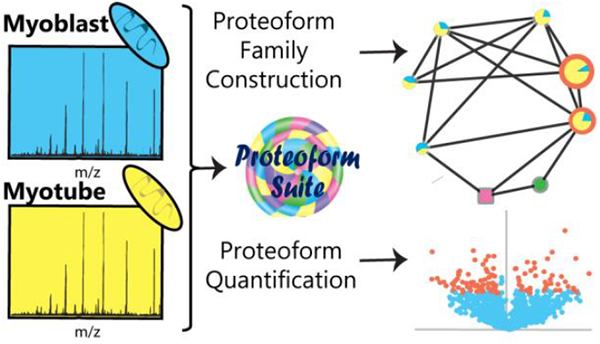
Abstract
The development of effective strategies for the comprehensive identification and quantification of proteoforms in complex systems is a critical challenge in proteomics. Proteoforms, the specific molecular forms in which proteins are present in biological systems, are the key effectors of biological function. Thus, knowledge of proteoform identities and abundances is essential to unraveling the mechanisms that underlie protein function. We recently reported a strategy that integrates conventional top-down mass spectrometry with intact-mass determinations for enhanced proteoform identifications and the elucidation of proteoform families and applied it to the analysis of yeast cell lysate. In the present work, we extend this strategy to enable quantification of proteoforms, and we examine changes in the abundance of murine mitochondrial proteoforms upon differentiation of mouse myoblasts to myotubes. The integrated top-down and intact-mass strategy provided an increase of ~37% in the number of identified proteoforms compared to top-down alone, which is in agreement with our previous work in yeast; 1779 unique proteoforms were identified using the integrated strategy, compared to 1301 using top-down analysis alone. Quantitative comparison of proteoform differences between the myoblast and myotube cell types showed 129 observed proteoforms exhibiting statistically significant abundance changes (fold change > 2 and false discovery rate < 5%).
Keywords: Top-down proteomics, proteoform, quantification, proteoform family, software, mitochondria
Graphical abstract
INTRODUCTION
Cells require a high level of proteomic complexity to perform a vast array of biological functions. One important aspect of proteome complexity comes from the diversity of proteoforms present. Here, the term proteoform refers to the different forms of a protein, arising from sources such as genetic variation, RNA splicing, and post-translational modification (PTM).1 The set of all proteoforms derived from the same gene make up a proteoform family.2 The identification of proteoforms is possible with top-down proteomic analysis, where intact proteins are analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS), and identifications are made using precursor mass (MS1) and fragment (MS2) information. Proteoforms are usually identified in top-down software by the precursor monoisotopic mass and fragment information.3–7 A top-down identification of a proteoform means the protein from which is it derived has also been identified, and the protein inference problem of bottom-up proteomics has been avoided.8 However, challenges remain in sample preparation and separation of intact proteins, in optimization of sequence coverage to localize modifications, and in the data analysis of complex top-down mass spectra.9
Although top-down proteomics has been used successfully to identify proteoforms in complex samples such as yeast10, E. coli11,12, and human13,14 lysates, major challenges remain when attempting to analyze intact proteins by MS/MS. One such limitation is that many more proteoforms are observed in MS1 spectra than may be subjected to fragmentation analysis15–17, due to both time constraints and signal-to-noise limitations.18,19 We recently reported a strategy that integrates conventional top-down mass spectrometry with intact-mass determinations for enhanced proteoform identifications and the elucidation of proteoform families, and we applied this strategy to the analysis of yeast cell lysate.20 In the present study, we explore the utility of this approach for proteoform analysis in a mammalian system of reduced scope - the mitochondrial proteoform-ome from the C2C12 murine myoblast cell line before and after differentiation into myotubes, which is a well-studied model system for skeletal muscle myocyte development.21–23 In addition, we augmented the analysis software to provide label-free quantification of intact proteoforms, enabling quantitative comparison of proteoform expression levels between these two cellular states.
Protein quantification in bottom-up analyses of both labeled and unlabeled samples is well established.24–27 However, in bottom-up quantitative analyses, the overall amount of each protein present is estimated by analyzing intensities from a limited subset of individual peptides that are thought to be derived from that protein. As such, quantitative analysis in bottom-up proteomics is generally limited to reporting bulk changes in the abundance of a particular peptide or given protein (while not discriminating among the various proteoforms of that protein). Proteoform-level abundance information can have important biological consequences, as different proteoforms often vary dramatically with respect to function.28 Label-free quantification at the intact proteoform level enables individual proteoforms to be quantified.29–32 In this study, proteoforms were identified, quantified, and assembled into proteoform families using the program Proteoform Suite33, which is open-source, freely available (https://smith-chem-wisc.github.io/ProteoformSuite/), and readily integrated into current top-down proteomic workflows.
METHODS
Data Acquisition
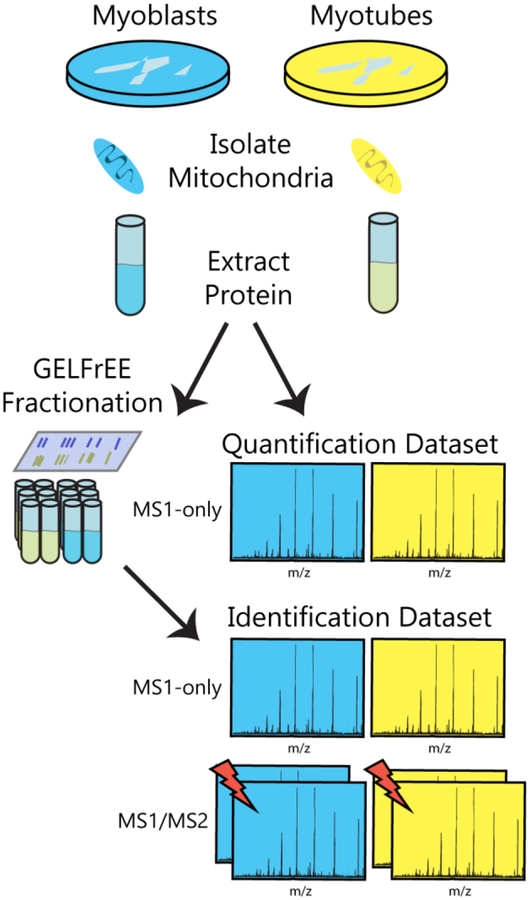
We analyzed both size-fractionated and unfractionated mitochondrial proteins from C2C12 myoblasts and myotubes to generate various datasets used for proteoform identification and quantification. The fractionated samples yielded deeper proteome coverage for identification, while the unfractionated samples provided more accurate proteoform intensity measurements for quantification (as fractionation introduces variability in MS intensity measurements). An outline of the data acquisition workflow is depicted in Figure 1. All mass spectrometry raw files are freely available on the MassIVE platform (https://massive.ucsd.edu; ID: MSV000082366). The sample preparation and mass spectrometric analyses are described in detail in the Materials and Methods section in the Supporting Information.
Figure 1.
Overview of data acquisition workflow. Proteins were extracted from mitochondria of myoblast and myotube cells. GELFrEE size-based prefractionation was performed in the case of integrated top-down (MS1/MS2) and intact-mass (MS1-only) measurements used for proteoform identifications, whereas no such pre-fractionation was employed for MS1-only measurements used for quantification. Lightning bolts denote fragmentation processes.
Data Acquisition for Identification Dataset
Undifferentiated C2C12 mouse myoblast cells and differentiated myotube cells were pelleted, and mitochondria were isolated, flash-frozen in liquid nitrogen, and stored at −80 °C until use. Cells from three biological replicates for each condition were lysed by heat, precipitated with acetone, and resuspended in 1% sodium dodecyl sulfate (SDS). Samples were fractionated using gel-eluted liquid entrapment electrophoresis (GELFrEE).34 Approximately 165 μg of each biological replicate was loaded onto a 12% GELFrEE cartridge (Expedeon) and separated by molecular weight into twelve fractions in accordance with the manufacturer’s recommendations. Two adjacent fractions were combined to produce six fractions for each biological replicate, and methanol-chloroform precipitation was performed to remove SDS. For each fraction, two replicates of top-down with top-2 data-dependent acquisition were acquired on a QE-HF Orbitrap (Thermo Fisher Scientific), as well as two MS1-only replicates, totaling 144 mass spectrometry raw files (Identification Dataset).
Data Acquisition for Quantification Dataset
An unfractionated dataset for accurate quantification was also generated. Three biological replicates for each condition were prepared as described above. After resuspension in 1% SDS, an aliquot was set aside for tryptic digestion and bottom-up analysis (see Supporting Information). Methanol-chloroform precipitation was performed on each sample (no fractionation was performed). Two MS1-only replicates of each intact sample were analyzed to produce 12 mass spectrometry raw files (Quantification Dataset). Two bottom-up top-10 data-dependent acquisition runs were performed on each digested sample, producing 12 bottom-up mass spectrometry raw files (Bottom-Up Dataset).
Data Analysis
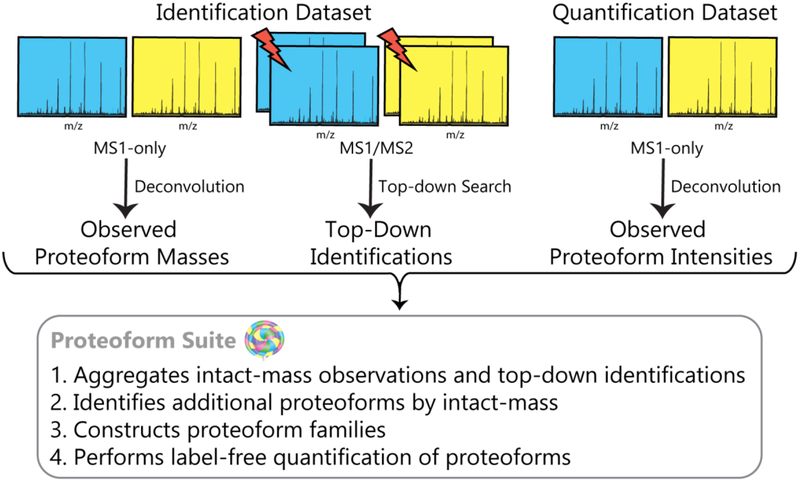
An outline of the data analysis workflow, including the steps automated in Proteoform Suite, is depicted in Figure 2. Version 0.3.3 of Proteoform Suite was used for all analyses (https://github.com/smith-chem-wisc/ProteoformSuite/releases). A schematic of the data processing steps in Proteoform Suite is shown in Figure S-1. All files used for the analysis, including method (.xml) files to automatically perform end-to-end analyses and reproduce the results in the software, are available in the Vignette folder in the release.
Figure 2.
Overview of data analysis workflow. Lightning bolts denote fragmentation processes.
Top-Down Data Analysis
Top-down analysis of the raw files from the Identification Dataset was performed using TDPortal from the National Resource for Translational and Developmental Proteomics (NRTDP, Northwestern University, Evanston, IL). TDPortal is available for academic collaborators at http://nrtdp.northwestern.edu/tdportal-request/. A shotgun annotated database from the May 2016 Swiss-Prot release of the Mus musculus proteome was created containing theoretical proteoforms for each protein with the sequence, potential sequence variations, and potential site-specific modifications.3,35 TDPortal performed a search with two modes, as defined for ProSight PTM 2.03: tight absolute mass search (2.2 Da tolerance for MS1 and 10 ppm tolerance for MS2); and biomarker search (10 ppm tolerance for both MS1 and MS2). TDPortal results were analyzed in TDViewer (http://topdownviewer.northwestern.edu), and top-down hits (protein spectrum matches) were exported to a Microsoft Excel file. These hits were filtered to satisfy a 1% protein-level false discovery rate (FDR) cutoff.
Deconvolution of MS1 Scans
All MS1-only raw files (for both the Identification Dataset and Quantification Dataset) were charge-state deconvoluted and deisotoped using Thermo Protein Deconvolution 4.0. The following parameters were utilized: fit factor = 70%, minimum S/N = 2, remainder threshold = 10%, minimum detected charge states = 3, charge range of +5 to +30. For the Quantification Dataset, the minimum of detected charge states was set to 2 and a charge range of +2 to +30 was utilized. A sliding window of 0.20 min and 34% offset was used to deconvolute the retention time range of 40 – 100 min, which is when most proteins eluted, in multiple ranges (the data export format of .xls has a maximum row allowance). The resulting Microsoft Excel files contained a list of the “raw experimental components,” which are the monoisotopic masses and corresponding integrated intensities for detected proteoforms.
Proteoform Suite Calibration
Mass calibration was performed in Proteoform Suite to improve mass accuracy using the software lock-mass concept developed for bottom-up proteomics.36,37 This algorithm has been described previously.20,37 Top-down hits with a minimum C-score of 40 (corresponding to well-characterized identifications38) were used to determine mass error as a function of retention time, m/z value, scan total ion current, and scan injection time for each individual raw file using a random forest machine-learning algorithm. For MS1-only files from both the Identification Dataset and the Quantification Dataset, components from deconvolution were selected as calibration points if within 10 ppm and 5 minutes of a top-down identification from the same condition (myoblast or myotube). In the Identification Dataset, the component from deconvolution also had to be from the same fraction and biological replicate as the corresponding top-down identification. A minimum of 5 top-down hits and 10 calibration points were required for each raw file; files failing these criteria were not analyzed further. Calibrated top-down hits (77 874 hits) and calibrated deconvolution results (485 835 raw components from Identification Dataset and 159 640 from Quantification Dataset) were used for subsequent analyses.
Proteoform Suite Identification Analysis
Proteoform Suite was used to identify additional proteoforms observed in MS1 spectra but not identified by top-down.20 Top-down hits with a minimum C-score of 3 (corresponding to identified proteoforms, but not necessarily well-characterized38) were imported and deconvolution errors were automatically corrected. The 1301 unique proteoform identifications became 1586 top-down experimental proteoforms during the “aggregation” step because if the same proteoform identification eluted at different retention times (>5 min), it was separated into different top-down experimental proteoforms in Proteoform Suite (Supporting Table S-1). We used a retention time tolerance of 5 minutes because top-down proteoforms from different LC-MS runs were aggregated; as a result, a wider tolerance is needed than the expected chromatographic peak width for a given proteoform in a single run. Figure S-2 shows a histogram of the maximum retention time difference for hits of the same top-down identification for each individual .raw file and for all .raw files. As seen in the figure, while top-down identified proteoforms of the same .raw file typically elute over the course of less than 1 or 2 minutes, this retention time difference increases to between 5 and 10 minutes when many .raw files are included. Future versions of Proteoform Suite will perform retention time calibration, similar to the mass calibration already performed by this software, in order to reduce proteoform retention time differences across MS runs.
The deconvolution results from the MS1-only raw files of the Identification Dataset (fractionated by GELFrEE) were imported, deconvolution errors were automatically corrected, and observations were aggregated by mass (allowing deviations of up to 5 ppm and up to 3 missed monoisotopic mass units) and retention time (deviations allowed of up to 5 min) to create a list of unique intact-mass experimental proteoforms. Only intact-mass experimental proteoforms present in at least three biological replicates of a single condition were selected for further analysis. The top-down experimental proteoforms were added to the intact-mass experimental proteoforms list, replacing intact-mass proteoforms with the same mass and retention time tolerances used for aggregation. The final list of 3794 experimental proteoforms thus contained top-down identified experimental proteoforms and intact-mass observations that were not identified by top-down fragmentation. A theoretical proteoform database was generated from the Mus musculus UniProt XML database (downloaded March 2017) with reviewed sequences and a database with common contaminants. The theoretical database contained unmodified proteoforms and proteoforms with combinations of up to two annotated modifications. Additional theoretical proteoforms were added, corresponding to proteoforms identified by TDPortal that were not already present in the database.
The process of constructing proteoform families has been described previously.2,20,33,39 Experimental proteoforms first were compared to the theoretical database, yielding experimental-theoretical pairs (ET pairs). To identify additional proteoforms not in the theoretical database, experimental proteoform masses were compared with masses of other experimental proteoforms that eluted within 2.5 minutes of one another to form experimental-experimental pairs (EE pairs). Pairs with mass differences that corresponded to known sets of PTMs or amino acid differences were accepted (Supporting Table S-2). Finally, proteoforms joined by accepted mass differences were formed into proteoform families. Proteoform Suite calculated FDR by dividing the number of identifications in target families by the average number of identifications in ten sets of created decoy families, as previously described.20,33 In this work, the FDR for proteoform identification was calculated to be 3.8%.
Proteoform Suite Quantitative Analysis
Previous work used Proteoform Suite with isotopic labeling to determine proteoform abundance changes in a yeast salt-stress response.33 In the current study, we have enabled label-free quantification in Proteoform Suite using the log2 fold-change analysis commonly used in bottom-up proteomic quantification.40 Deconvolution components from the MS1-only raw files of the Quantification Dataset (unfractionated) were imported and assigned to experimental proteoforms from the Identification Dataset with the same mass and retention time tolerances used for aggregation. Experimental proteoforms from the Identification Dataset with quantitative deconvolution components from a minimum of three biological replicates of a single condition were accepted for quantification analysis. For each quantified experimental proteoform, integrated ion intensities reported by Thermo Protein Deconvolution from all assigned quantitative components were summed across technical replicates for each biological replicate of each condition and normalized to total intensity of each biological replicate. Missing intensities for each biological replicate of each condition were imputed by selecting random values from a background distribution.40 The background intensity distribution was calculated from the Gaussian distribution of log2-quantified proteoform intensities with an intensity distribution width of 0.7σ and shift of −1.5σ from the population mean. A log2 fold-change analysis (Student’s t-test) was performed on the Quantification Dataset after adding these imputed intensity values. Changes in the abundances of experimentally observed proteoforms were defined as significant if they exhibited a minimum fold-change of 2.0 and a 5% FDR controlled for multiple testing by applying the Benjamini-Hochberg procedure41 to the calculated p-values.
Visualization of Proteoform Families
Finally, the results from both the identification and quantification analyses were visualized as proteoform family networks.33 Each unique proteoform is represented as a node (circle), and mass differences corresponding to modifications are represented as edges (lines) connecting “related” proteoforms. The area of each intact-mass experimental proteoform node is proportional to the integrated ion intensity reported by Thermo Protein Deconvolution. For each quantified proteoform, the intensity ratios between conditions are displayed as a pie chart, and proteoforms with significant changes have an orange annulus. Proteoform Suite outputs scripts that can be run by the visualization program Cytoscape42,43 to automatically visualize proteoform families.
Bottom-Up Quantitative Analysis
Bottom-up raw files were analyzed by MaxQuant44 version 1.6.1.0 with a Mus musculus UniProt FASTA database with reviewed sequences downloaded December 2017. Precursor ion mass tolerance was set to 4.5 ppm, and product ion mass tolerance was set to 20 ppm. Cysteine carbamidomethylation was included as a fixed modification, and oxidation of methionine and N-terminal protein acetylation were included as variable modifications. Two missed cleavages were allowed, and two unique peptides per protein were required. The MaxQuant label-free quantification algorithm with “match between runs” enabled was used to quantify peptides; only unique and razor peptides (which are peptides that are shared between protein groups8) were used. Perseus software (version 1.6.1.3)40 was used to perform statistical analysis with the protein groups output file. We required a protein to have been observed in at least three biological replicates of one condition. Intensities were log2 transformed, and imputation was used to replace missing intensities values with a background intensity distribution width of 0.3σ and shift of −1.8σ. We performed a Student’s t-test, and proteins with abundance changes that met the 5% Benjamini-Hochberg FDR cutoff were considered to have statistically significant abundance changes.
RESULTS AND DISCUSSION
The analysis of mitochondrial proteoforms from C2C12 myoblasts and myotubes presented here was performed in two parts: Identification, employing searches of the Identification Dataset (see Figure 1 and Methods Section); and Quantification, in which the proteoforms observed in the Identification Dataset were then quantified using the Quantification Dataset. Any proteoform quantified thus had to have been present in both datasets. The two datasets differ in that the Identification Dataset was generated from GELFrEE-fractionated samples, whereas the Quantification Dataset was from unfractionated samples.
Identification of Proteoforms
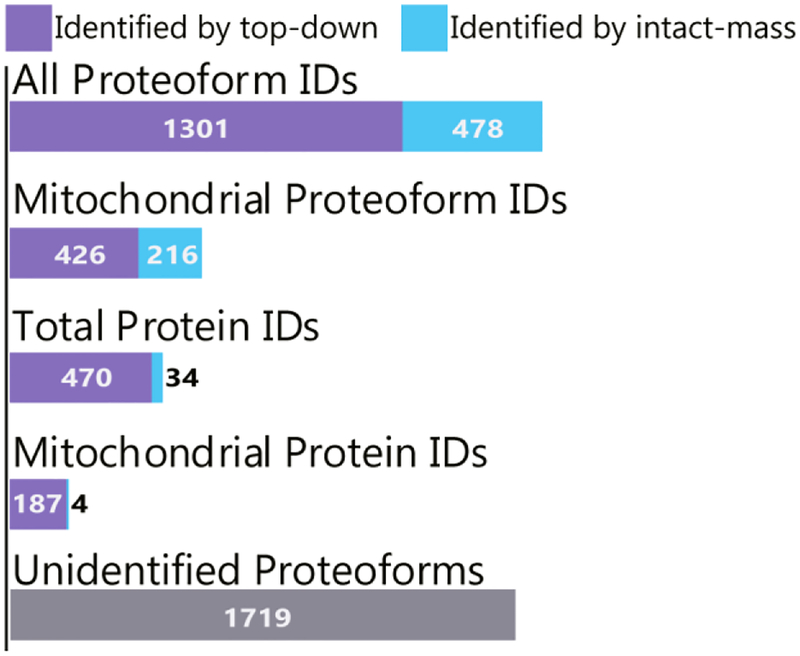
In top-down analyses, many proteoforms are observed in the MS1 spectra that are not identified by top-down fragmentation analysis17; we used Proteoform Suite to analyze the accurate intact-masses and make additional identifications of these observed but unidentified proteoforms. Figure 3 depicts the ~37% increase in number of proteoform identifications obtained by using Proteoform Suite to analyze intact masses in this study. There were 1301 unique identifications made by top-down fragmentation and 478 additional unique proteoform identifications made by intact-mass analysis in Proteoform Suite. The FDR for Proteoform Suite analysis was calculated to be 3.8%, whereas the TDPortal FDR was set to 1%. TDPortal was only able to identify 8 of the 478 intact-mass identifications, even when the TDPortal FDR was relaxed to 5%. Importantly, many of the additional proteoform identifications are from mitochondrial proteins, defined here as proteins that are present in the mitochondrial inventory MitoCarta.45 Specifically, 426 unique mitochondrial proteoforms were identified by top-down analysis in TDPortal, and 216 additional unique mitochondrial proteoforms were identified by intact-mass analysis in Proteoform Suite (Figure 3).
Figure 3.
Proteoform and protein identification results. Proteoform Suite increased the number of proteoform identifications by 37% overall and by 51% for mitochondrial proteoforms. Additionally, the number of unique protein IDs (each corresponding to a particular gene) increased by 7% overall and by 2% for mitochondrial proteins. There were 1719 intact-mass experimental proteoforms observed in at least three biological replicates of a single condition that were unidentified by either top-down or intact-mass analysis.
Additionally, the number of protein identifications (each corresponding to a specific gene, or proteoform family) increased by ~7% using the intact-mass analysis. There were 470 unique proteins identified by top-down analysis, and 34 additional proteins were identified by intact-mass; thus intact-mass analysis enabled entirely new proteoform families to be identified. Of the new protein identifications, there were 4 mitochondrial proteins identified by intact-mass analysis. These results illustrate how Proteoform Suite may be integrated into a top-down proteomics workflow to increase the number of proteoform and protein identifications. All intact-mass identifications are shown in Supporting Table S-3.
The top-down identified experimental proteoforms also undergo intact-mass analysis in Proteoform Suite; these Proteoform Suite identification assignments are shown in Supporting Table S-1, which displays top-down experimental proteoforms. Of the 1586 top-down experimental proteoforms, 932 were assigned the same identification by intact-mass analysis. There were 172 top-down experimental proteoforms where the identification assigned by Proteoform Suite differed from the identification assigned by TDPortal; the TDPortal identification determined with fragmentation was utilized for subsequent descriptions of the quantitative analysis. The majority of these proteoforms were histones, which are challenging for both top-down and intact-mass analysis due to sequence similarities and number of modifications. Finally, 482 top-down experimental proteoforms were not assigned an identification in Proteoform Suite because the precursor mass error was too large.
In total, our analysis of mitochondrial extracts from C2C12 myoblasts and myotubes identified 642 mitochondrial proteoforms from 191 unique mitochondrial proteins. We identified 259 proteoforms and 55 proteins from oxidative phosphorylation complexes. A previous top-down study that also used GELFrEE separation of mitochondrial extracts identified 107 unique annotated mitochondrial proteins46, and a study that used deeper sample fractionation by employing two orthogonal separation modes (GELFrEE and isoelectric focusing) identified 347 mitochondrial proteins at 1% FDR13. While more extensive pre-fractionation is able to provide deeper proteome coverage, it also requires increased sample amounts and instrument time; the intact-mass strategy employed here thus comprises a valuable strategy to provide increased proteome coverage at little cost. For studies where depth of proteome coverage is paramount, the present top-down/intact-mass strategy used in conjunction with multiple orthogonal separation modalities would likely provide the deepest coverage presently possible. This is because the orthogonal separations lead to more proteoforms being observed in the MS1, which are then available for identification by intact-mass, and the extra instrument time utilized for additional fractions allows more proteoforms to be selected for top-down fragmentation and thereby potentially identified in the MS2.
A number of identified mitochondrial proteoforms contained post-translational modifications. Top-down fragmentation analysis in TDPortal identified 224 mitochondrial proteoforms with at least one post-translational modification, including 2 proteoforms with the lipid modification N-myristoyl glycine. Proteoform Suite identified an additional 211 mitochondrial proteoforms containing modifications. A limitation of intact-mass analysis is that modifications on identified proteoforms are not localized; when necessary, targeted top-down analysis may be used to fragment proteoforms of interest and localize modifications of interest. Alternatively, in many cases PTM localization may be inferred from bottom-up results obtained using the global PTM discovery strategy (GPTM-D).37,47,48
There were many oxidized proteoforms identified by the intact-mass analysis - 306 total proteoforms, of which 154 were from mitochondrial proteins (these were not identified in the top-down analysis, as TDPortal does not include oxidation as a modification). Mitochondrial proteins are susceptible to oxidation because mitochondria are the main source of endogenous reactive oxygen species.49 Oxidation is both a naturally occurring regulatory modification50,51 and a sample handling artifact; we required an intact-mass experimental proteoform to be observed in three biological replicates, however this does not exclude the possibility of a given oxidation modification being an artifact from sample preparation.
Mass Calibration Improves Mass Accuracy
Because Proteoform Suite identifies additional proteoforms by experimental mass alone, mass accuracy is of utmost importance. We used mass calibration to increase the mass accuracy of both deconvoluted intact-mass observations and the top-down identification precursors by using high-scoring top-down identifications as calibration points for each raw file.20,36,37 A histogram of the mass error of precursor mass for top-down hits before and after calibration (Figure S-3) shows how mass accuracy improved for the top-down precursor masses. The number of identified intact-mass experimental proteoforms increased from 229 to 489 identifications over uncalibrated data analyzed with the same parameters, and the FDR decreased from 7.1% to 3.8%. These results demonstrate how mass calibration significantly improves intact-mass analysis. Mass calibration is performed in Proteoform Suite with the graphical user interface to produce calibrated deconvolution results (.xlsx), top-down results (.xlsx), and mass spectra files (.mzML).
Quantification of Proteoforms
We implemented a label-free quantitative analysis in Proteoform Suite to analyze myogenesis using a C2C12 mouse cell model, comparing undifferentiated myoblasts and differentiated myotubes. This analysis quantified proteoforms that were observed in the Identification Dataset. The MS intensities utilized for quantification analysis were measured in the Quantification Dataset. We required that an experimental proteoform be observed in at least three biological replicates of a single condition from the Quantification Dataset to be eligible for quantification, yielding 936 experimental proteoforms from the Identification Dataset (Supporting Table S-4). These quantified experimental proteoforms included 554 observed masses that were intact-mass experimental proteoforms, of which 194 were identified by Proteoform Suite. The remaining 382 quantified experimental proteoforms were identified by top-down fragmentation.
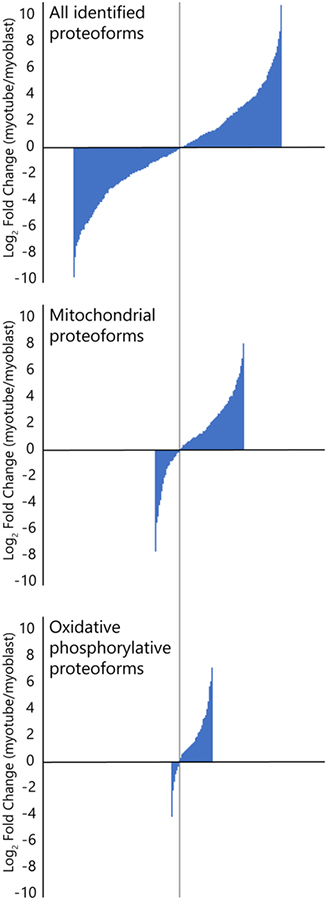
We analyzed the relative abundance changes across myoblast and myotube conditions for identified experimental proteoforms, as shown in Figure 4. For the set of all identified proteoforms, the distribution of positive and negative abundance changes is similar, which indicates that normalization was effective and loading amounts were similar for each condition; 50.8% of abundance changes favored myoblasts, and 49.2% of abundance changes favored myotubes. For mitochondrial proteoforms, 71.4% of the abundances increased in myotubes relative to myoblasts. This difference is even more pronounced for proteoforms of oxidative phosphorylation complexes, with 80.5% of abundances increasing in myotubes. These results are consistent with previous bottom-up quantitative analyses of mitochondrial protein abundance changes following C2C12 myoblast differentiation21,23,52, as myotubes have a greater reliance on mitochondrial metabolism and oxidative phosphorylation compared to myoblasts.21,22
Figure 4.
Bar graphs showing a comparison of relative abundance changes between myoblasts and myotubes for all identified and quantified proteoforms (top), mitochondrial proteoforms (middle), and mitochondrial proteoforms involved with oxidative phosphorylation (bottom). The height of each bar corresponds to an identified proteoform’s fold change value. The bars were ordered (left to right) by size from smallest (most negative fold change) to largest (most positive fold change), and the three plots were aligned by the bar corresponding to the fold change closest to 0 (vertical grey line). While the distribution of positive and negative changes in abundance are similar for the set of all proteoforms, positive changes are more frequent than negative changes for mitochondrial proteoforms and much more frequent for proteoforms involved with oxidative phosphorylation.
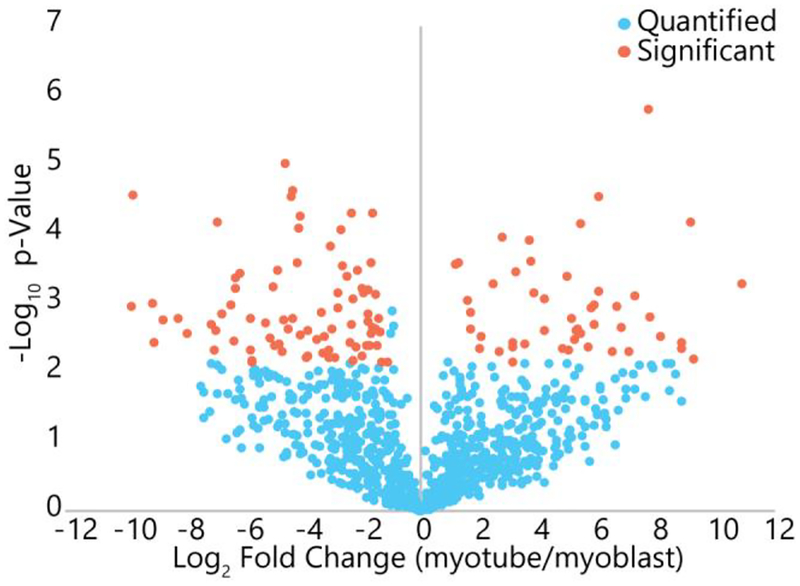
The volcano plot of log10 p-value versus the log2 fold-change for each quantified experimental proteoform is displayed in Figure 5, and orange points represent experimental proteoforms with statistically significant fold-changes. There were 129 experimental proteoforms with a significant abundance change across myoblast and myotube conditions. Of these, 84 were identified: 25 were identified by intact-mass analysis in Proteoform Suite, and 59 were identified by top-down fragmentation analysis. The remaining 45 observed intact-mass experimental proteoforms with statistically significant abundance changes were unidentified. Because Proteoform Suite performs the quantification analysis on all observed experimental proteoforms, unidentified observed proteoforms with significant abundance changes can still be determined, enabling a potential follow-up study where these changing proteoforms are targeted for fragmentation and identified.
Figure 5.
Volcano plot of changes in quantified proteoforms between myoblasts and myotubes. Blue represents proteoforms that did not change significantly, while orange represents those that did change significantly (fold change greater than 2 and p-value satisfying a Benjamini-Hochberg adjusted 5% FDR threshold). Of the quantified proteoforms, 13.7% showed significant changes between the two cell types.
We utilized intensity values from the unfractionated dataset for quantification because we found that fractionation introduced intensity variations which resulted in fewer statistically significant abundance changes. When using fractionated intensity values with the same parameters in Proteoform Suite, only 19 proteoforms were found to have statistically significant abundance changes across conditions. Of these, 9 proteoforms were identified, three of which were mitochondrial. A proteoform from the mitochondrial gene NDUFV3 was found to have statistically significant abundance changes in the unfractionated dataset, which agrees with bottom-up results (discussed below); however, this proteoform in the fractionated dataset did not have a statistically significant change, which we attribute to the increased intensity variations that occur for fractionated data. The standard deviation in log2 intensity across biological replicates for myoblasts and myotubes were both 0.37 in the unfractionated dataset, but 4.32 and 2.03 in the fractionated dataset. These results indicate that more reproducible separation is needed for pre-fractionation of intact proteins if reliable quantification is needed. Additionally, better normalization could reduce intensity variation, such as the MaxQuant normalization procedure used in bottom-up quantitative analyses.24
We performed a bottom-up protein quantification analysis with the software program MaxQuant24 (quantitative results in Supporting Table S-5) to compare with our proteoform quantification results. MaxQuant calculates a protein abundance value for each condition from the median of peptide intensity ratios for those peptides observed in both samples.24 From our intact proteoform results, we highlighted 26 proteoforms with statistically significant abundance changes that are known to be mitochondrial, and these correspond to 16 unique proteins. Bottom-up quantitative analysis determined significant abundance changes in 8 of these 16 proteins (discussed below) and non-significant abundance changes in 4 of these proteins. The remaining 4 proteins were not identified by bottom-up analysis. While it is interesting to compare the results between bottom-up and proteoform-level quantification, it is important to note that bottom-up and top-down quantification are comparing different molecular entities (peptides vs. proteoforms).53 When Proteoform Suite determines a statistically significant change in a given proteoform, bottom-up analysis does not necessarily determine this same change for the overall abundance levels. We attribute this to the fact that bottom-up analysis quantifies peptides from a mixture of different proteoforms in the sample, thus overall protein amount is being quantified as opposed to specific proteoform amount. In theory, a proteoform-specific peptide could be used to determine a proteoform abundance change across conditions; however, it can be difficult to confidently determine a peptide as proteoform-specific. In many cases, this requires that a PTM-containing or terminal peptide be identified.
Various mitochondrial proteoforms showed abundance changes from myogenesis (see Supporting Table S-4). Notably, a di-phosphorylated proteoform of COX4I1 (a subunit of complex IV of the electron transport chain) consisting of amino acid positions 23 to 168 significantly decreased in myotubes, whereas the unmodified form non-significantly increased in myotubes. The phosphorylated COX4I1 proteoform, identified by top-down fragmentation, contained phosphorylation on amino acids S56 and S58. Interestingly, previous work has determined that COX4I1 S58 phosphorylation regulates metabolic activity and decreased phosphorylation of this site is associated with inhibition of COX activity.54 However, as mitochondrial biogenesis increases during myotube formation, our result could suggest that decreased phosphorylation at one or both of these sites promotes mitochondrial activity. Future studies are needed to determine the significance of this decreased di-phosphorylated proteoform in myotubes, where the shift in metabolism is towards oxidative phosphorylation. Our MaxQuant analysis did not determine significant changes for the overall COX4I1 protein amount; however, an individual peptide with amino acid positions 160 to 168 decreased in abundance in myotubes.
Proteoforms from the genes NDUFA7, NDUFA12, and NDUFV3, which are members of complex I of the electron transport chain, were more abundant in myotubes, consistent with previous observations.52 MaxQuant reported that each of these proteins have statistically significant greater abundances in myotubes. While bottom-up analysis did identify similar abundance changes of these proteins, top-down analysis and Proteoform Suite identified the specific proteoforms from these genes that showed abundance changes across conditions. In Proteoform Suite, we observed that the significantly changing proteoform from NDUFA7 was acetylated and oxidized. In the bottom-up search, oxidation and protein N-terminal acetylation were included as variable modifications; three different oxidized peptides were identified from the NDUFA7 gene, but no acetylated peptides from either protein were identified by bottom-up analysis.
Proteoforms from ATP5F1 and ATP5I (subunits of mitochondrial ATP synthase complex V) and from USMG5 (involved in maintaining ATP synthases) were found at statistically significant increased abundances in myotubes, which is consistent with the MaxQuant results. Proteoforms from the genes HSPE1, PAM16, and PET117 with a single acetylation were detected at higher abundances in myoblasts than myotubes. MaxQuant also determined a significant abundance decrease in myotubes for HSPE1 (although the acetylation was not identified), did not identify any peptides from PET117, and did not determine statistically significant abundance changes for PAM16. Proteoform Suite also determined that a proteoform with four acetylations from HSPE1 increased in abundance in myotubes. Thus, in this HSPE1 example, different proteoforms from the same gene were shown to either decrease or increase in abundance under myogenesis, but this interesting circumstance was not detected by bottom-up analysis.
TDPortal and Proteoform Suite identified several proteoforms from the ATPIF1 gene, a mitochondrial ATPase inhibitor, with statistically significant abundance changes between myoblast and myotube cell types. While the proteoform with a sequence corresponding to amino acid positions 26 to 106 was present at lower levels in the myotubes than in the myoblasts, smaller proteoforms with sequences from amino acid positions 29 to 106, 30 to 106, 33 to 106, and 35 to 106 were present at higher levels in the myotubes. Truncated proteoforms are of great interest because they can have important biological consequences; for example, histone H3 clipping has been found to regulate gene expression.55 A semi-specific enzyme search can be performed in bottom-up analyses to identify these truncated forms, but these searches increase the search space which can negatively impact the FDR.56 Additionally, the identification of a truncated protein by bottom-up analysis requires the identification of a peptide at the N or C terminus. Future studies could determine whether the differences in the abundances of these truncated ATPIF1 proteoforms are in fact biologically significant or a result of sample handling. Bottom-up quantification reported an increase in overall ATPIF1 protein levels for myotubes (log2 fold-change = 1.07) and did not detect these subtle differences in specific truncated proteoform abundances; i.e. the proteoform-level information was lost. In summary, Proteoform Suite enabled a label-free quantification analysis that revealed individual mitochondrial proteoform abundance changes that were not revealed by bottom-up quantitative analysis.
Construction and Visualization of Proteoform Families
Proteoform Suite constructs proteoform families from accepted experimental-experimental (EE) and experimental-theoretical (ET) pairs, as explained in the Methods section. From this process, 669 proteoform families were constructed: 413 proteoform families corresponded to one gene, 29 proteoform families corresponded to more than one gene, and 227 proteoform families remained unidentified. In the families containing more than one gene, 9 identified intact-mass experimental proteoforms were potentially ambiguous identifications, meaning the proteoform had equal numbers of connections from theoretical proteoforms of different genes. Finally, 1024 intact-mass experimental proteoforms were orphans, as no relations were formed with another proteoform, neither theoretical nor experimental. Future targeted top-down analysis could help identify orphans and unidentified proteoforms; there were 1719 intact-mass experimental proteoforms observed that remained unidentified by top-down or intact-mass analysis.
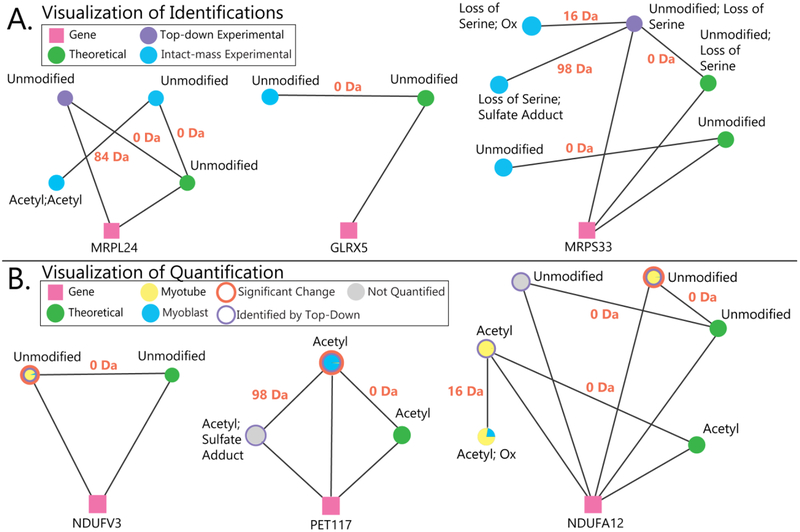
Proteoform Suite also enables the visualization of proteoform families as a network of masses related by PTMs, amino acid differences, chemical adducts, or by relation to the same gene, shown in Figures 6 and S-4. The visualization of proteoform families lets the user view all gene products and their modifications in a single graphic. Several examples of visualized mitochondrial proteoform families from identification results are shown in Figure 6A. Top-down fragmentation analysis was able to identify an unmodified form of a proteoform from the gene MRPL24; Proteoform Suite identified a diacetylated form of this protein by intact-mass analysis. Proteoform Suite also identified an unmodified proteoform from GLRX5; this protein was not identified in the top-down analysis, so an entire proteoform family was revealed by intact-mass analysis. In a third example, Proteoform Suite identified additional proteoforms from the MRPS33 proteoform family by intact-mass analysis.
Figure 6.
A) Examples of visualized proteoform families. Pink squares represent genes, and green circles represent theoretical proteoforms. Experimental proteoforms identified by top-down fragmentation are purple circles, and experimental proteoforms observed in MS1 spectra but not identified by top-down analysis are blue circles. The lines between circles are labeled with mass differences corresponding to modifications between proteoforms. In the left family, top-down analysis identified the unmodified form of MRPL24, and Proteoform Suite enabled the identification of the acetylated proteoform by intact-mass analysis. In the middle family, the mitochondrial proteoform family GLRX5 was identified by intact-mass alone; this protein would have remained unidentified if only a typical top-down analysis had been performed. The right family shows how Proteoform Suite identified three additional proteoforms by intact-mass analysis. B) Examples of quantified proteoform families. For quantified experimental proteoforms, a pie chart of yellow and blue depicts the abundance ratio in myoblasts and myotubes, an orange annulus indicates a significant change, and a purple annulus indicates that the experimental proteoform was identified through top-down fragmentation; unquantified experimental proteoforms are illustrated as grey circles. In the left family, an unmodified proteoform from the mitochondrial gene NDUFV3 was more abundant in myotubes, and in the middle family, an acetylated form of PET117 was more abundant in myoblasts. In the right family, different oxidized and acetylated proteoforms were identified from the NDUFA12 gene; an unmodified form was significantly more abundant in myotubes.
Proteoform Suite also provides a script that allows the user to visualize quantification results in Cytoscape. In these diagrams, quantified proteoforms show relative proteoform abundances in myoblasts (blue) and myotubes (yellow) as a pie chart in the circles representing experimental proteoforms. Several examples of quantified mitochondrial proteoform families are shown in Figure 6B, from the NDUFV3, PET117, and NDUFA12 proteoform families, which were discussed previously in the Quantification of Proteoforms section of the Results and Discussion.
CONCLUSIONS
This study employed an integrated intact-mass and top-down strategy to analyze murine mitochondrial proteoforms during myogenesis. We identified additional proteoforms by their intact masses, quantified unlabeled experimental proteoforms across undifferentiated myoblast and differentiated myotube cell types, and visualized the results as networks of related proteoforms. This strategy was implemented in the software program Proteoform Suite, which is open-source, freely available (https://smith-chem-wisc.github.io/ProteoformSuite/), and can be integrated into current top-down proteomic workflows to identify, quantify, and visualize proteoform families.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Smith lab who provided continual help and feedback on Proteoform Suite, including Robert Millikin, Zach Rolfs, Yunxiang Dai, Rachel Knoener, and Rachel Miller. We also thank Ryan Fellers, Richard LeDuc, Joseph Greer, Bryan Early, and AJ van Nispen at the NRTDP, who developed TDPortal.
Funding Sources
This work was supported by National Institutes of Health grants R01GM114292 (to L.M.S.) and R01GM115591 (to D.J.P.) from the National Institute of General Medical Sciences (NIGMS) and R01DK098672 from the National Institute of Diabetes and Digestive and Kidney Diseases. L.V.S. was supported by the NIGMS Biotechnology Training Program, T32GM008349. A.J.C was supported by the Computation and Informatics in Biology and Medicine Training Program, T15LM007359.
ABBREVIATIONS
- PTM
post-translational modification
- MS
mass spectrometry
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- SDS
sodium dodecyl sulfate
- GELFrEE
gel-eluted liquid entrapment electrophoresis
- NRTDP
National Resource for Translational and Developmental Proteomics
- FDR
false discovery rate
- XML
extensible markup language
- ET
experiment-theoretical
- EE
experiment-experiment
- G-PTM-D
global PTM discovery
Footnotes
ASSOCIATED CONTENT
Supporting Information. Materials and Methods, Raw Data File Summary, Proteoform Identification; Figure S-1: Proteoform Suite Overview; Figure S-2: Proteoform Retention Time Differences; Figure S-3, Mass Error Histogram; Figure S-4, Visualized Proteoform Families; Table S-1, Top-Down Experimental Proteoforms; Table S-2, Accepted EE and ET Pair Mass Differences; Table S-3, Intact-Mass Experimental Proteoform Identifications; Table S-4, Quantified Experimental Proteoforms; Table S-5, Bottom-Up Quantification Results.
REFERENCES
- (1).Smith LM; Kelleher NL Proteoform: A Single Term Describing Protein Complexity Lloyd. Nat. Methods 2013, 10 (3), 186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Shortreed MR; Frey BL; Scalf M; Knoener RA; Cesnik AJ; Smith LM Elucidating Proteoform Families from Proteoform Intact-Mass and Lysine-Count Measurements. J. Proteome Res. 2016, 15 (4), 1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Zamdborg L; LeDuc RD; Glowacz KJ; Kim Y. Bin; Viswanathan V; Spaulding IT; Early BP; Bluhm EJ; Babai S; Kelleher NL ProSight PTM 2.0: Improved Protein Identification and Characterization for Top down Mass Spectrometry. Nucleic Acids Res. 2007, 35, 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kou Q; Wu S; Tolic N; Pasa-Tolic L; Liu Y; Liu X A Mass Graph-Based Approach for the Identification of Modified Proteoforms Using Top-down Tandem Mass Spectra. Bioinformatics 2016, 33 (9), 1309–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sun RX; Luo L; Wu L; Wang RM; Zeng WF; Chi H; Liu C; He SM pTop 1.0: A High-Accuracy and High-Efficiency Search Engine for Intact Protein Identification. Anal. Chem 2016, 88 (6), 3082–3090. [DOI] [PubMed] [Google Scholar]
- (6).Frank AM; Pesavento JJ; Mizzen CA; Kelleher NL; Pevzner PA Interpreting Top-down Mass Spectra Using Spectral Alignment. Anal. Chem 2008, 80 (7), 2499–2505. [DOI] [PubMed] [Google Scholar]
- (7).Karabacak NM; Li L; Tiwari A; Hayward LJ; Hong P; Easterling ML; Agar JN Sensitive and Specific Identification of Wild Type and Variant Proteins from 8 to 669 kDa Using Top-down Mass Spectrometry. Mol. Cell. Proteomics 2009, 8 (4), 846–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Nesvizhskii AI; Aebersold R Interpretation of Shotgun Proteomic Data. Mol. Cell. Proteomics 2005, 4 (10), 1419–1440. [DOI] [PubMed] [Google Scholar]
- (9).Chen B; Brown KA; Lin Z; Ge Y Top-down Proteomics: Ready for Prime Time? Anal. Chem 2018, 90 (1), 110–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kellie JF; Catherman AD; Durbin KR; Tran JC; Tipton JD; Norris JL; Witkowski CE; Thomas PM; Kelleher NL Robust Analysis of the Yeast Proteome under 50 kDa by Molecular-Mass-Based Fractionation and Top-down Mass Spectrometry. Anal. Chem 2012, 84 (1), 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).McCool E; Lubeckyj RA; Shen X; Chen D; Kou Q; Liu X; Sun L Deep Top-down Proteomics Using Capillary Zone Electrophoresis-Tandem Mass Spectrometry: Identification of 5700 Proteoforms from the Escherichia Coli Proteome. Anal. Chem 2018, 90, 5529–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wang Z; Ma H; Smith K; Wu S Two-Dimensional Separation Using High-pH and Low-pH Reversed Phase Liquid Chromatography for Top-down Proteomics. Int. J. Mass Spectrom. 2018, 427, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Catherman AD; Durbin KR; Ahlf DR; Early BP; Fellers RT; Tran JC; Thomas PM; Kelleher NL Large-Scale Top-down Proteomics of the Human Proteome: Membrane Proteins, Mitochondria, and Senescence. Mol. Cell. Proteomics 2013, 12 (12), 3465–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Anderson LC; DeHart CJ; Kaiser NK; Fellers RT; Smith DF; Greer JB; LeDuc RD; Blakney GT; Thomas PM; Kelleher NL; Hendrickson CL Identification and Characterization of Human Proteoforms by Top-Down LC-21 Tesla FT-ICR Mass Spectrometry. J. Proteome Res. 2017, 16 (2), 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhao Y; Sun L; Zhu G; Dovichi NJ Coupling Capillary Zone Electrophoresis to a Q Exactive HF Mass Spectrometer for Top-down Proteomics: 580 Proteoform Identifications from Yeast. J. Proteome Res. 2016, 15 (10), 3679–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Tran JC; Zamdborg L; Ahlf DR; Lee JE; Catherman AD; Durbin KR; Tipton JD; Vellaichamy A; Kellie JF; Li M; Wu C; Sweet SMM; Early BP; Siuti N; LeDuc RD; Compton PD; Thomas PM; Kelleher NL Mapping Intact Protein Isoforms in Discovery Mode Using Top-down Proteomics. Nature 2011, 480 (7376), 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Durbin KR; Tran JC; Zamdborg L; Sweet SMM; Adam D; Lee JE; Li M; Kellie JF; Kelleher NL Intact Mass Detection, Interpretation, and Visualization to Automate Top-Down Proteomics on a Large Scale. Proteomics 2011, 10 (20), 3589–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Riley NM; Mullen C; Weisbrod CR; Sharma S; Senko MW; Zabrouskov V; Westphall MS; Syka JEP; Coon JJ Enhanced Dissociation of Intact Proteins with High Capacity Electron Transfer Dissociation. J. Am. Soc. Mass Spectrom. 2016, 27 (3), 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Compton PD; Zamdborg L; Thomas PM; Kelleher NL On the Scalability and Requirements of Whole Protein Mass Spectrometry. Anal. Chem 2011, 83 (17), 6868–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Schaffer LV; Shortreed MR; Cesnik AJ; Frey BL; Solntsev SK; Scalf M; Smith LM Expanding Proteoform Identifications in Top-Down Proteomic Analyses by Constructing Proteoform Families. Anal. Chem 2018, 90, 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kislinger T; Gramolini AO; Pan Y; Rahman K; MacLennan DH; Emili A Proteome Dynamics during C2C12 Myoblast Differentiation. Mol. Cell. Proteomics 2005, 4 (7), 887–901. [DOI] [PubMed] [Google Scholar]
- (22).Casadei L; Vallorani L; Gioacchini AM; Guescini M; Burattini S; D’Emilio A; Biagiotti L; Falcieri E; Stocchi V Proteomics-Based Investigation in C2C12 Myoblast Differentiation. Eur. J. Histochem 2009, 53 (4), 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Cui Z; Chen X; Lu B; Park SK; Xu T; Xie Z; Xue P; Hou J; Hang H; Yates JR; Yang F Preliminary Quantitative Profile of Differential Protein Expression between Rat L6 Myoblasts and Myotubes by Stable Isotope Labeling with Amino Acids in Cell Culture. Proteomics 2009, 9 (5), 1274–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Cox J; Hein MY; Luber CA; Paron I Accurate Proteome-Wide Label-Free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteomics 2014, 13 (9), 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Veenstra TD; Martinovic S; Anderson GA; Pasa-Tolic L; Smith RD Proteome Analysis Using Selective Incorporation of Isotopically Labeled Amino Acids. J. Am. Soc. Mass Spectrom. 2000, 11 (1), 78–82. [DOI] [PubMed] [Google Scholar]
- (26).Old WM; Meyer-Arendt K; Aveline-Wolf L; Pierce KG; Mendoza A; Sevinsky JR; Resing KA; Ahn NG Comparison of Label-Free Methods for Quantifying Human Proteins by Shotgun Proteomics. Mol. Cell. Proteomics 2005, 4 (10), 1487–1502. [DOI] [PubMed] [Google Scholar]
- (27).Wiese S; Reidegeld KA; Meyer HE; Warscheid B Protein Labeling by iTRAQ: A New Tool for Quantitative Mass Spectrometry in Proteome Research. Proteomics 2007, 7 (3), 340–350. [DOI] [PubMed] [Google Scholar]
- (28).Yang X; Coulombe-Huntington J; Kang S; Sheynkman GM; Hao T; Richardson A; Sun S; Yang F; Shen YA; Murray RR; Spirohn K; Begg BE; Duran-Frigola M; MacWilliams A; Pevzner SJ; Zhong Q; Trigg SA; Tam S; Ghamsari L; Sahni N; Yi S; Rodriguez MD; Balcha D; Tan G; Costanzo M; Andrews B; Boone C; Zhou XJ; Salehi-Ashtiani K; Charloteaux B; Chen AA; Calderwood MA; Aloy P; Roth FP; Hill DE; Iakoucheva LM; Xia Y; Vidal M Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell 2016, 164 (4), 805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ntai I; Kim K; Fellers RT; Skinner OS; Smith AD; Early BP; Savaryn JP; Leduc RD; Thomas PM; Kelleher NL Applying Label-Free Quantitation to Top down Proteomics. Anal. Chem 2014, 86 (10), 4961–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Durbin KR; Fornelli L; Fellers RT; Doubleday PF; Narita M; Kelleher NL Quantitation and Identification of Thousands of Human Proteoforms Below 30 kDa. J. Proteome Res. 2016, 15, 976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Mazur MT; Cardasis HL Quantitative Analysis of Apolipoproteins in Human HDL by Top-down Differential Mass Spectrometry. Methods Mol. Biol 2013, 1000 (17), 115–137. [DOI] [PubMed] [Google Scholar]
- (32).Zhang J; Guy MJ; Norman HS; Chen YC; Xu Q; Dong X; Guner H; Wang S; Kohmoto T; Young KH; Moss RL; Ge Y Top-down Quantitative Proteomics Identified Phosphorylation of Cardiac Troponin I as a Candidate Biomarker for Chronic Heart Failure. J. Proteome Res. 2011, 10 (9), 4054–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Cesnik AJ; Shortreed MR; Schaffer LV; Knoener RA; Frey BL; Scalf M; Solntsev SK; Dai Y; Gasch AP; Smith LM Proteoform Suite: Software for Constructing, Quantifying, and Visualizing Proteoform Families. J. Proteome Res. 2018, 17 (1), 568–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tran JC; Doucette AA Gel-Eluted Liquid Fraction Entrapment Electrophoresis: An Electrophoretic Method for Broad Molecular Weight Range Proteome Separation. Anal. Chem 2008, 80 (5), 1568–1573. [DOI] [PubMed] [Google Scholar]
- (35).LeDuc RD; Taylor GK; Kim Y. Bin; Januszyk TE; Bynum LH; Sola JV; Garavelli JS; Kelleher NL ProSight PTM: An Integrated Environment for Protein Identification and Characterization by Top-down Mass Spectrometry. Nucleic Acids Res. 2004, 32, 340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Cox J; Michalski A; Mann M Software Lock Mass by Two-Dimensional Minimization of Peptide Mass Errors. J. Am. Soc. Mass Spectrom. 2011, 22 (8), 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Solntsev SK; Shortreed MR; Frey BL; Smith LM Enhanced Global Post-Translational Modification Discovery with MetaMorpheus. J. Proteome Res. 2018, 17, 1844–1851. [DOI] [PubMed] [Google Scholar]
- (38).Leduc RD; Fellers RT; Early BP; Greer JB; Thomas PM; Kelleher NL The C-Score: A Bayesian Framework to Sharply Improve Proteoform Scoring in High-Throughput Top down Proteomics. J. Proteome Res. 2014, 13 (7), 3231–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Dai Y; Shortreed MR; Scalf M; Frey BL; Cesnik AJ; Solntsev S; Schaffer LV; Smith LM Elucidating Escherichia Coli Proteoform Families Using Intact-Mass Proteomics and a Global PTM Discovery Database. J. Proteome Res. 2017, 16 (11), 4156–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Tyanova S; Temu T; Sinitcyn P; Carlson A; Hein MY; Geiger T; Mann M; Cox J The Perseus Computational Platform for Comprehensive Analysis of (Prote)omics Data. Nat. Methods 2016, 13 (9), 731–740. [DOI] [PubMed] [Google Scholar]
- (41).Benjamini Y; Hochberg Y Controlling the False Discovery R ate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57 (1), 289–300. [Google Scholar]
- (42).Shannon P; Markiel A; Ozier O; Baliga NS; Wang JT; Ramage D; Amin N; Schwikowski B; Ideker T Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13 (11), 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Smoot ME; Ono K; Ruscheinski J; Wang P; Ideker T Cytoscape 2.8: New Features for Data Integration and Network Visualization. Bioinformatics 2011, 27 (3), 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Cox J; Mann M MaxQuant Enables High Peptide Identification Rates, Individualized P.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol 2008, 26 (12), 1367–1372. [DOI] [PubMed] [Google Scholar]
- (45).Calvo SE; Clauser KR; Mootha VK MitoCarta2.0: An Updated Inventory of Mammalian Mitochondrial Proteins. Nucleic Acids Res. 2015, 44, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Catherman AD; Li M; Tran JC; Durbin KR; Compton PD; Early BP; Thomas PM; Kelleher NL Top down Proteomics of Human Membrane Proteins from Enriched Mitochondrial Fractions. Anal. Chem 2013, 85 (3), 1880–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Shortreed MR; Wenger CD; Frey BL; Sheynkman GM; Scalf M; Keller MP; Attie AD; Smith LM Global Identification of Protein Post-Translational Modifications in a Single-Pass Database Search. J. Proteome Res. 2015, 14 (11), 4714–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Li Q; Shortreed MR; Wenger CD; Frey BL; Schaffer LV; Scalf M; Smith LM Global Post-Translational Modification Discovery. J. Proteome Res. 2017, 16 (4), 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Bulteau AL; Szweda LI; Friguet B Mitochondrial Protein Oxidation and Degradation in Response to Oxidative Stress and Aging. Exp. Gerontol 2006, 41 (7), 653–657. [DOI] [PubMed] [Google Scholar]
- (50).Hamanaka RB; Chandel NS Mitochondrial Reactive Oxygen Species Regulate Cellular Signaling and Dictate Biological Outcomes. Trends Biochem. Sci 2010, 35 (9), 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Sena LA; Chandel NS Physiological Roles of Mitochondrial Reactive Oxygen Species. Mol. Cell 2012, 48 (2), 158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Vincent CE; Rensvold JW; Westphall MS; Pagliarini DJ; Coon JJ Automated Gas-Phase Purification for Accurate, Multiplexed Quantification on a Stand-Alone Ion-Trap Mass Spectrometer. Anal. Chem 2013, 85 (4), 2079–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Ntai I; LeDuc RD; Fellers RT; Erdmann-Gilmore P; Davies SR; Rumsey J; Early BP; Thomas PM; Li S; Compton PD; Ellis MJC; Ruggles KV; Fenyö D; Boja ES; Rodriguez H; Townsend RR; Kelleher NL Integrated Bottom-Up and Top-Down Proteomics of Patient-Derived Breast Tumor Xenografts. Mol. Cell. Proteomics 2016, 15 (1), 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Acin-Perez R; Gatti DL; Bai Y; Manfredi G Protein Phosphorylation and Prevention of Cytochrome Oxidase Inhibition by ATP: Coupled Mechanisms of Energy Metabolism Regulation. Cell Metab. 2011, 13 (6), 712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Santos-Rosa H; Kirmizis A; Nelson C; Bartke T; Saksouk N; Cote J; Kouzarides T Histone H3 Tail Clipping Regulates Gene Expression. Nat. Struct. Mol. Biol 2009, 16 (1), 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Gupta N; Bandeira N; Keich U; Pevzner PA Target-Decoy Approach and False Discovery Rate: When Things May Go Wrong. J. Am. Soc. Mass Spectrom. 2011, 22 (7), 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.