S. aureus and E. faecium strains, exceeding the potency of control antibiotics oxacillin and vancomycin. Surprisingly and unlike the other compounds, 9 and 11 also inhibited the growth of E. coli with MICs of 3.1 and 12.5 μg/mL, respectively. Though previously reported to have broad-spectrum antibacterial activity24 in our antimicrobial assays, compound 10 did not inhibit the growth of E. coli. None of the compounds were active against P. aeruginosa or C. albicans.
Table 3.
Minimum Inhibitory Concentrations and Cytotoxicity of Compounds 1–14, 3a–6a, 8a, and 15a
| S. aureus ATCC | S. aureus ATCC | E.faecium ATCC | E.faecium ATCC | E. coli ATCC | P. aeruginosa | C. albicans ATCC | ||
|---|---|---|---|---|---|---|---|---|
| 29213 | 43300 | 29212 | 51299 | 25922 | ATCC 27853 | 28517 | Bsc-1 | |
| 1 | 100 | >50 | 12.5 | 25 | >100 | >50 | >50 | >50 |
| 2 | 1.6 | 1.25 | 3.1 | 1.6 | 50 | >50 | >50 | >50 |
| 3 | 1.6 | 0.31 | 3.1 | 3.1 | 100 | >50 | >50 | >50 |
| 4 | 0.78 | 0.16 | 1.6 | 0.78 | 50 | >50 | >50 | >50 |
| 5 | 0.31 | 0.16 | 1.6 | 0.78 | 50 | 50 | >50 | >50 |
| 6 | 0.31 | 0.078 | 0.39 | 0.39 | 25 | 50 | >50 | >50 |
| 7 | 0.39 | 0.16 | 0.78 | 0.39 | 50 | 50 | 50 | >50 |
| 8 | 0.78 | 0.39 | 3.1 | 1.56 | >50 | >50 | >50 | >50 |
| 3a | >50 | >50 | 50 | >50 | >50 | >50 | >50 | >50 |
| 4a | >50 | >50 | 50 | 50 | >50 | >50 | >50 | >50 |
| 5a | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 6a | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 8a | 3.1 | 3.1 | 13 | 13 | >50 | >50 | >50 | >50 |
| 9 | 0.042 | 0.08 | 1.2 | 1.2 | 3.1 | >50 | >50 | 7.0 |
| 10 | 0.78 | 0.19 | 0.8 | 0.8 | >100 | 32 | ||
| 11 | 0.14 | 0.015 | 0.4 | 0.4 | 12.5 | 8.8 | ||
| 12 | 3.7 | 0.4 | 1.2 | 1.2 | >100 | 15 | ||
| 13 | 1.6 | 0.8 | 33 | 33 | >100 | 29 | ||
| 14 | 3.7 | 0.4 | 11 | 11 | >100 | 35 | ||
| 15 | 50 | 50 | >50 | >50 | ||||
| oxacillin | 0.13 | >32 | 32 | |||||
| gentamicin | 0.5 | 1 | ||||||
| vancomycin | 1 | 2 | ||||||
| amphotericin B | <7.8 |
MICs and cytotoxity data are reported in μg/mL.
Compounds 1–14, 3a–6a, and 8a were tested for cytotoxicity against a monkey kidney cell line (BSC-1) and a human colorectal tumor cell line (HCT-116).25 Compounds 1–8, 3a–6a, and 8a were nontoxic at the maximum tested concentration (50 μg/mL). Compounds 9–14 showed some toxicity against the kidney cell line BSC-1 with IC50’s between 7 and 35 μg/mL. Although their ring B structures are similar, rings A of compounds 9–14 lack a hydroxy and contain bromine atoms ortho and para to the ether compared to compounds 2–8. This difference in conjunction with the cell-based screens suggests that the lack of the hydroxy group on ring A and/or the bromine substitution pattern leads to increased cytotoxicity. To explore whether halogens are required for these activities, we tested the commercially available compound 2,2′-dihydroxydiphenyl ether (15) for cytotoxity and antibacterial activity; it was inactive in all assays, as were the permethylated derivatives 3a–6a and 8a. The observed structure−activity relationship suggests that ring B needs two bromine atoms and a C-1′ hydroxy group for antibacterial activity. Further, the presence of two phenolic hydroxy groups at C-1′ and C-2 in PBDEs decreases cytotoxity, but also corresponds to a loss in activity against the Gram-negative bacterium E. coli.
In summary, we isolated 14 polybrominated and hydroxylated diphenyl ethers from related Dysideidae sponges and demonstrated a simple method employing methylation and associated changes in 13C chemical shifts to assign complex substitution patterns in proton-deficient structures. Because it can be used on compounds of this class that contain both hydroxy and methoxy substitutents, this method augments previous NMR approaches that have been used in PBDE analyses.23 Although superficially compounds 1–14 appear to be highly similar to one another, their antibacterial spectrum and potencies, as well as cellular cytotoxicities, differed greatly as a function of the presence and pattern of hydroxy and bromine substituents. Given the difficulty of identifying small molecules that inhibit the growth of Gram-negative organisms, these results may provide insights into features that lead to this activity.
EXPERIMENTAL SECTION
General Experimental Procedures.
UV−vis spectra were recorded on an Agilent 8453 spectrophotometer, and IR spectra on a PerkinElmer Frontier FT-IR spectrophotometer (KBr). NMR spectra were recorded at 298 K on a Bruker Avance500 spectrometer equipped with a triple resonance cryoprobe and z gradients. Transient NOE spectra were acquired with a 1 s mixing time using a selective 1D double pulsed field gradient spin echo pulse program incorporating perfect-echo excitation, alternating gradient polarity, and zero quantum suppression.26–29 LRESIMS was carried out on an Agilent 1100 LC system and a 6310 MSD. HRESIMS was carried out on a Waters time-of-flight mass spectrometer model LCT Premier. Semipreparative HPLC (Agilent 1100) was performed using a Waters XBridge preparative C18 column (10 × 250 mm, S-5 μm, 12 nm). All solvents (HPLC grade) and Diaion HP20SS gel were obtained from Sigma-Aldrich.
Animal Material.
Extracts from the three Dysideidae sponge samples used in this study were obtained from the NCI Open Repository. Lamellodysidea sp. (NPID no. C024121) was collected at a depth of 43 m in Papua New Guinea in June 2003. Dysidea granulosa (NPID no. C024075) was collected in Papua New Guinea at a depth of 10 m, and a separate collection of D. granulosa (NPID no. C031381) was collected in June 2010 in the Palau Islands. Taxonomic identifications were made by Michelle Kelly or one of the authors, L.J.B. Voucher specimens are available at the NCI, Frederick, MD, USA, and at the Smithsonian Museum, Washington, DC.
Extraction and Isolation.
Organic extracts from the above three sponges were prepared by extraction with MeOH−CH2Cl2 and stored at −50 °C until shipment. The organic extract of Lamellodysidea sp. (5 g) was chromatographed on a Diaion HP20SS-gel column equilibrated in H2O and was eluted with a 10% stepwise gradient from H2O to MeOH. Fractions were combined into three groups, A–C, on the basis of antimicrobial activity. Fraction C (4.5 g) was purified by semipreparative HPLC (5 mL/min, 0–15 min, 50–85%; 15–40 min 85–95% MeCN−H2O gradient elution) to yield compounds 1 (0.4 mg, tR 14.5 min), 2 (1.1 mg, tR 24.0 min), 8 (3.3 mg, tR 24.5 min), a mixture of compounds with a tR of 27 min, 4 (10 mg, tR 27.5 min), 5 (4.2 mg, tR 29.5 min), and 7 (114 mg, tR 31.0 min). The mixture eluting at 27 min was resubjected to semipreparative HPLC (5 mL/min, 30 min, 50–70% MeCN−H2O gradient elution) to give compounds 6 (6.4 mg, tR 23.5 min) and 3 (15 mg, tR 25.0 min).
The organic extract (2.19 g) of D. granulosa (NPID no. C031381) was dissolved in MeOH and chromatographed on a Diaion HP20SS-gel column eluting with water to MeOH, followed by acetone, to give seven fractions (A–G), which were combined on the basis of their LCMS profiles. The acetone fraction G (1.44 g) was subjected to reversed-phase HPLC (XBridge prep, Waters C18, 5 μm) eluting with a linear gradient of 0.1% TFA in H2O and 0.05% TFA in MeCN (60–100% in 40 min, flow rate 5 mL/min) to yield compounds 9 (38 mg, tR 14.5 min), 10 (1.2 mg, tR 17.3 min), 11 (38.6 mg, tR 18.0 min), 12 (13.4 mg, tR 19.0 min), and 13 (1.7 mg, tR 22.7 min).
The organic extract (2 g) of D. granulosa (NPID no. C024075) was dissolved in MeOH and chromatographed on a Diaion HP20SS-gel column as described above. The acetone fraction G (0.35 g) was subjected to reversed-phase HPLC (5 mL/min, 60–80% H2O−MeCN with 0.01% TFA, gradient elution in 40 min) to yield compounds 9 (20.2 mg, tR 18.5 min), 10 (6.0 mg, tR 18.5 min), and 14 (2.1 mg, tR 37.5 min).
1-(3′,5′,6′-Tribromo-4′-methoxy-1′-hydroxyphenoxy)-3,5-dibromo-2-phenol (8): white solid; UV (MeOH) λmax (log ε) 213 (5.64), 297 (4.57) nm; IR (film) νmax 2918, 2865, 1711, 1415, 1234 cm−1; 1H and 13C NMR data, Table 1; LRESIMS m/z 620.6 [M – H]−; HRESIMS m/z 620.6187 [M – H]− (calcd for C13H6Br5O4, 620.6183).
1-(3′,4′,6′-Tribromo-1′-methoxyphenoxy)-3,5-dibromo-2-methoxybenzene (3a): white solid; UV (MeOH) λmax (log ε) 214 (5.12) nm; IR (film) νmax 2917, 2864, 1701, 1413, 1232 cm−1; 1H and 13C NMR data, Supporting Information; HRESIMS m/z 619.6458 [M + H]+ (calcd for C14H9Br5O3, 619.6468).
1-(4′,5′,6′-Tribromo-1′-methoxyphenoxy)-3,5-dibromo-2-methoxybenzene (5a): white solid; UV (MeOH)0020λmax (log ε) 216 (5.34) nm; IR (film) νmax 2917, 2865, 1701, 1510, 1493, 1482 cm−1; 1H and 13C NMR data, Supporting Information; HRESIMS m/z 619.6473 [M + H]+ (calcd for C14H9Br5O3, 619.6468).
1-(3′,5′,6′-Tribromo-1′,4′-dimethoxy-1′-phenoxy)-3,5-dibromo-2-methoxybenzene (8a): UV (MeOH) λmax (log ε) 214 (5.68), 295 (4.44) nm; IR (film) νmax 2917, 2865, 1711, 1415, 1234 cm−1; 1H and 13C NMR data, Table 1; HRESIMS m/z 649.6567 [M + H]+ (calcd for C15H11Br5O4, 649.6574).
Methylation of Compounds 3–6 and 8.
Compound 3 (2.0 mg, 3 μmol) was added to a solution of acetone (500 μL) in the presence of K2CO3 (0.1 mg) and MeI (400 μL, 6 μM) at rt. The solution was stirred for 1 h, dried under a stream of N2, and extracted with CHCl3 to give 3a as a white solid (2.3 mg, 100%). By the same method compounds 4a (1.7 mg, 95%), 5a (2.0 mg, 84%), 6a (3.3 mg, 95%), and 8a (0.6 mg, 100%) were obtained from 4 (1.5 mg), 5 (2.0 mg), 6 (3.0 mg), and 8 (0.5 mg), respectively.
MIC and Cytotoxicity Determination.
MICs were determined by testing all compounds in duplicate in at least two independent experiments for their antimicrobial activity against laboratory strains listed in Table 3 and as described in the Clinical and Laboratory Standards Institute (CLSI) guidelines.30 All laboratory bacterial strains were obtained from the American Type Culture Collection. Experimental details for MIC and cytotoxicity data are provided in the Supporting Information.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Lloyd for HRMS data and the NCI Open Repository for organic extracts. This work was supported by the NIH Intramural Research Program (NIDDK). T.P. acknowledges a Park Scholarship.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jnat-prod.6b00229.
IR, HRMS, and 1H, 13C, and 2D NMR spectra of new compounds and protocols for biological testing (PDF)
REFERENCES
- (1).Walsh C Antibiotics: Actions, Origins, Resistance, 1st ed.; ASM Press: Washington, DC, 2003. [Google Scholar]
- (2).Payne DJ; Gwynn MN; Holmes DJ; Pompliano DL Nat. Rev. Drug Discovery 2007, 6, 29–40. [DOI] [PubMed] [Google Scholar]
- (3).Silver LL Expert Opin. Drug Discovery 2008, 3, 487–500. [DOI] [PubMed] [Google Scholar]
- (4).Silver LL Future Microbiol 2015, 20, 1711–1718. [DOI] [PubMed] [Google Scholar]
- (5).Boonlarppradab C; Faulkner DJ J. Nat. Prod 2007, 70, 846–848. [DOI] [PubMed] [Google Scholar]
- (6).De Almeida Leone P; Redburn J; Hooper JNA; Quinn RJ J. Nat. Prod 2000, 63, 694–697. [DOI] [PubMed] [Google Scholar]
- (7).Milkova TS; Mikhova BP; Nikolov NM; Popov SS; Andreev SN J. Nat. Prod 1992, 55, 974–978. [Google Scholar]
- (8).Costantino V; Fattorusso E; Imperatore C; Mangoni A J. Nat. Prod 2002, 65, 883–886. [DOI] [PubMed] [Google Scholar]
- (9).Kernan MR; Cambie RC; Bergquist PR J. Nat. Prod 1991, 54, 265–268. [Google Scholar]
- (10).Du L; Zhou YD; Nagle DG J. Nat. Prod 2013, 76, 1175–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Carroll AR; Buchanan MS; Edser A; Hyde E; Simpson M; Quinn RJ J. Nat. Prod 2004, 67, 1291–1294. [DOI] [PubMed] [Google Scholar]
- (12).Carroll AR; Pierens GK; Fechner G; De Almeida Leone P; Ngo A; Simpson M; Hyde E; Hooper J. N.a.; Boström SL; Musil D; Quinn RJ J. Am. Chem. Soc 2002, 124, 13340–13341. [DOI] [PubMed] [Google Scholar]
- (13).Harrigan GG; Goetz GH; Luesch H; Yang S; Likos J J. Nat. Prod 2001, 64, 1133–1138. [DOI] [PubMed] [Google Scholar]
- (14).Carté B; Faulkner DJ Tetrahedron 1981, 37, 2335–2339. [Google Scholar]
- (15).Norton RS; Croft KD; Wells RJ Tetrahedron 1981, 37, 2341–2349. [Google Scholar]
- (16).Sharma GM; Vig B Tetrahedron Lett 1972, 13, 1715–1718. [Google Scholar]
- (17).Agarwal V; Li J; Rahman I; Borgen M; Aluwihare LI; Biggs JS; Paul VJ; Moore BS Environ. Sci. Technol 2015, 49, 1339–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Faulkner DJ Nat. Prod. Rep 1984, 1, 551–598. [Google Scholar]
- (19).Malmvärn A; Marsh G; Kautsky L; Athanasiadou M; Bergman Å; Asplund L Environ. Sci. Technol 2005, 39, 2990–2997. [DOI] [PubMed] [Google Scholar]
- (20).Wang HS; Chen ZJ; Ho KL; Ge LC; Du J; Lam MHW; Giesy JP; Wong MH; Wong CKC Environ. Int 2012, 47, 66–72. [DOI] [PubMed] [Google Scholar]
- (21).Agarwal V; El Gamal A.a.; Yamanaka K; Poth D; Kersten RD; Schorn M; Allen EE; Moore BS Nat. Chem. Biol 2014, 10, 640–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Hanif N; Tanaka J; Setiawan A; Trianto A; De Voogd NJ; Murni A; Tanaka C; Higa T J. Nat. Prod 2007, 70, 432–435. [DOI] [PubMed] [Google Scholar]
- (23).Calcul L; Chow R; Oliver AG; Tenney K; White KN; Wood AW; Fiorilla C; Crews P J. Nat. Prod 2009, 72, 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Shridhar DM; Mahajan GB; Kamat VP; Naik CG; Parab RR; Thakur NR; Mishra PD Mar. Drugs 2009, 7, 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Alley MC; Scudiero D.a.; Monks A; Assay MT; Scudiere D. a.; Hursey ML; Czerwinski MJ; Fine DL; Abbott BJ; Mayo JG; Shoemaker RH; Boyd MR Cancer Res 1988, 48, 589–601. [PubMed] [Google Scholar]
- (26).Stott K; Keeler J; Van QN; Shaka AJ J. Magn. Reson 1997, 125, 302–324. [Google Scholar]
- (27).Takegoshi K; Ogura K; Hikichi K J. Magn. Reson 1989, 84, 611–615. [Google Scholar]
- (28).Thrippleton MJ; Keeler J Angew. Chem., Int. Ed 2003, 42, 3938–3941. [DOI] [PubMed] [Google Scholar]
- (29).Butts CP; Jones CR; Towers EC; Flynn JL; Appleby L; Barron NJ Org. Biomol. Chem 2011, 9, 177. [DOI] [PubMed] [Google Scholar]
- (30).CLSI. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard-ninth ed CLSI document M07-A9; CLSI: Wayne, PA, 2012; Vol. M07-A9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


