Abstract
Mycobacterium tuberculosis (Mtb) Rv2671 is annotated as a 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate (AROPP) reductase (RibD) in the riboflavin biosynthetic pathway. Recently, a strain of Mtb with a mutation in the 5′ untranslated region of Rv2671, which resulted in its overexpression, was found to be resistant to dihydrofolate reductase (DHFR) inhibitors including the anti-Mtb drug para-aminosalicylic acid (PAS). In this study, a biochemical analysis of Rv2671 showed that it was able to catalyze the reduction of dihydrofolate (DHF) to tetrahydrofolate (THF), which explained why the overexpression of Rv2671 was sufficient to confer PAS resistance. We solved the structure of Rv2671 in complex with the NADP+ and tetrahydrofolate (THF), which revealed the structural basis for the DHFR activity. The structures of Rv2671 complexed with two DHFR inhibitors, trimethoprim and trimetrexate, provided additional details of the substrate binding pocket and elucidated the differences between their inhibitory activities. Finally, Rv2671 was unable to catalyze the reduction of AROPP, which indicated that Rv2671 and its closely related orthologues are not involved in riboflavin biosynthesis.
Graphical Abstract

One of the first drugs developed to treat tuberculosis (TB) was para-aminosalicylic acid (PAS), and over 70 years later, it is still in use. While PAS has been replaced as a first line drug by isoniazid, pyrazinamide, and rifampin, it has found new usefulness as a second line drug to treat drug-resistant TB.1
PAS is a structural analogue of p-aminobenzoic acid (pABA), the substrate of dihydropteroate synthase (DHPS), and for years DHPS was thought to be the target. However, PAS was recently shown to be a prodrug,2,3 which is incorporated into the folate pathway through the conversion to hydroxy intermediates. Mycobacterium tuberculosis (Mtb) cytotoxicity is due to either a general poisoning of the folate-dependent pathway by hydroxy intermediates2 or the inhibition of dihydrofolate reductase (DHFR) by 12-hydroxy-7,8-dihydrofolate (hydroxy dihydrofolate).3 In the previously referenced study, a laboratory-derived Mtb mutant strain resistant to a DHFR inhibitor (NITD344,3 a WR992104-analogue) was found to also be cross-resistant to PAS. Most notably, this same mutation has been reported in 10 out of 208 PAS resistant clinical isolates of Mtb, plainly demonstrating its association with PAS resistance in vivo.5 This resistant strain (R7) contained a mutation in a noncoding region upstream of the Mtb Rv26713 and produced increased levels of Mtb RibD (Rv2671). Mtb Rv2671 is currently annotated as RibD, an enzyme in the riboflavin biosynthetic pathway;6 therefore, it was surprising to find that increased expression of an enzyme involved in riboflavin biosynthesis would confer resistance to PAS.
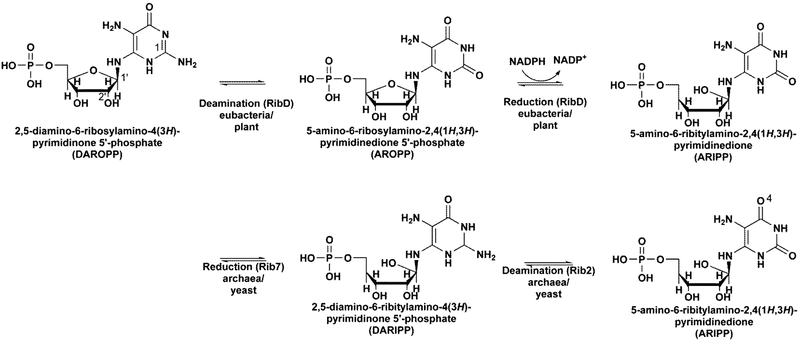
In eubacteria RibD, also referred to as RibG in some organisms, is typically a bifunctional enzyme which catalyzes the second and third reactions of the riboflavin biosynthetic pathway, that is, the conversion of 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate (DAROPP) to 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate (ARIPP). The RibD catalyzes the reaction that involves the deamination of the 2-amino group of the pyrimidine ring followed by a reduction of the ribose ring using either NADPH or NADH.7 In some eubacteria, the two activities are on separate enzymes. Mtb Rv3752c is annotated as the deaminase that would convert DAROPP to 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate (AROPP), and Mtb Rv2671 is annotated as an AROPP reductase converting AROPP to ARIPP. In fungi and some archaea, the reduction precedes the deamination, and the reactions are catalyzed by two monofunctional enzymes: the reductase Rib7 and the deaminase Rib2, respectively8,9 (Scheme 1). Other than Rv2671 and Rv3572c, Mtb RibG (Rv1409) encodes for a bifunctional RibD containing a DAROPP deaminase domain and an AROPP reductase domain. This is particularly unusual as the reactions, substrates, and products of Rv1409 should be identical to that of Rv2671 and Rv3571c.
Scheme 1.
In order to gain insights into the molecular mechanisms of PAS resistance and define the catalytic activity of Rv2671, we have completed a biochemical and structural characterization of Mtb Rv2671. The biochemical assays indicated that Mtb Rv2671 catalyzes the reduction of dihydrofolate (DHF), and the structural studies assisted in defining the molecular basis for the high Km for DHF. Our results also explained the mechanism for the resistance to PAS in the R7 and clinical strains. Furthermore, activity assays using purified recombinant Mtb Rv2671 revealed that it is neither an AROPP nor a DAROPP reductase, suggesting that Rv2671 and other related orthologues are not involved in riboflavin biosynthesis.
■ EXPERIMENTAL PROCEDURES
Materials.
All chemical reagents used in buffers, protein purification, and enzymatic assays were purchased from Sigma-Aldrich (St. Louis, MO). Dihydropteroic acid was purchased from Schircks Laboratories (Switzerland).
Cloning, Expression, and Purification of Rv2671.
The sequence of the full length Rv2671 gene was amplified from the Mtb H37Rv genome by PCR Primers were 5′-GGACCTCCATATGCCCGACTCTGGTCAGCTCG-3′ and 5′-C C C A A G C T T T C A G G T C - T T G A C G T A G C - GGGTGTACAGG-3′. The amplified gene was inserted into a pET28b expression vector (Novagen) containing an N-terminal 6x-His tag with the TEV cleavage site using the NdeI and HindIII restriction sites. The plasmid was transformed into BL21(DE3) Escherichia coli (E. coli) cells (Novagen) for Rv2671 expression. The cells with the plasmid were grown at 37 °C to OD600 ~0.6–0.8 in LB medium with 50 μg/mL kanamycin followed by induction with 1 mM IPTG and grown overnight at 18 °C.
The cells were lysed via a Microfluidizer M-100P (Microfluidics, Worcestershire, UK) in lysis buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 5% glycerol, 1 mM PMSF, 10 μg/mL DNase, and 2 mM MgCl2) and centrifuged at 27,216g for 1 h. The supernatant was purified over a nickel column (GE Healthcare) with a 0 to 300 mM imidazole gradient, and the 6x-His tag was cleaved using tobacco etch virus protease. The protein was >95% pure, as observed by SDS–PAGE, and was concentrated to 7.5 mg/mL, flash frozen, and stored in dialysis buffer (50 mM Tris, pH 7, 50 mM NaCl, 5% glycerol, and 1 mM DTT) at −80 °C. The concentration of the protein was determined by measuring A280 using a NanoDrop 2000 UV–vis spectrophotometer with an extinction coefficient of the enzyme of 17,545 M−1cm-1.
Crystallization.
In order to obtain cocrystal structures, Rv2671 was incubated with substrates or inhibitors and screened for crystallization conditions with a Mosquito liquid dispenser (TTP Labtech, Cambridge, MA, USA) using the sitting drop vapor diffusion technique in the CrystalMation Intelli-Plate 96-3 low-profile crystallization plate (Hampton Research). For each condition, 1 μL of protein (7.5 mg/mL) and 1 μL of crystallization formulation were mixed, and the mixture was equilibrated with 50 μL of crystallization solution in the reservoir well. Full length Rv2671 protein crystals were further optimized via hanging drop vapor diffusion by incubating 1.5 μL of purified protein solution with 1.5 μL of crystallization solution (0.2 M NaCl, 0.1 M Bis-Tris, pH 6.5, and 25–27% PEG 3350) at 18 °C for 2 d. Crystals were cryo-protected with 30% glycerol and flash frozen prior to data collection. For the NADP+ complex crystals, 2 mM NADP+ was cocrystallized with Rv2671 using the above procedure. The crystals of the Rv2671 with NADP+ and trimetrexate (TMQ) bound were also obtained from cocrystallization with 2 mM NADP+ and 2 mM TMQ. The Rv2671 crystal in complex with tetrahydrofolate (THF)-NADP+, trimethoprim (TMP)-NADP+, and dihydropteroic acid (DHP)-NADP+ were obtained by soaking 5 mM of compounds into the NADP+ binary crystals.
Data Collection and Structure Determination.
Data were collected at Argonne National Laboratory using the Advanced Photon Source (APS) beamline 19ID or 23ID-B. All data were processed and reduced using HKL3000.10 The structure of the copurified NADP(H) bound Rv2671 was solved by molecular replacement using MOLREP11 in CCP412 with the protein atomic coordinates for the truncated pyrimidine reductase-like protein of Corynebacterium diphtheria from the Protein Data Bank (accession code 2P4G, deposited by Joint Center for Structural Genomics). The exact search model contains residues 36–86 and residues 174–258 of 2P4G. The complex structures were solved by molecular replacement using MOLREP11 in CCP412 with the solved copurified NADP(H) bound Rv2671 structure. These crystals belong to the C2221 space group and contained one molecule in the asymmetric unit. Refinement and manual model building were performed with Phenix13 and COOT,14 respectively, and TLS was not used in the refinement.
Enzyme Kinetics.
The DHFR activity of Rv2671 was measured in a 1 mL solution containing 100 mM HEPES, 50 mM KCl, 200 μM NADPH, 50 mM 2-mercaptoethanol, and 11.1 μg of Rv2671 at pH 7.5, which was adapted from a similar assay to what has been previously reported for E. coli DHFR.15 Assays were started by adding DHF in concentration ranging from 0 μM to 250 μM. Enzyme activity was monitored using a Cary 100 spectrophotometer at 340 nm. Initial velocity data were fitted with the Michaelis–Menten equation16 using the SigmaPlot program (Systat Software, San Jose, CA).
The dihydropteroic acid reductase (DHPR) activity15 of Rv2671 was measured in 200 μL of enzyme solution containing 100 mM HEPES, 50 mM KCl, 200 μM NADPH, 50 mM 2-mercaptoethanol, and 1.7 μg of Rv2671 at pH 7.5. Assays were started by adding 10 μL of DHP from 0 μM to 530 μM. Enzyme activity was monitored using a Cary 100 spectrophotometer at 340 nm. Initial velocity data were fitted with the Michaelis–Menten equation16 using the SigmaPlot program (Systat Software, San Jose, CA).
The DHFR activity inhibition assay was performed under the same conditions described above but with the addition of 5 μL of inhibitor in 100% DMSO. The inhibitor was preincubated with the protein for 10 min. After preincubation, 5 μL of DHF was added to a final concentration of 200 μM in order to start the reaction. The reaction activity with 5 μL of DMSO alone served as a negative control. Activity data were plotted and fitted into the four parameter logistic equation (eq 1) by the SigmaPlot program (Systat Software, San Jose, CA).
| (1) |
The pyrimidine reductase activity of Rv2671 was followed by monitoring NADPH oxidation at 340 nm in a solution containing 50 mM HEPES, 5 mM MgCl2, 200 μM NADPH, 30 mM DTT, and 1.32–26.50 μg/mL Rv2671 at pH 8. The substrates, DAROPP and AROPP were synthesized enzymatically. DAROPP was generated by mixing 5 mM GTP, and 2 mg/mL E. coli GCH-II and AROPP was synthesized by incubating 1 mg/mL E. coli RibD with DAROPP in the absence of NADPH.17,18 The enzymes were removed by filtering through a 10 kDa cutoff concentrator (Vivaspin 500 centrifugal concentrator). DAROPP was analyzed as its diacetyl derivative and detected by fluorometric HPLC. AROPP was analyzed by LC-MS using an SHIMADZU LCMS-2010 spectrometer.
HPLC Analysis of Tetrahydrofolate Formation.
Rv2671 (400 nM) was incubated with 500 μM NADPH and 200 μM DHF in the aforementioned buffer (100 mM HEPES, pH 7.5, 50 mM KCl, and 50 mM 2-mercaptoethanol). Aliquots were removed every 2 min over a 10 min period and were then added to a solution of 0.1 M TCA and 0.55 M ascorbic acid to stop the reaction.19 To extend the stability of THF, the pH of the reaction mixture was immediately adjusted to pH 4 by adding NaOH.19 The denatured protein was removed by centrifuging at 16,200g for 10 min at 4 °C. An Agilent technologies HPLC (1200 infinity) with a reversed-phase C18 Atlantis T3, 5 μm, 4.6 × 250 mm column (Waters) was used to separate the components in the reaction mixtures.19 The column was eluted by the following optimized gradient at a 1 mL/min flow rate: solvent A is water, solvent B is 100 mM KPi pH 6.6, and solvent C is methanol; 0 min 100% B, 4 min 25% A 60% B 15% C, 8 min 20% A 40% B 40% C, 14 min 10% A 10% B 80%C, 18 min 10% A 10% B 80% C, 20 min 25% A 50% B 25% C, 22 min 100% B, and 25 min 100% B. The absorbance at 290 nm was monitored. The fluorescent chromatograph was monitored with excitation at 290 nm and emission at 360 nm.
Evolutionary Tree Construction.
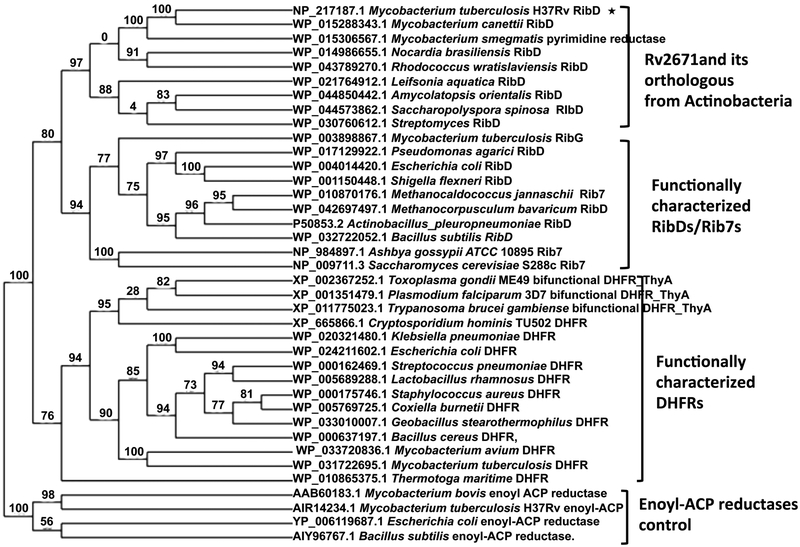
The amino acid sequences of thirty-eight representative homologues/orthologues were selected to generate the phylogenetic tree including 9 pyrimidine reductases from Actinobacteria, 15 DHFRs, 10 RibDs or Rib7s, and 4 enoyl ACP reductases used as the reference genes. Nine pyrimidine reductases from Actinobacteria were selected from the NCBI reference protein sequence database,20–22 which showed >50% sequence similarity with Mtb Rv2671. The DHFRs and RibD/Gs were selected from functionally characterized proteins. The phylogenetic tree was generated by the Phylogeny.fr server,23 which used MUSCLE in order to carry out the multiple alignments of protein sequences,24 PhyML to build the tree using the marginal likelihood method,25 and TreeDyn in order to generate tree rendering.26 The sequences used to generate the tree are listed in Supporting Information.
■ RESULTS AND DISCUSSION
Determination of Dihydrofolate Reductase Activity of Rv2671.
The genome sequence of Mtb has Rv2671 annotated as an AROPP reductase (RibD) which is part of the riboflavin biosynthetic pathway that catalyzes the reduction of AROPP to ARIPP.6 Its annotation was most likely due to the sequence similarity analysis showing that Rv2671 has a significantly better sequence similarity to the reductase domains of prokaryotic RibDs (AROPP reductases) (22% identity with E. coli RibD) and Rib7s (DAROPP reductases) (22% identity with Methanocaldococcus jannaschii Rib7), compared to that of any other enzyme family. The next closest was the dihydrofolate reductase family (14% identity with E. coli DHFR). Therefore, it was surprising that the overexpression of Rv2671 was able to replace Mtb DHFR function in vitro.3 Specifically, a mutant strain of Mtb with increased levels of Rv2671 was shown to be resistant to the DHFR inhibitor NITD344,3 an analogue of a well-known DHFR inhibitor WR99210.4 In addition, a multicopy plasmid that expressed Rv2671 was sufficient to allow for the production of a knockout of the dfrA gene,3 which codes for DHFR and is essential in Mtb based on transposon hybridization.27 Together, these studies suggested the possibility that Rv2671 was a bifunctional enzyme capable of catalyzing the reduction of DHF (Figure 1A) as well as AROPP.
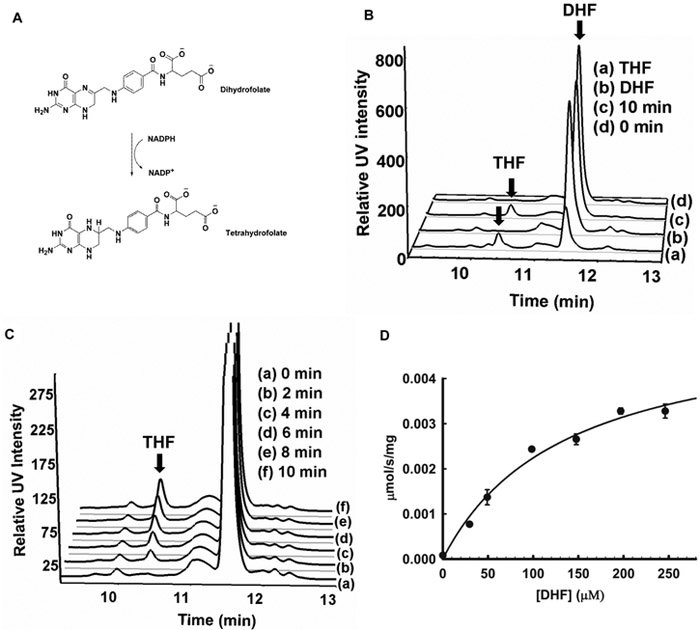
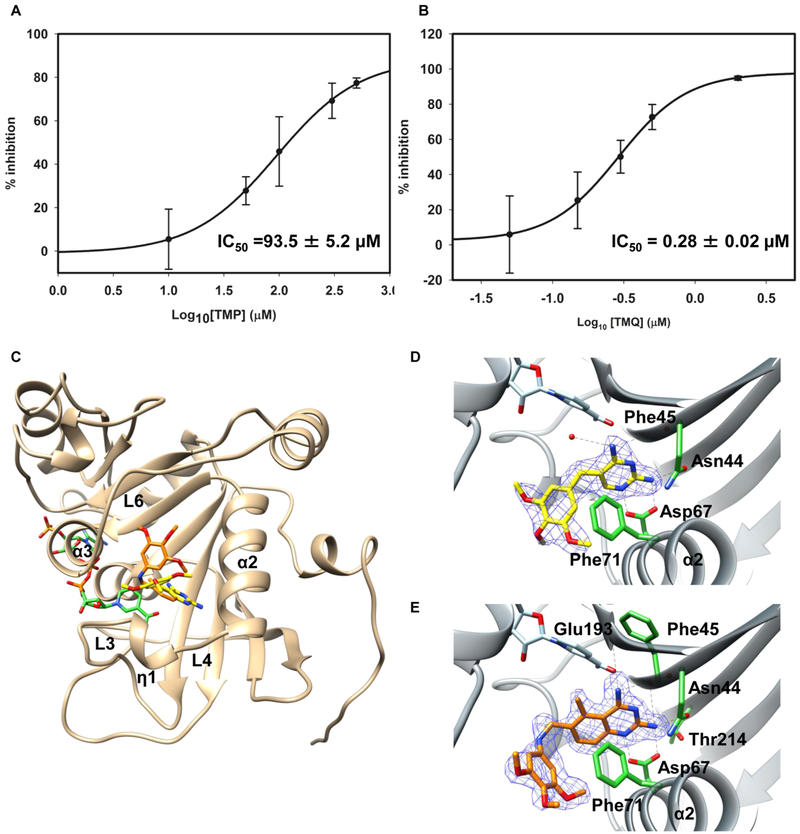
Figure 1. Characterization of Rv2671 as a DHFR.
(A) Scheme of the DHF reduction reaction. (B) HPLC analysis of the in vitro reaction and the relevant control. (C) Time course for the reaction shows that the amount of THF increases with time. (D) Steady state kinetic reaction measured in the presence of Rv2671 and saturated NADPH. Reaction velocity plotted versus DHF concentration (Km = 136 ± 32 μM; kcat = 0.15 ± 0.016 s−1). Each DHF concentration was performed in triplicate, and the data represent mean values ± SD.
We implemented a direct DHFR in vitro assay with pure recombinant Mtb Rv2671 to determine if Rv2671 could catalyze the reversible reduction of DHF to THF in the presence of NADPH. Tetrahydrofolate, the product of the reaction, was observed by HPLC spectrometry after the addition of DHF and NADPH to Rv2671 (Figure 1B and C). We detected an increase in a single species over time, and the HPLC peak comigrated with a tetrahydrofolate standard (rt = 10.4 min). The identity was further confirmed to be tetrahydrofolate by MS. The Km and kcat of Rv2671 for DHF were determined to be 136 ± 32 μM and 0.15 ± 0.016 s−1, respectively (Figure 1D). A comparison of the kinetic parameters with DHFRs from different organisms is listed in Table 1. Briefly, the kcat of the DHFRs ranged from 2 to 30 s−1, at least 1 order of magnitude faster than the kcat of Rv2671. Moreover, studied DHFRs have a lower KM for DHF, in the range of 0.7 to 4 μM, which is at least 25 times lower than that of Rv2671 (136 ± 32 μM). The Km of DHF for Mtb DHFR is 4.2 ± 6 μM with a kcat of 2.3 s−1.28 The Km of DHF is about 30 times lower than that of Rv2671, and the reaction rate is 15 times faster. Therefore, Rv2671 can indeed catalyze the DHF reduction reaction but with less efficiency than the DHFRs of Mtb and other organisms.
Table 1.
Kinetic Parameters of Rv2671 and DHFRs
Overall Structures of Rv2671 with Cofactor.
The X-ray crystal structure of recombinant E. coli-derived Rv2671 was determined to 1.9 Å resolution. The crystal belongs to the orthorhombic space group C2221 with one molecule in the asymmetric unit. In the final refined structure (Table 2), a total of 246 out of 258 residues were built into the electron density map. Residues 1–9 and 91–95 were disordered in the structure. The structure of Rv2671 is a variant Rossmann fold, composed of eight parallel β-strands (β8, β7, β1, β6, β2, β3, β4, and β5) and one antiparallel β-strand, β9. This mixed β-sheet is surrounded by seven α-helices and one 310-helix (η1). Helices α1 to α5 are on one side of the central sheet and α6, α7, and η1 are on the opposite side (Figure S1A). Weak electron density for NADP(H) was observed in the active site suggesting NADP(H) copurified with the protein. The electron density of the NADP(H) indicates an occupancy of less than 50% in the crystal.
Table 2.
Data Collection and Refinement Statistics
| copurified NADP(H) bound Rv2671 |
NADP+ bound Rv2671 |
THF bound Rv2671 |
|
|---|---|---|---|
| PDB ID | 4XRB | 4XT5 | 4XT6 |
| Data collection | |||
| space group | C2221 | C2221 | C2221 |
| cell dimensions | |||
| a, b, c (Å) | 71.4, 96.6, 75.7 | 69.7, 95.3, 73.9 | 70.5, 95.9,76.1 |
| α, β, γ (deg) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| resolution (Å)a | 27.4–1.9 (1.93–1.9) | 44.75–2.13 (2.17–2.13) | 36.61–1.85 (1.88–1.85) |
| Rsym | 0.045 (0.290) | 0.069 (0.418) | 0.053 (0.274) |
| I/σ(I) | 31.0 (8.3) | 26.8 (7.02) | 34.1(9.7) |
| completeness (%) | 99.36 (100) | 99.96 (99.71) | 99.34 (97.58) |
| redundancy | 7.1 (7.3) | 9.0 (8.9) | 8.7(9.0) |
| refinement | |||
| no. of reflections | 149934 | 127251 | 194557 |
| Rwork/Rfree | 0.18/0.20 | 0.19/0.24 | 0.20/0.24 |
| no. of atoms | 1974 | 1975 | 1919 |
| protein | 1851 | 1863 | 1799 |
| ligand/ion | 55 | 48 | 44 |
| water | 68 | 64 | 73 |
| B-factors | 36.4 | 37.9 | 30.9 |
| protein | 36.0 | 37.9 | 30.6 |
| ligand/ion | 48.3 | 35.6 | 35.8 |
| water | 38.9 | 38.9 | 33.7 |
| rms deviations | |||
| bond lengtds (Å) | 0.018 | 0.013 | 0.015 |
| bond angles (deg) | 1.96 | 1.36 | 1.24 |
| Ramachandran favored (%) | 99 | 97 | 97 |
| Ramachandran allowed (%) | 1 | 3 | 3 |
| Ramachandran outliers (%) | 0 | 0 | 0 |
| TMP bound Rv2671 |
TMQ bound Rv2671 |
DHP bound Rv2671 |
|
|---|---|---|---|
| PDB ID | 4XT7 | 4XT8 | 4XT4 |
| Data collection | |||
| space group | C2221 | C2221 | C2221 |
| cell dimensions | |||
| a, b, c (Å) | 69.0, 93.2, 74.6 | 69.4, 93.6, 74.3 | 70.3, 94.4, 75.8 |
| α, β, γ (deg) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| resolution (Å)a | 46.56–2.3 (2.34–2.3) | 46.81–1.95 (1.98–1.95) | 45.24–1.89 (1.92–1.89) |
| Rsym | 0.081 (0.331) | 0.08 (0.288) | 0.070 (0.585) |
| I/σ(I) | 21.3 (6.5) | 34.8 (12.9) | 21.7 (2.4) |
| completeness (%) | 99.94 (99.36) | 99.89 (99.72) | 99.43 (94.66) |
| redundancy | 7.3 (7.4) | 8.6 (8.8) | 9.3 (6.6) |
| refinement | |||
| no. reflections | 81136 | 155567 | 189305 |
| Rwork/Rfree | 0.17/0.22 | 0.18/0.22 | 0.19/0.22 |
| no. of atoms | 2006 | 2072 | 2002 |
| protein | 1858 | 1885 | 1864 |
| ligand/ion | 82 | 103 | 56 |
| water | 66 | 84 | 82 |
| B-factors | 24.4 | 23.9 | 33.8 |
| protein | 24.3 | 23.6 | 33.5 |
| ligand/ion | 25.9 | 24.6 | 36.1 |
| water | 25.5 | 28.2 | 38.2 |
| rms deviations | |||
| bond lengths (Å) | 0.054 | 0.051 | 0.011 |
| bond angles (deg) | 1.43 | 1.43 | 1.36 |
| Ramachandran favored (%) | 98 | 98 | 97 |
| Ramachandran allowed (%) | 2 | 2 | 3 |
| Ramachandran outliers (%) | 0 | 0 | 0 |
Resolution values for collection and refinement are the same.
Rv2671 is a homodimer in solution as determined by size-exclusion chromatography (Figure S1B). In the crystal, the homodimer is generated by 2-fold crystallographic symmetry (Figure 2A). The free energy of assembly dissociation (ΔGdiss) is 44.3 kcal/mol; the positive value suggests that this dimer is thermodynamically stable. The buried surface area of the dimer interface accounts for 37% of the total surface area.29 This large dimer interface contains residues from three loops (L1, L3, and L15) and four β-strands (β1, β7, β8, and β9). The intersubunit interactions are dominated by hydrogen bonds and electrostatic interactions, with roughly 56 hydrogen bonds and 8 electrostatic interactions across the interface. Each subunit of the homodimer contains one active site. The oligomeric state of Rv2671 is consistent with that of other RibD/Gs and Rib7s, except Bacillus subtilis RibG (Bs RibG), which is reported to be a tetramer in solution.30–33 In contrast, most DHFRs are monomeric.34
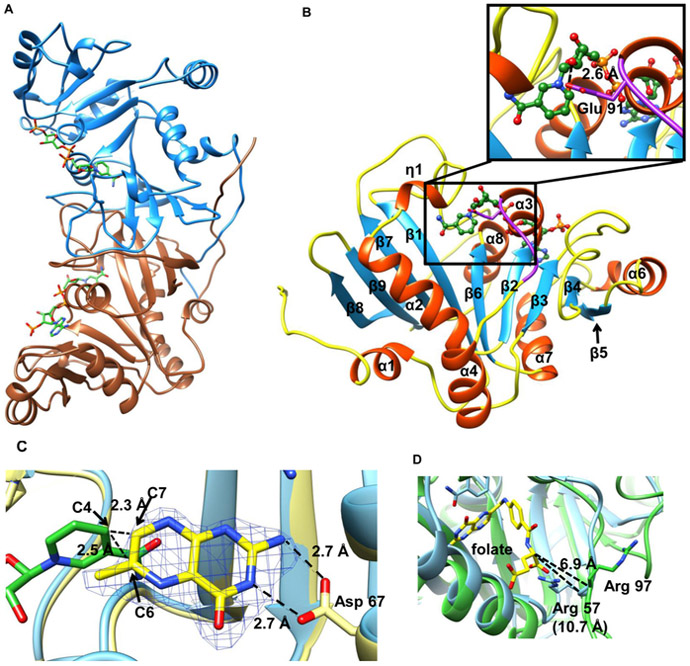
Figure 2. Crystal structure of Rv2671 and the THF binding mode.
(A) Ribbon representation of the dimeric Rv2671. (B) Ribbon representation of the Rv2671 in complex with NADP+. The secondary structural elements are labeled sequentially. The loop (residue 91–95) disordered in the native purified structure is shown in purple, and NADP+ is shown in the green ball and stick model. The hydrogen bond interaction between the ribose group of NADP+ and Glu 91 in the loop (residue 91–95) is shown in the zoomed-in figure. (C) Superimposition of the THF bound Rv2671 structure (yellow) with the NADP+ bound Rv2671 structure (blue). NADP+ and the pteridine ring of THF are colored in green and yellow. The simulated annealing omit map contoured at 2.5σ around the pteridine ring is shown in blue. Distances between the C4 of nicotinamide and the C6 and C7 of the pteridine are labeled. (D) Superimposition of the NADP+ bound Rv2671 structure (green) with the folate-NADPH bound E. coli RibD (4pdj, blue). Folate is colored in yellow. The side chain of the Arg 97 in the Rv2671 complex structure points to the other direction against the folate. The distances between α-carbon of the glutamate group in folate and the main chain carbon of the arginines are 6.9 and 10.7 Å, respectively.
The crystals of Rv2671 preincubated with NADP+ were nearly isomorphous with the natively purified Rv2671 crystals and the structure was refined with diffraction data extending to 2.1 Å resolution (Table 2). The overall fold of the protein is highly conserved, with the partial NADP(H) bound structure having a root-mean-square deviation (rmsd) of 0.42 Å over 243 Cα pairs. The disordered segment (residues 91–95) of the partial NADP(H) bound structure was clear in the electron density map and formed a loop in the NADP+ bound structure. With the exception of the loop, NADP+ bound Rv2671 exhibits no significant structural differences to the partial NADP(H) bound structure. This loop is near the NADP+ binding site and interacts directly with the cofactor and is stabilized by both a hydrogen bond between the side chain oxygen of Glu91 and the 2′ hydroxyl group of the nicotinamide ribose group (2.6 Å) and multiple van der Waals interactions (Figure 2B). The NADP+ binds at the C-terminal edge of the central β-sheet (β2, β3, and β6) and the pyrophosphate group locates between the α3 and α6 adjacent to the putative substrate binding cleft formed by α2, α3, β2, and β3 (Figure 2B). The conformation of NADP+ and its binding site is very similar to that of RibD/G structures, as well as DHFR structures.32
Complex Structure of Rv2671 with THF.
In order to understand the molecular basis for the relatively low DHFR activity of Rv2671, the structure of Rv2671 ternary complex with the products, NADP+ and THF, was solved. The crystals diffracted to about 2.1 Å resolution (Table 2). The backbone Cα rmsd between 236 atom pairs is 0.49 Å with the NADP+ bound structure. The electron density was not visible for residues 90–95, residues 56–57, or the nicotinamide ribose moiety of the NADP+ in the THF bound structure. Notably, the disordering of the nicotinamide ribose moiety of the NADP+ was also observed in a recent Mtb DHFR structural report35 which showed that the density for the nicotinamide ribose moiety of NADPH was missing in the open conformation of the Mtb DHFR-NADPH complex structure.35 We observed the same phenomenon in Rv2671. After soaking NADP+ bound Rv2671 crystals with THF, the density for the nicotinamide ribose moiety disappeared along with the density for the adjacent loop residues 90–95. These residues correspond to loop2 (L2), a part of the ligand access control segment in Mtb DHFR,35 suggesting that residues 90–95 serve the same function in Rv2671.
Unambiguous electron density for the tetrahydropteridine ring of THF was observed and built into the electron density (Figure 2C). However, electron density for the pABG group (pABA and the glutamate tail) of THF was not visible. NA2 and N3 of the pteridine ring interact with the side chain of Asp67 (2.7 and 2.7 Å), which is typically observed in DHF-complexed DHFR structures. Although the nicotinamide ribose moiety is disordered in the ternary structure, after we overlaid it with the NADP+ Rv2671 structure the distances between the nicotinamide C4 and the pteridine C6 and C7, where the hydride transfer would occur, were measured to be 2.5 and 2.3 Å (Figure 2C). Similar positions for these groups have been observed in the E. coli DHFR-DHF complex structure.36 Upon the basis of the theoretical calculation, 2.6 Å is the optimal distance between the donor and acceptor atoms, while the hydride transfer occurs via a bent geometry.37,38
The missing density for the pABG group in the ternary structure suggests that Rv2671 does not have the optimal amino acids to mediate its binding. In all DHF–DHFR complex structures, a conserved arginine forms an electrostatic interaction with the α-carboxyl group of the glutamate tail. In the E. coli folate-DHFR structure (4pdj),39 Arg57 corresponds to the conserved arginine in DHFRs. Superimposition of Rv2671 and E. coli folate-DHFR structures put E-coli folate-DHFR Arg57 at the cleft of loop 6 (L6) and loop 7 (L7) in Rv2671 (Figure 2D). The main chain of Arg97 at L6 in Rv2671 is in close proximity to that of E. coli DHFR Arg57. However, Arg97 is 3 Å closer to the α-carboxyl group and points in the opposite direction against the DHF (Figure 2D). Therefore, it is unlikely that this Arg97 can fulfill the role of stabilizing the α-carboxyl group of DHF. The position of Arg97 may be responsible for the 30 times higher Km for Rv2671 compared to that of other DHFRs. The potential pABG binding cavity is also larger in Rv2671 (16 Å long, 12.5 A wide) than it is in Mtb DHFR (17 Å long, 7.5 A wide). This could reduce the affinity of pABG for Rv2671 but may also provide Rv2671 with the ability to bind another ligand.
Dihydropteroic acid (DHP) was tested as a potential substrate for Rv2671 since DHP contains a dihydropteridine ring and a benzoic acid group like THF, but does not have the glutamate tail. The Km of Rv2671 for DHP was 180.2 ± 25.5 μM with a kcat of 0.32 ± 0.016 s−1, unexpectedly similar to the Km and kcat for DHF. Therefore, the crystal structure of DHP complexed to Rv2671 was solved and refined to 1.89 Å resolution (Table 2) to investigate the binding mode of DHP. The overall structure was nearly identical to the THF bound structure with an rmsd of 0.36 Å over 236 Cα pairs. The location of the dihydropteridine ring of DHP was also identical to the tetrahydropteridine ring of THF. The benzoic acid in the DHP and the nicotinamide ribose moiety of NADP+ are disordered, as in the THF bound structure. The structural similarity and the missing benzoic acid density explain the similar steady state kinetic parameters for DHF and DHP. This result also suggests that Rv2671 can bind and reduce a broad spectrum of dihydropteridine containing analogues.
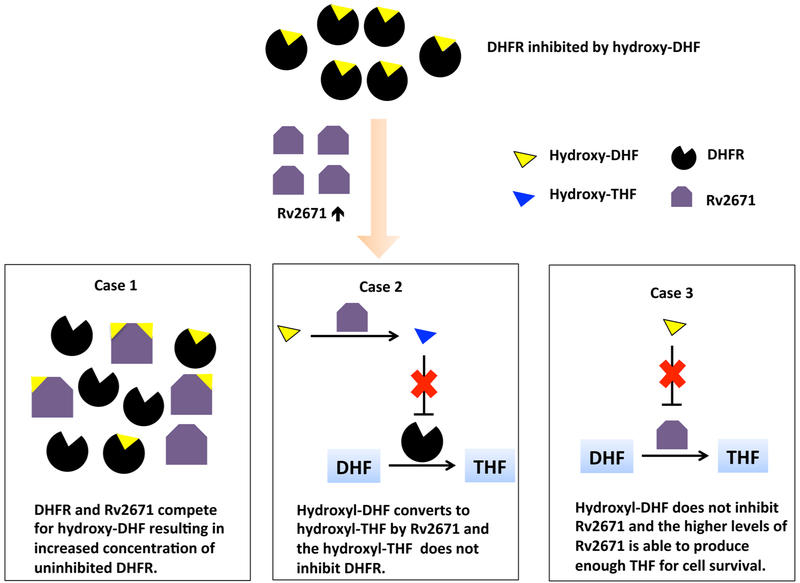
As previously stated, the drug PAS is converted in the cell to hydroxylated intermediates by enzymes in the folate biosynthetic pathway.2,3 The ability of Rv2671 to catalyze the reduction of DHF suggests several potential mechanisms for the observed resistance to PAS (Scheme 2). One scenario is that Rv2671, like DHFR, binds and is inhibited by the 12-hydroxy-7,8-dihydrofolate (hydroxy dihydrofolate) (Scheme 2, Case 1). In this case, the higher levels of Rv2671 would serve to sequester the inhibitor that would otherwise bind to DHFR within the cell. However, due to the high Km and inability to see in the structure, our studies indicate that this scenario is unlikely to be the reason for resistance since the lack of affinity of Rv2671 for DHF suggests that hydroxy dihydrofolate would likely prefer binding with Mtb DHFR instead of Rv2671. A second and more likely scenario is that the hydroxy dihydrofolate is a substrate of Rv2671 based on the less specific dihydropteridine reductase activity of Rv2671 (Scheme 2, Case 2). Thus, the higher levels of Rv2671 would effectively reduce the levels of DHFR inhibitor (hydroxy dihydrofolate) in the cell by converting it to the product hydroxy tetrahydrofolate. A third possibility is that Rv2671 does not bind to the hydroxy dihydrofolate (Scheme 2, Case 3). For this scenario, Rv2671 serves as the functional homologue of DHFR and overrides DHFR’s inhibition by hydroxy dihydrofolate. It has been shown that Mtb can survive in a strain where >97% of the DHFR enzyme had been depleted.40 Therefore, the higher levels of Rv2671 would be able to sustain folate biosynthesis at a level sufficient to support Mtb even though Rv2671 has only about 10% of the wild-type DHFR activity.
Scheme 2.
Another outstanding question is whether PAS’s cytotoxicity results from a general poisoning of folate-dependent pathways2 or if it acts specifically on DHFR through the conversion of PAS to hydroxy dihydrofolate.3 The nonspecific dihydropteridine reductase activity of Rv2671 indicates that the hydroxy dihydrofolate is likely a substrate of Rv2671 capable of converting a hydroxy dihydrofolate into a hydroxy tetrahydrofolate. This would mean that higher levels of Rv2671 in the cell would lead to the production of more hydroxy-folate intermediates which would in turn cause higher levels of sensitivity to PAS, if indeed PAS’s cytotoxicity results from the assumption of general poison. Since this was not the case, and the cells were resistant to PAS when Rv2671 was at higher levels, the data suggest that hydroxy dihydrofolate inhibition of DHFR is the event that leads to cell death.
Potential of Other Riboflavin Biosynthesis Reductases to Carry out DHFR’s Function.
Our discovery that Rv2671 has DHFR activity led us to test to see if DHFR activity is a general feature of the AROPP reductase domains of RibD/Gs. DHFR activity was not detected for the recombinant E. coli RibD (the AROPP reductase domain shows 22% sequence identity to Rv2671). This is not surprising since overexpression of Mtb RibG with both DAROPP deaminase domain and AROPP reductase domain was unable to replace the function of Mtb DHFR.3 In all known AROPP reductase sequences, there is a conserved lysine which interacts with the keto group of the pyrimidine ring of the AROPP in the active site.30–33 In the Bs RibG complex structure, the side chain of Lys151 forms a hydrogen bond with the keto group of the AROPP (3.4 Å).41 Superimposing E. coli RibD (2OBC)32 with Rv2671 shows that Asn44 in Rv2671 is in place of the aforementioned lysine (Lys152 in E. coli RibD) (Figure S2). The side chain carbonyl group of Asn44 forms a hydrogen bond with the pteridine NA2 of THF (3.1 Å) in the Rv2671 structure. A clash of the lysine side chain with the THF pteridine ring was observed when superimposing the structures. Our results indicate that bacterial AROPP reductases are unable to catalyze the reduction of DHF. This is consistent with the inability of bifunctional Mtb RibG (Rv1409) to compensate for a loss in DHFR activity.3
DHFR Inhibitors, Trimethoprim (TMP) and Trimetrexate (TMQ), Bind and Inhibit the DHFR Activity of Rv2671 in Vitro.
Two FDA approved DHFR inhibitors, TMP and TMQ, were tested for inhibitory activity against Rv2671. TMP is a selective DHFR inhibitor which typically shows about 2,500 times more potency against bacterial DHFR over human DHFR.42 TMQ is a DHFR inhibitor used in cancer treatment43 and does not show selectivity between bacterial and mammalian DHFRs.44 A 350-fold difference was observed in the IC50 values of TMP (Figure 3A) and TMQ (Figure 3B) to Rv2671 (93.5 ± 5.2 μM and 0.28 ± 0.02 μ;M, respectively). Although TMP is usually selective for bacterial DHFRs, it shows a low inhibition of not only Rv2671 but also Mtb DHFR (IC50 for TMP is 16.5 μM45).
Figure 3. Crystal structure of Rv2671 in complex with TMP or TMQ.
(A) The mean % inhibition values of Rv2671 are plotted against the log10 concentration of the DHFR inhibitor TMP. The calculated IC50 value is 93.5 ± 5.2 μM. (B) The mean % inhibition values of Rv2671 are plotted against the log10 concentration of the DHFR inhibitor TMQ. The calculated IC50 value is 0.28 ± 0.02 μM. (C) Ribbon representation of Rv2671 (tan) in complex with TMP (yellow) and TMQ (orange). (D) Binding of the TMP with Rv2671. The simulated annealing omit map contoured at 2.5σ around the inhibitor TMP is shown in blue. The key residues interacting with TMP are labeled and shown as sticks in green. The hydrogen bonds are shown as dashed lines. (E) Binding of TMQ with Rv2671. The simulated annealing omit map contoured at 2.5 σ around the inhibitor TMQ is shown in blue. The main residues interacting with TMP are labeled and shown as green sticks. Different binding modes of the trimethoxybenzyl rings of TMP and TMQ in Rv2671 can be observed in D and E. The trimethoxybenzyl ring of TMP in the complex structure is perpendicular to Phe71. The trimethoxybenzyl ring of TMQ in the complex structure is parallel to Phe71.
The structures of Rv2671 in complex with NADP+ and either TMP or TMQ (Figure 3C) revealed the molecular basis for the differences in inhibitory activity. The overall structures of the protein with inhibitors are similar to the NADP+ bound structure. For both inhibitors, hydrogen bonding interactions with Rv2671 appear to be the dominant binding interaction for the 2,4-diaminopyrimidine and the quinazoline group. The 2,4-diaminopyrimidine group of TMP forms six hydrogen bonds with residues of Rv2671 including Asn44, Asp67, Thr214, and a water molecule (Figure 3D). However, the quinazoline ring of TMQ forms seven hydrogen bonds with residues of Rv2671 including Asn44, Phe45, Asp67, Glu193, Thr214, and Glu91 (Figure 3E). When the two structures are superimposed, the 2,4-diaminopyrimidine group of TMP overlaps and is nearly coplanar (about 5 degrees out of plane) with the 2,4-diaminopyrimidine part of TMQ. The slight rotation results in one extra hydrogen bond between Rv2671 and TMQ, but the binding site of the two rings is identical. A more detailed analysis of the hydrogen bonding interactions between Rv2671 and the ligands is provided in Table S1.
The trimethoxybenzyl groups of TMP and TMQ were found to be in different binding orientations with Rv2671 (Figure 3C) due to the extra secondary amide in the linker region of TMQ which allows the trimethoxybenzyl ring to flip and move away from α2. The ring plane of the trimethoxybenzyl group of TMP is perpendicular to that of Phe71 in Rv2671 (Figure 3D), but the trimethoxybenzyl ring of TMQ in Rv2671 is parallel to the ring plane of Phe71 (Figure 3E). Different trimethoxybenzyl ring binding conformations result in different binding cavities, and the trimethoxybenzyl groups hold only via the van der Waals interactions with Rv2671, especially the hydrophobic effects. Since the hydrophobic interactions are the major contacts between the trimethoxybenzyl ring and the protein, the hydrophobicity of the ligand and its binding cavity was analyzed using the PLATINUM server, which calculates the hydrophobic properties of molecules and evaluates their match or mismatch in the protein–ligand complexes.46 The calculated hydrophobic match surface (SLL) between TMP and the surrounding residues is 0.06 Å2 compared with 34.24 Å2 between TMQ and its environment, and the fraction of lipophilic match (Match2) of TMP is 0.0007 compared with 0.2175 of TMQ. The higher match numbers of TMQ indicate better hydrophobic contacts between TMQ and Rv2671. The additional hydrogen bond and better hydrophobic interactions with the trimethoxybenzyl ring results in significantly better inhibition by TMQ compared with that of TMP.
Structural Comparison with Known RibDs/Rib7s from Other Organisms Reveals Differences in the Active Sites.
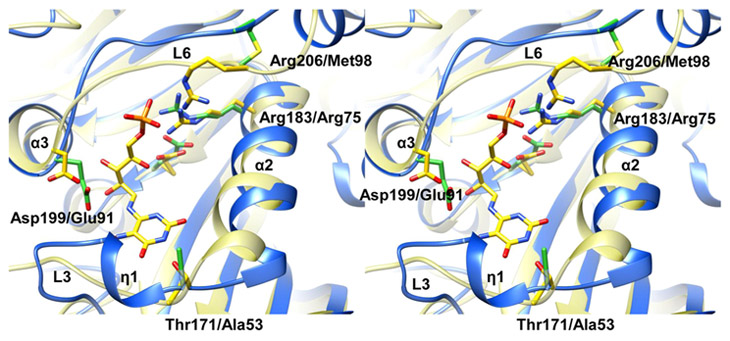
A structural similarity analysis using DALI47 identified a number of closely related homologues of Rv2671, and over 700 entries were identified with a Z-score above 2. Out of the 700 entries, 7 pyrimidine reducatases were found to be the closest structural homologues with Z-scores ranging from 16 to 27 and rmsd of Cα ranging from 2.1 to 2.6 Å, 4 of which were RibD/Gs, 1 was a Rib7, and 2 were putative pyrimidine reductases from different species. DHFRs also exhibited a significant match with 34 dihydrofolate reductases or bifunctional dihydrofolate reductase–thymidylates with Z-scores ranging from 11.4 to 18.7 and rmsd ranging from 1.8 to 2.9 Å. A pyrimidine reductase-like protein from C. diphtheria (2p4g) belonging to the monofunctional RibD_C family is the closest structural homologue of Rv2671 (Z-score of 27.1 and rmsd of 2.6 Å).
The AROPP reductase domain of RibD/Gs and Rib7s (DAROPP reductases) are often classified as pyrimidine reductases with identical reaction mechanism, overall structure, and substrate binding residues.48 2,5-Diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate (DAROPP) is the Rib7′s substrate and 5-amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate (AROPP) is the substrate of the reductase domain of RibD/Gs (Scheme 1).8,9 DAROPP and AROPP differ at the C2 position of the pyrimidine ring; DAROPP has an amino group in place of AROPP’s keto group. The distinct difference of RibD/Gs and Rib7s is a conserved lysine which interacts with the keto group of AROPP in the RibD/Gs. Pyrimidine reductases and Rv2671 share a common core structure including the central β-sheet and the flanking α-helices. The N-terminal region and α6 (in Rv2671) show the most structural diversity. For RibD/Gs, the N-terminus of the reductase domain connects to the deaminase domain.32,33 For Rib7s and Rv2671, the N-terminal region participates in the dimerization of two subunits. Helix α6 in Rv2671 is a loop in RibD/Gs, and in Rib7s the α6 helices are located in different spatial positions depending on the length of the adjacent cofactor binding loop.
Bs RibG in complex with ([(2R,3S,4R,5E)-5-[(5-amino-2,6- dioxo-3H-pyrimidin-4-yl)imino]-2,3,4-trihydroxy-pentyl] dihydrogen phosphate (AIF) is the only structure available in the protein databank (PDB) of a RibG with a substrate (analogue in this case) of a pyrimidine reductase.41 AIF is a proposed intermediate of the AROPP reduction reaction. The structures of Rv2671 and the Bs RibG-AIF complex were superimposed using the Matchmaker function of Chimera.49 The two structures were very similar to an rmsd of 1.9 Å over 202 Cα pairs. AIF was located at the cleft created by L3-η1-L4, L6, α2, α3, and the central β-sheet; α2, α3, and the central β-sheets were in similar spatial positions between the two structures, but the L6 loop of Rv2671 was 2.8 Å away from the phosphate group of AIF, and the region between L3-η1-L4 was 4.7 Å closer to α2 compared to the analogous regions of Bs RibG.
The enzymatic reaction of pyrimidine reductases is initiated by a conserved ionizable residue to abstract the proton from the amine group next to C1 of DAROPP/AROPP to form a Schiff base intermediate at C1, followed by a direct hydride transfer to C1 from the nicotinamide group of NADPH.31,32,41 The amino acids proposed for the proton abstraction are Asp200 in E. coli RibD and Glu290 in Bs RibG. In the structure of Rv2671, Glu193 corresponds to Bs RibG Glu290, and Glu91 corresponds to Asp200 of E. coli RibD. Although the length of the side chains of Glu and Asp differ by one carbon, the distance between the side chain oxygen of Glu91 and the amine nitrogen beside the C1 of the modeled AIF is 2.8 Å away, within proton transfer distance. However, based upon the superimposed structures, two differences in the active site could interfere with the binding of DAROPP/AROPP to Rv2671. First, there could be a clash between the pyrimidine ring and the backbone of Rv2671. As stated earlier, the location of L3-η1-L4 is different between Rv2671 and Bs RibG, and the backbone of L3-η1 in Rv2671 showed the potential for overlap with the pyrimidine ring of AIF in the superimposed structure. Although the possibility of main chain conformational changes cannot be ruled out, the distance between the main chain carbonyl group of Ser59 of Rv2671 and the O4 of the AIF from the Bs RibG structure is 1.14 A. Second, several conserved residues that are involved in substrate binding among functionally characterized pyrimidine reductases are not present in Rv2671 (Figure 4); for example, Thr171, Arg183, Asp199, and Arg206 interact directly with AIF in Bs RibG and are not conserved in Rv2671. The backbone of Ala53 in Rv2671 overlaps with the side chain of Thr171 in Bs RibG. The pyrimidine ring of AIF forms hydrogen bonds with the hydroxyl group of the side chain (2.1 Å) and the main chain nitrogen (3.1 Å) of Thr171 in Bs RibG. The main chain of Glu91 in Rv2671 is in the position of the main chain of Asp199 in Bs RibG; the side chain oxygen of Asp199 forms hydrogen bonds with two ribosyl hydroxyl groups of AIF (2.8 and 3.1 Å). The side chains of Arg183 and Arg206 in Bs RibG interact with the phosphate of AIF by hydrogen bonds and electrostatic interactions. Arg75 in Rv2671 was on top of Arg183 in Bs RibG. Met98 of Rv2671 corresponds to Arg206 in Bs RibG, but the distance between the two main chain carbons was 3.3 A since Met98 is located on L6. The differences between the substrate binding residues, especially missing Thr171 and Arg206, may cause Rv2671 not to bind to DAROPP or AROPP.
Figure 4.
Residue differences between the substrate binding sites of Rv2671 and Bs RibG. Superposition of NADP+ bound Rv2671 (blue) with the AIF bound Bs RibG (yellow) is shown in a stereo view. AIF and the residues directly interacting with AIF in Bs RibG are shown as yellow sticks, and the corresponding residues in Rv2671 are shown as green sticks.
Rv2671 Is Not a DAROPP or AROPP Reductase.
As discussed above, the annotation and structural similarity suggest two possible biological substrates for Rv2671: DAROPP or AROPP. Because of the instability of DAROPP and AROPP, these substrates were prepared enzymatically by E. coli RibA and E. coli RibD for the characterization of the pyrimidine reductase activity of Rv2671. Rv2671 showed no detectable reductase activity for either DAROPP or AROPP with different enzyme concentrations as well as substrate concentrations (Figure S3). As a positive control, pure recombinant E. coli RibD showed both deaminase and reductase activity under the reaction condition described in the Experimental Procedures section. Rv2671’s inability to catalyze the reduction of either DAROPP or AROPP under the tested conditions suggests that it is not a pyrimidine reductase and therefore not involved in the riboflavin biosynthetic pathway.
The evolutionary relationship between Rv2671, pyrimidine reductases, and DHFRs are apparent in the phylogenetic tree (Figure 5) constructed with 9 pyrimidine reductases from Actinobacteria which showed >50% sequence similarity to Rv2671, 15 DHFRs, 10 functionally identified RibDs or Rib7s, and 4 enoyl ACP reductases used as the out-group control. The full tree contained three well-supported monophyletic groups, enoyl ACP reductases, DHFRs, and RibDs/Rib7s. Although Rv2671 is part of the RibDs/Rib7s superfamily, the Rv2671-like pyrimidine reductases from Actinobacteria formed an independent clade within the RibD/Rib7 family. These enzymes have a conserved Asp (Asp67 in Rv2671) for pteridine recognition and a Glu (Glu91 in Rv2671) in place of the catalytically critical Asp of the pyrimidine reductases (Figure S4). This indicates that this group of enzymes, with high sequence similarity to Rv2671 from Actinobacteria, is likely misannotated. The ability of Rv2671 to catalyze the reduction of dihydrofolate to tetrahydrofolate or dihydropteroic acid to tetrahydropteroic acid further suggests that this particular group of proteins appears to have a fairly nonspecific dihydropteridine reductase activity. Moreover, the inability of Mtb Rv2671 to function as an AROPP reductase suggests that Mtb RibG (Rv1409) is likely to be the true AROPP reductase that generates ARIPP in the riboflavin biosynthetic pathway of Mtb.
Figure 5.
Phylogenetic tree of pyrimidine reductases and DHFRs. Sequences were aligned with MUSCLE; the tree was generated by the approximation of the standard Likelihood Ratio Test with PhyML. The numbers at the nodes indicate the level of confidence for the branches.
Supplementary Material
■ ACKNOWLEDGMENTS
We thank the beamline staff of APS-19 ID and APS-23 ID-B for data collection, Tracey Musa, Stephanie Courtright, and Dr. Inna Krieger for critical reading and editing of the manuscript, and Rung-Yi Lai for the HPLC experiment.
Funding
This work is supported in part by the TB structural genomics grant by the National Institutes of Health (Grant no. P01AI095208) and in part by the R.J. Wolfe-Welch Foundation (Grant A-0015 to J.C.S.). This work was supported by the National Institutes of Health [U19 AI107774].
■ ABBREVIATIONS
- AIF
([(2R,3S,4R,5E)-5-[(5-amino-2,6-dioxo-3H-pyrimidin-4-yl)imino]-2,3,4-trihydroxy-pentyl] dihydrogen phosphate
- ARIPP
5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate
- AROPP
5-amino-6-ribosylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate
- DAROPP
2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate
- DHF
dihydrofolate
- DHFR
dihydrofolate reductase
- DHP
dihydropteroic acid
- DHPR
dihydropteroic acid reducatase
- Mtb
Mycobacterium tuberculosis
- MTX
methotrexate
- PAS
para-aminosalicylic acid
- rmsd
rootmeansquare deviation
- TB
tuberculosis
- THF
tetrahydrofolate
- TMP
trimethoprim
- TMQ
trimetrexate
Footnotes
ASSOCIATED CONTENT
Supporting Information
Supporting Information includes one method section, one table, four figures and their associated legends and citations. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.5b00993.
Sequences used in the phylogenetic tree; hydrogen bond interactions between Rv2671 and TMP/TMQ figure legends; and references (PDF)
Figures showing the ribbon representation of Rv2671 and the gel filtration chromatography of Rv2671; superimposition of the THF bound Rv2671 structure with the E. coli RibD structure; pyrimidine reductase activity assays; and multiple sequence alignment of Rv2671 with its close orthologues and functionally characterized pyrimidine reductases (PDF)
Notes
The authors declare no competing financial interest.
■ REFERENCES
- (1).CDC. Update: Availability of Streptomycin and para-Amino-salicylic Acid—United States, Morbidity and Mortality Weekly Report; 41, 26; Center for Disease Control and Prevention: Atlanta, GA, 1992; 482. [Google Scholar]
- (2).Chakraborty S, Gruber T, Barry CE 3rd, Boshoff HI, and Rhee KY (2013) Para-aminosalicylic acid acts as an alternative substrate of folate metabolism in Mycobacterium tuberculosis. Science, 339, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Zheng J, Rubin EJ, Bifani P, Mathys V, Lim V, Au M, Jang J, Nam J, Dick T, Walker JR, Pethe K, and Camacho LR (2013) para-Aminosalicylic acid is a prodrug targeting dihydrofolate reductase in Mycobacterium tuberculosis. J. Biol. Chem 288, 23447–23456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Rieckmann KH (1973) The in Vitro Activity of Experimental Antimalarial Compounds against Strains of Plasmodium falciparum with Varying Degrees of Sensitivity to Pyrimethamine and Chloroquine, in Chemotherapy of Malaria and Resistance to Antimalarials, 529th ed., p 58, WHO Tech. Rep. Ser., World Health Organization, Geneva, Switzerland. [Google Scholar]
- (5).Zhang X, Liu L, Zhang Y, Dai G, Huang H, and Jin Q (2015) Genetic determinants involved in p-aminosalicylic acid resistance in clinical isolates from tuberculosis patients in northern China from 2006 to 2012. Antimicrob. Agents Chemother 59, 1320–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, and Barrell BG (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. [DOI] [PubMed] [Google Scholar]
- (7).Bacher A, Eberhardt S, Fischer M, Kis K, and Richter G (2000) Biosynthesis of vitamin b2 (riboflavin). Annu. Rev. Nutr 20, 153–167. [DOI] [PubMed] [Google Scholar]
- (8).Graupner M, Xu H, and White RH (2002) The pyrimidine nucleotide reductase step in riboflavin and F(420) biosynthesis in archaea proceeds by the eukaryotic route to riboflavin. J. Bacteriol 184, 1952–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Behm-Ansmant I, Grosjean H, Massenet S, Motorin Y, and Branlant C (2004) Pseudouridylation at position 32 of mitochondrial and cytoplasmic tRNAs requires two distinct enzymes in Saccharomyces cerevisiae. J. Biol. Chem 279, 52998–53006. [DOI] [PubMed] [Google Scholar]
- (10).Otwinowski Z, and Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- (11).Vagin A, and Teplyakov A (1997) MOLREP: an Automated Program for Molecular Replacement. J. Appl. Crystallogr 30, 1022–1025. [Google Scholar]
- (12).Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, and Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr., Sect. D: Biol. Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Emsley P, and Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- (15).Stone SR, and Morrison JF (1982) Kinetic mechanism of the reaction catalyzed by dihydrofolate reductase from Escherichia coli. Biochemistry 21, 3757–3765. [DOI] [PubMed] [Google Scholar]
- (16).Michaelis L, and Menten ML (1913) Die kinetik der invertinwirkung. Biochem. Z. 49, 352. [Google Scholar]
- (17).Magalhães ML, Argyrou A, Cahill SM, and Blanchard JS (2008) Kinetic and mechanistic analysis of the Escherichia coli ribD-encoded bifunctional deaminase-reductase involved in riboflavin biosynthesis. Biochemistry 47, 6499–6507. [DOI] [PubMed] [Google Scholar]
- (18).Hasnain G, Frelin O, Roje S, Ellens KW, Ali K, Guan JC, Garrett TJ, de Crécy-Lagard V, Gregory JF, McCarty DR, and Hanson AD (2013) Identification and characterization of the missing pyrimidine reductase in the plant riboflavin biosynthesis pathway. Plant Physiol. 161, 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Bailey SW, and Ayling JE (2009) The extremely slow and variable activity of dihydrofolate reductase in human liver and its implications for high folic acid intake. Proc. Natl. Acad. Sci. U. S. A 106, 15424–15429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Pruitt KD, Tatusova T, Brown GR, and Maglott DR (2012) NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, Murphy MR, O’Leary NA, Pujar S, Rajput B, Rangwala SH, Riddick LD, Shkeda A, Sun H, Tamez P, Tully RE, Wallin C, Webb D, Weber J, Wu W, DiCuccio M, Kitts P, Maglott DR, Murphy TD, and Ostell JM (2014) RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 42, D756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Tatusova T, Ciufo S, Fedorov B, O’Neill K, and Tolstoy I (2014) RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 42, D553–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, and Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, and Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol 59, 307–321. [DOI] [PubMed] [Google Scholar]
- (26).Chevenet F, Brun C, Banuls AL, Jacq B, and Christen R (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinf. 7, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Sassetti CM, Boyd DH, and Rubin EJ (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol 48, 77–84. [DOI] [PubMed] [Google Scholar]
- (28).Czekster CM, Vandemeulebroucke A, and Blanchard JS (2011) Kinetic and chemical mechanism of the dihydrofolate reductase from Mycobacterium tuberculosis. Biochemistry 50, 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Krissinel E, and Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- (30).Lv Z, Sun J, and Liu Y (2013) Structural and functional insights into Saccharomyces cerevisiae riboflavin biosynthesis reductase RIB7. PLoS One 8, e61249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Chatwell L, Krojer T, Fidler A, Romisch W, Eisenreich W, Bacher A, Huber R, and Fischer M (2006) Biosynthesis of riboflavin: structure and properties of 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate reductase of Methanocaldococcus jannaschii. J. Mol. Biol 359, 1334–1351. [DOI] [PubMed] [Google Scholar]
- (32).Stenmark P, Moche M, Gurmu D, and Nordlund P (2007) The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism. J. Mol. Biol 373, 48–64. [DOI] [PubMed] [Google Scholar]
- (33).Chen SC, Chang YC, Lin CH, and Liaw SH (2006) Crystal structure of a bifunctional deaminase and reductase from Bacillus subtilis involved in riboflavin biosynthesis. J. Biol. Chem 281, 7605–7613. [DOI] [PubMed] [Google Scholar]
- (34).Wilquet V, Gaspar JA, van de Lande M, Van de Casteele M, Legrain C, Meiering EM, and Glansdorff N (1998) Purification and characterization of recombinant Thermotoga maritima dihydrofolate reductase. Eur. J. Biochem 255, 628–637. [DOI] [PubMed] [Google Scholar]
- (35).Dias MV, Tyrakis P, Domingues RR, Paes Leme AF, and Blundell TL (2014) Mycobacterium tuberculosis dihydrofolate reductase reveals two conformational states and a possible low affinity mechanism to antifolate drugs. Structure 22, 94–103. [DOI] [PubMed] [Google Scholar]
- (36).Reyes VM, Sawaya MR, Brown KA, and Kraut J (1995) Isomorphous crystal structures of Escherichia coli dihydrofolate reductase complexed with folate, 5-deazafolate, and 5,10-dideazate-trahydrofolate: mechanistic implications. Biochemistry 34, 2710–2723. [DOI] [PubMed] [Google Scholar]
- (37).Wu Y, and Houk K (1987) Transition structures for hydride transfers. J. Am. Chem. Soc 109, 906–908. [Google Scholar]
- (38).Wu Y, and Houk K (1987) Theoretical transition structures for hydride transfer to methyleniminium ion from methylamine and dihydropyridine - on the nonlinearity of hydride transfers. J. Am. Chem. Soc 109, 2226–2227. [Google Scholar]
- (39).Wan Q, Bennett BC, Wilson MA, Kovalevsky A, Langan P, Howell EE, and Dealwis C (2014) Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography. Proc. Natl. Acad. Sci. U. S. A 111, 18225–18230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Wei JR, Krishnamoorthy V, Murphy K, Kim JH, Schnappinger D, Alber T, Sassetti CM, Rhee KY, and Rubin EJ (2011) Depletion of antibiotic targets has widely varying effects on growth. Proc. Natl. Acad. Sci U. S. A 108, 4176–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Chen SC, Lin YH, Yu HC, and Liaw SH (2009) Complex structure of Bacillus subtilis RibG: the reduction mechanism during riboflavin biosynthesis. J. Biol. Chem 284, 1725–1731. [DOI] [PubMed] [Google Scholar]
- (42).Margosiak SA, Appleman JR, Santi DV, and Blakley RL (1993) Dihydrofolate reductase from the pathogenic fungus Pneumocystis carinii: catalytic properties and interaction with antifolates. Arch. Biochem. Biophys 305, 499–508. [DOI] [PubMed] [Google Scholar]
- (43).Jackson RC, Fry DW, Boritzki TJ, Besserer JA, Leopold WR, Sloan BJ, and Elslager EF (1984) Biochemical pharmacology of the lipophilic antifolate, trimetrexate. Adv. Enzyme Regul 22, 187–206. [DOI] [PubMed] [Google Scholar]
- (44).Gangjee A, Jain HD, Phan J, Guo X, Queener SF, and Kisliuk RL (2010) 2,4-Diamino-5-methyl-6-substituted arylthio-furo[2,3-d]pyrimidines as novel classical and nonclassical antifolates as potential dual thymidylate synthase and dihydrofolate reductase inhibitors. Bioorg. Med. Chem 18, 953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).White EL, Ross LJ, Cunningham A, and Escuyer V (2004) Cloning, expression, and characterization of Mycobacterium tuberculosis dihydrofolate reductase. FEMS Microbiol. Lett 232, 101–105. [DOI] [PubMed] [Google Scholar]
- (46).Pyrkov TV, Chugunov AO, Krylov NA, Nolde DE, and Efremov RG (2009) PLATINUM: a web tool for analysis of hydrophobic/hydrophilic organization of biomolecular complexes. Bioinformatics 25, 1201–1202. [DOI] [PubMed] [Google Scholar]
- (47).Holm L, and Rosenström P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Romisch-Margl W, Eisenreich W, Haase I, Bacher A, and Fischer M (2008) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5′-phosphate synthases of fungi and archaea. FEBS J. 275, 4403–4414. [DOI] [PubMed] [Google Scholar]
- (49).Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- (50).Argyrou A, Vetting MW, Aladegbami B, and Blanchard JS (2006) Mycobacterium tuberculosis dihydrofolate reductase is a target for isoniazid. Nat. Struct. Mol. Biol 13, 408–413. [DOI] [PubMed] [Google Scholar]
- (51).Bock RA, Soulages JL, and Barrow WW (2007) substrate and inhibitor specificity of Mycobacterium avium dihydrofolate reductase. FEBS J 274, 3286–3298. [DOI] [PubMed] [Google Scholar]
- (52).Maglia G, Javed MH, and Allemann RK (2003) Hydride transfer during catalysis by dihydrofolate reductase from Thermotoga maritima. Biochem. J 374, 529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Fierke CA, Johnson KA, and Benkovic SJ (1987) Construction and evaluation of the kinetic scheme associated with dihydrofolate reductase from Escherichia coli. Biochemistry 26, 4085–4092. [DOI] [PubMed] [Google Scholar]
- (54).Lee J, Yennawar NH, Gam J, and Benkovic SJ (2010) Kinetic and structural characterization of dihydrofolate reductase from Streptococcus pneumoniae. Biochemistry 49, 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Thillet J, Adams JA, and Benkovic SJ (1990) The kinetic mechanism of wild-type and mutant mouse dihydrofolate reductases. Biochemistry 29, 5195–5202. [DOI] [PubMed] [Google Scholar]
- (56).Appleman JR, Beard WA, Delcamp TJ, Prendergast NJ, Freisheim JH, and Blakley RL (1990) Unusual transient- and steady-state kinetic behavior is predicted by the kinetic scheme operational for recombinant human dihydrofolate reductase. J. Biol. Chem 265, 2740–2748. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.