Abstract
Multi-agent chemotherapeutic regimes remain the cornerstone treatment for Ewing sarcoma, the second most common bone malignancy diagnosed in pediatric and young adolescent populations. We have reached a therapeutic ceiling with conventional cytotoxic agents, highlighting the need to adopt novel approaches that specifically target the drivers of Ewing sarcoma oncogenesis. As KDM1A/LSD1 (Lysine Specific Demethylase 1) is highly expressed in Ewing sarcoma cell lines and tumors, with elevated expression levels associated with worse overall survival (P=0.033), this study has examined biomarkers of sensitivity and mechanisms of cytotoxicity to targeted KDM1A inhibition using SP-2509 (reversible KDM1A inhibitor). We report, that innate resistance to SP-2509 was not observed in our Ewing sarcoma cell line cohort (n=17)(IC50 range 81nM-1593nM), in contrast resistance to the next generation KDM1A irreversible inhibitor GSK-LSD1 was observed across multiple cell lines (IC50>300μM). Although TP53/STAG2/CDKN2A status and basal KDM1A mRNA and protein levels did not correlate with SP-2509 response, induction of KDM1B following SP-2509 treatment was strongly associated with SP-2509 hyper-sensitivity. We show that the transcriptional profile driven by SP-2509 strongly mirrors KDM1A genetic depletion. Mechanistically, RNA-seq analysis revealed that SP-2509 imparts robust apoptosis through engagement of the endoplasmic reticulum (ER) stress pathway. In addition, ETS1/HIST1H2BM were specifically induced/repressed respectively following SP-2509 treatment only in our hypersensitive cell lines. Together, our findings provide key insights into the mechanisms of SP-2509 cytotoxicity as well as biomarkers that can be used to predict KDM1A inhibitor sensitivity in Ewing sarcoma.
Keywords: KDM1A/LSD1, SP-2509, Ewing sarcoma, GSK-LSD1, Endoplasmic reticulum stress response
Introduction
Ewing sarcoma (ES), is a highly aggressive pediatric bone tumor that has only seen incremental improvements in survival rates for patients diagnosed with primary metastatic or relapsed disease in recent years (<20% event-free survival). At the genome level, the sole distinguishing feature of ES is the presence of the reciprocal EWS/ETS translocation, with the resulting fusion protein, EWS/FLI in 85% of cases, acting as the molecular driver for tumor development (1,2). Although this poorly differentiated tumor has a strict dependence on EWS/ETS fusion proteins for oncogenesis, therapeutic targeting of EWS/FLI remains unrealized due to its intrinsically disordered protein nature and lack of enzymatic activity (3). Current chemotherapy backbones for the treatment of primary ES have historically consisted of regiments based on alkylating and anthracycline agents. However, extensive protocol-driven clinical research evaluating dose intensification and schedule optimization of the current five drug regiment (Vincristine, Adriamycin, Cyclophosphamide, Ifosfamide and Etoposide) suggests that further modification will unlikely produce additional benefit. This coupled with the high risk of anthracycline-induced cardiomyopathy and development of secondary neoplasms (4), clearly underscores the urgent requirement to incorporate new therapeutic agents with lower toxicity profiles to improve overall survival for these patients.
For the past several decades, the overwhelming emphasis in cancer biology has been defining and targeting DNA aberrations. However, there is growing appreciation that dysregulation of epigenetic machinery and chromatin modifications are important mechanisms utilized by tumors to favorably modulate DNA repair, cell cycle control, and apoptosis-promoting genes. This is particularly pertinent for ES, as recent high throughput screening efforts have shown that this malignancy possesses one of the lowest mutation rates amongst all cancers (0.15 mutations/Mb) (5–7), yielding a paucity of pharmacologically actionable mutations.
Histone methylation is a major determinant of chromatin structure and function and has shown to play critical roles in transcriptional regulation and genomic stability (8). KDM1A (LSD1/BHC110), was the first flavine adenine dinucleotide (FAD)-dependent lysine specific demethylase identified to regulate chromatin states through the removal of mono and dimethyl groups (H3K4 or H3K9) (9). KDM1A is overexpressed in several solid and hematological malignancies including breast, colorectal, prostate, ovarian cancer and sarcomas with KDM1A inhibition significantly reducing cell proliferation, migration and invasion in vitro (10). Notably, elevated KDM1A expression is associated with poor clinical outcome (11–14). These observations have led to the active development of several small molecule KDM1A inhibitors, with Tranylcypomine (NCT02273102, NCT02261779), GSK-LSD1 (NCT02177812, NCT02034123) and ORY-1001 (2013-002447-29) currently undergoing phase I/II clinical evaluation in patients with acute myeloid leukemia (AML) and small cell lung carcinoma (SCLC).
In 2012 Bennani-Baiti et al., documented KDM1A overexpression in ES patient tumors (15) highlighting the therapeutic utility of targeted KDM1A inhibition for this disease. We previously demonstrated that directed KDM1A inhibition in ES with the use of SP-2509 (previously HCI-2509), a non-competitive, phenylethylidene-benzohydrazide reversible KDM1A inhibitor (Ki: 31nM, KDM1A IC50:13nM) (16), reverses the EWS/ETS transcriptional signature, impairs several EWS/ETS-associated oncogenic phenotypes, and shows single-agent efficacy in multiple xenograft models of ES (17). However the precise mechanism by which SP-2509 induces apoptosis remains unknown. In addition, it is imperative that biomarkers of ES sensitivity and resistance to this exciting epigenetic therapeutic are elucidated to guide clinical trial patient stratification. In this report, we have built on our previous work and conducted a series of studies to define mechanisms of ES SP-2509 sensitivity as well as the biological pathways that govern apoptotic cytotoxicity.
Materials and Methods
Cell lines and compounds
Cell lines were sourced and cultured as detailed in Supplementary Table S1. Cell cultures were not used beyond 2 months from initial thawing with culture supernatants tested yearly for Mycoplasma infection using a PCR based detection kit (Southern Biotech, USA). Cell lines were authenticated by STR profiling (Genetica LabCorp, USA). SP-2509 was provided by Dr Sunil Sharma (TGen Clinical Sciences), Doxorubicin hydrochloride and GSK-LSD1 (18) were purchased from Cayman Chemical, Thapsigargin and AraC (Cytosine Arabinoside) were sourced from Sigma-Aldrich. [3H]GSK-LSD1 (specific activity, 9 Ci/mmol) was purchased from ViTrax Radiochemicals.
Immunodetection
Whole cell lysate (35μg) was run on 4-15% Tris-Glycine polyacrylamide gels and transferred onto nitrocellulose membranes using a iBlot2 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Membranes were blocked in Odyssey Blocking Buffer (PBS) (LI-COR Bioscience) for 1hr at room temperature followed by overnight 4°C incubation with primary antibodies. Immunodetection was achieved after incubation with infrared (IR)-dye-conjugated 800CW secondary antibodies (LiCor) with bands visualized using the Odyssey Imaging System. Antibodies provided in Supplementary Table S1. Densitometry analysis was performed using ImageJ software (V1.51).
IncuCyte cell proliferation and caspase 3/7 assay
ES cells were seeded (4000-10000 cells/well) in 96 clear micro-titer plates (triplicate wells per condition) and left to adhere overnight. If required, compounds of interest were added to each well (SP-2509, GSK-LSD1, Doxorubicin) 18hrs post seeding. Real-time apoptosis assays were assessed through the addition of IncuCyte Caspase-3/7 Apoptosis Assay Reagent (Essen BioSciences), final concentration of 5μM. Phase contest and/or green fluorescent images were taken in the IncuCyte ZOOM Kinetic Imaging System (Essen BioScience) at 3hr intervals for a minimum of 96hrs. Cell confluence (Phase contrast) or Green Fluorescence (Green Object Count per well) was evaluated using IncuCyte ZOOM 2016A software (Essen BioScience). Data represents mean ± SEM from a minimum of three independent experiments.
qRT-PCR
Total RNA was extracted using the RNeasy kit (Qiagen) with on-column DNase digestion. cDNA synthesis and subsequent qRT-PCR was performed with 50ng of total RNA using iTaq Universal SYBR Green 1 Step Reaction Mix (Bio-Rad), according to the manufacturer’s protocol. Primer specificity was determined using melting curve analysis with all amplicons sequenced verified during primer optimization studies. Reactions were processed on a CFX connect Real-Time System (Bio-Rad) with subsequent gene expression quantified using the ΔΔCT method from triplicate reactions. Gene expression was normalized to the internal housekeeping gene RPL19. Primer sequences are listed in Supplementary Table S1.
shRNA generation
shRNA’s targeting the 3’UTR of KDM1A, KDM1B and ERN1 were designed using sirna.wi.mit.edu (Whitehead Institute at MIT). shRNA sequences are listed in Supplementary Table S1. To create shRNA expression retroviral constructs, oligonucleotides were annealed and cloned into Age1 and EcoR1 sites of pMKO.1. To generate retroviruses, 293 EBNA cells were co-transfected (Mirus Bio TransIT-LT1) with retroviral expression plasmids (10μg), vesicular stomatitus virus-G glycoprotein and gag/pol packaging plasmids, with filtered viral supernatant collected over a 48hr period. Retroviral infected ES cell lines were selected in puromycin (0.05-2mg/ml) (Sigma) for a minimum of 48hrs.
Soft agar assays
Cells were seeded at a density of 7500-12500 cells per 6cm plate in 0.8% SeaPlaque GTG agarose (Lonza), in media containing 20% FCS, Iscove’s modification of Eagle’s media, penicillin/streptomycin/glutamine and puromycin. Duplicate plates per condition were seeded. Colonies were quantified using ImageJ software (V1.51) a minimum of 16 days post seeding.
Viability assays
ES cells were seeded (4000-10000 cells/well) in 96 white micro-titer plates (triplicate wells per condition) and left to adhere overnight. Cells were treated with vehicle control, media control or serial dilutions of SP-2509 18hrs post seeding (0.1% final DMSO concentration). Cell viability was assessed 72hrs post treatment using Cell Titer-Glo (Promega) according to the manufacturer’s instructions with luminescence read on a GloMax 96 Microplate Luminometer (Promega). Viability was calculated relative to vehicle control cells with IC50 values calculated using GraphPad Prism (Version 7.00).
XBP1 splice variant PCR
To amplify unspliced and spliced XBP1 variants, 50ng of total RNA was reverse transcribed and amplified using iTaq Universal SYBR Green 1 Step Reaction Mix (Bio-Rad) and the following primers (19): Forward: 5’ TTACGAGAGAAAACTCATGGCC’3, Reverse 5’ GGGTCCAAGTTGTCCAGAATGC ’3. PCR conditions were as follows; 95°C for an initial 1min followed by 50 cycles of denaturation at 95°C for 10secs, annealing at 57°C for 30secs and extension at 72°C for 30secs. PCR products were electrophoresed on a 2.5% agarose gel. Size difference between unspliced XBP1 (289bp) and spliced XBP1 (263bp) is 26 nucleotides.
Statistically analysis
P-values were calculated using Student t-test using Graph Pad Prism (Version 7). Additional methods provided in Supplementary Table S1.
RESULTS
KDM1A is highly expressed in ES tumors and cell lines
KDM1A represents an ideal therapeutic target for the treatment of ES for two reasons; KDM1A is highly expressed in ES tumors (15) and secondly no mutations in KDM1A have ever been documented in ES tumor samples (10). To confirm elevated KDM1A expression in ES compared to other malignancies, we examined the Broad Institute Cancer Cell Line panel database (1035 cell lines, 36 distinct cancer subtypes). ES was the second highest KDM1A expressing malignancy after T-cell AML (Fig. 1A), with expression levels significantly higher than other solid bone sarcomas including osteosarcoma (P=0.0206) and chondrosarcoma (P=0.0497). These findings were further supported through immuno-histochemical analysis of ES tumor microarray’s which demonstrated moderate-strong KDM1A expression in the majority of tumors (62.1%) (Fig. 1B).
Fig. 1. KDM1A is highly expressed in Ewing sarcoma cell lines and tumors.
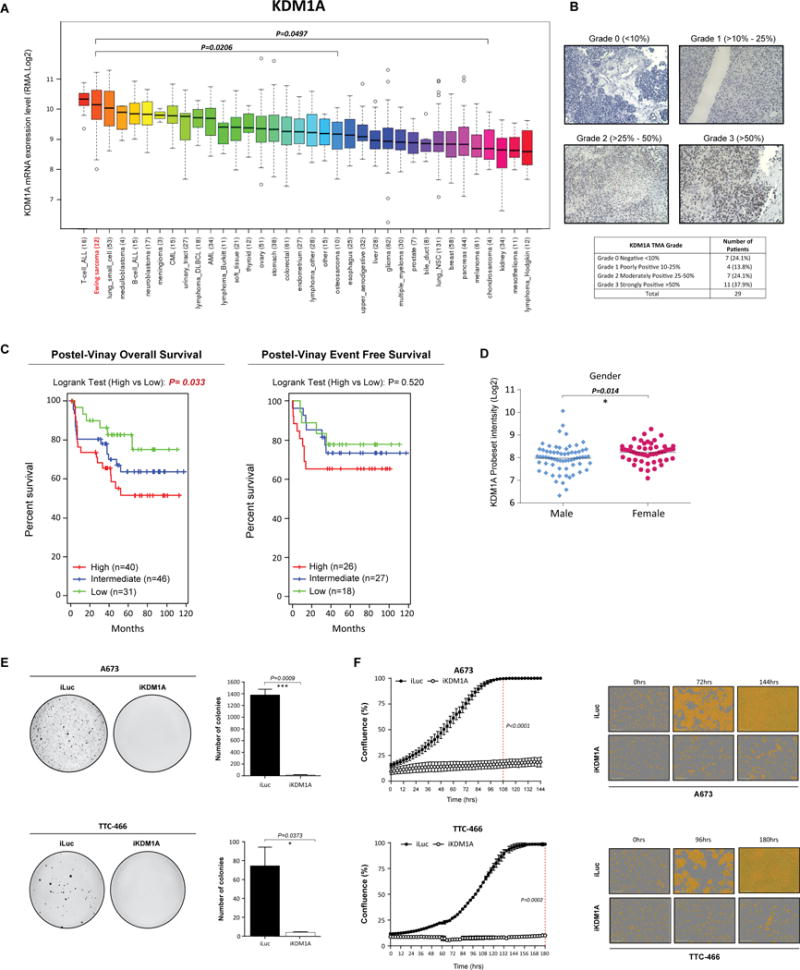
(A) BROAD Institute Cancer Cell line expression data of KDM1A across 36 distinct cancer entities. (B) Representative immuno-histochemical analysis of KDM1A in Ewing sarcoma patient tumors. (C) Event free and overall survival of Ewing sarcoma patients stratified by high (median +0.5xMAD), intermediate and low (median -0.5xMAD) KDM1A expression. Expression data obtained from Postel-Vinay et al., 2012. (D) Correlation between KDM1A expression and patient gender. Expression data obtained as described in (D). Log-rank (Mantel Cox Test) used to determine survival significance. (E) Representative images and quantification of soft agar colonies of A673 and TTC-466 cells stably transduced with iKDM1A or iLuc control shRNA constructs following Puromycin selection. (F) Real-time live cell imaging (IncuCyte ZOOM) of A673 and TTC-466 cells transduced with iKDM1A or iLuc shRNA constructs. Cell proliferation measured for 144 and 180hrs respectively. Dashed line represents 100% confluency (iLuc). Representative phase contrast images are also depicted. Data represents mean ± SEM from three independent experiments for A673 cells and mean ± STDEV from two independent experiments for TTC-466 cells. Asterisks denote statistical significance (*P<0.05, ***P <0.001).
We next elevated whether KDM1A expression correlates with ES clinical outcome or pathological features. Analysis of KDM1A expression in six ES micro-array/RNA-seq expression platform studies was conducted (6,20–24) with no correlation between KDM1A expression and pathological criteria including age, tumor location, EWS/ETS translocation or presence of metastatic disease observed in any study (Supplementary Table S2). Although females displayed significantly higher KDM1A expression compared to males (P=0.014) in one large cohort of 117 patients (Fig. 1D), this association was not observed in four smaller cohort studies (n=14-46 patients) (Supplementary Table S2). Notably, high KDM1A expression was significantly correlated with worse overall survival (Postel-Vinay 2012, P=0.033) and a worse overall survival trend in two additional small studies (Ohali 2004, P=0.054 and Volchenboum 2015, P=0.052) (Fig. 1C, Supplementary Fig. 1A). In contrast high KDM1A expression was significantly correlated with poor event free survival only in 1/4 studies (Ohali 2004, P=0.016) (Supplementary Fig. 1A). Data described in Postel-Vinay is the only study that assessed KDM1A expression in chemotherapy/radiotherapy naïve primary tumor samples. The remaining 3 studies all included patient samples from heavily treated and relapsed patients which could account for the discrepancy in survival findings. Together, although the clinical ES cohorts are small, our findings suggest that high KDM1A expression levels are associated worse overall survival, and that targeting KDM1A has broad utility for this aggressive malignancy.
KDM1A is required for ES tumorigenesis
We previously demonstrated targeted inhibition of KDM1A with SP-2509 significantly impairs the tumorigenic growth properties of ES cell lines (17). As such we sought to investigate whether direct loss of KDM1A recapitulates these findings in vitro. shRNA retroviral knockdown of KDM1A significantly impaired the anchorage-independent growth of A673 and TTC-466 (95.2/18.5 fold reduction in colony number respectively) ES cell lines in soft agar (Fig. 1E, Supplementary Fig 1B/C). Correspondingly, silencing of KDM1A significantly reduced the proliferative capacity of both cell lines as shown through IncuCyte live imaging analysis. Following 108hrs of growth, confluency of A673 iLuc shRNA control cells was 100% versus 16.8% for KDM1A knockdown cells (P<0.0001) (Fig. 1F). Analogous results were observed for TTC-466 cells suggesting that both small molecule inhibition (SP-2509) and direct depletion of KDM1A impairs EWS/ETS driven oncogenic transformation.
Targeted KDM1A inhibition and EWS/FLI knockdown induces similar transcriptional signatures
As we previously established that SP-2509 comprehensively disrupts the transcriptional signature of EWS/ETS (17), we next investigated whether genetic depletion of KDM1A mirrors the gene expression profiles of both SP-2509 treatment and EWS/FLI knockdown. RNA-seq analysis following KDM1A shRNA knockdown in A673 cells revealed 880 genes significantly induced (KDM1A repressed genes) (>2Log2 fold) and 952 genes significantly down-regulated (KDM1A activated genes) (Supplementary Fig. 2A). Ingenuity Pathway Analysis (IPA) revealed the top canonical enriched pathways associated with KDM1A activated and repressed genes were ethanol degradation IV (P=1.89×10−9) and hepatic stellate cell activation (P=7.51×10−9) respectively. We next assessed the similarity between the KDM1A and EWS/FLI driven transcriptional signature. Using our previous published A673 EWS/FLI knockdown RNA-seq data set (17), significant overlap (χ2) (P<0.0001) between these two transcriptional profiles was observed (Supplementary Fig. 2B/C). In total 195 repressed genes (11.2%) were commonly induced upon KDM1A-EWS/FLI knockdown, which were highly enriched for hepatic stellate cell activation (P=3.56×10−11) and HMGB1 signaling (P=7.69×10−08). Having established a similarity between EWS/FLI and KDM1A transcriptional profiles, we next compared the global signature of small molecule KDM1A blockade against KDM1A shNRA knockdown. χ2 analysis revealed a statistically significant overlap (P<0.0001) between genes activated/repressed following A673 treatment with SP-2509 (2μM) and KDM1A knockdown. In total 162 genes were commonly induced upon SP-2509/KDM1A knockdown (KDM1A repressed genes) with 107 genes commonly down-regulated (KDM1A activated genes) (Supplementary Fig. 2D/E). IPA of these cohorts established that granulocyte/agranulocyte adhesion and diapedesis, HMGB1 signaling, hepatic stellate cell activation, and Il-8 signaling were highly enriched for KDM1A knockdown/SP-2509 induced genes, with genes significantly repressed following SP-2509/KDM1A treatment associated with G2/M DNA damage checkpoint regulation (P=1.26×10−04). We alternatively compared SP-2509 and KDM1A transcriptional profiles using GSEA with a more stringent cutoff of a 3-fold for the KDM1A knockdown gene set (Supplementary Fig. 2F). GSEA showed significant correlation (P<0.001) for genes both up- and downregulated upon KDM1A knockdown in the SP-2509-regulated gene list. Together, these results suggest that genes regulated by KDM1A in ES correlate with EWS/FLI transcriptional function, with SP-2509 expression profiles strongly mimicking KDM1A genetic depletion.
ES cell lines are hypersensitive to reversible KDM1A inhibition
We next sought to elucidate the molecular basis of SP-2509 induced apoptotic cytotoxicity. Seventeen ES cell lines with varying STAG2 and TP53 mutational status were treated with SP-2509 for 72hrs with viability determined through Cell Titer Glo (CTG). Notably, innate resistance to SP-2509 was not observed, with all cell lines sensitive to the cytotoxic effects of KDM1A inhibition, IC50 range 81nM-1593nM (Mean IC50 0.621 ± 0.095μM) (Table 1). Importantly primary hMSC cells, a putative ES cell of origin and TIP5 fibroblasts remained unaffected at these low concentrations, SP-2509 IC50 of >4μM and 18.2μM respectively. To confirm the superior cytotoxicity of SP-2509 in ES, sensitivity of osteosarcoma, rhabdomyosarcoma, prostate (25) and neuroblastoma (26) cell lines to SP-2509 was also assessed. ES cell lines were significantly more sensitive (range: 2.1-11.4 fold greater) to SP-2509 compared to all other cancer subtypes (Supplementary Fig. 3A).
Table 1.
Genomic characteristics and SP-2509 sensitivity of the Ewing sarcoma cell line cohort
| Cell line | Histology | Fusion Gene | TP53 Status | STAG2 Status#^ | KDM1A Status# | KDM1A mRNA Expression | 72hr SP-2509 IC50 (μM) | 144hr GSK-LSD1 IC50 (μM) |
|---|---|---|---|---|---|---|---|---|
| SK-N-MC | ES | EWS/FLI Type I | Truncation | Wild-type | UN | 2.80 ± 0.04 | 0.081 ± 0.002 | ND |
| A673 | ES | EWS/FLI Type I | Q119fs | Wild-type | Wild-type | 8.49 ± 1.29 | 0.109 ± 0.030 | >330 |
| TC252 | ES | EWS/FLI Type I | Wild-type | UN | Wild-type | 2.23 ± 0.07 | 0.183 ± 0.017 | 266.23 ± 12.99 |
| TC32 | ES | EWS/FLI Type I | Wild-type | I636fs | Wild-type | 2.32 ± 0.23 | 0.226 ± 0.032 | 221.67 ± 18.03 |
| SK-ES-1 | ES | EWS/FLI Type II | C176F | Q735* | UN | 7.08 ± 1.47 | 0.418 ± 0.017 | ND |
| ES-4 | ES | EWS/FLI Type II | Wild-type | Wild-type | UN | 2.15 ± 0.30 | 0.502 ± 0.101 | >330 |
| TTC-466 | ES | EWS/ERG | R175G | Null exp | Wild-type | 7.10 ± 2.27 | 0.536 ± 0.078 | >330 |
| CHLA-9 | ES | EWS/FLI Type I | Wild-type | V628insTDI | UN | 2.75 ± 0.27 | 0.547 ± 0.029 | ND |
| VH64 | ES | EWS/FLI Type II | Wild-type | UN | UN | 1.18 ± 0.19 | 0.561 ± 0.043 | ND |
| EW-8 | ES | EWS/FLI Type I | Y220C | N475fs | Wild-type | 3.52 ± 0.20 | 0.568 ± 0.031 | ND |
| CADO-ES1 | ES | EWS/ERG | Wild-type | Wild-type | Wild-type | 3.92 ± 1.21 | 0.593 ± 0.068 | ND |
| CHLA-258 | ES | EWS/FLI Type III | Wild-type | Wild-type | Wild-type | 0.67 ± 0.02 | 0.810 ± 0.126 | 300.14 ± 43.82 |
| RD-ES | ES | EWS/FLI Type II | R273C | Null exp | Wild-type | 5.66 ± 0.75 | 0.857 ± 0.068 | ND |
| EWS-502 | ES | EWS/FLI Type I | C135F | Null exp | Wild-type | 4.99 ± 0.17 | 0.902 ± 0.078 | >330 |
| CHLA-10 | ES | EWS/FLI Type I | NF | UN | UN | 2.94 ± 0.26 | 0.952 ± 0.060 | >330 |
| ES-2 | ES | EWS/FLI Type III | R175H | E523* | UN | 1.98 ± 0.22 | 1.128 ± 0.126 | >330 |
| TC-71 | ES | EWS/FLI Type I | R213* | Wild-type | Wild-type | 3.14 ± 0.50 | 1.593 ± 0.036 | ND |
| hMSC | hMSC | - | UN | UN | UN | - | >4 | ND |
| TIP5 | HUFF | - | UN | UN | UN | - | 18.20 ± 2.131 | ND |
ES: Ewing sarcoma, hMSC: Primary human mesenchymal stem cells, HUFF: Human foreskin fibroblasts, ND: Not determined, NF: Non Functional, UN: Unknown, Null exp: No expression
Fusion Gene: Type I (Exon 7 EWS/Exon 6 of FLI1), Type II (Exon 7 EWS/Exon 5 FLI1) and Type III (Exon 10 EWS/Exon 6 FLI-1)
Crompton et al (2014),
Brohl et al (2014)
To determine the antitumor activity associated with catalytic inhibition of KDM1A, a panel of ES cell lines was also treated with the irreversible KDM1A inhibitors tranylcypromine (72hrs) and its more specific next generation analogue derivative GSK-LSD1 (144hrs). Unlike SP-2509, tranylcypromine and GSK-LSD1 both covalently bind the FAD pocket of KDM1A. Strikingly, the low dose rapid cytotoxicity profile of SP-2509 was not recapitulated with either compound (Supplementary Fig. 3B). GSK-LSD1 concentrations exceeding 300μM were required to achieve 50% anti-proliferative activity in 7/9 cell lines and >400μM for tranylcypromine in 6/6 cell lines.
To address whether the pronounced disparity in cellular activity between irreversible and reversible KDM1A inhibitors were due in part to intracellular drug accumulation, whole cell and nuclear uptake of tritiated [3H] GSK-LSD1 was assessed. GSK-LSD1 resistant ES cell lines (TC252, A673) and two additional cell lines HL60 and H1417 (AML and SCLC respectively) previously shown to be hypersensitive to the cytotoxic effects of GSK-LSD1 both in vitro (EC50 <20nM) and in vivo (18) were treated with [3H] GSK-LSD1 (1μM). No significant difference in both whole cell lysate and nuclear uptake between ES and hypersensitive GSK-LSD1 control cell lines was observed following incubation with [3H] GSK-LSD1 (5min and 2hr treatment) (Supplementary Fig. 3C). Our data suggests that failure of GSK-LSD1 to impart a cytostatic effect in ES cell lines cannot be attributed to the inability of this KDM1A inhibitor to accumulate within the cell.
Finally, in order to assess whether irreversibly blocking KDM1A enzymatic activity has any effect on SP-2509 sensitivity, A673 and TC252 cells were pretreated with GSK-LSD1 for 48hrs prior to treatment with SP-2509 alone or in combination with GSK-LSD1 for an additional 72hrs. No significant change in SP-2509 sensitivity was observed following pre-treatment with GSK-LSD1 in both A673 and TC252 cells, suggesting that blockade of KDM1A enzymatic activity is not required for SP-2509 to induce cytotoxicity in Ewing sarcoma (Supplementary Fig. 3D). We propose that ES cells are more susceptible to disruption of the KDM1A-EWS/ETS-chromatin regulatory complex (17) rather than enzymatic irreversible inactivation through direct covalent modification of the KDM1A FAD pocket.
SP-2509 but not GSK-LSD1 significantly impairs the proliferative growth of ES cell lines
To confirm that GSK-LSD1 has no effect on the proliferative growth of ES cell lines, A673, TC252 and ES-2 cells were treated with SP-2509, GSK-LSD1 or the ES chemotherapeutic agent Doxorubicin for 96hrs. As expected, Doxorubicin significantly impaired the proliferative growth of all three cell lines as early as 18hrs post treatment (ES-2, P=0.020) (Fig. 2) with strong induction of apoptosis observed as quantified through caspase 3/7 activity. Similarly SP-2509 concentrations exceeding 0.5μM in hypersensitive cell lines (A673, TC252) and 1μM in the sensitive cell line ES-2, significantly reduced proliferative capacity as early as 42hrs (A673 and ES-2). In contrast no significant difference in cellular growth or caspase 3/7 induction between GSK-LSD1 and vehicle control treatment was observed for all three cell lines across all time-points, underscoring the superiority of reversible KDM1A inhibitors for the treatment of ES.
Fig 2. SP-2509 but not GSK-LSD1 significantly reduces the proliferative capacity of Ewing sarcoma cell lines.
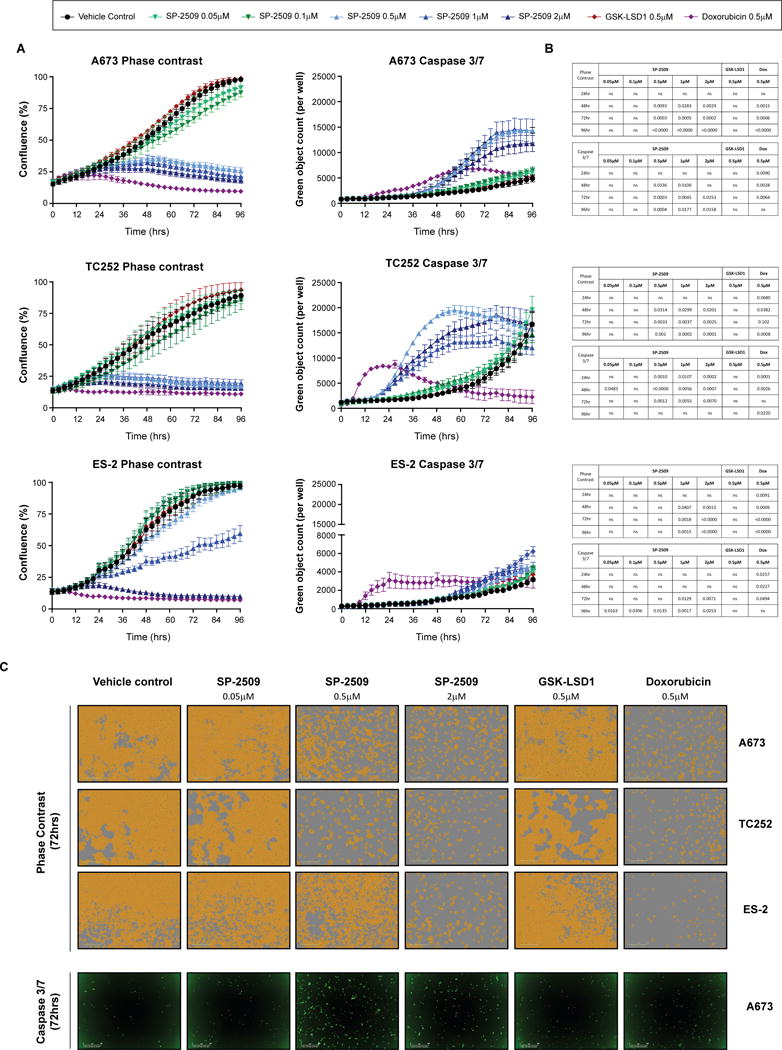
(A) SP-2509 hypersensitive (A673, TC252) and sensitive (ES-2) Ewing sarcoma cell lines treated with the indicated concentrations of SP-2509, GSK-LSD1, Doxorubicin or vehicle control (DMSO) for 96hrs. Proliferative capacity and induction of apoptosis (caspase 3/7 activity) was measured in real-time through IncuCyte ZOOM live cell imaging. Data represents mean ± SEM confluency or green object count, from three independent experiments. (B) Statistical analysis of proliferation or caspase 3/7 induction compared to vehicle control cells (ns: not significant). (C) Representative phase contrast and green fluorescence (caspase 3/7) images following 72hrs of treatment with the indicated agents.
TP53/STAG2/CDKN2A status and basal KDM1A expression levels do not confer SP-2509 cytotoxicity
Several recent large scale sequencing studies have confirmed the low mutational burden in ES tumors, with only recurrent deleterious mutations reported in STAG2 (21.5%) and TP53 (6.2%) and focal deletions at the CDKN2A locus (5,6,27). As such, we investigated whether STAG2, TP53 and CDKN2A status correlate with SP-2509 sensitivity. Despite that 3/5 hypersensitive SP-2509 cells lines (IC50 <500nM) (SK-N-MC, A673 and SK-ES-1) harbor null/mutant TP53, no correlation between TP53 mutational status and SP-2509 sensitivity was observed (P=0.256). This TP53 finding was additional confirmed in our expanded panel of neuroblastoma, osteosarcoma, prostate and rhabdomyosarcoma cell lines (Supplementary Fig. 3E). Similarly, neither STAG2, CDKN2A status nor EWS/ETS translocation partner correlated with SP-2509 sensitivity (P=0.887, P=0.884 and P=0.866 respectively) (Fig. 3A). We next investigated whether basal KDM1A or KDM1B mRNA expression levels correlate with SP-2509 sensitivity. Although overexpression of KDM1A (>2 fold) was observed in 14/17 cell lines, no correlation between basal KDM1A or KDM1B mRNA levels was demonstrated (R2=0.039 and R2=0.013 respectively) (Fig. 3B/C). In addition, although strong protein expression of KDM1A was evident across the entire cell line cohort, no significant association between SP-2509 sensitivity and KDM1A, KDM1B, FLI and p53 protein levels as well as cell doubling time (28) or whether the cell line was obtained from chemotherapy-naïve or treated tumors, was observed (Fig. 3D and Supplementary Fig. 3F/G).
Fig. 3. Basal KDM1A mRNA and protein levels do mediate SP-2509 sensitivity in Ewing sarcoma.
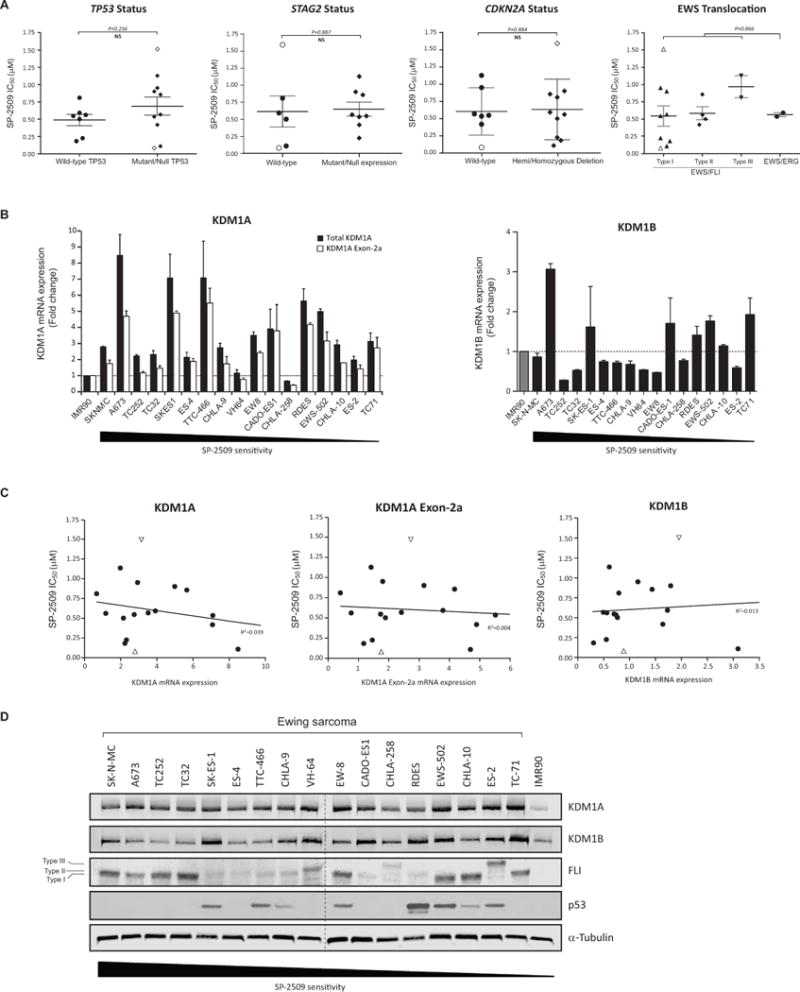
(A) Lack of correlation between TP53/STAG2/CDKN2A status, EWS/ETS translocation partner and SP-2509 sensitivity (IC50). (B) Relative mRNA expression (fold change) of KDM1A, KMD1A Exon-2a and KDM1B in the Ewing sarcoma cell line cohort. Expression normalized to IMR90 cells. Data represents mean mRNA expression ± STDEV from two independent experiments. (C) Lack of correlation between KDM1A, KMD1A Exon-2a and KDM1B mRNA expression levels and SP-2509 sensitivity (IC50). (D) Western blot analysis of KDM1A, KDM1B, FLI and TP53 protein expression in the cell line cohort. Cell lines are ranked in order of SP-2509 sensitivity (IC50). Data represents mean SP-2509 IC50 ± SEM from three independent experiments. Open symbols denote most (SK-N-MC) and least (TC-71) SP-2509 sensitive cell lines.
Four unique KDM1A isoforms generated through either single or double insertion of two alternatively spliced exons (2a and 8a) in the unstructured amino-terminus and amino oxidase domains of KDM1A respectively have been identified (29). As retention of the exon 8a splice variant is restricted to cells of neuronal lineage, mRNA expression levels of exon 2a (60bp) was evaluated in our cell line panel. Overexpression of exon 2a was observed in 7/17 cell lines and strongly correlated with total KDM1A mRNA levels (R2=0.902) (Fig. 3B, Supplementary Fig. 3H). Similarly to total KDM1A, no association between basal KDM1A exon 2a mRNA expression levels and SP-2509 sensitivity was observed (R2=0.004) (Fig. 3C). Our data demonstrates that regardless of KDM1A expression levels, ES cells are susceptible to the cytotoxic effects of SP-2509, thereby strengthening the broad utility of targeted KDM1A inhibition for the treatment of ES.
KDM1B expression levels post SP-2509 treatment strongly correlate with SP-2509 sensitivity
To assess whether SP-2509 treatment directly modulates KDM1A, KDM1B and EWS/FLI mRNA and protein levels, a panel of eight ES cell lines of varying sensitivity were treated with SP-2509 (2μM) for 48hrs. Although KDM1A mRNA levels were significantly down-regulated in 7/8 cell lines, maximum of 52.0% (A673), the degree of KDM1A mRNA reduction did not correlate with SP-2509 sensitivity (Supplementary Fig. 3I) (R2=0.022). In contrast KDM1B mRNA levels were significantly induced following SP-2509 treatment in 7/8 cell lines with the greatest induction observed in the hypersensitive SP-2509 cell line A673 (2.6 fold) (Supplementary Fig. 3J). KDM1B mRNA levels were strongly predictive of SP-2509 sensitivity (R2=0.562), suggesting that hypersensitive ES cell lines positively induce KDM1B following SP-2509 compared to sensitive cell lines. Similarly, KDM1B protein levels were significantly induced (>25%) only in hypersensitive cell lines 44 and 55hrs post SP-2509 treatment (maximum 1.72 fold induction, TC252) (Supplementary Fig. 6A). EWS/FLI mRNA levels were modulated in 4/5 cell lines, with a maximum reduction of 80.5% observed in TC32 cells (Supplementary Fig. 3K). However, this striking effect was only observed at the transcript level, as only a significant reduction in KDM1A but not EWS/FLI protein levels was observed across multiple cell lines, maximum reduction of 26.1% (A673) (Supplementary Fig. 3L).
To exclude the possibility that SP-2509’s modulation of the KDM1A, KDM1B and EWS/FLI signature was simply a nonspecific response to treatment with cytotoxic agents, we sought to determine whether other compounds such as GSK-LSD1 and Doxorubicin also induce similar gene expression patterns (Supplementary Fig. 4A/B). No consistent changes (mRNA and protein) in KDM1A, KDM1B or EWS/FLI were observed across multiple ES cell lines following treatment with Doxorubicin or GSK-LSD1. Finally to elucidate whether small molecule modulation of EWS/FLI protein levels also affects KDM1A and KDM1B expression, A673, TC252 and ES-2 cells were also treated with the nucleoside analog ARA-C (cytosine arabinoside), a known modulator of EWS/FLI protein but not transcript levels (30). Although a significant reduction in EWS/FLI protein levels was achieved, no consistent change across all three cell lines was observed for KDM1A and KDM1B protein levels (Supplementary Fig. 4C/D). Together, these findings suggest that the ability of SP-2509 to modulate KDM1A, KDM1B and EWS/FLI expression levels is a direct affect, and not simply a cellular stress response to cytotoxic agents.
KDM1B mediates SP-2509 sensitivity in hypersensitive ES cell lines
On the premise that induction of KDM1B following SP-2509 treatment is significantly correlated with SP-2509 cytotoxicity, we next addressed the role of KDM1B in mediating SP-2509 sensitivity. Stable retroviral knockdown of KDM1B in the hypersensitive SP-2509 cell line A673 and sensitive cell line EWS-502 was achieved with two KDM1B shRNAs (shRNA #3 and shRNA #7). KDM1B targeting shRNA effectively reduced endogenous KDM1B mRNA expression by a maximum of 76.1% and 86.1%, A673 and EWS-502 cells respectively (Fig. 4A). Importantly, no significant reduction in KDM1A protein or mRNA levels were observed for both KDM1B hairpins, (Fig. 4B/C). Interestingly, knockdown of KDM1B significantly reduced the proliferative capacity of both A673 and EWS-502 cells compared to non-targeting control (iLuc). Following five day (120hrs) IncuCyte proliferation analysis, A673 confluency for iLuc, KDM1B shRNA #3 and KDM1B shRNA #7 generated cell lines were as follows, 99.4%, 27.9% and 41.7%. Analogous results were observed for EWS-502 (Fig. 4D). Finally, to address whether KDM1B has any effect on SP-2509 sensitivity, non-targeting and KDM1B shRNA A673 and EWS-502 cell lines were treated with SP-2509 (0-4μM). Following 72hrs of treatment, KDM1B knockdown significantly reduced the sensitivity of A673 cells to SP-2509 compared to controls by 2.3 and 4.3 fold, shRNA #3 and shRNA #7 respectively. In contrast, KDM1B knockdown had no effect on EWS-502 sensitivity (IC50: 1.386μM iLuc, 1.250μM KDM1B shRNA #3, 1.370μM KDM1B shRNA #7) (Fig. 4E), advocating a potential role for KDM1B driving SP-2509 hypersensitivity.
Fig. 4. KDM1B mediates SP-2509 sensitivity in hypersensitive Ewing sarcoma cell lines.
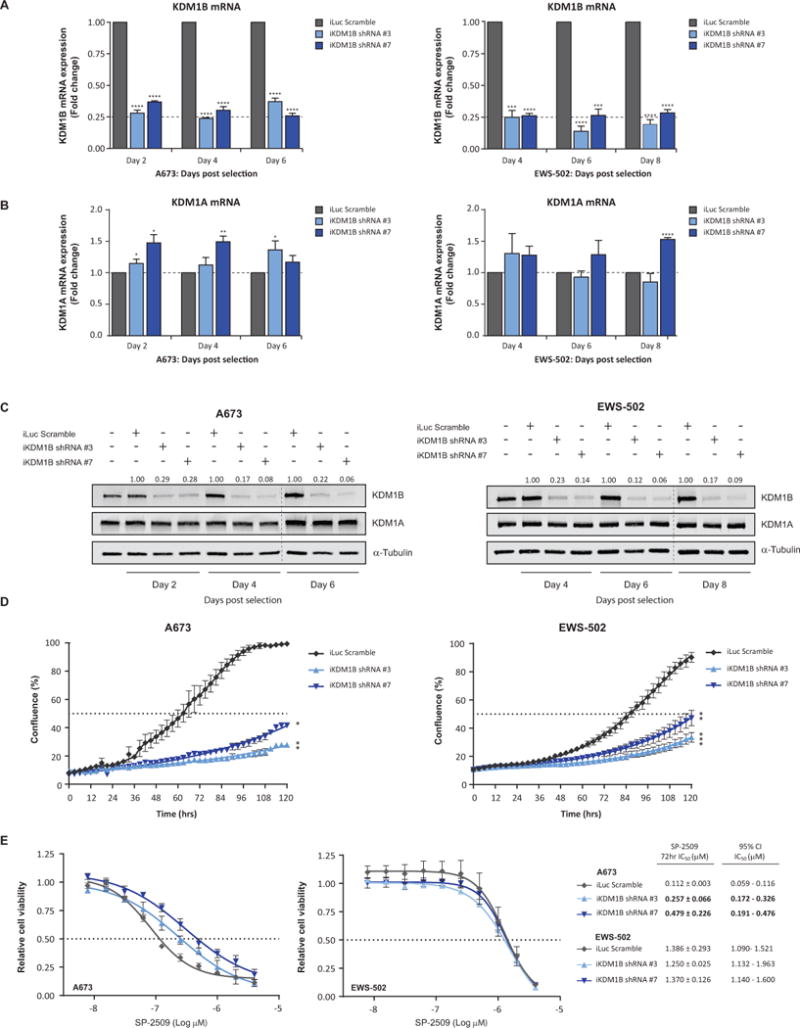
Relative (A) KDM1B and (B) KDM1A mRNA levels in A673 and EWS-502 cells following transduction with two unique KDM1B knockdown shRNA constructs (#3 and #7) or iLuc shRNA control. (C) Representative western blot analysis of KDM1B and KDM1A protein levels in A673 and EWS-502 cells following KDM1B knockdown. α-Tubulin was used a loading control. KDM1B densitometry normalized to α-Tubulin and relative to iLuc shRNA control is depicted. (D) Real-time live cell imaging (IncuCyte ZOOM) of A673 and EWS-502 cells transduced with KDM1B and iLuc shRNA constructs. Cell proliferation measured for 120hrs. (E) Cell viability analysis (Cell Titer Glo) of A673 and EWS-502 transduced with the indicated shRNA constructs and treated with SP-2509 (0-4μM) for 72hrs. Data represents mean ± SEM from three-four independent experiments. Asterisks denote statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
SP-2509 hypersensitive cell lines share similar basal transcriptomic profiles
To gain additional insight into the molecular mechanisms and pathways that mediate SP-2509 cytotoxicity and sensitivity, three hypersensitive (IC50<300nM: A673, TC252, TC32) and three sensitive (IC50>900nM: EWS-502, ES-2, TC-71) ES cell lines treated with either vehicle control (DMSO) or SP-2509 (2μM) were submitted for RNA-seq analysis. Unsupervised hierarchical clustering analysis of these transcriptomes showed that those cell lines that were highly sensitive to SP-2509 shared similar basal gene expression profiles and clustered separately from the sensitive cell lines (Supplementary Fig. 5A/B). In all, 424 genes were significantly differentially expressed between the two groups with HIST1H2AJ (Histone H2A type 1-J) being the most significantly differentially expressed gene (P=1.156×10−20). IPA revealed that the top four canonical pathways associated with this core subset of genes included actin cytoskeleton signaling (P=0.002), CdC42 signaling (P=0.0161), D-mannose degradation (P=0.020), and RhoA signaling (P=0.035).
To complement and extend these analyses, we next sought to identify which core subset of genes were specifically induced and repressed in our hypersensitive but not sensitive ES cell lines following SP-2509 treatment (Supplementary Fig. 5C/D). In total 44 genes were uniquely induced only in the hypersensitive cohort (2 fold) with ETS1, HIF1A-AS2, RASEF, KCNK9 and CRIM1 being the top five most highly expressed genes. Interestingly, 14/44 (31.8%) of these genes overlapped with known EWS/FLI regulated targets (17). Similarity, 53 genes were specifically repressed in our hypersensitive cell lines (2 fold) with HIST1H2BM, HIST1H2AJ, CEACAM5, ICAM3 and MORC1 being the top five most highly repressed genes following SP-2509 treatment. Of these genes 43.4% (23/53) overlapped with known EWS/FLI regulated targets.
SP-2509 induces endoplasmic reticulum stress in ES cell lines
On the basis that SP-2509 resistance was not observed in our ES cell line cohort, we next examined which genes are commonly induced and repressed across all cell lines to mediate SP-2509 cytotoxicity. In total 103 genes were commonly induced (>2 fold) with 82 genes universally repressed following SP-2509 treatment (Fig. 5A). Interestingly 30/103 (29.1%) and 33/82 (40.2%) of these genes overlapped with known EWS/FLI regulated targets, corroborating our previous findings that SP-2509 reverses the EWS/ETS-driven transcriptional program in ES (Supplementary Table S2). In addition, the majority of genes modulated by SP-2509 were protein coding (80.6% and 69.5%), followed by long coding RNA’s (17.5% and 15.9% up/down-regulated respectively) (Fig. 5B). Notably tRNA’s (11.0%) were also only specifically repressed following SP-2509 treatment.
Fig. 5. SP-2509 cytotoxicity is mediated through the ER-stress response pathway.
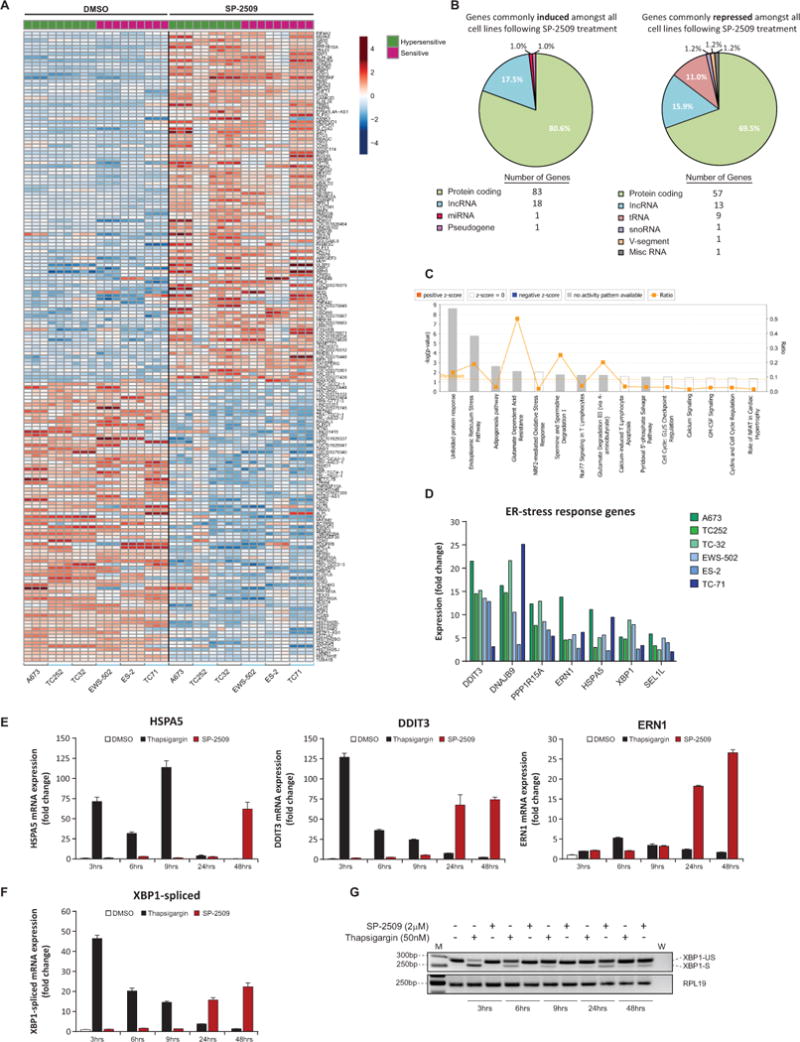
(A) Heatmap representation of genes commonly induced/repressed following SP-2509 treatment (2μM, 48hrs) across 6 ES cell lines compared to vehicle control (DMSO). Three independent replicates per treatment and cell line are depicted. Scale: Mean-centered rlog-transformed expression. (B) Distribution of SP-2509 regulated genes according to class. (C) IPA of the top canonical pathways associated with SP-2509 induced genes (>2 fold increase from vehicle control). (D) Normalized RNA-seq mRNA expression of ER-stress response genes following SP-2509 treatment across the Ewing sarcoma cell line cohort. (E/F) Relative mRNA expression levels of HSPA5, DDIT3, ERN1 and spliced XBP1 in TC252 cells following treatment with DMSO, Thapsigargin (50nM) or SP-2509 (2uM) for the indicated time periods. Data represents mean expression ± SEM from triplicate reactions. (G) PCR analysis of unspliced (XPB1-US) and spliced XBP1 (XPB1-S) in TC252 cells following treatment as in (E). “M” and “W” denote marker and water control respectively. RPL19 housekeeper was used as loading control.
IPA of SP-2509 induced genes uncovered the following five highly enriched pathways: unfolded protein response (P=2.15×10−9), endoplasmic reticulum (ER) stress pathway (P=1.56×10−6), EIF2 signaling (P=1.86×10−3), adipogenesis pathway (P=2.23×10−3), and NRF2-mediated oxidative stress response (P=8.09×10−3) (Fig. 5C). Indeed the following genes associated with the ER-stress response pathway; DDIT3, DNAJBP, PPP1R15A, ERN1, HSPA5, XBP1 and SEL1L, were significantly induced across all cell lines following SP-2509 treatment (maximum 25.2 fold increase from vehicle control) (Fig. 5D). For this reason, we focused on delineating the role of ER-stress in mediating SP-2509 cytotoxicity.
To confirm that SP-2509 activates the ER-stress response pathway, TC252 cells were treated with SP-2509 (2μM), vehicle control or thapsigargin (50nM), a chemical agent known to activate the UPR stress response by blocking ER calcium ATPase pumps, leading to depletion of ER calcium stores (31,32). Significant induction of HSPA5 (BIP) (71.4 fold) and the terminal UPR pro-apoptotic transcription factor DDIT3 (CHOP) (126.9 fold) occurred rapidly after thapsigargin treatment compared to vehicle control (3hrs) (Fig. 5E) with expression weakening (>10 fold induction) after 24hrs of treatment. In contrast, a time dependent increase in HSPA5 and DDIT3 following SP-2509 treatment was observed, with maximum thapsigargin comparable transactivation of HSPA5 and DDIT3 detected at 48hrs (62.2 fold and 74.3 fold respectively). Regardless of SP-2509 sensitivity, similar protein induction of ERN1, PERK and DDIT3 was observed across A673, TC252, ES-2 and EWS-502 cell lines following both thapsigargin and SP-2509 treatment. In contrast, marked protein induction of BiP/HSPA5 (>25% from DMSO control) 48 and 55hrs post SP-2509 treatment was only observed in our hypersensitive cell lines (Supplementary Fig. 6A). As BiP is a major ER chaperone protein critical for protein quality control of the ER and initiation of the UPR, the lack of protein induction in sensitive SP-2509 cell lines may account for the divergent SP-2509 IC50s documented across our cell line panel. Finally, mRNA levels of 12 known UPR pathway genes was assessed to determine whether hypersensitive cell lines are basally primed for greater robust apoptotic responses following SP-2509 treatment. No significant difference in any UPR mediators was observed between hypersensitive and sensitive SP-2509 cell lines (Supplementary Fig. 6B).
It is well established that during the initiation of the UPR, dissociation of HSPA5 from ERN1 (IRE1α) results in dimerization and activation of ERN1. The endoribonuclease domain of activated ERN1 subsequently cleaves a 26-nucleotide intron from XBP1, causing a frame shift enabling translation and generation of a basic leucine zipper family transcription factor, spliced XBP1. Spliced XBP1 consequently binds numerous UPR and ER-stress response genes leading to transcription of ER-chaperone proteins. Following thapsigargin treatment in TC252 cells, rapid induction of XBP1 spliced mRNA was observed (46.5 fold) as early as 3hrs post treatment which rapidly declined over a 48hr period to near pre-treatment levels (Fig. 5F). In contrast, similar to HSPA5, DDIT3 and ERN1, significant induction of spliced XBP1 was only observed 24hrs post SP-2509 treatment with peak induction observed 48hrs post drug exposure (22.3 fold increase from vehicle control). PCR analysis of the spliced (263bp) and un-spliced (289bp) XBP1 variants following thapsigargin and SP-2509 corroborated these findings. The slow kinetic splicing of XBP1 was only visualized 24 and 48hrs post SP-2509 treatment. In contrast spliced XBP1 was observed as early as 3hrs post thapsigargin treatment, with strong diminution of splicing observed with prolonged drug exposure (Fig. 5G). Taken together these results reveal that SP-2509 induces the UPR and ER-stress response, although at a slower kinetic rate than known UPR inducing agents.
Knockdown of ERN1 significantly attenuates the cytotoxic activity of SP-2509
The UPR comprises of three parallel signaling branches: EIF2AK3/PERK, ATF6α and ERN1/IRE1α. It is well accepted that the ER localized transmembrane receptor ERN1 is one of the first pro-survival arms activated upon ER stress, with subsequent attenuation of activity facilitating apoptosis in response to chronic unresolved stress. As ERN1 expression was notably induced across all six ES cell lines following SP-2509 treatment (Fig. 5A/D) and is responsible for the cleavage of XBP1, we next investigated whether stable retroviral knockdown of ERN1 mitigates SP-2509 cytotoxicity. Both ERN1 targeting shRNAs (shRNA #1 and #2) effectively reduced endogenous ERN1 mRNA and protein levels (maximum of 80.7% and 61.5% respectively) (Supplementary Fig. 6C-E). Following 72hr treatment with SP-2509 (0-4μM), shRNA knockdown of ERN1 (shRNA #2) significantly increased the concentration of SP-2509 required to reduce viability by 50% (2.06 fold increase). These findings corroborate our RNA-seq study and validate the role of the UPR response in instigating SP-2509 cytotoxicity in ES cells.
Both oxidative stress and nutrient deprivation are well established stimulators of the UPR response. In order to further evaluate how SP-2509 initiates this adaptive pathway, reactive oxygen species (ROS), HIF1α protein levels and 2-deoxyglucose-6-phosphate (2DG6P) uptake following SP-2509 treatment were assessed (Supplementary Fig. 6F-H). No induction of ROS or HIF1α was observed following SP-2509 treatment. In contrast, a significant dose dependent reduction in 2-deoxyglucose-6-phosphate (2DG6P) uptake was observed in both A673 and TC252 cells following treatment with SP-2509 (48/72hrs) suggesting that SP-2509 may engage the accumulation of misfolded proteins through nutrient deprivation.
Discussion
Despite global efforts, only minor improvement in overall survival rates for patients afflicted with relapsed and metastatic ES has been achieved over the past 40 years, emphasizing the need to move away from traditional non-specific agents to drugs that specifically target the underlying drivers of Ewing sarcomagenesis. We clearly show that ES cell lines and tumors express elevated levels KDM1A, with shRNA depletion abolishing the proliferative and tumorigenic properties of ES cells in vitro. Elevated KDM1A expression was associated with poor overall survival, consistent with several other solid cancers including ovarian (14), lung (11), esophageal squamous cell carcinoma (13), and neuroblastoma (12). Thus in addition to EWS/FLI, KDM1A may represent an additional Achilles’ heel of ES, which unlike EWS/FLI can be easily targeted through small molecule blockade.
In an effort to favorably impact patient outcome, the premise of this study was to elucidate mechanisms of sensitivity to the KDM1A reversible small molecule SP-2509, a non-competitive KDM1A inhibitor we have previously shown to reverse the EWS/ETS transcriptional signature and impair multiple EWS/ETS-associated oncogenic phenotypes both in vitro and in vivo (17). We reveal that proposed biomarkers such as TP53/STAG2/CDKN2A status in addition to basal KDM1A mRNA/protein levels do not correlate with SP-2509 sensitivity. However, our results uncovered a previously unappreciated role for KDM1B/LSD2, the only mammalian homologue of KDM1A, in SP-2509 hypersensitivity. In addition we demonstrate that induction of the UPR response mediates SP-2509 cytotoxicity in ES, a pathway previously unlinked with KDM1A blockade.
Notably, shRNA targeting of KDM1B significantly reduced the proliferative capacity of ES cell lines and mitigated the sensitivity of hypersensitive SP-2509 cells to small molecule KDM1A inhibition (Fig. 4). Furthermore, SP-2509 specific induction of KDM1B following treatment was also predictive of SP-2509 hypersensitivity (Supplementary Fig. 3). The reported SP-2509 IC50 for KDM1B is 11.2μM, well above the SP-2509 IC50 for our entire ES cell line cohort, validating that changes in KDM1B mRNA levels post treatment are not due to direct inhibition of KDM1B. In contrast no consistent upregulation of KDM1B was observed following 2μM treatment with GSK-LSD1 (KDM1B IC50 93.7μM).
Although KDM1A and KDM1B only share 33% amino acid sequence homology in the amine oxidase domain, they are both capable of demethylating the same enzymatic substrate, H3K4me1/2 (33). Emerging evidence has also supported that these two histone demethylases possess distinct functions, primarily due to the fact that KDM1B contains an N-terminal CW-type zinc finger domain which is absent in KDM1A and lacks a tower domain which is instrumental for binding to the co-repressor protein CoREST (33). Genome wide mapping has also revealed that KDM1B predominantly associates with the gene bodies of actively transcribed genes, but is markedly absent from promoters, which is in direct contrast to KDM1A which largely associates with promoter regions of genes (34). In addition functional classification of KDM1B-associated polypeptides suggests that the composition of the KDM1B complex is notably distinct from KDM1A, with KDM1B-associated proteins linked to splicing, DNA replication and damage repair, nucleosome remodeling and histone modification. Despite this knowledge, the precise role of KDM1B in cancer remains relatively unexplored. Amplification of KDM1B has been observed in breast (35), bladder urothelia carcinoma (36), and neuroendocrine prostate cancer (37), with stable shRNA silencing of KDM1B significantly reducing the colony formation capacity of breast cancer cell lines (38). Although high expression of KDM1B is associated with shorter overall survival in prostate cancer (39), we show that KDM1B expression is not predictive of event free or overall survival in ES (6,24) (Supplementary Fig. 6I/J).
In 2010, Fang et al., employed MS/MS proteomic analysis to identify KDM1B associated proteins, one of which was PARP-1 (34). It is well established that the EWS/FLI fusion present in ES drives and maintains PARP-1 expression, which in turn further promotes transcriptional activation by EWS/FLI (40,41). It may be plausible that induction of KD1MB in hypertensive SP-2509 cell lines, directly affects the interaction between KDM1B/PARP-1 thereby enhancing the execution of PARP-1 driven apoptosis following SP-2509 treatment. Indeed our results demonstrated that knockdown of KDM1B only in the hypersensitive SP-2509 cell line A673 but not EWS-502 (sensitive) significantly reduced the cytotoxic effects of SP-2509. Together, our findings lay the foundation for future studies to elucidate the specific role of KDM1B in mediating SP-2509 sensitivity.
To date, the precise mechanism by which SP-2509 drives ES cells towards an apoptotic fate remains unanswered. Significant induction of caspase 3/7 activity, a marker for apoptosis was clearly evident across multiple cell lines as early as 15hrs post SP-2509 treatment (Fig. 2). Previous studies have reported that KDM1A is required for cell cycle progression as it co-localizes with centrosomes in interphase and spindle poles during mitosis to permit proper chromosome segregation (42). Notably, down-regulation of KDM1A leads to abnormal centrosome duplication and significantly impairs nuclear pore complex assembly subsequently leading extended telophase (43). In contrast, our RNA-seq analysis of six Ewing cell lines post SP-2509 treatment revealed a strong enrichment for genes associated with the unfolded protein (UPR) and endoplasmic reticulum (ER) stress response (Fig. 5).
A number of pathophysiological extracellular perturbations (disturbed Ca2+ homeostasis, oxidative stress, metabolic changes, acidosis and nutrient deprivation), lead to the accumulation of unfolded/misfolded proteins in the ER subsequently triggering ER stress (44). The UPR is a double edge sword, initially promoting coordinated cytoprotective cascades to re-establish cellular homeostasis and preserve ER protein folding fidelity. However, under conditions of chronic unmitigated stress, UPR signaling shifts away from pro-survival adaptive programs with ER-stress-induced apoptosis ensued. The transcription factor C/EBP homologous protein CHOP (also known as DDIT3/GADD153) is a common point of convergence for all three arms of the UPR as evidenced by the discovery that the DDIT3 promoter contains binding sites for the major UPR induces ATF4, ATF6 and XBP1. In addition, it is well established that DDIT3 promotes UPR apoptotic cell death following unresolved ER stress (45). Indeed strong induction of DDIT3 was observed across all six ES cell lines; approximately 24hrs post SP-2509 treatment.
Our A673 KDM1A knockdown RNA-seq data revealed that several UPR genes including DDIT3, DNAJB9 and PPP1R15A are upregulated following KDM1A depletion. Similarly, DNAJB9, XBP1 and HSPA5 were identified as direct EWS/ETS repressed targets (17). In addition, to classify regulated proteins associated with EWS/FLI1, recent network i-TRAQ analysis of ES cell lines following EWS/FLI knockdown revealed XBP1 as the highest ranked pathway (46). These findings demonstrate that UPR targets are directly repressed by both KDM1A and EWS/ETS. Interestingly we show that SP-2509 significantly attenuates glucose uptake in ES cell lines. This observation is supported by multiple lines of evidence advocating the essential role of KDM1A in coordinating glycolytic and mitochondrial metabolism in cancerous cells. Inactivation of KDM1A (chemical and genetic depletion) has been shown to suppress glucose uptake and glycolytic activity, with concurrent activation of mitochondrial respiration (47–50). Collectively, our preliminary findings clearly show for the first time an association between the UPR and the ability of SP-2509 to induce apoptosis in ES cell lines.
In summary, our findings add to the growing body of evidence highlighting the oncogenic role of KDM1A in tumors such as ES, and validate the broad therapeutic utility of targeted KDM1A inhibition for the treatment of this aggressive pediatric malignancy. We demonstrate that ES cells are exceptionally dependent upon KDM1A for growth and are exquisitely sensitive to small molecule reversible but not irreversible KDM1A blockade (SP-2509). Epigenetic regulators such as SP-2509 represent an exciting class of pharmacologic agents for the treatment of ES, with clinical formulations of SP-2509 (SP-2577/Seclidemstat) entering phase I clinical testing in 2018. For this reason, further studies are required to define mechanism(s) of SP-2509 resistance, a key unanswered question which will be essential to guide combinatorial treatment methods to circumvent patient drug resistance and define more selective inclusion criteria required for KDM1A inhibitor clinical trials.
Supplementary Material
Acknowledgments
We acknowledge experimental support from Iftekhar Showpnil and Julia Selich-Anderson. This study is dedicated to the memory of Tom Wood and Brad Neilsen.
Financial Support: Stephen L. Lessnick was supported by the NIH/NCI grants R01 CA140394 and R01 CA183776, the NCTN Grant U10 CA180886, Human Specimen Banking Grant U24 CA114766, and the NCTN Statistics & Data Center Grant U10 CA 180899 of the Children’s Oncology Group from the NCI/NIH (Bethesda, MD, USA). Additional support for research for S. Lessnick was provided by a grant from the WWWW (QuadW) Foundation, Inc. (www.QuadW.org) to the Children’s Oncology Group. Kathleen I. Pishas acknowledges financial support from the University of Adelaide Florey Medical Research Foundation Clinical Cancer Research Fellowship, NHMRC CJ Martin Overseas Biomedical Fellowship (APP1111032) and the Alex’s Lemonade Stand Young Investigators Grant (APP37138). M. C. Beckerle acknowledges financial support from CureSearch for Children’s Cancer and the Huntsman Cancer Institute Cancer Center Support Grant (P30-CA04214).
Footnotes
Conflict of interest: Stephen L. Lessnick is a consultant/advisory board member for Salarius Pharmaceuticals. Sunil Sharma is a founder and has equity in Salarius Pharmaceuticals
References
- 1.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359(6391):162–5. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 2.Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT. A second Ewing’s sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nature genetics. 1994;6(2):146–51. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- 3.Todorova R. Structure-function based molecular relationships in Ewing’s sarcoma. BioMed research international. 2015;2015:798426. doi: 10.1155/2015/798426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marina NM, Liu Q, Donaldson SS, Sklar CA, Armstrong GT, Oeffinger KC, et al. Longitudinal follow-up of adult survivors of Ewing sarcoma: A report from the Childhood Cancer Survivor Study. Cancer. 2017 doi: 10.1002/cncr.30627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS genetics. 2014;10(7):e1004475. doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer discovery. 2014;4(11):1326–41. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Putiri EL, Robertson KD. Epigenetic mechanisms and genome stability. Clinical epigenetics. 2011;2(2):299–314. doi: 10.1007/s13148-010-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Theisen ER, Pishas KI, Saund RS, Lessnick SL. Therapeutic opportunities in Ewing sarcoma: EWS-FLI inhibition via LSD1 targeting. Oncotarget. 2016;7(14):17616–30. doi: 10.18632/oncotarget.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X, et al. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PloS one. 2012;7(4):e35065. doi: 10.1371/journal.pone.0035065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer research. 2009;69(5):2065–71. doi: 10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 13.Yu Y, Wang B, Zhang K, Lei Z, Guo Y, Xiao H, et al. High expression of lysine-specific demethylase 1 correlates with poor prognosis of patients with esophageal squamous cell carcinoma. Biochemical and biophysical research communications. 2013;437(2):192–8. doi: 10.1016/j.bbrc.2013.05.123. [DOI] [PubMed] [Google Scholar]
- 14.Chen C, Ge J, Lu Q, Ping G, Yang C, Fang X. Expression of Lysine-specific demethylase 1 in human epithelial ovarian cancer. Journal of ovarian research. 2015;8:28. doi: 10.1186/s13048-015-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennani-Baiti IM, Machado I, Llombart-Bosch A, Kovar H. Lysine-specific demethylase 1 (LSD1/KDM1A/AOF2/BHC110) is expressed and is an epigenetic drug target in chondrosarcoma, Ewing’s sarcoma, osteosarcoma, and rhabdomyosarcoma. Human pathology. 2012;43(8):1300–7. doi: 10.1016/j.humpath.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Sorna V, Theisen ER, Stephens B, Warner SL, Bearss DJ, Vankayalapati H, et al. High-throughput virtual screening identifies novel N’-(1-phenylethylidene)-benzohydrazides as potent, specific, and reversible LSD1 inhibitors. Journal of medicinal chemistry. 2013;56(23):9496–508. doi: 10.1021/jm400870h. [DOI] [PubMed] [Google Scholar]
- 17.Sankar S, Theisen ER, Bearss J, Mulvihill T, Hoffman LM, Sorna V, et al. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20(17):4584–97. doi: 10.1158/1078-0432.CCR-14-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, et al. A DNA Hypomethylation Signature Predicts Antitumor Activity of LSD1 Inhibitors in SCLC. Cancer cell. 2015;28(1):57–69. doi: 10.1016/j.ccell.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Cawley K, Deegan S, Samali A, Gupta S. Assays for detecting the unfolded protein response. Methods in enzymology. 2011;490:31–51. doi: 10.1016/B978-0-12-385114-7.00002-7. [DOI] [PubMed] [Google Scholar]
- 20.Volchenboum SL, Andrade J, Huang L, Barkauskas DA, Krailo M, Womer RB, et al. Gene Expression Profiling of Ewing Sarcoma Tumors Reveals the Prognostic Importance of Tumor-Stromal Interactions: A Report from the Children’s Oncology Group. The journal of pathology Clinical research. 2015;1(2):83–94. doi: 10.1002/cjp2.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaefer KL, Eisenacher M, Braun Y, Brachwitz K, Wai DH, Dirksen U, et al. Microarray analysis of Ewing’s sarcoma family of tumours reveals characteristic gene expression signatures associated with metastasis and resistance to chemotherapy. Eur J Cancer. 2008;44(5):699–709. doi: 10.1016/j.ejca.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Ohali A, Avigad S, Zaizov R, Ophir R, Horn-Saban S, Cohen IJ, et al. Prediction of high risk Ewing’s sarcoma by gene expression profiling. Oncogene. 2004;23(55):8997–9006. doi: 10.1038/sj.onc.1208060. [DOI] [PubMed] [Google Scholar]
- 23.Ferreira BI, Alonso J, Carrillo J, Acquadro F, Largo C, Suela J, et al. Array CGH and gene-expression profiling reveals distinct genomic instability patterns associated with DNA repair and cell-cycle checkpoint pathways in Ewing’s sarcoma. Oncogene. 2008;27(14):2084–90. doi: 10.1038/sj.onc.1210845. [DOI] [PubMed] [Google Scholar]
- 24.Postel-Vinay S, Veron AS, Tirode F, Pierron G, Reynaud S, Kovar H, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nature genetics. 2012;44(3):323–7. doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 25.Gupta S, Weston A, Bearrs J, Thode T, Neiss A, Soldi R, et al. Reversible lysine-specific demethylase 1 antagonist HCI-2509 inhibits growth and decreases c-MYC in castration- and docetaxel-resistant prostate cancer cells. Prostate cancer and prostatic diseases. 2016;19(4):349–57. doi: 10.1038/pcan.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta S, Doyle K, Mosbruger TL, Butterfield A, Weston A, Ast A, et al. Reversible LSD1 inhibition with HCI-2509 induces the p53 gene expression signature and disrupts the MYCN signature in high-risk neuroblastoma cells. Oncotarget. 2018;9(11):9907–24. doi: 10.18632/oncotarget.24035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer discovery. 2014;4(11):1342–53. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.May WA, Grigoryan RS, Keshelava N, Cabral DJ, Christensen LL, Jenabi J, et al. Characterization and drug resistance patterns of Ewing’s sarcoma family tumor cell lines. PloS one. 2013;8(12):e80060. doi: 10.1371/journal.pone.0080060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, et al. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30(7):2521–32. doi: 10.1523/JNEUROSCI.5500-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stegmaier K, Wong JS, Ross KN, Chow KT, Peck D, Wright RD, et al. Signature-based small molecule screening identifies cytosine arabinoside as an EWS/FLI modulator in Ewing sarcoma. PLoS medicine. 2007;4(4):e122. doi: 10.1371/journal.pmed.0040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Schadewijk A, van’t Wout EF, Stolk J, Hiemstra PS. A quantitative method for detection of spliced X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic reticulum (ER) stress. Cell stress & chaperones. 2012;17(2):275–9. doi: 10.1007/s12192-011-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price BD, Mannheim-Rodman LA, Calderwood SK. Brefeldin A, thapsigargin, and AIF4- stimulate the accumulation of GRP78 mRNA in a cycloheximide dependent manner, whilst induction by hypoxia is independent of protein synthesis. Journal of cellular physiology. 1992;152(3):545–52. doi: 10.1002/jcp.1041520314. [DOI] [PubMed] [Google Scholar]
- 33.Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, et al. A novel mammalian flavin-dependent histone demethylase. The Journal of biological chemistry. 2009;284(26):17775–82. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Molecular cell. 2010;39(2):222–33. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518(7539):422–6. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nature medicine. 2016;22(3):298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katz TA, Vasilatos SN, Harrington E, Oesterreich S, Davidson NE, Huang Y. Inhibition of histone demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases sensitivity to DNMT inhibitor-induced apoptosis in breast cancer cells. Breast cancer research and treatment. 2014;146(1):99–108. doi: 10.1007/s10549-014-3012-9. [DOI] [PubMed] [Google Scholar]
- 39.Crea F, Sun L, Mai A, Chiang YT, Farrar WL, Danesi R, et al. The emerging role of histone lysine demethylases in prostate cancer. Molecular cancer. 2012;11:52. doi: 10.1186/1476-4598-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenner JC, Feng FY, Han S, Patel S, Goyal SV, Bou-Maroun LM, et al. PARP-1 inhibition as a targeted strategy to treat Ewing’s sarcoma. Cancer research. 2012;72(7):1608–13. doi: 10.1158/0008-5472.CAN-11-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pishas KI, Lessnick SL. Recent advances in targeted therapy for Ewing sarcoma. F1000Research. 2016:5. doi: 10.12688/f1000research.8631.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lv S, Bu W, Jiao H, Liu B, Zhu L, Zhao H, et al. LSD1 is required for chromosome segregation during mitosis. European journal of cell biology. 2010;89(7):557–63. doi: 10.1016/j.ejcb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Schooley A, Moreno-Andres D, De Magistris P, Vollmer B, Antonin W. The lysine demethylase LSD1 is required for nuclear envelope formation at the end of mitosis. Journal of cell science. 2015;128(18):3466–77. doi: 10.1242/jcs.173013. [DOI] [PubMed] [Google Scholar]
- 44.Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO reports. 2006;7(9):880–5. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell death and differentiation. 2004;11(4):381–9. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 46.Adhikari S, Toretsky JA, Yuan L, Roy R. Magnesium, essential for base excision repair enzymes, inhibits substrate binding of N-methylpurine-DNA glycosylase. The Journal of biological chemistry. 2006;281(40):29525–32. doi: 10.1074/jbc.M602673200. [DOI] [PubMed] [Google Scholar]
- 47.Castex J, Willmann D, Kanouni T, Arrigoni L, Li Y, Friedrich M, et al. Inactivation of Lsd1 triggers senescence in trophoblast stem cells by induction of Sirt4. Cell death & disease. 2017;8(2):e2631. doi: 10.1038/cddis.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kosumi K, Baba Y, Sakamoto A, Ishimoto T, Harada K, Nakamura K, et al. Lysine-specific demethylase-1 contributes to malignant behavior by regulation of invasive activity and metabolic shift in esophageal cancer. International journal of cancer Journal international du cancer. 2016;138(2):428–39. doi: 10.1002/ijc.29714. [DOI] [PubMed] [Google Scholar]
- 49.Qin Y, Zhu W, Xu W, Zhang B, Shi S, Ji S, et al. LSD1 sustains pancreatic cancer growth via maintaining HIF1alpha-dependent glycolytic process. Cancer letters. 2014;347(2):225–32. doi: 10.1016/j.canlet.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 50.Sakamoto A, Hino S, Nagaoka K, Anan K, Takase R, Matsumori H, et al. Lysine Demethylase LSD1 Coordinates Glycolytic and Mitochondrial Metabolism in Hepatocellular Carcinoma Cells. Cancer research. 2015;75(7):1445–56. doi: 10.1158/0008-5472.CAN-14-1560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


